Abstract
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide. SAR1 gene homolog B (SAR1B) is a GTPase that has been reported to have a central role in the regulation of lipid homeostasis and is associated with numerous diseases. However, its role in cancer, particularly in CRC, remains unclear. The present study revealed that SAR1B was overexpressed in CRC samples and this was associated with shorter overall survival time in patients with CRC. Colony formation, cell proliferation and flow cytometry assays were conducted to evaluate the functions of SAR1B in CRC. It was reported that SAR1B may be associated with tumorigenesis of CRC. Knockdown of SAR1B suppressed cell proliferation and induced significant apoptosis of RKO cells. Furthermore, microarray analysis was performed to identify the potential targets of SAR1B in CRC. Bioinformatics analysis revealed that SAR1B was significantly involved in regulating ‘TGF-β signaling’, ‘paxillin signaling’, ‘cell cycle regulation by BTG family proteins’ and ‘IGF-1 signaling’. These results suggested that SAR1B may be considered a potential prognostic biomarker and therapeutic target for CRC.
Keywords: colorectal cancer, SAR1 gene homolog B, cell growth, cell apoptosis
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second leading cause of cancer-related death among men and women worldwide (1). Surgery and chemotherapy are the main treatment options for CRC. A cytotoxic combination of chemotherapy drugs, such as fluorouracil and irinotecan, and the medication leucovorin is the first line approach to treat CRC (2,3). However, as cancer cells usually develop mechanisms to evade cell death induced by chemotherapy (4–6), the effects of these treatments are still limited. Therefore, there is an urgent need to explore novel tumor-related genes and potential therapeutic approaches to improve the clinical outcomes of CRC.
SAR1 gene homolog B (SAR1B), a coat protein II (COPII) component, serves a central role in the lipid metabolism involved in vesicular COPII-dependent transport of proteins from the endoplasmic reticulum (ER) to the Golgi apparatus (7,8). Mutations of the SAR1a gene homolog 2 that encodes SAR1B protein cause the rare recessive disorder chylomicron retention disease (CMRD) (9), which is associated with developmental problems in infancy. The characteristics of CMRD include fat malabsorption, steatorrhea, deficiency in fat-soluble vitamins, low blood cholesterol and a selective absence of chylomicrons in the blood (9–11). A Previous study demonstrated that patients with CMRD exhibit diffuse enterocyte vacuolization, large cytosolic lipid droplets in intestinal histological tests and accumulation of chylomicron-like particles in enterocytes (12). As lipid metabolism is crucial in cancer cell proliferation, survival and other biochemical processes (12,13), SAR1B may be associated with physiological functions in CRC. However, limited data are available to determine the involvement and physiological role of SAR1B in CRC.
The present study demonstrated that the mRNA expression levels of SAR1B were the highest in SW620 cells among three CRC cell lines. In addition, using lentivirus-mediated infection with short hairpin (sh)RNA targeting SAR1B (shSAR1B), the expression of SAR1B was effectively knocked down in RKO cells. The Celigo system, MTT and colony formation assays identified the inhibitory effects of SAR1B on proliferation of RKO cells. Molecular mechanism analysis results suggested that knockdown of SAR1B with shSAR1B induced significant apoptosis of RKO cells compared with cells infected with control short hairpin (sh)RNA (shCtrl). Together, the results of the present study indicated a tumorigenic function and a potential mechanism of SAR1B in CRC.
Materials and methods
Public datasets analysis
The expression of SAR1B in CRC was analyzed using GEPIA datasets (http://gepia.cancer-pku.cn/) (14). Moreover, Kaplan-Meier method (14) was used to analyze the association between SAR1B expression levels and overall survival (OS) time in patients with CRC using two public datasets, GSE17536 (15) and GSE17537 (15). In order to explore the potential functions of SAR1B in CRC, the DepMap database was analyzed (https://depmap.org/rnai/index) (16). The function of SAR1B in cancer cell survival was analyzed using DepMap portal (https://depmap.org/rnai/index) based on the results of genome-wide shRNA (or small interfering RNA) inhibition of SAR1B from the Broad (17) and Novartis (18) datasets. A PPI network regulated by SAR1B was constructed using the STRING database (https://string-db.org/cgi/network.pl) (19). The bioinformatics analysis was conducted using the Ingenuity Pathway Analysis system (https://www.nihlibrary.nih.gov/resources/tools/ingenuity-pathways-analysis-ipa).
Patients
A total of 15 patients with CRC were recruited for the present study. Patients from Taizhou People's Hospital (Taizhou, China) were pathologically diagnosed with CRC and underwent surgical resection between July 2019 and October 2019. No patient received chemotherapy or radiotherapy treatment prior to surgery. The clinical information of these patients is included in Table I. All patients provided written informed consent, and the present study was approved by the Committees of Taizhou People's Hospital.
Table I.
Clinical information of patients with colorectal cancer used in this study.
| Patient | Age | Sex | Tumor type | Lymph node metastasis |
|---|---|---|---|---|
| Patient 1 | 51 | Female | Rectal protuberance-type differentiated adenocarcinoma | Negative |
| Patient 2 | 38 | Female | Rectal ulcer type of moderately poorly differentiated mucinous adenocarcinoma | Positive |
| Patient 3 | 46 | Male | Rectal ulcer type moderately differentiated adenocarcinoma | Positive |
| Patient 4 | 53 | Male | Rectal protuberance-type differentiated adenocarcinoma | Positive |
| Patient 5 | 61 | Female | Colonic ulcer type moderately differentiated adenocarcinoma | Negative |
| Patient 6 | 68 | Female | Rectal ulcer type moderately differentiated adenocarcinoma | Positive |
| Patient 7 | 71 | Female | Colonic ulcer type moderately differentiated adenocarcinoma | Positive |
| Patient 8 | 29 | Male | Rectal ulcer type moderately differentiated adenocarcinoma | Negative |
| Patient 9 | 38 | Male | Colonic bulging type of differentiated adenocarcinoma | Positive |
| Patient 10 | 46 | Female | Rectal ulcer mucinous adenocarcinoma | Negative |
| Patient 11 | 55 | Female | Rectal ulcer type moderately poorly differentiated adenocarcinoma | Negative |
| Patient 12 | 51 | Male | Rectal ulcer type moderately differentiated adenocarcinoma | Negative |
| Patient 13 | 63 | Female | Rectal prolapse type moderately differentiated adenocarcinoma | Negative |
| Patient 14 | 65 | Female | Rectal ulcer type moderately differentiated adenocarcinoma | Positive |
| Patient 15 | 68 | Male | Sigmoid colonic hypertrophic adenocarcinoma | Negative |
Cell lines and culture
CRC cell lines HCT116, SW620 and RKO cells were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). Penicillin (100 U/ml) and streptomycin (100 U/ml) were added to the medium to prevent contamination. Cells were incubated at 37°C in a humidified atmosphere containing 95% air and 5% CO2.
