Abstract
The pathogenesis of intervertebral disc degeneration (IDD) is complex, and a better understanding of IDD pathogenesis may provide a better method for the treatment of IDD. Exosomes are 40–100 nm nanosized vesicles that are released from many cell types into the extracellular space. We speculated that exosome-transported circular RNAs (circRNAs) could regulate IDD. Exosomes from different degenerative grades were isolated and added to nucleus pulposus cells (NPCs), and indicators of proliferation and apoptosis were detected. Based on the previous circRNA microarray results, the top 10 circRNAs were selected. PCR was performed to determine the circRNA with the maximum upregulation. Competing endogenous RNA (ceRNA) analysis was carried out, and the sponged microRNA (miRNA) was identified. Further functional verification of the selected circRNA was carried out in vivo and in vitro. NPCs of different degenerative grades secreted exosomes, which could regulate IDD. circRNA_0000253 was selected as having the maximum upregulation in degenerative NPC exosomes. ceRNA analysis showed that circRNA_0000253 could adsorb miRNA-141-5p to downregulate SIRT1. circRNA_0000253 was confirmed to increase IDD by adsorbing miRNA-141-5p and downregulating SIRT1 in vivo and in vitro. Exosomal circRNA_0000253 owns the maximum upregulation in degenerative NPC exosomes and could promote IDD by adsorbing miRNA-141-5p and downregulating SIRT1.
Keywords: circular RNAs, exosome, microRNA, SIRT1, intervertebral disc degeneration
Graphical Abstract
Intervertebral disc degeneration (IDD) disease may cause substantial medical costs. Song et al. report that circRNA_0000253 could adsorb microRNA-141-5p to downregulate SIRT1, which may increase IDD. The study provides insight into the IDD and great help to prevent the occurrence of IDD.
Introduction
The incidence of spine musculoskeletal disorders mainly caused by intervertebral disc degeneration (IDD) disease is increasing in the population year by year as society ages, which may seriously affect patient quality of life and cause substantial medical costs.1,2 The extracellular matrix (ECM) consists of type II collagen (collagen II), which helps to maintain the disc height and respond to external mechanical stress.3 ECM metabolism during IDD is regulated by growth factors, inflammatory factors, and noncoding RNAs,4, 5, 6, 7, 8, 9 and it is important to clarify the metabolic mechanism of ECM catabolism during IDD.
Circular RNAs (circRNAs) are a family of covalently closed noncoding RNA molecules produced by the backsplicing of exons in precursor mRNAs in eukaryotes.10,11 Current studies have shown that circRNAs are closely involved in the occurrence and development of IDD.3,9,12,13 Exosomes are microvesicles (MVs) with a diameter of 40–100 nm, and they can carry various proteins, lipids, and nucleic materials, such as DNA, RNA, messenger RNA (mRNA), and noncoding RNAs.14, 15, 16 Exosomes originate from multivesicular endosomes by inverse budding and are released into the extracellular space when a multivesicular body fuses with the plasma membrane to regulate the occurrence and development of various diseases.17 A recent study showed that delivery of mesenchymal stem cell (MSC)-derived exosomes in vivo modulated endoplasmic reticulum (ER) stress-related apoptosis and retarded IDD progression in a rat tail model.18
Our previous study showed that circRNAs were highly expressed in intervertebral discs and played important roles in regulating IDD.9 However, it is still unclear where circRNAs are distributed in nucleus pulposus (NP) tissues and how they are transported to regulate IDD. In this study, we hypothesized that circRNAs are abundant in NP cell (NPC) exosomes and are transferred from transformed NPCs into neighboring, normal NPCs via exosomes to regulate IDD.
Results
Exosomes Were Abundant in NPCs and Differentially Expressed in Degenerative and Normal NPCs
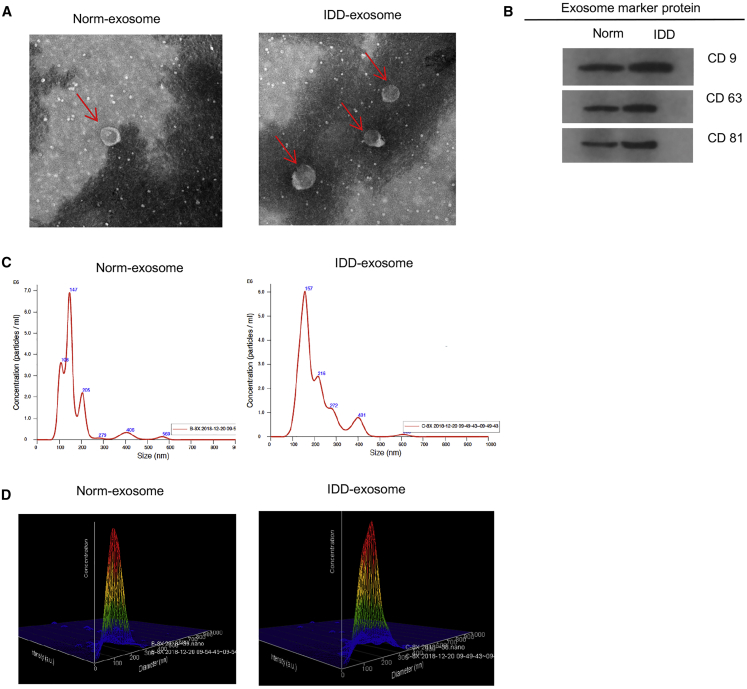
The detection and classification of exosomes are challenging.19 To investigate whether exosomes can be secreted from NPCs, the NPC culture medium was collected, and exosomes were isolated. Electron microscopy revealed that the exosomes of normal and degenerative NPCs showed typical rounded shapes, and the degenerative NPCs secreted more exosomes than the normal NPCs (Figure 1A). Western blot (WB) analysis confirmed the presence of exosome markers CD9, CD63, and CD81 in exosomes derived from normal and degenerative NPCs (Figure 1B). The particle sizes and number of exosomes showed that no significant differences were detected between exosomes derived from degenerative and normal NPCs (Figures 1C and 1D). The exosomes with the highest concentration were 147 nm in normal NPCs, and the exosomes with the highest concentration were 157 nm in degenerative NPCs.
Figure 1.
Exosomes Were Abundant in NPCs and Differently Distributed in Degenerative and Normal Nucleus Pulposus Cells (NPCs)
(A) Electron microscopy showed the shape of exosomes isolated from normal and degenerative NPCs. Scale bar, 100 nm. (B) WB shows the exosomal marker protein expression of normal and degenerative NPCs. (C) The particle size and number of the exosomes are shown by ZetaView nanoparticle tracker. (D) The particles and number of the exosomes are shown at the three dimensional.
