Abstract
Joubert syndrome is characterized by unique malformation of the cerebellar vermis. More than thirty Joubert syndrome genes have been identified, including ARL13B. However, its role in cerebellar development remains unexplored. We found that knockdown or knockout of arl13b impaired balance and locomotion in zebrafish larvae. Granule cells were selectively reduced in the corpus cerebelli, a structure homologous to the mammalian vermis. Purkinje cell progenitors were also selectively disturbed dorsomedially. The expression of atoh1 and ptf1, proneural genes of granule and Purkinje cells, respectively, were selectively down-regulated along the dorsal midline of the cerebellum. Moreover, wnt1, which is transiently expressed early in cerebellar development, was selectively reduced. Intriguingly, activating Wnt signaling partially rescued the granule cell defects in arl13b mutants. These findings suggested that Arl13b is necessary for the early development of cerebellar granule and Purkinje cells. The arl13b-deficient zebrafish can serve as a model organism for studying Joubert syndrome.
Electronic supplementary material
The online version of this article (10.1007/s12264-020-00554-y) contains supplementary material, which is available to authorized users.
Keywords: Joubert syndrome, arl13b, Cerebellum, Development, Granule cell, Purkinje cell, Wnt
Introduction
Joubert syndrome (JS) is an autosomal-recessive neurodevelopmental disorder, which is characterized morphologically by the unique molar tooth sign, a complex malformation of the cerebellar vermis and brainstem, with abnormalities of axonal decussation affecting the corticospinal tract and superior cerebellar peduncles [1]. JS is clinically characterized by impaired motor functions and intellectual disability. Most cases of JS are variably associated with impairments of additional organs, including the retina, kidney, skeleton, and liver. More than 30 genes have been identified to cause JS (213300, Online Mendelian Inheritance in Man). Most of the proteins encoded by these genes are located in or near the primary cilium, an organelle found in eukaryotic cells, and this makes JS a typical ciliopathy [1]. Although malformation of the cerebellar vermis is common in JS, the role and mechanisms of the causative genes in cerebellar development have only been reported recently [2, 3].
Mutations of ARL13B (HGNC ID 25419) lead to the classical form of JS [4]. The ARL13B gene encodes an enzyme belonging to the small GTPase superfamily and this makes it unique among the known causative genes of JS. It has been demonstrated that Arl13b is critical for processes of neural development, such as interneuron migration and placement [5], polarized radial glial scaffold formation [6], and neural tube patterning [7]. Arl13b might also be involved in photoreceptor degeneration and kidney cysts [8, 9]. However, the role of Arl13b in cerebellar development remains a mystery.
The zebrafish has been established as a model organism in studying JS [9, 10]. Here, we explored the role of arl13b in early development of the cerebellum and we hope use this powerful model to investigate the pathological mechanisms of JS and help to screen for therapeutic targets.
Materials and Methods
Zebrafish Maintenance and Embryo Collection
All zebrafish lines were raised and maintained under a photoperiod of 14 h/10 h (light/dark) at 28.5°C in our facility supplied with filtered fresh circulating water. Wild-type zebrafish of the AB strain and the arl13b mutants were kindly provided by Dr. Ying Cao (Tongji University, Shanghai, China) and the Tg(neurod1:EGFP) transgenic zebrafish were a gift from Dr. Jing-Wei Xiong (Peking University, Beijing, China). arl13b homozygous mutants only survive up to 10 days, so heterozygous mutants were mated to produce homozygous embryos. The homozygous embryos were picked according to the curved tail since this phenotype is almost fully penetrant. The developmental stages of zebrafish were characterized following previously-described morphological criteria [11]. Fish embryos and larvae for in situ hybridization and immunostaining were raised in E3 supplemented with 0.003% phenylthiourea from 24 hpf onward to prevent pigment formation. All handling procedures were approved by the Ethics Review Committee at Nanchang University.
Morpholino Oligonucleotide Microinjection
The morpholino (MO) antisense oligonucleotide blocking the translation of arl13b (5’-TTTCCCCCCTAAATGCTTTCACTGG-3’) described previously [9] was purchased from Gene Tools LLC (Philomath, OR, USA). The MO was microinjected into zebrafish embryonic yolk at the one- to two-cell stage.
Imaging of Zebrafish and Behavior
The morphology of the zebrafish larvae (otolith and body curvature) was imaged at 4 dpf using a Nikon AZ100 microscope (Nikon, Tokyo, Japan). The larvae were raised in Petri dishes and transferred to a new dish at 5 dpf for behavioral analysis. After allowing adaptation to the new environment for 5 min, locomotion was video-recorded for 3 min using a Nikon DS-Fil1 digital camera and processed with NIS-Elements F3.0 (Nikon).
Whole-Mount In Situ Hybridization
We made RNA probes from different templates: PCR products and linearized plasmid DNA. For PCR-based in situ templates, we designed PCR primers (listed in Supplemental Table 1) to amplify gene-specific products that contained the T3 promoter sequence, and RNA probes were transcribed in vitro using T3-RNA-polymerase. For the linearized plasmid DNA-based in situ templates, shh (HindIII/T7), atoh1a (Nco I /SP6), ptf1a (Nco I/SP6), reelin (NcoI/SP6), and roraa (ApaI/SP6), the last four plasmids were kindly provided by Dr. Sheng-Ping L. Hwang (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, China). The antisense RNA probes were synthesized with T7 or SP6 RNA polymerase after plasmid DNA linearization. Whole-mount in situ hybridization was performed using digoxigenin-labeled antisense RNA probes and alkaline phosphatase-conjugated anti-digoxigenin antibodies (Roche, Mannheim, Germany), as described previously [12]. Embryos were mounted in glycerol, and images were captured using a Nikon AZ100 microscope.
RNA Isolation and Quantitative Real-Time PCR (qPCR)
Total RNA was extracted from the embryos using RNAiso Plus following the manufacturer’s protocol (Takara, Shiga, Japan). Reverse-transcription reactions were carried out with M-MLV reverse transcriptase (Takara). qPCR assays were performed with SYBR Premix Ex Taq II (Takara) on the Abi-Step-One plus Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). All the primer sequences used for qPCR are listed in Supplemental Table 2. All experiments were conducted at least three times. Student’s t-test was applied to analyze the data and P < 0.05 was indicated a statistically significant difference.
Immunostaining
For tubulin antibody staining, we fixed larvae with 2% trichloroacetic acid in PBS for 3 h at room temperature. For other antibodies, we fixed larvae overnight at 4°C in 4% paraformaldehyde (PFA) in PBS supplemented with sucrose (4% w/v) [13]. The larvae were rinsed 3 times in 1×PBST (1×PBS and 0.8% Triton) for 10 min each. Afterward, larvae were dehydrated and rehydrated through graded methanols (50% MeOH once, 100% twice, and 50% once, 10 min each), and then rinsed 3 times in 1×PBST for 5 min each. The larvae were immersed in ice-cold acetone for 20 min at –20 °C, rinsed 3 times in 1×PBST for 5 min each and digested with proteinase K for permeabilization. The concentration and treatment time of proteinase K was determined by the developmental stages of the larvae. After digestion, the larvae were re-fixed with fresh 4% PFA in 1×PBS for 20 min at room temperature (RT), and then rinsed 3 times in 1×PBST for 10 min each. The larvae were blocked with buffer containing 10% serum and 1% dimethylsulfoxide in 1×PBST for 3 h at RT and incubated with primary antibody overnight at 4°C. After rinsing 4 times in 1×PBST for 30 min each, the larvae were incubated with secondary antibody for 4 h at RT. The larvae were rinsed and counterstained with Hoechst 33342 (10 μg/mL) for nuclear staining. The following primary antibodies were used: mouse anti-tubulin (1:1000, T6793, Sigma), mouse anti-parvalbumin (1:1000, MAB1572, Chemicon), goat 3A10 (1:1000, DSHB), and goat anti-GFP (1:500, 600-101-215, Rockland). For fluorescence detection, we used Alexa Fluor 488 donkey anti-goat IgG (H+L) (1:500, A11055, Invitrogen) and Alexa Fluor 488 donkey anti-mouse IgG (H+L) (1:500, A21202, Invitrogen). Fluorescence images of larvae were acquired using an Olympus FV1000 confocal microscope, and Z-series stacks were shown as two-dimensional projections.