Construction of SAR1B-knockdown lentivirus
The target DNA sequence (5′-TGGCACAGTACATTGATTA-3′) of SAR1B was selected from the full-length sequence (NM_016103) by Shanghai GeneChem Co., Ltd. According to the sequence of SAR1B, two vectors, shRNA S1 and shRNA S2, were designed. The sequences were as follows: shRNA S1, 5′-CCGGGATGGCACAGTACATTGATTACTCGAGTAATCAATGTACTGTGCCATCTTTTTG-3′; shRNA S2, 5′-AATTCAAAAAGATGGCACAGTACATTGATTACTCGAGTAATCAATGTACTGTGCCATC-3′. The shRNAs were annealed and ligated to the linearized GV115-GFP lentivirus vector (Shanghai GeneChem Co., Ltd.). The plasmid was extracted from the DH5α cells (Shanghai GeneChem Co., Ltd.) and verified by restriction endonuclease digestion followed by Sanger sequencing. The plasmid was extracted and verified by enzymatic digestion and sequencing. RKO cells (ATCC) at a density of 8,000 cells/well were infected with the lentivirus. RKO Cells infected with an empty lentiviral vector were used as a control (shCtrl). Cells at a density of 8,000 cells/well were cultured in RPMI-1640 medium with lentiviruses at a multiplicity of infection of 10 for 24 h at 37°C. A stable SAR1B-knockdown RKO cell line was constructed using the GV115-GFP lentivirus vector system (Shanghai GeneChem Co., Ltd.), which could express GFP. After 72 h of infection, the fluorescence and infection efficiency were determined using an inverted fluorescence microscope at 200× magnification (IX-71; Olympus Corporation). When the infection efficiency was 80%, the expression of SAR1B was analyzed by reverse transcription-quantitative PCR (RT-qPCR) and western blotting. The strong and positive EGFP signals in RKO cells following infection with a lentivirus recombined with shSAR1B or shCtrl indicated that the efficiency of infection in RKO cells and were determined using an inverted fluorescence microscope (magnification, ×200; Olympus).
RT-qPCR
Total RNA from HCT116, SW620 and RKO cells and 15 paired CRC samples were extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. A total of 2 µg RNA RNA samples were reverse transcribed using a Reverse Transcription kit (Roche Diagnostics GmbH) to synthesize cDNA, according to the manufacturer's protocol. The reverse transcription temperature protocol was as follows: 65°C for 10 min, 25°C for 10 min, 50°C for 1 h and 85°C for 5 min. cDNA (1 µl) was used as a template for PCR using the SYBR® Green qPCR kit (Takara Biotechnology Co., Ltd.), using the ViiA™ 7 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers used were as follows: SAR1B, forward 5′-TTTTCCTACGATGGAATCTGGC-3′, reverse 5′-CAGCCTGTCTCCTCGCTTTC-3′; and GAPDH, forward 5′-TGACTTCAACAGCGACACCCA-3′ and reverse 5′-CACCCTGTTGCTGTAGCCAAA-3′. The thermocycling conditions were as follows: Initial denaturation at 95°C for 15 sec, followed by 45 cycles at 95°C for 5 sec and 60°C for 30 sec. The PCR products of SAR1B and GAPDH were 241 and 121 bp, respectively. All samples were analyzed by using the 2−ΔΔCq method in triplicate (20).
Colony formation assay
shSAR1B-infected and shCtrl RKO cells were digested with trypsin and resuspended in standard medium after reaching the logarithmic growth phase. Cells were seeded into 6-well plates at a density of 500 cells/well, incubated at 37°C in a 5% CO2 incubator and observed for 10 days with half of the medium changed every 3 days. Cells were washed with PBS and fixed with 4% paraformaldehyde (1 ml/well; Shanghai Sangong Pharmaceutical Co., Ltd.) for 30–60 min at room temperature. Cells were washed with PBS, stained with 500 µl Giemsa (cat. no. ECM550; Chemicon; Thermo Fisher Scientific, Inc.) for 20 min at room temperature and washed with deionized water three times. Images of cell colonies were captured using inverted light microscope (200× magnification; IX71; Olympus, Tokyo, Japan) and counted using ImageJ software (version 4.0; National Institutes of Health).
Cell proliferation assay
An MTT assay (Sigma-Aldrich; Merck KGaA) was performed to assess cell proliferation following after knockdown of SAR1B as previously described. Briefly, lentivirus-infected RKO cells were seeded in 96-well plates at 3,000 cells/well. At 1, 2, 3, 4 and 5 days following incubation, 20 µl MTT solution (1 mg/ml) was added to each well and incubated at 37°C for 4 h. Subsequently, the medium was carefully removed and 100 µl acidic isopropanol (10% SDS, 5% isopropanol and 0.01 mol/l HCl) was added to each well. The plates were read using an automated microplate reader (Molecular Devices, LLC) at 490 nm.
Plate analysis with the adherent cell cytometry system Celigo®
Briefly, shSAR1B- or shCtrl-infected RKO cells were digested with trypsin and resuspended in standard medium after achieving the logarithmic growth phase. Cells were seeded into 96-well plates at a density of 2,000 cells/well. Fluorescence was detected every day using the Celigo system (Nexcelom Bioscience LLC) for 5 days continuously. Following adjustments in the parameters in analysis settings, the number of cells by scanning green fluorescence daily for 5 days at room temperature according to the manufacturer's instructions.
Analysis of apoptosis using flow cytometry
Apoptosis assessment was performed using an Annexin-V/FITC Apoptosis Detection kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Briefly, cells were infected with shSAR1B or shCtrl. Cells were harvested and stained with Annexin-V/FITC and propidium iodide (PI) in binding buffer for 15 min at room temperature in the dark. The samples were analyzed using a BD FACScan system (BD Biosciences) to determine the percentage of cells exhibiting Annexin-V and PI staining. Apoptotic cells were subsequently analyzed via flow cytometry, using MoFlo XDP (Beckman Coulter, Inc.).
Western blot analysis
Cells were infected with shSAR1B or shCtrl. Protein was extracted using RIPA lysis buffer with proteinase inhibitor (Beyotime Institute of Biotechnology) and western blotting was performed as previously described (12). Protein concentrations of cell lysates were determined using the Bradford method. Equal amounts of protein (20 µg/lane) were separated using 12% SDS-PAGE and transferred to a PVDF membrane. The membranes were blocked using 5% BSA, at 25°C for 2 h. After blocking at room temperature, the membrane was incubated with anti-SAR1B (1:1,000; cat. no. ab155278, Abcam) and anti-GAPDH (1:1,000; cat. no. sc-32233, Santa-Cruz Biotechnology, Inc.) primary antibodies at 4°C for 12 h. Membranes were washed three times with Tris-buffered saline Tween-20 buffer (TBST; 10 mM Tris, 150 mM NaCl, 0.05% Tween-20; Beijing Solarbio Science & Technology Co., Ltd.). Following the primary incubation, membranes were incubated with the horseradish peroxidase-conjugated (HRP) anti-mouse secondary antibody (1:5,000; cat. no. sc-2005; Santa-Cruz, Inc.) and anti-rabbit secondary antibody (1:5,000; cat. no. sc-2004; Santa-Cruz Biotechnology, Inc.) at 25°C for 2 h. Membranes were re-washed three times with TBST buffer. Electrochemiluminescence (cat. no. 6883; Cell Signaling Technology, Inc.) was used for visualization of the protein bands. GAPDH was used as the loading control and protein expression was quantified using ImageJ Software version 1.47 (National Institutes of Health).
