Erasing Rubisco small subunit nuclear synthesis in tobacco provides a new plant chassis for investigating novel Rubisco complexes in a whole plant context via chloroplast transformation of both its large and small subunits.
Abstract
Engineering improved Rubisco for the enhancement of photosynthesis is challenged by the alternate locations of the chloroplast rbcL gene and nuclear RbcS genes. Here we develop an RNAi-RbcS tobacco (Nicotiana tabacum) master-line, tobRrΔS, for producing homogenous plant Rubisco by rbcL-rbcS operon chloroplast transformation. Four genotypes encoding alternative rbcS genes and adjoining 5′-intergenic sequences revealed that Rubisco production was highest (50% of the wild type) in the lines incorporating a rbcS gene whose codon use and 5′ untranslated-region matched rbcL. Additional tobacco genotypes produced here incorporated differing potato (Solanum tuberosum) rbcL-rbcS operons that either encoded one of three mesophyll small subunits (pS1, pS2, and pS3) or the potato trichome pST-subunit. The pS3-subunit caused impairment of potato Rubisco production by ∼15% relative to the lines producing pS1, pS2, or pST. However, the βA-βB loop Asn-55-His and Lys-57-Ser substitutions in the pS3-subunit improved carboxylation rates by 13% and carboxylation efficiency (CE) by 17%, relative to potato Rubisco incorporating pS1 or pS2-subunits. Tobacco photosynthesis and growth were most impaired in lines producing potato Rubisco incorporating the pST-subunit, which reduced CE and CO2/O2 specificity 40% and 15%, respectively. Returning the rbcS gene to the plant plastome provides an effective bioengineering chassis for introduction and evaluation of novel homogeneous Rubisco complexes in a whole plant context.
INTRODUCTION
There is an escalating need to improve agricultural photosynthetic efficiency and yield to support the food requirements of a growing global population (Bailey-Serres et al., 2019; Simkin et al., 2019). An enzyme with considerable influence on crop productivity and resource use is Rubisco, the CO2-fixing enzyme in photosynthesis (Parry et al., 2012). Rubisco is considered an inefficient catalyst, due to its slow carboxylation rate. Rubisco is also competitively inhibited by O2, where 2-phosphoglycolate is produced and recycled via photorespiration, a metabolic cycle that consumes energy and releases fixed CO2 (Sharwood, 2017). To circumvent Rubisco-related losses in photosynthetic efficiency, significant efforts have been devoted to the integration of CO2-concentrating mechanisms in the chloroplasts of C3-crops to maximize their carbon fixation efficiency (Long et al., 2016). Another engineering focus has been to improve the carboxylation properties of plant Rubisco itself (Sharwood, 2017). While such endeavors have proven difficult, improving plant Rubisco function may not be an immutable challenge. We have demonstrated that more efficient Rubisco isoforms have naturally evolved in some red algae (Whitney et al., 2001; Andrews and Whitney, 2003) and can be artificially generated in the laboratory by directed evolution (Wilson et al., 2016, 2018; Zhou and Whitney, 2019).
Current evidence indicates that step improvements in plant Rubisco catalysis will require coordinated changes in both the large (L) and small (S) subunits. Transgenic modification of plant Rubisco is, however, hindered by the disparate locations of the rbcL gene in the chloroplast genome (the plastome, which encodes the ∼50-kD L-subunit) and multiple RbcS genes in the nucleus (coding for the ∼14.5-kD S-subunits). Whereas mutagenic studies have primarily focused on the L-subunits, because they house the catalytic site, Rubisco bioengineering studies in the unicellular green alga Chlamydomonas (Chlamydomonas reinhardtii; Spreitzer et al., 2005; Genkov et al., 2010), in Escherichia coli (Wilson et al., 2018; Zhou and Whitney, 2019) and in plants (Ishikawa et al., 2011; Fukayama et al., 2015; Whitney et al., 2015) have demonstrated how amino acid changes in the S-subunits can considerably influence catalysis. For example, the largest improvements to the carboxylation properties of the plant-like “green” Rubisco in Chlamydomonas required compatible sequence changes in both the L- and S-subunits (Spreitzer et al., 2005). Achieving this feat required independent transgenic events in the nucleus and the plastome of Chlamydomonas mutants either lacking full-length sequences of both nuclear RbcS genes or having no plastome rbcL (which can only survive if provided with a source of reduced carbon such as acetate). A comparable gene deletion approach in plants is challenged by their higher RbcS gene copy number and debilitated growth of progeny lacking Rubisco (Kanevski and Maliga, 1994; Whitney and Sharwood, 2008; Sharwood et al., 2016). A potential alternative approach toward engineering multiple RbcS genes in plants might be to exploit their high sequence similarity to introduce desired nucleotide changes into most, if not all, nuclear copies by gene editing (McCarty et al., 2020). Unfortunately, the incorporation of accompanying and matching L-subunit mutations would require additional gene editing of rbcL by plastome transformation, a technology only established in a limited range of plant species (Bock, 2014).
An alternative, although untested, approach to engineer Rubisco would rely on the cointroduction of rbcL and rbcS genes into the nucleus using new-generation multigene nucleus transformation capabilities (Engler et al., 2014). To date, such recombinant L-subunit synthesis via nuclear rbcL transformation has proven inadequate, producing between only 2% (Kanevski and Maliga, 1994) and 60% (Wostrikoff et al., 2012) of wild-type Rubisco content. By contrast, relocating an RbcS gene back into its pre-endosymbiotic location within the chloroplast genome might allow for its coordinated plastome bioengineering with rbcL. Although prior transplastomic rbcS integration studies have shown different efficiencies in plastid S-subunit incorporation into the L8S8 holoenzyme, they have also demonstrated the requirement for silencing of the nucleus-encoded RbcS gene to avoid the preferential incorporation of S-subunits made in the cytosol (Whitney and Andrews, 2001; Zhang et al., 2002; Dhingra et al., 2004).
A problem encountered in plastome transformation studies is the inability to predict the amount of recombinant protein accumulation in chloroplasts (Bock, 2014). This often requires several transformation trials to optimize transgene expression, especially when introducing polycistrons that encode multiple protein components (Bock, 2015). Protein production is highly influenced by chloroplast mRNA structure, stability, and capacity to engage with RNA regulatory elements and protein translation factors encoded by genes in both the chloroplast and the nucleus (Trösch et al., 2018). Key RNA regulatory components that modulate chloroplast mRNA translation are primarily located in the 5′ untranslated region (UTR). These include the Shine–Dalgarno sequence, as well as secondary stem-loop structures that determine mRNA stability and translation (Zoschke and Bock, 2018). For genes such as rbcL, the Shine–Dalgarno sequence shows strong complementarity to the anti-Shine–Dalgarno recognition sequence at the 3′ end of the 16S ribosomal RNA, possibly helping to bolster the large amount of L-subunit made in plants (Scharff et al., 2017).
New capabilities to express plant Rubisco in E. coli offer great hope to discover L- and S-subunit structural changes with potential to improve catalysis more rapidly (Aigner et al., 2017; Conlan and Whitney, 2018) especially if successfully adapted to suit synthetic laboratory evolution approaches (Aigner et al., 2017; Conlan and Whitney, 2018). However, as indicated above, identifying a genetic approach to efficiently transplant these changes into plants remains a challenge. To address this, we report on the development of a next-generation tobacco (Nicotiana tabacum) line called “tobRrΔS,” generated from blocking S-subunit production by RNA interference (RNAi) in the transplastomic cmtrL genotype (herein called “tobRr)” in which tobacco L8S8 Rubisco production has been substituted with the bacterial Rhodospirillum rubrum L2 Rubisco (Figure 1A; Whitney and Sharwood, 2008). We used tobRrΔS to produce a range of rbcL- and rbcS-cotransformed transplastomic lines. These lines were analyzed both in vitro and in vivo to assess how modifications to the L- and/or S-subunit influence tobacco and potato (Solanum tuberosum) L8S8 biogenesis, enzyme kinetics, leaf photosynthesis, and plant growth. Our findings provide novel insights into (1) the pervasive effects of the S-subunit on Rubisco catalysis, (2) the identity of amino acids governing the carboxylation properties of potato Rubisco, and (3) the critical role of 5′UTR and codon use on plastid gene translation.
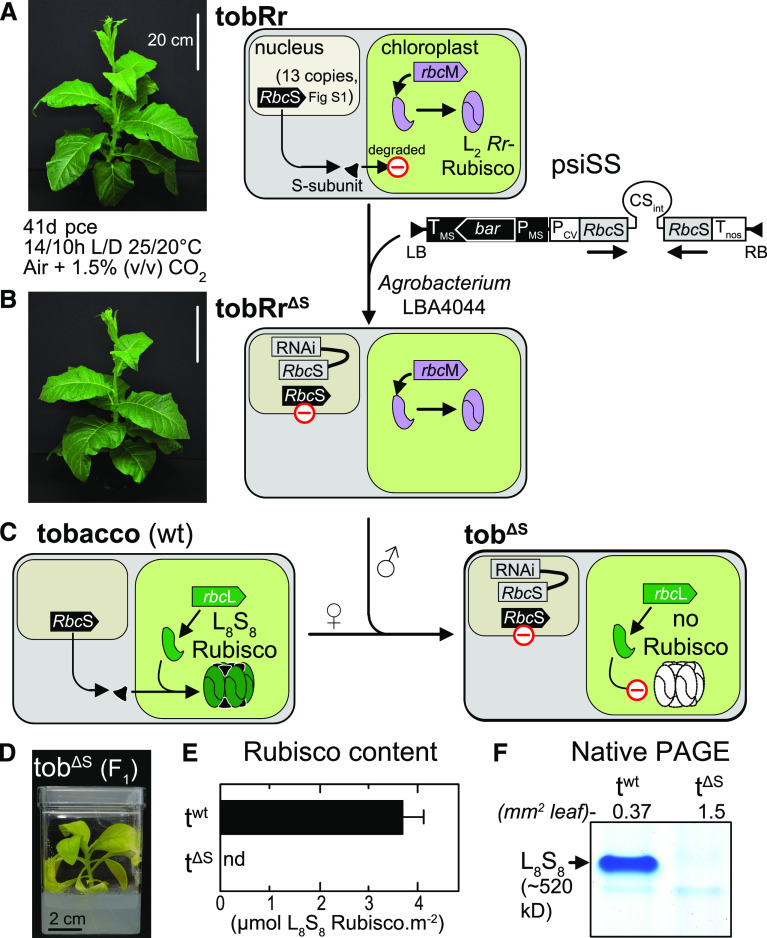
Figure 1.
RNAi Silencing of S-Subunit Production in the New tobRrΔS Genotype.
(A) Schematic summary showing how the RNAi-rbcS silencing plasmid psiSS (Supplemental Figure 1A; Wostrikoff and Stern, 2007) was introduced into tobacco genotype tobRr, producing the bacterial L2-Rubisco (Whitney and Sharwood, 2008), via Agrobacterium-mediated transformation.
(B) and (C) The resulting tobRrΔS plants produce no S-subunits. The phenotype of homozygous tobRrΔS1 plants (in [B], and see Supplemental Figure 2) matched tobRr (showing both plants 41-d post-cotyledon emergence, pce). Flowers from homozygous T2, single RNAi-RbcS insertion tobRrΔS plants identified by segregation analysis (Supplemental Figure 2) were crossed with wild-type (wt) tobacco (C) to produce heterozygous F1 tobΔS progeny.
(D) tobΔS plants can only be grown in tissue culture.
(E) and (F) 14C-CABP binding from n = 3 independent tissue-cultured tobΔS plants or glasshouse-grown wild-type tobacco (twt), shown in (E), and native PAGE from leaf area of soluble protein separated by PAGE, shown in (F), indicated that the tobΔs lines produced no L8S8 Rubisco (nd, not detected).
RESULTS
Silencing Rubisco S-Subunit Production in Tobacco
Reducing Rubisco production in plants via anti-RbcS methods produce photosynthetically impaired progeny that require elevated CO2 to grow in soil (Andrews and Whitney, 2003; Sharwood, 2017). Fully eliminating Rubisco production is more problematic, as the resulting plants are only viable in tissue culture (Whitney and Sharwood, 2008) or when grafted onto wild-type stock (Kanevski and Maliga, 1994). To circumvent these limitations, we blocked tobacco S-subunit production by RNA interference of RbcS (RNAi-RbcS) in the transplastomic tobacco genotype tobRr (previously called “cmtrL”; Whitney and Sharwood, 2008), where the tobacco rbcL gene had been replaced with a rbcM gene encoding R. rubrum L2 Rubisco, which does not require S-subunits (Figure 1A). We achieved silencing of the RbcS gene encoding the S-subunit in tobRr by Agrobacterium (Agrobacterium tumefaciens)-mediated transformation of the inverted hairpin RNAi-rbcS plasmid, psiSS (Supplemental Figure 1A). The 13 RbcS gene copies in tobacco share high sequence similarity (Supplemental Figure 1B, Supplemental Table 1; Gong et al., 2014) and are predicted to encode nine different mature S-subunit isoforms (NtS1 to NtS9) that differ by one to seven polymorphic residues (Table 1). Due to this high sequence conservation, transient psiSS expression effectively silences Rubisco production in tobacco (Wostrikoff and Stern, 2007).
Table 1. Tobacco Rubisco S-Subunit Sequence Variation.
| S-Subunit Isoform | RbcS Genea | Amino Acid Position in the Mature S-Subunit | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 8 | 23 | 26 | 30 | 31 | 48 | 70 | 88 | 96 | 121 | ||
| NtS1b | NtS1a, NtS1b | I | N | Q | S | V | E | H | W | E | Q | E |
| NtS2 | NtT1 | Y | G | E | — | I | — | R | — | G | E | — |
| NtS3 | NtS2 | — | — | R | — | D | — | — | — | — | — | |
| NtS4 | NtT2, NtS, NtT3, NtS4 | — | — | E | R | — | — | — | — | — | — | — |
| NtS5 | NtT3b | — | — | E | R | — | — | — | — | — | R | — |
| NtS6 | NtT4a | — | — | E | R | — | — | — | R | — | — | A |
| NtS7 | NtT4b | — | — | E | R | — | — | — | — | — | — | A |
| NtS8 | NtS5 | — | — | E | R | — | D | — | — | — | — | — |
| NtS9 | NtT5 | — | — | V | R | — | — | — | — | — | — | — |
Dashes indicate no data.
