Abstract
The management of food allergy is complicated by the lack of highly predictive biomarkers for diagnosis and prediction of disease course. Measurement of food-specific IgE is a useful tool together with clinical history, but is an imprecise predictor of clinical reactivity. The gold standard for diagnosis and clinical research is a double-blind placebo-controlled food challenge. Improvement in our understanding of immune mechanisms of disease, development of high-throughput technologies, and advances in bioinformatics have yielded a number of promising new biomarkers of food allergy. In this review, we will discuss advances in immunoglobulin measurements, the utility of the basophil activation test, T cell profiling, and the use of -omic technologies (transcriptome, epigenome, microbiome, and metabolome) as biomarker tools in food allergy.
Keywords: Double blind placebo controlled food challenge (DBPCFC), components, epitopes, basophil activation test, T cell receptor repertoire, transcriptome, microbiome, epigenome, metabolome, biomarker
Introduction
The gold standard for diagnosis of food allergy is the double-blind placebo-controlled food challenge (DBPCFC). In the past decade, there has been considerable effort dedicated to standardizing the DBPCFC, from procedure 1 to assessment and interpretation 2. The DBPCFC can be unpleasant for the patient and carries a burden of time and resources to perform in a safe manner which can make it difficult to offer in private practice. The DBPCFC is also the standard for determining efficacy in treatment studies for food allergy and is used at a minimum before and after treatment to assess the impact of treatment on threshold of reactivity to the food. This is a barrier to conducting clinical trials in food allergy. Furthermore, as studies move from single allergen immunotherapy to treatments for multi-food allergy, this requirement of entry and exit food challenges becomes a major burden, limiting participant recruitment and ability of clinical sites to participate in treatment trials. We are currently at a time of rapid acceleration of treatment options, including novel biologics, for testing in food allergy. There is a clear unmet need for biomarkers to reduce the need for DBPCFC for clinical assessment. There is also a need for biomarkers informative of key clinical parameters of food allergy, including threshold of reactivity, risk of severe reactions, probability of natural resolution, and probability of successful treatment response to immunotherapy. Figure 1 provides an overview to the biomarker approaches discussed in this review.
Figure 1. Biomarkers in Food Allergy.
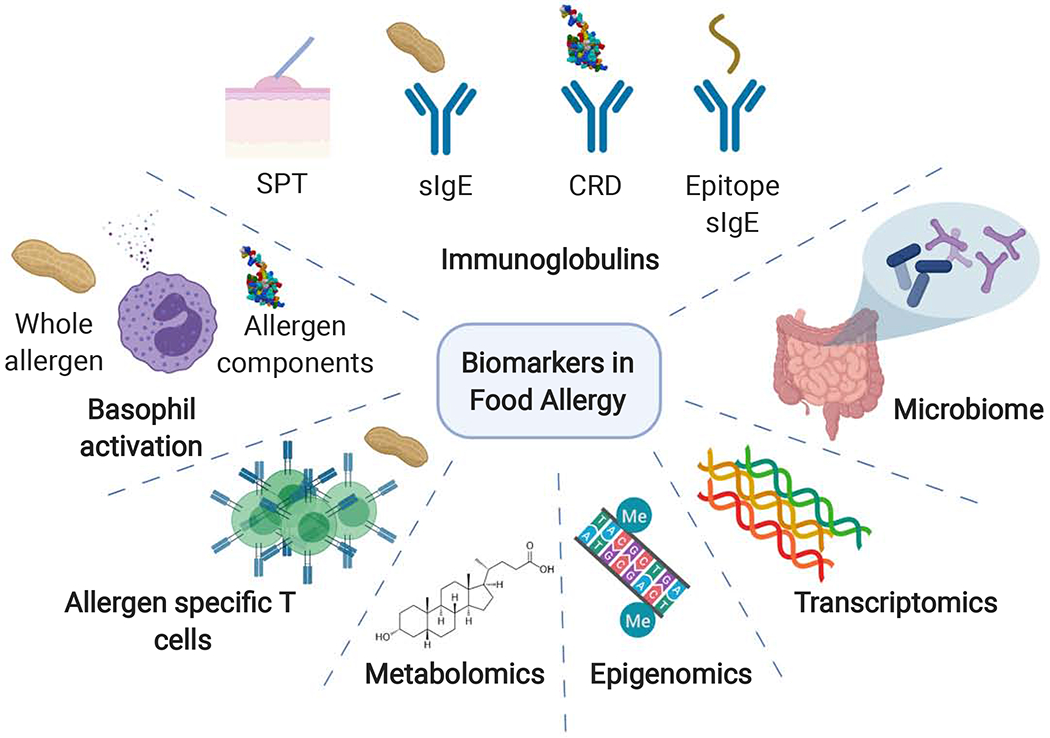
Recent innovations have led to the discovery and integration of multiple biomarkers for both diagnosis and prognosis for the management of IgE-mediated food allergies. Biomarkers include skin prick testing (SPT), specific immunoglobulins (sIgE), component resolved diagnostics (CRD), epitope specific IgE, basophil activation, allergen-specific T cells, transcriptomics, epigenomics, microbiome, and metabolomics (Created with Biorender.com).
Immunoglobulins
Immunoglobulins and food allergy diagnosis
Food-specific IgE:
The blood biomarker currently in use as a guide to clinical decision-making is allergen-specific IgE. The most commonly used IgE lab test is ImmunoCAP™. Food-specific IgE is measured in serum and values are reported as kilounits of antibody per liter (kU/L) based on World Health Organization IgE standards. Range of ImmunoCAP is 0.1 – 100 kU/L, and sera above 100 kU/L can be diluted to obtain an accurate measure of food-specific IgE. The level of food-specific IgE can be a useful tool for estimating the probability of clinical reactivity to some foods some foods 3–7. However, the predictive value of specific IgE measurement varies by geography and age and is not predictive for all foods.
Allergen-specific IgE:
Component resolved diagnostics, or the measurement of IgE binding to food allergens (components) provide improved predictive performance compared to IgE levels to whole allergens. The best example of this is for peanut, where binding to the Bet v 1-cross reactive protein Ara h 8 can contribute significantly to the peanut-specific IgE level in birch pollen allergic patients. Ara h 8 is not digestion stable, and elicits reactions primarily limited to the oral cavity. Ara h 9 is a lipid transfer protein and, in many parts of the world, can be identified more often in peanut-sensitized but tolerant individuals. In contrast, presence of IgE specific to the seed storage proteins (Ara h 2, Ara h 1, Ara h 3, and Ara h 6) are more predictive of peanut allergy,8 with Ara h 2-specific IgE documented to have greater predictive performance than peanut-specific IgE. 9, 10 Similarly, binding of IgE to the Cor a 14 allergen of hazelnut was a better predictor of hazelnut allergy than IgE to hazelnut. 11 There are significant age and geographic factors that contribute to sensitization to food components with differing clinical significance, and therefore influence the performance of component and food-specific IgE levels in prediction of clinical reactivity.
