Abstract
A prevalent developmental mechanism for the assignment of cell identities is the production of spatiotemporal concentration gradients of extracellular signaling molecules that are interpreted by the responding cells. One of such signaling systems is the Shh gradient that controls neuronal subtype identity in the ventral spinal cord. Using loss and gain of function approaches in chick and mouse embryos, we show here that the fibroblast growth factor (FGF) signaling pathway is required to restrict the domains of ventral gene expression as neuroepithelial cells become exposed to Shh during caudal extension of the embryo. FGF signaling activates the expression of the Shh receptor and negative pathway regulator Patched 2 (Ptch2) and therefore can enhance a negative feedback loop that restrains the activity of the pathway. Thus, we identify one of the mechanisms by which FGF signaling acts as a modulator of the onset of Shh signaling activity in the context of coordination of ventral patterning and caudal axis extension.
Keywords: chick embryo, mouse embryo, neural tube, gene regulatory network, FGF, Shh, Ptch
INTRODUCTION
Development of the nervous system involves the control of proliferation, cell cycle exit and cell fate assignment which depend on the position of neural progenitors and neurons at their birth time. The spinal cord of amniotes derives from a caudal primordium located in the vicinity of the node, which regresses with the primitive streak, progressively leaving in its wake spinal cord cells (Brown and Storey, 2000; Mathis et al., 2001). Although the signaling pathways important for cell fate assignment along the dorso-ventral axis and for caudal extension of the spinal cord are known, their interactions and temporal control have not been fully elucidated yet.
Ventral patterning genes encode transcription factors (i.e. FoxA2, Nkx2.2, Nkx6.1, and Olig2) that, in combination, allow the specification of motor neurons and different types of interneurons in a spatially and temporally ordered way (Briscoe et al., 2000). The restricted expression pattern of these genes along the dorso-ventral axis depends on Sonic Hedgehog (Shh) signaling that is activated in a ventral to dorsal gradient in spinal cord progenitor cells exposed to Shh, emanating from the notochord and the floor plate (Jessell, 2000). The core Shh signaling pathway involves the receptors Patched 1 and Patched 2 (Ptch1, Ptch2) which, in the absence of Shh, block the activity of the transmembrane protein Smoothened (Smo). With Smo active, an intracellular cascade leads to the accumulation of activator forms of Gli transcription factors and the reduction of Gli repressive forms that, in turn, act on appropriate target genes (Dessaud et al., 2008).
The strict control of Shh concentration, timing of exposure and response is achieved, in part, by several positive and negative feedback loops, as well as by modulators of the pathway (Dessaud et al., 2008; Ribes and Briscoe, 2009). One important negative feedback loop results from the ability of Shh to activate transcription of Ptch1 and Ptch2 which, in turn, can limit Smo function and Shh spreading across the tissue (Goodrich et al., 1996, 1997; Marigo and Tabin, 1996; Pearse et al., 2001; Jeong and McMahon, 2005; Vokes et al., 2007; Holtz et al., 2013; Alfaro et al., 2014). Thus, for instance, overexpression of Ptch1 or Ptch2 in the chick spinal cord downregulates expression of Nkx6.1, which is one of the factors induced by Shh (Holtz et al., 2013).
Neural progenitors become exposed to Shh from the notochord when they leave the spinal cord primordium region. However, the expression of ventral specification transcription factors that require Shh signaling only starts as cells progress into the neural tube flanked by somites (Diez del Corral et al., 2003; Novitch et al., 2003). This delay has been related to changes in FGF signaling (Diez del Corral et al., 2003; Novitch et al., 2003), which is specifically required for the maintenance of cells within the spinal cord primordium (Mathis et al., 2001). Downregulation of FGF signaling is critical for the transition from an immature spinal cord primordium to a more mature spinal cord state where neuronal differentiation (Diez del Corral et al., 2002), neural crest cell specification and emigration (Martinez-Morales et al., 2011) and expression of a number of ventral patterning genes can occur (Bertrand et al., 2000; Diez del Corral et al., 2003; Novitch et al., 2003).
Here, we demonstrate that FGF signaling is required for the proper dorso-ventral restriction of progenitor domains and for the expression of the Shh receptor and pathway inhibitor Ptch2. This regulation of a negative feedback loop at the core of the Shh pathway by FGF signaling would modulate the onset of Shh signaling in the ventral spinal cord.
MATERIALS AND METHODS
DNA Constructs
PCIGs (pCAGGS-IRES–nuclear EGFP (Megason and McMahon, 2002) or pCAGGS-IRES–EGFP) were used as expression vectors, unless stated otherwise, and were the control DNA in electroporation experiments. DN-FGFR encodes a truncated chick FGFR1c (aa 1–425) provided by C. Weijer (University of Dundee, Dundee, Scotland, UK; Yang et al., 2002). Mouse Etv4 was obtained from C. Tabin (Brent and Tabin, 2004). Human GliA encodes a truncated version of Gli3 (amino acids 468–1580 of hGli3) that behaves as a constitutively activator form of Gli targets (Stamataki et al., 2005). Mouse FGF8b was obtained from T. Schimmang (Dominguez-Frutos et al., 2009).
Mouse Ptch2 was obtained by PCR amplification of mouse cDNA and ligated in frame to Cherry at its 5′ end and the HA-epitope at its 3′ end and inserted in the PCAGGS expression vector. For the luciferase reporter construct, a fragment from genomic chick Ptch2 was obtained by PCR amplification with primers (FWD: TTTAAGCTTTCTCGGTCGGTTG TGG and REV: TTTGTGACAGGGATGGCTGCGGA CA) and cloned in PGL3-promoter.
In Ovo Electroporation
Chick embryos were electroporated with purified plasmid DNA (1–2 μg/μL) as previously described (Martinez-Morales et al., 2011) and they were analysed 24 h postelectroporation.
Explant Cultures and Pharmacological Treatments
Explants were dissected from HH9–13 chicken embryos. Three different types of explants including the region of the neural plate or tube adjacent to presomitic mesoderm were isolated (as depicted in Fig. 4): (1) pNT explants correspond to neural tissue caudal to the last formed somite and rostral to the node [Figs. 2(A) and 4(K), and Supporting information Fig. (2)], (2) one side pNT without floor plate is a similar explant containing only one hemitube and devoid of the floor plate region [Fig. 4(F)], and (3) pNT explants with mesoderm, endoderm and node extend more caudally to include the node and adjacent tissue [Fig. 4(A)]. For neural only explants, mesoderm was removed with trypsin treatment (Diez del Corral et al., 2002). Explants were cultured in collagen beds as previously described (Martinez-Morales et al., 2011) in medium containing either BSA only, 330 to 660 ng/mL of human FGF4 (Sigma) or mouse FGF8b (Sigma) (both supplemented with 1–10 ng/μL heparin) or containing 10 μM PD184352 (Axon Medchem), 10 μM PD173074 (Sigma), 1 to 10 μM SANT1 (Calbiochem), 0.1% DMSO, or combinations of the different compounds as indicated.
Figure 4.
