Abstract
The use of the smartphone is an ideal platform to realize the future point-of-care (POC) diagnostic system. Herein, we propose an integrated smartphone-based genetic analyzer. It consists of a smartphone and an integrated genetic analysis unit (i-Gene), in which the power of the smartphone was utilized for heating the gene amplification reaction, and the camera function was used for imaging the colorimetric change of the reaction for quantitative and multiplex foodborne pathogens. The housing of i-Gene was fabricated by using a 3D printer, which was equipped with a macro lens, white LEDs, a disposable microfluidic chip for loop-mediated isothermal amplification (LAMP), a thin-film heater, and a power booster. The i-Gene was installed on the iPhone in alignment with a camera. The LAMP mixture for Eriochrome Black T (EBT) colorimetric detection was injected into the LAMP chip to identify Escherichia coli O157:H7, Salmonella typhimurium, and Vibrio parahaemolyticus. The proportional–integral–derivative controller-embedded film heater was powered by a 5.0 V power bank to maintain 63 °C for the LAMP reaction. When the LAMP proceeded, the color was changed from violet to blue, which was real-time monitored by the smartphone complementary metal oxide semiconductor camera. The images were transported to the desktop computer via Wi-Fi. The quantitative LAMP profiles were obtained by plotting the ratio of green/red intensity versus the reaction time. We could identify E. coli O157:H7 with a limit of detection of 101 copies/μL within 60 min. Our proposed smartphone-based genetic analyzer offers a portable, simple, rapid, and cost-effective POC platform for future diagnostic markets.
Introduction
Foodborne disease have been one of the major public health problems, and the latest news by the World Health Organization (WHO) in 2019 reported that 550 million people suffer from diarrheal diseases, and 33 million people lose healthy life over the world due to food safety violation.1 To handle this issue, the Food and Drug Administration (FDA) announced three important elements for managing food safety including (i) preventing the outbreak of foodborne pathogens from the infected area, (ii) intervening at a critical point in the food supply chain, and (iii) rapidly responding when a foodborne pathogen is detected.2 To keep the abovementioned three elements should be premised on the development of the early diagnostic tools.
The diagnostic methods for the foodborne pathogen are commonly divided into immunoassay and molecular diagnosis. Of the two, the molecular-based analysis is highly accurate, so it has been widely used for the definite diagnosis of the pathogens. Recent advancement of the genetic amplification technology as well as the miniaturized hardware and devices moves toward point-of-care (POC) testing for early diagnostics of pathogens.3 In terms of the amplification technology, a variety of isothermal amplification methods have been developed including loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA), nucleic acid sequence-based amplification (NASBA), and rolling circle amplification. These techniques require only one constant temperature for gene amplification, making the whole system simple for the POC DNA testing. For example, Park et al. used the LAMP to amplify the target gene of Salmonella typhimurium and Vibrio parahaemolyticus at 65 °C for 60 min.4 Ahn et al. utilized the RPA for gene amplification at 37 °C for 20 min to identify Escherichia coli,Staphylococcus aureus, and S. typhimurium.(5) NASBA has been applied for detecting S. aureus and E. coli, which was carried out at 41 °C for 60 min.6
Regarding the miniaturized hardware, lab-on-a-chip technology, smartphone-based diagnostic tools, and a variety of hand-held devices have been developed.7−16 In particular, the smartphone-based method has recently garnered great attention due to the popularity to the general public, the ideal platform for U-healthcare monitoring, and the tremendous progress in the now and future. Liao et al. reported a smartphone-based device that used the smartphone camera to detect the LAMP reaction and produced an amplification curve by plotting the fluorescence intensity of the captured image along with the LAMP reaction time.17 Kong et al. reported a smartphone-based fluorescence reader for detecting qPCR products at the end.18 Gou et al. introduced a smartphone-based digital PCR device that used the smartphone camera for detecting a fluorescence signal.19 Yamanaka et al. presented a smartphone-based colorimetric reader to monitor the color change of an LAMP/hydroxy naphthol blue (HNB) assay.20 Until now, most of the smartphone-based diagnostics have focused on using the camera function of the smartphone for the fluorescence detection, which requires expensive optical accessories such as a blue LED as a light source and long/short pass filters for eliminating interference light.
In this study, we propose an advanced integrated genetic analysis platform to realize the POC DNA testing on the smartphone. We utilized the diverse functions of the smartphone such as the high-quality optical and imaging sensor, long-lasting battery, cloud-based data storage, and data transfer via Wi-Fi or Bluetooth. The integrated smartphone-based genetic analyzer (i-Gene) contains multiple components including a film heater for isothermal heating, optical accessories for colorimetric monitoring, and an LAMP chip with multiplex pathogen detection. We adopted an EBT-mediated LAMP reaction for real-time colorimetric qualitative and quantitative analyses of three pathogenic bacteria (Escherichia coli O157:H7 (E. coli O157:H7), Salmonella typhimurium (S. typhimurium), and Vibrio parahaemolyticus (V. parahaemolyticus)). Our proposed i-Gene analyzer is mobile, cost effective, and user friendly, demonstrating the high potential of the promising POC platform for the molecular diagnostics of pathogens in the future.