NPC Medium Could Regulate IDD by Secreted Exosomes
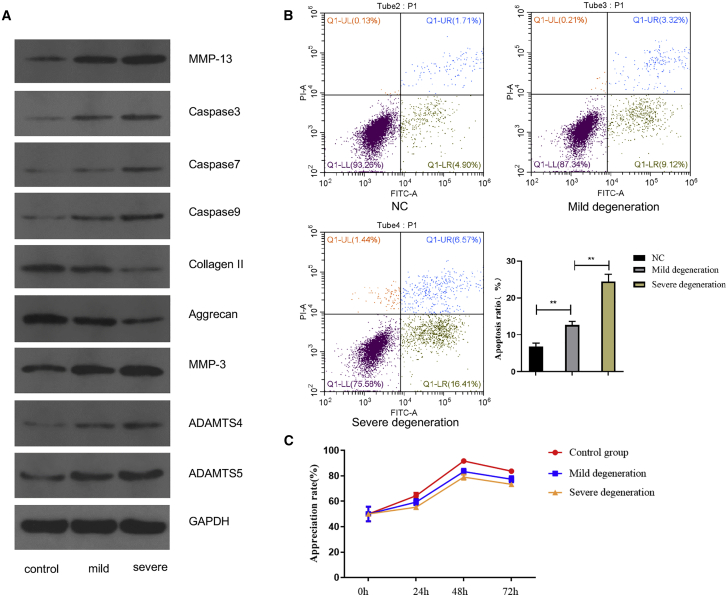
Mild- and severe-degeneration exosomes were isolated from NPCs9 The proteins caspase-3, caspase-7, caspase-9, matrix metallopeptidase (MMP)-3, MMP-13, a disintegrin and metalloproteinase thrombospondin type I motifs 4 (ADAMTS4), and ADAMTS5 were regarded as the degeneration-promoting indexes, whereas the protein levels of aggrecan and collagen II were regarded as the degeneration-inhibitory indexes.3 The results showed that the expression levels of caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, and ADAMTS5 were higher in NPCs with degenerative exosomes than in NPCs without exosomes, and the expression level was higher in NPCs treated with severely degenerated exosomes than in those treated with mildly degenerated exosomes (Figure 2A). The flow cytometry results showed that the NPC apoptotic rate in the negative control (NC) group was much lower than that in the mild-degeneration exosome group, and the NPC apoptotic rate in the severe-degeneration exosome group was much higher than that in the mild-degeneration exosome group (Figure 2B). The Cell Counting Kit 8 (CCK-8) results showed that the mild-degeneration exosome group had a lower NPC proliferative capacity when compared with the control group, and the NPC proliferative capacity in the severe-degeneration exosome group was lower than that in the mild-degeneration exosome group (Figure 2C). The results of flow cytometry and CCK-8 confirmed that exosomes secreted by degenerative NPCs could promote apoptosis and inhibit the proliferation of NPCs.
Figure 2.
NPC-Conditioned Medium Regulates IDD by Secreting Exosomes
(A) WB showing the differentially expressed proteins between the mild and severe groups. (B) Flow cytometry showing the apoptosis of NPCs between the mild and severe groups. (C) CCK-8 assay showing the proliferation of NPCs between the mild and severe groups.
circRNA_0000253 Could Competitively Adsorb microRNA (miRNA)-141-5p
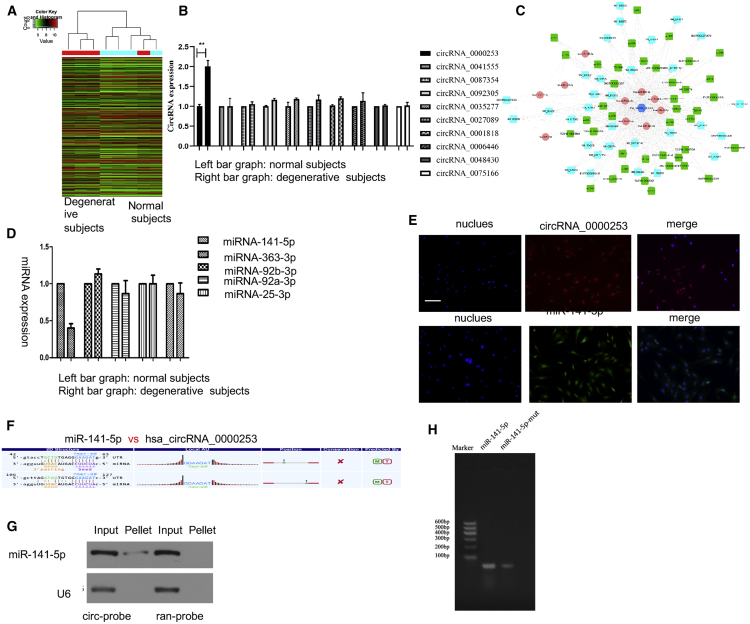
To explore the possible roles of circRNAs in IDD, PCR was used to detect circRNA expression between normal and degenerative NPC exosomes based on the results of the circRNA microarray (Figure 3A).9 The top 10 upregulated circRNAs were selected, and the PCR results showed that circRNA_0000253 was the most upregulated circRNA in the degenerative NPC exosomes when compared with the normal NPC exosomes (Figure 3B). Competing endogenous RNA (ceRNA) analysis showed that 10 miRNAs were identified that were related to circRNA_0000253, and the top 5 miRNAs with the most binding sites were miRNA-141-5p, miRNA-363-3p, miRNA-92b-3p, miRNA-92a-3p, and miRNA-25-3p (Figure 3C). PCR was used to detect the expression levels of the miRNAs between the normal and degenerative NPC exosomes, and the results showed that miRNA-141-5p was the most strongly upregulated miRNA in the degenerative NPC exosomes compared with the normal NPC exosomes (Figure 3D). Fluorescence in situ hybridization (FISH) analysis showed that circRNA_0000253 and miRNA-25-3p were distributed in the cytoplasm and nucleus of NPCs (Figure 3E). The binding sequence diagram of circRNA_0000253 and miRNA-141-5p is shown in Figure 3F. Northern blot analysis revealed that circRNA_0000253 could reverse the pull-down of miRNA-141-5p (Figure 3G). Pull-down assay analysis confirmed a greater enrichment of circRNA_0000253 in the miRNA-141-5p-captured fraction compared with the fraction with the miRNA-141-5p mutation, which disrupted the binding site of miRNA-141-5p on circRNA_0000253 (Figure 3H).
Figure 3.
circRNA_0000253 Competitively Adsorbs microRNA (miRNA)-141-5p
(A) The circRNAs differentially expressed in the nucleus pulposus (NP) tissue. (B) The top 10 upregulated circRNAs were verified by qPCR in degenerative and normal NP exosomes. (C) The possible sponged miRNAs of circRNA_0000253. (D) The possible sponged miRNAs were verified by qPCR in degenerative and normal NP exosomes. (E) RNA fluorescence in situ hybridization (FISH) was used to detect the colocalization of circRNA_0000253 and miRNA-141-5p in NPCs. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 100 μm. (F) The binding sequence diagram of circRNA_0000253 and miRNA-141-5p. (G) miRNA-141-5p was pulled down by the circular probe (circ-probe) but not the random probe (ran-probe), and the levels of miRNA-141-5p were detected by northern blot. Input, 20% samples were loaded; pellet, all samples were loaded. (H) Pull-down assay analysis confirmed a greater enrichment of circRNA_0000253 in the miRNA-141-5p-captured fraction compared with the introduction of the miRNA-141-5p mutation that disrupted the binding site of miRNA-141-5p in circRNA_0000253.
