Abstract
Fungal bicyclo[2.2.2]diazaoctane indole alkaloids represent an important family of natural products with a wide-spectrum of biological activities. Although biomimetic total syntheses of representative compounds have been reported, the details of their biogenesis, especially the mechanisms for assembly of diastereomerically distinct and enantiomerically antipodal metabolites, have remained largely uncharacterized. Brevianamide A represents a basic form of the sub-family bearing a dioxopiperazine core and a rare 3-spiro-ψ-indoxyl skeleton. Here, we identified the Brevianamide A biosynthetic gene cluster from Penicillium brevicompactum NRRL 864 and elucidated the metabolic pathway. BvnE was revealed to be an essential isomerase/semi-pinacolase that specifies selective production of the natural product. Structural elucidation, molecular modeling, and mutational analysis of BvnE, and quantum chemical calculations provided mechanistic insights into the diastereoselective formation of the 3-spiro-ψ-indoxyl moiety in Brevianamide A. This occurs through a BvnE-controlled semi-pinacol rearrangement and a subsequent spontaneous intramolecular [4+2] hetero-Diels-Alder cycloaddition.
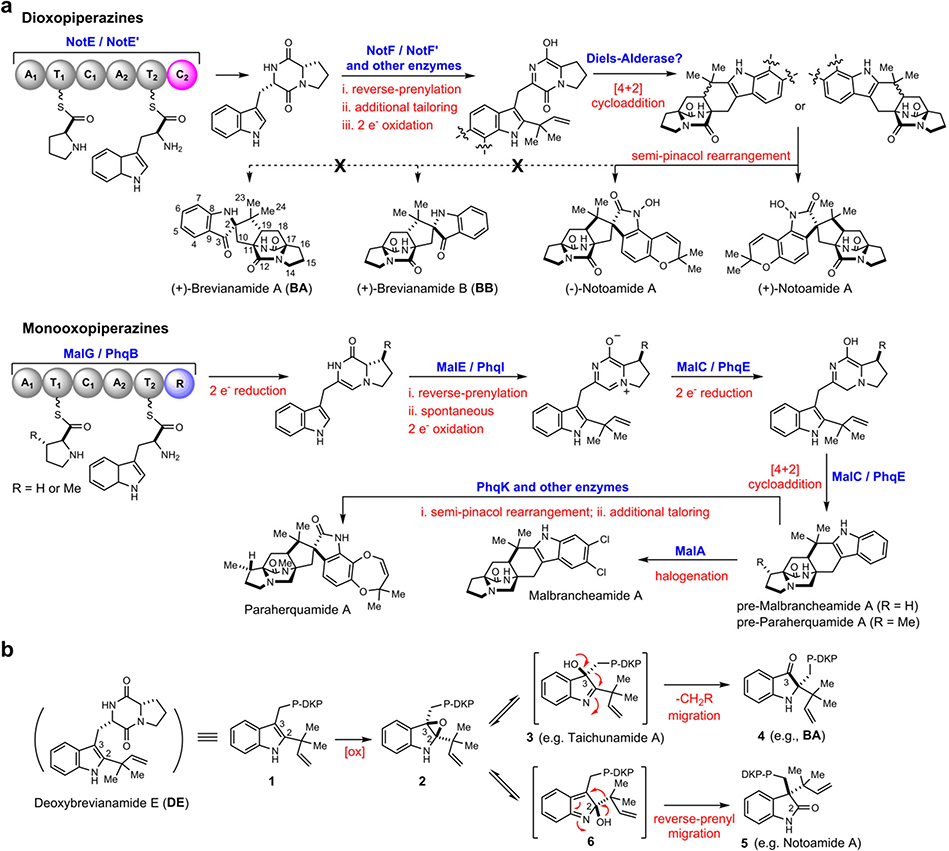
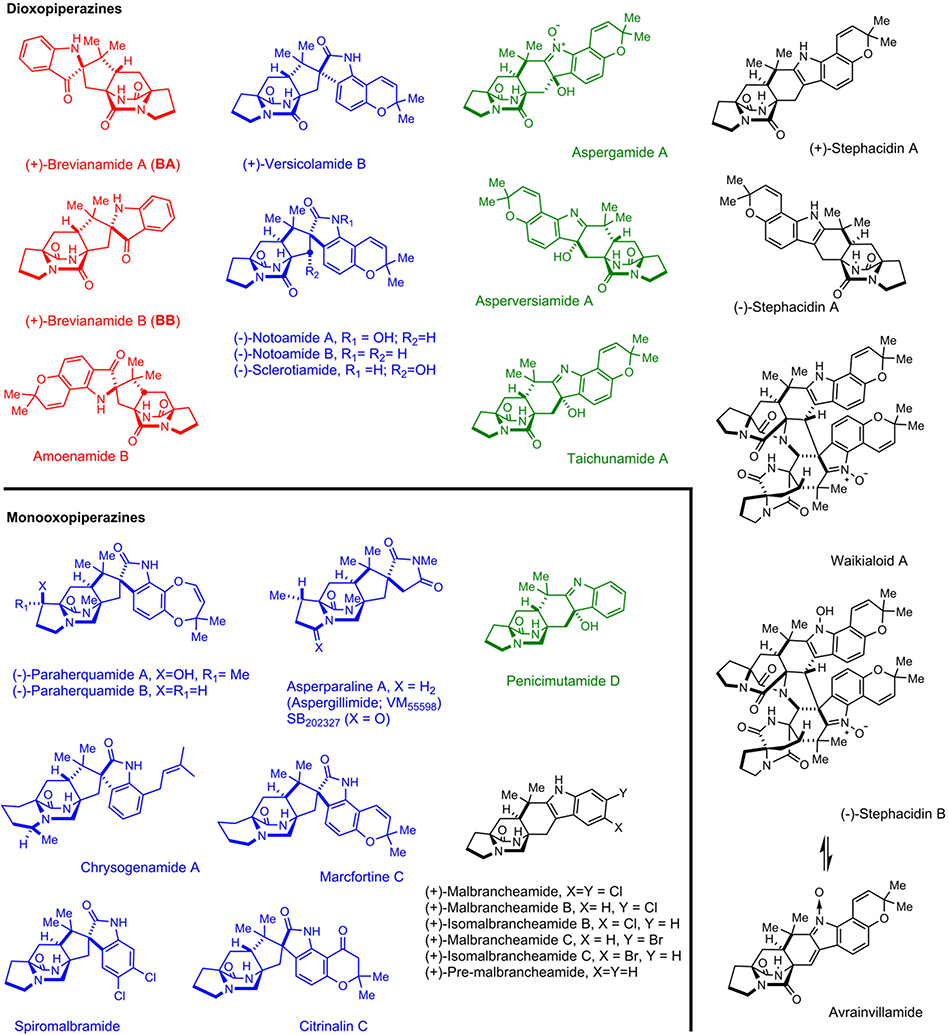
Fungal indole alkaloids bearing the unusual bicyclo[2.2.2]diazaoctane core have drawn considerable attention from natural product, synthetic and biological chemists for decades. A wealth of studies on the discovery of analogs (including semi-synthetic, synthetic and natural), biological activities and biosynthetic mechanisms have been conducted.1 The prominent representatives of this structural family have been recognized by our laboratories and others as belonging to two main biogenetic subfamilies (Extended Data Fig. 1): (1) the dioxopiperazines that includes: the insecticidal Brevianamide A and B (BA/BB) from Penicillium brevicompactum,2 the anticancer agents (−)-Notoamide A isolated from Aspergillus protuberus (formerly Aspergillus sp. MF297–2), and (+)-Notoamide A from A. amoenus (formerly A. versicolor NRRL 35600), the anti-cancer agents Stephacidins A and B from A. ochraceus, the Sclerotiamides, Versicolamides, Taichunamides, anti-fungal Waikialoids, Amoenamide B, Speramides, and Asperochramides; and (2) the monooxopiperazines that includes: the anti-parasitic Paraherquamides from Penicillium spp., the Asperparalines, Marcfortines, calmodulin-inhibiting Malbrancheamides, Chrysogenamide, Mangrovamides, Penioxalamine, Penicimutamides, and Asperversiamides.1,3,4
The bicyclo[2.2.2]diazaoctane core structure has long been proposed to arise from an intramolecular [4+2] hetero-Diels-Alder (IMDA) construction.5–10 In our continuing pursuit of biocatalysts responsible for the [4+2] cycloaddition that is essential for assembly of diverse diastereomerically distinct and enantiomerically antipodal metabolites, we realised that dioxopiperazines and monooxopiperazines adopt distinct biosynthetic strategies for building their respective bicyclo[2.2.2]diazaoctane scaffold (Fig. 1a). For instance, we recently revealed that the bifunctional NADPH-dependent reductase/Diels-Alderase MalC (and PhqE) catalyses diastereo- and enantioselective IMDA cyclization via a zwitterionic intermediate for assembly of the monooxopiperazines Malbrancheamides (and Paraherquamides).11 However, the biosynthetic gene clusters for dioxopiperazines such as Notoamides lack of a MalC/PhqE homologue (Extended Data Fig. 2), indicating alternative yet uncharacterised mechanisms to generate their bicyclo[2.2.2]diazaoctane system.
Fig. 1. Present knowledge on bicyclo[2.2.2]diazaoctane assembly and semi-pinacol rearrangements of indole- 2,3-epoxide systems.
a, Distinct biosynthetic strategies for assembly of the bicyclo[2.2.2]diazaoctane core structure in dioxopiperazines and monooxopiperazines. b, Disparate fates of indole-2,3-epoxide moieties (2) and semi-pinacol rearrangements leading to indoxyls (4), oxindoles (5), and stable 3-hydroxyindolenines (3). P-DKP refers to prolyl- DKP moiety of DE.
