A unified 4-clade phylogenetic tree of CCDs highlights ZmCCD10a, a previously unknown CCD enzyme in terms of cleavage sites, that facilitates plant tolerance to phosphate limitation.
Abstract
Carotenoid cleavage dioxygenases (CCDs) drive carotenoid catabolism to produce various apocarotenoids and immediate derivatives with particular developmental, ecological, and agricultural importance. How CCD genes evolved with species diversification and the resulting functional novelties in cereal crops have remained largely elusive. We constructed a unified four-clade phylogenetic tree of CCDs, revealing a previously unanchored basal clade CCD10. CCD10 underwent highly dynamic duplication or loss events, even in the grass family. Different from cleavage sites of CCD8 and ZAXINONE SYNTHASE (ZAS), maize (Zea mays) ZmCCD10a cleaved differentially structured carotenoids at 5, 6 (5′, 6') and 9, 10 (9', 10') positions, generating C8 (6-methyl-5-hepten-2-one) and C13 (geranylacetone, α-ionone, and β-ionone) apocarotenoids in Escherichia coli. Localized in plastids, ZmCCD10a cleaved neoxanthin, violaxanthin, antheraxathin, lutein, zeaxanthin, and β-carotene in planta, corroborating functional divergence of ZmCCD10a and ZAS. ZmCCD10a expression was dramatically stimulated in maize and teosinte (Z. mays ssp. parviglumis, Z. mays ssp. huehuetenangensis, Zea luxurians, and Zea diploperennis) roots by phosphate (Pi) limitation. ZmCCD10a silencing favored phosphorus retention in the root and reduced phosphorus and biomass accumulation in the shoot under low Pi. Overexpression of ZmCCD10a in Arabidopsis (Arabidopsis thaliana) enhanced plant tolerance to Pi limitation by preferential phosphorus allocation to the shoot. Thus, ZmCCD10a encodes a unique CCD facilitating plant tolerance to Pi limitation. Additionally, ZmCCD10a silencing and overexpression led to coherent alterations in expression of PHOSPHATE STARVATION RESPONSE REGULATOR 1 (PHR1) and Pi transporters, and cis-regulation of ZmCCD10a expression by ZmPHR1;1 and ZmPHR1;2 implies a probable ZmCCD10a-involved regulatory pathway that adjusts Pi allocation.
Carotenoids are fat-soluble secondary metabolites in plants and microbes (Nisar et al., 2015). In plants, carotenogenesis starts with phytoene production and continues with sequential desaturation, isomerization, and lycopene cyclization, giving rise to various carotenes such as δ-carotene, ε-carotene, α-carotene, and β-carotene; further oxygenation of cyclized carotenes produces a series of xanthophylls, including lutein, zeaxanthin, antheraxanthin, violaxanthin, and neoxanthin (Tian et al., 2003; Farré et al., 2011). They serve as photoprotectants, antioxidants, and accessory pigments, or further undergo carotenoid cleavage dioxygenase (CCD)–mediated catabolism to generate apocarotenoids (Schwartz et al., 1997; Auldridge et al., 2006a; Auldridge et al., 2006b; Ballottari et al., 2014; Park et al., 2019).
CCDs cleave carotenoids at their double bonds, and Viviparous 14 is the first cloned CCD family gene regulating abscisic acid (ABA) biosynthesis in maize (Zea mays; Schwartz et al., 1997). Similar to Viviparous 14, five NCEDs (nine-cis epoxycarotenoid dioxygenases; NCED2, NCED3, NCED5, NCED6, and NCED9) cleave the 11, 12 (11', 12') double bond of 9-cis-violaxanthin or 9-cis-neoxanthin toward ABA production in Arabidopsis (Arabidopsis thaliana; Tan et al., 2003); ABA plays crucial roles in plant senescence, fruit and seed development, stoma closure, and environmental adaptation (Zeevaart and Creelman, 1998; Cheng et al., 2014). Five CCDs, including CCD1, CCD2, CCD4, MORE AXILLARY GROWTH3 (MAX3; CCD7), and MAX4 (CCD8), have divergent cleavage properties (Auldridge et al., 2006a; Huang et al., 2009; Frusciante et al., 2014). The cytosol specifically localized CCD1 cleaves a wide range of carotenoids at varying positions 5, 6 (5′, 6'); 7, 8 (7', 8'); or 9, 10 (9', 10'; Schwartz et al., 2001; Vogel et al., 2008). CCD2 drives zeaxanthin catabolism at positions 7, 8, or 7', 8' in Crocus sativus (Frusciante et al., 2014). CCD4 acts on carotenoids at positions 7, 8 (7', 8') or 9, 10 (9', 10') in plastids (Ahrazem et al., 2016). CCD1- and CCD4-mediated carotenoid cleavage produces a variety of color, flavor, and aroma compounds, enabling flowers and fruits to better attract pollinators (Ohmiya et al., 2006; Gonzalez-Jorge et al., 2013; Ahrazem et al., 2016). CCD1 also promotes root colonization of arbuscular mycorrizhal fungi (AMF) in Medicago truncatula (Floss et al., 2008b). Carotenoids can be sequentially cleaved by plastidic MAX3 (at positions 9, 10, or 9', 10') and MAX4 (at positions 13, 14), generating carlactone as the strigolactone precursor (Alder et al., 2012). Strigolactones coordinate shoot and root development, promote germination of parasitic seeds, and trigger AMF branching (Umehara et al., 2008). Loss-of-function mutation of MAX3 or MAX4 leads to dramatically more axillary branches in Arabidopsis. OsCCD8b also regulates rice (Oryza sativa) tillering, whereas ZmCCD8 plays essential roles in root and shoot development, with smaller roots, shorter internodes, and longer tassels in Zmccd8 mutant plants (Arite et al., 2007; Guan et al., 2012). Recent findings reveal that ZAXINONE SYNTHASE (ZAS) cleaves apo-10'-zeaxanthinal at the 13, 14 double bond to produce zaxinone to reduce strigolactone levels in rice (Wang et al., 2019). With increasingly more genomes sequenced, a growing body of CCD genes form a large family across bacteria, plants, and animals (Kloer and Schulz, 2006; Ahrazem et al., 2016; Wang et al., 2019), including CCD1, CCD4, CCD7, CCD8, CCD-like, ZAS, and five NCED clades (Vallabhaneni et al., 2010; Wei et al., 2016; Wang et al., 2017; Chen et al., 2018; Wang et al., 2019). However, the evolutionary relationship of CCD, CCD-like, ZAS, and NCED genes remains unresolved, which, to a certain extent, hinders their functional prediction and characterization in different species.
A particularly interesting feature of a number of CCD genes is their active involvement in phosphate (Pi) stress responses. Upon Pi limitation, enhanced MAX4 expression boosts strigolactone production, reducing tiller proliferation and stimulating AMF-mediated Pi acquisition (Umehara et al., 2008; Czarnecki et al., 2013). ZmCCD7 transcription is greatly up-regulated, especially in crown roots of Pi-limited seedlings (Pan et al., 2016). PHOSPHATE STARVATION RESPONSE REGULATOR 1 (PHR1), a MYB family transcription factor, is a crucial regulator of plant responses to Pi limitation via binding to the P1BS cis-element (GNATATNC) of target genes (Chiou and Lin, 2011). Heterologous expression of ZmPHR1 in Arabidopsis increases the shoot biomass and inorganic Pi content (Wang et al., 2013). Auxin response factors ARF7 and ARF19 function upstream of PHR1 to modulate its transcription in Arabidopsis roots (Huang et al., 2018). PHOSPHATE1 (PHO1) mediates Pi loading into the xylem and root-to-shoot translocation (Poirier et al., 1991). Regulated by PHR1 and miRNA399 at the transcription level, PHO2 degrades PHO1 and Pi transporters to down-scale Pi absorption and translocation; pho2 mutant leaves show Pi toxicity due to Pi over-accumulation (Delhaize and Randall, 1995; Bari et al., 2006; Liu et al., 2012). The family of PHOSPHATE TRANSPORTER 1 (PHT1) plays crucial roles in Pi uptake from the soil and translocation within the plant (Nussaume et al., 2011). Knockout of AtPHT1;1 significantly reduces Pi uptake, and AtPHT1;5 regulates Pi translocation from source to sink tissues to maintain Pi homeostasis (Shin et al., 2004; Nagarajan et al., 2011).
Maize is an evolutionarily, genetically, and economically important crop promoting global food and feed security (Kellogg, 1997; Coe, 2001; Kumar and Jhariya, 2013). Ancient polyploidization and mobilization of highly abundant transposable elements has led to a 2.5-gigabase maize genome (Lai et al., 2004; Schnable et al., 2009). Maize grains contain highly concentrated phosphorus (P) to nourish embryo and seedling development (Nadeem et al., 2011); however, Pi limitation frequently occurs due to the low bioavailabilty of soil Pi (Shen et al., 2011). It is particularly necessary to explore evolutionary novelties of CCDs in maize and their potential relationship with P nutrition. Here, we constructed a unified CCD phylogenetic tree and found that CCD10 represents a previously unanchored basal clade adding to CCD1, CCD7, CCD8 clades in the entire gene family. ZmCCD10a encoded a new CCD in terms of cleavage sites. Down-regulation of ZmCCD10a expression made plants susceptible to Pi deficiency, whereas its heterologous expression in Arabidopsis enhanced plant tolerance to Pi limitation.
RESULTS
ZmCCD10 was a Distinct Member of the CCD Family
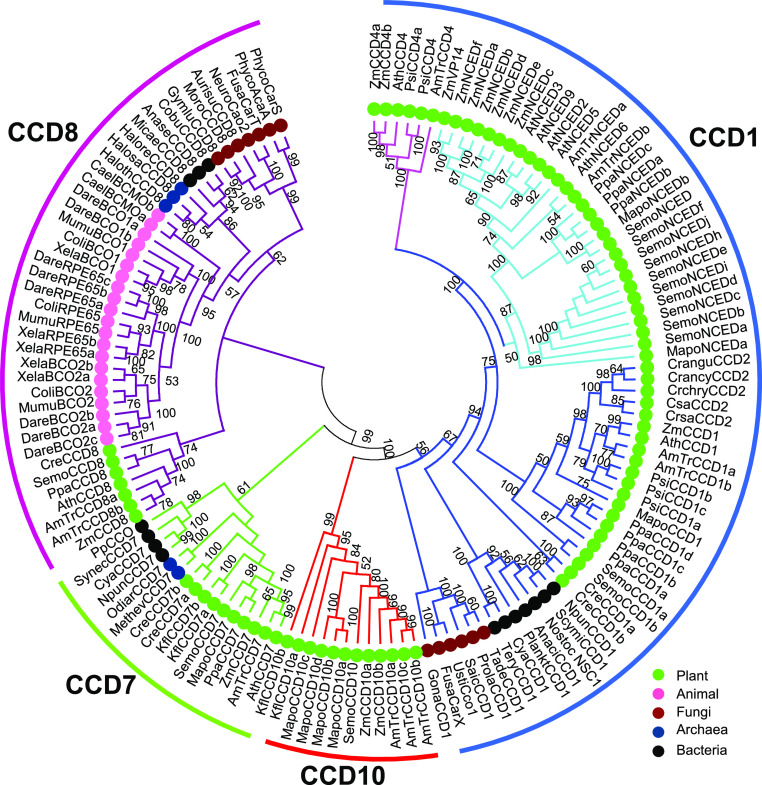
To analyze CCD genes in maize, 14 CCDs were retrieved from the maize genome and classified into four basal clades according to systemic phylogenetic analysis: CCD1, CCD7, CCD8, and CCD10 (Fig. 1). ZmCCD1, two ZmCCD4s, and seven ZmNCEDs were members of the CCD1 clade, which existed in bacteria, fungi, and plants, and underwent two rounds of duplication along with plant adaptation to land, giving rise to NCED and CCD4 subclades. ZmCCD7 belonged to a second basal clade persisting in bacteria, archaea, and plants. ZmCCD8 appeared as a single-copy gene in the CCD8 clade spreading across five fundamental kingdoms. CCD10 was named as CCD-like or more recently ZAS in different contexts (Wei et al., 2016; Wang et al., 2017; Chen et al., 2018; Wang et al., 2019); here, we renamed it as CCD10 according to three criteria: (1) reflecting its fundamental functions in carotenoid cleavage; (2) following CCD1, CCD4, CCD7, CCD8, NCED2, NCED3, NCED5, NCED6, and NCED9; and (3) accommodating wide functional divergence across species beyond mediation of zaxinone synthesis in rice. Thus, we constructed a unified CCD phylogenetic tree with four basal clades (Fig. 1). ZmCCD10a and ZmCCD10b represented a previously unanchored basal clade containing only plant species, most likely due to their loss in bacteria, archaea, and fungi. Notably, basal clade genes CCD1, CCD7, CCD8, and CCD10 contained 1 to 14, 7 and 8, 4 to 10, and 9 to 15 exons in land plants, respectively, indicating large variations in gene structure across four CCD clades (Supplemental Fig. S1). Together, our analysis suggested that ZmCCD10 encodes a distinct CCD in a basal clade.
Figure 1.
Phylogenetic analysis of the CCD gene family. Four basal clades are indicated by blue (CCD1), green (CCD7), purple (CCD8), and red (CCD10) curves. Within the CCD1 clade, CCD4 and NCED branches are in pink and light blue, respectively. Numbers on branches indicate bootstrap values above 50.
