The MYB transcription factor FUSED LEAVES1 regulates and abscisic acid influences cuticle biosynthesis and cuticle-mediated drought response during the juvenile phase of maize plant growth.
Abstract
In all land plants, the outer surface of aerial parts is covered by the cuticle, a complex lipid layer that constitutes a barrier against damage caused by environmental factors and provides protection against nonstomatal water loss. We show in this study that both cuticle deposition and cuticle-dependent leaf permeability during the juvenile phase of plant development are controlled by the maize (Zea mays) transcription factor ZmFUSED LEAVES 1 (FDL1)/MYB94. Biochemical analysis showed altered cutin and wax biosynthesis and deposition in fdl1-1 mutant seedlings at the coleoptile stage. Among cutin compounds, ω-hydroxy fatty acids and polyhydroxy-fatty acids were specifically affected, while the reduction of epicuticular waxes was mainly observed in primary long chain alcohols and, to a minor extent, in long-chain wax esters. Transcriptome analysis allowed the identification of candidate genes involved in lipid metabolism and the assembly of a proposed pathway for cuticle biosynthesis in maize. Lack of ZmFDL1/MYB94 affects the expression of genes located in different modules of the pathway, and we highlighted the correspondence between gene transcriptional variations and biochemical defects. We observed a decrease in cuticle-dependent leaf permeability in maize seedlings exposed to drought as well as abscisic acid treatment, which implies coordinated changes in the transcript levels of ZmFDL1/MYB94 and associated genes. Overall, our results suggest that the response to water stress implies the activation of wax biosynthesis and the involvement of both ZmFDL1/MYB94 and abscisic acid regulatory pathways.
In all land plants, the outer surface of aerial parts, including vegetative organs, flowers, fruits, seeds and pollen grains, is constituted by a continuous hydrophobic layer termed the cuticle, which consists of two major components, the polymer cutin and cuticular waxes. Cutin is a polymer of C16 to C18 hydroxylated fatty acids that are cross-esterified to each other either directly or via a glycerol backbone (Fich et al., 2016). Cutin monomers are synthetized in the endoplasmic reticulum (ER) through different reactions, including the esterification of fatty acids to CoA, ω-hydroxylation and further oxidation, glycerol-3-phosphate transacylation, and export through the cell wall to the surface, where polymerization occurs (Fich et al., 2016). Cuticular waxes are constituted by a complex mixture of very-long-chain fatty acids (VLCFAs) with >20 carbon atoms, and their derivatives, which include alcohols, aldehydes, alkanes, ketones, and wax esters (WEs). They also include variable amounts of cyclic compounds, such as triterpenoids and phenylpropanoids (Bernard and Joubès, 2013; Lee and Suh, 2015b). The first phase of wax biosynthesis consists in the elongation of C16 and C18 fatty acids produced in the plastids VLCFAs with a chain length between C22 and C38 by elongase complexes in the ER (Haslam and Kunst, 2013). Wax components are then produced through two different pathways. Primary alcohols and esters are produced by the alcohol-forming pathway, also termed the acyl-reduction pathway (Rowland et al., 2006; Li et al., 2008), while alkanes, secondary n-alcohols, and ketones are produced by the alkane-forming pathway, also called the decarbonylation pathway (Bernard et al., 2012). All wax compounds are synthesized in the epidermal cell layer (L1) and are polarly secreted to cover the outer cell wall of epidermal cells, where they are embedded in the cutin and deposited on the cuticle surface as films or wax crystals. Waxes forming the outer layer of plant tissues are called epicuticular waxes.
Cuticle formation prevents postgenital fusions among organs that grow very tightly appressed to each other when enclosed in vegetative shoots or within buds (Ingram and Nawrath, 2017). In addition, cuticle constitutes a constant barrier against damage caused by environmental abiotic and biotic factors, including UV light, temperature changes, pests, and pathogens, as well as a primary waterproof barrier that controls nonstomatal water loss and gas exchange from leaves (Yeats and Rose, 2013). Several studies in various plants have shown that changes in the amount and/or distribution of cuticular waxes lead to alterations in cuticular permeability. In tomato (Solanum licopersicum), mechanical and genetic manipulations of the cuticular components show that aliphatic constituents of the intracuticular wax layer have a key role in limiting the transpiration rate across the epidermis (Vogg et al., 2004). In sorghum (Sorghum bicolor), a bloomless mutant with a high reduction in epicuticular waxes appears to have a higher rate of epidermal permeability and night-time water loss (Burow et al., 2008). In rice (Oryza sativa), impairments in the organization of crystal waxes on the leaf surfaces of the wilted dwarf and lethal 1 (wdl1) mutant are correlated with a 2.3-fold increase in transpiration rates and higher rates of water loss (Park et al., 2010). A positive correlation between an increase in wax amount and tolerance to drought stress has been reported in Arabidopsis (Arabidopsis thaliana; Aharoni et al., 2004; Kosma et al., 2009; Cui et al., 2016). Similarly, alkanes and primary alcohols are increased by drought treatment in two Australian wheat (Triticum aestivum) cultivars. This study allowed the characterization of two MYB transcription factors that were activated by drought and in turn stimulated the expression of cuticle biosynthesis-related genes (Bi et al., 2016). In rice, the Glossy1-3 (OsGL1-3) gene, highly similar to Arabidopsis ECERIFERUM1 (AtCER1) and involved in cuticular wax accumulation, is activated by drought (Zhou et al., 2015). Finally, in maize (Zea mays) seedlings, both cuticular permeability and drought sensitivity are increased by a mutation in the ZmGL6 gene, the product of which is putatively involved in intracellular trafficking of cuticular waxes (Li et al., 2019).
In Arabidopsis, deposition of cuticular waxes in both leaves and stems is regulated by the AtMYB96, AtMYB30, and AtMYB94 transcription factors, which collectively modulate the expression of wax biosynthetic enzymes (Raffaele et al., 2008; Seo et al., 2011; Lee and Suh, 2015b). Abscisic acid (ABA), drought, and high salinity activate the expression of AtMYB96 (Seo et al., 2009), which in turn mediates the activation of cuticle biosynthetic genes to increase drought tolerance (Seo et al., 2011). Similarly, the expression level of AtMYB94 is also increased by drought, and transgenic Arabidopsis lines overexpressing AtMYB94 show increased accumulation of cuticular waxes and, under drought conditions, a reduction in the rate of cuticular transpiration in leaves (Lee and Suh, 2015a). We showed in a previous work that the maize ZmFDL1/MYB94 transcription factor is specifically required for cuticle-mediated postgenital organ separation during embryo development and early phases of seedling growth. Lack of ZmFDL1/MYB94 activity in the fused leaves1-1 (fdl1-1) recessive mutant specifically affects seedling development at early developmental stages and results in organ fusions due to the lack of cuticular material in the boundary between organs and irregular distribution of wax crystals on young leaf epidermal surfaces (La Rocca et al., 2015).
To gain further insight into the role of ZmFDL1/MYB94, we compared in this study the cuticle composition of mutant and wild-type seedlings and analyzed the impact of the mutation on the transcriptome during early phases of seedling development. We also investigated ZmFDL1/MYB94 involvement in controlling cuticular permeability and in mediating drought stress response in maize seedlings. The isolation of genes involved in the biosynthesis and transport of cuticular waxes, which are responsive to drought, is of particular interest for crop breeding. The detailed functional characterization of the maize ZmFDL1/MYB94 regulatory gene proposed in this work will contribute to unravelling the genetic-molecular mechanisms at the basis of the cuticle-mediated drought stress tolerance in the important cereal crop maize.
RESULTS
ZmFDL1/MYB94 Regulates Cuticle Deposition in a Phase-Dependent Manner
The most evident defects observed in fdl1-1 homozygous mutants were the irregular coleoptile opening and the presence of fusions between coleoptile and first leaf (Fig. 1A; La Rocca et al., 2015). To characterize the effects of the fdl1-1 mutation on cuticle composition, we extracted cutin and epicuticular waxes from mutant and wild-type seedlings and analyzed them by gas chromatography (Domergue et al., 2010; Bourdenx et al., 2011).
Figure 1.
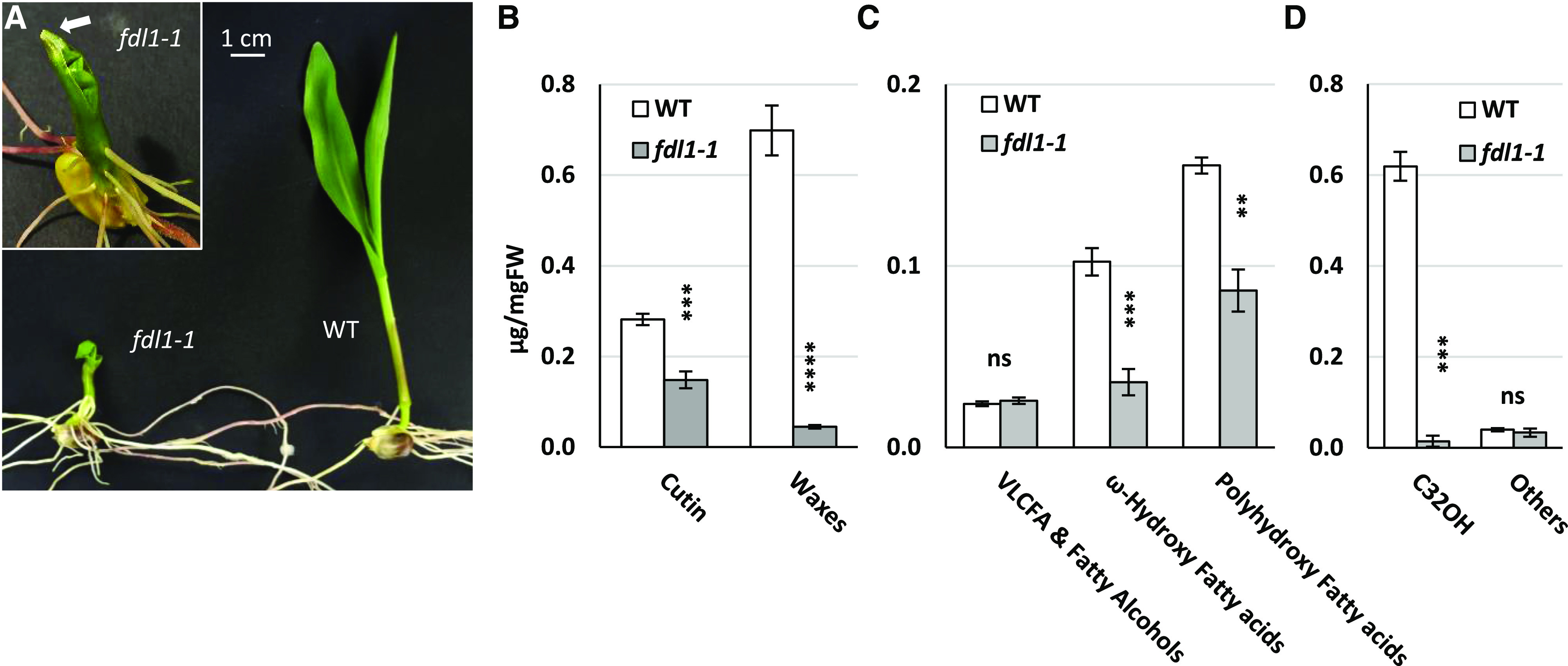
Cuticle-related defects in the fdl1-1 homozygous mutant. A, Representative phenotype of 10-d-old wild-type (WT) and fdl1-1 mutant seedlings. Inset, The white arrow indicates the fusion between the coleoptile and the first leaf of a fdl1-1 plant. B to D, Total cutin and wax loads (B), cutin composition (C), and wax composition (D) in fdl1-1 and wild-type seedlings at the coleoptile developmental stage. The data represent the means ± se of five biological replicates. Significant differences between the wild type and the mutant were assessed by Student’s t test (**P < 0.01, ***P < 0.001, and ****P < 0.0001). FW, Fresh weight.
