Overexpression of OsPAP10c driven by its native promoter enhances the growth and yield of rice in comparison both with the wild-type and with plants where overexpression is driven by a ubiquitin promoter.
Keywords: Acid phosphatase, native promoter, organic phosphate, overexpression, Pi deficiency, rice, root
Abstract
Whilst constitutive overexpression of particular acid phosphatases (APases) can increase utilization of extracellular organic phosphate, negative effects are frequently observed in these transgenic plants under conditions of inorganic phosphate (Pi) sufficiency. In this study, we identified rice purple acid phosphatase 10c (OsPAP10c) as being a novel and major APase that exhibits activities associated both with the root surface and with secretion. Two constructs were used to generate the OsPAP10c-overexpression plants by driving its coding sequence with either a ubiquitin promoter (UP) or the OsPAP10c-native promoter (NP). Compared with the UP transgenic plants, lower expression levels and APase activities were observed in the NP plants. However, the UP and NP plants both showed a similar ability to degrade extracellular ATP and both promoted root growth. The growth performance and yield of the NP transgenic plants were better than the wild-type and UP plants in both hydroponic and field experiments irrespective of the level of Pi supply. Overexpression of APase by its native promoter therefore provides a potential way to improve crop production that might avoid increased APase activity in untargeted tissues and its inhibition of the growth of transgenic plants.
Introduction
With the increasing demands on agricultural production worldwide, phosphorus (P) is receiving more and more attention as a non-renewable resource (Gilbert, 2009). Although P is abundant in the Earth’s crust, plants can only take up the water-soluble inorganic phosphate (Pi) in the soil. As P diffuses slowly and is highly fixed in the soil, the concentration of Pi is usually very low, and not enough to support the farming systems in many countries. Consequently, to obtain high crop yields, chemical P fertilizer is widely used on agricultural land and has increased costs and caused environmental damage (Shen et al., 2011). Developing crop varieties that are adapted to conditions of low-Pi stress therefore has an important role to play in developing sustainable agriculture and maintaining the global food supply.
Pi starvation has been reported to induce the synthesis of intra- and extracellular acid phosphatase (APase) isozymes in plants (Bozzo et al., 2006). More specifically, intracellular acid phosphatases have been shown to be up-regulated after 2 d of Pi starvation, which coincides with a reduction in the levels of intracellular free Pi, and they are believed to be involved in recycling Pi from intracellular P metabolites (Bozzo et al., 2006; Pratt et al., 2009). In contrast, under prolonged conditions of Pi starvation, secreted acid phosphatases accumulate in the growing medium in order to degrade extracellular organic P (Po) compounds (Bozzo et al., 2006). The major intracellular and secreted acid phosphatases have been determined in several plant species, and the majority of them are encoded by purple acid phosphatase genes (Miller et al., 2001; Bozzo et al., 2004; Veljanovski et al., 2006; Liang et al., 2010; Tran et al., 2010b).
Purple acid phosphatases (PAPs; EC 3.1.3.2) are a family of binuclear metalloenzymes that show a purple or pink colour in solution (Olczak et al., 2003). A wide range of phosphate esters and anhydrides can be hydrolysed by different PAPs (Tran et al., 2010a). PAPs were first isolated from mammalian species and play a role in bone metabolism (Feder et al., 2019a). Serum concentrations of PAPs are significantly increased in patients suffering from osteoporosis, and hence several inhibitors have been developed as therapeutic agents to treat this condition (Hussein et al., 2018; Feder et al., 2019b). Since their discovery in mammals, many PAP isoforms have also been purified from plant tissues (del Pozo et al., 1999; Miller et al., 2001; Bozzo et al., 2004), and the physiological functions of a number of them have been characterized, most of which are related to P nutrition (Tran et al., 2010b; Wang et al., 2011; Robinson et al., 2012; Lu et al., 2016; Li et al., 2017; Mehra et al., 2017; Wu et al., 2018). In Arabidopsis, AtPAP12 and AtPAP26 have been shown to be the predominant PAP isozymes that are secreted into the medium under Pi starvation (Tran et al., 2010b; Ghahremani et al., 2019). Mutations of AtPAP12 and AtPAP26 result in a significant reduction in the activity of secreted APases and hence in the ability to utilize extracellular Po (Robinson et al., 2012). AtPAP12 and AtPAP26 can also be secreted into the cell wall and account for a substantial proportion of the APase activity localized to the root surface (Robinson et al., 2012). However, mutation of AtPAP10 results in a ‘white root’ phenotype and impaired growth, indicating that AtPAP10 is the major APase isoform associated with the roots in Arabidopsis (Wang et al., 2011; Zhang et al., 2014). Recently, two root-associated APase isozymes induced by Pi starvation in soybean, GmPAP1-like and GmPAP21, have been identified using a proteomic analysis of cell wall proteins (Wu et al., 2018), and they have been shown to be involved in the adaptation of soybean roots to Pi starvation (Li et al., 2017; Wu et al., 2018). In addition to utilization of Po, NtPAP12 and OsPAP21b have been reported to regulate cell wall biosynthesis and root development (Kaida et al., 2009; Mehra et al., 2017).
As Po can account for 30–65% of the total P in soils, a number of acid phosphatases have been used to generate transgenic crops that have an increased utilization ability in order to improve growth performance under low-Pi conditions. In rice, overexpression of OsPAP10a, OsPAP10c, and OsPAP21b significantly increase APase activity and Po degradation (Tian et al., 2012; Lu et al., 2016; Mehra et al., 2017). The extracellular utilization of ATP is significantly enhanced in hairy roots of transgenic beans with overexpression of PvPAP3 or GmPAP1-like (Liang et al., 2010; Wu et al., 2018). In addition, in transgenic Arabidopsis with overexpression of AtPAP10, AtPAP12, and AtPAP26, growth is improved when plants are supplied with ADP and Fru-6-P (Wang et al., 2011, 2014). Overexpression of AtPAP15 and SgPAP23 (Stylosanthes guianensis), which show high phytase activities, facilitates the utilization of phytate-P and significantly improves the growth of transgenic plants (Wang et al., 2009; Liu et al., 2018). Transgenic soybean lines with overexpression of AtPAP15 also exhibit improved yields when grown on acid soils (Wang et al., 2009).
Although the overexpression of these PAP genes successfully increases the ability to degrade Po and improves plant growth under Pi-deficient conditions, the transgenic plants usually show either no response or inhibited growth performance under Pi-sufficient conditions (Wang et al., 2014; Lu et al., 2016; Liu et al., 2018; Wu et al., 2018). To avoid the negative effects of constitutive overexpression of PAP genes, in this study we increased the expression of OsPAP10c by means of its own promoter. It has previously been reported that OsPAP10c is a novel monocotyledon PAP gene that is specifically induced by Pi starvation in roots (Lu et al., 2016). We found that mutants of OsPAP10c had significantly decreased activities of both root surface-associated and secreted APase, indicating that OsPAP10c is one of the major APase isoforms in rice. Compared with plants with constitutive overexpression of OsPAP10c, transgenic plants with OsPAP10c driven by the native promoter showed lower transcripts levels and APase activity in untargeted tissues. The growth performance and yield of the transgenic plants driven by the native promoter were better than those of the wild-type and of plants with constitutive overexpression in both hydroponic and field experiments irrespective of the level of Pi supply.
Materials and methods
Plant materials and growth conditions
Seeds of rice (Oryza sativa) cv Nipponbare were surface-sterilized with 1% nitric acid and germinated at 37 °C in the dark for 2 d. Uniform germinated seeds were cultured hydroponically using a solution containing 1.425 mM NH4NO3, 0.513 mM K2SO4, 0.998 mM CaCl2, 1.643 mM MgSO4, 0.25 mM NaSiO3, 0.009 mM MnCl2, 0.075 μM (NH4)6Mo7 O24, 0.019 μM H3BO3, 0.155 μM CuSO4, 0.152 μM ZnSO4, and 0.125 mM EDTA-Fe(II), pH 5.5. For the Pi-sufficient treatment (+Pi) the nutrient solution also contained 0.323mM NaH2PO4; the solution without NaH2PO4 represented the Pi-deficient treatment (–P).The seedlings were grown in a greenhouse under a 12/12-h photoperiod (200 µmol photons m–2 s–1) at 32/24 °C and 60% relative humidity.
