Abstract
Healthy function of the gut microenvironment is dependent on complex interactions between the bacteria of the microbiome, epithelial and immune (host) cells, and the surrounding tissue. Misregulation of these interactions is implicated in disease. A range of tools have been developed to study these interactions, from mechanistic studies to therapeutic evaluation. In this Digest, we highlight select tools at the cellular and molecular level for probing specific cell-microenvironment interactions. Approaches are overviewed for controlling and probing cell-cell interactions, from transwell and microfluidic devices to engineered bacterial peptidoglycan fragments, and cell-matrix interactions, from three-dimensional scaffolds to chemical handles for in situ modifications.
Introduction
The innate immune system is the first line of defense against invading pathogens; as the name implies, the system is ancient and has been evolutionarily conserved in some form from plants to humans.1–3 Originally, the innate immune system was thought to arbitrarily keep infection at bay until the activation of the adaptive immune system. However, Charles Janeway and Nobel prize winning work accurately predicted that the innate immune system is much more complex.1 A key first step in recognition of pathogenic bacteria is their engulfment by macrophages, which patrol tissues throughout the body engulfing and digesting pathogens, dead cells, and cell debris.4 Macrophages express a variety of receptors on their surface that identify pathogens and initiate phagocytosis for appropriate uptake into the cell for degradation. This immune recognition can be described in three major strategies: recognition of “microbial nonself,” the ability of the host to recognize products of microbial metabolism unique to these organisms and not the host; recognition of “missing self,” which is the ability to detect markers of normal self, such as macrophage surface markers; and recognition of “induced or altered self,” the ability to detect markers of abnormal self during infection that mark cells for elimination by the immune system.1 In particular during this process, pathogen-associated molecular patterns (PAMPs) including lipopolysaccharides (LPS) and peptidoglycans (PG) are detected by pattern recognition receptors (PRRs), allowing the immune system to distinguish self from microbes.5 Misregulation of this process is thought to lead to a variety of autoimmune diseases.6,7 For example, irritable bowel disease (IBD) arises from acute and chronic inflammation of the gut mucosa without the presence of specific pathogens, indicating misregulation of immune response to commensal or uncharacterized pathogenic bacteria in the gut.8 Innovative tools are needed to dissect these complex host-bacteria interactions for understanding and ultimately targeting critical regulators of these diseases at the molecular, cellular, and tissue levels. In this Digest, we will highlight essential aspects of the host-bacteria interactions in the gut microenvironment, current techniques for studying these interactions, and applications of and opportunities for the use of innovative organic and bioorganic chemical tools in parsing these interactions.
Host-bacteria interactions in the gut
Cell-cell interactions: bacteria, epithelial, and select immune cells.
Consideration of interactions between host-bacteria cells and the context in which they occur are both important in the design of model systems and molecular tools for understanding these complex systems. At the tissue and cellular levels in the gut, bacteria are in contact with the intestinal epithelium, the most rapidly self-renewing tissue where high turnover aids in prevention of pathogenic bacterial infections.9 The epithelium in the small intestine is composed of crypts and villi, which facilitate nutrient absorption, and in the colon flat crypts, with multiple stem cells per crypt that facilitate self-renewal (Figure 1).10 These cells are protected by a thick layer, roughly 100 microns, of mucus classified into adherent and non-adherent layers, which bacterial cells must invade to gain access to the intestinal epithelial cells (IECs). While an important and critical part of the gut microenvironment, the scope of modeling this mucosal layer is very complex and will not be discussed in this Digest; however, for excellent reviews and discussion on the mucosal layer, please see these references.11–13
Figure 1: Overview of key host-bacteria interactions in healthy and diseased intestinal epithelium.
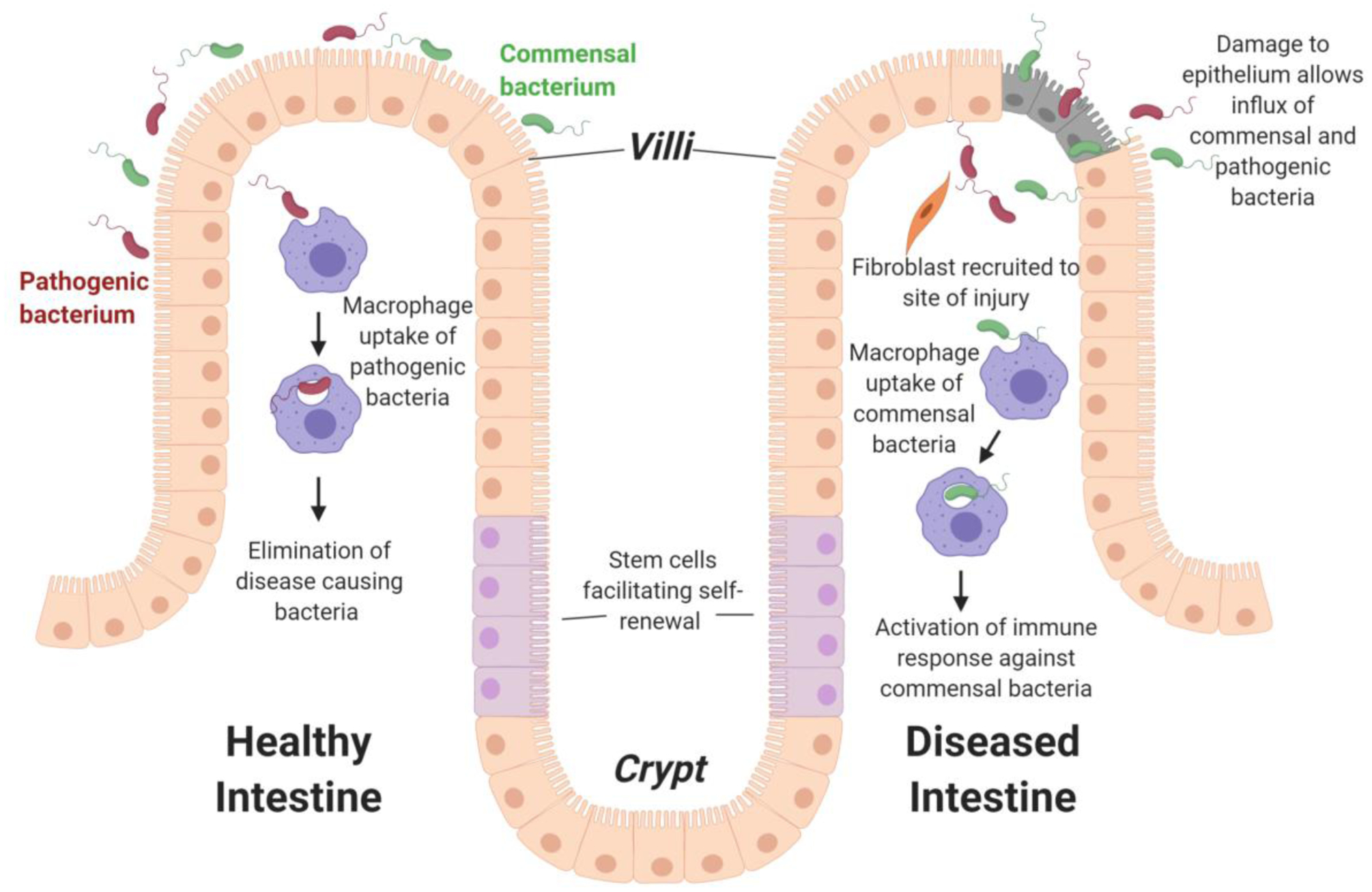
In healthy intestines, macrophages clear any pathogenic bacteria that cross over the epithelium layer. However, damage to the epithelium layer, such as in IBD, leads to an influx of both commensal and pathogenic bacteria, which can cause misrecognition of commensal bacteria, improper activation of immune system, and initiation of tissue remodeling responses (e.g., recruitment and activation of fibroblasts that leads to the production of scar-like tissue). Note, not shown in this schematic for simplicity is the protective mucosal layer that separates the epithelium layer and the bacterial microbiome and the lamina propria (LP) connective tissue that underlies the epithelium.
IECs are connected by tight junctions, forming an effective barrier within the healthy gut, and secrete a variety of factors that regulate interactions with bacteria and other host cells, including antimicrobial peptides that influence microbe colonization and penetration of the epithelium and chemokines and cytokines to recruit and activate host immune cells.14 Macrophages reside in the lamina propria (LP), the large layer of connective tissue that underlies the epithelium, as well as in the deeper layers of the gut wall, such as the submucosa and muscularis, and are well positioned to sense bacteria and identifying features (PAMPs) that cross the epithelium.15 Macrophages play a key role in maintaining the integrity of epithelial layer and managing the balance of the microbiota.16 However, when macrophages begin to elicit improper responses, such as aggressive responses to LPS present on some bacteria, the epithelial integrity can be destroyed and requires mucosal healing in the case of IBD.17 Probing these cell-cell interactions at the cellular and molecular level is pivotal in understanding IBD development and progression.
