Summary
The aggregation and joint analysis of large numbers of exome sequences has recently made it possible to derive estimates of intolerance to loss-of-function (LoF) variation for human genes. Here, we demonstrate strong and widespread coupling between genic LoF intolerance and promoter CpG density across the human genome. Genes downstream of the most CpG-rich promoters (top 10% CpG density) have a 67.2% probability of being highly LoF intolerant, using the LOEUF metric from gnomAD. This is in contrast to 7.4% of genes downstream of the most CpG-poor (bottom 10% CpG density) promoters. Combining promoter CpG density with exonic and promoter conservation explains 33.4% of the variation in LOEUF, and the contribution of CpG density exceeds the individual contributions of exonic and promoter conservation. We leverage this to train a simple and easily interpretable predictive model that outperforms other existing predictors and allows us to classify 1,760 genes—which are currently unascertained in gnomAD—as highly LoF intolerant or not. These predictions have the potential to aid in the interpretation of novel variants in the clinical setting. Moreover, our results reveal that high CpG density is not merely a generic feature of human promoters but is preferentially encountered at the promoters of the most selectively constrained genes, calling into question the prevailing view that CpG islands are not subject to selection.
Keywords: CpG islands, haploinsufficiency, gnomAD, loss-of-function, dosage sensitivity, promoters, GC content, CpG density, epigenetics, selection
Introduction
A powerful way of gaining insight into a gene’s contribution to organismal homeostasis is by studying the fitness effect exerted by loss-of-function (LoF) variants in that gene. Fully characterizing this effect is challenging, as it requires estimation of both the selection coefficient for individuals with bi-allelic LoF variants as well as the dominance coefficient.1,2 However, recent studies based on the joint processing and analysis of large numbers of exome sequences have developed metrics which serve as approximations to genic LoF intolerance in humans.3, 4, 5 These metrics correlate with several properties indicative of LoF intolerance (such as enrichment for known haploinsufficient genes4,5) and can substantially help in the assignment of pathogenicity to novel variants encountered in individuals as recommended by the American College of Medical Genetics and Genomics.6
At the core of all these metrics is a comparison of the observed to the expected number of LoF variants. Hence, genes where the latter is small (e.g., due to small coding sequence length or low mutation rate) will not be amenable to this approach until the sample sizes become much larger than they presently are. Currently in gnomAD, the largest such effort with publicly available constraint data based on 125,748 exomes, approximately 28% of genes are unascertained with respect to their LoF intolerance.5 It has been estimated that even with 500,000 individuals, the discovery of LoF variants will remain far from saturation, with potentially a sizeable fraction of genes still difficult to ascertain.7
The cardinal feature of highly LoF-intolerant genes, i.e., genes depleted of even monoallelic LoF variants in healthy individuals, is dosage sensitivity; a gene copy containing one or more LoF variants produces mRNAs that are typically degraded via nonsense-mediated decay.8,9 Therefore, the deleterious effects of LoF variants in these genes are often mediated through a reduction of the normal amount of mRNA used for protein production. This in turn, implies that studying the characteristics of regulatory elements controlling the expression of highly LoF-intolerant genes has the potential to yield two important benefits.10,11 First, it can highlight the features of the most functionally important regulatory elements in the human genome. Second, such features can then provide the basis for predictive models of LoF intolerance, which can be applied to unascertained genes.
In promoters, one sequence feature that has been extensively studied is CpG density. A large number of mammalian promoters harbor CpG islands,12,13 which typically remain constitutively unmethylated in all cell types.14,15 Recently, it has been shown that clusters of unmethylated CpG dinucleotides are recognized by CxxC-domain-containing proteins,16,17 thereby facilitating the deposition of transcription-associated marks such as H3K4me3.18, 19, 20 Additionally, there is now evidence that unmethylated CpGs surrounding transcription factor (TF) motifs may contribute to promoter activity by also increasing the probability that the cognate TFs will bind.21,22
Material and Methods
Selecting Transcripts with High-Confidence Loss-of-Function Intolerance Estimates
In total, gnomAD5 provides LoF intolerance estimates for 79,141 human protein-coding transcripts (hereafter referred to as transcripts) labeled with ENSEMBL identifiers, of which 19,172 are annotated as canonical. For each transcript, these LoF intolerance estimates consist of the point estimate of the observed/expected number of LoF variants, as well as a 90% confidence interval around it. The upper bound of this confidence interval (LOEUF) is the suggested metric of LoF intolerance.5 For any given transcript, the ability to reliably estimate LOEUF is directly related to the expected number of LoF variants; when that expected number is small, there is uncertainty around the point estimate (and thus a large LOEUF value), because it is not possible to determine whether an observed depletion of LoF variants is due to negative selection against these variants in the population or due to inadequate sample size. Therefore, for transcripts with high-confidence LOEUF values, there should be a strong positive correlation between the point estimate and LOEUF; in contrast, low-confidence LOEUF transcripts will have LOUEF values substantially larger than their point estimates.
Based on this assessment, and consistent with Karczewski et al.,5 we determined that for transcripts with ≤10 expected LoF variants, there is inadequate power for LOEUF estimation (34,232 out of 79,141 total transcripts; 5,413 out of 19,172 canonical transcripts; Figure S1). Throughout the text, we refer to the genes encoding for these transcripts as “unascertained.”
Even though in Karczewski et al.5 most of the analyses were performed using transcripts with >10 expected LoF variants, we saw that, with increasing expected number of LoF variants, there was a non-negligible increase in the probability (conditional on a given point estimate) of a transcript belonging in the highly LoF-intolerant category (LOEUF < 0.35), even for genes with expected LoF variants between 10 and 20. We thus adopted a more stringent threshold, and considered transcripts with ≥20 expected LoF variants (25,474 out of 79,141 total transcripts; 8,506 out of 19,172 canonical transcripts; Figure S1) to have high-confidence LOEUF. The genes encoding for these transcripts form the “well-ascertained” set, which, after further filtering based on promoter annotation (see the section Selecting Transcripts with High-Confidence Annotations in GENCODE v.19), we used to establish the association between promoter CpG density and LOEUF and to train predLoF-CpG.
Selecting Transcripts with High-Confidence Annotations in GENCODE v.19
gnomAD supplies LOEUF estimates for 79,141 transcripts in GENCODE v.19. However, we conducted our analyses at the gene level, based on the following reasoning: typically, transcripts from the same gene have overlap in their coding sequence, which makes it hard to disentangle their LOEUF estimates. For example, a transcript whose loss does not have severe phenotypic consequences, and therefore its promoter does not contain informative features, may still have low LOEUF merely because it overlaps with a different transcript of the same gene.
For each gene, GENCODE labels a single transcript as canonical and recognizes the difficulty of accurately annotating transcriptional start sites (TSSs).23 We manually inspected GENCODE’s choices of canonical transcripts and found some problematic cases. An illustrative example is KMT2D (Figure S2). First, even though this gene is broadly expressed across tissues in GTEx, its canonical promoter shows POLR2A (the major subunit of RNA PolII complex24,25) ChIP-seq peaks in only 4 ENCODE experiments (out of 74 total). Even though there does exist a non-canonical transcript whose promoter has POLR2A signal in 59 experiments (as would be expected for a broadly expressed gene since binding of the RNA PolII complex is the main hallmark of transcriptional initiation at protein-coding gene promoters), that non-canonical transcript has an unusually short coding sequence, which does not even encode for the catalytic SET domain. In this particular case, we reasoned that the 5′ UTR of the canonical transcript needs to be extended up until the TSS of the non-canonical transcript. Such an annotation would also be consistent with the annotation of the mouse ortholog. Importantly, if this annotation error is ignored, it is impossible to select a KMT2D transcript with accurate estimates of both LOEUF and promoter CpG density.