circRNA_0000253 Could Promote IDD by Competitively Adsorbing miRNA-141-5p
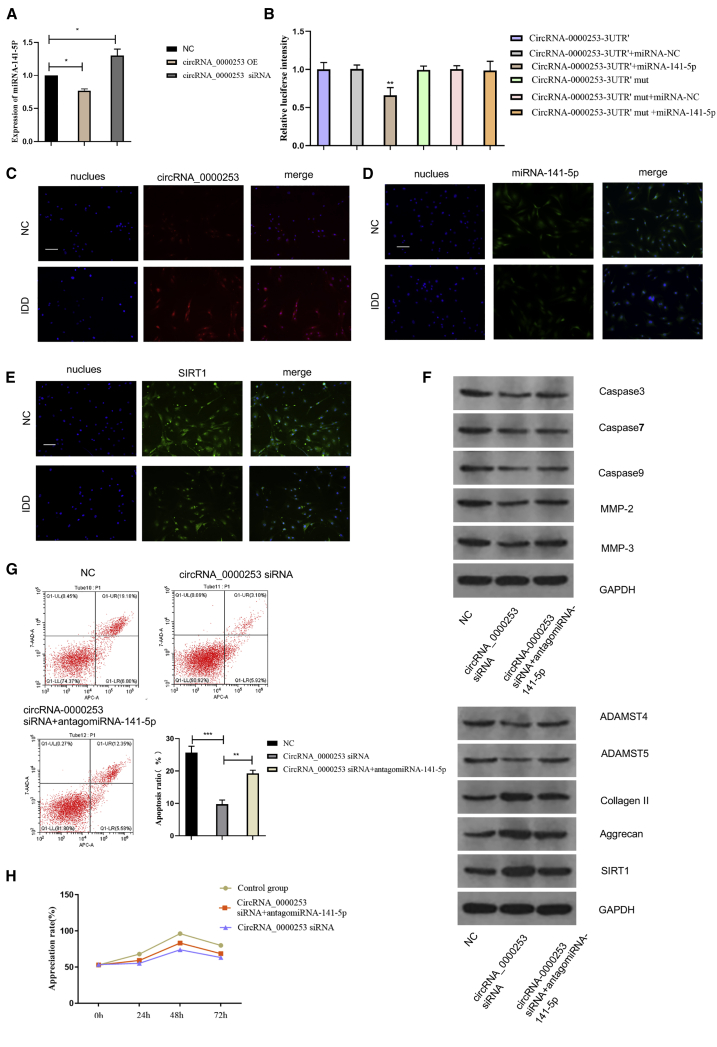
Overexpression and inhibition of circRNA_0000253 were achieved. The results showed that overexpression of circRNA_0000253 could downregulate miRNA-141-5p expression compared with the NC group, and interfering circRNA_0000253 upregulates miRNA-141-5p expression. The results confirm that circRNA_0000253 could competitively adsorb miRNA-141-5p (Figure 4A). The dual-luciferase reporter assay showed that the luciferase signal of the circRNA_0000253 reporter was suppressed by miRNA-141-5p, whereas the introduction of mutations in the circRNA abolished the inhibitory effect of miRNA-141-5p. The results confirmed that circRNA_0000253 could directly absorb miRNA-141-5p to regulate IDD (Figure 4B). FISH assay was carried out to compare the gene expression between IDD NPCs and normal NPCs, and the results showed that circRNA_0000253, miRNA-141-5p, and SIRT1 were expressed in both the nucleus and cytoplasm of NPCs (Figures 4C–4E). The results also confirmed that circRNA_0000253 levels were markedly increased in IDD NPCs compared with normal NPCs, and miRNA-141-5p and SIRT1 levels were markedly reduced in IDD NPCs compared with normal NPCs (Figures 4C–4E). To further confirm that circRNA_0000253 could competitively absorb miRNA-141-5p and downregulate SIRT1, the protein levels of caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, ADAMTS5, collagen II, aggrecan, and SIRT1 were measured by WB and compared among the NC, circRNA_0000253 small interfering RNA (siRNA), and circRNA_0000253 siRNA + antagomiRNA-141-5p groups. The results showed that circRNA_0000253 could promote the expression of caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, and ADAMTS5 and inhibit collagen II and aggrecan by competitively absorbing miRNA-141-5p and downregulating SIRT1 (Figure 4F). Proliferation and apoptosis assays in NPCs were also performed, and the results showed that circRNA_0000253 could promote apoptosis and inhibit proliferation of NPCs by competitively absorbing miRNA-141-5p and downregulating SIRT1 (Figures 4G and 4H). Blockade of circRNA_0000253 may serve as a potential therapeutic approach in the treatment of IDD.
Figure 4.
circRNA_0000253 Upregulates IDD by Competitively Adsorbing miRNA-141-5p
(A) qPCR showed the miRNA-141-5p expression in the NC, circRNA_0000253 siRNA, and circRNA_0000253 siRNA + antagomiRNA-141-5p groups; ∗p < 0.05, ∗∗p < 0.01. (B) Dual-luciferase assay showed the binding effect between circRNA_0000253 and miRNA-141-5p; ∗∗p < 0.01. (C–E) The expression of circRNA_0000253 (C), miRNA-141-5p (D), and SIRT1 (E) was detected in NP samples from patients with or without IDD by RNA FISH. circRNA_0000253, miRNA-141-5p, and SIRT1 probes were labeled with Alexa 488. Nuclei were stained with DAPI. Scale bars, 100 μm. (F) WB shows the protein expression of caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, ADAMTS 5, collagen II, aggrecan, and SIRT1 in the NC, circRNA_0000253 siRNA, and circRNA_0000253 siRNA + antagomiRNA-141-5p groups. (G) Flow cytometry showed the apoptosis of NPCs in the NC, circRNA_0000253 siRNA, and circRNA_0000253 siRNA + antagomiRNA-141-5p groups. (H) CCK-8 assay showed the proliferation of NPCs in the NC, circRNA_0000253 siRNA, and circRNA_0000253 siRNA+ antagomiRNA-141-5p groups.
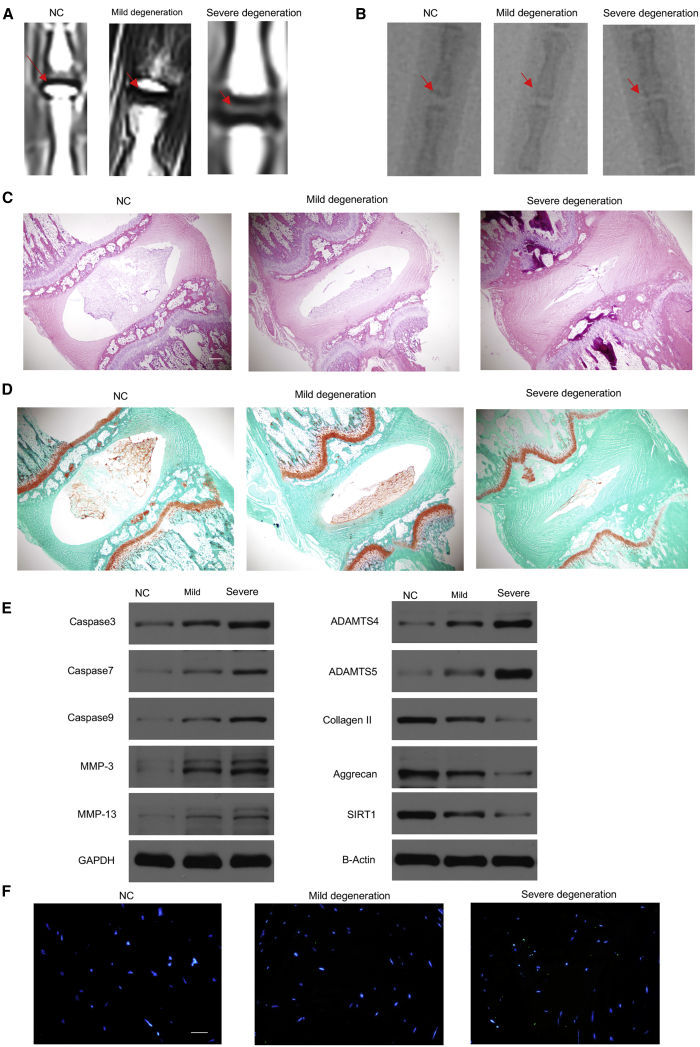
Exosomes from NPCs of Different Degeneration Levels Could Regulate Rat IDD
It is necessary to think about whether all concentrations of exosomes used in the experiments have any relevance to the conditions in vivo. Based on a modified Thompson classification, the rats injected with exosomes from the severe IDD group had a higher degeneration grade compared with the rats injected with exosomes from the mild IDD and NC groups, and the rats injected with exosomes from the NC group had a lower degeneration grade compared with the exosomes from the mild IDD groups (Figure 5A). The rats injected with exosomes from the severe IDD group had a lower disc height index (%DHI) compared with the exosomes from the mild IDD and NC groups, and rats injected with exosomes from the mild IDD group had a lower %DHI compared with exosomes from the NC groups (Figure 5B). Hematoxylin and eosin (H&E) and SafraninO-Fast Green Staining were used to evaluate the degeneration grade of rat NP. Rats injected with exosomes from the severe IDD group had a higher degree of IDD than the exosomes from the mild IDD and NC groups, and the rats injected with exosomes from the NC group had a lower degree of IDD than the exosomes from the mild IDD groups (Figures 5C and 5D). WB indicators were used to evaluate rat IDD, including caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, ADAMTS 5, collagen II, aggrecan, and SIRT1. The results showed that rats injected with exosomes from the severe IDD group had increased expression of caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, and ADAMTS 5 and decreased expression of collagen II, aggrecan, and SIRT1 compared with the exosomes from the mild IDD groups, whereas rats injected with exosomes from the mild IDD group had increased expression of caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, and ADAMTS 5 and decreased expression of collagen II, aggrecan, and SIRT1 compared with the exosomes from the NC groups (Figure 5E). Terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate (dUTP) nick end labeling (TUNEL) staining showed that apoptosis of the NP in the severe-degeneration groups owns a higher apoptosis rate than the mild- and NC-degeneration groups, whereas the mild-degeneration group owns a higher apoptosis rate than the NC group (Figure 5F).
