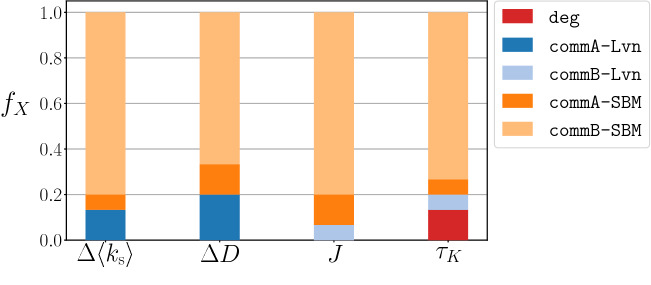Abstract
The organisation of a network in a maximal set of nodes having at least k neighbours within the set, known as -core decomposition, has been used for studying various phenomena. It has been shown that nodes in the innermost -shells play a crucial role in contagion processes, emergence of consensus, and resilience of the system. It is known that the -core decomposition of many empirical networks cannot be explained by the degree of each node alone, or equivalently, random graph models that preserve the degree of each node (i.e., configuration model). Here we study the -core decomposition of some empirical networks as well as that of some randomised counterparts, and examine the extent to which the -shell structure of the networks can be accounted for by the community structure. We find that preserving the community structure in the randomisation process is crucial for generating networks whose -core decomposition is close to the empirical one. We also highlight the existence, in some networks, of a concentration of the nodes in the innermost -shells into a small number of communities.
Subject terms: Statistical physics, thermodynamics and nonlinear dynamics; Complex networks
Introduction
Whenever a system can be abstracted as a set of units (nodes) interacting in pairs (edges), we can describe it as a network (also called a graph). Network analysis has proven to be a valuable framework to aid us to understand a plethora of phenomena taking place in many complex systems. Examples include cascades and collective behaviour in socio-technical systems, the emergence of cognitive functions in neural systems, the stability of chemical/biological systems, and the shape of spatially embedded systems, to cite a few1–3.
One of the advantages of the network representation is the possibility to probe the system in a coarse-grained manner, going beyond dyadic interactions by identifying high-order structures of the network4,5. Examples include tightly connected groups of nodes, i.e., communities6, multiscale coarse-grained structures7, core-periphery structure8–10, nested assembly of nodes11, rich clubs12,13, and the -core14,15.
The -core decomposition of a network is the maximal set of nodes that have at least k neighbours within the set14,15. The algorithm to extract the -core consists in recursively removing the nodes having less than k connections. A -shell is defined as the set of nodes belonging to the kth core but not to the th core15. The -core decomposition has proven to be useful in a variety of domains such as identifying and ranking the most influential spreaders in networks, identifying keywords used for classifying documents, and in assessing the robustness of mutualistic ecosystem and protein networks16,17.
Models to generate random networks with specific features should help us to understand how the mechanisms governing the establishment of edges account for properties of empirical networks. Despite the vast range of applications of the -core decomposition, to the best of our knowledge, there have been only few attempts to build models to generate networks with a given -core structure. One indirect attempt to generate networks with a given -core decomposition is the so-called brite model18. Originally, this model sought to replicate the features (including the -core) of the Internet network at the Autonomous System (AS) level by mixing the mechanism of growth with preferential attachment19,20 and that of adding edges between already existing nodes. Another model aimed at generating networks with a -core structure akin to an empirical one by leveraging the information stored in the so-called core fingerprint21. The core fingerprint corresponds to knowing the number of nodes in each -shell, the number of intra-shell edges (i.e., those connecting nodes belonging to the same -shell), and the number of inter-shell edges (i.e., those connecting nodes belonging to different -shells) of a given network. Moreover, the authors qualitatively compared the Internet AS networks and synthetic networks preserving the core fingerprint of the original networks using several indicators21. More recently, models based on modified versions of the so-called configuration model have been proven to be effective in generating networks with -core structure akin to that of empirical networks22,23. In a nutshell, in these models the edge stubs attached to each node are divided into two groups: red and blue. Red stubs can create any edges regardless of the -shell structure. Blue stubs only form edges connecting nodes belonging to distinct -shells. Among the possible pairs of stubs’ colours, only the blue-blue pair is forbidden.
As mentioned above, another type of mesoscale structure is communities. Although there is not a univocal definition of what a community is, in general the community refers to a group of nodes that are more tightly connected between each other than with the other nodes of the network6. Communities are also defined by the concept of stochastic equivalence, i.e., nodes in the same group/community interact, on average, with nodes in other groups in the same way24. Methods based on different definitions of communities may return different partitions of the node set. However, there is often some consistency between those partitions, which indicates the presence of groups of nodes acting like the building blocks of communities25. The presence of communities is an important large-scale characteristic of many empirical networks because a system’s different functions tend to be located in different communities (e.g., in functional brain networks26 and protein-protein interaction networks27). Moreover, it has been proven that communities play a role in the resilience of the system28 and the presence of triangles29, as well as in the emergence of collective behaviour including synchronisation30, the emergence of cooperation31,32, spreading of a pandemic33, and the attainment of consensus34,35.
Although -core and communities are two ways of decomposing the same network, there may be overlaps or intricate relationships between them. In the present paper, we study the relation between the -core decomposition and the community structures of several empirical and synthetic networks. In particular, we leverage the work of Alvarez-Hamelin et al.36 and confirm that the nodes’ degrees (i.e., their number of edges) alone are not capable of reproducing the network’s -shell structure. We find that one has to include information about the community structure to obtain networks whose -core decomposition looks sufficiently close to the empirical one. We also highlight the existence of a concentration-like phenomenon of the innermost -shells into a small number of communities, which is stronger in some data sets than others.
Results
Degree-based reconstruction of the -core
As stated above, various studies on networks leverage the -core decomposition to extract insightful information from networks. However, less studies have asked which mechanisms are sufficient for explaining generation of networks having empirically observed patterns of -core decomposition. More specifically, Alvarez-Hamelin et al. found that networks generated using the configuration model37 having a Poisson or power-law degree distribution do not display a -core structure similar to the one displayed by the AS network36. Using the results of Alvarez-Hamelin et al. as a starting point, given an empirical network G with N nodes, we check whether its -core decomposition can be reproduced solely from the degree of each node i (i.e., the number of edges that node i has), denoted by . We generated random networks by a standard configuration model preserving the degree of each node of G, which we denote by deg (see “Methods” section for details).