With this example in mind, we developed an empirical approach to only retain transcripts with high-confidence GENCODE annotations in our analysis. First, we defined promoters as 4 kb elements centered around the TSS. We then leveraged data from ENCODE26 on the genome-wide binding locations of POLR2A from 74 ChIP-seq experiments on several cell lines, originating from diverse human tissues (see POLR2A ENCODE ChIP-Seq Data section below).
As expected, we observed that genes that are broadly expressed across the 53 different tissues in GTEx (; see GTEx Expression Data section below) tend to have promoters with POLR2A ChIP-seq peaks in multiple experiments, while the opposite is true for genes expressed in a restricted number of tissues (, Figures S3A–S3C). However, as in the KMT2D example above, we also observed genes with broad expression and very low binding of POLR2A at their canonical promoter (Figure S3C) and a few genes with restricted expression but POLR2A peaks at their canonical promoter in multiple experiments (Figure S3C), raising our suspicion that these reflect inaccurate annotation of the canonical TSS.
Therefore, we required that the canonical promoter of a broadly expressed gene exhibits POLR2A peaks in multiple ENCODE experiments and that the canonical promoter of a gene with restricted expression exhibits POLR2A peaks only in a small number of ENCODE experiments. As additional layers of evidence for canonical promoters, we used the presence of CpG islands, which are known markers of promoters in mammalian genomes,12,13 as well as the concordance between the human TSS coordinate and the TSS coordinate of a mouse ortholog transcript (when the latter is mapped onto the human genome).
Specifically, we first excluded genes on the sex chromosomes, since, due to X-inactivation in females and hemizygosity in males, LoF intolerance estimates have different interpretation in these cases. This gave us 17,657 genes with at least one canonical transcript, of which 17,359 had expression measurements in GTEx. We then applied the following criteria (when none of the criteria were satisfied, we entirely discarded the gene):
Criterion 1: The gene is broadly expressed () and the canonical promoter has a POLR2A peak in more than 35 ENCODE experiments.
We found 7,250 cases satisfying this criterion and therefore kept the canonical promoter annotation.
Criterion 2: The gene is broadly expressed (), the canonical promoter has a POLR2A peak in less than 10 ENCODE experiments, and there is an alternative promoter with POLR2A peaks in more than 35 experiments.
We found 218 cases satisfying this criterion (Figure S3D) and therefore classified the alternative promoter as the canonical (all such cases are provided in Table S1). When there were more than one alternative promoter satisfying our requirement, we distinguished the following subcases:
-
(a)
If none of these alternative promoters overlapped a CpG island, we classified the promoter corresponding to the transcript with the greater number of expected LoF variants as the canonical.
-
(b)
If exactly one of these alternative promoters overlapped a CpG island, we classified that promoter as the canonical.
-
(c)
If more than one of these alternative promoters overlapped a CpG island, we classified the promoter that, among the CpG-island-overlapping promoters, had the greatest number of expected LoF variants as the canonical.
For our subsequent analyses, we used the LOEUF value of the newly annotated canonical promoter.
Criterion 3: The gene is not broadly expressed () and the canonical promoter has a POLR2A peak in fewer than 10 ENCODE experiments and overlaps a CpG island.
We found 1,862 cases satisfying this criterion and therefore kept the canonical promoter annotation.
Criterion 4: The gene is not broadly expressed (), the canonical promoter has a POLR2A peak in fewer than 10 ENCODE experiments, none of the promoters corresponding to the gene overlap a CpG island, and there is a mouse ortholog TSS in RefSeq no more than 500 bp away from the canonical human TSS.
We found 3,049 cases satisfying this criterion and therefore kept the canonical promoter annotation.
Criterion 5: The gene is not broadly expressed (), the canonical promoter has a POLR2A peak in fewer than 10 ENCODE experiments, none of the promoters corresponding to the gene overlap a CpG island, there is no mouse ortholog TSS in RefSeq, and there are no alternative transcripts with different TSS coordinates.
We found 1,411 cases satisfying this criterion and therefore kept the canonical promoter annotation.
The promoters selected from the above five criteria along with their coordinates are provided in Table S2.
Finally, regarding coding sequence annotations, errors such as the one in KMT2D described at the beginning of the section are difficult to systematically detect and correct, and our manual inspection suggested that they are also less frequent. We chose to entirely discard cases where:
-
(a)
the transcript we had selected after promoter filtering had ≤10 expected LoF variants (placing the gene into the unascertained category) and
-
(b)
there was an alternative transcript that had longer coding sequence and ≥20 more expected LoF variants compared to the one our procedure selected.
This approach removes KMT2D and 14 more potentially problematic cases such as ZNF609.
Overlapping Promoters
When defining the set of genes with high-confidence LOEUF estimates, we excluded genes whose promoters overlapped promoters of genes with fewer than 20 expected LoF variants, with an observed/expected LoF point estimate for the latter suggestive of LoF intolerance (<0.5). In cases of overlapping promoters with both genes having ≥20 expected LoF variants, we kept the promoter corresponding to the gene with the lowest LOEUF. In cases of overlapping promoters with both genes having ≤10 expected LoF variants, we kept the promoter with the highest CpG density. Finally, when defining the set of unascertained genes, we excluded genes whose promoters overlapped promoters of genes with more than 10 expected LoF variants, unless there was strong evidence that these were LoF tolerant (observed/expected LoF point estimate >0.8 and at least 20 expected LoF variants).
We recognize, however, that in cases where promoters overlap, the predictions are potentially informative not only for the gene whose promoter was ultimately used, but also for the genes with overlapping promoters. In addition, in cases of genes predicted as highly LoF intolerant, these predictions might also have been influenced by the overlapping promoter (there are only three such potential cases). With that in mind, in Tables S3, S4, and S5, we provide such information under the column “other_genes_with_overlapping_promoter.”
Promoters in Subtelomeric Regions
It is known that subtelomeric regions are rich in CpG islands, which are however different than those in the rest of the genome, in that they appear in clusters and their CpG richness is driven mainly by GC-biased gene conversion.27 We thus excluded promoters residing in subtelomeric regions (defined as 2 Mb on each of the two chromosomal ends of each chromosome) from our analyses.
A schematic of our overall approach to partitioning genes, based on this and the previous three sections, is shown in Figure S4.
Calculating the CpG Density of a Promoter
For a given promoter, we defined its CpG density as the observed-to-expected (o/e) CpG ratio of the 4 kb interval centered around the TSS. To calculate the o/e CpG ratio, we used the definition in Gardiner-Garden and Frommer,28 applied to the entire 4 kb sequence (that is, without using sliding windows). Specifically, we used the formula
with p(CG) being the proportion of CpG dinucleotides observed in the sequence (and similarly for p(C), p(G)). The sequence of each promoter was obtained using the BSgenome.Hsapiens.UCSC.hg19 R package.
Given that CpG density is a ratio, it is theoretically possible that it becomes an unreliable metric when the expected number of CpGs is small. We therefore asked whether the association with LOEUF persists if, instead of the CpG density, we use the observed count of CpGs in a promoter. We found this to be true, with the results being almost the same quantitatively (Figures 1A, 1B, and S5).