Microarray analysis
The stable RKO cells after shRNA infection were used for the microarray analysis. Whole-genome gene expression levels of stable RKO cells following SAR1B knockdown were examined using the gene chip PrimeView Human Gene Expression Array (Thermo Fisher Scientific, Inc.). The sample labeling, microarray hybridization, washing and gene normalization were performed according to the manufacturer's protocols. Differentially expressed genes were subsequently identified via fold-change analysis and P-values; the threshold set for upregulated and downregulated genes was fold-change ≥1.5 and P<0.05. Cluster 3.0 software was used to analyze the differentially expressed genes, using hierarchical clustering method, and gene expression correlation coefficient as the distance, average connection. Then the Treeview software was used to draw the DEGs cluster map.
Bioinformatics analysis
The present study used the Ingenuity Pathway Analysis system (https://www.nihlibrary.nih.gov/resources/tools/ingenuity-pathways-analysis-ipa) to perform Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (21) and Gene Ontology (GO) (21,22) analyses of potential targets of SAR1B.
Statistical analysis
All experiments were performed in triplicate and were repeated at least three times. Data are presented as the mean ± SD. Statistical analysis was performed using unpaired Student's t-test. The probability of survival was estimated using the Kaplan-Meier method. The log-rank test was used to statistically compare the differences in survival times. P<0.05 was considered to indicate a statistically significant difference.
Results
SAR1B is upregulated in CRC
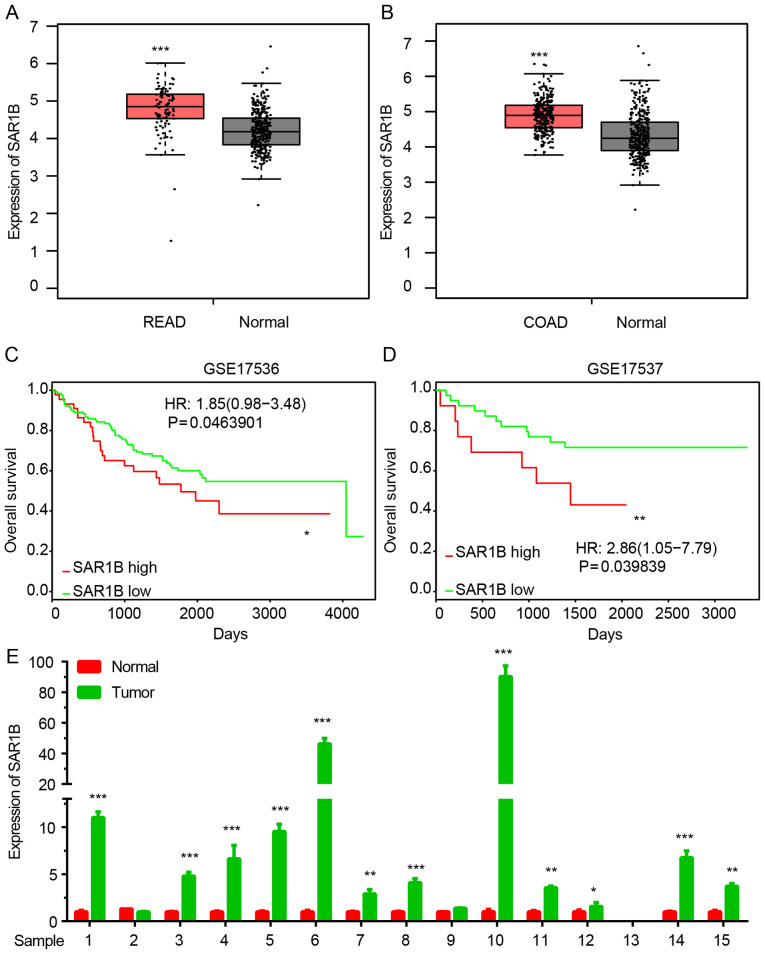
The expression of SAR1B in CRC was analyzed using GEPIA datasets. As demonstrated in Fig. 1A and B, SAR1B expression was significantly upregulated in colon and rectum cancer samples compared with normal tissues.
Figure 1.
SAR1B is upregulated in CRC. (A and B) SAR1B was upregulated in CRC compared with normal samples. ***P<0.001 vs. normal. (C and D) High SAR1B expression was associated with a shorter overall survival time in patients with CRC. *P<0.05, **P<0.01 vs. SAR1B low. (E) SAR1B was upregulated in CRC samples compared with in adjacent normal tissues. *P<0.05, **P<0.01, ***P<0.001 vs. normal. COAD, colon adenocarcinoma; READ, rectum adenocarcinoma; CRC, colorectal cancer; SAR1B, SAR1 gene homolog B.
Kaplan-Meier method was used to analyze the association between SAR1B expression levels and overall survival (OS) time in patients with CRC using two public datasets, GSE17536 and GSE17537. The results revealed that high expression of SAR1B was significantly associated with shorter OS time in patients with CRC (Fig. 1C and D).
In order to further validate the above analysis, 15 pairs of CRC samples were collected. As shown in Fig. 1E, RT-qPCR confirmed that SAR1B was significantly upregulated in CRC samples compared with in normal tissues (Fig. 1E).