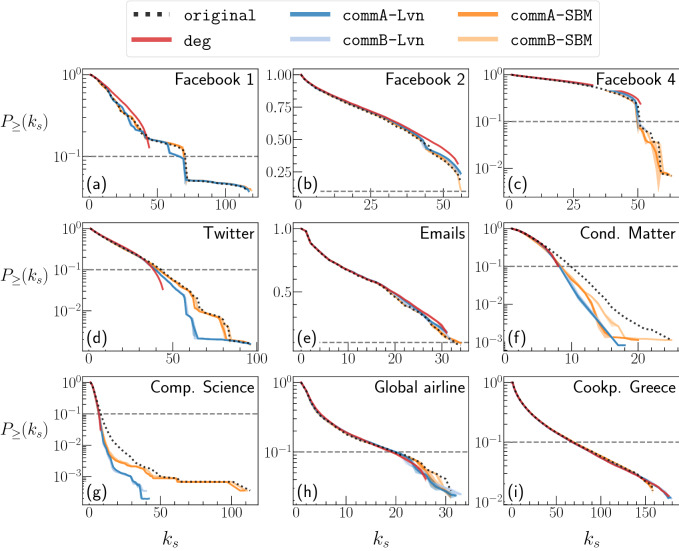
We have analysed several empirical networks encompassing social, technological, linguistic, and transportation systems whose main properties are summarised in Table 1. In Fig. 1, we show the survival function of the probability distributions of the -shell index, (i.e., fraction of nodes whose -shell index is larger than or equal to ), for a selection of data sets, compared across the original networks and their synthetic counterparts (see Supplementary Fig. S1 in SM for the other data sets). Figure 1 indicates that the degree of each node is not sufficient for reproducing the -core structure of the original networks because for deg considerably deviates from that for the original networks. This result is consistent with the previous results36. In fact, we find that fixing the degree of each node is sufficient to recover the -core profile in some networks. For these networks the empirical and deg networks are not too different in terms of (e.g., Facebook 2 and Cookpad networks). We point out two main differences in between the empirical and deg networks. First, for most data sets, the largest value, which is denoted by D and called the degeneracy, is considerably smaller for the networks generated by deg than the original networks. Second, the of some empirical networks have plateaus and abrupt drops in . The plateaus imply that some of the -shells are completely or almost empty, whereas the abrupt drops indicate that some -shells are more densely populated than those adjacent to them. In contrast, for the deg networks does not have a notable plateau or drop in . Therefore, in the deg networks, all the -shells up to are populated, and there is no -shell that is substantially more populated than its adjacent -shells.
Table 1.
Main properties of the data sets used in the present study. N: number of nodes, L: number of edges, : average degree, : maximum degree, : average value of the -shell index, D: maximum value of the -shell index, , : number of communities determined by the Louvain method and the corresponding modularity, respectively, , : number of communities determined by the SBM and the corresponding modularity, respectively.
| Data set | N | L | D | References | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Facebook 1 | 4039 | 88234 | 43.691 | 1045 | 26.880 | 115 | 16 | 0.835 | 62 | 0.551 | 55,74 |
| Facebook 2 | 6386 | 217662 | 68.168 | 930 | 35.712 | 56 | 19 | 0.419 | 198 | 0.158 | 56–58,75 |
| Facebook 3 | 2235 | 90954 | 81.391 | 467 | 44.508 | 63 | 8 | 0.436 | 87 | 0.139 | 56–58,76 |
| Facebook 4 | 11247 | 351358 | 62.480 | 415 | 32.413 | 63 | 10 | 0.438 | 274 | 0.193 | 56–58,77 |
| Facebook 5 | 27737 | 1034802 | 74.615 | 2555 | 38.681 | 81 | 18 | 0.470 | 547 | 0.172 | 56–58,78 |
| 81306 | 1342296 | 33.018 | 3383 | 17.762 | 96 | 73 | 0.808 | 510 | 0.511 | 55,79 | |
| Web-blogs | 1490 | 16715 | 22.436 | 351 | 12.154 | 36 | 275 | 0.426 | 17 | 0.076 | 60,80 |
| Emails | 1005 | 16064 | 31.968 | 345 | 17.063 | 34 | 26 | 0.410 | 33 | 0.232 | 81–83 |
| Cond. Matter | 23133 | 93439 | 8.078 | 279 | 4.900 | 25 | 619 | 0.730 | 203 | 0.633 | 83,84 |
| Comp. Science | 317080 | 1049866 | 6.622 | 343 | 4.215 | 113 | 209 | 0.822 | 676 | 0.726 | 59,85,86 |
| Global airline | 3376 | 19179 | 11.362 | 248 | 6.123 | 31 | 26 | 0.665 | 40 | 0.311 | 61 |
| Words | 146005 | 656999 | 9.000 | 1008 | 5.289 | 31 | 378 | 0.759 | 548 | 0.583 | 59,87,88 |
| Cookpad Greece | 32235 | 745178 | 46.234 | 8196 | 23.709 | 158 | 40 | 0.166 | 76 | 0.020 | – |
| Cookpad Spain | 122158 | 1749751 | 28.647 | 12637 | 14.547 | 162 | 262 | 0.270 | 90 | 0.035 | – |
| Cookpad UK | 13758 | 47525 | 6.909 | 1880 | 3.558 | 33 | 199 | 0.350 | 8 | 0.114 | – |
Figure 1.
Survival function of the probability distributions of the -shell index, i.e., as a function of for the original network (dotted line) and shuffled networks (solid line). Each panel corresponds to a data set, i.e., Facebook 1 (panel a), Facebook 2 (b), Facebook 4 (c), Twitter (d), Emails (e), Cond. Matter (f), Comp. Science (g), Global airline (h), and Cookpad Greece (i). The horizontal dashed lines indicate that . Results are averaged over 10 different runs of each shuffling method, and the shaded areas (when visible) represent the standard deviations.
A more quantitative comparison of distribution between the empirical and deg networks may be done by, for example, the Kolmogorov-Smirnov (KS) test38. However, because a majority of the nodes usually belongs to outer -shells, (i.e., set of nodes with small values) and Fig. 1 shows that the strongest discrepancies between the two distributions tend to occur at large values, the KS test fails to grasp the differences at large values that we are mostly interested in. Therefore, we compare the -core decomposition of the empirical and deg networks using four indicators, i.e., the relative difference in the average -shell index, , the relative difference in the network’s degeneracy, , the Jaccard score, J, and Kendall’s, of the nodes belonging to the top 10% (i.e., innermost -shells) of the distribution. The average of each indicator over all the data sets for the networks obtained with the deg shuffling method is equal to , , , and . The value of indicates that is only different between the original and deg networks on average. However, their degeneracy differs by on average. The and values inform us that innermost -shells of the original networks and those of the deg networks tend to share approximately half of the nodes, albeit their ranking seems to be fairly preserved. Supplementary Table S1 reports the values of each indicator.