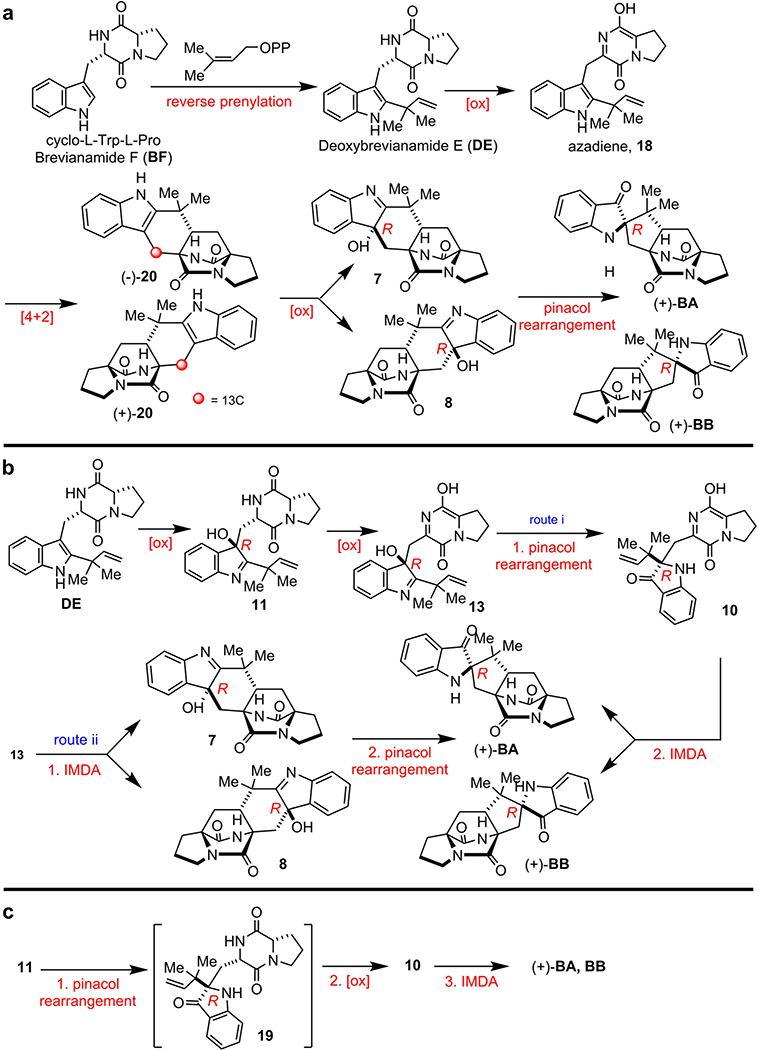
BA and BB are among the original natural alkaloids isolated that comprise the bicyclo[2.2.2]diazaoctane core (Fig. 1a).2 After we established the correct absolute configuration of natural (+)-BB through total synthesis,12,13 it became evident that the biogenesis of BA/BB must accommodate assembly of the core tricyclic ring systems that are pseudo-enantiomeric. Based on the pioneering proposal first suggested by Porter and Sammes in 197014, several hypotheses for their biogenesis were proposed to accommodate the stereochemistry, presumably installed by a [4+2] IMDA cycloaddition, and the unique 3-spiro-ψ–indoxyl moiety. Specifically (Fig. 1b, Extended Data Fig. 3), a highly modified, reverse-prenylated indole (1, e.g., Deoxybrevianamide E (DE)) undergoes an apparent indole 2,3-epoxidation either before or after the IMDA cyclization. Ring-opening of epoxide 2 can form 3-hydroxyindolenines (3), which in some instances are stable (e.g., Taichunamide A15) or, suffers a spontaneous semi-pinacol rearrangement to generate 3-spiro-ψ–indoxyls (4, e.g., BA), or spiro-2-oxindoles (5, e.g., Notoamide A16) via an alternative ring-opening intermediate 6.1,3,4
While the spiro-2-oxindoles are more frequently observed in the prenylated indole alkaloid family, only a small number of 3-spiro-ψ-indoxyl species have been isolated so far, and very rarely are natural spiro-2-oxindole species and 3-spiro-ψ-indoxyls co-produced by a single microbe.17,18 These co-metabolite profiles suggest that the semi-pinacol rearrangement might be differentially controlled in the partitioning of the putative indole-2,3-epoxides and implies a specific biocatalyst-controlled mechanism. Recently, the chemical synthesis of BA, the typical structure that contains the dioxopiperazine and 3-spiro-ψ–indoxyl moieties, has been accomplished and suggested a Diels-Alderase-free biosynthesis.19 However, the biocatalytic details and the sequential timing for the putative indole oxidation, semi-pinacol rearrangement, and IMDA cycloaddition (Extended Data Fig. 3) remain unresolved experimentally due to lack of access to the key enzymes.
In this study, we addressed these outstanding biosynthetic questions by elucidating the BA biosynthetic pathway through gene disruption, heterologous expression, precursor incorporation experiments, and in vitro biochemical analysis. Particularly, an isomerase/semi-pinacolase BvnE was revealed to catalyse an essential semi-pinacol rearrangement, thereby directing diastereoselective assembly of BA via a spontaneous [4+2] IMDA cycloaddition. Resolution of this 50-year old mechanistic mystery together with our recent characterization of the Diels-Alderase-mediated biogenesis of monooxopiperazines11 highlight the diversified biosynthetic strategies deployed by fungi for creating structurally diverse spiro-cyclized indole alkaloids.
Results and discussion
Identification of the Brevianamide A biosynthetic gene cluster
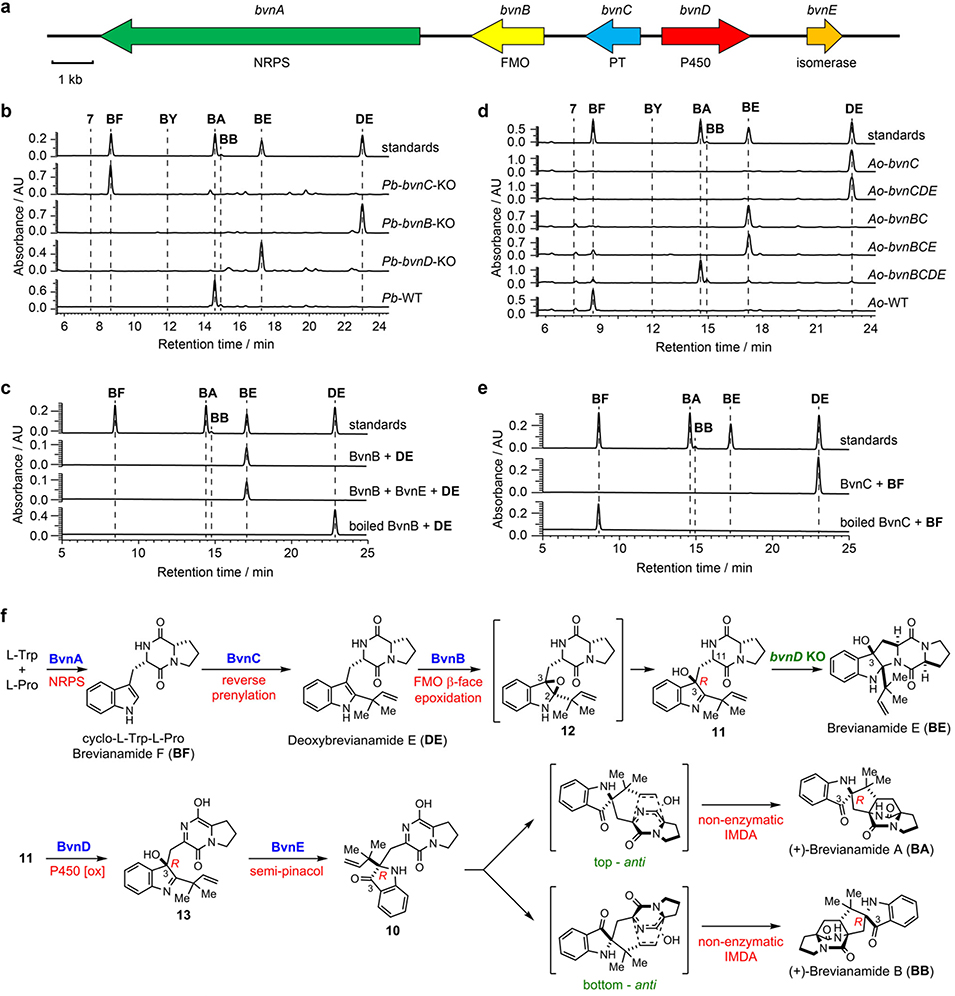
Initially, genome mining of a BA producing strain P. brevicompactum NRRL 864 (Pb) was conducted using the Notoamide nonribosomal peptide synthetase (NRPS) gene notE as a probe to search for its homolog responsible for assembling the cyclodipeptide Brevianamide F (BF).3 A putative 16 kb BA biosynthetic gene cluster (bvn, GenBank accession number: MN401751) was revealed (Fig. 2a) that contains bvnA (NRPS), bvnB (flavin monooxygenase, FMO), bvnC (prenyltransferase, PT), and bvnD (P450 monooxygenase, P450), all of which have homologues in the (+)-/(−)-Notoamide A biosynthetic gene clusters3. The bvnE gene (465 bp) that encodes a putative isomerase shows low homology to Trt14, AusH and PrhC involved in meroterpenoid biosynthetic pathways.20 Interestingly, the biosynthetic gene clusters of the spiro-2-oxindole containing Notoamides and Paraherquamides3 lack a bvnE homologue (Extended Data Fig. 2), and further mining of these corresponding genomes failed to identify homologous genes. Thus, we surmised that BvnE might play an important role in 3-spiro-ψ-indoxyl formation.