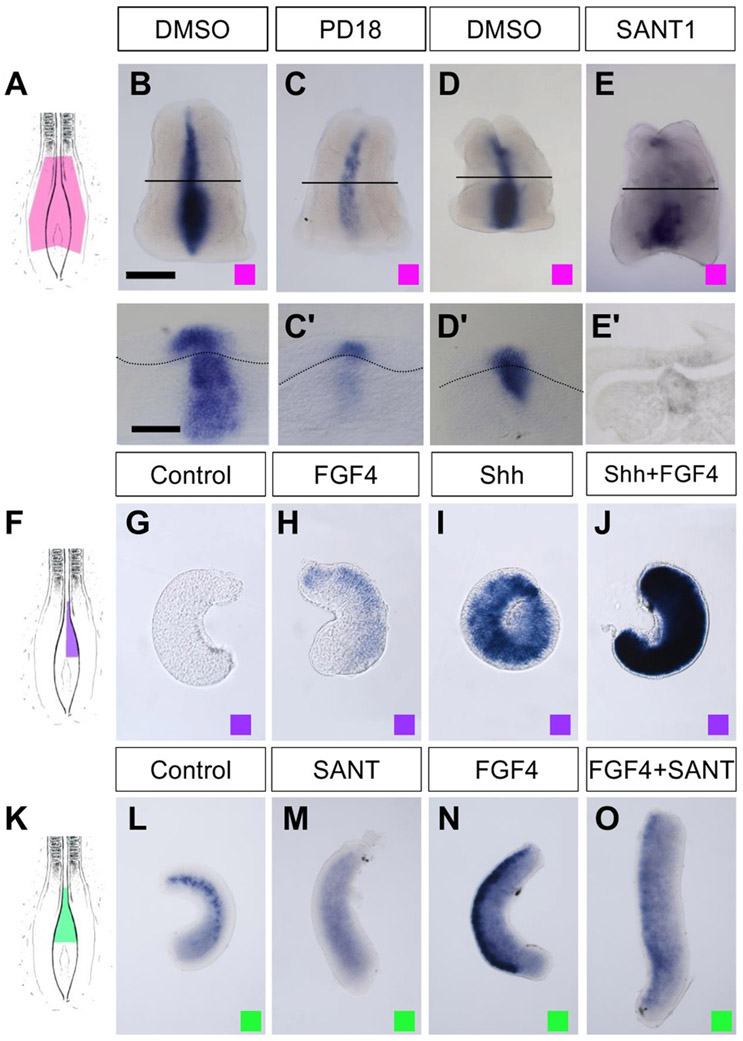
FGF and Shh signaling control the onset of Ptch2 expression. (A, F, K) Diagrams showing the regions used for explant cultures in the experiments shown in (B–E, G–J, L–O) as indicated with colour coded squares. Explants include either three germinal layers and node (A) or only ectoderm (F and K). (B–E) Expression of Ptch2 in chick explants cultured for 4 h in control conditions (B, D) and in the presence of PD184352 (C) or SANT-1 (E). (B′–E′) are transverse sections. (G–J) Expression of Ptch2 in chick neural explants devoid of floor plate cultured for 4 h in control conditions (G) or in the presence of the indicated factor (H–J). (L–O) Expression of Ptch2 in chick neural explants cultured for 4 h in control conditions (L) or in the presence of the indicated molecule (M–O). Scale bars: 200 μm in B (for B–E, G–J, and L–O) and 100 μm in B′ for (B′–E′).
Figure 2.
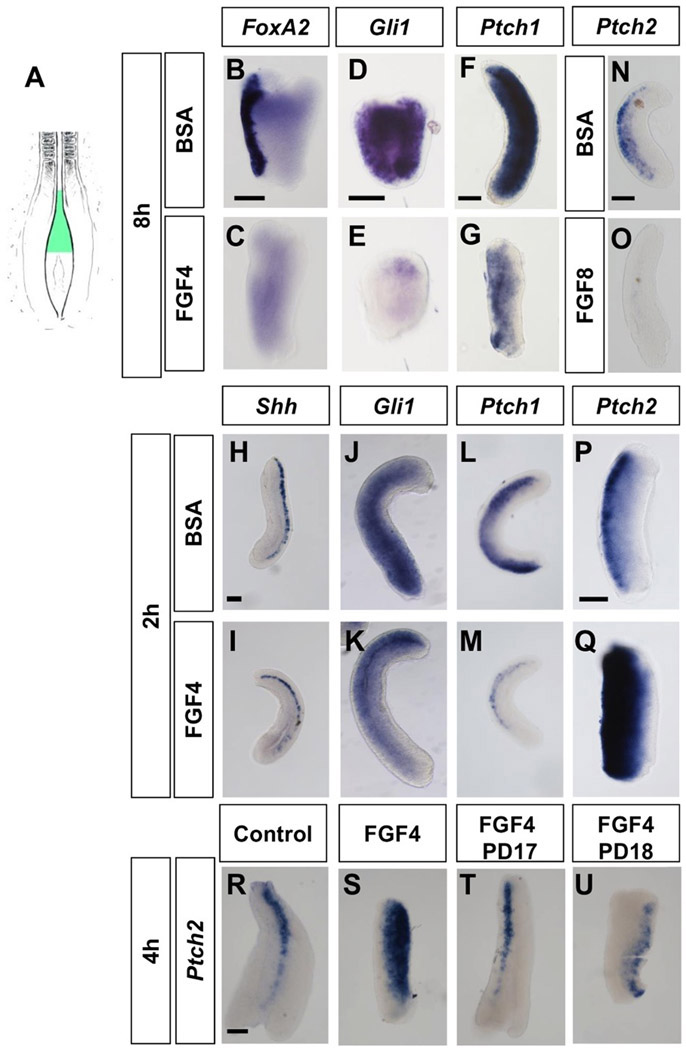
FGF signaling upregulates Ptch2 expression through the FGFR-MAPK pathway. (A) Diagram showing the ectoderm region used for the chick neural explant cultures shown in (B–U). (B–G) Expression of the Shh downstream targets FoxA2, Gli1 and Ptch1 in explants cultured for 8 h either in control conditions (B, D, F) or in the presence of FGF4 (C, E, G). (H–M) Expression of the Shh signaling components Shh, Gli1, Ptch1 in explants cultured for 2 h either in control conditions or in the presence of FGF4 as indicated. (N, O) Expression of Ptch2 in explants cultured for 8 h either in control conditions (N) or in the presence of FGF8 (O). (P, Q) Expression of Ptch2 in explants culture for 2 h either in control (P) or in the presence of FGF8 (Q). (R–U) Expression of Ptch2 in explants cultured for 4 h in control conditions (R) or the presence of FGF4 (S), FGF4 and the FGFR antagonist PD173074 (T) or FGF4 and the MEK antagonist PD184352 (U). Scale bars: 100 μm (in B for B–C, in D for D-E., in F for F–G, in H for H–M, in N for N–O, in P for P–Q, and in R for R–U).
Down-regulation of the FGF signaling target Sprouty2 in node region explants (Minowada et al., 1999) and of Pax6 in neural explants (Diez del Corral et al., 2003) were monitored to confirm down-regulation or activation of FGF signaling, respectively.
Mouse Mutants
Female mice homozygous for a floxed allele of Fgfr1 (Xu et al., 2002) were crossed to males homozygous for TCre (Perantoni et al., 2005) and heterozygous for the Fgfr1 null allele (the Cre- mediated derivative of the floxed allele). The progeny of this cross are 50% mutant (TCre; Fgfr1flox/null) and 50% control (T-Cre; Fgfr1flox/+). The first few litters were identified by PCR-genotyping, and after that via phenotyping as the abnormal caudal extension defect was obvious (Wahl et al., 2007).
In Situ Hybridization
Embryos were fixed overnight at 4°C and in situ hybridisation with digoxigenin labelled probes was performed following standard methods. Embryos were embedded in 5% agarose/10% sucrose and sectioned on a vibratome (40 μm; VT1000S; Leica). Chick Smo probe was synthesized from EST clone pgn1c.pk009.m19 (Carre et al., 2006) (University of Delaware collection) and mouse Ptch2 probe corresponds to a 2440 bp fragment (nucleotide 1110 to 3549 of NM_008958) which is longer than the probe previously described in Motoyama et al. (1998) and Takabatake et al. (1997) corresponding to a 766 bp cDNA fragment (AB000847). Other probes used have been described in the literature: chick FoxA2, Gli1, Gli2, Gli3, Ptch1, Ptch2, Shh, and Spry2.
Immunohistochemistry
Embryos were fixed for 2 to 4 h at 4°C with 4% PFA in PBS, and they were immersed in 30% sucrose solution, embedded in 7.5% gelatin/15% sucrose and sectioned on a cryostat (15 μm; CM1900; Leica). For immunohistochemistry, 15 μm cryostat were permeabilised with 0.5% Triton X-100, blocked with 10% FBS, and incubated overnight at 4°C with the primary antibody. After washing, the cryostat sections were incubated for 1 h with secondary antibodies. Polyclonal primary antibodies against Olig2 (Millipore: AB9610) were used. Monoclonal antibodies against FoxA2 (4C7), Nkx2.2 (74.5A5), Nkx6.1 (F55A10), and Shh (5E1) were all obtained from the Developmental Studies Hybridoma Bank (developed under the auspices of the National Institute of Child Health and Development and maintained by the University of Iowa). Alexa Fluor 488, Alexa Fluor 647, and Cy3-conjugated antimouse or rabbit secondary antibodies (Invitrogen and Jackson Immunoresearch) were used for detection and nuclei were stained with bisbenzimide. Sections were mounted in Mowiol and photographed using a confocal scanner microscope (SP5; Leica). Cell counting was carried out in four to eight sections of three to six different embryos from each experimental condition.