Cell-matrix interactions: key insoluble cues within the epithelium and LP.
Pathogen/host cell interactions (Figure 1) are directed and influenced by interactions with the mucosal layer that lies above the epithelium, as noted earlier,11–13 and the insoluble extracellular matrix (ECM) of the gut tissue that lies below the epithelial cells in both healthy and diseased states. The ECM provides both structure and bioactivity through hierarchical organization and an array of receptor and soluble factor binding sites.8 Here, we will focus on the epithelium and LP and highlight key insoluble cues and compositional changes that occur from healthy to diseased states.
ECM proteins such as laminins (e.g., laminin-511 and -111 at the base of villus cells; laminin-211 in the crypts), collagens (e.g., collagen IV), and different proteoglycans comprise the basement membrane that sequesters secreted factors and supports and separates the intestinal epithelium from the underlying connective tissue in the LP, influencing the morphology, adhesion, and function of epithelial cells.18–20 Engagement of specific integrins by the ECM is necessary for the appropriate cell function; for example, engagement of αVβ5 is required for macrophage engulfment of apoptotic cells, where mutations in it lead to accumulation of apoptotic cells in the colon and colitis.17,21 During IBD such as ulcerative colitis and Crohn’s disease, enrichment of different laminin isoforms (e.g., laminin-511 and -111) occurs in the crypts. Further, excessive deposition and accumulation of collagens (e.g., collagens V, III, and I) occurs throughout the mucosa and submucosa, resulting from activation of immune cells and mesenchymal cells (e.g., fibroblasts, smooth muscle cells) that remodel the local ECM to produce fibrotic scar-like tissue and ultimately thickening of the bowel wall and loss of tissue function.22 These compositional and structural changes in the ECM during disease correlate with and are thought to play a role in the loss of integrity of the epithelium during IBD, leading to increased migration of bacteria, accumulation of lymphocytes, and both a local and systemic immune response. Model systems and tools are needed that allow not only control of cell-cell interactions, but also cell-matrix interactions over relevant time and size scales for hypothesis testing and therapeutic screening.
Select tools for studying host-bacteria interactions at the tissue and cell level
A range of traditional tools for studying cell biology have been applied to the study of host-bacteria interactions in IBD.23 For example, at the whole organism level, several mouse models have been established that allow investigation of the role of genetic factors in the spontaneous development of mucosal inflammation or exogenous agents in inducing inflammation.24 At the tissue level, human mucosa explants have been cultured ex vivo, where the apical to basolateral polarity is preserved during stimulation.25 These model systems replicate organ complexity, yet challenges remain in parsing individual cell-microenvironment interactions owing to the limited handles available for modifying these systems without unintended downstream effects. To address this, a range of in vitro culture platforms have been developed and applied for probing cell-cell and cell-matrix interactions over different length and time scales, from the design of devices for microphysiological models to synthetic ECMs for molecular control of cell-microenvironment interactions, as overviewed below (Figure 2). Here, we overview select tools for probing interactions of bacteria with host cells (e.g., epithelial, stromal, and immune cells), as well as interactions between host cells, and with the ECM of the epithelium and LP.
Figure 2: Devices used for intestinal mono- and co-cultures in well-defined microenvironments.
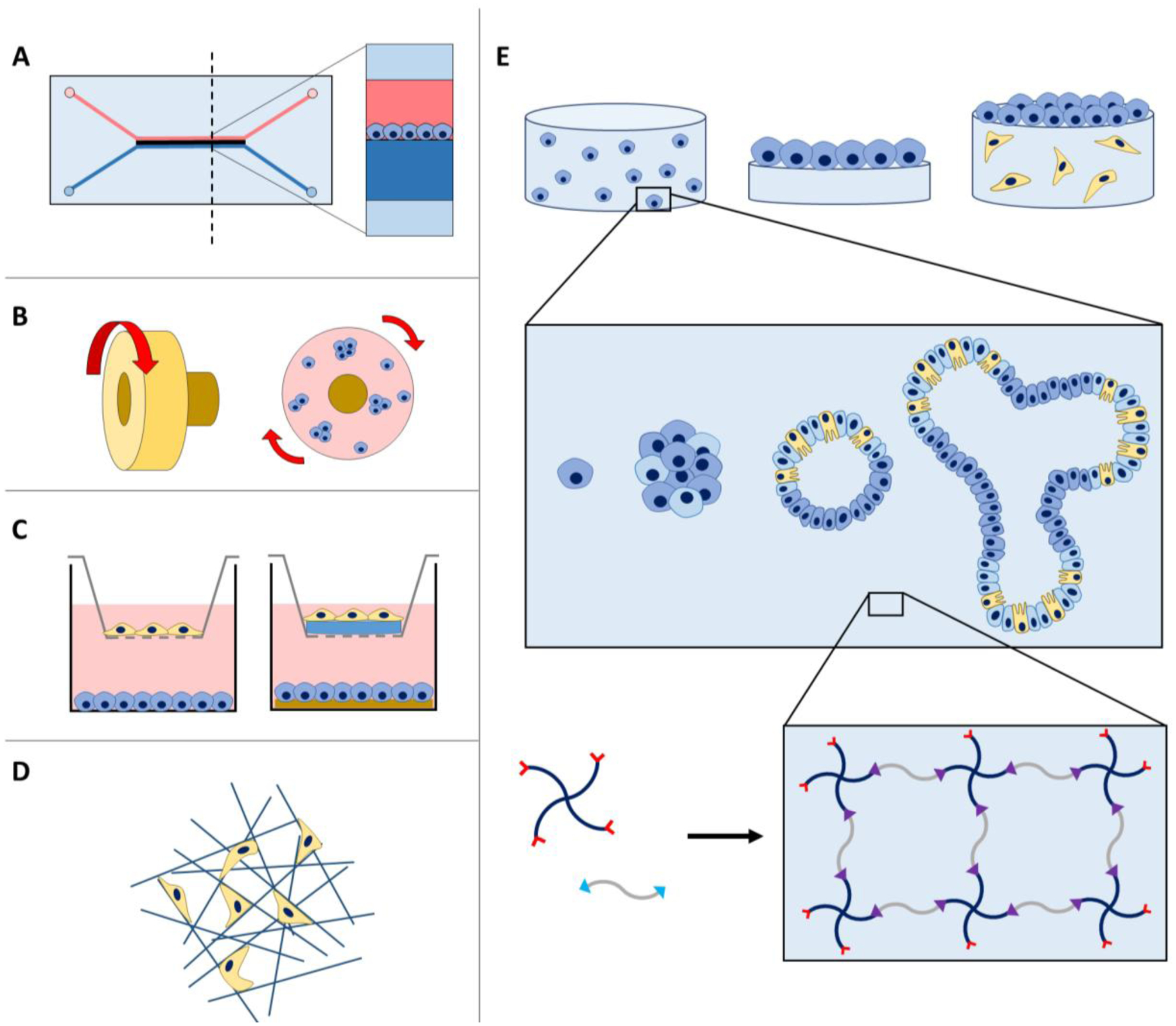
A) Microfluidic devices provide fluid flow over cell monolayers for the construction of ‘organs on a chip’. B) Rotating wall vessels contain media and suspended cells, where rotation encourages spheroid formation. C) Transwell inserts with cells seeded on and below the insert provide dynamic direct or indirect cell-cell interactions. ECM mimics such as hydrogels formed with harvested proteins (e.g., Matrigel, collagen I) have been integrated within these transwell systems to control matrix composition or dimensionality. D) Fibrous porous scaffolds also allow seeding of cells within the voids of these structures to enable multidimensional culture in more well-defined environments. E) Further, hydrogels can be utilized for cell culture in multiple geometries, including in 3D culture (top left), 2D culture (top center), or a combination (top right). Single cells, spheroid clusters, or established organoids have been encapsulated within hydrogels for 3D culture (middle), where crosslinking of multiarm polymers and peptides functionalized reactive handles is a robust and modular approach for the formation of well-defined hydrogel-based synthetic extracellular matrices (bottom).
Devices for micro-physiological models.
For increased control of the microenvironment while retaining features of whole organisms or tissues, multiscale devices have been designed to create microphysiological models with physiologically relevant fluid flow and cell polarization (Figure 2A). For example, model IECs (Caco-2, a heterogeneous human epithelial colorectal adenocarcinoma cell line) have been cultured on hard or soft membranes within different device designs for mono-culture under flow to probe cell function or response to soluble factors or co-culture with bacteria.26–28 Fluid flow influences mucus secretion and allows the long-term co-culture with microbes of interest with appropriate oxygenation and in morphologically relevant configurations with the aim of creating human guts-on-a-chip for evaluation of therapeutics.29,30
Transwell plate co-cultures.
