Significance
cGAS is a protein which senses the infected viral DNA in the cytosol. Sensing of viral DNA by cGAS signals an innate immune response to virus. How cGAS is regulated by posttranslational modifications is still not fully understood. In this study, we found that the acetyltransferase KAT5 mediated acetylation of the N terminus of cGAS after infection of DNA virus, such as herpes simplex virus 1. This modification of cGAS causes its higher affinity to viral DNA and therefore promotes innate antiviral response. This study helps to understand the delicate regulatory mechanisms of innate antiviral response. Manipulation of these mechanisms may help to develop novel therapeutics for infectious and inflammatory diseases.
Keywords: DNA virus, cGAS, KAT5, acetylation, innate immune response
Abstract
The DNA sensor cGMP-AMP synthase (cGAS) senses cytosolic microbial or self DNA to initiate a MITA/STING-dependent innate immune response. cGAS is regulated by various posttranslational modifications at its C-terminal catalytic domain. Whether and how its N-terminal unstructured domain is regulated by posttranslational modifications remain unknown. We identified the acetyltransferase KAT5 as a positive regulator of cGAS-mediated innate immune signaling. Overexpression of KAT5 potentiated viral-DNA–triggered transcription of downstream antiviral genes, whereas a KAT5 deficiency had the opposite effects. Mice with inactivated Kat5 exhibited lower levels of serum cytokines in response to DNA virus infection, higher viral titers in the brains, and more susceptibility to DNA-virus–induced death. Mechanistically, KAT5 catalyzed acetylation of cGAS at multiple lysine residues in its N-terminal domain, which promoted its DNA-binding ability. Our findings suggest that KAT5-mediated cGAS acetylation at its N terminus is important for efficient innate immune response to DNA virus.
Upon infection of pathogenic microbes, the host innate immune system employs germline-encoded pattern-recognition receptors to detect pathogen-associated molecular patterns (PAMPs), which triggers a series of immune signaling cascades for ultimate elimination of the pathogens and clearance of infected cells (1–5). Upon DNA virus infection, viral DNA acts as a major PAMP to trigger antiviral innate immune response. In mammalian cells, the most widely expressed and utilized cytosolic viral DNA sensor is cGMP-AMP (cGAMP) synthase (cGAS), which forms a “cGAS-DNA” liquid droplet upon viral infection and mediates synthesis of cGAMP (6, 7). cGAMP in turn binds to the endoplasmic reticulum (ER)-associated protein MITA (also called STING), which then translocates from the ER via the ER–Golgi intermediate complex to perinuclear punctate structures. During these processes, the kinase TBK1 and the transcriptional factor IRF3 are recruited to the MITA-associated complex, leading to phosphorylation of IRF3 by TBK1 and its activation to induce transcription of hundreds of downstream antiviral genes (8–11).
cGAS consists of two domains, including the N-terminal unstructured domain ([NUD]: amino acid [aa] 1 to 160) and the C-terminal catalytic domain ([CCD]: aa 161 to 521) which binds to its substrates GTP and ATP. The CCD of cGAS is heavily regulated by different posttranslational modifications. In resting cells, the CCD of cGAS is (poly)glutamylated by TTLL4/6 at multiple residues to keep its enzymatic inactive before viral infection, whereas viral infection causes its deglutamylation and activation by CCP5/6 (12). The CCD of cGAS is also modified by K48-linked polyubiquitin chains at multiple lysine residues to limit its protein level in resting cells, whereas viral infection induces its sumoylation by TRIM38 and deubiquitination by USP14, leading to its stability and activation for a fast and robust innate immune response at the early phase of infection. At the late phase of infection, cGAS undergoes desumoylation by SENP2 and subsequent K48-linked polyubiquitination, leading to termination of the innate immune response (13, 14). Recently, it has been shown that the CCD of cGAS is acetylated to suppress its enzymatic activity (15). It has also been shown that the NUD of cGAS is critically involved in its optimal DNA-binding (16), phase-separation (7), and subcellular locations (17). However, whether and how the NUD of cGAS is regulated remains unknown.
The lysine acetyltransferase 5 (KAT5) is a catalytic subunit of the highly conserved NuA4 acetyltransferase complex, which plays critical roles in DNA damage repair, p53-mediated apoptosis, HIV-1 transcription, and autophagy (18–21). Although KAT5 has been investigated mostly as a transcriptional regulator, there is increasing evidence that KAT5 also acts as a key regulator in signal transduction pathways by targeting nonhistone proteins such as ULK1, PRAK, and Lipin1 for acetylation (20, 22, 23).
In this study, we identify KAT5 as a positive regulator of cGAS in response to cytosolic double-stranded DNA (dsDNA) and DNA viral infection. KAT5 deficiency or impairment of its acetyltransferase activity inhibits cytosolic dsDNA- or DNA-virus–triggered transcription of downstream antiviral genes and innate antiviral response in vitro and in vivo. Mechanistically, KAT5 catalyzes acetylation of cGAS at four lysine residues in its NUD, which promotes its DNA-binding ability and activation. Our findings suggest that KAT5 acts as an important regulator of cGAS-mediated innate antiviral response by promoting acetylation of its N terminus.
Results
KAT5 Positively Regulates cGAS-Mediated Innate Immune Signaling.