Epitope-specific IgE:
Just as there are allergens with more or less clinical relevance, there are epitopes (regions on the allergen that bind IgE) for which IgE binding appears to have more clinical relevance. Most of this data is derived from studies of linear epitopes, in which IgE binding to sequential 15-20 amino acid peptides along the allergens is tested. Microarrays of epitopes bound to glass slides were initially used 12, 13, while next generation formats use epitopes bound to beads for efficient multiplexing capacity and advanced bioinformatics for identification of informative epitopes. Epitope-specific IgE measured in multiplexed bead-based assay format has been found to outperform food-specific and component-specific IgE in prediction of phenotype of milk allergy 14 and predicting reactivity versus sensitization to peanut 15. Epitope-specific binding has also been used to parse out clinically meaningful binding to shellfish epitopes from cross-reactivity to inhaled mite and cockroach allergens 16. Studies are needed to determine if predictive epitopes are conserved across geographic regions and age groups.
Food-specific IgG and IgA:
Measurements of food-specific IgG, IgG4, or IgA are not useful for the diagnosis of food allergy and do not predict clinical reactivity17–19
Immunoglobulins and prediction of threshold and severity
Beyond diagnosis of food allergy, a prediction of threshold of clinical reactivity (i.e. risk of reacting to contaminating amounts of allergen) is useful for assessing risk. There is a wide range of severity of clinical reactions. Therefore, identifying those at risk of severe symptoms would be beneficial. Rolinck-Werninghaus et al reported on immune parameters associated with 1,671 oral food challenges (OFCs) to milk, egg, soy, and wheat. Elevated milk and egg-specific IgE were associated with reactions at lower threshold of clinical reactivity, and were weakly associated with reaction severity 20. Specific IgE to the peanut component Ara h 2 was associated with a lower threshold of clinical reactivity to peanut on DBPCFC 21. Measurement of IgE binding to milk epitopes 15 and peanut epitopes 22, 16 has also been identified as predicting threshold of clinical reactivity on DBPCFC. In addition to threshold, measurement of IgE binding to epitopes in milk can effectively predict the milk allergy phenotype (tolerance of fermented or baked forms of milk) 14. Thus, higher resolution of IgE binding is predictive of the clinical presentation of food allergy.
Immunoglobulins and prediction of natural resolution and response to treatment
Individuals who go on to outgrow their food allergy have lower levels of IgE than those who have persistent food allergy. In the CoFAR2 longitudinal study and the HealthNuts study, low IgE was the only immunoglobulin predictive of resolution to milk, egg, and peanut 23–25. Predictive calculators were generated to estimate the probability of milk and egg allergy resolution based on clinical features and food-specific IgE. Low levels of food-specific IgE is also predictive of successful response to allergen immunotherapy 26, 27.
As with diagnosis, information on the specificity of IgE binding is predictive of natural history and response to therapy. The HealthNuts study found that the diversity of the IgE response to egg predicted persistence of allergy 28. IgE binding to all 4 egg components measured (ovalbumin, ovomucoid, ovotransferrin, and egg yolk) was associated with 4-fold elevated risk of persistence. A greater diversity of IgE binding to linear epitopes in caseins within milk was associated with natural milk allergy persistence 29, and milk epitope binding is predictive of response to milk oral immunotherapy (OIT) 30.
Food-specific IgG and IgA are elevated in response to allergen immunotherapy 31, 32. Both isotypes can function as blocking antibodies, and IgG can also provide inhibitory signaling to basophils and mast cells through the IgG receptor FcγRIIb 33, 8 An early rise in egg-specific IgG4 was shown to be predictive of later treatment success, defined as sustained unresponsiveness to egg after a 2 week discontinuation of treatment34, however others have shown no significant difference in IgG4 levels and long-term treatment success 26.
Basophil Activation Tests
Activation of basophils, a rare circulating allergen effector cells, has been used as an ex vivo functional surrogate of in vivo allergen reactivity 35. The high affinity Fc epsilon receptor I (FcεRI) on the surface of basophils in allergic individuals is occupied by allergen specific IgE. Exposure of basophils to allergen cross-links the surface IgE, leading to the release of inflammatory mediators, including histamine. Histamine release has been correlated to the bimodal expression of surface CD63 36, later determined to be a lysosomal associated membrane protein. The cell surface marker CD203c 37 is also upregulated with basophil activation, albeit with more rapidity. Basophil activation is typically assessed using a dose response curve to increasing antigen doses38. Antigen stimulation with whole allergen extract or specific allergen protein components have been studied in food allergy.
Two measures of basophil activation based on the dose-response can be assessed. Basophil reactivity is measured by the maximal response of the dose-response or by the area-under-the-curve of the dose-response curve. Basophil sensitivity is measured as the ED50 (median effective dose) or CD-sens of the dose response curve, reflecting the antigen dose that corresponds to 50% of the maximal degranulation response.
Basophil activation testing in the diagnosis of food allergy
Basophil activation testing (BAT) has been used in the diagnosis of IgE-mediated food allergies. In peanut allergy, basophil activation to whole peanut extract has been shown to effectively identify clinically reactive versus tolerant individuals.39 The use of BAT in a step-wise approach improves the diagnostic accuracy in children with equivocal SPT and specific IgE to peanut and its components.39, 40 Basophil reactivity to whole peanut extract was associated with severity of reactions on OFC to peanut whereas the basophil sensitivity was associated with the threshold of peanut allergen that elicited a reaction on OFC.41 Subsequent work suggests that basophil activation to the immunodominant allergen Ara h 2 is also an effective predictor of peanut allergy in children.
Similarly in milk allergy, increased basophil activation to whole milk extract provided additional sensitivity and specificity for the diagnosis of IgE-mediated cow’s milk allergy in young children.42 Basophil reactivity to milk protein has been shown to be higher in milk allergic children who react to baked milk versus those who tolerate it.43 Furthermore, basophil activation to cow’s milk protein was found to be suppressed in those individuals who passed an oral milk challenge, suggesting that basophil activation may be a useful tool for monitoring development of natural tolerance over time.44
Basophil activation testing in food allergen immunotherapy
Many clinical trials of food allergen immunotherapy to many allergens, including egg45 and peanut,26, 46–50 have robustly demonstration that immunotherapy modulates basophil activation tests over time. Basophil suppression occurs in oral,26, 46–50 sublingual,51, 52 and epicutaneous53 immunotherapy. During active immunotherapy, basophil reactivity is suppressed in peanut26, 46–49 and egg45 immunotherapy. This suppression occurs in both high and low dose peanut immunotherapy54 and in multi-allergen immunotherapy as well. However, studies examining the kinetics of basophil activation during OIT found that suppression is partially transient during the maintenance phase of OIT.55 The reproducibility of basophil suppression from baseline across multiple forms and routes of active immunotherapy, during chronic antigenic delivery, has made this test a particularly useful biomarker in immunotherapy. Currently, the clinical applications of BAT testing have been assessed in research settings.
Basophil activation testing has been shown to be a biomarker of tolerance in immunotherapy. The decrease in basophil sensitivity to Ara h 2 within the first 3 months was found to predict sustained unresponsiveness after peanut OIT. Basophil reactivity to Ara h 2 and whole peanut correlates with the return of clinical reactivity after cessation of peanut OIT. Therefore, serial monitoring with basophil activation testing may be a useful non-invasive and safe monitoring tool in food allergen immunotherapy.