S-subunit sequence used in the transgenic tobacco lines reported in this study. Only amino acids that differ from NtS1 are shown. Dashes indicate identical amino acids to NtS1.
NtS:RbcS genes were maternally derived from Nicotiana sylvestris; NtT:RbcS genes were paternally derived from Nicotiana tomentosiformis. GenBank accession numbers for each RbcS gene are listed in Supplemental Table 1.
We obtained 11 independent glufosinate-resistant T0 tobRr plants transformed with the RNAi-RbcS construct. We subjected four of these plants (tobRrΔS1 to tobRrΔS4) to glufosinate resistance/sensitivity segregation analyses on their T1 and T2 progeny (Supplemental Figure 2). All four corresponding tobRrΔS lines contained single RNAi-RbcS transfer DNA insertions and produced no RbcS mRNA, as determined by RNA gel blotting (Supplemental Figure 2). Homozygous T2 progeny of the tobRrΔS1 line (subsequently called “tobRrΔS”) was used for all subsequent transformation and crossing analyses (Figure 1B). The growth and phenotypes of both the tobRrΔS and tobRr plants were indistinguishable (Figures 1A and 1B). This is consistent with the lack of the S-subunit requirement of R. rubrum L2-Rubisco.
We confirmed the efficiency of RNAi-mediated RbcS silencing in tobRrΔS by genetic crossing with wild-type tobacco (Figure 1C). The resulting heterozygous F1 tobΔS progeny were all resistant to glufosinate (encoded by the nuclear bar transgene from tobRrΔS) with no RbcS mRNA evident by RNA gel blotting (Supplemental Figure 2). The phenotype exhibited by tobΔS plants matches that seen in other tobacco genotypes making no Rubisco: highly chlorotic, and unable to survive outside of tissue-culture–growth conditions (Figure 1D; Whitney and Sharwood, 2008; Sharwood et al., 2016). We failed to detect L8S8 Rubisco in the tobΔS plants by [14C]-CABP binding (Figure 1E), native PAGE (Figure 1F), or by SDS PAGE (Supplemental Figure 2). These data are consistent with RbcS mRNA being effectively silenced and S-subunit synthesis undetectable in the tobRrΔS line and the tobΔS progeny.
Comparing Chloroplast rbcL-rbcS Operon Design on Rubisco Production
To develop a robust transplastomic strategy capable of simultaneously engineering both rbcL and rbcS, we stably introduced four differing synthetic tobacco rbcL-rbcS-aadA operons into the tobRrΔS plastome to replace the rbcM gene (Figure 2A). All four transformation plasmids were derived from pLEV4 Whitney et al., 2011 and all incorporated an rbcL-rbcS-aadA operon comprising the native tobacco rbcL promoter, 5′UTR, coding sequence, and the same rbcL 3′UTR-aadA-rbcL 3′UTR sequence (compare Figure 2B with 2C). Each operon varied only in their intergenic region (IR)-rbcS sequences. To test the effect of codon use on S-subunit translation, the synthetic rbcS gene within the rbcL-rbcS-aadA operon was either chloroplast-optimized (corbcSH) or nonoptimized (norbcSH). We altered the codon use of corbcSH to match the tobacco rbcL gene (Supplemental Figure 4). We modified the codon use of norbcSH to limit its sequence similarity to either corbcSH or the native NtS1 RbcS gene (Supplemental Figure 3B). Accordingly, the corbcSH and norbcSH genes share only 70% sequence identity with the NtS1 gene, and ∼80% with each other (Supplemental Figure 3B). Both corbcSH and norbcSH genes encoded the NtS1-subunit with a C-terminal 6× His tag that increased the size of the protein from 14.7 to 15.4 kD (Supplemental Figure 3A).
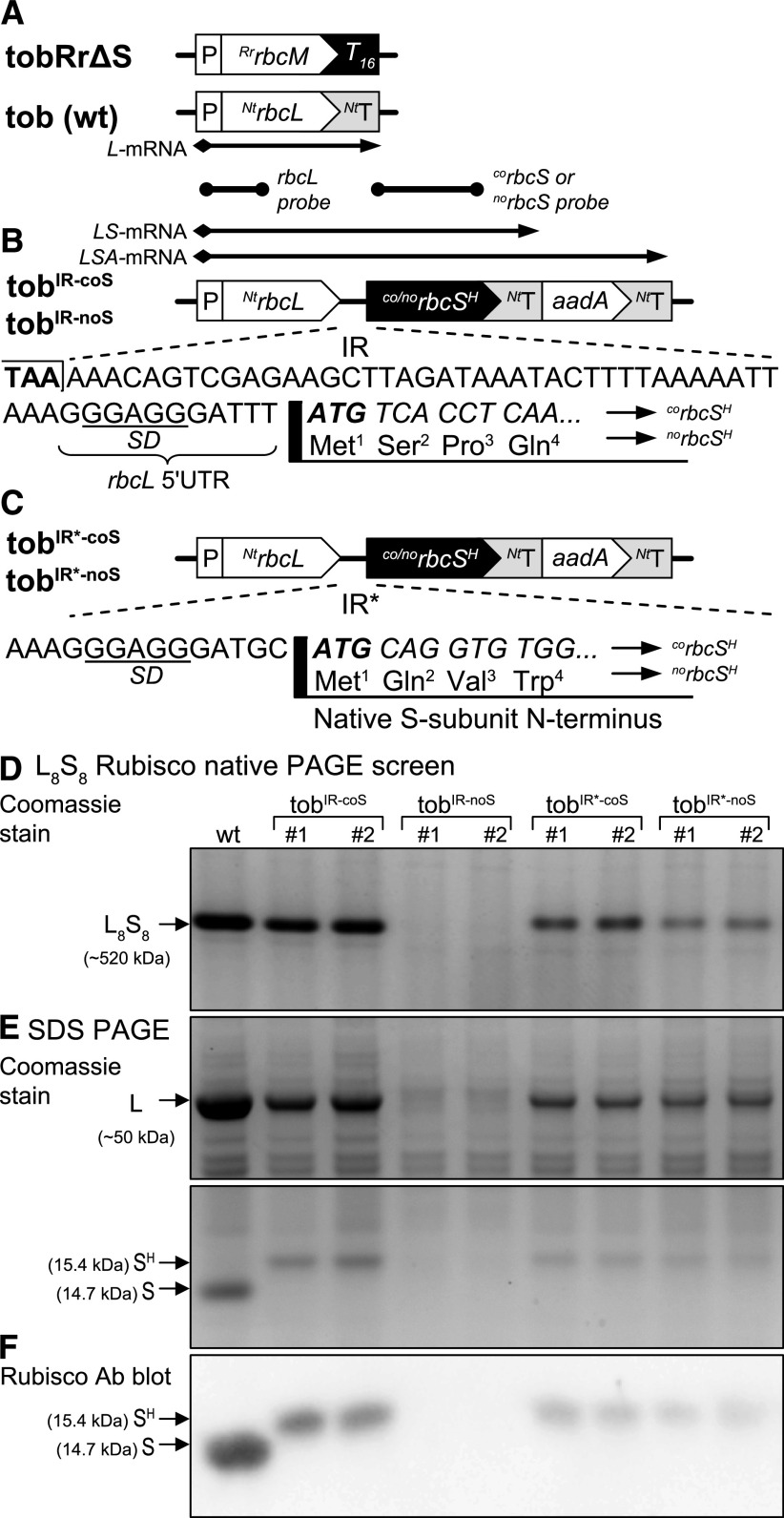
Figure 2.
Co-Engineering Plant Rubisco L- and S-Subunits into tobRrΔS Chloroplasts.
(A) Comparative plastome detail around the rbcL gene in wild-type (wt) tobacco and the rbcM gene in the tobRrΔS line that expresses R. rubrum L2 Rubisco.
(B) Corresponding plastome sequence in the tobIR-coS and tobIR-noS genotypes after transformation of the tobacco rbcL, an aadA gene (confers spectinomycin resistance) and either a corbcSH or norbcSH gene encoding a 6× His C-terminal tag (H). Shown is the sequence of the IR introduced into both the tobIR-coS and tobIR-noS genotypes whose corbcSH and norbcSH genes encode the same N termini (MSPQTWPP; see Supplemental Figure 3 for full sequence information and Supplemental Figure 4 for codon use comparison). The arrows in (A) and (B) indicate the expected mRNA transcripts produced, with the gray shading indicating which fragment will hybridize with the rbcL probe and either the corbcS or norbcS probes. NtT and T16, 3′UTR (terminator) sequences for the tobacco rbcL and rps16 genes, respectively.
(C) Comparisons in the IR and the corbcS and norbcS 5′sequences (that encodes a shorter MQVWPP N terminus that matches the native S-subunits) transformed into the tobIR*-coS and tobIR*-noS genotypes relative to that shown in (B).
(D) and (E) Native PAGE (D) and SDS PAGE (E) analysis of the soluble protein from leaf no. 5 (the youngest, near-fully expanded leaf, 13 ± 1 cm in diameter) of glasshouse-grown T0 plants (when 45 ± 3 cm in height) or tissue-culture–dependent tobIR-noS and tobIR*-noS plants. Here, numbers 1 and 2 represent samples from independently transformed lines for each genotype.
(F) Rubisco antibody immunoblots verify that the Rubisco produced in transplastomic plants only comprises chloroplast-made 15.4-kD SH-subunits and no apparent endogenous (nucleus-encoded) 14.7-kD S-subunits.
We tested the effects of translation initiation on chloroplast S-subunit synthesis by modifying the sequence between the Shine–Dalgarno and the rbcS initiator ATG codon in the IR of the rbcL-rbcS-aadA operon. In the tobIR-coS and tobIR-noS lines, the 3′ end of the 52-bp IR incorporated the native rbcL Shine–Dalgarno-5′UTR sequence (GGGAGGGATTT) and appended the L-subunit N-terminal MSPQT coding sequence onto the rbcS genes (Figure 2B). By contrast, the corresponding IR sequence in the tobIR*-coS and tobIR*-noS lines included the sequence GGGAGGGATGC, with the substituted 3′-GC nucleotides forming a SphI restriction site (and an out-of-frame AUG codon 5′ to the AUG initiator codon; Figure 2C). The S-subunit N terminus (MQVWPP) encoded in tobIR*-coS and tobIR*-noS matched the native tobacco S-subunit N terminus (Supplemental Figure 3).
We passed two independently transformed lines for each genotype through two rounds of selection on regeneration media of plants (RMOP) supplemented with 500 µg/mL of spectinomycin. We confirmed homoplasmy by native PAGE testing for total replacement of R. rubrum L2 Rubisco by L8S8 Rubisco from tobacco (Whitney and Sharwood, 2008). The native PAGE analyses indicated that each genotype produced differing levels of L8S8 Rubisco (Figure 2D). From Coomassie staining, we clearly observed that the tobIR-coS and tobIR*-coS lines expressing the corbcSH gene produced more L8S8 Rubisco than the lines expressing the norbcSH gene, highlighting the necessity of codon optimization to improve expression. Of particular note, we detected no L8S8 Rubisco in any of the tobIR-noS plants produced (Figure 2D).
We confirmed the differing levels of Rubisco production by Coomassie staining (Figure 2E) and immunoblots with an anti-tobacco Rubisco antibody (Figure 2F) after SDS PAGE. These analyses also confirmed that the Rubisco produced in each transplastomic genotype only comprised the 15.4-kD SH subunit (made in the chloroplast) and no 14.7-kD S-subunits (made in the cytosol). This observation confirmed that the native RbcS mRNA, and corresponding S-subunit synthesis, remained effectively silenced in each transplastomic line.