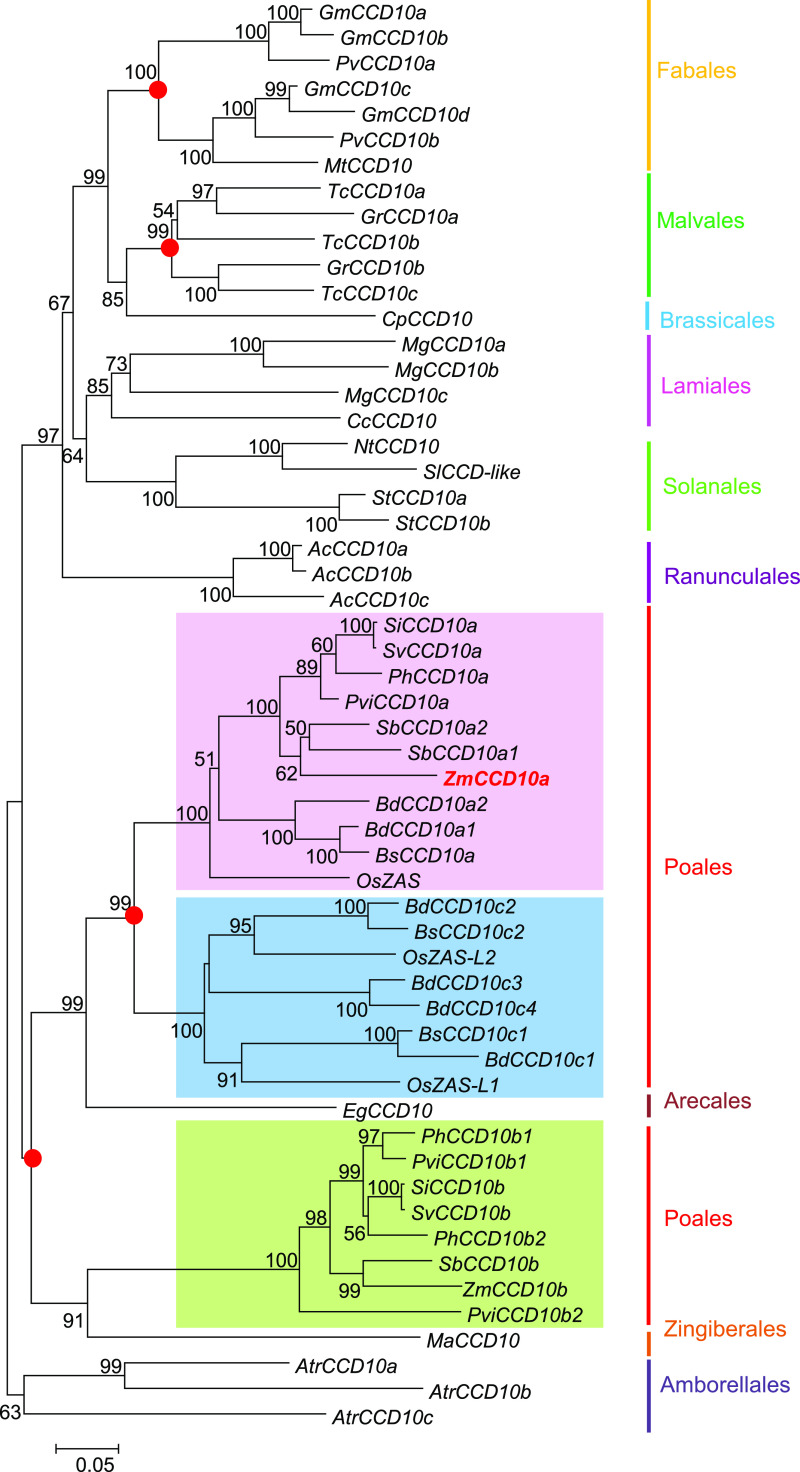
To trace CCD10 footprints over plant evolution in angiosperms, we constructed a phylogenetic tree with 24 representative species from 10 orders, revealing that CCD10 was highly dynamic and underwent recurrent gain and loss events during genome expanding and restructuring (Fig. 2). Its copy number dramatically varied from 0 (i.e. in the Brassicaceae family) to 6 in the examined species (Fig. 2; Supplemental Table S1). In dicots, CCD10 duplicated independently before Fabales and Malvales occurrence, and CCD10 doubled before monocot emergence. Different from maintenance of CCD10 duplicates in Poales, only one copy was retained in Elaeis guineensis in subclade a or in Musa acuminate in subclade b. The a subclade further duplicated before Poales split and gave rise to the c subclade. Particularly in the Poaceae family (Figs. 2 and 3), CCD10a remained a single copy in maize and rice and gained an extra copy in Brachypodium distachyon and sorghum (Sorghum bicolor). CCD10b persisted in maize and sorghum although it was eliminated in rice and B. distachyon. CCD10c was lost in maize and sorghum while maintaining two paralogs in rice and four in B. distachyon, indicative of a unique evolutionary path. Of the CCD10 triplicate, only CCD10a orthologs survived across maize, sorghum, rice, and B. distachyon in the grass family (Fig. 2).
Figure 2.
Phylogenetic analysis of the CCD10 gene family in angiosperms. The pink, green, and blue rectangles represent the CCD10a, CCD10b, and CCD10c subclades, respectively. Red solid circles indicate typical duplication events. ZmCCD10a is in red and bold. CCD10s of amborellales serve as the outgroup. Numbers on branches indicate bootstrap values above 50.
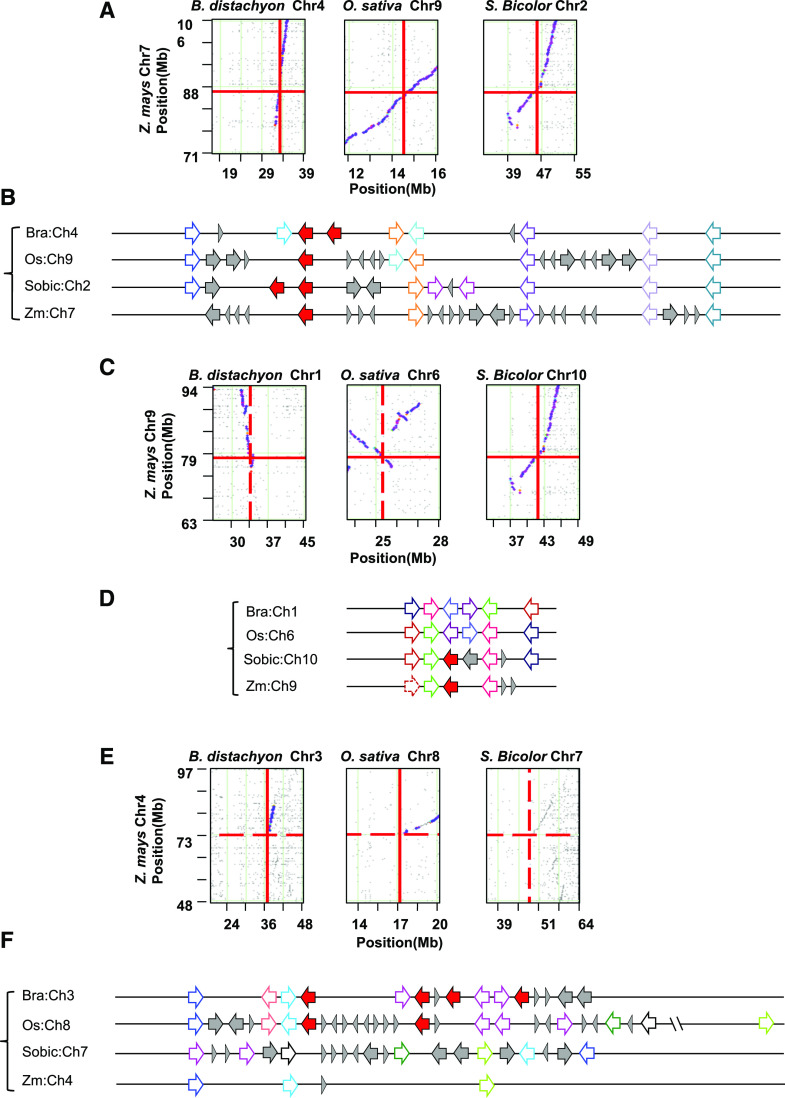
Figure 3.
CCD10 collinearity across four representative cereal species. Collinearity in the CCD10a (A), CCD10b (C), and CCD10c (E) regions across maize, sorghum, rice, and B. distachyon. Dot plots were drawn based on CoGe (https://genomevolution.org/coge/SynMap.pl) BlastP analysis and putative homologous pairs were plotted as purple dots corresponding to their genomic positions. Red lines cross at the CCD10 locus. Local collinearity of CCD10a (B), CCD10b (D), and CCD10c (F) across maize, sorghum, rice, and B. distachyon are shown. Solid red arrows represent CCD10 genes. Open arrows in the same color represent homologous genes across species, except for solid gray arrows and triangles to indicate nonhomologous genes or elements.
In maize, CCD10a was previously reported as ZmCCD8b, a CCD8 closely related name requiring further functional justification (Vallabhaneni et al., 2010). To verify our evolutionary classification, ZmCCD10a and ZmCCD8 were constitutively expressed in max4 (Auldridge et al., 2006a). Heterologous expression of ZmCCD8 functionally complemented the mutant and fully suppressed its over-branching, whereas ZmCCD10a transgenic plants still had as many (6–8, n = 20) bolts as the max4 mutant (Fig. 4A). Dramatically different phenotypes of these two transgenic lines provided functional evidence to favorably position ZmCCD10a into a separate clade rather than the ZmCCD8 clade in this context.
Figure 4.
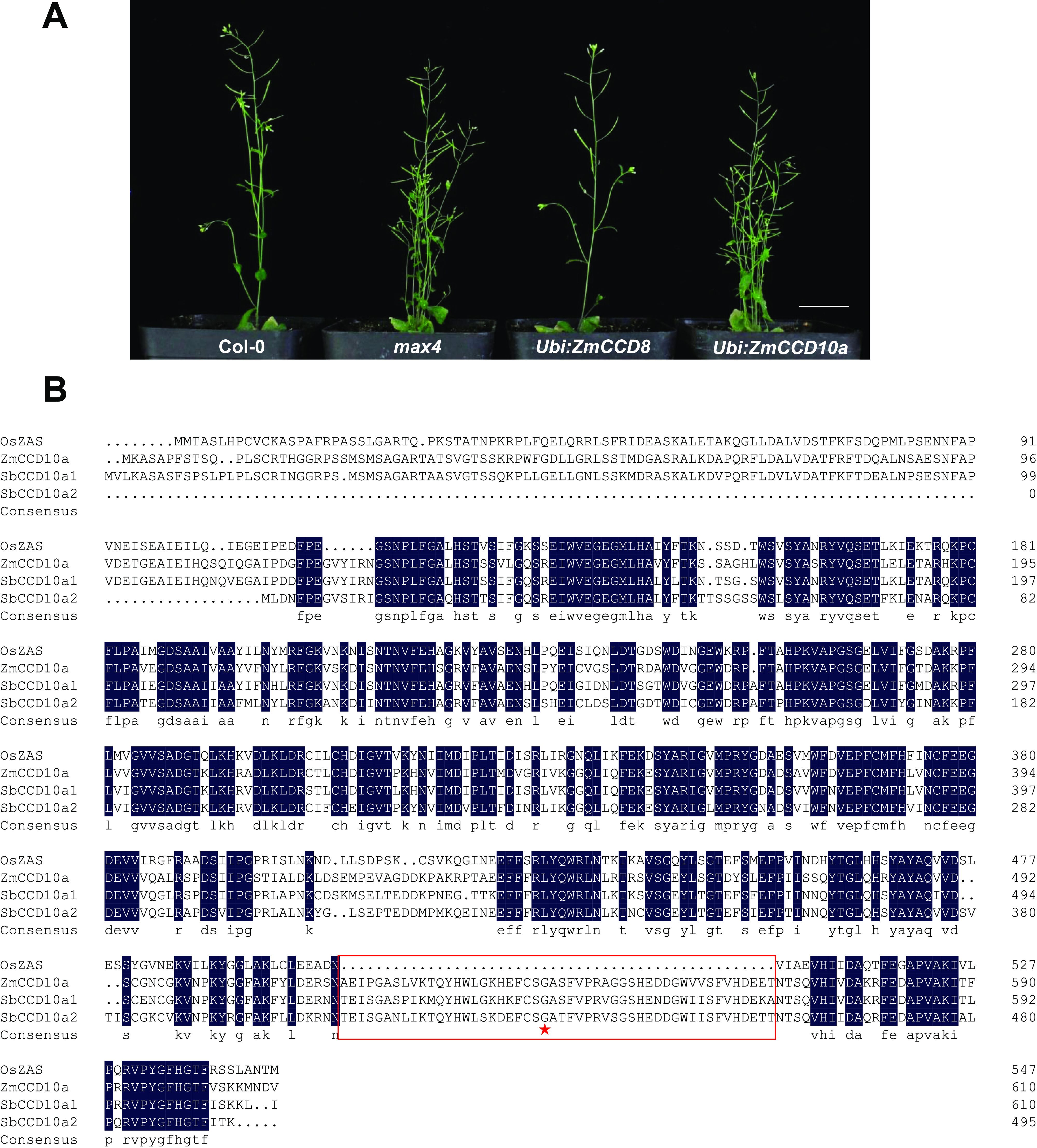
Heterologous expression of ZmCCD10a in the max4 mutant and protein alignment of CCD10a in rice, maize, and sorghum. A, Transformation of ZmCCD10a (Ubi:ZmCCD10a) and ZmCCD8 (Ubi:ZmCCD8) into the max4 mutant and bolts of homozygous transgenic lines. Wild-type (Col-0), max4 mutant, and transgenic plants were grown in the soil for 4 weeks. Scale bar = 3 cm. B, Protein alignment with conserved amino acids blue shaded, 50 amino acids lost in OsZAS shown in the red box, and the conserved Gly-541 in ZmCCD10a marked by the red star.
