Abstract
Single cell MS (SCMS) techniques are under rapid development for molecular analysis of individual cells among heterogeneous populations. Lipids are basic cellular constituents playing essential functions in energy storage and the cellular signaling processes of cells. Unsaturated lipids are characterized with one or multiple carbon–carbon double (C=C) bonds, and they are critical for cell functions and human diseases. Characterizing unsaturated lipids in single cells allows for better understanding of metabolomic biomarkers and therapeutic targets of rare cells (e.g., cancer stem cells); however, these studies remain challenging. We developed a new technique using a micropipette needle, in which Paternò-Büchi (PB) reactions at C=C bond can be induced, to determine locations of C=C bonds in unsaturated lipids at the single-cell level. The micropipette needle is produced by combining a pulled glass capillary needle with a fused silica capillary. Cell lysis solvent and PB reagent (acetone or benzophenone) are delivered into the micropipette needle (tip size ≈ 15 um) through a fused silica capillary. The capillary needle plays multiple functions (i.e., single cell sampling probe, cell lysis container, microreactor, and nano-ESI emitter) in the experiments. Both regular (no reaction) and reactive (with PB reaction) SCMS analyses of the same cell can be achieved. C=C bond locations were determined from MS scan and MS/MS of PB products assisted by Python programs. This technique can potentially be used for other reactive SCMS studies to enhance molecular analysis for broad ranges of single cells.
Graphical Abstract

Among all known organisms, cells are the smallest unit of life. The majority of current studies of cells are based on ensemble measurement. However, each individual cell has unique genomic and phenotypic traits that can distinguish it from other adjacent cells, causing cell to cell heterogeneity in any population.1 Numerous studies indicate that small subpopulations of cells are overlooked using population measurements, resulting in the loss of important biology information on rare cells.1 Therefore, molecular analysis of single cells is an inevitable choice to understand cellular mechanisms that cannot be studied using traditional bulk analysis.1 Among all cellular molecules, lipids are crucial components of cell membrane and other cellular compartments, including the endoplasmic reticulum, Golgi apparatus, and nuclear membrane. In addition, lipids play important roles such as regulation of cell metabolism and signal transport.2,3 Unsaturated lipids are a subclass of lipids containing one or more carbon–carbon double (C=C) bonds. Identification of unsaturation sites is critical for understanding lipid biochemistry. For example, previous studies indicate that cancer stem cells have relatively higher levels of unsaturated lipids compared with nonstem cancer cells,4,5 and higher abundances of unsaturated lipids in cancer stem cells are related to the upregulation of de novo fatty acid (FA) synthesis pathway.4,6,7 The unsaturation level of lipids influences many cell physiological properties such as membrane fluidity,8,9 neuro-transmitter release,10,11 and cardiolipin remodeling.12 The location of C=C bond in unsaturated lipids is critical for their biological functions. For example, lipid isomers with different C=C bond positions are related to numerous diseases, including cancer, cardiovascular disease, type II diabetes, Barth syndrome, Alzheimer’s disease, and Parkinson’s disease.12–14 Thus, determining the C=C bond locations in unsaturated lipids is needed in studies of fundamental cell biology and wide types of diseases.
Mass spectrometry (MS) has become one of the most effective tools for lipid profiling and quantification.15,16 MS based methods have been widely used for targeted and nontargeted lipidomics studies, including identification of specific lipid classes using shotgun MS17 and analysis of complex lipids (e.g., glycerolipids, glycerophospholipids, and glycolipids).18–21 Particularly, combined with chemical reactions, MS has been used to pinpoint C=C bond sites in unsaturated lipids based on three different reactions. (1) Paternò–Büchi (PB) reaction. PB reaction is a classical photochemical derivatization that can specifically form adducts at C=C bonds under UV irradiation. Both acetone and benzophenone have been used as the PB reagents to study unsaturated lipids. For example, Xia20,22 and Liu21 have utilized tandem MS (MS/MS) to analyze adducts formed in the PB reactions to identify the structure of unsaturated lipids. (2) Ozone-induced dissociation (OzID). This technique can directly utilize ozonolysis, an organic reaction allowing for the cleavage of alkene double bonds using ozone, or combine it with CID (collision induced dissociation) to elucidate C=C bonds in unsaturated lipids.23 (3) meta-Chloroperoxy benzoic acid (m-CPBA) epoxidation reaction. m-CPBA is an oxidant that can convert alkene to epoxide. Both Li24 and Hsu12 have used m-CPBA in reactions with unsaturated lipids to form products with triatomic rings, which generated two diagnostic ion pairs (with 16 Da mass difference) to determine the C=C bond positions.
Using PB, m-CPBA, and ozonolysis reactions as mentioned above, MS determination of C=C bonds in unsaturated lipids has been carried out in bulk analysis (e.g., lipids prepared solutions and lipids extractions form cells, tissues, and plasma).20,22,24,25 However, the corresponding studies at the single-cell level remain unexplored due to extremely small amount analytes in a single cell (e.g., a few pLs)26 and the absence of appropriate techniques. Single cell MS (SCMS) methods have been rapidly developed for metabolomics and proteomics studies of a broad range of cells, including plant cells, mammalian cells, and yeasts.27–29 Under vacuum conditions, matrix-assisted desorption/ionization (MALDI) MS30 and secondary ion MS (SIMS) are commonly used for single cell analysis. In recent years, a variety of ambient based MS methods have been developed for single cell analysis. The representative examples include single-cell video-MS,31 induced nanoESI (InESI) MS,32 probe electrospray ionization (PESI),33 the Single-probe,34–37 and the T-probe.38 Cell attachment (e.g., onto a substrate) is generally required for most SCMS techniques. However, the requirement of cell attachment prior to MS analysis largely limits the type of cells in studies. In addition, cell attachment may alter cell status and their molecular compositions.39 A number of ambient MS techniques have been developed to analyze single nonadherent cells, such as mass cytometry,40 drop-on-demand inkjet printing coupled with PESI,41 the redesigned T-probe,26 integrated cell manipulation platform (ICMP)/Single-probe secondary ion MS (SIMS),42 and techniques coupled with microfluidic chips.43,44 However, none of the existing SCMS methods has been used for studies of C=C bond positions in unsaturated lipids at the single-cell level.
