Abstract
Patients with the pre-leukemia bone marrow failure syndrome called severe congenital neutropenia (CN) have an approximately 15% risk of developing acute myeloid leukemia (AML; called here CN/AML). Most CN/AML patients co-acquire CSF3R and RUNX1 mutations, which play cooperative roles in the development of AML. To establish an in vitro model of leukemogenesis, we utilized bone marrow lin− cells from transgenic C57BL/6-d715 Csf3r mice expressing a CN patient–mimicking truncated CSF3R mutation. We transduced these cells with vectors encoding RUNX1 wild type (WT) or RUNX1 mutant proteins carrying the R139G or R174L mutations. Cells transduced with these RUNX1 mutants showed diminished in vitro myeloid differentiation and elevated replating capacity, compared with those expressing WT RUNX1. mRNA expression analysis showed that cells transduced with the RUNX1 mutants exhibited hyperactivation of inflammatory signaling and innate immunity pathways, including IL-6, TLR, NF-kappaB, IFN, and TREM1 signaling. These data suggest that the expression of mutated RUNX1 in a CSF3R-mutated background may activate the pro-inflammatory cell state and inhibit myeloid differentiation.
Electronic supplementary material
The online version of this article (10.1007/s00277-020-04194-0) contains supplementary material, which is available to authorized users.
Keywords: Severe congenital neutropenia, Pre-leukemia bone marrow failure syndrome, G-CSFR mutations, RUNX1 mutations
Introduction
Patients with the inborn pre-leukemia bone marrow failure syndrome called severe congenital neutropenia (CN) have a very low level (less than 500 cells/μl blood) or even a complete lack of mature neutrophilic granulocytes in the peripheral blood which is caused by blockade of the terminal differentiation of bone marrow myeloid progenitor cells at the promyelocytes/myelocyte stage [1, 2]. In most CN patients, this granulocyte differentiation defect can be successfully treated by daily subcutaneous administration of recombinant human granulocyte colony–stimulating factor (G-CSF). Approximately 15% of CN patients develop myelodysplastic syndrome or acute myeloid leukemia (MDS or AML). Inherited mutations in the ELANE, HAX1, G6PC3, SRP54, GFI1, and JAGN1 genes cause CN, and leukemic progression has been seen in CN patients of all genetic groups [1].
CSF3R mutations resulting in the production of truncated G-CSFR proteins that lack from one to four phospho-tyrosine residues and exhibit defective receptor internalization were reported in a majority of CN patients with overt AML or MDS [3–9]. However, transgenic d715 Csf3r mice lacking three tyrosines do not develop AML or MDS [3–9], suggesting that additional genetic alterations in combination with CSF3R mutation are needed for the progression of AML. We recently examined a large cohort of CN/AML patients (31 patients) and found cooperative acquired mutations of CSF3R and RUNX1 (runt-related transcription factor 1) in 55% of CN patients with overt AML or MDS [10]. However, the detailed mechanism underlying the leukemogenic transformation downstream of CSF3R and RUNX1 mutations remained unknown.
Acquired mutations in RUNX1 occur in AML, mostly secondary to MDS, radiation therapy, or chemotherapy [11–16]. Most RUNX1 mutations are acquired heterozygous point mutations; they are predominantly located in the Runt homology/DNA-binding (RHD) or transactivation (TAD) domains. Interestingly, a majority of patients with familial platelet disorder (FPD) and a predisposition for AML have germline RUNX1 mutations [17]. Some FPD patients with overt AML gain additional RUNX1 mutations [17]. Among the described groups of AML patients, the incidence of acquired RUNX1 mutations is the highest in CN/AML patients. RUNX1 mutations in CN/AML patients are distributed throughout the RHD (primarily) and TAD of the RUNX1 protein, and some hot spot positions have been noted [10]. For example, amino acid residues 139 and 174 of the RUNX1 protein were found to be mutated in four and three CN/AML patients, respectively [10] (data not shown). The functional outcomes of RUNX1 mutations at different positions have not yet been clearly defined, but we speculate that they may affect the DNA binding of RUNX1 to target genes or the protein–protein interactions, intracellular localization, protein stability, and/or post-translational modification(s) of RUNX1.
The role of inflammation in cancer was first mentioned in 1863 by Virchow [18]. A growing body of research suggests that pro-inflammatory signaling acts through diverse mechanisms to increase the proliferation rate of hematopoietic stem and progenitor cells (HSPCs), which induces genotoxicity, increases survival, and produces pre-leukemia stem cells (pre-LSCs) with a high likelihood of leukemic transformation [19].
In the present study, we sought to establish an in vitro experimental model to study the intracellular mechanisms of leukemia development downstream of CSF3R and RUNX1 mutations. Using this model, we identified upregulation of an inflammatory signature signaling in mouse HSPCs expressing mutated CSF3R and RUNX1. This expression signature may predispose CN patients toward leukemic transformation.
Material and methods
Mice
Male d715 Csf3r mice on the C57BL/6J background have been described previously [9]. Mice were housed under pathogen-free conditions in the animal facility of Tübingen University.
Cell purification and separation
Mouse bone marrow cells were isolated by flushing the long bones with ice-cold PBS. Bone marrow mononuclear cells were isolated by Ficoll–Hypaque gradient centrifugation (Amersham Biosciences) and positively selected bone marrow lin− cells by immunomagnetic labeling with corresponding MACS beads (Miltenyi Biotec). Cells were counted, and viability was assessed by Trypan blue dye exclusion.
Generation of the lentiviral vectors expressing WT or mutant RUNX1 cDNA
To generate RUNX1 mutants, we performed site-directed mutagenesis. As a template, we used Lego-iG/Puro-RUNX1-wt-CTAP plasmid expressing human wild type (WT) RUNX1 generously provided by Dr. Boris Fehse and Dr. Carol Stocking. Specific primers to introduce mutations p.Arg139Gly (R139G) and p.Arg174Leu (R174L) in wild type RUNX1 nucleotide sequence were designed using the QuickChange Primer Design tool (https://www.agilent.com/store/primerDesignProgram.jsp). Primer sequences are available upon request. Lego-iG/Puro-RUNX1-R139G-CTAP and Lego-iG/Puro-RUNX1-R174L-CTAP plasmids were generated using the QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Inc.) according to the manufacturer’s instruction. RUNX1 cDNAs (WT and two mutants) were subsequently re-cloned into lentiviral pRRL.PPT.CBX3.SFFV.hRUNX1.i2.EBFP.puro.pre vector. Transgene expression was controlled by the spleen focus–forming virus promoter (SFFV) juxtaposed to the minimal ubiquitous chromatin opening element (CBX3). EBFP translation was initiated by an internal ribosomal entry side (i2).
Transduction of cells
Lin− cells were cultured in a hematopoietic stem cell expansion medium consisting of Stemline II medium supplemented with Pen/Strep, 10% FCS (Sigma-Aldrich), 1 μm dexamethasone, 100 ng/ml of mSCF, 4 ng/ml of mIL-3, 10 ng/ml of hIL-6, 40 ng/ml of murine IGF-1, and 20 ng/ml of human Flt-3L at 2 × 105 cells/ml for 2 days. Cells were transduced at MOIs of 5–10 in the presence of polybrene (5 μg/ml) and re-transduced after 12–24 h. Percentage of EBFP+ cells was assessed by FACS.