Figure 5.
Exosomes from NPCs of Different Degeneration Levels Could Regulate Rat IDD
(A) MRI showed rat disc degeneration in the NC, mild-degeneration, and severe-degeneration groups. (B) X-ray showing the disc height index (%DHI) of the NC, mild-degeneration, and severe-degeneration groups. (C and D). H&E (C) and SafraninO-Fast Green (D) staining compared rat NP among the NC, mild-degeneration, and severe-degeneration groups. Scale bar, 500 μm. (E) WB showed caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, ADAMTS 5, collagen II, aggrecan, and SIRT1 expression in the NC, mild-degeneration, and severe-degeneration groups. Nuclei were stained with DAPI. (F) Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining showed apoptosis of the NP in the NC, mild-degeneration, and severe-degeneration groups. Scale bar, 50 μm.
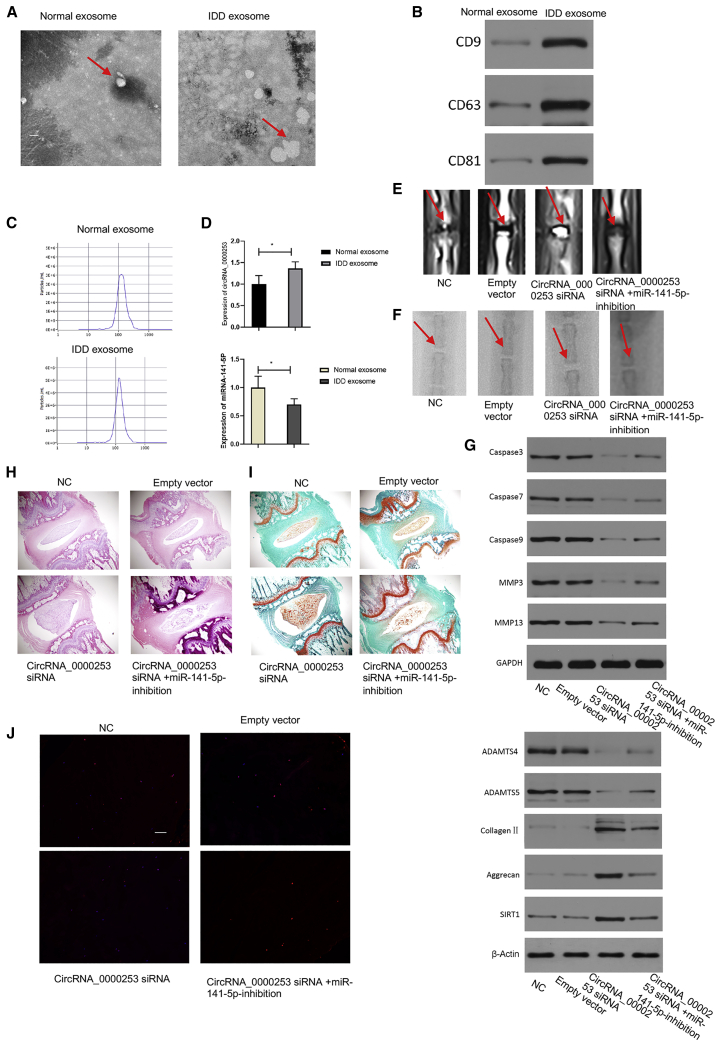
circRNA_0000253 Could Promote IDD in Rats by Competitively Adsorbing miRNA-141-5p
Previous in vitro studies on NPCs yielded good results. It is necessary to think about whether all concentrations of exosomal circRNA_0000253 used in these experiments are relevant to the conditions in vivo. Electron microscopy revealed that the exosomes of normal and degenerative rat NPCs showed typically rounded shapes, and the degenerative NPCs secreted more exosomes than the normal NPCs (Figure 6A). WB analysis confirmed the presence of the exosomal markers CD9, CD63, and CD81 in exosomes derived from normal and degenerative rat NPCs (Figure 6B). The particle sizes and numbers of exosomes showed that no significant differences were detected between exosomes derived from degenerative and normal NPCs, as determined by a ZetaView nanoparticle tracker (Figure 6C). To confirm further that exosomal circRNA_0000253 could regulate IDD, PCR was used to compare the expression of circRNA_0000253 and miRNA-141-5p between normal and degenerative rat exosomes, and the results revealed that circRNA_0000253 had higher expression in degenerative rat exosomes than in normal rat exosomes, and miRNA-141-5p had lower expression in degenerative rat exosomes than in normal rat exosomes (Figure 6D). In this study, adeno-associated virus 2 (AAV-2), carrying a gene-interference sequence, was synthesized and injected into selected rat discs. The rats were randomly divided into the following 4 groups: NC group (normal saline), AAV-2-empty vector group, AAV-2-circRNA_0000253-siRNA group, and AAV-2-circRNA_0000253-siRNA + AAV-2-miRNA-141-5p-inhibitor group. MRI and X-ray were used to evaluate the degenerative disc grade in the rats. Based on a modified Thompson classification, AAV-2-circRNA_0000253-siRNA could significantly alleviate the degree of IDD when compared with the NC and empty vector groups, and the AAV-2-circRNA_0000253-siRNA + AAV-2-miRNA-141-5p-inhibitor group could aggravate the degree of IDD when compared with the AAV-2-circRNA_0000253-siRNA group, whereas no significant differences in the IDD degree were detected between the NC and empty vector groups (Figure 6E). AAV-2-circRNA_0000253-siRNA had a higher %DHI compared with the NC and empty vector groups, and the AAV-2-circRNA_0000253-siRNA + AAV-2-miRNA-141-5p-inhibitor group had a lower %DHI compared with the AAV-2-circRNA_0000253-siRNA group, whereas no significant differences in the %DHI were detected between the NC and empty vector groups (Figure 6F). WB showed that the AAV-2-circRNA_0000253-siRNA group could decrease caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, and ADAMTS 5 and increase collagen II, aggrecan, and SIRT1 compared with the NC and empty vector groups, whereas the AAV-2-circRNA_0000253-siRNA + AAV-2-miRNA-141-5p-inhibitor group could increase caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, and ADAMTS 5 and decrease collagen II, aggrecan, and SIRT1 compared with the AAV-2-circRNA_0000253-siRNA group (Figure 6G). H&E (Figure 6H) and SafraninO-Fast Green (Figure 6I) staining showed similar results to MRI. The TUNEL assay results demonstrated that AAV-2-circRNA_0000253-siRNA could significantly reduce apoptosis of NPCs compared with the NC and empty vector groups, and the AAV-2-circRNA_0000253-siRNA + AAV-2-miRNA-141-5p-inhibitor could increase apoptosis of NPCs when compared with the AAV-2-circRNA_0000253-siRNA group, whereas no significant differences in NPC apoptosis were detected between the NC and empty vector groups (Figure 6J).