Community-aware reconstruction of the -core
We have seen that the degree distribution by itself does not reproduce main features of the -shell index distribution. An alternative feature that may explain the -shell index distribution is the community structure. For this reason, we generated synthetic networks that preserve both the degree of each node and the community structure, , where is the number of communities of the original network. To account for the multiple definitions of what a community is, we identified the communities of each network using two methods: the Louvain method39, denoted by Lvn, and the degree-corrected stochastic block model40, denoted by SBM. In combination with each of the two community detection methods, we considered two rewiring methods preserving and the degree of each node, denoted by commA and commB. Method commA preserves the exact number of inter- and intra-community edges at the level of single communities. Method commB preserves the number of inter- and intra-community edges for each node.
Figure 1 indicates that preserving the community structure in addition to the degree of each node improves the similarity in between the empirical and synthetic networks, especially at large values, which correspond to inner -shells. In particular, commA and commB generate networks whose D value tends to be closer to the empirical value than deg does. Furthermore, for commA and commB tends to have plateaus and abrupt drops at similarly to the empirical networks. Overall, synthetic networks preserving the SBM community structure have a -core decomposition more akin to the empirical one than those preserving the Lvn community structure. This observation is quantitatively supported by the values of the four indices reported in Supplementary Table S1.
To obtain an overview of the performances of different network randomisation methods, in Fig. 2 we show the fraction of data sets, , for which a certain shuffling method generates a -core decomposition that is the most similar to that of the empirical network according to each indicator. The figure indicates that commB-SBM (i.e., the commB shuffling method that preserves the community structure determined by SBM) performs the best in mimicking the -shell index features for approximately 65–80% of the data sets, depending on the indicator. Detailed results for the performance of each method for each empirical network are shown in Supplementary Fig. S2 and Supplementary Table S1.
Figure 2.
Performances of different shuffling methods in terms of four indicators. We report the fraction of data sets for which a given combination of the shuffling method and the community detection method yields an indicator’s value closest to that for the original network. Each bar refers to an indicator, i.e., average -shell’s difference, , degeneracy’s difference, , Jaccard score, J, and Kendall’s tau, .
One issue of Lvn is that it cannot discover small communities6,41. One way to mitigate this limitation is to introduce in Lvn a resolution parameter, , regulating the resolution scale. It is possible to detect small communities when r is small, whereas the original Lvn corresponds to 42. We denote by LvnR the Louvain method with , i.e., with a resolution higher than that used by Lvn. In Sect. 2 of SM we report whether preserving the communities found using LvnR instead of Lvn improves our ability to reproduce the -core decomposition of the original network. We found that LvnR performs better than Lvn (Supplementary Figs. S3 and S4) but worse than SBM in general (Supplementary Fig. S5).
Imposing the simultaneous conservation of each node’s degree and community structure may result in synthetic networks that are not substantially different from the original ones. To exclude this possibility, we computed the Jaccard score, , (see Eq. (3)) for the sets of edges, and , of the original and shuffled networks, respectively. The values of J approximately fall between 0.01 and 0.5, confirming that the set of edges – hence, the networks – are considerably different.
The results presented so far suggest that preserving the community structure improves the preservation of the -core decomposition of the original network. Therefore, the mere presence of a community structure may be enough to preserve the main features of the -core decomposition of the original networks. To test this possibility, we applied the -core decomposition to networks with communities generated using the LFR model43 (see Sect. 3 of SM). The plots of shown in Supplementary Figs. S7–S10 indicate that the presence of a community structure alone is not sufficient for producing major features of the -core structure of the empirical networks. Specifically, the of the networks generated by the LFR model is always smooth and shows neither plateaus nor abrupt drops as increases. Moreover, with the LFR, is narrowly distributed, i.e., . These differences between the -core structure of the LFR model and that of empirical networks are not sensitive to the value of the mixing parameter, , of the LFR model, which controls how distinct the communities are. It should also be noted that for the LFR model, as for the empirical network, the commB-SBM generates networks that are the most similar to the original LFR networks among the different shuffling methods in terms of .
Overlap between communities and -core
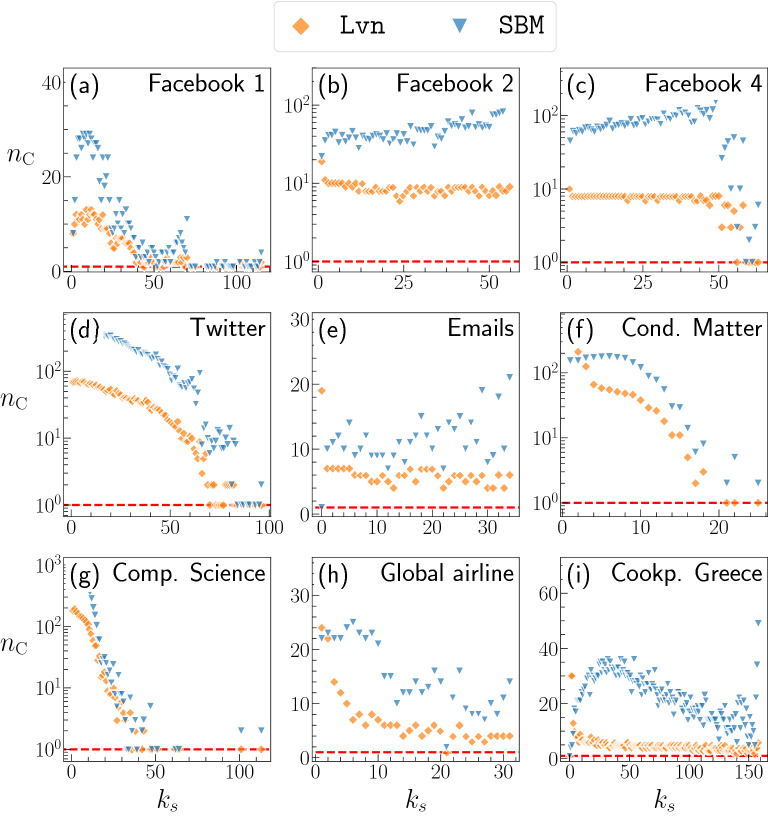
Preserving the community structure in addition to the node’s degree can lead to preservation of features of the -core structure possibly because nodes with high values of form a -core which tend to belong to the same community. To examine this possibility, we show the number of communities to which the nodes of a given -shell belong, , in Fig. 3 (see Supplementary Fig. S11 for the other data sets). Although each data set shows a distinct pattern, for many data sets, inner -shells (i.e., nodes with large values) are concentrated into one or a few communities. The concentration effect is particularly noticeable for some data sets, e.g., Facebook 1 and Twitter. To check whether the number of communities per -shell is merely a byproduct of the random combinatorial effect owing to the number of communities, the distribution of the community size, and the distribution of , we computed a random assignments of the nodes to communities and then calculated for each value (see Sect. 4 and Supplementary Fig. S12 of the SM). We have found that the nodes in each -shell are almost always more concentrated into a smaller number of communities than what is expected by the random assignment of the nodes to communities for all the data sets and community detection methods, with the only exception of SBM for Cookpad’s data sets. This finding is in agreement with the previous result that nodes with high tend to belong to the same community, which has been observed in networks embedded into hyperbolic spaces44,45. In particular, we observe a strong concentration of the -shells into a few communities for the Facebook 1, Twitter, Cond. Matter, Comp. Science, and Words networks, which are those showing a more pronounced difference in the values of D between the original and deg networks.