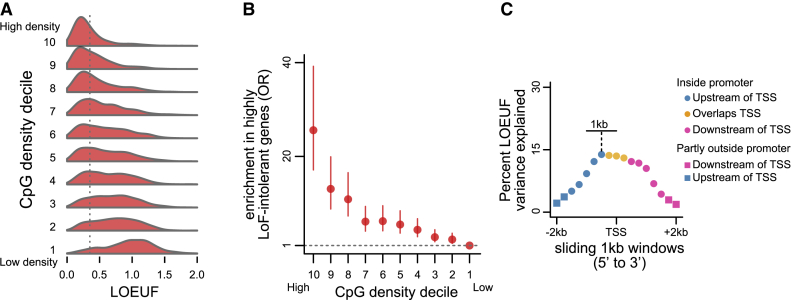
Figure 1.
The Relationship between Promoter CpG Density (o/e CpG Ratio) and Downstream Gene Loss-of-Function Intolerance
(A) The distribution of genic LOEUF (as provided by gnomAD) in each decile of promoter CpG density. The vertical line corresponds to the cutoff for highly LoF-intolerant genes (LOEUF < 0.35).
(B) Odds ratios and the corresponding 95% confidence intervals, quantifying the enrichment for highly LoF-intolerant genes (LOEUF < 0.35) that is exhibited by the set of genes in each decile of promoter CpG density. For each of the other deciles, the enrichment is computed against the 10th decile. The horizontal line corresponds to zero enrichment.
In both (A) and (B), CpG density deciles are labeled from 1-10 with 1 being the most CpG-poor and 10 the most CpG-rich decile.
(C) The percentage of LOEUF variance (adjusted r2) explained by CpG density, computed in 1 kb windows. Each point corresponds to a window. We start with a window centered at 2 kb upstream of the TSS, and slide it in 250 bp steps in the 5′-to-3′ direction, until the final window is centered at 2 kb downstream. Red and pink points correspond to intervals entirely upstream or downstream, respectively, of the TSS, with squares indicating intervals extending beyond 2 kb. Orange points correspond to intervals containing both upstream and downstream sequence.
The Impact of Promoter Definition
There is currently no single accepted definition of a promoter in terms of the size of the interval around the TSS. Our main motivation behind the choice of 4 kb was that CpG density has been mechanistically linked to the presence of histone marks such as H3K4me3,18,20 which are typically detected in that interval. However, since using 4 kb around the TSS often leads to the inclusion of some exonic sequence, we sought to compare the contribution of promoter CpGs to that of CpGs in the N-terminal part of the encoded protein. We used 1 kb windows, starting with a window centered at 2 upstream of the TSS, and slid these windows (in 250 bp steps in the 5′-to-3′ direction) until the final window was centered at 2 downstream of the TSS. In each window, we computed the CpG density and asked how much LOEUF variance (adjusted r2) it explains. This clearly revealed that the association between CpG density and LOEUF is driven by the CpGs proximal to the TSS, with the maximal explained variance attained with a window centered at 500 bp upstream of the TSS (Figure 1C). As these sliding windows move away from the TSS and into the coding sequence, the explained variance drops to almost 0 (Figure 1C). This result can be interpreted in two ways. One is that the CpGs proximal to the TSS (both upstream and downstream) are driving the association with LOEUF, because they are part of the promoter region. This is the interpretation we favor and is consistent with the aforementioned experiments which suggest causal links between high CpG density and histone mark recruitment, as well as TF binding. The alternative interpretation is that there is an independent contribution of the CpGs upstream and those downstream of the TSS, with the downstream ones having a different biological role related to their presence within the exonic sequence. We find this interpretation less plausible, especially in light of the fact that exonic sequence has no contribution once we start moving away from the TSS.
ENCODE ChIP-Seq Data
We used the rtracklayer R package to download the “wgEncodeRegTfbsClusteredV3” table from the “Txn Factor ChIP” track, part of the “Regulation” group as provided by the UCSC Table Browser for the hg19 human assembly. We then restricted to peak clusters corresponding to our factor of interest. For POLR2A, for example, this gave us a set of genomic intervals, each of which has been derived from uniform processing of 74 POLR2A ChIP experiments on 32 distinct cell lines (some cell lines were represented by more than one experiments). Each genomic interval was associated with a single number, which ranged from 0 to 74 and indicated the number of ChIP experiments where a peak was detected at that interval. The EZH2 and CTCF data were downloaded in an identical manner.
The EZH2 ChIP experiments were performed on the following cell lines: H1-hESC (embryonic stem cells), HeLa-S3 (cervical carcinoma), HMEC (mammary epithelial cells), HSMM (skeletal muscle myoblasts), NH-A (astrocytes), NHDF-Ad (dermal fibroblasts), NHEK (epidermal keratinocytes), NHLF (lung fibroblasts), Dnd41 (T cell leukemia with Notch mutation), GM12878 (lymphoblastoid), HepG2 (hepatocellular carcinoma), HSMMtube (skeletal muscle myotubes differentiated from the HSMM cell line), HUVEC (umbilical vein endothelial cells), and K562 (lymphoblasts). The cell lines on which the POLR2A and CTCF ChIP experiments were performed are too numerous to list here and can be found on the UCSC genome browser.
GTEx Expression Data
We used the GTEx portal to download a matrix with the gene-level TPM expression values from the v7 release, derived from RNA-seq expression measurements from 714 individuals, spanning 53 tissues.29
As the metric of tissue specificity for a given gene, we used τ, which has been shown to be the most robust such measure when benchmarked against alternatives.30 To calculate τ, we first computed the gene’s median expression across individuals, within each tissue. Since it has been shown that the transcriptomic profiles of the different brain regions are very similar, with the exception of the two cerebellar tissues,31 which are similar to one another, we aggregated the median expression of each gene in the different brain regions into two “meta-values.” One meta-value corresponded to the median of its median expression in the two cerebellar tissues, and the other to the median of its median expression in the other brain regions. We then formed a matrix where rows corresponded to genes and columns to tissues, with one column for the across-brain-regions meta-value and another for the across-cerebellar-tissues meta-value; the entries in the matrix were log2(TPM+1) median expression values. Finally, for each gene, τ was calculated as described in Kryuchkova-Mostacci and Robinson-Rechavi.30
For our analyses of the association between promoter CpG density and expression level, we used the median (across individuals) expression (log2(TPM+1)), computed for the tissue where the gene had the maximum median expression.
TSS Coordinates of Mouse Orthologs
We used the biomaRt R package to obtain a list of mouse-human homolog pairs, using the human Ensembl gene IDs as the input. For this query, we set the “mmusculus_homolog_orthology_confidence” parameter equal to 1 (indicating high-confidence homolog pairs). Then, for each of the mouse homolog Ensembl IDs, we retrieved the RefSeq mRNA IDs, again with biomaRt. We discarded cases where the same RefSeq mRNA ID was associated with more than one Ensembl gene ID. We then used the rtracklayer R package to download the “xenoRefGene” UCSC table, from the “Other RefSeq” track, containing the TSS coordinates for each of the mouse RefSeq transcripts.