Lentivirus-mediated knockdown of SAR1B in CRC RKO cells
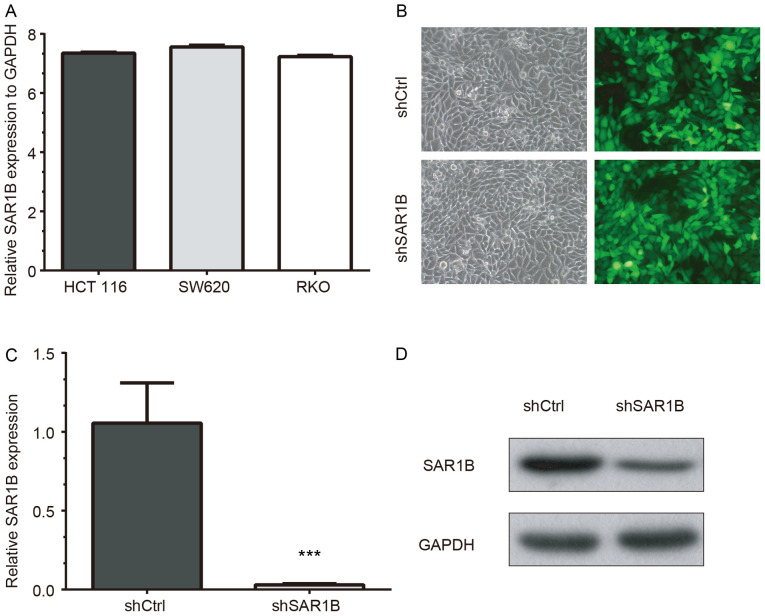
RT-qPCR was used to determine the mRNA expression levels of SAR1B in CRC cell lines HCT116, SW620 and RKO. The results demonstrated that SAR1B was expressed in the three cell lines. The highest SAR1B mRNA expression was observed in SW620 cells, which was 7.56-fold higher than GAPDH expression (Fig. 2A). SAR1B mRNA expression levels were also high in HCT116 cells (7.35-fold higher compared with GAPDH). In addition, SAR1B mRNA expression was 7.23-fold higher than the level of GADPH in RKO cells, which was consistent with HCT116 and SW620 cells. The present study selected RKO cells for the validation of functional importance of SAR1B in CRC, which has been widely used in previous CRC studies (23–26).
Figure 2.
Successful knockdown of SAR1B using lentivirus-mediated shSAR1B in CRC RKO cells. (A) SAR1B was expressed in three cell lines, including HCT116, SW620 and RKO CRC cells. (B) White light and fluorescence images of shCtrl and shSAR1B lentivirus-infected RKO cells. Magnification, ×200. (C) mRNA and (D) protein expression levels of SAR1B were suppressed after lentivirus-mediated knockdown of SAR1B. Each experiment was independently performed in triplicate. ***P<0.001 vs. shCtrl. CRC, colorectal cancer; Ctrl, control; SAR1B, SAR1 gene homolog B; sh, short hairpin RNA.
To further explore the function of SAR1B in CRC, SAR1B was silenced in CRC RKO cells using lentivirus-mediated gene knockdown. As demonstrated in Fig. 2B, the strong and positive EGFP signals in RKO cells following infection with a lentivirus recombined with shSAR1B or shCtrl indicated that the efficiency of infection in RKO cells was high and thus qualified for subsequent experiments.
RT-qPCR and western blot analysis results revealed that knockdown of SAR1B was efficient at the mRNA and protein levels in shSAR1B-infected RKO cells (Fig. 2C and D). The RT-qPCR results demonstrated that the reduction efficiency reached 97% following infection with shSAR1B, which was significantly different compared with the shCtrl group (P<0.05; Fig. 2C). The western blotting results as also showed SAR1B protein was reduced in shSAR1B-infected cells (Fig. 2D). These results indicated that recombinant lentivirus targeting SAR1B effectively suppressed mRNA and protein expression of endogenous SAR1B in CRC cells.
Knockdown of SAR1B inhibits RKO cell proliferation
In order to explore the potential functions of SAR1B in CRC, the DepMap database was used. The effects of SAR1B on cancer cell survival was analyzed using the DepMap portal. The results of genome-wide shRNA (or small interfering RNA) inhibition of SAR1B from the Broad (Fig. S1A), and Novartis (Fig. S1B) datasets showed the dependency of SAR1B for cancer cell survival, thus suggesting that SAR1B may promote the proliferation of CRC cells.
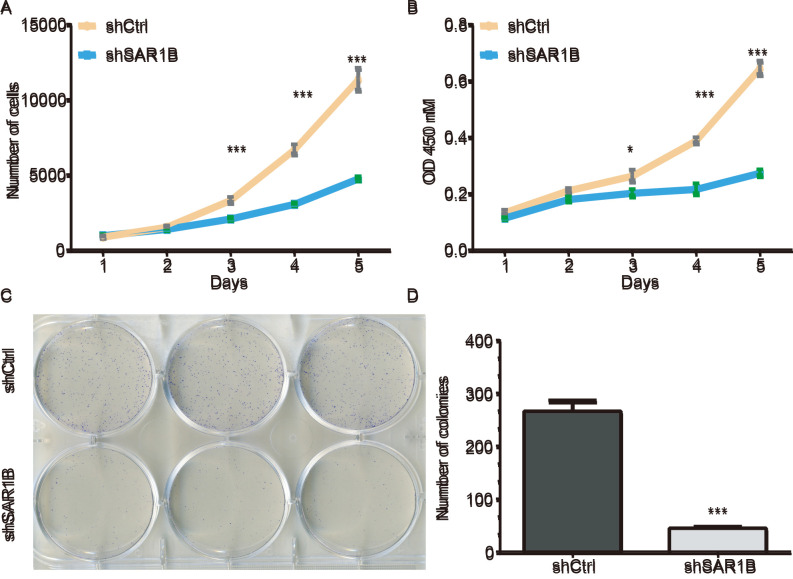
A Celigo cell counting application, which allows for direct imaging and counting of cells, was used to detect RKO cell proliferation following SAR1B knockdown. The data were collected for 5 days continually, and the results are presented in Fig. 3A. The results demonstrated that compared with cells infected with shCtrl, the green signals in cells infected with shSAR1B were significantly inhibited (P<0.05). These results suggested that the downregulation of SAR1B may inhibit cell proliferation (Fig. 3A). The average cell number was 12.76-fold higher on day 5 compared with that on day 1 in cells infected with shCtrl, whereas the average cell number was 4.74-fold higher on day 5 compared with that on day 1 in cells infected with shSAR1B, which was significantly different (P<0.001).
Figure 3.
Knockdown of SAR1B using shSAR1B led to inhibition of RKO cell growth. (A) Number of RKO cells expressing shCtrl lentivirus and shSAR1B lentivirus on days 1, 2, 3, 4 and 5, as measured by Celigo cell counting assay. (B) Cell proliferation of RKO cells expressing shCtrl and shSAR1B on days 1, 2, 3, 4 and 5, as measured by an MTT assay. *P<0.05, ***P<0.001 vs. shSAR1B. (C) Microscope images showing colonies of RKO cells that have been stained with Giemsa. (D) Representative histogram showing the number of colonies in RKO cells. Each experiment was independently performed in triplicate. ***P<0.001 vs. shCtrl. CRC, colorectal cancer; Ctrl, control; OD, optical density; SAR1B, SAR1 gene homolog B; sh, short hairpin RNA.
MTT and colony formation assays were used to further investigate the potential effects of SAR1B on the proliferation of RKO cells. A total of 2,000 cells were seeded and the cell numbers were analyzed using an MTT assay daily for 5 days (Fig. 3B). The proliferation was reduced in cells infected with shSAR1B compared with that in cells infected with shCtrl (P<0.05), which suggested that the rate of cell proliferation was significantly inhibited.