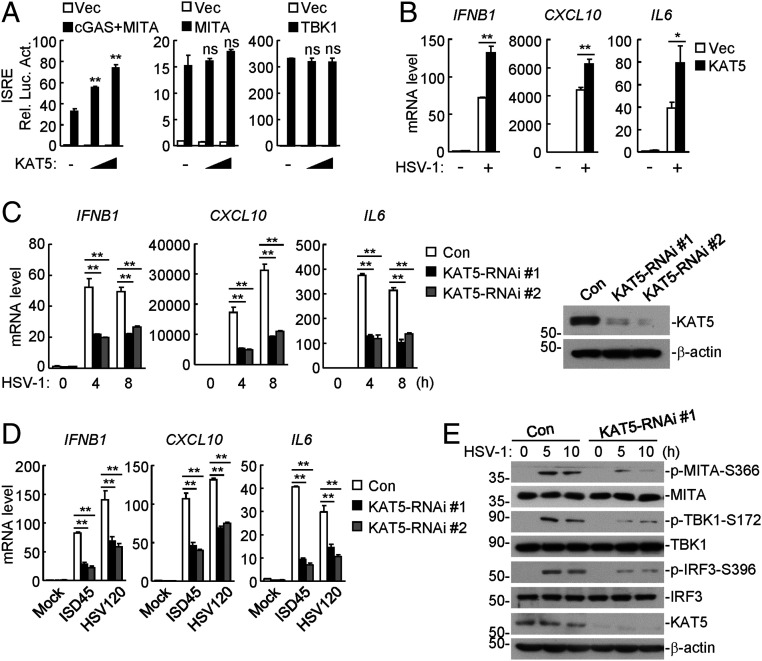
To identify potential acetyltransferases that regulate cGAS-mediated innate immune response, we examined 13 KAT family acetyltransferases for their abilities to regulate cGAS-mediated activation of the interferon-stimulated response element (ISRE) by reporter assays. As shown in SI Appendix, Fig. S1A, overexpression of KAT5 but not the other members of the KAT family markedly potentiated cGAS-mediated ISRE activation in HEK293 cells stably expressing MITA. In similar experiments, overexpression of KAT5 dose-dependently and specifically promoted cGAS-, but not MITA- or TBK1-mediated activation of ISRE (Fig. 1A). Quantitative PCR (qPCR) analysis indicated that overexpression of KAT5 potentiated transcription of downstream antiviral genes including IFNB1, CXCL10, and IL6 induced by the DNA virus herpes simplex virus 1 (HSV-1) (Fig. 1B). To investigate the roles of endogenous KAT5 in cGAS-mediated innate immune response, we constructed two human KAT5-RNAi plasmids which were able to markedly down-regulate the KAT5 level (Fig. 1C). qPCR analysis indicated that knockdown of KAT5 markedly inhibited HSV-1– and transfected dsDNA-induced transcription of downstream IFNB1, CXCL10, and IL6 genes in both human monocytic THP-1 (Fig. 1 C and D) and human foreskin fibroblast cells (SI Appendix, Fig. S1 B and C). Knockdown of KAT5 also markedly inhibited HSV-1–induced phosphorylation of MITA S366, TBK1 S172, and IRF3 S396, which are hallmarks for activation of virus-triggered innate immune signaling (Fig. 1E). As a control, knockdown of KAT5 potentiated SeV- and poly(I:C) (low- and high-molecular-weight)–induced transcription of the IFNB1 gene (SI Appendix, Fig. S1 D and E). These results suggest that KAT5 positively regulates DNA- virus– and cytosolic dsDNA-triggered innate immune signaling.
Fig. 1.
KAT5 positively regulates DNA-virus–triggered signaling. (A) KAT5 promotes cGAS- but not MITA- or TBK-mediated ISRE activation. HEK293 cells were transfected with the indicated plasmids for 20 h before luciferase assays were performed. (B) Overexpression of KAT5 promoted HSV-1–induced transcription of downstream genes. The stable KAT5-overexpressing and control THP-1 cell lines were left uninfected or infected with HSV-1 (multiplicity of infection [MOI] = 1) for 10 h before qPCR analysis. (C) Effects of KAT5 knockdown on HSV-1–induced transcription of downstream genes. The KAT5 knockdown and control THP-1 cell lines were left uninfected or infected with HSV-1 (MOI = 1) for the indicated times before qPCR analysis. Knockdown efficiency of KAT5 was confirmed by immunoblots (Right). (D) Effects of KAT5 knockdown on transcription of downstream genes induced by transfected DNA. The KAT5-RNAi stable knockdown and control THP-1 cell lines were transfected with the indicated dsDNA for 4 h before qPCR analysis. (E) Effects of KAT5 knockdown on HSV-1–induced phosphorylation of MITA-S366, TBK1-S172, and IRF3-S396. The KAT5 knockdown and control THP-1 cell lines were left uninfected or infected with HSV-1 (MOI = 1) for the indicated times before immunoblotting analysis with the indicated antibodies. ns, no significance. *P < 0.05, **P < 0.01 (unpaired t test). Data shown are mean ± SD from one representative experiment performed in triplicates (A–D).
KAT5 Mediates Acetylation of cGAS NUD.