Lithium Treatment
Embryos at 30–37 hpf were incubated in E3 containing LiCl (A100416-0025, Sangon) at a final concentration of 50 mmol/L. Age-matched untreated embryos served as controls.
Results
Depletion of arl13b Impairs Posture and Locomotion in Zebrafish Larvae
In order to explore the function of ARL13B in vivo, we started to investigate whether the neurological features of JS are mimicked in arl13b mutant zebrafish, an established model organism [4, 9]. The null mutation of arl13b (arl13b-/-), identified in a retroviral insertion screen, led to body axis curvature (Fig. S1) [4, 14]. The curved tail phenotype is almost fully penetrant in homozygous embryos while it is rarely found in wild-type and heterozygous embryos [4]. We took advantage of this readily-recognizable morphological phenotype to pick homozygous mutants for further experiments. The picked embryos were genotyped by PCR which confirmed retroviral insertion into the first exon of the arl13b gene. Furthermore, qPCR results demonstrated that the expression of arl13b mRNA was almost undetectable in the picked embryos, while it was normally-expressed in wild-type embryos (Fig. S3). The arl13b-/- mutant larvae [5 days post-fertilization (dpf)] were usually motionless and occasionally moved by trembling and circling. These abnormal movements were not found in wild-type larvae, which swam around freely and elegantly (Movie 1).
To assess the phenotype specificity and confirm the locomotor defects, we performed transient knockdown experiments with an antisense morpholino oligonucleotide (MO) designed to specifically block the translation of Arl13b [9]. After injecting 9.7 ng arl13b MO into embryos, we observed body curvature and abnormal locomotion like that in arl13b-/- mutants (Movie 1). Considering that the curvature might interfere with locomotion, we injected a subthreshold dose of MO (7.3 ng) and found that the larvae showed no apparent body axis defects while they still exhibited impairments in posture and locomotion (Movie 1). Wild-type larvae maintained a normal posture (Fig. 1A). In contrast, the subthreshold-dose morphants usually laid on their sides, i.e., lost posture (Fig. 1B, C). As for the swimming patterns, wild-type larvae often exhibited spontaneous swimming characterized by small bending angles (Fig. 1D). However, the subthreshold-dose morphants swam with trembling and exhibited large bending angles (Fig. 1E). The subthreshold-dose results suggested that the posture and locomotion defects in arl13b mutants and morphants are due to the depletion of arl13b rather than body curvature.
Fig. 1.
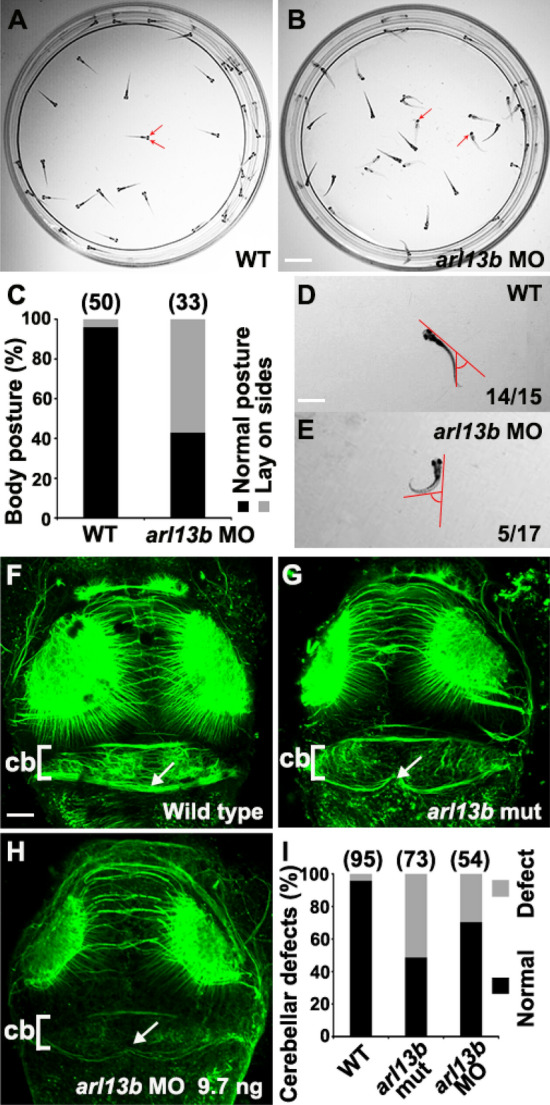
Knockdown of arl13b impairs posture, locomotion, and cerebellar morphology in zebrafish larvae. A Wild-type sibling larvae remain vertically oriented at 5 days post-fertilization (dpf), with both eyes visible from a top view (arrows). B In contrast, larvae injected with arl13b MO (subthreshold dose, 7.3 ng) often lie on their side at the bottom of the dish, with only one eye visible (arrows). Note that the body of the subthreshold-dose arl13b morphants are relatively straight and only slightly curved. C Statistics of the posture of zebrafish larvae at 5 dpf. D Wild-type larvae perform stereotyped spontaneous swimming with small bending angles. E The arl13b morphants (subthreshold dose, 7.3 ng) swim slower and exhibit greater bending angles. F–H Immunostaining with acetylated tubulin antibody outlines the cerebellum of larvae at 3 dpf. Comparing the dorsal view of wild-type embryos (F) with arl13b mutants (G) reveals morphological defects of the cerebellum (arrows). The cerebellar defects were also present in embryos injected with arl13b MO (H) (cb, cerebellum). I Statistics of the embryos with morphological defects of the cerebellum. The number of embryos examined in each condition is indicated above each column. A–E, Scale bar 1 cm. F–H, Scale bar 100 μm.
In addition, the sensation of arl13b mutants and morphants was compromised; they were not sensitive to a needle poke, while wild-type embryos were sensitive and swam away quickly (Movie 1).
Taken together, the above results suggested that zebrafish deficient in arl13b exhibit impairments in posture and swimming pattern, reminiscent of the signs of JS.