Analysis of Mouse Mutant Embryos
In order to identify alterations in the onset of ventral patterning we analyzed transverse sections of the caudal regions of mouse embryos. As somites in the TCre; Fgfr1flox/null embryos were not correctly formed or were misshaped (Wahl et al., 2007), we could not use them as a reference of the rostrocaudal level and thus, focused our attention on the notochord. The caudal-most notochord appears as a dispersed group of cells, that aggregate in a large bulge and then becomes rod-shaped at more rostral presomitic regions. For each embryo we performed double immunohistochemistry and identified the most caudal sections that included the rod-shaped notochord, cells expressing Nkx6.1 or cells expressing Olig2. This allowed us to determine the number of sections and therefore the distance that separated them (each section is 15 μm thick). Data correspond to mean±SEM.
Luciferase Reporter Assay
Transcriptional activity assays were performed in embryos electroporated with the indicated expression DNA constructs, together with the luciferase reporter to be tested and two Renilla luciferase constructs carrying either the cytomegalovirus or the simian virus 40 promoter (Promega, Southampton, UK) for normalization. Luciferase activities were measured by the Dual Luciferase Reporter Assay System (Promega). Both sides of the tube were electroporated by changing the electrode polarity. Eggs were further incubated for 6 h and were assayed for EGFP expression in the neural tube and the electroporated region was dissected (only levels adjacent to paraxial mesoderm and last four somites where considered), frozen and processed for luciferase assays. Experiments were performed two times with four individual samples per treatment and measures were normalized to the same day mean value of control samples. Data were analysed using IBM SSPS software to perform one-way ANOVA followed by the Tamhane post hoc test for pairwise comparison when variances are not equal. Data correspond to mean±SEM.
Sequence Analysis, Comparison, and Motif Identification
Available chick genomic sequences at public databases (UCSC genome browser galGal4 assembly) present gaps at the predicted 5′ end and intron 2 of the Ptch2 gene. We have PCR amplified and sequenced genomic DNA corresponding to the gaps using primers from the flanking regions (Genebank accession numbers: KJ433682 and KJ433683) to reconstitute the complete Ptch2 genomic DNA sequence.
Genomic regions of Ptch2 from different species were compared with the ERC browser (http://ecrbrowser.dcode.org/) to identify conserved regions that were subsequently analysed to identify possible Gli and ETS binding sites. For GBS, we searched for the sequence TGGGTGGTC with one mismatch allowed, (Kinzler and Vogelstein, 1990; Hallikas et al., 2006; Vokes et al., 2007) and for ETS consensus we searched for GGA(A/T) (Wei et al., 2010).
RESULTS
FGF Signaling is Required for Correct Ventral Patterning
In order to determine whether FGF signaling is required for correct ventral patterning of the neural tube, we electroporated a dominant-negative form of the FGF receptor 1 (DN-FGFR1-pCIG) (Martinez-Morales et al., 2011) in the caudal presomitic neural tube (pNT) of Hamburger and Hamilton (HH) stages 11-13 chick embryos and analysed the expression of ventral neural identity markers (Briscoe et al., 2000; Novitch et al., 2001; Ribes et al., 2010) after 20 h.
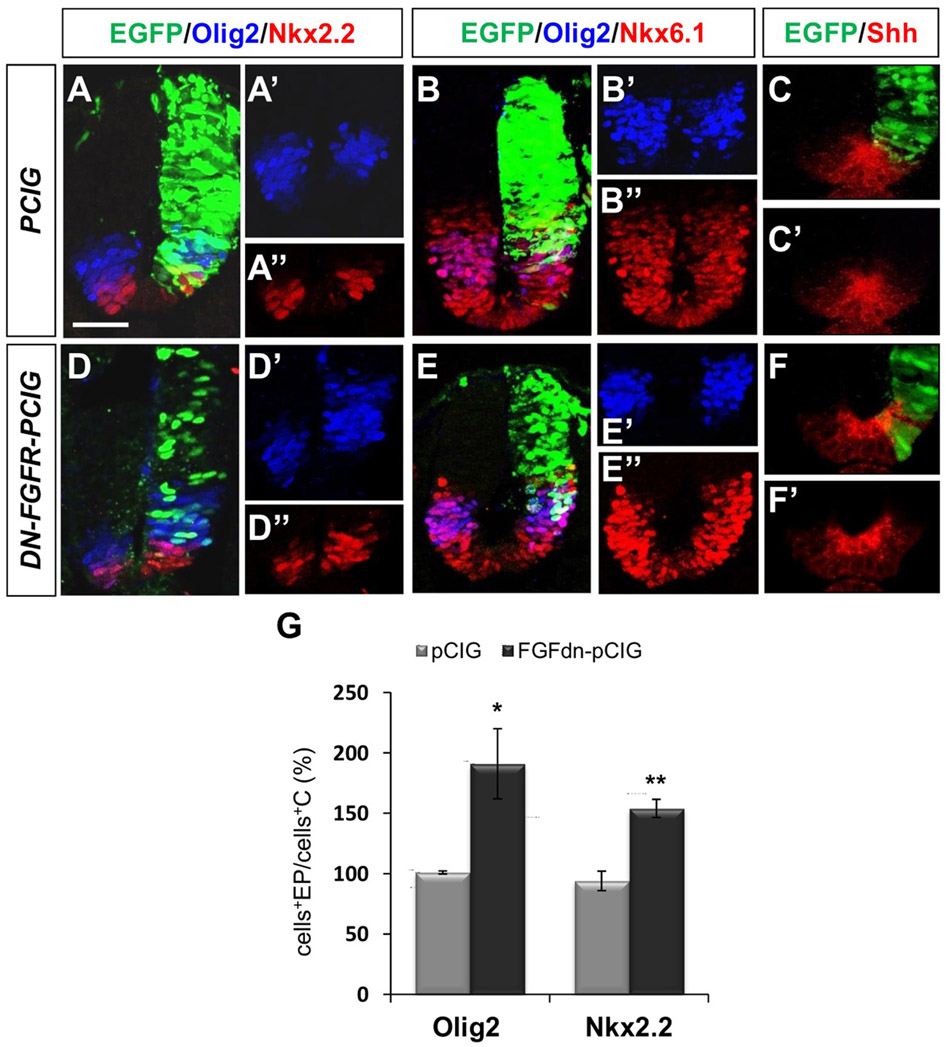
Blockade of FGF signaling caused a broadening and dorsal shifting of the domains of the motor neuron and ventral interneuron type 3 progenitors, identified by Olig2 and Nkx2.2, respectively, when compared with the non-electroporated hemitube [Fig. 1(D,G)]. The Nkx6.1 expression domain (from progenitors of ventral interneuron type 2 to floor plate) was only slightly increased [n = 4/6; Fig. 1(E)] and no clear change in the extent of the FoxA2 expressing domain (floor plate) was observed in comparison with the non electroporated side (n = 5/6; Supporting information Fig. 1). By contrast, no side to side differences were observed in embryos electroporated with the control expression vector (pCIG) for any of the markers [Fig. 1(A,B,G)].
Figure 1.
FGF signaling is required for proper ventral patterning specification. (A–C) Control (PCIG) or (D–F) DN-FGFR electroporated neural tube sections showing the electroporated region in green (EGFP) and immunostaining for Olig2, Nkx2.2, Nkx6.1, and Shh as indicated. (A′–F′, A″–B″, D″–E″) Non-EGFP channel images. (G) Quantitation of the number of Olig2 and Nkx2.2 expressing cells in PCIG (n = 6 and 3, respectively) and DN-FGFR (n = 5 and 4, respectively) electroporated hemitubes with respect to corresponding control hemitubes. Each bar represents mean±SEM (see methods). Scale bar: 30 μm. *p < 0.05 and ** p < 0.01 (Student’s t-test).
As Nkx.2.2, Nkx6.1, and Olig2 expression depends on the Shh signal (Briscoe et al., 2000; Cai et al., 2000; Lu et al., 2000), changes in their expression domains could be due to an increase in Shh expression. However, there were no apparent changes in Shh levels following electroporation of DN-FGFR1 [n = 4/4; Fig. 1(F)] compared with control pCIG electroporated embryos [n = 4/4; Fig. 1(C)] by immunostaining and also by Western blot analysis of the dissected neural tube (without notochord) after both-side-electroporation (see Supporting Information Materials and Methods and Supporting Information Fig. 2). These data indicate that a decrease in FGF signaling can alter the ventral pattern, without changing noticeably the amount of Shh produced by floor plate cells.