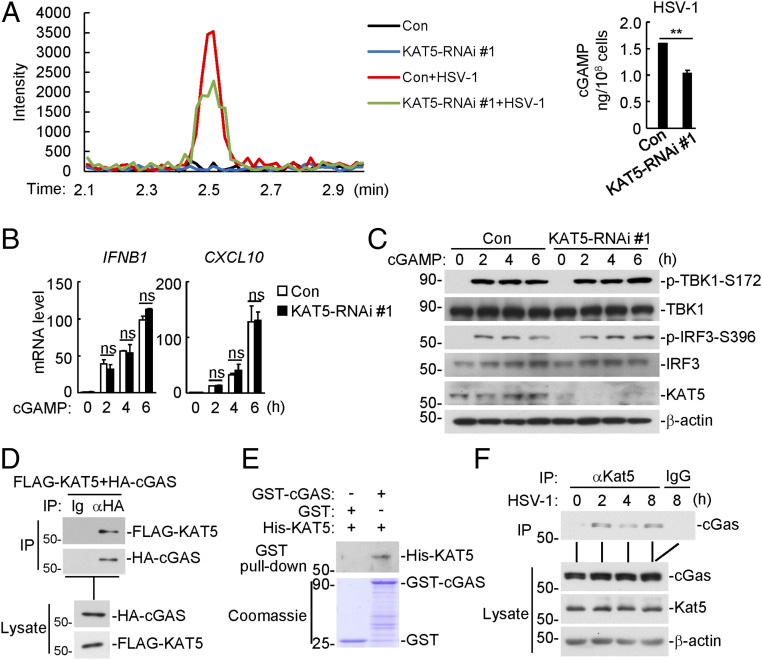
To determine the targets of KAT5, we examined the effects of KAT5 deficiency on synthesis of cGAMP in response to HSV-1 infection as well as cGAMP-triggered signaling. As shown in Fig. 2A, knockdown of KAT5 markedly reduced HSV-1–induced cGAMP production, but had no marked effects on cGAMP-induced transcription of downstream IFNB1 and CXCL10 genes as well as phosphorylation of TBK1 and IRF3 (Fig. 2 B and C). These results suggest that KAT5 functions at the cGAS level. Transient transfection and coimmunoprecipitation experiments indicated that cGAS interacted with KAT5 (Fig. 2D). Confocal microscopy showed that cGAS and KAT5 partially colocalized in the cytoplasm (SI Appendix, Fig. S2A). In vitro GST-pulldown assays further confirmed that cGAS directly interacted with KAT5 (Fig. 2E). Endogenous coimmunoprecipitation experiments indicated that KAT5 was constitutively associated with cGAS in resting cells, and viral infection further enhanced their association at 2 and 4 h post infection (Fig. 2F). Domain mapping experiments indicated that KAT5 interacted with all of the examined cGAS truncations, but the middle fragment aa 212 to 382 of cGAS could mediate stronger interaction with KAT5. Similar experiments indicated that cGAS interacted with the MYST domain of KAT5 (aa 262 to 471) (SI Appendix, Fig. S2B). These results suggest that KAT5 directly targets cGAS.
Fig. 2.
KAT5 functions at the cGAS level. (A) Effects of KAT5 knockdown on cGAMP synthesis induced by HSV-1 infection. The KAT5-knockdown and control THP-1 cells (1 × 108) were left uninfected or infected with HSV-1 (MOI = 2) for 3 h, and then the cell extracts containing cGAMP were separated by chromatography using a C18 column. The abundance of cGAMP was quantitated by mass spectrometry. Quantification of cGAMP was shown in the histograph (Right). **P < 0.01 (unpaired t test). Data shown are mean ± SD of three technical repeats. (B) Effects of KAT5 knockdown on cGAMP-induced transcription of downstream genes. The KAT5-knockdown and control THP-1 cells were left untreated or treated with 2′3′-cGAMP (40 nM) for the indicated times before qPCR analysis. ns, no significance (unpaired t test). Data shown are mean ± SD from one representative experiment performed in triplicate. (C) Effects of KAT5 knockdown on cGAMP-induced phosphorylation of TBK1-S172 and IRF3-S396. The KAT5-knockdown and control THP-1 cells were left untreated or treated with 2′3′-cGAMP (40 nM) for the indicated times before immunoblotting analysis with the indicated antibodies. (D) Interaction of KAT5 and cGAS in mammalian overexpression system. HEK293 cells were transfected with the indicated plasmids for 20 h. Cell lysates were immunoprecipitated with control IgG or anti-HA. The lysates and immunoprecipitates were subjected to immunoblotting analysis with the indicated antibodies. (E) Interaction of KAT5 and cGAS in vitro. GST, GST-cGAS, and His-KAT5 proteins were purified from Escherichia coli. Recombinant GST or GST-cGAS was mixed with His-KAT5. The GST or GST-cGAS protein was pulled down with glutathione Sepharose beads and then analyzed by immunoblotting with the indicated antibodies. (F) Association of endogenous KAT5 with cGAS. The mouse lung fibroblasts were left uninfected or infected with HSV-1 (MOI = 1) for the indicted times before cells were harvested for immunoprecipitation with control IgG or anti-KAT5. The lysates and immunoprecipitates were subjected to immunoblotting analysis with the indicated antibodies.
It has been reported that simultaneous mutation of Q377 and G380 of KAT5 to glutamate (E) impairs its acetyltransferase activity (24). In reporter assays, wild-type KAT5 but not KAT5(QG/EE), in which Q377 and G380 are mutated to glutamate, potentiated cGAS-mediated activation of ISRE (SI Appendix, Fig. S2C). Furthermore, MG149, a potent and selective inhibitor of KAT5 enzymatic activity (25), markedly inhibited HSV-1–induced transcription of downstream effector genes (SI Appendix, Fig. S2D). These results suggest that KAT5 promotes a cGAS-mediated innate immune response in an acetyltransferase-activity–dependent manner.