At the coleoptile stage, the total cutin and wax loads of fdl1-1 mutants were considerably reduced compared with those of wild-type seedlings, but not completely abolished (Fig. 1B). In fdl1-1 coleoptiles, the reduction in total waxes was more severe than that observed for cutin (94% and 47% decreases, respectively; Fig. 1B). These differences progressively decreased during seedling development and at the third leaf stage, total amounts of cutin (Supplemental Fig. S1A) and waxes (Supplemental Figs. S1, B and C, and S2, M–T) were similar in fdl1-1 and wild-type plants. Reduction in cutin content was mainly due to decreases in ω-hydroxy fatty acids and polyhydroxy-fatty acids (Fig. 1C; Supplemental Fig. S2, E–L). The ω-hydroxy fatty acid content was more impaired than the polyhydroxy-fatty acids content (65% and 44% decrease, respectively) in the coleoptiles of the homozygous fdl1-1 mutant (Fig. 1C). Such differences were detected up to the second leaf stage of seedling development (Supplemental Fig. S2, E, F, I, and J). Loss of ZmFDL1/MYB94 activity had no substantial effect on the VLCFAs and fatty alcohols (Fig. 1C) as only a few monomers were significantly different from the wild type at the early stage of seedling development (Supplemental Fig. S2, A and B).
The decrease in epicuticular wax load (Fig. 1B; Supplemental Figs. S1 and S2) was mainly due to the reduction in primary long-chain alcohols (Fig. 1D; Supplemental Figs. S1 and S2), which represent the major components of maize seedling waxes and are dominated by the C32 primary alcohol isomer. In coleoptiles, the C32 primary alcohol content was decreased by 98% in fdl1-1 compared to wild type (Fig. 1D). The amounts of other minor primary long-chain alcohols with 30 or more carbons (C31 to C34OH) were also lower in fdl1-1, whereas the amounts of fatty alcohols with <30 carbons (C28 and C26OH) showed the opposite trend and were increased in fdl1-1 with respect to wild-type seedlings (Supplemental Fig. S2, Q and R). These differences disappeared with seedling development, and in the third leaf, the amounts of primary alcohol were similar in fdl1-1 mutant and wild-type seedlings (Supplemental Figs. S1C and S2, Q–T). Alkanes and aldehydes were also analyzed (Supplemental Fig. S2, M–P) and were reduced only in the early stages of seedling development (Supplemental Fig. S2, M and N). In particular, the C32 n-aldehyde was nearly absent in fdl1-1 coleoptiles, whereas alkanes were less affected (Supplemental Fig. S2M). Finally, minor long-chain WEs were detected at the coleoptile stage and their content was much lower in the homozygous fdl1-1 mutant than in wild-type seedlings (Supplemental Fig. S1D). Altogether, these data suggested that ZmFDL1/MYB94 is a regulatory component of both cutin and wax biosynthesis and deposition. Moreover, as previously observed for visual mutant traits, ZmMYB94-related biochemical defects undergo a progressive reversion during seedling development, further confirming that ZmMYB94 activity occurs in a specific developmental phase (La Rocca et al., 2015).
Cuticular Permeability Is Altered in the fdl1-1 Mutant
Reduced cuticle load is frequently accompanied by increased cuticle permeability (Aharoni et al., 2004; Park et al., 2010), and changes in this parameter can be assessed by measuring the efflux of chlorophyll (Lolle et al., 1997; Kurdyukov et al., 2006b). The chlorophyll leaching assay showed that the second leaf of fdl1-1 homozygous seedlings released chlorophyll faster than the second leaf of nonmutant B73 siblings (Fig. 2A).
Figure 2.
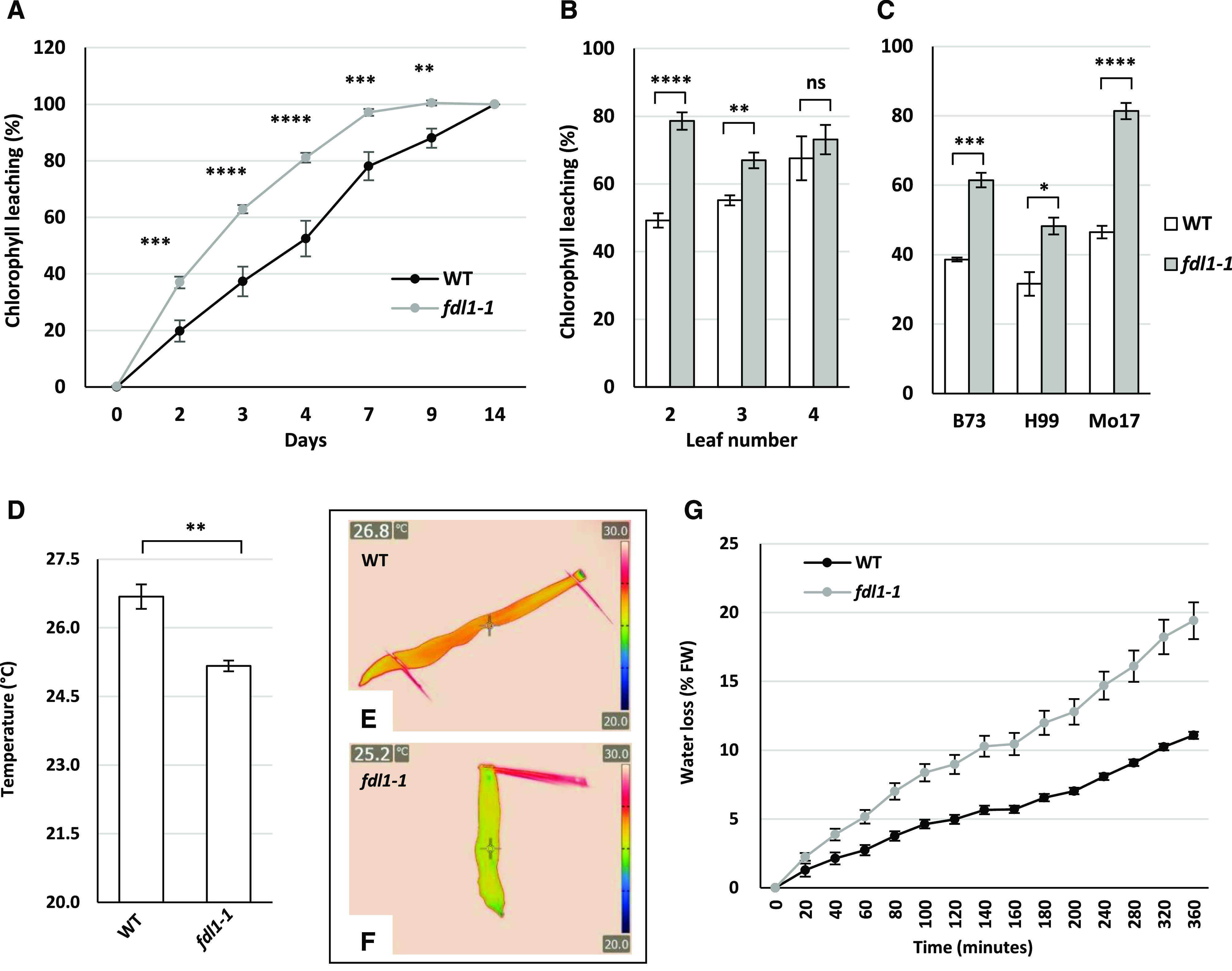
Cuticle-dependent leaf permeability in the fdl1-1 mutant. A to C, The chlorophyll leaching assay was performed on the second fully expanded leaf of 14-d-old fdl1-1 and B73 wild-type (WT) control plants (A), the second, third, and fourth fully expanded leaves of fdl1-1 and wild-type control plants (B), and the second fully expanded leaf of fdl1-1 and wild-type plants in different genetic backgrounds (C). The data in B and C are from day 3 of the chlorophyll leaching assay. Values represent the mean ± se of a minimum of five replicates. D, Temperature of the second fully expanded leaf in 14-d-old wild-type and fdl1-1 homozygous plants. Values represent the mean ± se of five biological replicates. E and F, Representative images of the leaf temperature acquired with the thermographic camera in wild-type (E) and homozygous fdl1-1 (F) plants. G, Percentage of water loss in detached 10-d-old homozygous fdl1-1 and wild-type seedlings. Values represent the mean ± se of 13 and 21 biological replicates for wild-type and fdl1-1 homozygous plants, respectively. Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001; ns, not significant). FW, Fresh weight.
Differences observed between mutant and wild-type chlorophyll leaching progressively decreased in the second, third and fourth leaves (Fig. 2B). The differences were maintained in different genetic backgrounds, as observed in F2 progenies obtained after introgressing the mutation in the H99 and Mo17 inbred lines (Fig. 2C).
To further understand the impact of the increased leaf surface permeability, thermography images of the second fully expanded leaves of wild-type (Fig. 2E) and fdl1-1 homozygous mutant (Fig. 2F) seedlings were analyzed. The fdl1-1 presented a reduced leaf temperature of ∼1.5°C compared to its control (Fig. 2D). Moreover, a water loss time course experiment was performed on 10-d-old seedlings by estimating the loss of weight with respect to the initial seedling fresh weight. The resulting profiles showed that homozygous fdl1-1 had a higher water loss rate compared to the wild-type plants (Fig. 2G).
Transcriptome Profile of the fdl1-1 Mutant
To investigate the regulatory network associated with ZmFDL1/MYB94, cDNA libraries were produced from coleoptiles of fdl1-1 and wild-type seedlings and sequenced on a HiSeq 2000 platform (Supplemental Table S1). A total of 2,213 and 2,399 genes were considered differentially expressed by Limma and DESeq2, respectively, when a significance cutoff false discovery rate (FDR) of 0.05 was applied (Supplemental Table S2). Importantly, 1,639 genes were differentially expressed according to both methods and were considered for subsequent analyses (Supplemental Table S3). Of these, 612 were downregulated and 1,027 were upregulated (Fig. 3A).
Figure 3.
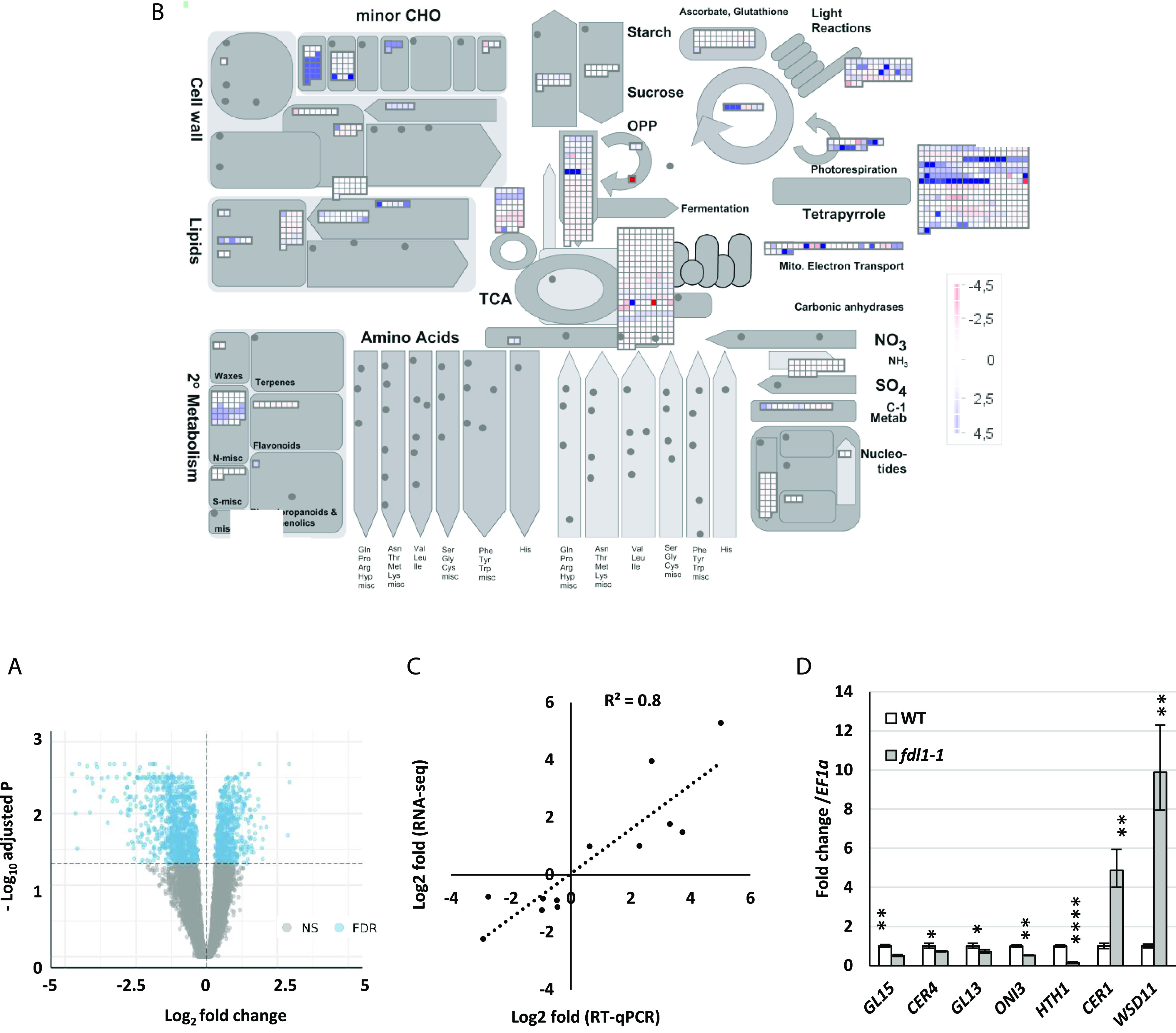
Functional enrichment of metabolic pathways and cuticle-related genes. A, Volcano plot of DEGs in fdl1-1 compared to wild-type control plants. Blue dots denote upregulated and downregulated DEGs. Gray dots represent genes with no significant P value. B, Functional enrichment of metabolic pathways. The DEGs have been assigned to different cellular components through GO analysis. MapMan was used to provide a general overview of differentially expressed transcripts involved in different metabolic pathways and cellular processes. ZmFDL1/MYB94-mediated changes in different metabolic processes in seedlings are illustrated, with blue indicating induced genes and red repressed genes. C, RT-qPCR validation of DEGs characterized by RNA-Seq. Correlation of log2 fold change data obtained using RT-qPCR (y axis) and with RNA-Seq analysis (x axis). D, Gene expression level of putative cuticle-related genes, analyzed by RT-qPCR, in fdl1-1 and wild-type control seedlings at the coleoptile developmental stage. Values represent the mean fold change variations ± sd of four biological replicates. Comparison is made between wild-type and homozygous fdl1-1 genotypes. Significant difference was assessed by Student’s t test (*P < 0.05; **P < 0.01; and ****P < 0.0001; ns, not significant).