Here, we used the glass micropipette needles to perform PB reactions of single cells followed by MS determination of C=C bonds’ positions in unsaturated lipids (Figure 1). The micropipette needle is produced by combing a fused silica capillary with a pulled glass micropipette (Figures 1B and 2D). The fused silica capillary is connected with a syringe to provide solution containing cell lysis solvent and the PB reagent. The micropipette needle plays multiple functions, including cell selection, cell lysis, microreactor for PB reaction, and nano-ESI emitter for ionization (Figure 2). To conduct the PB reactions of single cells, the reagent (i.e., acetone or benzophenone solution) was drawn into the glass micropipette. Using an integrated cell manipulation system,44 which contains two Eppendorf cell manipulation systems, an inverted microscope (Nikon Eclipse TE300, Tokyo, Japan), and a syringe pump, a target cell was selected and sucked into the glass micropipette, in which the cell underwent a rapid lysis process (Figure 1). A mercury lamp was placed next to the micropipette needle to provide UV irradiation and induce PB reactions with unsaturated lipids (Figure 2). For MS analysis, the ionization voltage was applied onto the conductive union to generate nano-ESI at the tip of the micropipette needle. Two types of SCMS experiments, including the regular (no UV irradiation) and reactive (after 15 min of UV irradiation) methods, were conducted for the same cell to acquire comprehensive information for studying unsaturated lipids.
Figure 1.
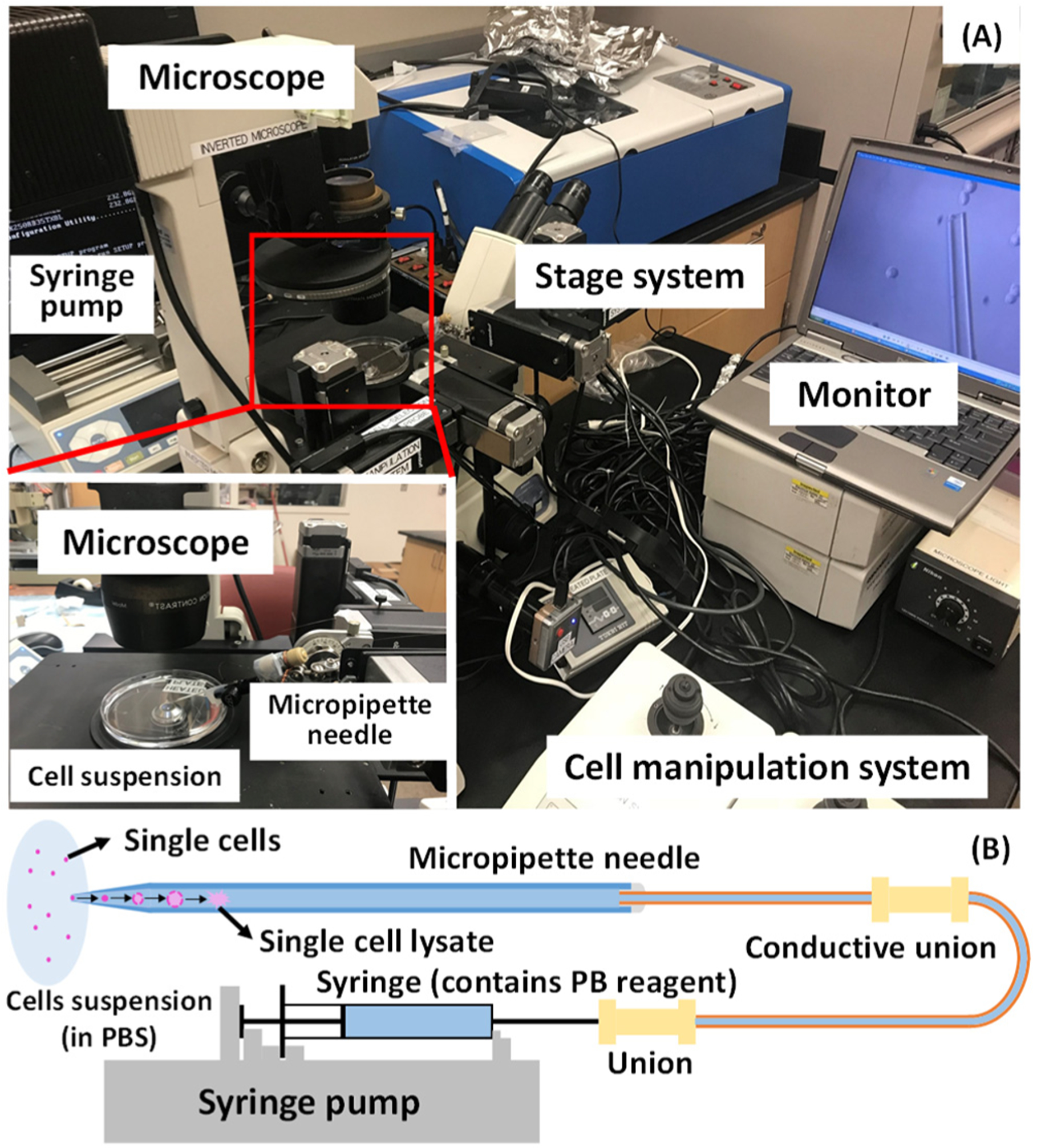
(A) Experimental setup of the micropipette needle for single cell sampling. An inverted microscope was used to provide visual guide during the experiment, and cell manipulation system was used to control the micropipette needle to aim the targeted cell. The micropipette needle was connected with a syringe using capillary and unions. A syringe pump controlled the syringe to suck the targeted cell into the pipet. (B) Sketch of the single-cell sampling using a micropipette needle.
Figure 2.