Figure 3.
Number of different communities, , that the set of nodes of a given -shell value, , overlaps. The horizontal dashed line is a guide to the eyes showing . Each panel accounts for a different data set (see the caption of Fig. 1 for the details). For each data set, we show the results corresponding to the community structure obtained using either Lvn or SBM.
Discussion
The information encoded in the degree of each node is not sufficient for generating networks with a -core structure that is similar to those of empirical networks36. This gap of knowledge calls for the design of generative models of networks beyond the configuration model. Such models are expected to be useful to generate benchmark networks and to understand the mechanisms behind the emergence of the -core. To the best of our knowledge, few models are available to generate networks with a given -core decomposition21–23.
In the present study, we investigated how much the combination of the nodes’ degrees and community structure accounts for -core structure of empirical networks. Given a network G, we randomly shuffled G’s edges to generate its synthetic counterparts preserving each node’s degree and/or community structure of G. We found that randomised networks preserving the community structure obtained through a stochastic block model showed a -shell index distribution that was reasonably similar to the distribution for the original networks. The success of the stochastic block model in mimicking the features of the -core decomposition might be due to its ability to approximate the mesoscale structures of networks with a good accuracy24,46, including communities. We also sought to understand more the relationship between -core and communities by studying networks generated by the LFR model which enables us to control the extent to which the communities are distinguished from each other. However, regardless of whether or not different communities are relatively distinguished from each other in a network, the -shell index distribution of LFR networks does not show the same features as those observed in the empirical networks. Finally, we have investigated the overlap between communities and -shells and found that, in some empirical networks, the nodes in inner -shells are concentrated into a small number of communities much more so than a randomised counterpart. This result is in agreement with the observations made for networks embedded in hyperbolic spaces44,45. Up to our numerical efforts, the concentration is observed if and only if the empirical network and its deg counterpart are substantially different in terms of their -core decomposition. The concentration suggests that inner -shells may perform specific functions in such networks, corresponding to the functions of the communities they belong to as observed in, for instance, functional brain networks26 and protein-protein interaction networks27.
The “community aware” rewiring mechanisms introduced in this paper can be used for assessing whether or not a given property of a network is a direct expression of its community structure. One example of such an approach is given in47, where the authors have improved the robustness against attacks on a network while keeping its community structure. In that case, the method only preserves the communities and alters the connectivity pattern by increasing the density of intra-community edges as well as changing the edges between communities. It may be interesting, instead, to check whether the robustness of the network can be improved even when one also preserves the degree of the nodes using our community-aware rewiring mechanisms.
One viable extension of our work is to the case of k-peak graph decomposition method48. In Ref.48, the authors argue that for networks with communities, the -core decomposition should be performed locally rather than globally, thus returning the k-peak decomposition of each of the system’s regions. The rationale behind this approach is to avoid that, if the network contains regions with different densities of edges, the standard -core decomposition would fail to recognise local core nodes in sparser regions. Studying the evolution of the k-peak decomposition in response to the rewiring of the connections may unveil salient features of complex systems. Another possible direction of research is to concatenate the information encoded into the -shell index, , with the one provided by the so-called onion decomposition (OD)49. The OD is an extension of the -core decomposition where a node is labelled with both its and its layer index. The layer of a node i represents the iteration number with which node i is removed in the recursive pruning process of the -core decomposition. The OD provides a further characterisation of the structure of the network than the -core, revealing, for example, how tree-like the network is.
Summing up, in this work we have analysed the interplay between the -core decomposition and community structure of networks. Understanding such a relationship is useful not only owing to the broad range of applications of -core decomposition, but also to inform the design of models capable of generating networks with both a community structure and -core’s features beyond those explainable by the degree distribution. Such models may stand on, for instance, the stochastic block model40, the enhanced configuration model based on maximum entropy50, or the hierarchical extension of the LFR model51. Alternatively, models based on microscopic growth mechanisms such as triadic closure52,53 or modified preferential attachment54 may deserve further investigation.
Methods
Data
We have considered networks corresponding to systems of different types: from social to technological, from semantic to transportation. Table 1 summarises main properties of such networks. Except for Cookpad networks, all the data sets are publicly available and have been retrieved from the Stanford Large Network data set Collection55 (Facebook 1, Twitter, Emails, and Cond. Matter), the Network Repository56–58 (Facebook 2, 3, 4, and 5), the Koblenz Network Collection (KONECT)59 (Comp. Science, and Words), Mark E. J. Newman’s personal network data repository60 (Web-blogs), and the OpenFlights data repository61 (Global airline). In the following text, we provide a brief description of each data set.
Facebook and Twitter These networks describe social relationships. Nodes are people. Edges represent their friendship relations.
Web-blogs This network is composed of the hyperlinks (edges) between weblogs on US politics (nodes) recorded in 2005.
Emails This is a network of email data from a large European research institution. Nodes are people. Edges connect pairs of individuals who have exchanged at least one e-mail.
Cond. Matter and Comp. Science The former network is the co-authorship network of the authors of preprint manuscripts submitted to the Condensed Matter Physics arXiv e-print archive from January 1993 to April 2003. The latter network is similarly defined using manuscripts appearing in the DBLP computer science bibliography, using a comprehensive list of research papers in computer science. The submission time of the papers of the DBLP collection is unavailable. A node is an author. An edge represents the existence of at least one manuscript co-authored by two authors.
Global airline In this network nodes are airports across the globe. An edge indicates direct commercial flights between two airports.
Words This network accounts for the lexical relationships among words extracted from the WordNet data set. Nodes are English words. Edges are relationships (synonymy, antonymy, meronymy, etc.) between pairs of words.
Cookpad These networks are extracted from the Cookpad online recipe sharing platform62. Users can post and browse recipes, as well as interact with other users through recipes in multiple ways including liking, sharing, and posting a comment. The platform is present in many countries (e.g., Japan, Indonesia, United Kingdom, and Italy). Here, we consider the data collected from September to November of 2018 in Greece, Spain, and the United Kingdom, separately for each country. In the three networks, nodes are users. An edge between a pair of users exists if one or more of the following types of events takes place: like or follow a user, viewing, bookmarking, commenting, or making a cooksnap of another user’s recipe.
