Abstract
Purpose
PSA has limited performance in detecting prostate cancer (PCa). The transcription factor GATA2 is expressed in aggressive PCa. Here we analyzed the predictive value of urine extracellular vesicle (EV) GATA2 mRNA alone and in combination with a multigene panel to improve detection of PCa and high-risk disease.
Materials and Methods
GATA2 mRNA was analyzed in matched EVs isolated from urines pre- and post-prostatectomy (n=16) and paired urine and tissue prostatectomy samples (n=19). EV GATA2 mRNA performance to distinguish PCa and high-grade disease was tested in training (n=52) and validation (n=165) cohorts. The predictive value of a multigene score including GATA2, PCA3 and TMPRSS2-ERG (GAPT-E) was tested in both cohorts.
Results
Confirming its prostate origin, urine EV GATA2 mRNA levels significantly dropped after prostatectomy and correlated with PCa tissue GATA2 mRNA levels. In the training and validation cohort, GATA2 discriminated PCa (area under the curve (AUC), 0.74 and 0.66) and high-grade disease (AUC 0.78 and 0.65), respectively. Notably, the GAPT-E score improved discrimination of PCa (AUC 0.84 and 0.72) and high-grade cancer (AUC 0.85 and 0.71), in both cohorts when compared with each biomarker alone and PT-E (PCA3 and TMPRSS2-ERG). A GAPT-E score for high-grade PCa would avoid 92.1% of unnecessary prostate biopsies, compared to 61.9% when a PT-E score is used.
Conclusions
Urine EV GATA2 mRNA analysis improves the detection of high-risk PCa and may reduce the number of unnecessary biopsies.
Keywords: Prostate cancer, extracellular vesicles, GATA2, TMPRSS2-ERG, PCA3
INTRODUCTION
Prostate cancer (PCa) is the most common solid malignant disease and fifth leading cause of cancer death in men worldwide 1. In the 1990s, widespread adoption of prostate specific antigen (PSA) for PCa screening resulted in a spike in the incidence of localized disease followed by a decrease in mortality 2. However, the positive effects of PSA screening are overshadowed by its low specificity which compromises its ability to distinguish benign conditions and low-risk from high-risk (Gleason score, GS≥7) tumors and subjects patients to unnecessary biopsies 3–5. Thus, better biomarkers are needed to predict indolent tumors prior to invasive procedures.
Importantly, non-invasive liquid biopsy approaches for the detection of cancer have been developed 6. These novel urine- and blood-based liquid biopsies may add complementary information since they possibly reflect the molecular signature of the entire tumor while conventional methods such as tissue biopsies inform about the obtained sample 7. This tumor circulating material is mainly composed of circulating tumor cells, extracellular vesicles (EVs), DNA, RNA, and proteins 8. In particular, EVs which range from approximately 30 to 1,000 nm in diameter and contain RNA, DNA, and protein that are preserved from degradation, are secreted by most cell types into nearly all bodily fluids 9. EV content can be examined to determine the health of the cell from which they originate. Thus, taking into consideration all these characteristics, detailed molecular analysis of EVs may represent a valuable source to develop novel liquid biopsy biomarkers to study disease conditions, such as PCa.
In recent years several urine- and blood-based PCa biomarkers have been developed to better predict PCa10–17. However, there continues to be an unmet clinical need to develop novel liquid biopsy biomarkers that are not only prostate specific, but also define the aggressiveness of PCa. In this context, we and others have described that the pioneer transcription factor GATA2 plays a key role in the development of the prostate and in enhancing PCa aggressiveness 18. GATA2 contributes to the metastatic dissemination of PCa 19, as well as the survival of tumor cells to standard therapy 20–24. In tissue specimens, GATA2 mRNA and protein expression are increased in patients with high-risk primary PCa and correlate to clinical outcome 19, 20, 25. Patients with high GATA2 mRNA and protein expression have a higher GS, decreased disease free survival, and lower overall survival.
In this study, we investigated GATA2 mRNA expression levels in EVs from non-digital rectal exam (DRE) urine obtained from males with suspicion of PCa prior to biopsy to evaluate its predictive value for detecting any cancer and high-risk PCa. Moreover, based on previous studies which have shown that PCA3 and TMPRSS2-ERG discriminate PCa 13, 14, we evaluated whether a multiple gene score index that included GATA2 would further improve the detection of this disease.