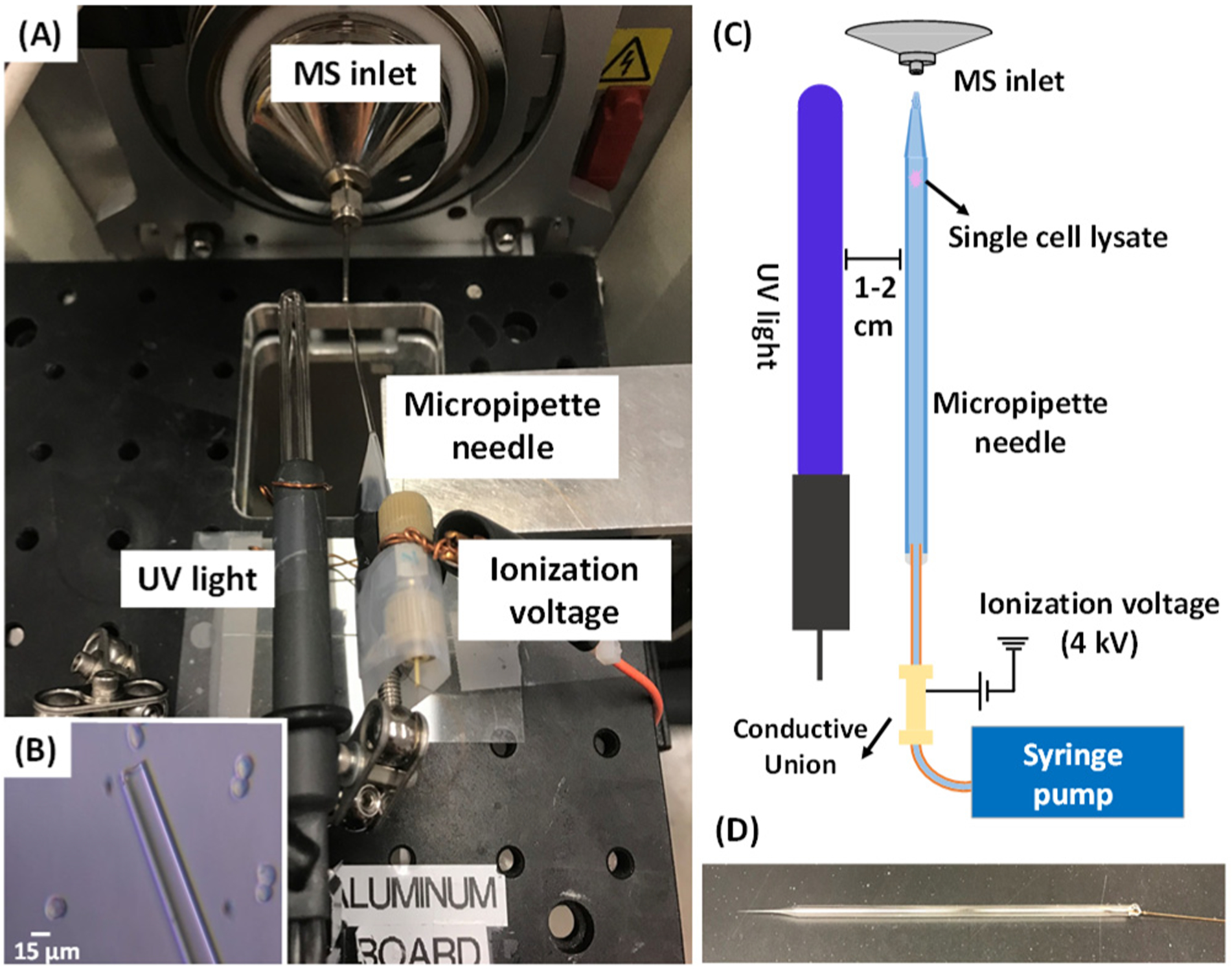
(A) Experimental setup of the micropipette needle for C=C bond identification at the single-cell level. The mercury UV light was placed next to the micropipette needle, and the ionization voltage was applied on the micropipette needle. (B) Sampling a suspended HCT-116 cell under the microscope. (C) Schematics of single cell sampling and SCMS analysis. (D) Photo of a micropipette needle.
METHODS
Fabrication of the Micropipette Needle.
The micropipette needle (tip size ≈ 15 um) was pulled from a glass capillary tube (size: 0.8 × 90 mm2, Kimble Chase Life Science and Research Products, Rockwood, TN) using a pipet puller (KOPF, Tujunga, CA). UV epoxy (Prime-Dent, Chicago, IL) was used to connect the micropipette needle to a fused silica capillary (OD: 150 μm, ID: 75 μm, Polymicro Technologies, Phoenix, AZ). A syringe was connected to the fused silica capillary via a conductive union (IDEX Health & Science LLC, Oak Harbor, WA).
Preparation of SCMS Solutions.
In the reactive SCMS studies, the solvent has three major functions: inducing cell lysis, performing PB reactions, and playing the role as the ionization solvent. First, deoxygenation of solvents was performed to minimize side reactions while promoting PB reactions.21,45 An Erlenmeyer flask (with stopper) containing pure acetone or ACN (acetonitrile) was placed on the ice and vacuumed for 30 min, followed by bubbling nitrogen for 30 min.21 This process was repeated three times to deplete oxygen from solvents. Next, the solution containing PB reagents was prepared and added in the micropipette needle. Two different reagents, benzophenone solution (5 mM benzophenone in ACN with 0.1% formic acid) and acetone have been tested in our studies to compare their performance.
Reactive SCMS Experiments.
Using an Eppendorf cell manipulation system and a syringe pump, a target cell was sucked into the glass micropipette (flow rate 10 μL/min) containing prefilled acetone or benzophenone solution (Figure 1). Additional solution was drawn into the micropipette needle to ensure cell lysis. The syringe pump was turned on to deliver (flow rate 0.2 μL/min) the single cell lysate toward the nano ESI emitter. A DC ionization voltage (+4 kV in the positive ion mode or −4 kV in the negative ion mode) was applied on a conductive union and transmitted through the solvent to induce ionization of cell lysis at the tip of the micropipette for MS analysis. Due to the long signal duration time of a single cell (20–30 min), both the regular (no UV irradiation) and the reactive (after UV irradiation) SCMS experiments can be conducted for the same single cell. Specifically, after accomplishing data acquisition of the regular SCMS experiment, the ionization energy was turned off and the syringe pump was paused. The UV lamp (BHK, Ontario, CA) was then turned on to generate UV radiation and initiate PB reactions between the reagents and unsaturated cellular lipids. After 15 min of reaction, the UV light was turned off, and then the reactive SCMS experiment was started by turning on the ionization voltage and resuming the syringe pump. Products from the PB reactions were analyzed using both MS scan (to obtain accurate m/z values of all ions) and tandem MS (MS/MS) analysis (to acquire fragments of selected ions). As illustrated in Figure 3, the PB reaction at each C=C bond can produce two isomers (i.e., Isomers I and II), which then produce one pair of diagnostic ions during CID, i.e., aldehyde and alkene ions can be generated from Isomer I and Isomer II, respectively.
Figure 3.

Mechanism of PB reactions and the formation of diagnostic ion pairs (aldehyde and alkene formats) in CID.
Cells Culture and Sample Preparation.