Depletion of arl13b Results in Morphological Defects of the Cerebellum
The above behavioral defects suggested the arl13b-deficient zebrafish might serve as a good model for studying JS. We first investigated the development of the cerebellum since it is the main cause of the characteristic molar tooth sign in JS. The outline of the cerebellum can easily be distinguished by immunostaining with an anti-tubulin antibody (Fig. 1F). Obvious morphological defects were found in the cerebella of both arl13b mutants and morphants, and the antibody-labeled fibers were globally reduced (Fig. 1G, H). The posterior outline of the cerebellum was thinner than that of the wild-type and invaginated anteriorly at the midline, while the anterior outline was relatively normal (Fig. 1F–I). Moreover, the acetyl-tubulin-positive parallel fibers connecting the cerebellar hemispheres were disturbed in arl13b-deficient larvae while the commissural axons between the optic tecta remained largely unaffected (Fig. 1F–H). The midline cerebellar defects in arl13b mutants and morphants are reminiscent of the midline cerebellar defects in JS patients [2].
Besides the cerebellum, we also checked other neural tissues. arl13b is highly expressed in the ventricle and otic vesicle at early developmental stages [9]; the ciliated cells in the inner ear are important for the formation of otoliths [15]. Usually, there are two otoliths in zebrafish (Fig. S1). We found that the otoliths displayed defects in number and size in both arl13b mutants and morphants (Fig. S1). The otoliths are critical for proper balance and hearing, and their impairment might contribute to the postural and locomotor defects in arl13b-depleted larvae. We also checked motor axons and Mauthner axons, which are involved in locomotion. No morphological defect was observed in either acetylated tubulin-positive motor axons or 3A10-positive Mauthner axons (Fig. S2). We focused on the cerebellum in the subsequent experiments since cerebellar malformation is a major hallmark of JS.
Depletion of arl13b Impairs Granule Cell Progenitors in the Developing Cerebellum
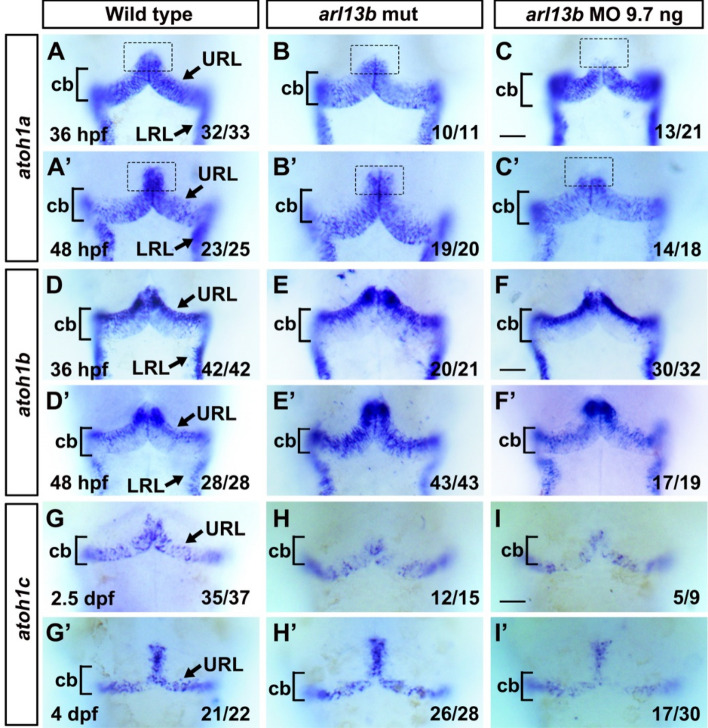
The morphological defects in arl13b-deficient embryos prompted us to further investigate the role of arl13b in the development of cerebellar neural circuits. Like mammals, the zebrafish cerebellum is derived from the dorsal part of the anterior hindbrain [16]. The cerebellum is composed of several types of neurons, which are categorized according to their major neurotransmitter, glutamate or GABA. The glutamatergic granule neurons are derived from granule cell progenitors located in the upper rhombic lip (URL). Granule cells are the most abundant type of neuron in the cerebellum. We first examined the expression of the granule progenitor cell marker atoh1 using whole-mount in situ hybridization. In zebrafish, there are three paralogues of atoh1 – 1a, 1b, and 1c – which are expressed sequentially in overlapping but distinct granule cell progenitors within the rhombic lip [17, 18]. In wild-type embryos, atoh1a was strongly expressed in the URL and the lateral rhombic lip (Fig. 2A). In some arl13b morphants, atoh1a was absent from the anterior dorsomedial URL along the midline (Fig. 2C). The specific absence of atoh1a was maintained at 48 hours post-fertilization (hpf) (Fig. 2A’–C’), suggesting that the phenotype could not be due to developmental delay. No detectable change of atoh1b was found (Fig. 2D–F’), indicating that the absence of atoh1a dorsomedially was specific and not due to defects in cerebellar structure. The expression level of atoh1c was decreased in arl13b morphants at 2.5 dpf and 4 dpf (Fig. 2G–I’). In arl13b mutants, atoh1c was decreased slightly at 2.5 dpf (Fig. 2H). The expression defects of atoh1a and atoh1c were more apparent in arl13b morphants than in mutants. This could be due to the maternally-deposited arl13b mRNA in null mutants which might mask the early defects caused by Arl13b deficiency [9]. The MO blocks the translation of arl13b mRNA, including the maternally-derived mRNA, so the phenotypes in morphants are more penetrant than in mutants [9]. However, potential off-target effects of the MO could not be excluded although it seems unlikely since most phenotypes have been reported in arl13b mutants [9] (and our data).
Fig. 2.
The expression of markers of cerebellar granule cell progenitors is impaired in arl13b-deficient embryos. A–I’ Representative images of in situ hybridization illustrate that the three paralogues of atoh1 (atoh1a, 1b and 1c) are expressed in distinct populations of cerebellar granule cell progenitors. atoh1a is expressed in the URL and LRL. Similar expression patterns of atoh1a occur in wild-type embryos (A) and arl13b mutants (B) while its expression is absent from the oral dorsomedial URL (dashed box) in arl13b morphants (C) at 36 hpf and 48 hpf (A’–C’). The expression patterns of atoh1b remained unaffected in arl13b mutants and morphants (D–F’). The expression level of atoh1c was decreased in the URL in arl13b morphants (I and I’) (cb, cerebellum; URL, upper rhombic lip; LRL, lower rhombic lip). A–I’, Scale bar 100 μm.
We performed quantitative PCR (qPCR) using whole body tissues to verify the above results. atoh1c was consistently down-regulated in arl13b mutants from 36 hpf to 4 dpf (Fig. S3). However, atoh1a and 1b were up-regulated at 36 hpf but down-regulated at later stages, when comparing arl13b mutants with the wild-type (Fig. S3). It should be noted that we used the whole zebrafish body and the qPCR results represented the mixed expression of all tissues. According to our in situ hybridization results and those of others [17, 18], atoh1c is mainly expressed in the URL (Fig. 2). The decrease of atoh1c in arl13b mutants revealed by qPCR indicated that atoh1c is reduced mainly in the URL and confirmed our in situ hybridization results.
atoh1c and 1a are critical for the full complement of granule cells in the corpus cerebelli (CCe), a structure homologous to the mammalian cerebellar vermis [17, 19]. The decrease of atoh1a and particularly atoh1c revealed by in situ hybridization and qPCR in arl13b-deficient embryos might interfere with the development of granule cells in the CCe.