Differences in Leaf Rubisco Content alter Photosynthetic Potential and Plant Growth
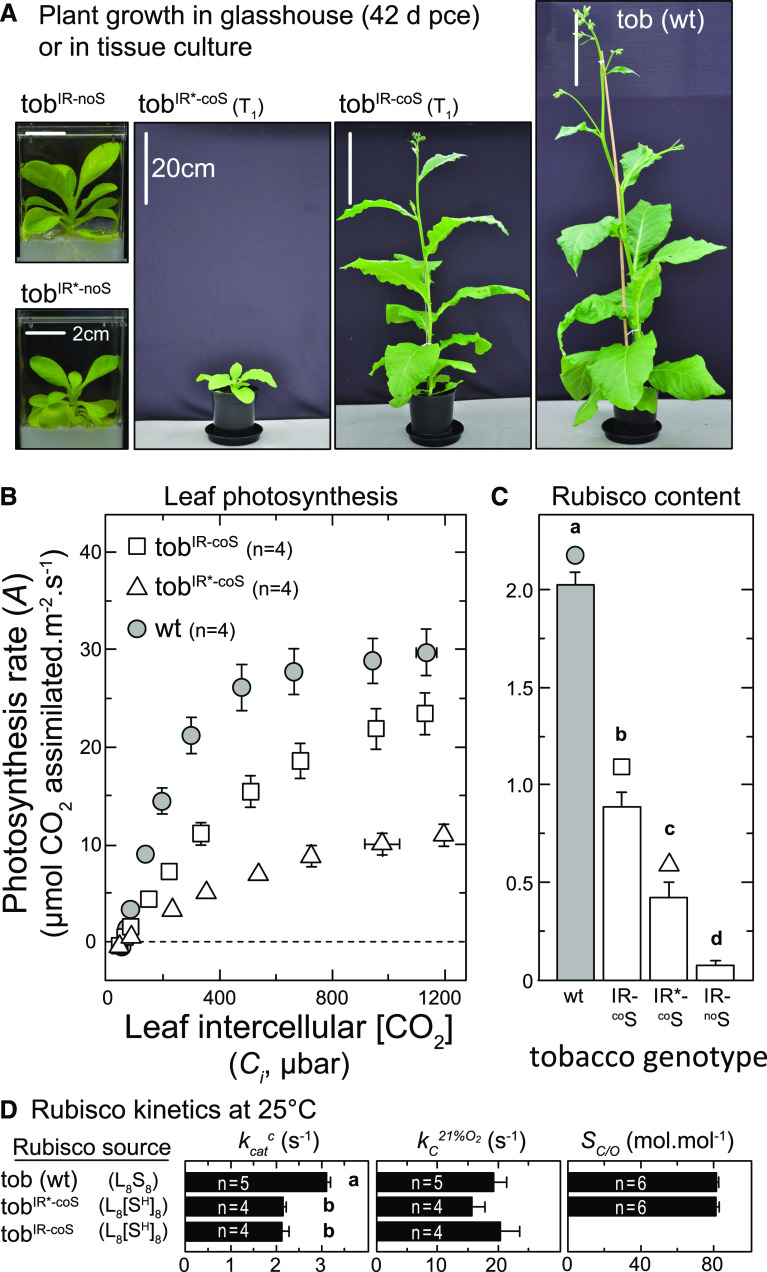
Consistent with the low amounts of Rubisco produced in the tobIR*-noS plants, and the absence of Rubisco in tobIR-noS, these genotypes failed to grow in soil and could only be maintained in tissue culture (Figure 3A). By comparison, the tobacco Rubisco quantities produced in the tobIR-coS and tobIR*-coS plants supported plant growth to fertile maturity in soil in the glasshouse, albeit at a slower rate than wild type (Figure 3A). Leaf gas exchange measurements performed on leaf no. 5 of T1 plants at 45 ± 3 cm in height showed genotype-dependent variations in their photosynthetic rates (Figure 3B). The range of photosynthetic capacity among genotypes correlated with the Rubisco content measured in the same leaves (Figure 3C) and accounted for the observed differences in growth rate (Figure 3A). For example, the response of the photosynthetic CO2 assimilation rate (A) to varying intercellular CO2 concentrations (Ci) in the wild type followed a characteristic biphasic A-Ci curve (Figure 3B). Below ∼400 μbar Ci, Rubisco carboxylase activity primarily limited the photosynthetic rate in wild-type leaves, while the electron transport rate-dependent regeneration of Ribulose 1,5-bisphosphate (RuBP) constituted the main limiting factor at higher Ci levels (Farquhar et al., 1980). By comparison, the 50% decrease in Rubisco content in tobIR-coS (Figure 3C) lowered the A-Ci response, with A remaining limited by Rubisco activity over the Ci range tested (Figure 3B). In tobIR*-coS leaves, the A-Ci response was further compromised by their 4-fold–lower Rubisco content (relative to wild type; Figure 3C), which accordingly markedly slowed their growth in soil (Figure 3A).
Figure 3.
Variation in Plant Growth, Leaf Photosynthesis, Rubisco Content, and Catalysis among Each Tobacco Genotype.
(A) Representative phenotype and growth stage of glasshouse-grown (25 ± 3°C, natural illumination) T1 tobIR-coS and tobIR*-coS plants relative to the wild-type (wt) 42-d post-cotyledon emergence (pce). The tobIR-noS and tobIR*-noS grew on Suc-containing tissue-culture medium, but were unable to survive growth on soil.
(B) and (C) Average (±sd) A-Ci response measured in leaf 5 of n = 4 plants (when 45 ± 3 cm in height) of each genotype (B) and the corresponding Rubisco contents measured in the same leaves by 14C-CABP binding (C); note that the tobIR*-noS Rubisco measures were made on leaves grown in tissue culture.
(D) Mean (±sd) of the CO2-fixation rate (kcatc), the apparent Michaelis–Menten constant for CO2 under ambient O2 (KC21%O2), and the CO2/O2 specificity (Sc/o) for L8S8 Rubisco from tobacco and L8(SH)8 Rubiscos (that comprise 6× His-tagged chloroplast-made SH-subunits) from tobIR-coS (encodes native S-subunit N terminus) and tobIR*-coS (N terminus encodes an MSP fusion; Figure 2B). Statistical differences after a one-way ANOVA and post hoc Tukey test are indicated by lowercase letters (P < 0.05).
As the initial slope of the A-Ci response is influenced by both the content and kinetics of Rubisco (Farquhar et al., 1980), we compared the carboxylation properties of the L8(SH)8 complex produced in both tobIR-coS and tobIR*-coS (comprising chloroplast-made tobacco L- and SH-subunits) with that of wild-type tobacco L8S8 Rubisco (Figure 3D). The maximal CO2-fixation rate (kcatc) by both L8(SH)8 holoenzymes were reduced ∼one-third, with no significant effect on CO2-affinity under ambient O2 (Kc21%O2) or the CO2/O2 specificity (Sc/o) of Rubisco in tobIR*-coS (Figure 3D). This suggests that the reduced kcatc of the L8(SH)8 complexes resulted from the C-terminal 6× His tag on the SH-subunits, rather than the additional MSP N-terminal residues in tobIR-coS. The resulting impairment to the carboxylation efficiency of L8(SH)8 Rubisco (i.e., the ratio of kcatc to Kc21%O2) would therefore have contributed to the weakened A-Ci response in tobIR*-coS and tobIR-coS leaves.
Rubisco Production in tobIR-coS and tobIR*-coS Is Constrained by Chloroplast rbcLS mRNA Abundance and rbcS Translation Initiation
We used samples taken from the leaves analyzed by gas exchange to compare Rubisco transcript abundance with the Rubisco content in tobIR-coS, tobIR*-coS, and wild-type leaves. We separated equivalent amounts of total leaf RNA under denaturing conditions (Figure 4A) before blotting onto a nylon membrane and hybridization with 32P-labeled probes to rbcL (Figure 4B), corbcS (Figure 4C), or norbcS (Figure 4D). Both the rbcL and corbcS probes identified an abundant dicistronic rbcL-rbcS mRNA, as well as an ∼6-fold–less abundant rbcL-rbcS-aadA tricistronic mRNA in both tobIR-coS and tobIR*-coS plants. By contrast, we only detected the native rbcL mRNA in the wild type (Figure 4B). The norbcS probe identified the rbcL-rbcS and rbcL-rbcS-aadA transcripts in tobIR-noS or tobIR*-noS but not in the tobIR-coS or tobIR*-coS lines (Figure 4C), indicating that the 70% sequence identity between norbcS and corbcS (Supplemental Figure 3B) precluded their cross hybridization. The absence of an rbcL mRNA in the transplastomic lines matches prior observations that inclusion of analogous short IR sequences in rbcL-rbcS mRNA prevented their processing into separate rbcL and rbcS transcripts (Whitney and Sharwood, 2008).
Figure 4.
Comparative Leaf Rubisco mRNA Analysis.
(A) Ethidium-bromide staining of total leaf RNA (3 μg) purified from the same leaves analyzed in Figure 3 separated by denaturing agarose gel electrophoresis. wt, wild type.
(B) to (D) Nitrocellulose membrane blots of the separated RNA hybridized with 32P-labeled rbcL (B), corbcS (C), or norbcS (D) probes (refer to Figure 2A). The differing Rubisco transcripts recognized are annotated L (rbcL), LS (dicistronic rbcL-rbcS mRNA), or LSA (tricistronic rbcL-rbcS-aadA mRNA), as depicted in Figure 2A. The sequence diversity between corbcS and norbcS (see Supplemental Figure 3B) precluded cross hybridization between their respective LS and LSA mRNAs and with the endogenous rbcS mRNA.
(E) Rubisco transcript abundance (relative to the rbcL mRNA content in the wild-type controls) determined by densitometry from (B) and the relative mRNA levels indicated in gray (L), black (LS), or white (LSA). Data (±sd) are the average of n = 2 samples from independent tobacco (wt), tobIR-coS (IR-coS), tobIR*-coS (IR*-coS), tobIR-noS (IR-noS), and tobIR*-noS (IR*-noS) plants.
Densitometry analysis of the rbcL-probed blots showed that the tobIR-coS and tobIR*-coS lines produced comparable amounts of rbcL-rbcS and rbcL-rbcS-aadA mRNA, which together equated to ∼60% of the rbcL mRNA content in the wild type (Figure 4E). This reduced Rubisco mRNA pool was in agreement with the 50% decrease in Rubisco content in tobIR-coS leaves (Figure 3C), suggesting that chloroplast rbcL-rbcS and rbcL-rbcS-aadA mRNA abundance may primarily limit Rubisco production in tobIR-coS. By contrast, the Rubisco content in tobIR*-coS leaves was ∼25% that of the wild type (Figure 3C), despite producing comparable rbcL-rbcS and rbcL-rbcS-aadA mRNA amounts to tobIR-coS. This suggests that constraints to S-subunit translation impair Rubisco production in the tobIR*-coS lines. A comparison of the tobIR-coS and tobIR*-coS engineered plastomes show that they only differ by nucleotide changes adjacent to the corbcS ATG initiator codon (Figures 2B and 2C). That is, TTATGCAGGTGTGGACT in tobIR-coS compared with GCATGTCACCT in tobIR*-coS. This suggests that translation initiation of corbcS impaired Rubisco production in tobIR*-coS. This effect on translation initiation likely arises from changes in mRNA secondary structure in the rbcL-rbcS and rbcL-rbcS-aadA transcripts, which will influence anti-Shine–Dalgarno/Shine–Dalgarno interactivity. Consistent with this hypothesis, we determined that the rates of 35S-L8S8 holoenzyme formation in tobIR-coS and tobIR*-coS were ∼40% and ∼30%, respectively, compared with the wild type, as measured by 35S-Met labeling in young intact leaves and native PAGE separation (Figures 5A and 5B). We tested the stability of the assembled 35S-L8S8 complexes by infiltrating the leaves with saturating amounts of unlabeled Met (Whitney et al., 2015). The 35S-Rubisco signal during this methionine “chase” remained unchanged over 7 h in all the tobacco genotypes (Figure 5C). This result indicates that differences in S-subunit translation, not the rate of L8S8 proteolysis, accounted for the variation in leaf Rubisco content between tobIR-coS and tobIR*-coS.
Figure 5.
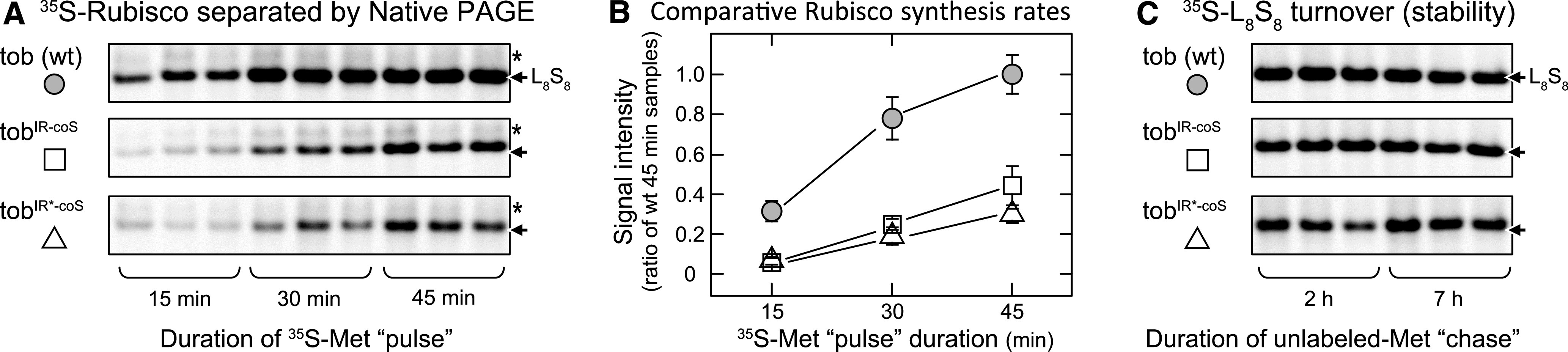
Rubisco Synthesis and Degradation.
(A) to (C) L8S8 Rubisco synthesis and turnover in leaf 5 of 35 cm in height tobIR-coS, tobIR*-coS, and wild-type tob (wt) control plants using 35S-Met “pulse” then unlabeled-Met “chase” analyses at 25°C under constant illumination (∼500 μmol quanta m−2 s−1).
(A) Autoradiography signals of native PAGE-separated soluble protein from 6 mm2 of attached leaves taken 15, 30, and 45 min after 35S-Met infiltration.
(B) The average L8S8 Rubisco densitometry signals at each time point (n = 3 ± sd) relative to the averaged 45-min wild-type sample signal.
(C) Native PAGE analyses of soluble protein from the same leaves 2 h and 7 h after infiltrating with 10 mM of unlabeled-Met.
The Codon Use of the norbcS Gene Impairs its Translation in Plastids
We detected both rbcL-rbcS and rbcL-rbcS-aadA mRNA when using the rbcL and norbcS probes against leaf RNA isolated from tobIR-noS and tobIR*-noS plants grown in tissue culture (Figures 4B and 4D). Densitometry analyses estimated the Rubisco mRNA levels in tobIR-noS and tobIR*-noS to be between 30% and 40% of the rbcL content measured in the wild type (Figure 4E). Despite these appreciable Rubisco transcript levels, both transplastomic lines produced very little Rubisco (tobIR*-noS; <1 µmol catalytic sites.m−2, Figure 3C) or no Rubisco (tobIR-noS; Figure 2D). This result implies that a post-transcriptional process, such as translation initiation and/or elongation of the norbcS transgene, impaired S-subunit synthesis, and Rubisco production in both tobIR-noS and tobIR*-noS.