ZmCCD10a was also quite different from ZAS, its ortholog in rice (Wang et al., 2019); the latter lost 50 amino acids in a conserved region on the C-terminal (Fig. 4B). Within this conserved region, Gly-541 in ZmCCD10a was highly conserved across all angiosperms, corresponding to a Glu residue in lycophytes, liverworts, and charophytes (Fig. 4B; Supplemental Fig. S2). The Glu/Gly residue is involved in binding of the Fe2+ cofactor to condition enzymatic functions of CCDs (Takahashi et al., 2005; Kloer and Schulz, 2006).
Expression of ZmCCD10a was Upregulated by Low Phosphate
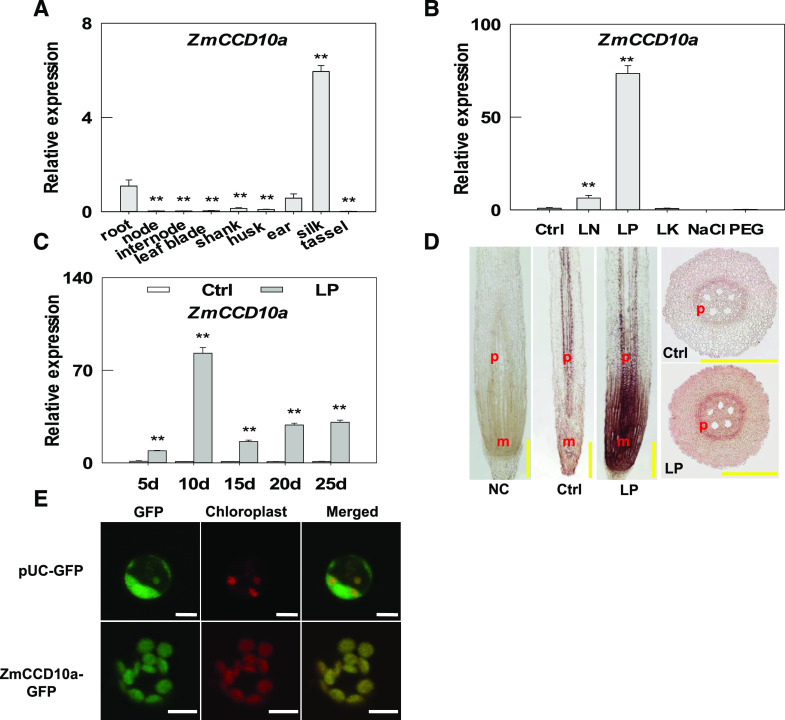
To investigate the fundamental function of ZmCCD10a, we characterized its expression pattern by reverse transcription quantitative PCR (RT-qPCR). At the silking stage, transcript levels of CCD10a were highest in the silk tissue, less abundant in the ear and root, very low in the husk, shank, and leaf blade, and hardly detectable in the tassel, node, and internode (Fig. 5A). Interestingly, CCD10a expression was dramatically stimulated by ∼73-fold in the root under low phosphate (LP) and ∼6-fold under low nitrogen, but remained at a similar level compared with the control under low potassium (Fig. 5B). Time course analysis revealed that CCD10a expression in the root was nearly 10-fold on day 5, increased to 82-fold on day 10, and decreased to ∼20- to ∼30-fold from day 15 to 25 under LP compared with the control (Fig. 5C). We also quantified CCD10a expression in the root in another maize inbred line Kansas Yellow Saline and four teosinte lines (Z. mays ssp. parviglumis, Z. mays ssp. huehuetenangensis, Zea luxurians, and Zea diploperennis) on day 10 under LP and found ∼55-, 11-, 23-, 16-, and 34-fold upregulation, respectively (Supplemental Fig. S3, A–E), indicating conserved upregulation of CCD10a expression in the root by LP despite intensive artificial selection and dramatic habitat changes over domestication.
Figure 5.
ZmCCD10a expression and localization. A, Relative expression of ZmCCD10a in different maize organs/tissues. The gene expression level in the root was normalized to one for comparison with that in the other organs/tissues. B, Relative abundance of ZmCCD10a transcripts under LN (low nitrogen), LP, LK (low potassium), 100 mm NaCl, and PEG (5% [w/v] PEG6000) 10 d after the treatment compared with the control (Ctrl). C, Stimulation of ZmCCD10a expression during the 25-d LP treatment. Error bars represent the se of four biological replicates. Asterisks indicate significant difference as determined by one-way ANOVA (**P < 0.01). D, ZmCCD10a expression in the primary root tip in maize by in situ hybridization. Negative control (NC) was hybridized with the sense probe. ZmCCD10a antisense probes were applied to the longitudinal and cross section of the maize root. P, Pericycle; M, meristem. Scale bars = 500 μm. E, Subcellular localization of GFP-tagged ZmCCD10a. GFP protein (plasmid University of California [pUC]-GFP) and GFP-tagged ZmCCD10a (ZmCCD10a-GFP) were transiently expressed in maize mesophyll protoplasts. GFP signals in green, auto-fluorescent signals of chloroplasts in red, and merged signals in yellow. Scale bars = 5 μm.
We next performed in situ hybridization to examine tissue expression patterns of ZmCCD10a within the root. As shown in Figure 5D, no signal was detected in the root tip of the negative control. ZmCCD10a signals were mostly present in the pericycle under the Pi sufficient condition. Under LP, strong ZmCCD10a signals were observed in cell division and elongation zones in addition to pericycle signals. When transiently expressed in maize leaf protoplasts, GFP-only signals diffused within the entire cell, whereas GFP-tagged CCD10a mirrored auto-fluorescent signals of chloroplasts (Fig. 5E), indicating that, like CCD7 and CCD8 (Auldridge et al., 2006a), CCD10a localized to plastids.
ZmCCD10a Cleaved Carotenoids in Escherichia coli
To identify potential substrates of ZmCCD10a, the coding sequence of ZmCCD10a was expressed into E. coli strains capable of synthesizing individual carotenoids with distinct colors, including phytoene, lycopene, δ-carotene, ε-carotene, β-carotene, and zeaxanthin (Cunningham and Gantt, 1998; Booker et al., 2004; Pan et al., 2016). Upon ZmCCD10a expression, color intensities of E. coli pellets were significantly reduced compared with the negative control without ZmCCD10a expression, suggesting efficient cleavage of corresponding carotenoids by ZmCCD10a (Fig. 6A). Carotenoids were extracted from E. coli cultures and quantitatively analyzed by high performance liquid chromatography (HPLC). When ZmCCD10a expression was induced, phytoene, lycopene, δ-carotene, ε-carotene, β-carotene, and zeaxanthin accumulation in the corresponding strain significantly decreased by 82.48%, 51.34%, 81.81%, 55.31%, 48.53%, and 59.66%, respectively (Fig. 6, B–G).
Figure 6.
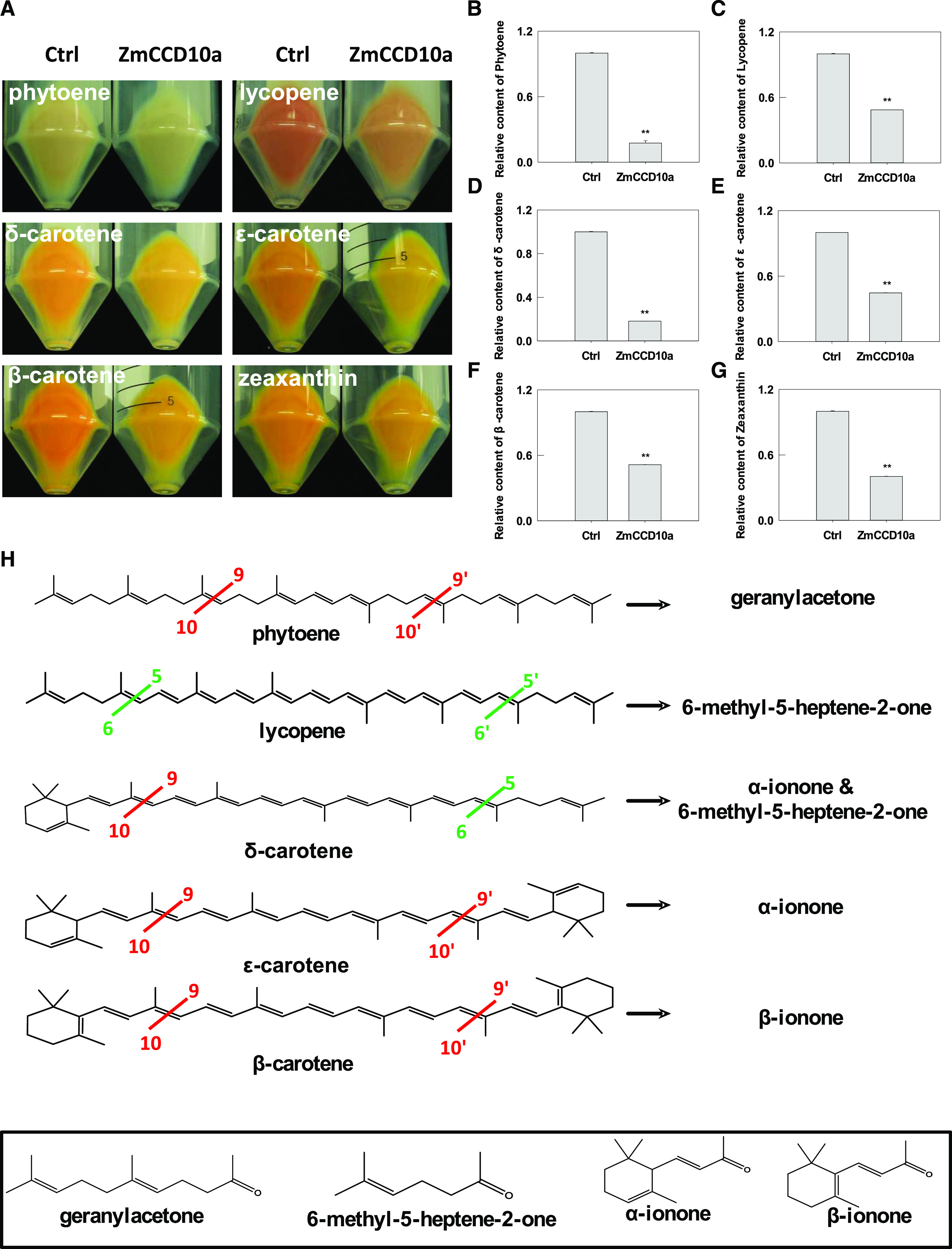
Carotenoid cleavage by ZmCCD10a in E. coli. A, Visualization of carotenoid cleavage without or with ZmCCD10a in E. coli. Six panels indicate accumulation of phytoene, lycopene, δ-carotene, ε-carotene, β-carotene, and zeaxanthin in E. coli without or with ZmCCD10a cleavage. Carotenoid accumulation in the culture was quantified by HPLC analysis: phytoene (B), lycopene (C), δ-carotene (D), ε-carotene (E), β-carotene (F), and zeaxanthin (G). The carotenoid content in the negative control (Ctrl) was normalized to 1 for comparison. Error bars represent the se of three biological replicates. Asterisks indicate significant difference as determined by one-way ANOVA (**P < 0.01). H, Cleavage sites of CCD10a and resulting volatiles. Green and red lines indicate the 5, 6 (5′, 6') and 9, 10 (9', 10') double bonds cleaved by ZmCCD10a. Resulting volatiles from individual carotenoid cleavage are listed to the right of black arrows, and their structures are illustrated in the bottom rectangle.
We performed headspace solid-phase microextraction coupled with gas chromatography (GC)-mass spectrometry analysis to qualitatively investigate products of ZmCCD10a-mediated carotenoid metabolism in E. coli and found geranylacetone in the E. coli culture expressing phytoene, 6-methyl-5-hepten-2-one (MHO) in the culture expressing lycopene, MHO and α-ionone in the culture expressing δ-carotene, α-ionone in the culture expressing ε-carotene, and β-ionone in the culture expressing β-carotene (Fig. 6H; Supplemental Figs. S4, A–F, and S5, A–F). No volatile was identified in the culture expressing zeaxanthin, probably due to low concentrations of resulting products. Thus, ZmCCD10a was able to cleave the above carotenoids at 5, 6 (5′, 6') and 9, 10 (9', 10') positions.
Heterologous Expression of CCD10a in Arabidopsis Conferred LP Tolerance
ZmCCD10a was constitutively expressed in Arabidopsis driven by the Ubiqutin promoter (Desfeux et al., 2000), and three independent transgenic lines were selected for further analysis (Fig. 7, A and B). With sufficient Pi, the total carotenoid levels in siliques of the transgenic plants significantly decreased compared with those of wild-type plants, including neoxanthin, violaxanthin, antheraxanthin, lutein, zeaxanthin, and β-carotene (Table 1); no such significant change occurred in leaves and roots (Table 1).