For ATP degradation and utilization experiments, plants were grown in +P media for 15 d, after which half of the seedlings were treated with 0.1 mM ATP for a further 10 d.
For analysis of agronomic traits, plants were grown in an experimental field where they received either 80 kg ha–1 phosphate fertilizer (high-P treatment, HP) or no fertilizer (low-P treatment, LP). The properties of the soil were as follows: pH 7.01; organic matter, 9.03 g kg–1; available nitrogen, 76.38 mg kg–1; available potassium, 195.33 mg kg–1; and effective phosphorus, 6.11 mg kg–1. The following agronomic traits were determined at maturity: plant height, tiller number, seed-setting rate, 1000-grain weight, grain number per plant, and grain yield per plant. Seed-setting rate was calculated as the ratio of solid grains to total grains on the spikes.
Vector construction and plant transformation
To create OsPAP10c CRISPR lines, a 20-bp sgRNA targeting the first exon of OsPAP10c was cloned into the CRISPR-Cas9 expression vector pRGEB31 according to the method described by Xie et al. (2014). OsPAP10c-overexpression plants driven by a ubiquitin promoter (UP) were obtained as described by Lu et al. (2016). In addition, the full-length cDNA of OsPAP10c was cloned into the vector pCAMIA1300 using the OsPAP10c-native promoter (NP) to drive its coding sequence (CDS). Transgenic plants were generated by Agrobacterium-mediated transformation as reported previously (Tian et al., 2012).
To construct the CaMV 35S::OsPAP10c transformation vector, the CDS of OsPAP10c was amplified using a High-Fidelity PCR Kit (CWBIO) with the forward primer 5´-CGGAGCTCGGTACCCATGGGGATGCTGCGGTGG-3´ and reverse primer 5´-CTCTAGAGGATCCCCCTACATCGTCGTTGGTGGGC-3´. The amplified CDS was subsequently cloned into the pCAM1300-mod vector using a GBclonart Seamless Cloning Kit (GBI, Suzhou, China) according to the manufacturer’s instructions. The construct was transferred into Agrobacterium tumefaciens strain GV3101 and transformed into the Arabidopsis pap10 mutant using the floral dip method (Clough and Bent, 1998). The resultant transgenic plants were confirmed by their resistance to hygromycin and PCR analysis.
Southern blot analysis of transgenic plants
For Southern blot hybridization analysis, 10 μg of genomic DNA was digested with the EcoR I restriction enzyme, separated on a 0.8% agarose/TAE gel, and capillary-blotted onto a nylon membrane. The hygromycin phosphotransferase (hpt) probe and OsPAP10c probe were labeled with digoxigenin and the membranes were hybridized with each probe separately. The membranes were visualized using a multifunctional image analyser (FLA-1500, FUJIFILM).
Quantitative real-time PCR
Total RNA was extracted from the tissues of rice plants using TRIzol reagent (ThermoFisher Scientific). Samples of 2 μg of total RNA were used to synthesize cDNA using a High-Capacity cDNA reverse-transcription kit (ThermoFisher Scientific). qRT-PCR assays were performed with SYBR Green Real-Time PCR Master Mix reagent (Takara) on an Applied Biosystems QuantstudioTM Real-Time PCR System. The rice actin gene (OsACTIN) was used as the internal reference and relative expression levels were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001). The sequences of the OsACTIN primers were as follows: 5´-GACGGACGCACCCTGGCTGA-3´ and 5´-TGCTGCCAATTACCATATACC-3´. The sequences of the OsPAP10c primers were as follows: 5´-AGCCGTTCTGGTACTCCGTCAAG-3´ and 5´-CGCGCATCGTCTCCCCCTCCAT-3´. The sequences of the OsPAP10a primers were as follows: 5´-GAGATAGATTTTGCCCCAGAACT-3´ and 5´-AGCTTCAAGCCACTTGTACTGAG-3´.
Protein extraction and quantification of APase activity in plant tissues
Total protein was extracted from root and leaf tissues in frozen extraction buffer as described by Lu et al. (2016). The protein content was quantified using Coomassie Blue in assay reagent. For quantification of tissue APase activity, 1 µg (roots) or 10 µg (leaves) of total protein were incubated with 600 µl of 10 mM p-nitrophenyl phosphate (pNPP) in a reaction mixture containing 50 mM sodium acetate, pH 5.5. Reactions were incubated at 25 ºC for 30 min and were then stopped by the addition of 1.2 ml of 1 M NaOH. The absorbance was determined at 410 nm using a microplate assay (Tecan). The APase activity was expressed as μmol pNP min–1 mg–1 protein.
Protein extraction from culture media and in-gel APase profiling
Rice calli were liquid-cultured according to the method described in detail by Lu et al. (2016). In brief, the calli were cultured with Pi-sufficient and -deficient R2S solutions for 7 d, and the liquid culture media were then collected. Proteins were concentrated and isolated using ultrafiltration centrifuge tubes (Millipore) with a 10-kDa cut-off at 4 °C according to the manufacturer’s instructions. The protein contents were quantified using Coomassie Plus (Bradford) Assay Kit (ThermoFisher Scientific). In-gel APase profiling was carried out as described by Wang et al. (2011). The proteins were separated on a 10% non-reducing PAGE gel at 4 °C. The gels were gently washed in cold distilled water to remove salt (10 min per wash and a total of six washes). The gels were then washed twice (15 min each) in a buffer containing 50 mM sodium acetate (pH 4.9) and 10 mM MgCl2. After equilibrium with the buffer, the gels were stained for APase activity in buffer containing 0.5 mg l–1 Fast Black potassium salt and 0.5 mg l–1 ß-naphthyl acid phosphate at 37 °C. The gels were then washed in distilled water and imaged.
Activity assays for root-associated APase
In vivo root activity staining of acid phosphatase was performed as previously described (Tian et al., 2012). Briefly, the roots of seedlings were excised and incubated with a 5-bromo-4-chloro-3-indolyl-phosphate (BCIP)-agar overlay solution containing 50 mM Na-acetate, 10 mM MgCl2, 0.5% agar, 0.05 mg ml−1 BCIP, pH 5.3, for 2 h at 25 °C for the visualization of the activity of root secreted acid phosphatases. The BCIP solution is a colorless acid phosphatase substrate and the activity is visualized as the blue color that results from its hydrolysis.
For quantification of root-associated APase activities, individual seedling was rinsed in distilled water and transferred to centrifuge tubes containing culture medium with 10 mM pNPP (pH 5.5). The seedlings were then placed in an incubator at 37 °C for 30 min. The reaction was stopped with 1 M NaOH and the absorbance was determined at 410 nm using a microplate assay (Tecan). APase activity was normalized to the fresh weigh of the roots (units g–1 FW) or expressed per seedling of Arabidopsis (nmol pNP min–1 per seedling).
Measurements of tissue Pi concentration and total P content
Pi concentrations were measured using the methods described previously by Lu et al. (2016). Briefly, 25 mg of fresh tissue samples were homogenized with 25 µl 5 M H2SO4 and 1.5 ml distilled water. After centrifugation at 11 180 g at 4 °C the supernatant was collected. The supernatant was diluted and mixed with malachite green reagent (19.4 mM H3BO3, 27.64 mM (NH4)6MO7O24.4H2O, 2.38 M H2SO4, 627.5 μM malachite green, and 0.1% polyvinyl alcohol) at a 3:1 ratio and absorbance at 650 nm was measured after 30 min using a microplate assay (Tecan). The Pi concentration was calculated based on a standard curve generated using varying concentrations of KH2PO4. Free Pi released from ATP in the nutrient solutions was measured directly using the malachite green reagent described above.