Experimental Section
Schematics of the i-Gene
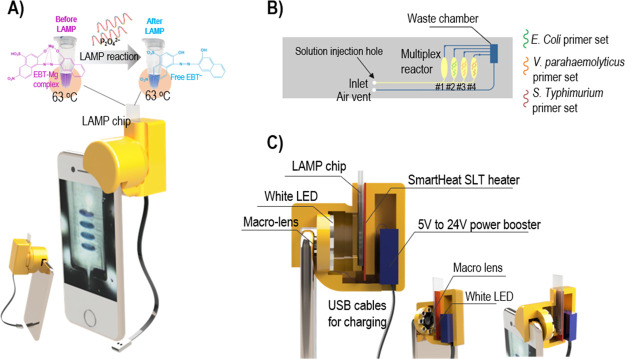
The i-Gene, whose size was 4.5 cm (length) × 6.3 cm (width) × 6.0 cm (height), was attached on the top of the smartphone in alignment with the camera as shown in Figure 1A. Inside the genetic analyzer that was fabricated by a 3D printer, there were a macro lens, eight white LEDs, a microfluidic LAMP chip, a film heater, and a power booster (Figure 1C). The disposable LAMP chip (6 cm in length × 1.5 cm in width) was made of a poly(methyl methacrylate) (PMMA) sheet through a CNC milling and could be inserted and pulled out by hand. The structure of the microfluidic chip consists of four reaction chambers for multiplex pathogen detection (Figure 1B).
Figure 1.
(A) Schematic illustration of the i-Gene attached on the smartphone. (B) Microfluidic LAMP reaction chip (yellow line channels: vistex coating, blue line channels: no coating). (C) Inside view of the genetic analyzer.
Design and Fabrication of the LAMP Chip
The microfluidic LAMP chip was fabricated from a 2 mm-thick (PMMA) sheet (Acrytal, Korea) using a CNC machine (Tinyrobo, Korea). The chip size was 6.0 cm (length) × 1.5 cm (width). On the surface of the reaction chambers, the primer mixture (FIB, BIP, F3, and B3) to amplify the target gene of bacteria was manually coated by pipetting and dried overnight in the dark at room temperature. The chamber #1 has no primer sets for a negative control, and the chambers #2, #3, and #4 were coated with 2 μL of the primer sets for targeting E. coli O157:H7, S. typhimurium, and V. parahaemolyticus, respectively. Then, the chip was sealed with a pressure-sensitive olefin film (PSA) (HJ-Bioanalytik GmbH, Germany) and stored in the refrigerator at 4 °C before use. The inlet channel and the four reaction chambers were coated with a hydrophilic reagent, Vistex 111-50 (FSI Coating Technologies, USA), while other channels and the waste chamber were left uncoated. The Vistex solution was diluted to a 10 wt % concentration by isopropanol, injected from the inlet hole for coating, and was incubated at 70 °C for 30 min to stabilize the Vistex layer.
Design and Fabrication of the i-Gene
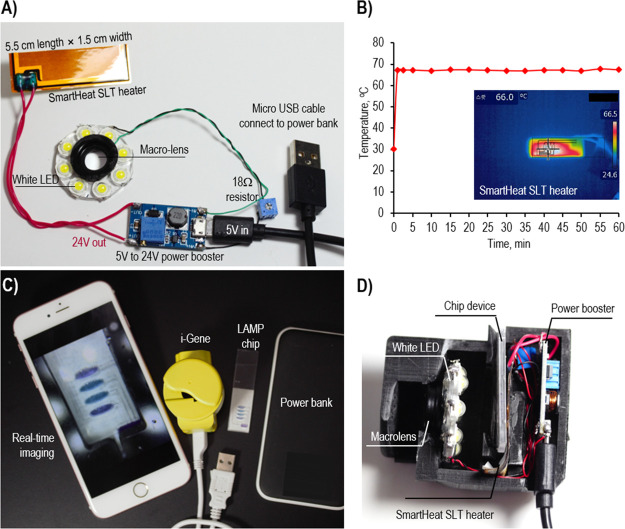
For POC genetic analysis on a smartphone, we designed a unique compact functional unit, which can perform heating, imaging, and data analysis. The genetic analyzer was designed using Autodesk Fusion360 software (Autodesk, USA) and was fabricated using a 3D printer (Cubicon, Korea). The total weight of the genetic analyzer was 60 g, and the overall size was 4.5 cm (length) × 6.3 cm (width) × 6.0 cm (height) (Figure S1). Since we utilized the EBT-mediated colorimetric detection of the LAMP, eight white LEDs (Tongyifang, China) and an iPhone 6S plus (CMOS image sensor, 12 MP color camera with F/2.2 aperture and 29 mm focal length) were used as the detector unit. In order to tune the focal length from the complementary metal oxide semiconductor (CMOS) camera sensor to the reaction chamber on a chip, a macro lens (Shanghai SKINA Digital Technology, China) was installed in front of the camera window of the smartphone for 1.5 cm close-up. For the LAMP reaction, a constant temperature of 63 °C was provided by a SmartHeat SLT thin-film heater (Minco, USA) that contains an embedded proportional–integral–derivative (PID) controller. To power the heater, 5 V derived from the smartphone battery was increased to 24 V via a micro-USB power booster DC–DC (Tuozhanteng Hong Kong Technology, China) (Figure 1C). The temperature of the heater was recorded and determined by using an IR camera (FLIR system, USA). The electronic circuits are off-the-shelf, easy to buy, and easy to connect with less effort. Those miniaturized electrical components enable us to construct a portable integrated genetic analyzer, so that the smartphone can be applied for disease diagnostics.