FGF Promotes Transient Induction of the Shh Signaling Modulator Ptch2
The effect of FGF on ventral patterning was further examined in pNT explants [Fig. 2(A)]. Exposure to FGF4 for 8 h has been shown to impair the expression of ventral patterning genes such as Nkx6.1 (Diez del Corral et al., 2003) and we extended this analysis to examine FoxA2, another neural target of the pathway, and generic readouts such as Gli1 (Marigo and Tabin, 1996) and Ptch1 (Lee et al., 1997; Vokes et al., 2007). A clear down-regulation of FoxA2 (n = 8; Fig. 2(C)] and both Gli1 [n = 11; Fig. 2(E)] and Ptch1 [n = 7; Fig. 2(G)] expression was observed in the presence of FGF4 in comparison with BSA treated explants [n = 7, 11, and 8, respectively; Fig. 2(B,D,F)], consistent with a decrease in Shh signaling pathway activity in the presence of FGF. These results indicate that the Shh pathway is susceptible of attenuation by FGF in the presomitic neural tube, a region in which key components of the pathway (Smo, Ptch1 and Ptch2, and Gli1-3) are present [Supporting Information Fig. 3(A-G)] (Marigo and Tabin, 1996; Quirk et al., 1997; Pearse et al., 2001; Diez del Corral et al., 2003; Novitch et al., 2003).
To determine possible immediate changes in the expression of core Shh components, we analysed pNT explants exposed to FGF4 for 2 or 4 h (Fig. 2 and Supporting Information Fig. 4). Short-term exposure (2 h and 4 h) did not affect Shh expression [n = 4 for each time period; Fig. 2(I); Supporting Information Fig. 4(C)] in comparison with BSA treated explants [n = 4 for each time period; Fig. 2(H) and Supporting Information Fig. 4(B)]. By contrast, a similar treatment reduced Gli1 [n ≥ 4 for each time period; Fig. 2(K) and Supporting Information Fig. 4(E)] and more dramatically Ptch1 levels [n ≥ 4; Fig. 2(M) and Supporting Information Fig. 4(G)] with respect to BSA treated explants [(n ≥ 4 for each time period; Fig. 2(J, L) and Supporting information Fig. 4(D,F)]. However, the observed Ptch1 and Gli1 reductions in chicken explants are more likely consequences and not causes of the Shh pathway attenuation (Marigo and Tabin, 1996; Lee et al., 1997; Vokes et al., 2007) as Gli1 mutant embryos do not exhibit ventral patterning defects (Park et al., 2000) and the Ptch1 mutation is associated with an expansion and not a reduction of ventral gene expression (Goodrich et al., 1997).
The expression of the other Shh receptor Ptch2 [highly present in the caudal pNT; Supporting information Fig. 3(D)] displayed instead the most interesting response. It has been previously shown that Ptch2 expression is down-regulated in explants after 8 h culture in the presence of FGF4 (Diez del Corral et al., 2003) and we have also confirmed this down-regulation after 8 h treatment with FGF8 (n = 10; Fig. 2(N,O)]). However, Ptch2 was strongly upregulated after 2 h and 4 h in the presence of either FGF4 or FGF8 [n ≥ 7 for each time period and treatment; Fig. 2(P,Q), Supporting Information Fig. 4(H-K)]. Moreover, this up-regulation was dependent on the FGFR-MAPK pathway as Ptch2 levels did not increase in explants treated with FGF in the presence of either the FGFR antagonist PD173074 or the MAPK kinases MEK1 and MEK5 antagonist PD184352 (Mohammadi et al., 1998; Sebolt-Leopold et al., 1999) [n = 3 to 4 for each treatment, Fig. 2(R-U)]. These results indicate that Ptch2 is an early transcriptional response target of FGF signaling which may be responsible for the FGF mediated repression of the Shh pathway and ventral patterning genes.
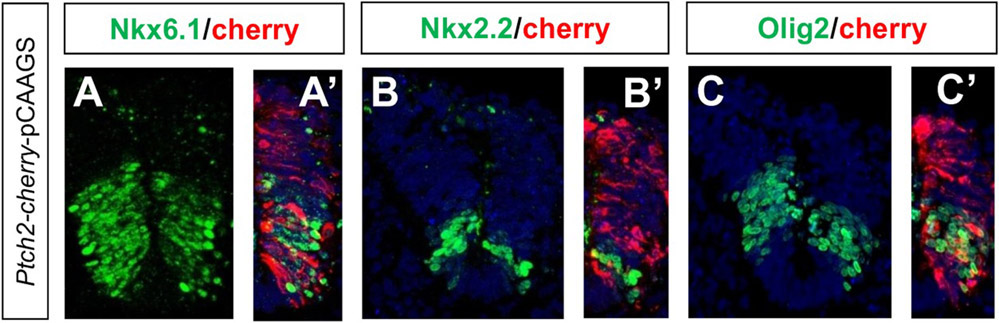
Overexpression of Ptch2 in the chick spinal cord has been shown to affect the expression of Shh responsive genes, down-regulating Nkx6.1 and upregulating Pax7 (Holtz et al., 2013). We have electroporated mouse Ptch2 in the chick spinal cord and confirm a clear reduction of Nkx6.1 [n = 5/5; Fig. 3(A-A′)] and show a reduction of the more ventral progenitor domains identified by Nkx2.2 [n = 3/3; Fig. 3(B-B′)] and Olig2 [n = 4/5; Fig. 3(C-C’)], as expected for a Shh pathway inhibitor.
Figure 3.
Ptch2 overexpression reduces expression of ventral genes. (A–C) mPtch2-Cherry electroporated neural tube sections showing immunostaining for Nkx6.1, Nkx2.2, and Olig2 as indicated. (A’-C’) Merge images showing the cherry channel.
These results reveal the Shh pathway modulator Ptch2 as a possible mediator of FGF function in the control of neural tube ventral patterning.
FGF and Shh are Cooperatively Required to Activate Ptch2
To examine whether FGFR-MAPK signaling can not only promote Ptch2 expression but it is also necessary for its expression, explants containing the node and the surrounding neural plate, mesoderm and endoderm [Fig. 4(A)] were treated with the MAPK kinase antagonist PD184352. In this condition, Ptch2 expression was severely downregulated after 4 h in comparison with DMSO treated explants [n ≥ 12 for each treatment; Fig. 4(B,C)], supporting an important role for the FGF-MAPK pathway on the onset of Ptch2 expression.
Expression of Ptch2 in the midline of early embryos depends on Shh signaling (Pearse et al., 2001). Interestingly, Ptch2 expression in caudal tissue explants [Fig. 4(A)] was down-regulated in the presence of the blocker of Smo activation SANT-1 (Chen et al., 2002) [n ≥ 3 for each treatment; Fig. 4(D,E)], confirming that its expression requires Shh signaling at these later stages of development as well.
To examine whether the FGF and Shh pathways could act in concert to activate Ptch2 expression, we cultured neural tube explants without the floor plate [and therefore without a source of Shh; Fig. 4(F)] in the presence of FGF4 and Shh separately or in combination. After 4 h exposure to FGF4, Ptch2 expression was only slightly increased with respect to control BSA treated explants [n = 7 for each treatment; Fig. 4(G,H)], whereas Shh activated Ptch2 expression to moderate levels [n = 7; Fig. 4(I)]. By contrast, the levels of Ptch2 expression were dramatically increased in the presence of both Shh and FGF, suggesting a synergistic effect [n = 8; Fig. 4(J)].
The limited increase in Ptch2 expression with the FGF4 treatment in the absence of a Shh source suggested that FGF requires Shh signaling to promote efficiently Ptch2 transcription. As expected, Ptch2 mRNA levels in pNT explants that included the floor plate [Fig. 4(K)] were decreased upon 4 h exposure to the Smo blocker SANT-1 [n = 8 for each treatment; Fig. 4(L,M)] and its induction by FGF was blocked in the presence of the SANT-1 inhibitor [n = 8, Fig. 4(N,O)]. In summary, these results indicate that the strong caudal expression of Ptch2 requires the concerted action of both Shh and FGF signaling.