To identify biological processes and pathways differentially modulated in the fdl1-1 mutant, functional enrichment analyses of differentially expressed genes (DEGs) were executed. Observed patterns of functional enrichment were highly consistent with phenotypic traits of fdl1-1 mutants. Ontology terms associated with water deprivation, wax biosynthesis and lipid metabolism were consistently associated with genes dysregulated in fdl-1, irrespective of the type of ontology used for the functional annotation.
The Gene Ontology (GO) terms “GO:0010025: wax biosynthetic process,” “GO:0005618: cell wall,” “GO:0009414: response to water deprivation,” and “GO:0019216: regulation of lipid metabolic process” were highly enriched in both sets of up and downregulated genes (Supplemental Fig. S3, B and C; Supplemental Tables S4 and S5), suggesting that DEGs identified by our analyses are likely to be involved in the modulation of wax/cutin biosynthesis and deposition in maize. Consistent with these observations, MapMan ontology (Fig. 3B; Supplemental Table S6) analyses of metabolic pathways suggested a strong enrichment of terms related to cell wall organization and lipid biosynthesis, such as “.Cell wall organisation.cutin and suberine” and “.Lipid metabolism”. Similarly, functional enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Supplemental Tables S7 and S8) identified that pathways related to cutin and fatty acid metabolism were among the most enriched pathways in genes both significantly downregulated (zma01212 [fatty acid metabolism], zma00061 [fatty acid biosynthesis], and zma00073 [cutin, suberin, and wax biosynthesis]), and significantly upregulated (zma00561 [glycerolipid metabolism]).
The high levels of agreement here are highly consistent with the idea that genes differentially modulated in the fdl1-1 mutant are likely to mediate the phenotypic traits observed in this developmental stage, as demonstrated by the systematic enrichment of ontology terms associated with wax, cutin, and lipid metabolism in general.
Accordingly, we performed a careful manual annotation of our list of DEGs to identify candidate genes directly involved in cutin metabolism. Based on a combination of GO, KEGG pathway, and orthology predictions with Arabidopsis and rice, as available from the phytozome annotation of the ZmB73_RefGen_v4 gene models and the ARALIP Web site (Li-Beisson et al., 2013), we identified 79 candidate DEGs that were tentatively assigned to nine distinct lipid- and cuticle-related biosynthetic processes, defined by expert manual curation (Supplemental Table S9). These genes were mainly related to the biosynthesis and transport of secondary metabolites, but also to the regulation of gene expression. According to our manual annotation, 15 DEGs were assigned generically to lipid metabolism, 9 DEGs to glycerolipid metabolism, and 5 DEGs to glycerophospholipid metabolism. The 50 remaining genes were assigned to the putative biosynthetic pathways as described below. Five genes (ZmLPD1, ZmACLB-1, ZmACLA-3, ZmACC2, and ZmAAE13) were associated with the formation of acetyl-CoA and Malonyl-CoA precursors. Six genes (ZmACP4, ZmSSI2/ZmFAB2, ZmFAB1.1, ZmFAB1.2, ZmALDH3F1, and ZmFAD7) were associated with the biosynthesis and desaturation of fatty acid, and seven genes (ZmGL26, ZmPAS2, ZmKCS24, ZmKCS17, ZmKCS22/ZmCER60, ZmKCS15, and ZmELO1) with the fatty acid elongation pathway. Fourteen genes (ZmCER8, ZmLACS2, ZmLACS4, ZmAAE16, ZmCER1, ZmMSH1/ZmCER4, ZmWSD11, ZmHTH1, ZmONI3, ZmCUS2, ZmGPAT8, ZmFAH1, ZmFAAH, and ZmLDAP3) were assigned to the cutin and wax biosynthesis pathway and three genes (ZmHCT12/ZmDCR, ZmECH1, ZmFAO4) to the β-oxidation pathway. In addition, 10 genes (ZmABCG11, ZmCER5, ZmCTS, ZmACBP4, ZmGL13, ZmCHI4/ZmFAP2, ZmLTPG6, ZmLTPG2, ZmPLT3, and ZmDIR1) might be involved in binding and transport of fatty acid, cutin, and wax monomers. Finally, five genes (ZmOCL1, ZmMYB30, ZmGL15, ZmSHN2.1, and ZmSHN2.2) codify for regulatory proteins (Fig. 4; Supplemental Table S9). The data obtained provided an overview and valuable information for investigating ZmFDL1/MYB94-dependent cuticle regulation, biosynthesis, and transport in maize (Fig. 4; Supplemental Table S9).
Figure 4.
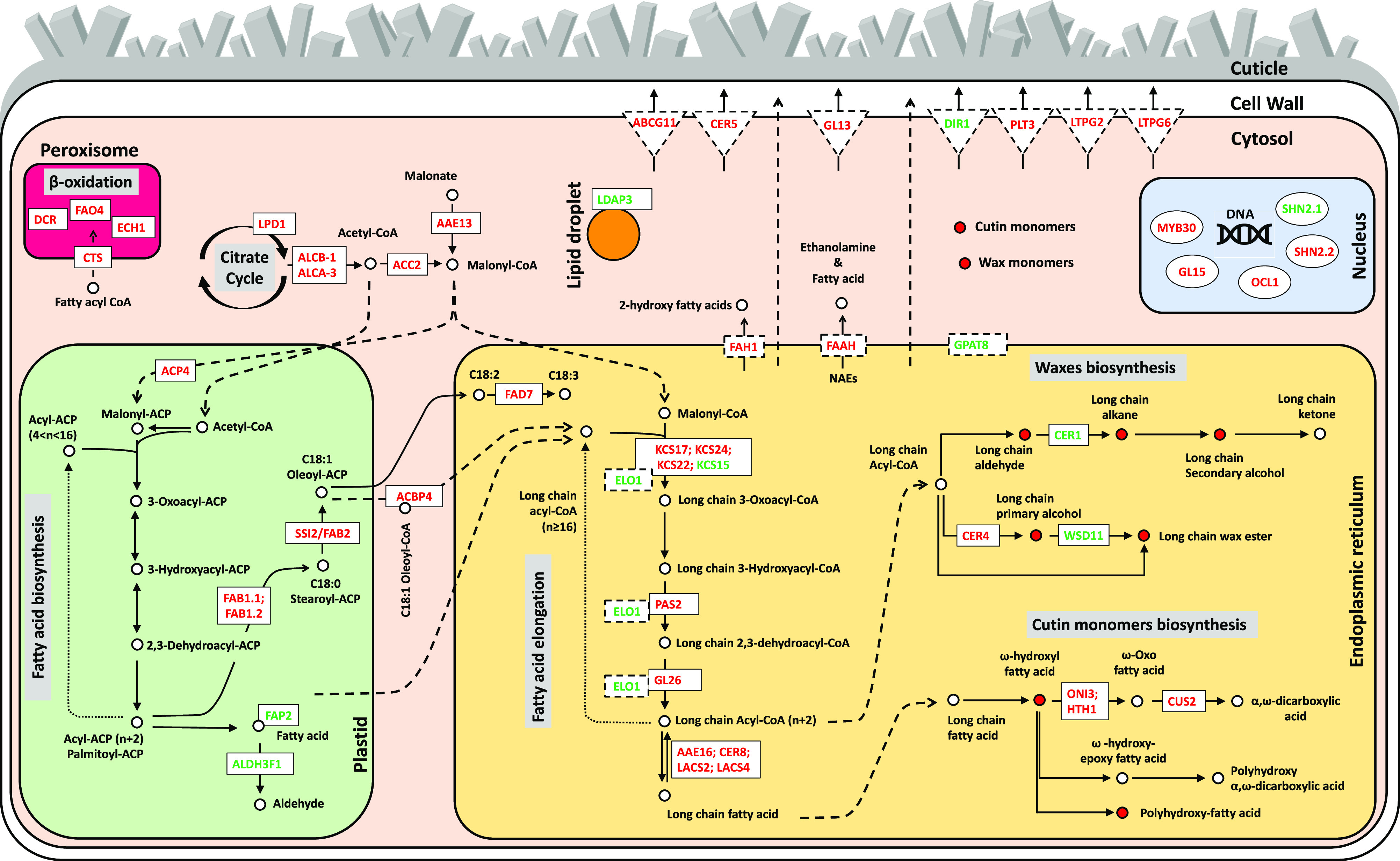
Pathways involved in cuticle biosynthesis. Schemes were designed using pathways available through the KEGG database. Transcript regulation of the citrate cycle and acetyl-CoA biosynthesis, fatty acid biosynthesis, fatty acid elongation, fatty acid β-oxidation, wax biosynthesis, cutin biosynthesis, and cuticle monomer transport are represented in the corresponding cell compartments. Solid black arrows indicate enzymatic reaction steps and gene symbols refer to DEGs in specific steps. Intermediates are shown as circles that are red when their amount is lower in fdl1-1 compared to the wild-type plants. Dashed black arrows indicate the movement of specific compounds into or toward a cell compartment. Products of down- and upregulated genes (Supplemental Table S9) are represented with red and green colored symbols, respectively. Genes encoding soluble enzymes are represented in rectangles with a continuous line, whereas genes encoding transmembrane enzymes are represented in rectangles with a dashed line. Transporters of cuticle monomers are represented in triangles.
To validate the reproducibility of the gene expression data obtained by the RNA-sequencing (RNA-Seq) analysis (Supplemental Table S2), expression patterns of 12 DEGs were investigated by reverse transcription quantitative PCR (RT-qPCR). Notably, RNA-Seq expression data were confirmed for all the DEGs considered and displayed high levels of correlation with RT-qPCR data (R2 = 0.8; Fig. 3C). Among the 12 DEGs selected for validation, seven genes differentially expressed in fdl1-1 compared to wild-type seedlings at the coleoptile developmental stage were putative cuticle-related genes (Fig. 3D).
