Significance
The cancer-dependent metabolic rewiring is mainly associated with the synthesis of building blocks that are needed to fulfill the proliferating cell metabolic requirements. However, the proliferation-independent instructive role of metabolic enzymes in tumor plasticity is still unclear. Here, we introduce glutathione peroxidase 8 (GPX8) as a metabolic enzyme that regulates cancer aggressiveness. We found that lack of GPX8 suppresses the aggressive phenotype and stemness features of the tumor cells. Mechanistically, these cells express a nonfunctional IL-6 receptor, which fails to interact with IL-6. This impaired binding hinders the activation of the downstream JAK/STAT3 signaling pathway, thereby inhibiting cancer cells transition to aggressive phenotypes. Thus, we present this GPX8/IL-6/STAT3 axis as a prototype of metabolic enzymes regulating cancer aggressiveness-associated signaling pathways.
Keywords: cancer metabolism, epithelial–mesenchymal transition, GPX8, JAK/STAT3 signaling
Abstract
One of the emerging hallmarks of cancer illustrates the importance of metabolic reprogramming, necessary to synthesize the building blocks required to fulfill the high demands of rapidly proliferating cells. However, the proliferation-independent instructive role of metabolic enzymes in tumor plasticity is still unclear. Here, we provide evidence that glutathione peroxidase 8 (GPX8), a poorly characterized enzyme that resides in the endoplasmic reticulum, is an essential regulator of tumor aggressiveness. We found that GPX8 expression was induced by the epithelial–mesenchymal transition (EMT) program. Moreover, in breast cancer patients, GPX8 expression significantly correlated with known mesenchymal markers and poor prognosis. Strikingly, GPX8 knockout in mesenchymal-like cells (MDA-MB-231) resulted in an epithelial-like morphology, down-regulation of EMT characteristics, and loss of cancer stemness features. In addition, GPX8 knockout significantly delayed tumor initiation and decreased its growth rate in mice. We found that these GPX8 loss-dependent phenotypes were accompanied by the repression of crucial autocrine factors, in particular, interleukin-6 (IL-6). In these cells, IL-6 bound to the soluble receptor (sIL6R), stimulating the JAK/STAT3 signaling pathway by IL-6 trans-signaling mechanisms, so promoting cancer aggressiveness. We observed that in GPX8 knockout cells, this signaling mechanism was impaired as sIL6R failed to activate the JAK/STAT3 signaling pathway. Altogether, we present the GPX8/IL-6/STAT3 axis as a metabolic-inflammatory pathway that acts as a robust regulator of cancer cell aggressiveness.
During recent decades there is substantial progress in cancer treatment due to a better understanding of tumor biology. However, despite these advances in therapy, the disease can relapse, as it acquires more aggressive traits, including resistance to chemotherapeutic drugs (1). This plasticity is achieved through significant alterations in the tumor physiology, such as its ability to transdifferentiate into a mesenchymal-like state (2), orchestrated by the epithelial–mesenchymal transition (EMT) program (3). The execution of the EMT program induces significant changes in the cellular phenotype as cells acquire chemoresistance, lose their polarity, eliminate their interactions with neighboring cells, and gain invasive properties (4). These shifts in the tumor characteristics provide a model describing how tumors gain the ability to detach from the primary site and promote the metastatic cascade (5).
The EMT program is regulated by a core set of transcription factors (EMT-TFs), which includes Twist family BHLH transcription factor 1 (TWIST1), zinc finger E-box binding homeobox 1 and 2 (ZEB1 and ZEB2), Snail family transcriptional repressor 1 and 2 (SNAI1 and SNAI2 [SLUG]) (6). The EMT-TFs’ mechanism of action is directed by intracellular signaling pathways, including Wnt and Notch or by specific ligands such as chronic transforming growth factor-beta (TGFβ), mitogenic growth factors (1), and inflammatory cytokines, such as IL-6 (7). In recent years, many studies demonstrated a link between the EMT program and cancer stem cells (CSC) (8, 9). For example, induction of the EMT program in epithelial cells results in the expression of stemness markers such as CD44high/CD24low and their ability to form mammospheres (9–11). However, these CSCs are not the outcome of the full execution of the EMT program but instead are at an intermediate state along the epithelial–mesenchymal spectrum (1).
Tumors exhibit a distinct metabolic pattern relative to nonproliferative cells in order to satisfy the metabolic demands of the rapidly proliferating cells (reviewed in refs. 12, 13). This metabolic remolding is mediated via the expression of a unique metabolic gene signature (14). However, cancer-dependent metabolic rewiring is more complicated than initially described (15) as there are metabolic processes essential only for particular tumor types (16, 17). These findings indicate that the cancer-dependent metabolic rewiring is not only limited to support cell proliferation but is also required to satisfy other proliferation-independent cellular needs, such as acquiring traits associated with high-grade malignancy.
To methodically identify the metabolic enzymes regulating tumor dynamics, we generated MERAV, a web-based tool to analyze human gene expression between different cancer types and normal tissues (http://merav.wi.mit.edu/, ref. 18). By analyzing MERAV, we characterized a set of 44 metabolic genes that are selectively present in high-grade tumors bearing mesenchymal markers, which we designated as the “mesenchymal metabolic signature” (MMS) (11). To systematically determine the role of MMS in tumor progression, we developed a fluorescence-activated cell sorting (FACS)-based shRNA screen that identified 16 genes as essential for the EMT program (11). Among them is dihydropyrimidine dehydrogenase (DPYD), the rate-limiting enzyme of the pyrimidine degradation pathway (19), whose activity has been demonstrated to be vital for the proper execution of the EMT program (11). Having verified the essential role of DPYD in the EMT, we were intrigued as to whether the second hit in the screen, glutathione peroxidase 8 (GPX8), is also critical for tumor aggressiveness.
The primary function of the glutathione peroxidase (GPx) family of proteins is to limit the cellular accumulation of the reactive oxygen species (ROS) (20). These enzymes use glutathione (GSH) as a reducing agent (21) to catabolize peroxides to the corresponding alcohols. The specific activity of GPx is determined by its particular amino acid composition, as most members contain the nonstandard amino acid selenocysteine in their active site (GPX1-4 and GPX6), whereas the others (GPX5, GPX7-8) have a cysteine (21). GPX8, the last member of this family to be identified (21), is a type II transmembrane protein with high sequence similarity to the soluble GPX7 (NPGPx). These two enzymes share many characteristics, as they contain a KDEL-like endoplasmic reticulum (ER) retrieval motif and are localized in the ER (22). Despite their name and their similarity to the other members of the GPx family, both GPX7 and GPX8 demonstrate low GPx activity (22), as they lack the GSH-binding domain (23). Thus, the proposed function of both GPX7 and GPX8 is associated with protein disulfide isomerase (PDI) peroxide-mediated oxidative protein folding (22). Specifically, they bind and clear the peroxides generated in the ER by the endoplasmic reticulum oxidoreductase 1 Alpha (ERO1α) enzyme, which introduces disulfide bonds into PDI (24). The physiological function of GPX8 is still unclear; however, it has been reported to be involved in diverse physiological processes. For example, GPX8 protects against colitis (25), serves as a cellular substrate to the hepatitis C virus NS3-4A protease (26), induces ER stress in rat pancreatic β-cells (27), and regulates calcium flux in HeLa cells (28).
Our knowledge about GPX8 regulation is still limited, although its expression was found to be regulated by hypoxia-inducible factor (HIF1α) (29) and repressed by insulin-like growth factor 1 receptor (IGF1R) in the lung (30). These studies are only starting to reveal some of the physiological functions of GPX8, but its role in tumor biology is still unclear. Here, we report that GPX8 robustly maintains cancer cells at their aggressive state via regulation of the IL-6/JAK/STAT3 signaling pathway.
Results
GPX8 Expression Is Up-Regulated during the EMT Program.