Human colon cancer cell line, HCT-116, was chosen as a model system in the current study. Cells were cultured using standard protocol as briefly described following.34 Cells were cultured in McCoy’s 5A medium, detached from Petri dish using trypsin, rinsed by PBS (phosphate-buffered saline), centrifuged (1000 rmp, 10 min, three times), and then resuspended in PBS. Cell density was controlled to be around 5 × 104 cells/mL, and ~7 mL of the cell suspension solution was transferred to a culture dish for the following SCMS analysis. Cell lysis sample was prepared using standard protocols for comparative studies.46 The detailed procedure is shown in the Supporting Information (SI).
RESULTS AND DISCUSSION
Sampling Solvent Selection.
Previous studies utilized both acetone and benzophenone as the PB reagents to analyze unsaturated lipids in bulk samples.20,21 Acetone and acetonitrile are common organic solvents generally used to prepare lysate. Thus, acetone and benzophenone solution (5 mM in acetonitrile) were selected in the current studies to induce cell lysis and PB reactions. Because a small amount of PBS is inevitably drawn into the micropipette needle during single cell sampling, a dilution of cell lysis solution can occur, potentially resulting in reduced cell lysis efficiency. For example, our previous studies show that acetonitrile can induce rapid cell lysis (<15 s) when its concentration is >80%, whereas lower concentrations result in slower lysis processes.26 Thus, we prepared cell lysis solutions using acetone and acetonitrile without adding other solvents commonly used in MS studies such as water and methanol. Because benzophenone concentration can affect the yield of products from the PB reactions,21 we prepared a series of acetonitrile solutions containing benzophenone (i.e., 0.5, 2.0, 5.0, and 10.0 mM) to optimize the PB reaction conditions in SCMS studies. Our experiments indicated that PB products were not observed using 0.5 and 2 mM benzophenone solutions. Although both 5 and 10 mM benzophenone induced PB reactions, the later one generated more undesired side products. Thus, 5 mM benzophenone was selected as the optimum concentration for both the regular and reactive SCMS experiments. To generate PB products in the reactive SCMS experiments, UV irradiation (~15 min) was necessary for both PB reagents. Relatively abundant ions (e.g., intensity >104) were selected for MS/MS analysis.
Characterization of the Micropipette Needle.
To evaluate the sensitivity of the micropipette needle, we measured the limit of detection (LOD) of a number of standard compounds relevant to our studies. The LODs were determined as 1.0, 0.1, and 0.1 pM, for irinotecan, verapamil, and a phosphatidylcholine (PC(16:0/18:1)), respectively, which are comparable with the results obtained using standard nano-ESI source (Table S1).26,38
Workflow of Data Analysis.
To efficiently analyze the experimental data, we wrote two different Python scripts to determine the locations of C=C bonds in lipids through three steps: screening the potential lipids and their corresponding PB products, predicting the fragmentation of PB products, and identifying C=C bonds in lipids. First, we screened the potential lipids and their corresponding PB products using the Script A as shown in Figure S1. Briefly, relatively abundant ions (intensity >104) were retained from SCMS data in both the “regular” (without UV irradiation) and “reactive” (15 min UV irradiation) groups. Script A was used to search for the m/z values with mass difference of 58.0418 (using acetone reagent) or 182.0731 (using benzophenone reagent) between the “regular” and “reactive” groups ((m/z)reactive – (m/z)regular = 58.0418 or 182.0731, within 20 ppm), and to generate a list of m/z pairs. In each m/z pair, the (m/z)regular was regarded as a candidate of a potential lipid, whereas the (m/z)reactive was considered as the candidate of the corresponding PB product. Then, METLIN47 was used to find all potential lipids for each (m/z)regular, whereas those (m/z)regular values that cannot be found in METLIN were removed from the list along with their corresponding (m/z)reactive. An updated list of m/z pairs with potential identifications was then generated, and MS/MS experiments were conducted for ions in this list (intensity >104). Second, we predicted the diagnostic fragments of all potential PB products obtained from the previous step using the Script B (Figure S2). All featured fragments (i.e., m/z values) representing the head groups, tails, and adducts (i.e., H+, Na+, and K+) of each PB product were predicted according to the potential lipids generated from METLIN database searching. Last, we determined locations of C=C bonds in unsaturated lipids by comparing the predicted fragments in the second step and experimental MS/MS spectra, which were measured from selected ions using both CID (for diagnostic ions) and HCD (for lipids head groups) modes.
According to the METLIN database searching, all lipids detected in our experiments potentially belong to phospholipids. Phospholipids contain two fatty acid tails and a hydrophilic head with a phosphate group. For ions with the same m/z value, differences in these two fatty acid tails can generate multiple lipid isomers that can produce the same diagenetic ions in MS/MS (Table S2). To further elucidate the structure of each fatty acid tail, SCMS experiments were also conducted in the negative ion mode, in which ammonium acetate (10 mM) was added in the solvent to enhance the ion intensities. Acetate adducts of phospholipids were detected, and their fragments were used to determine the fatty acid chains in these phospholipids20 (Figure S3).
Determination of C=C Bond Locations in Unsaturated Lipids of Single Cells.
Determining the exact structures of large numbers of unsaturated lipids in single cells is challenging due to multiple factors, including extremely complex compositions of cellular species, small sample amounts, and the lack of complete structure information on all lipids in the current database. Nevertheless, our technique can be used as an analytical tool to identify structures or confine the detected species to limited numbers of isomers. We conducted comprehensive data analysis and tentatively determined 16 unsaturated lipids at the single-cell level in the current study (Table 1). Among them, PB products of three ions (m/z 760.5821, 253.2162, and 281.2485) were detected using both acetone and benzophenone as the reagents, whereas the rest species were only observed using acetone in the experiment. The presence of benzophenone (5 mM) likely affected the detection sensitivity of lipids (Figure S4). Our results may indicate that acetone is a more effective PB reagent to identify lipids C=C bond at the single-cell level.
Table 1.