Preparation of Bacterial Genomic DNA
E. coli O157:H7, (KCCM 11835), V. parahaemolyticus (KCCM 11965), and S. typhimurium (KCCM 11806) were purchased from the Korean Culture Center of Microorganisms (Korea). The bacteria were grown in the media following the manufacturer’s guidelines. Genomic DNAs of the three target bacteria were extracted by a QIAamp DNA mini kit (Qiagen, UK). The quantity of the purified DNA was measured by the NanoDrop One (Thermo Fisher Scientific, USA) based on the absorbance at 260 nm. The purified DNAs were diluted by DNase/RNase water into the concentration ranging from 101 to 106 copies/μL for further experiments.
Preparation of the LAMP Reaction Mixture
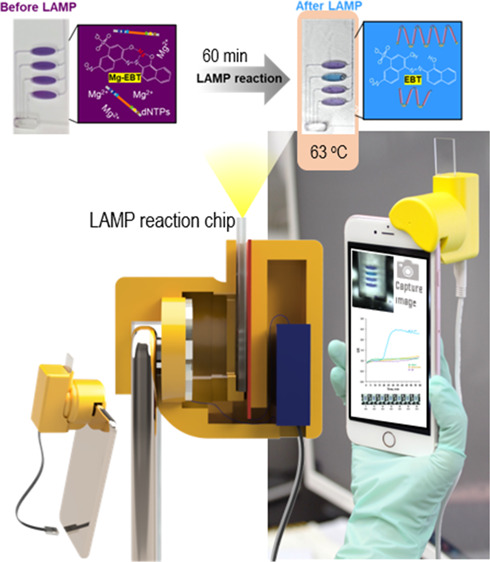
To amplify the target gene of the three bacteria via LAMP, the primer sets including FIB, BIP, F3, and B3 were designed by PrimerExplorerV5 software (Eiken Chemical Co., Tokyo, Japan) and synthesized by Macrogen (Korea). The information of the primer sequence is shown in Table S1. The Bst 2.0 WarmStart DNA polymerase enzyme for the LAMP reaction was purchased from New England Biolabs (USA). Each reaction chamber was designed with a defined volume of 20 μL containing 1× isothermal amplification buffer, 8.20 mM Mg2+, 6.11 mM dNTPs, 1220 unit/mL Bst 2.0 WarmStart DNA polymerase, 0.20 pmol/μL FIP, 0.20 pmol/μL BIP, 0.02 pmol/μL F3, 0.02 pmol/μL B3, and 0.10 mM EBT. The temperature was kept at 63 °C for 60 min. The success of the gene amplification was judged by observing the color change from violet to blue. The monitoring for the color change and the real-time quantification during the amplification process was performed by the smartphone camera.
Operation Procedure of the Smartphone-Based Genetic Analyzer
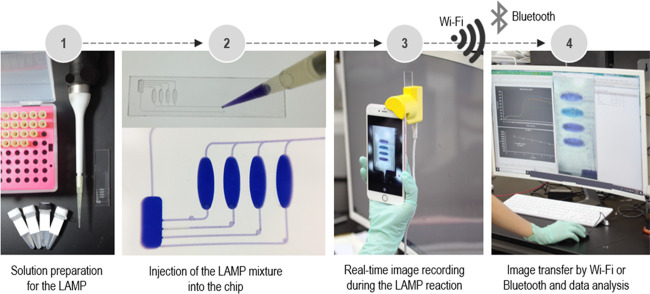
The overall procedure for the pathogen detection by the portable i-Gene on the smartphone is presented in Figure 2. First, the purified genomic DNAs were mixed with the LAMP cocktail (Figure 2, first step). Then, the mixture was injected into the microfluidic LAMP chip from the inlet hole. The solution was equally aliquoted into the four reaction chambers. After filling the chambers, the air vent hole and the inlet hole were sealed with the PSA film to prevent the evaporation of the LAMP reaction mixture during the LAMP reaction (Figure 2, second step). Then, the backside of the chip, which is located against the heater surface, was covered with a white label paper (Avery Dennison, USA). The chip was inserted into the chip-mounting slit of the i-Gene. The USB cable was connected to a 5.0 V power bank to power up the LAMP reaction. During the reaction for 60 min, the smartphone camera automatically captured the chip image at an interval of 1 min by using an iOS time-lapse app (Figure 2, third step). The 60 captured photographs were then sent to a laptop computer by Wi-Fi or Bluetooth. The color intensity of the reaction chambers was measured by ImageJ software (LOCI, University of Wisconsin, USA), and the qualitative and quantitative analyses for the pathogens were conducted. The whole process is shown in Video S1.
Figure 2.
Procedure of the molecular diagnostics by the portable smartphone-based genetic analyzer.
Image Processing
The captured images of the reaction chambers were processed using ImageJ software. The region of interest (ROI) (50 × 50 pixels) was cropped from the original image. The average intensity value of the red (R), green (G), and blue (B) color of the ROI was then determined. We chose the intensity ratio of the G and R as an indicator for color change. The real-time LAMP curve was plotted by the LAMP reaction time versus the G/R ratio. The threshold time (Ct) was determined with a value of the G/R ratio of 1.4. For the quantitative analysis, the real-time LAMP reaction was performed with the amount of genomic DNA of E. coli O157:H7 ranging from 101 to 106 copies/μL. At each concentration of genomic DNA, the Ct value was obtained, and then the quantitative calibration curve between log(DNA) and the Ct value was produced.