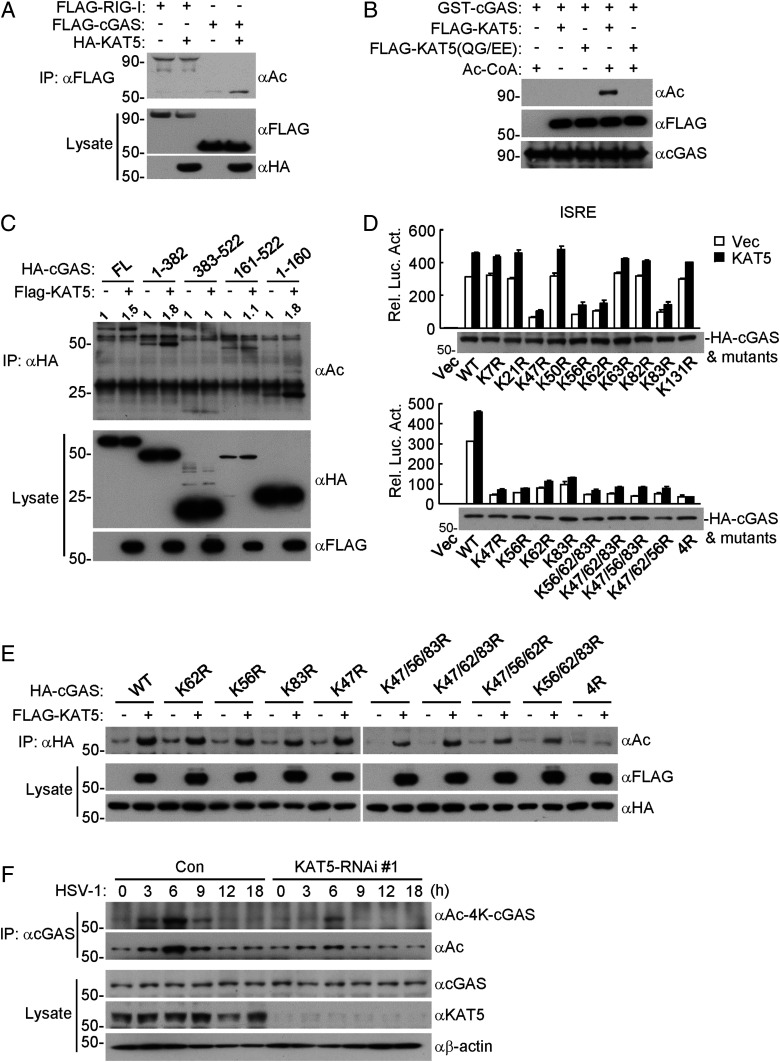
We next examined whether KAT5 could acetylate cGAS. Overexpression of KAT5 increased acetylation of cGAS, but not acetylation of RIG-I (Fig. 3A). In vitro acetylation assays showed that KAT5 but not KAT5(QG/EE) catalyzed acetylation of cGAS (Fig. 3B). Further experiments indicated that KAT5 increased the acetylation of cGAS truncation mutants containing the N-terminal 160 aa but not those lacking the N-terminal 160 aa. Notably, both the N terminus and C terminus of cGAS were basally acetylated in non-KAT5– overexpressing cells (Fig. 3C).
Fig. 3.
KAT5 mediates acetylation of the N-terminal domain of cGAS. (A) Overexpression of KAT5 increased acetylation of cGAS but not RIG-I. HEK293 cells were transfected with the indicated plasmids for 20 h, and then the cells were treated with the deacetylase inhibitors TSA and NAM for 8 h before coimmunoprecipitation and immunoblotting analysis were performed with the indicated antibodies. (B) Effects of KAT5 and its mutant on acetylation of cGAS in vitro. GST-cGAS was purified from E. coli. FLAG-KAT5 and FLAG-KAT5(QG/EE) were purified from HEK293. The indicated purified proteins were incubated for 2 h, followed by immunoblotting analysis with the indicated antibodies. (C) Effects of KAT5 on acetylation of cGAS and its truncated mutants. The experiments were similarly performed as in A. (D) Effects of KAT5 on ISRE activation mediated by cGAS and its mutants. The HEK293 cells stably expressing MITA were transfected with the indicated plasmids for 24 h before luciferase assays were performed. (E) Effects of KAT5 on acetylation of cGAS and its N-terminal lysine mutants. The experiments were similarly performed as in A. (F) Effects of KAT5 knockdown on HSV-1–induced acetylation of cGAS. The KAT5-knockdown or control THP-1 cell lines were infected with HSV-1 (MOI = 2) for the indicated times before coimmunoprecipitation and immunoblotting analysis were performed with the indicated antibodies.
To determine which lysine residues of cGAS are targeted by KAT5, we transfected cGAS and KAT5 into HEK293 cells and immunoprecipitated cGAS for mass spectrometry analysis. These experiments identified seven acetylation sites at the N terminus of cGAS, including K21, K47, K50, K62, K63, K82, and K83 (SI Appendix, Table S1). We mutated all of the lysine residues at the N terminus of cGAS individually or in different combinations and then examined the abilities of these mutants to activate ISRE in reporter assays as well as their acetylation by KAT5. As shown in Fig. 3D, mutation of K47, K56, K62, or K83 to arginine reduced the ability of cGAS to activate ISRE, whereas simultaneous mutation of all four of these lysine residues of cGAS to arginine residues (4R) further impaired cGAS's ability to activate ISRE and to be potentiated by KAT5 for ISRE activation (Fig. 3D). Furthermore, acetylation assays indicated that mutation of each of the four lysine residues of cGAS had minimal effects on its acetylation by KAT5, whereas simultaneous mutation of all four lysine residues abolished its acetylation by KAT5 (Fig. 3E). These results suggest that KAT5-mediated acetylation of K47, K56, K62, and K83 of cGAS is important for its functions in innate immune response.