Genes with Developmentally Specific Expression
We obtained mouse genes expressed at specific time points during embryogenesis (see Web Resources). Specifically, these genes were identified as differentially expressed across 5 time points during mouse embryogenesis (E9.5 to E13.5) using single-cell RNA-seq.32 For each of the 10 main developmental trajectories provided, we kept genes with a q-value < 0.01 and absolute fold change ≥2. We then pooled the resulting mouse ENSEMBL gene IDs from all 10 trajectories and obtained their human homologs using the biomaRt R package. We restricted the human-mouse homolog pairs to those where the “mmusculus_homolog_orthology_confidence” was equal to 1. Intersecting these genes with our list of 4,743 well-ascertained genes and reliable promoter annotation yielded 559 genes, which we used for our analysis. Genes encoding for human key developmental regulators (defined as such on the basis of their regulation by arrays of highly conserved non-coding elements) were obtained from the supplemental material of Akalin et al.33 (where they were labeled as “target genes”).
Across-Species Conservation Quantification
For each nucleotide, we quantified conservation across 100 vertebrate species using the PhastCons score,34 obtained with the phastCons100way.UCSC.hg19 R package. The PhastCons score ranges from 0 to 1 and represents the probability that a given nucleotide is conserved. As the promoter PhastCons score for a given gene, we computed the average PhastCons of all nucleotides in the 4 kb region centered around the TSS. As the exonic PhastCons for a given gene, we pooled all nucleotides belonging to the coding sequence of the gene (that is, excluding the 5′ and 3′ UTRs), and computed their average PhastCons.
Previously Published LoF Intolerance Predictions
The updated version of the score of Huang et al.35 was downloaded from the DECIPHER database (see Web Resources). The scores of Steinberg et al.36 and Han et al.10 were downloaded from the supplemental materials of the respective publications. In our comparison we did not include HIPred,37 since it provides binary haploinsufficiency predictions for only a small number of genes.
Structural Variation Data
We used the gnomAD browser to download a bed file containing the coordinates and characteristics of structural variants in gnomAD v.2 (see Web Resources). We then restricted to deletions that passed quality control (“FILTER” column value equal to “PASS”). Subsequently, we excluded deletions that overlapped more than one of our high-confidence promoters (n = 499), in order to avoid ambiguous links between deletions and genes.
Gene Catalogs
The following gene catalogs were used for Figure 5D.
-
(a)
404 heterozygous lethal genes in mouse (see Web Resources, and see the supplemental material of Karczewski et al.5 for details on obtaining this set). We mapped these genes to their human homolog ensembl IDs with the biomaRt R package using the “mgi_symbol” filter, keeping only pairs with the “mmusculus_homolog_orthology_confidence” parameter equal to 1. This yielded a total of 390 human homologs.
-
(b)
1,254 high-confidence transcription factor genes from Barrera et al.38
-
(c)
371 olfactory receptor genes (see Web Resources).
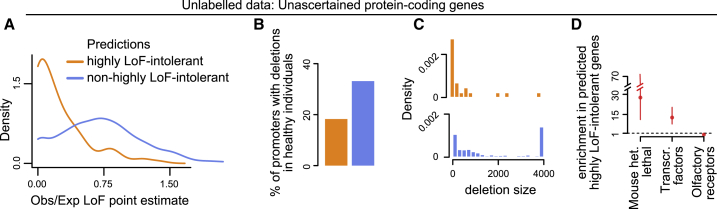
Figure 5.
Using predLoF-CpG to Classify Currently Unascertained Genes as Highly Loss-of-Function Intolerant or Not
(A) The distribution of point estimates of the observed/expected proportions of LoF variants. Genes are stratified according to their classification as highly LoF intolerant or not.
(B) The proportion of promoters which harbor deletions in a sample of 14,891 healthy individuals. Promoters are stratified according to downstream gene classification as highly LoF intolerant or not.
(C) The distribution of the size of deletions harbored by promoters in a sample of 14,891 healthy individuals. Promoters are stratified according to downstream gene classification as highly LoF intolerant or not.
(D) Odds ratios and the corresponding 95% confidence intervals quantifying the enrichment for genes in each of the x axis groups that is exhibited by genes predicted as highly LoF intolerant by predLoF-CpG. The enrichment is computed against genes predicted as non-highly LoF intolerant. The horizontal line at 1 corresponds to zero enrichment.
Enrichment Quantification
All enrichment point estimates in the text correspond to odds ratios, and the associated p values were calculated using Fisher’s exact test (two-sided) with the “fisher.test” function in R.
Results
Promoter CpG Density Is Strongly and Quantitatively Associated with Downstream Gene LoF Intolerance
We discovered a strong relationship between the observed-to-expected CpG ratio (hereafter referred to as CpG density) of a promoter and LoF intolerance of the downstream gene (Figures 1A and 1B); high CpG density is associated with high LoF intolerance. To establish this, we used the LOEUF metric provided by gnomAD, an updated and more accurate measure of genic LOF intolerance compared to pLI.5 In contrast to pLI, which is essentially a binary metric with limited resolution,4 LOEUF places human genes on a 0-to-2 continuous scale, with lower values indicating higher LoF intolerance. Following previous work,39 we classified genes with LOEUF < 0.35 as highly LoF intolerant.
In Karczewski et al.,5 genes with ≤10 expected LoF variants were found to be insufficiently powered for LOEUF estimation in gnomAD. We refer to these genes as unascertained. Based on additional assessment (Figure S1; Material and Methods), we here adopted an even more stringent threshold and considered 8,506 genes with ≥20 expected LoF variants, which we refer to as “well-ascertained.” We refer to genes in the intermediate category (expected LoF variants between 10 and 20) as “ascertained.” We then further restricted our analysis to those genes for which we could reliably determine the canonical promoter (4,743 well ascertained, 2,772 ascertained, and 2,430 unascertained genes; Material and Methods; Figure S4 contains a schematic of our approach to partitioning genes).
When ranked according to the CpG density of their promoter, genes in the top 10% have a 67.2% probability of being highly LoF intolerant. This in contrast to 7.4% for genes in the bottom 10%, yielding a 25.6-fold enrichment (p < 2.2 × 10−16; Figure 1B). We note that there is a continuous gradient of enrichment across CpG density deciles (Figure 1B). When splitting genes into just two groups, consisting of those with CpG island-overlapping promoters and those without, we found that the enrichment for highly LoF-intolerant genes in the CpG-island-overlapping group is markedly weaker (odds ratio = 3.71, p < 2.2 × 10−16), showing that this dichotomy masks the more continuous nature of CpG density. Finally, regression modeling revealed that CpG density alone can explain 19.3% of the variation in LOEUF (p < 2.2 × 10−16; β = −1.02) (Figure S6; Material and Methods) and that its effect on LOEUF is unchanged when accounting for coding sequence length (p < 2.2 × 10−16; β = −1.00).
We emphasize that our result remains pronounced even when we omit the filtering for high-confidence promoters and merely consider all canonical promoters with ≥20 expected LoF variants (p < 2.2 × 10−16; Figure S7). However, the association becomes weaker (14.6-fold enrichment of highly LoF-intolerant genes in the top CpG density decile), underscoring the importance of accurate promoter annotation. We also found that the relationship between CpG density and LOEUF is mostly driven by the CpGs in the TSS-proximal region (Figure 1C; Material and Methods) and that the exact definition of the promoter (in terms of the size of the interval around the TSS) has only a small impact on the strength of this relationship (Figure S8).