Analysis of Unsaturated Lipids in Single Cells
| Ion mode | (m/z)Expa | (m/z)Calb | Error (ppm)c | reagentd | (m/z)Prode | identified lipids | diagnostic ions (m/z) |
|---|---|---|---|---|---|---|---|
| + | 760.5899 | 760.5856 | 5.7 | Ph2CO | 942.6542 | PC (16:0/18:1 (9)f | 650.4362, 800.5195 |
| + | 760.5821 | 760.5856 | −4.6 | ACE | 818.6579 | PC (16:0/18:1 (9)f | 650.4359, 676.4872 |
| + | 732.5506 | 732.5543 | −5.1 | ACE | 790.5902 | PC (16:0/16:1 (9)f | 650.4357, 676.4882 |
| + | 729.5892 | 729.5910 | −2.5 | ACE | 787.6395 | SM(d 18:1/18:1 (9))f | 645.4260, 729.3963 |
| + | 756.5475 | 756.5500 | −3.3 | ACE | 814.5931 | PC (16:1(7)/18:2(9,12))f | 646.6411, 672.4830, 728.5173 |
| + | 780.5482 | 780.5543 | −7.8 | ACE | 838.5899 | PC (16:1(9)/20:4(5,8,11,14))f | 712.5525, 714.5693, 738.8998, 780.4312 |
| + | 784.5746 | 784.5855 | −13.9 | ACE | 842.6530 | PC (16:0/20:3(11,14,17))f | 700.4511, 728.4319, 784.4609 |
| + | 786.5971 | 786.6012 | −5.2 | ACE | 844.7043 | PC (18:1(15)/18:1(15))f | 660.4500, 662.4649, 786.4759 |
| + | 787.6745 | 787.6693 | 6.6 | ACE | 845.6319 | SM(d 16:1/24:0) | 661.462 |
| + | 782.5654 | 782.5699 | −5.8 | ACE | 840.6057 | PC (16:0/18:1 (9)f | 656.5559, 672.4178, 698.4372, 782.5636 |
| + | 754.5334 | 754.5387 | −7.0 | ACE | 812.5792 | PC (16:0/18:4(9,11,1,115))f | 566.3104, 628.5224, 644.4230, 687.4790, 728.6622, 754.4146 |
| + | 810.5944 | 810.6012 | −8.5 | ACE | 868.6410 | PC (16:0/20:1 (11))f | 700.4489, 726.5560, 756.5484, 782.5640 |
| − | 253.2162 | 253.2173 | −4.3 | Ph2CO | 435.1712 | FA (16:1 (9)) | 171.1232 |
| − | 281.2485 | 281.2486 | −0.4 | Ph2CO | 463.3290 | FA (18:1 (9)) | 171.3439, 321.0381 |
| − | 253.2174 | 253.2173 | 0.4 | ACE | 311.1686 | FA (16:1 (9)) | 197.0279 |
| − | 281.2495 | 281.2486 | 3.2 | ACE | 339.1998 | FA (18:1 (9)) | 197.0279 |
Here, we presented an example, in which m/z 760.5821 was identified as PC(16:0/18:1(9)), with details to illustrate the workflow of locating C=C bonds in unsaturated lipids through comprehensive data analysis. First, we obtained all potential species with the m/z of 760.5821 ((m/z)regular = 760.5821). This peak is commonly detected in the regular SCMS experiment, and its potential PB adducts with acetone ((m/z)reactive 818.6579) and benzophenone ((m/z)reactive 942.6542) were extracted from experimental data using the Script A (Figure S1). We then searched for the potential species with the m/z of 760.5821 obtained from METLIN searching, and discovered that among all 36 potential lipids, 20 of them are phosphatidylcholines (PCs) and the rest 16 species are phosphatidylethanolamines (PEs) (Table S3). Second, structure identification of lipids and corresponding PB products were performed based on their MS/MS fragments. The MS/MS spectra of m/z 760.5821 in both HCD and CID modes are shown in Figure S5B, D. Because m/z 184.0724 is the headgroup of PC or sphingomyelin (SM),48,49 we excluded PEs from the list of potential candidates (Table S3). We then used the Script B to predict the featured fragments of the potential PB products based on the type of lipids, and the results are listed in Table S2. Last, we determined the position of C=C bond in the unsaturated lipid. The comparison between the experimental MS/MS of m/z 818.6579 and the predicted list led to the discovery of diagnostic ionpairs of m/z 650.4359 and m/z 676.4872 (i.e., Δ26) using acetone as the PB reagent (Figure 4A, Table S2)20 However, five lipids (Table S2) can generate the same diagnostic ion pair (m/z 650/676). To narrow down the potential candidates, we performed MS/MS analysis of m/z 818.59 (acetate adduct of m/z 760.5821) in the negative mode, and determined the fatty acid tails of m/z 760.5821 (Table S4). Combining all above results, the ion m/z 760.5821 was identified as PC (16:0/18:1(9)) (Table S5). This identification was further confirmed by comparing its MS/MS fragments with those obtained from standard compound PC (16:0/18:1(9)) measured in our experiments (Figure S5).
Figure 4.