Moving to the cellular level, bacterial-epithelium co-cultures have been established, where some of the first systems used were rotating-wall culture vessels for studies of viral infection amongst other applications (Figure 2B).31,32 Building upon this, traditional approaches for co-culture of different cell types in a multi-well plate format have been investigated: cell types of interest are separated by transwell inserts allowing dynamic exchange of soluble factors between them or studies of cell migration between the top and bottom chambers that each cell type occupies based on the respective pore size of the transwell membrane (Figure 2C). Such transwell systems allow facile replication for hypothesis testing and screening assays and have been applied for investigations of migration, drug penetration, and paracrine signaling in co-cultures of IECs, fibroblasts or other stromal cells, or immune cells (often RAW 264.7 cells or THP-1 differentiated into macrophages), as well as bacteria.33,34 For example, model IECs (Caco-2) have been grown on transwell inserts to create a model epithelium with relevant barrier functions, often assessed by immunocytochemistry, model compound transport, and trans-epithelial electrical resistance (TEER), and used in co-culture studies, such as with non-pathogenic bacteria to investigate their activation of cytokine secretion by IECs.33,34
Scaffolds for three-dimensional mono- and co-cultures.
These more traditional in vitro culture methods take a reductionist approach to both mono- and co-culture of relevant cell types. However, use of flat, hard culture substrates (e.g., tissue culture polystyrene) may limit insights that can be gleaned about specific cellular functions that require cell interaction with, degradation of, and response to the ECM in multiple dimensions, including bacteria invasion. Indeed, culture of primary cells on such traditional substrates leads to loss of phenotype during long term propagation.35 In this context, different mimics of the ECM have been applied to allow the multidimensional culture of relevant cell types. Traditional, stiff porous scaffolds formed with natural materials such as silk, harvested proteins such as covalently-crosslinked collagen I or synthetic polymers such as poly(lactic acid-co-glycolic acid) (PLGA) have be utilized for three-dimensional (3D) culture of epithelial cells and their co-culture with bacteria (Figure 2D), although most studies to date have focused on mono-cultures.33,36,37 While these materials mimic aspects of the multidimensional nature of native ECM, they are more rigid and different in composition from the soft epithelium and LP of the healthy gut.
Water swollen polymer networks, known as hydrogels, have been formed with harvested proteins (e.g., physically crosslinked Matrigel, collagen I, alginate) to better mimic the soft tissue of the gut.38 Broadly, Matrigel™ and collagen I hydrogels have been long-used tools for 3D culture owing to their accessibility and utility in promoting the viability and function of a range of cell types including epithelial cells, although with batch-to-batch variability given their harvest from animal tissue.39 For example, Matrigel™ is a heterogeneous mixture of ECM proteins, proteoglycans, and growth factors secreted by Engelbreth-Holm-Swarm mouse sarcoma cells with compositional and structural variance between isolations.40 Collagen I typically is harvested from rat tail tendon or bovine skin or tendon and solubilized in acid for hydrogel formation over minutes upon neutralization with base.41 In these types of soft materials formed with harvested proteins, immune cells and epithelial cells have been co-cultured, from the study of IBD to the study of cancer.42,43 For example, model macrophage or dendritic cells (differentiated THP-1 human monocytes or MUTZ-3 dendritic cells) have been embedded in collagen I hydrogels and co-cultured with model IECs (Caco-2) seeded on top for investigating toxicity of common nanoparticles.42
For increased control of matrix properties, hydrogels formed with synthetic or biological polymers modified with reactive handles also have been investigated (Figure 2E). For example, comparing between naturally-derived and synthetic systems, culture of model IECs (Caco-2 and mucus-secreting human colorectal cancer cell line HT29-MTX) on and within alginate hydrogels led to spheroid formation, whereas culture on poly(N-isopropylacrylamide) (pNIPAM) led to the formation of villus-like structures.44 Deploying the temporal and spatial resolution of property control afforded by light-based chemistries, photolithographic techniques have been used to form poly(ethylene glycol) (PEG) diacrylate (PEGDA) based synthetic hydrogels that mimic the shape of the intestinal microvilli, and these micropatterned soft materials further were modified with additional functional handles for incorporation of collagen I and laminin proteins.45 IECs (Caco-2) seeded on these structures are capable of proliferating, aligning more similarly to epithelial cells in vivo, and forming tight junctions. These findings highlight the opportunity that chemical approaches provide for directing formation of tissue-mimicking structures and for controlling the cell microenvironment more generally, whether for the creation of improved model systems as done here or future studies testing hypotheses about specific interactions between host and bacteria.
Organoid cultures.
Organoid culture is an emerging approach for bridging the gap between whole organisms and traditional in vitro culture of models that use immortalized cell lines. Organoids often are formed with patient-derived cells, such as absorptive enterocytes from the small intestines or induced pluripotent stem cells. These organoid cultures also often move beyond the culture of such cells as spheroids by embedding them in matrix to encourage the formation of structures similar in morphology and function to native tissues, including the luminal structure of the gut. The resulting 3D clusters of progenitor cells embedded in matrix are capable of self-organization and self-renewal and can exhibit similar functions to in vivo organs (Figure 2E).46 A range of approaches are being explored for the creation of organoid microtissues, including encapsulation of tissue fragments, different single cells from niche of interest, or spheroids formed from cells cultured in droplets in Matrigel or collagen I hydrogels.47–49 Such 3D gastrointestinal model systems are being used for drug discovery, toxicology, and disease modeling.50 However, challenges remain in these cultures: these include the lengthy timescale to fully generate these organoids systems (circa 1 to 3 months); the artful and variable nature of their preparation that includes the use of harvested proteins in addition to primary cells with batch to batch variation; and often the absence of immune cells which can be key in studying host-bacteria interactions in IBD. For the study of immune cell-bacteria interactions, spheroid cultures,51 or transwell co-cultures, as described above, still remain the current ‘gold standard’ in this context.52
Table 1 below summarizes the application of these different tools and approaches for the study of cell-cell and cell-matrix interactions in both mono- and co-cultures.
Table 1:
Application of different in vitro culture platforms to mono- and co-culture of relevant cell types, including integration of different harvested and synthetic ECMs for 2D and 3D controlled cell culture.
| Cell Types | Matrix Properties | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Culture Platform | Epithelial (E) | Immune (I) | Stromal (S) | Bacteria (B) | Co-culture | Synthetic | Naturally Derived | 2D | 3D |
| Microfluidics | 27,28,53 | 53 | E/B53 | 27,28,53 | 27,28,53 | ||||
| Rotating wall vessel | 31,32 | 31 | E/B31 | ||||||
| Transwell | 33,34,54,55 | 34,54 | 55 | 33,34,54 | E/I/B34,54 E/S55 E/B33 | 54,55 | 33,34,54,55 | ||
| Porous scaffolds | 33,37 | 37 | 33,37 | E/S/B37 E/B33 | 33 | 37 | 33,37 | ||
| Hydrogels | 44,56–59 | 56,57,60–74 | E/S56 | 44,56,57,60,62,64–74 | 44,63,65 | 44 | 44,56,57,60,62–72,74 | ||
| Spheroids | 58,59,75–78 | 77 | E/S77 | 58,59,76,78 | 75,76 | 58,59,75,76,78 | |||
Innovative approaches for probing cell-microenvironment interactions with opportunities for the integration of new chemical tools in the study of host-bacteria interactions in the gut
Cell-cell interactions: organotypic model systems.
Aspects of well-defined co-cultures and chemical approaches for engineering the matrix are being integrated with organoid approaches for improved control of cell-microenvironment interactions in patient-derived systems.49 For example, focusing on cell-cell interactions, human small intestine cells, enterocytes, from donors have been cultured in transwell inserts with and without fibroblasts. Other systems have been designed to recapitulate the air-liquid interface (ALI) to create organotypic models that better mimic the villus structure and barrier function of the human gut and allow co-culture with macrophages and E. coli.54,55 Similar strategies of pre-mixing critical niche cell types with patient-derived intestinal cells also are being developed for shortening the timeline for organoid formation.77
Cell-matrix interactions: well-defined synthetic ECM mimics.