Results and Discussion
Design of the LAMP Reaction Chip
The design of the LAMP reaction chip for multiplex pathogenic bacterial detection is illustrated in Figure 1B. Four reaction chambers were patterned on a chip. One negative control (NC) chamber was uncoated with the primer set to serve as a standard color. The other three chambers were coated with the primer sets for targeting E. coli O157:H7 (chamber #2), V. parahaemolyticus (chamber #3), and S. typhimurium (chamber #4). The LAMP/EBT reaction mixture was injected through the inlet hole and was automatically separated into the four chambers. The reaction chamber was designed in an angle-free oval shape in order to smoothly fill the chamber without any bubble formation. The inlet channels and the reaction chambers were coated with Vistex 10% w/w to reduce the back pressure due to the hydrophobic property of the PMMA material. The static contact angle (N = 10) of the reaction buffer on the Vistex-coated polycarbonate polymer is 19.3 ± 1.4°, while the contact angle on the untreated polymer is 53.8 ± 1.1°, increasing the wettability dramatically.21 On the other hand, the outlet channels and the waste chamber remained uncoated, so the hydrophobic surface plays a role as a passive valve to prevent the LAMP mixture filled in one reaction chamber from overflowing to the waste chamber before occupying all other chambers. The hydrophilic and hydrophobic surface treatments provide a robust aliquoting of the reaction mixture into the four chambers by one injection. The waste chamber was connected to an air vent hole for venting the air during the injection. Finally, the injection hole and the air vent hole were sealed prior to the LAMP reaction to avoid an evaporation issue at 63 °C.
Design of the i-Gene
When we developed the i-Gene, the size of the platform was considered as a critical factor to apply for the POC diagnostics. Among the electronic components, the SmartHeat SLT thin-film heater provides a miniaturized heating system without an external bulky PID controller. The eight white LEDs were aligned in a circle toward the LAMP chip for homogeneous lighting during the image recoding by a camera. The LAMP chip was inserted between the camera and the heater, and the distance between the camera window and the chip was 2.3 cm. To capture a clear image over a short distance, it is necessary to place the macro lens in front of the camera window to focus the image. This macro lens allows the capture of a clear picture in a very close distance to the iPhone camera. Without the macro lens, the minimal focus distance of the camera is about 6 cm, which would make the platform bigger. The power of the heater and the LED lights was supplied by a 5.0 V, 2.0 A power bank. However, to run the heater, a high voltage around 24.0 V is required. So, a power booster was utilized to convert DC 5.0 V of the power bank to 24.0 V. On the other hand, the LED lighting ring was operated with a 3.2 V power. Therefore, the input 5.0 V was directly connected to the LED ring circuit through an 18.0 Ω resistor, supplying 3.2 V (Figure 3A, C, D). In the proposed configuration, the illumination of the white LED was absorbed by the LAMP solution on a chip. The color change and the reduced intensity of the light were recorded. The capture images were stored in a .jpg format, and the color intensity was digitally scaled from 0 to 255 for the R, G, and B channels.22 These color intensities corresponded to the light intensity entering the CMOS camera sensor after passing through a Bayer filter.
Figure 3.
(A) Assembly of the electrical circuit to run the heater and the LED lighting system. (B) Temperature profile for 60 min at 24.0 V input voltage. (C) Full set for the molecular diagnostics consisting of an i-Gene, a smartphone, an LAMP chip, and a power bank. (D) Cross-sectional digital image of the i-Gene.
Temperature Calibration of the PID Controller-Embedded Film Heater
The SmartHeat SLT thin-film heater utilizes a unique patented polymer inner-lay material to maintain a set temperature without the need for an external PID controller. The temperature of the heater was varied according to the applied voltage, which is in the range of 6.0–24.0 V. The device was powered by a power bank with a fixed output voltage of 5.0 V. Thus, a USB power booster circuit was equipped to amplify 5.0 to 24 V to run the heater. Several values of the input voltages from 12.0 to 24.0 V were tested for heating. As shown in Figure S2A, the output temperature proportionally increased with the input voltages. As the input voltage increased from 12 to 24 V, the temperature was augmented from 50.7 to 66.5 °C with the linear regression equation of y = 2.56x + 48.8 (R2 = 0.99) (x: input voltage, y: temperature). The input voltage at 24.0 V produces a temperature of 66 °C, which was an optimal condition for the LAMP reaction. Under these conditions, the ramping rate for heating was 2.0 °C/s, and the ramping rate for natural cooling was 0.72 °C/s (Figure S2B). As shown in Figure 3B, the heater provides a stable temperature profile during the LAMP reaction for 60 min.
Data Processing
The captured images were processed using ImageJ software. The ROI was cropped at the center of the chamber from the original image with a size of 50 × 50 pixels as shown in Figure 4. Since the reaction chamber was designed in an oval shape, the bubbles that formed during the LAMP reaction were gathered at the border of the chamber. Thus, the cropped image took the homogeneous area in the reactor, avoiding the error derived from the bubble. The average value of the R, G, and B color intensity of the ROI was then determined.
Figure 4.