All the networks considered in this work are treated as undirected and unweighted, even when the original data contains more information. Finally, we also consider synthetic networks, generated using the LFR (Lancichinetti–Fortunato–Radicchi) model43 (see Sect. 3 of SM for details).
Network shuffling
Given a network, G, with N nodes and L edges, we generate a randomised counterpart, , that has the same nodes and the same number of edges by shuffling the edges of G. We consider three shuffling methods denoted by deg, commA, and commB; each shuffling method preserves different properties of G. The shuffling consists in selecting uniformly at random two edges (a, b) and (c, d), and replacing them with, e.g., (a, c) and (b, d), if the swapping of the edges is accepted. An attempt to swap edges is accepted, in which case we call the swapping effective, if and only if it respects the rule of the specific shuffling method and the swapping does not generate self-loops or multiple edges. We continued the shuffling until we carried out 2L effective swaps, such that an edge was swapped four times on average.
In the following text, we provide the details of each shuffling method. Assume that network G partitions into communities such that the set of the communities is , where is the number of communities. Furthermore, let , be the community to which the ith node belongs and be the degree of node i. We have:
Degree-preserving shuffling (deg) This method preserves degree of each node i and is equivalent to the configuration model37.
Community-preserving shuffling of type A (commA) On top of the degree of each node, this method preserves the total number of edges within each community and between each pair of communities. In attempts to swap edges, we replace two randomly selected edges (a, b) and (c, d) by (a, c) and (b, d) if and only if an end node of edge (a, b) and an end node of edge (c, d) belong to the same community (i.e., if or ).
Community-preserving shuffling of type B (commB) Like commA, this method preserves the degree of each node and the number of edges within each community and between each pair of communities. In contrast with commA, the commB method preserves the numbers of edges within and across communities for each node, and not only for each community or pairs of communities. Given two selected edges (a, b) and (c, d), we replace them with (a, c) and (b, d) if and only if the two new edges connect the same community pairs as before the swapping (i.e., and ).
Comparison of the -core decomposition
To assess the similarity between the -core decomposition of the original network, G, and of its shuffled counterpart, , we used four indicators: the average -shell index, , the network’s degeneracy, D, the Jaccard score, J, and the generalised Kendall’s tau, . The indicator explicitly depends on all the nodes in the network, whereas D, J and only depend on the nodes belonging to the innermost -shell(s). We use the latter three indicators because, although a majority of nodes tends to belong to outer -shells, it is a difference in the tails of the distributions that often affect functions of networks such as the impact of influencers in contagion processes63. The four indicators are defined as follows.The average of the -shell index, , is equal to
| 1 |
where is the -shell index of node i. The degeneracy, D, of a network G is given by64
| 2 |
Rather than using these raw indicators, to compare across the different data sets, we compute their relative difference between the empirical network and its shuffled counterpart given by , where .
To compute J and , we need to define a criterion to select nodes belonging to the innermost -shells. We decided to confine the comparison to the nodes whose falls within the top among the N nodes. The horizontal lines in Fig. 1 indicate the threshold values of such that . In the same manner, we define such that in network . To calculate J and , we use the nodes belonging to -shells with in G and the nodes belonging to -shells with in without duplication of the nodes. There are several remarks. First, it may hold that . Second, the value of varies from one combination of a run of shuffling and community detection to another. Third, as in the case of the Facebook 2 data set, sometimes does not even exist. In such a case, we set and select all the nodes belonging to the innermost -shell although they constitute more than 10% of the nodes in the network. Fourth, additional tests using different threshold percentages, 5% and 20%, instead of 10%, did not qualitatively change the results. Fifth, while the Jaccard score simply compares the nodes belonging to two sets, the generalised Kendall’s tau, compares ranked sets. In our case, the node’s rank is equivalent to the value.
Given two sets and , the Jaccard score quantifies their overlap and is given by
| 3 |
The Jaccard score ranges between 0 and 1. A value of 1 indicates the complete overlap between the two sets (i.e., the sets are the same), whereas a value of 0 indicates that the sets are completely different.
The generalised Kendall’s tau, , measures the consistency between two rankings by assigning penalties to pairs of elements on which the two rankings disagree65,66. Given two sets and having and elements, respectively, consider their associated ranking functions and . We denote with an arbitrary pair of elements of . We assign a penalty to if (a) the rankings of the two elements within each set are different (i.e., and ), (b) the element with the higher rank in one set is missing in the other set, i.e., and (or and ), or (c) both elements belong to one set each, which is not the same set, i.e., and (and vice-versa). In all the other cases , such that we do not penalise the pair. Finally, we sum the penalties over all the possible pairs of elements and normalise it, thus obtaining the generalised Kendall’s tau:
| 4 |
Index ranges between 0 and 1. If , the two rankings are completely coherent. If , the two sets and have no pair of elements on which rankings and are coherent. The above formulation of the Kendall’s tau is the so-called optimistic approach65. This means that we do not penalise the case in which a pairs of elements is present in one set and not in the other set.
Community detection methods
We considered two methods for community detection. The first is the Louvain method (Lvn)39, which is a heuristic greedy multiscale method that approximately maximises the modularity function. Given a network with N nodes distributed among communities, the modularity, Q, reads
| 5 |
where is the element of the network’s adjacency matrix A; g(i) is the community to which the i-th node belongs (), and is the Kronecker delta. A large value of Q implies a good partitioning. The Louvain method seeks the partitioning that maximises the modularity. Note that we obtain for random assignment of nodes to communities and that we obtain when the network is made of perfectly disjoint communities.
The other community detection method that we used is the stochastic block model67. It uses the probabilities with which there exists an edge (a, b) connecting an arbitrarily selected node a in community (i.e., ) and an arbitrarily selected node b in community (i.e., ). Different instances of probabilities allow the description of different mixing patterns. When the diagonal entries of predominate, we obtain the most usual community structure, whereas other instances yield other structures such as bipartite or core-periphery structure.
To find the optimal partition, one maximises the likelihood function with respect to corresponding to the partitioning , where . The unnormalised log-likelihood, , with which a partition of network G into communities, , is reproduced reads
| 6 |
where is the number of edges connecting community and community , and is the number of nodes belonging to .
The above formulation, however, has one major limitation: it assumes that the degrees of the nodes are distributed according to a Poisson-like function. To account for the degrees’ heterogeneity, Karrer et al. have implemented the so-called degree corrected stochastic block model, in which the expected degree of each node is kept constant via the introduction of additional parameters40. Let be the sum of the node’s degree over all nodes in community . Then, the unnormalised log-likelihood for the degree-corrected stochastic block model reads
| 7 |
Equations (6) and (7) depend on the number of communities . Because the value of is not known a priori, it is inferred through the minimisation of a quantity called the description length. The minimum description length principle describes how much a model compresses the data and allows us to find the optimal number of communities while avoiding overfitting68. In the present work we use the degree-corrected stochastic block model and its implementation available in the Python Graph-tool package69, which we refer to as SBM for brevity.