For measurement of total P content, 150 mg of dried plant tissue was pre-digested in glass tubes with concentrated sulfuric acid overnight. The tubes were then heated to 120 °C for 1 h and 4–5 drops of 30% H2O2 were added every 30 min until the solution turned colourless. The digestion was continued for an additional 30 min, and then the total P concentration was determined by molybdenum blue colorimetry. In this study, we define the P-use efficiency as the amount of biomass produced by a given amount of P content in the tissue, according to Liao and Yan (1999)
Statistical analysis
SPSS Statistics Base software (version 22) was used for statistical analysis. Significant differences were evaluated using one-way ANOVA and Turkey’s test.
Results
Expression of OsPAP10c is specifically induced by Pi starvation in roots
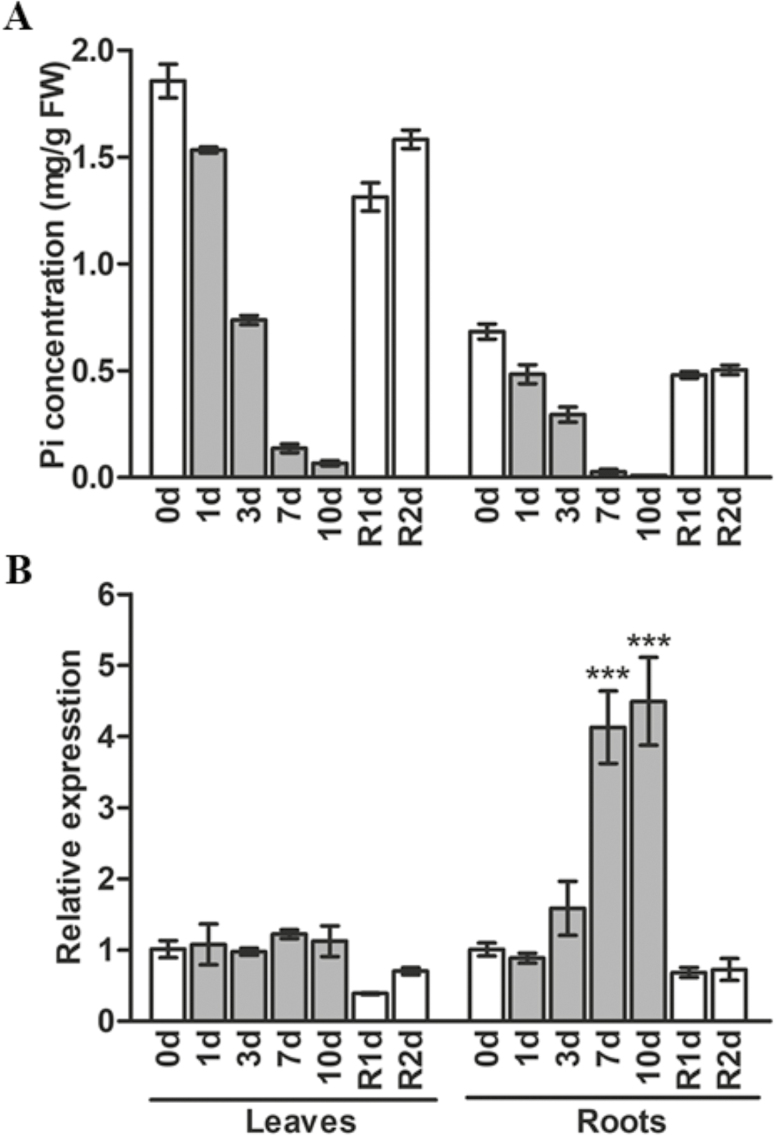
To determine the expression of OsPAP10c in rice under Pi-starvation conditions, a time-course of transcript levels was extracted from published RNA-seq data (Secco et al., 2013), which showed that transcripts increased in roots within 3 d of a Pi-deficient treatment (Supplementary Fig. S1 at JXB online). By 7 d of treatment the transcript level had increased by more than 10-fold, and this was maintained at 21 d. However, after a recovery period of resupply of Pi for only 1 d the expression of OsPAP10c had decreased back to the same level as the control roots. In contrast to the roots, there was no effect on transcript levels in response to Pi starvation even after 21 d in the shoots. To confirm these RNA-seq results, we used qRT-PCR to determine the expression of OsPAP10c at different time-points during Pi starvation. The concentrations of Pi in both the roots and leaves steadily decreased with time in response to the lack of supply (Fig. 1A). Resupply of Pi in the nutrient solution significantly increased the concentrations in both the roots and shoots. The decrease in Pi concentration in the roots was accompanied by a significant increase in the expression of OsPAP10c after 7 d and 10 d of the starvation treatment, but expression returned to the control level after 1 d of resupply (Fig. 1B). In contrast, the expression of OsPAP10c was maintained at a low level in the shoots irrespective of the Pi supply and the changes in internal concentration. These results therefore showed that the expression of OsPAP10c was specifically induced by Pi starvation only in the roots.
Fig. 1.
(A) Pi concentration and (B) expression of OsPAP10c in leaves and roots of rice during a period of P starvation followed by resupply. Germinated seeds were grown in normal nutrient solution for 10 d and then transferred to solution without Pi for 10 d, followed by 2 d recovery (R) in normal solution. RNA was extracted for quantitative RT-PCR and OsPAP10c expression was normalized to that of OsACTIN. Data are means (±SEM) of three replicates. Significant differences compared with the control (0 d) were determined using Turkey’s test (***P<0.001).
Mutations of OsPAP10c decrease the activities of root-associated and secreted acid phosphatase
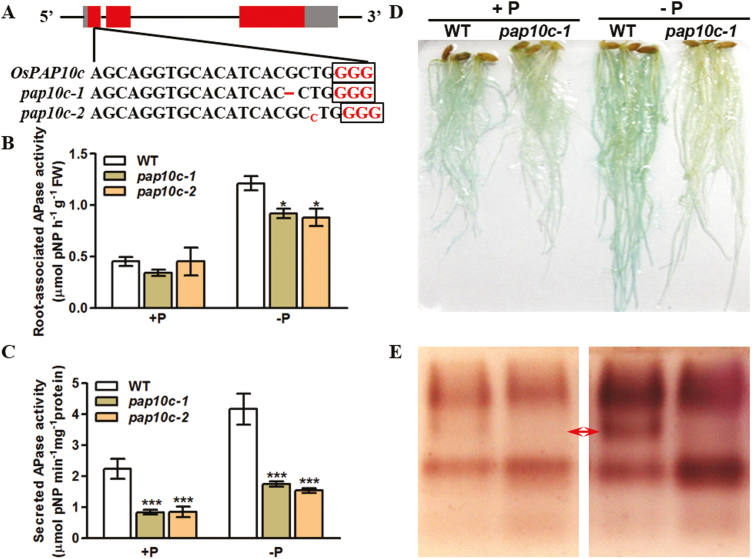
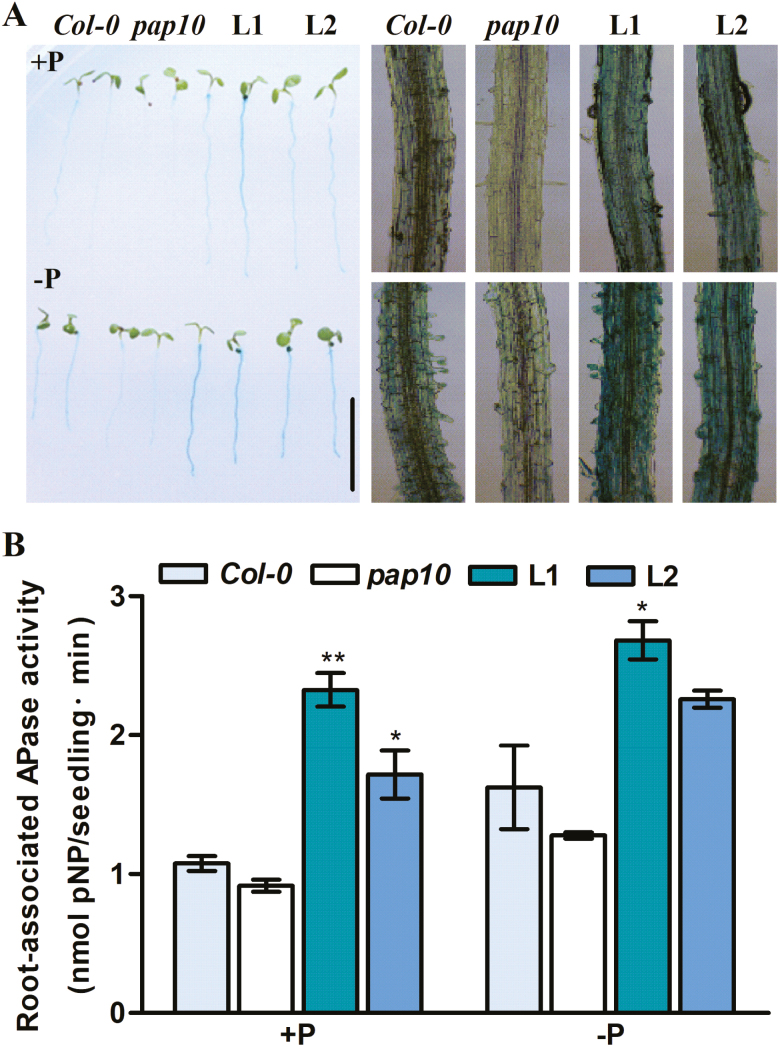
To examine whether OsPAP10c is a major secreted acid phosphatase in rice, we transformed a CRISPR-Cas9 construct that targeted the first exon of OsPAP10c into the Nipponbare wild-type (WT) and identified two knock-out plants with frame shifts (pap10c-1 and pap10c-2, Fig. 2A). Different patterns of acid phosphatase (APase) activity were observed in the mutants compared with the WT. The APase activities in the leaves and roots were similar between the mutants and the WT plants under both Pi-sufficient and -deficient conditions (Supplementary Fig. S2); however, the activities of both the root surface-associated and secreted APase were significantly decreased in the mutants under deficient conditions (Fig. 2B, C). To further determine the surface-associated APase activity, roots of pap10c-1 were stained with BCIP. Pi starvation induced less staining in the roots of the mutant than in those of the WT under both Pi-sufficient and -deficient conditions (Fig. 2D). The white roots of Ospap10c-1 were reminiscent of the Arabidopsis pap10 mutant, encoding a root-associated PAP. We therefore transformed OsPAP10c into the Atpap10 mutant and obtained two OsPAP10c-overexpression lines in the mutant background. As previously reported, the Atpap10 mutant showed white roots under BCIP staining and had decreased root-associated APase activity compared with the WT under both Pi-sufficient and -deficient conditions (Fig. 3A, B). Overexpression of OsPAP10c in Atpap10 fully complemented the white root phenotype and significantly increased the root-associated APase activity. The secreted proteins from suspension cells were purified and analysed by in-gel APase activity assays in order to determine whether OsPAP10c was present as a major APase isoform. Pi starvation induced a specific APase isoform in the WT that was absent in the Ospap10c mutant (Fig. 2E). Taken together, the results indicated that OsPAP10c is not only a major root surface-associated APase in rice but also that it is a major secreted isoform.