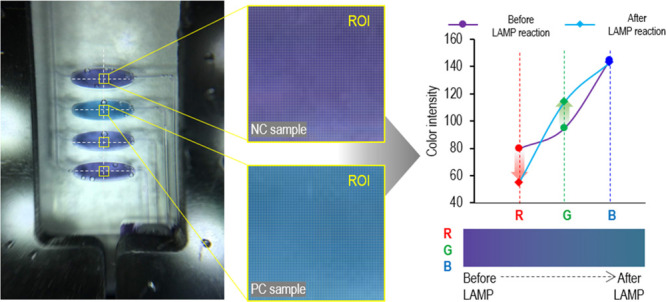
RGB colorimetric assay for quantifying the EBT-mediated LAMP reaction.
In principle, the LAMP/EBT reaction produces a color change from violet to blue for a positive control (PC) sample. In the NC sample, the color tone remained violet. This phenomenon relies on the fact that the EBT–Mg complex in the initial LAMP/EBT mixture reveals a violet color. During the amplification of the target gene, the released P2O74– takes up Mg2+ from the EBT–Mg complex and combines with Mg2+ to form white precipitates. As a result, the free EBT– ion shows a sky blue color (Figure 1A).23,24 This colorimetric assay can be estimated by naked eyes to identify the positive and negative results, but the visual decision cannot avoid a human error. In this study, we propose a numerical method for quantifying the DNA templates by an RGB analysis. As shown in Figure 4, the color intensity of the R, G, and B was changed as the LAMP reaction proceeded. The R intensity decreased, and the G intensity increased, while the B intensity remained unchanged during the LAMP reaction. Therefore, the G/R ratio would be the best indicator to monitor the success of the LAMP reaction as well as the quantification for the initial DNA templates since the value of the G/R would increase as the LAMP reaction is successful.
Monoplex, Duplex, and Triplex Identification of Foodborne Pathogens
The LAMP chip was designed to detect three target bacteria in one sample in a single run. For the monoplex test, the LAMP mixture only contained the genomic DNA of E. coli O157:H7. In the duplex test, the LAMP mixture contained the genomic DNA of E. coli O157:H7 and V. parahaemolyticus. In the triplex test, the LAMP mixture contained the genomic DNA of E. coli O157:H7, V. parahaemolyticus, and S. typhimurium. The number of purified DNAs was 105 copies/μL for all studies. Figure 5 shows the digital images for the chip and the real-time LAMP profiles. The first-row chamber is an NC, the second-row chamber is for E. coli O157:H7, the third-row chamber is for V. parahaemolyticus, and the last row chamber is for S. typhimurium. Figure 5A shows the monoplex result to detect E. coli O157:H7. Only the chamber in the second row displayed a blue color, while others were violet. The color change was initiated after 20 min. For the duplex test, the second and third rows show the color change, meaning that the sample contained E. coli O157:H7 and V. parahaemolyticus (Figure 5B). The LAMP amplificiation for V. parahaemolyticus started around 42 min. When triplex pathogens were