To further explore acetylation of endogenous cGAS at these lysine residues, we generated site-specific antibodies by using synthetic peptides that bear acetylation modifications at K47, K56, K62, and K83 of human cGAS (SI Appendix, Fig. S3 A and B). As shown in Fig. 3F, cGAS acetylation increased with the specific antibody as well as with a pan-acetylation antibody after HSV-1 infection for 3 to 9 h, which was markedly reduced in KAT5-knockdown cells. Recently, cGAS has been reported to be acetylated at K384, K394, and K414 by unknown acetyltransferases for its inactivation in resting cells, and viral infection or DNA stimulation triggers deacetylation of cGAS at these residues, which was also observed in our experiments (SI Appendix, Fig. S3C). Knockdown of KAT5 had no marked effects on acetylation of cGAS at K384, K394, and K414 (SI Appendix, Fig. S3C), indicating that acetylation of cGAS at these residues is catalyzed by other acetyltransferases but not KAT5. These results suggest that KAT5 catalyzes acetylation of cGAS at multiple residues including K47, K56, K62, and K83 in the N-terminal domain of cGAS.
Acetylation of cGAS by KAT5 Promotes Its DNA-Binding Activity.
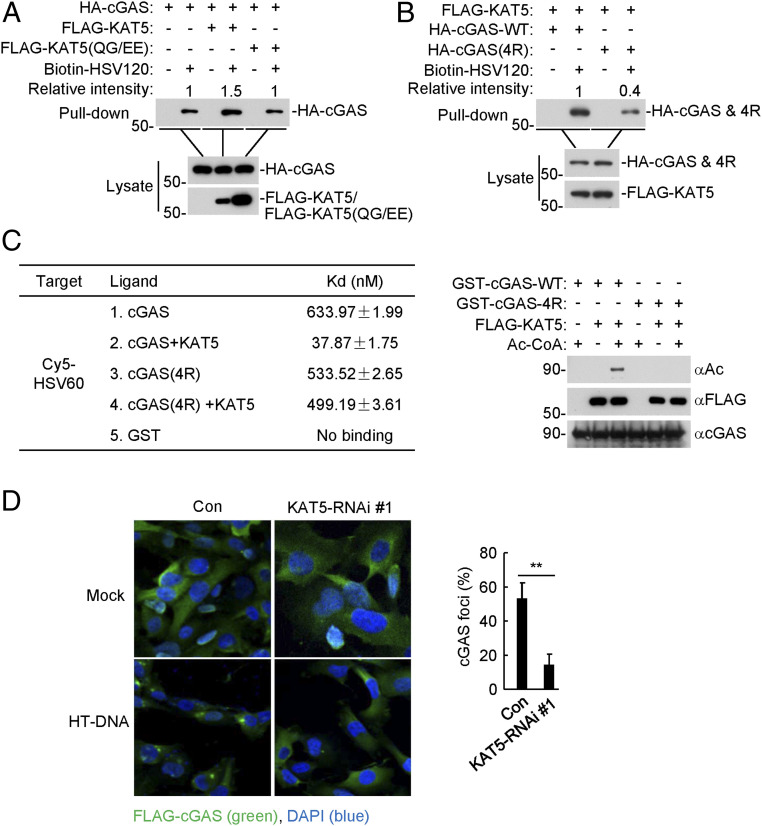
We next investigated how KAT5-mediated acetylation of cGAS regulates its activity. Overexpression of KAT5 but not KAT5(QG/EE) increased the binding of cGAS to the DNA HSV120, which is a synthetic 120-bp dsDNA representing the genome of HSV-1 (Fig. 4A). Consistently, cGAS(4R) had markedly reduced ability in binding to HSV120 in comparison to wild-type cGAS (Fig. 4B). We next examined the binding affinities of recombinant cGAS and cGAS(4R) to dsDNA. Recombinant GST-cGAS or GST-cGAS(4R) was acetylated by KAT5 in vitro and then mixed with the synthetic Cy5-labeled HSV60 DNA to measure their affinities by microscale thermophoresis technology (MST). As shown in Fig. 4C, the KAT5-acetylated cGAS bound to Cy5-HSV60 with an affinity of Kd = 37.87 ± 1.75 nM, which was ∼17-fold higher than that of unacetylated cGAS with HSV60 (Kd = 633.97 ± 1.99 nM). The DNA-binding affinity of cGAS(4R) treated with KAT5 or left untreated was comparable (Fig. 4C). It has been reported that binding of cGAS to DNA triggers liquid phase separation of cGAS-DNA and formation of cGAS foci in the cytoplasm (7). As shown in Fig. 4D, the ratio of cells containing cGAS foci was significantly reduced in KAT5-knockdown cells (Fig. 4D). These results suggest that KAT5-mediated acetylation of cGAS promotes its DNA-binding ability and activity.
Fig. 4.
Acetylation of cGAS by KAT5 increases its DNA-binding affinity. (A) Effects of KAT5 and KAT5(QG/EE) on the binding of cGAS to dsDNA. HEK293 cells were transfected with the indicated plasmids for 20 h. The cell lysates were then incubated with biotinylated HSV120 and streptavidin-Sepharose beads. The bead-bound proteins were analyzed by immunoblotting with the indicated antibodies. (B) Binding of dsDNA by cGAS and cGAS(4R). The experiments were similarly performed as in A. (C) Binding affinities of acetylated or unacetylated cGAS to dsDNA. GST-cGAS and GST-cGAS(4R) were purified from E. coli. FLAG-KAT5 was purified from HEK293. GST-cGAS or GST-cGAS(4R) were subjected to acetylation reaction with FLAG-KAT5. The reactions were then mixed with Cy5-labeled HSV60 before MST measurements. Aliquots of the reactions were also analyzed by immunoblotting with the indicated antibodies. (D) Effects of KAT5 knockdown on dsDNA-induced cGAS aggregation. The KAT5-knockdown and control HT1080-FLAG-cGAS cell lines were transfected with HT-DNA for 5 h before immunofluorescent analysis of cGAS (green) (Left). A total of more than 180 cells from each sample were quantified for cGAS foci (Right). **P < 0.01 (unpaired t test). Data shown are representative of three experiments with similar results.