MATERIAL AND METHODS
Study population
The study population consisted of men with suspicious DRE and/or PSA serum levels scheduled for initial prostate needle biopsy. Non-DRE urine samples prior to biopsy were tested in a training cohort of 52 men and an independent validation cohort of 165 men. Non-DRE urines were collected prospectively in three institutions (Hospital Calella, Barcelona, Spain; Thomas Jefferson University Hospital, Philadelphia, PA, USA; and Mount Sinai Hospital, New York, NY, USA) from September 2017 to April 2019. Pathological analysis of biopsies was performed blinded to the urine EV biomarker result. Paired urine samples from 16 patients pre- and post-prostatectomy and 19 patients matched formalin-fixed paraffin-embedded (FFPE) prostate tissues from radical prostatectomies (RPs) and urine samples pre-prostatectomy were analyzed. Magnetic resonance imaging (MRI) was not required to participate in this study. The study was approved by the Ethics Committee of each institutional review boards and conducted satisfying the principles of the Helsinki Declaration. All patients provided written informed consent.
PCa cell culture and generation of PCa cells stably expressing GATA2 cDNA and shRNAs Human PCa cell lines, DU145 and 22Rv1, were obtained from American Type Culture Collection (ATCC HTB-81 and CRL-2505, respectively) and maintained in RPMI 1640 medium (GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO) ultracentrifuged 120,000 g for 18 hours at 4°C to remove FBS-contained EVs. Cells were grown at 37°C in a humidified atmosphere with 5% CO2 and conditioned media for EV isolation collected after 48 hours of cell culture. GATA2 overexpression and short hairpin RNA (shRNA) stable expressing cells were generated as previously reported 23.
Sample processing and EV isolation
Non-digital rectal exam (DRE) urines samples (15 – 40 ml) were collected and stored in sterile urine collection vessels at 2–8°C no more than 14 days before analysis. Urines from patients with clinical symptomatology of infection and positive leukocyte-nitrite dipstick result at the time of collection were not included in the study. EVs were isolated by ultracentrifugation. Briefly, sequential centrifugation was performed at 4°C first at 300 g for 10 min, followed by 16,000 g for 20 min. Finally, the supernatant was ultracentrifuged at 120,000 g for 2 hours at 4°C in L8–70M Ultracentrifuge (Beckman) with 50.2 Ti Rotor (Beckman).
RNA extraction and PCR analysis
EV RNA was obtained using the Total Exosome RNA and Protein Isolation Kit (Invitrogen), FFPE RNA using the Recover All Total Nucleic Acid Isolation Kit (Invitrogen), and cellular RNA using the RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. cDNA was synthesized using SuperScript III First-Strand Synthesis System (Invitrogen) according to manufacturer’s instructions. RT-qPCR reactions were performed with the QuantStudio3 Real-Time PCR System (Thermo Fisher) using SYBR Green Master Mix (Applied Biosystems). A Ct (cycle threshold) value was derived for GATA2, PCA3, TMPRSS2-ERG, and ACTIN. Relative gene expression was calculated using the 2−ΔΔCt method. Primer sequences are described in Supplementary Table 1. Samples with RNA concentration <1.25 ng/μL and/or ACTIN Ct value >34 did not fulfill quality control and were excluded from the analysis. GATA2 RT-qPCR linearity and reproducibility were assessed using a standard curve constructed with GATA2 cDNA (Fig. S1).
Statistical analysis
PCA3, GATA2, PSA, and age were modeled as continuous variables while race, family history of PCa, and TMPRSS2-ERG were modeled as categorical variables. For all models the logarithm of PCA3 and GATA2 was used. TMPRSS2-ERG was dichotomized as 0 vs greater than 0. The three categories used for race were Caucasian, African-American, and other.