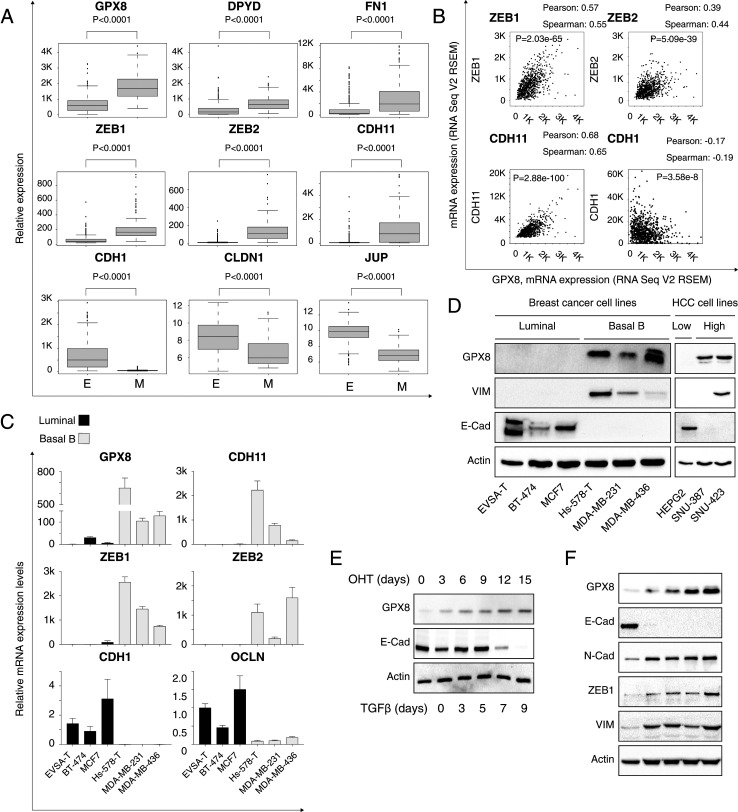
Previous unsupervised hierarchical clustering analysis of cancer cell lines’ metabolic gene expression profile generated by the MERAV web portal (http://merav.wi.mit.edu/) (18) classified five distinct groups (11). These include the epithelial group comprising 378 cell lines originating from the epithelial tissues and the mesenchymal group consisting of 150 cell lines expressing a shared mesenchymal signature. We compared the expression profile between these two groups and identified significant up-regulation of GPX8 in the mesenchymal group. We found this GPX8 expression pattern to be similar to that of other known mesenchymal markers such as DPYD (11), fibronectin (FN1) (10), ZEB1 (31), ZEB2 (32), and cadherin 11 (CDH11) (33) in contrast to the epithelial markers cadherin 1 (CDH1 [E-cadherin]) (31), claudin (CLDN1) (10), and junction plakoglobin (JUP [ɣ-catenin]) (Fig. 1A).
Fig. 1.
GPX8 expression is up-regulated in mesenchymal-like cells. (A) Elevated GPX8 gene expression in mesenchymal cell lines. Cancer cell lines were divided into epithelial (n = 378 cell lines) and mesenchymal (n = 150 cell lines) groups based on the expression of known mesenchymal markers. Box plots represent the expression levels of the indicated genes in each group. The P value was determined by Student’s t test. (B) GPX8 expression correlates with mesenchymal markers. Patients’ gene expression data were generated by the TCGA project and analyzed using the cBioportal web tool (https://www.cbioportal.org). GPX8 expression positively and significantly correlated with known mesenchymal markers (ZEB1, ZEB2, and CDH11) and negatively with the epithelial marker CDH1. The Pearson, Spearman correlation, and the P value, were calculated by the analysis tool. (C) GPX8 mRNA level is up-regulated in basal B breast cancer cells. The relative level of GPX8, as well as other indicated EMT markers in breast cancer cell lines, were determined by qPCR. The expression level of all cell lines is relative to that of the EVSA-T cell line. Each value represents the mean ± SD for n = 3. (D) GPX8 protein levels are up-regulated in high-grade breast and HCC cancer cell lines. Cells were lysed and subjected to immunoblotting using the indicated antibodies. (E) GPX8 expression is up-regulated during the EMT program. HMLE-Twist-ER cells were treated with OHT to induce EMT for a total of 15 d. Every 3 d, cells were collected, lysed, and subjected to immunoblotting using the indicated antibodies. (F) TGFβ1 induces GPX8 expression. A549 cells were treated with 5 ng/mL TGFβ1 to induce EMT for a total of 9 d. On each indicated day, cells were collected and subjected to immunoblotting using the indicated antibodies.
We then wanted to determine whether the GPX8 expression pattern in patient-derived samples is similar to that seen in cancer cell lines. To this end, we analyzed the GPX8 expression profile in breast cancer samples derived from the cancer genome atlas (TCGA) project (34) available on the cBioportal web-based tool [https://www.cbioportal.org (34)]. Specifically, we correlated GPX8 expression to every gene in the genome and subjected the obtained Spearman’s rank correlation coefficients to gene set enrichment analysis (GSEA) (35, 36) which revealed a significantly high correlation with the EMT markers (“hallmark epithelial–mesenchymal transition” [SI Appendix, Fig. S1A]). This analysis was then individually validated by the mesenchymal markers ZEB1, ZEB2, and CDH11 which showed a significantly high positive correlation with GPX8 expression (Spearman’s rank correlation coefficients = 0.55, 0.44, and 0.65, respectively) and a negative correlation with the epithelial markers CDH1 (Spearman’s rank correlation coefficients = −0.19) (Fig. 1B). Together, we systematically confirmed that GPX8 expression correlates with the more aggressive mesenchymal-like characteristics of cancer cell lines and breast cancer patients.
To validate these bioinformatics results, we determined GPX8 expression at both messenger RNA (mRNA) and protein levels in different cancer cell lines. We confirmed elevated GPX8 expression in basal B (mesenchymal-like) relative to luminal (epithelial)-derived cancer cell lines (Fig. 1 C and D). This GPX8 expression pattern is similar to other known mesenchymal markers (CDH11, ZEB1, VIM, and ZEB2) and different to epithelial markers (E-Cad [E-cadherin] [CDH1] and epithelial cell junction protein Occludin [OCLN]) (37) (Fig. 1 C and D). We also found up-regulation of GPX8 expression in high-grade hepatocellular carcinoma cell lines (HCC) (SNU-387 and SNU-423) (38) relative to the low-grade cell line (HepG2) (Fig. 1D). Additionally, GPX8 expression in melanoma was up-regulated in the highly metastatic, mesenchymal-like cell line (A375-MA2) (39) as compared to its less aggressive parental cell line (A375) (SI Appendix, Fig. S1B). Altogether, these results confirm relatively high GPX8 expression in mesenchymal-like cancer samples, indicating its biological role in cancer cell aggressiveness.
The elevated GPX8 expression in mesenchymal-like cells indicates that the EMT program regulates its expression. To investigate this further, we exploited the engineered human mammary epithelial (HMLE) cells expressing Twist conjugated to the estrogen receptor. Upon 4-hydroxytamoxifen (OHT) treatment, this ectopically expressed Twist translocates to the nucleus and gradually induces the EMT-promoting factors (9). We found that OHT treatment stimulated GPX8 expression and simultaneously down-regulated the epithelial marker E-Cad (Fig. 1E). In addition, GPX8 expression was up-regulated in the lung carcinoma cell line A549 treated with transforming growth factor-beta 1 (TGFβ1) for 9 d (40) (Fig. 1F). This treatment induced the expression of other mesenchymal markers (N-Cad [N-cadherin], ZEB1, and vimentin [VIM]) but repressed the epithelial marker (E-Cad). Together, these results confirm that the EMT program regulates GPX8 expression, verifying its biological role in cancer cell aggressiveness.
GPX8 Expression Is Associated with Poor Patient Prognosis.