Figure 6.
circRNA_0000253 Promotes Rat IDD by Competitively Adsorbing miRNA-141-5p
(A) Electron microscopy showed the shape of exosomes isolated from normal and degenerative rat NPCs. Scale bar, 50 nm. (B) WB showed the expression of marker proteins in exosomes isolated from normal and degenerative rat NPCs. (C) The particle sizes and number of exosomes were determined by a ZetaView nanoparticle tracker. (D) PCR showed the expression of circRNA_0000253 and miRNA-141-5p in normal and degenerative rat NPC exosomes. (E) MRI compared rat disc degeneration among the NC, empty vector, AAV-2-circRNA_0000253-siRNA, and AAV-2-circRNA_0000253-siRNA + AAV-2-miRNA-141-5p-inhibitor groups. (F) X-ray showing the %DHI among the NC, empty vector, circRNA_0000253 siRNA, and circRNA_0000253 siRNA + antagomiRNA-141-5p groups. (G) WB shows the caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, ADAMTS 5, collagen II, aggrecan, and SIRT1 expression among the NC, empty vector, circRNA_0000253 siRNA, and circRNA_0000253 siRNA + antagomiRNA-141-5p groups. (H and I) H&E (H) and SafraninO-Fast Green (I) staining compared the rat NP among the NC, empty vector, AAV-2-circRNA_0000253-siRNA, and AAV-2-circRNA_0000253-siRNA + AAV-2-miRNA-141-5p-inhibitor groups. Scale bar, 500 μm. (J) TUNEL staining showed apoptosis of the NP in the NC, empty vector, circRNA_0000253 siRNA, and circRNA_0000253 siRNA + antagomiRNA-141-5p groups. Scale bar, 50 μm.
Discussion
Exosomes are known as transporters that deliver cargoes from parental cells to recipient cells to regulate pathophysiological processes, including immune responses, inflammation, tumor growth, and infection in a paracrine manner, and healthy subjects and patients with different diseases release exosomes with different nucleic acid and protein contents into the circulation.20,21 Exosomes have been demonstrated to play important roles in regulating IDD, and accumulating studies have confirmed that MSC transplantation could regulate IDD progression through paracrine effects. Exosomes are a critical bioactive component of MSC secretion and may serve as an alternative to MSC-based therapy.22 However, the regulation of IDD by NPC autocrine exosomes remains unknown. In this study, exosomes were found to be abundant in the medium of NPCs, and NPCs of different degenerative grades secreted different amounts of exosomes, with NPCs of high-degenerative grade secreting more exosomes than normal NPCs. The results confirmed that exosomes were differentially expressed in NPCs and may regulate or provide biological markers for IDD. Further studies were carried out to explore the effect of exosomes on IDD, and the results showed that exosomes secreted by NPCs with a higher degeneration level could promote apoptosis and inhibit the proliferation of NPCs compared with those secreted by NPCs with a low-degeneration level or normal NPCs. It has been confirmed that exosomes can regulate IDD by releasing nucleic acids or proteins. Thus, it is important to clarify the molecules contained in NPC exosomes and their regulation of IDD as well as the potential mechanism.
Studies regarding the molecular drivers of IDD have been performed over the past few years, and circRNAs have been regarded as key players in the pathogenesis of IDD.3,23 circRNAs regulate IDD by acting as post-transcriptional regulators, and they can interact with miRNAs as miRNA sponges and ceRNAs in the NPC cytoplasm.24,25 Our previous research results show that circRNAs are abundant in IDD NPCs and play important roles in regulating IDD. Further studies were carried out to explore the role of exosomal circRNAs in regulating IDD. The 10 most upregulated circRNAs were selected for exosome PCR verification, and circRNA_0000253 was confirmed to be the most upregulated. ceRNA and PCR results showed that circRNA_0000253 may competitively absorb miRNA-141-5p to regulate IDD.
SIRT1 is an Nicotinamide adenine dinucleotide (NAD)+-dependent deacetylase that can reduce apoptosis in several different cells. SIRT1 has also been proven to play a protective role in the survival of degenerative human NPCs26, 27, 28. The underlying mechanism regarding SIRT1 regulation of IDD remains controversial. A study showed that SIRT1 exerts anti-inflammatory effects against interleukin (IL)-1β-mediated degeneration of NPCs through the Toll-like receptor 2 (TLR2)/SIRT1/nuclear factor κB (NF-κB) pathway,29 whereas another study showed that SIRT1 ameliorates oxidative stress-induced senescence of rat NPs regulated by the Akt/FoxO1 pathway.26 miRNAs are highly conserved molecules that can post-transcriptionally regulate protein expression levels. Aberrant miRNA expression has been detected in various musculoskeletal disorders, including osteoporosis, osteoarthritis, rheumatoid arthritis, and IDD.30,31 A large number of miRNAs are differentially expressed in IDD tissues and cells. Among these, some of the miRNAs have been shown to be involved in multiple pathological processes during IDD progression, including apoptosis, ECM degradation, cell proliferation, and inflammatory response.32 In this study, we confirmed that circRNA_0000253 could downregulate SIRT1 to promote IDD, and a previous study showed that miRNA-141 could directly target SIRT1 to promote IDD progression.33 Since miRNA-141-3p is the commonly detected strand (miRNA-141), whereas miRNA-141-5p∗ is the rare strand, we speculated that miRNA-141-5p could inhibit IDD by downregulating miRNA-141-3p (miRNA-141) in IDD. As described above, we demonstrated that circRNA_0000253 regulates IDD by sponging miRNA-141-5p. Competitive adsorption of miRNA by circRNA to regulate IDD was also confirmed by our previous study.9 More studies are needed to confirm the direct interaction between circRNA_0000253 and miRNA-141-5p. PCR showed that circRNA_0000253 could inhibit miRNA-141-5p expression in NPCs. The dual-luciferase reporter assay confirmed that circRNA_0000253 could directly act on miRNA-141-5p. The FISH results showed that circRNA_0000253 and miRNA-141-5p were expressed in both the nucleus and cytoplasm of NPCs. Experiments on NCP proliferation and apoptosis showed that circRNA_0000253 could promote apoptosis and inhibit proliferation of NPCs by competitive adsorption of miRNA-141-5p.
The AAV vector is well suited for gene transfer in vivo due to its low immunogenicity and high safety.34,35 The observed rapid and prolonged upregulation of protein expression and increased matrix synthesis indicated that AAV-mediated therapeutic gene transfer could be a promising treatment for IDD in vivo.34 Another study showed that AAV-mediated BMP-7 and SOX9 cotransfection in vitro can promote the synthesis of collagen II in NPCs in a human-degenerative intervertebral disc.36 In this study, a degenerative spine model in rats was generated by acupuncture of the intervertebral disc, and AAV-2 carrying specific genes was injected into rat intervertebral disc tissue. The rats were randomly divided into the following four groups: NC (normal saline), AAV-2-empty vector, AAV-2-circRNA_0000253 siRNA, and AAV-2- circRNA_0000253 siRNA + AAV-2-miRNA-141-5p-inhibitor. The results showed that circRNA_0000253 could promote rat IDD by competitively inhibiting miRNA-141-5p and downregulating SIRT1. Therefore, the role of circRNA_0000253 in regulating IDD was confirmed in vitro and in vivo.