Supplementary information
Acknowledgements
The authors thank A. Barrat for helpful comments on a preliminary version of this work. A.C. acknowledges the support of the Spanish Ministerio de Ciencia e Innovacion (MICINN) through Grant IJCI-2017-34300. I.M., A.C., and N.M. acknowledge the support of Cookpad Limited. This work was carried out using the computational facilities of the Advanced Computing Research Centre, University of Bristol – http://www.bristol.ac.uk/acrc/. Numerical analysis has been carried out using the NumPy, NetworkX, and Graph-tool Python packages69–72. Graphics have been prepared using the Matplotlib Python package73.
Author contributions
I.M. analysed data, designed the experiments, and performed simulations, A.C. analysed the results and designed the experiments. All authors discussed the methods and results, and wrote and reviewed the manuscript.
Data availability
The data sets on Cookpad™analysed in the current study are not publicly available due to exclusive ownership of Cookpad Limited. All the other data sets are available from the corresponding repositories listed in the bibliography.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alessio Cardillo, Email: alessio.cardillo@urv.cat.
Naoki Masuda, Email: naokimas@buffalo.edu.
Supplementary information
is available for this paper at 10.1038/s41598-020-71426-8.
References
- 1.Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang D. Complex networks: Structure and dynamics. Phys. Rep. 2006;424:175–308. doi: 10.1016/j.physrep.2005.10.009. [DOI] [Google Scholar]
- 2.Barabási A-L. The network takeover. Nat. Phys. 2011;8:14–16. doi: 10.1038/nphys2188. [DOI] [Google Scholar]
- 3.Vespignani A. Modelling dynamical processes in complex socio-technical systems. Nat. Phys. 2012;8:32–39. doi: 10.1038/nphys2160. [DOI] [Google Scholar]
- 4.Lambiotte R, Rosvall M, Scholtes I. From networks to optimal higher-order models of complex systems. Nat. Phys. 2019;15:313–320. doi: 10.1038/s41567-019-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson AR, Gleich DF, Leskovec J. Higher-order organization of complex networks. Science. 2016;353:163–166. doi: 10.1126/science.aad9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fortunato S, Hric D. Community detection in networks: a user guide. Phys. Rep. 2016;659:1–44. doi: 10.1016/j.physrep.2016.09.002. [DOI] [Google Scholar]
- 7.Gfeller D, De Los Rios P. Spectral coarse graining of complex networks. Phys. Rev. Lett. 2007;99:038701. doi: 10.1103/PhysRevLett.99.038701. [DOI] [PubMed] [Google Scholar]
- 8.Borgatti SP, Everett MG. Models of core/periphery structures. Soc. Netw. 2000;21:375–395. doi: 10.1016/S0378-8733(99)00019-2. [DOI] [Google Scholar]
- 9.Csermely P, London A, Wu L-Y, Uzzi B. Structure and dynamics of core/periphery networks. J. Complex Netw. 2013;1:93–123. doi: 10.1093/comnet/cnt016. [DOI] [Google Scholar]
- 10.Rombach P, Porter MA, Fowler JH, Mucha PJ. Core-periphery structure in networks (revisited) SIAM Rev. 2017;59:619–646. doi: 10.1137/17M1130046. [DOI] [Google Scholar]
- 11.Mariani MS, Ren Z-M, Bascompte J, Tessone CJ. Nestedness in complex networks: observation, emergence, and implications. Phys. Rep. 2019;813:1–90. doi: 10.1016/j.physrep.2019.04.001. [DOI] [Google Scholar]
- 12.Zhou S, Mondragon RJ. The rich-club phenomenon in the Internet topology. IEEE Commun. Lett. 2004;8:180–182. doi: 10.1109/LCOMM.2004.823426. [DOI] [Google Scholar]
- 13.Colizza V, Flammini A, Serrano MÁ, Vespignani A. Detecting rich-club ordering in complex networks. Nat. Phys. 2006;2:110–115. doi: 10.1038/nphys209. [DOI] [Google Scholar]
- 14.Erdős P, Hajnal A. On chromatic number of graphs and set-systems. Acta Math. Acad. Sci. Hung. 1966;17:61–99. doi: 10.1007/BF02020444. [DOI] [Google Scholar]
- 15.Seidman SB. Network structure and minimum degree. Soc. Netw. 1983;5:269–287. doi: 10.1016/0378-8733(83)90028-X. [DOI] [Google Scholar]
- 16.Kong Y-X, Shi G-Y, Wu R-J, Zhang Y-C. k-core: theories and applications. Phys. Rep. 2019;832:1–32. doi: 10.1016/j.physrep.2019.10.004. [DOI] [Google Scholar]
- 17.Malliaros FD, Giatsidis C, Papadopoulos AN, Vazirgiannis M. The core decomposition of networks: theory, algorithms and applications. VLDB J. 2020;29:61–92. doi: 10.1007/s00778-019-00587-4. [DOI] [Google Scholar]
- 18.Medina A, Lakhina A, Matta I, Byers J. Proceedings ninth international symposium on modeling. Anal. Simul. Comput. Telecommun. Syst. 2001;346–353:2001. doi: 10.1109/MASCOT.2001.948886. [DOI] [Google Scholar]
- 19.de Solla Price D. Networks of scientific papers. J. Sci. 1965;149:510–515. doi: 10.1126/science.149.3683.510. [DOI] [PubMed] [Google Scholar]
- 20.Barabàsi A, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 21.Baur M, Gaertler M, Görke R, Krug M, Wagner D. Augmenting -core generation with preferential attachment. Netw. Heterogeneous Media. 2008;3:277–294. doi: 10.3934/nhm.2008.3.277. [DOI] [Google Scholar]
- 22.Hébert-Dufresne L, Allard A, Young J-G, Dubé LJ. Percolation on random networks with arbitrary -core structure. Phys. Rev. E. 2013;88:062820. doi: 10.1103/PhysRevE.88.062820. [DOI] [PubMed] [Google Scholar]
- 23.Allard A, Hébert-Dufresne L. Percolation and the effective structure of complex networks. Phys. Rev. X. 2019;9:011023. doi: 10.1103/PhysRevX.9.011023. [DOI] [Google Scholar]
- 24.Young J-G, St-Onge G, Desrosiers P, Dubé LJ. Universality of the stochastic block model. Phys. Rev. E. 2018;98:032309. doi: 10.1103/PhysRevE.98.032309. [DOI] [Google Scholar]
- 25.Riolo MA, Newman MEJ. Consistency of community structure in complex networks. Phys. Rev. E. 2020;101:052306. doi: 10.1103/PhysRevE.101.052306. [DOI] [PubMed] [Google Scholar]
- 26.Meunier D, Lambiotte R, Bullmore ET. Modular and hierarchically modular organization of brain networks. Front. Neurosci. 2010;4:200. doi: 10.3389/fnins.2010.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huttlin E, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilarranz LJ, Rayfield B, Liñán-Cembrano G, Bascompte J, Gonzalez A. Effects of network modularity on the spread of perturbation impact in experimental metapopulations. Science. 2017;357:199–201. doi: 10.1126/science.aal4122. [DOI] [PubMed] [Google Scholar]
- 29.Orman, K., Labatut, V. & Cherifi, H. Complex Networks, vol. 424 of Studies in Computational Intelligence, chap. An empirical study of the relation between community structure and transitivity, 99–110 (Springer, Berlin, Heidelberg, 2013).