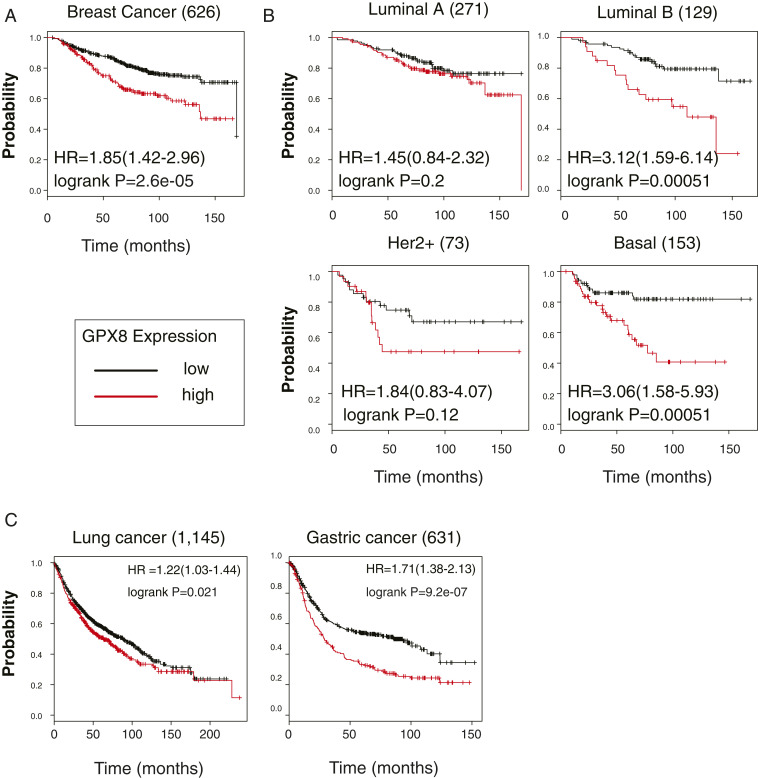
To further assess GPX8 role in tumor aggressiveness, we analyzed whether GPX8 up-regulation is associated with poor patient outcomes. By utilizing the Kaplan–Meier (KM) Plotter tool (http://kmplot.com/analysis/) (41), we found that high GPX8 expression was associated with reduced breast cancer patient overall survival (OS) (Fig. 2A), distance metastasis-free survival (DMFS), and relapse-free survival (RFS) (41) (SI Appendix, Fig. S2A). Moreover, when we classified the breast cancer samples to tumor subtypes according to their aggressiveness, we found that high GPX8 expression level correlates with poor patient outcomes. For example, the effect of high GPX8 levels on the OS of patients with the less aggressive breast cancer subtype (luminal A) was insignificant (P = 0.2), in contrast to the effect on those with the aggressive breast cancer subtype (basal) where it was significant (P = 0.00051) (Fig. 2B). Similarly, high GPX8 expression significantly affected the DMS and RFS in basal breast cancer samples (P = 0.0049 and 0.00059, respectively) relative to luminal A (P = 0.37 and 0.052, respectively) (SI Appendix, Fig. S2B). This correlation between GPX8 expression level and patient outcome was not restricted to breast cancer, as it was also found in lung and gastric cancers (Fig. 2C). To reinforce our findings regarding the direct role of GPX8 in cancer aggressiveness, we demonstrate that other mesenchymal markers (ZEB1, DPYD, and VIM) did not show any significant effect on patient outcome (SI Appendix, Fig. S2C). Therefore, our findings emphasize that association with the poor patient outcome is a unique feature of GPX8 expression and not a common characteristic of the EMT markers. Collectively, patient outcome data support the clinical significance of GPX8 function in aggressive cancer subtypes.
Fig. 2.
GPX8 expression is associated with poor patient prognosis. (A) KM survival plots for patients with breast cancer divided into high GPX8 expression (“high” [red]) and low (“low” [black]). The columns represent overall survival data of all breast cancers (Breast Cancer). Number in parentheses indicates the total number of patients. These plots were generated in the KM plotter website. The GPX8 228141_at Affymetrix ID symbol was used for all of the analyses. The P value (P), the hazard ratio (HR), and the number at risk were determined by the analysis tool. (B) GPX8 expression effects on patient overall survival are more profound in the aggressive breast cancers subtypes. KM plots represent the overall survival data from breast cancers; Luminal A, Luminal B, Her2+, and Basal subtypes. (C) Overall survival data of GPX8 expression levels in lung (Lung Cancer) and gastric cancers (Gastric Cancer).
GPX8 Loss in MDA-MB-231 Induces Epithelial-Like Phenotype.
We wanted to explore GPX8’s function in cancer cell aggressiveness. To this end, we knocked out GPX8 in the mesenchymal-like basal B breast cancer cell line MDA-MB-231, by applying the CRISPR-Cas9 gene knockout system. GPX8 knockout (KO) in two different clones (GPX8-KO-1 and GPX8-KO-2) resulted in significant changes in cell morphology relative to wild-type (WT) cells. These KO cells became smaller, rounder, and clustered into an island-like morphology (Fig. 3A), phenocopying epithelial cell characteristics. Moreover, despite the differences in their DNA deletion pattern (SI Appendix, Fig. S3A), both clones lost GPX8 expression (Fig. 3B) without affecting cell proliferation (SI Appendix, Fig. S3B). To eliminate the possibility of off-target effects, we restored GPX8 expression in GPX8-KO-1 background by ectopically expressing GPX8 (GPX8-KO-1+GPX8-OE). This rescue GPX8 construct is mutated in its PAM and guide recognition sites but translates the same amino acid sequence. We found that GPX8 overexpression rescued the cellular morphology changes induced by GPX8 loss (Fig. 3 A and B), thus supporting GPX8’s role in maintaining the cells’ mesenchymal morphology.
Fig. 3.
GPX8 loss results in epithelial-like characteristics. (A) Silencing of GPX8 expression in MDA-MB-231 cells, using the CRISPR-Cas9 system, induces epithelial-like morphology. (Scale bar, 100 µm.) (B) GPX8 protein levels in the different clones. Cells were separated into single clones, and for each, GPX8 levels were measured by immunoblot using a specific antibody against GPX8. GPX8-KO-1+GPX8-OE: GPX8 was reintroduced in the background of GPX8-KO-1. (C) GPX8 loss leads to reduced gene expression of EMT markers. MDA-MB-231 WT cells and GPX8-KO-1 were subjected to RNA-Seq analysis. The expression ratio between all genes (∼22,000) was calculated and ranked based on the relative expression between the GPX8 WT and KO. The samples were subjected to GSEA. GSEA computed FDR q-value. (D) KO of GPX8 in MDA-MB-231 cells reduces the expression of known mesenchymal markers. The RNA was isolated from the different clones as described above (A) and subjected to qPCR analysis. Each value represents the mean ± SD for n = 9. (E) GPX8 KO inhibits the EMT program. HMLE-Twist-ER cells were infected with the indicated hairpins. The cells were either left untreated or treated with OHT for 15 d, followed by FACS analysis of the cell-surface markers CD24 and CD44 to separate the epithelial and mesenchymal populations. The percentage of cells in each gate is presented. Each value represents the mean ± SD for n = 3.
Next, we subjected WT and GPX8-KO-1 cells to RNA-Seq analysis (Dataset S1) to systematically determine the differential gene expression profile (SI Appendix, Fig. S3C). GSEA confirmed that GPX8 loss caused a significant reduction in “the hallmarks of the epithelial–mesenchymal transition” gene set (false discovery rate [FDR] q-value < 0.0001; Fig. 3C). Specifically, GPX8-KO-1 resulted in the down-regulation of the known mesenchymal markers (FN1, SNAI2 [SLUG], and CDH11) along with the up-regulation of the epithelial marker OCLN (Fig. 3D). This induction of the mesenchymal–epithelial transition (MET) program by GPX8 loss was inhibited by reintroducing GPX8, implying that this gene has a role as a guardian of the mesenchymal state.