The Association between CpG Density and LoF Intolerance Is Not Mediated through Tissue/Developmental Specificity or Expression Level
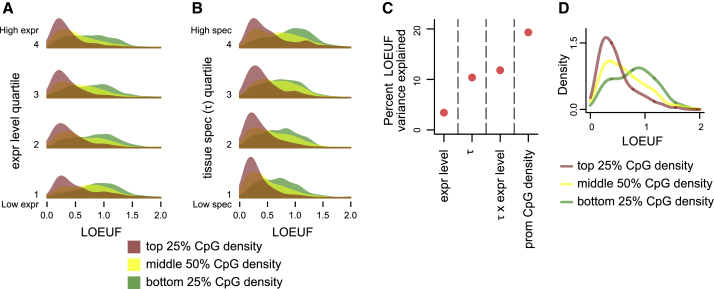
It is established that promoter CpG islands are associated with genes that exhibit broad, housekeeping-like expression,40,41 genes whose expression is developmentally regulated,41 and genes expressed at high levels.22,42 However, we found that these associations are not sufficient to explain the relationship with LoF intolerance. First, after stratifying genes according to either expression level or tissue specificity (using RNA-seq data from the GTEx consortium; Material and Methods), we saw a clear relationship between promoter CpG density and LOEUF within each stratum (Figures 2A and 2B). Second, the effect of CpG density on LOEUF is almost equally strong when adjusting for either expression level or tissue specificity (regression and , respectively, p < 2.2 × 10−16 for both regression models; Figure S9). Third, even the combination of the two expression properties explains less LOEUF variance than CpG density by itself (Figure 2C). Finally, when restricting to 559 genes whose mouse homologs are differentially expressed at specific time points during embryogenesis32 (Material and Methods), the relationship between CpG density and LOEUF is still pronounced (Figure 2D); the same is true when focusing on 46 key human developmental regulator genes33 (Figure S10, Material and Methods), even though these genes overall have very high promoter CpG density (25th percentile = 0.58).
Figure 2.
The Relationship between Promoter CpG Density (o/e CpG Ratio) and Loss-of-Function Intolerance Conditional on Downstream Gene Expression Level and Tissue/Developmental Specificity (τ)
(A) The distribution of LOEUF, stratified by promoter CpG density, in each quartile of downstream gene expression level, computed using the GTEx dataset (Material and Methods).
(B) The distribution of LOEUF, stratified by promoter CpG density, in each quartile of downstream tissue specificity. For each gene, tissue specificity is quantified by τ, and is computed using the GTEX dataset (Material and Methods).
For both (A) and (B) quartiles are labeled from 1-4, with 1 being the quartile with the lowest and 4 the quartile with the highest expression/tissue specificity, respectively.
(C) The percentage of LOEUF variance (adjusted r2) that is explained by downstream gene expression level, τ, the interaction between the two, and promoter CpG density.
(D) The distribution of LOEUF, stratified by promoter CpG density, for 559 genes whose mouse homologs are differentially expressed at specific time points during embryogenesis (Material and Methods). The stratification was done based on the CpG density quartiles calculated for all 4,743 genes, as in (A) and (B).
Regulatory Factor Binding at Promoters Can Provide Information about LoF Intolerance which Adds to CpG Density
We next turned our attention to the fraction of LOEUF variation (80.7%) that remains unexplained by CpG density. We hypothesized that part of it might be explained by preferential binding of specific regulatory factors at LoF-intolerant gene promoters. Since a comprehensive assessment of this is currently out of reach (due to the lack of extensive genome-wide binding data for most regulatory factors), we focused on two such factors, EZH2 and CTCF, as a proof-of-principle. EZH2 is a relatively well-characterized histone methyltransferase that specifically localizes to CpG islands of non-transcribed genes43,44 (Figures 3A and S11); CTCF is a transcription factor with diverse roles in gene activation, repression, and 3D-contact regulation.45,46
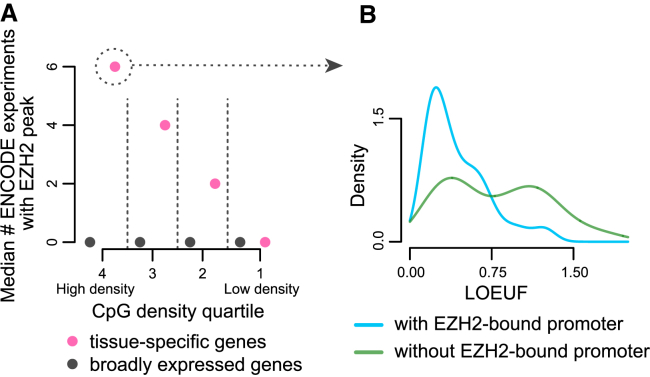
Figure 3.
The Loss-of-Function Intolerance of Tissue-Specific Genes Conditional on High Promoter CpG-Density (o/e CpG Ratio) and Promoter EZH2 Binding
(A) The median number of ENCODE ChIP-seq experiments (out of 14 total) where an EZH2 peak is detected, shown separately for tissue-specific () and broadly expressed () genes, within each quartile of promoter CpG density. The quartiles are labeled from 1-4, with 1 being the most CpG-poor and 4 the most CpG-rich.
(B) The LOEUF distributions of tissue-specific genes with high-CpG-density (top 25%) promoters, stratified according to whether their promoters show EZH2 peaks in at least 2 ENCODE experiments, or in less than 2 experiments.
We discovered that tissue-specific genes with CpG-dense and EZH2-bound promoters (EZH2 binding in at least two ENCODE experiments) have lower LOEUF compared to their EZH2-unbound counterparts (Figure 3B; regression , p < 5.21 × 10−8, for the interaction between CpG density and EZH2 binding, conditional on tissue specificity ). In this subset of promoters, the interaction of EZH2 binding with CpG density explains an additional 27.1% of LOEUF variance on top of what CpG density explains (2.1%). In contrast to EZH2, however, we saw that CTCF binding has no effect on LOEUF on top of CpG density (Figure S12). Together, these results illustrate that regulatory factor binding can indeed modify the relationship between CpG density and LoF intolerance, but this is not universally true even for factors with established importance.
Promoter CpG Density with Promoter and Exonic Across-Species Conservation Can Collectively Predict LoF Intolerance with High Accuracy
We then sought to develop a predictive model for LoF intolerance, with the goal of providing high-confidence predictions for the unascertained genes. Specifically, we aimed to classify genes as highly LoF intolerant (LOEUF < 0.35) or not.
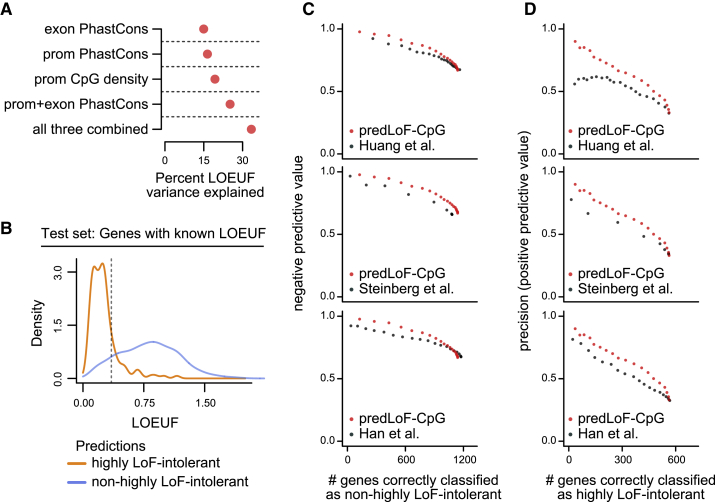
To build our model, we first separately computed the promoter and exonic across-species conservation for each gene (using the PhastCons score; Material and Methods) and asked whether they provide information about LOEUF complementary to CpG density. We found this to be true (Figure S13); notably, CpG density explains at least as much LOEUF variance as exonic or promoter conservation (Figure 4A). When all three metrics are combined, 33.4% of the total LOEUF variation is explained (Figure 4A). We note that while EZH2 explains a substantial amount of LOEUF variance when considering tissue-specific genes with high CpG-density promoters, these are a small subset. Hence, inclusion of this feature only minimally increases the overall explained variance (0.4% increase). We therefore settled on training a logistic regression model with CpG density, and promoter/exonic conservation as three linear predictors. As our training set we used 3,000 genes, randomly selected from the 4,743 well-ascertained genes.