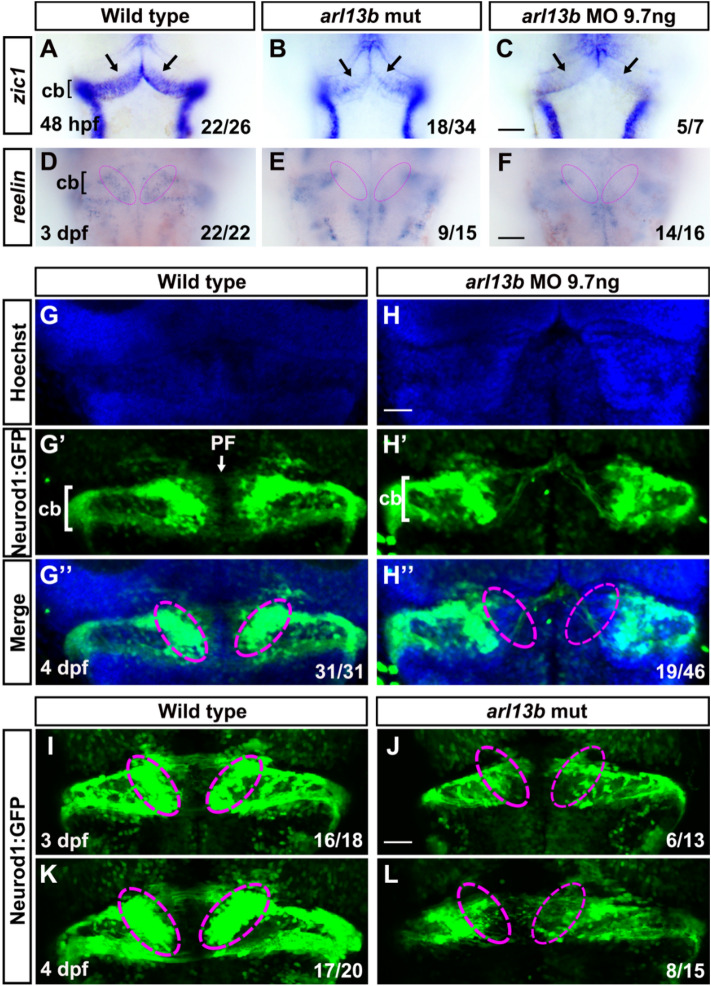
Zic1 has been shown to be involved in mouse granule cell proliferation [20]. We found that zic1 was expressed in the URL cells in wild-type zebrafish at 48 hpf while it was dramatically down-regulated in arl13b morphants and mutants (Fig. 3A–C).
Fig. 3.
Disruption of arl13b impairs the development of cerebellar granule cells. A Expression of the granule cell progenitor marker, zic1, in the cerebellum of wild-type embryos at 48 hpf revealed by in situ hybridization. B, C Expression of zic1 is reduced in the URL of both arl13b mutant and morphant embryos compared with wild-type embryos. Note that zic1 expression is severely reduced in the dorsomedial subregions of the URL (arrows). D The expression of the differentiated granule cell marker, reelin, in the cerebellum of wild-type embryos at 3 dpf. E, F Expression of reelin is reduced in the cerebellum of both arl13b mutant and morphant embryos. Note that in some embryos reelin expression is almost absent in the dorsal medial subregions of the cerebellum (colored ovals). G–G’’ In Tg(neurod1:eGFP) transgenic embryos, GFP+ granule cells are grouped into three clusters, the dorsomedial (dashed ovals), dorsoposterior, and ventrolateral subdivisions. H–H’’ The pattern of GFP+ granule cells is dramatically altered in the cerebellum, and particularly in the dorsomedial cerebellar subdivisions (dashed ovals) are severely affected in arl13b morphants. The parallel fibers connecting the two hemispheres are disrupted in arl13b morphants. Dorsal views of the embryos are shown. I–L Malformations of the dorsomedial cerebellar subdivisions (dashed ovals) and parallel fibers are also present in arl13b mutants both at 3 dpf and 4 dpf. cb, cerebellum; PF, parallel fiber. A–F, Scale bar 100 μm. G–L, Scale bar 50 μm.
These results suggested that Arl13b participates in cerebellar development by regulating the development of granule cell progenitors.
Depletion of arl13b Impairs Granule Neurons Specifically in the Corpus Cerebelli
The impairment of granule cell progenitors might interfere with the subsequent development of granule neurons, so we next examined the differentiation of granule neurons. We found that reelin, a marker of differentiated granule cells [21], was markedly decreased in the cerebellum of arl13b-deficient embryos, particularly in the dorsomedial subregions (Fig. 3D–F). It has been demonstrated that NEUROD1 is expressed in immature and mature cerebellar granule neurons in both mammals and zebrafish [18, 22]. We found that neurod1 was absent from the dorsomedial cerebella of arl13b morphants (Fig. S4) while the dorsolateral neurons were not affected, similar to the expression of reelin. We further examined the development of cerebellar granule cells using the transgenic line Tg(neurod1:EGFP) [23]. In these embryos, GFP was expressed in three main clusters: two close to the midline, the dorsomedial and dorsoposterior granule cells, and one distant from the midline, the ventrolateral granule cells (Fig. 3G–G’’). The patterns of the Tg(neurod1:EGFP) GFP+ granule cells resembled the granule cells labeled by Tg(gata1:GFP) [19]. Knocking down arl13b in Tg(neurod1:EGFP) embryos caused a global reduction of GFP+ granule neurons compared to the control transgenic embryos. Particularly, the dorsomedial clusters were severely affected and, in some embryos, these clusters were totally absent (Fig. 3H–H’’). The ventrolateral and dorsoposterior clusters were still present though with reduced numbers of GFP+ neurons (Fig. 3H–H’’). These results were consistent with the expression pattern of neurod1 detected by in situ hybridization (Fig. S4). The dorsomedial subdivision of granule neurons populate the CCe [17, 19]. The defects of CCe granule cells in arl13b-deficient zebrafish resembled the defects in the cerebellar vermis found in JS patients. Tg(neurod1:EGFP) labeled some parallel fibers at the midline of cerebellum (Fig. 3G’). Upon arl13b knockdown, the parallel fibers were dramatically disrupted (Fig. 3H’), similar to those found by anti-tubulin immunostaining (Fig. 1F–I). These phenotypes were also found in arl13b mutants (Fig. 3I–L). The arl13b mutants were crossed with Tg(neurod1:EGFP) fish and transgenic labeled arl13b homozygotes were picked. The selective malformation of CCe granule cells and disruption of parallel fibers were frequently observed in arl13b mutants both at 3 dpf (Fig. 3I, J) and 4 dpf (Fig. 3K, L).
The qPCR results revealed that the markers of differentiated granule cells calbindin 2a (calretinin, calb2a) and GABA receptor alpha 6a (gabra6a) were dramatically reduced in arl13b mutants (Fig. S3). These results suggested that not only the proliferation and migration but also the differentiation of granule cells is impaired in arl13b-deficient embryos.