Next, we determined whether GPX8 plays a critical role in the proper execution of the EMT program. Thus, we induced this program in HMLE-Twist-ER cells and found that as opposed to control cells (shGFP), where OHT shifted the cell-surface markers from an epithelial (CD24high/CD44low) to a mesenchymal (CD24low/CD44high) profile (42), GPX8-silenced cells (SI Appendix, Fig. S3D) maintained their epithelial profile (Fig. 3E). In addition, knocking down GPX8 or DPYD (another MMS gene) resulted in high expression of the epithelial marker CDH1 and low expression of the mesenchymal markers ZEB1 and CDH2 (SI Appendix, Fig. S3D). Collectively, these findings indicate that GPX8 is a critical component of the EMT program and an essential factor in maintaining the cell’s mesenchymal properties.
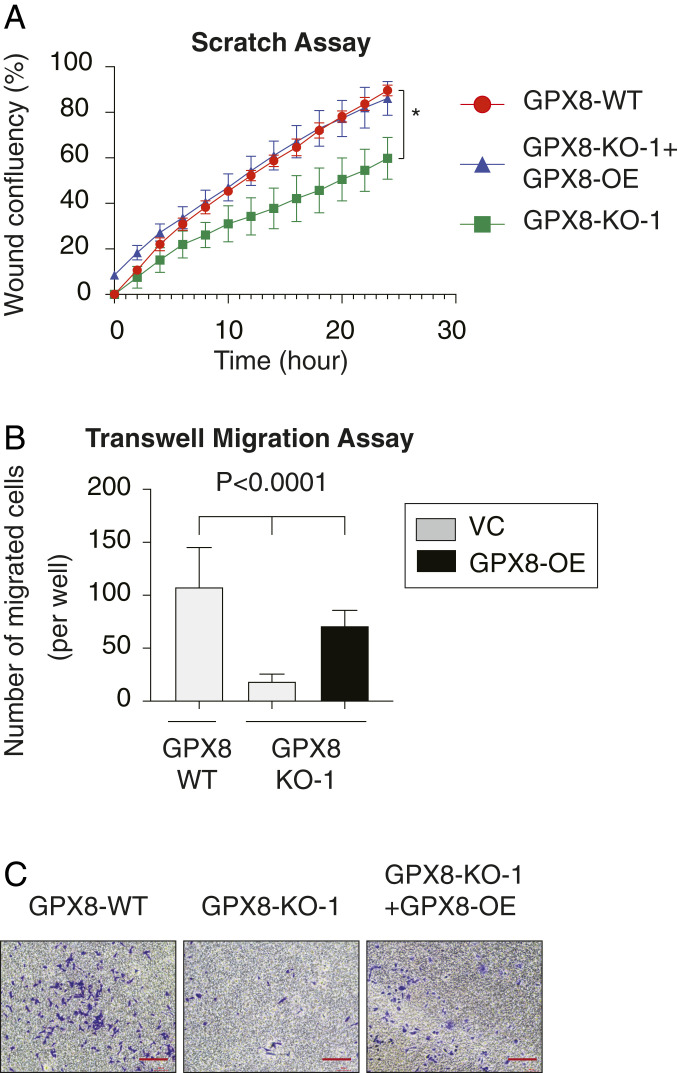
As an in vitro functional readout for the EMT program, we determined the consequence of GPX8 loss on the migratory capabilities of MDA-MB-231 cells using two methods, the Incucyte Live-Cell Analysis System to monitor in real time the rate of cell migration in a scratch assay (SI Appendix, Fig. S4A) and the determination of the number of migrating cells in Boyden chamber with the transwell migration assay (SI Appendix, Fig. S4B). We found that GPX8 loss in two different colonies resulted in a significant reduction in migration efficiency (SI Appendix, Fig. S4 A and B). Furthermore, restoring GPX8 expression levels in clone-1(GPX8-KO-1+GPX8-OE) rescued these observed effects in both the scratch assay and the transwell assay (Fig. 4 A and B). These results demonstrate that GPX8 has a vital role in maintaining cellular mesenchymal features, such as the ability to migrate.
Fig. 4.
GPX8 loss inhibits cell migration in the MDA-MB-231 breast cancer cell line. (A) Quantification of scratch confluence during 24 h for the indicated WT, GPX8-KO-1, and GPX8-KO-1+ GPX8-OE cells. Each value represents the mean ± SD for n = 3. The P value = 0.01 (*) was determined by Student’s t test. (B) GPX8 loss inhibits the MDA-MB-231 migration capabilities. The migration capability of the different samples was determined in a transwell assay. The data are reported as the number of migrated cells per 10,000 seeded cells; each value represents the mean ± SD for n = 6. The P value was determined by Student’s t test. (C) Representative cell migration images of each sample. (Scale bar, 200 µm.)
GPX8 Regulates the Stemness Properties of Cancer Cells.
Previous studies connected the EMT program and the elevation of CSC markers (9, 43). Indeed, several of the basal B breast cancer cell lines, including MDA-MB-231, have stem-like properties as they express specific markers and can initiate tumors in mice (44, 45). We identified that in addition to the EMT markers, GPX8 loss resulted in the down-regulation of the CSC markers such as CD44 (42) (Fig. 5A), [NGFR, p75NTR (46)] (SI Appendix, Fig. S5A), and integrin-β4 (ITGB4) (47) (Fig. 5B and SI Appendix, Fig. S5B). To further determine the role of GPX8 as a regulator of the stemness state, we ectopically expressed GPX8 (GPX8-OE) both in GPX8-KO-1 cells and in the basal epithelial breast cancer cell line MDA-MB-468 (48), in which GPX8 expression is relatively low (SI Appendix, Fig. S5C). We found that GPX8-OE in GPX8-KO-1 cells rescued both CD44 and ITGB4 expression levels (Fig. 5 A and B). In addition, introducing GPX8 in MDA-MB-468 resulted in CD44 up-regulation (Fig. 5C), demonstrating GPX8 sufficiency for inducing this CSC/mesenchymal marker expression.
Fig. 5.
GPX8 loss in MDA-MB-231 cells affects cancer stemness. (A) Loss of GPX8 results in CD44 cell surface expression reduction. The different indicated samples were subjected to FACS analysis of the cell-surface markers CD44. The histogram represents CD44 fluorescence intensity values. n = 3. (B) GPX8 expression levels correlate with the stem cell markers. Cells were subjected to immunoblotting with the indicated antibodies. (C) GPX8 overexpression in MDA-MB-468 cells induces CD44 expression. Cells were subjected to immunoblotting with the indicated antibodies. VC, vector control; GPX8-OE, GPX8 overexpression. (D) GPX8 expression in MDA-MD-231 cells correlates with the cells’ ability to form mammospheres. Quantification of in vitro mammosphere formation by cells from the different clones was performed. The data are reported as the number of mammospheres formed per 600 seeded cells; each value represents the mean ± SD for n = 5. The P value was determined by Student t test. (E) GPX8 loss affects tumor formation in mice. Female NOD-SCID mice were injected with 106 cells generated from the different clones. After 7 wk, the proportion of animals bearing tumors was assessed and presented. The P value was determined by Fisher’s exact test. (F) GPX8 expression affects tumor formation and growth rate in mice. During the in vivo time course, the tumor volume of each group was measured weekly and presented in a graph; each value represents the mean ± SD. For GPX8-WT, n = 8; GPX8-KO-1, n = 3; GPX8-KO-1+GPX8-OE, n = 7. The P value was determined by Student t test.
As an in vitro functional readout for the stemness properties of the cells, we determined the effect of GPX8 loss on mammosphere formation ability in MDA-MB-231 cells. We found that in comparison to the WT cells, GPX8 KO clones (GPX8-KO-1 and GPX8-KO-2) formed significantly fewer mammospheres (Fig. 5D and SI Appendix, Fig. S5 D and E). In contrast, GPX8-OE in GPX8-KO-1 cells resulted in a significant increase in the number of mammospheres (Fig. 5D and SI Appendix, Fig. S5E), thus demonstrating the specific role of GPX8 in mammosphere formation capabilities.