Figure 7.
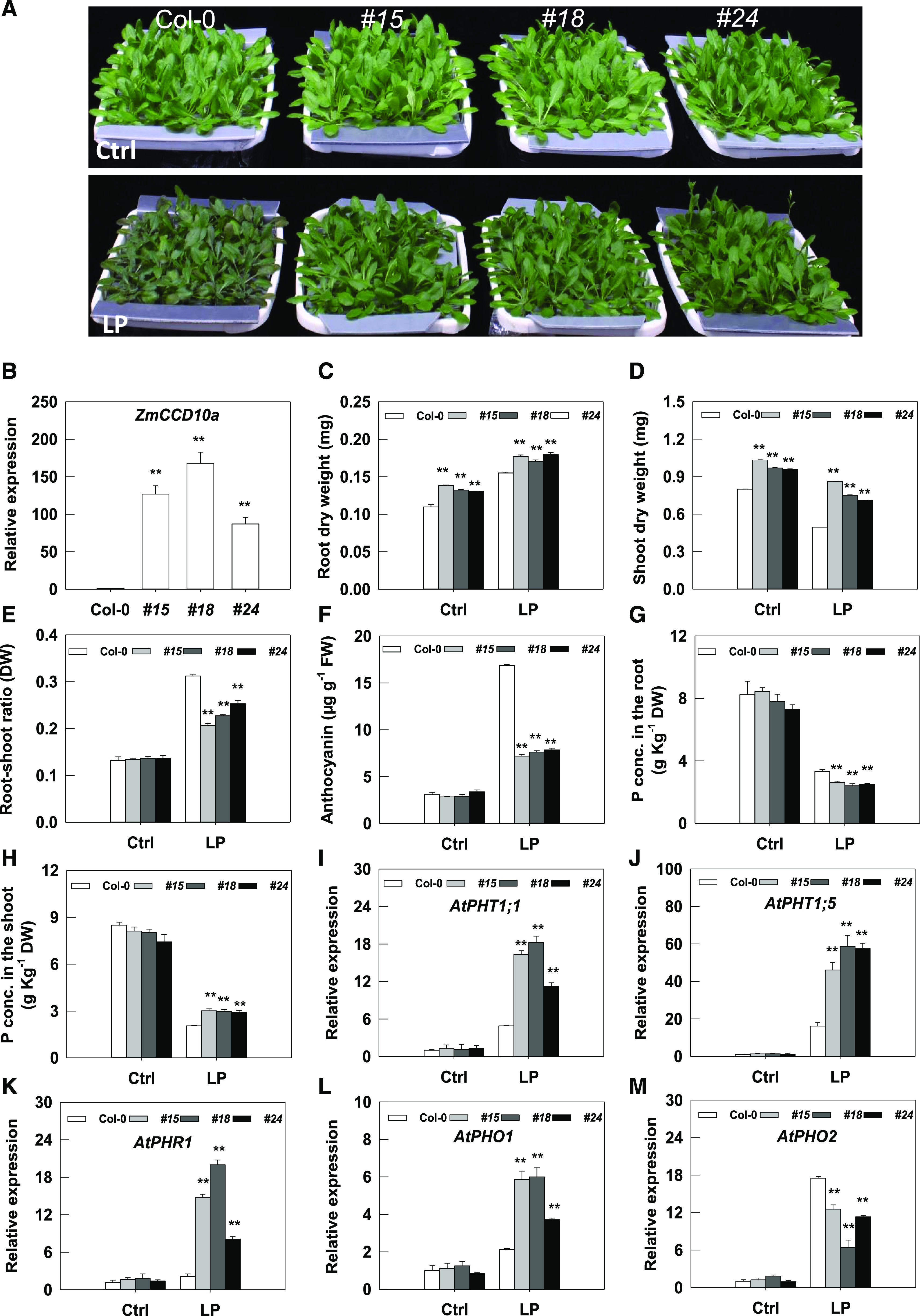
Tolerance of ZmCCD10a-overexpressing Arabidopsis plants to LP. A, ZmCCD10a-overexpressing (nos. 15, 18, 24) and wild-type (Col-0) plants under sufficient (Ctrl) and LP. B, Relative expression of ZmCCD10a in three overexpression lines. C to E, Root (C) and shoot (D) biomass, and the root-shoot ratio (E) of ZmCCD10a-overexpressing lines under control (Ctrl) and LP conditions. F, Anthocyanin accumulation in ZmCCD10a-overexpressing leaves. P concentrations (conc.) in roots (G) and shoots (H) of ZmCCD10a-overexpressing lines compared with wild-type plants. Relative expression of P starvation-induced genes AtPHT1;1 (I), AtPHT1;5 (J), AtPHR1 (K), AtPHO1 (L), and AtPHO2 (M) in ZmCCD10a-overexpressing lines in comparison to wild-type plants. Error bars represent the se of four biological replicates. Asterisks indicate significant difference as determined by one-way ANOVA (**P < 0.01). DW, Dry weight.
Table 1. CCD10a-mediated differential accumulation of carotenoids in plant tissues.
Arabidopsis, including wild-type (Col-0) and three transgenic lines (nos. 15, 18, and 24), with heterologous expression of ZmCCD10a, including control (vector control [V/C]) and ZmCCD10a silencing line ccd10a. Ctrl, Control. Asterisks indicate significant difference as determined by one-way ANOVA (*P < 0.05 and **P < 0.01).
| Treatment | Genotype | Neoxanthin | Violaxanthin | Antheraxanthin | Lutein | Zeaxanthin | β-Carotene | Total Carotenoids |
|---|---|---|---|---|---|---|---|---|
| μg g−1 | ||||||||
| Siliques_At | Col-0 | 63.53 ± 6.59 | 9.17 ± 0.72 | 25.69 ± 1.19 | 282.64 ± 10.05 | 102.37 ± 2.76 | 27.15 ± 2.66 | 510.55 ± 19.47 |
| 15 | 13.32 ± 2.14** | 2.53 ± 0.49** | 11.20 ± 1.83** | 190.94 ± 6.42** | 77.72 ± 2.20** | 12.86 ± 1.74** | 308.57 ± 13.01** | |
| Ctrl | 18 | 32.88 ± 1.48** | 2.36 ± 0.36** | 9.10 ± 2.10** | 169.16 ± 3.30** | 73.84 ± 8.73** | 17.43 ± 2.36** | 302.77 ± 12.31** |
| 24 | 39.96.0 ± 1.81** | 4.60 ± 0.70** | 15.75 ± 1.99** | 210.88 ± 11.44** | 82.14 ± 3.55** | 15.05 ± 0.76** | 368.39 ± 14.69** | |
| Col-0 | 85.85 ± 7.11 | 11.26 ± 1.34 | 31.31 ± 2.50 | 307.32 ± 9.20 | 129.42 ± 5.27 | 27.22 ± 3.13 | 592.38 ± 18.62 | |
| 15 | 84.30 ± 10.39 | 10.57 ± 0.86 | 31.22 ± 3.02 | 300.79 ± 4.09 | 117.54 ± 3.81 | 26.82 ± 3.83 | 571.25 ± 6.50 | |
| LP | 18 | 81.56 ± 11.45 | 8.13 ± 1.59 | 28.38 ± 4.74 | 305.32 ± 10.33 | 120.84 ± 7.74 | 22.67 ± 3.50 | 566.91 ± 21.89 |
| 24 | 79.54 ± 17.04 | 9.84 ± 1.40 | 29.59 ± 4.57 | 305.70 ± 12.85 | 124.31 ± 5.94 | 24.15 ± 3.68 | 573.13 ± 25.36 | |
| Leave_At | Col-0 | 33.26 ± 1.57 | 10.66 ± 0.47 | 5.89 ± 0.41 | 487.64 ± 15.18 | 20.81 ± 0.78 | 44.70 ± 1.94 | 602.96 ± 19.01 |
| 15 | 33.01 ± 1.33 | 9.10 ± 0.31 | 6.03 ± 0.66 | 479.76 ± 7.46 | 23.41 ± 1.65 | 47.88 ± 2.27 | 599.19 ± 12.87 | |
| Ctrl | 18 | 35.07 ± 1.68 | 9.30 ± 0.17 | 5.95 ± 0.41 | 473.25 ± 5.65 | 24.03 ± 2.82 | 45.83 ± 2.63 | 593.42 ± 9.27 |
| 24 | 34.10 ± 3.37 | 10.31 ± 0.67 | 6.27 ± 0.73 | 487.38 ± 13.70 | 21.16 ± 3.40 | 48.29 ± 2.79 | 607.52 ± 19.75 | |
| Col-0 | 52.58 ± 0.57 | 15.92 ± 0.11 | 10.33 ± 0.30 | 608.81 ± 17.60 | 27.09 ± 3.09 | 82.47 ± 4.81 | 797.20 ± 13.04 | |
| 15 | 52.58 ± 0.30 | 15.59 ± 0.31 | 9.80 ± 0.35 | 766.01 ± 5.85** | 25.73 ± 2.35 | 79.55 ± 3.29 | 949.26 ± 11.55** | |
| LP | 18 | 51.02 ± 2.18 | 13.51 ± 0.90 | 10.79 ± 0.28 | 771.96 ± 7.33** | 25.96 ± 5.35 | 81.26 ± 8.28 | 953.51 ± 19.57** |
| 24 | 50.76 ± 1.10 | 15.90 ± 0.61 | 10.18 ± 0.20 | 767.67 ± 11.51** | 26.81 ± 2.78 | 75.90 ± 4.11 | 947.22 ± 18.49** | |
| Root_At | Col-0 | 0.02 ± 0.01 | — | — | 1.13 ± 0.16 | 0.08 ± 0.01 | 0.11 ± 0.02 | 1.34 ± 0.20 |
| 15 | 0.03 ± 0.00 | — | — | 1.21 ± 0.06 | 0.10 ± 0.01 | 0.13 ± 0.01 | 1.48 ± 0.05 | |
| Ctrl | 18 | 0.02 ± 0.00 | — | — | 1.03 ± 0.06 | 0.09 ± 0.00 | 0.11 ± 0.00 | 1.25 ± 0.05 |
| 24 | 0.02 ± 0.00 | — | — | 1.17 ± 0.02 | 0.10 ± 0.01 | 0.13 ± 0.01 | 1.42 ± 0.03 | |
| Col-0 | 0.04 ± 0.01 | — | — | 0.98 ± 0.00 | 0.10 ± 0.01 | 0.13 ± 0.02 | 1.24 ± 0.01 | |
| 15 | 0.02 ± 0.00 | — | — | 0.73 ± 0.01** | 0.08 ± 0.00 | 0.10 ± 0.01 | 0.93 ± 0.01** | |
| LP | 18 | 0.02 ± 0.00 | — | — | 0.70 ± 0.02** | 0.08 ± 0.00 | 0.10 ± 0.00 | 0.89 ± 0.01** |
| 24 | 0.02 ± 0.00 | — | — | 0.77 ± 0.02** | 0.08 ± 0.00 | 0.12 ± 0.01 | 0.99 ± 0.03** | |
| Leave_Zm | ||||||||
| Ctrl | V/C | 79.55 ± 1.99 | 64.20 ± 4.11 | 42.20 ± 5.02 | 582.10 ± 43.28 | 14.61 ± 0.78 | 28.85 ± 3.25 | 811.51 ± 47.51 |
| ccd10a | 67.76 ± 1.17* | 54.46 ± 0.73* | 35.56 ± 3.00 | 447.71 ± 13.75* | 11.71 ± 0.47* | 22.71 ± 0.56 | 639.92 ± 14.96* | |
| LP | V/C | 77.22 ± 4.30 | 67.55 ± 4.46 | 44.93 ± 5.23 | 597.19 ± 7.28 | 14.41 ± 0.76 | 30.23 ± 1.00 | 831.55 ± 21.11 |
| ccd10a | 60.85 ± 2.45* | 55.39 ± 2.92* | 37.24 ± 3.35 | 447.36 ± 17.96* | 10.98 ± 0.48* | 23.59 ± 1.53* | 635.42 ± 19.04* | |
| Root_Zm | ||||||||
| Ctrl | V/C | 0.01 ± 0.00 | 0.00 ± 0.00 | 0.03 ± 0.00 | 0.87 ± 0.19 | 0.1 ± 0.04 | 0.06 ± 0.01 | 1.07 ± 0.22 |
| ccd10a | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.04 ± 0.01 | 1.22 ± 0.55 | 0.06 ± 0.01 | 0.10 ± 0.04 | 1.43 ± 0.60 | |
| LP | V/C | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.19 ± 0.04 | 0.04 ± 0.01 | 0.03 ± 0.00 | 0.27 ± 0.05 |
| ccd10a | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.03 ± 0.00** | 0.57 ± 0.13* | 0.07 ± 0.01** | 0.05 ± 0.01** | 0.73 ± 0.13** | |
CCD10a was dramatically upregulated under Pi limitation in maize (Fig. 5B); here, we took advantage of CCD10a transgenic lines to analyze biological consequences of CCD10a upregulation in planta. Both CCD10a transgenic plants and wild-type plants showed robust growth under nutrient-sufficient conditions (Fig. 7A); however, CCD10a transgenic plants had significantly greater shoot and root dry weights and similar root/shoot ratios compared with the wild-type plants (Fig. 7, C–E). Transgenic plants remained normal even 10 d after Pi limitation (Fig. 7A); by sharp contrast, a typical Pi-deficient phenotype of reddish-purple discoloration occurred in leaves of wild-type plants (Jiang et al., 2007). The anthocyanin concentration in leaves of wild-type plants (16.9 μg g−1 fresh weight) was approximately two times that of the transgenic plants under LP (Fig. 7F). Importantly, when compared with substantial decreases in biomass accumulation in the shoot of wild-type plants under Pi limitation, transgenic plants had relatively lower reductions (Fig. 7D). As a result, the root/shoot ratio of transgenic plants was significantly lower than that of wild-type plants under Pi limitation (Fig. 7E). Therefore, ZmCCD10a transgenic plants were less stressed by LP.