Figure 4.
Training and Assessing predLoF-CpG: A Predictor of Loss-of-Function Intolerance Based on CpG Density
(A) The percentage of LOEUF variance (adjusted r2) that is explained by CpG density (o/e CpG ratio), exonic or promoter conservation, and their combinations.
(B) The out-of-sample performance of predLoF-CpG. Shown are the LOEUF distributions of 1,743 genes belonging to the holdout test set (which consists of well-ascertained genes with respect to LOEUF), stratified according to their classification as highly LoF-intolerant or not. The dashed vertical line corresponds to the cutoff for highly LoF-intolerant genes (LOEUF < 0.35).
(C and D) The negative predictive value (y axis in C) and precision (y axis in D) plotted against the number of correctly classified genes (x axis), for different predictors of loss-of-function intolerance. Predictors are from Han et al.,10 Huang et al.,35 and Steinberg et al.36 Each point corresponds to a threshold. The thresholds span the [0,1] interval, with a step size of 0.05. We note that because we are using two classification thresholds, a ROC curve would not be an appropriate evaluation metric here.
Our predictor, which we called predLoF-CpG (predictor of LoF intolerance based on CpG density) showed strong out-of-sample performance on the test set of the remaining 1,743 genes. The precision (positive predictive value) was 82.6% at the 0.75 prediction probability cutoff, and the negative predictive value was 88.4% at the 0.25 cutoff (Figure 4B); 144 genes were predicted to be highly LoF intolerant, 753 were predicted as non-highly LoF intolerant, and 806 (47.3%) were left unclassified. We chose to use two thresholds instead of one, at the expense of leaving a fraction of genes unclassified, since this endows our predictor with precision and negative predictive value high enough to be useful in the clinical setting. We note that our predictive accuracy is comparable to that of widely adopted tools for predicting damaging missense variants.47,48 Further examining our out-of-sample classifications, we found that (1) the genes falsely predicted as highly LoF intolerant had a median observed-to-expected LoF point estimate of 0.29, indicating that at least half of them are very LoF intolerant even though their LOEUF values do not exceed the 0.35 cutoff, and (2) 25% of the genes correctly predicted as non-highly LoF intolerant had LOEUF greater than 1.1, and a lower confidence interval bound for their observed-to-expected LoF point estimates greater than 0.56, suggesting that they are likely to be relatively tolerant of bi-allelic inactivation as well (Figure 4B).
Regardless of the choices for the two classification thresholds, predLoF-CpG outperforms all of the previously published predictors of LoF intolerance (Figure 4C). Specifically, all models have comparable and high negative predictive value, with ours being slightly superior (Figure 4C). However, within a range of thresholds that yield high precision, as would be required for use in clinical decision making, predLoF-CpG provides clear gain versus the rest (Figure 4D, upper left area of the plots). As an additional evaluation, we found that predLoF-CpG is capable of explaining a greater proportion of out-of-sample LOEUF variance compared to the other three (Figure S14).
Finally, we mention GeVIR, a recently developed metric (primarily for intolerance to missense, but also useful for LoF variation49) which identifies regions depleted of protein-altering variation50 and weights these regions by conservation within each gene. As expected given its dependency on observed variation, GeVIR exhibits substantial correlation with the expected number of LoF variants (Spearman correlation = 0.42 versus 0.26 for predLoF-CpG). This limits its applicability to unascertained genes, even though the weighting step slightly alleviates this issue compared to LOEUF (Spearman correlation = 0.49).
32.5% of Currently Unascertained Genes in gnomAD Receive High-Confidence Predictions by predLoF-CpG
We applied predLoF-CpG to genes unascertained in gnomAD. After filtering for these with high-confidence promoter annotation, we retained 2,430 (out of 5,413). Of these, 104 were classified as highly LoF intolerant, 1,656 as non-highly LoF intolerant, and 670 were left unclassified (Tables S3 and S4). We first examined the ratio of observed-to-expected LoF variants in these genes. Even though these point estimates are uncertain, there is a clear difference in the distribution of the point estimates between genes we classify as highly LoF intolerant (median = 0.14) and those as not (median = 0.70), with the difference being in the expected direction (Figure 5A; Wilcoxon test, p < 2.2 × 10−16).
Next, to provide orthogonal support for our predictions, we leveraged a set of 175,716 deletions detected in 14,891 healthy individuals using whole-genome sequencing (Material and Methods).51 We reasoned that LoF-intolerant gene promoters should be depleted of such deletions; when they do harbor deletions, these should be small. By considering only promoters, we ensured that our assessment is not dependent on gene length, which confounds LOEUF estimation. Using the 4,743 genes with high-confidence LOEUF (from the training and test sets), we first observed that low LOEUF is indeed associated with the presence of both fewer (p <=2.39 × 10−15) and smaller (p < 2.2 × 10−16) promoter deletions (Figures S15A and S15B), showing that this is a legitimate assessment strategy. Turning to our predictions, we found the same: genes predicted to be highly LoF intolerant are less likely to contain deletions in their promoters compared to genes classified as non-highly LoF intolerant (Figure 5B; probability of overlapping at least one deletion = 0.18 versus 0.33, permutation one-sided p < 4 × 10−4 after 10,000 permutations); when such deletions are observed, they tend to be much smaller (Figure 4C; median size = 129 versus 1,092; Wilcoxon test, p < 4.49 × 10−5).
Finally, we found that our predictions are in strong agreement with what would be expected based on known mouse phenotypes and membership in specific gene classes (Figure 5D). First, the predicted highly LoF-intolerant genes show a 27.6-fold enrichment for genes heterozygous lethal in mouse (p < 1.03 × 10−12), when compared against those predicted as non-highly LoF intolerant. Second, they exhibit a 12.7-fold enrichment for transcription factors (p < 2.2 × 10−16), consistent with the known dosage sensitivity of these genes.52, 53, 54 Third, they show a total depletion (odds ratio = 0) of olfactory receptor genes (p < 2.5 × 10−5).
predLoF-CpG Classifies 101 Genes with Expected LoF Variants between 10 and 20 as Highly LoF Intolerant
In our analyses so far, we have ignored the set of ascertained genes (3,440 genes with expected LoF variants between 10 and 20). Even though in Karczewski et al.5 these were treated as well powered, our assessment suggests that lack of power can affect whether they are categorized as highly LoF intolerant or not (Figure S1, Material and Methods). After filtering for reliable promoter annotation, we applied predLoF-CpG to 2,772 genes and obtained high-confidence classifications for 1,675. For the great majority (93.9%), we agree with the classification obtained by purely considering whether their LOEUF is <0.35. However, we observed 101 genes that were classified as highly LoF intolerant by predLoF-CpG but had LOEUF ≥ 0.35, a number not explained by the false positive rate of our predictor (Table S5). 75% of these genes have an observed/expected LoF point estimate less than 0.31, suggesting that they are indeed highly LoF intolerant, but do not exceed the required LOEUF threshold because of inadequate power. Therefore, when interpreting LoF variants in these genes, we suggest that both LOEUF as well as predLoF-CpG are taken into account.