We then determined the effect of GPX8 loss on the ability of the MDA-MB-231 to form tumors in mice. Accordingly, we injected cells originating from WT, GPX8-KO-1, or GPX8-KO-1+GPX8-OE lines into the mammary fat pad of female NOD-SCID mice and monitored weekly the number and size of the generated tumors for up to 7 wk. Interestingly, only three out of seven mice injected with GPX8-KO-1 developed tumors, whereas tumors formed in all mice injected with WT or GPX8-KO-1+GPX8-OE cells (Fig. 5E). Moreover, tumors generated from GPX8-KO-1 cells weighed significantly less (P < 0.005) (SI Appendix, Fig. S5F) and were smaller in size (Fig. 5F and SI Appendix, Fig. S5G). Altogether, we found that GPX8 regulates the stemness characteristics of these cancer cells as it plays an essential role in their initiation ability and tumor growth.
GPX8 Regulates the Secretion of Cytokines.
Our next goal was to define the cellular mechanism by which GPX8 regulates the aggressiveness of cancer cells. Analysis of our RNA-Seq data followed by GSEA revealed that GPX8 loss does not only affect the hallmark of the EMT (Fig. 3C) but also modulates the cellular inflammatory response (“hallmark inflammatory response”) (SI Appendix, Fig. S6A). To experimentally verify these global changes in the production of secreted factors, we cultured GPX8-KO-1 cells for 3 d in the growth media from WT cells (WT-conditioned media), which includes all of the desired cytokines/chemokines. While GPX8-KO-1 cells in regular media demonstrated a reduction in CD44, ITGB, and fibronectin (FN1) expression, the addition of WT-conditioned media rescued the expression of these EMT and CSC markers (Fig. 6A).
Fig. 6.
GPX8 KO impairs cytokine production in MDA-MB-231 cells. (A) Conditioned media from WT cells can rescue the expression of CSC and EMT markers. Conditioned media from the WT were added to GPX8-KO-1cells for 3 d. The cells were lysed and subjected to immunoblot using the indicated Abs. (B) GPX8 loss induces a global reduction in cytokine expression. The expression level in WT and GPX8 KO cells is presented in a volcano plot. Cytokines and chemokines are presented as a blue triangle apart from IL-6 expression, which is presented in red. (C) GPX8 loss in MDA-MB-231 cells reduces the expression of selected cytokines. The RNA was isolated from WT, GPX8-KO-1, and GPX8-KO-1+GPX8-OE cells, and the expression of the selected genes was determined by qPCR. Each value represents the mean ± SD for n = 3. (D) IL-6 level is reduced in GPX8 KO cells growth media. Cell growth media were collected from each of the indicated samples after 24 h. The level of IL-6 was determined using a specific ELISA kit (n = 3). The P value was determined by Student t test.
We then aimed to systematically identify the GPX8-regulated secreted factors that modulate cancer cell aggressiveness. Using the “Human Protein Atlas” database (https://www.proteinatlas.org), we generated a list of all of the predicted secreted proteins (secretome). From the 2,249 listed genes, we chose 78 genes encoding for cytokines and 44 for chemokines. Then, by analyzing our RNA-Seq data, we limited our secretome only to genes that are expressed in MDA-MB-231 cells. These restrictions limited our secretome to 50 genes, which we designated as the cytokines gene set (CGS, Dataset S2). By further analyzing our RNA-Seq data, we found a significant down-regulation of the CGS in GPX8-KO-1 relative to the WT cells (Fig. 6B and SI Appendix, Fig. S6B). To validate this GPX8 loss-dependent expression pattern, we focused on interleukine-15 (IL-15), interleukine-1β (IL1B), C-X-C motif chemokine ligand 10 (CXCL10), colony-stimulating factor 3 (CSF3), and interleukine-6 (IL-6) as they were all significantly down-regulated in the RNA-Seq analysis by twofold (SI Appendix, Fig. S6C). This GPX8 loss-dependent expression pattern was then validated by qPCR. Moreover, the down-regulation of these selected cytokines in KO cells was rescued by GPX8 overexpression (Fig. 6C).
Among these selected cytokines, we then focused our study on IL-6, as it has been reported to play a crucial role in cancer aggressiveness and the EMT program (49, 50). We validated IL-6’s selective up-regulation expression pattern in mesenchymal relative to epithelial cell lines using the MERAV database (SI Appendix, Fig. S6D). Accordingly, GPX8-KO-1 cells demonstrated a significant reduction in the IL-6 secretion relative to WT cells. This effect on cytokine secretion was rescued by GPX8 overexpression (Fig. 6D). Together, these findings indicate that GPX8 is a global regulator of fundamental cancer-associated cytokines such as IL-6.
GPX8 Regulates IL6R, an Essential Player in the IL-6/STAT3 Signaling Pathway.
The Janus kinase (JAK)-signal transducer and activator of transcription 3 (STAT3) signaling pathway provides a critical link between inflammation and cancer (51). In tumors, STAT3 activation is associated with the induction of the EMT program (52) and enhancing stem-like properties (53). Activation of the JAK/STAT3 signaling pathway induces the expression of multiple cancer aggressiveness promoting factors such as IL-6 (54). IL-6 is stimulated by an autocrine loop, whereby the cytokine interacts with the IL6 receptor (IL6R), resulting in JAK/STAT3 signal transduction (54). Therefore, IL-6 levels serve as a readout for IL6R function and the downstream JAK/STAT3 signal activation.
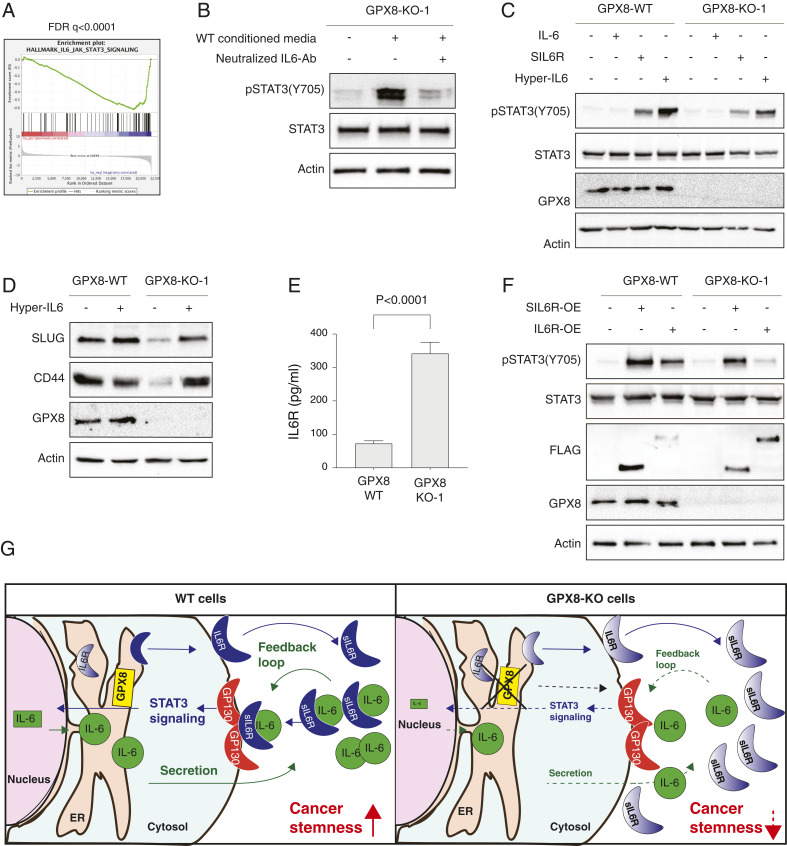
Comparative gene expression analysis followed by GSEA revealed that GPX8 loss resulted in a significant reduction in “hallmark of IL-6/JAK/STAT3 signaling” (Fig. 7A), indicating that GPX8 is a key regulator of this signaling pathway. In addition, we performed an mRNA stability assay that demonstrated that IL-6 down-regulation was due to reduced transcription rate in GPX8-KO-1 cells (SI Appendix, Fig. S7A) and not due to posttranscriptional regulation by mRNA stability factors (55).