Depletion of arl13b Impairs the Development of Cerebellar Purkinje Cells
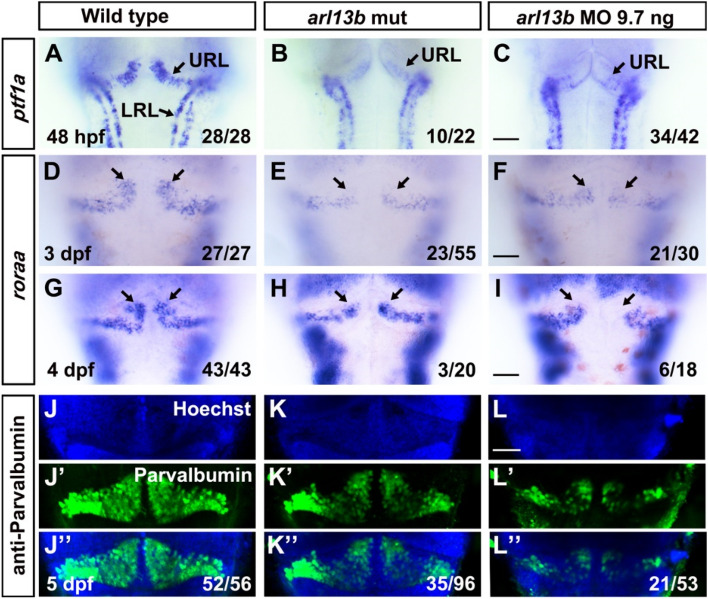
Both the function and development of the cerebellum are dependent on the cooperation between granule neurons and Purkinje cells, two major neuronal types in the cerebellum, so we further investigated the role of arl13b in the development of Purkinje cells. ptf1a was used as a marker to label the precursors of Purkinje cells by in situ hybridization and it was found to be expressed in the ventricular zone of wild-type embryos at 48 hpf (Fig. 4A). In arl13b mutant and morphant embryos, the expression level of ptf1a was dramatically and selectively reduced in the dorsomedial ventricular zone, while it was relatively normal in the ventrolateral regions (Fig. 4B, C). The phenotype of morphants was more penetrating than that of mutants, as found in granule cells. These results demonstrated that the progenitors of Purkinje cells are selectively reduced dorsomedially in arl13b-deficient embryos. We further examined differentiating Purkinje cells using roraa as a marker [24]. This revealed that the dorsomedial clusters of differentiating Purkinje cells were dramatically reduced, while the ventrolateral clusters were only mildly reduced in arl13b-deficient embryos at 3 dpf (Fig. 4D–F). This is reminiscent of the selective reduction of dorsomedial granule cell clusters (Fig. 3). A similar decrease of dorsomedial clusters was observed at 4 dpf (Fig. 4G–I). As Purkinje cells differentiate and mature, early distinct clusters of Purkinje cells merge together, become unified, and form a continuous layer spanning the mediolateral width of the cerebellum [25]. Immunostaining with an antibody against parvalbumin was used to label differentiated Purkinje cells and demonstrated that they were dramatically reduced globally in the cerebellum at 5 dpf, including both the dorsomedial and ventrolateral clusters (Fig. 4K–L’’). qPCR results revealed that the markers of immature and mature Purkinje cells roraa, aldoca, and pvalb7 were reduced (Fig. S3D), consistent with the immunostaining results.
Fig. 4.
Disruption of arl13b reduces both precursor and differentiated cerebellar Purkinje cells. A–I Representative images of in situ hybridization using anti-sense probes against ptf1a and roraa to label Purkinje precursors and differentiated cells, respectively. The dorsomedial clusters of precursor of Purkinje cells and differentiated Purkinje neurons are selectively reduced (arrows). J–L’’ Immunostaining of mature Purkinje neurons using anti-parvalbumin antibody reveals that the dorsomedial Purkinje neurons are reduced in arl13b-deficient embryos. A–I, Scale bar 100 μm. J–L’’ Scale bar 50 μm.
The global reduction of mature Purkinje cells differed from the specific reduction of dorsomedial clusters of granule cells at the late larval stage. The early specific dorsomedial defects of both Purkinje progenitors and granule progenitors are likely due to unknown common defects in the mechanisms required and partly shared by the two types of neural progenitor.
Depletion of arl13b Reduces wnt1 Expression in the Developing Cerebellum
Arl13b has been demonstrated to be associated with Hedgehog signaling pathways [7, 26, 27], so we examined the expression of Hedgehog signaling components by in situ hybridization and qPCR. shh was not detected in the cerebellar anlage at 24 hpf although it was ventrally expressed along the neural tube (Fig. S5). No detectable change of shh expression in the cerebellum was observed in arl13b morphants, although its expression in the zona limitans intrathalamica was reduced dorsally (Fig. S5). The expression level of the hedgehog receptor patched1 was relatively low (Fig. S5). It was hard to tell whether it was expressed in the cerebellum in wild-type embryos at 30 hpf as well as whether its expression level or pattern changed. We then used qPCR to assess the expression levels of Shh signaling components. The expression of the receptors smoothen (smo) and patched1 (ptch1), and the Hedgehog signaling components gli2a, gli2b, and gli3 were dynamically regulated in the early stages of cerebellar development (Fig. S5). These results suggested that Hedgehog signaling components are altered globally in arl13b mutants while no detectable change occurs early in cerebellar development.
Wnt signaling is also critical for cerebellar development [28] and cooperates with Hedgehog signaling to regulate cerebellar olig2+ cell development [29]. Intriguingly, recessive mutations of human WNT1 result in hypoplasia of the cerebellum, particularly of the vermis, suggesting a conserved role of WNT1 in vermis development [30]. Besides being mainly expressed at the midbrain–hindbrain boundary, in mice wnt1 is also transiently expressed in the URL [31], from which many cerebellar neurons are derived. The transient expression of wnt1 in the URL is conserved in zebrafish [29]. We found that wnt1 was transiently expressed in the dorsal cerebellum at 30 hpf in wild-type embryos (Fig. 5A). Its expression was strikingly reduced and almost absent from the cerebellum of some arl13b mutants (Fig. 5B), indicating that Wnt signaling is disrupted early in cerebellar development.
Fig. 5.
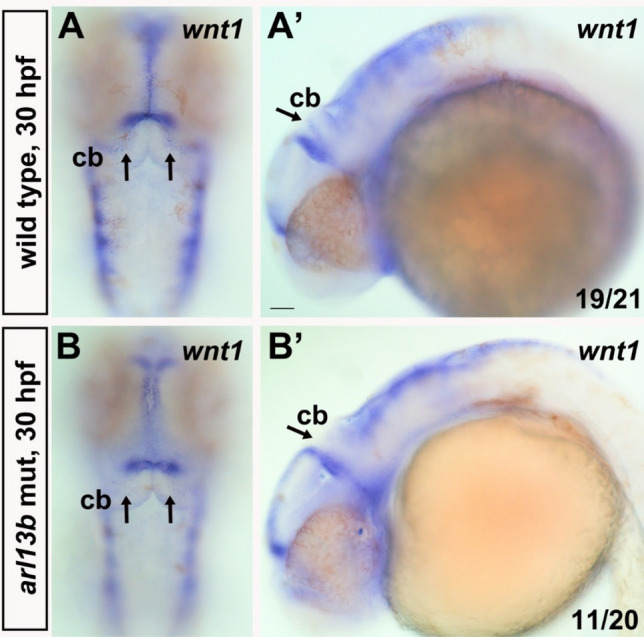
Wnt1 is selectively down-regulated in the cerebellum of arl13b mutants. wnt1 is transiently expressed in the cerebellum of WT embryos (A, A’) (arrows) while it is dramatically reduced in arl13b mutants (B, B’). A and B, dorsal view; A’ and B’, lateral view. Note that the expression of wnt1 is selectively decreased in the cerebellum while its expression at the midbrain–hindbrain boundary is intact. A–B’, Scale bar 100 μm.