Focusing on cell-matrix interactions, chemical approaches for control of the matrix have been deployed for improved control of the formation of intestinal organoids in 3D culture and their delivery in vivo for regeneration of the gut.58,59,78 For example, well-defined biomimetic synthetic hydrogels were formed with multi-arm biologically-inert polymer (4-arm PEG) functionalized with maleimide reactive end groups (Figure 3A), linked with dithiol cell-degradable peptides, and modified with pendant monothiol integrin-binding peptides to control the mechanical and biochemical properties of the matrix.59 Properties of the synthetic matrix were tuned to promote the viability and growth of encapsulated stem-cell-derived spheroids and the formation of intestinal organoids (e.g., Figure 2E). Transitioning spheroids initially grown in Matrigel to this synthetic matrix minimized potential immunogenicity issues with Matrigel upon transplantation in vivo and allowed consistent and reproducible growth of spheroids and their delivery. In a separate study, utilizing the capability to tune both matrix degradation and composition afforded by such synthetic matrices, murine intestinal crypts were cultured within synthetic matrices formed by FXIIIa crosslinking of 8-arm PEG functionalized with vinyl sulfone and acrylate groups and peptide functionalized with acrylate or transglutaminase (TG) factor XIII handles (Figure 3A). These studies revealed both matrix modulus and cell-matrix interactions are critical to organoid formation. These careful investigations with temporally modulating specific synthetic matrix properties demonstrated the influence of yes associated protein (YAP) signaling and high matrix modulus on increased iPSC propagation and the need for a laminin-enriched, soft matrix modulus for differentiation and organoid development. This synthetic alternative to Matrigel streamlines and decreases variability of previous protocols by allowing for both propagation of iPSC colonies, and through dynamic changes to matrix properties, sequential generation of intestinal organoids.76
Figure 3: Chemical handles for the formation of synthetic matrices with molecular-level property control.
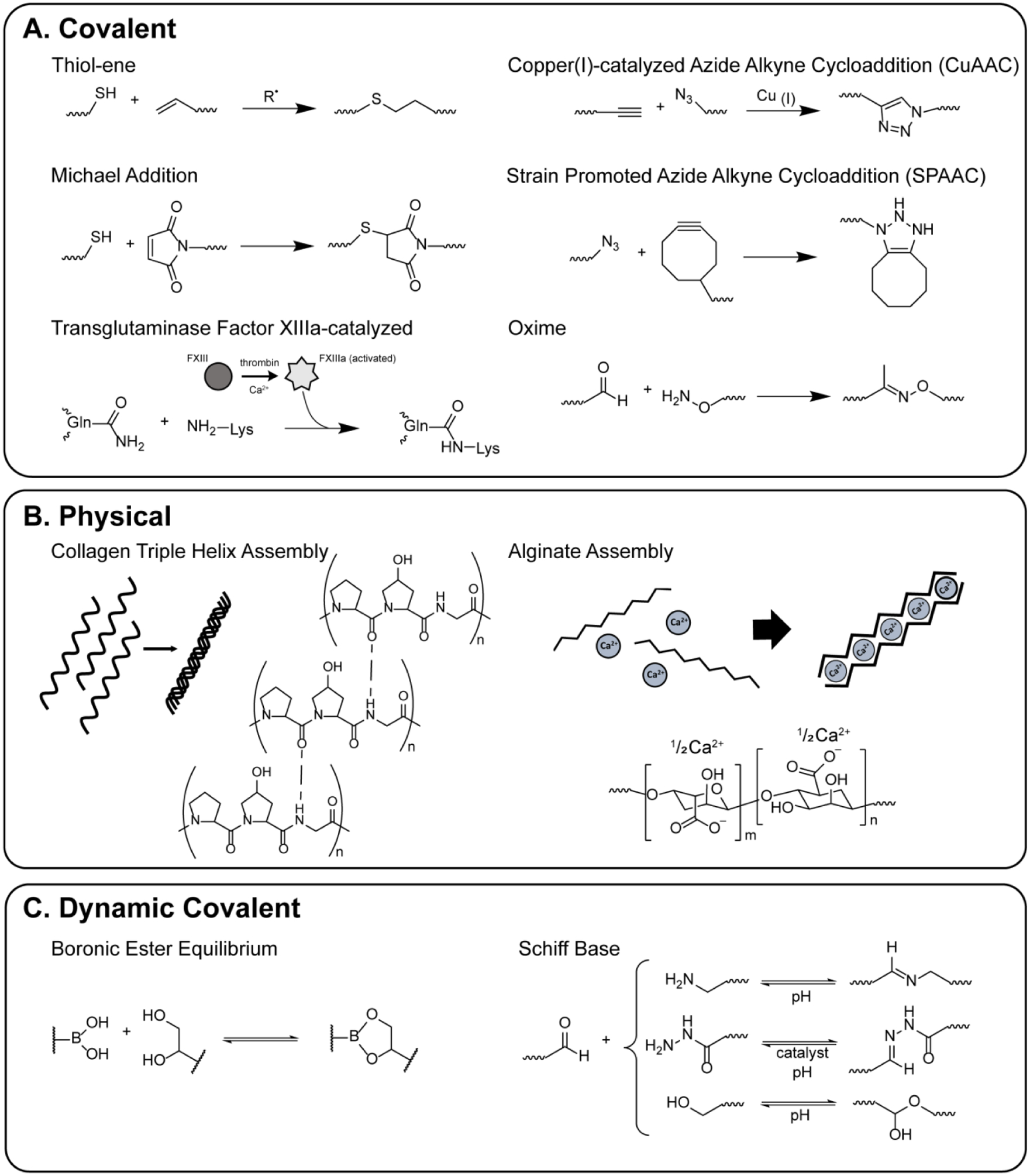
Here, A) covalent, B) physical, and C) dynamic covalent chemistries utilized in the encapsulation of cells for 3D culture are highlighted.
Chemical approaches for controlling synthetic ECM properties.
These seminal works highlight the opportunity that well-defined multidimensional cultures that integrate tunable synthetic matrices present for studies of host and bacteria cells. For example, such approaches using chemical tools can be deployed for innovative studies of key cell-cell and cell-matrix interactions for new insights into pivotal regulators in IBD. A range of new chemical reactive handles have been and are being established to further control and parse the influence of matrix mechanics, composition, and structure in spatially defined or dynamic fashion, including bio-orthogonal click chemistries, stimuli responsive (pH, light, temperature) chemistries, and dynamic covalent compositions that impart viscoelastic behavior to the synthetic matrix, all toward better mimicking native tissue properties and enabling hypothesis testing.
Synthetic and hybrid hydrogels composed of (bio)polymers (e.g., PEG, poly(vinyl alcohol), hyaluronic acid) decorated with different reactive functional groups have been formed with a variety of ‘click’ chemistries, including strain-promoted azide-alkyne cycloadditions (SPAAC), copper-catalyzed azide-alkyne cycloadditions (CuAAC), thiol-Michael addition, and oxime ligation (Figure 3A).79 Well-defined and robust soft materials have been fabricated with this approach to achieve a range of matrix moduli relevant for applications across a variety of tissue types, which is not possible with traditional physically-crosslinked harvested protein hydrogels. Further, these macromolecular building blocks and resulting hydrogels provide a modular platform for presentation of covalently conjugated biochemical cues. Photo-triggered chemistries (e.g., photoinitiated thiol-ene ‘click’ chemistry,57,67–69 photolabile nitrobenzyl groups70–74) allow formation and modification when and where desired, where photomasks or focused light have been used to control matrix modulus or presentation of specific biochemical cues, such as pendant peptides for promoting binding of desired integrins. For example, stromal cells (fibroblasts) have been encapsulated in synthetic hydrogels formed with SPAAC reactions between 8-arm PEG dibenzylcyclooctyne, a 4-arm PEG azide, and azido-RGDS integrin binding peptide; cell-matrix interactions then were modulated dynamically with reversible spatially-defined protein conjugation (thiolated TGFβ1) using allyl sulfide chemistry, and fibroblast activation was only observed in regions where protein was tethered.60 Thiol-ene chemistry has been used to spatially photopattern nanofibrous structures: for example, norbornene functionalized hyaluronic acid was electrospun and swollen with thiolated RGDS peptide, which was patterned in regions of interest utilizing photolithographic techniques.57 In another work, light-mediated degradation of a nitrobenzyl crosslinker was utilized to spatially pattern aligned softened channels into the surface of a gelatin-methacrylate hydrogel, where patterned gels directed cell alignment.72
More recently, the importance of both hierarchical structure and matrix viscoelasticity in addition to matrix mechanical properties has been demonstrated.80,81 This level of property control can be achieved through integration of assembling or stress-relaxing chemistries that utilize physical or dynamic covalent interactions within synthetic matrices, respectively (Figure 3B and C). For example, boronic acid (BA)-based moieties reversibly react with 1,2- and 1,3 diols to provide stress relaxation and self-healing capabilities.82 BA-based hydrogels have been used to culture human mesenchymal stem cells (hMSCs), which demonstrate morphological and mechanotransduction (YAP/TAZ signaling) changes when cultured in viscoelastic BA-based environments compared to control hydrogels with purely elastic properties.64 The dynamic self-healing nature of BA-based cultures also provides opportunity for facile co-culture construction.56 Additionally, other dynamic covalent chemistries, such as modified hydrazone and imine approaches,66,83,84 diels alder,69 bio-inspired62 and ionic based crosslinks,63,85 provide innovative platforms for investigations of the impact of more complex matrix properties on cell behavior.