analyzed, the violet color was turned to blue in all the rows, except the first row in which the NC was performed (Figure 5C). Accordingly, the LAMP profiles show that the G/R value gradually increased in the LAMP reaction chambers as the reaction time went on. Since the LAMP efficiency could depend on the target gene and primer design, the amplification profile for each pathogen was different. These results demonstrated high performance of the i-Gene for high multiplexity, high specificity, and no contamination problem between the chambers. The amplified products were confirmed by the gel electrophoresis data as shown in Figure S3.
Figure 5.
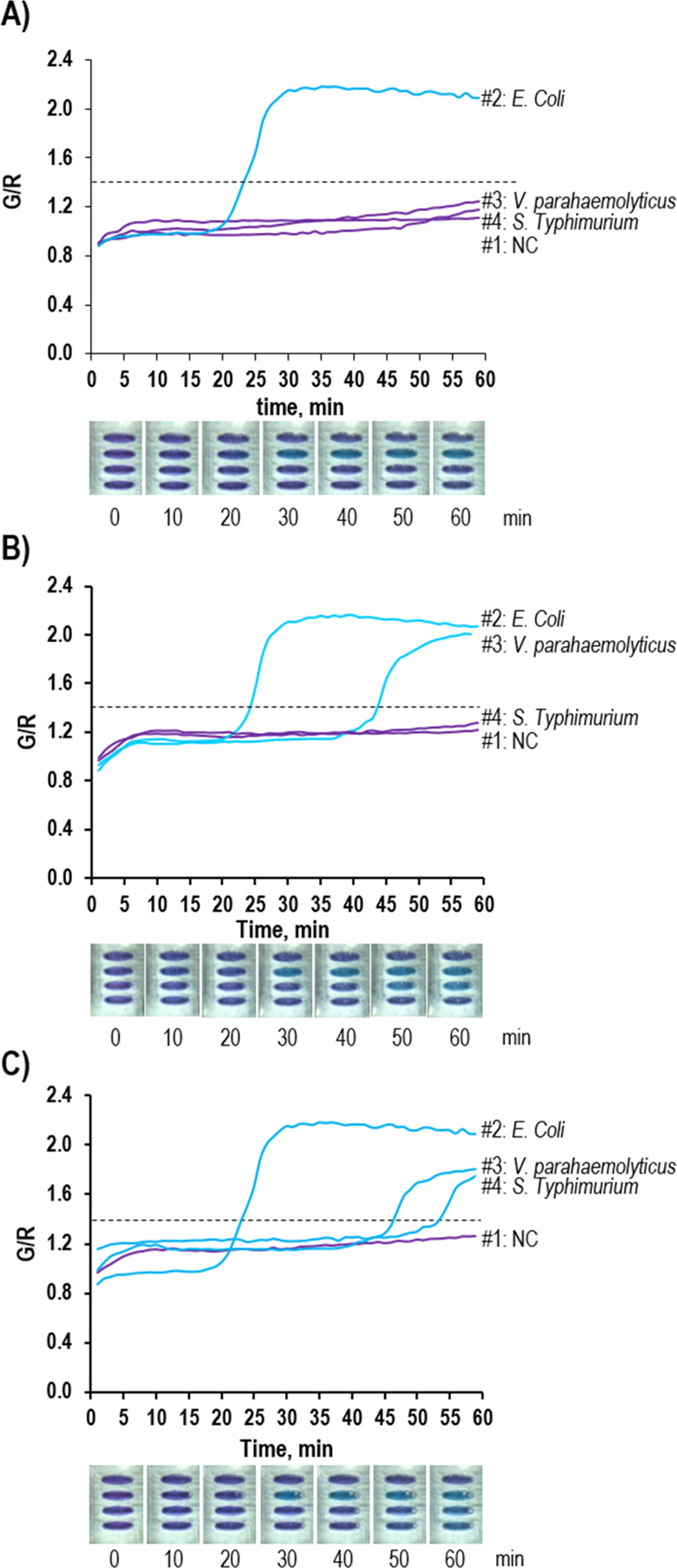
Colorimetric detection and the real-time LAMP profiles by the i-Gene for the (A) monoplex target (E. coli O157:H7), (B) duplex target (E. coli O157:H7 and V. parahaemolyticus), and (C) triplex target (E. coli O157:H7, V. parahaemolyticus, and S. typhimurium).
Generation of a Quantification Calibration Curve on the Integrated Smartphone-Based Genetic Analyzer
We, for the first time, performed not only qualitative analysis for the pathogen but also quantitative analysis to determine the initial copy number of pathogens. To do the quantitative analysis, it is necessary to generate a quantification curve. By choosing E. coli O157:H7 as a model, a variety of concentrations of the genomic DNA in the range of 101–106 copies/μL were prepared, and the real-time LAMP profiles were obtained by plotting the G/R ratio versus the reaction time (Figure 6A). Similar to the real-time PCR, we set the value of 1.40 of the G/R ratio to find out the threshold time (Ct) in the real-time LAMP. The Ct values were 30.2, 28.0, 25.7, 24.4, 23.2, and 22.3 at the concentrations of 101, 102, 103, 104, 105, and 106, respectively. From these data, we could generate the quantitative calibration curve by plotting the Ct value versus log(DNA) (Figure 6B), which shows a good correlation between the initial DNA amount and the Ct value (R2 = 0.97). Thus, the limit-of-detection (LOD) of our proposed system can reach up to 101 copies/μL (2 copies/reaction chamber), which was in a good agreement with the previous results.23,25 Chen et al. reported the LOD of 5 copies/reaction chamber by using the fluorescence-labeled LAMP on the smartphone-based detector.26
Figure 6.
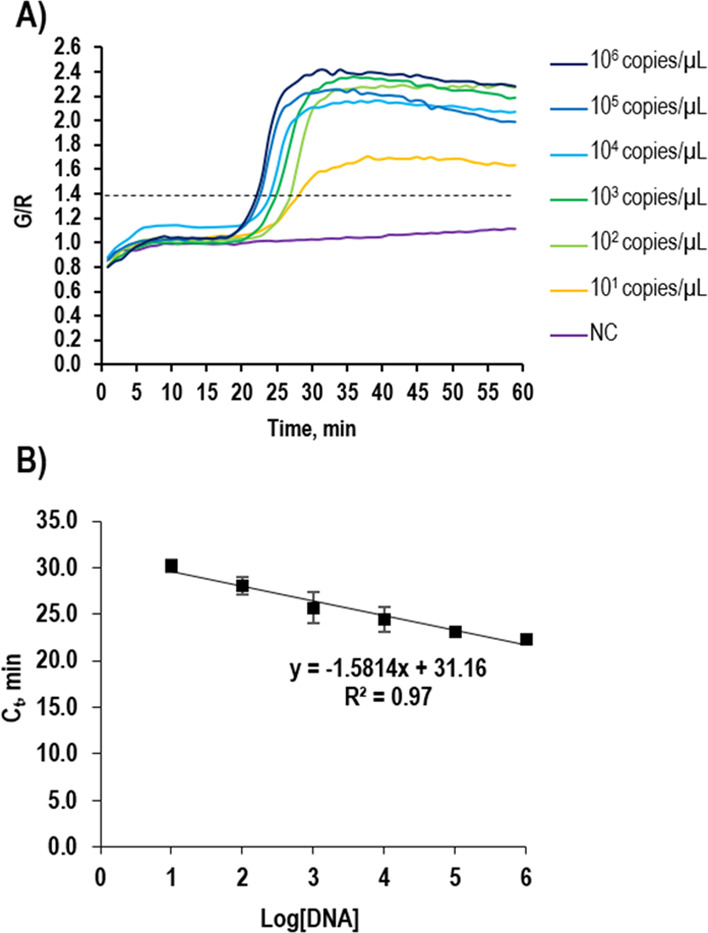
Quantification calibration curve. (A) LAMP profiles using various amounts of genomic DNAs of E. coli O157:H7 ranging from 101 to 106 copies/μL. (B) Linear regression plot of the Ct versus log(the amount of DNA).