Allergen-specific T cells
Phenotype of allergen-responsive T cells in food allergy
Food-responsive T cells have been measured primarily by re-stimulating peripheral blood mononuclear cells (PBMCs) with food extract or purified allergens. Proliferation was initially measured by thymidine incorporation, and then the advent of carboxyfluorescein succinimidyl ester (CFSE) to identify proliferated cells allowed for simultaneous phenotypic analysis of food-responsive T cells. More recently, the use of activation markers such as CD154 (CD40L), CD69, and CD137 have enabled the detection of allergen responsive cells without extensive time in culture (minimizing culture artifact). Finally, tetramers comprised of epitopes from food allergens have enabled identification of antigen-specific T cells without the need for culture at all. All of these approaches have determined that allergic individuals have a greater frequency of food-responsive T cells than non-allergic individuals, and the food-responsive T cells are predominantly composed of Th2 cells expressing IL-4 and IL-13 56–60. Peanut allergy is associated with a population of highly differentiated Th2 cells expressing IL-5 and IL-9, and expressing receptors for IL-25 and IL-33 59, 61–63. Highly differentiated memory Th2 cells expressing CRTH2, CD161, and lacking CD27 expression have been termed Th2A cells 62. These are common to a range of allergic disorders, and therefore antigen specificity is important to their use as a biomarker for food allergy.
The association of allergen-responsive T cells in different phenotypes of food allergy have been studied. Egg-responsive Th2 cells were identified in egg allergic individuals who were reactive or tolerant to heated forms of egg 64. Although heated egg reactive individuals tended to have higher Th2 responses, this was not sufficient to discriminate between the two phenotypes of egg allergy 64, 65. In peanut allergy, individuals with differing thresholds of reactivity to peanut have been studied 59, 63. Ruiter et al studied peanut-sensitized individuals with a threshold of reactivity above or below a cumulative dose of 443 mg of peanut 63. Those with a lower threshold of reactivity had more peanut-responsive T effector cells, no difference in peanut-responsive T regulatory cells (Tregs), a greater T cell receptor (TCR) diversity in T effector cells, more Th2 cytokine production, and a unique transcriptional profile consistent with Th2A cells. They also identified a population of cells with a transcriptome consistent with Th17 cells that were enriched in those with a low threshold of reactivity.
Utility of T cells as biomarkers of food allergy
Studies examining allergen-responsive T cells have primarily been used to elucidate mechanisms underlying food allergy and development of tolerance. Low levels of allergen-responsive T cells expressing IL-4 were found to be predictive of natural resolution of milk allergy 23. Those with resolved allergy have a greater frequency of allergen-responsive Tregs than those with active food allergy 66. Allergen immunotherapy is generally associated with a reduced frequency of Th2 cells 49, 55, 62 and a small study showed an expansion of a subset of cells with an anergic phenotype 67. The approaches used to identify and phenotype T cells currently require a high level of laboratory expertise and are labor and time intensive. Thus it is unlikely that the type of assays that have been used to study T cells in food allergy could be readily translated to biomarkers. However, the identification of TCR clones that are shared between peanut-allergic individuals, and the lack of shared clones between effector and regulatory cells, suggests that it is possible to envision a simple PCR-based blood test to quantify “pathogenic” clones of T cells in blood 63.
Transcriptomic markers of food allergy
Transcriptomic profiling of food allergic individuals has been used to uncover other molecular underpinnings of food allergy. Using an in vivo approach where transcriptomic profiling was performed on peripheral blood samples obtained before, during, and after reaction from peanut allergic children undergoing DBPCFC to peanut, investigators identified genes with significant changes in expression induced by peanut but not placebo during acute allergic reactions.68 These identified peanut reaction genes were uniquely enriched in a gene co-expression module for acute phase response and pro-inflammatory pathways, for which 6 key driver genes (LTB4R, PADI4, IL1R2, PPP1R3D, KLHL2, ECHDC3) were predicted by probabilistic causal network analysis to causally modulate the state of this peanut reaction module at the most upstream level.68 The results replicated in an independent cohort, and the identified key drivers could be targeted as biomarkers of peanut allergic reactions.68
In a subsequent study of the same children, Do et al. identified transcriptomic markers of reaction severity.69 The investigators examined the symptoms experienced and reaction eliciting doses for the peanut allergic children undergoing DBPCFC to peanut to calculate threshold-weighted reaction severity scores.69 They identified and replicated 318 genes with expression level changes associated with reaction severity.69 The genes upregulated with increasing reaction severity were associated with neutrophil degranulation and neutrophil-mediated immunity, and interaction networks for these severity-associated genes revealed NFKBIA and ARG1 as hubs.69
Gene expression studies have also identified signatures of immature neonatal T-cell function in children who subsequently develop food allergy. Specifically, peripheral blood CD4+ T cells collected at birth and at age 1 year in children with and without food allergy by age 1 year have been transcriptionally examined.70 Compared to the non-allergic group, the allergic group at birth had fewer genes upregulated in response to anti-CD3 treatment, suggesting reduced capacity for T cell proliferation.70 This differential response to polyclonal activation was not seen at age 1 year, suggesting transience in this suboptimal neonatal T-cell activation.70
Epigenomic markers of food allergy
As food allergy is mediated by genetic and environmental risk factors, there has also been interest in epigenomic markers of food allergy. Using methylation profiles from blood mononuclear cells obtained from 29 egg or peanut allergic and 42 non-food allergic subjects, a supervised learning approach was used to identify a classifier based on 96 CpG sites to classify OFC outcomes.71 Testing of this classifier in an independent cohort of 12 food allergic and 12 nonallergic controls assayed at two time points demonstrated accurate classification of OFC result for 79.2% of the outcomes.71
In addition to identifying peripheral blood transcripts associated with reaction severity in peanut allergic children as discussed above in “Transcriptomic Markers of Food Allergy”, Do et al. performed parallel genome-wide DNA methylation profiling of peripheral blood CD4+ lymphocytes from these children.69 From these methylome data, they identified 203 CpG sites with differential methylation associated with reaction severity.69 The identified CpG sites were replicated in an independent cohort. Integrated analyses of the parallel transcriptional and methylation signatures of reaction severity revealed that the correlations between severity-associated CpG methylation and gene expression were mostly negative (i.e. lower CpG methylation at baseline was associated with increased gene expression associated with severity).69 Network analyses incorporating the peanut severity genes and peanut severity CpGs highlighted biological processes for chemotaxis, immune response, and regulation of macroautophagy.69 Causal mediation analyses revealed that the associations between two CpGs and reaction severity were each causally mediated by gene expression changes of their respective genes (PHACTR1 and ZNF121), suggesting that CpG methylation may serve as an anchor upon which gene expression modulates reaction severity.69
Also employing genome-wide DNA methylation and transcriptional profiling, another group investigated naïve CD4+ T cell activation in egg-allergic children and controls at age 12 months and at follow-up at age 2 or 4 years, when 59% had resolved allergy.72.65 In line with the group’s prior gene expression finding of reduced capacity for T cell proliferation in neonates who later develop food allergy70, the investigators found reduced lymphoproliferative responses to polyclonal activation of naive CD4+ T cells in children with food allergy.72 The reduced response was associated with lower expression of E2F and MYC transcription factor networks and remodeling of DNA methylation at metabolic and inflammatory genes.72
Microbiome markers of food allergy
Accumulating findings about the gut microbiome and food allergy suggest potential roles for microbial markers of food allergy.73–76 Observational human cohort studies have demonstrated differences in gut microbiota in individuals with and without food allergy. For example, a multi-center study of infants with egg allergy vs. non-food allergic controls revealed compositional differences in their gut microbiota, with genera from Lachnospiraceae and Streptococcaceae significantly more abundant, and Leuconostoceae significantly reduced, in the gut microbiota of egg allergic infants.77 Separately, studies of milk allergic infants have shown that their gut microbial community structures are dominated by Lachnospiraceae and Ruminococcaceae compared to healthy controls.78
Other studies have reported associations between gut microbiota and food allergy trajectory over time, suggesting the possibility of microbial markers of food allergy course. 73–76 A multi-center study of 226 infants with milk allergy who were followed up to age 8 years identified taxa associated with the eventual resolution of milk allergy.79 Specifically, among infants age 3-6 months, taxa from the Firmicutes phylum, including Clostridia, were enriched in the infant gut microbiota of subjects whose milk allergy later resolved by age 8 years, whereas taxa from Bacteroidetes were more abundant in the infant gut microbiota of subjects with persistent milk allergy.79 In another population-based study of 166 infants, each quartile increase in the ratio of Enterobacteriaceae/Bacteroidaceae at 3 months was associated with two-fold risk of sensitization to at least one common food allergen by age 1 year.80 Separately, other investigators found that lower relative abundances of Haemophilus, Dialister, Dorea, and Clostridium in gut microbiota at age 3-6 months was associated with sensitization to at least 1 food allergen at age 3 years.81
Metabolomics
Metabolomics is the untargeted measurement of small molecule end products (metabolites) of cellular processes. This encompasses measurement of a range of physically distinct categories of molecules by liquid chromatography-mass spectrometry. Measurement of metabolites is a way to assess the biological consequences of gene expression. Metabolomics is a relatively young field, and the information pertaining to food allergy remains sparse.