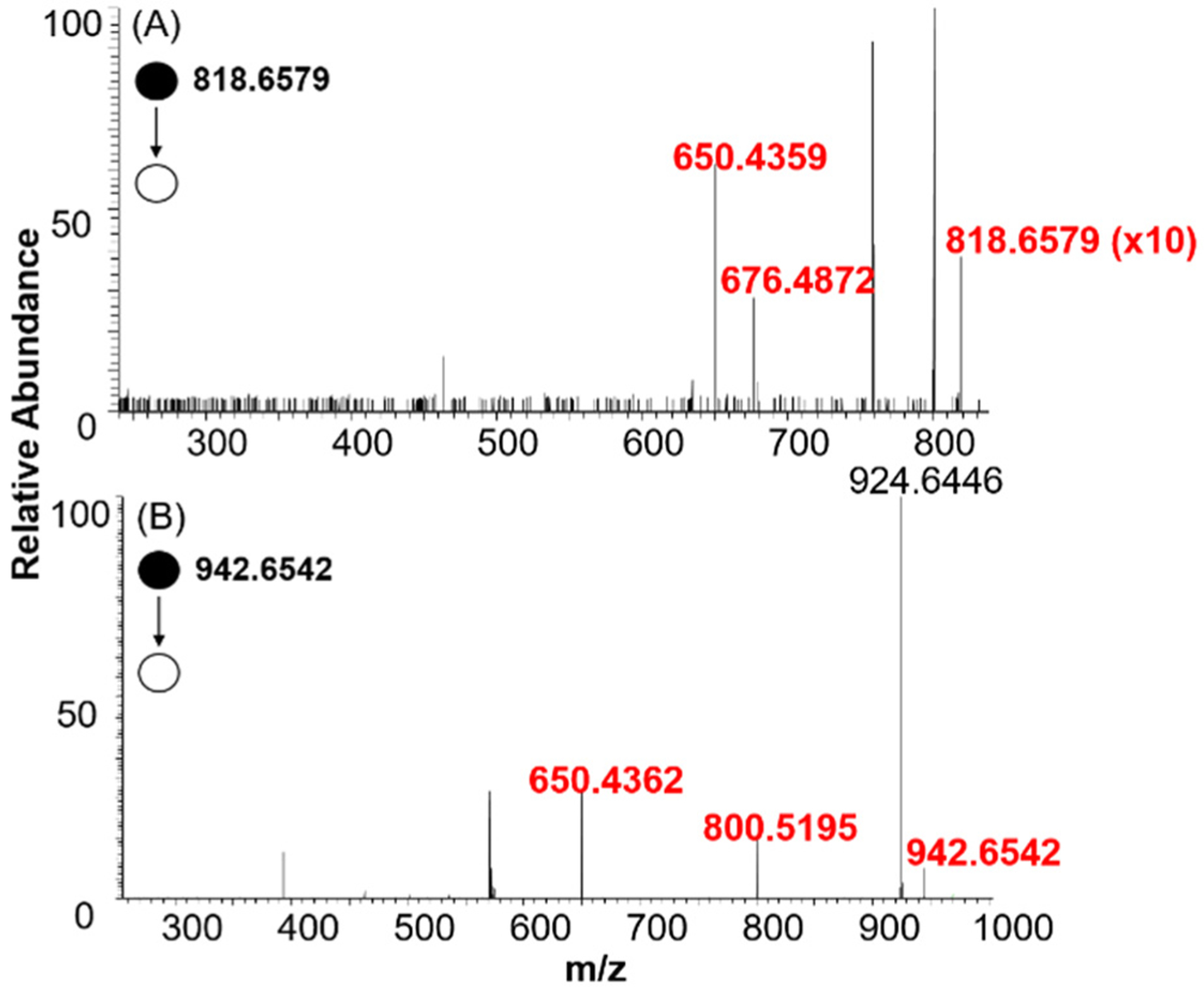
(A) MS/MS spectra of PB product of m/z 760.5899 (m/z 818.6579) using acetone as the PB reagent detected at single-cell level. Ions labeled in red font are diagnostic ions (m/z 650.4359 and m/z 676.4872). (B) MS/MS spectra of the PB product of m/z 760.5821 (m/z 942.6542) using benzophenone as the PB reagent detected from a single cell. Ions labeled in red font are diagnostic ions (m/z 650.4362 and m/z 800.5195).
As illustrated in Figure 3, the PB products at one C=C bond can produce a pair of diagnostic ions during fragmentation. In our experiments, nine different peaks of the PB products (Tables S5–S13) generated one or two pairs of diagnostic ions, with a mass difference of Δ26 (acetone) or Δ150 (150.0836, benzophenone21), in MS/MS analysis (Figures 4, S6, and S7). The production of two pairs of diagnostic ions likely due to the coexistence of isomeric lipids with C=C bond at different locations. For example, MS/MS experiment of m/z 868.6410 (the acetone PB product of m/z 810.5944) produced two pairs of diagnostic ions with a mass difference of Δ26: m/z 700.4489/726.5560 and 756.5484/782.5640 (Figure S6B). Previous studies found that a featured ion m/z 146.9807 was produced from the head-groups of sodiated PCs, SMs, or PEs in CID.50,51 MS/MS spectra of the corresponding lipid(s) (m/z 810.5944) in the regular SCMS experiment also contained the peak of m/z 146.9807 (Figure S6A), supporting the prediction that the ion m/z 810.5944 belongs to one or multiple Na+ adducts of PCs, SMs, or PEs. By searching for the potential species in METLIN database, we were able to exclude SMs and PEs from the list while keeping the rest PCs as potential candidates. Using the Script B, we predicted the diagnostic ions of the PB products of all potential PCs (Table S6) and compared with experimental observation (Figure S6B), in which two pairs of diagnostic ions (m/z 700.4489/726.5560 and 756.5484/782.5640) were detected. Because all potential PCs acquired from METLIN contain only one C=C bond, our results may indicate the coexistence of isomers with different locations of a C=C bond. Combining the results from PB reactions (positive ion mode) and information on fatty acid tails (negative ion mode, Table S4), these potential isomers were determined as seven unsaturated PCs as listed in Table S6: PC (16:0/20:1(11)), PC (18:0/18:1(13)), PC(18:0/18:1(9)), PC(18:1(9)/18:0), PC(20:1(11)/16:0), PC(22:0/14:1(9)), and PC(14:1(9)/22:0).
Interestingly, our experimental results indicate that the relative signal intensities of two diagnostic ions in a pair are different: more than half of the aldehyde ions are more abundant than the alkene ions. These differences are likely attributed to the relatively higher abundances of Isomer I than Isomer II produced in the PB reactions. Similar results have been reported in previous studies of bulk samples.20 In addition to paired diagnostic ions, we observed that the PB products of four lipids (m/z 814.5931, 838.5899, 840.6057, and 812.5792) produced unpaired diagnostic ions (Tables S9–S12). This is likely due to the relatively low concentrations of these lipids in single cells, and the abundances of their PB products were insufficient to produce detectable diagnostic ion pairs. The remaining seven lipids PB products (m/z 787.6395, 842.6530, 844.7043, 845.6319, 435.1712 (negative ion mode), 311.1686 (negative ion mode), and 339.1998 (negative ion mode)) also produced unpaired diagnostic ions. We totally analyzed 17 and 9 single cells in the positive and negative ion modes, respectively. The results of these ions are summarized in Tables S14–S20, and MS/MS spectra of these PB products and their corresponding lipids are shown in Figure S8.
Comparison Studies of Cell Lysates and Single Cells.