Materials and Methods
Patient Samples and circRNA Microarray Analysis
NP specimens for microarray analysis were obtained from 8 patients: 4 NP specimens were obtained from Hirayama disease (HD) patients (3 males and 1 female; age 17.3 ± 4.1 years; Pfirrmann grade, 1),37 whereas another 4 NP specimens were obtained from patients with cervical spondylosis undergoing discectomy (3 males and 1 female; age 62.1 ± 15.6 years; average Pfirrmann grade, 3.4).9 The study protocol was approved by the Huashan Hospital Ethics Committee Affiliated with Fudan University. Informed consent was obtained from each donor. The detailed processes of the microarray analysis are described in our previous study.9 The NP specimens collected for microarray analysis were obtained from the patients enrolled in our previously published study.9
Isolation and Identification of Exosomes
NP tissues were harvested from the intervertebral disc and then incubated with 0.25 mg/mL collagenase II for 4 h at 37°C (Invitrogen, Carlsbad, CA, USA). The NP tissues for NPC isolation were from the patients enrolled in the present study. We filtered the digested tissue with 70 μm pore-size mesh and cultured the NPCs in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA), supplemented with 10% fetal bovine serum (FBS; Invitrogen), 1% penicillin-streptomycin (Sigma, USA), 2 mM glutamine (Sigma, USA), and 50 μg/mL L-ascorbic acid (Sigma) in a humidified 37°C incubator with 5% CO2. Second-passage NPCs were cultured in a monolayer for further experiments, including exosome isolation.
To extract exosomes from NPCs, the cell culture medium was replaced with medium containing 10% FBS free of exosomes (d-FBS; FBS was centrifuged at 100,000 × g at 4°C for 16 h to remove the exosomes, overnight), and 10 mL of culture medium was collected after centrifugation at 3,000 × g for 15 min. Exosomes were extracted from the NPC culture medium using a Total Exosome Isolation ExoQuick PLUS Exosome Purification Kit (System Biosciences [SBI], USA), according to the manufacturer’s protocol.38,39 The exosomes were purified and then stored in 50–100 μL of particle-free PBS (pH 7.4) at −80°C. Total RNA was extracted from the exosomes by using TRIzol reagent (Life Technologies, USA), followed by RT-PCR as described below. Exosomes were further analyzed by electron microscopy to verify their presence, by a nanoparticle characterization system to measure their size and concentration, and by WB to detect marker proteins (CD9, CD63, and CD81).
Quantitative Real-Time PCR
Total RNA was extracted from the exosomes using TRIzol reagent (Life Technologies, USA). The expression level of upregulated genes was assessed by quantitative real-time PCR according to the manufacturer’s guidelines. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize circRNA and mRNA expression levels, and U6 was used to normalize miRNA expression levels. The primers used for PCR are provided in Table S1.
ceRNA Analysis
The circRNA with the highest upregulation was selected based on exosome real-time PCR. ceRNA analysis was carried out. A circRNA-miRNA network was constructed to predict the possible interactions between the selected circRNA and sponged miRNAs by using Arraystar software, which was developed based on TargetScan and miRanda.
Luciferase Reporter Assay
The 3′ UTR of circRNA_0000253 was inserted into the luciferase vector. NPCs were cotransfected with the vector and miRNA-141-5p. The specific primers are provided in Table S1. miRNA-141-5p or NC oligonucleotides were cotransfected with the pmiR-RB-Report vector, with or without the 3′ UTR sequence of circRNA_0000253, using Opti-MEM (Invitrogen) and Lipofectamine 3000. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) after cotransfection. A histogram was constructed to visualize the experimental results.
Northern Blot and Pull-Down Assay
Northern blotting was carried out based on the manufacturer’s protocol.3 The blot was visualized using the ChemiDoc XRS system. A digoxin-labeled 18S or U6 probe was used as a control. For the pull-down assay, a biotinylated DNA probe complementary to circRNA_0000253 was synthesized and dissolved in 500 μL of lysis buffer. The probes were incubated with streptavidin-coated magnetic beads at 25°C for 3 h. The cell lysates were incubated with probe-coated beads. The RNA mixture was eluted and extracted for northern blot analysis. The probe sequences are provided in Table S2.
FISH
The FISH assay was performed using RNA samples and SIRT1 FISH Probe and Kit (Roche, Indianapolis, IN, USA), according to the manufacturer’s guidelines. NPCs were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. After prehybridization for 30 min, NPCs were hybridized with specific probes at 37°C overnight (Fluorescent In Situ Hybridization Kit; RiboBio). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). RNA and the SIRT1 gene appeared as red signals, and DAPI appeared as a blue signal. Fluorescence signals were captured by an OLYMPUS laser confocal microscope FV1000 (Olympus, Tokyo, Japan). The probe sequences are provided in Table S3.
Synthesis of siRNAs and Cell Transfection
siRNAs were synthesized that targeted the back-splice junction of circRNA_0000253 (circRNA_0000253 siRNA, circRNA_0000253 siRNA) and miRNA-141-5p (antagomiRNA-141-5p) (GenePharma, Shanghai). NPCs were transfected with circRNA_0000253 siRNA, antagomiRNA-141-5p, and NC sequences using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. The synthesized gene primers are described in Table S4.
Western Blotting
WB was carried out based on standard methods. The proteins were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and then transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked in 5% milk for 30 min and incubated with primary antibody (dilution 1:200; anti-GAPDH dilution 1:1,000; Santa Cruz Biotechnology) for 12 h. The blots were rinsed several times with Tris-buffered saline containing 0.05% Tween-20 (TBST). The PVDF membranes were then incubated for 2 h with a secondary goat anti-rabbit antibody (1:1,000; Santa Cruz Biotechnology).
NPC Proliferation Assay
Flow cytometry and CCK-8 analyses were carried out to detect NPC proliferation. For the flow cytometry assay, NPCs were plated into 6-well plates at a density of 1 × 105 cells per well. NPC apoptosis rates were assessed by flow cytometry using the Annexin V/propidium iodide (PI) apoptosis detection kit (BD Biosciences, NJ, USA). NPCs were washed twice with PBS, resuspended in binding buffer, and incubated with 5 μL fluorescein isothiocyanate (FITC)-Annexin V and 5 μL PI for 15 min at room temperature (RT). NPC staining was then observed and analyzed with a FACScan system (Becton Dickinson, CA, USA).
For the CCK-8 analysis, NPCs were inoculated into 96-well plates at 104 cells per well and incubated overnight at 37°C in 5% CO2 incubators. Then, 20 μL CCK-8 (Dojindo Molecular Technologies, Kyushu, Japan) was added to each plate, and the NPCs were cultured in a humidified 37°C incubator with 5% CO2 for 4 h. Absorbance was determined using a microplate reader (Bio-Rad, Hercules, CA, USA) at 450 nm.
Sprague-Dawley (SD) Rat In Vivo Studies
SD rats were purchased from Shanghai Slake Laboratory Animal (Shanghai, China) and housed at the Animal Laboratory Centre, Basic Medical College, Fudan University (8 weeks old, 290–310 g). All enrolled SD rats were male. All animal procedures were approved by the Animal Care and Use Committee of Fudan University and were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Exosomes from Different Degenerative SD Rats Were Injected into SD Rat Discs
The SD rats were anaesthetized with pentobarbital by intraperitoneal injection (35 mg/kg). A rat-tail model of degenerative vertebrae was established by acupuncture of selected intervertebral discs using a 24G syringe needle. The NP of rats was separated from degenerative and normal rat discs and cultured in medium. The rats were randomly divided into 3 groups and injected with saline, normal rat NPC medium, or degenerative rat NPC medium (1 mL). MR images and X-rays were taken 4 weeks after the injection. The MR images were obtained using a Siemens Trio Tim 3.0T MR scanner (Siemens Medical Solutions, Erlangen, Germany) with a quadrature extremity coil receiver at the scheduled time. The degree of disc degeneration was evaluated by MR based on a modified Thompson classification40 and by X-ray (Philips, Netherlands) based on the disc height. The NP tissues were collected for H&E, SafraninO-Fast Green staining, and TUNEL assays, and the stained slides were viewed under a fluorescence microscope. The staining procedure was carried out based on standard methods.41 WB was carried out to detect the expression of caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, ADAMTS 5, collagen II, aggrecan, and SIRT1 in NP tissues.