The property control afforded by the presented systems amongst others is applicable to not only the culture of stromal cells, but to all cell types broadly. For the generation of a range of cell types, synthetic matrices with controlled integrin binding sequences have been used to direct the proliferation and differentiation of iPSCs and the formation of organoids.58,76 Further, changes in the mechanical properties of the matrix are known to alter the behavior of many cell types, from fibroblasts to hMSCs to cancer, and have been implicated in many diseases, including various IBDs.47,68,86 The complex material platforms described here provide modular multidimensional systems for the investigation of cell-cell and cell-matrix interactions, with future opportunities to investigate a myriad of diseases, including IBD, for a more comprehensive understanding of key cell-microenvironment interactions, host-bacteria cell interactions, and the development of relevant disease models.
Molecular and imaging-based tools for probing host-bacteria interactions on the cellular and sub-cellular level
Moving to the molecular level, studies between host and bacteria cells are revealing the importance of not only whole cell-cell interactions, but also how fragments of cells (e.g., debris from bacterial cells) are important in both local and systemic immune response in IBD and other diseases. Specifically, the bacterial cell wall is composed of peptidogylcan (PG), a glycan strand of N-acetyl glucosamine (NAG) and N-acetyl muramic acid (NAM) crosslinked by D-amino acids containing short peptides.87,88 Although this glycan macromolecule remains generally conserved, the stitching together of the polymer creates diversity between bacteria species.88 As both commensal and pathogenic bacteria produce PG fragments with the core building block NAM, the innate immune system has developed molecular mechanisms to sense and respond to bacteria based on their PG fragments.89,90 Although the PG polymer has incredible importance, a gap in the field has existed until recently with the development of strategies for direct imaging and labeling of PG.
Initial work to label PG used a chemical biology approach to fluorescently label D-amino acids in order to visualize cell wall synthesis in real time in a variety of bacterial species as well as in live cells.87 Small fluorophores, 7-hydroxycoumarin-3-carboxylic acid (HCC-OH) and 4-chlro-7-nitrobenzofuran (NBD-Cl), were coupled to the D-amino acid backbone, 3-amino-D-alanine, to visualize peripheral and septal labeling of entire cell populations of a wide spectrum of bacterial species, including Escherichia coli, Bacillus subtilis, and Agrobacterium tumefaciens (Figure 4A).87 This was accomplished in one doubling time and did not affect the growth rate of the species.87 These probes and others more recently developed can be used to track PG at sites of active synthesis for a comprehensive analysis of bacterial growth.91,92
Figure 4: Structure of various bacterial labeling probes.
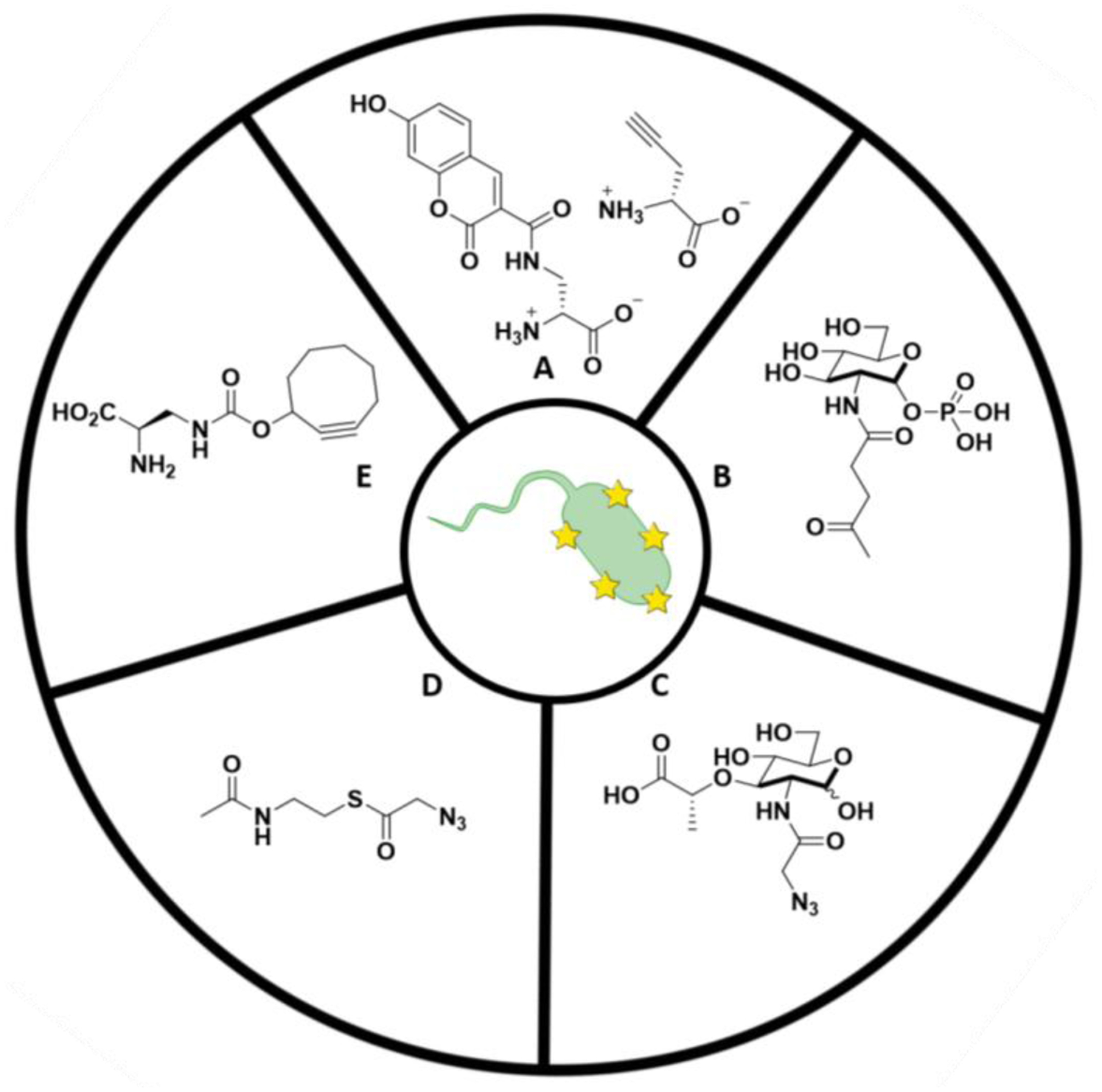
A) Small fluorophores used to couple to D-amino acids in bacterial PG. B) NAG sugar probe for PG visualization. C) NAM sugar probe for metabolic labeling of PG. D) SNAc derivative example for post-synthetic modification of PG. E) NIR fluorogenic probes for no-wash labeling of PG.
Besides labeling the amino acid residues of the PG, specifically for lactic acid bacteria, the NAG unit in PG can be labeled at the 2-N acetyl position.93 Using N-acetylglucosamine-1-phosphate (GlcNAc-1-phosphate) as a precursor, derivatives were synthesized with non-native reactive groups for incorporation into the bacteria cell wall (Figure 4B).93 Using the lactic acid bacteria Lactobacillus plantarum, incorporation was observed of the GlcNAc-1-phosophate derivative that was acetylated and contained a ketone group at cell-wall biosynthesis.93 This opened the door for further investigation in bacterial probes on the NAG sugar that could potentially be utilized for PG visualization.
Bacterial probe development has even extended to probes that hijack the natural biosynthetic pathway.94,95 Currently, the information about biological identity and generation of N-acetyl muramic acid-containing PG fragments is limited. Based on natural modifications of the 2-N acetyl position of NAM glycan to a N-glycolyl with a NAM hydroxylase by mycobacteria and actinomycetes, a set of NAM probes has been developed around a synthesis of 2-azido muramyl dipeptide.94,95 The NAM probes contain an azide or alkyne at the 2-N position which allow for fluorophore labeling though azide-alkyne Huisegen cycloaddition (CuAAC), allowing for robust labeling and visualization of the NAM carbohydrate backbone in both Gram-positive B. subtilis and Gram-negative E. coli modified with recycling enzymes AmgK and MurU (Figure 4C).94 These remodeled bacteria have been utilized for invasion applications with mouse macrophages, showing the versatility of these probes for future application within in vitro invasion models.