Fig. 2. Functional analyses of Brevianamide A biosynthetic genes and the proposed biosynthetic pathway.
a, Brevianamide A biosynthetic gene cluster (bvn) from Penicillium brevicompactum NRRL 864 (Pb). b. HPLC analysis (230 nm) of Pb KO mutants. c, HPLC analysis (230 nm) of the in vitro assays of BvnB and BvnE. d, HPLC analysis (230 nm) of the Brevianamide F (BF) feeding experiments with recombinant Aspergillus oryzae M-2–3 (Ao) strains expressing different bvn gene(s). e, HPLC analysis (230 nm) of in vitro assays using BvnC. f. Revised biosynthetic pathway for BA and BB.
Functional analysis of Brevianamide A biosynthetic genes
In the bvn gene cluster, the bimodular NRPS encoded by bvnA is predicted to catalyze BF formation. This was confirmed by its heterologous expression in Aspergillus oryzae NSAR1 (Ao) (Supplementary Fig. 1). To investigate the function of bvnB-E, four individual single gene knockout (KO) strains of Pb were constructed. The resulting Pb-bvnC-KO strain accumulated BF (Fig. 2b) as the only product. The Pb-bvnB-KO and Pb-bvnD-KO strains accumulated DE and Brevianamide E (BE) (Fig. 2b), respectively. BE is proposed to be a rearranged shunt product resulting from initial 2,3-indole epoxidation of DE by BvnB FMO (Fig. 2f). Indeed, the recombinant N-His6-tagged BvnB efficiently converted DE into BE in vitro (Fig. 2c) likely through a mechanism previously demonstrated for NotB21 (a BvnB homolog with 62%/75% identity/similarity) in Notoamide biosynthesis. Since Pb-bvnD-KO did not produce a bicyclo[2.2.2]diazaoctane structure, we reasoned that BvnD is a key enzyme for the proposed IMDA reactions.
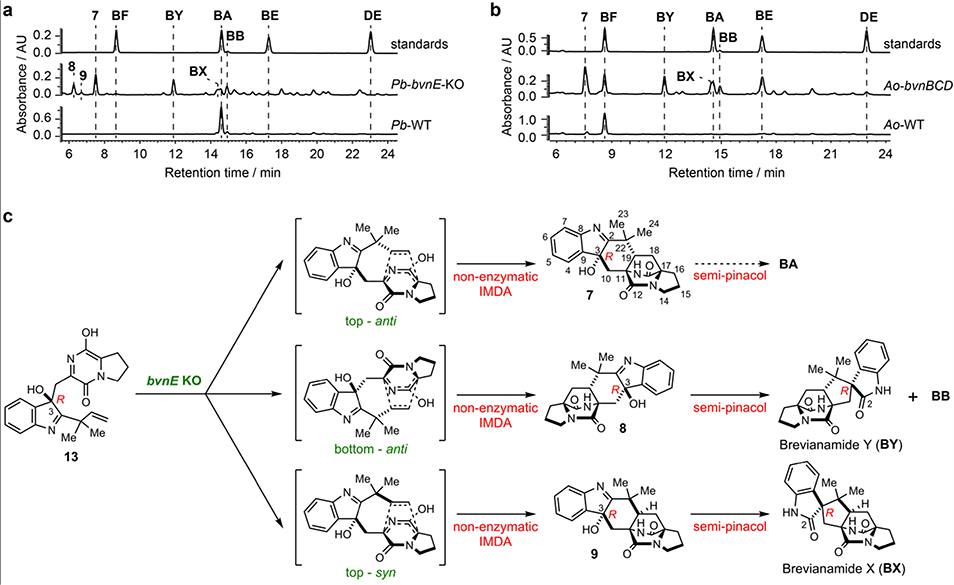
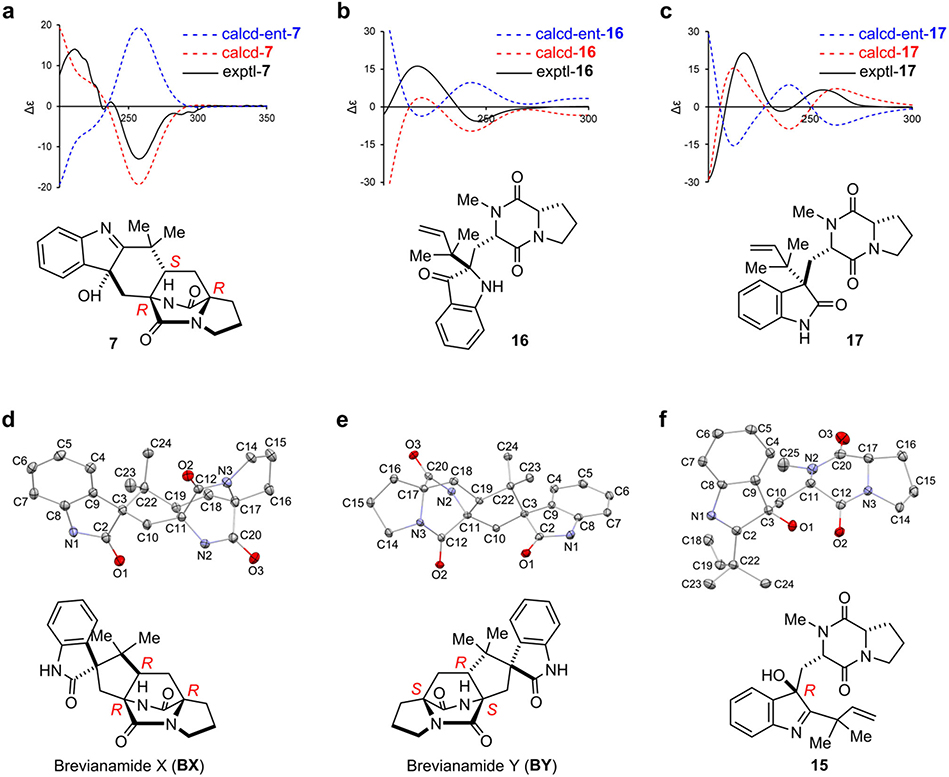
When bvnE was deleted in Pb, five detectable substances were observed in addition to low levels of BA (Supplementary Table 1, Supplementary Figs.2–3) and BB (Fig. 3a), all showing the same molecular weight of 365 Da (Supplementary Fig. 4). These include three previously unknown 3-hydroxyindolenine derivatives 7–9 and the two reported spiro-2-oxindoles Brevianamide X (BX) and Brevianamide Y (BY). The planar structure of 7 was constructed based on 1D and 2D nuclear magnetic resonance (NMR) analyses (Supplementary Table 2, Supplementary Figs. 5–10): the key heteronuclear multiple bond correlation (HMBC) correlations of H3-23/24 to C-2/19/22, H2-10 to C-2/3/11/19, and H-4 to C-3 (Supplementary Fig. 9) readily built the hexatomic ring neighboring the indole base; the C-2=N double bond was deduced from the chemical shift of C-2 (189.29 ppm); and the α-configuration of 3-OH was determined from the nuclear Overhauser effect spectroscopy (NOESY) analysis (Supplementary Fig. 10). The absolute configuration of 7 was determined by comparison of the experimental and computational electronic circular dichroism (ECD) spectra (Extended Data Fig. 4). The structures of BX and BY were determined through high resolution mass spectrometry (HRMS) (Supplementary Fig. 4), NMR analyses (Supplementary Table 1, Supplementary Figs. 11–22), and comparison with the corresponding synthesized authentic standards (see Supplementary Methods). Their absolute configurations were established by single-crystal X-ray diffraction (CCDC No. BX: 1973959, BY: 1973957; Extended Data Fig. 4, Supplementary Table 3). The ratio of BA:BB:7:BX:BY was determined to be 1:1:7:5:9 (Supplementary Table 4). Instability of 8 and 9 prevented direct structural determination of these isomers. Nonetheless, the observations that 8 and 9 quickly collapsed to BY/BB and BX (Supplementary Fig. 23), respectively, and their similar UV spectra to that of 7 suggested the structures of these two unstable metabolites being 3-hydroxyindolenines (Fig. 3c, Supplementary Fig. 24). To determine the structures of 8 and 9, we chemically synthesized their hypothetical structures (see Supplementary Methods,Supplementary Figs. 25–28). The matched UV spectra and retention times on HPLC, and the same decomposition behaviors of the synthetic standards to the isolated samples (Supplementary Fig. 29) confirmed their structures.
Fig. 3. Functional analyses of bvnE.
a, HPLC analysis (230 nm) of the Pb-bvnE-KO mutant. b, HPLC analysis (230 nm) of the Ao-bvnBCD mutant. c, the putative spontaneous transformations in absence of BvnE.