Activating Wnt Signaling Partially Rescues Cerebellar Defects in arl13b Mutants
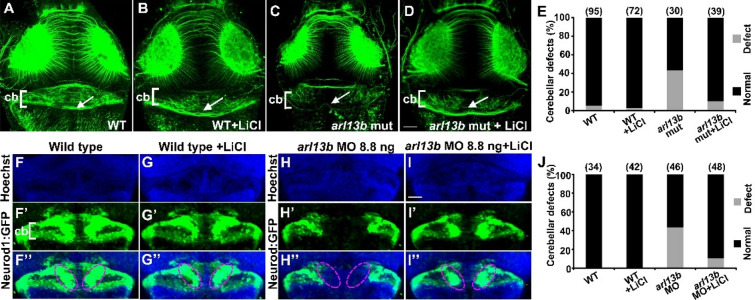
We started to explore whether activating Wnt signaling with lithium, an agonist of Wnt signaling, can mitigate the cerebellar defects in arl13b-deficient embryos. The dose of lithium chloride (LiCl) was optimized for different developmental stages. We found that treating the embryos with 50 mmol/L LiCl at 30–37 hpf reduced the cerebellar structural defects in arl13b mutants. The wild-type embryos treated with LiCl at the same time showed no detectable cerebellar defects (Fig. 6A–E). We further examined the rescue effects of LiCl with Tg(neurod1:EGFP) embryos and found that the dorsomedial reduction of granule cells caused by arl13b knockdown was mitigated by LiCl treatment (Fig. 6F–J).
Fig. 6.
Treating the arl13b mutants with lithium mitigates the morphological defects in the cerebellum. A–D Representative images of embryos treated with 50 mmol/L LiCl at 30–37 hpf, fixed at 3 dpf, and immunostained with anti-tubulin antibody to reveal cerebellar morphology. Treatment of wild-type embryos with Li+ does not affect the cerebellar morphology (A, B) (arrows). The morphological defects of the cerebellum in arl13b mutants treated with Li+ are partially rescued (C, D) (arrow). A–D, Scale bar 100 μm. E Statistics revealing that the proportion of arl13b mutant embryos with cerebellar defects is dramatically decreased by LiCl treatment. F–I’’ Representative images of Tg(neurod1:EGFP) transgenic embryos used to label granule cells. Treating wild-type transgenic embryos with Li+ causes no defect (F–G’’) (dashed ovals). Treating arl13b morphant transgenic embryos restores the dorsomedial cluster of granule cells (H–I’’) (dashed ovals). F–I’’, Scale bar 100 μm. J The proportion of arl13b morphant embryos with cerebellar defects in the dorsomedial clusters is dramatically reduced by LiCl treatment.
Discussion
Most published papers study the role of JS genes during late embryonic or postnatal development of the cerebellum, though the molar tooth sign can be detected early in the first trimester [32]. Moreover, these studies mainly focus on cerebellar granule cells. In this study, we found that the disruption of arl13b in zebrafish larvae leads to early cerebellar malformations and defects in both granule and Purkinje cell progenitors. The early developmental patterning of cerebellar granule neurons is compromised particularly in the dorsomedial subregions of the CCe, a structure homologous to the mammalian cerebellar vermis. This phenotype is reminiscent of hypoplasia of the cerebellar vermis in JS. Molecular and cellular studies revealed that the early development of the progenitors of both granule cells and Purkinje cells are selectively altered in arl13b-deficient embryos. Arl13b may be involved in regulating a network of signaling pathways, including Wnt and Atoh1. Treating the arl13b mutants with Li+, an agonist of Wnt signaling, partially rescued the cerebellar defects.
Our finding that granule neurons are reduced specifically in the dorsomedial subregions of the CCe in arl13b-deficient zebrafish larvae is consistent with the enrichment of arl13b expression in the ventricle at early stages, from 25 hpf to 40 hpf and later [9, 33]. This finding is also consistent with reports that granule neurons are reduced along the anteroposterior but not the mediolateral axis in mouse mutants of the cilia genes Kif3a, Ift8, or Rpgrip1l [34, 35]. The reduction of granule neurons in these mutants could be caused by proliferation defects in granule progenitors [34, 35]. Although the cerebellar granule neurons in Ahi1- or Cep290- knockout mice are only slightly affected, there are significant proliferation defects of granule cells at an early developmental stage (E16.5)[2]. Proliferation defects of granule progenitors seem to be common in these cilia gene mutants [3, 34, 35]. The reduction of granule neurons in arl13b-deficient zebrafish was likely caused by proliferation defects in granule progenitors since the expression pattern of atoh1a was altered and atoh1c was dramatically down-regulated (Fig. 2 and S3). Atoh1 is known to be critical for cerebellar granule neurogenesis in the mouse [36] and for the proliferation of granule cells in zebrafish [18]. atoh1c-derived cells contribute to the majority of granule neurons in the CCe, while a minority is derived from atoh1a progenitors [17, 18]. The dramatic down-regulation of atoh1c and the selective hypoplasia of the CCe in arl13b-deficient embryos are consistent with the critical role of atoh1c in populating granule neurons in the CCe [17]. The dramatically decreased expression of zic1 and neurod1 specifically in the dorsomedial subregions of the cerebellum (Figs. 2 and S4) is also consistent with the proliferation defects of granule progenitors since both genes are implicated in granule cell proliferation [18, 20]. We immunostained the embryos using anti-pH3 antibody, a marker of cell proliferation, and found reduced fluorescence intensity of pH3-positive cells in the cerebellum of arl13b mutants (Fig. S6). These results are consistent with potential proliferation defects of granule cells in the cerebellum.
It has been shown that Arl13b regulates the migration and location of interneurons [5]. The specific hypoplasia of the dorsomedial subregions of the CCe in arl13b-deficient zebrafish could also be due to migration defects in granule cell precursors. It has been demonstrated that the URL generates different populations of granule cell precursors along its mediolateral axis. These precursors migrate along different routes and form different functional compartments of the mature cerebellum: the eminentia granularis and the CCe [19]. Atoh1 has been shown to be critical for the migration of granule cell precursors out of the URL in mice and particularly Atoh1c in zebrafish [17, 36]. The reduction of atoh1c expression in arl13b-deficient zebrafish could lead to migration defects of the granule cell precursors. The migration defects might impair the differentiation of granule neurons since several markers of granule neurons are altered, particularly neurod1. The absence of Tg(neurod1:EGFP)+ cells in the dorsomedial domain of the cerebellum in arl13b mutants differs from that seen in atoh1c mutants [17], in which neurod1 is expressed in most of the Tg(atoh1c:kaede)+ granule cell progenitors, including the dorsomedial cells along the midline. These results suggest that depletion of arl13b also disrupts other signaling pathways besides Atoh1.