Detection of the human RUNX1 protein in transduced bone marrow lin− cells from d715 Csf3r mice using western blotting
A total of 1 × 106 transduced bone marrow lin− cells from d715 Csf3r mice were lysed in 200 μl 3 × Laemmli buffer, and protein was denatured for 10 min at 95 °C. Five microliter of cell lysate in Laemmli buffer was loaded per lane. Proteins were separated on a 12.5% polyacrylamide gel and transferred on a nitrocellulose membrane (GE Healthcare) (1 h, 100 V, 4 °C). The membrane was blocked for 1 h in 5% milk and incubated with primary rabbit anti-human and anti-mouse RUNX1-specific (Cell Signaling Technology #4334) or GAPDH (Cell Signaling Technology, #2118) antibody overnight (at 4 °C). After that, membranes were washed and incubated with secondary HRP-conjugated antibody (Cell signaling, #7074) for 1 h at room temperature. Pierce ECL solution (Thermo Fisher) and Amersham Hyperfilms were used to detect chemiluminescence signal of proteins.
Liquid culture differentiation of transduced lin− cells
A total of 2 × 105 of transduced cells/ml were incubated for 7 days in RPMI 1640 GlutaMAX supplemented with 10% FCS, 10 ng/ml hIL-6, 5 ng/ml IL-3, 5 ng/ml GM-CSF, and 10 ng/ml G-CSF. The medium was exchanged every second day. On day 7, the medium was changed to RPMI 1640 GlutaMAX supplemented with 10% FBS, 1% penicillin/streptomycin, and 10 ng/ml G-CSF. The medium was exchanged every second day until day 11. On day 11, cells were analyzed by FACS using the following antibody: rat anti-mouse Gr-1 (BD 553128) and rat anti-mouse CD11b (BD 553312) on FACSCanto II.
Colony-forming unit assay
A total of 1000 transduced EBFP+ cells were plated directly after transduction in 1 ml methylcellulose medium (MethoCult GF M3434; StemCell Technologies) supplemented with 10 ng/ml of G-CSF. After 14 days of culture, the numbers of CFU-G, CFU-GM, and BFU-E colonies were counted. Cells were collected for the colony replating experiments, washed 3 times with PBS, and plated 1000 cells/dish in new methylcellulose for an additional 2 weeks (1st replating). The procedure was repeated one more time (2nd replating).
Microarray-based mRNA expression analysis
After 48 h of lentiviral transduction, lin− cells were starved for 24 h and subsequently treated with 10 ng/ml of G-CSF for 24 h. After that, transduced cells were sorted, and mRNA was isolated using the RNeasy Mini Kit (Qiagen, #74106) according to the manufacturer’s instructions.
RNA from transduced lin− cells was subjected to microarray analysis using the Affymetrix Microarray Platform. The GeneChip WT cDNA Synthesis and Amplification Kit was used to make double-stranded cDNA from total RNA, which was then labeled with biotin (GeneChip WT Terminal Labeling Kit). After chemical fragmentation of the biotin-labeled cDNA targets, they were hybridized to the GeneChip Mouse Gene 2.0 ST Array using the Fluidics Station 450 and scanned using the Affymetrix GeneChip Scanner 3000 with the GeneChip Operating Software 1.4 (Affymetrix, Santa Clara, CA). Data analysis was performed using Affymetrix Expression Console Version 1.1 for invariant set normalization, and the Ingenuity Pathway Analysis (IPA) software (Qiagen) was used for identification of differentially expressed genes. Motif activity response analysis (MARA) was conducted using the Integrated System for Motif Activity Response Analysis (ISMARA); CEL files were uploaded to the server using the web interface.
Cytospin preparation, staining, and microscopic image acquisition
After sorting, 1 × 104 cells were centrifuge onto microscope slides at 250 rpm for 3 min. The slides were air-dried, and subsequent Wright–Giemsa staining was carried out. Images where acquired on a Nikon Eclipse TS100. Cells were covered with oil, and images were collected at × 630 magnification.
qRT-PCR
RNA isolation was performed using the RNeasy Micro Kit (Qiagen). cDNA was synthesized from 1 μg total RNA with the Omniscript RT Kit (Qiagen). qRT-PCR was conducted with SYBR Green qPCR master mix (Roche) on a Light Cycler 480 (Roche). Target genes were normalized to ACTB. Primers are available upon request.
LSK cell analysis
A total of 2 × 105 cells were incubated with FcR blocking antibody (BioLegend #101320), 7AAD, and lineage cocktail biotin–conjugated antibody (BioLegend #79750, #79748, #79752, #79748, #79749), and stained with APC-Cy7-conjugated streptavidin (BioLegend #405208), anti-mouse Sca-1 BV510–conjugated antibody (BioLegend, #108129), and anti-mouse-c-KIT APC–conjugated antibody (BioLegend, #105812). After staining and washing, cells were analyzed on a CANTO II (BD) flow cytometer.
Statistics
Statistical analysis was performed using a two-sided unpaired Student t test for the analysis of differences in mean values between groups.
Results
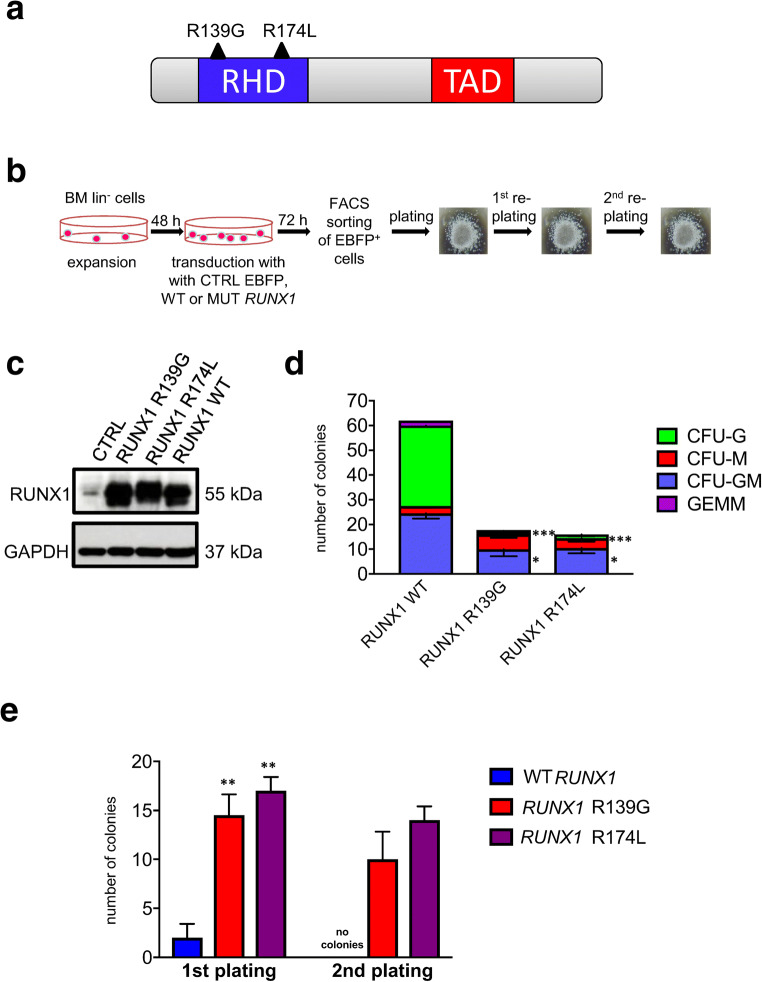
Diminished formation of myeloid colonies and elevated replating capacity of d715 Csf3r lin− cells transduced with mutated RUNX1
We isolated bone marrow lin− cells from d715 Csf3r mice and transduced these cells with lentiviral vectors encoding EBFP and RUNX1 wild type (RUNX1-WT), or mutants, RUNX1-R139G, or RUNX1-R174L. Amino acids 174 and 139 were found to be “hot spots” for mutation in the RHD domain of the RUNX1 protein, as they were detected in four and three CN-AML patients, respectively (Fig. 1a) [10] (data not shown). At 72 h post-transduction, we performed colony-forming units and replating experiments of sorted EBFP+ cells (Fig. 1b). We observed comparable expression of human WT and mutant RUNX1 proteins in transduced cells (Fig. 1c). Low levels of endogenous murine runx1 protein were detected in BFP+ control transduced cells (Fig. 1c). Interestingly, d715 Csf3r lin− cells transduced with mutated RUNX1 had markedly diminished capacities to form myeloid colonies, including CFU-G and CFU-GM, as compared with cells transduced with WT RUNX1 (Fig. 1d). In line with these findings, CFU replating experiments showed that cell transduced with each RUNX1 mutant had markedly higher replating capacities than RUNX1 WT–transduced cells (Fig. 1e).