To validate the function of bvn genes and also to clarify the order of biosynthetic steps, we conducted heterologous expression with different gene combinations in Ao. As expected, BF was converted into DE by either Ao-bvnC in vivo (Fig. 2d) or purified BvnC in vitro in the presence of DMAPP and Mg2+ (Fig. 2e). Thus, PT BvnC is a Deoxybrevianamide E synthase as previously demonstrated for NotF.22 Ao-bvnCDE produced DE exclusively in the feeding experiment, while Ao-bvnBC and Ao-bvnBCE both accumulated BE exclusively (Fig. 2d). These results indicate that the presumed indole epoxidation catalyzed by FMO BvnB occurs prior to the P450 BvnD-mediated step and preceding the BvnE-catalyzed step. Upon introduction of BF to the Ao-bvnBCD culture, five products with identical m/z 366 ([M+H]+) were observed (Fig. 3b, Supplementary Fig. 4). Their relative abundance (BA:BB:7:BX:BY = 1:1:5:2:6) was qualitatively consistent with the observations in Pb-bvnE-KO (Fig. 3a, Supplementary Table 4) except that 8 and 9 were not detected probably due to their instability. Finally, when bvnE was incorporated, and the Ao-bvnBCDE strain grown in the presence of exogenous BF, we observed BA and BB in ~10:1 ratio (Fig. 3b, Supplementary Table 4), which is consistent with the product profile of wild-type Pb (Pb-WT) (Fig. 3a).
These results demonstrated that the metabolites generated through Ao biotransformations and individual Pb knockout mutants correlated directly with one another. The reaction sequence of each enzyme in the pathway can now be deduced as BvnA→BvnC→BvnB→BvnD→BvnE (Fig. 2f). Both systems showed a BvnE-dependent change of product profile, suggesting that BvnE might function to mediate formation of the 3-spiro-ψ-indoxyl species 10, and directly impacts diastereo-outcome of the presumed IMDA reactions. With respect to BvnD activity, intensive attempts to produce this P450 enzyme in E. coli and Saccharomyces cerevisiae were unsuccessful, thus preventing direct functional analysis. Nonetheless, the results that Ao-bvnBCD transformed BF into multiple IMDA products (Fig. 3b) and that Ao-bvnCDE was unable to recognize DE as a substrate (Fig. 2d), and the principles for P450 enzymes and Diels-Alder reactions together suggest that BvnD might oxidize a non-isolable intermediate 11 (derived from ring opening of the indole epoxide 12) to 13, thereby generating the diene moiety required for IMDA cyclization (Fig. 2f). In the absence of BvnD, compound 11 would collapse to BE via an energetically favored N-C ring closure (Supplementary Fig. 30).
Although we cannot exclude the possibility that BvnD might directly desaturate the N-C bond in the dioxopiperazine ring of 11,23 it is more likely that BvnD first catalyzes hydroxylation of 11 and then undergoes spontaneous dehydration/tautomerization to yield 13 (Fig. 2f). Regarding the position on 11 likely oxidized by BvnD, bond dissociation energy calculations (Supplementary Fig. 31) indicated that the most probable hydroxylation site is the tertiary C-H bond at C-11, which is supported by (1) FtmG, a BvnD homologue with 47%/64% protein sequence identity/similarity in the Fumitremorgin biosynthetic pathway, has been experimentally confirmed to catalyze hydroxylation of an analogous position;23 and (2) a fungal dioxopiperazine structure Asperversiamide I was recently discovered to contain the C-11 hydroxylation, which might be installed by an unidentified BvnD homologue.24
Probing the catalytic mechanism of the isomerase/pinacolase BvnE
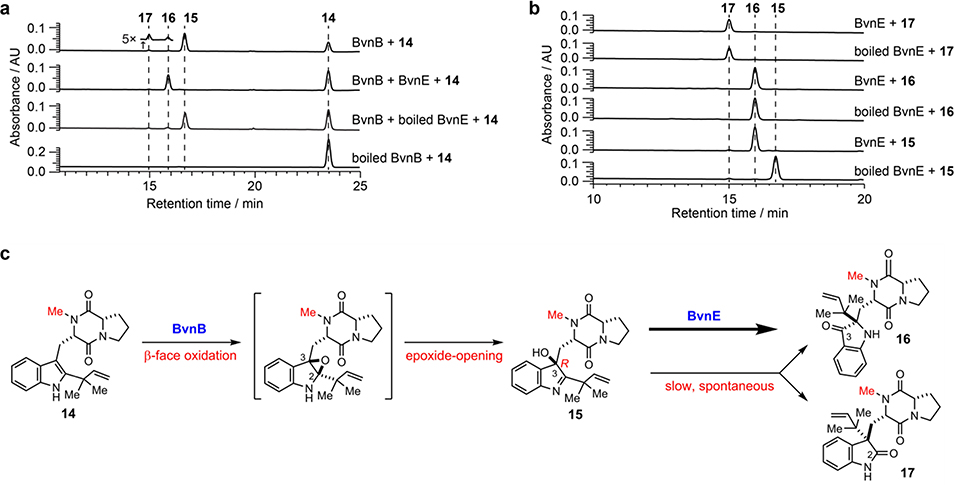
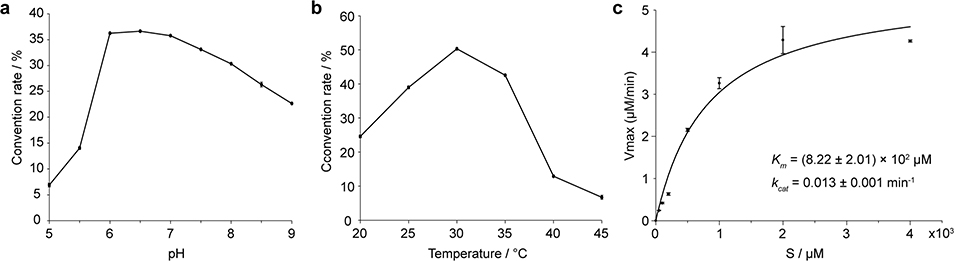
Based on the results of Pb-bvnE-KO and Ao-bvnBCD (Fig. 3), BvnE appeared to be a central enzyme for controlling the product profile in Brevianamide biogenesis. To assess this unique isomerase in vitro, the N-His6-tagged BvnE was produced in E. coli BL21(DE3) and purified to homogeneity (Supplementary Fig. 32). Next, we sought to identify the potential natural substrate of BvnE from Pb-bvnE-KO. Compounds 7, 8, 9, BX, BY, and BE were tested as substrates, but failed to be transformed (Supplementary Figs. 33–34). Thus, we hypothesized that 13 might be the native substrate of BvnE. Considering the inaccessibility of this unstable intermediate, we selected to chemoenzymatically synthesize 15 (see Supplementary Methods), a stable analogue of 13 (Fig. 4c) in order to block the dioxopiperazine nitrogen from N-C ring closure, and prevent formation of the unstable azadiene intermediate upon spontaneous dehydration. The mechanistic probe 15 was prepared through BvnB-catalysed in vitro conversion of the chemically synthesized N-methyl-Deoxybrevianamide E (14) (Supplementary Figs. 35–43); and the absolute configuration of 15 was determined by single-crystal X-ray diffraction (CCDC No. 1973958, Extended Data Fig. 4). Interestingly, two minor products 16 and 17 were also generated from 14 along with the predominant product 15 (Fig. 4a). Structural determination indicated that 16 and 17 are 3-spiro-ψ-indoxyl and spiro-2-oxindole isomers of 15, respectively (Supplementary Table 5, Supplementary Figs. 35, 44–55). Their absolute configurations were determined by comparison of their experimental and calculated ECD spectra (Extended Data Fig. 4), which were consistent with the structures arising from the semi-pinacol rearrangement of 15. Moreover, when 15 was incubated with BvnE, it was completely converted into 16 (Fig. 4b). These results indicated that BvnE functions as a semi-pinacolase responsible for the selective formation of the 3-spiro-ψ-indoxyl sub-structure (in 16) from the 3-hydroxy-pyrrole moiety (in 15). Spontaneous conversion from 15 to 16 and 17 at room temperature could also occur slowly (Fig. 4c, Supplementary Fig. 56). Loss of stereocontrol of IMDA cycloaddition in terms of top/bottom (7/9/BA/BX vs 8/BY/BB) and anti/syn selectivity (7/8/BA/BB/BY vs 9/BX) in the absence of BvnE strongly suggests that these isomers are generated from non-enzymatic IMDA reactions. Moreover, the observed product profile (Fig. 3, Supplementary Table 6) is quantitatively consistent with the calculated product distribution (see below). With respect to the catalytic properties of BvnE (Extended Data Fig. 5), the apparent kcat and Km values of BvnE towards 15 were determined to be 0.013 min−1 and 822 μM, respectively, under the optimal pH (6.5) and temperature (30 °C). The low catalytic efficiency (kcat/Km = 1.58 × 10−5 min−1 μM −1) was probably because 15 is not the native substrate.
Fig. 4. Probing the functionality of BvnE using substrate mimics.
a, HPLC analysis (230 nm) of the in vitro assays of BvnB/BvnE. b, HPLC analysis (230 nm) of the in vitro assays of BvnE with the N-methylated substrates. c, the reaction scheme.
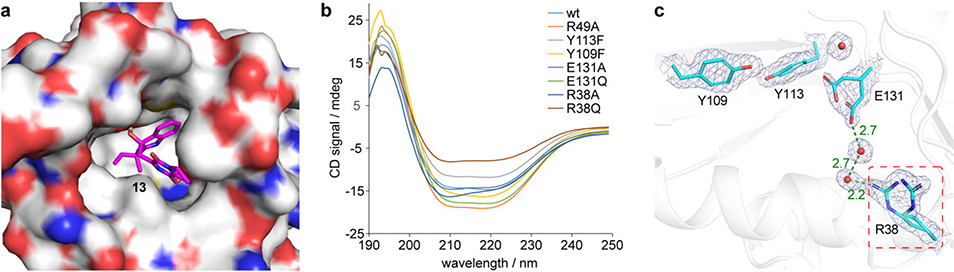
As a key component in controlling product outcome in Brevianamide biosynthesis, we sought to determine how BvnE catalyzes formation of the 3-spiro-ψ-indoxyl species. BvnE is related to NTF2-like superfamily enzymes that have been studied in fungal meroterpenoid biogenesis.20 We solved the crystal structure of BvnE at 1.8 Å resolution (Fig. 5a, Supplementary Table 7, PDB ID: 6U9I) by molecular replacement using PrhC (PDB ID: 5X9J) as a search model (Supplementary Fig. 57).25 BvnE is a symmetric homodimer that adopts an α+β-barrel fold with the presumed active sites at the end of each barrel. This cavity has a hydrophobic interior and also contains several polar residues, which could be involved in the acid-base chemistry reported for this family of enzymes (Fig. 5a, Supplementary Fig. 58).