Wnt1 is transiently expressed in the cerebellum of zebrafish [29] (Fig. 5) and mouse [31] and its mutations in humans result in hypoplasia of the cerebellar vermis [30], indicating that Wnt signaling is a potential mechanism underlying the pathology of arl13b mutant zebrafish and humans. Besides wnt1, other wnt genes have also been detected in the cerebellum, such as wnt3, wnt7a and wnt10b [37], which might be involved in early cerebellar development [3, 38, 39]. Further study is required to investigate whether they are also regulated by Arl13b. The Wnt downstream signaling components β-catenin and Gsk3β regulate the transcription and protein stability of Atoh1 [40, 41]. Wnt1 has been proposed to regulate Atoh1 expression in the developing cerebellum [31]. These reports are consistent with our findings that atoh1c and 1a expression is reduced in arl13b-deficient embryos (Figs. 2 and S3). Moreover, wnt1 is also expressed at the midbrain–hindbrain boundary and it has been speculated to contribute to the development of the cerebellum. Whether the boundary Wnt1 also contributes to the cerebellar defects in arl13b-deficient embryos needs further experiments. Lithium treatment partially rescues the cerebellar defects in both zebrafish arl13b (Fig. 6) and mouse Ahi1 mutants [2], suggesting that Wnt signaling is a conserved key pathway regulating early cerebellar development and could serve as a common potential therapy target in JS. Atoh1 and Wnt signaling might only partially contribute to the mechanisms of Arl13b signaling since Arl13b has been found to be distributed subcellularly in the cilium and cytoplasm and is expressed in many tissues and cells.
Most JS research has focused on granule cells and has seldom dealt with Purkinje cells. In human JS samples, the migration of cerebellar Purkinje cells is faulty, with heterotopic and locally-interrupted Purkinje cell layers [42]. The malformation of the Purkinje cell monolayer has also been found in JS gene mutant mice [3, 43]. These late Purkinje cell defects could be due to an early developmental deficiency. It has been demonstrated that mutations of the JS gene Zfp423/ZNF423 in the mouse impair the early development of Purkinje cell progenitors [44]. Our results also demonstrated that the progenitors of Purkinje cells were selectively disrupted in the dorsomedial cluster in arl13b-defecient embryos (Fig. 4), like the early defects in granule cell progenitors. The coincident selective defects of Purkinje and granule cell progenitors suggest that these two populations interact or share common mechanisms. Radial glia and the Bergmann glial cells derived from them provide a scaffold for the migration of both Purkinje cells and granule cells early in cerebellar development [45, 46]. Ciliary Arl13b has been shown to be critical for the polarization of the radial glial scaffold and the laminar organization of neurons in mouse cerebral cortex [6]. The available evidence and our current results suggest that the role of Arl13b in neural development is conserved between species and Arl13b participates in the polarization of the radial glial scaffold as well as coordinating the proliferation and migration of both Purkinje cells and granule cells early in cerebellar development. The migration of Purkinje cells and granule cells also shares molecular mechanisms, such as Reelin signaling [45], the expression level of which was also selectively reduced in the dorsomedial cerebellum in arl13b-deficient embryos (Fig. 3).
Conclusions
We have established a JS model using arl13b-deficient zebrafish, which recapitulate some of the signs of JS, such as locomotor and cerebellar developmental defects. The disruption of arl13b resulted in a dramatic reduction of granule cells specifically in the CCe, a structure homologous to the human cerebellar vermis. The expression of the proneural gene atoh1 in a subpopulation of granule cells was down-regulated in the cerebellum. The early development of Purkinje cells was also selectively disrupted in the dorsomedial cerebellum. wnt1, a gene transiently expressed early in cerebellar development, was dramatically down-regulated. Furthermore, activating Wnt signaling mitigated the granule cell defects caused by arl13b-disruption. Our results reveal the critical role of arl13b in the early development of cerebellar granule and Purkinje cells. We propose that the arl13b-deficient zebrafish can serve as a powerful tool to investigate the pathological mechanisms underlying JS.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31171044, 81160144, and 81760216) and the Young Scientist Project of Jiangxi Province, China (20122BCB23007). We thank Dr. Zhao-Xia Sun (Yale University) for kindly providing the arl13b mutants, Dr. Jing-Wei Xiong (Peking University) for the Tg(neurod1:EGFP) transgenic fish, and Dr. Sheng-Ping L. Hwang (Institute of Cellular and Organismic Biology, Academia Sinica, Taipei, China) for kindly providing plasmids. We specially thank Professor Shi-Wen Luo (Nanchang University) for critical discussion and valuable advice.
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Jian Zhu and Han-Tsing Wang have contributed equally to this work.
References
- 1.Romani M, Micalizzi A, Valente EM. Joubert syndrome: congenital cerebellar ataxia with the molar tooth. Lancet Neurol. 2013;12:894–905. doi: 10.1016/S1474-4422(13)70136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lancaster MA, Gopal DJ, Kim J, Saleem SN, Silhavy JL, Louie CM, et al. Defective Wnt-dependent cerebellar midline fusion in a mouse model of Joubert syndrome. Nat Med. 2011;17:726–731. doi: 10.1038/nm.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashford AL, Subramanian V. Mice with a conditional deletion of Talpid3 (KIAA0586)—a model for Joubert syndrome. J Pathol. 2019;248:396–408. doi: 10.1002/path.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, et al. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higginbotham H, Eom TY, Mariani LE, Bachleda A, Hirt J, Gukassyan V, et al. Arl13b in primary cilia regulates the migration and placement of interneurons in the developing cerebral cortex. Dev Cell. 2012;23:925–938. doi: 10.1016/j.devcel.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higginbotham H, Guo J, Yokota Y, Umberger NL, Su CY, Li J, et al. Arl13b-regulated cilia activities are essential for polarized radial glial scaffold formation. Nat Neurosci. 2013;16:1000–1007. doi: 10.1038/nn.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspary T, Larkins CE, Anderson KV. The graded response to sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Song P, Dudinsky L, Fogerty J, Gaivin R, Perkins BD. Arl13b interacts with Vangl2 to regulate cilia and photoreceptor outer segment length in zebrafish. Invest Ophthalmol Vis Sci. 2016;57:4517–4526. doi: 10.1167/iovs.16-19898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duldulao NA, Lee S, Sun Z. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development. 2009;136:4033–4042. doi: 10.1242/dev.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu L, Chen L, Yan L, Perkins BD, Li S, Li B, et al. Mutant Ahi1 affects retinal axon projection in zebrafish via toxic gain of function. Front Cell Neurosci. 2019;13:81. doi: 10.3389/fncel.2019.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 12.Liu ZZ, Zhu J, Wang CL, Wang X, Han YY, Liu LY, et al. CRMP2 and CRMP4 are differentially required for axon guidance and growth in zebrafish retinal neurons. Neural Plast. 2018;2018:8791304. doi: 10.1155/2018/8791304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mu Z, Zhang S, He C, Hou H, Liu D, Hu N, et al. Expression of SoxC transcription factors during zebrafish retinal and optic nerve regeneration. Neurosci Bull. 2017;33:53–61. doi: 10.1007/s12264-016-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 15.Riley BB, Zhu C, Janetopoulos C, Aufderheide KJ. A critical period of ear development controlled by distinct populations of ciliated cells in the zebrafish. Dev Biol. 1997;191:191–201. doi: 10.1006/dbio.1997.8736. [DOI] [PubMed] [Google Scholar]