The colony formation assay also exhibited consistent inhibition of cell proliferation in shSAR1B-infected RKO cells. As demonstrated in Fig. 3C, the colonies were relatively smaller and lower in number in the shSAR1B group compared with the shCtrl group in RKO cells. Statistical analysis further revealed that knockdown of SAR1B significantly reduced the number of colonies formed by RKO cells (P<0.001; Fig. 3D). These results suggested that knockdown of SAR1B inhibited RKO cell proliferation.
Knockdown of SAR1B induces apoptosis of RKO cells
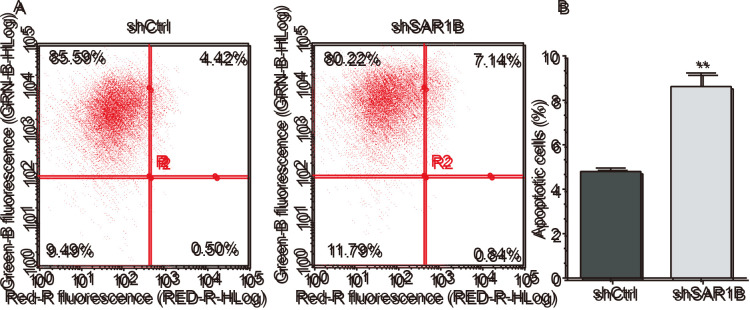
To explore the role of SAR1B in apoptosis, flow cytometry was performed to determine the apoptotic rates in RKO cells infected with shCtrl or shSAR1B. As demonstrated in Fig. 4A and B, apoptosis was induced following shCtrl or shSAR1B infection. Further analysis revealed that the apoptotic rate was significantly reduced in the shCtrl group compared with the shSAR1B group, which was significantly different (P<0.01). These results suggested that SAR1B knockdown induced apoptosis of RKO cells, indicating the tumorigenic potential of SAR1B.
Figure 4.
Knockdown of SAR1B induces apoptosis of RKO cells. (A) Cell apoptosis was analyzed by flow cytometry. Cells in the left upper and left lower quadrants represent living cells. The cells in right upper and right lower quadrants represent apoptotic cells. (B) Statistical analysis of the number of apoptotic cells after SAR1B knockdown. Each experiment was independently performed in triplicate. **P<0.01 vs. shCtrl. Ctrl, control; PI, propidium iodide; SAR1B, SAR1 gene homolog B; sh, short hairpin RNA.
Microarray analysis of the potential roles of SAR1B in CRC
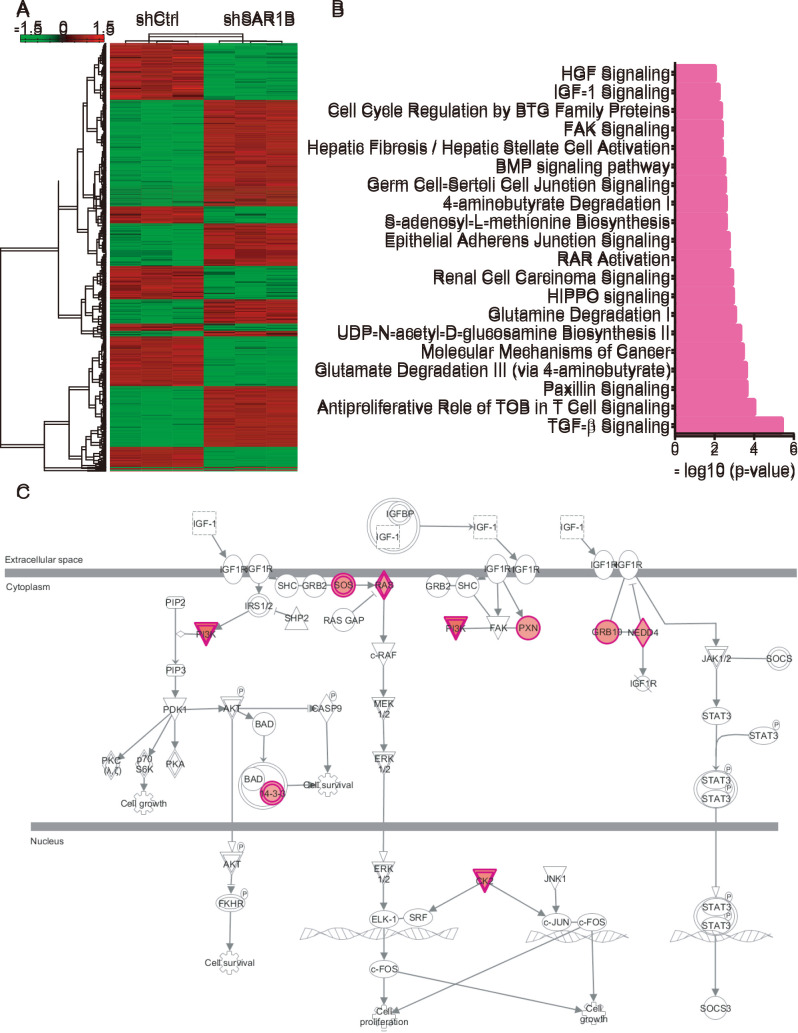
As the mechanisms underlying the effects of SAR1B on CRC progression remain largely unknown, microarray analysis was conducted. As presented in Fig. 5A, a total of 333 genes were upregulated and 369 genes were downregulated following SAR1B knockdown in RKO cells (Table SI).
Figure 5.
Microarray analysis showing the potential roles of SAR1B in CRC. (A) Heatmap showing differentially expressed genes after SAR1B knockdown in RKO cells. (B) Bioinformatics analysis revealed the potential functions of SAR1B in CRC. (C) Analysis showed that ‘IGF-1 signaling’ was significantly activated, >9 regulators of this pathway were upregulated after SAR1B knockdown in CRC. CRC, colorectal cancer; SAR1B, SAR1 gene homolog B; sh, short hairpin RNA.
KEGG pathway analysis revealed that SAR1B was significantly enriched in ‘TGF-β signaling’, ‘antiproliferative role of TOB in T cell signaling’, ‘paxillin signaling’, ‘glutamate degradation III (via 4-aminobutyrate)’, ‘molecular mechanisms of cancer’, ‘UDP-N-acetyl-D-glucosamine biosynthesis II’, ‘glutamine degradation I’, ‘HIPPO signaling’, ‘renal cell carcinoma signaling’, ‘RAR activation’, ‘epithelial adherens junction signaling’, ‘S-adenosyl-L-methionine biosynthesis’, ‘4-aminobutyrate degradation I’, ‘germ cell-sertoli cell junction signaling’, ‘BMP signaling pathway’, ‘hepatic fibrosis/hepatic stellate cell activation’, ‘FAK signaling’, ‘cell cycle regulation by BTG family proteins’ and ‘IGF-1 signaling’ (Fig. 5B). Of note, ‘IGF-1 signaling’ was significantly activated; >9 regulators of this pathway were upregulated following SAR1B knockdown in CRC by using Ingenuity Pathway Analysis system (https://www.nihlibrary.nih.gov/resources/tools/ingenuity-pathways-analysis-ipa) (Fig. 5C).