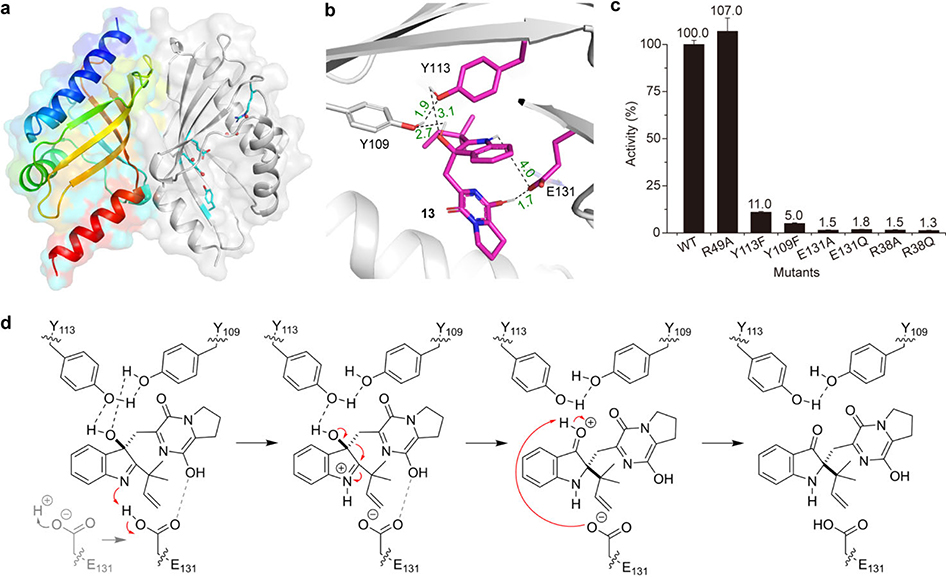
Fig. 5. BvnE crystal structure, docking and the proposed catalytic mechanism.
a, Structure overview of BvnE: left chain shown in rainbow spectrum from N-terminus (blue) to C-terminus (red); the right chain in grey cartoon with putative catalytic acid-basic residues (cyan) and water molecules in the active site (red spheres). b, BvnE-13 complex with key residues and hydrogen bonding shown. c, Site-directed mutagenesis results; Data represent the average of triplicated independent experiments (centre values, means; error bars, standard deviations; n = 3). d, The proposed BvnE reaction mechanism for isomerization of 13. Glu131-mediated proton transfer could be assisted by 540 Arg38, which interacts with Glu131 via an ordered water network (Supplementary Fig. 62c). Indirect deprotonation of the oxonium intermediate via bound solvent molecules rather than a direct interaction with Glu131 cannot be excluded.
Docking of the presumed native substrate 13 (Fig. 5b, Extended Data Fig. 6a) helped reveal the BvnE active site. We identified Arg38, Tyr109, Tyr113 and Glu131 as candidates for site-directed mutagenesis and in vitro enzyme assays with 15. In each case, mutation of these residues caused severely attenuated activity (Fig. 5c). All CD spectra of BvnE mutants matched wild-type protein (Extended Data Fig. 6b), indicating that these point mutations did not significantly impact protein folding. The electron density indicated that Arg38, Tyr113 and Glu131 adopt multiple conformations in the BvnE crystal lattice. Therefore, these residues were designated as flexible in molecular docking (Extended Data Fig. 6c). In the lowest energy output of the BvnE-13 docking complex, the substrate reverse prenyl group packed into a hydrophobic pocket (Extended Data Fig. 6a), and Tyr109 and Tyr113 interacted with the 3–OH of 13 (Fig. 5b). Glu131 is positioned to interact with the indole nitrogen and the C-18 oxygen of 13, suggesting its key role in the semi-pinacol rearrangement. (Fig. 5d). On the basis of these docking studies, we propose that the reaction mechanism of BvnE includes: (1) Glu131-mediated proton transfer to initiate the semi-pinacol rearrangement and to provide charge stabilization of the intermediate, and (2) hydrogen bond-assisted activation of the semi-pinacol rearrangement through Tyr109/Try113 (Fig. 5d). Arg38, which was essential for activity, does not directly interact with the docked ligands or active site residues. However, the structure reveals two ordered water molecules in a hydrogen bonding network between Arg38 and Glu131(Extended Data Fig. 6c), suggesting that it may assist in proton transfer by regenerating the carboxylate form of E131.
Quantum chemical calculations
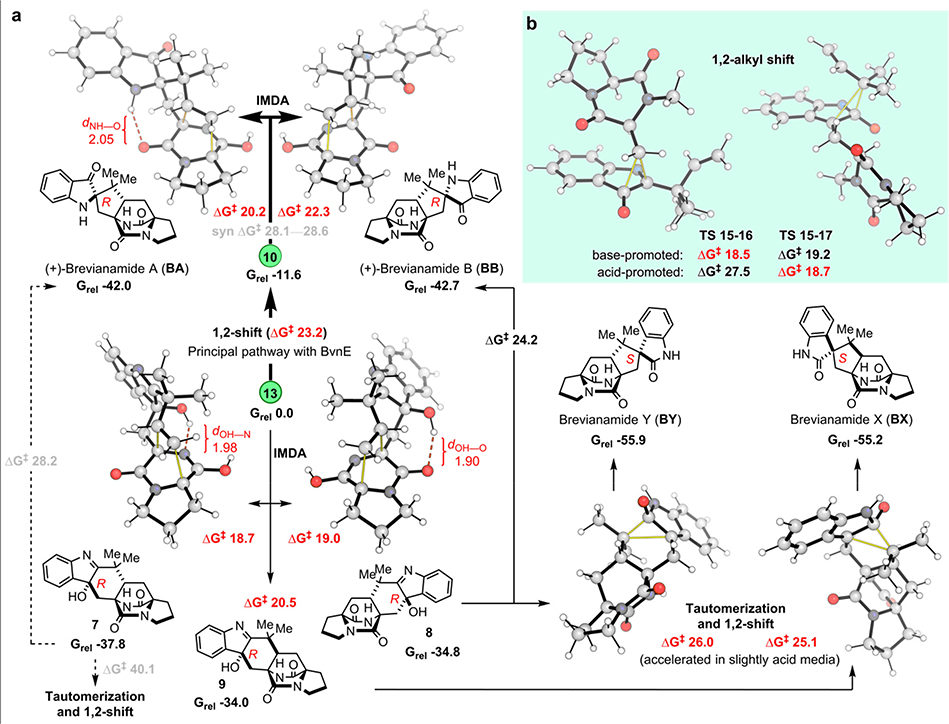
Next, we used density functional theory and coupled cluster theory calculations (SMD-DLPNO-CCSD(T)//M06–2X-D3/6–31+G(d,p)) (see Supplementary Methods) to evaluate the intermediate and transition state (TS) structures in the revised Brevianamide biosynthetic pathway (Figs. 2f and 3c), and to explore innate regio- and stereochemical preferences associated with key steps (Fig. 6a). Accordingly, under Pb-bvnE-KO conditions, the IMDA reactions that transform 13 into 7-9 are favored. In the calculated mechanism, only two diastereomers of 7 (8 and 9) undergo a subsequent 1,2-alkyl shift to irreversibly generate BY/BB (from 8), and BX (from 9). Before the shifts that lead to ring system formation, an initial tautomerization that switches the position of the OH group between C3 and C2 proceeds quickly (Supplementary Fig. 59). The reason 7 fails to undergo subsequent transformation is likely due to the high energy required for the tautomerization process (35.5 kcal·mol-1 between 7 and its tautomer) (Supplementary Fig. 60). In the computed kinetic profile, the initial irreversible IMDA (from 13 to 7-9) and CH2-dioxopiperazine shift (from 13 to 10) determine the selectivity of the process and their activation barriers are consistent with room-temperature reactivity over the course of minutes/hours. Considering that BY and BX are the major products obtained from 8 and 9, respectively, the calculated results agree qualitatively with the experimental results: the two most favorable products are 7 and BY, followed by BX; and BA and BB are minor products (experimental ratio, BY:7:BX:BA:BB = 9:7:5:1:1, Supplementary Table 4).
Fig. 6. Quantum chemical calculation results.
a, anti-Selective IMDA transition structures and products accessible from intermediates 13 and 10. b, 1,2-alkyl shift (semi-pinacol) transition structures for the migration of CH2-dioxopiperazine (LHS) and reverse-prenyl (RHS) groups. Computed Gibbs energies are in kcal/mol and highlighted distances in Å. Normal lines follow the major pathways obtained under Pb-bvnE-KO conditions. Pathways in bold correspond to the major reactivity obtained when using BvnE (switch in reactivity). Dashed lines represent pathways with prohibitively high activation energies under the reaction conditions.