Fig. 1.
In HSPCs of d715 Csf3r mice, RUNX1 mutations decrease CFU-G and CFU-GM formation but increase the replating capacity. a Schematic of RUNX1 protein showing the location and amino acid changes of the mutations, which are indicated by black triangles. The functionally important Runt homology DNA-binding domain (RHD) is shown in blue, and the transactivation domain (TAD) is shown in red. b Schematic of CFU experiments performed using transduced bone marrow lin− cells of C57BL/6-d715 Csf3r mice. c Representative WB images of lin− cells of C57BL/6-d715 Csf3r mice transduced with corresponding lentiviral constructs. GAPDH was used as loading control. d CFU assay and e replating CFU assay of transduced C57BL/6-d715 Csf3r lin− cells. Data represent means ± SD from triplicates of two independent experiments; *p < 0.05, **p < 0.01,*** p < 0.001
Reduced liquid culture myeloid differentiation of d715 Csf3r lin− cells transduced with mutated RUNX1
We next compared the G-CSF-triggered myeloid differentiation of transduced d715 Csf3r lin− cells in vitro. Transduced cells were cultured in liquid culture myeloid differentiation medium for 11 days (Fig. 2a). FACS was used to count the absolute numbers of myeloid CD11b+ and Gr-1+ cells on day 11 of liquid culture, and the results showed that d715 Csf3r lin− cells transduced with each of the RUNX1 mutants exhibited reduced myeloid differentiation, compared with WT RUNX1–overexpressing samples (Fig. 2b). At the same time, no marked difference was observed in the absolute numbers of BFP+ cells between groups transduced with WT RUNX1 or with each of RUNX1 mutants on days 3 and day 7 of liquid culture differentiation. On day 11 of culture, numbers of BFP+ cells transduced with RUNX1-R139G mutant were significantly (p < 0.05) increased, as compared with RUNX1 WT– or RUNX1-R174L–transduced samples (Fig. 2c).
Fig. 2.
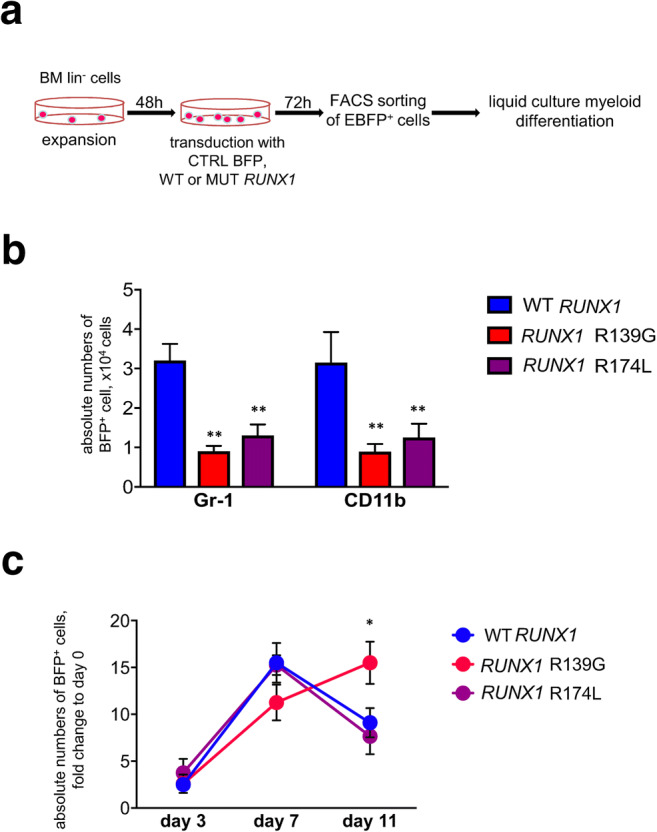
Liquid culture differentiation of transduced d715 Csf3r HSPCs. a Schematic of the workflow for liquid culture myeloid differentiation of transduced bone marrow lin− cells obtained from C57BL/6-d715 Csf3r mice. b Transduced cells were subjected to liquid culture myeloid differentiation (see “Material and methods” for details). FACS was used to count the myeloid and granulocytic cells on day 11 of culture. Graph bars represent absolute cell counts of Gr-1+ or CD11b+ cells. Data represent means ± SD from triplicates of two independent experiments; **p < 0.01
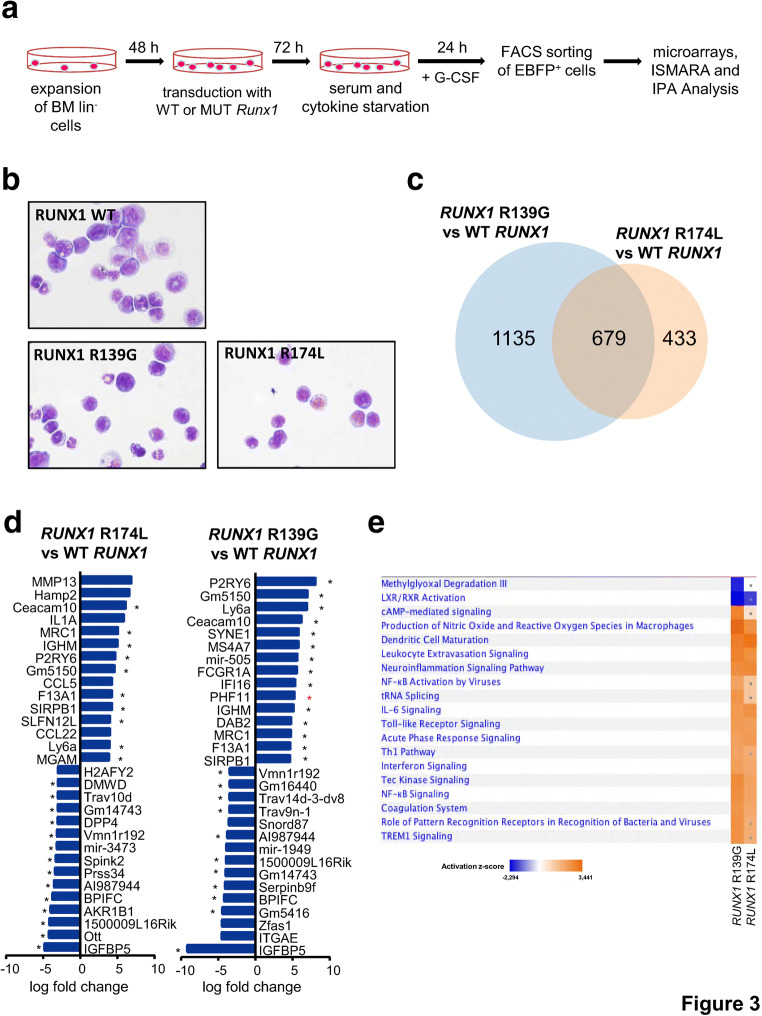
RUNX1 missense mutations induce inflammatory signaling pathways in G-CSF-treated d715 Csf3r hematopoietic cells
We further aimed to identify intracellular signaling pathways downstream of csf3r and RUNX1 mutations that may affect the in vitro proliferation and myeloid differentiation of mouse HSPCs. The d715 Csf3r lin− cells were expanded in HSPC expansion medium and transduced with lentiviral vectors carrying RUNX1 WT, RUNX1-R139G, or RUNX1-R174L. Transduced cells were starved for 24 h and then treated with G-CSF for 24 h. EBFP+ cells were sorted in RLT buffer, and mRNA expression was evaluated using an Affymetrix MoGene 2.0 Chip (Fig. 3a). Representative images of Wright–Giemsa–stained cytospins prepared from sorted cells show no difference in the cell morphology between studied groups: all samples show immature cell morphology (Fig. 3b). These data suggest that the differences in mRNA expression should not contribute to a strong diversity in the cell composition between studied groups.
Fig. 3.