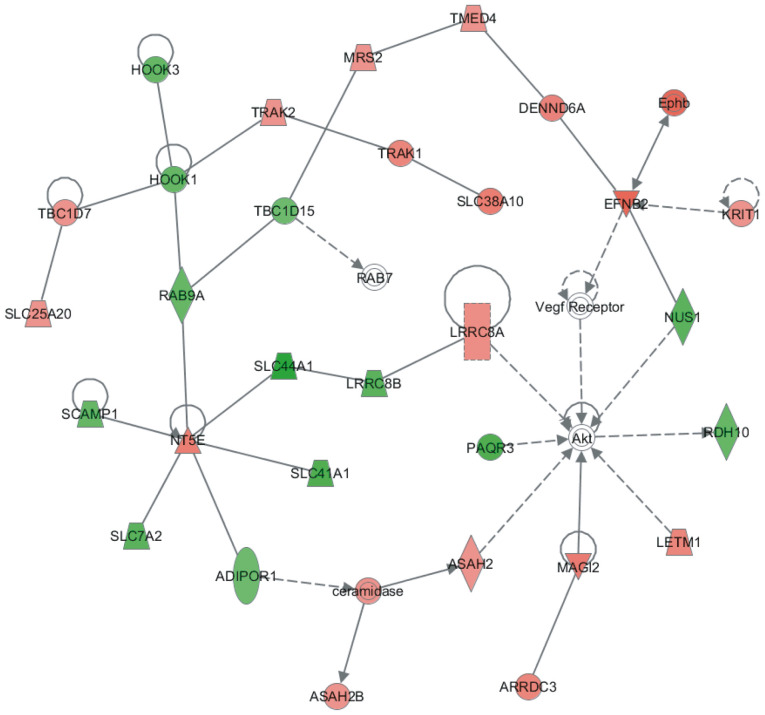
Construction of a protein-protein interaction (PPI) network regulated by SAR1B in CRC
A PPI network regulated by SAR1B was constructed using the STRING database. This network contained 35 nodes. Among them, HOOK1, HOOK3, RAB9A, TBC1D15, SLC44A1, LRRC8B, SLC41A1, SCAMP1, SLC7A2, ADIPOR1, PAQR3, NUS1 and RDH10 were downregulated following SAR1B knockdown in RKO cells. Conversely, Akt, ARRDC3, ASAH2, ASAH2B, ceramidase, DENND6A, EFNB2, Ephb, KRIT1, LETM1, LRRC8A, MAGI2, MRS2, NT5E, RAB7, SLC25A20, SLC38A10, TBC1D7, TMED4, TRAK1, TRAK2 and vascular endothelial growth factor receptor were upregulated following SAR1B knockdown in RKO cells (Fig. 6).
Figure 6.
Construction of protein-protein interaction network regulated by SAR1B in CRC. Red node, upregulated genes after SAR1B knockdown in CRC; green node, downregulated genes after SAR1B knockdown in CRC; Akt, Vegf receptor and RAB7 were labeled with white circles. CRC, colorectal cancer; SAR1B, SAR1 gene homolog B.
Discussion
The results of the present study indicated that SAR1B may be closely associated with the tumorigenesis of CRC. SAR1B expression was determined in CRC cell lines, including HCT116, SW620 and RKO. Functional analysis demonstrated that knockdown of SAR1B suppressed CRC cell proliferation. Furthermore, flow cytometry results revealed that knockdown of SAR1B induced apoptosis of RKO cells. In addition, SAR1B downstream targets and pathways in CRC were determined using microarray and bioinformatics analyses.
Low molecular weight GTPases are involved in vesicular transport associated with multiple signaling transduction pathways that affect the regulation of cell proliferation, differentiation and transformation (7,8). It has been demonstrated that SAR1B GTPase is central to lipid metabolism and regulates chylomicron secretion by the small intestine (27,28). According to previous reports, the expression of SAR1B was identified in cell lines (including Caco-2/15 and McArdle-RH7777 cells), tissues and blood samples from patients (12,29,30). In the present study, SAR1B expression was upregulated in CRC patient samples. By using RT-PCR, we found SAR1B was expressed in HCT116, SW620 and RKO cell lines. These data suggested that SAR1B may be associated with CRC tumorigenesis.
Previous metabolic studies have revealed that when nutrients, such as lipids, proteins and nucleic acids, are abundant, oncogenic signaling pathways directly enhance nutrient acquisition, and facilitate cancer cell proliferation (31–33). These results were reproduced in cultures of clear cell renal cell carcinoma, glioblastoma and bladder cancer, suggesting that increased lipid levels may contribute to cancer cell proliferation (11) (9,34,35). Considering the regulatory role of SAR1B in lipid transport, it could be hypothesized that SAR1B may serve a role in tumorigenesis. A lentiviral system with shSAR1B was used to knock down SAR1B in RKO CRC cells. The effects of SAR1B on RKO proliferation were determined by Celigo cell counting application, MTT and colony formation assays. shSAR1B effectively suppressed mRNA and protein expression levels of endogenous SAR1B in RKO cells. In addition, RKO cell proliferation was reduced in cells infected with shSAR1B compared with those infected with shCtrl. The colony formation assay also demonstrated that knockdown of SAR1B significantly reduced the number of colonies formed by RKO cells. The results of the present study supported the involvement of SAR1B in CRC cell function and demonstrated that the suppression of SAR1B inhibited RKO cell proliferation.
Accumulating evidence has demonstrated that genes encoding certain endocytosis-related proteins are the targets of chromosomal rearrangement in human hematopoietic malignancies, and abnormal expression or mutations of these proteins have been reported in human cancer, suggesting a potential link between endocytosis proteins and cancer (36). SAR1B is a GTPase that serves a key role in the exportation of proteins from the ER to the Golgi apparatus by recruiting β-COP to the ER and promoting vesicular transport (10,37,38). Considering the essential function of lipids in cancer, it is possible that SAR1B may affect the molecular mechanisms involved in cell proliferation or apoptosis. To further explore the role of SAR1B in CRC in the present study, RKO cells were infected with shSAR1B, and apoptosis was detected by flow cytometry. The apoptotic rate of RKO cells infected with shSAR1B was significantly higher compared with that in cells infected with shCtrl. These results suggested that SAR1B knockdown induced apoptosis of RKO cells.