Fig. 7.
GPX8-KO cellular effect is mediated by IL-6/JAK/STAT3 signaling pathway. (A) GPX8 loss leads to a reduction in the gene expression pattern for “hallmark IL-6/JAK/STAT3 signaling.” MDA-MB-231 WT cells and GPX8-KO-1 were subjected to RNA-Seq analysis. The expression ratio of all genes was calculated and ranked based on the relative expression in GPX8 WT and KO. The samples were subjected to GSEA. The FDR q-value was computed by GSEA. (B) STAT3 signaling is reduced by the addition of neutralizing IL-6 antibodies to the conditioned media. Conditioned media from the WT cultured for 3 d were added to GPX8-KO-1 cells in the absence or presence of neutralizing IL-6 antibodies (800 ng/mL) for 1 h. The cells were lysed and subjected to immunoblot using the indicated antibodies. (C) GPX8 WT and KO cells respond to soluble IL6 receptor and Hyper IL-6 but not to IL-6 treatment. GPX8-WT and GPX8-KO-1 cells were starved with 0.1% FBS medium for 24 h and treated with 20 ng/mL IL-6, 300 ng/mL soluble IL-6 receptor, and 50 ng/mL Hyper IL-6 for 1 h. Cells were subjected to immunoblot using the indicated Abs. (D) Hyper-IL6 induces EMT markers expression in GPX8-KO cells. WT and GPX8 KO cells were stimulated with Hyper IL-6 (50 ng/mL) for 3 d. The cells were then lysed and subjected to immunoblot using the indicated antibodies. (E) The soluble IL-6 receptor level is affected by the expression level of GPX8 in cell growth media. Cell growth media were collected from each of the indicated samples after 24 h starvation. The level of sIL6R was determined using a specific ELISA kit (n = 4). The P value was determined by Student t test. (F) Ectopic expression of full-length IL-6 receptor activates the STAT3 signaling pathway only in WT cells. The soluble IL6R and the full-length IL6R were overexpressed (SIL6R-OE, IL6R-OE, respectively, both FLAG-tag) in both GPX8-WT and GPX8-KO-1 cells, were lysed after 24 h, and subjected to immunoblot using the indicated antibodies. (G) A scheme representing the role of GPX8 in IL-6/JAK/STAT3 activation. Green arrows represent IL-6 secretion, and dark blue arrows represent intracellular IL-6 signaling. Dashed arrows and reduced font size represent impaired secretion and intracellular signaling. In WT cells, IL-6 and IL6R are processed in the ER; gradient color represents proteins before maturation and solid color after maturation.
We then determined whether GPX8 regulation of cytokine production (Fig. 6) is mediated by the IL6/JAK/STAT3 signaling cascade. We found that treating GPX8-KO-1 cells with WT growth media (WT-conditioned media) resulted in a dramatic increase in STAT3 phosphorylation on tyrosine 705 (Fig. 7B). However, supplementing this conditioned media with neutralizing IL-6 antibodies, which specifically blocked the activity of this cytokine (SI Appendix, Fig. S7B), resulted in JAK/STAT3 signaling pathway inhibition (Fig. 7B). These findings highlight the importance of GPX8 as a regulator of IL-6 production and its subsequent impact on the downstream JAK/STAT3 cascade.
The proper protein folding and maturation of both IL-6 and IL6R is mediated by disulfide bonds (56, 57) that can be potentially regulated by GPX8. Thus, we wanted to understand the exact cellular mechanisms by which GPX8 regulates JAK/STAT3 signaling. Since IL-6 failed to induce STAT3 phosphorylation (Fig. 7C) in both WT and GPX8-KO-1 cells, we speculated that GPX8 function is mediated through the IL-6 receptor (IL6R) regulation. IL6R can induce its cellular effect by two mechanisms; the “IL-6 classic signaling”, which is composed of secreted IL-6 and transmembrane IL6R, or the alternative IL-6 trans-signaling (58). In this alternative mechanism, activation of the coreceptor IL-6 signal transducer (IL6ST or GP130) is mediated via a complex of IL-6 with the soluble form of IL6R (sIL6R). We found that WT, GPX8-KO-1, and GPX8-KO-1+GPX8-OE cells expressed GP130 at similar levels (SI Appendix, Fig. S7C); however, IL6R expression is absent from the surface of all these cells (SI Appendix, Fig. S7D). Accordingly, we propose that GPX8 regulates IL6R, which consequently activates the JAK/STAT3 signaling through the IL-6 trans-signaling mechanism.
To provide further support for this IL-6 trans-signaling mechanism, we treated both WT and GPX8-KO-1 cells with IL-6, soluble IL-6 receptor (sIL6R), or IL-6/sIL6R fusion protein called Hyper-IL6 (59). In contrast to recombinant IL-6 treatment, sIL6R and Hyper-IL6 induced STAT3 phosphorylation in WT cells and, to a lesser extent, in the GPX8-KO-1 cells (Fig. 7C). Moreover, long-term treatment with Hyper-IL-6 rescued the expression of the EMT markers SLUG and CD44 in the GPX8-KO-1 cells (Fig. 7D). Since sIL6R was sufficient to induce STAT3 phosphorylation, these results indicate the endogenous IL-6 is active and thus not affected by the GPX8 loss. Therefore, these results reveal that GPX8 mediates the expression of mesenchymal markers by regulating the IL-6 receptor and not the cytokine itself.
Next, we determined the cellular mechanisms by which GPX8 regulates IL6R activity. Surprisingly, we found that when compared to WT, KO cells up-regulate IL6R expression (SI Appendix, Fig. S7E) and secrete higher levels of sIL6R (Fig. 7E). Importantly, as demonstrated in Fig. 7F, IL6R produced in GPX8-KO cells cannot activate the JAK/STAT3 signaling cascade, and therefore its elevation can be presumably attributed to compensation machinery. Moreover, the high secretion levels of sIL6R in GPX8-KO-1 cells indicates that the cellular trafficking machinery is unaffected by GPX8 activity.
Soluble IL6R (sIL6R) can be generated by two mechanisms; either through alternative splicing, which skips the exon encoding the transmembrane domain or by shedding of the full-length receptor by ADAM metallopeptidase domain 17 (ADAM17) (58). We excluded the first possibility as we detected a reduction in the transcript levels of the sIL6R form in the GPX8-KO-1 cells relative to WT cells (SI Appendix, Fig. S7F). However, we found ADAM17 to be an active metalloprotease in all of the samples, including GPX8-KO (SI Appendix, Fig. S7G). We conclude that the shedding of the full-length receptor in GPX8-KO-1 is the central mechanism in the generation of sIL6R.
To gain insight into GPX8 activity, we overexpressed sIL6R and IL6R in WT and GPX8-KO-1 cells and assessed the impact on STAT3 signaling. We found that overexpression of the sIL6R induced STAT3 phosphorylation in both WT and GPX8-KO-1 cells. Remarkably, overexpression of the full-length receptor in GPX8-KO-1 cells failed to activate this signaling cascade (Fig. 7F). Thus, GPX8 preferentially regulates full-length IL6R prior to its cleavage, as the function of the ectopically expressed sIL6R was unaffected.
Discussion
We have found that the MMS gene GPX8 is a guardian of the mesenchymal state in aggressive cancer cells. GPX8 demonstrated a significant correlation with known mesenchymal markers in both cell lines and patients. Moreover, high GPX8 expression was associated with poor patient prognosis in aggressive breast cancer samples. We show that GPX8 expression was up-regulated during the EMT program, and its loss conferred epithelial characteristics in the mesenchymal-like breast cancer cells, MDA-MB-231. Specifically, knocking out GPX8 resulted in a profound inhibition of cytokine production, including the EMT-promoting factor, IL-6. In these cells, IL-6 is produced through an autocrine loop, which activates the JAK/STAT3 signaling pathway through a trans-signaling mechanism. We demonstrated that GPX8 modulated the ability of IL-6 and its receptor to activate this signaling cascade. Thus, we propose a model in which GPX8 regulates the full-length IL6R activity, probably by affecting its interaction with its ligand, resulting in impaired JAK/STAT3 signaling (Fig. 7G).