Fig. 2.
APase activity in rice Ospap10c mutants. (A) Schematic diagram of deletion mutations at the target sites in two representative knock-out lines generated by CRISPR/Cas9 technology. The sgRNA target sequence is shown and the protospacer adjacent motifs are indicated by the boxes. (B) Root surface-associated APase activity of the wild-type (WT) and pap10c mutants under +P and –P conditions. (C) Secreted APase activity of the WT and pap10c mutants under +P and –P conditions. (D) BCIP staining of the WT and the pap10c-1 mutant under +P and –P conditions. (E) In-gel APase activity assay of secreted proteins extracted from suspension cells of the WT and pap10c-1 mutant. The arrows indicate that the absence of a specific APase isoform in the mutant under Pi-deficient conditions.
Fig. 3.
Root-associated APase activity in Arabidopsis Col-0, the Atpap10 mutant, and OsPAP10c complementary lines in the Atpap10 background (L1, L2) under +P and –P conditions. (A) BCIP staining of roots of 7-d-old plants. The scale bar is 1 cm. (B) Quantification of the APase activity. Data are means (±SEM) of three replicates. Significant differences compared with Col-0 were determined using Turkey’s test (*P<0.05; **P<0.01).
The native promoter increases expression of OsPAP10c
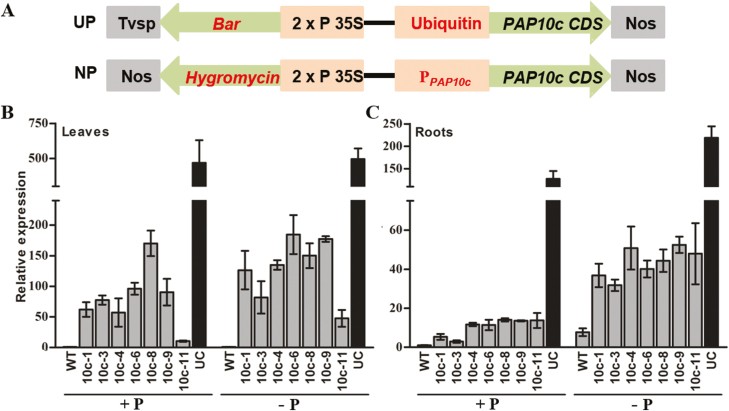
Although overexpression lines of OsPAP10c driven by a ubiquitin promoter (UP) have significantly enhanced APase activity in leaves and roots, the growth of the transgenic plants is not stimulated under both Pi-sufficient and -deficient conditions (Lu et al., 2016). Because the expression of OsPAP10c is specifically induced by Pi starvation in roots, we made another construct using the OsPAP10c native promoter (NP) to drive its coding sequence and generated seven positive lines by Agrobacterium-mediated transformation (Fig. 4A). Southern blot analysis using the hygromycin phosphotransferase (hpt) probe detected one band in the lines NP-1, NP-6, and NP-9, and two or three bands in the other lines (Supplementary Fig. S3A). To further confirm the copy numbers in these transgenic lines, the OsPAP10c probe was used for Southern blotting. Interestingly, four bands were detected in the WT plants, indicating that there are several homologues of OsPAP10c in the rice genome (Supplementary Fig. S3B). Compared with the WT, an additional band was detected in the lines NP-1, NP-3, and NP-9, while the others contained more bands of OsPAP10c. Combining the information obtained from the two probes, it appeared that lines NP-1 and NP-9 were single-copy insertion events while the other lines were multiple-copy events.
Fig. 4.
Relative expression levels of OsPAP10c in rice wild-type (WT) and transgenic plants. (A) Schematic diagram of the vectors with the constitutive ubiquitin promoter (UP) and the native promoter (NP). (B, C) Relative expression levels of OsPAP10c in the leaves and roots of the genotypes. The expression levels are relative to that of the wild-type in the +P treatment. Data are means (±SEM) of three replicates.
The expression levels of OsPAP10c were measured in the leaves and roots of WT and the two types of transgenic plants under both Pi-sufficient and -deficient conditions. As expected, Pi starvation significantly induced the expression of OsPAP10c in the roots of all the genotypes, but the relative increase in the UP plants was less and not significant (Fig. 4C). Although expression was significantly increased in both the UP and NP plants compared with the WT, the level was much lower in the NP plants than in the UP plants in both the leaves and roots (Fig. 4B, C). The expression of OsPAP10c in the UP plants under both Pi-sufficient and -deficient conditions was nearly 500-fold and 127-fold that of the leaves and roots, respectively, of the WT plants. The mean expression level of OsPAP10c in the NP plants was only ~10% of that of the UP plants.