Canonical pathway analysis of microarray data obtained from transduced d715 Csf3r HSPCs. a Schematic of the experimental procedure performed for microarray analysis. The experiment was conducted in duplicate. After quality control analysis, one sample from the RUNX1-R139G group was excluded from the final analysis. b Wright–Giemsa–stained cytospin preparations of Csf3r lin− cells transduced with RUNX1 WT and mutants and sorted for fluorescent protein expression. Images were acquired at × 630 magnification. c Venn diagram depicting the overlay of significantly up- or downregulated transcripts in each RUNX1 mutant group, as compared with WT RUNX1–transduced cells. d Canonical pathways that were significantly (−log(p value) > 1.3) enriched and significantly predicted (z-value > 2 and < − 2) to be up- or downregulated in each RUNX1 mutant group compared with WT RUNX1–transduced cells. Shared pathways are marked with an asterisk (*). e IPA analysis of the significantly regulated pathways shared by lin− cells transduced with each RUNX1 mutant, as compared with WT RUNX1–overexpressing samples. Upregulated pathways are shown in orange, and downregulated pathways are shown in blue
After invariant set normalization, the expression levels of the RUNX1 mutants were normalized to those obtained from WT RUNX1–transduced cells. The fold change expression table (Suppl. Table 1) was uploaded to the IPA software, and the cutoff values were set to − 1.7 and + 1.7 for log fold change, with the goal of identifying differentially expressed genes. This analysis showed that 1113 and 1814 genes were differentially expressed in the RUNX1-R174L and RUNX1-R139G mutant groups compared with the WT RUNX1 group (Suppl. Table 2). Overlap analysis revealed that 679 genes (37.4% for the RUNX1-R139G mutant and 61% for the RUNX1-R174L mutant) were co-regulated in both RUNX1 mutant groups (Fig. 3c). The 15 most highly up- and downregulated genes were very similar between the RUNX1 mutant groups. Of the top upregulated genes, 10/15 from the RUNX1-R174L group and 14/15 from the RUNX1-R139G group were co-activated in both groups. Of the top downregulated genes, 14/15 from the RUNX1-R174L group and 11/15 from the RUNX1-R139G group were co-downregulated (Fig. 3d). A comparison analysis performed using the IPA software revealed that 46 and 79 canonical pathways were significantly enriched (log(p value) ≥ 1.3 corresponding to p ≤ 0.05) in d715 Csf3r lin− cells transduced with RUNX1-R174L or RUNX1-R139G, respectively. Of these pathways, 11 in the RUNX1-R174L group and 18 in the RUNX1-R139G group were significantly activated (z-value ≥ 2) or inhibited (z-value ≤ − 2). Most of the regulated pathways belonged to members of the activated innate immune pathways category; these included NF-kappaB signaling, toll-like receptor signaling (TLR), acute-phase response signaling, production of nitric oxide and reactive oxygen species, pattern-recognizing receptors, Trem 1 signaling, and IL-1 signaling. Activation was also seen among members of the inflammatory signaling pathways, such as IL-6 signaling, interferon signaling, Tec kinase signaling, and leukocyte extravasation signaling (Fig. 3e, Suppl. Table 3).
Representative pathways of the innate immune system activated in d715 Csf3r lin− cells transduced with RUNX1 mutants
As examples of the activated canonical inflammatory and innate immune signaling pathways, canonical TLR signaling and IL-6 pathways are depicted in Suppl. Figs. 2 and 3. TLR signaling was upregulated in d715 Csf3r lin− cells transduced with the R139G or R174L RUNX1 mutants; this resulted in activation of transcription factors through Janus kinase 1 (JAK1) and p38 MAPK, promoting the expression of IL-1, TNF-alpha, and IL-12 (Suppl. Fig. 2A, B). Interferon signaling acted through JAK1 and JAK2 to activate the transcription factors, STAT1 and STAT2, thereby upregulating interferon response factors and other pro-inflammatory genes (Suppl. Fig. 2A, B).
The upregulation of IL-6 signaling–dependent genes in d715 Csf3r lin− cells transduced with each RUNX1 mutant was also mediated through JAK signaling, but relied on the transcription factor, STAT3, to confer translational regulation in the nucleus. Additional IL-6-dependent activation of ERK1 induced the NF-kappaB-NF-IL-6 transcription factor complex (Suppl. Fig. 3A, B).