Drought Affects Cuticular Permeability and the Expression of Cuticle-Related Genes
We previously showed that the fdl1-1 mutation causes an increase of the cuticle-mediated leaf permeability in young maize seedlings (Fig. 2G). Since we could exclude that the observed differences in water loss values might be due to alterations in the stomatal index (Supplemental Fig. S4, A and B), we suggest that they are attributable to alterations in cuticular transpiration and not stomatal conductance. We further investigated the role of cuticle and cuticle-related genes in mediating water stress response. Wild-type B73 seedlings were grown under either low watering/drought stress (LW), which was applied by withholding irrigation and maintaining relative soil water content (RSWC) at 40%, or normal watering (NW) conditions. After 14 d, the morphological parameters of seedlings grown under restricted water were similar to those of control plants (Fig. 5A), indicating that the applied stress was mild. We performed chlorophyll leaching assays and observed that in treated seedlings (LW) the permeability of the cuticle to chlorophyll was lower compared to that in control plants grown under the NW condition (Fig. 5B). The same response was observed in wild-type seedlings treated by applying a 1 μm ABA solution directly to the roots up to 14 d after sowing (DAS). Chlorophyll leaching was lower in the second leaf of ABA-treated plants compared to that of mock-treated control seedlings (Fig. 5C).
Figure 5.
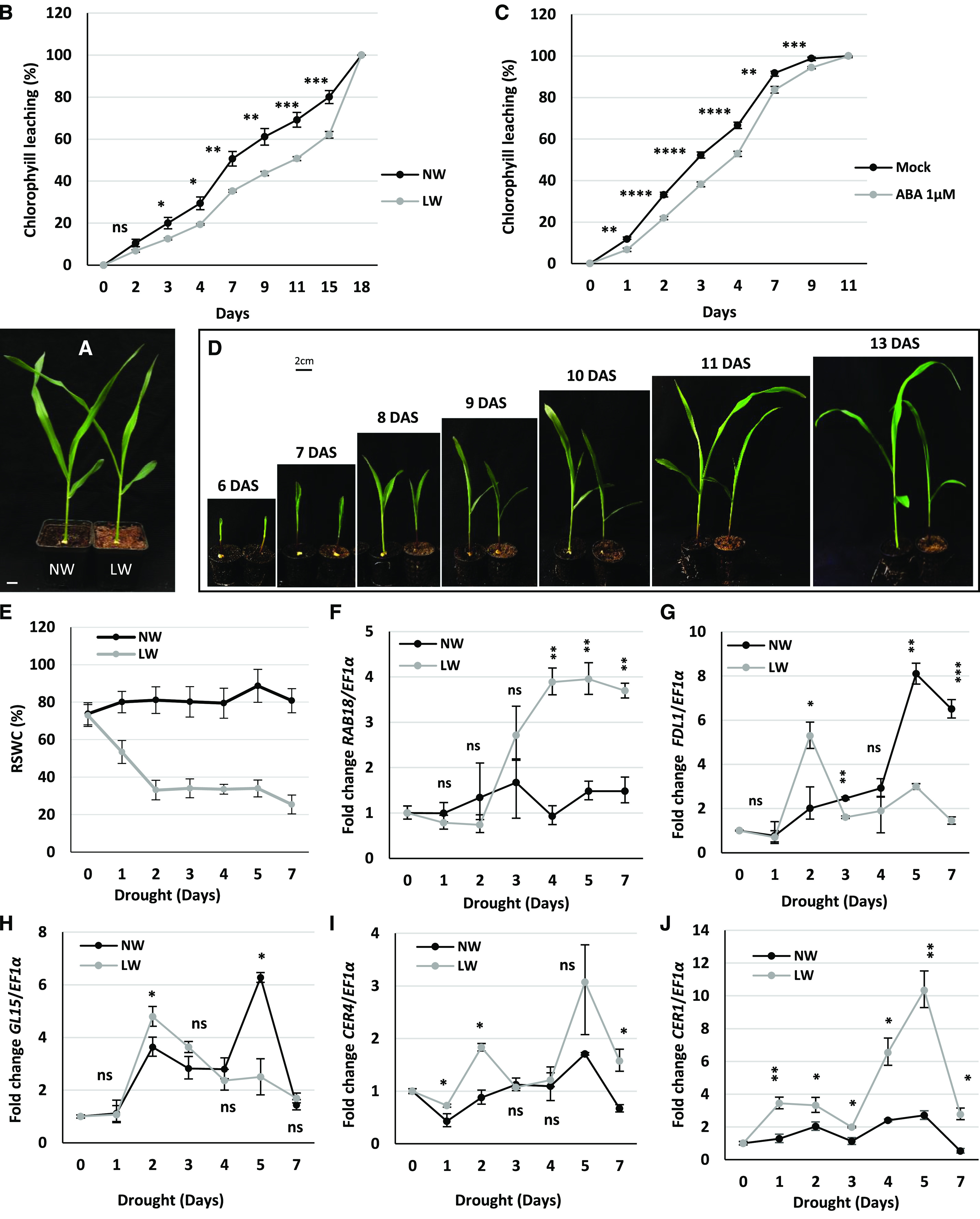
Drought modulates cuticle-dependent leaf permeability and the expression of cuticle-related genes. A, Representative images of 18-d-old B73 wild-type seedlings grown under NW or LW conditions for 13 d. B and C, Chlorophyll leaching from leaves of B73 wild-type plants grown for 13 d under LW or NW conditions (B) and B73 wild-type plants treated with control (Mock) or 1 μm ABA solution (C). Values represent the mean ± se of 10 replicates. Comparison was made at each time point between LW (B) or ABA-treated (C) and control plants. D, Representative images of B73 wild-type seedlings grown under NW (left) or LW (right) conditions. E and F, Water stress was applied to 6-d-old seedlings and was monitored through measurement of RSWC (E) and expression analysis of the drought stress marker gene ZmRAB18 (F). Values in E represent the mean ± se of 10 replicates. G to J, Patterns of ZmFDL1/MYB94 (G), ZmGL15 (H), ZmCER4 (I), and ZmCER1 (J) transcript accumulation analyzed by RT-qPCR in the second leaves of wild-type plants grown under LW or NW conditions. In F to J, values represent the mean fold change ± sd of three independent biological replicates. Comparison was made at each time point between LW and control (NW) plants. Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001; ns, not significant). Scale bar = 1 cm (A) and 2 cm (D).
To investigate transcriptomic changes induced by the drought stress stimuli, maize seedlings were grown under the NW condition until 5 DAS and subjected to drought stress the next day (Fig. 5D). To apply moderately stronger stress, RSWC was maintained at 30% of the total (Fig. 5E) for a shorter period. As a consequence, after 7 d (13 DAS), treated plants were visibly stunted compared to control seedlings (Fig. 5D). Every day, the second leaf from LW-treated and NW control seedlings was sampled and gene expression analysis was performed at every time point.
To test the effectiveness of the drought treatment, we analyzed changes in the expression of the ZmRAB18 gene, a known drought stress marker (Mao et al., 2015), which showed an increased transcript level in seedlings grown under LW compared to those grown under NW conditions (Fig. 5F), thus confirming that the plants were perceiving the stress.
The expression profile of ZmFDL1/MYB94, analyzed in the second leaf of seedlings grown under the NW condition, revealed that ZmFDL1/MYB94 transcripts accumulated during leaf expansion to reach a peak around 11 DAS (Fig. 5G, black line). In the LW condition (Fig. 5G, gray line), the ZmFDL1/MYB94 transcript profile was different, since the peak of maximum expression was detected earlier, after 2 d of drought stress (Fig. 5D, 8 DAS), exactly when the RSWC reached 30% of the total (Fig. 5E). At subsequent time points, the ZmFDL1/MYB94 transcript level decreased to levels lower in LW than in NW growth conditions.
The ZmGL15 transcription factor expression profile mirrored what we observed for ZmFDL1/MYB94 (Fig. 5H). An increase in transcript accumulation was detected during leaf growth (Fig. 5H, black line). Compared to the control condition, ZmGL15 expression was upregulated after 2 d of drought stress and then downregulated after 5 d of stress (Fig. 5H, gray line). On the other hand, the expression pattern of the ZmCER4, ZmCER1, and ZmWSD11 genes showed a trend of upregulation under LW.
ZmCER4 expression was significantly upregulated compared to NW after 2 and 5 d of stress (Fig. 5I, gray line), whereas the transcript levels of ZmCER1 and ZmWSD11 were consistently higher in seedlings grown under the LW compared to the NW condition and reached a peak at 5 and 7 d, respectively, after the initiation of stress (Fig. 5J; Supplemental Fig. S4C, gray line).
The expression profiles of ZmGL13, ZmONI3, and ZmHTH1 in control conditions generally decreased throughout leaf development (Supplemental Fig. S4, D–F, black line). The ZmGL13 gene was slightly upregulated after 2 d of drought (Supplemental Fig. S4D, gray line), at the same time point when ZmFDL1, ZmGL15, ZmCER4, ZmCER1, and ZmWSD11 were upregulated (Fig. 5, G–J, Supplemental Fig. S4C). Instead, the transcript levels of ZmONI3 and ZmHTH1 were less abundant in seedlings grown in the LW condition compared to control plants (Supplemental Fig. S, E and F, gray line).
ABA Treatment Affects ZmFDL1/MYB94 and Wax-Related Gene Expression
We previously showed that plants responded to ABA treatment by reducing the permeability of the cuticle to chlorophyll (Fig. 5C). We therefore analyzed a maize mutant in the VIVIPAROUS1 (ZmVP1) gene (vp1-1). The transcription factor VP1 is the ortholog of the Arabidopsis ABA INSENSITIVE3 (AtABI3) gene (Giraudat et al., 1992), and mutants in this gene have been reported to have a reduced sensitivity to ABA (McCarty et al., 1991; Suzuki et al., 2003; Cao et al., 2007).
Interestingly, we noticed that the vp1-1 mutant seedlings showed increased chlorophyll leaching compared with wild-type controls (Fig. 6A). Subsequently, to investigate the possible interplay between ABA and ZmFDL1/MYB94-regulated pathways, we evaluated the effect of exogenous application of the ABA hormone on the fdl1-1 mutant. After 14 d of a treatment consisting in the application of a 1 μm ABA solution (directly to the root, as for the wild type; Fig. 5C), no significant reduction in the permeability of the cuticle to chlorophyll was observed in fdl1-1 mutant seedling compared to the mock-treated control (Fig. 6B).
Figure 6.