Discussion
Our study reveals that (1) there exists a strong, widespread coupling between promoter CpG density and downstream gene LoF intolerance in the human genome and (2) this coupling can be exploited to predict LoF intolerance for almost 1,800 genes that are otherwise largely intractable with current sample sizes. Our predictions for these genes (which we make available in Table S3) can inform research into novel disease candidates and now become incorporated in the clinical genetics laboratory setting. Similarly to existing tools for missense variants,47,48 they can provide corroborating evidence during the evaluation of the pathogenicity of LoF variants harbored by individuals with disease phenotypes, as recommended by the American College of Medical Genetics and Genomics.6
In terms of understanding the regulatory architecture of the genome, our findings extend decades of work12,13 to show that high CpG density is not just a prevalent feature of many promoters but is preferentially marking the promoters of the most selectively constrained genes. We believe this casts doubt on the prevailing view that CpG islands are not under selection,27 as constrained genes are typically paired with constrained promoters.55 However, we note that our current results are correlative in nature.
If promoter CpG density is indeed under selection, its presence at LoF-intolerant gene promoters has to be advantageous, which raises the question of the underlying biological mechanism. Our findings suggest that this mechanism is not related to the high and constitutive expression that LoF-intolerant genes typically exhibit. An intriguing possibility has been recently raised by single-cell expression measurements showing that promoter CpG islands are associated with reduced expression variability.56 We hypothesize that this decreased variability is beneficial for many processes where LoF-intolerant genes are known to play central roles, such as neurodevelopment.57
Our work represents an attempt at deciphering the link between regulatory element characteristics and the LoF intolerance of the genes they control. The fact that taking promoter EZH2 binding into account improves our ability to recognize LoF-intolerant genes on top of CpG density implies that this mapping can be learned with even greater accuracy by incorporating information about other regulatory factors as well. However, a current barrier to employing this approach, and understanding its limits, is the relative paucity of genome-wide binding data across the full repertoire of transcription factors: the human genome encodes approximately 1,500 transcription factors38,58,59 and at least 295 epigenetic regulators.54 In contrast to these numbers, currently ENCODE has profiled only ~330 regulatory factors in K562 cells, the most extensively characterized cell line.
It is also natural to consider moving beyond promoters to other regulatory elements. An initial step in this direction has recently been taken in Wang and Goldstein,11 motivated by work in Drosophila showing that developmentally important genes can have multiple redundant enhancers.60,61 While this “enhancer domain score” was not designed to capture LoF intolerance and has poor association with LOEUF (adjusted r2 = 0.03), it has been shown to have some predictive capacity for human disease genes, especially those with a developmental basis.
In summary, our study shows the existence of a strong and widespread association between promoter CpG density and genic LoF intolerance and leverages this relationship to predict LoF intolerance for unascertained genes.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM121459. L.B. was partly supported by the Maryland Genetics, Epidemiology, and Medicine (MD-GEM) training program, funded by the Burroughs Wellcome Fund. H.T.B. received support from the Louma G. Foundation.
Published: August 14, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.07.014.
Contributor Information
Hans T. Bjornsson, Email: hbjorns1@jhmi.edu.
Kasper D. Hansen, Email: khansen@jhsph.edu.
Data and Code Availability
Code described in this paper can be found at https://github.com/hansenlab/lof_prediction_paper_repro.
Web Resources
Mouse developmental scRNA-seq data, https://oncoscape.v3.sttrcancer.org/atlas.gs.washington.edu.mouse.rna/downloads.
Mouse lethal genes, https://github.com/macarthur-lab/gnomad_lof/blob/master/R/ko_gene_lists/list_mouse_het_lethal_genes.tsv.
Olfactory genes, https://github.com/macarthur-lab/gene_lists/blob/master/lists/olfactory_receptors.tsv.
DECIPHER (accessed November 2019), https://decipher.sanger.ac.uk/files/downloads/HI_Predictions_Version3.bed.gz.
gnomAD structural variants, https://storage.googleapis.com/gnomad-public/papers/2019-sv/gnomad_v2.1_sv.sites.bed.gz.tbi.
Supplemental Data
The table contains the promoter coordinates and transcript ENSEMBL ids of both the canonical, as well as the alternative transcript that was selected. All coordinates refer to hg19.
Prediction probabilities are provided in the ”prediction probability of high LoF intolerance by predLoF-CpG” column. Probabilities > 0.75 correspond to genes predicted as highly LoF-intolerant, and probabilities < 0.25 to genes predicted as non-highly LoF-intolerant. ENSEMBL gene/transcript ids and coordinates of the promoters used for prediction are also provided; all coordinates refer to hg19.
These prediction probabilities are provided in the ”prediction probability of high LoF intolerance by predLoFCpG” column. We provide this table for completeness, but do not recommend using these probabilities for clinical decision making.