Upstream pathway analysis points toward inflammatory dysregulation
Upstream analysis performed using the IPA software identified potential upstream regulators responsible for the gene expression signatures observed in the studied groups. We detected 768 upstream regulators in the RUNX1-R139G mutant group and 875 regulators in the RUNX1-R174L mutant group (Suppl. Fig. 4A, Suppl. Table 4). Five hundred upstream regulators were commonly found in both groups: 65.1% in the RUNX1-R139G mutant group and 57.1% in the RUNX1-R174L mutant group (Suppl. Fig. 4A). Among the 15 top activated upstream regulators, 9/15 for RUNX1-R174L and 11/15 for RUNX1-R139G were shared between the groups. Among the 15 top downregulated regulators, overlaps were seen in 6/15 for RUNX1-R174L and 9/15 for RUNX1-R139G (Suppl. Fig. 4B). Selected up- and downregulated candidate genes were validated by qRT-PCR in the independent set of experiments (Suppl. Fig. 4C). Also, staining of transduced G-CSF-treated lin− cells with Sca-1 and c-kit antibody followed by flow cytometry analysis revealed an increase of lin−Sca-1+c-KIT+ cell population in RUNX1-R139G– and RUNX1-R174L–transduced groups, compared with cells expressing RUNX1 WT, or GFP CTRL (Suppl. Fig. 4D).
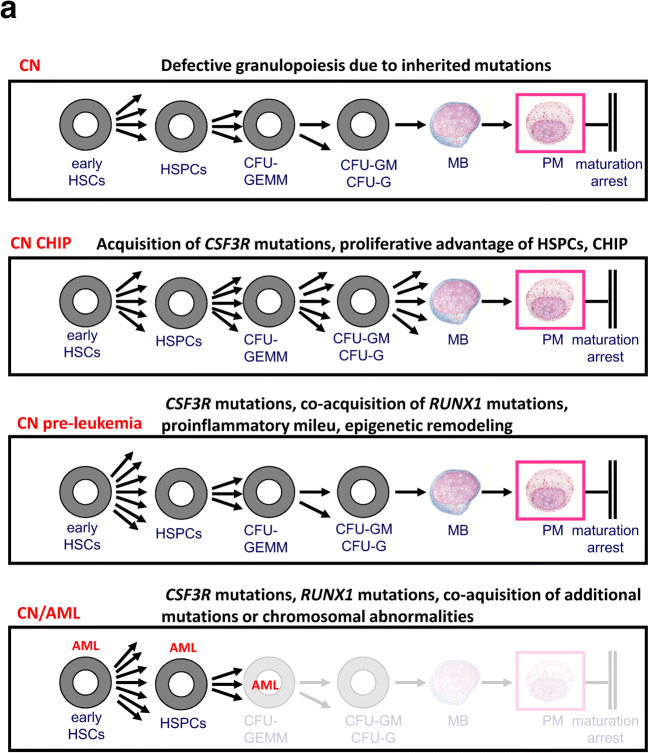
Fig. 4.
Proposed model for leukemia development in CN. a CN, CN-associated germline mutations cause maturation arrest of granulopoiesis at the stage of promyelocytes/myelocyte. CN CHIP, HSPCs that acquire CSF3R mutations gain a proliferative advantage that may mimic the CHIP phenotype. CN pre-leukemia, the co-acquisition of RUNX1 mutation induces an inflammatory milieu, leading to genotoxicity, additional defects of myeloid differentiation, and elevated proliferation that constitute the pre-leukemia stage. CN/AML, the acquisition of additional leukemia-associated gene mutations or chromosomal abnormalities results in AML or MDS
The top overlapping upstream regulators included a number of inflammatory cytokines, such as IL-15 and IL-18. IL-15 has been described as a responsible driver for chronic inflammation in autoimmune diseases and hematological malignancies [20]. Both IL-15 and IL-18 are confirmed targets for antitumor activity [21, 22]. Another factor that promotes survival of leukemogenic cells, Bcl2a1 [23], was also highly expressed, as were Ly6a and Sca-1, which are known as stemness factors for HSPCs.24 The top downregulated genes included anti-inflammatory genes, such as Il-10 [25], and the cytokine-processing factor, Dpp4, which can increase proliferation by prolonging cytokine signaling [26]. Interestingly, Csf3r was among the highest expressed genes in both groups. This is in accordance with the literature showing that G-CSF sensitivity is increased in AML blasts with missense RUNX1 mutations.
Motif activity response analysis confirms the central role of inflammatory and innate immunity signaling and the presence of early molecular changes related to MDS/AML downstream of Csf3r and RUNX1 mutations
We used the ISMARA [27] web tool to analyze the transcription factor binding motifs enriched among the differentially expressed genes. We found that the most highly enriched motif was Irf2_Irf1_Irf8_Irf9_Irf7 (z-value 4.564), which corresponds to the interferon regulatory factors. This indicates that an interferon-related change in inflammation signaling is responsible for at least some of the observed gene expression differences. The Stat2 motif was significantly correlated with the expression signature (z-value 2.707), and we observed significant activation of the Klf4_Sp3 (z-value 2.349) and Mecp-2 (z-value 2.627) transcription factor binding motifs known to be associated with AML or MDS, but significant downregulation of the Max–Mycn motif (z-value 2.016), upon transduction of d715 Csf3r lin− cells with RUNX1 mutants (Suppl. Fig. 5A). Finally, our inferred activity analysis of each motif revealed that there was a high degree of similarity in the significantly activated motifs associated with the two different RUNX1 mutants (Suppl. Fig. 5B).
Discussion
The identification of cooperative CSF3R and RUNX1 mutations in a majority of CN patients with overt MDS or AML brought us one step closer to understanding leukemia development [3]. Truncated CSF3R mutations are a very rare event in de novo AML, while RUNX1 mutations are most frequent among patients with secondary AML (sAML) after chemotherapy or radiation therapy, which represents about 30% of adult AML cases. Acquired cooperative CSF3R and RUNX1 mutations in approximately 55% of CN/AML patients are a unique feature and may reflect the inherited background of patients with CN-associated gene mutations, such as those in ELANE or HAX1. Mechanistically, ELANE mutations induce unfolded protein response (UPR) [28, 29] and endoplasmic reticulum (ER) stress, while mutations in HAX1 have pro-apoptotic functions [30, 31]. The role of inherited CN-associated mutations in the process of leukemogenesis needs to be investigated. The presence of these mutations may predispose to acquisition of secondary mutations in CSF3R and RUNX1 and the development of leukemia. Similar to secondary leukemias, wherein HSPCs are damaged by chemotherapeutic agents or radiation therapy, inherited CN-specific mutations may induce DNA damage or stress responses in HSPCs. Indeed, using CN patient–derived iPSCs as an experimental model, we recently showed that an elevation of DNA damage in CN HSPCs precedes the leukemic transformation [32]. In the present study, we found that csf3r and RUNX1 mutations in HSPCs induce reduced myeloid differentiation and enhanced clonogenic potential. Our findings demonstrated that cells transduced with WT RUNX1 differentiated and lost the capacity to proliferate, making no colonies in replating experiments. In contrast, cells transduced with RUNX1 mutants differentiate less, but retain proliferative potential. Leukemogenic activity of csf3R and RUNX1 mutations should be further elaborated in vivo using a mouse model.