Obeso et al studied PBMC transcriptomics together with plasma metabolomics in a European cohort of patients with severe allergy to profilin on oral challenge 82. Pathway analysis of differentially expressed genes in those with severe versus mild, or severe versus moderate allergy identified platelet activation pathways. Metabolomics identified a number of differentially expressed molecules related to platelet biology, including sphingosine and lysophospholipids. This study is an example of a hypothesis generating use of -omics in food allergy, pointing to an important role of platelets in food allergy severity. This is consistent with previous findings, such as a positive association between anaphylaxis severity and platelet activating factor (PAF) 83, 84, and findings of basophil-platelet interactions in peanut allergy 85.
Crestani et al recently compared the metabolomic profile of 70 individuals with food allergy +/− asthma 86. The well-characterized cohort allowed for the analysis of food allergy, asthma, anaphylaxis, multi-food allergy, and food allergy severity (through history of anaphylaxis). Comparing food allergics to controls (healthy or asthmatic controls), sphingolipids, lysophospholipids, and ceramides were significantly different in abundance 86. Multi-food allergy was associated with tryptophan, a key metabolite in the production of serotonin. A metabolite upstream of PAF, 1-arachidonoyl-GPD, was found to be significantly and inversely associated with food allergy.
From these two independent studies on diverse cohorts of food allergy, we can conclude that pathways related to sphingolipids, lysophospholipids, and platelet biology are altered in food allergy. Furthermore, the consistent findings between the two studies support the idea that measurement of a panel of candidate metabolites could be a useful biomarker of food allergy.
Multifactorial prediction models
Combinatorial use of multiple biomarkers has been used to design approaches for both reducing the need for oral food challenges and predicting the severity of reactions. When basophil activation is used in a step-wise approach along with specific IgE measurement, the need for diagnostic challenges can be theoretically reduced 39, 40.
Clinical tools to predict the severity of reactions on oral challenge have used multivariate analyses for multiple foods, including hazelnut 87, peanut88, cow’s milk89, 90, and wheat91. In these approaches, the severity of clinical reactions were grouped into a categorical variable, followed by the use of multiple biomarkers and allergic comorbidities, such as atopic dermatitis, asthma, specific IgE levels, skin prick testing, component IgE testing, and basophil activation. In particular, this approach increases the diagnostic accuracy with foods such as hazelnut, for which individual biomarker performance is considerably less accurate87.
Conclusions and Future Directions
As reviewed here and summarized in Table 1 (Figure 2), there has been a remarkable expansion in candidate biomarkers for use in food allergy. Of these biomarkers, the basophil activation test has been most widely tested in diverse clinical cohorts and has the strongest supporting evidence for the potential to reduce the clinical need for oral food challenges. At this point, there needs to be a shift to testing the utility of this biomarker for use in guiding clinical care rather than remaining a research tool. Epitope binding is similarly close to testing as a biomarker for clinical setting.
Table 1:
Summary of the utility of biomarkers in prediction of clinical reactivity and key clinical parameters
| Biomarker | Allergy Y/N | Threshold | Severity | Natural Resolution | Response to IT |
|---|---|---|---|---|---|
| Food-specific IgE | ++ | + | +/− | + | + |
| Component-specific IgE | ++ | − | − | + | + |
| Food-specific IgG | − | − | − | − | + |
| Food-specific IgA | − | − | − | − | + |
| Epitope-specific IgE | ++ | ND | ND | ND | ++ |
| Basophil Activation Test | +++ | ++ | ++ | + | ++ |
| Food-specific T cell | + | + | ND | + | + |
| Transcriptomics | + | ND | + | ND | ND |
| Epigenomics | + | ND | + | ND | ND |
| Microbiome | + | ND | ND | + | ND |
| Metabolomics | −/+ | ND | ND | ND | ND |
ND = not done. − not predictive, + some evidence, +++ strong evidence, −/+ data for and against
Figure 2.
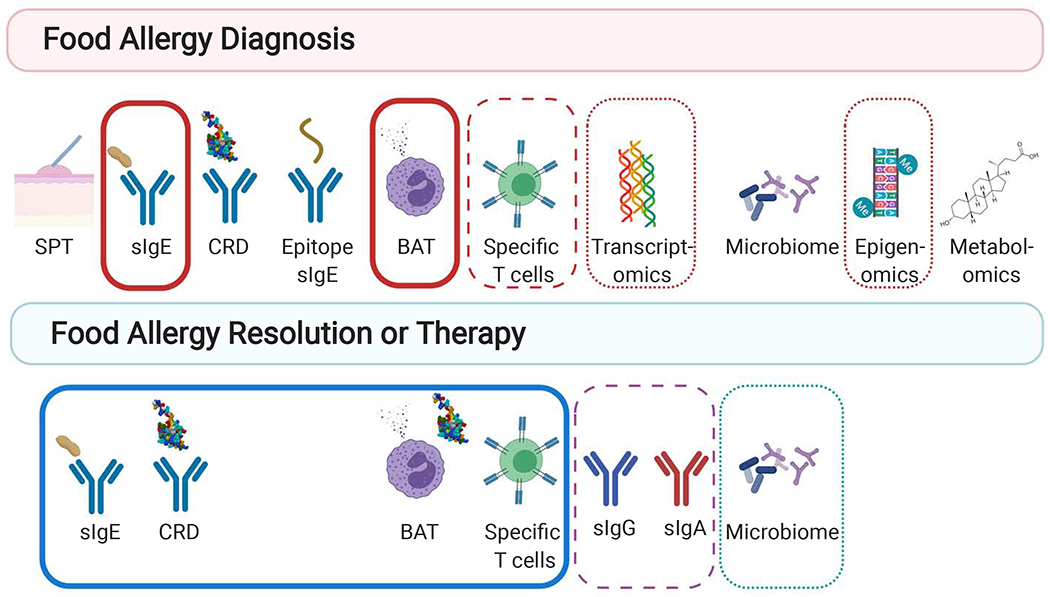
Utility of Biomarkers in Food Allergy. Diagnostic biomarkers of food allergy include include skin prick testing (SPT), specific immunoglobulins (sIgE), component resolved diagnostics (CRD), epitope specific IgE, basophil activation, allergen-specific T cells, transcriptomics, epigenomics, microbiome, and metabolomics. Biomarkers correlate with both the threshold and severity of reactions on oral food challenge (OFC) (solid red outline) or have been correlated with only threshold (dashed red outline) or severity (dotted red outline). Biomarkers of tolerance due to either natural tolerance of therapeutic intervention include sIgE, CRD, BAT, allergen-specific T cells, allergen specific IgG and IgA, and microbiome. Biomarkers of both types of tolerance are outlined in blue, biomarkers of therapeutic intervention are outlined in a purple dashed line, and biomarkers of natural resolution are outlined in a green dotted line (Created with Biorender.com).