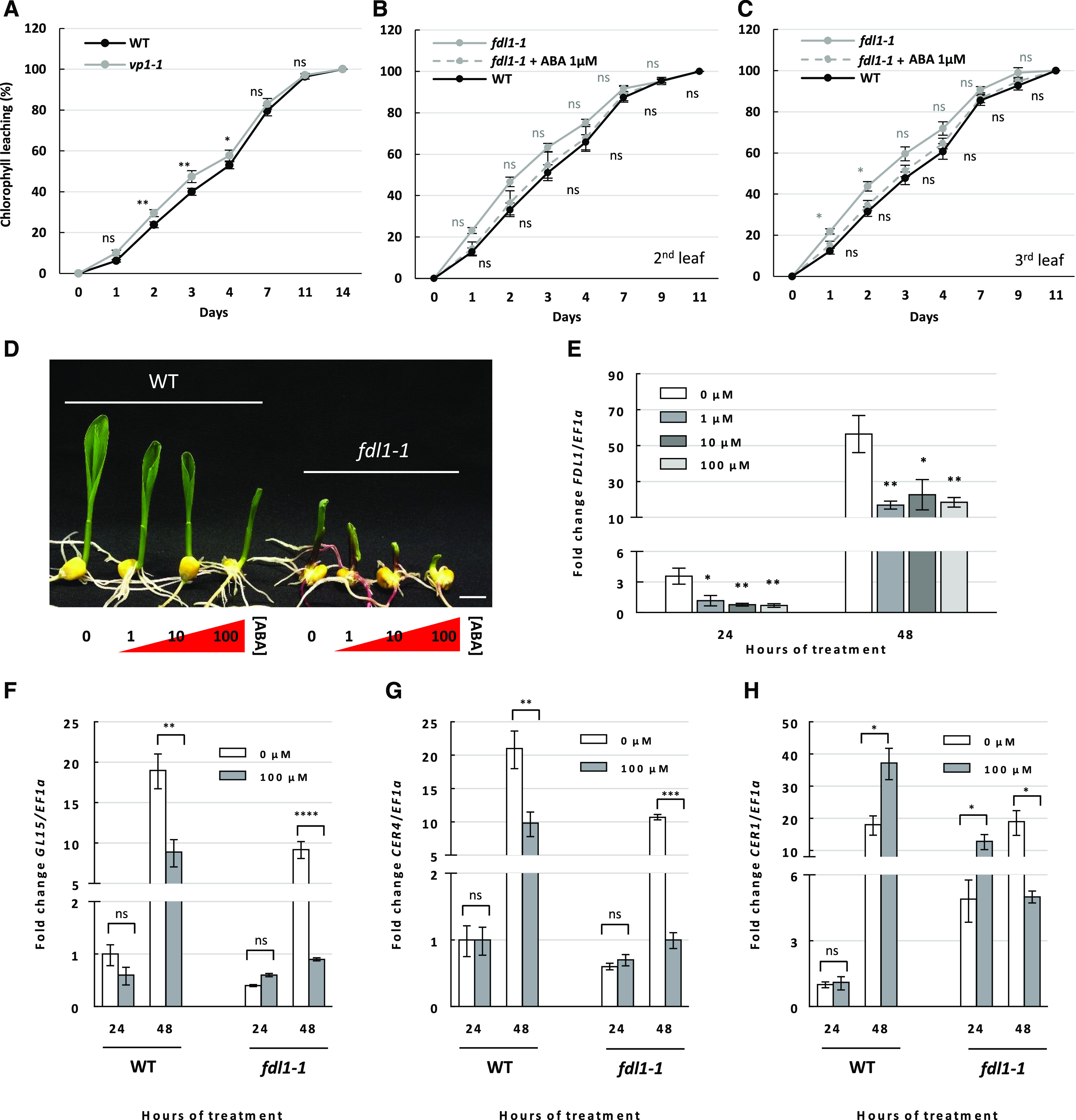
Effect of ABA treatment on leaf permeability and gene expression. A to C, Kinetics of chlorophyll leaching using the second fully expanded leaf of homozygous vp1-1 and B73 wild-type (WT) control plants (A) and the second (B) and third (C) leaves of fdl1-1 plants treated with a mock or 1 μm ABA solution and wild-type plants. Values represent the mean ± se of 10 biological replicates. D, Representative images of B73 wild-type and homozygous fdl1-1 mutant seedlings grown for 48 h with the root apparatus in liquid solutions with increasing ABA concentration (0, 1, 10, and 100 μm). Scale bar = 1 cm. E, Expression profile of ZmFDL1/MYB94 in wild-type plants after treatment with control (0 μm ABA) or 1, 10, and 100 μm ABA solution for 24 and 48 h. F to H, RT-qPCR analysis of the cuticle-related ZmGL15 (F), ZmCER4 (G), and ZmCER1 (H) genes in mock (0 μm ABA) or 100 μm ABA-treated wild-type and homozygous fdl1-1 mutant seedlings for 24 and 48 h. Values in E to G represent the mean fold change ± sd for a minimum of three independent biological replicates. Comparisons were made at each time point between wild-type and vp1-1 genotypes (A); between ABA and mock treatments in fdl1-1 (gray) and wild-type plants (black); or between control and ABA-treated plants (E–H). Significant differences were assessed by one-way ANOVA or Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001; ns, not significant).
Comparisons of expression profiles in ABA- and control-treated wild-type plants showed a consistent downregulation of ZmFDL1/MYB94 upon treatment with increasing concentrations of the hormone after both 24 and 48 h of treatment (Fig. 6E). Importantly, seedlings of both fdl1-1 and wild-type control plants responded to ABA with a decrease in growth, suggesting that the perception of ABA was not altered (Fig. 6D).
Expression levels of cuticle-related genes were investigated in fdl1-1 and wild-type plants treated with ABA and in corresponding untreated controls. The transcript levels of both ZmGL15 and ZmCER4 genes were reduced upon ABA treatment in both fdl1-1 and wild-type plants after 48 h of ABA treatment (Fig. 6, F and G). The ZmCER1 gene (Fig. 6H) was upregulated in fdl1-1 after 24 h of ABA treatment (164%) and in wild-type plants after 48 h (102%), while it was downregulated in mutant plants after 48 h (−74%).
Consistent with our previous observations based on RNA-Seq data (Supplemental Table S9) and quantitative expression analysis (Fig. 3D), the ZmGL15 and ZmCER4 genes were downregulated in untreated fdl1-1 seedlings compared to untreated control wild-type plants, while ZmCER1 was upregulated (Fig. 6, F–H). The same trends were also observed upon ABA treatment. It is remarkable that after 48 h of ABA treatment, the reduction in expression of ZmGL15 and ZmCER4 compared to their untreated controls was more pronounced. The difference between ABA-treated and control plants was ∼90% and 53% for ZmGl15 (Fig. 6F) and 91% and 52% for ZmCER4 (Fig. 6G) in fdl1-1 mutant and wild-type plants, respectively. Similarly, the upregulation of ZmCER1 was higher in ABA-treated fdl1-1 mutant plants (164%) than in wild-type plants (102%) and the transcript changes occurred earlier in fdl1-1 (24 h) than in wild-type (48 h; Fig. 6H).
DISCUSSION
As shown in our previous study (La Rocca et al., 2015), maize plants lacking the ZmFDL1/MYB94 transcription factor are defective in seedling development, since fusions, which occur specifically in embryonic primordia and seedling organs, impair coleoptile opening as well as leaf expansion. Such fusions were attributed to the irregular deposition of intervening cuticular material between juxtaposed cell walls and to the patchy distribution of epicuticular waxes on the epidermis of the young leaves. This primary phenotypic characterization suggested that ZmFDL1/MYB94 may have an important role in controlling cuticle biosynthesis in maize.
ZmFDL1/MYB94 Is a Key Regulator of Cuticle Biosynthesis and Deposition in Maize
In this work, a more thorough characterization, including biochemical as well as transcriptional analyses, has been undertaken to gain further insight into the role of ZmFDL1/MYB94 in cuticle formation. The data herein obtained strongly confirm that ZmFDL1/MYB94 is a key regulatory component of both cutin and wax biosynthesis and deposition in maize seedlings. Among cutin compounds, ω-hydroxy fatty acids and polyhydroxy fatty acids were specifically affected in fdl1-1 coleoptiles. In epicuticular waxes, the reduction was mainly observed in primary long-chain alcohols, although a reduction of long-chain WEs was also detected (Supplemental Fig. S1D).
It is conceivable that the defects observed in both cuticle components, i.e. waxes and cutin, were causative for the organ fusions in the fdl1-1 mutant seedlings. In contrast, fusions do not occur in maize mutants carrying defects in GL genes specifically controlling wax biosynthesis, such as GL1 and GL2, which are involved in the alkane-forming pathway and the elongation of VLCFAs to C30 (Sturaro et al., 2005; Tacke et al., 1995), or GL4 and GL8, which belong to the fatty acid elongase complex (Liu et al., 2009; Dietrich et al., 2005). These gl mutants display glossy, bright green leaves but normal seedling morphology. A similar observation applies to mutants in ZmGL3, which is involved in the control of wax deposition and closely related to the Arabidopsis AtMYB60 (Liu et al., 2012), and in GL6, the product of which is responsible for the intracellular trafficking of cuticular waxes (Li et al., 2019). In this context, one exception might be represented by mutants in the ADHERENT1 (AD1) gene encoding the 3-ketoacyl-coa synthase (KCS) required for cuticular wax biosynthesis (Liu et al., 2020), which display organ fusions in seedlings as well as in adult organs and tassel branches. However, the nature of fusions seems different between the two mutants, since an intact cuticular layer is maintained in ad1 adherent regions (Sinha and Lynch, 1998), while the cuticle layer is completely absent in fdl1-1 (La Rocca et al., 2015).
A whole picture of the complex biosynthetic network underlying cuticle formation is proposed in Figure 4, in which DEGs, as identified from our transcriptome analysis, are also reported. Lack of ZmFDL1/MYB94 has an effect on the activities of genes located in different modules of the proposed pathway. With the absence of publicly available large-scale data regarding possible in vivo direct targets of ZmFDL1/MYB94, it is difficult to speculate whether this effect is direct or indirect. Nevertheless, we notice that lack of ZmFDL1/MYB94 activity affects the expression levels of several waxes and cutin-related genes, including, for example, transporters of cuticle components such as the ZmGL13 wax transporter (Li et al., 2013). Interestingly, ZmGL13 was also consistently downregulated by quantitative expression analysis (Fig. 3D).
The decrease in very-long-chain C32 primary alcohols (C32OH; Fig. 1D) observed in mutant coleoptile waxes associates well with the downregulation of ZmCER4 (Fig. 3D; Supplemental Table S9), whose closest homolog is AtCER4, which is involved in the alcohol-forming pathway (Fig. 4; Rowland et al., 2006).
ZmWSD11 (Fig. 4), which corresponds to the Arabidopsis FOLDED PETAL 1 (AtFOP1/AtWSD11) that encodes a bifunctional WE synthase/diacylglycerol acyltransferase (Takeda et al., 2013), was upregulated in the fld1-1 mutant. We also observed upregulation of the ZmCER1 gene (Fig. 3E), a homolog of AtCER1 that was shown to be involved in the production of very-long-chain alkanes (Bourdenx et al., 2011). However, the increased expression of ZmCER1 in fdl1-1 did not agree with the content of these compounds observed in mutant seedlings (Supplemental Fig. S2). A general downregulation of the genes involved in the biosynthesis of long-chain fatty acids, such as ZmKCS17, ZmKCS22, ZmKCS24, ZmPAS2, and ZmGL26 (Fig. 4; Supplemental Table S9), and a strong decrease in the content of the very-long-chain aldehydes (Supplemental Fig. S2M), precursors of the very-long-chain alkanes, could possibly explain the discrepancy between the transcript levels of ZmCER1 and ZmWSD11 (Fig. 3D) and their respective products (Supplemental Figs. S1D and S2M).
The reduction of ω-hydroxy fatty acids and polyhydroxy fatty acids (Supplemental Fig. S2, I and E) was consistent with the low expression of genes involved in the biosynthesis of cutin precursors and cutin monomers, such as ZmONI3, ZmHTH1, ZmCUS2, ZmAAE16, ZmCER8, ZmLACS2, and ZmLACS4 (Fig. 4; Supplemental Table S9).
ZmONI3 and ZmHTH1 share high levels of similarity with the rice HOTHEAD (HTH) proteins OsONI3 and Os08g0401500 (close homolog of OsONI3; Akiba et al., 2014), respectively, and also with Arabidopsis HTH/EDA17 (Yephremov et al., 1999; Krolikowski et al., 2003), which were suggested to encode an ω-hydroxy dehydrogenase catalyzing the reaction from long-chain ω-hydroxy fatty acids to ω-oxo fatty acids in the β-oxidation pathway (Kurdyukov et al., 2006a). Interestingly, the seedling morphology of the OsONI3 mutant highly resembles that of fdl1-1, showing regions of fusion involving embryo leaf primordia as well as shoot leaves. This may suggest that the proper synthesis of cutin-related compounds is required to prevent postgerminative adhesion among organs.
The results of a recent genome-wide analysis based on DAP-Seq (Liu et al., 2020) identified six of our selected genes as possible direct targets of ZmFDL1/MYB94 (P-value hypergeometric 1.6E−07), confirming the robustness of our approach. These direct targets are Zm00001d004125 (ZmACC2), Zm00001d044579 (ZmKCS22/ZmCER60), Zm00001d039053 (ZmKCS15), Zm00001d053127 (ZmLACS2), Zm00001d017251 (ZmCER1), and Zm00001d043853 (ZmWSD11).