Lipidomics studies are generally conducted using lysates prepared from bulk biological samples (e.g., populations of cells, tissues, and plasma extraction) through multiple steps (e.g., cell lysis, centrifuging, supernatant transfer, drying, and reconcentration), which may affect their molecular compositions due to the potential variance of sample preparation protocols.1 In contrast, entire cellular contents from individual cells are retained in our SCMS experiments, minimizing the influence of sample preparation variance on composition analysis.34 To evaluate the difference between these two approaches to the identification of C=C bond, we conducted MS measurements of cell lysates and compared the results with those obtained at the single-cell level. Cell lysates were prepared and loaded into the micropipette needle for MS analysis, and data were collected before (no UV) and after the PB reactions (after 15 min of UV irradiation). Data analysis was performed using the same procedures as those used to process single cell results (see details in the SI). In general, MS/MS spectra of PB products obtained from the cell lysates were more complex than those from single cells (Figures S9 and S10), likely due to larger amounts of cellular contents in cell lysates. However, more cellular contents allowed for the analysis of additional PB products, including those with relatively lower abundances. For example, m/z 790.5902 (the PB product of m/z 732.5506) produced two pairs of diagnostic ions with a difference of m/z 26 (m/z 650.4354/676.4868 and 622.4042/648.4544) using cell lysate (Figure S9B, Table S21), whereas only one pair of diagnostic ions (m/z 650.4357/676.4882) were detected from single cells. According to MS/MS spectra (Figure S9A) and METLIN database, the PB product m/z 732.5506 was produced from PCs containing one C=C bond, indicating that the peak detected in single cells is attributed to multiple isomers. Therefore, traditional MS analysis of lysate can provide complementary information for single cell studies. Similar analyses of other ions using both cell lysates and single cells were conducted, as summarized in Table S22 and Figure S10.
Although enhanced ion signal intensities were obtained from cell lysates, larger amounts of cellular contents resulted in more undesired products from PB side reactions. Previous studies indicate that abundant lipids can react with PB reagents to generate side products, which may have very similar m/z values as the regular PB products or induce the retro-PB reactions, i.e., decomposition of PB products back into reactants in CID,20,25 resulting in interference for C=C bond analysis.20,52 For example, paired diagnostic ions in CID analysis of both m/z 868.6410 and 838.5899 were not observed using cell lysate, but they were detected at the single-cell level, providing structural information for the C=C bond analysis (Tables S6 and S11, Figures S6 and S7D).
CONCLUSIONS
We report a simple SCMS analysis device, the micropipette needle, that can accommodate PB reactions to determine C=C bond positions in unsaturated lipids at the single-cell level. HCT-116 cell line was used as a model, and individual cells were drawn into a micropipette needle to induce rapid cell lysis after mixing with acetone or acetonitrile solution containing benzophenone. To determine C=C bond locations in unsaturated lipids, the micropipette needle was then used as a nano-ESI emitter and coupled to a mass spectrometer to conduct both regular and reactive SCMS analyses of the same single cell. The regular SCMS measurement provided molecular analysis of single cell lysate without PB reactions. When conducting reactive SCMS experiments, the lysis solution also played a role as the PB reagent by reacting with unsaturated lipids at C=C bonds under UV irradiation. Python scripts were used to analyze data obtained from both regular and reactive SCMS experiments to screen potential lipids and their corresponding PB products. Assisted by MS/MS analysis of candidate PB product ions, C=C bond locations were determined to identify unsaturated lipids at the single-cell level. Experiments were conducted in both positive and negative ion modes to obtain comprehensive structure information. Comparative studies between cell lysates and single cells were performed, and we found that bulk sample analysis can provide complementary information for single cell studies. However, limited by a number of factors (e.g., complex compositions of cellular contents with limited amounts, lack of separation, and requirement of abundant PB product ions for MS/MS analysis), our current method is more effective for structure identification of unsaturated lipids and fatty acids with relatively high abundances and simple structures (e.g., with one or two C=C bonds). In addition, to generate abundant PB products, the throughput of reactive SCMS is primarily restricted by the time (15 min) needed for UV irradiation. The reaction time can likely be shortened using micropipette needles produced from thin-wall glass capillaries or other materials with higher UV transmission (e.g., quartz). Although a common cancer cell line was used as a model system in the current proof-of-concept studies, other systems (e.g., rare cells and heterogeneous cells) can be studied using this technique to answer specific biological questions that are intractable from bulk analysis. In addition, our techniques can potentially be used for broad ranges of reactive SCMS studies, in which other chemical reactions can be utilized to enhance the molecular analysis of single cells or trace amounts of biological samples.
SAFETY CONSIDERATIONS
Needles are hazardous (e.g., glass needles and fused silica capillaries) and should be handled with care. Cell culture and chemical handling need to follow standard safety protocols.
Supplementary Material
ACKNOWLEDGMENTS
This research project is partially supported by grants from National Institutes of Health (R01GM116116 and R21CA204706) and National Science Foundation (OCE 1634630)
ABBREVIATIONS USED
- SCMS
Single cell MS
- C=C
carbon–carbon double-bonds
- PB
Paternò–Büchi
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- SM
sphingomyelin
- FA
fatty acid
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c02245.
Cell lysis preparation, lipids C=C bond identification using cell lysate, MS/MS spectra of lipids, and their corresponding PB products (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.analchem.0c02245