It has been proposed that mutation of an acetylation targeting lysine residue to glutamine (Q) in a protein mimics its acetylation state. We mutated the four N-terminal lysine residues of cGAS to glutamine, cGAS(4Q), and examined its activity. Pull-down assays indicated that the binding ability of cGAS(4Q) to HSV120 DNA was higher than wild-type cGAS (SI Appendix, Fig. S4A). MST assays indicated that cGAS(4Q) bound to HSV60 DNA with an affinity of Kd = 118.24 ± 10.47 nM, which was approximately six-fold higher than that of wild-type cGAS (Kd = 714.01 ± 17.52 nM) or cGAS(4R) (Kd = 721.41 ± 46.25 nM) (SI Appendix, Fig. S4B). In addition, DNA-induced formation of foci increased for cGAS(4Q), whereas it decreased for cGAS(4R) in comparison to wild-type cGAS (SI Appendix, Fig. S4C). To further evaluate the function of cGAS-4K acetylation, KAT5 was depleted by retroviral-mediated transduction of KAT5-RNAi in HT1080 cells stably expressing wild-type cGAS, cGAS(4Q), or cGAS(4R). The cells were then infected with HSV-1 for 9 h or left uninfected before qPCR analysis. The results indicated that cGAS(4Q) had increased ability in mediating HSV-1–induced transcription of downstream genes, while cGAS(4R) had reduced ability in comparison to wild-type cGAS (SI Appendix, Fig. S4D).
KAT5 Is Important for cGAS-Mediated Antiviral Response In Vivo.
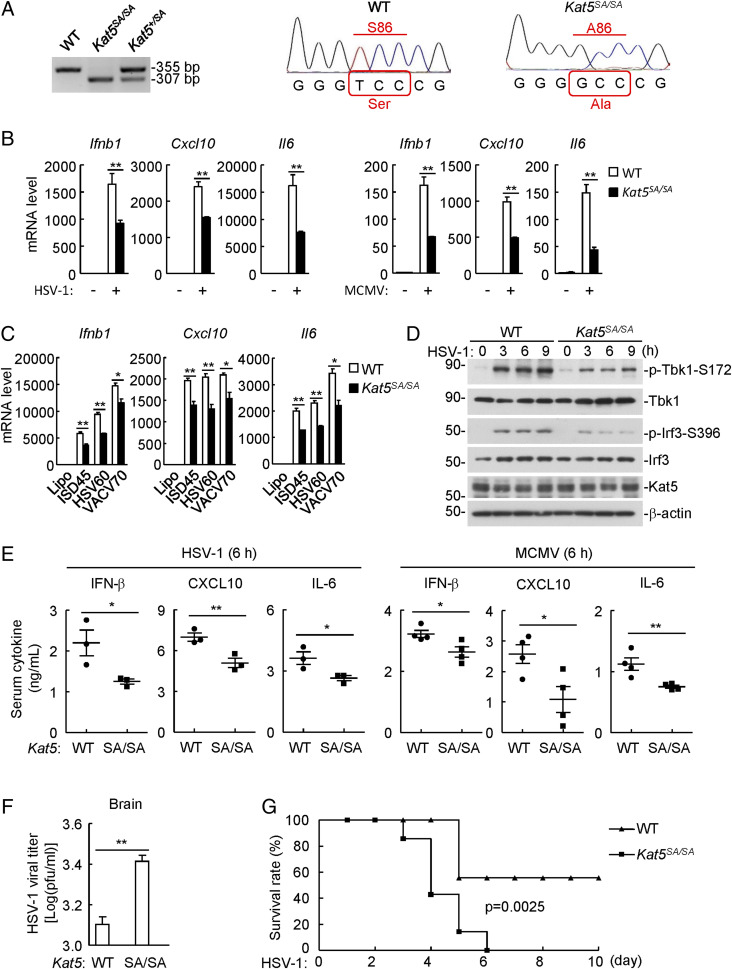
To investigate the roles of KAT5 in cGAS-mediated antiviral response in vivo, we obtained a knock-in mouse strain in which the wild-type Kat5 allele is replaced by the Kat5-S86A mutant (Ser86 altered to alanine), and thus the expressed KAT5S86A protein could not be phosphorylated by glycogen synthase kinase 3 (GSK3), leading to attenuation of KAT5 acetyltransferase activity (23, 26) (SI Appendix, Fig. S5A). The genotypes of mice were confirmed by PCR analysis and sequencing of genomic DNAs from wild-type and Kat5SA/SA mice (Fig. 5A). qPCR analysis indicated that transcription of the downstream Ifnb1, Cxcl10, and Il6 genes induced by HSV-1 and the DNA virus murine cytomegalovirus (MCMV) or transfected dsDNA was markedly inhibited in Kat5SA/SA bone-marrow–derived macrophages (BMDMs) (Fig. 5 B and C) and bone-marrow–derived dendritic cells (SI Appendix, Fig. S5 B and C) compared to their wild-type counterparts. In these experiments, transcription of the downstream Ifnb1, Cxcl10, and Il6 genes induced by Sendai virus (SeV) and vesicular stomatitis virus was comparable between Kat5SA/SA and wild-type BMDMs (SI Appendix, Fig. S5D). Consistently, phosphorylation of TBK1 S172 and IRF3 S396 induced by HSV-1 was markedly inhibited in Kat5SA/SA BMDMs (Fig. 5D). Secretion of IFN-β, CXCL10, and IL-6 was significantly inhibited in Kat5SA/SA BMDMs following HSV-1 or MCMV infection (SI Appendix, Fig. S5E). HSV-1–induced cGAMP production was reduced in Kat5SA/SA BMDMs (SI Appendix, Fig. S5F), but cGAMP-induced transcription of downstream antiviral genes and phosphorylation of Tbk1 S172 and Irf3 S396 were comparable in Kat5SA/SA and wild-type BMDMs (SI Appendix, Fig. S5 G and H). Collectively, these results suggest that KAT5 potentiates cGAS-mediated antiviral response in mouse immune cells.