AtMYB30, AtMYB96, and AtMYB94, which are the genes in Arabidopsis most closely related to ZmFDL1/MYB94, are similarly involved in the regulation of cuticular wax biosynthesis and transport (Raffaele et al., 2008; Seo et al., 2011; Lee and Suh, 2015b). AtKCS1, AtKCS2, AtFDH, AtHCD1, AtPAS2, and AtCER10 are activated by AtMYB30 (Raffaele et al., 2008). Moreover, AtKCS1 and AtKCS2, along with AtKCS6, AtKCR1, and AtCER3, are direct targets of AtMYB96 (Seo et al., 2011). Similarly, AtKCS2, AtCER2, AtECR/AtCER10, AtFAR3/AtCER4, and AtWSD1 have been identified as direct target genes of AtMYB94 (Lee and Suh, 2015a). Furthermore, AtMYB94 and AtMYB96 additively activate the wax biosynthetic genes AtKCS1, AtKCS2, AtKCS6, AtKCR1, AtCER2, AtCER1, AtCER3, and AtWSD1 under drought stress conditions (Lee et al., 2016b). No evidence has been provided that these AtMYBs are also involved in the regulation of cutin-related genes. Most maize homologs of these Arabidopsis genes were found to be differentially expressed in our RNA-Seq experiment, as highlighted in Supplemental Table S9.
Interestingly, the strong enrichment of downregulated DEGs in the KEGG “zma04712: Circadian rhythm” pathway (Supplemental Table S10) could reflect a conserved function between ZmFDL1/MYB94 and its Arabidopsis homolog AtMYB96, which is connected with the clock to shape the circadian gating of ABA responses (Lee et al., 2016a).
ZmFDL1/MYB94 Regulates Cuticle Deposition in the Juvenile Phase of the Plant
In maize, cuticle properties are different in juvenile and adult leaves. Maize juvenile leaves have a thin cuticle and are covered with epicuticular wax crystals, whereas adult leaves have a thick cuticle and an amorphous wax layer on their surfaces (Sylvester et al., 1990). Interestingly, long-chain alcohols (69%) are the main components of cuticular waxes in seedling leaves, followed by aldehydes (25%), alkanes (4%), and esters (2%; Javelle et al., 2010). In adult leaves, on the other hand, alkanes and alkyl esters are the main components (Bourgault et al., 2020). Cutin composition has been characterized in adult leaves and is mainly composed of dihydroxyhexadecanoic acid and typical members of the C18 family of cutin acids, including hydroxy and hydroxy-epoxy acids (Espelie and Kolattukudy, 1979).
In agreement with the pattern observed for the morphological traits (La Rocca et al., 2015), the biochemical defects appeared transiently in germinating fdl1-1 mutant seedlings (up to the second leaf stage), and a progressive shift toward control values was observed in subsequent developmental stages for all examined compounds (Supplemental Figs. S1 and S2). These data, along with the previous finding that the level of the ZmFDL1/MYB94 transcript showed a progressive decrease in the second and third leaves (La Rocca et al., 2015), provide further proof that the action of ZmFDL1/MYB94 is confined to a precise developmental window delimited by the third leaf stage. After the third leaf stage, ZmFDL1/MYB94 may act redundantly with its paralogue ZmMYB70 (GRMZM2G139284; La Rocca et al., 2015) to control cuticle deposition. This may explain the absence of phenotypic effects at later stages in fdl1-1 mutant plants, although future studies are required to confirm this hypothesis. The absence of evident morphological alterations, including organ fusions, in adult fdl1-1 mutant plants confirms that ZmFDL1/MYB94 action is dispensable at later stages, in which a different regulatory network might be involved in controlling cuticle deposition.
An intact cuticle is clearly important throughout the plant cycle to prevent organ fusion; therefore, other genes must fulfill a ZmFDL1/MYB94-like role at later stages. Relatively limited information is available in this regard. However, CRINKLY4, the product of which belongs to a family of receptor kinases, constitutes an interesting candidate for the control of epidermis differentiation as well as cuticle deposition throughout the maize life cycle, including the adult phase (Becraft et al., 2001; Jin et al., 2000). Moreover, a recent genome-wide association study aimed at the identification of genes involved in controlling the water barrier function of the maize leaf cuticle identifies various candidates for the regulation of cuticle deposition, among which are genes involved in membrane trafficking and deposition of cutin lipids. It also identifies an ortholog of the AtCER7 genes, which regulates the biosynthesis of alkanes (Hooker et al., 2007), one of the major classes of cuticle waxes in Arabidopsis and also in adult maize leaves (Bourgault et al., 2020).
The expression profile of ZmGL3, encoding another R2R3-MYB transcription factor that controls cuticle deposition, is also confined to the maize juvenile phase (Liu et al., 2012). Both ZmGL3 and ZmFDL1/MYB94 might, in turn, interact with additional regulatory factors involved either in the maintenance of the juvenile phase or in promoting the transition from juvenile to adult phase. In this context, an interesting candidate is ZmGL15 (Moose and Sisco, 1994), which encodes a transcription factor of the AP2-domain family, showing high similarity to the Arabidopsis APETALA2 gene (AtAP2; Jofuku et al., 1994). ZmGL15 is required for the maintenance of juvenile traits in the leaf epidermis, and its transcriptional level is controlled by miR172, which, by downregulating ZmGL15, promotes the transition from juvenile to adult phase (Lauter et al., 2005). The gl15 mutant exhibits a decrease in juvenile waxes within the third leaf and a glossy phenotype beginning with the fourth leaf (Moose and Sisco, 1994), when the transition from juvenile to adult phase takes over, indicating that another factor is sufficient to promote wax synthesis in the first two leaves. Therefore, our hypothesis is that ZmFLD1 is sufficient to ensure a correct cuticle biosynthesis up to the third leaf stage, whereas in subsequent juvenile leaves its role is achieved by ZmGl15.
ZmFDL1/MYB94 and Associated Cuticle-Related Genes Mediate Leaf Permeability under Drought and ABA Treatments
We have shown in the current study that ZmFDL1/MYB94, in addition to promoting organ separation, is required in maize seedling leaves to control cuticle-dependent leaf permeability, to reduce cuticular water loss, and eventually to preserve leaf water status (Fig. 2).
Several studies have shown that cuticle formation responds to environmental conditions, suggesting that the cuticle retains an active role in the plant response to environmental stress conditions, among them water scarcity (Xue et al., 2017). Our study provides support for these findings and further indicates that in maize seedlings this response is mediated by ZmFDL1/MYB94. It also provides evidence that additional regulators, such as ZmGL15, as well the ABA hormone signaling pathway, are involved. Cuticle-dependent leaf permeability, as measured through chlorophyll leaching analysis on B73 maize seedlings, decreased under limited water supply, as well as under exogenous administration of the ABA hormone (Fig. 5, A and B). Moreover, the application of a drought stress treatment (LW) to developing seedlings caused a change in the expression profile of ZmFDL1/MYB94 and associated genes (Figs. 5 and 7; Supplemental Fig. S4).
Figure 7.
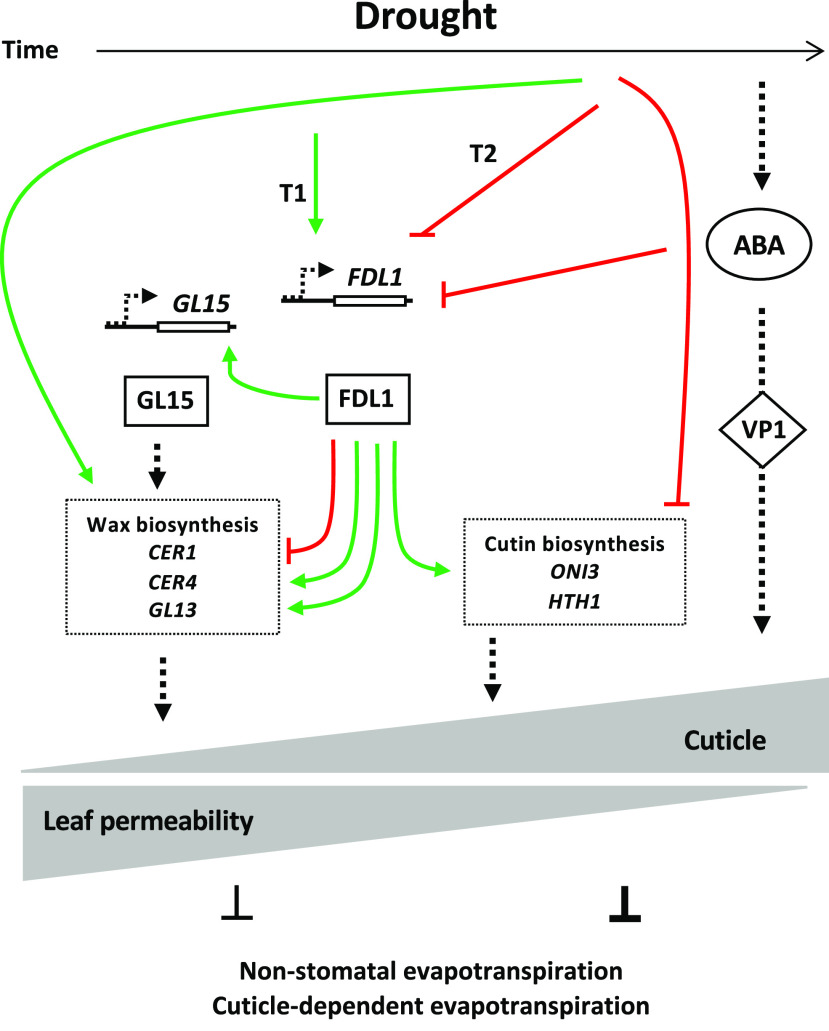
Proposed roles of ZmFDL1/MYB94 and ABA in the regulation of cuticle-dependent leaf permeability under drought. ZmFDL1/MYB94 is drought sensitive and promotes an early (T1) response. Its action, which is accompanied by the activation of the regulatory gene ZmGL15, allows plants to rapidly cope with water scarcity conditions by stimulating, in young tissues, the transcription of genes involved in cuticle biosynthesis, and more specifically in wax deposition. At later stages (T2), ABA and ZmFDL1/MYB94 pathways interact to integrate the leaf developmental program and external cues. This leads to proper modulation of cuticle composition that reduces the cuticle-dependent leaf evapotranspiration and prevents massive water loss. Green arrows indicate transcriptional activation and red T-shaped lines indicate transcriptional repression. Black dotted arrows indicate a putative activation, although the mode of interaction is unknown.
A peak in the expression profile of both ZmFDL1/MYB94 and ZmGL15 regulatory genes was observed, which appeared earlier in the LW condition than in the NW condition (Fig. 5, G and H). This observation applies also to ZmCER4 (Fig. 5I), while the expression profiles of ZmCER1, ZmWSD11, and ZmGL13 showed a neat peak under LW that was not observed in the NW condition (Fig. 5J; Supplemental Fig. S4, C and D). We may speculate that the effect of drought on the expression profile of ZmCER4 is modulated to a greater extent by the positive action exerted by ZmFDL1/MYB94, which might also mediate the positive effect exerted by drought on the ZmGL13 transcript level. On the other hand, if we assume from our previous data that ZmFDL1/MYB94 is a repressor of ZmCER1 and ZmWSD11, the upregulation of these genes observed under drought might be independent of the action of this MYB factor. Overall, our data indicate that genes involved in wax biosynthesis (ZmCER1, ZmCER4, and ZmWSD11), and eventually the total amount of cuticular wax, are increased in response to drought stress conditions, thus improving plant protection from water loss. This finding is in agreement with a number of studies conducted for wax-related Arabidopsis genes. One example is constituted by AtCER1, which controls the biosynthesis of very-long-chain alkanes (Bernard et al., 2012) and the expression of which correlates with drought stress responses (Bourdenx et al., 2011). Its overexpression in plants significantly increases the production of alkanes and leads to increased drought tolerance (Bourdenx et al., 2011). Moreover, the expression of AtWAX2, which is involved in both cutin and wax biosynthesis, is induced by drought, ABA, low temperature, and salinity in Arabidopsis (Chen et al., 2003).
Our analysis suggests that by increasing their transcriptional activity, ZmFDL1/MYB94 and other genes involved in wax deposition provide a rapid response to the drought stimulus. This response seems transient, since at later stages, when the stress condition is more severe, as indicated by the high expression level of the ZmRAB18, their transcript levels are diminished. Negative feedback might be produced, which causes repression of wax biosynthesis. The observed gene pattern might also reflect the pattern of leaf growth, meaning that in leaves approaching an advanced developmental stage, cuticle biosynthesis is diminished.