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Altschuler SJ; Wu LF Cell 2010, 141 (4), 559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Muro E; Atilla-Gokcumen GE; Eggert US Mol. Biol. Cell 2014, 25 (12), 1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Sunshine H; Iruela-Arispe ML Curr. Opin. Lipidol 2017, 28 (5), 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sun M; Yang Z Anal. Chem 2019, 91 (3), 2384–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Li J; Condello S; Thomes-Pepin J; Ma X; Xia Y; Hurley TD; Matei D; Cheng J-X Cell Stem Cell 2017, 20 (3), 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mukherjee A; Kenny HA; Lengyel E Cell Stem Cell 2017, 20 (3), 291–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Tirinato L; Pagliari F; Limongi T; Marini M; Falqui A; Seco J; Candeloro P; Liberale C; Di Fabrizio E Stem Cells Int. 2017, 2017, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Murata N; Wada H Biochem. J 1995, 308, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cybulski LE; Del Solar G; Craig PO; Espinosa M; De Mendoza DJ Biol. Chem 2004, 279 (38), 39340–39347. [DOI] [PubMed] [Google Scholar]
- (10).Antonny B; Vanni S; Shindou H; Ferreira T Trends Cell Biol. 2015, 25 (7), 427–436. [DOI] [PubMed] [Google Scholar]
- (11).Lesa GM; Palfreyman M; Hall DH; Clandinin MT; Rudolph C; Jorgensen EM; Schiavo GJ Cell Sci. 2003, 116 (24), 4965–4975. [DOI] [PubMed] [Google Scholar]
- (12).Ting H-C; Chen L-T; Chen J-Y; Huang Y-L; Xin R-C; Chan J-F; Hsu Y-HH Lipids Health Dis. 2019, 18 (1), 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kelley NS; Hubbard NE; Erickson KL J. Nutr 2007, 137 (12), 2599–2607 [DOI] [PubMed] [Google Scholar]
- (14).Hrelia S; Biagi PL; Lorenzini A; Jimenez JAL; Horrobin DF; Bordoni A IUBMB Life 1997, 41 (2), 423–430. [DOI] [PubMed] [Google Scholar]
- (15).Hu T; Zhang JL J. Sep. Sci 2018, 41 (1), 351–372. [DOI] [PubMed] [Google Scholar]
- (16).Köfeler HC; Fauland A; Rechberger GN; Trötzmüller M Metabolites 2012, 2 (1), 19–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Han X; Yang K; Gross RW Mass Spectrom. Rev 2012, 31 (1), 134–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Quehenberger O; Armando AM; Brown AH; Milne SB; Myers DS; Merrill AH; Bandyopadhyay S; Jones KN; Kelly S; Shaner RL; et al. J. Lipid Res 2010, 51 (11), 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Andreyev AY; Fahy E; Guan Z; Kelly S; Li X; McDonald JG; Milne S; Myers D; Park H; Ryan A; et al. J. Lipid Res 2010, 51 (9), 2785–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ma X; Chong L; Tian R; Shi R; Hu TY; Ouyang Z; Xia Y Proc. Natl. Acad. Sci. U. S. A 2016, 113 (10), 2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Xu T; Pi Z; Song F; Liu S; Liu Z Anal. Chim. Acta 2018, 1028, 32–44 [DOI] [PubMed] [Google Scholar]
- (22).Ma X; Zhao X; Li J; Zhang W; Cheng J-X; Ouyang Z; Xia Y Anal. Chem 2016, 88 (18), 8931–8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Thomas MC; Mitchell TW; Harman DG; Deeley JM; Nealon JR; Blanksby SJ Anal. Chem 2008, 80 (1), 303–311. [DOI] [PubMed] [Google Scholar]
- (24).Feng Y; Chen B; Yu Q; Li L Anal. Chem 2019, 91 (3), 1791–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ma X; Xia Y Angew. Chem., Int. Ed 2014, 53 (10), 2592–2596. [DOI] [PubMed] [Google Scholar]
- (26).Zhu Y; Liu R; Yang Z Anal. Chim. Acta 2019, 1084, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Yang Y; Huang Y; Wu J; Liu N; Deng J; Luan T. TrAC, Trends Anal. Chem 2017, 90, 14–26. [Google Scholar]
- (28).Sun M; Yang Z; Wawrik B Front. Plant Sci 2018, 9, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Duncan KD; Fyrestam J; Lanekoff I Analyst 2019, 144 (3), 782–793. [DOI] [PubMed] [Google Scholar]
- (30).Dueñas ME; Essner JJ; Lee YJ Sci. Rep 2017, 7 (1), 14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Tsuyama N; Mizuno H; Tokunaga E; Masujima T Anal. Sci 2008, 24 (5), 559–561. [DOI] [PubMed] [Google Scholar]
- (32).Zhu H; Zou G; Wang N; Zhuang M; Xiong W; Huang G Proc. Natl. Acad. Sci. U. S. A 2017, 114 (10), 2586–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gong X; Zhao Y; Cai S; Fu S; Yang C; Zhang S; Zhang X Anal. Chem 2014, 86 (8), 3809–3816. [DOI] [PubMed] [Google Scholar]
- (34).Pan N; Rao W; Kothapalli NR; Liu R; Burgett AW; Yang Z Anal. Chem 2014, 86 (19), 9376–9380. [DOI] [PubMed] [Google Scholar]
- (35).Rao W; Pan N; Yang ZJ Visualized Exp. 2016, No. 112, No. e53911. [Google Scholar]
- (36).Sun M; Yang Z; Wawrik B Front. Plant Sci 2018, 9, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Liu R; Zhang G; Yang Z Chem. Commun 2019, 55 (5), 616–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Liu R; Pan N; Zhu Y; Yang Z Anal. Chem 2018, 90 (18), 11078–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Katz B-Z; Zamir E; Bershadsky A; Kam Z; Yamada KM; Geiger B Mol. Biol. Cell 2000, 11 (3), 1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bandura DR; Baranov VI; Ornatsky OI; Antonov A; Kinach R; Lou X; Pavlov S; Vorobiev S; Dick JE; Tanner SD Anal. Chem 2009, 81 (16), 6813–6822. [DOI] [PubMed] [Google Scholar]
- (41).Chen F; Lin L; Zhang J; He Z; Uchiyama K; Lin J-M Anal. Chem 2016, 88 (8), 4354–4360. [DOI] [PubMed] [Google Scholar]
- (42).Passarelli MK; Newman CF; Marshall PS; West A; Gilmore IS; Bunch J; Alexander MR; Dollery CT Anal. Chem 2015, 87 (13), 6696–6702. [DOI] [PubMed] [Google Scholar]
- (43).Mao S; Zhang W; Huang Q; Khan M; Li H; Uchiyama K; Lin JM Angew. Chem., Int. Ed 2018, 57 (1), 236–240. [DOI] [PubMed] [Google Scholar]
- (44).Standke SJ; Colby DH; Bensen RC; Burgett AW; Yang Z Anal. Chem 2019, 91 (3), 1738–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Stinson CA; Xia Y Analyst 2016, 141 (12), 3696–3704. [DOI] [PubMed] [Google Scholar]
- (46).Floch JJ Biol. Chem 1957, 226, 497–509. [PubMed] [Google Scholar]
- (47).Smith CA; O’Maille G; Want EJ; Qin C; Trauger SA; Brandon TR; Custodio DE; Abagyan R; Siuzdak G Ther. Drug Monit 2005, 27 (6), 747–751. [DOI] [PubMed] [Google Scholar]
- (48).Karnezis T; Fisher HC; Neumann GM; Stone BA; Stanisich VA J. Bacteriol 2002, 184 (15), 4114–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Brügger B; Erben G; Sandhoff R; Wieland FT; Lehmann WD Proc. Natl. Acad. Sci. U. S. A 1997, 94 (6), 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Zemski Berry KA; Hankin JA; Barkley RM; Spraggins JM; Caprioli RM; Murphy RC Chem. Rev 2011, 111 (10), 6491–6512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chughtai K; Jiang L; Greenwood TR; Glunde K; Heeren RM J. Lipid Res 2013, 54 (2), 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Zhang W; Zhang D; Chen Q; Wu J; Ouyang Z; Xia Y Nat. Commun 2019, 10 (1), 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