Microarray analysis was performed to identify the potential targets of SAR1B in CRC. Bioinformatics analysis revealed that SAR1B was significantly enriched in ‘TGF-β signaling’, ‘paxillin signaling’, ‘cell cycle regulation by BTG family proteins’ and ‘IGF-1 signaling’. Of note, >9 regulators of ‘IGF-1 signaling’ were upregulated following SAR1B knockdown in CRC. Additionally, a SAR1B-related PPI network was constructed; several key regulators of cancer were included in this network, such as Akt, HOOK1 and HOOK3. HOOK1 has been demonstrated to inhibit hepatocellular carcinoma progression and epithelial-mesenchymal transition (EMT) (39), and high expression of HOOK3 is associated with poor prognosis of prostate cancer (39). These reports, together with the present results, suggested that SAR1B may serve crucial roles in CRC by regulating these targets.
A recent study from Huang and Wang (40) revealed that SAR1B knockdown promoted the migration and invasion of CRC cells. While this previous study focused on the effects of SAR1B on CRC migration, invasion and EMT, the present study focused on the effects of SAR1B on CRC proliferation, colony formation and apoptosis. By combining these previous results and the current findings, it can be concluded that SAR1B may promote cancer proliferation, while suppressing cancer metastasis in CRC. Of note, emerging studies have revealed that a number of genes serve different roles in regulating cancer proliferation and metastasis. For example, Wan et al (41) revealed that patched 1 suppressed lung cancer proliferation via its coding protein; however, it also promoted cell migration, invasion and adhesion in non-small cell lung cancer via its 3′ untranslated region. Although further validation into the roles of SAR1B in CRC is still required, the present study could provide novel insights into the functions of SAR1B.
It should be noted that there are several limitations of the current study. Firstly, an in vivo model should be used to further validate the roles of SAR1B in CRC, which would be helpful to gain an understanding into the functional importance of SAR1B in CRC. Secondly, the current study explored the potential mechanisms of SAR1B in CRC using microarray analysis; however, the microarray analysis was not validated in this study.
In conclusion, the results of the present study demonstrated that SAR1B was overexpressed in CRC cells. Inhibition of SAR1B suppressed CRC cell proliferation and induced apoptosis of RKO cells. Moreover, it was found that SAR1B may be involved in CRC cell proliferation and apoptosis. These results suggested that SAR1B could be used in the prognosis and treatment of CRC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
YL and TB designed the present study. YL, SZ and RC performed the majority of the experiments and drafted the initial manuscript. YL and SZ performed the statistical analysis. LJ helped perform the experiments and critically revised the manuscript for important intellectual content. LY interpreted the data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
All patients provided written informed consent, and the present study was approved by The Ethics Committee of Taizhou Hospital of Zhejiang Province Affiliated to Wenzhou Medical University (approval no. 2019068).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fakih MG. Metastatic colorectal cancer: Current state and future directions. J Clin Oncol. 2015;33:1809–1824. doi: 10.1200/JCO.2014.59.7633. [DOI] [PubMed] [Google Scholar]
- 2.Engelhardt EG, Revesz D, Tamminga HJ, Punt CJA, Koopman M, Onwuteaka-Philipsen BD, Steyerberg EW, Jansma IP, De Vet HCW, Coupé VMH. Clinical usefulness of tools to support decision-making for palliative treatment of metastatic colorectal cancer: A systematic review. Clin Colorectal Cancer. 2018;17:e1–e12. doi: 10.1016/j.clcc.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Advani SM, Advani P, DeSantis SM, Brown D, VonVille HM, Lam M, Loree JM, Mehrvarz Sarshekeh A, Bressler J, Lopez DS, et al. Clinical, pathological, and molecular characteristics of CpG island methylator phenotype in colorectal cancer: A systematic review and meta-analysis. Transl Oncol. 2018;11:1188–1201. doi: 10.1016/j.tranon.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Safa AR. Resistance to cell death and its modulation in cancer stem cells. Crit Rev Oncog. 2016;21:203–219. doi: 10.1615/CritRevOncog.2016016976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gebremeskel S, Johnston B. Concepts and mechanisms underlying chemotherapy induced immunogenic cell death: Impact on clinical studies and considerations for combined therapies. Oncotarget. 2015;6:41600–41619. doi: 10.18632/oncotarget.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letai A. Cell death and cancer therapy: Don't forget to kill the cancer cell! Clin Cancer Res. 2015;21:5015–5020. doi: 10.1158/1078-0432.CCR-15-1204. [DOI] [PubMed] [Google Scholar]
- 7.Fryer LG, Jones B, Duncan EJ, Hutchison CE, Ozkan T, Williams PA, Alder O, Nieuwdorp M, Townley AK, Mensenkamp AR, et al. The endoplasmic reticulum coat protein II transport machinery coordinates cellular lipid secretion and cholesterol biosynthesis. J Biol Chem. 2014;289:4244–4261. doi: 10.1074/jbc.M113.479980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loftus AF, Hsieh VL, Parthasarathy R. Modulation of membrane rigidity by the human vesicle trafficking proteins Sar1A and Sar1B. Biochem Biophys Res Commun. 2012;426:585–589. doi: 10.1016/j.bbrc.2012.08.131. [DOI] [PubMed] [Google Scholar]
- 9.Charcosset M, Sassolas A, Peretti N, Roy CC, Deslandres C, Sinnett D, Levy E, Lachaux A. Anderson or chylomicron retention disease: Molecular impact of five mutations in the SAR1B gene on the structure and the functionality of Sar1b protein. Mol Genet Metab. 2008;93:74–84. doi: 10.1016/j.ymgme.2007.08.120. [DOI] [PubMed] [Google Scholar]
- 10.Sane AT, Seidman E, Peretti N, Kleme ML, Delvin E, Deslandres C, Garofalo C, Spahis S, Levy E. Understanding chylomicron retention disease through sar1b gtpase gene disruption: Insight from cell culture. Arterioscler Thromb Vasc Biol. 2017;37:2243–2251. doi: 10.1161/ATVBAHA.117.310121. [DOI] [PubMed] [Google Scholar]
- 11.Silvain M, Bligny D, Aparicio T, Laforêt P, Grodet A, Peretti N, Ménard D, Djouadi F, Jardel C, Bégué JM, et al. Anderson's disease (chylomicron retention disease): A new mutation in the SARA2 gene associated with muscular and cardiac abnormalities. Clin Genet. 2008;74:546–552. doi: 10.1111/j.1399-0004.2008.01069.x. [DOI] [PubMed] [Google Scholar]