The role of ABA in mediating the response to water deficit has been known for many years (Bartels and Sunkar, 2005; Shinozaki and Yamaguchi-Shinozaki, 2007), and the effect of ABA treatment on cuticle composition and related gene expression has already been reported in Arabidopsis (Kosma et al., 2009) and tomato (Martin et al., 2017). We observed in this study that the absence of ABA perception in vp1 mutant seedlings resulted in increased leaf permeability to chlorophyll (Fig. 7), while application of exogenous ABA treatment resulted in a slight decrease (Fig. 7), thus showing for the first time in maize the involvement of ABA in cuticle-dependent leaf permeability.
We also observed that exogenous ABA application to B73 wild-type seedlings caused severe repression of ZmFDL1/MYB94 after both 24 and 48 h of treatment. As expected, repression at the transcriptional level was also observed for both ZmGL15 and ZmCER4, although only after 48 h of ABA treatment (Fig. 6, F and G). A different pattern was observed for ZmCER1, the expression of which was upregulated in the ABA-treated plants (Fig. 6E). The early response observed in the fdl1-1 mutant plants might be due to the absence of the repressive role exerted by ZmFDL1/MYB94 on the transcription of this gene. Accordingly, in Arabidopsis, the level of waxes, and in particular of alkanes, increases following ABA application (Kosma et al., 2009).
The unexpected reduction of the expression of cuticle-related genes observed in this experiment, which resembles the reduction observed at later stages in seedlings grown under limited watering (Fig. 5, E–J, days 5 and 7), might be due to the high concentrations of the applied exogenous ABA, which are considerably higher than physiological levels. These experimental conditions, with increasing ABA concentration, plausibly mimic severe drought stress (Dallmier and Stewart, 1992). It is remarkable that the changes in gene expression of ZmGL15 and ZmCER4 caused by ABA treatment, although qualitatively similar in both wild-type and fdl1-1 mutant plants, were considerably more pronounced in the latter. Taken together the data from the expression analysis strongly suggest the existence of two signaling pathways promoted by the ABA hormone for the regulation of cuticular wax biosynthesis, of which one is independent and the other requires the action of ZmFDL1/MYB94. The two pathways may act additively in controlling the transcription of biosynthetic genes.
In summary (Fig. 7), we showed in this study that the ZmFDL1/MYB94 transcription factor is a key regulator of cuticle deposition and mediates the active response of the cuticle to water scarcity conditions. ZmFDL1/MYB94 sensitivity to drought stress, consisting in the upregulation of its transcript (2d; Fig. 5G), occurred very early during seedling leaf development, as observed in the second not yet emerged leaf (7 DAS; Fig. 5D). This early ZmFDL1/MYB94 response, accompanied by the activation of the regulatory gene ZmGL15, may allow plants to rapidly cope with water scarcity conditions in young tissues by stimulating the activity of genes involved in cuticle biosynthesis, and more specifically in wax deposition. A reduction in nonstomatal evapotranspiration is thus achieved to counteract water deficit. It appeared, however, that the FDL1-mediated response is less effective at later developmental stages, as well as when the severity of the imposed stress is high. We also showed that in maize seedlings, ABA signaling influences cuticle-dependent leaf permeability, and that ABA has a negative effect on ZmFDL1/MYB94 transcription. Although mechanisms of interaction between the hormone signaling pathway and the ZmFDL1/MYB94 regulatory pathway remain to be elucidated, our model suggests that the two pathways interact to modulate the expression of cuticle-related genes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The maize (Zea mays) fdl1-1 mutant, first described by La Rocca et al. (2015), was introgressed into B73, H99, and Mo17 inbred lines. In all the experiments performed, homozygous mutants and their wild-type control plants were from the same F2 segregating progeny. Plants were grown under a long-day photoperiod (16 h light/8 h dark) in a growth chamber with controlled temperature (25°C night/30°C day) and with photon fluence of 270 µmol m−2 s−1 in pots containing S.Q.10 substrate (peat, sand, compost; Vigorplant).
For drought treatment, maize seedlings were germinated and grown in soil under NW conditions (RSWC = 80%) until 5 DAS and the next day (6 DAS, day 0 of LW) the drought stress was imposed by withholding irrigation. To monitor the drought stress level, the RSWC was measured (Janeczko et al., 2016), as previously described (Castorina et al., 2018). To keep the plants under NW or LW conditions, the RSWC was measured every day and the pots were watered as necessary until the end of the experiments. For the chlorophyll leaching assay the drought (RSWC = 40%) was imposed up to 18 DAS (Fig. 5, A and B), and for each treatment, seven pots were used. For the expression analysis of cuticle-related genes, the drought (RSWC = 30%) was imposed up to 13 DAS (Fig. 5, D–J; Supplemental Fig. S4), and for each time point, the second leaf was sampled from four/five independent plants per treatment.
To mimic drought stress, plants were treated with exogenous application of the ABA hormone (Duchefa), as previously described (Riboni et al., 2016). The fdl1-1 and B73 wild-type seedlings at the coleoptile developmental stage were treated with 1 mL ABA (1 μm) or mock control (0.001% [v/v] ethanol) solution. Starting from 4 DAS, the treatments were dispensed, every other day, watering the roots directly up to 14 DAS. For each treatment (Mock and ABA) the second leaf was taken from a minimum of seven independent plants per genotype (Fig. 2E; Supplemental Fig. S3).
To analyze the transcript regulation effect of ABA on ZmFDL1/MYB94 and other cuticle-related genes, B73 wild-type and fdl1-1 seedlings were treated by dipping only the root apparatus into an ABA (1, 10, and 100 μm) or mock-control solution. After 24 and 48 h of treatment, only the green tissues (coleoptile and leaves) from a minimum of six independent plants per genotype were sampled for subsequent total RNA extraction.
Cuticular Cutin and Wax Analysis
Cuticle composition was analyzed in fdl1-1 and wild-type seedlings at successive developmental stages: coleoptile, first emerging leaf, second emerging leaf, and third emerging leaf stages, at 2, 6, 10, and 14 DAS, respectively. Cutin and epicuticular waxes were extracted and identified by the combination of gas chromatography on-column injection and gas chromatography-mass spectrometry performed according to previously described methods (Domergue et al., 2010; Bourdenx et al., 2011).
Chlorophyll Leaching Assay, Leaf Temperature, and Water Loss
For the chlorophyll leaching analysis, second, third, and fourth fully expanded leaves were taken from a minimum of five independent plants per genotype and dissected into pieces of 8 cm in length measured from the apex. Leaf sectors were weighed, immersed in 80% [v/v] ethanol and incubated in the dark at room temperature. Absorbance was measured every day at 647 and 664 nm with a spectrophotometer (Cary 60 UV-Vis, Agilent Technologies) to quantify the chlorophyll released in the solution, and measurements were performed until chlorophyll extraction was complete. The micromolar concentration of chlorophyll was calculated using the equation total micromoles chlorophyll = 7.93 × A664 + 19.53 × A647 (Lolle et al., 1997). The data obtained were normalized per gram of fresh weight and area of the 8-cm-long leaf pieces to be expressed as a percentage of the total chlorophyll.
Thermal images of the second fully expanded leaves were taken from 14-d-old fdl1-1 and wild-type seedlings with a semiautomated long-wave infrared camera system (FLIR T650sc) in a growth chamber at a constant temperature of 25°C and with photon fluence of 270 μmol m−2 s−1. The temperature of the leaves was measured using the FLIR ResearchIR Max software.
To determine the time course of the seedling water loss, 10-d-old seedlings were detached and weighed immediately. Seedling weight was then estimated at designated time intervals and water loss was calculated as the percentage of fresh weight based on the initial weight. Several biological replicates were measured for each genotype.
Cell Density and Stomatal Index Analysis
To measure stomatal density and stomatal index, a leaf surface imprint method was used as previously described (Castorina et al., 2018). We analyzed second, third, and fourth fully expanded leaves of fdl1-1 mutant and wild-type plants. Stomatal index was determined as (number of stomata/[number of epidermal cells + number of stomata]) × 100. A one-way ANOVA test was performed with the statistical package SPSS 21.0 (IBM; P > 0.05).
RNA, cDNA Preparation, and Quantitative Gene Expression Analysis
Total RNA was extracted from maize tissues using the TRIzol Reagent (Life Technologies) and treated with RQ1 RNase-Free DNase (Promega) according to the manufacturer’s instructions. First-strand cDNA was synthetized with the SuperScript III First-Strand Synthesis System (Invitrogen) from 1,000 ng of total RNA, according to the manufacturer's instructions. RT-qPCR was performed with the 7300 Real-Time PCR System (Applied Biosystems), using GoTaq qPCR Master Mix (Promega), in a final volume of 10 μL. The following cycle was used: 10 min preincubation at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The relative transcript level of each gene was calculated by the 2−ΔΔCt method (Livak and Schmittgen, 2001) using expression of the ZmEF1a gene as a reference. The gene-specific primers are listed in Supplemental Table S2.
The RNA extractions and purifications for RNA-Seq were performed on seedlings at the coleoptile stage of fdl1-1 and wild-type plants 2 d after germination, collecting three plants per genotype (n = 3), and homogenized in liquid nitrogen. The total RNA extraction was performed with the Pure Link RNA Mini Kit (Invitrogen). Four micrograms of total RNA with an RNA integrity number ≥8 (Bioanalyzer 2100, Agilent Technologies) was sent to the IGA Technology Services4 for library preparation (TruSeq Stranded mRNA, Illumina) and sequencing on a HiSeq 2000 platform (single-read 50 bp, 6-plex, ∼20 million reads/sample; Supplemental Table S1).
Differential Expression Analysis and Functional Enrichment Analyses
Six biological replicates, three for the fdl1-1 mutant and three for the wild-type background, were analyzed. Reads were mapped on the Zm00001d.2 gene model annotation of the B73 reference assembly (ZmB73_RefGen_v4) of the maize genome, as obtained from http://plants.ensembl.org/info/website/ftp/index.html, using the bowtie2 program (Langmead and Salzberg, 2012). Statistics concerning the total number of reads produced and the proportion of reads assigned to gene models according to the Zm00001d annotation of the ZmB73_RefGen_v4 reference assembly (Jiao et al., 2017) are reported in Supplemental Data S1. Estimation of gene expression levels was performed using RSEM (Li and Dewey, 2011). Identification of DEGs was performed applying the latest versions of DESeq2 (Love et al., 2014) and Limma (Ritchie et al., 2015) to RSEM-estimated read counts. Only genes with a median read count of 10 or more were considered (10 reads in at least three samples). Genes showing a false discovery rate <0.05 according to both tools were considered differentially expressed. GO annotation of maize genes according to Wimalanathan et al. (2018) was obtained from http://datacommons.cyverse.org/browse/iplant/home/shared/commons_repo/curated/Carolyn_Lawrence_Dill_maize-GAMER_July_2017_V.1. Functional enrichment analyses of DEGs were performed according to a collection of ontologies for the functional annotation of maize gene models and biosynthetic pathways, including GO (Wimalanathan et al., 2018), MapMan (Schwacke et al., 2019), and KEGG (Moriya et al., 2007; Okuda et al., 2008), by means of a custom script implementing a Fisher exact test and the Benjamini Hochberg procedure for the correction of multiple testing.
Graphical representation of the results of Gene Ontology functional enrichment was performed by means of the REViGO tool (Supek et al., 2011; http://revigo.irb.hr/) using default parameters. GO terms of interest were highlighted manually.
Annotation of maize metabolic pathways according to the Mercator MapMan ontology (Schwacke et al., 2019) was obtained from https://mapman.gabipd.org/mapmanstore. Functional enrichment of metabolic pathways was performed using MapMan version 3.5.1.
Publicly available KEGG pathways for maize were obtained directly from the KEGG PATHWAY repository, listed at https://www.genome.jp/dbget-bin/get_linkdb?-t+pathway+gn:T01088.
Accession Numbers
The gene sequences from this article can be found in the Maize Genetics and Genomics Database (MaizeGDB) or GenBank/EMBL databases under the following accession numbers: Zm00001d022227 or GRMZM2G056407 (ZmFDL1/MYB94), Zm00001d046449 or GRMZM2G153541 (ZmEF1a), Zm00001d045321 or GRMZM2G098750 (ZmRAB18). Accession numbers are available in the Supplemental Data.