The Ptch2 Genomic Region Contains a Regulatory Element Responsive to Gli and Etv4 Factors Combination
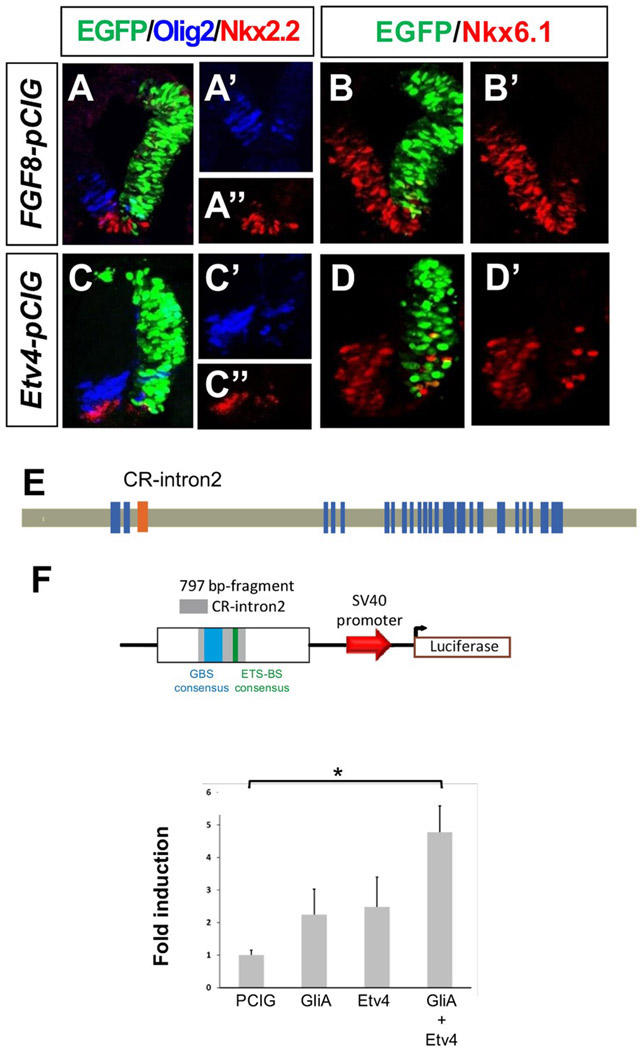
A number of MAPK dependent FGF signaling events are mediated by transcription factors of the ETS family such as Etv3, Etv4 and Etv5 (Raible and Brand, 2001; Roehl and Nusslein-Volhard, 2001; Brent and Tabin, 2004). To examine whether FGF action on Shh signaling could be mediated through these effectors, we electroporated either Fgf8b-pCIG or Etv4-pCIG and examined the expression of ventral genes. Both constructs caused a severe down-regulation of Nkx2.2, Olig2, and Nkx6.1 expression in the ventral neural tube 20 h postelectroporation [Fgf8: n = 5/6, 7/7, and 7/7, respectively; Etv4: n = 6/6, 7/7, and 7/7, respectively; Fig. 5(A-D)] whereas no changes were observed in control electroporated embryos [Fig. 1(A,B)]. In some Etv4-pCIG electroporated embryos, we also observed disorganization of dorsal cells accompanied with ectopic intermediate and dorsal activation of Olig2 and Nkx2 (n = 4/6; Supporting Information Fig. 5) but this may reflect the interaction of Etv4 with dorsal specific pathways and is not explored here.
Figure 5.
The Etv4 transcription factor mimics the effect of FGF8 on ventral patterning genes and in combination with GliA can activate a regulatory region from the Ptch2 locus. (A, B) FGF8 or (C-D) Etv4 electroporated neural tube sections showing the extent of electroporation in green (EGFP) and immunostaining for Olig2, Nkx2.2, Nkx6.1. (E) Diagram showing chick Ptch2 genomic structure (blue rectangles indicate exons) and highlighting the localization of the conserved region in intron 2 (CR-intron2) that includes a Gli binding site consensus sequence. (F) Quantitative analysis of the transcriptional activity of the 797 bp-fragment-luciferase plasmid (shown schematically above) following electroporation of PCIG, GliA, Etv4, or a combination of GliA and Etv4 expression plasmids, as indicated. Each bar represents mean±SEM (see methods). *p < 0.05 (ANOVA followed by Tamhane post hoc test).
These results, indicating that Etv4 may be one of the effectors of the FGF signal in the neural tube and the rapid activation of Ptch2 by the Shh and FGF pathways, suggest that Ptch2 transcription may be directly regulated by the combined activity of Gli and ETS factors. We thus searched the chick Ptch2 locus for the presence of conserved cis-regulatory sites that could respond to these transcription factors. Ptch2 regulatory regions controlled by Gli activity have been described in mouse (Vokes et al., 2007; Holtz et al., 2013) and in zebrafish (Wang et al., 2013). Comparison of the chick Ptch2 locus with that of other vertebrates revealed a conserved region (CR) with a putative Gli binding site (GBS) and a consensus ETS binding site [Fig. 5(E)] located in intron 2 [CR-intron2; present in fish (Wang et al., 2013), bird and reptile Ptch2 genes; Supporting Information Fig. 6(A)].
To determine whether this region may mediate the activation of Ptch2 transcription in response to combined FGF and Shh exposure, we examined its activity using a luciferase reporter vector where we inserted a 787 bp fragment containing CR-Intron 2, [Fig. 5(F)]. Embryos electroporated with a Gli activator (GliA) or with Etv4 did not change luciferase levels significantly (ANOVA followed by Tamhane posthoc test) with respect to control PCIG [Fig. 5(F)]. Interestingly, combined electroporation of GliA and Etv4 did increase the levels of luciferase activity driven by this fragment up to (4.8±0.8) fold with respect to control PCIG [Fig. 5(F)].
These results suggest that the genomic fragment located in the second intron of Ptch2 contains a cis-regulatory module that drives Ptch2 expression in response to combined Shh-Gli and FGF/Etv4 signaling. This region may therefore be responsible for the expression of Ptch2 in the transition zone region where both Shh and FGF signals coincide.
FGF Signaling is Required for Ptch2 Expression and Ventral Patterning in Mouse Embryos
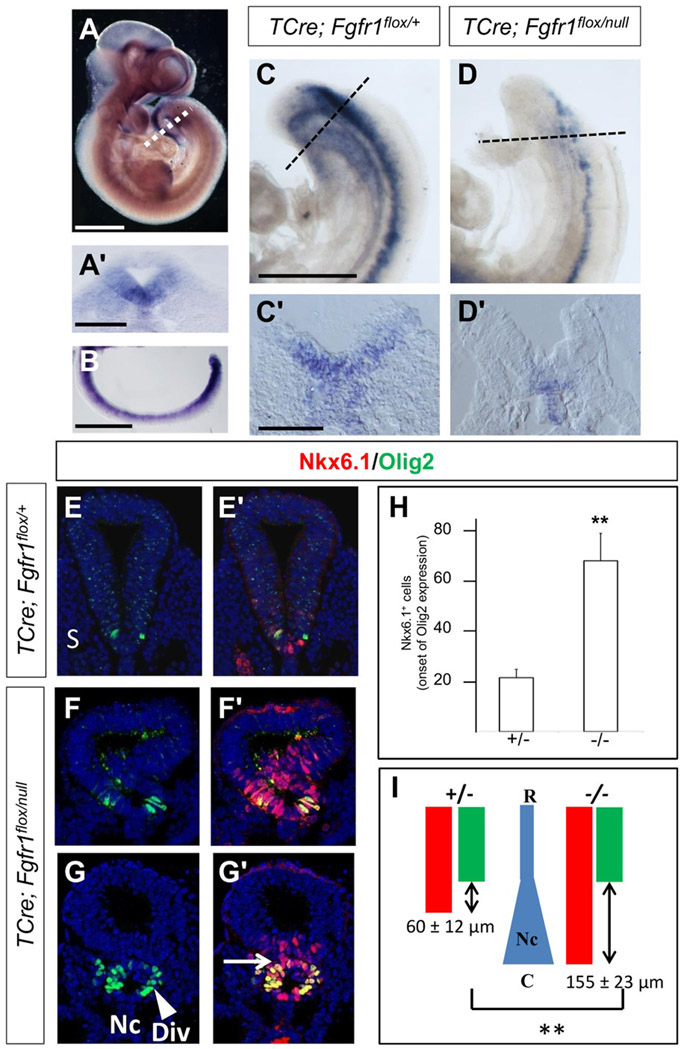
Caudal FGF signaling is also active in mammalian embryos suggesting that the regulation of Ptch2 expression by FGF signaling could be conserved among vertebrates. Previous reports have described only low levels of mouse Ptch2 mRNA expression in the developing ventral spinal cord (Motoyama et al., 1998) but with an improved Ptch2 in situ hybridization probe (see Methods), we observed strong gene expression in the ventral neural tube, especially at the level of the caudal neural tube at embryonic stages E9.5-E10.5 [Fig. 6(A,B)].