We speculated that ZmCCD10a expression affected the uptake, accumulation, or allocation of Pi in transgenic plants. Under Pi-sufficient conditions, no significant difference in the P concentration was observed between wild-type and transgenic plants (Fig. 7, G and H). However, under Pi limitation, the P concentration in the root of transgenic plants was 24.48% lower than that of wild-type plants (Fig. 7G), and the P concentration in the shoot of the transgenic plants was 30.82% higher than that of wild-type plants (Fig. 7H). We further analyzed related gene expression in the root. CCD10a heterologous expression significantly stimulated expression of AtPHT1;1 and AtPHT1;5 under Pi limitation (Fig. 7, I and J). Pi limitation also caused up-regulation of AtPHR1 and AtPHO1 and down-regulation of AtPHO2 in the transgenic plants (Fig. 7, K–M). Thus, ZmCCD10a transgenic plants were more LP resistant by allocating more P into the shoot. Signaling- and stress-related genes were also involved in such regulation as indicated by their significant enrichment and overall up-regulation in the transcriptome profile (Supplemental Fig. S6, A–C).
Different from downregulation of PHO2 expression in Arabidopsis under LP (Lin et al., 2008), we found its upregulation at a late time point (Fig. 7M; Supplemental Fig. S7A), implying a potentially dynamic functional mode of PHO2 in the Arabidopsis response to Pi limitation. As positive regulators of PHR1 (Huang et al., 2018), transcript levels of ARF19 in the root of CCD10a overexpression lines under LP were significantly higher than those in the wild-type root, whereas ARF7 expression in the root did not respond to LP (Supplemental Fig. S7, B and C).
Pi limitation also affects carotenoid accumulation. The total carotenoid concentration in leaves of transgenic plants was significantly higher than that of wild-type plants under Pi limitation, with a particular increase in lutein accumulation and others remaining unchanged (Table 1). However, when compared with wild type, concentrations of lutein and total carotenoids in the transgenic root decreased under LP (Table 1). We detected no significant difference in carotenoid accumulation in siliques between wild-type and transgenic plants under Pi limitation (Table 1).
ZmCCD10a silencing made maize seedlings more sensitive to LP
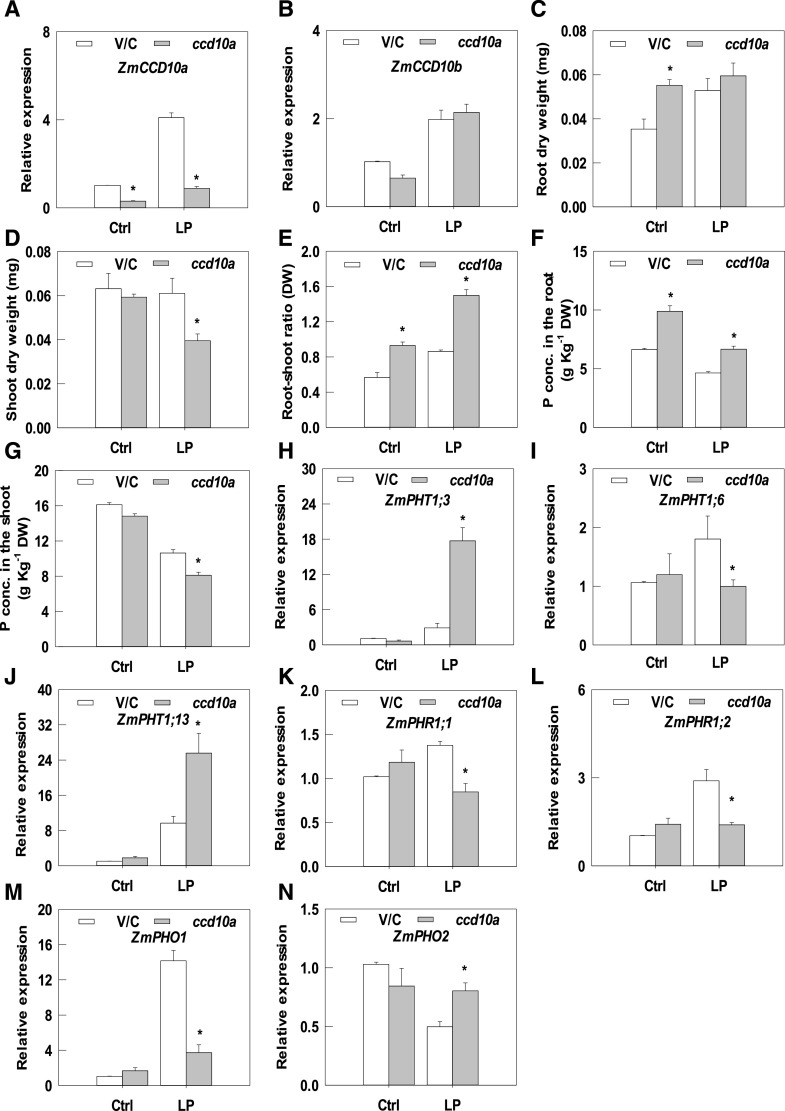
To investigate in vivo functions of ZmCCD10a, ZmCCD10a knockdown lines were generated by virus-mediated gene silencing (Benavente et al., 2012; Wang et al., 2016). Most positive plants showed 15% to 85% reductions in CCD10a expression compared with the vector control under the Pi-sufficient condition, whereas ZmCCD10b expression remained the same (Fig. 8, A and B). Seedlings with 68% to 78% reductions in CCD10a expression were then used for phenotypic characterization. When compared with the control seedlings, ccd10a seedlings had similar biomass accumulation in the shoot under Pi-sufficient conditions; however, the shoot dry weight of ccd10a knockdown lines significantly decreased by 35.25% under LP (Fig. 8D). Notably, the P concentration decreased by 23.79% in the shoot by contrast with an increase of 43.54% in the root of ccd10a lines under Pi limitation (Fig. 8, F and G), suggesting that more P retention in the root negatively affected shoot growth. ccd10a also caused down-regulation of expression of ZmPHT1;6 and up-regulation of ZmPHT1;3 and ZmPHT1;13 under Pi limitation (Fig. 8, H–J). Expression of ZmPHR1;1, ZmPHR1;2, ZmPHO1, and ZmARF27 was down-regulated and that of ZmPHO2 was up-regulated in ccd10a lines under Pi limitation (Fig. 8, K–N; Supplemental Fig. S7D).
Figure 8.
ZmCCD10a silencing in maize. Expression levels of ZmCCD10a (A) and ZmCCD10b (B) in control (vector control [V/C]) and ZmCCD10a silencing (ccd10a) plants under sufficient (Ctrl) and LP conditions are shown. Root (C) and shoot (D) biomass and the root-shoot ratio (E) of control and ccd10a plants are shown. P concentrations (conc.) in roots (F) and shoots (G) are also shown. Expression of phosphate starvation-induced genes ZmPHT1;3 (H), ZmPHT1;6 (I), ZmPHT1;13 (J), ZmPHR1;1 (K), ZmPHR1;2 (L), ZmPHO1 (M), and ZmPHO2 (N) in control and ccd10a plants. Error bars represent the se of four biological replicates. Asterisks indicate significant difference as determined by one-way ANOVA (*P < 0.05). DW, Dry weight.
Interestingly, accumulation of neoxanthin, violaxanthin, lutein, zeaxanthin, and total carotenoids significantly decreased in leaves of ccd10a seedlings with sufficient Pi compared with that in control leaves. In addition to reduction in accumulation of the above carotenoids, β-carotene accumulation also significantly decreased in the leaf of ccd10a transgenic lines under LP (Table 1). However, the ccd10a root accumulated significantly more antheraxanthin, lutein, zeaxanthin, β-carotene, and total carotenoids than the control root under Pi limitation (Table 1). We also tried to qualify aromatic volatiles derived from CCD10a functioning in the maize root with the authentic standard and bacterial culture as positive controls. When compared with the obvious peak of the standard, we observed the fairly low peak from the bacteria culture and were unable to detect meaningful peaks from root samples, likely due to extremely low levels of aromatic metabolites in the root (Supplemental Fig. S8).
One incompatible result derived from CCD10a overexpression and knockdown was that both transgenic lines promoted biomass accumulation in the root (Figs. 7C and 8C). Monocots and dicots have quite different developmental patterns (Atkinson et al., 2014). A given gene may have primarily conserved functions across monocots and dicots (Kim et al., 2006); however, it may confer divergent phenotypes on some other perspectives (Guan et al., 2012; Huang et al., 2017). The root dry weight is programmed by a wide array of endogenous developmental and exogenous environmental cues (Zhang and Forde, 1998; Remans et al., 2006; Di Mambro et al., 2017). Inconsistent effects of CCD10a expression on the root dry weight in Arabidopsis and maize may be attributed to accumulative evolutionary divergence of these two species or other yet unknown mechanisms to be further investigated in the future.
In addition, we found no visible difference in the root system between the control and ccd10a lines. CCD10a knockdown did not affect the total root length (V/C, 286.06 ± 16.30 cm; ccd10a, 297.22 ± 12.93 cm, n = 4, P < 0.05), although Pi limitation led to significantly longer root systems (V/C, 453.92 ± 38.51 cm; ccd10a, 444.62 ± 26.86 cm, n = 4, P < 0.05).
ZmPHR1;1 and ZmPHR1;2 Bound to the P1BS Element of ZmCCD10a
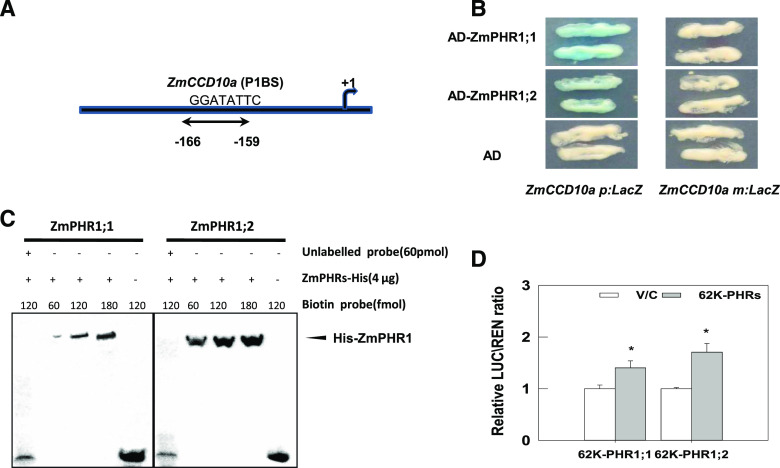
We next analyzed ZmCCD10a’s cis-elements to identify its upstream regulators. ZmCCD10a harbors a P1BS (−166GGATATTC−159) element in its promoter region (Fig. 9A). As shown in Figure 9B, both PHR1;1 and PHR1;2 bind to the P1BS element of ZmCCD10a by yeast one hybrid assay. Such binding activities were verified by interaction of ZmPHR1;1 and ZmPHR1;2 with the biotin-labeled P1BS oligonucleotide of ZmCCD10a and obviously reduced mobility of ZmPHR1;1 and ZmPHR1;2 in the electrophoresis mobility shift assay (EMSA) analysis (Fig. 9C). We further tested whether ZmPHR regulated ZmCCD10a in planta. In Nicotiana benthamiana leaves, relative luciferase (LUC)/Renilla luciferase (REN) expression, driven by the ZmCCD10a promoter, significantly increased when the ZmCCD10a promoter was coexpressed with ZmPHR1;1 or ZmPHR1;2, indicating up-regulation of ZmCCD10 expression mediated by both ZmPHRs (Fig. 9D).
Figure 9.
Regulation of ZmCCD10a by ZmPHR1s. A, The P1BS element in the ZmCCD10a promoter. B, ZmPHR1;1 and ZmPHR1;2 binding to ZmCCD10a p:LacZ, rather than ZmCCD10a m:LacZ with a mutated P1BS element in the yeast one-hybrid assay. C, Binding of PHR1;1 and PHR1;2 to the biotin-labeled P1BS-containing oligonucleotide derived from the ZmCCD10a promoter in the EMSA assay. D, Transcriptional enhancement of ZmCCD10a by transactivation of PHR1;1 (62K-PHR1;1) and PHR1;2 (62K-PHR1;2) compared to the vector control (V/C), as indicated by the relative LUC/REN ratio. Error bars represent the se of six biological replicates. Asterisks indicate significant difference as determined by one-way ANOVA (*P < 0.05).