Secreted APase activity and ATP degradation are increased in NP plants
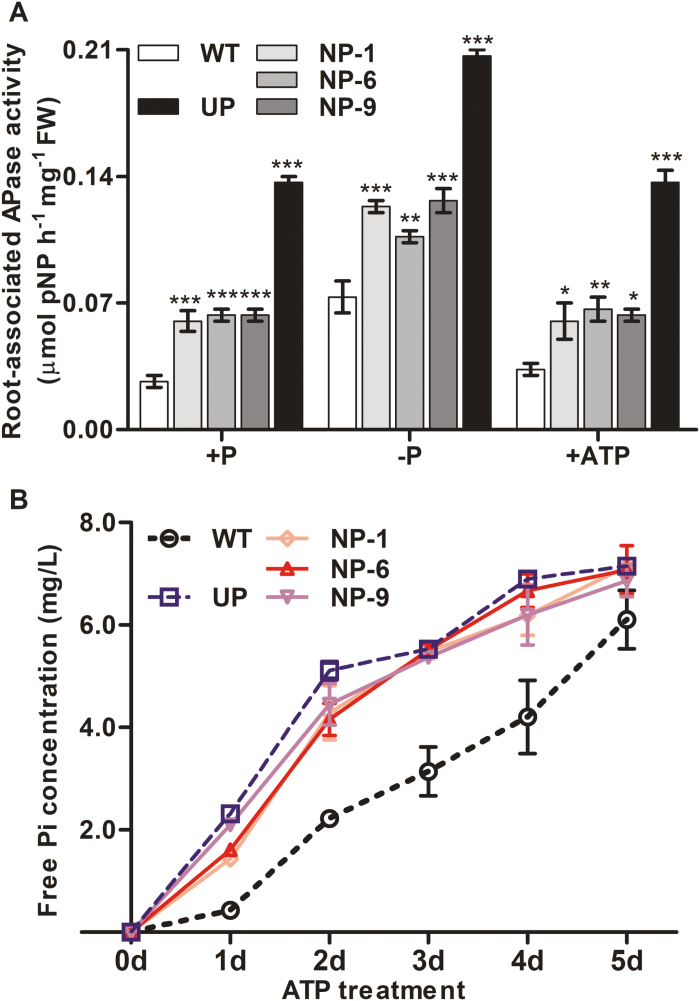
To evaluate the effects of the increased copy number of OsPAP10c in rice, the transgenic plants were grown under Pi-sufficient and -deficient conditions. Compared with the WT, the growth of the UP plants was only enhanced under Pi-deficient conditions (Supplementary Fig. S4). In contrast, the growth of all the NP lines was better than the WT and the UP plants under both Pi-sufficient and -deficient conditions. Lines NP-1, NP-6, and NP-9 showed the highest FW and were selected for further analysis. APase activities were determined under different Pi supply conditions. APase activities were significantly increased in the leaves and roots in the UP plants under both Pi-sufficient and -deficient conditions (Supplementary Fig. S5). In contrast, under both conditions the APase activity of leaves was similar between the NP plants and the WT plants. Whilst the activity in the roots was higher in the NP plants than in the WT, it was lower than that of the UP plants, especially under Pi-sufficient conditions.
With regards to the activity of secreted APase, overexpression of OsPAP10c in the UP plants increased the activity by 7-fold in comparison to the WT under Pi sufficient-conditions (Fig. 5A), whilst there was only a 2-fold increase in the NP plants. Pi starvation significantly induced APase activity in all the genotypes, and levels were 3- and 1.5-fold greater in the UP and NP plants, respectively, compared with the WT. When ATP was used as the only P source, the activity of secreted APase was 2- and 5-fold greater in the UP and NP plants, respectively, compared with the WT. As previously reported, the UP plants released more Pi from ATP during the first 4 d of treatment compared with the WT (Fig. 5B; Lu et al., 2016). Interestingly, a similar pattern for ATP degradation was observed between the NP and UP plants.
Fig. 5.
(A) Root-associated APase activity of the rice wild-type (WT) and of transgenic plants with OsPAP10c driven by the constitutive ubiquitin promoter (UP) or the native promoter (NP) grown either with or without Pi, or with ATP as the only source of P. (B) Concentration of free Pi in the growth solution for plants grown with ATP. Germinated seeds were grown in normal nutrient solution for 15 d and then transferred to solution either with or without Pi, or containing 0.1 mM ATP as the only source of P. Root-associated APase activities were measured after 10 d treatments. Data are means (±SEM) of three replicates. Significant differences compared with the WT were determined using Turkey’s test (*P<0.05; **P<0.01; ***P<0.001).
Plant growth and accumulation of P are increased in NP plants
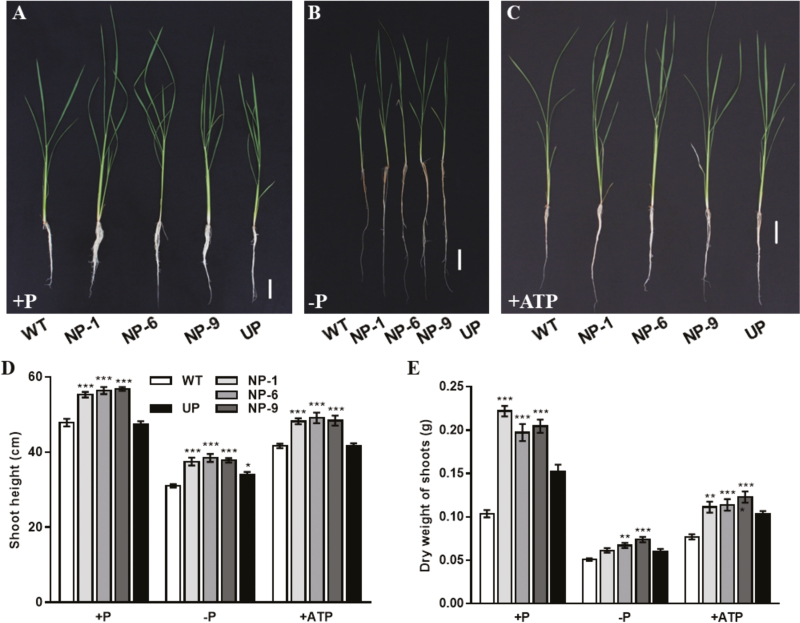
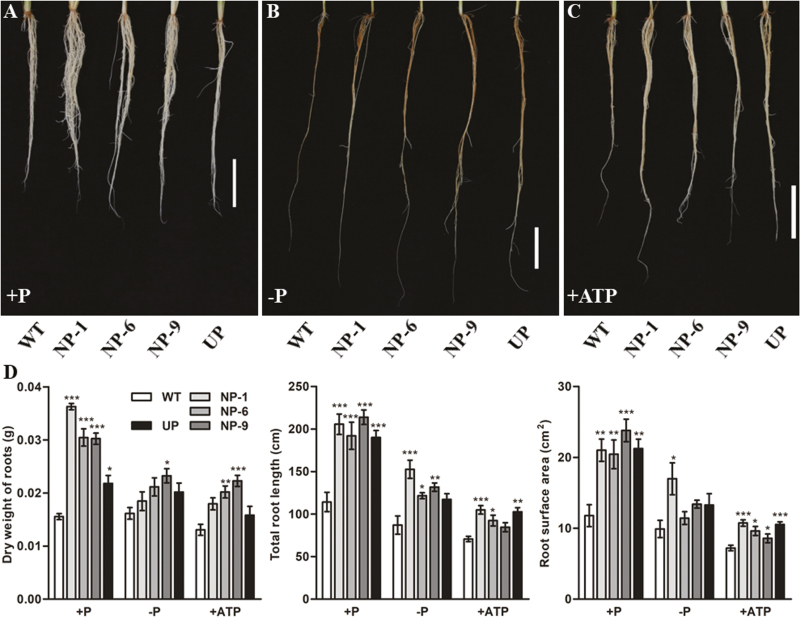
Growth parameters were measured in the WT, NP and UP transgenic plants under different P conditions. The shoot length and dry weight were significantly increased in the NP plants compared with the WT irrespective of the P supply (Fig. 6). The root dry weight, total root length, and root surface area were all significantly greater in the NP and UP plants compared with the WT under Pi-sufficient conditions (Fig. 7A, D), and a similar pattern was observed in the ATP treatment. Under Pi-deficient conditions, total root length was significantly increased in the NP plants but not in the UP plants. The root dry weight and surface area were only significantly affected in individual NP lines under Pi deficit (Fig. 7B, D).