Figure 6.
FGF signaling is required for the onset of mouse Ptch2 expression and for the initial ventral patterning of the neural tube in mouse embryos. (A, A′, B) Expression of Ptch2 in stage E10.5 (A, A′) and E9.5 (B) mouse embryos in whole mount (A), in a transverse section (A′) at the level of the dotted line in (A) and in a dissected caudal neural tube (B). (C–C′, D–D′) Expression of Ptch2 in E9.5 TCre; Fgfr1flox/+ (C–C′) or in TCre; Fgfr1flox/null (D–D′) mouse embryos in whole mount (C, D) and in transverse sections (C′, D′). (E–E′, F–F′, G–G′) Transverse sections of E10.5 stage spinal cord of the indicated genotypes, showing the expression of Olig2 and Nkx6.1 at caudal levels. (E–E′, F–F′) Sections corresponding to the region where the first Olig2 expressing cells appear in a TCre; Fgfr1flox/+ embryo or in a TCre; Fgfr1flox/null embryo. (G–G′) Section showing the abnormal morphology of the neural tube of a TCre; Fgfr1flox/null embryo. (H) Quantification of the number of Nkx6.1+ cells at the spinal cord levels where the first Olig2 expressing cells appear in control and TCre; Fgfr1flox/null embryos. Each bar represents mean±SEM. (I) Diagram depicting the domains of Olig2 and Nkx6.1 along the rostral (R) to caudal (C) axis in TCre; Fgfr1flox/+ and TCre; Fgfr1flox/null embryos. The rostrocaudal distance separating the initiation of Olig2 and Nkx6.1 expression is shown with a double arrowed symbol and mean±SEM are indicated. The changing shape of the notochord is also depicted in blue. Nc: Notochord, S: somite, Div: diverticulum, arrow points at the separation of the tube and the diverticulum, R: Rostral, C: Caudal. Scale bars: 500 μm (A), 100 μm (A′), 500 μm (B), 500 μm (C for C and D), 100 μm (C′ for C′ and D′). ** p ≤ 0,01 (Student’s t test).
To address the possibility that FGF signaling could control Ptch2 expression in mouse embryos, we deleted Fgfr1 specifically in the caudal neural tissue and underlying mesoderm using a transgenic TCre line (Perantoni et al., 2005; Wahl et al., 2007). Stage E9.5-10 TCre; Fgfr1flox/null mutant embryos showed no or very low levels of Ptch2 expression in the caudal neural tube [n = 4/6; Fig. 6(D,D′)] in comparison with their control TCre; Fgfr1flox/+ siblings [n = 7; Fig. 6(C,C′)], indicating that FGF signaling is required for Ptch2 expression during mouse neural development.
At caudal levels of E10-10.5 mutant embryos, we observed a neuroepitelial diverticulum as a ventral invagination of the neural tube or even a separate tube between the neural tube and the notochord [n = 12/13; Fig. 6(F,G)], which was never observed in their control siblings [n = 0/7; Fig. 6(E)]. This diverticulum formed by ventral neural progenitors expressing Nkx6.1 [Fig. 6(F′,G′)] is reminiscent of the ectopic neural tube described for a conditional double Fgf4/Fgf8 mutant (Boulet and Capecchi, 2012) and may be related to changes in cell proliferation and morphogenesis or in mesoderm versus neural cell fate assignment and has not been further explored here.
To identify possible differences in ventral specification in mutant embryos with respect to controls, we analysed expression of Nkx6.1 and Olig2 focusing in the caudal spinal cord. Changes in ventral progenitor domains are difficult to assess in caudal regions as the borders of the domains are still dynamic and depend on the precise rostro-caudal level. However, the analysis of Nkx6.1 expression at a rostro-caudal level corresponding to onset of Olig2 expression (where only 2–11 Olig2+ cells are present) showed an enlarged Nkx6.1 domain in mutants [69.2±11.2 Nkx6.1+ cells; n = 9; Fig. 6(F′,G′H)] in comparison with controls [21.8±6.9 Nkx6.1+ cells; n = 5; Fig. 6(E′,H)]. These changes in the number of Nkx6.1+ neural progenitors are compatible with a broader initial activation of Shh signaling in the caudal neural tube owing to the reduced Ptch2 expression in the caudal neural tube of mutant embryos.
Furthermore, to examine whether precocious activation of Shh signaling occurred in the mutants, we measured the distance that separates the rostro-caudal level where the ventral markers Olig2 and Nkx6.1 first appear and the level at which the notochord displays its characteristic rod shape (see methods for measurement details). Olig2 expression started at the level of the rod-shaped notochord in both mutant (n = 6) and control embryos (n = 6). By contrast, Nkx6.1 expression started around 200±25 μm caudal to the beginning of the rod-shaped notochord in mutant embryos whereas this distance was only about 60±27 μm in controls. As a result, the distance that separates the onset of Nkx6.1 and Olig2 was 60±12 μm in control embryos (n = 5) and increased to 155±23 μm in mutants [n = 12; Fig. 6(I)]. These data suggest that the ventral marker Nkx6.1 is expressed precociously in Fgfr1 mutants.
Overall, these data support the conclusion that there is an evolutionary conservation in the control of the onset of ventral gene expression in the neural tube, governed by caudal FGF signaling regulation of Ptch2 expression.
DISCUSSION
FGF Signaling Promotes a Negative Feedback Loop of the Shh Pathway at the Onset of Ventral Patterning
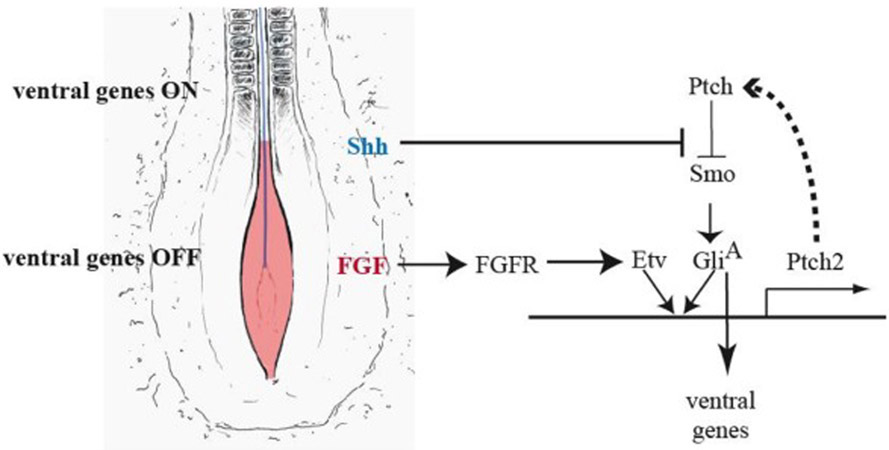
Ventral patterning is sustained by a complex gene regulatory network governed by Shh signaling and modulated by other signaling pathways. Here we have reported the novel observation that the FGF signaling pathway, acting at the initial stages of Shh signaling, is required for the proper restriction of ventral progenitor identity. We have also shown that FGF enhances the ability of Shh to promote the expression of Ptch2, a receptor and negative regulator of the Shh pathway (Briscoe et al., 2001; Holtz et al., 2013) and may thus attenuate the response to Shh and delay the onset of ventral patterning (summary in Fig. 7).
Figure 7.
Proposed model of FGF-mediated regulation of the Shh pathway in the spinal cord. Neural tube/plate cells adjacent to the presomitic mesoderm are exposed to FGF4/8 (red) and Shh (blue, produced by the underlying notochord). Shh triggers a gene regulatory network that activates downstream genes, including Ptch2, a component of a negative feedback loop of the pathway. In this region, FGF signaling further promotes Ptch2 expression and therefore reinforces the negative feedback loop in the Shh pathway, delaying the onset of ventral gene expression.