Logistic regression was used to develop two scores for the outcome cancer/no cancer in the training set. The variables used here were GATA2, PCA3, and TMPRSS2-ERG (GAPT-E) or PCA3, and TMPRSS2-ERG (PT-E). The parameters from fitting this regression model in the training set were then used to generate a predicted probability of cancer which was multiplied by 100 to the get the proposed score for each patient. The scoring model developed in the training set was used to generate scores for each patient in the validation set. Similarly, a SOC score was developed using age, PSA, and family history in the training set. A cutoff for diagnosing cancer vs no cancer was determined by finding the score in the training set that corresponded to approximately 90% sensitivity. This sensitivity level was chosen under the assumption that high sensitivity was a minimum requirement of a diagnostic test in this context. Performance of these cutoffs was evaluated in the validation cohort by calculating the sensitivity, specificity, negative predictive value (NPV), and positive predictive value (PPV). ROC curves were developed to assess the accuracy of the GAPT-E and PT-E scores, component variables, and a multivariable model incorporating standard of care (SOC) variables (race [validation cohort only], age, PSA, and family history) and other models for predicting any cancer and high-grade cancer in both the training and validation cohorts. To analyze correlations between variables, we used Spearman’s correlation tests when the two variables were assessed as continuous, t test when one variable was assessed as continuous and the other as qualitative and χ2 test (Fisher exact test) when the two variables were qualitative. Statistical tests were conducted at the two-sided 0.05 level of significance. All analyses were completed using SAS 9.4 and SAS/STAT 14.1 (SAS Institute, Cary, NC).
RESULTS
Urine EV GATA2 mRNA a biomarker of PCa aggressiveness
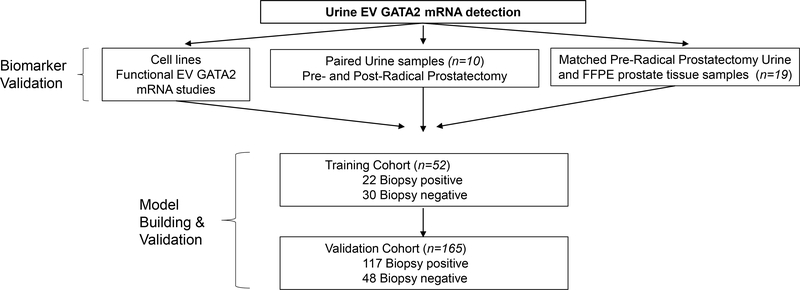
The usefulness of EV GATA2 mRNA to discriminate PCa was studied in PCa cells, urine pre- and post- radical prostatectomy and paired urine and tissue samples. In addition the capacity of urine EV GATA2 mRNA to predict biopsy result was studied in a training and validation cohorts of men with suspicion of PCa. Study flow diagram is presented in Fig. 1.
Fig. 1.
Scheme of study design. EV= extracellular vesicle, FFPE= formalin-fixed paraffin embedded.
To test whether GATA2 mRNA is expressed in EVs secreted from PCa cells we performed RT-qPCR analysis of GATA2 mRNA in EVs isolated from conditioned media of two PCa cell lines (DU145 and 22Rv1) and observed that EVs contained GATA2 mRNA (Fig. S2A). To study if mRNA GATA2 levels detected in EVs mimic the GATA2 mRNA levels expressed in PCa cells, we generated stable cell lines (DU145 and 22Rv1) that expressed high and low levels of GATA2 by either transducing cells with a GATA2 vector or two independent shRNAs, respectively. EV GATA2 mRNA levels secreted into conditioned media increased (Fig. S2B) or decreased (Fig. S2C) in a similar magnitude to those observed in the parental corresponding cells. Thus, these results demonstrate that GATA2 mRNA is expressed in EVs secreted by PCa cells and mirror the GATA2 mRNA levels of the PCa cells from which they originate.