Looking to Nature for inspiration for how bacteria naturally modify their PG, post synthetic strategies also have arisen for pathogenic bacteria species Staphylococcus aureus and Neisseria gonorrheae, which O-acetylate their PG to avoid degradation by lysozyme.96 The enzymes peptidoglycan O-acetyltransferase B (PatB) and peptidoglycan acetyltransferase A (PatA) are believed to work in conjunction in Gram-negative bacteria to O-acetylate the PG using acetyl-CoA.96 Through the development of synthetic pathways for N-acetylcysteamine-thioesters (SNAc), truncated CoA thioesters, and purified PatB, a modified strain of B. subtilis (Gram-positive) was co-labeled with wheatgerm agglutinin (WGA), providing a post-synthetic modification strategy to label NAG sugars in the PG (Figure 4D).96 While robust kinetic analysis was performed on purified PatB using model tri-saccharide substrates for the SNAcs, these data do not report on the efficiency of labeling whole cells with fluorescent probe.96 The labeling methodology was extended to Gram-negative bacteria in in vitro purified PG assays; this methodology will most likely not be useful for Gram-negative bacteria in whole cells due the presence of the outer membrane. In the in vitro assay, PatB, was able to decorate a series of Gram-negative bacterial PG providing protection from lysosomal degradation, thus providing a mechanism to produce modified PG materials.96
Biorthogonal chemistry has proven extremely useful in developing a wide variety of bacterial labeling probes. Recent strategies have developed near-infrared (NIR) fluorogenic probes that are activated by biorthogonal chemistry for tissue studies.97 Fluorogenic probes activated through biorthogonal reactions can have minimized fluorescence from excess probe.97 Longer wavelength, NIR fluorogenic probes are desirable for interrogating biological systems due to higher tissue penetrance and reduced background autofluorescence.97 For example, fluorogenic azido Si-rhodamine probes with emission maxima near 670 nm have been synthesized and optimized, providing opportunities for “no wash” labeling with terminal alkynes displayed on mammalian cell surfaces (Figure 4E).97 In addition, bacterial species such as M. smegmatis and C. glutamicum are grown for one doubling time with new cyclooctyne d-alanine analogs which react with the new fluorogenic azido Si-rhodamine probes though copper-catalyzed conditions, allowing for no-wash visualization of PG for in vivo imaging of bacterial pathogens97,98.
These PG labeling strategies, ranging from amino acid derivatives to metabolic incorporation and post-synthetic modification of glycans, coupled to invasion models, will be critical in conducting vital studies to inform how these bacterial fragments are interacting with the body’s innate immune system.
Conclusion and opportunities for future investigations
With the advent of microbiome studies, the models discussed above become much more complex. The microbiome which comprises over 30 trillion bacteria cells from a variety of bacterial species is an integral player in human health. However, these bacterial cells contain many of the same patterns as pathogens. Now the question becomes how does the human immune system differentiate between these two types of microbial signatures – one pathogenic and the other commensal? The ability to properly investigate the interaction of human innate immune cells with pathogenic and commensal bacterial will be critical in understanding this differentiation. An array of well-defined and accessible chemical tools for controlling host-bacteria interactions, from the bacteria cell wall to the multiscale extracellular matrix, will enable these investigations and provide new insights into these complex systems and ultimately improved therapeutic strategies for enhancing human health.
Acknowledgements
K.A.W. would like to thank NIH for support through CBI training grant (5T32GM008550) and the NIH U01 Common Fund project (U01CA221230-01). C.L.G. is a Pew Biomedical Scholar and Sloan Scholar, and would like to thank the Pew Foundation, Sloan Foundation, and the Camille and Henry Dreyfus Foundation. S.E.C. and A.M.K. would like to thank Pew Charitable Trusts (00026178) and NIH Director’s New Innovator Award (DP2HL152424) for their funding support.
References
- 1.Janeway CA, Medzhitov R. Innate Immune Recognition. Annu Rev Immunol. 2002. doi: 10.1146/annurev.immunol.20.083001.084359 [DOI] [PubMed] [Google Scholar]
- 2.Tosi MF. Innate immune responses to infection. J Allergy Clin Immunol. 2005. doi: 10.1016/j.jaci.2005.05.036 [DOI] [PubMed] [Google Scholar]
- 3.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016. doi: 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: Macrophages, mast cells and neutrophils. Nat Immunol. 2011. doi: 10.1038/ni.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medzhitov R, Janeway CA. Decoding the patterns of self and nonself by the innate immune system. Science (80- ). 2002. doi: 10.1126/science.1068883 [DOI] [PubMed] [Google Scholar]
- 6.Geddes K, Magalhães JG, Girardin SE. Unleashing the therapeutic potential of NOD-like receptors. Nat Rev Drug Discov. 2009. doi: 10.1038/nrd2783 [DOI] [PubMed] [Google Scholar]
- 7.Thaiss CA, Levy M, Itav S, Elinav E. Integration of Innate Immune Signaling. Trends Immunol. 2016. doi: 10.1016/j.it.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 8.Melmed G, Thomas LS, Lee N, et al. Human Intestinal Epithelial Cells Are Broadly Unresponsive to Toll-Like Receptor 2-Dependent Bacterial Ligands: Implications for Host-Microbial Interactions in the Gut. J Immunol. 2003. doi: 10.4049/jimmunol.170.3.1406 [DOI] [PubMed] [Google Scholar]
- 9.Kim M, Ashida H, Ogawa M, Yoshikawa Y, Mimuro H, Sasakawa C. Bacterial interactions with the host epithelium. Cell Host Microbe. 2010. doi: 10.1016/j.chom.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 10.Goodlad RA, Plumb JA, Wright NA. Simultaneous measurement of intestinal crypt cell production rate and water absorption. Gut. 1987. doi: 10.1136/gut.28.Suppl.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner CE, Wheeler KM, Ribbeck K. Mucins and Their Role in Shaping the Functions of Mucus Barriers. Annu Rev Cell Dev Biol. 2018. doi: 10.1146/annurev-cellbio-100617-062818 [DOI] [PubMed] [Google Scholar]
- 12.Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012. doi: 10.1016/j.mib.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommer F, Bäckhed F. The gut microbiota-masters of host development and physiology. Nat Rev Microbiol. 2013. doi: 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- 14.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011. doi: 10.1038/nature10208 [DOI] [PubMed] [Google Scholar]
- 15.Mowat AM, Bain CC. Mucosal macrophages in intestinal homeostasis and inflammation. J Innate Immun. 2011. doi: 10.1159/000329099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortha A, Chudnovskiy A, Hashimoto D, et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science (80- ). 2014. doi: 10.1126/science.1249288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bain CC, Schridde A. Origin, differentiation, and function of intestinal macrophages. Front Immunol. 2018. doi: 10.3389/fimmu.2018.02733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lussier C, Basora N, Bouatrouss Y, Beaulieu JF. Integrins as mediators of epithelial cell-matrix interactions in the human small intestinal mucosa. Microsc Res Tech. 2000. doi: [DOI] [PubMed] [Google Scholar]
- 19.Nikulin SV, Knyazev EN, Gerasimenko TN, et al. Non-Invasive Evaluation of Extracellular Matrix Formation in the Intestinal Epithelium. Bull Exp Biol Med. 2018. doi: 10.1007/s10517-018-4283-7 [DOI] [PubMed] [Google Scholar]
- 20.Li ACY, Thompson RPH. Basement membrane components. J Clin Pathol. 2003. doi: 10.1136/jcp.56.12.885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schridde A, Bain CC, Mayer JU, et al. Tissue-specific differentiation of colonic macrophages requires TGFβ receptor-mediated signaling. Mucosal Immunol. 2017. doi: 10.1038/mi.2016.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spenlé C, Lefebvre O, Lacroute J, et al. The laminin response in inflammatory bowel disease: Protection or malignancy? PLoS One. 2014. doi: 10.1371/journal.pone.0111336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrila J, Crabbé A, Yang J, et al. Modeling host-pathogen interactions in the context of the microenvironment: Three-dimensional cell culture comes of age. Infect Immun. 2018;86(11):1. doi: 10.1128/IAI.00282-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blumberg RS, Saubermann LJ, Strober W. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr Opin Immunol. 1999. doi: 10.1016/S0952-7915(99)00032-1 [DOI] [PubMed] [Google Scholar]