As is the case for other biomarker candidates, the clinical translation of biomarkers identified via -omics approaches will involve development work to ensure that the particular features sets identified via -omic approaches (e.g. selected sets of genes, CpG loci, microbiota, and/or metabolites) can be measured and interpreted in clinical settings.
Learning objectives:
To identify and evaluate biomarkers relevant for diagnosis, prognosis and treatment of food allergy.
To evaluate emerging biomarkers for the diagnosis and treatment of food allergy.
Questions:
1. Which of the following is true of component resolved diagnostics (CRD) in the diagnosis of food allergy?
It is the measurement of IgE to allergenic epitopes.
It improves predictive performance in the diagnosis of peanut allergy.
It does not have any geographic diversity.
It does not distinguish sensitization to pollen cross-reactive allergens.
Answer: B
Explanation: The clinical utility of CRD in the diagnosis of peanut allergy has been established by numerous studies, which have find that Ara h 2 specific IgE has a better predictive performance for the diagnosis of peanut allergy when compared to peanut specific IgE alone. Epitope specific IgE, not CRD, measures IgE specific to epitopes within allergenic proteins. CRD varies by geography and can be used to identify sensitization to crossreactive pollen proteins.
2. Which of the following describes the clinical utility of basophil activation testing?
Basophil activation to allergenic components is not a predictor of peanut allergy.
Basophil activation testing improves diagnostic accuracy in peanut allergy but not milk allergy.
A decrease in basophil sensitivity early in peanut oral immunotherapy correlates with tolerance.
Suppression of basophil activation occurs only in immunotherapy but not in natural resolution of milk allergy.
Answer: C
Explanation: Previous studies have shown that a decrease in basophil sensitivity early in peanut oral immunotherapy is a biomarker of tolerance. Basophil activation to Ara h 2 has been shown to be an effector predictor of peanut allergy. Basophil activation testing improves diagnostic accuracy in both peanut and milk allergy. Suppression of basophil activation occurs both in immunotherapy and the natural resolution of milk allergy.
3. Which of the following is true about the T cells in food allergy?
Peanut allergy is not correlated with highly differentiated, allergen-specific Th2 cells that also express receptors for IL-25 and IL-33.
Peanut allergic individuals with a low threshold of reactivity (<443 mg peanut protein) have been found to have an increased frequency of peanut-responsive T effector cells.
Highly differentiated TH2A cells are specific to food allergy.
Egg-responsive Th2 cells are a robust biomarker of tolerance to heated forms of egg.
Answer: b
Explanation: Peanut allergic individuals with a low threshold of reactivity (<443 mg of peanut) have been found to have an increased frequency of peanut-responsive T effector cells. Previously, studies have demonstrated the presence of highly differentiated, allergen-specific Th2 cells that also express receptors for IL-25 and IL-33 in peanut allergy. Highly differentiated TH2A cells are not specific to food allergy as they also can be identified in other allergic diseases. Although heated egg reactive individuals tended to have higher Th2 responses, this was not sufficient to discriminate between the two phenotypes of egg allergy
4. Which of the following is true about the microbiome as a biomarker in food allergy?
The gut microbiome is identical in those with and without current food allergy.
Early gut microbiome composition does not influence food allergy outcomes.
No relationships between gut microbiome and food allergen sensitization have been identified.
Gut microbiota differences have been found in milk-allergic individuals and in those with persistent versus transient milk allergy.
Answer: d
Explanation: Two studies have found differences in gut microbiota in milk allergic versus nonallergic individuals and in those with persistent versus transient milk allergy (answer D is correct). The other answers are incorrect because gut microbiome has been associated with current food allergy, and outcomes for food sensitization and allergy.
Acknowledgments
Funding sources: S.U.P. is supported through National Institutes of Health NIAID grant K23AI121491. M.C.B. is supported by National Institutes of Health NIAID grant U19AI136053.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: S. U. Patil is supported through National Institutes of Health NIAID grant K23AI121491. S. Bunyavanich declares no relevant conflicts of interest. M. C. Berin serves on the Scientific Advisory Board of Prota Therapeutics; and is supported by National Institutes of Health NIAID grant U19AI136053.
CME EXAM
Issue: September 2020 (Diagnostic Methods in Allergy theme)
Review series: Clinical Management
MS# & Article title: Emerging food allergy biomarkers
Author(s): Drs. Sarita U. Patil, Supinda Bunyavanich, M. Cecilia Berin
Editor: Scott H. Sicherer, MD
Editor disclosure: S. H. Sicherer declares no conflicts to disclose.