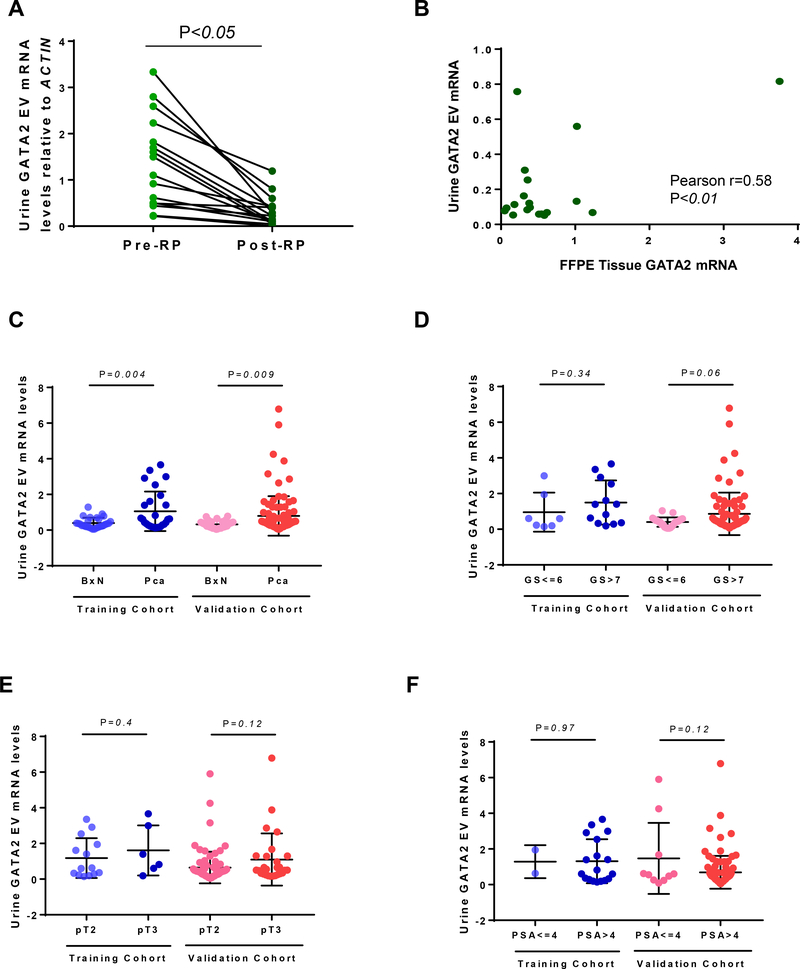
Based on these results, we conducted a series of studies to assess whether urine EV GATA2 mRNA expression derived from the patient’s prostate. Analysis of paired urine of 16 PCa patients pre- and post-RP showed that EV GATA2 mRNA levels significantly (p<0.05) decreased after surgery (Fig. 2A). Moreover, analysis of GATA2 mRNA in 19 paired prostate tissue samples and urine EVs showed a significant (r= 0.58, p<0.01) positive correlation (Fig. 2B). Collectively, these results indicate that urine EV GATA2 mRNA levels inform on the levels of GATA2 in PCa tumors and provide the rational to further study its use as a urine biomarker for detecting PCa.
Fig. 2.
Analysis of EV GATA2 mRNA in A) paired pre- and post-RP urine samples (n=16, t-test); B) matching urine and RP FFPE tissue (n=19, Pearson correlation); C) biopsy-negative and -positive (t-test); D) low- versus high-grade PCa (t-test); E) pT2 compared to pT3 (t-test) and; F) PSA<4ng/ml comparted to PSA≥4ng (t-test) in the training and validation cohorts.
EV = extracellular vesicle; BxN = biopsy negative; PCa = prostate cancer; GS = Gleason score; pT = pathological stages RP = radical prostatectomy; FFPE = formalin-fixed paraffin-embedded.
Next, we addressed whether urine EV GATA2 mRNA would associate with PCa and established clinico-pathologic variables of aggressive disease, such as Gleason score, stage and PSA levels. Detailed information of each patient cohort is provided in Table 1.
Table 1.