The semi-pinacol rearrangement to form 3-spiro-ψ-indoxyl 10 is favorably exergonic and irreversible (ΔG −11.6 kcal·mol−1). Competition between the two possible non-enzymatic semi-pinacol rearrangements (CH2-dioxopiperazine vs. reverse-prenyl shifts) was found computationally to depend strongly on whether the reaction undergoes basic activation of the hydroxyl group or acidic activation of the imine group (Fig. 6b): whereas general/specific acid activation at nitrogen results in a buildup of positive charge in the migrating group in the TS, favoring 1,2-prenyl migration (ΔΔG‡ 8.8 kcal·mol−1), basic activation of the hydroxyl group has the opposite effect, inverting the selectivity to favor migration of the dioxopiperazine-containing group (ΔΔG‡ 0.7 kcal·mol−1). The contrast between regioselective biocatalytic conversion of 13 into 10 and the spontaneous conversion of 15 to 16/17 under acid-base conditions corroborates the involvement of hydrogen-bond acceptors Tyr113 or Tyr109 as a likely candidate for activation of the 3-hydroxyl group in BvnE (Fig. 5d, Supplementary Fig. 61) during this key step.
Next, we investigated distinct IMDA cyclizations from 10 and 13 (Fig. 6a). In each case, there are four possible stereochemical outcomes. The selectivity for anti- over syn-cycloadducts is controlled by the balance stabilizing intramolecular H-bonds and unfavorable steric interactions (Supplementary Fig. 62). Whereas syn-pathways from 10 are highly disfavored relative to anti-pathways, those from 13 form C–H···O interactions26,27, making them more competitive (Supplementary Fig. 63). In this regard, the levels of IMDA diastereoselectivity of 10 are exceptional, displaying high selectivity for the formation of BA over pseudo-enantiomeric bicyclo[2.2.2]diazaoctane BB by 2 kcal·mol−1 due to an intramolecular N–H···O hydrogen bond, with a very large (ca. 6 kcal·mol−1) preference for the two anti-configured products over their syn-diastereomers (Fig. 6a). The stereospecific conversion of 7 into BA, via a ring-contraction was found to be more challenging than the earlier 1,2-rearrangements (activation barrier of 28.2 kcal·mol−1); however, this process is exergonic by 4 kcal·mol−1. Accordingly, this ring-contraction can be accomplished under laboratory conditions by treating 7 with sodium hydroxide in water (Fig. 3c, Supplementary Fig. 64).
Conclusion
We have characterized and fully reconstituted BA/BB biosynthesis and identified a key semi-pinacolase BvnE that mediates diastereocontrol for subsequent spontaneous [4+2] cycloaddition for biogenesis of the Brevianamides. Formation of the dioxopiperazine bicyclo[2.2.2]diazaoctane core construction has also been demonstrated and the computational data support a spontaneous [4+2] pericyclic reaction. Resolution of this mechanistic mystery together with our recent characterization of the Diels-Alderase-mediated biogenesis of monooxopiperazines11 highlight the diversified biosynthetic strategies deployed by fungi for creating structurally diverse spiro-cyclized indole alkaloids. However, several important mechanistic questions remain, including: 1) what are the mechanistic details of the BvnD-catalyzed two-electron oxidation required for azadiene formation to enable the IMDA, and 2) what, if any, biogenetic relationships exist between Brevianamides and other dioxopiperazine families comprised of the bicyclo[2.2.2]diazaoctane system, such as (+)/(−)-Notoamide A whose producer genomes lack apparent bvnE homologs?
Methods
Strains and culture conditions
All Escherichia coli strains were grown at 37 °C unless otherwise specified. E. coli DH5α strain was used for vector construction and plasmid isolation using Luria-Bertani (LB) agar plates or LB liquid media. E. coli BL21 (DE3) strain was used for protein production and purification, and cultured in Terrific Broth (TB, tryptone 1.2%, yeast extract 2.4%, K2HPO3 0.94%, KH2PO3 0.22%, glycerol 4%) media. Penicillium brevicompactum NRRL 864 (Pb) was obtained from the Agricultural Research Service Culture Collection (NRRL) collection. Pb was cultured on Potato dextrose agar (PDA) plates at 28 °C for 7 d before spores collection and preservation. For Brevianamides fermentation, solid Czapek-Dox (CD) agar media were inoculated with spore suspension and incubated at 28 °C for 6 d. Aspergillus oryzae NSAR1 (Ao) was a gift from Professor Oikawa’s laboratory. Ao and its different transformants were grown on DPY (dextrin-polypeptone-yeast extract: 2% dextrin, 1% polypeptone, 0.5% yeast extract, 2% agar) agar plates at 30 °C for 7 d. For metabolites production, CMP liquid media (CD broth supplemented with 3% maltose and 1% peptone) were used. Upon inoculation, cultures were incubated at 28 °C for 5 d with shaking at 200 rpm. All strains and vectors used in this study are listed in Supplementary Table 8.
Protein expression and purification of BvnB, BvnC and BvnE
For protein expression, the E. coli BL21 (DE3) strain carrying a certain gene expression vector was used. A single colony was picked, inoculated and overnight cultured at 37 °C in LB broth with appropriate selective antibiotics (commonly 50 μg/mL kanamycin unless otherwise specified). The overnight seed culture was inoculated (1:100) into 1 L TB medium containing 4% glycerol, selective antibiotics, and 0.1% rare salt solution (FeCl3 0.68%, ZnCl2 0.05%, CoCl2 0.05%, Na2MoO4 0.05%, CaCl2 0.025%, CuSO4 0.047%, and H3BO3 0.013%). The culture was then shaking incubated at 37 °C, 220 rpm. Protein expression was induced by adding IPTG to the final concentration of 0.2 mM, when OD600 reached ~0.6 (typically ~3 h after inoculation). The cells were grown for an additional 20 h at 18 °C, 220 rpm. After cell harvesting by centrifugation (4,000 ×g, 15 min) at 4 °C, cells were frozen at −20 °C for 30 min and then melted at ambient temperature. All procedures of protein purification were carried out at 4 °C. Briefly, 40 mL of lysis buffer (NaH2PO4 50 mM, NaCl 300 mM, glycerol 10%, imidazole 5 mM, pH 7.5) was used to re-suspend the cell pellet by vortexing, and a Model 500 Sonic Dismembrator was used for ultrasonication-based cell lysis (5 sec on; 5 sec off, 1 h in total). After centrifugation at 12,000 ×g for 30 min, the supernatant was collected, to which 1 mL Ni-NTA resin (Qiagen, Germany) was added and incubated on a shaker at 30 rpm, 4 °C for 30 min. The slurry was then loaded to an empty column. Then, 100–300 mL of wash buffer (NaH2PO4 50 mM, NaCl 300 mM, glycerol 10%, imidazole 10 mM, pH 7.5) was used to wash the protein-bound resin until no protein was detectable in flow-through by Coomassie Brilliant Blue G-250 assay. Target proteins were eluted by 5 mL elution buffer (NaH2PO4 50 mM, NaCl 300 mM, glycerol 10%, imidazole 250 mM, pH 7.5) and concentrated using an Amicon Ultra centrifugal filter of appropriate molecular weight cutoff (Merck KGaA, Darmstadt, Germany) at 5,000 ×g for 30 min. The concentrated fraction (< 1 mL) was loaded onto a PD-10 column (GE Healthcare, Buckinghamshire, UK) pre-equilibrated with 25 mL desalting buffer (NaH2PO4 50 mM, glycerol 10%, pH 7.5) for buffer exchange. The aliquots of desalted fraction (100 μL per tube) were flash-frozen by liquid N2 and stored at −80 °C for later use.
Gene knockout in Pb
For targeted gene disruption in Pb, two specific homologous arms (~1 kb) were designed and amplified from Pb genomic DNA. Knockout vectors were constructed using common ligation or recombinase strategies, assembling the two arms respectively into the two multi-cloning sites (MCS) of the plasmid pRSF-hyg (Supplementary Fig. 65), which contains a hygromycin resistance marker for selection of knockout mutants. Primers used in this study are listed in Supplementary Table 9. Gene knockout was performed using the split-marker strategy28. Briefly, two knockout cassette fragments were PCR amplified using the primers x-up-F/Hyg-R (x indicates the target gene to be knocked out) and Hyg-F/x-down-R, and subsequently concentrated to >200 ng/μL. About 10 μL of the two concentrated samples were mixed and transformed into Pb protoplasts. The protoplast preparation and PEG-mediated transformation were carried out by following the procedures developed by Heneghan et al.29 with some minor modifications. Specifically, PDA and potato dextrose broth (PDB) were used as solid and liquid media. Protoplasts were collected immediately after lysing enzyme digestion. The PEG-mediated protoplast transformation was conducted on PDA plates containing 1 M sorbitol and 100 μg/mL hygromycin as the selection antibiotic. The genotypes of the knockout mutants derived from homologous recombination were verified through PCR with appropriate primers listed in Supplementary Table 9.
Heterologous gene expression in Ao
A spore suspension of Ao-WT or an Ao mutant (1.0 × 106 cells) was inoculated into 100 mL DPY medium (dextrin-polypeptone-yeast extract: 2% dextrin, 1% polypeptone, 0.5% yeast extract, 100 mL) supplemented with appropriate nutrients. After a 3-d incubation at 30 °C, 200 rpm, mycelia were collected by filtration and washed with ddH2O. Protoplasting was performed using Yatalase (Takara; 5.0 mg/mL) in Solution 1 (0.8 mM of NaCl, 10 mM of NaH2PO4, pH 6.0) at 30 °C for 2 h. Protoplasts were centrifuged at 900 ×g (Beckman JLA10.500) for 5 min and washed with 0.8 M of sterile NaCl solution. Then, protoplasts were adjusted to 2.0 × 108 cells/mL by adding Solution 2 (0.8 M of NaCl, 10 mM of CaCl2, 10 mM of Tris-HCl, pH 8.0) and Solution 3 (40% (w/v) of PEG4000, 50 mM of CaCl2, 50 mM of Tris-HCl, pH 8.0) in a 4:1 volume ratio. To the protoplast solution (200 μL) appropriate plasmid (13 μg) was added. The aliquot was incubated on ice for 20 min, to which Solution 3 (1 mL) was added. After 20 min incubation at room temperature, Solution 2 (10 mL) was added to the mixture and the mixture was centrifuged at 900 ×g (Beckman JLA10.500) for 5 min. The transformation mixture was poured onto CD agar plates supplemented with 0.8 M of NaCl and appropriate nutrients, and then overlaid with the soft-top agar (1.2 M of sorbitol, 3.5% of CD broth, 0.6% of agar). The plates were incubated at 30 °C for 3–7 d until colonies showed up.