Conclusions
In summary, we have demonstrated an advanced portable POC platform using a smartphone for the molecular diagnostics of pathogen. The i-Gene consists of a PID controller-embedded heater, an inexpensive macro lens, a white LED ring, and a power booster to execute the functions of heating for gene amplification, colorimetric detection, and wireless data transfer. The proposed platform was developed with a miniaturized size (4.5 × 6.3 × 6.0 cm) and lightweight (60 g), which is suitable for POC DNA testing for U-healthcare monitoring. The overall cost of the i-Gene is estimated to be about 120.0 USD (Table S2), and the disposable chip is about 0.1 USD, so it can be applied for resource-limited environments. The high performance of the proposed platform was proven by evaluating the multiplex as well as monoplex identification of the pathogen with excellent specificity. In addition, we have shown a possibility for the quantitative analysis of the pathogen on the smartphone. We believe that our platform would accelerate the advancement of the smartphone-based diagnostic tools to be used anywhere at any time.
Acknowledgments
This work was supported by the Engineering Research Center of Excellence Program of Korea Ministry of Science, ICT and Future Planning (MSIP)/National Research Foundation of Korea (NRF) (2014R1A5A1009799); National Research Foundation of Korea (NRF), The Ministry of Science and ICT (MSIT) (2020R1A2C1003960); and Korea Health Technology R&D Project/Korea Health Industry Development Institute (KHIDI), Ministry of Health and Welfare of South Korea (HI20C0644).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c02317.
(Figure S1) Integrated smartphone-based genetic analyzer with different views, (Figure S2) temperature profile according to the nominal voltage, (Figure S3) gel electrophoresis results, (Table S1) primer sequence information for the LAMP reaction, and (Table S2) cost of each component and the overall cost of the i-Gene (PDF)
Video S1 Whole process (MP4)
The authors declare no competing financial interest.
Supplementary Material
References
- WHO Food safety; https://www.who.int/news-room/fact-sheets/detail/food-safety.
- FDA Food protection plan : an integrated strategy for protecting the nation’s food supply; Food and Drug Administration: 2007.
- Nguyen H. V.; Nguyen V. D.; Lee E. Y.; Seo T. S. Point-of-care genetic analysis for multiplex pathogenic bacteria on a fully integrated centrifugal microdevice with a large-volume sample. Biosens. Bioelectron. 2019, 136, 132–139. 10.1016/j.bios.2019.04.035. [DOI] [PubMed] [Google Scholar]
- Park B. H.; Oh S. J.; Jung J. H.; Choi G.; Seo J. H.; Kim D. H.; Lee E. Y.; Seo T. S. An integrated rotary microfluidic system with DNA extraction, loop-mediated isothermal amplification, and lateral flow strip based detection for point-of-care pathogen diagnostics. Biosens. Bioelectron. 2017, 91, 334–340. 10.1016/j.bios.2016.11.063. [DOI] [PubMed] [Google Scholar]
- Ahn H.; Batule B. S.; Seok Y.; Kim M.-G. Single-Step Recombinase Polymerase Amplification Assay Based on a Paper Chip for Simultaneous Detection of Multiple Foodborne Pathogens. Anal. Chem. 2018, 90, 10211–10216. 10.1021/acs.analchem.8b01309. [DOI] [PubMed] [Google Scholar]
- Ma Y.; Dai X.; Hong T.; Munk G. B.; Libera M. A NASBA on microgel-tethered molecular-beacon microarray for real-time microbial molecular diagnostics. Analyst 2017, 142, 147–155. 10.1039/C6AN02192A. [DOI] [PubMed] [Google Scholar]
- Ahrberg C. D.; Ilic B. R.; Manz A.; Neužil P. Handheld real-time PCR device. Lab Chip 2016, 16, 586–592. 10.1039/C5LC01415H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia M.; Iemmolo R.; Petralia S.; Conoci S.; Cavallaro S. Miniaturized Real-Time PCR on a Q3 System for Rapid KRAS Genotyping. Sensors 2017, 17, 831. 10.3390/s17040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulberry G.; White K. A.; Vaidya M.; Sugaya K.; Kim B. N. 3D printing and milling a real-time PCR device for infectious disease diagnostics. PLoS One 2017, 12, e0179133 10.1371/journal.pone.0179133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S.; Gou T.; Hu J.; Wu W.; Ding X.; Fang W.; Hu Z.; Mu Y. A highly integrated real-time digital PCR device for accurate DNA quantitative analysis. Biosens. Bioelectron. 2019, 128, 151–158. 10.1016/j.bios.2018.12.055. [DOI] [PubMed] [Google Scholar]