Identification of Four Different Rubisco Small Subunits in Potato
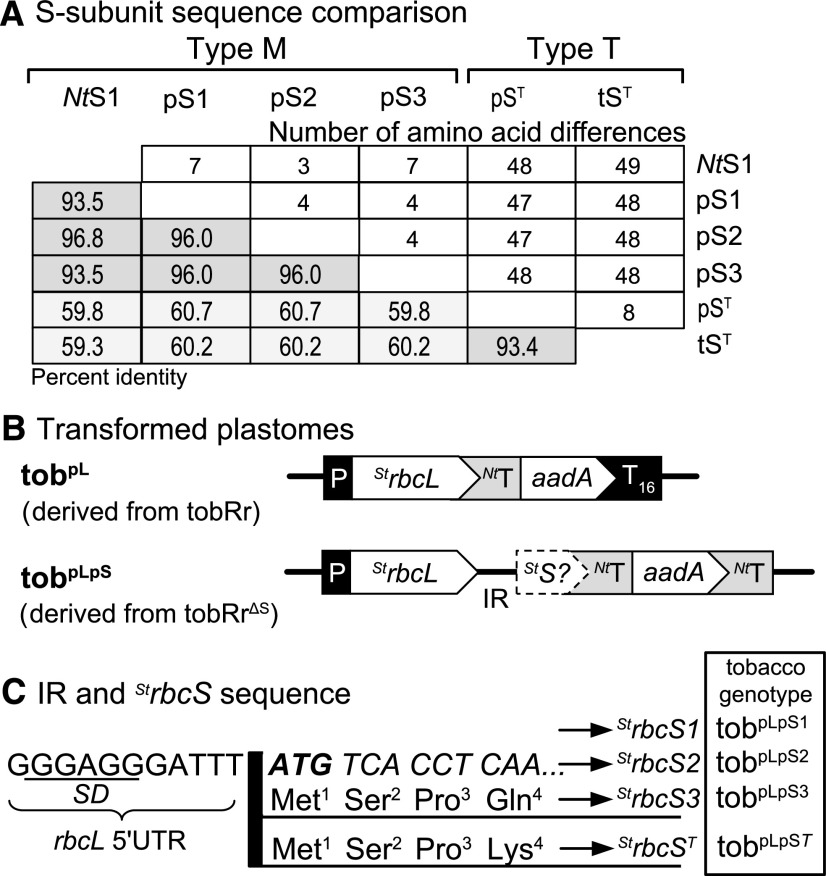
We tested the utility of tobRrΔS as a surrogate for crop Rubisco bioengineering via plastome transformation using Rubisco from the related Solanaceae plant potato. That the tobacco and potato Rubisco L-subunits share 97.5% identity (differing only in 10 out of 477 amino acids; Supplemental Figure 5A) engendered a hope that the biogenesis requirements of potato Rubisco would be readily met in tobacco. Analysis of the annotated potato genome in Phytozome identified four different mature S-subunits (after transit peptide removal; Supplemental Figure 5B), three of which (here termed “pS1,” “pS2,” and “pS3”) shared >93% identity with each other and the tobacco NtS1 subunit (Figure 6A). A phylogenetic analysis of S-subunits revealed that the fourth potato S-subunit (called “pST”) segregated in a distinct trichome-specific (Type T) S-subunit lineage (Supplemental Figure 6), as previously observed in Laterre et al. (2017). The pST shared only ∼60% identity with the mesophyll-cell–specific (Type M) pS1-, pS2-, and pS3-subunits and 93.4% identify with the tobacco trichome tST-subunit (Figure 6A).
Figure 6.
A Transplastomic Approach to Study how S-Subunit Diversity Influences Potato Rubisco Catalysis.
(A) Summary of the amino acid identity between mesophyll-cell–located (Type M) SM-subunits from tobacco (NtS1) and potato (pS1, pS2, and pS3) and the Type-T trichome-expressed ST-subunits in potato (pST) and tobacco (tST, GenBank:XM_016595419). See Supplemental Figure 5 for alignments of the potato and tobacco Rubisco L-, SM-, and ST-subunits and Supplemental Figure 6 for the phylogenetic relatedness of plant Type-M and Type-T Rubisco S-subunits.
(B) Schematic plastome maps of the tobpL genotype (after transformation of the potato StrbcL into tobRr) and differing tobpLpS genotypes produced by transforming differing StrbcL-S synthetic operons into the tobRrΔS plastome. T16, rps16 terminator sequence; P and NtT native tobacco rbcL promoter/5′UTR and terminator sequences.
(C) The StrbcL-S IR matched the tobIR-coS line (Figure 2B). The differing codon-optimized StrbcS genes coded pS1 (tobpLpS1), pS2 (tobpLpS2), pS3 (tobpLpS3), or a pST-subunit (tobpLpST).
Expressing Potato Rubisco in Tobacco
We assembled a set of chloroplast transformation plasmids similar to those made to generate tobIR-coS (Figure 2B), by incorporating a StrbcL gene coding for the potato L-subunit (pL) and a synthetic StrbcS gene encoding either the pS1-, pS2-, pS3-, or pST-subunits (nucleotide sequences, when possible, altered to match the tobacco corbcS gene). We transformed the plasmids into tobRrΔS to produce the tobacco genotypes tobpLpST, tobpLpS1, tobpLpS2, and tobpLpS3 (Figures 6B and 6C). The potato Rubisco produced in each tobacco genotype consisted of the same potato L-subunit and one of the four pS-subunits. We also transformed the StrbcL gene alone (i.e., without a cognate pS-subunit) into the plastome of tobRr (where tobacco S-subunits are still produced; Figure 1A) to generate tobpL plants that produced hybrid L8S8 Rubisco-comprising potato L-subunits and endogenous tobacco S-subunits (Figure 6B).
We identified transformed lines producing L8S8 Rubisco by native PAGE and determined homoplasmy based on ceased production of R. rubrum L2 Rubisco (Whitney and Sharwood, 2008). We grew at least two independent transformed lines for each genotype to maturity, crossed them with pollen from tobRrΔS (or from tobRr for tobpL), and used their progeny for all subsequent analyses.
We observed differences in the glasshouse growth of the tobacco lines producing the different potato Rubiscos. The growth rate and phenotype of the tobpL and wild-type tobacco plants were highly comparable, while the tobpLpST plants producing the trichome pST subunit grew the slowest (Figure 7A). The tobpLpS1 and tobpLpS2 plants grew at matching rates and slightly faster than the tobpLpS3 plants (Figure 7A). Gas exchange measures using leaf no. 5 of plants ∼33 cm in height showed differences in the A-Ci photosynthetic responses that correlated with the growth rate for each genotype (Figure 7B). While A in the wild type transitioned between being limited by carboxylase activity and electron transport rate at ∼400 to 600 μbar Ci, A remained limited by Rubisco activity across the entire Ci range tested in all the potato Rubisco-producing genotypes. The similar A-Ci responses in tobpLpS1 and tobpLpS2 leaves and marginally lower rates in tobpLpS3 correlated with their associated leaf Rubisco contents: equivalent in tobpLpS1 and tobpLpS2 leaves and ∼15% less in tobpLpS3 (Figures 7C and 7D). By contrast, the rates of photosynthesis and growth were lowest in tobpLpST (Figure 7B) despite having an L8S8 Rubisco content equivalent to tobpLpS1 and tobpLpS2 (Figure 7D).
Figure 7.

The Potato S-Subunit Sequence Influences Potato Rubisco Biogenesis, Leaf Photosynthesis, and Plant Growth.
(A) Comparative growth and phenotype 30-d post-cotyledon emergence of the potato Rubisco-producing tobacco lines grown in soil in a glasshouse at 25°C under natural illumination.
(B) Leaf A-Ci responses made on leaf no. 5 of glasshouse-grown plants (n = 3 ±sd) of each genotype when at 33 ± 3 cm in height.
(C) and (D) Native PAGE (n = 2, with nos. 1 and 2 representing independently transformed lines) (C) and 14C-CABP binding analysis (n = 5 or 6 ±sd) of Rubisco content (D) in wild-type (wt) and each potato Rubisco producing tobacco genotype at the time of gas-exchange. Different letters show significant differences to P < 0.05 using a Tukey multiple comparison test.
The pS3-subunit Differentially Affects the Kinetics of Potato Rubisco
In accordance with the high sequence similarity between the tobacco and potato Rubisco subunits (Supplemental Figure 5), both enzymes had equivalent CO2-fixation rates (kcatc) and apparent Michaelis–Menten constants for CO2 under ambient O2 (KC21%O2) and carboxylation efficiencies (kcatc/KC21%O2), although their specificity for CO2 over O2 (Sc/o) varied by ∼7% at 25°C (Table 2). Moreover, the highly similar S-subunits in both species (Figure 6A) meant that the kinetics of the hybrid Rubisco produced in tobpL also matched that of potato and tobacco Rubisco (Table 2). The kcatc and KC21%O2 of the Rubisco produced in tobpLpS1 and tobpLpS2 matched wild-type potato Rubisco, while the tobpLpS3 Rubisco showed a modest, but significant, 9% increase in kcatc and an unchanged KC21%O2. This kinetic variation mapped to the unique His-55-Asn (H55N) and Ser-57-Lys (S57K) substitutions at the apex of the βA-βB loop in the potato pS3-subunit produced in tobpLpS3 (Figure 8A). Within the L8S8 holoenzyme, H55N and S57K locate within the opening of the central solvent channel that traverses the hollow core (Figures 8B and8C). These data indicate that pS-subunits encoding N55 and/or K57 enhanced the potato Rubisco carboxylation rate, but they also impaired the biogenesis of these enzymes by ∼15% relative to Rubisco production in tobpLpS1 (Figure 7D), whose plastome differs only by 4 nt in the StRbcS gene encoded in tobpLpS3.
Table 2. Rubisco Kinetics at 25°C.
| Rubisco Source | kcatc (s−1) n = 5–6 | KC21%O2 (μM) n = 5–6 | kcatc/KC21%O2 (mM−1.s−1) n = 5–6 | Sc/o (mol.mol−1) n = 10–11 |
|---|---|---|---|---|
| Tobacco | 3.1 ± 0.1* | 18.5 ± 1.4* | 167 ± 11* | 80.6 ± 1.2* |
| Potato | 3.2 ± 0.1* | 18.8 ± 0.8* | 170 ± 10* | 84.4 ± 1.7**,*** |
| tobpL | 3.1 ± 0.1* | 18.7 ± 1.1* | 168 ± 13* | 80.7 ± 3.0* |
| tobpLpS1 | 3.0 ± 0.2* | 20.1 ± 1.8* | 149 ± 14* | 84.5 ± 1.4**,*** |
| tobpLpS2 | 3.0 ± 0.2* | 19.6 ± 1.6* | 155 ± 18* | 82.6 ± 3.6*,** |
| tobpLpS3 | 3.5 ± 0.1** | 19.5 ± 1.5* | 180 ± 12** | 87.3 ± 2.0*** |
| tobpLpST | 4.4 ± 0.2*** | 50.6 ± 2.9** | 86 ± 8*** | 70.6 ± 2.0**** |
Values are the average (±sd) of the indicated number of biological samples analyzed (n); values of kcatC/KC21%O2 were derived from the averaged values of kcatC and KC21%O2. The asterisks *, **, ***, and **** indicate significant differences to P < 0.05 using a Tukey multiple comparison test.
Figure 8.
Mapping the Amino Acid Diversity in the Potato Rubisco Mesophyll S-Subunits.
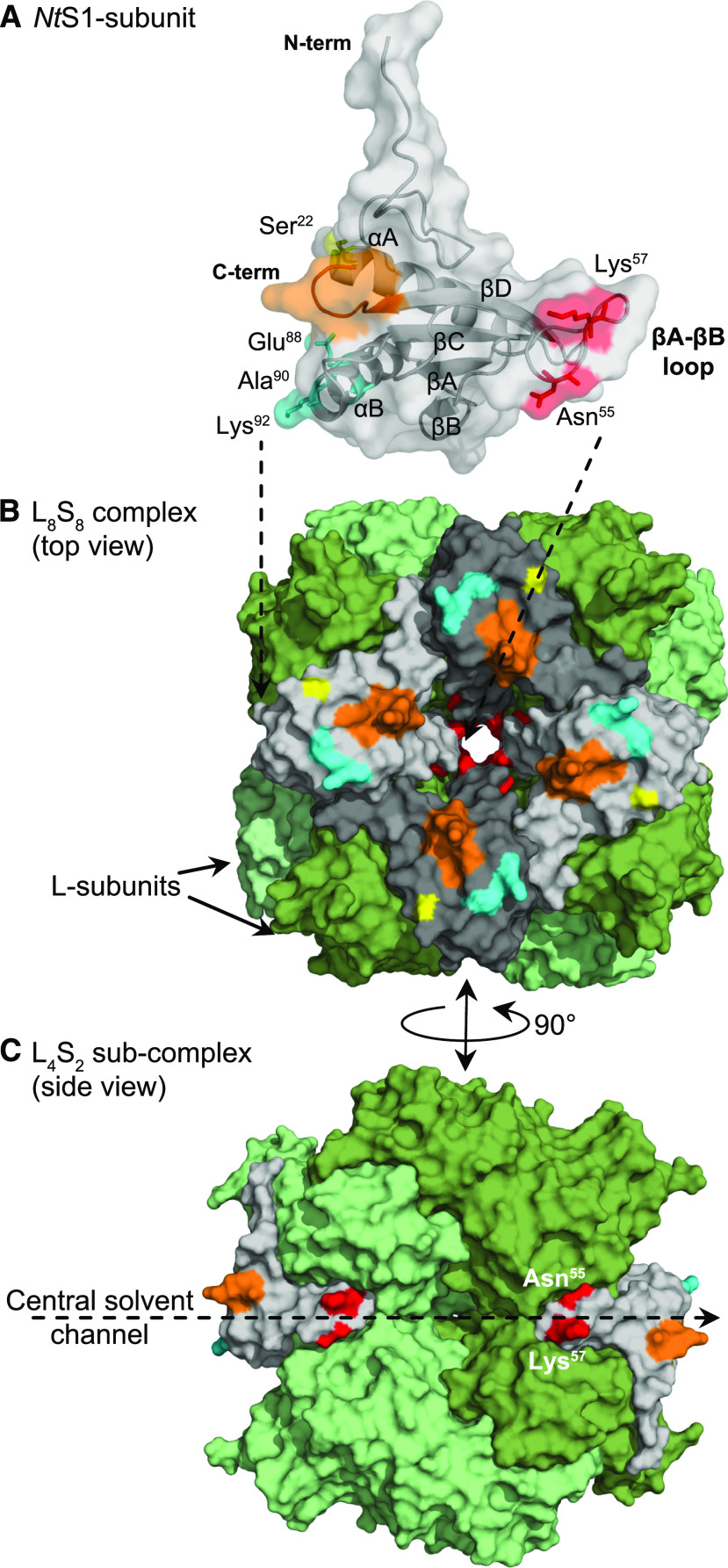
(A) Transparent surface rendering (performed in the software PyMol, https://pymol.org/2/) over the peptide backbone of a single NtS1-subunit depicted as a ribbon structure, with the side chains shown for the six variant amino acids between the pS1-, pS2-, and pS3-subunits, colored relative to Supplemental Figure 5B (and see below). The βA-βB loop, two α-helices, and the four β-strands are indicated.