In vitro enzyme assays
Unless otherwise specified, all enzymatic assays were carried out in 100 μL of 50 mM Tris·HCl buffer (pH 7.5) at 30 °C for 2 h. The enzyme reaction mixture contained 1 mM substrate(s) (BF and DMAPP for BvnC, DE or 14 for BvnB, 15 for BvnE (or its mutants), necessary cofactors (10 mM Mg2+ for BvnC, 2 mM NADPH for BvnB), and 10 μL of each purified enzyme.
Mutagenesis of BvnE
The mutagenesis PCR reaction contained 50 ng pET28b-bvnE as template, 0.2 μM primer (in our system, only one primer is enough for successful mutagenesis), 2.5 μL 10× Pfu buffer, 0.5 μL dNTPs mix (25 mM of each dNTP), 1 mL DMSO and 0.5 μL PfuUltra II fusion HS DNA polymerase (Agilent, USA) in a total volume of 25 μL. The PCR cycles were set as follows: 1) 95°C for 3 min, 2) 95°C for 30 s, 3) 53°C for 1 min, 4) 65°C for 2 min, and steps 2–4 repeated for 30 cycles. DpnI digestion contained 1 μL 2 U/μL DpnI and 25 μL PCR reaction solution for 2 h at 37°C. From the digested solution, 1 μL was taken to transform 30 μL DH10β Electrocompetent Competent cell (Takara, Japan). The mutant plasmids were purified and subject to sequencing at the University of Michigan DNA Sequencing Core (Michigan, USA).
BvnE protein expression and purification for crystallization
E. coli BL21(DE3) pRARE2 were transformed with the plasmid (BvnE_pET28H8-MBP-T) and grown in 1 L Terrific Broth medium with 50 μg/mL kanamycin and 100 μg/mL spectinomycin at 37 °C to OD600 ~1. The culture was shifted to 20 °C over 1 h, induced with 0.25 mM IPTG, and incubated for 24 h (20 °C, 225 rpm shaking). Cells were harvested by centrifugation (4 °C, 5,000 ×g) and stored at −20 °C. All BvnE purification steps were performed at 4 °C. The cell pellet was resuspended in lysis buffer (50 mM Tris pH 7.5, 300 mM NaCl, 20 mM imidazole, 10 % glycerol, 0.5 mg/mL lysozyme, 0.05 mg/mL DNase, 5 mM MgCl2) and lysed by sonication followed by high speed centrifugation (4 °C, 60,000 ×g, 25 min). The lysate was filtered (0.45 μm) and loaded on a Fast Protein Liquid Chromatography (FPLC) Ni-NTA Histrap 5mL column and washed with 20 CV Ni-NTA buffer (50 mM Tris pH 7.5, 300 mM NaCl, 20 mM imidazole, 10 % glycerol) at 3 mL/min. BvnE was eluted with elution buffer (50 mM Tris pH 7.5, 300 mM NaCl, 500 mM imidazole, 10 % glycerol). Pooled fractions were incubated with His-tagged tobacco etch virus (TEV) protease in a 1:20 molar ratio to remove the His-maltose binding protein (MBP)-tag. The protein was dialyzed overnight against 50 mM Tris pH 7.5, 300 mM NaCl, 2 mM DTT, and passed through a Ni-NTA column to remove TEV protease and MBP. Further purification was accomplished by size-exclusion chromatography using a Superdex 75 10/300 GL column equilibrated with 50 mM Tris pH 7.5, 300 mM NaCl. SDS-PAGE was used to assess protein homogeneity; BvnE was >95 % pure. Protein was concentrated using the Amicon Ultra Centrifugal Concentrator (10,000 NMWL) to 29 mg/mL. The concentration was determined by UV using the calculated molar extinction coefficient.
Crystallization and structure determination
BvnE was screened for initial crystallization conditions using the Midwest Center Structural Genomics (MCSG) screen at 25 and 4 °C. Single, diffraction-quality crystals of BvnE were grown by sitting drop vapor diffusion at 4 °C by mixing 29 mg/mL BvnE with well solution (26% PEG 3350, 0.1 M Tris pH 8.5, 4% v/v Pentaerythritol ethoxylate (3/4 EO/OH)) in a 1:1 ratio. Sitting droplets were nucleated from an earlier spontaneous crystallization using a cat whisker. 8 μL of cryo-protectant (12.75 % glycerol, 26% PEG 3350, 0.1 M Tris pH 8.5, 4% v/v Pentaerythritol ethoxylate (3/4 EO/OH), 50 mM Tris pH 7.5, 300 mM NaCl) was added directly to the sitting drops, and the crystals were harvested using nylon loops and vitrified by rapid plunging into liquid nitrogen. Diffraction data were collected at beamline 23-ID-B at the Advanced Photon Source (APS) using an X-ray wavelength of 1.033 Å (360° of data, 100 K, 0.2° image width). Data were integrated and scaled with XDS30. The structure was solved by molecular replacement using PrhC (PDB id: 5X9J) as a search model (Phaser-MR)31. The final model was completed using alternate cycles of manual model building using Coot32 and refinement using PHENIX.refine33. The structure was validated with MolProbity34. Data collection and refinement statistics are listed in Supplementary Table 7.
Molecular docking of BvnE with compound 13
Compound 13 was docked into the BvnE structure with the automated docking software Autodock435. The conformational heterogeneity of key active site residues Arg38, Tyr113 and Glu131 experimentally observed in the electron density/crystal structure was incorporated into the docking model. Docking was performed with Arg38, Tyr113 and Glu131 as flexible residues in effort to obtain lowest energy conformations.
Data Availability
Sequence data referenced in this study are available in GenBank with the accession number MN401751. Coordinates and associated structure factors have been deposited in Protein Data Bank (PDB) with the PDB ID 6U9I (BvnE) and in the Cambridge Crystallographic Data Centre (CCDC) with the CCDC numbers 1973959 (BX), 1973957 (BY), and 1973958 (15). Molecular coordinates as well as absolute and relative thermochemistry data have been provided for the computational studies. All other data are available from corresponding author on reasonable request.
Extended Data
Extended Data Figure 1. Representative fungal bicyclo[2.2.2]diazaoctane indole alkaloids.
Compounds with 3-spiroψ–indoxyls, spiro-2-oxindoles and 3-hydroxyindolenines are presented in red, blue and green, respectively.
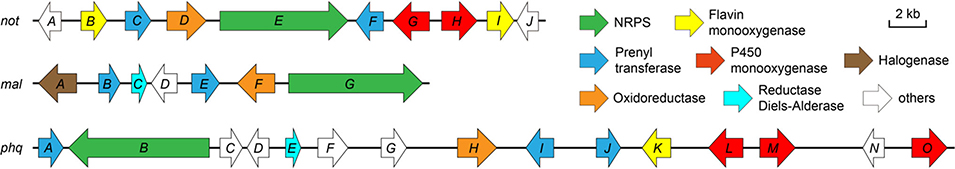
Extended Data Figure 2. The biosynthetic gene clusters for representative fungal bicyclo[2.2.2]indole alkaloids.
The gene clusters not, mal and phq are responsible for biosynthesis of Notoamides, Malbrancheamides and Parapherquamides, respectively.
Extended Data Figure 3. Biogenetic proposals for Brevianamide A (BA) and Brevianamide B (BB).
a, Early biosynthetic proposal suggested and interrogated by Williams et. al.13 b, Original biogenesis proposed by Porter and Sammes (via 7).14 c, More recent biosynthetic proposals suggested by Williams et al..5,6,10 Several biogenetic hypotheses based on the pioneering proposal first suggested by Porter and Sammes in 197014 reasoned that the bicyclo[2.2.2]diazaoctane core arises via an intramolecular [4+2] hetero-Diels-Alder (IMDA) construction.5–10,36 We experimentally interrogated the biogenetic proposal a, through the synthesis of 13C-labeled putative Diels-Alder products (±)-20, but could not detect incorporation into either BA or BB in cultures of Pb. Based on these results, we then suggested the biosynthetic pathways illustrated in b and c. The fundamental difference between the biogenesis described in a, and that in b and c, is the sequential timing of the indole oxidation, the semi-pinacol rearrangement and the crucial IMDA reactions. Thus, it remained conceivable that oxidation of DE to the (R)-hydroxyindolenine provides species 11, which can suffer several fates. One is N-C ring closure to co-metabolite Brevianamide E (BE); a second possibility shown in b is oxidation and tautomerization to the azadiene species 13, which can suffer IMDA cyclization providing 7 and 8, then undergo a final semi-pinacol rearrangement to furnish BA and BB (route i). Alternatively, azadiene 13 could first suffer semi-pinacol rearrangement to 10, which then undergoes the IMDA construction to generate BA and BB (route ii). Another possibility c involves (R)-hydroxyindolenine 11, which proceeds through a semi-pinacol rearrangement to the indoxyl species 19, followed by further oxidation to the azadiene species 10 and subsequent IMDA to furnish BA and BB. Experimental supports to distinguish between these proposed pathways, all of which embrace the relative and absolute stereochemistry of BA and BB, have remained unresolved until this work.