- 30.Lotfi N, Rodrigues FA, Darooneh AH. The role of community structure on the nature of explosive synchronization. Chaos. 2018;28:033102. doi: 10.1063/1.5005616. [DOI] [PubMed] [Google Scholar]
- 31.Fotouhi B, Momeni N, Allen B, Nowak MA. Evolution of cooperation on large networks with community structure. J. R. Soc. Interface. 2019;16:20180677. doi: 10.1098/rsif.2018.0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giatsidis, C., Thilikos, D. M. & Vazirgiannis, M. Evaluating cooperation in communities with the k-core structure. In 2011 International Conference on Advances in Social Networks Analysis and Mining, 87–93, 10.1109/ASONAM.2011.65 (IEEE, 2011).
- 33.Salathé M, Jones JH. Dynamics and control of diseases in networks with community structure. PLoS Comput. Biol. 2010;6:1–11. doi: 10.1371/journal.pcbi.1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mistry D, Zhang Q, Perra N, Baronchelli A. Committed activists and the reshaping of status-quo social consensus. Phys. Rev. E. 2015;92:042805. doi: 10.1103/PhysRevE.92.042805. [DOI] [PubMed] [Google Scholar]
- 35.Masuda N. Voter model on the two-clique graph. Phys. Rev. E. 2014;90:012802. doi: 10.1103/PhysRevE.90.012802. [DOI] [PubMed] [Google Scholar]
- 36.Alvarez-Hamelin JI, DallAsta L, Barrat A, Vespignani A. K-core decomposition of internet graphs: hierarchies, self-similarity and measurement biases. Netw. Heterogeneous Media. 2008;3:371. doi: 10.3934/nhm.2008.3.371. [DOI] [Google Scholar]
- 37.Fosdick BK, Larremore DB, Nishimura J, Ugander J. Configuring random graph models with fixed degree sequences. SIAM Rev. 2018;60:315–355. doi: 10.1137/16M1087175. [DOI] [Google Scholar]
- 38.Massey FJ., Jr The Kolmogorov–Smirnov test for goodness of fit. J. Am. Stat. Assoc. 1951;46:68–78. doi: 10.1080/01621459.1951.10500769. [DOI] [Google Scholar]
- 39.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech: Theory Exp. 2008;2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 40.Karrer B, Newman MEJ. Stochastic blockmodels and community structure in networks. Phys. Rev. E. 2011;83:016107. doi: 10.1103/PhysRevE.83.016107. [DOI] [PubMed] [Google Scholar]
- 41.Fortunato S, Barthélemy M. Resolution limit in community detection. Proc. Nat. Acad. Sci. U.S.A. 2007;104:36–41. doi: 10.1073/pnas.0605965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambiotte R, Delvenne JC, Barahona M. Random walks, Markov processes and the multiscale modular organization of complex networks. IEEE Trans. Net. Sci. Eng. 2014;1:76–90. doi: 10.1109/TNSE.2015.2391998. [DOI] [Google Scholar]
- 43.Lancichinetti A, Fortunato S, Radicchi F. Benchmark graphs for testing community detection algorithms. Phys. Rev. E. 2008;78:046110. doi: 10.1103/PhysRevE.78.046110. [DOI] [PubMed] [Google Scholar]
- 44.Faqeeh A, Osat S, Radicchi F. Characterizing the analogy between hyperbolic embedding and community structure of complex networks. Phys. Rev. Lett. 2018;121:098301. doi: 10.1103/PhysRevLett.121.098301. [DOI] [PubMed] [Google Scholar]
- 45.Osat S, Radicchi F, Papadopoulos F. -core structure of real multiplex networks. Phys. Rev. Res. 2020 doi: 10.1103/PhysRevResearch.2.023176. [DOI] [Google Scholar]
- 46.Olhede SC, Wolfe PJ. Network histograms and universality of blockmodel approximation. Proc. Nat. Acad. Sci. U.S.A. 2014;111:14722–14727. doi: 10.1073/pnas.1400374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mozafari M, Khansari M. Improving the robustness of scale-free networks by maintaining community structure. J. Complex Netw. 2019;7:838–864. doi: 10.1093/comnet/cnz009. [DOI] [Google Scholar]
- 48.Govindan, P., Wang, C., Xu, C., Duan, H. & Soundarajan, S. The k-peak decomposition: Mapping the global structure of graphs. In Proceedings of the 26th International Conference on World Wide Web, 1441–1450, 10.1145/3038912.3052635 (International World Wide Web Conferences Steering Committee, 2017).
- 49.Hébert-Dufresne L, Grochow JA, Allard A. Multi-scale structure and topological anomaly detection via a new network statistic: the onion decomposition. Sci. Rep. 2016;6:31708. doi: 10.1038/srep31708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mastrandrea R, Squartini T, Fagiolo G, Garlaschelli D. Enhanced reconstruction of weighted networks from strengths and degrees. New J. Phys. 2014;16:043022. doi: 10.1088/1367-2630/16/4/043022. [DOI] [Google Scholar]
- 51.Yang Z, Perotti JI, Tessone CJ. Hierarchical benchmark graphs for testing community detection algorithms. Phys. Rev. E. 2017;96:052311. doi: 10.1103/PhysRevE.96.052311. [DOI] [PubMed] [Google Scholar]
- 52.Kumpula JM, Onnela J-P, Saramäki J, Kaski K, Kertész J. Emergence of communities in weighted networks. Phys. Rev. Lett. 2007;99:228701. doi: 10.1103/PhysRevLett.99.228701. [DOI] [PubMed] [Google Scholar]
- 53.Bianconi G, Darst RK, Iacovacci J, Fortunato S. Triadic closure as a basic generating mechanism of communities in complex networks. Phys. Rev. E. 2014;90:042806. doi: 10.1103/PhysRevE.90.042806. [DOI] [PubMed] [Google Scholar]
- 54.Shang, K.-k., Yang, B., Moore, J. M., Ji, Q. & Small, M. Growing networks with communities: a distributive link model. Chaos: An Interdiscip. J. Nonlinear Sci.30, 041101, 10.1063/5.0007422 (2020). [DOI] [PMC free article] [PubMed]
- 55.McAuley, J. & Leskovec, J. Learning to discover social circles in ego networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems—Volume 1, NIPS’12, 539–547 (Curran Associates Inc., Red Hook, NY, USA, 2012).