References
- 1.Sampson HA, Gerth van Wijk R, Bindslev-Jensen C, Sicherer S, Teuber SS, Burks AW, et al. Standardizing double-blind, placebo-controlled oral food challenges: American Academy of Allergy, Asthma & Immunology-European Academy of Allergy and Clinical Immunology PRACTALL consensus report. J Allergy Clin Immunol 2012; 130:1260–74. [DOI] [PubMed] [Google Scholar]
- 2.Grabenhenrich LB, Reich A, Bellach J, Trendelenburg V, Sprikkelman AB, Roberts G, et al. A new framework for the documentation and interpretation of oral food challenges in population-based and clinical research. Allergy 2017; 72:453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sampson HA, Ho DG. Relationship between food-specific IgE concentrations and the risk of positive food challenges in children and adolescents. J Allergy Clin Immunol 1997; 100:444–51. [DOI] [PubMed] [Google Scholar]
- 4.Sampson HA. Utility of food-specific IgE concentrations in predicting symptomatic food allergy. J Allergy Clin Immunol 2001; 107:891–6. [DOI] [PubMed] [Google Scholar]
- 5.Perry TT, Matsui EC, Kay Conover-Walker M, Wood RA. The relationship of allergenspecific IgE levels and oral food challenge outcome. The Journal of Allergy and Clinical Immunology 2004; 114:144–9. [DOI] [PubMed] [Google Scholar]
- 6.Celik-Bilgili S, Mehl A, Verstege A, Staden U, Nocon M, Beyer K, et al. The predictive value of specific immunoglobulin E levels in serum for the outcome of oral food challenges. Clin Exp Allergy 2005; 35:268–73. [DOI] [PubMed] [Google Scholar]
- 7.Komata T, Soderstrom L, Borres MP, Tachimoto H, Ebisawa M. The predictive relationship of food-specific serum IgE concentrations to challenge outcomes for egg and milk varies by patient age. J Allergy Clin Immunol 2007; 119:1272–4. [DOI] [PubMed] [Google Scholar]
- 8.Nicolaou N, Poorafshar M, Murray C, Simpson A, Winell H, Kerry G, et al. Allergy or tolerance in children sensitized to peanut: prevalence and differentiation using component-resolved diagnostics. J Allergy Clin Immunol 2010; 125:191–7 e1–13. [DOI] [PubMed] [Google Scholar]
- 9.Dang TD, Tang M, Choo S, Licciardi PV, Koplin JJ, Martin PE, et al. Increasing the accuracy of peanut allergy diagnosis by using Ara h 2. J Allergy Clin Immunol 2012; 129:1056–63. [DOI] [PubMed] [Google Scholar]
- 10.Klemans RJ, Broekman HC, Knol EF, Bruijnzeel-Koomen CA, Otten HG, Pasmans SG, et al. Ara h 2 is the best predictor for peanut allergy in adults. J Allergy Clin Immunol Pract 2013; 1:632–8 e1. [DOI] [PubMed] [Google Scholar]
- 11.Masthoff LJ, Mattsson L, Zuidmeer-Jongejan L, Lidholm J, Andersson K, Akkerdaas JH, et al. Sensitization to Cor a 9 and Cor a 14 is highly specific for a hazelnut allergy with objective symptoms in Dutch children and adults. J Allergy Clin Immunol 2013; 132:393–9. [DOI] [PubMed] [Google Scholar]
- 12.Shreffler WG, Beyer K, Chu T- HT, Burks AW, Sampson HA. Microarray immunoassay: association of clinical history, in vitro IgE function, and heterogeneity of allergenic peanut epitopes. J Allergy Clin Immunol 2004; 113:776–82. [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Bruni FM, Fu Z, Maloney J, Bardina L, Boner AL, et al. A bioinformatics approach to identify patients with symptomatic peanut allergy using peptide microarray immunoassay. J Allergy Clin Immunol 2012; 129:1321–8 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sackesen C, Suarez-Farinas M, Sillva R, Lin J, Schmidt S, Getts R, et al. A new luminexbased peptide assay to identify reactivity to baked, fermented and whole milk. Allergy 2018. [DOI] [PubMed] [Google Scholar]
- 15.Suprun M, Grishina G, Henning A, Sicherer S, Wood R, Jones S, et al. Peanut epitopespecific IgE binding can predict clinical peanut allergy. Allergy 2018; 73:116. [Google Scholar]
- 16.Nugraha R, Kamath SD, Johnston E, Karnaneedi S, Ruethers T, Lopata AL. Conservation Analysis of B-Cell Allergen Epitopes to Predict Clinical Cross-Reactivity Between Shellfish and Inhalant Invertebrate Allergens. Front Immunol 2019; 10:2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panel NI-SE, Boyce JA, Assa’ad A, Burks AW, Jones SM, Sampson HA, et al. Guidelines for the diagnosis and management of food allergy in the United States: report of the NIAIDsponsored expert panel. J Allergy Clin Immunol 2010; 126:S1–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stapel SO, Asero R, Ballmer-Weber BK, Knol EF, Strobel S, Vieths S, et al. Testing for IgG4 against foods is not recommended as a diagnostic tool: EAACI Task Force Report. Allergy 2008; 63:793–6. [DOI] [PubMed] [Google Scholar]
- 19.Husby S, Oxelius VA, Teisner B, Jensenius JC, Svehag SE. Humoral immunity to dietary antigens in healthy adults. Occurrence, isotype and IgG subclass distribution of serum antibodies to protein antigens. Int Arch Allergy Appl Immunol 1985; 77:416–22. [DOI] [PubMed] [Google Scholar]
- 20.Rolinck-Werninghaus C, Niggemann B, Grabenhenrich L, Wahn U, Beyer K. Outcome of oral food challenges in children in relation to symptom-eliciting allergen dose and allergen-specific IgE. Allergy 2012; 67:951–7. [DOI] [PubMed] [Google Scholar]
- 21.Elegbede CF, Papadopoulos A, Just J, Moneret-Vautrin DA, Deschildre A, Crepet A. Gender, prick test size and rAra h 2 sIgE level may predict the eliciting dose in patients with peanut allergy: Evidence from the Mirabel survey. Clin Exp Allergy 2019; 49:677–89. [DOI] [PubMed] [Google Scholar]
- 22.Dreskin SC, Germinaro M, Reinhold D, Chen X, Vickery BP, Kulis M, et al. IgE binding to linear epitopes of Ara h 2 in peanut allergic preschool children undergoing oral Immunotherapy. Pediatr Allergy Immunol 2019; 30:817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood RA, Sicherer SH, Vickery BP, Jones SM, Liu AH, Fleischer DM, et al. The natural history of milk allergy in an observational cohort. J Allergy Clin Immunol 2013; 131:805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sicherer SH, Wood RA, Vickery BP, Jones SM, Liu AH, Fleischer DM, et al. The natural history of egg allergy in an observational cohort. J Allergy Clin Immunol 2014; 133:492–9 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters RL, Allen KJ, Dharmage SC, Koplin JJ, Dang T, Tilbrook KP, et al. Natural history of peanut allergy and predictors of resolution in the first 4 years of life: A population-based assessment. J Allergy Clin Immunol 2015; 135:1257–66 e1–2. [DOI] [PubMed] [Google Scholar]
- 26.Vickery BP, Scurlock AM, Kulis M, Steele PH, Kamilaris J, Berglund JP, et al. Sustained unresponsiveness to peanut in subjects who have completed peanut oral immunotherapy. J Allergy Clin Immunol 2014; 133:468–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chinthrajah RS, Purington N, Andorf S, Long A, O’Laughlin KL, Lyu SC, et al. Sustained outcomes in oral immunotherapy for peanut allergy (POISED study): a large, randomised, double-blind, placebo-controlled, phase 2 study. Lancet 2019; 394:1437–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang TD, Peters RL, Koplin JJ, Dharmage SC, Gurrin LC, Ponsonby AL, et al. Egg allergen specific IgE diversity predicts resolution of egg allergy in the population cohort HealthNuts. Allergy 2019; 74:318–26. [DOI] [PubMed] [Google Scholar]
- 29.Caubet JC, Lin J, Ahrens B, Gimenez G, Bardina L, Niggemann B, et al. Natural tolerance development in cow’s milk allergic children: IgE and IgG4 epitope binding. Allergy 2017; 72:1677–85. [DOI] [PubMed] [Google Scholar]