(B) and (C) Surface representation of the tobacco L8S8 holoenzyme complex (PDB:1EJ7) (B) and a cross-section view of a L4S2 subcomplex (C). The two L-subunits assembled head-to-tail in each L2-unit are colored in differing shades of green. The S-subunits are colored in two shades of gray. The six variant amino acids include the surface-exposed amino acid 22 (yellow, Ser22 in NtS1, pS2, pS3; Thr22 in pS1); 55 and 57 (red, His55, Ser57 in pS3; Asn55, Lys57 in NtS1, pS1, pS2); and 88, 90, and 92 (cyan, Gln88, Cys90, and Asn92 in pS1; Gln88, Ala90, and Asn92 in pS3; Glu88, Ala90, and Lys92 in NtS1 and pS2; see alignment in Supplemental Figure 5B). The C-terminal residues Ala117 to Tyr123 are colored orange. Residues 55 and 57 (in red) are located in the βA-βB loop that lines the opening of the central solvent channel, a hollow core that traverses the length of the L8S8 complex; refer to (C).
The kinetic similarity of the potato Rubisco in tobpLpS1 and tobpLpS2 indicates that the differing amino acids at residues 22, 88, 90, and 92 in the pS1- and pS2-subunits (Supplemental Figure 5B) are catalytically neutral. Consistent with this hypothesis, all four residues are exposed to solvent and distant from the catalytic pockets that are located at the adjoining interface of each L-subunit pair (Figure 8).
The Trichome pST-subunit Confers C4-like Kinetics to Potato Rubisco
The kcatc for Rubisco from tobpLpST was ∼40% higher than potato Rubisco (Table 2). The improvement in kcatc was offset by a reduction in CO2-affinity (i.e., an ∼2.3-fold increase in KC21%O2) that reduced carboxylation efficiency of the enzyme by ∼2-fold and impaired the enzyme Sc/o by ∼15%. These perturbations to the carboxylation properties of tobpLpST Rubisco are consistent with the reduced growth (Figure 7A) and lower photosynthetic potential of this line (i.e., slow A-Ci response; Figure 7B), despite producing equivalent levels of Rubisco as the tobpLpS1 and tobpLpS2 genotypes (Figures 7C and 7D).
DISCUSSION
This study demonstrates that the modest amino acid variation among the S-subunits encoded by the RbcS multigene family in plants can differentially influence both the catalytic properties and biogenesis potential of plant Rubisco. Mutagenic analyses have underpinned decades of Rubisco structure-function studies to demonstrate how residues in both the L- and S-subunits can affect catalysis. Plant Rubisco mutagenic studies are underrepresented in the data due to the historical necessity for using protracted genome transformation approaches (Andrews and Whitney, 2003; Sharwood, 2017). While new capabilities to produce plant Rubisco in E. coli may enable plant Rubisco mutagenic studies at a higher throughput (Aigner et al., 2017), identifying a strategy for simultaneously transplanting rbcL and rbcS sequence alterations into the plant is challenged by their differing genome locations and the multiple RbcS copies. Here, we address this limitation using tobRrΔS, a tobacco plant producing R. rubrum L2 Rubisco, where the superfluous cytosolic S-subunit synthesis is efficiently, and stably, blocked by RNA interference of the encoding genes. Using chloroplast transformation, we show that introducing rbcL-rbcS operons into the tobRrΔS allows for the production of Rubisco containing a homogeneous population of L- and S-subunits—analogous to that from non-green algae that maintain the Rubisco operon in their plastome (Reith, 1995). Here we exploited this capacity and generated tobacco genotypes producing homogeneous potato Rubisco that incorporated either a mesophyll pS1-, pS2-, or pS3-subunit or the trichome pST-subunit. Biochemical and physiological phenotyping of each genotype showed how changes to residues 55 and 57 in the mesophyll S-subunit βA-βB loop and the 40%-sequence–divergent pST-subunit can influence Rubisco catalysis, L8S8 biogenesis, leaf photosynthesis, and plant growth.
Catalytic Switches in the βA-βB Loop of Plant Rubisco S-subunits
Our understanding of Rubisco structure-function has insufficient resolution to predict how S-subunit replacement or targeted amino acid changes will affect Rubisco catalysis. Heterologous RbcS expression studies in plants have produced conflicting outcomes. Arabidopsis Rubisco incorporating either pea (Pisum sativum; Getzoff et al., 1998) or tobacco (Whitney et al., 2015) S-subunits showed impaired catalysis, while rice (Oryza sativa) Rubisco incorporating a S-subunit from sorghum (Sorghum bicolor, a C4-plant) showed a kinetic transition toward C4-like catalysis (Ishikawa et al., 2011). By contrast, hybrid Rubisco enzymes consisting of tobacco S-subunits and the L-subunits from sunflower (Helianthus annuus), tomato (Solanum lycopersicum), or various marigold (Flaveria spp) species maintain equivalent catalytic properties to the native enzymes from which the L-subunits were sourced (Sharwood et al., 2008; Sharwood and Whitney, 2010; Whitney et al., 2011; Zhang et al., 2011). A limitation in interpreting the structure-function associations of these studies is that the heterologous Rubisco enzymes produced in these transgenic plants integrate more than one S-subunit. However, plastome rbcL-rbcS engineering in tobRrΔS circumvents this obstacle to produce homogenous plant Rubisco that comprises a single L- and S-subunit. By this approach, we uniquely show alterations in the size and charge of the side chains accompanying H55N or/and S57K substitutions at the apex of the βA-βB loop in the pS3-subunit of potato Rubisco stimulated kcatc and carboxylation efficiency (kcatc/KC21%O2) by 15% (Table 2). By contrast, the similar Rubisco kinetics in tobpLpS1 and tobpLpS2 demonstrated that substitutions to pS-subunit surface amino acids 22, 88, 90, and 92 are catalytically neutral to potato Rubisco (Table 2; Figure 8).
Structurally, the βA-βB loop interacts with L-subunit residues within the openings of the Rubisco solvent channel, the central cavity traversing the holoenzyme (Figure 8C). While the βA-βB loop is structurally remote to the catalytic sites (>16 Å in distance), its pervasive influence on catalysis is well understood in the context of Chlamydomonas Rubisco (Flachmann et al., 1997; Genkov and Spreitzer, 2009; Genkov et al., 2010). The intersubunit interactivity associated with βA-βB loop meant that improving Chlamydomonas Rubisco Sc/o necessitated complimentary amino acids changes at the adjoining L-subunit interface (Spreitzer et al., 2005). Crystal structure inter-subunit modeling suggested that the βA-βB loop constitutes one of three regions in the S-subunit whose L-subunit interactions have a pervasive effect on substrate interaction within the catalytic sites of the L8S8 holoenzyme (van Lun et al., 2011). Followup, in silico modeling of S-L inter-subunit interactions to mechanistically understand how S-subunit sequence changes can modulate catalysis, has not been forthcoming.
Rubisco Evolution: Evidence for a Compromise Between Catalysis and Biogenesis
Mounting evidence indicates that the reliance of plant Rubisco biogenesis on multiple chaperones has impeded its catalytic evolution. For example, amino acid changes in the L-subunit that perturb structural complementarity with its co-evolved Rubisco accumulation factor1 can perturb plant Rubisco production (Whitney et al., 2015). Inclusion of the Rubisco-specific assembly chaperone RbcX also affected the population of cyanobacteria Rubisco mutants selected by directed evolution (Durão et al., 2015). Inexplicably, amino acid substitutions S112F or G332S in the L-subunit prevent tobacco Rubisco biogenesis entirely (Avni et al., 1989; Shikanai et al., 1996). Conversely, the biogenesis of differing cyanobacteria Rubiscos in E. coli can be stimulated 2- to 11-fold by introducing a F345I substitution in the L-subunit (Wilson et al., 2018), in addition to a variety of other single S- and L-subunit mutations (see summary in Zhou and Whitney, 2019). The consequences arising from point mutations on Rubisco biogenesis have also been seen in tobacco rbcL transplastomic studies, where the D149A substitution in Flaveria L-subunits impaired hybrid L8S8 formation with tobacco S-subunits (Whitney et al., 2011). As shown in Figure 7, the H55N and S57K substitutions in the pS3-subunit also influence potato Rubisco biogenesis, reducing L8S8 formation by ∼15% and resulting in a slower photosynthetic rate and growth of the tobpLpS3 plants relative to tobpLpS1 and tobpLpS2. Mutagenic studies of the βA-βB loop in the Rubisco S-subunit from pea using isolated chloroplast 35S-Met-peptide feeding studies similarly found that an R53E substitution slowed the rate of S-subunit incorporation into the L8S8 holoenzyme by >10-fold (Flachmann and Bohnert, 1992; Adam, 1995). More significant structural changes to the surface alpha-helices of Arabidopsis S-subunits were also shown to compromise Rubisco biogenesis, albeit with little kinetic consequence (Atkinson et al., 2017). This suggests that the accessible sequence landscape for plant Rubisco catalytic evolution may not only be modulated by the requirement to maintain L-subunit/assembly chaperone structural complementarity (Whitney et al., 2011), but also by the need to preserve structural compatibility with the S-subunits to sustain sufficient L8S8 assembly. Arguably, the plant RbcS multigene family may therefore ensure adequate S-subunit production (Ogawa et al., 2011) as well as provide plasticity in Rubisco catalytic evolution. For example, current evidence indicates that RbcS transcript accumulation patterns vary over the course of organ development and environmental acclimation (Sugita and Gruissem, 1987; Dedonder et al., 1993; Yoon et al., 2001). Whether this biochemically translates to S-subunit–induced changes in catalysis and/or L8S8 biogenesis potential remains unclear.
Avoiding the Catalytic Constraints of a C-Terminal S-Subunit Epitope Tag
The use of epitope and marker protein fusion tags is commonplace in transgenic studies that seek to monitor accumulation and localization of a protein of interest, as well as purify the recombinant protein. Limitations in epitope tag uses include their ability to destabilize protein structure and perturb protein interactions or/and alter protein function. How epitope tags influence such property changes are often overlooked or are too problematic to be experimentally resolved (Saiz-Baggetto et al., 2017). Our data show that incorporating a C-terminal 6× His tag onto the S-subunit can perturb Rubisco catalysis. While the SH-subunit had no significant effect on tobacco Rubisco CO2 affinity or Sc/o, it reduced kcatc by 30%. As depicted in Figure 8, the S-subunit C terminus is exposed to solvent (residues 117 to 123 shown in orange) and forms no inter-subunit connections. Nevertheless, our findings suggest that inclusion of additional sequence to the S-subunit C terminus may induce conformational changes in holoenzyme structure that affect catalytic function.
The C4-Like Kinetics and Low Abundance of Trichome Rubisco
The pervasive influence of an ST-subunit on Rubisco kinetics was demonstrated previously by overexpressing the OsRbcS1 isoform in rice (Morita et al., 2013) and assembling tobacco tST with Chlamydomonas L-subunits (Laterre et al., 2017). In both studies, the trichome S-subunits increased kcatc and KC. Similarly, we showed that the kcatc of potato Rubisco incorporating pST-subunits increased ∼40%, while its carboxylation efficiency (kcatc/KC21%O2) decreased by 50% and Sc/o dropped by ∼15% (Table 2). Existing hypotheses suggest that the faster activity and lower CO2 affinity of trichome Rubisco reflects an evolutionary adaptation of the enzyme to the specialized metabolic functions of trichome chloroplasts (Laterre et al., 2017; Pottier et al., 2018). Underscoring these hypotheses is how trichome Rubisco kinetics reflect those evolved by C4-plant Rubisco in response to the acquisition of a CO2-concentrating mechanism. Accordingly, it is proposed that CO2 released during the production of pathogen-deterring diterpenes and sucrose esters within trichomes, along with a possibly lower stroma pH, culminates in an elevated CO2-environment within their chloroplasts (Laterre et al., 2017; Pottier et al., 2018). As seen in C4-plants, such a microenvironment would lessen the need for a high CO2-affinity Rubisco, thereby allowing a higher kcatc enzyme to form and be maintained.