References
- 1.Falconer D.S., Mackay T.F.C. Pearson; 1996. Introduction to Quantitative Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuller Z.L., Berg J.J., Mostafavi H., Sella G., Przeworski M. Measuring intolerance to mutation in human genetics. Nat. Genet. 2019;51:772–776. doi: 10.1038/s41588-019-0383-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abou Tayoun A.N., Pesaran T., DiStefano M.T., Oza A., Rehm H.L., Biesecker L.G., Harrison S.M., ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI) Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat. 2018;39:1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou J., Valiant G., Valiant P., Karczewski K., Chan S.O., Samocha K., Lek M., Sunyaev S., Daly M., MacArthur D.G. Quantifying unobserved protein-coding variants in human populations provides a roadmap for large-scale sequencing projects. Nat. Commun. 2016;7:13293. doi: 10.1038/ncomms13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 9.Lindeboom R.G.H., Vermeulen M., Lehner B., Supek F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat. Genet. 2019;51:1645–1651. doi: 10.1038/s41588-019-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han X., Chen S., Flynn E., Wu S., Wintner D., Shen Y. Distinct epigenomic patterns are associated with haploinsufficiency and predict risk genes of developmental disorders. Nat. Commun. 2018;9:2138. doi: 10.1038/s41467-018-04552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Goldstein D.B. Enhancer domains predict gene pathogenicity and inform gene discovery in complex disease. Am. J. Hum. Genet. 2020;106:215–233. doi: 10.1016/j.ajhg.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird A.P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- 13.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Straussman R., Nejman D., Roberts D., Steinfeld I., Blum B., Benvenisty N., Simon I., Yakhini Z., Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 16.Lee J.H., Voo K.S., Skalnik D.G. Identification and characterization of the DNA binding domain of CpG-binding protein. J. Biol. Chem. 2001;276:44669–44676. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- 17.Long H.K., Blackledge N.P., Klose R.J. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem. Soc. Trans. 2013;41:727–740. doi: 10.1042/BST20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson J.P., Skene P.J., Selfridge J., Clouaire T., Guy J., Webb S., Kerr A.R.W., Deaton A., Andrews R., James K.D. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clouaire T., Webb S., Skene P., Illingworth R., Kerr A., Andrews R., Lee J.-H., Skalnik D., Bird A. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wachter E., Quante T., Merusi C., Arczewska A., Stewart F., Webb S., Bird A. Synthetic CpG islands reveal DNA sequence determinants of chromatin structure. eLife. 2014;3:e03397. doi: 10.7554/eLife.03397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White M.A., Myers C.A., Corbo J.C., Cohen B.A. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc. Natl. Acad. Sci. USA. 2013;110:11952–11957. doi: 10.1073/pnas.1307449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartl D., Krebs A.R., Grand R.S., Baubec T., Isbel L., Wirbelauer C., Burger L., Schübeler D. CG dinucleotides enhance promoter activity independent of DNA methylation. Genome Res. 2019;29:554–563. doi: 10.1101/gr.241653.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wintzerith M., Acker J., Vicaire S., Vigneron M., Kedinger C. Complete sequence of the human RNA polymerase II largest subunit. Nucleic Acids Res. 1992;20:910. doi: 10.1093/nar/20.4.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mita K., Tsuji H., Morimyo M., Takahashi E., Nenoi M., Ichimura S., Yamauchi M., Hongo E., Hayashi A. The human gene encoding the largest subunit of RNA polymerase II. Gene. 1995;159:285–286. doi: 10.1016/0378-1119(95)00081-g. [DOI] [PubMed] [Google Scholar]
- 26.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen N.M., Kenigsberg E., Tanay A. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell. 2011;145:773–786. doi: 10.1016/j.cell.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 28.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 29.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 30.Kryuchkova-Mostacci N., Robinson-Rechavi M. A benchmark of gene expression tissue-specificity metrics. Brief. Bioinform. 2017;18:205–214. doi: 10.1093/bib/bbw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akalin A., Fredman D., Arner E., Dong X., Bryne J.C., Suzuki H., Daub C.O., Hayashizaki Y., Lenhard B. Transcriptional features of genomic regulatory blocks. Genome Biol. 2009;10:R38. doi: 10.1186/gb-2009-10-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinberg J., Honti F., Meader S., Webber C. Haploinsufficiency predictions without study bias. Nucleic Acids Res. 2015;43:e101. doi: 10.1093/nar/gkv474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shihab H.A., Rogers M.F., Campbell C., Gaunt T.R. HIPred: an integrative approach to predicting haploinsufficient genes. Bioinformatics. 2017;33:1751–1757. doi: 10.1093/bioinformatics/btx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrera L.A., Vedenko A., Kurland J.V., Rogers J.M., Gisselbrecht S.S., Rossin E.J., Woodard J., Mariani L., Kock K.H., Inukai S. Survey of variation in human transcription factors reveals prevalent DNA binding changes. Science. 2016;351:1450–1454. doi: 10.1126/science.aad2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cummings B.B., Karczewski K.J., Kosmicki J.A., Seaby E.G., Watts N.A., Singer-Berk M., Mudge J.M., Karjalainen J., Satterstrom F.K., O’Donnell-Luria A.H., Genome Aggregation Database Production Team. Genome Aggregation Database Consortium Transcript expression-aware annotation improves rare variant interpretation. Nature. 2020;581:452–458. doi: 10.1038/s41586-020-2329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saxonov S., Berg P., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenhard B., Sandelin A., Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 42.Agarwal V., Shendure J. Predicting mRNA abundance directly from genomic sequence using deep convolutional neural networks. Cell Rep. 2020;31:107663. doi: 10.1016/j.celrep.2020.107663. [DOI] [PubMed] [Google Scholar]
- 43.Riising E.M., Comet I., Leblanc B., Wu X., Johansen J.V., Helin K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Berrozpe G., Bryant G.O., Warpinski K., Spagna D., Narayan S., Shah S., Ptashne M. Polycomb responds to low levels of transcription. Cell Rep. 2017;20:785–793. doi: 10.1016/j.celrep.2017.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filippova G.N. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 46.Ong C.-T., Corces V.G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452-W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;Chapter 7:20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abramovs N., Brass A., Tassabehji M. GeVIR is a continuous gene-level metric that uses variant distribution patterns to prioritize disease candidate genes. Nat. Genet. 2020;52:35–39. doi: 10.1038/s41588-019-0560-2. [DOI] [PubMed] [Google Scholar]
- 50.Havrilla J.M., Pedersen B.S., Layer R.M., Quinlan A.R. A map of constrained coding regions in the human genome. Nat. Genet. 2019;51:88–95. doi: 10.1038/s41588-018-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins R.L., Brand H., Karczewski K.J., Zhao X., Alföldi J., Francioli L.C., Khera A.V., Lowther C., Gauthier L.D., Wang H., Genome Aggregation Database Production Team. Genome Aggregation Database Consortium A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jimenez-Sanchez G., Childs B., Valle D. Human disease genes. Nature. 2001;409:853–855. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- 53.Seidman J.G., Seidman C. Transcription factor haploinsufficiency: when half a loaf is not enough. J. Clin. Invest. 2002;109:451–455. doi: 10.1172/JCI15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boukas L., Havrilla J.M., Hickey P.F., Quinlan A.R., Bjornsson H.T., Hansen K.D. Coexpression patterns define epigenetic regulators associated with neurological dysfunction. Genome Res. 2019;29:532–542. doi: 10.1101/gr.239442.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.di Iulio J., Bartha I., Wong E.H.M., Yu H.-C., Lavrenko V., Yang D., Jung I., Hicks M.A., Shah N., Kirkness E.F. The human noncoding genome defined by genetic diversity. Nat. Genet. 2018;50:333–337. doi: 10.1038/s41588-018-0062-7. [DOI] [PubMed] [Google Scholar]
- 56.Morgan M.D., Marioni J.C. CpG island composition differences are a source of gene expression noise indicative of promoter responsiveness. Genome Biol. 2018;19:81. doi: 10.1186/s13059-018-1461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: postnatal malleability and therapeutic prospects. Hum. Mol. Genet. 2019;28(R2):R254–R264. doi: 10.1093/hmg/ddz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 59.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. The human transcription factors. Cell. 2018;175:598–599. doi: 10.1016/j.cell.2018.09.045. [DOI] [PubMed] [Google Scholar]
- 60.Perry M.W., Boettiger A.N., Bothma J.P., Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frankel N., Davis G.K., Vargas D., Wang S., Payre F., Stern D.L. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table contains the promoter coordinates and transcript ENSEMBL ids of both the canonical, as well as the alternative transcript that was selected. All coordinates refer to hg19.
Prediction probabilities are provided in the ”prediction probability of high LoF intolerance by predLoF-CpG” column. Probabilities > 0.75 correspond to genes predicted as highly LoF-intolerant, and probabilities < 0.25 to genes predicted as non-highly LoF-intolerant. ENSEMBL gene/transcript ids and coordinates of the promoters used for prediction are also provided; all coordinates refer to hg19.
These prediction probabilities are provided in the ”prediction probability of high LoF intolerance by predLoFCpG” column. We provide this table for completeness, but do not recommend using these probabilities for clinical decision making.
Data Availability Statement
Code described in this paper can be found at https://github.com/hansenlab/lof_prediction_paper_repro.