DISCUSSION
CCD10 Represents a Previously Unanchored Basal Clade of CCDs Within the Unified Four-Clade CCD Family Tree
CCDs play a wide array of fundamental roles in plant development, secondary metabolism, stress biology, and interspecies interaction (Schwartz et al., 1997; Akiyama et al., 2005; Auldridge et al., 2006b; Guan et al., 2012; Liu et al., 2015). How CCDs originated and evolved over species diversification and sophistication has been under intensive investigation. Four CCD and five NCED clades are most frequently placed on the phylogenetic trees as parallel clades (Cui et al., 2012; Helgeland et al., 2014; Wei et al., 2016; Wang et al., 2017), and inclusion of CCD-like and ZAS clades (Wei et al., 2016; Wang et al., 2019) makes a more complicated and yet unresolved CCD tree. However, the primary evolutionary path of CCDs remains unknown. More and more functional characterization of CCDs also calls for a better understanding of CCD evolution history. We found that all CCDs and NCEDs formed a unified phylogenetic tree with CCD1, CCD7, CCD8, and CCD10 as four basal clades (Fig. 1). First, ZmCCD10 represented an unexpectedly basal clade within the large CCD family spreading across bacterial, archaeal, fungal, plant, and animal kingdoms (Fig. 1). The CCD10 clade accommodated CCD-like and ZAS genes (Wei et al., 2016; Wang et al., 2019). Second, CCD4 and NCEDs were not independent basal clades, but subclades of the large CCD1 clade (Fig. 1). CCD2 belonged to the CCD1 clade (Frusciante et al., 2014). Thus, our work resolved the evolutionary puzzle of the large CCD gene family, providing a fundamental frame for in-depth analysis of evolution patterns of each subclade and informative guidance for further functional investigation of CCDs.
As a plant-specific clade, CCD10s (9 to 15 exons) mostly have more exons than CCD7s (7 and 8 exons) and CCD8s (4 to 10 exons) and display less variation in exon number than CCD1s (1 to 14 exons) in land plants (Fig. 1; Supplemental Fig. S1), indicating unique gene structure of CCD10s. Duplication is an essential driving force of gene and species evolution by providing genetic materials for selection (Lynch and Conery, 2000; Bowers et al., 2003); selection and adaptation may cause gene losses on the other hand (Lai et al., 2004; Koskiniemi et al., 2012; Albalat and Cañestro, 2016). Copy number of CCD10 neither followed a general ascending path from algae to land plants, nor was proportional to genome size across species (Fig. 2; Supplemental Table S1). CCD10 expands from one copy in lycophyte Selaginella moellendorffii to three copies in basal angiosperm Amborella trichopoda and shrinks back to one copy in monocots M. acuminate or E. guineensis (Fig. 2; Supplemental Table S1). Two rounds of CCD10 duplication predates Poales origination (Fig. 2), continues with asymmetric evolution in the Poaceae family, or further expands in B. distachyon (6 CCD10s) in contrast to two copies in maize (Fig. 3). Together, CCD10 is a previously unanchored basal clade of the CCD family illustrating highly dynamic evolutionary features.
ZmCCD10a Is a Distinct Broad-Spectrum CCD
Maize, a genetically and evolutionarily important crop (Doebley et al., 2006; Schnable et al., 2009), contains two CCD10 genes, with ZmCCD10a as the orthologous gene across examined cereal crops (Fig. 2). ZmCCD10a shows higher levels of expression in the silk, ear, and root at the reproductive stage, with concentrated expression in pericycle in the root (Fig. 5, A and D). GFP-tagged ZmCCD10a is localized to plastids—the organelle containing carotenoids (Fig. 5E). When coexpressed in E. coli, ZmCCD10a significantly cleaves differentially structured carotenoid substrates including phytoene, lycopene, δ-carotene, ε-carotene, and β-carotene at the 5, 6 (5′, 6') and 9, 10 (9', 10') positions and generates C8 (MHO) and C13 apocarotenoids (geranylacetone, α-ionone, and β-ionone; Fig. 6, A–H; Supplemental Fig. S5, A–F). When compared with that, ZAS cleaves the 13, 14 bond of apo-10'-zeaxanthinal (C27 carotenoids) to generate zaxinone (Wang et al., 2019); ZmCCD10a is a functionally new CCD enzyme in terms of cleavage sites of C40.
Importantly, ZmCCD10a knockdown leads to overaccumulation of lutein, antheraxanthin, zeaxanthin, β-carotene, and total carotenoids in the root under LP in comparison with wild-type seedlings; consistently, heterologous expression of ZmCCD10a in Arabidopsis significantly reduces accumulation of lutein and total carotenoids in the LP root (Table 1), suggesting that ZmCCD10a is a functional CCD in planta. ZmCCD10a expression in Arabidopsis also reduces accumulation of neoxanthin, violaxanthin, antheraxathin, lutein, zeaxanthin, β-carotene, and total carotenoids in siliques (Table 1). Similarly, knockout of related CCDs (CCD1, CCD4, or NCED2) significantly promote accumulation of total and other individual carotenoid in siliques in Arabidopsis (Gonzalez-Jorge et al., 2013). In addition, leaves, when compared with nongreen tissues (fruits and roots), have very different metabolic and accumulation patterns of carotenoids (Shewmaker et al., 1999; Ye et al., 2000; Lätari et al., 2015). Under LP, accumulation of lutein and total carotenoids increases in leaves of Arabidopsis with ZmCCD10a expression (Table 1); ZmCCD10a knockdown reduces accumulation of neoxanthin, violaxanthin, lutein, zeaxanthin, β-carotene, and total carotenoids in maize leaves. Such positive association of carotenoid accumulation in leaves with ZmCCD10a expression in two contrasting biological contexts needs to be further investigated (Table 1).
Together, our in vivo and in vitro results suggested that ZmCCD10 functions in a highly divergent manner in terms of substrate preference and cleavage positions compared with its counterpart ZAS in rice (Wang et al., 2019). Our conclusion was supported by another line of evidence that OsZAS lost a conserved 50-amino acid domain at its C terminus (Fig. 4B; Supplemental Fig. S2; Takahashi et al., 2005; Kloer and Schulz, 2006); such domain loss may result in substantial functional divergence from ZmCCD10 and other CCDs (Wang et al., 2019).
ZmCCD10a Is a Positive Regulator of Plant Tolerance to LP
Dramatic and persistent upregulation of CCD10a expression in the maize and teosinte root in response to LP strongly suggests association of CCD10 with Pi nutrition (Fig. 5B; Supplemental Fig. S3, A–E). Heterologous expression of ZmCCD10a in Arabidopsis promotes P accumulation in the shoot, resulting in higher shoot dry weight under Pi limitation (Fig. 7, D and H). PHT1;1 and PHT1;5 mediate Pi acquisition and translocation from source to sink organs, respectively (Shin et al., 2004; Nagarajan et al., 2011). Enhanced expression of PHR1, PHO1, PHT1;1, and AtPHT1;5 and reduced expression of PHO2 in roots of ZmCCD10a-expressing plants favors Pi uptake and translocation toward the LP shoot (Fig. 7, H–M). In opposition to preferential P retention in the shoot of LP-stressed Arabidopsis transgenic lines, ZmCCD10a silencing causes more P retention in the root and less P allocation to the shoot under LP, which hampers shoot growth and biomass accumulation (Figs. 7H and 8, D–G). ZmPHT1;6 knockout dramatically reduces aboveground biomass in the field (Willmann et al., 2013). Lower PHT1;6 expression is in agreement with decreases in shoot biomass of ZmCCD10a silencing plants (Fig. 8, D and I). Elevation of ZmPHT1;3 and ZmPHT1;13 expression in the ZmCCD10a silencing lines may be a feedback outcome of more severe P shortage in the shoot, which probably facilitates Pi absorption or translocation in the root (Fig. 8, F, H, and J; Liu et al., 2016). Together with up-regulation of PHO2 expression and downregulation of ZmPHR1;1, ZmPHR1;2, and PHO1 expression tend to reduce P allocation to the shoot under LP (Fig. 8, G and K–N). Therefore, CCD10a modulates relative Pi allocation between the root and shoot depending on its transcript abundance; enhanced transcription, probably under regulation of ZmPHR1;1 and ZmPHR1;2 (Fig. 9D), favors Pi flow to the shoot to support aboveground functioning under LP.
As a putative loop linker, ZmCCD10a overexpression and silencing leads to contrasting expression of PHR1, PHO2, PHO1, and related Pi transporters. How does ZmCCD10a transact on the PHR1 pathway? In Arabidopsis, ARF19 positively regulates PHR1 expression specifically in the root (Huang et al., 2018). Here, transcript levels of ARF19 in Arabidopsis roots with heterologous CCD10a expression significantly increase under LP compared with those in wild type, whereas transcript abundance of ZmARF27 (ARF19’s ortholog) in the CCD10a knockdown root is significantly reduced upon Pi limitation (Supplemental Fig. S7, C–E). Thus, one possibility is that CCD10a may modulate PHR1 expression in an ARF19-dependent manner. Alternatively, ZmCCD10a triggers the LP signaling cascade via potential protein partners or yet to be validated metabolites, which is supported by surprising enrichment and upregulation of signaling related genes in Arabidopsis roots with heterologous ZmCCD10a expression (Supplemental Fig. S6, A–C). Together, ZmCCD10a functions as a positive regulator of plant tolerance to Pi limitation, and the presence of CCD10a in cereal ancestors probably facilitates their adaptation to Pi limited habitats. Finally, given that only mycorrhizal plants contain CCD10 (Wang et al., 2019) and that C13 apocarotenoids are strongly associated with colonization and functional maintenance of mycorrhizas (Walter et al., 2000; Floss et al., 2008a; Floss et al., 2008b), elevated expression of CCD10a under LP accelerates carotenoid cleavage to boost C13 apocarotenoid provision, potentially favoring Pi acquisition via the mycorrhiza fungal pathway.
MATERIALS AND METHODS
Sequence Blast, Alignment, and Phylogenetic Construction
Bacterial, archaeal, fungal, animal, and plant CCD protein sequences were identified and retrieved from phytozome (https://phytozome.jgi.doe.gov), National Center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and Cyanobase (http://genome.microbedb.jp/cyanobase/) using nine Arabidopsis (Arabidopsis thaliana) AtCCDs (CCD1, NCED2, NCED3, CCD4, NCED5, NCED6, CCD7, CCD8, and NCED9) as the primary query (Supplemental Datasets S1 and S2). These protein sequences were aligned, manually adjusted, and scored sequentially using MUSCLE 3.6 (http://www.drive5.com/muscle/), GeneDoc 3.2 (https://www.softpedia.com/get/Science-CAD/GeneDoc.shtml), and CLUSTALX (http://www.clustal.org/clustal2/). Sites with quality scores higher than five were selected to construct the CCD family tree. The coding sequence matrix was generated on the basis of the protein matrix by PAL2NAL (http://www.bork.embl.de/pal2nal/), and sites with quality scores higher than 12 were chosen for construction of the CCD10 tree. Phylogenetic trees were generated by the MEGA 6.0 (https://www.megasoftware.net/history.php) following the neighbor joining method, and the bootstrap was analyzed with 1000 replications.
Plant Materials and Growth Conditions
Hydroponic culture of maize (Zea mays; inbred lines B73 and Kansas Yellow Saline) and teosinte seedlings (Z. mays ssp. parviglumis; Z. mays ssp. huehuetenangensis; Zea luxurians; Zea diploperennis) followed our well-established protocol (Pan et al., 2016). The full nutrient solution comprised of 2 mm NH4NO3, 2 mm CaCl2, 0.75 mm K2SO4, 0.1 mm KCl, 0.65 mm MgSO4, 0.25 mm KH2PO4, 0.2 mm Fe-EDTA, 1 × 10−3 mm ZnSO4, 1 × 10−3 mm MnSO4, 1 × 10−4 mm CuSO4, 5 × 10−6 mm (NH4)6Mo7O24, and 1 × 10−3 mm H3BO3. For the low nitrogen treatment, the NH4NO3 concentration was reduced from 2 mm to 40 μm; 2.5 μm KH2PO4 and 0.1 mm KCl were adopted for the LP and low potassium treatment, respectively, whereas other nutrient concentrations remained unchanged or balanced. To simulate salt or drought stress, 100 mm NaCl or 5% (w/v) polyethylene glycol (PEG) 6000 was applied to the full nutrient solution. Every treatment had three technical and three biological replicates, and the nutrient solution (pH = 6.0) was changed every 2 d. Plants were grown in a controlled growth chamber having continuous aeration, 14/10-h light/dark photoperiod, 28°C/22°C day/night temperature, and 60% relative humidity. Seedlings were harvested on 5, 10, 15, 20, and 25 d after treatments. Field experiments with four biological replicates were conducted at the Shangzhuang Experimental Station, Beijing (Liao et al., 2012; Pan et al., 2015; Pan et al., 2016). Maize samples at silking were harvested as the root (five randomly selected crown roots), node (next to the ear), internode (immediately next to the ear), leaf blade of the ear leaf, shank, ear, husk, silk, and tassel. All samples were immediately frozen in liquid nitrogen and kept in the −80°C freezer for further analysis.
Arabidopsis (Columbia-0; wild-type and transgenic lines) was grown in hydroponic solution (pH = 5.8) containing 0.5 mm Ca(NO3)2, 2 mm K2SO4, 2 mm KH2PO4, 1 mm MgSO4, 100 μm Fe-EDTA, 1 μm H3BO3, 0.2 μm ZnSO4, 0.2 μm MnSO4, 0.05 μm CuSO4, and 0.05 μm Na2MoO4 and replaced daily. Five-week-old plants were treated with the LP nutrient solution containing 20 μm KH2PO4 (the K+ was balanced by KCl, and other nutrient concentrations remained unchanged), and both control and LP plants were grown under long-day conditions (16-h light/8-h dark, light 100–120 μmol m−2 s−1) at 22°C. Root and shoot tissues were harvested after 10 d of treatment. Leaves and siliques were harvested after 25 d of treatment for carotenoid quantification.