As indicated above, the catalytic evolution of the S-subunit has likely been influenced by a need to sustain its L8S8 biogenesis potential. Our finding of matching Rubisco contents in the tobpLpS1, tobpLpS2, and tobpLpST genotypes (Figure 7D) therefore suggests that the potato L8 core binds the mesophyll pS1-, pS2-, and trichome pST-subunits with comparable avidity, despite only sharing 60% amino acid identity (Figure 6A). That is, these phylogenetically distinct pS-subunit families (Figure 6) appear to have independently evolved equivalent assembly compatibilities with the same L8 core. The resulting L8S8 complexes differ in their native PAGE mobilities (Figure 7C), akin to the slower mobility seen for native tobacco trichome Rubisco (Laterre et al., 2017). This difference in holoenzyme quaternary structure questions whether structural incompatibilities at adjoining S/S- and L/S-subunit interfaces might preclude co-integration of mesophyll and trichome subunits into the same L8S8 complex. Notably, we failed to detect the slower migrating trichome Rubisco in protein extracts from wild-type potato separated by native PAGE (Figure 7C) or tobacco (Laterre et al., 2017). This indicates that trichome Rubisco is produced in finite amounts that would have no discernible effects on the kinetics of Rubisco isolated from whole leaf extract.
Codon Use and the 5′UTR Impacted Plastid S-Subunit Translation
The twofold difference in Rubisco content between the tobIR-coS and tobIR*-coS lines producing a codon-optimized corbcS and the paucity, or absence, of Rubisco in the corresponding tobIR-noS and tobIR*-noS lines producing a nonoptimized norbcS, provide a compelling demonstration of how both the 5′UTR sequence adjacent to the ATG initiator codon and the codon use of the mRNA itself can dramatically influence plastid gene translation. Appealing features of plastome transgenics are the high genome copy number, the corresponding high mRNA abundance, and the potential for high-level protein production (Bock, 2015; Daniell et al., 2016). Contrary to these expectations, however, the production of transplastomic recombinant protein is often inadequate, or totally lacking, despite abundant transgene mRNA levels (Bock, 2014). The tobIR-noS line produced here, with abundant rbcLS but lacking Rubisco, provides an example of this phenomenon.
A key challenge to chloroplast bioengineering is designing transformation plasmids that can reliably produce abundant recombinant protein(s). Exacerbating this challenge is the limited range of available gene-expression parts (i.e., choice of promoter 5′UTR and 3′UTR regulatory/stabilizing sequences) and lack of reproducible transgene expression using a common regulatory element (Maliga, 2002; Bock, 2015). To advance our mechanistic understanding of chloroplast protein biosynthesis, ribosome profiling tools can provide in vivo measures of mRNA translation activity (Fujita et al., 2019). Such measures have discovered that protein synthesis is affected by codon usage, regions of ribosome pausing, the Shine–Dalgarno sequence, and its positioning relative to the ATG initiation codon (Bock, 2015; Scharff et al., 2017; Trösch et al., 2018; Zoschke and Bock, 2018). Our finding that the tobacco lines encoding the corbcS gene produced more Rubisco than those coding for the norbcS gene is in line with prior transplastomic studies, where incorporating the synonymous codons used by highly expressed chloroplast genes enhanced the expression of R. rubrum Rubisco (Whitney and Sharwood, 2008), Bacillus thuringiensis cryIA(b) (Perlak et al., 1991), and Clostridium tetani TetC (Tregoning et al., 2003) in tobacco chloroplasts. Incorporating a portion of the rbcL translational control region (Kuroda and Maliga, 2001) onto the corbcS (Figure 2B) also appeared to increase S-subunit translation, and thus Rubisco content, ∼2-fold in tobIR-coS relative to the tobIR*-coS genotype (Figure 3C). This increase possibly stemmed from improvements in corbcS translation initiation through the functional complementarity between the rbcL mRNA Shine–Dalgarno sequence and the anti-Shine–Dalgarno sequence located at the 3′ end of the ribosomal 16S ribosomal RNA (Scharff et al., 2017). By contrast, appending the same rbcL translational-control–region sequence to the norbcS gene precluded S-subunit production in the tobIR-noS lines, while some Rubisco production was still feasible in the tobIR*-noS lines (Figure 3C). This result implies that translation initiation was impeded in tobIR-noS, possibly due to unfavorable mRNA secondary structure folding around the norbcS Shine–Dalgarno sequence—a hypothesis for future testing by ribosome profiling. Similarly, ribosome fingerprinting would also help decipher how the 8-nt differences between the tobIR*-coS and tobIR-coS lines (Figured 2B and 2C) around the AUG initiator codon of corbcS differentially influence translation initiation (Figure 5).
Enhancing Plastid S-Subunit Translation
While the tobRrΔS line successfully allows for simultaneous co-engineering of rbcL and rbcS genes in the rbcL coding region of the plastome, our strategy of using a polycistronic mRNA regulated by the native rbcL promoter likely requires modification to increase S-subunit translation. One possibility is to equip the rbcS gene (and aadA gene) with independent regulatory elements. An alternative approach is to emulate the native process of some polycistronic plastid mRNAs that employ RNA binding proteins to cleave and stabilize these transcripts into single gene units to promote their translation (Bock, 2014). The intercistronic expression element (IEE) constitutes one such native sequence that harbors the binding site of the RNA binding protein HIGH CHLOROPHYLL FUORESCENT 107A (Zhou et al., 2007). Incorporation of the IEE between individual genes within a cistron can facilitate mRNA processing into stable monocistronic mRNAs with the ensuing potential for enhanced translation (Lu et al., 2013). Notably. the IEE was incorporated into Synechococcus elongatus PCC7942 and Cyanobium marinum PCC7001 rbcL-rbcS polycistrons introduced into the tobacco plastome to enhance S-subunit translation (Occhialini et al., 2015; Long et al., 2018). Unfortunately, the biogenesis requirements of cyanobacterial Rubisco are poorly met in leaf chloroplasts, limiting L8S8 production such that altering cyanobacterial Rubisco mRNA abundance had little effect on enzyme biogenesis, which reached at best ∼15% of the tobacco controls (Occhialini et al., 2015). As discussed below, limitations in plant Rubisco production in chloroplasts might also arise if the import and processing of cytosolic-made S-subunits (by auxiliary proteins located at the inner envelope membrane) comprise a critical component of the Rubisco biogenesis process. To address this potential limitation, future objectives are to test whether Rubisco production can be increased in variants of tobIR-coS where the corbcS gene contains its own regulatory elements or the rbcL-rbcS-aadA polycistron incorporates IEE sequences to produce a stable, more translatable, corbcS monocistron mRNA.
Increasing Plastid Rubisco mRNA Abundance
In addition to translational processing deficiencies, the >2-fold–lower tobacco rbcL-rbcS mRNA abundance in all the transplastomic lines generated in this study likely contributed to their lower Rubisco contents (Figure 4E). Plausible causes of this reduction are variations in mRNA synthesis and/or stability—both of which are likely sensitive to cellular homeostasis. This appears more compromised in the tobIR-noS and tobIR*-noS lines lacking Rubisco, even when grown in tissue culture (Figure 4B).
Prior tobacco S-subunit transplastomic studies have primarily focused on inserting the tobacco native NtS1a gene encoding NtS1 (Table 1) into the duplicated inverted repeat regions (IRA, IRB) of the plastome. When inserted between the trnV gene and rps12/7 operon and when its expression is regulated by the tobacco psbA regulatory sequences, approximately only one in 10 L8S8 complexes contained a plastid-made S-subunit (Whitney and Andrews, 2001). Greater success was achieved by inserting a comparable rbcS cassette between the trnI-trnA genes of the IRA and IRB in an anti-RbcS tobacco line (Dhingra et al., 2004). A consideration to the perceived advantage of increasing Rubisco mRNA abundance by transforming into the IRA and IRB is the objective of co-engineering both rbcL and rbcS simultaneously in a genotype where the RbcS genes are silenced by RNAi. Using tobRrΔS this could be achieved by first transforming a rbcL-rbcS operon into the IRA and IRB of the plastome then subsequently deleting the rbcM gene from the large single-copy region. Alternatively, it might be feasible to generate the desired transgenic lines using a co-transformation strategy of the tobRrΔS plastome (Ye et al., 2003). For example, the strategy might include co-introduced transgenic events that replace rbcM with a selectable marker gene and integrate rbcLS and a second marker gene into the IR. Lines carrying both transgenic events would be readily identifiable as those producing L8S8 Rubisco, and no longer the L2 Rubisco.
Cytosolic SSU Import, an Unbeatable Evolutionary Advantage?
A fundamental question is whether the limited capacity to incorporate stroma-synthesized S-subunits into L8S8 complexes stems from their spatial segregation of their natural chaperone-facilitated import through the chloroplast envelope translocon. The processing of imported cytosol-made precursor S-subunits involves a range of compatible chloroplast envelope and stromal molecular partners. By contrast, engineered S-subunits made in the chloroplast are likely synthesized by thylakoid-bound or stroma-located chloroplast polysomes (Thomson et al., 2020), which may place these S-subunits spatially far from compatible envelope-associated molecular partners that would otherwise provide proteolytic stability or/and facilitate their passage to L8S8 assembly. As hypothesized by Whitney and Andrews (2001), the relocation of RbcS genes to the nucleus appears to have led to adaptive changes in S-subunit evolution that influenced the biogenesis pathway of Rubisco and may possibly now limit the incorporation of chloroplast-made S-subunits. Better understanding of the natural import and chaperoning processes of cytosol-made S-subunits may provide clues as to why they may be preferentially incorporated during L8S8 biogenesis in leaf chloroplasts.
Future Challenges for Bioengineering Plant Rubisco
The potential for high-resolution mutagenic profiling of plant Rubisco now appears feasible using new E. coli expression capabilities (Wilson and Hayer-Hartl, 2018; Wilson et al., 2019). Further study into the development of versatile E. coli plant Rubisco expression systems is critical to (1) exploring how catalytic diversity is influenced by L- and S-subunit sequences, and (2) using this information to bioengineer improved crop Rubisco (Conlan and Whitney, 2018). What remains unclear, however, is the extent to which E. coli can be used as a surrogate for assessing the biogenesis potential of Rubisco in chloroplasts. To date, the capacity for cyanobacterial Rubisco expression in E. coli has not correlated with its biogenesis potential in tobacco chloroplasts (Wilson et al., 2018). Plant L- and S-subunits produced in E. coli also lack the series of post-translational modifications that occur during L8S8 biogenesis in the stroma (Houtz et al., 2008), the functions of which remain enigmatic and their effect on catalysis poorly understood. Current evidence, however, indicates that trimethylation of Lys-14 in plant L-subunits has no discernible effect on Rubisco catalysis or CO2-assimilation rate in transgenic tobacco lacking this post-translational modification (Dirk et al., 2006).
A key challenge facing Rubisco bioengineering in plants is being able to meet the Rubisco content required. For example, Rubisco L- and S-subunits in tobacco typically constitute ∼19% and 6% (w/w) of the leaf soluble protein, respectively (Whitney and Sharwood, 2014) and this investment is doubled in rice (Suzuki et al., 2009). Increasing Rubisco production together with plant productivity is achievable in rice through co-expressing an additional RbcS gene (Yoon et al., 2020) and in maize (Zea mays) through nuclear RbcL, RbcS, and Rubisco accumulation factor1 overexpression (Salesse-Smith et al., 2018). However, transgenic RbcS substitution attempts in plants remain unsuccessful in producing recombinant S-subunits in amounts equivalent to the wild-type controls (Ishikawa et al., 2011). Indeed, even the tobIR-coS lines produce only approximately half the Rubisco content of the wild type (Figure 3C). As discussed above, identifying a plastome transformation strategy that can produce 2-fold–higher amounts of Rubisco relative to tobIR-coS is a significant future objective.
METHODS
RbcS Phylogenetic Reconstruction
RbcS sequences were retrieved from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) and Phytozome (phytozome.jgi.doe.gov) websites (see Supplemental Table and Supplemental Data Sets 1 and 2 for species and gene accession numbers). Introns were removed and exons translated into proteins, which were then aligned using the software MUSCLE (Edgar, 2004). The software MODELTEST 3.7 (Posada and Buckley, 2004) was used to check for the best model before running phylogenetic analyses using maximum-likelihood inference conducted with the program RAxML v.7.2.6 (Stamatakis, 2014).
Plant Tissue Culture, Growth Conditions, and Transformation
All genetic transformations were performed in tobacco (Nicotiana tabacum cv Petit Havana N,N). Plant tissue culture was handled on RMOP (Whitney and Sharwood, 2014) containing 3% (w/v) Suc. The plant tissue was grown at 25°C in air supplemented with 2% (v/v) CO2 under 50- to 100-μmol photons.m−2.s−1 artificial white fluorescent light (Philips) illumination on a 14-h/10-h L/D cycle. Transformed T0 plants with established roots were transferred from tissue culture into 2-L pots in soil and grown at 25°C, with 300 to 450 μmol photons m−2 s−1 illumination (14-h/10-h L/D cycle, Warm Deluxe Metal Halide Lamps; Venture Lighting International) in air (0.04% [v/v] CO2) or in air with 1% or 1.5% (v/v) CO2. Plants were fertilized every two weeks with Osmocote (Scotts).
The RNAi-RbcS nucleus-transformation plasmid psiSS (Supplemental Figure 1A, a gift from Katia Wostrikoff; Wostrikoff and Stern, 2007) was transformed into the Rhodospirillum rubrum L2 Rubisco-producing tobacco genotype tobRr (Figure 1A) using Agrobacterium (Agrobacterium tumefaciens) strain LB4404. Transformed tissue was grown on RMOP containing 10 μg mL−1 glufosinate at 25°C in air with 2.5% (v/v) CO2, and 50- to 100-μmol photons m−2 s−1 illumination. Glufosinate-resistant calli from different leaf sections were regenerated twice on RMOP-glufosinate before growing plantlets in 750-mL pots containing 150 mL of RMOP before being transplanted to soil and allowed to grow to maturity. Their flowers were appropriately pollinated for segregation analysis and identification of homozygous tobRrΔS1, tobRrΔS2, tobRrΔS3, and tobRrΔS4 lines (see Figure 1 and Supplemental Figure 2 for details).