Fig. 5.
KAT5 potentiates viral DNA-triggered innate immune response in vivo. (A) Genotyping of Kat5-mutated mice. The genomic DNA was extracted from wild-type, Kat5+/SA, or Kat5SA/SA knock-in mice and analyzed by PCR and DNA sequencing. (B) Inhibition of DNA-virus–induced transcription of downstream antiviral genes in Kat5SA/SA BMDMs. Wild-type and Kat5SA/SA BMDMs were left uninfected or infected with HSV-1 or MCMV for 6 h before qPCR analysis. **P < 0.01 (unpaired t test). Data shown are mean ± SD from one representative experiment performed in triplicate. (C) Inhibition of transfected DNA-induced transcription of downstream antiviral genes in Kat5SA/SA BMDMs. Wild-type and Kat5SA/SA BMDMs were transfected with the indicated dsDNA for 4 h before qPCR analysis. *P < 0.05, **P < 0.01 (unpaired t test). Data shown are mean ± SD from one representative experiment performed in triplicate. (D) Inhibition of DNA-virus–induced phosphorylation of Tbk1-S172 and Irf3-S396 in Kat5SA/SA BMDMs. Wild-type and Kat5SA/SA BMDMs were left uninfected or infected with HSV-1 for the indicated times before immunoblotting analysis with the indicated antibodies. (E) Serum levels of IFN-β, CXCL10, and IL-6 following DNA virus infection in wild-type and Kat5SA/SA mice. The mice (n = 3 or 4 per strain, 8 wk old, male) were infected with HSV-1 (3 × 107 plaque-forming units [PFU]/mouse) or MCMV (1 × 104 PFU/mouse) for 6 h before enzyme-linked immunosorbent assay. *P < 0.05, **P < 0.01 (unpaired t test). Data shown are mean ± SD from one representative experiment. (F) Viral titers in brain following HSV-1 infection in wild-type and Kat5SA/SA mice. Wild-type and Kat5SA/SA mice (n = 3 per strain, 8 wk old, male) were infected with HSV-1 at 3 × 107 PFU per mouse for 5 d. HSV-1 viral titers in the brains of infected mice were quantified by plaque assays. **P < 0.01 (unpaired t test). Data shown are mean ± SD from one representative experiment. (G) Effects of Kat5 mutation on HSV-1–induced death of mice. Wild-type and Kat5SA/SA mice (n = 7 per strain, 8 wk old, female) were infected intraperitoneally with HSV-1 at 5 × 107 PFU per mouse, and the survival of mice was monitored daily for 10 d. P = 0.0025 (log-rank test).
To evaluate the importance of KAT5 in host defense against viral infection in vivo, the age- and sex-matched wild-type and Kat5SA/SA mice were infected intraperitoneally with HSV-1 or MCMV. As shown in Fig. 5E, HSV-1– and MCMV-induced productions of IFN-β, CXCL10, and IL-6 were severely impaired in the sera of Kat5SA/SA compared to wild-type mice. The viral titers in the brains of Kat5SA/SA mice were markedly higher than those of wild-type mice at 5 d after HSV-1 infection (Fig. 5F). In addition, Kat5SA/SA mice were more susceptible to HSV-1–induced death in comparison to wild-type mice (Fig. 5G). These results suggest that KAT5 plays an important role in host defense against DNA virus in vivo.
Discussion
Posttranslational modifications act as regulatory signals for control of the stability, localization, activity, and function of proteins. As a major DNA sensor in the cytoplasm, cGAS is tightly regulated by posttranslational modifications on its CCD. In this study, we reveal a critical role of the acetyltransferase KAT5 in innate antiviral response by mediating acetylation of the NUD of cGAS.
In an expression screen, KAT5 was identified as a positive regulator of cGAS-mediated signaling. Overexpression of KAT5 promoted cGAS-mediated transcription of downstream antiviral genes, whereas knockdown of KAT5 or treatment of cells with KAT5 specific inhibitor had opposite effects. In vivo experiments indicated that KAT5-inactive mice produced fewer serum cytokines, had higher viral titers in the brain, and were more susceptible to HSV-1–induced death. These results suggest that KAT5 plays an important role in efficient innate immune response to DNA virus.
Several experiments suggest that KAT5 functions by targeting cGAS for acetylation. Knockdown of KAT5 inhibited viral-DNA–induced cGAMP production, but had no marked effects on cGAMP-induced transcription of downstream antiviral genes. KAT5 was constitutively associated with cGAS, although their association increased following viral infection. Furthermore, KAT5 catalyzed acetylation of cGAS in vitro and in vivo. Mass spectrometry and mutagenesis indicated that KAT5 catalyzed cGAS acetylation at multiple lysine residues including K47, K56, K62, and K83 in the NUD. DNA pulldown and MST assays indicated that KAT5-acetylated cGAS or cGAS(4Q) had a higher affinity to DNA. In addition, cGAS(4Q) had an increased, whereas cGAS(4R) had a decreased, ability to mediate HSV-1–triggered activation of ISRE in comparison to wild-type cGAS. These results suggest that KAT5 promotes innate antiviral response to DNA virus by acetylating the N terminus of cGAS, leading to its higher affinity to viral DNA. Previously, it was demonstrated that another DNA sensor, IFI16, is also regulated by acetylation, which results in its cytoplasmic localization and detection of cytoplasmic DNA (27). Whether KAT5 is involved in acetylation of IFI16 or other proteins in innate antiviral response needs to be investigated in future studies. Interestingly, we found that KAT5 could negatively regulate an innate immune response to RNA virus, which will be further investigated in a separate study.