- 56.Rossi, R. A. & Ahmed, N. K. The network data repository with interactive graph analytics and visualization. In Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence, AAAI’15, 4292–4293 (AAAI Press, 2015).
- 57.Traud AL, Mucha PJ, Porter MA. Social structure of Facebook networks. Phys. A. 2012;391:4165–4180. doi: 10.1016/j.physa.2011.12.021. [DOI] [Google Scholar]
- 58.Traud AL, Kelsic ED, Mucha PJ, Porter MA. Comparing community structure to characteristics in online collegiate social networks. SIAM Rev. 2011;53:526–543. doi: 10.1137/080734315. [DOI] [Google Scholar]
- 59.Kunegis, J. KONECT – The Koblenz Network Collection. In Proceedings of the International Conference on World Wide Web Companion, 1343–1350 (2013).
- 60.Newman, M. E. J. Network data repository: Political blogs dataset. http://www-personal.umich.edu/~mejn/netdata/. Accessed on 01/10/2019.
- 61.The OpenFlights database. https://openflights.org/data.html. Accessed on 01/10/2019.
- 62.Cookpad: Make everyday cooking fun! https://cookpad.com/. Accessed on 01/10/2019.
- 63.Kitsak M, et al. Identification of influential spreaders in complex networks. Nat. Phys. 2010;6:888–893. doi: 10.1038/nphys1746. [DOI] [Google Scholar]
- 64.Bollobás, B. Modern graph theory. Graduate Texts in Mathematics 184 (Springer-Verlag New York, 1998).
- 65.Fagin R, Kumar R, Sivakumar D. Comparing top k lists. SIAM J. Discrete Math. 2003;17:134–160. doi: 10.1137/S0895480102412856. [DOI] [Google Scholar]
- 66.McCown, F. & Nelson, M. L. Agreeing to disagree: search engines and their public interfaces. In Proceedings of the 7th ACM/IEEE-CS Joint Conference on Digital Libraries, 309–318, 10.1145/1255175.1255237 (2007).
- 67.Holland PW, Laskey KB, Leinhardt S. Stochastic blockmodels: first steps. Soc. Netw. 1983;5:109–137. doi: 10.1016/0378-8733(83)90021-7. [DOI] [Google Scholar]
- 68.Peixoto TP. Nonparametric bayesian inference of the microcanonical stochastic block model. Phys. Rev. E. 2017;95:012317. doi: 10.1103/PhysRevE.95.012317. [DOI] [PubMed] [Google Scholar]
- 69.Peixoto TP. The graph-tool python library. figshare. 2014 doi: 10.6084/m9.figshare.1164194. [DOI] [Google Scholar]
- 70.Oliphant, T. Guide to NumPy (Trelgol Publishing, 2006).
- 71.van der Walt S, Colbert SC, Varoquaux G. The numpy array: a structure for efficient numerical computation. Comput. Sci. Eng. 2011;13:22–30. doi: 10.1109/MCSE.2011.37. [DOI] [Google Scholar]
- 72.Hagberg, A. A., Schult, D. A. & Swart, P. J. Exploring network structure, dynamics, and function using networkx. In Proceedings of the 7th Python in Science Conference (eds. Varoquaux, G., Vaught, T. & Millman, J.) 11 – 15 (Pasadena, CA USA, 2008).
- 73.Hunter JD. Matplotlib: a 2d graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 74.Stanford Network Analysis Project (SNAP): “social circles: Facebook” dataset. http://snap.stanford.edu/data/ego-Facebook.html. Accessed on 01/10/2019.
- 75.Network Repository: “American75” dataset. http://networkrepository.com/socfb-American75.php. Accessed on 01/10/2019.
- 76.Network Repository: “Amherst41” dataset. http://networkrepository.com/socfb-Amherst41.php. Accessed on 01/10/2019.
- 77.Network Repository: “Cal65” dataset. http://networkrepository.com/socfb-Cal65.php. Accessed on 01/10/2019.
- 78.Network Repository: “FSU53” dataset. http://networkrepository.com/socfb-FSU53.php. Accessed on 01/10/2019.
- 79.Stanford Network Analysis Project (SNAP): “social circles: Twitter” dataset. http://snap.stanford.edu/data/ego-Twitter.html. Accessed on 01/10/2019.
- 80.Adamic, L. A. & Glance, N. The political blogosphere and the 2004 us election: divided they blog. In Proceedings of the 3rd International Workshop on Link discovery, 36–43, 10.1145/1134271.1134277 (ACM, 2005).
- 81.Stanford Network Analysis Project (SNAP): “email-EU-core network” dataset. http://snap.stanford.edu/data/email-Eu-core.html. Accessed on 01/10/2019.
- 82.Yin, H., Benson, A. R., Leskovec, J. & Gleich, D. F. Local higher-order graph clustering. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 555–564, 10.1145/3097983.3098069 (ACM, 2017). [DOI] [PMC free article] [PubMed]
- 83.Leskovec J, Kleinberg J, Faloutsos C. Graph evolution: densification and shrinking diameters. ACM Trans. Knowl. Discov. Data (TKDD) 2017;1:2. doi: 10.1145/1217299.1217301. [DOI] [Google Scholar]
- 84.Stanford Network Analysis Project (SNAP): “Condensed Matter collaboration network” dataset. https://snap.stanford.edu/data/ca-CondMat.html. Accessed on 01/10/2019.
- 85.The KONECT Project: DBLP co-authorship network dataset. http://konect.cc/networks/com-dblp (2017). Accessed on 01/10/2019.
- 86.Yang, J. & Leskovec, J. Defining and evaluating network communities based on ground-truth. In Proceedings of the ACM SIGKDD Workshop on Min. Data Semant., 3, 10.1007/s10115-013-0693-z (2012).
- 87.The KONECT Project: WordNet network dataset. http://konect.cc/networks/wordnet-words (2017). Accessed on 01/10/2019.
- 88.Fellbaum C, editor. WordNet: an Electronic Lexical Database. Cambridge: MIT Press; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets on Cookpad™analysed in the current study are not publicly available due to exclusive ownership of Cookpad Limited. All the other data sets are available from the corresponding repositories listed in the bibliography.