- 25.Tsilingiri K, Barbosa T, Penna G, et al. Probiotic and postbiotic activity in health and disease: Comparison on a novel polarised ex-vivo organ culture model. Gut. 2012. doi: 10.1136/gutjnl-2011-300971 [DOI] [PubMed] [Google Scholar]
- 26.Shah P, Fritz JV, Glaab E, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun. 2016;7(May). doi: 10.1038/ncomms11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pocock K, Delon LC, Khatri A, et al. Uptake of silica particulate drug carriers in an intestine-on-a-chip: Towards a better: In vitro model of nanoparticulate carrier and mucus interactions. Biomater Sci. 2019;7(6):2410. doi: 10.1039/c9bm00058e [DOI] [PubMed] [Google Scholar]
- 28.Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (United Kingdom). 2013;5(9):1130. doi: 10.1039/c3ib40126j [DOI] [PubMed] [Google Scholar]
- 29.Bein A, Shin W, Jalili-Firoozinezhad S, et al. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cmgh. 2018;5(4):659–668. doi: 10.1016/j.jcmgh.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Choi JH, Kim HJ. Human gut-on-a-chip technology: will this revolutionize our understanding of IBD and future treatments? Expert Rev Gastroenterol Hepatol. 2016;10(8):883. doi: 10.1080/17474124.2016.1200466 [DOI] [PubMed] [Google Scholar]
- 31.Nickerson CA, Goodwin TJ, Terlonge J, et al. Three-dimensional tissue assemblies: Novel models for the study of Salmonella enterica serovar typhimurium pathogenesis. Infect Immun. 2001;69(11):7106. doi: 10.1128/IAI.69.11.7106-7120.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long JP, Pierson S, Hughes JH. Rhinovirus replication in HeLa cells cultured under conditions of simulated microgravity. Aviat Space Environ Med. 1998;69(9):851. [PubMed] [Google Scholar]
- 33.Costello CM, Sorna RM, Goh Y, Cengic I, Jain NK, March JC. 3‑D Intestinal Scaffolds for Evaluating the Therapeutic Potential of Probiotics. Mol Pharm. 2014;11:2030. doi: 10.1021/mp5001422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haller D, Bode C, Hammes WP, Pfeifer AMA, Schi EJ V, Blum S. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell / leucocyte co-cultures. Gut. 2000;47:79. doi: 10.1136/gut47.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Battiston KG, Cheung JWC, Jain D, Santerre JP. Biomaterials in co-culture systems: Towards optimizing tissue integration and cell signaling within scaffolds. Biomaterials. 2014;35(15):4465. doi: 10.1016/j.biomaterials.2014.02.023 [DOI] [PubMed] [Google Scholar]
- 36.Roh TT, Chen Y, Paul HT, Guo C, Kaplan DL. 3D bioengineered tissue model of the large intestine to study inflammatory bowel disease. Biomaterials. 2019;225(September):119517. doi: 10.1016/j.biomaterials.2019.119517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardenas D, Bhalchandra S, Lamisere H, et al. Two- and Three-Dimensional Bioengineered Human Intestinal Tissue Models for Cryptosporidium BT - Cryptosporidium: Methods and Protocols. In: Mead JR, Arrowood MJ, eds. New York, NY: Springer New York; 2020:373. doi: 10.1007/978-1-4939-9748-0_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dosh RH, Jordan-Mahy N, Sammon C, Le Maitre CL. Long-term in vitro 3D hydrogel co-culture model of inflammatory bowel disease. Sci Rep. 2019. doi: 10.1038/s41598-019-38524-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caliari SR, Burdick JA. A practical guide to hydrogels for cell culture. Nat Methods. 2016. doi: 10.1038/nmeth.3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes CS, Postovit LM, Lajoie GA. Matrigel : A complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886. doi: 10.1002/pmic.200900758 [DOI] [PubMed] [Google Scholar]
- 41.Caliari SR, Burdick JA. A practical guide to hydrogels for cell culture. Nat Methods. 2016;13(5). doi: 10.1038/nmeth.3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Susewind J, De Souza Carvalho-Wodarz C, Repnik U, et al. A 3D co-culture of three human cell lines to model the inflamed intestinal mucosa for safety testing of nanomaterials. Nanotoxicology. 2016;10(1):53–62. doi: 10.3109/17435390.2015.1008065 [DOI] [PubMed] [Google Scholar]
- 43.Ham SL, Joshi R, Thakuri PS, Tavana H. Liquid-based three-dimensional tumor models for cancer research and drug discovery. Exp Biol Med. 2016. doi: 10.1177/1535370216643772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dosh RH, Essa A, Jordan-Mahy N, Sammon C, Le Maitre CL. Use of hydrogel scaffolds to develop an in vitro 3D culture model of human intestinal epithelium. Acta Biomater. 2017;62:128. doi: 10.1016/j.actbio.2017.08.035 [DOI] [PubMed] [Google Scholar]
- 45.Castano AG, Garcia-Diaz M, Torras N, Altay G, Comelles J, Martinez E. Dynamic photopolymerization produces complex microstructures on hydrogels in a moldless approach to generate a 3D intestinal tissue model. Biofabrication. 2019;11:025007. doi: 10.1088/1758-5090/ab0478 [DOI] [PubMed] [Google Scholar]
- 46.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18(3):246. doi: 10.1038/ncb3312 [DOI] [PubMed] [Google Scholar]
- 47.Pastuła A, Middelhoff M, Brandtner A, et al. Three-Dimensional Gastrointestinal Organoid Culture in Combination with Nerves or Fibroblasts: A Method to Characterize the Gastrointestinal Stem Cell Niche. Stem Cells Int. 2016;2016. doi: 10.1155/2016/3710836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khalil HA, Lei NY, Brinkley G, et al. A novel culture system for adult porcine intestinal crypts. Cell Tissue Res. 2016;365(1):123. doi: 10.1007/s00441-016-2367-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kratochvil MJ, Seymour AJ, Li TL, Paşca SP, Kuo CJ, Heilshorn SC. Engineered materials for organoid systems. Nat Rev Mater. 2019;4(9):606. doi: 10.1038/s41578-019-0129-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartfeld S, Clevers H. Stem cell-derived organoids and their application for medical research and patient treatment. J Mol Med. 2017;95(7):729. doi: 10.1007/s00109-017-1531-7 [DOI] [PubMed] [Google Scholar]
- 51.Sebrell TA, Hashimi M, Sidar B, et al. A Novel Gastric Spheroid Co-culture Model Reveals Chemokine-Dependent Recruitment of Human Dendritic Cells to the Gastric Epithelium. Cmgh. 2019;8(1):157.e3. doi: 10.1016/j.jcmgh.2019.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology. 2016;150(5):1098. doi: 10.1053/j.gastro.2015.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12(12):2165. doi: 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- 54.Noel G, Baetz NW, Staab JF, et al. A primary human macrophage- enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Nat Publ Gr. 2017; (September 2016):1. doi: 10.1038/srep45270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ayehunie S, Landry T, Stevens Z, Armento A, Hayden P, Klausner M. Human Primary Cell-Based Organotypic Microtissues for Modeling Small Intestinal Drug Absorption. Pharm Res. 2018;35(4). doi: 10.1007/s11095-018-2362-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smithmyer ME, Deng CC, Cassel SE, Levalley PJ, Sumerlin BS, Kloxin AM. Self-Healing Boronic Acid-Based Hydrogels for 3D Co-cultures. ACS Macro Lett. 2018;7(9):1105. doi: 10.1021/acsmacrolett.8b00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wade RJ, Bassin EJ, Gramlich WM, Burdick JA. Nanofibrous hydrogels with spatially patterned biochemical signals to control cell behavior. Adv Mater. 2015;27(8). doi: 10.1002/adma.201404993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cruz-Acuña R, Quirós M, Farkas AE, et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat Cell Biol. 2017;19(11):1326. doi: 10.1038/ncb3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cruz-Acuña R, Quirós M, Huang S, et al. PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery. Nat Protoc. 2018;13(9):2102. doi: 10.1038/s41596-018-0036-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grim JC, Brown TE, Aguado BA, et al. A Reversible and Repeatable Thiol-Ene Bioconjugation for Dynamic Patterning of Signaling Proteins in Hydrogels. ACS Cent Sci. 2018;4(7):909. doi: 10.1021/acscentsci.8b00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W, Ma H, Zhang J, Zhu L, Wang C, Yang Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci Rep. 2017;7(1):1. doi: 10.1038/s41598-017-14364-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu L, Shadish JA, Arakawa CK, Shi K, Davis J, DeForest CA. Cyclic Stiffness Modulation of Cell-Laden Protein–Polymer Hydrogels in Response to User-Specified Stimuli Including Light. Adv Biosyst. 2018;2(12):1. doi: 10.1002/adbi.201800240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaudhuri O, Gu L, Klumpers D, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater. 2016;15(3):326. doi: 10.1038/nmat4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang S, Ma H, Tu HC, Wang HR, Lin PC, Anseth KS. Adaptable Fast Relaxing Boronate-Based Hydrogels for Probing Cell–Matrix Interactions. Adv Sci. 2018;5(9):1. doi: 10.1002/advs.201800638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang X, Liu G, Peng L, et al. Highly Efficient Self-Healable and Dual Responsive Cellulose-Based Hydrogels for Controlled Release and 3D Cell Culture. Adv Funct Mater. 2017;27(40):1. doi: 10.1002/adfm.201703174 [DOI] [Google Scholar]