- Hung T. Q.; Chin W. H.; Sun Y.; Wolff A.; Bang D. D. A novel lab-on-chip platform with integrated solid phase PCR and Supercritical Angle Fluorescence (SAF) microlens array for highly sensitive and multiplexed pathogen detection. Biosens. Bioelectron. 2017, 90, 217–223. 10.1016/j.bios.2016.11.028. [DOI] [PubMed] [Google Scholar]
- Rane T. D.; Chen L.; Zec H. C.; Wang T.-H. Microfluidic continuous flow digital loop-mediated isothermal amplification (LAMP). Lab Chip 2015, 15, 776–782. 10.1039/C4LC01158A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez N. M.; Wong W. S.; Liu L.; Dewar R.; Klapperich C. M. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip 2016, 16, 753–763. 10.1039/C5LC01392E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Gallegos R. A.; Rios A.; Garcia-Cordero J. L. An Affordable and Portable Thermocycler for Real-Time PCR Made of 3D-Printed Parts and Off-the-Shelf Electronics. Anal. Chem. 2018, 90, 5563–5568. 10.1021/acs.analchem.7b04843. [DOI] [PubMed] [Google Scholar]
- Papadakis G.; Pantazis A. K.; Ntogka M.; Parasyris K.; Theodosi G. I.; Kaprou G.; Gizeli E. 3D-printed Point-of-Care Platform for Genetic Testing of Infectious Diseases Directly in Human Samples Using Acoustic Sensors and a Smartphone. ACS Sens. 2019, 4, 1329–1336. 10.1021/acssensors.9b00264. [DOI] [PubMed] [Google Scholar]
- Choi G.; Jung J. H.; Park B. H.; Oh S. J.; Seo J. H.; Choi J. S.; Kim D. H.; Seo T. S. A centrifugal direct recombinase polymerase amplification (direct-RPA) microdevice for multiplex and real-time identification of food poisoning bacteria. Lab Chip 2016, 16, 2309–2316. 10.1039/C6LC00329J. [DOI] [PubMed] [Google Scholar]
- Liao S.-C.; Peng J.; Mauk M. G.; Awasthi S.; Song J.; Friedman H.; Bau H. H.; Liu C. Smart cup: A minimally-instrumented, smartphone-based point-of-care molecular diagnostic device. Sens. Actuators, B 2016, 229, 232–238. 10.1016/j.snb.2016.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J. E.; Wei Q.; Tseng D.; Zhang J.; Pan E.; Lewinski M.; Garner O. B.; Ozcan A.; Di Carlo D. Highly Stable and Sensitive Nucleic Acid Amplification and Cell-Phone-Based Readout. ACS Nano 2017, 11, 2934–2943. 10.1021/acsnano.6b08274. [DOI] [PubMed] [Google Scholar]
- Gou T.; Hu J.; Wu W.; Ding X.; Zhou S.; Fang W.; Mu Y. Smartphone-based mobile digital PCR device for DNA quantitative analysis with high accuracy. Biosens. Bioelectron. 2018, 120, 144–152. 10.1016/j.bios.2018.08.030. [DOI] [PubMed] [Google Scholar]
- Yamanaka E. S.; Tortajada-Genaro L. A.; Pastor N.; Maquieira Á. Polymorphism genotyping based on loop-mediated isothermal amplification and smartphone detection. Biosens. Bioelectron. 2018, 109, 177–183. 10.1016/j.bios.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Focke M.; Stumpf F.; Roth G.; Zengerle R.; von Stetten F. Centrifugal microfluidic system for primary amplification and secondary real-time PCR. Lab Chip 2010, 10, 3210–3212. 10.1039/c0lc00161a. [DOI] [PubMed] [Google Scholar]
- Bueno D.; Muñoz R.; Marty J. L. Fluorescence analyzer based on smartphone camera and wireless for detection of Ochratoxin A. Sens. Actuators, B 2016, 232, 462–468. 10.1016/j.snb.2016.03.140. [DOI] [Google Scholar]
- Oh S. J.; Park B. H.; Choi G.; Seo J. H.; Jung J. H.; Choi J. S.; Kim D. H.; Seo T. S. Fully automated and colorimetric foodborne pathogen detection on an integrated centrifugal microfluidic device. Lab Chip 2016, 16, 1917–1926. 10.1039/C6LC00326E. [DOI] [PubMed] [Google Scholar]
- Seo J. H.; Park B. H.; Oh S. J.; Choi G.; Kim D. H.; Lee E. Y.; Seo T. S. Development of a high-throughput centrifugal loop-mediated isothermal amplification microdevice for multiplex foodborne pathogenic bacteria detection. Sens. Actuators, B 2017, 246, 146–153. 10.1016/j.snb.2017.02.051. [DOI] [Google Scholar]
- Oh S. J.; Seo T. S. Combination of a centrifugal microfluidic device with a solution-loading cartridge for fully automatic molecular diagnostics. Analyst 2019, 144, 5766–5774. 10.1039/C9AN00900K. [DOI] [PubMed] [Google Scholar]
- Chen W.; Yu H.; Sun F.; Ornob A.; Brisbin R.; Ganguli A.; Vemuri V.; Strzebonski P.; Cui G.; Allen K. J.; Desai S. A.; Lin W.; Nash D. M.; Hirschberg D. L.; Brooks I.; Bashir R.; Cunningham B. T. Mobile Platform for Multiplexed Detection and Differentiation of Disease-Specific Nucleic Acid Sequences, Using Microfluidic Loop-Mediated Isothermal Amplification and Smartphone Detection. Anal. Chem. 2017, 89, 11219–11226. 10.1021/acs.analchem.7b02478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.