Elucidation of the deregulated intracellular signaling pathways downstream of CSF3R and RUNX1 mutations will help to identify druggable targets, with the goal of eliminating leukemia-predisposed HSPCs and/or specifically targeting CN/AML blasts. Interestingly, mRNA expression analysis revealed that HSPCs harboring mutations in Csf3R and RUNX1 exhibit activation of the inflammatory and innate immunity pathways, including interferon signaling; IL-1, IL-6, IL-8, and TLR signaling; and TREM1 signaling. Additionally, we also found marked upregulation of Ly6a (also known as Sca-1), a marker of stemness in HSPCs [24] that is also upregulated upon inflammation [33–35]. IL-6 and IL-8 were previously known to be hyper-activated in MDS and AML [19, 36, 37]. Moreover, the increased genotoxic stress in HSPCs of 40% de novo MDS cases has been associated with elevated TLRs/Myd88-triggered intracellular signaling and IL-8 expression [19]. Most probably, the presence of truncated CSF3R and RUNX1 mutations in HSPCs of CN patients alters the pro-inflammatory cell state, enhances proliferation, and increases the susceptibility of HSPCs to genomic toxicity. Recent reports revealed that pre-leukemic HSPCs carrying an ETV6-RUNX1 fusion gene or having Pax5 haploinsufficiency evolved to precursor B cell acute lymphocytic leukemia upon activation of the pro-inflammatory pathways [38, 39]. The role of activated innate immunity and inflammatory pathways in the leukemogenesis of CN HSPCs was not studied yet. It would be interesting to investigate the susceptibility and expression kinetics of TLRs and the receptors for IL-1 and IL-8 on HSPCs from CN/AML patients during the development of leukemia.
We found that the Spi1/PU.1 transcription factor motif is activated in HSPCs downstream of Csf3r and RUNX1 mutations. PU.1 is upregulated in hematopoietic cells of CN patients [40] and is an essential transcription factor for monocytic differentiation [41, 42]. PU.1 also acts as a maintenance factor for pre- and leukemia-initiating cells [43]. The leukemogenic function of upregulated PU.1 in HSPCs of CN patients has not been studied yet.
Taken together, our results support the following proposed mechanism for leukemia development in CN: CSF3R mutations represent a state of clonal hematopoiesis of indeterminate potential (CHIP) in CN patients in that they confer a clonal advantage. Co-acquisition of RUNX1 mutation further increases proliferation and genotoxicity, including hypersensitivity to pro-inflammatory signaling. This combination may lead to acquisition of additional somatic mutations (e.g., in SUZ12 or ASXL1) or chromosomal abnormalities (e.g., monosomy 7 or trisomy 21), which finally overt to MDS or AML (Fig. 4).
Electronic supplementary material
Selected significantly enriched canonical pathways detected by IPA core analysis. (PNG 610 kb)
TLR and IFN pathways are hyper-activated in d715 Csf3r HSPCs transduced with RUNX1 mutants, as compared to WT RUNX1 transduced cells. Pathway analysis of differentially expressed genes between WT RUNX1-overexpressing d715 Csf3r lin- cells and RUNX1-R139G- (A) or RUNX1-R174L (B) transduced cells was conducted using IPA software. Selected canonical signaling pathways (left: TLR signaling, right: Interferon signaling) that are activated in the presence of mutated RUNX1, in comparison to WT RUNX1 are depicted. The overlay of all matching molecules of selected pathways in the IPA dataset is shown. Genes marked in red are upregulated in RUNX1-mutant groups, as compared to WT-RUNX1 sampels, while green coloured genes are downregulated. (PNG 3476 kb)
IL-6 signaling is upregulated in d715 Csf3r HSPCs transduced with RUNX1 mutants. Overlays for IL-6 signaling pathway of all matching molecules contained in the IPA dataset demonstrate pathway activation in A, RUNX1-R139G and B, RUNX1-R174L overexpressing d715 Csf3r lin- cells compared to WT RUNX1 overexpressing cells. Green represents negative expression fold change while red marks positive expression fold change. Genes marked in red are upregulated in RUNX1-mutant groups, as compared to WT-RUNX1 sampels, while green coloured genes are downregulated. (PNG 2433 kb)
Analysis of the upstream regulators responsible for the described phenotypes. A, Venn diagram depicting the overlay of all significantly enriched upstream regulators. B, Among all significantly enriched upstream regulators found by IPA core analysis, the 15 top up- and downregulated upstream regulators are shown. Black stars mark genes that are differentially expressed in the same direction in both RUNX1 mutants; red stars mark genes that are expressed oppositely. C, qRT-PCR of the selected candidate genes that were identified in the microarray analysis. The fold change of expression differences of cells transduced with RUNX1 mutants normalized to RUNX1 wildtype transduced samples is shown. D, Flow cytometry analysis of LSK cells in d715 Csf3r lin- cell population transduced with RUNX1-WT, or RUNX1 mutants and treated with G-CSF, as described in the Material and Methods section. Representative results of one donor mouse are shown. (PNG 599 kb)
Motif Activity Response Analysis performed using the ISMARA webtool for d715 Csf3r lin- cells transduced with WT RUNX1 or the RUNX1 mutants A, Significant active motifs found to be responsible for the observed differential gene expression patterns in d715 Csf3r lin- cells transduced with RUNX1 mutants compared to WT RUNX1-overexpressing cells (z-value >2). B, Activity scores (arbitrary unit) for each motif in the variously transduced cells. (PNG 253 kb)
Gene lists used for IPA Ingenuity. (XLSX 2914 kb)
List of genes used in core analysis by IPA comparing expression profiles of RUNX1 mutant transduced cells normalized to RUNX1 WT transduced cells. (XLSX 114 kb)
List of significantly enriched canonical pathways detected by IPA core analysis. (XLSX 18 kb)
List of significantly enriched upstream regulators detected by IPA core analysis. (XLSX 166 kb)
Acknowledgments
We would like to thank C. Stocking for providing RUNX1 expression plasmids.
Authorship contributions
JS and MR made initial observations, designed the experiments, analyzed the data, supervised experimentation, and wrote the manuscript; MR performed the main experiments; MK generated RUNX1 mutants; A Scham. and DH re-cloned RUNX1 cDNAs into pRRL.PPT.CBX3SF hRUNX1.i2.EBFP.puro.pre backbone; OK assisted with the transduction of mouse lin− cells and differentiation experiments; A Schm. and DCL provided bone marrow of C57BL/6-d715 Csf3r mice; and KW and LK provided insightful comments.
Funding information
Open Access funding provided by Projekt DEAL. This study was funded by the Fritz Thyssen Stiftung and DFG. We would like to acknowledge the financial support of the study by the German Jose Carreras Leukemia foundation (M.R. and J.S.).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All animal studies included in this manuscript were reviewed and approved by the Regional Board of the City of Tübingen.
Human and animal rights
This article does not contain any study with humans participants performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/16/2021
The financial support by the German Jose Carreras Leukemia foundation (M.R. and J.S.) was missed by the author to be included in this papper, thus, this correction was made.