- 66.McKinnon DD, Domaille DW, Cha JN, Anseth KS. Biophysically defined and cytocompatible covalently adaptable networks as viscoelastic 3d cell culture systems. Adv Mater. 2014;26(6):865. doi: 10.1002/adma.201303680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sawicki LA, Kloxin AM. Design of thiol-ene photoclick hydrogels using facile techniques for cell culture applications. Biomater Sci. 2014. doi: 10.1039/c4bm00187g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mabry KM, Lawrence RL, Anseth KS. Dynamic stiffening of poly(ethylene glycol)-based hydrogels to direct valvular interstitial cell phenotype in a three-dimensional environment. Biomaterials. 2015;49:47. doi: 10.1016/j.biomaterials.2015.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu F, Cao X, Li Y, Chen X. Diels-Alder Click-Based Hydrogels for Direct Spatiotemporal Postpatterning via Photoclick Chemistry. ACS Macro Lett. 2015;4(3):289. doi: 10.1021/mz5007427 [DOI] [PubMed] [Google Scholar]
- 70.Azagarsamy MA, Marozas IA, Spaans S, Anseth KS. Photoregulated Hydrazone-Based Hydrogel Formation for Biochemically Patterning 3D Cellular Microenvironments. ACS Macro Lett. 2016;5(1):19. doi: 10.1021/acsmacrolett.5b00682 [DOI] [PubMed] [Google Scholar]
- 71.Kirschner CM, Alge DL, Gould ST, Anseth KS. Clickable, photodegradable hydrogels to dynamically modulate valvular interstitial cell phenotype. Adv Healthc Mater. 2014;3(5):649. doi: 10.1002/adhm.201300288 [DOI] [PubMed] [Google Scholar]
- 72.Tsang KMC, Annabi N, Ercole F, et al. Facile one-step micropatterning using photodegradable gelatin hydrogels for improved cardiomyocyte organization and alignment. Adv Funct Mater. 2015;25(6):977. doi: 10.1002/adfm.201403124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Musoke-Zawedde P, Shoichet MS. Anisotropic three-dimensional peptide channels guide neurite outgrowth within a biodegradable hydrogel matrix. Biomed Mater. 2006;1:162. doi: 10.1088/1748-6041/1/3/011 [DOI] [PubMed] [Google Scholar]
- 74.Lunzer M, Shi L, Andriotis OG, et al. A Modular Approach to Sensitized Two-Photon Patterning of Photodegradable Hydrogels Angewandte. 2018:15122. doi: 10.1002/anie.201808908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panek M, Grabacka M, Pierzchalska M. The formation of intestinal organoids in a hanging drop culture. Cytotechnology. 2018;70(3):1085. doi: 10.1007/s10616-018-0194-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gjorevski N, Sachs N, Manfrin A, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539(7630):560. doi: 10.1038/nature20168 [DOI] [PubMed] [Google Scholar]
- 77.Zoetemelk M, Rausch M, Colin DJ, Dormond O, Nowak-Sliwinska P. Short-term 3D culture systems of various complexity for treatment optimization of colorectal carcinoma. Sci Rep. 2019;9(1):1. doi: 10.1038/s41598-019-42836-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hernandez-Gordillo V, Kassis T, Lampejo A, et al. Niche-inspired synthetic matrices for epithelial organoid culture. bioRxiv. 2019:806919. doi: 10.1101/806919 [DOI] [Google Scholar]
- 79.Kharkar PM, Kiick KL, Kloxin AM. Designing degradable hydrogels for orthogonal control of cell microenvironments. Chem Soc Rev. 2013. doi: 10.1039/c3cs60040h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lou J, Stowers R, Nam S, Xia Y, Chaudhuri O. Stress relaxing hyaluronic acid-collagen hydrogels promote cell spreading, fiber remodeling, and focal adhesion formation in 3D cell culture. Biomaterials. 2018;154:213. doi: 10.1016/j.biomaterials.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 81.Baker BM, Trappmann B, Wang WY, et al. Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments. Nat Mater. 2015;14(12). doi: 10.1038/nmat4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dufort BM, Tibbitt MW. Design of moldable hydrogels for biomedical applications using dynamic covalent boronic esters. Mater Today Chem. 2019;12:16. doi: 10.1016/j.mtchem.2018.12.001 [DOI] [Google Scholar]
- 83.Escobar F, Anseth KS, Schultz KM. Dynamic Changes in Material Properties and Degradation of Poly(ethylene glycol)-Hydrazone Gels as a Function of pH. Macromolecules. 2017;50(18):7351. doi: 10.1021/acs.macromol.7b01246 [DOI] [Google Scholar]
- 84.Yang X, Liu G, Peng L, Guo J, Tao L, Yuan J. Highly Efficient Self-Healable and Dual Responsive Cellulose-Based Hydrogels for Controlled Release and 3D Cell Culture. 2017;1703174:1. doi: 10.1002/adfm.201703174 [DOI] [Google Scholar]
- 85.Barrett DG, Fullenkamp DE, He L, Holten-andersen N, Lee KYC, Messersmith PB. pH-Based Regulation of Hydrogel Mechanical Properties Through Mussel-Inspired Chemistry and Processing. 2013:1111. doi: 10.1002/adfm.201201922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee JY, Chaudhuri O. Regulation of Breast Cancer Progression by Extracellular Matrix Mechanics: Insights from 3D Culture Models. ACS Biomater Sci Eng. 2018;4(2):302. doi: 10.1021/acsbiomaterials.7b00071 [DOI] [PubMed] [Google Scholar]
- 87.Kuru E, Hughes HV, Brown PJ, et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chemie - Int Ed. 2012. doi: 10.1002/anie.201206749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rogers HJ. PEPTIDOGLYCANS (MUCOPEPTIDES): STRUCTURE, FUNCTION, AND VARIATIONS. Ann N Y Acad Sci. 1974. doi: 10.1111/j.1749-6632.1974.tb43255.x [DOI] [PubMed] [Google Scholar]
- 89.Walsh C Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000. doi: 10.1038/35021219 [DOI] [PubMed] [Google Scholar]
- 90.Strominger JL. Bacterial cell walls, innate immunity and immunoadjuvants. Nat Immunol. 2007. doi: 10.1038/ni1207-1269 [DOI] [PubMed] [Google Scholar]
- 91.Hsu YP, Hall E, Booher G, et al. Fluorogenic d-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays. Nat Chem. 2019. doi: 10.1038/s41557-019-0217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Silva AM, Otten C, Biboy J, et al. The fluorescent D-Amino Acid NADA as a tool to study the conditional activity of transpeptidases in Escherichia coli. Front Microbiol. 2018. doi: 10.3389/fmicb.2018.02101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sadamoto R, Matsubayashi T, Shimizu M, et al. Bacterial surface engineering utilizing glucosamine phosphate derivatives as cell wall precursor surrogates. Chem - A Eur J. 2008. doi: 10.1002/chem.200801734 [DOI] [PubMed] [Google Scholar]
- 94.Liang H, DeMeester KE, Hou CW, Parent MA, Caplan JL, Grimes CL. Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat Commun. 2017. doi: 10.1038/ncomms15015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Demeester KE, Liang H, Jensen MR, et al. Synthesis of Functionalized N-Acetyl Muramic Acids to Probe Bacterial Cell Wall Recycling and Biosynthesis. J Am Chem Soc. 2018. doi: 10.1021/jacs.8b03304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Lazor KM, DeMeester KE, Liang H, Heiss TK, Grimes CL. Postsynthetic modification of bacterial peptidoglycan using bioorthogonal n-acetylcysteamine analogs and peptidoglycan o-acetyltransferase B. J Am Chem Soc. 2017. doi: 10.1021/jacs.7b06820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shieh P, Siegrist MS, Cullen AJ, Bertozzi CR. Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proc Natl Acad Sci. 2014. doi: 10.1073/pnas.1322727111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. D-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 2013. doi: 10.1021/cb3004995 [DOI] [PMC free article] [PubMed] [Google Scholar]