We determine GPX8 up-regulation in mesenchymal-like aggressive cancer cells and in two different EMT-induction system models (TGFβ and Twist) (Fig. 1). HIF1α plays a regulatory role in the EMT program (60) as it represses E-cadherin expression (61) or activates the expression of the key EMT transcription factors such as TWIST (62), ZEB1, and ZEB2 (3). Moreover, this factor in triple-negative breast cancer promotes tumorigenesis and adaptation to hypoxia (63). A previous study has shown that GPX8 expression is transcriptionally regulated by HIF1α in HeLa cells (29), as GPX8 promotor contains two hypoxia-response elements. Thus, we suggest HIF1α as a key regulator of GPX8 expression in breast cancer.
The EMT program was first reported in embryonic development when selected cells change their epithelial identity and gain mesenchymal-like characteristics (3). Further studies expanded the role of the EMT program to be included in wound healing, fibrosis, and cancer. Fibrosis is a complex disease associated with reduced organ function (64), which is caused by cellular mechanisms reminiscent of oncogenesis (65). Interestingly, fibrosis, like cancer aggressiveness, is driven by ROS (65) and selected cytokines such as IL-6 (66). Since both of these factors are associated with GPX8, this enzyme is a potential druggable candidate to inhibit cancer aggressiveness and fibrosis.
The eight-member GPx family of enzymes is divided into two groups based on the presence of the nonstandard amino acid, selenocysteine (GPX1-4, and GPX6) or of cysteine (GPX5, GPX7-8) in their active site. Unambiguous phylogeny analysis further subdivides this family into three groups, whereby GPX4, GPX7, and GPX8 belong to the same evolutionary branch (67). GPX4 was found to be one of the primary regulators of ferroptosis (68), a form of apoptotic cell death (69). A selective GPX4 inhibitor, RSL3, induces cell death in epithelial cancer-derived cell lines expressing mesenchymal markers such as ZEB1 (70). In addition, persistent cancer cells, which are resistant to lapatinib treatment and up-regulate mesenchymal as well as stem cell markers, are vulnerable to GPX4 inhibition by RSL3 (71). This mutual mesenchymal cell-dependent function of GPX4 and GPX8 suggests a conserved role for this subfamily of GPx in aggressive cancer cells.
The maturation process of cytokines and their respective receptors, such as IL-6 and IL6R, occurs in the ER. In this organelle, these proteins acquire their proper structural conformation via modifications such as disulfide bond formation (56, 57). Specifically, the extracellular soluble domain of IL6R, which interacts with the ligand, contains four conserved cysteines that form disulfide bonds (72). Here, we determined that GPX8 modulates IL6R, as the addition of recombinant IL6R rescues the GPX8 loss cellular phenotype. Thus, we suggest a potential mechanism whereby the ER-localized GPX8 and its function in protein disulfide bond formation (22) regulate the maturation of IL6R, essential for its ability to transmit the signaling downstream.
We suggest an overall model in which the EMT program elevates GPX8 expression central to the activity of cytokines and their receptors. These fully functional autocrine cytokines are secreted and induce the aggressive mesenchymal-like properties in these tumors via the activation of signaling cascade such as JAK/STAT3 (54) (Fig. 7G). Finally, we have revealed a GPX8/IL-6/STAT3 axis, essential for the maintenance of the aggressive mesenchymal-like state in cancer cells. We hypothesize that further studies evaluating GPX8 function in posttranslational modification, such as disulfide bond formation, would identify new mechanisms essential for the regulation of cancer cell aggressiveness.
Materials and Methods
Cell Lysis and Immunoblotting.
Cells were rinsed once with ice-cold PBS and lysed with RIPA lysis buffer (20 mM Tris [pH 7.4]), 137 mM NaCl, 10% glycerol (vol/vol), 1% Triton X-100, 0.5% (wt/vol) deoxycholate, 0.1% (wt/vol) SDS, 2.0 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0), and one tablet of EDTA-free protease inhibitor (Roche) and phosphatase inhibitor mixture mixes A and B (100X) (Bimake). The protein concentration was determined by Bradford (BioRad, 500-0006). The supernatants were separated by a 10% SDS/PAGE, transferred onto a 0.45 µm PVDF membrane (Merck), and probed with the appropriate antibodies.
Antibodies.
Antibodies were obtained from the following sources: GPX8 (HPA036720) from Sigma Aldrich, CDH1 (3195), CDH2 (13116), β-actin (4970)\(3700), ZEB1 (3396), VIM (5741), ITGB4 (14803), CD44 (for immunoblotting [3570]), p-sSTAT3-Tyr705 (9145), STAT3 (9139), from Cell Signaling Technology; FITC-labeled anti-CD24 (555427), APC-labeled anti-CD44 (559942) from BD Bioscience; IL6-R anti-mouse (sc-373708) HRP-labeled anti-mouse and anti-rabbit secondary antibodies from Santa Cruz Biotechnology. GP130 anti-rabbit (06-291) from Upstate Biotechnology. For IL-6 neutralization assay, we used goat anti-human IL-6 (500-P26G; Peprotech).
Cancer Sample Analysis.
KM analysis of the data from breast and other cancer samples were analyzed and generated by the KM Plotter website (http://kmplot.com/analysis/) (73). GPX8 was searched as a gene symbol, and the 228141_at Affymetrix ID was chosen. The autoselect best cutoff was selected as well. The obtained KM plots are presented. The statistics were generated by the website itself.
Animal Studies.
MDA-MB-231 WT, GPX8-KO-1, and GPX8 rescue cells were injected into the mammary fat pad of female NOD-SCID mice (1 × 106 cells per mouse). The tumors were measured weekly. After 7 wk, the tumors were removed and weighed. All mouse experiments were carried out under Hebrew University Institutional Animal Care and Use Committee-approved protocol MD-16-14939-5. Hebrew University is Association for Assessment and Accreditation of Laboratory Animal Care-approved.
Cytokine and Receptor Measurement.
Measurement of IL-6 levels was performed by sandwich ELISA using the human IL-6 mini ABTS ELISA development kit (Peprotech). Human soluble IL-6 receptor levels in the medium were measured by the in vitro ELISA kit (Abcam, ab46029). WT and GPX8-KO-1 cells were starved with serum-free media for 36 h. The media were collected and concentrated by centrifugation using 10 KDa cutoff Spin filter (Amicon).
Supplementary Material
Acknowledgments
We thank the members of the Y.D.S. laboratory. This work was supported by the Israel Science Foundation (Grant 1816/16), the Israel Cancer Association (Grant 20180062, Abraham Rutstein funds), and the Hebrew University start-up funds. B.S. is supported by the Lady Davis Fellowship for postdoctoral researchers at The Hebrew University of Jerusalem. The Genomic Applications Laboratory of the Core Research Facility, The Faculty of Medicine, The Hebrew University of Jerusalem, Israel, performed the RNA-Seq data analysis. Prof. Rotem Karni, Hebrew University, designed the primers for IL6R alternative splice isoform analysis. Prof. Irit Sagi, Weizmann Institute of Science, assisted the ADAM17 activity assay.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010275117/-/DCSupplemental.
Data Availability.
All relevant data in the paper are entirely available through both text and figures, in the main text and SI Appendix.