Fig. 6.
Phenotypes and growth of wild-type (WT) rice and of transgenic plants with OsPAP10c driven by the constitutive ubiquitin promoter (UP) or the native promoter (NP) grown either with or without Pi, or with ATP as the only source of P. Germinated seeds were grown in normal nutrient solution for 15 d and then transferred to different solutions for 10 d. (A–C) Phenotypes of seedlings under the different treatments. The scale bars are 5 cm. (D) Shoot height and (E) dry weight of the genotypes under the different treatments. Data are means (±SEM) of six replicates. Significant differences compared with the WT were determined using Turkey’s test (*P<0.05; **P<0.01; ***P<0.001).
Fig. 7.
Root phenotypes and growth of wild-type (WT) rice and of transgenic plants with OsPAP10c driven by the constitutive ubiquitin promoter (UP) or the native promoter (NP) grown either with or without Pi, or with ATP as the only source of N. Germinated seeds were grown in normal nutrient solution for 15 d and then transferred to different solutions for 10 d. (A–C) Phenotypes of the roots under the different treatments. The scale bars are 5 cm. (D) Root dry weight, total root length, and root surface area under the different treatments. Data are means (±SEM) of six replicates. Significant differences compared with the WT were determined using Turkey’s test (*P<0.05; **P<0.01; ***P<0.001).
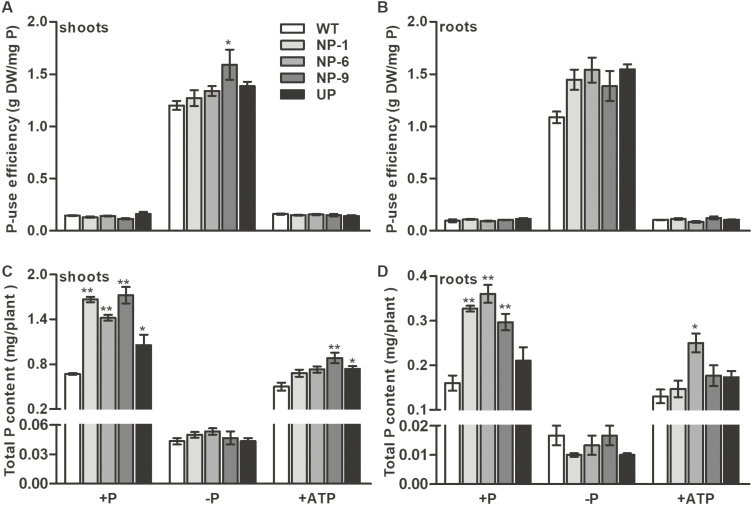
We also determined Pi concentration, P-use efficiency, and P content under the different P supply conditions. Similar Pi concentrations were observed in all the genotypes in both shoots and roots irrespective of the Pi supply (Supplementary Fig. S6). The ATP treatment significantly increased the Pi concentration of the shoots in the UP plants and in one line of the NP lines. With the exception of one NP line, no significant differences in P-use efficiency were observed between the genotypes in either of the Pi treatments or the ATP treatments (Fig. 8A, B). The total P content was significantly increased in the shoots and roots of NP plants under Pi-sufficient conditions compared with the WT, but no differences were observed under Pi-deficient conditions (Fig. 8C, D). Compared with the WT, the P content of the shoots of UP plants was significantly increased under P-sufficient conditions and in the ATP treatment. The P content of the line NP-9 was also increased in the shoots and roots in the ATP treatment.
Fig. 8.
P-use efficiency of wild-type (WT) rice and of transgenic plants with OsPAP10c driven by the constitutive ubiquitin promoter (UP) or the native promoter (NP) grown either with or without Pi, or with ATP as the only source of P. Germinated seeds were grown in normal nutrient solution for 15 d and then transferred to different solutions for 10 d. (A, B) P-use efficiency ratio of the genotypes under the different treatments. P-use efficiency was defined as the amount of biomass produced by a given amount of P content in the tissue. (C, D) Total P content of the genotypes under the different treatments. Data are means (±SEM) of three replicates. Significant differences compared with the WT were determined using Turkey’s test (*P<0.05; **P<0.01).
Growth and yield of NP plants are increased under field conditions
Given the results from the hydroponic experiments, we further evaluated the performance of NP transgenic plants grown in the field with or without application of P fertilizer. Overall shoot growth and panicle size were increased in the NP plants compared with the WT (Supplementary Fig. S7). In terms of agronomical traits, similar values for tiller number, seed-setting rate, and 1000-grain weight were observed between the WT and NP plants under both low and normal P conditions (Table 1). Plant height, grain number, and grain yield per plant were significantly increased in the NP plants compared with the WT under both P treatments.
Table 1.
Agronomic traits of wild-type and transgenic plants driven by the native promoter grown in the field under different Pi supply
| Pi supply | Line | Plant height (cm) | Tiller number | Seed-setting rate (%) | 1000-grain weight (g) | Grain number per plant | Grain yield per plant (g) |
|---|---|---|---|---|---|---|---|
| LP | WT | 87.36±2.53e | 26.33±3.50a | 72.51±7.56b | 22.56±0.36ab | 1593.49±137.68d | 35.95±3.26e |
| NP-1 | 115.21±2.15bc | 23.83±5.19a | 79.91±3.04ab | 23.91±2.09ab | 2121.44±321.34bc | 50.76±8.92bcd | |
| NP-6 | 124.64±4.62a | 24.67±3.61a | 79.20±5.76ab | 23.47±1.17ab | 1930.03±270.43bcd | 45.48±8.16cde | |
| NP-9 | 113.39±3.97c | 28.17±3.13a | 81.50±3.12ab | 25.01±1.95ab | 1994.97±230.48bcd | 49.67±5.12bcd | |
| HP | WT | 95.46±2.70d | 30.17±5.49a | 74.82±8.57ab | 22.16±1.16b | 1783.01±223.28cd | 39.31±2.77de |
| NP-1 | 119.36±5.00b | 31.00±4.69a | 83.28±2.52a | 24.69±2.36ab | 2818.01±433.94a | 69.00±7.65a | |
| NP-6 | 124.89±4.06a | 26.67±4.59a | 84.37±5.77a | 24.56±1.54ab | 2394.53±235.01ab | 59.00±8.42ab | |
| NP-9 | 113.46±4.31c | 25.33±3.67a | 82.58±5.58ab | 25.49±1.61a | 2133.21±207.66bc | 54.22±4.24bc |
LP, low Pi (without P fertilizer); HP, high Pi (80 kg P ha–1). WT, wild-type; NP, native promoter. Data are means (±SD). Different letters indicate significant differences between the WT and transgenic plants as determined by one-way ANOVA followed by Tukey’s test (P<0.05).