- 12.Levy E, Harmel E, Laville M, Sanchez R, Emonnot L, Sinnett D, Ziv E, Delvin E, Couture P, Marcil V, Sane AT. Expression of Sar1b enhances chylomicron assembly and key components of the coat protein complex II system driving vesicle budding. Arterioscler Thromb Vasc Biol. 2011;31:2692–2699. doi: 10.1161/ATVBAHA.111.233908. [DOI] [PubMed] [Google Scholar]
- 13.Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, Krill-Burger JM, Green TM, Vazquez F, Boehm JS, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun. 2018;9:4610. doi: 10.1038/s41467-018-06916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu C, Mannan AM, Yvone GM, Ross KN, Zhang YL, Marton MA, Taylor BR, Crenshaw A, Gould JZ, Tamayo P, et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat Biotechnol. 2016;34:419–423. doi: 10.1038/nbt.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald ER, III, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, Belur D, Castelletti D, Frias E, Gampa K, et al. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, Deep RNAi screening. Cell. 2017;170:577–592.e10. doi: 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Gene Ontology Consortium, corp-author. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang CY, Zhang LJ, Lu ZC, Ma CY, Ye Y, Rahman K, Zhang H, Zhu JY. Antitumor activity of diterpenoids from jatropha gossypiifolia: Cell cycle arrest and apoptosis-inducing activity in RKO colon cancer cells. J Nat Prod. 2018;81:1701–1710. doi: 10.1021/acs.jnatprod.7b01036. [DOI] [PubMed] [Google Scholar]
- 24.Garufi A, Ricci A, Trisciuoglio D, Iorio E, Carpinelli G, Pistritto G, Cirone M, D'Orazi G. Glucose restriction induces cell death in parental but not in homeodomain-interacting protein kinase 2-depleted RKO colon cancer cells: Molecular mechanisms and implications for tumor therapy. Cell Death Dis. 2013;4:e639. doi: 10.1038/cddis.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tansuwanwong S, Hiroyuki Y, Kohzoh I, Vinitketkumnuen U. Induction of apoptosis in RKO colon cancer cell line by an aqueous extract of Millingtonia hortensis. Asian Pac J Cancer Prev. 2006;7:641–644. [PubMed] [Google Scholar]
- 26.Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. Expression of the gut-enriched Kruppel-like factor (Kruppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sane A, Ahmarani L, Delvin E, Auclair N, Spahis S, Levy E. SAR1B GTPase is necessary to protect intestinal cells from disorders of lipid homeostasis, oxidative stress, and inflammation. J Lipid Res. 2019;60:1755–1764. doi: 10.1194/jlr.RA119000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy E, Spahis S, Garofalo C, Marcil V, Montoudis A, Sinnet D, Sanchez R, Peretti N, Beaulieu JF, Sane A. Sar1b transgenic male mice are more susceptible to high-fat diet-induced obesity, insulin insensitivity and intestinal chylomicron overproduction. J Nutr Biochem. 2014;25:540–548. doi: 10.1016/j.jnutbio.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Magnolo L, Najah M, Fancello T, Di Leo E, Pinotti E, Brini I, Gueddiche NM, Calandra S, Slimene NM, Tarugi P. Novel mutations in SAR1B and MTTP genes in Tunisian children with chylomicron retention disease and abetalipoproteinemia. Gene. 2013;512:28–34. doi: 10.1016/j.gene.2012.09.117. [DOI] [PubMed] [Google Scholar]
- 30.Marcil V, Seidman E, Sinnett D, Sanchez R, Spahis S, Sané A, Levy E. Tissue distribution and regulation of the small Sar1b GTPase in mice. Cell Physiol Biochem. 2014;33:1815–1826. doi: 10.1159/000362960. [DOI] [PubMed] [Google Scholar]
- 31.Okada T, Miyashita M, Fukuhara J, Sugitani M, Ueno T, Samson-Bouma ME, Aggerbeck LP. Anderson's disease/chylomicron retention disease in a Japanese patient with uniparental disomy 7 and a normal SAR1B gene protein coding sequence. Orphanet J Rare Dis. 2011;6:78. doi: 10.1186/1750-1172-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cefalu AB, Calvo PL, Noto D, Baldi M, Valenti V, Lerro P, Tramuto F, Lezo A, Morra I, Cenacchi G, et al. Variable phenotypic expression of chylomicron retention disease in a kindred carrying a mutation of the Sara2 gene. Metabolism. 2010;59:463–467. doi: 10.1016/j.metabol.2009.07.042. [DOI] [PubMed] [Google Scholar]
- 33.Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treepongkaruna S, Chongviriyaphan N, Suthutvoravut U, Charoenpipop D, Choubtum L, Wattanasirichaigoon D. Novel missense mutations of SAR1B gene in an infant with chylomicron retention disease. J Pediatr Gastroenterol Nutr. 2009;48:370–373. doi: 10.1097/MPG.0b013e318183188f. [DOI] [PubMed] [Google Scholar]
- 35.Barbe L, Lundberg E, Oksvold P, Stenius A, Lewin E, Björling E, Asplund A, Pontén F, Brismar H, Uhlén M, Andersson-Svahn H. Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics. 2008;7:499–508. doi: 10.1074/mcp.M700325-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, Svrzikapa N, Hirozane-Kishikawa T, Rietman E, Yang X, et al. Next-generation sequencing to generate interactome datasets. Nat Methods. 2011;8:478–480. doi: 10.1038/nmeth.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen JH, Hsieh CJ, Huang YL, Chen YC, Chen TF, Sun Y, Wen LL, Yip PK, Chu YM. Genetic polymorphisms of lipid metabolism gene SAR1 homolog B and the risk of Alzheimer's disease and vascular dementia. J Formos Med Assoc. 2016;115:38–44. doi: 10.1016/j.jfma.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 38.Sane A, Seidman E, Spahis S, Lamantia V, Garofalo C, Montoudis A, Marcil V, Levy E. New insights in intestinal Sar1B GTPase regulation and role in cholesterol homeostasis. J Cell Biochem. 2015;116:2270–2282. doi: 10.1002/jcb.25177. [DOI] [PubMed] [Google Scholar]
- 39.Sun X, Zhang Q, Chen W, Hu Q, Lou Y, Fu QH, Zhang JY, Chen YW, Ye LY, Wang Y, et al. Hook1 inhibits malignancy and epithelial-mesenchymal transition in hepatocellular carcinoma. Tumour Biol. 2017;39:1010428317711098. doi: 10.1177/1010428317711098. [DOI] [PubMed] [Google Scholar]
- 40.Huang M, Wang Y. Targeted quantitative proteomic approach for probing altered protein expression of small GTPases associated with colorectal cancer metastasis. Anal Chem. 2019;91:6233–6241. doi: 10.1021/acs.analchem.9b00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan X, Kong Z, Chu K, Yi C, Hu J, Qin R, Zhao C, Fu F, Wu H, Li Y, Huang Y. Co-expression analysis revealed PTCH1-3′UTR promoted cell migration and invasion by activating miR-101-3p/SLC39A6 axis in non-small cell lung cancer: Implicating the novel function of PTCH1. Oncotarget. 2018;9:4798–4813. doi: 10.18632/oncotarget.23219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.