Recently, it has been reported that cGAS is acetylated at multiple lysine residues including K384, K394, and K414 in its C terminus by unknown acetyltransferases in resting cells. Acetylation of these C-terminal lysine residues of cGAS impairs its enzymatic activity, whereas viral infection triggers HDAC3-mediated deacetylation of these residues for its activation to synthesis cGAMP (15). In our experiments, we confirmed these observations by using the acetylated K384/394- and K414-specific antibodies. Interestingly, by using both the pan-acetylation and acetylated 4K-specific antibodies, we noted that the acetylation levels of cGAS increased at 3 to 9 h post HSV-1 infection, which were markedly suppressed in KAT5 knockdown cells. In addition, viral-infection–triggered deacetylation of cGAS at K384/394 and K414 was not markedly affected in KAT5-deficient cells, suggesting that certain unknown acetyltransferases but not KAT5 are responsible for cGAS acetylation on cGAS's C-terminal lysine residues. In our experiments, we observed that cGAS and KAT5 were associated with each other in uninfected cells, although their association increased at 2 h after HSV-1 infection. However, KAT5-mediated acetylation of cGAS N terminus was mostly induced at 6 h after HSV-1 infection, at which time point the acetylation of cGAS C terminus diminished. It is possible that deacetylation of cGAS at its C terminus is a prerequisite step for the acetylation of the cGAS N terminus by KAT5. In light of our and other previous studies, we propose a model for the regulation of innate antiviral response by acetylation and deacetylation of cGAS. In uninfected cells, cGAS is acetylated at K384/394/414 by unknown acetyltransferases to keep cGAS inactive. Upon DNA virus infection, HDAC3 deacetylates cGAS at these residues to unlock the inhibition of cGAS. This enables KAT5 to acetylate cGAS at its N-terminal K47/52/62/83 residues, leading to its increased affinity to viral DNA and efficient induction of innate antiviral response. Since the N-terminal domain of cGAS is unstructured and its structural properties are unknown, it is unclear how the acetylation of cGAS N terminus affects its DNA-binding affinity. Nevertheless, our study reveals an additional regulatory mechanism in the cGAS-mediated innate immune response to DNA virus.
Materials and Methods
All animal experiments were performed in accordance with the Wuhan University Animal Care and Use Committee guidelines. The information on reagents, antibodies, cells, constructs, PCR primers, RNA interference target sequences, mice, and various other methods are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Tian Xia, Wei-Wei Luo, Mi Li, Lu Zhou, Ru Zang, and Xuan Zhong for technical help and Cao-Qi Lei for critical reading of the manuscript. This work was supported by grants from the State Key R&D Program of China (2017YFA0505800, 2016YFA0502102) and by the National Natural Science Foundation of China (31830024, 31630045).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922330117/-/DCSupplemental.
Data Availability Statement.
All study data are included in the article and SI Appendix.
References
- 1.Akira S., Uematsu S., Takeuchi O., Pathogen recognition and innate immunity. Cell 124, 783–801 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Hu M. M., Shu H. B., Cytoplasmic mechanisms of recognition and defense of microbial nucleic acids. Annu. Rev. Cell Dev. Biol. 34, 357–379 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Wu J., Chen Z. J., Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Barbalat R., Ewald S. E., Mouchess M. L., Barton G. M., Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 29, 185–214 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi O., Akira S., Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Sun L., Wu J., Du F., Chen X., Chen Z. J. J., Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du M., Chen Z. J., DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa H., Barber G. N., STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong B. et al., The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Jin L. et al., MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28, 5014–5026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun W. et al., ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia P. et al., Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol. 17, 369–378 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Hu M. M. et al., Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity 45, 555–569 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Chen M. et al., TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol. Cell 64, 105–119 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Dai J. et al., Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell 176, 1447–1460.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao J. et al., Nonspecific DNA binding of cGAS N terminus promotes cGAS activation. J. Immunol. 198, 3627–3636 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Barnett K. C. et al., Phosphoinositide interactions position cGAS at the plasma membrane to ensure efficient distinction between self- and viral DNA. Cell 176, 1432–1446.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y., Jiang X., Chen S., Fernandes N., Price B. D., A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 102, 13182–13187 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z. et al., The KAT5-Acetyl-Histone4-Brd4 axis silences HIV-1 transcription and promotes viral latency. PLoS Pathog. 14, e1007012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin S. Y. et al., GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336, 477–481 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Sykes S. M. et al., Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng H. et al., A posttranslational modification cascade involving p38, Tip60, and PRAK mediates oncogene-induced senescence. Mol. Cell 50, 699–710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T. Y. et al., Tip60-mediated lipin 1 acetylation and ER translocation determine triacylglycerol synthesis rate. Nat. Commun. 9, 1916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikura T. et al., Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Ghizzoni M. et al., 6-alkylsalicylates are selective Tip60 inhibitors and target the acetyl-CoA binding site. Eur. J. Med. Chem. 47, 337–344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brauns-Schubert P. et al., CDK9-mediated phosphorylation controls the interaction of TIP60 with the transcriptional machinery. EMBO Rep. 19, 244–256 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T., Diner B. A., Chen J., Cristea I. M., Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. U.S.A. 109, 10558–10563 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.