The plastome-transformation plasmids were derived from pLEV4 (Whitney et al., 2015) and biolistically transformed as described by Svab and Maliga (1993) into leaves of a homozygous T2 tobRrΔS1 line (renamed tobRrΔS) or the tobRr genotype. The transformation plasmids pLEVIR-coS (GenBank:MT596796), pLEVIR*-coS, pLEVIR-noS, pLEVIR*-noS, pLEVpL (GenBank:MT596797), pLEVpLpS1 (GenBank:MT596798), pLEVpLpS2, pLEVpLpS3, and pLEVpLpST (GenBank:MT596799) allow the targeted integration of the rbcL (±rbcS) and aadA transgenes into the tobacco plastome around rbcL (Figures 2 and 6B; Whitney and Sharwood, 2008). The plasmids were assembled by standard restriction enzyme cloning from commercially synthesized genetic elements cloned into pUC57 (GenScript). Each plasmid was fully sequenced using a BigDye Terminator Cycle Sequencing Kit (Thermo Fisher Scientific) before bombarding into five leaf sections, from which five to 15 spectinomycin-resistant plants were obtained for each genotype. After two rounds of regeneration on selective RMOP-spec (0.5 mg mL−1 of spectinomycin), homoplastomic-transformed lines were identified by native PAGE screening for total replacement of L2 Rubisco (100 kD) with L8S8 Rubisco (520 kD) production. Two independent lines of each genotype were grown to maturity in soil and either self-pollinated (to maintain RNAi-RbcS homozygosity) or crossed with wild-type pollen (heterozygous RNAi-RbcS progeny).
RNA Extraction and Blot Analyses
Leaf samples (0.5-cm2 discs) from plants grown in tissue culture or soil were snap-frozen in liquid nitrogen and stored at –80°C until analyzed for RNA, protein, or Rubisco content and activity. Leaf total RNA was purified with the RNAeasy RNA Extraction Kit (Qiagen) and either separated by denaturing formaldehyde agarose gel electrophoresis and blotted onto a Hybond-N+ nylon membrane (Bio-Rad), or blotted directly onto the membrane using a Bio-Dot SF Cell (Bio-Rad). The latter process involved assembling a membrane pre-wet with sterile water into the Bio-Dot SF Cell and adding the RNA samples diluted 1:100 with sterile water under gentle vacuum. The wells were then washed with 3× 0.2-mL RNA wash solution (10 mM of NaOH and 1 mM of EDTA) under vacuum before immobilizing the RNA onto the membrane using a UV Stratalinker 1800 (Stratagene). The RNA gel blots were hybridized in AlkPhos Direct Hybridization Buffer (GE Healthcare) at 55°C for >16 h with heat-denatured [32P]-labeled DNA probes prepared using the Prime-a-Gene Labeling System (Promega). The DNA probes were amplified by PCR using the primer pairs S1For (5′-ATGTCACCTCAAACTTGGCC-3′)/SRev (5′-GGCTTATATGCGATAAA3′; probe corbcS1), S2For (5′ATGCAGGTGTGGCCTCCT3′)/SRev (probe norbcS), and LSa/LSb (probe rbcL; Whitney et al., 1999). The membranes were rinsed twice for 20 min with 200 mL of 0.5× SSC buffer (75 mM of NaCl, 7.5 mM of trisodium citrate at pH 7.0, and 0.1% [w/v] SDS) at 55°C, then patted dry with toweling and exposed to a Storage Phosphor Screen GP (Kodak). The hybridization signals were visualized using a PharosFX Plus Molecular Imager (Bio-Rad) and quantified using the software Quantity One (Bio-Rad).
Leaf Protein and Rubisco Quantification and PAGE Analysis
Total leaf protein was isolated in 0.5 or 1 mL of ice-cold extraction buffer (50 mM of EPPS-NaOH at pH 8.0, 1 mM of EDTA, 10 mM of MgCl2, 2 mM of DTT, 1% [w/v] polyvinylpolypyrrolidone, and 0.1% [v/v] plant protease inhibitor; Sigma-Aldrich) using 2 mL of glass tissue homogenizers (Wheaton). The soluble protein fraction was recovered by centrifugation (16,400g, 5 min, 4°C). Protein content was determined against BSA using the Coomassie Plus Protein Assay Reagent (Pierce). The Rubisco content was determined by [14C]-CABP binding (Whitney and Sharwood, 2014). Soluble proteins were separated by native PAGE (4 to 12% Tris-Gly gels; Invitrogen) or SDS PAGE (4 to 12% Bis-Tris gel; Invitrogen) electrophoresis. The separated protein bands were either visualized using Gelcode Blue Stain Reagent (Invitrogen) or blotted onto Hybond-C Extra nitrocellulose membrane using an XCell II Blot Module (Invitrogen) as described by Whitney and Sharwood (2014). The membranes were blocked in tris buffered saline (TBS) buffer (50 mM of Tris-Cl and 150 mM of NaCl at pH 7.6) containing 5% (w/v) skim milk powder before probing with polyclonal antibodies (diluted 1:5,000 in TBS) raised in a rabbit against purified tobacco Rubisco (WEHI Antibody Facility, Bundoora, Australia) for 30 min. After three 5-min washes in TBS, the membranes were incubated for 30 min with AP-conjugated anti-Rabbit secondary antibody (diluted 1:3,000 in TBS). After three 5-min washes in TBS, the immuno-reactive proteins were visualized with AttoPhos AP Fluorescent Substrate (Promega). The chemi-fluorescent bands were imaged using a Pharos FX Plus Molecular imager and analyzed using the software Quantity One (Bio-Rad).
Rubisco Kinetics
RuBP was synthesized and purified as described by Kane et al. (1998) and used in RuBP-dependent 14CO2 fixation assays to measure the Rubisco Michaelis–Menten constants for CO2 under ambient O2 concentrations (KC21%O2) as well as the maximal carboxylation turnover rate (kcatc) at 25°C (Sharwood et al., 2016). The assays were initiated by adding 20 μL of rapidly extracted soluble leaf protein whose Rubisco catalytic sites had been fully CO2-Mg2+–activated by first incubating with an equal volume of 2× Mg−14CO2 buffer (50 mM of EPPS-OH at pH 8.0, 20 mM of MgCl2, 1 mM of EDTA, and 20 mM of NaH14CO3) at 25°C for 7 or 11 min. The assays were performed in septum-sealed 8-mL glass scintillation vials containing 480 μL of 14CO2 fixation assay buffer (50 mM of EPPS-OH pH 8.2, 1 mM of EDTA, 20 mM of MgCl2, 10 μg mL−1 of carbonic anhydrase, and 0.2 mM of RuBP) containing five differing CO2 concentrations (from 0 to 37.4 μM of 14CO2 [i.e., ∼4.1 mM of NaH14CO3], specific activity ∼25 to 30 Bq nmol−1). The CO2 (C) concentrations were calculated using the Henderson–Hasselbalch equation:
where Ct is total organic carbon, V/v is the ratio of reaction vial headspace (V) to solution volume (v), q is the solubility of CO2 in water at 1 atm at 25°C (0.03292 mol L−1 atm−1), R is the universal gas constant, and T is the reaction temperature (298 K). The pK1 and pK2 values of 6.25 and 10.33, respectively, were used in the calculations. RuBP-independent 14CO2 fixation control assays were run for each sample and contained water in place of RuBP. After adding each sample, the assays were stopped after 1 min by adding 100 μL of 20% (v/v) formic acid. The mixture was dried at 90°C, then the residue dissolved in 0.5 mL water and mixed with 1 mL of scintillant (Ultima-Gold; Perkin Elmer). Acid stable radioactivity was measured in a scintillation counter. The data were fitted to the Michaelis–Menten equation to derive values for Kc21%O2 and substrate-saturated carboxylation rate (Vcmax), the latter divided by the Rubisco active site content assayed (quantified by [14C]-CABP binding) to derive kcatc.
Measurements of CO2/O2 specificity (Sc/o) were measured with Rubisco purified by ion-exchange chromatography (Sharwood et al., 2016) using the method of Kane et al. (1994).
Leaf Gas Exchange
Plants were transferred from their growth facility to a laboratory and left overnight before measuring the photosynthetic gas exchange properties of leaf no. 5, the youngest, near-fully expanded leaf of plants of similar height (see legens to Figures 3 and 7), using a model no. 6400-02B (Li-COR) fitted with a red/blue (10%) LED light source. Measurements of how leaf CO2 assimilation rates (A) responded to changes in intercellular CO2 concentrations (Ci; i.e., A-Ci response) were performed at a leaf temperature of 25°C and with 1,500 μmol photons m−2 s−1 illumination.
35S-Labeling of L8S8 Rubisco Synthesis and Turnover
Leaf no. 5 of plants 35 ± 2 cm in height were placed under 480 ± 50 µmol photons m2 s−1 white fluorescent illumination for 0.5 h before infiltrating with 2 mL of Trans35S-label (ICN Biomedicals) diluted to 0.25 mCi mL−1 (9.25 MBq mL−1) with infiltration buffer (10 mM of MES-NaOH at pH 5.5 and 10 mM of MgSO4) as described by Whitney et al. (2015). Leaf discs (0.5 cm2) were collected after 15, 30, and 45 min and frozen in liquid nitrogen. Infiltration buffer containing 10 mM of Met was then infiltrated into the same leaves and leaf disc samples then taken after 2 h and 7 h. The incorporation of 32P into L8S8 Rubisco after separation by native PAGE was undertaken and analyzed as described by Whitney et al. (2015).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under accession numbers listed in the Supplemental Table and in Supplemental Data Set 1.
Supplemental Data
Supplemental Figure 1. Tobacco Rubisco S-subunit sequence diversity and silencing by RNAi.
Supplemental Figure 2. Overview of tobRrΔS generation, segregation analysis, and crossing to generate the F1 tobΔS progeny.
Supplemental Figure 3. The tobacco S-subunit and RbcS genes examined in this study.
Supplemental Figure 4. The codon use of rbcL, NtS1a, and the corbcS and norbcS transgenes.
Supplemental Figure 5. Potato and tobacco Rubisco subunit alignments.
Supplemental Figure 6. The phylogenetic relatedness of plant Type-M and Type-T Rubisco S-subunits.
Supplemental Table. GenBank accession numbers for each tobacco RbcS gene. Data used to construct Table 1.
Supplemental Data Set 1. GenBank accession numbers for genes used to construct the RbcS phylogenetic tree in Supplemental Figure 6.
Supplemental Data Set 2. Raw data and ANOVA results of data.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This research was supported by the Australian Government through the Australian Research Council Centre of Excellence for Translational Photosynthesis (grant CE140100015) and the Discovery Program (grant DP130103825).
AUTHOR CONTRIBUTIONS
S.M.W. and E.M.-A. designed the research; E.M.-A., Y.L.-L., R.B., L.M.A.D., R.E.S., and S.M.W. performed the research; T.R. and M.V.K. contributed analytic and computational tools; E.M.-A., S.B., S.M.W., and R.B. analyzed the data; all authors helped write the article.