- 30.Wood RA, Kim JS, Lindblad R, Nadeau K, Henning AK, Dawson P, et al. A randomized, double-blind, placebo-controlled study of omalizumab combined with oral immunotherapy for the treatment of cow’s milk allergy. J Allergy Clin Immunol 2016; 137:1103–10 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulis M, Saba K, Kim EH, Bird JA, Kamilaris N, Vickery BP, et al. Increased peanut-specific IgA levels in saliva correlate with food challenge outcomes after peanut sublingual immunotherapy. J Allergy Clin Immunol 2012; 129:1159–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright BL, Kulis M, Orgel KA, Burks AW, Dawson P, Henning AK, et al. Componentresolved analysis of IgA, IgE, and IgG4 during egg OIT identifies markers associated with sustained unresponsiveness. Allergy 2016; 71:1552–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burton OT, Tamayo JM, Stranks AJ, Koleoglou KJ, Oettgen HC. Allergen-specific IgG antibody signaling through FcgammaRIIb promotes food tolerance. J Allergy Clin Immunol 2018; 141:189–201 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burks AW, Jones SM, Wood RA, Fleischer DM, Sicherer SH, Lindblad RW, et al. Oral immunotherapy for treatment of egg allergy in children. The New England journal of medicine 2012; 367:233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann HJ, Santos AF, Mayorga C, Nopp A, Eberlein B, Ferrer M, et al. The clinical utility of basophil activation testing in diagnosis and monitoring of allergic disease. Allergy 2015; 70:1393–405. [DOI] [PubMed] [Google Scholar]
- 36.Knol EF, Mul FP, Jansen H, Calafat J, Roos D. Monitoring human basophil activation via CD63 monoclonal antibody 435. J Allergy Clin Immunol 1991; 88:328–38. [DOI] [PubMed] [Google Scholar]
- 37.Buhring HJ, Simmons PJ, Pudney M, Muller R, Jarrossay D, van Agthoven A, et al. The monoclonal antibody 97A6 defines a novel surface antigen expressed on human basophils and their multipotent and unipotent progenitors. Blood 1999; 94:2343–56. [PubMed] [Google Scholar]
- 38.Patil SU, Shreffler WG. Immunology in the Clinic Review Series; focus on allergies: basophils as biomarkers for assessing immune modulation. Clin Exp Immunol 2012; 167:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos AF, Douiri A, Becares N, Wu SY, Stephens A, Radulovic S, et al. Basophil activation test discriminates between allergy and tolerance in peanut-sensitized children. J Allergy Clin Immunol 2014; 134:645–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos AF, Shreffler WG. Road map for the clinical application of the basophil activation test in food allergy. Clin Exp Allergy 2017; 47:1115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santos AF, Du Toit G, Douiri A, Radulovic S, Stephens A, Turcanu V, et al. Distinct parameters of the basophil activation test reflect the severity and threshold of allergic reactions to peanut. J Allergy Clin Immunol 2015; 135:179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruinemans-Koerts J, Schmidt-Hieltjes Y, Jansen A, Savelkoul HFJ, Plaisier A, van Setten P. The Basophil Activation Test reduces the need for a food challenge test in children suspected of IgE-mediated cow’s milk allergy. Clin Exp Allergy 2019; 49:350–6. [DOI] [PubMed] [Google Scholar]
- 43.Ford LS, Bloom KA, Nowak-Wegrzyn AH, Shreffler WG, Masilamani M, Sampson HA. Basophil reactivity, wheal size, and immunoglobulin levels distinguish degrees of cow’s milk tolerance. J Allergy Clin Immunol 2013; 131:180–6 e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubio A, Vivinus-Nebot M, Bourrier T, Saggio B, Albertini M, Bernard A. Benefit of the basophil activation test in deciding when to reintroduce cow’s milk in allergic children. Allergy 2011; 66:92–100. [DOI] [PubMed] [Google Scholar]
- 45.Vila L, Moreno A, Gamboa PM, Martinez-Aranguren R, Sanz ML. Decrease in antigenspecific CD63 basophil expression is associated with the development of tolerance to egg by SOTI in children. Pediatr Allergy Immunol 2013; 24:463–8.23682931 [Google Scholar]
- 46.Jones SM, Pons L, Roberts JL, Scurlock AM, Perry TT, Kulis M, et al. Clinical efficacy and immune regulation with peanut oral immunotherapy. J Allergy Clin Immunol 2009; 124:292–300, e1–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patil SU, Steinbrecher J, Calatroni A, Smith N, Ma A, Ruiter B, et al. Early decrease in basophil sensitivity to Ara h 2 precedes sustained unresponsiveness after peanut oral immunotherapy. J Allergy Clin Immunol 2019; 144:1310–9 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thyagarajan A, Jones SM, Calatroni A, Pons L, Kulis M, Woo CS, et al. Evidence of pathway-specific basophil anergy induced by peanut oral immunotherapy in peanutallergic children. Clin Exp Allergy 2012; 42:1197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varshney P, Jones SM, Scurlock AM, Perry TT, Kemper A, Steele P, et al. A randomized controlled study of peanut oral immunotherapy: clinical desensitization and modulation of the allergic response. J Allergy Clin Immunol 2011; 127:654–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai M, Mukai K, Chinthrajah RS, Nadeau KC, Galli SJ. Sustained successful peanut oral immunotherapy associated with low basophil activation and peanut-specific IgE. J Allergy Clin Immunol 2020; 145:885–96 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim EH, Bird JA, Kulis M, Laubach S, Pons L, Shreffler W, et al. Sublingual immunotherapy for peanut allergy: clinical and immunologic evidence of desensitization. J Allergy Clin Immunol 2011; 127:640–6 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim EH, Yang L, Ye P, Guo R, Li Q, Kulis MD, et al. Long-term sublingual immunotherapy for peanut allergy in children: Clinical and immunologic evidence of desensitization. J Allergy Clin Immunol 2019; 144:1320–6 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones SM, Sicherer SH, Burks AW, Leung DY, Lindblad RW, Dawson P, et al. Epicutaneous immunotherapy for the treatment of peanut allergy in children and young adults. J Allergy Clin Immunol 2017; 139:1242–52 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kulis M, Yue X, Guo R, Zhang H, Orgel K, Ye P, et al. High-and low-dose oral immunotherapy similarly suppress pro-allergic cytokines and basophil activation in young children. Clin Exp Allergy 2019; 49:180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorelik M, Narisety SD, Guerrerio AL, Chichester KL, Keet CA, Bieneman AP, et al. Suppression of the immunologic response to peanut during immunotherapy is often transient. J Allergy Clin Immunol 2015; 135:1283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turcanu V, Maleki SJ, Lack G. Characterization of lymphocyte responses to peanuts in normal children, peanut-allergic children, and allergic children who acquired tolerance to peanuts. J Clin Invest 2003; 111:1065–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeLong JH, Simpson KH, Wambre E, James EA, Robinson D, Kwok WW. Ara h 1-reactive T cells in individuals with peanut allergy. J Allergy Clin Immunol 2011; 127:1211–8 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wisniewski JA, Commins SP, Agrawal R, Hulse KE, Yu MD, Cronin J, et al. Analysis of cytokine production by peanut-reactive T cells identifies residual Th2 effectors in highly allergic children who received peanut oral immunotherapy. Clin Exp Allergy 2015; 45:1201–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiang D, Chen X, Jones SM, Wood RA, Sicherer SH, Burks AW, et al. Single-cell profiling of peanut-responsive T cells in patients with peanut allergy reveals heterogeneous effector TH2 subsets. J Allergy Clin Immunol 2018; 141:2107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weissler KA, Rasooly M, DiMaggio T, Bolan H, Cantave D, Martino D, et al. Identification and analysis of peanut-specific effector T and regulatory T cells in children allergic and tolerant to peanut. J Allergy Clin Immunol 2018; 141:1699–710 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brough HA, Cousins DJ, Munteanu A, Wong YF, Sudra A, Makinson K, et al. IL-9 is a key component of memory TH cell peanut-specific responses from children with peanut allergy. J Allergy Clin Immunol 2014; 134:1329–38 e10. [DOI] [PubMed] [Google Scholar]