RT-qPCR, Gene Cloning, In Situ Hybridization, and Subcellular Localization
RNA isolation, RT, real time qPCR with gene specific primers (Supplemental Table S2), and data analysis were carried out following standard protocols (Livak and Schmittgen, 2001; Pan et al., 2016). ZmUbi (for maize) and AtTub4 (for Arabidopsis) were used as the internal control. Each treatment for all RT-qPCR in this study had three biological replicates, and each biological replicate had three technical replicates.
The full CCD10a coding sequence was amplified with addition of restriction enzyme/cleavage sites (SalI and SpeI), cloned into the pMD19-T vector (Takara), and sequenced as previously described (Pan et al., 2016). The PCR fragments were listed in the Supplemental Table S2.
The root tips of maize seedlings were fixed and hybridized following previous protocols (Zhang et al., 2013). Probes were generated through PCR amplification using gene-specific primers (Supplemental Table S2). Images were captured by an Olympus BX53 microscope.
The CCD10a coding sequence lacking a termination codon was inserted into the pUC-GFP vector for GFP-fused protein expression driven by the 35S promoter. Maize protoplasts were cultured, isolated according to Yoo et al. (2007), and transformed with the 35S::CCD10a-GFP construct using the PEG–mediated method (Zuo et al., 2015). Briefly, 0.2 mL of suspended protoplasts (0.5 × 106 cells mL−1) with 20% (w/v) PEG4000 (Sigma) were transformed with 20 μg plasmids and incubated at 23°C for 15 min. Transformed protoplasts were cultured at 25°C for 16 h in the dark and observed under a Zeiss confocal microscope.
Analysis of CCD10a Activity
CCD10a was cloned into pDEST15 (Invitrogen) from CCD10a-pDONR221 by recombination. Plasmids containing individual carotenoid biosynthetic genes for phytoene, lycopene, δ-carotene, ε-carotene, β-carotene, and zeaxanthin were solely transformed (as the negative control) or cotransformed with pDEST15 harboring CCD10a into the Escherichia coli strain BL21-AI (Invitrogen) for l-arabinose induced expression; ZmCCD10a expression was analyzed by the standard SDS-PAGE method. Three individual colonies per carotenoid strain were used to separately inoculate 5 mL Luria-Bertani medium containing 100 μg mL−1 ampicillin and 34 μg mL−1 chloramphenicol (Sigma). The 5-mL cultures were grown overnight at 37°C. Four milliliters of overnight culture was used to inoculate 200 mL of Luria-Bertani medium containing 100 μg mL−1 ampicillin and 34 μg mL−1 chloramphenicol (Sigma) in a 500-mL flask.
For HPLC analysis, cultures were grown with vigorous shaking for 9 to 10 h at 30°C to an absorbance of 0.9 to 1 at 600 nm, then divided into two equal parts in two 250-mL flasks. One flask was induced with a final concentration of 0.1% (w/v) l-arabinose (Sigma), and another flask was added equal volume of water as control. The cultures were grown for additional 16 h at 25°C. Cells were pelleted at 5000 g for 5 min. Lycopene, phytoene, δ-carotene, and ε-carotene were extracted from cell pellets by vortexing in 12 mL acetone. β-Carotene was extracted from cell pellets by vortexing in 4-mL ethyl alcohol followed by extraction in 8-mL ethyl acetate. To collect zeaxanthin, cell pellets were extracted in 12 mL of 95% (v/v) ethanol. All solvents were purchased from Fischer Scientific, and all steps were carried out on ice and shielded from strong light. After filtration, 20 μL extract was injected onto the HPLC column. The Waters HPLC was equipped with a photodiode array detector and Agilent SB-C18 (150 mm × 4.6 mm; 5 μm; Waters). For lycopene, δ-carotene, ε-carotene, β-carotene, and zeaxanthin, mobile phases consisted of acetonitrile/water (90:10 [v/v]; A) and ethyl acetate (B). The linear gradient elution increased to 100% B from 0 in 15 min with a flow rate of 1 mL min−1, and the detection wavelength was 475 nm. For phytoene, the mobile phase was ethanol (1 mL min−1) at room temperature and the detection wavelength was 286 nm. Standard phytoene, lycopene, β-carotene, and zeaxanthin were from Aladdin, and δ-carotene and ε-carotene were from CaroteNature.
Volatile compounds were determined by headspace solid-phase microextraction coupled with GC-mass spectrometry (Zhang et al., 2011). Cultures were grown for 5 to 6 h at 30°C to an absorbance of 0.6 to 0.8 at 600 nm, divided, and induced at 16°C for an additional 16 h. Then 5 mL culture and 1 g NaCl were placed into a 15-mL sample vial tightly capped with a polytetrafluoroethylene-silicon septum and heated at 40°C for 30 min on a heating platform with agitation at 400 rpm. The solid-phase microextraction (85 μm polyacrylate, 100 μm polydimethylsiloxane, and 50/30 μm Divinylbenzene/Carboxen/Polydimethyl-siloxane [DVB/CAR/polydimethylsiloxane]; Supelco), preconditioned according to manufacturers' instructions, was then inserted into the headspace, where the extraction lasted for 30 min with heating at 40°C and agitation by a magnetic stirrer. The fiber was subsequently desorbed in the GC injector for 25 min. The Agilent 6890N GC was equipped with an Agilent 5975B mass spectrometer. The column was a HP-INNOWAX column (60 m × 0.25 mm internal diameter, 0.25 μm; J&W Scientific). Helium was used as carrier gas with a flow rate of 1 mL min−1. The GC oven was held at 40°C for 1 min, increased up to 220°C at a rate of 3°C min−1 and held at 220°C for 5 min. The mass spectrometer operated in the electron impact mode at 70 eV, and samples were scanned in the range of mass-to-charge ratio 50 to 500. Identification was based on retention indices of reference standards and mass spectra matching in the National Institute of Standards and Technology 08 library.
Constructs and Generation of Transgenic Plants
The CCD10a coding region was cloned into SalI and SpeI sites of the pSuper1300+ vector (Yang et al., 2010). The construct was transformed into Agrobacterium tumefaciens strain GV3101 for Arabidopsis transformation (Desfeux et al., 2000). Positive transformants were selected on Murashige and Skoog plates with 35 μg mL−1 hygromycin. The resulting positive lines (T1 generation) were transferred to soil (perlite:vermiculite, 1:1 [v/v]) in a growth chamber as described above for seed proliferation and homozygote screen on the selective growth media. Three independent T3 homozygous lines were selected for further analyses according to the expression level of ZmCCD10a.
To generate gene silencing lines, a 238-bp fragment from the CCD10a coding sequence (C1185 to C1422) was cloned into the pCMV201-2bN81 vector, transformed into the A. tumefaciens strain C58C1, coinfiltrated with C58C1 cultures harboring pCMV101 or pCMV301 (with a ratio of 1:1:1) into the leaves 5 and of 6-week Nicotiana benthamiana seedlings, and incubated for 3 d (Wang et al., 2016). CCD10a silencing constructs were introduced into excessive amount of maize seeds (0.15–0.16 g grain−1; 100 seeds per strain per treatment) by vascular puncture after virus extraction from infected leaves, with empty pCMV201-2bN81 as the negative control (Wang et al., 2016). Maize seeds were then germinated and grown in control and LP nutrient solutions for 10 d. The cotyledon tip (2 cm × 0.5 cm) was cut off to screen for CCD10a silencing seedlings using the virus specific primers; 2-cm long root tips were sampled and frozen in liquid nitrogen to analyze gene expression. The remaining tissue of individual plants was used for other phenotypic analysis with four biological replicates. The whole experiment was repeated three times. Dry mass was determined after oven drying at 120°C for 30 min and 65°C for 72 h using a BSA124S analytical balance (Sartorius). The samples were then ground into fine powder for P analysis (Soon and Kalra, 1995). Primers are listed in Supplemental Table S2.
RNA Isolation, Library Construction, and Transcriptome Sequencing
Total RNA was extracted from the root of Col-0 and ZmCCD10a-overexpressing plants grown in the full nutrient solution for 4 weeks, followed by library construction, RNA sequencing, and data analysis according to detailed protocols described previously (Pan et al., 2015). A representative transgenic line (no. 15) was selected for sequencing with three biological replicates.
Measurement of Anthocyanin Content
Pigments were extracted with 5 mL 99:1 (v/v) methanol:HCl at 4°C (Rabino and Mancinelli, 1986). The OD530 and OD657 for each sample were measured. The shoot anthocyanin content was calculated as (OD530 – 0.25 × OD657) × 5/fresh weight (g).
Elemental Assay
The appropriate amount of the ground plant material was digested by a modified Kjeldahl procedure using H2SO4 + H2O2 as a catalyst. Digests were analyzed for P concentrations by automated colorimetry using the molybdovanadate method (Soon and Kalra, 1995).
Isolation and Quantification of Carotenoids in the Plant Tissue
Fresh plant tissues (10 to 20 mg for the leaf and 400–500 mg for the root) were ground into powder and suspended in the 500 μL extraction buffer (acetone: ethyl acetate, 3:2 [v/v]) containing 0.1% (w/v) butylated hydroxytoluene. After 10 min of shaking, 100 μL HPLC-grade water was added followed by 10 min centrifugation at 2500g. The 20 μL top organic phase was used for the HPLC injection. Then 500 μL of extraction buffer (methanol: chloroform, 2:1 [v/v]) with butylated hydroxytoluene and 100 μL of HPLC-grade water/200 μL of chloroform was used for extraction of silique carotenoids. Carotenoids were identified by comparison of the retention time with that of the authentic standard. Standard curves were prepared using commercial standards, with antheraxanthin, neoxanthin, and violaxanthin from CaroteNature and lutein, zeaxanthin, and β-carotene from Aladdin.
Yeast One Hybrid Assay
Two 200-bp promoter fragments of ZmCCD10a containing the P1BS (GGATATGC) or mutated P1BS (TGATCTGA, negative control) element were synthesized and separately cloned into the pLacZi2u vector (reporter) and cotransformed into the EGY48 yeast (Saccharomyces cerevisiae) strain with the pB42AD plasmid fused with PHR1;1 or PHR1;2 (primers listed in Supplemental Table S2). Yeast transformation was performed following instructions in the Yeast Protocols Handbook (Clontech). Transformants were grown on (Trp/-Ura) dropout plates containing 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside for blue color development.
EMSA
Coding sequences of ZmPHR1;1 and ZmPHR1;2 were cloned into the PET-30a vector, transformed into the BL21 strain, cultured for 5 to 6 h at 30°C until an absorbance of 0.6 to 0.8 at 600 nm, and then induced with 1% (w/v) isopropylthio-β-galactoside for 16 h at 16°C. The recombinant protein was purified with Ni-NTA-Agarose. EMSA was carried out following the instructions of the LightShift Chemiluminescent EMSA Kit (Thermo Scientific). The ∼40-bp probe containing the P1BS element of ZmCCD10a was synthesized and labeled with biotin, with unlabeled biotin as the negative control. The primers and oligonucleotides were listed in Supplemental Table S2.
Transient Activation Assay in N. benthamiana Leaves
Coding sequences of PHR1;1 and PHR1;2 were cloned into pGreen 62-SK as effectors, and the 2054-bp promoter of CCD10a was fused to pGreen II 0800-LUC as the reporter. Both vectors were transformed into A. tumefaciens GV3101 (pSOUP 19). After construct verification, the culture mixture (9 effector:1 reporter) was infiltrated into N. benthamiana leaves and incubated for 3 d, with the empty effector vector as the negative control. Activities of firefly luciferase (LUC) and Renilla luciferase (REN) in leaves were measured with the Dual Luciferase Assay System (Promega) on a GLO-MAX 20/20 Luminometer (Promega). The ratio of LUC to REN was calculated with six biological replicates, and the value derived from the vector control was normalized to one. Primers are listed in Supplemental Table S2.
Statistical Analysis
Significant differences were analyzed using the one-way ANOVA in SAS (v8; SAS Institute Inc.). Means of different treatments were compared based on the lsd at a 0.05 or 0.01 level of probability.
Accession Numbers
Accession numbers of all genes or proteins analyzed in this study are listed in Supplemental Datasets S1 and S2 and Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The structure of CCD family genes in land plants.
Supplemental Figure S2. Partial alignment of CCD10 protein sequences.
Supplemental Figure S3. CCD10a expression in maize and teosinte lines.
Supplemental Figure S4. Coomassie Blue-stained SDS-PAGE gels showing His-tagged ZmCCD10a expression in E. coli.
Supplemental Figure S5. GC-mass spectrometry analysis of products derived from carotenoid cleavage by ZmCCD10a.
Supplemental Figure S6. Transcriptomic analysis of ZmCCD10a-overexpressing Arabidopsis plants.
Supplemental Figure S7. Gene expression analysis in wild-type Arabidopsis and ZmCCD10a-overexpressing and silencing seedlings.
Supplemental Figure S8. Aromatic metabolite analysis in roots of ZmCCD10a silencing lines by GC-MS.