The mechanism of Shh signaling regulation by FGF described here reveals a specific feature of the Shh receptor gene Ptch2 which is considered as a modulator of the Shh pathway activity [(Holtz et al., 2013; Alfaro et al., 2014) and our results]. Similarly to Ptch1, Ptch2 interacts with Shh and Smo (Carpenter et al., 1998) and blocks, albeit less efficiently than Ptch1, the activation of a Gli reporter in vitro (Rahnama et al., 2004; Nieuwenhuis et al., 2006; Holtz et al., 2013). Whereas mutations in mouse Ptch1 result in a dramatic upregulation of Shh pathway activity (Goodrich et al., 1997), the requirement of Ptch2 in mouse has so far been revealed when the levels of Ptch1 are compromised (Lee et al., 2006; Nieuwenhuis et al., 2006; Holtz et al., 2013; Zhulyn et al., 2015) suggesting that its role may be at play within a particular time window or when developmental conditions are suboptimal.
As FGF8 production is tightly linked to the elongation process, it may serve as a sensor of caudal elongation and we propose that the FGF-Ptch2 network may precisely coordinate the onset of ventral patterning to axis extension. Our results, thus, place Ptch2 at the centre of the coordination of axial extension and ventral patterning initiation. They also suggest a role of Ptch2 in conferring robustness in patterning under fluctuations in the elongation process.
FGF Signaling Modulates Several Signaling Pathways during Dorsoventral Patterning
Here we have focused on the role of FGF as a factor that limits Shh signaling activity. However, our results showing stronger ventral patterning defects associated to FGF8 or Etv4 overexpression in comparison with Ptch2 overexpression indicate that Ptch2 is not the sole mediator of the effect of FGF signaling on ventral specification. Indeed, FGF also affects the onset of other signaling pathways involved in spinal cord patterning such as BMP and Wnt dorsally and retinoic acid (RA) throughout the DV axis (Diez del Corral et al., 2003; Novitch et al., 2003; Olivera-Martinez and Storey, 2007; Martinez-Morales et al., 2011; Sasai et al., 2014).
RA signaling is active at the level of the somites and is required for the expression of a subset of ventral specification genes (Diez del Corral et al., 2003; Olivera-Martinez and Storey, 2007). As FGF restrains RA signaling in the neural tube, the alterations due to changes in FGF could be indirectly attributed to changes in RA. However, that is not the case as not all genes affected by FGF are dependent on RA (Diez del Corral et al., 2003; Novitch et al., 2003) and furthermore, RA signaling is not changed caudally in the conditional Fgfr mutant used in our work (Wahl et al., 2007).
FGF can also downregulate expression of intermediate genes normally repressed by Shh such as Pax6 and Irx3 (Bertrand et al., 2000; Diez del Corral et al., 2003; Novitch et al., 2003) through a mechanism that involves chromatin compaction and changes in nuclear position of the Pax6 and Irx3 loci (Patel et al., 2013). As these intermediate patterning genes inhibit Olig2 and Nkx2.2, respectively (Briscoe et al., 2000; Novitch et al., 2001), the manipulation of FGF levels may also affect Olig2 and Nkx2.2 indirectly through an effect on intermediate genes. This may explain some discrepancies between our FGF results and previous data showing that FGF8 overexpression in chicken does not reduce Olig2 and Nkx2.2 levels (Novitch et al., 2003). We propose that the results of misexpression experiments may differ depending on temporal and space windows used for FGF8 overexpression, earlier and more caudal in our case. Our loss-of-function experiments reveal that FGF is required for the dorso-ventral restriction of Olig2 or Nkx2.2 domains but these domains would be further refined by the repression activities of Irx3 and Pax6 (Briscoe et al., 2000; Novitch et al., 2001).
In addition, transient exposure to FGF signaling has been recently shown to be important for floor plate specification (Sasai et al., 2014) and thus the overall picture that emerges from these results is that of a highly interconnected gene regulatory network that sustains dorso-ventral patterning, with FGF signaling acting as a crucial modulator of the initiation of Shh dependent ventral patterning as well as of intermediate and dorsal patterning including neural crest specification.
FGF Signaling as a Modulator of the Shh Pathway in Multiple Contexts
Although we have identified a previously unreported molecular mechanism for FGF-mediated interference with the Shh pathway in the spinal cord, other interactions between these two pathways have been described in other contexts during development (Riobo et al., 2006). In most of the described cases, however, they display cooperative and not antagonistic functions (Ye et al., 1998; Storm et al., 2006; Blaess et al., 2008). In a few cases, the regulation of Shh components by FGF has been identified and for example FGF can activate Gli2 expression in early Xenopus mesoderm patterning (Brewster et al., 2000). One interesting situation is found during limb development where both cooperative and antagonistic relations have been described. In this case, FGF is required for initiation and maintenance of Shh expression but it also downregulates Shh transcription through a limb specific enhancer (Lettice et al., 2003; Mao et al., 2009; Zhang et al., 2009; Lettice et al., 2012).
FGF and Shh are also implicated in the control of proliferation during development and in cancer. Antagonism of the two pathways has been suggested in pathological situations such as medulloblastoma cells produced in Ptch1+/− mutant mice in which bFGF acts as a potent inhibitor of proliferation (Fogarty et al., 2007; Emmenegger et al., 2012). Considering that a reduction of Ptch2 can exacerbate the formation of medulloblastomas (Lee et al., 2006), it is tempting to propose that one possible mechanism for the anti-proliferative activity of FGF in this context could be an increase in Ptch2 levels.
Finally, a mechanism of negative feedback loop control, as the one we describe here to tune the onset of a critical signaling such as the Shh pathway, may be a common strategy used in other gene regulatory networks in development or in tissue homeostasis to provide robustness at the initial stages of signaling.
Supplementary Material
Acknowledgments
The authors are grateful to K. Storey for comments on the manuscript and to J.M. Frade for support during the initial stages of the project. The authors thank E. Martí, S. Martinez, T. Oosterveen, L. Puelles, L. Sánchez-Arrones, T Schimmang, C. Tabin, and C. Weijer for DNA templates and for expression constructs. The authors are grateful to C. Bahamon, M. Ciorraga, M.C. Escudero, C. Oueslati, and O. Parreño for technical assistance. The authors thank J. Briscoe and N. Sasai for sharing results prior to publication.
Contract grant sponsor: Spanish government; contract grant numbers: BFU2014-57494-R, BFU2011-29490, BFU2010-15665, BFU2005-02972 (to A. M. and R. D.) and BFU2007-61774, BFU2010-16031-P (to P. B.).
Contract grant sponsor: Madrid Local Government; contract grant number: CCG07-CSIC/SAL-1930 (to R. D.).
Contract grant sponsor: CIBERER (to P.B.).