References
- 1.Skokowa J, Dale DC, Touw IP, Zeidler C, Welte K. Severe congenital neutropenias. Nat Rev Dis Prim. 2017;3:17032. doi: 10.1038/nrdp.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skokowa J, Germeshausen M, Zeidler C, et al. Severe congenital neutropenia: inheritance and pathophysiology. Curr Opin Hematol. 2007;14:22–28. doi: 10.1097/00062752-200701000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Hermans MHA, Antonissen C, Ward AC, Mayen AEM, Ploemacher RE, Touw IP. Sustained receptor activation and hyperproliferation in response to granulocyte colony-stimulating factor (G-CSF) in mice with a severe congenital neutropenia/acute myeloid leukemia–derived mutation in the G-CSF receptor gene. J Exp Med. 1999;189:683–691. doi: 10.1084/jem.189.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong F, Brynes RK, Tidow N, Welte K, Löwenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333:487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 5.Dong F, Hoefsloot LH, Schelen AM, Broeders CA, Meijer Y, Veerman AJ, Touw IP, Lowenberg B. Identification of a nonsense mutation in the granulocyte-colony-stimulating factor receptor in severe congenital neutropenia. Proc Natl Acad Sci. 1994;91:4480–4484. doi: 10.1073/pnas.91.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong F, van Buitenen C, Pouwels K, Hoefsloot LH, Löwenberg B, Touw IP. Distinct cytoplasmic regions of the human granulocyte colony-stimulating factor receptor involved in induction of proliferation and maturation. Mol Cell Biol. 1993;13:7774–7781. doi: 10.1128/MCB.13.12.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward AC, van Aesch YM, Schelen AM, Touw IP. Defective internalization and sustained activation of truncated granulocyte colony-stimulating factor receptor found in severe congenital neutropenia/acute myeloid leukemia. Blood. 1999;93:447–458. doi: 10.1182/blood.V93.2.447. [DOI] [PubMed] [Google Scholar]
- 8.Liu F, Kunter G, Krem MM, Eades WC, Cain JA, Tomasson MH, Hennighausen L, Link DC. Csf3r mutations in mice confer a strong clonal HSC advantage via activation of Stat5. J Clin Invest. 2008;118:946–955. doi: 10.1172/JCI32704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLemore ML, Poursine-Laurent J, Link DC. Increased granulocyte colony-stimulating factor responsiveness but normal resting granulopoiesis in mice carrying a targeted granulocyte colony-stimulating factor receptor mutation derived from a patient with severe congenital neutropenia. J Clin Invest. 1998;102:483–492. doi: 10.1172/JCI3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skokowa J, Steinemann D, Katsman-Kuipers JE, Zeidler C, Klimenkova O, Klimiankou M, Ünalan M, Kandabarau S, Makaryan V, Beekman R, Behrens K, Stocking C, Obenauer J, Schnittger S, Kohlmann A, Valkhof MG, Hoogenboezem R, Göhring G, Reinhardt D, Schlegelberger B, Stanulla M, Vandenberghe P, Donadieu J, Zwaan CM, Touw IP, van den Heuvel-Eibrink MM, Dale DC, Welte K. Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood. 2014;123:2229–2237. doi: 10.1182/blood-2013-11-538025. [DOI] [PubMed] [Google Scholar]
- 11.Osato M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene. 2004;23:4284–4296. doi: 10.1038/sj.onc.1207779. [DOI] [PubMed] [Google Scholar]
- 12.Osato M, Asou N, Abdalla E, Hoshino K, Yamasaki H, Okubo T, Suzushima H, Takatsuki K, Kanno T, Shigesada K, Ito Y. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2alphaB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–1824. doi: 10.1182/blood.V93.6.1817.406k36_1817_1824. [DOI] [PubMed] [Google Scholar]
- 13.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations of AML1 are common in therapy-related myelodysplasia following therapy with alkylating agents and are significantly associated with deletion or loss of chromosome arm 7q and with subsequent leukemic transformation. Blood. 2004;104:1474–1481. doi: 10.1182/blood-2004-02-0754. [DOI] [PubMed] [Google Scholar]
- 14.Harada H, Harada Y, Tanaka H, Kimura A, Inaba T. Implications of somatic mutations in the AML1 gene in radiation-associated and therapy-related myelodysplastic syndrome/acute myeloid leukemia. Blood. 2003;101:673–680. doi: 10.1182/blood-2002-04-1010. [DOI] [PubMed] [Google Scholar]
- 15.Schnittger S, Dicker F, Kern W, Wendland N, Sundermann J, Alpermann T, Haferlach C, Haferlach T. RUNX1 mutations are frequent in de novo-AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117:2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 16.Gaidzik VI, Teleanu V, Papaemmanuil E, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2160–2168. doi: 10.1038/leu.2016.126. [DOI] [PubMed] [Google Scholar]
- 17.Preudhomme C, Renneville A, Bourdon V et al (2009) Brief report High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. 113:5583–5588 [DOI] [PubMed]
- 18.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 19.Hemmati S, Haque T, Gritsman K (2017) Inflammatory signaling pathways in preleukemic and leukemic stem cells. Front Oncol 7 [DOI] [PMC free article] [PubMed]
- 20.Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood. 2001;97:14–32. doi: 10.1182/blood.V97.1.14. [DOI] [PubMed] [Google Scholar]
- 21.Kim PS, Kwilas AR, Xu W et al (2016) IL-15 superagonist/IL-15RαSushi-Fc fusion complex (IL-15SA/IL-15RαSu-Fc; ALT-803) markedly enhances specific subpopulations of NK and memory CD8+ T cells, and mediates potent anti-tumor activity against murine breast and colon carcinomas. Oncotarget 7 [DOI] [PMC free article] [PubMed]
- 22.Senju H, Kumagai A, Nakamura Y, Yamaguchi H, Nakatomi K, Fukami S, Shiraishi K, Harada Y, Nakamura M, Okamura H, Tanaka Y, Mukae H. Effect of IL-18 on the expansion and phenotype of human natural killer cells: application to cancer immunotherapy. Int J Biol Sci. 2018;14:331–340. doi: 10.7150/ijbs.22809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogler M. BCL2A1: the underdog in the BCL2 family. Cell Death Differ. 2012;19:67–74. doi: 10.1038/cdd.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes C, Stanford WL. Concise review: stem cell antigen-1: expression, function, and enigma. Stem Cells. 2007;25:1339–1347. doi: 10.1634/stemcells.2006-0644. [DOI] [PubMed] [Google Scholar]
- 25.Kelly Á, Lynch A, Vereker E, Nolan Y, Queenan P, Whittaker E, O'Neill LAJ, Lynch MA. The anti-inflammatory cytokine, interleukin (IL)-10, blocks the inhibitory effect of IL-1β on long term potentiation. J Biol Chem. 2001;276:45564–45572. doi: 10.1074/jbc.M108757200. [DOI] [PubMed] [Google Scholar]
- 26.Deng L, Chan R, O’Leary HA, et al. DPP4 truncated GM-CSF and IL-3 manifest distinct receptor-binding and regulatory functions compared with their full-length forms. Leukemia. 2017;31:2468–2478. doi: 10.1038/leu.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 2014;24:869–884. doi: 10.1101/gr.169508.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grenda DS, Murakami M, Ghatak J, Xia J, Boxer LA, Dale D, Dinauer MC, Link DC. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood. 2007;110:4179–4187. doi: 10.1182/blood-2006-11-057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nustede R, Klimiankou M, Klimenkova O, Kuznetsova I, Zeidler C, Welte K, Skokowa J. ELANE mutant-specific activation of different UPR pathways in congenital neutropenia. Br J Haematol. 2016;172:219–227. doi: 10.1111/bjh.13823. [DOI] [PubMed] [Google Scholar]