Isolation and Identification of SD Rat Exosomes
The SD rats were anaesthetized with pentobarbital by intraperitoneal injection (35 mg/kg). A rat-tail model of degenerative vertebrae was established by acupuncture of selected intervertebral discs, and the intervertebral disc NP of rats was separated from degenerative and normal rat discs and cultured in medium. Exosomes were extracted from the NPC culture medium, according to the manufacturer’s protocol. Exosomes were further analyzed by electron microscopy to verify their presence, by a nanoparticle characterization system to measure their size and concentration, and by WB to detect marker proteins (CD9, CD63, and CD81). PCR was also carried out to explore the expression of circRNA_0000253 and miRNA-141-5p between normal and degenerative rat NPC exosomes.
Functional Verification of circRNA_0000253 In Vivo
The rats were randomly divided into 4 groups. A rat-tail model of degenerative vertebrae was also established by acupuncture of selected intervertebral discs (Co5/6, Co6/7, and Co7/8). AAV-2 was synthesized by Shanghai GeneChem (Shanghai, China). 4 weeks after the modeling, the 4 groups of rats were injected with normal saline, AAV-2-empty vector, AAV-2-circRNA_0000253-siRNA, or AAV-2-circRNA_0000253-siRNA + AAV-2-miRNA-141-5p-inhibitor (1.5 mL, 3.65E+12 vector genomes [v.g.]/mL). The synthesized targeting sequence is described in Table S5. SD rats were anaesthetized with 1% pentobarbital sodium at a dose of 45 mg/kg. MR images and X-rays were taken 4 weeks after the AAV-2 injection. The MR images were obtained using a Siemens Trio Tim 3.0T MR scanner (Siemens Medical Solutions, Erlangen, Germany) with a quadrature extremity coil receiver at the scheduled time. The degree of disc degeneration was evaluated by MR based on a modified Thompson classification40 and by X-ray (Philips, Netherlands) based on the disc height. The NP tissues were collected for H&E staining and TUNEL assays, and the stained slides were viewed under a fluorescence microscope. The staining procedure was carried out based on standard methods41. WB was carried out to detect the expression of caspase-3, caspase-7, caspase-9, MMP-3, MMP-13, ADAMTS4, ADAMTS 5, collagen II, aggrecan, and SIRT1 in NP tissues. The SD rats were sacrificed by cervical spine dislocation after the experiment.
Statistical Analyses
SPSS 21.0 was used for statistical analyses. Data are described as the mean ± SD. Student’s t test was used to compare two groups, and one-way analysis of variance (ANOVA), followed by Dunnett’s post hoc test, was used to compare more than two groups. Differences were considered statistically significant when p <0.05.
Author Contributions
J.S. and J.-Y.J. designed the experiments. J.S., K.-H.S., H.-L.W., G.-Y.X., S.X., and F.Z. performed the experiments and acquired the data. J.S., H.-L.W., Z.-H.C., and F.Z. analyzed the data. J.-Y.J., X.-S. M.,H.-L.W., and C.-J.Z. supervised the project and wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
Government funds were received in support of this work from the National Natural Science Foundation of China,China (81972109 and 81972093). The work was also sponsored by Shanghai Sailing Program, Shang Hai, China (20YF1429900 and 20YF1438100).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.07.039.
Contributor Information
Fei Zou, Email: zillion-faculty@126.com.
Xiao-Sheng Ma, Email: mxs893@sina.com.
Hong-Li Wang, Email: wanghongli0212@163.com.
Jian-Yuan Jiang, Email: jianyuanjiang@sina.com.
Supplemental Information
References
- 1.Nouri A., Tetreault L., Singh A., Karadimas S.K., Fehlings M.G. Degenerative Cervical Myelopathy: Epidemiology, Genetics, and Pathogenesis. Spine. 2015;40:E675–E693. doi: 10.1097/BRS.0000000000000913. [DOI] [PubMed] [Google Scholar]
- 2.Gille O., Bouloussa H., Mazas S., Vergari C., Challier V., Vital J.M., Coudert P., Ghailane S. A new classification system for degenerative spondylolisthesis of the lumbar spine. Eur. Spine J. 2017;26:3096–3105. doi: 10.1007/s00586-017-5275-4. [DOI] [PubMed] [Google Scholar]
- 3.Cheng X., Zhang L., Zhang K., Zhang G., Hu Y., Sun X., Zhao C., Li H., Li Y.M., Zhao J. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann. Rheum. Dis. 2018;77:770–779. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li B., Zheng X.F., Ni B.B., Yang Y.H., Jiang S.D., Lu H., Jiang L.S. Reduced expression of insulin-like growth factor 1 receptor leads to accelerated intervertebral disc degeneration in mice. Int. J. Immunopathol. Pharmacol. 2013;26:337–347. doi: 10.1177/039463201302600207. [DOI] [PubMed] [Google Scholar]
- 5.Wang K., Chen T., Ying X., Zhang Z., Shao Z., Lin J., Xu T., Chen Y., Wang X., Chen J., Sheng S. Ligustilide alleviated IL-1β induced apoptosis and extracellular matrix degradation of nucleus pulposus cells and attenuates intervertebral disc degeneration in vivo. Int. Immunopharmacol. 2019;69:398–407. doi: 10.1016/j.intimp.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Johnson Z.I., Schoepflin Z.R., Choi H., Shapiro I.M., Risbud M.V. Disc in flames: Roles of TNF-alpha and IL-1beta in intervertebral disc degeneration. Eur. Cell. Mater. 2015;30:104–116. doi: 10.22203/ecm.v030a08. discussion 116–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Yu X., Shen J., Chan M.T., Wu W.K. MicroRNA in intervertebral disc degeneration. Cell Prolif. 2015;48:278–283. doi: 10.1111/cpr.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W.-K., Yu X.-H., Yang W., Wang C., He W.-S., Yan Y.-G., Zhang J., Wang W.-J. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50:e12313. doi: 10.1111/cpr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song J., Wang H.L., Song K.H., Ding Z.W., Wang H.L., Ma X.S., Lu F.Z., Xia X.L., Wang Y.W., Fei-Zou, Jiang J.Y. CircularRNA_104670 plays a critical role in intervertebral disc degeneration by functioning as a ceRNA. Exp. Mol. Med. 2018;50:94. doi: 10.1038/s12276-018-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belousova E.A., Filipenko M.L., Kushlinskii N.E. Circular RNA: New Regulatory Molecules. Bull. Exp. Biol. Med. 2018;164:803–815. doi: 10.1007/s10517-018-4084-z. [DOI] [PubMed] [Google Scholar]
- 11.Li X., Yang L., Chen L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 12.Wang S., Sun J., Yang H., Zou W., Zheng B., Chen Y., Guo Y., Shi J. Profiling and bioinformatics analysis of differentially expressed circular RNAs in human intervertebral disc degeneration. Acta Biochim. Biophys. Sin. (Shanghai) 2019;51:571–579. doi: 10.1093/abbs/gmz036. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., He P., Pan H., Long J., Wang J., Li Z., Liu H., Jiang W., Zheng Z. Circular RNA circ-4099 is induced by TNF-α and regulates ECM synthesis by blocking miR-616-5p inhibition of Sox9 in intervertebral disc degeneration. Exp. Mol. Med. 2018;50:27. doi: 10.1038/s12276-018-0056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mousavi S., Moallem R., Hassanian S.M., Sadeghzade M., Mardani R., Ferns G.A., Khazaei M., Avan A. Tumor-derived exosomes: Potential biomarkers and therapeutic target in the treatment of colorectal cancer. J. Cell. Physiol. 2019;234:12422–12432. doi: 10.1002/jcp.28080. [DOI] [PubMed] [Google Scholar]