Extended Data Figure 4. Elucidation of absolute configurations.
a, Experimental and computational ECD spectra of 7. b, Experimental and computational ECD spectra of 16. c, Experimental and computational ECD spectra of 17. d, X-ray ORTEP diagram of BX. e, X-ray ORTEP diagram of BY. f, X-ray ORTEP diagram of 15.
Extended Data Figure 5. Enzyme properties of BvnE using compound 15 as a substrate.
a, pH dependency. b, temperature dependency. c, Michaelis-Menton kinetic analysis (centre values, means; error bars, standard deviations; n = 2).
Extended Data Figure 6. Docking results and CD spectra of BvnE.
a, Docking complex of BvnE with compound 13 (magenta). b, CD spectra of purified wild-type and mutant BvnE proteins. c, Electron density (2Fo-Fc, 1σ) for the active site residues of BvnE (PDB ID code: 6U9I) shows conformational flexibility at Arg38 and Glu131 suggestive of potential active site remodeling during catalysis. Arg38 (red box) which is essential for catalytic activity does not make direct interactions with the docked ligand or active site residues. However, the structure reveals two ordered water molecules (red spheres) in a hydrogen bonding network between Arg38 and Glu131. These binding poses reveal possible key interactions between the ligand and BvnE.
Supplementary Material
Acknowledgements
This work was supported by the National Key Research and Development Program (2019YFA0905100 and 2019YFA0706900), the National Natural Science Foundation of China (21472204 and 31872729 to S.L. and 31800041 to L.D.), the National Institutes of Health R35 GM118101, the Hans W. Vahlteich Professorship (to D.H.S.), R01 CA070375 (to R.M.W. and D.H.S.), the Shandong Provincial Natural Science Foundation (ZR2019ZD22 to S.L. and ZR201807060986 to L.D.), the National Postdoctoral Innovative Talents Support Program (BX20180325 to L.D.), China Postdoctoral Science Foundation (2019M652500 to L.D.), and funding from NSF grant CHE-0840456 (for X-ray instrumentation). GM/CA@APS is supported by the National Institutes of Health, National Institute of General Medical Sciences (AGM-12006) and National Cancer Institute (ACB-12002). We also thank Professor Janet L. Smith from University of Michigan for her assistance with the structure determination of BvnE. R.S.P. acknowledges computational resources from the RMACC Summit supercomputer supported by the National Science Foundation (ACI-1532235 and ACI-1532236), the University of Colorado Boulder and Colorado State University, and the Extreme Science and Engineering Discovery Environment (XSEDE) through allocation TG-CHE180056. XSEDE is supported by the National Science Foundation (ACI-1548562). The authors thank Ms. Xiaoju Li and Ms. Haiyan Sui from The State Key Laboratory of Microbial Technology, Shandong University for the assistance in X-ray single crystal diffractometer.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Klas KR et al. Structural and stereochemical diversity in prenylated indole alkaloids containing the bicyclo 2.2.2 diazaoctane ring system from marine and terrestrial. Nat. Prod. Rep 35, 532–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birch AJ & Wright JJ The brevianamides: a new class of fungal alkaloid. J. Chem. Soc. D, 644b-645 (1969). [Google Scholar]
- 3.Li S et al. Comparative analysis of the biosynthetic systems for fungal bicyclo 2.2.2 diazaoctane indole alkaloids: the (+)/(−)-notoamide, paraherquamide and malbrancheamide pathways. MedChemComm 3, 987–996 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finefield JM, Frisvad JC, Sherman DH & Williams RM Fungal origins of the bicyclo 2.2.2 diazaoctane ring system of prenylated indole alkaloids. J. Nat. Prod 75, 812–833 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanzcervera JF, Glinka T & Williams RM Biosynthesis of the brevianamides: quest for a biosynthetic Diels-Alder cyclization. J. Am. Chem. Soc 115, 347–348 (1993). [Google Scholar]
- 6.Sanzcervera JF, Glinka T & Williams RM Biosynthesis of brevianamides A and B: in search of the biosynthetic Diels-Alder construction. Tetrahedron 49, 8471–8482 (1993). [Google Scholar]
- 7.Williams RM, Sanz-Cervera JF, Sancenon F, Marco JA & Halligan K Biomimetic Diels-Alder cyclizations for the construction of the brevianamide, paraherquamide sclerotamide, and VM55599 ring systems. J. Am. Chem. Soc 120, 1090–1091 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Adams LA, Valente MWN & Williams RM A concise synthesis of d,l-brevianamide B via a biomimetically-inspired IMDA construction. Tetrahedron 62, 5195–5200 (2006). [Google Scholar]
- 9.Williams RM & Cox RJ Paraherquamides, brevianamides, and asperparalines: laboratory synthesis and biosynthesis. An interim report. Acc. Chem. Res 36, 127–139 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Stocking EM & Williams RM Chemistry and biology of biosynthetic Diels-Alder reactions. Angew. Chem. Int. Ed 42, 3078–3115 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Dan Q et al. Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels–Alderase. Nat. Chem 11, 972–980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams RM, Glinka T & Kwast E Facial selectivity of the intramolecular SN2’ cyclization: stereocontrolled total synthesis of brevianamide B. J. Am. Chem. Soc 110, 5927–5929 (1988). [Google Scholar]
- 13.Williams RM, Kwast E, Coffman H & Glinka T Remarkable, enantio-divergent biogenesis of brevianamide A and B. J. Am. Chem. Soc 111, 3064–3065 (1989). [Google Scholar]
- 14.Porter AEA & Sammes PG A Diels-Alder reaction of possible biosynthetic importance. J. Chem. Soc. D, 1103a (1970). [Google Scholar]
- 15.Li F et al. Determination of taichunamide H and structural revision of taichunamide A. Org. Lett 20, 1138–1141 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Greshock TJ et al. Isolation, structure elucidation, and biomimetic total synthesis of versicolamide B, and the isolation of antipodal (−)-stephacidin A and (+)-notoamide B from Aspergillus versicolor NRRL 35600. Angew. Chem. Int. Ed 47, 3573–3577 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kai A, Kato H, Sherman DH, Williams RM & Tsukamoto S Isolation of a new indoxyl alkaloid, Amoenamide B, from Aspergillus amoenus NRRL 35600: biosynthetic implications and correction of the structure of Speramide B. Tetrahedron Lett 59, 4236–4240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Zhang X, Nong X, Wang J & Qi S Brevianamides and mycophenolic acid derivatives from the deep-sea-derived fungus Penicillium brevicompactum DFFSCS025. Mar. Drugs 15, 43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfrey RC, Green NJ, Nichol GS & Lawrence AL Chemical synthesis of (+)-brevianamide A supports a Diels–Alderase-free biosynthesis (2019). Doi: 10.26434/chemrxiv.8224148.v1. [DOI] [Google Scholar]
- 20.Mori T et al. Molecular basis for the unusual ring reconstruction in fungal meroterpenoid biogenesis. Nat. Chem. Biol 13, 1066–1073 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Li S et al. Biochemical characterization of NotB as an FAD-dependent oxidase in the biosynthesis of notoamide indole alkaloids. J. Am. Chem. Soc 134, 788–791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Y et al. Genome-based characterization of two prenylation steps in the assembly of the stephacidin and notoamide anticancer agents in a marine-derived Aspergillus sp. J. Am. Chem. Soc 132, 12733–12740 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato N et al. Identification of cytochrome P450s required for fumitremorgin biosynthesis in Aspergillus fumigatus. ChemBioChem 10, 920–928 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Li H et al. HPLC-DAD-Directed Isolation of Linearly Fused Prenylated Indole Alkaloids from a Soil-Derived Aspergillus versicolor. J. Nat. Prod 82, 2181–2188 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Matsuda Y et al. Discovery of key dioxygenases that diverged the paraherquonin and acetoxydehydroaustin pathways in Penicillium brasilianum. J. Am. Chem. Soc 138, 12671–12677 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Sutor DJ The C–H… O Hydrogen Bond in Crystals. Nature 195, 68–69 (1962). [Google Scholar]
- 27.Schwalbe CH June Sutor and the C–H···O hydrogen bonding controversy. Crystallography Reviews 18, 191–206 (2012). [Google Scholar]
- 28.Goswami RS Targeted gene replacement in fungi using a split-marker approach. Methods Mol. Biol 835, 255–269 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Heneghan MN et al. First Heterologous Reconstruction of a Complete Functional Fungal Biosynthetic Multigene Cluster. ChemBioChem 11, 1508–1512 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W XDS. Acta Crystallogr. Sect. D. Biol. Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ et al. Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. Sect. D. Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D. Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Struct. Biol 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris GM et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem 30, 2785–2791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams RM, Sanz-Cervera JF, Sancenon F, Marco JA & Halligan KM Biomimetic Diels-Alder cyclizations for the construction of the brevianamide, paraherquamide, sclerotamide, asperparaline and VM55599 ring systems. Biorg. Med. Chem 6, 1233–1241 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data referenced in this study are available in GenBank with the accession number MN401751. Coordinates and associated structure factors have been deposited in Protein Data Bank (PDB) with the PDB ID 6U9I (BvnE) and in the Cambridge Crystallographic Data Centre (CCDC) with the CCDC numbers 1973959 (BX), 1973957 (BY), and 1973958 (15). Molecular coordinates as well as absolute and relative thermochemistry data have been provided for the computational studies. All other data are available from corresponding author on reasonable request.