- 30.Boztug K, Appaswamy G, Ashikov A, et al. A syndrome with congenital neutropenia and mutations in G6PC3. N Engl J Med. 2008;360:32–43. doi: 10.1056/NEJMoa0805051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein C, Grudzien M, Appaswamy G, Germeshausen M, Sandrock I, Schäffer AA, Rathinam C, Boztug K, Schwinzer B, Rezaei N, Bohn G, Melin M, Carlsson G, Fadeel B, Dahl N, Palmblad J, Henter JI, Zeidler C, Grimbacher B, Welte K. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nat Genet. 2007;39:86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- 32.Dannenmann B, Zahabi A, Mir P et al (2018) Human iPSC-based model of severe congenital neutropenia reveals elevated UPR and DNA damage in CD34+ cells preceding leukemic transformation. Exp Hematol [DOI] [PubMed]
- 33.Clapes T, Lefkopoulos S, Trompouki E. Stress and non-stress roles of inflammatory signals during HSC emergence and maintenance. Front Immunol. 2016;7:1–15. doi: 10.3389/fimmu.2016.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Essers MAG, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, Trumpp A. IFNα activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 35.Bujanover N, Goldstein O, Greenshpan Y, Turgeman H, Klainberger A, Scharff Y’, Gazit R. Identification of immune-activated hematopoietic stem cells. Leukemia. 2018;32:2016–2020. doi: 10.1038/s41375-018-0220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuett A, Rieger C, Perathoner D, et al. IL-8 as mediator in the microenvironment-leukaemia network in acute myeloid leukaemia. Sci Rep. 2015;5:1–11. doi: 10.1038/srep18411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharyya S, Shastri A, Bartenstein M, et al. IL8-CXCR2 pathway inhibition as a therapeutic strategy against MDS and AML stem cells. Blood. 2015;125:3144–3152. doi: 10.1182/blood-2014-12-612580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodríguez-Hernández G, Hauer J, Martín-Lorenzo A, Schäfer D, Bartenhagen C, García-Ramírez I, Auer F, González-Herrero I, Ruiz-Roca L, Gombert M, Okpanyi V, Fischer U, Chen C, Dugas M, Bhatia S, Linka RM, Garcia-Suquia M, Rascón-Trincado MV, Garcia-Sanchez A, Blanco O, García-Cenador MB, García-Criado FJ, Cobaleda C, Alonso-López D, de Las Rivas J, Müschen M, Vicente-Dueñas C, Sánchez-García I, Borkhardt A. Infection exposure promotes ETV6-RUNX1 precursor B-cell leukemia via impaired H3K4 demethylases. Cancer Res. 2017;77:4365–4377. doi: 10.1158/0008-5472.CAN-17-0701. [DOI] [PubMed] [Google Scholar]
- 39.Martín-Lorenzo A, Hauer J, Vicente-Dueñas C, et al. Infection exposure is a causal factor in B-cell precursor acute lymphoblastic leukemia as a result of Pax5-inherited susceptibility. Cancer Discov. 2015;5:1328–1343. doi: 10.1158/2159-8290.CD-15-0892. [DOI] [PubMed] [Google Scholar]
- 40.Skokowa J, Welte K. Dysregulation of myeloid-specific transcription factors in congenital neutropenia: rescue by namptnad+sirt1. Ann N Y Acad Sci. 2009;1176:94–100. doi: 10.1111/j.1749-6632.2009.04963.x. [DOI] [PubMed] [Google Scholar]
- 41.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 42.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 43.Staber PB, Zhang P, Ye M, Welner RS, Levantini E, di Ruscio A, Ebralidze AK, Bach C, Zhang H, Zhang J, Vanura K, Delwel R, Yang H, Huang G, Tenen DG. The Runx-PU.1 pathway preserves normal and AML/ETO9a leukemic stem cells. Blood. 2014;124:2391–2399. doi: 10.1182/blood-2014-01-550855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selected significantly enriched canonical pathways detected by IPA core analysis. (PNG 610 kb)
TLR and IFN pathways are hyper-activated in d715 Csf3r HSPCs transduced with RUNX1 mutants, as compared to WT RUNX1 transduced cells. Pathway analysis of differentially expressed genes between WT RUNX1-overexpressing d715 Csf3r lin- cells and RUNX1-R139G- (A) or RUNX1-R174L (B) transduced cells was conducted using IPA software. Selected canonical signaling pathways (left: TLR signaling, right: Interferon signaling) that are activated in the presence of mutated RUNX1, in comparison to WT RUNX1 are depicted. The overlay of all matching molecules of selected pathways in the IPA dataset is shown. Genes marked in red are upregulated in RUNX1-mutant groups, as compared to WT-RUNX1 sampels, while green coloured genes are downregulated. (PNG 3476 kb)
IL-6 signaling is upregulated in d715 Csf3r HSPCs transduced with RUNX1 mutants. Overlays for IL-6 signaling pathway of all matching molecules contained in the IPA dataset demonstrate pathway activation in A, RUNX1-R139G and B, RUNX1-R174L overexpressing d715 Csf3r lin- cells compared to WT RUNX1 overexpressing cells. Green represents negative expression fold change while red marks positive expression fold change. Genes marked in red are upregulated in RUNX1-mutant groups, as compared to WT-RUNX1 sampels, while green coloured genes are downregulated. (PNG 2433 kb)
Analysis of the upstream regulators responsible for the described phenotypes. A, Venn diagram depicting the overlay of all significantly enriched upstream regulators. B, Among all significantly enriched upstream regulators found by IPA core analysis, the 15 top up- and downregulated upstream regulators are shown. Black stars mark genes that are differentially expressed in the same direction in both RUNX1 mutants; red stars mark genes that are expressed oppositely. C, qRT-PCR of the selected candidate genes that were identified in the microarray analysis. The fold change of expression differences of cells transduced with RUNX1 mutants normalized to RUNX1 wildtype transduced samples is shown. D, Flow cytometry analysis of LSK cells in d715 Csf3r lin- cell population transduced with RUNX1-WT, or RUNX1 mutants and treated with G-CSF, as described in the Material and Methods section. Representative results of one donor mouse are shown. (PNG 599 kb)
Motif Activity Response Analysis performed using the ISMARA webtool for d715 Csf3r lin- cells transduced with WT RUNX1 or the RUNX1 mutants A, Significant active motifs found to be responsible for the observed differential gene expression patterns in d715 Csf3r lin- cells transduced with RUNX1 mutants compared to WT RUNX1-overexpressing cells (z-value >2). B, Activity scores (arbitrary unit) for each motif in the variously transduced cells. (PNG 253 kb)
Gene lists used for IPA Ingenuity. (XLSX 2914 kb)
List of genes used in core analysis by IPA comparing expression profiles of RUNX1 mutant transduced cells normalized to RUNX1 WT transduced cells. (XLSX 114 kb)
List of significantly enriched canonical pathways detected by IPA core analysis. (XLSX 18 kb)
List of significantly enriched upstream regulators detected by IPA core analysis. (XLSX 166 kb)