RNA sequencing data sets analyzed during this study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146905) under accession number GSE146905.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Developmental phase-dependent cuticle composition
Supplemental Figure S2. Detailed cuticle composition in seedlings at succeeding developmental stages.
Supplemental Figure S3. Venn diagram of DEGs and representation of enriched GO terms, according to the REViGO tool
Supplemental Figure S4. Stomatal index and additional information about drought stress experiments.
Supplemental Table S1. Number of reads and proportion of reads mapped.
Supplemental Table S2. Results of differential expression analyses.
Supplemental Table S3. Differentially expressed genes.
Supplemental Table S4. Significantly over-represented GO terms in down regulated genes.
Supplemental Table S5. Significantly over-represented GO terms in up regulated genes.
Supplemental Table S6. Significantly over-represented metabolic bins according to MapMan.
Supplemental Table S7. Significantly over-represented KEGG pathways in up regulated genes.
Supplemental Table S8. Significantly over-represented KEGG pathways in up regulated genes.
Supplemental Table S9. Differentially expressed genes involved in cuticle-related processes in the fdl1-1 mutant.
Supplemental Table S10. Differentially expressed genes involved in circadian rhythm in the fdl1-1 mutant.
Supplemental Table S11. Primers used for RT-qPCR analysis.
Acknowledgments
We acknowledge the Maize Genetics Cooperation Stock Center for providing the vp1 mutant.
Footnotes
This work was supported by the Università degli Studi di Milano (PSR project to G.Co and postdoctoral fellowship to G.Ca.).
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A(2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiba T, Hibara K, Kimura F, Tsuda K, Shibata K, Ishibashi M, Moriya C, Nakagawa K, Kurata N, Itoh J, et al. (2014) Organ fusion and defective shoot development in oni3 mutants of rice. Plant Cell Physiol 55: 42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Sunkar R(2005) Drought and salt tolerance in plants. CRC Crit Rev Plant Sci 24: 23–58 [Google Scholar]
- Becraft PW, Kang SH, Suh SG(2001) The maize CRINKLY4 receptor kinase controls a cell-autonomous differentiation response. Plant Physiol 127: 486–496 [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure JD, Haslam RP, Napier JA, Lessire R, Joubès J(2012) Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Joubès J(2013) Arabidopsis cuticular waxes: Advances in synthesis, export and regulation. Prog Lipid Res 52: 110–129 [DOI] [PubMed] [Google Scholar]
- Bi H, Luang S, Li Y, Bazanova N, Morran S, Song Z, Perera MA, Hrmova M, Borisjuk N, Lopato S(2016) Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J Exp Bot 67: 5363–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdenx B, Bernard A, Domergue F, Pascal S, Léger A, Roby D, Pervent M, Vile D, Haslam RP, Napier JA, et al. (2011) Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol 156: 29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgault R, Matschi S, Vasquez M, Qiao P, Sonntag A, Charlebois C, Mohammadi M, Scanlon MJ, Smith LG, Molina I(2020) Constructing functional cuticles: Analysis of relationships between cuticle lipid composition, ultrastructure and water barrier function in developing adult maize leaves. Ann Bot 125: 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow GB, Franks CD, Xin Z(2008) Genetic and physiological analysis of an irradiated bloomless mutant (epicuticular wax mutant) of sorghum. Crop Sci 48: 41–48 [Google Scholar]
- Cao X, Costa LM, Biderre-Petit C, Kbhaya B, Dey N, Perez P, McCarty DR, Gutierrez-Marcos JF, Becraft PW(2007) Abscisic acid and stress signals induce Viviparous1 expression in seed and vegetative tissues of maize. Plant Physiol 143: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castorina G, Persico M, Zilio M, Sangiorgio S, Carabelli L, Consonni G(2018) The maize lilliputian1 (lil1) gene, encoding a brassinosteroid cytochrome P450 C-6 oxidase, is involved in plant growth and drought response. Ann Bot 122: 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA(2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, Brosché M, Lehtonen MTT, Amiryousefi A, Xu E, Punkkinen M, Valkonen JPPT, Fujii H, Overmyer K(2016) Dissecting abscisic acid signaling pathways involved in cuticle formation. Mol Plant 9: 926–938 [DOI] [PubMed] [Google Scholar]
- Dallmier KA, Stewart CR(1992) Effect of exogenous abscisic acid on proline dehydrogenase activity in maize (Zea mays L.). Plant Physiol 99: 762–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich CR, Perera MADN, Yandeau-Nelson MD, Meeley RB, Nikolau BJ, Schnable PS(2005) Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J 42: 844–861 [DOI] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubès J, Ono J, Lee JA, Bourdon M, Alhattab R, Lowe C, Pascal S, Lessire R, et al. (2010) Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol 153: 1539–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelie KE, Kolattukudy PE(1979) Composition of the aliphatic component of “suberin” from the bundle sheaths of Zea mays leaves. Plant Sci Lett 15: 225–230 [Google Scholar]
- Fich EA, Segerson NA, Rose JKC(2016) The plant polyester cutin: Biosynthesis, structure, and biological roles. Annu Rev Plant Biol 67: 207–233 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM(1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam TM, Kunst L(2013) Extending the story of very-long-chain fatty acid elongation. Plant Sci 210: 93–107 [DOI] [PubMed] [Google Scholar]
- Hooker TS, Lam P, Zheng H, Kunst L(2007) A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19: 904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram G, Nawrath C(2017) The roles of the cuticle in plant development: Organ adhesions and beyond. J Exp Bot 68: 5307–5321 [DOI] [PubMed] [Google Scholar]
- Janeczko A, Gruszka S, Pociecha E, Dziurka M, Filek M, Jurczyk B, Kalaji HM, Kocurek M, Waligórski P(2016) Physiological and biochemical characterisation of watered and drought-stressed barley mutants in the HvDWARF gene encoding C6-oxidase involved in brassinosteroid biosynthesis. Plant Physiol Biochem 99: 126–141 [DOI] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Depège-Fargeix N, Arnould C, Oursel D, Domergue F, Sarda X, Rogowsky PM(2010) Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor OUTER CELL LAYER1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol 154: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Guo T, Becraft PW(2000) The maize CR4 receptor-like kinase mediates a growth factor-like differentiation response. Genesis 27: 104–116 [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK(1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lü S, Joubès J, Jenks MA(2009) The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol 151: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolikowski KA, Victor JL, Wagler TN, Lolle SJ, Pruitt RE(2003) Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J 35: 501–511 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bär S, Franke R, Efremova N, Tietjen K, Schreiber L, Saedler H, Yephremov A(2006a) Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224: 315–329 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Nawrath C, Bär S, Voisin D, Efremova N, Franke R, Schreiber L, Saedler H, Métraux JP, et al. (2006b) The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca N, Manzotti PS, Cavaiuolo M, Barbante A, Dalla Vecchia F, Gabotti D, Gendrot G, Horner DS, Krstajic J, Persico M, et al. (2015) The maize fused leaves1 (fdl1) gene controls organ separation in the embryo and seedling shoot and promotes coleoptile opening. J Exp Bot 66: 5753–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL(2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose SP(2005) microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA 102: 9412–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Mas P, Seo PJ(2016a) MYB96 shapes the circadian gating of ABA signaling in Arabidopsis. Sci Rep 6: 17754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Suh MC(2015b) Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep 34: 557–572 [DOI] [PubMed] [Google Scholar]
- Lee SB, Suh MC(2015a) Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol 56: 48–60 [DOI] [PubMed] [Google Scholar]
- Lee SB, Kim HU, Suh MC(2016b) MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis. Plant Cell Physiol 57: 2300–2311 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN(2011) RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, Jetter R, Kunst L(2008) Identification of the wax ester synthase/acyl-coenzyme A:diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol 148: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Du Y, He C, Dietrich CR, Li J, Ma X, Wang R, Liu Q, Liu S, Wang G, et al. (2019) Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J Exp Bot 70: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li D, Liu S, Ma X, Dietrich CR, Hu HC, Zhang G, Liu Z, Zheng J, Wang G, et al. (2013) The maize glossy13 gene, cloned via BSR-Seq and Seq-walking, encodes a putative ABC transporter required for the normal accumulation of epicuticular waxes. PLoS One 8: e82333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Dietrich CR, Schnable PS(2009) DLA-based strategies for cloning insertion mutants: Cloning the gl4 locus of maize using Mu transposon tagged alleles. Genetics 183: 1215–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Tang HM, Nettleton D, Schnable PS(2012) Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One 7: e36406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bourgault R, Strable J, Galli M, Chen Z, Dong J, Molina I, Gallavotti A (2020) The FUSED LEAVES1/ADHERENT1 regulatory module is required for maize cuticle development and organ separation. bioRxiv 943787, doi: 10.1101/2020.02.11.943787 [DOI] [PMC free article] [PubMed]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lolle SJ, Berlyn GP, Engstrom EM, Krolikowski KA, Reiter W, Pruitt RE(1997) Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: A role for the epidermal cell wall and cuticle. Dev Biol 189: 311–321 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S(2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Wang H, Liu S, Li Z, Yang X, Yan J, Li J, Tran LSP, Qin F(2015) A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun 6: 8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LBB, Romero P, Fich EA, Domozych DS, Rose JKC(2017) Cuticle biosynthesis in tomato leaves is developmentally regulated by abscisic acid. Plant Physiol 174: 1384–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK(1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- Moose SP, Sisco PH(1994) Glossy15 controls the epidermal juvenile-to-adult phase transition in maize. Plant Cell 6: 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M(2007) KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35: W182–W185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M(2008) KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res 36: W423–W426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JJ, Jin P, Yoon J, Yang JI, Jeong HJ, Ranathunge K, Schreiber L, Franke R, Lee IJ, An G(2010) Mutation in Wilted Dwarf and Lethal 1 (WDL1) causes abnormal cuticle formation and rapid water loss in rice. Plant Mol Biol 74: 91–103 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D(2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L(2016) ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J Exp Bot 67: 6309–6322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK(2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L(2006) CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol 142: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Ponce-Soto GY, Krause K, Bolger AM, Arsova B, Hallab A, Gruden K, Stitt M, Bolger ME, Usadel B(2019) MapMan4: A refined protein classification and annotation framework applicable to multi-omics data analysis. Mol Plant 12: 879–892 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park M-J, Go YS, Park C-M(2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM(2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K(2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Sinha N, Lynch M(1998) Fused organs in the adherent1 mutation in maize show altered epidermal walls with no perturbations in tissue identities. Planta 206: 184–195 [Google Scholar]
- Sturaro M, Hartings H, Schmelzer E, Velasco R, Salamini F, Motto M(2005) Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol 138: 478–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T(2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ketterling MG, Li QB, McCarty DR(2003) Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol 132: 1664–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M(1990) Division and differentiation during normal and liguleless-1 maize leaf development. Development 110: 985–1000 [DOI] [PubMed] [Google Scholar]
- Tacke E, Korfhage C, Michel D, Maddaloni M, Motto M, Lanzini S, Salamini F, Döring HP(1995) Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J 8: 907–917 [DOI] [PubMed] [Google Scholar]
- Takeda S, Iwasaki A, Matsumoto N, Uemura T, Tatematsu K, Okada K(2013) Physical interaction of floral organs controls petal morphogenesis in Arabidopsis. Plant Physiol 161: 1242–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg G, Fischer S, Leide J, Emmanuel E, Jetter R, Levy AA, Riederer M(2004) Tomato fruit cuticular waxes and their effects on transpiration barrier properties: Functional characterization of a mutant deficient in a very-long-chain fatty acid β-ketoacyl-CoA synthase. J Exp Bot 55: 1401–1410 [DOI] [PubMed] [Google Scholar]
- Wimalanathan K, Friedberg I, Andorf CM, Lawrence-Dill CJ(2018) Maize GO Annotation—Methods, Evaluation, and Review (maize-GAMER). Plant Direct 2: e00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Zhang X, Lu X, Chen G, Chen ZH(2017) Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front Plant Sci 8: 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC(2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H(1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li L, Xiang J, Gao G, Xu F, Liu A, Zhang X, Peng Y, Chen X, Wan X(2015) OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS One 10: e116676. [DOI] [PMC free article] [PubMed] [Google Scholar]