References
- Adam Z.(1995). A mutation in the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase that reduces the rate of its incorporation into holoenzyme. Photosynth. Res. 43: 143–147. [DOI] [PubMed] [Google Scholar]
- Aigner H., Wilson R.H., Bracher A., Calisse L., Bhat J.Y., Hartl F.U., Hayer-Hartl M.(2017). Plant Rubisco assembly in E. coli with five chloroplast chaperones including BSD2. Science 358: 1272–1278. [DOI] [PubMed] [Google Scholar]
- Andrews T.J., Whitney S.M.(2003). Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Arch. Biochem. Biophys. 414: 159–169. [DOI] [PubMed] [Google Scholar]
- Atkinson N., Leitão N., Orr D.J., Meyer M.T., Carmo-Silva E., Griffiths H., Smith A.M., McCormick A.J.(2017). Rubisco small subunits from the unicellular green alga Chlamydomonas complement Rubisco-deficient mutants of Arabidopsis. New Phytol. 214: 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni A., Edelman M., Rachailovich I., Aviv D., Fluhr R.(1989). A point mutation in the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase affects holoenzyme assembly in Nicotiana tabacum. EMBO J. 8: 1915–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J., Parker J.E., Ainsworth E.A., Oldroyd G.E.D., Schroeder J.I.(2019). Genetic strategies for improving crop yields. Nature 575: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R.(2014). Genetic engineering of the chloroplast: Novel tools and new applications. Curr. Opin. Biotechnol. 26: 7–13. [DOI] [PubMed] [Google Scholar]
- Bock R.(2015). Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 66: 211–241. [DOI] [PubMed] [Google Scholar]
- Conlan B., Whitney S.(2018). Preparing Rubisco for a tune up. Nat. Plants 4: 12–13. [DOI] [PubMed] [Google Scholar]
- Daniell H., Lin C.-S., Yu M., Chang W.-J.(2016). Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 17: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedonder A., Rethy R., Fredericq H., Van Montagu M., Krebbers E.(1993). Arabidopsis RbcS genes are differentially regulated by light. Plant Physiol. 101: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra A., Portis A.R., Daniell H.(2004). Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proc. Natl. Acad. Sci. USA 101: 6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirk L.M.A., Trievel R.C., Houtz R.L.(2006). 7 Non-histone protein lysine methyltransferases: Structure and catalytic roles In The Enzymes, Clarke S.G., and Tamanoi F., eds (Cambridge, MA: Academic Press; ), pp. 179–228. [DOI] [PubMed] [Google Scholar]
- Durão P., Aigner H., Nagy P., Mueller-Cajar O., Hartl F.U., Hayer-Hartl M.(2015). Opposing effects of folding and assembly chaperones on evolvability of Rubisco. Nat. Chem. Biol. 11: 148–155. [DOI] [PubMed] [Google Scholar]
- Edgar R.C.(2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Youles M., Gruetzner R., Ehnert T.-M., Werner S., Jones J.D.G., Patron N.J., Marillonnet S.(2014). A Golden Gate modular cloning toolbox for plants. ACS Synth. Biol. 3: 839–843. [DOI] [PubMed] [Google Scholar]
- Farquhar G.D., von Caemmerer S., Berry J.A.(1980). A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149: 78–90. [DOI] [PubMed] [Google Scholar]
- Flachmann R., Bohnert H.J.(1992). Replacement of a conserved arginine in the assembly domain of ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit interferes with holoenzyme formation. J. Biol. Chem. 267: 10576–10582. [PubMed] [Google Scholar]
- Flachmann R., Zhu G.H., Jensen R.G., Bohnert H.J.(1997). Mutations in the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase increase the formation of the misfire product xylulose-1,5-bisphosphate. Plant Physiol. 114: 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Kurihara Y., Iwasaki S.(2019). The plant translatome surveyed by ribosome profiling. Plant Cell Physiol. 60: 1917–1926. [DOI] [PubMed] [Google Scholar]
- Fukayama H., Koga A., Hatanaka T., Misoo S.(2015). Small subunit of a cold-resistant plant, timothy, does not significantly alter the catalytic properties of Rubisco in transgenic rice. Photosynth. Res. 124: 57–65. [DOI] [PubMed] [Google Scholar]
- Genkov T., Meyer M., Griffiths H., Spreitzer R.J.(2010). Functional hybrid Rubisco with plant small subunits and algal large subunits. J. Biol. Chem. 285: 19833–19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genkov T., Spreitzer R.J.(2009). Highly conserved small subunit residues influence Rubisco large subunit catalysis. J. Biol. Chem. 284: 30105–30112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getzoff T.P., Zhu G.H., Bohnert H.J., Jensen R.G.(1998). Chimeric Arabidopsis thaliana Rubisco containing a pea small subunit protein is compromised in carbamylation. Plant Physiol. 116: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L., Olson M., Wendel J.F.(2014). Cytonuclear evolution of Rubisco in four allopolyploid lineages. Mol. Biol. Evol. 31: 2624–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtz R.L., Magnani R., Nayak N.R., Dirk L.M.A.(2008). Co- and post-translational modifications in Rubisco: Unanswered questions. J. Exp. Bot. 59: 1635–1645. [DOI] [PubMed] [Google Scholar]
- Ishikawa C., Hatanaka T., Misoo S., Miyake C., Fukayama H.(2011). Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiol. 156: 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane H.J., Viil J., Entsch B., Paul K., Morell M.K., Andrews T.J.(1994). An improved method for measuring the CO2/O2 specificity of ribulosebisphosphate carboxylase-oxygenase. Aust. J. Plant Physiol. 21: 449–461. [Google Scholar]
- Kane H.J., Wilkin J.M., Portis A.R., John Andrews T.(1998). Potent inhibition of ribulose-bisphosphate carboxylase by an oxidized impurity in ribulose-1,5-bisphosphate. Plant Physiol. 117: 1059–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanevski I., Maliga P.(1994). Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 91: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H., Maliga P.(2001). Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol. 125: 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laterre R., Pottier M., Remacle C., Boutry M.(2017). Photosynthetic trichomes contain a specific Rubisco with a modified pH-dependent activity. Plant Physiol. 173: 2110–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long B.M., Hee W.Y., Sharwood R.E., Rae B.D., Kaines S., Lim Y.-L., Nguyen N.D., Massey B., Bala S., von Caemmerer S., Badger M.R., Price G.D.(2018). Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 9: 3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long B.M., Rae B.D., Rolland V., Förster B., Price G.D.(2016). Cyanobacterial CO2-concentrating mechanism components: Function and prospects for plant metabolic engineering. Curr. Opin. Plant Biol. 31: 1–8. [DOI] [PubMed] [Google Scholar]
- Lu Y., Rijzaani H., Karcher D., Ruf S., Bock R.(2013). Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc. Natl. Acad. Sci. USA 110: E623–E632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lun M., van der Spoel D., Andersson I.(2011). Subunit interface dynamics in hexadecameric Rubisco. J. Mol. Biol. 411: 1083–1098. [DOI] [PubMed] [Google Scholar]
- Maliga P.(2002). Engineering the plastid genome of higher plants. Curr. Opin. Plant Biol. 5: 164–172. [DOI] [PubMed] [Google Scholar]
- McCarty N.S., Graham A.E., Studená L., Ledesma-Amaro R.(2020). Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 11: 1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Hatanaka T., Misoo S., Fukayama H.(2013). Unusual small subunit that is not expressed in photosynthetic cells alters the catalytic properties of Rubisco in rice. Plant Physiol. 164: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhialini A., Lin M.T., Andralojc P.J., Hanson M.R., Parry M.A.J.(2015). Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO2. Plant J. 85: 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S., Suzuki Y., Yoshizawa R., Kanno K., Makino A.(2011). Effect of individual suppression of RBCS multigene family on Rubisco contents in rice leaves. Plant Cell Environ. 35: 546–553. [DOI] [PubMed] [Google Scholar]
- Parry M.A.J., Andralojc P.J., Scales J.C., Salvucci M.E., Carmo-Silva A.E., Alonso H., Whitney S.M.(2012). Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 64: 717–730. [DOI] [PubMed] [Google Scholar]
- Perlak F.J., Fuchs R.L., Dean D.A., McPherson S.L., Fischhoff D.A.(1991). Modification of the coding sequence enhances plant expression of insect control protein genes. Proc. Natl. Acad. Sci. USA 88: 3324–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Buckley T.R.(2004). Model selection and model averaging in phylogenetics: Advantages of Akaike Information Criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53: 793–808. [DOI] [PubMed] [Google Scholar]
- Pottier M., Gilis D., Boutry M.(2018). The hidden face of Rubisco. Trends Plant Sci. 23: 382–392. [DOI] [PubMed] [Google Scholar]
- Reith M.(1995). Molecular biology of rhodophyte and chromophyte plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 549–575. [Google Scholar]
- Saiz-Baggetto S., Méndez E., Quilis I., Igual J., Bañó M.(2017). Chimeric proteins tagged with specific 3×HA cassettes may present instability and functional problems. PLoS One 12: e0183067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salesse-Smith C.E., Sharwood R.E., Busch F.A., Kromdijk J., Bardal V., Stern D.B.(2018). Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat. Plants 4: 802–810. [DOI] [PubMed] [Google Scholar]
- Scharff L.B., Ehrnthaler M., Janowski M., Childs L.H., Hasse C., Gremmels J., Ruf S., Zoschke R., Bock R.(2017). Shine–Dalgarno sequences play an essential role in the translation of plastid mRNAs in tobacco. Plant Cell 29: 3085–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood R.E.(2017). Engineering chloroplasts to improve Rubisco catalysis: Prospects for translating improvements into food and fiber crops. New Phytol. 213: 494–510. [DOI] [PubMed] [Google Scholar]
- Sharwood R.E., Ghannoum O., Kapralov M.V., Gunn L.H., Whitney S.M.(2016). Temperature responses of Rubisco from Paniceae grasses provide opportunities for improving C3 photosynthesis. Nat. Plants 2: 16186. [DOI] [PubMed] [Google Scholar]
- Sharwood R.E., von Caemmerer S., Maliga P., Whitney S.(2008). The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol. 146: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood R.E., Whitney S.(2010). Engineering the sunflower Rubisco subunits into tobacco chloroplasts: New considerations In The Chloroplast: Advances in Photosynthesis and Respiration, Ca R., ed (Netherlands: Springer; ), pp. 285–306. [Google Scholar]
- Shikanai T., Foyer C.H., Dulieu H., Parry M.A.J., Yokota A.(1996). A point mutation in the gene encoding the Rubisco large subunit interferes with holoenzyme assembly. Plant Mol. Biol. 31: 399–403. [DOI] [PubMed] [Google Scholar]
- Simkin A.J., López-Calcagno P.E., Raines C.A.(2019). Feeding the world: Improving photosynthetic efficiency for sustainable crop production. J. Exp. Bot. 70: 1119–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer R.J., Peddi S.R., Satagopan S.(2005). Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco. Proc. Natl. Acad. Sci. USA 102: 17225–17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.(2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M., Gruissem W.(1987). Developmental, organ-specific, and light-dependent expression of the tomato ribulose-1,5-bisphosphate carboxylase small subunit gene family. Proc. Natl. Acad. Sci. USA 84: 7104–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Nakabayashi K., Yoshizawa R., Mae T., Makino A.(2009). Differences in expression of the RBCS multigene family and Rubisco protein content in various rice plant tissues at different growth stages. Plant Cell Physiol. 50: 1851–1855. [DOI] [PubMed] [Google Scholar]
- Svab Z., Maliga P.(1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90: 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S.M., Pulido P., Jarvis R.P.(2020). Protein import into chloroplasts and its regulation by the ubiquitin-proteasome system. Biochem. Soc. Trans. 48: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregoning J.S., Nixon P., Kuroda H., Svab Z., Clare S., Bowe F., Fairweather N., Ytterberg J., Wijk K.J., Dougan G., Maliga P.(2003). Expression of tetanus toxin Fragment C in tobacco chloroplasts. Nucleic Acids Res. 31: 1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trösch R., Barahimipour R., Gao Y., Badillo-Corona J.A., Gotsmann V.L., Zimmer D., Mühlhaus T., Zoschke R., Willmund F.(2018). Commonalities and differences of chloroplast translation in a green alga and land plants. Nat. Plants 4: 564–575. [DOI] [PubMed] [Google Scholar]
- Whitney S.M., Andrews T.J.(2001). The gene for the Rubisco small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into Rubisco. Plant Cell 13: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney S.M., Baldet P., Hudson G.S., Andrews T.J.(2001). Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J. 26: 535–547. [DOI] [PubMed] [Google Scholar]
- Whitney S.M., Birch R., Kelso C., Beck J.L., Kapralov M.V.(2015). Improving recombinant Rubisco biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proc. Natl. Acad. Sci. USA 112: 3564–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney S.M., Sharwood R.E.(2008). Construction of a tobacco master line to improve Rubisco engineering in chloroplasts. J. Exp. Bot. 59: 1909–1921. [DOI] [PubMed] [Google Scholar]
- Whitney S.M., Sharwood R.E.(2014). Plastid transformation for Rubisco engineering and protocols for assessing expression. Methods Mol. Biol. 1132: 245–262. [DOI] [PubMed] [Google Scholar]
- Whitney S.M., von Caemmerer S., Hudson G.S., Andrews T.J.(1999). Directed mutation of the Rubisco large subunit of tobacco influences photorespiration and growth. Plant Physiol. 121: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney S.M., Sharwood R.E., Orr D., White S.J., Alonso H., Galmés J.(2011). Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) carboxylation rate in Flaveria. Proc. Natl. Acad. Sci. USA 108: 14688–14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.H., Alonso H., Whitney S.M.(2016). Evolving Methanococcoides burtonii archaeal Rubisco for improved photosynthesis and plant growth. Sci. Rep. 6: 22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.H., Hayer-Hartl M.(2018). Complex chaperone dependence of Rubisco biogenesis. Biochemistry 57: 3210–3216. [DOI] [PubMed] [Google Scholar]
- Wilson R.H., Martin-Avila E., Conlan C., Whitney S.M.(2018). An improved Escherichia coli screen for Rubisco identifies a protein–protein interface that can enhance CO2-fixation kinetics. J. Biol. Chem. 293: 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.H., Thieulin-Pardo G., Hartl F.-U., Hayer-Hartl M.(2019). Improved recombinant expression and purification of functional plant Rubisco. FEBS Lett. 593: 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Clark A., Sato S., Clemente T., Stern D.(2012). Ectopic expression of Rubisco subunits in maize mesophyll cells does not overcome barriers to cell type-specific accumulation. Plant Physiol. 160: 419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Stern D.(2007). Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 104: 6466–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye G.-N., Colburn S.M., Xu C.W., Hajdukiewicz P.T.J., Staub J.M.(2003). Persistence of unselected transgenic DNA during a plastid transformation and segregation approach to herbicide resistance. Plant Physiol. 133: 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon D.-K., Ishiyama K., Suganami M., Tazoe Y., Watanabe M., Imaruoka S., Ogura M., Ishida H., Suzuki Y., Obara M., Mae T., Makino A.(2020). Transgenic rice overproducing Rubisco exhibits increased yields with improved nitrogen-use efficiency in an experimental paddy field. Nature Food 1: 134–139. [DOI] [PubMed] [Google Scholar]
- Yoon M., Putterill J.J., Ross G.S., Laing W.A.(2001). Determination of the relative expression levels of Rubisco small subunit genes in Arabidopsis by rapid amplification of cDNA ends. Anal. Biochem. 291: 237–244. [DOI] [PubMed] [Google Scholar]
- Zhang X.H., Ewy R.G., Widholm J.M., Portis A.R. Jr.(2002). Complementation of the nuclear antisense RbcS-induced photosynthesis deficiency by introducing an rbcS gene into the tobacco plastid genome. Plant Cell Physiol. 43: 1302–1313. [DOI] [PubMed] [Google Scholar]
- Zhang X.H., Webb J., Huang Y.H., Lin L., Tang R.S., Liu A.(2011). Hybrid Rubisco of tomato large subunits and tobacco small subunits is functional in tobacco plants. Plant Sci. 180: 480–488. [DOI] [PubMed] [Google Scholar]
- Zhou F., Karcher D., Bock R.(2007). Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 52: 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Whitney S.(2019). Directed evolution of an improved Rubisco: In vitro analyses to decipher fact from fiction. Int. J. Mol. Sci. 20: 5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Bock R.(2018). Chloroplast translation: Structural and functional organization, operational control, and regulation. Plant Cell 30: 745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]