- 15.Hon K.W., Abu N., Ab Mutalib N.S., Jamal R. Exosomes As Potential Biomarkers and Targeted Therapy in Colorectal Cancer: A Mini-Review. Front. Pharmacol. 2017;8:583. doi: 10.3389/fphar.2017.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skotland T., Sandvig K., Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H., Deng T., Ge S., Liu Y., Bai M., Zhu K., Fan Q., Li J., Ning T., Tian F. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Liao Z., Luo R., Li G., Song Y., Zhan S., Zhao K., Hua W., Zhang Y., Wu X., Yang C. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics. 2019;9:4084–4100. doi: 10.7150/thno.33638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y.T., Huang Y.Y., Zheng L., Qin S.H., Xu X.P., An T.X., Xu Y., Wu Y.S., Hu X.M., Ping B.H., Wang Q. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int. J. Mol. Med. 2017;40:834–844. doi: 10.3892/ijmm.2017.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barile L., Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 21.György B., Hung M.E., Breakefield X.O., Leonard J.N. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu. Rev. Pharmacol. Toxicol. 2015;55:439–464. doi: 10.1146/annurev-pharmtox-010814-124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung V.Y., Aladin D.M., Lv F., Tam V., Sun Y., Lau R.Y., Hung S.C., Ngan A.H., Tang B., Lim C.T. Mesenchymal stem cells reduce intervertebral disc fibrosis and facilitate repair. Stem Cells. 2014;32:2164–2177. doi: 10.1002/stem.1717. [DOI] [PubMed] [Google Scholar]
- 23.Guo W., Zhang B., Mu K., Feng S.Q., Dong Z.Y., Ning G.Z., Li H.R., Liu S., Zhao L., Li Y. Circular RNA GRB10 as a competitive endogenous RNA regulating nucleus pulposus cells death in degenerative intervertebral disk. Cell Death Dis. 2018;9:319. doi: 10.1038/s41419-017-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 26.He J., Zhang A., Song Z., Guo S., Chen Y., Liu Z., Zhang J., Xu X., Liu J., Chu L. The resistant effect of SIRT1 in oxidative stress-induced senescence of rat nucleus pulposus cell is regulated by Akt-FoxO1 pathway. Biosci. Rep. 2019;39 doi: 10.1042/BSR20190112. BSR20190112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Lin J., Tian N., Wu Y., Zhou Y., Wang C., Wang Q., Jin H., Chen T., Nisar M. Melatonin protects vertebral endplate chondrocytes against apoptosis and calcification via the Sirt1-autophagy pathway. J. Cell. Mol. Med. 2019;23:177–193. doi: 10.1111/jcmm.13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J., Shao M., Lu F., Jiang J., Xia X. Role of Sirt1 Plays in Nucleus Pulposus Cells and Intervertebral Disc Degeneration. Spine. 2017;42:E757–E766. doi: 10.1097/BRS.0000000000001954. [DOI] [PubMed] [Google Scholar]
- 29.Shen J., Fang J., Hao J., Zhong X., Wang D., Ren H., Hu Z. SIRT1 Inhibits the Catabolic Effect of IL-1β Through TLR2/SIRT1/NF-κB Pathway in Human Degenerative Nucleus Pulposus Cells. Pain Physician. 2016;19:E215–E226. [PubMed] [Google Scholar]
- 30.Zhou X., Chen L., Grad S., Alini M., Pan H., Yang D., Zhen W., Li Z., Huang S., Peng S. The roles and perspectives of microRNAs as biomarkers for intervertebral disc degeneration. J. Tissue Eng. Regen. Med. 2017;11:3481–3487. doi: 10.1002/term.2261. [DOI] [PubMed] [Google Scholar]
- 31.Wang C., Wang W., Yang W., Yu X., Yan Y., Zhang J., Jiang Z. [MicroRNAs: a type of novel regulative factor for intervertebral disc degeneration] Zhejiang Da Xue Xue Bao Yi Xue Ban. 2016;45:170–178. doi: 10.3785/j.issn.1008-9292.2016.03.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C., Wang W.J., Yan Y.G., Xiang Y.X., Zhang J., Tang Z.H., Jiang Z.S. MicroRNAs: New players in intervertebral disc degeneration. Clin. Chim. Acta. 2015;450:333–341. doi: 10.1016/j.cca.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 33.Ji M.L., Jiang H., Zhang X.J., Shi P.L., Li C., Wu H., Wu X.T., Wang Y.T., Wang C., Lu J. Preclinical development of a microRNA-based therapy for intervertebral disc degeneration. Nat. Commun. 2018;9:5051. doi: 10.1038/s41467-018-07360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H.F., Ning B., Zhang H., Wang D.C., Hu Y.L., Qiao G.X., Zhao Y.P., Hu Y.G. Effect of rAAV2-hTGFβ1 Gene Transfer on Matrix Synthesis in an In Vivo Rabbit Disk Degeneration Model. Clin. Spine Surg. 2016;29:E127–E134. doi: 10.1097/BSD.0b013e3182a26553. [DOI] [PubMed] [Google Scholar]
- 35.Mern D.S., Thomé C. Identification and characterization of human nucleus pulposus cell specific serotypes of adeno-associated virus for gene therapeutic approaches of intervertebral disc disorders. BMC Musculoskelet. Disord. 2015;16:341. doi: 10.1186/s12891-015-0799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren X.F., Diao Z.Z., Xi Y.M., Qi Z.H., Ren S., Liu Y.J., Yang D.L., Zhang X., Yuan S.L. Adeno-associated virus-mediated BMP-7 and SOX9 in vitro co-transfection of human degenerative intervertebral disc cells. Genet. Mol. Res. 2015;14:3736–3744. doi: 10.4238/2015.April.22.1. [DOI] [PubMed] [Google Scholar]
- 37.Urrutia J., Besa P., Campos M., Cikutovic P., Cabezon M., Molina M., Cruz J.P. The Pfirrmann classification of lumbar intervertebral disc degeneration: an independent inter- and intra-observer agreement assessment. Eur. Spine J. 2016;25:2728–2733. doi: 10.1007/s00586-016-4438-z. [DOI] [PubMed] [Google Scholar]
- 38.Li Z., Hu C., Jia J., Xia Y., Xie H., Shen M., Huang R., He L., Liu C., Wang S. Establishment and Evaluation of a Simple Size-Selective Method for Exosome Enrichment and Purification. J. Biomed. Nanotechnol. 2019;15:1090–1096. doi: 10.1166/jbn.2019.2768. [DOI] [PubMed] [Google Scholar]
- 39.Felicetti F., De Feo A., Coscia C., Puglisi R., Pedini F., Pasquini L., Bellenghi M., Errico M.C., Pagani E., Carè A. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J. Transl. Med. 2016;14:56. doi: 10.1186/s12967-016-0811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masuda K., Aota Y., Muehleman C., Imai Y., Okuma M., Thonar E.J., Andersson G.B., An H.S. A novel rabbit model of mild, reproducible disc degeneration by an anulus needle puncture: correlation between the degree of disc injury and radiological and histological appearances of disc degeneration. Spine. 2005;30:5–14. doi: 10.1097/01.brs.0000148152.04401.20. [DOI] [PubMed] [Google Scholar]
- 41.Chen J., Lin Z., Deng K., Shao B., Yang D. Tension induces intervertebral disc degeneration via endoplasmic reticulum stress-mediated autophagy. Biosci. Rep. 2019;39 doi: 10.1042/BSR20190578. BSR20190578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.