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Alfaro AC, Roberts B, Kwong L, Bijlsma MF, Roelink H. 2014. Ptch2 mediates the Shh response in Ptch1−/− cells. Development 141:3331–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Medevielle F, Pituello F. 2000. FGF signaling controls the timing of Pax6 activation in the neural tube. Development 127:4837–4843. [DOI] [PubMed] [Google Scholar]
- Blaess S, Stephen D, Joyner AL. 2008. Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development 135:2093–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet AM, Capecchi MR. 2012. Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev Biol 371:235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent AE, Tabin CJ. 2004. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development 131:3885–3896. [DOI] [PubMed] [Google Scholar]
- Brewster R, Mullor JL, Ruiz i Altaba A. 2000. Gli2 functions in FGF signaling during antero-posterior patterning. Development 127:4395–4405. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Chen Y, Jessell TM, Struhl G. 2001. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol Cell 7:1279–1291. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. 2000. A homeo-domain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101:435–445. [DOI] [PubMed] [Google Scholar]
- Brown JM, Storey KG. 2000. A region of the vertebrate neural plate in which neighbouring cells can adopt neural or epidermal cell fates. Curr Biol 10:869–872. [DOI] [PubMed] [Google Scholar]
- Cai J, Xu X, Yin H, Wu R, Modderman G, Chen Y, Jensen J, et al. 2000. Evidence for the differential regulation of Nkx-6.1 expression in the ventral spinal cord and foregut by Shh-dependent and -independent mechanisms. Genesis 27:6–11. [DOI] [PubMed] [Google Scholar]
- Carpenter D, Stone DM, Brush J, Ryan A, Armanini M, Frantz G, Rosenthal A, et al. 1998. Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc Natl Acad Sci USA 95:13630–13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre W, Wang X, Porter TE, Nys Y, Tang J, Bernberg E, Morgan R, et al. 2006. Chicken genomics resource: sequencing and annotation of 35,407 ESTs from single and multiple tissue cDNA libraries and CAP3 assembly of a chicken gene index. Physiol Genomics 25:514–524. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. 2002. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA 99:14071–14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. 2008. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development 135:2489–2503. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Breitkreuz DN, Storey KG. 2002. Onset of neuronal differentiation is regulated by paraxial mesoderm and requires attenuation of FGF signaling. Development 129:1681–1691. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey KG. 2003. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron 40:65–79. [DOI] [PubMed] [Google Scholar]
- Dominguez-Frutos E, Vendrell V, Alvarez Y, Zelarayan LC, Lopez-Hernandez I, Ros M, Schimmang T. 2009. Tissue-specific requirements for FGF8 during early inner ear development. Mech Dev 126:873–881. [DOI] [PubMed] [Google Scholar]
- Emmenegger BA, Hwang EI, Moore C, Markant SL, Brun SN, Dutton JW, Read TA, et al. 2012. Distinct roles for fibroblast growth factor signaling in cerebellar development and medulloblastoma. Oncogene 32:4181–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty MP, Emmenegger BA, Grasfeder LL, Oliver TG, Wechsler-Reya RJ. 2007. Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells. Proc Natl Acad Sci USA 104:2973–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. 1996. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev 10:301–312. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277:1109–1113. [DOI] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. 2006. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124:47–59. [DOI] [PubMed] [Google Scholar]
- Holtz AM, Peterson KA, Nishi Y, Morin S, Song JY, Charron F, McMahon AP, et al. 2013. Essential role for ligand-dependent feedback antagonism of vertebrate hedgehog signaling by PTCH1, PTCH2 and HHIP1 during neural patterning. Development 140:3423–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, McMahon AP. 2005. Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched 1 and Hhip1. Development 132:143–154. [DOI] [PubMed] [Google Scholar]
- Jessell TM. 2000. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet 1:20–29. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. 1990. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol 10:634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Platt KA, Censullo P, Ruiz i Altaba A. 1997. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 124:2537–2552. [DOI] [PubMed] [Google Scholar]
- Lee Y, Miller HL, Russell HR, Boyd K, Curran T, McKinnon PJ. 2006. Patched2 modulates tumorigenesis in patched1 heterozygous mice. Cancer Res 66:6964–6971. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, et al. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12:1725–1735. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Williamson I, Wiltshire JH, Peluso S, Devenney PS, Hill AE, Essafi A, et al. 2012. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev Cell 22:459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, et al. 2000. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25:317–329. [DOI] [PubMed] [Google Scholar]
- Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. 2009. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev Cell 16:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo V, Tabin CJ. 1996. Regulation of patched by sonic hedgehog in the developing neural tube. Proc Natl Acad Sci USA 93:9346–9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Morales PL, Diez Del Corral R, Olivera-Martinez I, Quiroga AC, Das RM, Barbas JA, Storey KG, et al. 2011. FGF and retinoic acid activity gradients control the timing of neural crest cell emigration in the trunk. J Cell Biol 194:489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis L, Kulesa PM, Fraser SE. 2001. FGF receptor signaling is required to maintain neural progenitors during Hensen’s node progression. Nat Cell Biol 3:559–566. [DOI] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. 2002. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development 129:2087–2098. [DOI] [PubMed] [Google Scholar]
- Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, et al. 1999. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development 126:4465–4475. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, Eliseenkova AV, et al. 1998. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J 17:5896–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama J, Heng H, Crackower MA, Takabatake T, Takeshima K, Tsui LC, Hui C. 1998. Overlapping and non-overlapping Ptch2 expression with Shh during mouse embryogenesis. Mech Dev 78:81–84. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis E, Motoyama J, Barnfield PC, Yoshikawa Y, Zhang X, Mo R, Crackower MA, et al. 2006. Mice with a targeted mutation of patched2 are viable but develop alopecia and epidermal hyperplasia. Mol Cell Biol 26:6609–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. 2001. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31:773–789. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Wichterle H, Jessell TM, Sockanathan S. 2003. A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron 40:81–95. [DOI] [PubMed] [Google Scholar]
- Olivera-Martinez I, Storey KG. 2007. Wnt signals provide a timing mechanism for the FGF-retinoid differentiation switch during vertebrate body axis extension. Development 134:2125–2135. [DOI] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, et al. 2000. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127:1593–1605. [DOI] [PubMed] [Google Scholar]
- Patel NS, Rhinn M, Semprich CI, Halley PA, Dolle P, Bickmore WA, Storey KG. 2013. FGF signaling regulates chromatin organisation during neural differentiation via mechanisms that can be uncoupled from transcription. PLoS Genet 9:e1003614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse RV II, Vogan KJ, Tabin CJ. 2001. Ptc1 and Ptc2 transcripts provide distinct readouts of Hedgehog signaling activity during chick embryogenesis. Dev Biol 239:15–29. [DOI] [PubMed] [Google Scholar]
- Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, et al. 2005. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development 132:3859–3871. [DOI] [PubMed] [Google Scholar]
- Quirk J, van den Heuvel M, Henrique D, Marigo V, Jones TA, Tabin C, Ingham PW. 1997. The smoothened gene and hedgehog signal transduction in Drosophila and vertebrate development. Cold Spring Harb Symp Quant Biol 62:217–226. [PubMed] [Google Scholar]
- Rahnama F, Toftgard R, Zaphiropoulos PG. 2004. Distinct roles of PTCH2 splice variants in Hedgehog signaling. Biochem J 378:325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F, Brand M. 2001. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech Dev 107:105–117. [DOI] [PubMed] [Google Scholar]
- Ribes V, Balaskas N, Sasai N, Cruz C, Dessaud E, Cayuso J, Tozer S, et al. 2010. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev 24:1186–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes V, Briscoe J. 2009. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb Perspect Biol 1:a002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riobo NA, Lu K, Emerson CP Jr. 2006. Hedgehog signal transduction: signal integration and cross talk in development and cancer. Cell Cycle 5:1612–1615. [DOI] [PubMed] [Google Scholar]
- Roehl H, Nusslein-Volhard C. 2001. Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol 11:503–507. [DOI] [PubMed] [Google Scholar]
- Sasai N, Kutejova E, Briscoe J. 2014. Integration of signals along orthogonal axes of the vertebrate neural tube controls progenitor competence and increases cell diversity. PLoS Biol 12:e1001907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebolt-Leopold JS, Dudley DT, Herrera R, Van Becelaere K, Wiland A, Gowan RC, Tecle H, et al. 1999. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat Med 5:810–816. [DOI] [PubMed] [Google Scholar]
- Stamataki D, Ulloa F, Tsoni SV, Mynett A, Briscoe J. 2005. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev 19:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, et al. 2006. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development 133:1831–1844. [DOI] [PubMed] [Google Scholar]
- Takabatake T, Ogawa M, Takahashi TC, Mizuno M, Okamoto M, Takeshima K. 1997. Hedgehog and patched gene expression in adult ocular tissues. FEBS letters 410:485–489. [DOI] [PubMed] [Google Scholar]
- Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJ, Davidson EH, et al. 2007. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134:1977–1989. [DOI] [PubMed] [Google Scholar]
- Wahl MB, Deng C, Lewandoski M, Pourquie O. 2007. FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development 134:4033–4041. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao Z, Muller J, Iyu A, Khng AJ, Guccione E, Ruan Y, et al. 2013. Targeted inactivation and identification of targets of the Gli2a transcription factor in the zebrafish. Biol Open 2:1203–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, et al. 2010. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J 29:2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Qiao W, Li C, Deng CX. 2002. Generation of Fgfr1 conditional knockout mice. Genesis 32:85–86. [DOI] [PubMed] [Google Scholar]
- Yang X, Dormann D, Munsterberg AE, Weijer CJ. 2002. Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev Cell 3:425–437. [DOI] [PubMed] [Google Scholar]
- Ye W, shimamura K, Rubenstein JLR, Hynes MA, Rosenthal A. 1998. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural ridge. Cell 93:755–766. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Verheyden JM, Hassell JA, Sun X. 2009. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev Cell 16:607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulyn O, Nieuwenhuis E, Liu YC, Angers S, Hui CC. 2015. Ptch2 shares overlapping functions with Ptch1 in Smo regulation and limb development. Dev Biol 397:191–202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.