- 16.Hibi M, Shimizu T. Development of the cerebellum and cerebellar neural circuits. Dev Neurobiol. 2012;72:282–301. doi: 10.1002/dneu.20875. [DOI] [PubMed] [Google Scholar]
- 17.Kidwell CU, Su CY, Hibi M, Moens CB. Multiple zebrafish atoh1 genes specify a diversity of neuronal types in the zebrafish cerebellum. Dev Biol. 2018;438:44–56. doi: 10.1016/j.ydbio.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kani S, Bae YK, Shimizu T, Tanabe K, Satou C, Parsons MJ, et al. Proneural gene-linked neurogenesis in zebrafish cerebellum. Dev Biol. 2010;343:1–17. doi: 10.1016/j.ydbio.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 19.Volkmann K, Rieger S, Babaryka A, Koster RW. The zebrafish cerebellar rhombic lip is spatially patterned in producing granule cell populations of different functional compartments. Dev Biol. 2008;313:167–180. doi: 10.1016/j.ydbio.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 20.Aruga J, Minowa O, Yaginuma H, Kuno J, Nagai T, Noda T, et al. Mouse Zic1 is involved in cerebellar development. J Neurosci. 1998;18:284–293. doi: 10.1523/JNEUROSCI.18-01-00284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costagli A, Kapsimali M, Wilson SW, Mione M. Conserved and divergent patterns of Reelin expression in the zebrafish central nervous system. J Comp Neurol. 2002;450:73–93. doi: 10.1002/cne.10292. [DOI] [PubMed] [Google Scholar]
- 22.Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H, Bu Y, Wu Q, Wang X, Chang N, Lei L, et al. Mecp2 regulates neural cell differentiation by suppressing the Id1 to Her2 axis in zebrafish. J Cell Sci. 2015;128:2340–2350. doi: 10.1242/jcs.167874. [DOI] [PubMed] [Google Scholar]
- 24.Katsuyama Y, Oomiya Y, Dekimoto H, Motooka E, Takano A, Kikkawa S, et al. Expression of zebrafish ROR alpha gene in cerebellar-like structures. Dev Dyn. 2007;236:2694–2701. doi: 10.1002/dvdy.21275. [DOI] [PubMed] [Google Scholar]
- 25.Hamling KR, Tobias ZJ, Weissman TA. Mapping the development of cerebellar Purkinje cells in zebrafish. Dev Neurobiol. 2015;75:1174–1188. doi: 10.1002/dneu.22275. [DOI] [PubMed] [Google Scholar]
- 26.Horner VL, Caspary T. Disrupted dorsal neural tube BMP signaling in the cilia mutant Arl13b hnn stems from abnormal Shh signaling. Dev Biol. 2011;355:43–54. doi: 10.1016/j.ydbio.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao J, Xu L, Chen L, Lu Q, Xie X, Shi W, et al. Arl13b promotes gastric tumorigenesis by regulating smo trafficking and activation of the Hedgehog signaling pathway. Cancer Res. 2017;77:4000–4013. doi: 10.1158/0008-5472.CAN-16-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hibi M, Matsuda K, Takeuchi M, Shimizu T, Murakami Y. Evolutionary mechanisms that generate morphology and neural-circuit diversity of the cerebellum. Dev Growth Differ. 2017;59:228–243. doi: 10.1111/dgd.12349. [DOI] [PubMed] [Google Scholar]
- 29.McFarland KA, Topczewska JM, Weidinger G, Dorsky RI, Appel B. Hh and Wnt signaling regulate formation of olig2+ neurons in the zebrafish cerebellum. Dev Biol. 2008;318:162–171. doi: 10.1016/j.ydbio.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aldinger KA, Mendelsohn NJ, Chung BH, Zhang W, Cohn DH, Fernandez B, et al. Variable brain phenotype primarily affects the brainstem and cerebellum in patients with osteogenesis imperfecta caused by recessive WNT1 mutations. J Med Genet. 2016;53:427–430. doi: 10.1136/jmedgenet-2015-103476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagan N, Zervas M. Wnt1 expression temporally allocates upper rhombic lip progenitors and defines their terminal cell fate in the cerebellum. Mol Cell Neurosci. 2012;49:217–229. doi: 10.1016/j.mcn.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quarello E. Enlarged intracranial translucency and molar tooth sign in the first trimester as features of Joubert syndrome and related disorders. Ultrasound Obstet Gynecol. 2016;48:532–534. doi: 10.1002/uog.15856. [DOI] [PubMed] [Google Scholar]
- 33.Song P, Perkins BD. Developmental expression of the zebrafish Arf-like small GTPase paralogs arl13a and arl13b. Gene Expr Patterns. 2018;29:82–87. doi: 10.1016/j.gep.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spassky N, Han YG, Aguilar A, Strehl L, Besse L, Laclef C, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chizhikov VV, Davenport J, Zhang Q, Shih EK, Cabello OA, Fuchs JL, et al. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. J Neurosci. 2007;27:9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 37.Duncan RN, Panahi S, Piotrowski T, Dorsky RI. Identification of Wnt genes expressed in neural progenitor zones during zebrafish brain development. PLoS One. 2015;10:e0145810. doi: 10.1371/journal.pone.0145810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lucas FR, Salinas PC. WNT-7a induces axonal remodeling and increases synapsin I levels in cerebellar neurons. Dev Biol. 1997;192:31–44. doi: 10.1006/dbio.1997.8734. [DOI] [PubMed] [Google Scholar]
- 39.Salinas PC, Fletcher C, Copeland NG, Jenkins NA, Nusse R. Maintenance of Wnt-3 expression in Purkinje cells of the mouse cerebellum depends on interactions with granule cells. Development. 1994;120:1277–1286. doi: 10.1242/dev.120.5.1277. [DOI] [PubMed] [Google Scholar]
- 40.Shi F, Cheng YF, Wang XL, Edge AS. Beta-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3’ enhancer. J Biol Chem. 2010;285:392–400. doi: 10.1074/jbc.M109.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuchiya K, Nakamura T, Okamoto R, Kanai T, Watanabe M. Reciprocal targeting of Hath1 and beta-catenin by Wnt glycogen synthase kinase 3beta in human colon cancer. Gastroenterology. 2007;132:208–220. doi: 10.1053/j.gastro.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 42.Aguilar A, Meunier A, Strehl L, Martinovic J, Bonniere M, Attie-Bitach T, et al. Analysis of human samples reveals impaired SHH-dependent cerebellar development in Joubert syndrome/Meckel syndrome. Proc Natl Acad Sci U S A. 2012;109:16951–16956. doi: 10.1073/pnas.1201408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdelhamed ZA, Abdelmottaleb DI, El-Asrag ME, Natarajan S, Wheway G, Inglehearn CF, et al. The ciliary Frizzled-like receptor Tmem67 regulates canonical Wnt/beta-catenin signalling in the developing cerebellum via Hoxb5. Sci Rep. 2019;9:5446. doi: 10.1038/s41598-019-41940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casoni F, Croci L, Bosone C, D’Ambrosio R, Badaloni A, Gaudesi D, et al. Zfp423/ZNF423 regulates cell cycle progression, the mode of cell division and the DNA-damage response in Purkinje neuron progenitors. Development. 2017;144:3686–3697. doi: 10.1242/dev.155077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schilling K. Moving into shape: cell migration during the development and histogenesis of the cerebellum. Histochem Cell Biol. 2018;150:13–36. doi: 10.1007/s00418-018-1677-6. [DOI] [PubMed] [Google Scholar]
- 46.Xu H, Yang Y, Tang X, Zhao M, Liang F, Xu P, et al. Bergmann glia function in granule cell migration during cerebellum development. Mol Neurobiol. 2013;47:833–844. doi: 10.1007/s12035-013-8405-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.