Discussion
OsPAP10c encodes a specific secreted phosphatase
It has been reported that Pi starvation induces extracellular APases, which degrade Po into Pi for plant growth and development (Tran et al., 2010a). The extracellular APase proteins can be secreted into the rhizosphere or the cell wall matrix. APase activities have been measured in mutants of 29 different PAP genes in Arabidopsis, with the results showing that only the Atpap12, Atpap15 and Atpap26 mutants have APase activity decreased by more than 30% compared with WT plants in roots under either Pi-sufficient or -deficient conditions (Wang et al., 2014). It is well established that AtPAP12 and AtPAP26 are the major APases that are secreted into the rhizosphere and cell wall in phosphate-starved Arabidopsis (Tran et al., 2010b; Robinson et al., 2012). However, mutations of the two corresponding genes decrease the activity of secreted APase rather than the activity APase associated with the root surface, as indicated by BCIP staining. In contrast, the Atpap10 mutant shows a ‘white root’ phenotype and AtPAP10 has been shown to be the major root surface-associated APase in Arabidopsis (Wang et al., 2011). OsPAP10c was previously identified as the rice homologue of AtPAP10 and AtPAP12, and it is specifically induced by Pi starvation in the roots (Lu et al., 2016). To determine whether OsPAP10c is one of the major secreted APase isoforms in rice, mutants were generated using CRISPR/Cas9 technology (Fig. 2A). Quantitative measurements of APase activity in the resulting mutants showed that the activity of APase secreted into the medium was decreased by more than 50% under both Pi-sufficient and -deficient conditions (Fig. 2B, C). The mutations also resulted a similar ‘white root’ phenotype in rice as seen in Atpap10 (Fig. 2D). Moreover, OsPAP10c could fully complement the decreased root surface-associated APase activity in the Atpap10 mutant (Fig. 3), indicating that OsPAP10c is an essential root surface-associated APase in rice. We then collected and analysed the proteins secreted into the medium by rice suspension cells in order to identify which APase isoforms were present (Fig. 2E). A specific isoform was absent in the samples from the pap10c mutant, and this might account for the significant decrease in the activity of secreted APase. Taken together, the results indicated that OsPAP10c is a major secreted APase in rice.
It has previously been shown that the major secreted APases from a variety of plant species belong to the PAP Ia family, and OsPAP10c is classified as being within a monocotyledon-specific PAP group (Lu et al., 2016). Four homologous genes of AtPAP10 and AtPAP12 have been found in rice, namely OsPAP10a–10d (Zhang et al., 2011; Lu et al., 2016). Corresponding with this, we detected four bands in the WT plants using OsPAP10c as the probe in Southern blot analysis (Supplementary Fig. S3), presumably representing the homologous genes. Interestingly, these four genes show different expression patterns in rice. The expressions of OsPAP10b and OsPAP10d are extremely low and show no response to Pi starvation, whereas in contrast OsPAP10a and OsPAP10c show relatively higher expression and are induced by Pi starvation, indicating that they might be involved the adaptation of rice to low P supply (Lu et al., 2016). However, expression of OsPAP10a is induced in both roots and leaves by Pi starvation while expression of OsPAP10c is only specifically induced in the roots. Although overexpression of OsPAP10a and OsPAP10c both significantly increase the utilization of Po in rice, overexpression lines of OsPAP10c exhibit higher activity of secreted APase compared with overexpression lines of OsPAP10a (Tian et al., 2012; Lu et al., 2016). In-gel APase activity assays have indicated that overexpression of OsPAP10a and OsPAP10c in rice increase different APase isoforms (Tian et al., 2012; Lu et al., 2016). We conducted a qRT-PCR analysis that ruled out the possibility that overexpression of OsPAP10c influenced the expression of OsPAP10a (Supplementary Fig. S8). Therefore, we assume that OsPAP10c represents a monocotyledon-specific APase isozyme, which can be secreted into the medium in a similar manner to AtPAP12 and can be associated with the root surface in a similar manner to AtPAP10.
Overexpression of OsPAP10c enhances root growth in rice
The expression of a number of PAP genes are induced by Pi starvation, indicating their important roles in Pi stress responses. It is generally recognized that PAPs are involved in the degradation of extracellular Po to convert it into plant-available Pi. However, there is a growing body of evidence to show that PAPs are also involved in regulating plant growth in addition to recycling Po. Increasing numbers of PAPs are being found to be secreted into the cell wall where limited Po exists. In tobacco, NtPAP12 has been identified as a cell wall-bound APase and it regulates the wall components by dephosphorylation of α-xylosidase and β-glucosidase. Although the physiological functions of NtPAP12 are unknown, overexpression of its homologue AtPAP12 in Arabidopsis significantly increases plant growth under Pi-deficient conditions without extracellular Po supply (Wang et al., 2014). Moreover, mutations of AtPAP10 and OsPAP21b inhibit root growth in Arabidopsis and rice, respectively (Wang et al., 2011; Mehra et al., 2017). It has been proposed that these PAPs may regulate growth by modifying the cell wall composition. Interestingly, we found that NP transgenic plants showed higher root-associated APase activity and increased root length and surface area compared with the WT (Figs 5, 7). OsPAP10c may play a key function in root growth under Pi-stress conditions, which would be in accordance with the induced expression that we observed (Fig. 1). It has been reported that increasing the root length and surface area can significantly increase Pi uptake and accumulation in rice (Gamuyao et al., 2012), and we found that the P content per plant was significantly increased in the NP transgenic plants compared with the WT, while the Pi concentration and P utilization efficiency did not significantly change (Fig. 8, Supplementary Fig. S6). Hence, the better performance of the NP plants may have been due to the stimulated root growth and increased nutrient uptake efficiency.
Increased expression of OsPAP10c driven by its native promoter enhances plant growth and yield
Although the overexpression lines of OsPAP10c driven by a ubiquitin promoter showed significantly enhanced APase activity in leaves and roots, the growth of these lines is inhibited in both pot and field experiments (Lu et al., 2016). Similarly, overexpression of GmPAP1-like and GmPAP21 in bean hairy roots also results in a reduction in fresh weight relative to control lines under Pi-sufficient conditions (Li et al., 2017; Wu et al., 2018). The expression of these PAP genes was induced by Pi starvation in specific tissues, whereas their constitutive overexpression would increase APase activity and degrade organic P compounds in all tissues included untargeted ones, and this might cause negative effects in the transgenic plants. To try to avoid this, we overexpressed OsPAP10c in rice by using its native promoter. As expected, the NP transgenic plants had increased expression of OsPAP10c to a moderate level, which was significantly lower than that in the UP transgenic plants (Fig. 4), and accordingly the APase activity was not changed in the leaves and was slightly increased in roots (Supplementary Fig. S5). Interestingly, the activities of both root surface-associated and secreted APase were significantly increased in the NP plants compared with the WT, although they were still lower than in the UP plants.
To evaluate the effects of overexpressing OsPAP10c by use of its native promoter, the transgenic plants were grown under different Pi supply conditions. Compared with the WT and UP plants, the growth of NP plants was significantly increased irrespective of the Pi supply (Fig. 6). This indicated that overexpression of OsPAP10c by its native promoter rather than a constitutive promoter was an effective way to stimulate plant growth. However, the expression levels and APase activity were not tightly correlated with the copy number of OsPAP10c. The three lines with the best performance (NP-1, NP-6, and NP-9) showed a level of expression level that was comparable with some of the other lines (Fig. 4). This variation in expression and phenotype between the different transgenic lines may have been be influenced by different insertion positions of the T-DNA or by random mutations during the tissue culture process in addition to copy number. Another possibility is that post-transcriptional regulation of OsPAP10c exists. Although the expression of PAP genes can be increased by a factor of tens or hundreds, in all the studies to date the activity of APase was always been increased only by a few fold (Wang et al., 2011, 2014; Tian et al., 2012; Li et al., 2017; Wu et al., 2018). Thus, the difference in APase activity is several magnitudes smaller than the difference in transcript levels in the plants, which would accounts for the similar APase activities in the different transgenic lines.
We conducted a field experiment to examine the agronomic traits of the NP transgenic plants, and found that plant growth and grain yield were significantly enhanced under both low and normal P conditions (Table 1). Overall, our results indicate that using the native promoter to overexpress PAPs that are induced by Pi starvation provides a potential means to improve future crop production
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Time-course of expression levels of OsPAP10c in leaves and roots under different P treatments, as extracted from previously published RNA-seq data.