- 62.Wambre E, Bajzik V, DeLong JH, O’Brien K, Nguyen QA, Speake C, et al. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med 2017; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruiter B, Smith NP, Monian B, Tu AA, Fleming E, Virkud YV, et al. Expansion of the CD4(+) effector T-cell repertoire characterizes peanut-allergic patients with heightened clinical sensitivity. J Allergy Clin Immunol 2020; 145:270–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berin MC, Grishin A, Masilamani M, Leung DYM, Sicherer SH, Jones SM, et al. Eggspecific IgE and basophil activation but not egg-specific T-cell counts correlate with phenotypes of clinical egg allergy. J Allergy Clin Immunol 2018; 142:149–58 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kosoy R, Agashe C, Grishin A, Leung DY, Wood RA, Sicherer SH, et al. Transcriptional Profiling of Egg Allergy and Relationship to Disease Phenotype. PLoS One 2016; 11:e0163831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qamar N, Fishbein AB, Erickson KA, Cai M, Szychlinski C, Bryce PJ, et al. Naturally occurring tolerance acquisition to foods in previously allergic children is characterized by antigen specificity and associated with increased subsets of regulatory T cells. Clin Exp Allergy 2015; 45:1663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ryan JF, Hovde R, Glanville J, Lyu SC, Ji X, Gupta S, et al. Successful immunotherapy induces previously unidentified allergen-specific CD4+ T-cell subsets. Proc Natl Acad Sci U S A 2016; 113:E1286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watson CT, Cohain AT, Griffin RS, Chun Y, Grishin A, Hacyznska H, et al. Integrative transcriptomic analysis reveals key drivers of acute peanut allergic reactions. Nat Commun 2017; 8:1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Do AN, Watson CT, Cohain AT, Griffin RS, Grishin A, Wood RA, et al. Dual transcriptomic and epigenomic study of reaction severity in peanut allergic children. J Allergy Clin Immunol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martino DJ, Bosco A, McKenna KL, Hollams E, Mok D, Holt PG, et al. T-cell activation genes differentially expressed at birth in CD4+ T-cells from children who develop IgE food allergy. Allergy 2012; 67:191–200. [DOI] [PubMed] [Google Scholar]
- 71.Martino D, Dang T, Sexton-Oates A, Prescott S, Tang ML, Dharmage S, et al. Blood DNA methylation biomarkers predict clinical reactivity in food-sensitized infants. J Allergy Clin Immunol 2015; 135:1319–28 e1–12. [DOI] [PubMed] [Google Scholar]
- 72.Martino D, Neeland M, Dang T, Cobb J, Ellis J, Barnett A, et al. Epigenetic dysregulation of naive CD4+ T-cell activation genes in childhood food allergy. Nat Commun 2018; 9:3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bunyavanich S Food allergy: could the gut microbiota hold the key? Nat Rev Gastroenterol Hepatol 2019; 16:201–2. [DOI] [PubMed] [Google Scholar]
- 74.Zhao W, Ho HE, Bunyavanich S. The gut microbiome in food allergy. Ann Allergy Asthma Immunol 2019; 122:276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ho HE, Bunyavanich S. Role of the Microbiome in Food Allergy. Curr Allergy Asthma Rep 2018; 18:27. [DOI] [PubMed] [Google Scholar]
- 76.Bunyavanich S, Berin MC. Food allergy and the microbiome: Current understandings and future directions. J Allergy Clin Immunol 2019; 144:1468–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fazlollahi M, Chun Y, Grishin A, Wood RA, Burks AW, Dawson P, et al. Early-life gut microbiome and egg allergy. Allergy 2018; 73:1515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J 2016; 10:742–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bunyavanich S, Shen N, Grishin A, Wood R, Burks W, Dawson P, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016; 138:1122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy 2015; 45:632–43. [DOI] [PubMed] [Google Scholar]
- 81.Savage JH, Lee-Sarwar KA, Sordillo J, Bunyavanich S, Zhou Y, O’Connor G, et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018; 73:145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Obeso D, Mera-Berriatua L, Rodriguez-Coira J, Rosace D, Fernandez P, Martin-Antoniano IA, et al. Multi-omics analysis points to altered platelet functions in severe foodassociated respiratory allergy. Allergy 2018; 73:2137–49. [DOI] [PubMed] [Google Scholar]
- 83.Vadas P, Perelman B, Liss G. Platelet-activating factor, histamine, and tryptase levels in human anaphylaxis. J Allergy Clin Immunol 2013; 131:144–9. [DOI] [PubMed] [Google Scholar]
- 84.Vadas P, Gold M, Perelman B, Liss GM, Lack G, Blyth T, et al. Platelet-activating factor, PAF acetylhydrolase, and severe anaphylaxis. N Engl J Med 2008; 358:28–35. [DOI] [PubMed] [Google Scholar]
- 85.Tordesillas L, Rahman AH, Hartmann BM, Sampson HA, Berin MC. Mass cytometry profiling the response of basophils and the complete peripheral blood compartment to peanut. J Allergy Clin Immunol 2016; 138:1741–4 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crestani E, Harb H, Charbonnier LM, Leirer J, Motsinger-Reif A, Rachid R, et al. Untargeted metabolomic profiling identifies disease-specific signatures in food allergy and asthma. J Allergy Clin Immunol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Datema MR, van Ree R, Asero R, Barreales L, Belohlavkova S, de Blay F, et al. Component-resolved diagnosis and beyond: Multivariable regression models to predict severity of hazelnut allergy. Allergy 2018; 73:549–59. [DOI] [PubMed] [Google Scholar]
- 88.Chinthrajah RS, Purington N, Andorf S, Rosa JS, Mukai K, Hamilton R, et al. Development of a tool predicting severity of allergic reaction during peanut challenge. Ann Allergy Asthma Immunol 2018; 121:69–76 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kawahara T, Tezuka J, Ninomiya T, Honjo S, Masumoto N, Nanishi M, et al. Risk prediction of severe reaction to oral challenge test of cow’s milk. Eur J Pediatr 2019; 178:181–8. [DOI] [PubMed] [Google Scholar]
- 90.Sugiura S, Sasaki K, Matsui T, Nakagawa T, Kando N, Ito K. Development of a prediction model for a severe reaction in cow’s milk challenges. Allergol Int 2017; 66:493–4. [DOI] [PubMed] [Google Scholar]
- 91.Sugiura S, Matsui T, Furuta T, Sasaki K, Kando N, Ito K. Development of a prediction model for severe wheat allergy. Pediatr Allergy Immunol 2018; 29:93–6. [DOI] [PubMed] [Google Scholar]