References
- 1.Dongre A., Weinberg R. A., New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Pastushenko I., Blanpain C., EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29, 212–226 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Nieto M. A., Huang R. Y.-J., Jackson R. A., Thiery J. P., EMT: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Yang J. et al.; EMT International Association (TEMTIA) , Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diepenbruck M., Christofori G., Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no, maybe? Curr. Opin. Cell Biol. 43, 7–13 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Stemmler M. P., Eccles R. L., Brabletz S., Brabletz T., Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 21, 102–112 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Bharti R., Dey G., Mandal M., Cancer development, chemoresistance, epithelial to mesenchymal transition and stem cells: A snapshot of IL-6 mediated involvement. Cancer Lett. 375, 51–61 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Pattabiraman D. R. et al., Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351, aad3680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mani S. A. et al., The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taube J. H. et al., Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A. 107, 15449–15454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaul Y. D. et al., Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell 158, 1094–1109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantor J. R., Sabatini D. M., Cancer cell metabolism: One hallmark, many faces. Cancer Discov. 2, 881–898 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee N., Kim D., Cancer metabolism: Fueling more than just growth. Mol. Cells 39, 847–854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rmaileh A. A. et al., Large-scale differential gene expression transcriptomic analysis identifies a metabolic signature shared by all cancer cells. Biomolecules 10, 701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vander Heiden M. G., DeBerardinis R. J., Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locasale J. W. et al., Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43, 869–874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Possemato R. et al., Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaul Y. D. et al., MERAV: A tool for comparing gene expression across human tissues and cell types. Nucleic Acids Res. 44, D560–D566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amstutz U., Froehlich T. K., Largiadèr C. R., Dihydropyrimidine dehydrogenase gene as a major predictor of severe 5-fluorouracil toxicity. Pharmacogenomics 12, 1321–1336 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Tosatto S. C. E. et al., The catalytic site of glutathione peroxidases. Antioxid. Redox Signal. 10, 1515–1526 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Brigelius-Flohé R., Maiorino M., Glutathione peroxidases. Biochim. Biophys. Acta 1830, 3289–3303 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Nguyen V. D. et al., Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J. Mol. Biol. 406, 503–515 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Chen Y.-I., Wei P.-C., Hsu J. L., Su F. Y., Lee W. H., NPGPx (GPx7): A novel oxidative stress sensor/transmitter with multiple roles in redox homeostasis. Am. J. Transl. Res. 8, 1626–1640 (2016). [PMC free article] [PubMed] [Google Scholar]
- 24.Ramming T., Hansen H. G., Nagata K., Ellgaard L., Appenzeller-Herzog C., GPx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic. Biol. Med. 70, 106–116 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Hsu J.-L. et al., Glutathione peroxidase 8 negatively regulates caspase-4/11 to protect against colitis. EMBO Mol. Med. 12, e9386 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa K. et al., Quantitative proteomics identifies the membrane-associated peroxidase GPx8 as a cellular substrate of the hepatitis C virus NS3-4A protease. Hepatology 59, 423–433 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Mehmeti I., Lortz S., Avezov E., Jörns A., Lenzen S., ER-resident antioxidative GPx7 and GPx8 enzyme isoforms protect insulin-secreting INS-1E β-cells against lipotoxicity by improving the ER antioxidative capacity. Free Radic. Biol. Med. 112, 121–130 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Yoboue E. D., Rimessi A., Anelli T., Pinton P., Sitia R., Regulation of calcium fluxes by GPX8, a type-II transmembrane peroxidase enriched at the mitochondria-associated endoplasmic reticulum membrane. Antioxid. Redox Signal. 27, 583–595 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Bosello-Travain V. et al., Glutathione peroxidase 8 is transcriptionally regulated by HIFα and modulates growth factor signaling in HeLa cells. Free Radic. Biol. Med. 81, 58–68 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Piñeiro-Hermida S. et al., IGF1R deficiency attenuates acute inflammatory response in a bleomycin-induced lung injury mouse model. Sci. Rep. 7, 4290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gröger C. J., Grubinger M., Waldhör T., Vierlinger K., Mikulits W., Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One 7, e51136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeisberg M., Neilson E. G., Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest. 119, 1429–1437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haakensen V. D. et al., Gene expression profiles of breast biopsies from healthy women identify a group with claudin-low features. BMC Med. Genomics 4, 77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao J. et al., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A. et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mootha V. K. et al., PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Aigner K. et al., The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 26, 6979–6988 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ku J.-L., Park J.-G., Biology of SNU cell lines. Cancer Res. Treat. 37, 1–19 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L. et al., Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol. Cancer Res. 6, 760–769 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J. et al., Suppression of SCARA5 by Snail1 is essential for EMT-associated cell migration of A549 cells. Oncogenesis 2, e73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Györffy B. et al., An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F., Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Aznar E., Wiesmüller L., Sainz B. Jr., Hermann P. C., EMT and stemness-key players in pancreatic cancer stem cells. Cancers 11, 1136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fillmore C. M., Kuperwasser C., Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 10, R25–R13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jonasson E. et al., Identification of breast cancer stem cell related genes using functional cellular assays combined with single-cell RNA sequencing in MDA-MB-231 cells. Front. Genet. 10, 500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehmann B. D. et al., Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bierie B. et al., Integrin-β4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 114, E2337–E2346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W. et al., Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci. Rep. 7, 13856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang G. X. et al., Interleukin-6 induces epithelial-mesenchymal transition in human intrahepatic biliary epithelial cells. Mol. Med. Rep. 13, 1563–1569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gyamfi J., Lee Y.-H., Eom M., Choi J., Interleukin-6/STAT3 signalling regulates adipocyte induced epithelial-mesenchymal transition in breast cancer cells. Sci. Rep. 8, 8859 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taniguchi K., Karin M., IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin. Immunol. 26, 54–74 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Rokavec M. et al., IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest. 124, 1853–1867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S.-Y. et al., Role of the IL-6-JAK1-STAT3-Oct-4 pathway in the conversion of non-stem cancer cells into cancer stem-like cells. Cell. Signal. 25, 961–969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson D. E., O’Keefe R. A., Grandis J. R., Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 234–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Masuda K. et al., Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc. Natl. Acad. Sci. U.S.A. 110, 9409–9414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clogston C. L., Boone T. C., Crandall B. C., Mendiaz E. A., Lu H. S., Disulfide structures of human interleukin-6 are similar to those of human granulocyte colony stimulating factor. Arch. Biochem. Biophys. 272, 144–151 (1989). [DOI] [PubMed] [Google Scholar]
- 57.Rock F. L., Li X., Chong P., Ida N., Klein M., Roles of disulfide bonds in recombinant human interleukin 6 conformation. Biochemistry 33, 5146–5154 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Rose-John S., The soluble interleukin 6 receptor: Advanced therapeutic options in inflammation. Clin. Pharmacol. Ther. 102, 591–598 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Fischer M. et al., I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 15, 142–145 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Rankin E. B., Giaccia A. J., Hypoxic control of metastasis. Science 352, 175–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krishnamachary B. et al., Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 66, 2725–2731 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Yang M.-H. et al., Direct regulation of TWIST by HIF-1α promotes metastasis. Nat. Cell Biol. 10, 295–305 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Briggs K. J. et al., Paracrine induction of HIF by glutamate in breast cancer: EglN1 senses cysteine. Cell 166, 126–139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shibue T., Weinberg R. A., EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14, 611–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morry J., Ngamcherdtrakul W., Yantasee W., Oxidative stress in cancer and fibrosis: Opportunity for therapeutic intervention with antioxidant compounds, enzymes, and nanoparticles. Redox Biol. 11, 240–253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fielding C. A. et al., Interleukin-6 signaling drives fibrosis in unresolved inflammation. Immunity 40, 40–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mariotti M. et al., Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS One 7, e33066 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang W. S. et al., Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dixon S. J., Stockwell B. B., The hallmarks of ferroptosis. Annu. Rev. Cancer Biol. 3, 35–54 (2019). [Google Scholar]
- 70.Viswanathan V. S. et al., Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hangauer M. J. et al., Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cole A. R. et al., Disulfide bond structure and N-glycosylation sites of the extracellular domain of the human interleukin-6 receptor. J. Biol. Chem. 274, 7207–7215 (1999). [DOI] [PubMed] [Google Scholar]
- 73.Lánczky A. et al., miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 160, 439–446 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data in the paper are entirely available through both text and figures, in the main text and SI Appendix.