Fig. S2. Acid phosphatase activities in leaves and roots of the wild-type and pap10c mutants under different Pi supply conditions.
Fig. S3. Southern blot analysis of the wild-type and transgenic plants using the hpt and OsPAP10c probes.
Fig. S4. Phenotypes and fresh weights of the wild-type and transgenic plants under different Pi supply conditions.
Fig. S5. Acid phosphatase activities in the leaves and roots of the wild-type and transgenic plants under different Pi supply conditions.
Fig. S6. Pi concentrations in the shoots and roots of the wild-type and transgenic plants under different Pi supply conditions, and with supply of ATP.
Fig. S7. Phenotypes of shoots and panicles of the wild-type and NP transgenic plants grown in the field with or without P fertilizer.
Fig. S8. The expression of OsPAP10a in the wild-type and OsPAP10c NP transgenic plants under different Pi supply conditions.
Acknowledgements
We thanked Professor Dong Liu of Tsinghua University for providing the Arabidopsis pap10 seeds. This work was supported by the National Science Foundation of China (31772378), the Transgenic Plant Research Special Program of China (2016ZX08003005), and the Fundamental Research Funds for the Central University of China (2016RC005, 2662017PY012, 2662019PY013).
Glossary
Abbreviations
- APase
acid phosphatase
- Pi
phosphate
- Po
organic phosphate
- NP
native promoter
- UP
ubiquitin promoter
Author contributions:
SD, JL, and TL performed the experiments with rice; LL constructed the transgenic lines of rice and Arabidopsis; ZD performed the experiments with Arabidopsis; CW, FX, and LS supervised the experiments; CW, SD, and WL analysed the data and wrote the manuscript; CW and HX designed the project; CW agreed to serve as the corresponding author.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
References
- Bozzo GG, Dunn EL, Plaxton WC. 2006. Differential synthesis of phosphate-starvation inducible purple acid phosphatase isozymes in tomato (Lycopersicon esculentum) suspension cells and seedlings. Plant, Cell & Environment 29, 303–313. [DOI] [PubMed] [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC. 2004. Structural and kinetic properties of a novel purple acid phosphatase from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. The Biochemical Journal 377, 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J. 1999. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. The Plant Journal 19, 579–589. [DOI] [PubMed] [Google Scholar]
- Feder D, Gahan LR, McGeary RP, Guddat LW, Schenk G. 2019a The binding mode of an ADP analogue to a metallohydrolase mimics the likely transition state. ChemBioChem 20, 1536–1540. [DOI] [PubMed] [Google Scholar]
- Feder D, Kan MW, Hussein WM, Guddat LW, Schenk G, McGeary RP. 2019b Synthesis, evaluation and structural investigations of potent purple acid phosphatase inhibitors as drug leads for osteoporosis. European Journal of Medicinal Chemistry 182, 111611. [DOI] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S. 2012. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488, 535–539. [DOI] [PubMed] [Google Scholar]
- Ghahremani M, Park J, Anderson EM, Marty-Howard NJ, Mullen RT, Plaxton WC. 2019. Lectin AtGAL1 interacts with high-mannose glycoform of the purple acid phosphatase AtPAP26 secreted by phosphate-starved Arabidopsis. Plant, Cell & Environment 42, 1158–1166. [DOI] [PubMed] [Google Scholar]
- Gilbert N. 2009. Environment: the disappearing nutrient. Nature 461, 716–718. [DOI] [PubMed] [Google Scholar]
- Hussein WM, Feder D, Schenk G, Guddat LW, McGeary RP. 2018. Purple acid phosphatase inhibitors as leads for osteoporosis chemotherapeutics. European Journal of Medicinal Chemistry 157, 462–479. [DOI] [PubMed] [Google Scholar]
- Kaida R, Satoh Y, Bulone V, Yamada Y, Kaku T, Hayashi T, Kaneko TS. 2009. Activation of beta-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiology 150, 1822–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li C, Zhang H, Liao H, Wang X. 2017. The purple acid phosphatase GmPAP21 enhances internal phosphorus utilization and possibly plays a role in symbiosis with rhizobia in soybean. Physiologia Plantarum 159, 215–227. [DOI] [PubMed] [Google Scholar]
- Liang C, Tian J, Lam HM, Lim BL, Yan X, Liao H. 2010. Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiology 152, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Yan X. 1999. Seed size is closely related to phosphorus use efficiency and photosynthetic phosphorus use efficiency in common bean. Journal of Plant Nutrition 22, 877–888. [Google Scholar]
- Liu P, Cai Z, Chen Z, Mo X, Ding X, Liang C, Liu G, Tian J. 2018. A root-associated purple acid phosphatase, SgPAP23, mediates extracellular phytate-P utilization in Stylosanthes guianensis. Plant, Cell & Environment 41, 2821–2834. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu L, Qiu W, Gao W, Tyerman SD, Shou H, Wang C. 2016. OsPAP10c, a novel secreted acid phosphatase in rice, plays an important role in the utilization of external organic phosphorus. Plant, Cell & Environment 39, 2247–2259. [DOI] [PubMed] [Google Scholar]
- Mehra P, Pandey BK, Giri J. 2017. Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnology Journal 15, 1054–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SS, Liu J, Allan DL, Menzhuber CJ, Fedorova M, Vance CP. 2001. Molecular control of acid phosphatase secretion into the rhizosphere of proteoid roots from phosphorus-stressed white lupin. Plant Physiology 127, 594–606. [PMC free article] [PubMed] [Google Scholar]
- Olczak M, Morawiecka B, Watorek W. 2003. Plant purple acid phosphatases – genes, structures and biological function. Acta Biochimica Polonica 50, 1245–1256. [PubMed] [Google Scholar]
- Pratt J, Boisson AM, Gout E, Bligny R, Douce R, Aubert S. 2009. Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells: an in vivo31P-nuclear magnetic resonance study using methylphosphonate as a Pi analog. Plant Physiology 151, 1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WD, Park J, Tran HT, Del Vecchio HA, Ying S, Zins JL, Patel K, McKnight TD, Plaxton WC. 2012. The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana. Journal of Experimental Botany 63, 6531–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J. 2013. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. The Plant Cell 25, 4285–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Yuan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F. 2011. Phosphorus dynamics: from soil to plant. Plant Physiology 156, 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Wang C, Zhang Q, He X, Whelan J, Shou H. 2012. Overexpression of OsPAP10a, a root-associated acid phosphatase, increased extracellular organic phosphorus utilization in rice. Journal of Integrative Plant Biology 54, 631–639. [DOI] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC. 2010a Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Science 179, 14–27. [Google Scholar]
- Tran HT, Qian W, Hurley BA, She YM, Wang D, Plaxton WC. 2010b Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant, Cell & Environment 33, 1789–1803. [DOI] [PubMed] [Google Scholar]
- Veljanovski V, Vanderbeld B, Knowles VL, Snedden WA, Plaxton WC. 2006. Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiology 142, 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li Z, Qian W, et al. 2011. The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiology 157, 1283–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lu S, Zhang Y, Li Z, Du X, Liu D. 2014. Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. Journal of Integrative Plant Biology 56, 299–314. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Y, Tian J, Lim BL, Yan X, Liao H. 2009. Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiology 151, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Lin Y, Liu P, Chen Q, Tian J, Liang C. 2018. Association of extracellular dNTP utilization with a GmPAP1-like protein identified in cell wall proteomic analysis of soybean roots. Journal of Experimental Botany 69, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. 2014. Targeted gene mutation in rice using a CRISPR-Cas9 system. Bio-Protocol 4, e1225. [Google Scholar]
- Zhang Q, Wang C, Tian J, Li K, Shou H. 2011. Identification of rice purple acid phosphatases related to phosphate starvation signalling. Plant Biology 13, 7–15. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Lu S, Liu D. 2014. A major root-associated acid phosphatase in Arabidopsis, AtPAP10, is regulated by both local and systemic signals under phosphate starvation. Journal of Experimental Botany 65, 6577–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.