Supplemental Table S1. Variations in copy number of CCD10 over evolution.
Supplemental Table S2. Primers used for gene cloning, expression, and vector construction.
Supplemental Dataset S1. Gene identifiers and related information for phylogenetic analysis of the CCD gene family.
Supplemental Dataset S2. Gene identifiers and related information for phylogenetic analysis of the CCD10 gene family in angiosperms.
Acknowledgments
We thank Dr. Francis Cunningham (University of Maryland) for donating bacterial plasmids for carotenoid production and Dr. Jun Fan (China Agricultural University) for insightful comments.
Footnotes
This work was supported by the Chinese Ministry of Science and Technology (grant no. 2016YFD0100707), the National Natural Science Foundation of China (grant nos. 31772385 and 31471928), the Chinese Ministry of Education (grant no. NCET–12–0521), and the Deutsche Forschungsgemeinschaft (grant no. 328017493/GRK 2366).
References
- Ahrazem O, Gómez-Gómez L, Rodrigo MJ, Avalos J, Limón MC(2016) Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: Features and functions. Int J Mol Sci 17: 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H(2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Albalat R, Cañestro C(2016) Evolution by gene loss. Nat Rev Genet 17: 379–391 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S(2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J(2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Atkinson JA, Rasmussen A, Traini R, Voß U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ(2014) Branching out in roots: uncovering form, function, and regulation. Plant Physiol 166: 538–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty DR, Klee HJ(2006a) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45: 982–993 [DOI] [PubMed] [Google Scholar]
- Auldridge ME, McCarty DR, Klee HJ(2006b) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol 9: 315–321 [DOI] [PubMed] [Google Scholar]
- Ballottari M, Alcocer MJ, D’Andrea C, Viola D, Ahn TK, Petrozza A, Polli D, Fleming GR, Cerullo G, Bassi R(2014) Regulation of photosystem I light harvesting by zeaxanthin. Proc Natl Acad Sci USA 111: E2431–E2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR(2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavente LM, Ding XS, Redinbaugh MG, Nelson RS, Balint-Kurti P(2012) Virus-induced gene silencing in diverse maize lines using the brome mosaic virus-based silencing vector. Maydica 57: 206–214 [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O(2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH(2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Chen H, Zuo X, Shao H, Fan S, Ma J, Zhang D, Zhao C, Yan X, Liu X, Han M(2018) Genome-wide analysis of carotenoid cleavage oxygenase genes and their responses to various phytohormones and abiotic stresses in apple (Malus domestica). Plant Physiol Biochem 123: 81–93 [DOI] [PubMed] [Google Scholar]
- Cheng ZJ, Zhao XY, Shao XX, Wang F, Zhou C, Liu YG, Zhang Y, Zhang XS(2014) Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26: 1053–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI(2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Coe EH., Jr.(2001) The origins of maize genetics. Nat Rev Genet 2: 898–905 [DOI] [PubMed] [Google Scholar]
- Cui H, Wang Y, Qin S(2012) Genomewide analysis of carotenoid cleavage dioxygenases in unicellular and filamentous cyanobacteria. Comp Funct Genomics 2012: 164690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E(1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 557–583 [DOI] [PubMed] [Google Scholar]
- Czarnecki O, Yang J, Weston DJ, Tuskan GA, Chen JG(2013) A dual role of strigolactones in phosphate acquisition and utilization in plants. Int J Mol Sci 14: 7681–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ(1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfeux C, Clough SJ, Bent AF(2000) Female reproductive tissues are the primary target of Agrobacterium-mediated transformation by the Arabidopsis floral-dip method. Plant Physiol 123: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, et al. (2017) Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci USA 114: E7641–E7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD(2006) The molecular genetics of crop domestication. Cell 127: 1309–1321 [DOI] [PubMed] [Google Scholar]
- Farré G, Bai C, Twyman RM, Capell T, Christou P, Zhu C(2011) Nutritious crops producing multiple carotenoids--a metabolic balancing act. Trends Plant Sci 16: 532–540 [DOI] [PubMed] [Google Scholar]
- Floss DS, Hause B, Lange PR, Küster H, Strack D, Walter MH(2008a) Knock-down of the MEP pathway isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J 56: 86–100 [DOI] [PubMed] [Google Scholar]
- Floss DS, Schliemann W, Schmidt J, Strack D, Walter MH(2008b) RNA interference-mediated repression of MtCCD1 in mycorrhizal roots of Medicago truncatula causes accumulation of C27 apocarotenoids, shedding light on the functional role of CCD1. Plant Physiol 148: 1267–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frusciante S, Diretto G, Bruno M, Ferrante P, Pietrella M, Prado-Cabrero A, Rubio-Moraga A, Beyer P, Gomez-Gomez L, Al-Babili S, et al. (2014) Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci USA 111: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jorge S, Ha SH, Magallanes-Lundback M, Gilliland LU, Zhou A, Lipka AE, Nguyen YN, Angelovici R, Lin H, Cepela J, et al. (2013) Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25: 4812–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JC, Koch KE, Suzuki M, Wu S, Latshaw S, Petruff T, Goulet C, Klee HJ, McCarty DR(2012) Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol 160: 1303–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeland H, Sandve SR, Torgersen JS, Halle MK, Sundvold H, Omholt S, Våge DI(2014) The evolution and functional divergence of the beta-carotene oxygenase gene family in teleost fish--exemplified by Atlantic salmon. Gene 543: 268–274 [DOI] [PubMed] [Google Scholar]
- Huang FC, Molnár P, Schwab W(2009) Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J Exp Bot 60: 3011–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KL, Ma GJ, Zhang ML, Xiong H, Wu H, Zhao CZ, Liu CS, Jia HX, Chen L, Kjorven JO, et al. (2018) The ARF7 and ARF19 transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis roots. Plant Physiol 178: 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Shi X, Wang W, Ryu KH, Schiefelbein J(2017) Diversification of root hair development genes in vascular plants. Plant Physiol 174: 1697–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Gao X, Liao L, Harberd NP, Fu X(2007) Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol 145: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA.(1997) Plant evolution: The dominance of maize. Curr Biol 7: R411–R413 [DOI] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT(2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloer DP, Schulz GE(2006) Structural and biological aspects of carotenoid cleavage. Cell Mol Life Sci 63: 2291–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi S, Sun S, Berg OG, Andersson DI(2012) Selection-driven gene loss in bacteria. PLoS Genet 8: e1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Jhariya AN(2013) Nutritional, medicinal and economical importance of corn: A mini review. Res J Pharmaceutical Sci 2: 7–8 [Google Scholar]
- Lai J, Ma J, Swigonová Z, Ramakrishna W, Linton E, Llaca V, Tanyolac B, Park YJ, Jeong OY, Bennetzen JL, et al. (2004) Gene loss and movement in the maize genome. Genome Res 14(10A): 1924–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lätari K, Wüst F, Hübner M, Schaub P, Beisel KG, Matsubara S, Beyer P, Welsch R(2015) Tissue-specific apocarotenoid glycosylation contributes to carotenoid homeostasis in Arabidopsis leaves. Plant Physiol 168: 1550–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C, Peng Y, Ma W, Liu R, Li C, Li X(2012) Proteomic analysis revealed nitrogen-mediated metabolic, developmental, and hormonal regulation of maize (Zea mays L.) ear growth. J Exp Bot 63: 5275–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ(2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Xu Y, Jiang H, Jiang C, Du Y, Gong C, Wang W, Zhu S, Han G, Cheng B(2016) Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int J Mol Sci 17: 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Shao Z, Zhang M, Wang Q(2015) Regulation of carotenoid metabolism in tomato. Mol Plant 8: 28–39 [DOI] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ(2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS(2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Nadeem M, Mollier A, Morel C, Vives A, Prud’homme L, Pellerin S(2011) Relative contribution of seed phosphorus reserves and exogenous phosphorus uptake to maize (Zea mays L.) nutrition during early growth stages. Plant Soil 346: 231–244 [Google Scholar]
- Nagarajan VK, Jain A, Poling MD, Lewis AJ, Raghothama KG, Smith AP(2011) Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol 156: 1149–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar N, Li L, Lu S, Khin NC, Pogson BJ(2015) Carotenoid metabolism in plants. Mol Plant 8: 68–82 [DOI] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC(2011) Phosphate import in plants: Focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K(2006) Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol 142: 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Hasan MM, Li Y, Liao C, Zheng H, Liu R, Li X(2015) Asymmetric transcriptomic signatures between the cob and florets in the maize ear under optimal- and low-nitrogen conditions at silking, and functional characterization of amino acid transporters ZmAAP4 and ZmVAAT3. J Exp Bot 66: 6149–6166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Zheng H, Zhao J, Xu Y, Li X(2016) ZmCCD7/ZpCCD7 encodes a carotenoid cleavage dioxygenase mediating shoot branching. Planta 243: 1407–1418 [DOI] [PubMed] [Google Scholar]
- Park S, Steen CJ, Lyska D, Fischer AL, Endelman B, Iwai M, Niyogi KK, Fleming GR(2019) Chlorophyll-carotenoid excitation energy transfer and charge transfer in Nannochloropsis oceanica for the regulation of photosynthesis. Proc Natl Acad Sci USA 116: 3385–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J(1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabino I, Mancinelli AL(1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A(2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103: 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JAD(2001) Characterization of a novel carotenoid cleavage dioxygenase from plants. J Biol Chem 276: 25208–25211 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JAD, McCarty DR(1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Shen J, Yuan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F(2011) Phosphorus dynamics: From soil to plant. Plant Physiol 156: 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY(1999) Seed-specific overexpression of phytoene synthase: Increase in carotenoids and other metabolic effects. Plant J 20: 401–412X [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ(2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Soon YK, Kalra YP(1995) A comparison of plant tissue digestion methods for nitrogen and phosphorus analyses. Can J Soil Sci 75: 243–254 [Google Scholar]
- Takahashi Y, Moiseyev G, Chen Y, Ma JX(2005) Identification of conserved histidines and glutamic acid as key residues for isomerohydrolase activity of RPE65, an enzyme of the visual cycle in the retinal pigment epithelium. FEBS Lett 579: 5414–5418 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR(2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tian L, Magallanes-Lundback M, Musetti V, DellaPenna D(2003) Functional analysis of β- and ε-ring carotenoid hydroxylases in Arabidopsis. Plant Cell 15: 1320–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Vallabhaneni R, Bradbury LM, Wurtzel ET(2010) The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch Biochem Biophys 504: 104–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Tan BC, McCarty DR, Klee HJ(2008) The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J Biol Chem 283: 11364–11373 [DOI] [PubMed] [Google Scholar]
- Walter MH, Fester T, Strack D(2000) Arbuscular mycorrhizal fungi induce the non-mevalonate methylerythritol phosphate pathway of isoprenoid biosynthesis correlated with accumulation of the ‘yellow pigment’ and other apocarotenoids. Plant J 21: 571–578 [DOI] [PubMed] [Google Scholar]
- Wang JY, Haider I, Jamil M, Fiorilli V, Saito Y, Mi J, Baz L, Kountche BA, Jia KP, Guo X, et al. (2019) The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat Commun 10: 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Yang X, Wang N, Liu X, Nelson RS, Li W, Fan Z, Zhou T(2016) An efficient virus-induced gene silencing vector for maize functional genomics research. Plant J 86: 102–115 [DOI] [PubMed] [Google Scholar]
- Wang XH, Bai JR, Liu HM, Sun Y, Shi XY, Ren ZQ(2013) Overexpression of a maize transcription factor ZmPHR1 improves shoot inorganic phosphate content and growth of Arabidopsis under low-phosphate conditions. Plant Mol Biol Report 31: 665–677 [Google Scholar]
- Wang Y, Ding G, Gu T, Ding J, Li Y(2017) Bioinformatic and expression analyses on carotenoid dioxygenase genes in fruit development and abiotic stress responses in Fragaria vesca. Mol Genet Genomics 292: 895–907 [DOI] [PubMed] [Google Scholar]
- Wei YP, Wan HQ, Wu ZM, Wang RQ, Ruan MY, Ye QJ, Li ZM, Zhou GZ, Yao ZP, Yang YJ(2016) A comprehensive analysis of carotenoid cleavage dioxygenases genes in Solanum lycopersicum. Plant Mol Biol Report 34: 512–523 [Google Scholar]
- Willmann M, Gerlach N, Buer B, Polatajko A, Nagy R, Koebke E, Jansa J, Flisch R, Bucher M(2013) Mycorrhizal phosphate uptake pathway in maize: Vital for growth and cob development on nutrient poor agricultural and greenhouse soils. Front Plant Sci 4: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Shi Y, Liu J, Guo L, Zhang X, Yang S(2010) A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J 63: 283–296 [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I(2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J(2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA(1998) Metabolism and physiology of abscisic acid. Annu Rev Plant Biol 39: 439–473 [Google Scholar]
- Zhang HM, Forde BG(1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang MX, Pan QH, Yan GL, Duan CQ(2011) Using headspace solid phase micro-extraction for analysis of aromatic compounds during alcoholic fermentation of red wine. Food Chem 125: 743–749 [Google Scholar]
- Zhang X, Zhou Y, Ding L, Wu Z, Liu R, Meyerowitz EM(2013) Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25: 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Chao Q, Zhang N, Ye J, Tan G, Li B, Xing Y, Zhang B, Liu H, Fengler KA, et al. (2015) A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet 47: 151–157 [DOI] [PubMed] [Google Scholar]