Subject clinical characteristics in the training and validation cohort
| Training | Validation | |||
|---|---|---|---|---|
| Median (range) or % | N | Median (range) or % | N | |
| QC passed | 92.3% | 48/52 | 88.5% | 146/165 |
| Age (years) | 67.8 (51 – 83) | 48 | 66.5 (47 – 82) | 146 |
| Serum PSA level (ng/ml) | 8.6 (1.5 – 39.2) | 48 | 7.9 (0.1 – 46.8) | 146 |
| Race | ||||
| Caucasian | 100% | 48 | 92.5% | 135 |
| African-American | - | - | 3.4% | 5 |
| Other | - | - | 4.1% | 6 |
| Family history of PCa | ||||
| Yes | 14.6% | 7 | 14.4% | 21 |
| No | 85.4% | 41 | 85.6% | 125 |
| Biopsy result positive | 41.6% | 20/48 | 63.2 % | 107/146 |
| Gleason score | ||||
| GS ≤ 6 (3+3) | 35% | 7 | 22.4% | 24 |
| GS ≥ 7 (3+4) | 65% | 13 | 77.6% | 83 |
| GS = 7 (3+4) | 25% | 5 | 40.2% | 43 |
| GS = 7 (4+3) | 20% | 4 | 20.6% | 22 |
| GS = 8 (4+4) | 10% | 2 | 9.3% | 10 |
| GS = 9 (4+5) | 10% | 2 | 5.6% | 6 |
| GS = 10 (5+5) | - | - | 1.9% | 2 |
| Pathological stages | ||||
| pT2 | 70% | 14 | 70.1% | 75 |
| pT3 | 30% | 6 | 29.9 % | 32 |
QC=quality control; PSA=prostate specific antigen; PCa=prostate cancer; GS=Gleason score
Indeed, we observed that urine EV GATA2 mRNA levels in patients that satisfied quality control criteria in a test (n=48) and validation cohort (n=146) prior to biopsy significantly increased in the biopsy-positive PCa patients, when compared to biopsy-negative patients (Fig. 2C). 75% of PCa in the training cohort and 63% in the validation cohort showed high levels of GATA2 (>0.32, the median in biopsy-negative patients in the training cohort). Of note, in biopsy-negative PCa, GATA2 mRNA levels did not increase in patients displaying benign prostatic hyperplasia (Fig. S3). Moreover, EV elevated GATA2 mRNA levels showed a trend towards being significantly associated to high Gleason Score (GS≥7) in the validation cohort (Fig. 2D) with 65.1% high-grade PCa expressing high GATA2, but did not associate with stage (Fig. 2E) and PSA levels (Fig. 2F). Of note, further detailed analysis between urine EV GATA2 levels and Gleason score showed no significant association with Gleason (3+4) or (4+3) and Gleason 7 or >7 (Fig. S4).
GATA2 alone and combined in a multigene score discriminates biopsy result
Mindful of the established role of PCA3 and TMPRSS2-ERG gene fusion as useful urine EV biomarkers to detect PCa 13, 14, we next extended our studies to explore if developing a multigene score that includes GATA2 improves the prediction of biopsy result. As observed with GATA2, urine PCA3 and TMPRSS2-ERG EV mRNA levels decreased after radical prostatectomy (Fig. S5A) and were significantly associated with the mRNA levels of paired tumor tissue samples (Fig. S5B), confirming its prostate origin. Of note, multivariable regression analysis showed that GATA2, PCA3, and TMPRSS2-ERG mRNA levels were independent predictive biomarkers for discriminating any cancer and high-grade PCa (Table 2).
Table 2.
Multivariate analysis of urine EV biomarkers and SOC variables for any cancer and high-grade PCa in the validation cohort.
| Predictor | Cancer vs. No Cancer OR (95% CI); p-value |
Gleason Score ≥7 vs <7 OR (95% CI); p-value |
|---|---|---|
| log_PCA3 | 1.96 (0.95 to 4.05); 0.069 | 1.63 (0.91 to 2.90); 0.099 |
| log_GATA2 | 1.80 (1.07 to 3.03); 0.027 | 2.15 (1.31 to 3.53); 0.002 |
| TE | ||
| TE>0 | 2.62 (1.09 to 6.26); 0.031 | 2.70 (1.23 to 5.92); 0.013 |
| PSA | 0.97 (0.92 to 1.04); 0.414 | 0.97 (0.91 to 1.03); 0.340 |
| Age | 0.98 (0.92 to 1.04); 0.456 | 0.98 (0.93 to 1.03); 0.469 |
| Race | ||
| African-American | 3.39 (0.19 to 83.98); 0.552 | 12.88 (0.49 to 294.24); 0.316 |
| Other | 2.19 (0.21 to 23.18); 0.950 | 5.40 (0.49 to 59.38); 0.780 |
| Family History of PCa | ||
| Yes | 0.49 (0.15 to 1.59); 0.236 | 0.41 (0.13 to 1.27); 0.123 |
EV=extracellular vesicles; SOC=standard of care; PCa=prostate cancer; TE=TMPRSS2-ERG; OR=odds ratio; CI=confidence interval
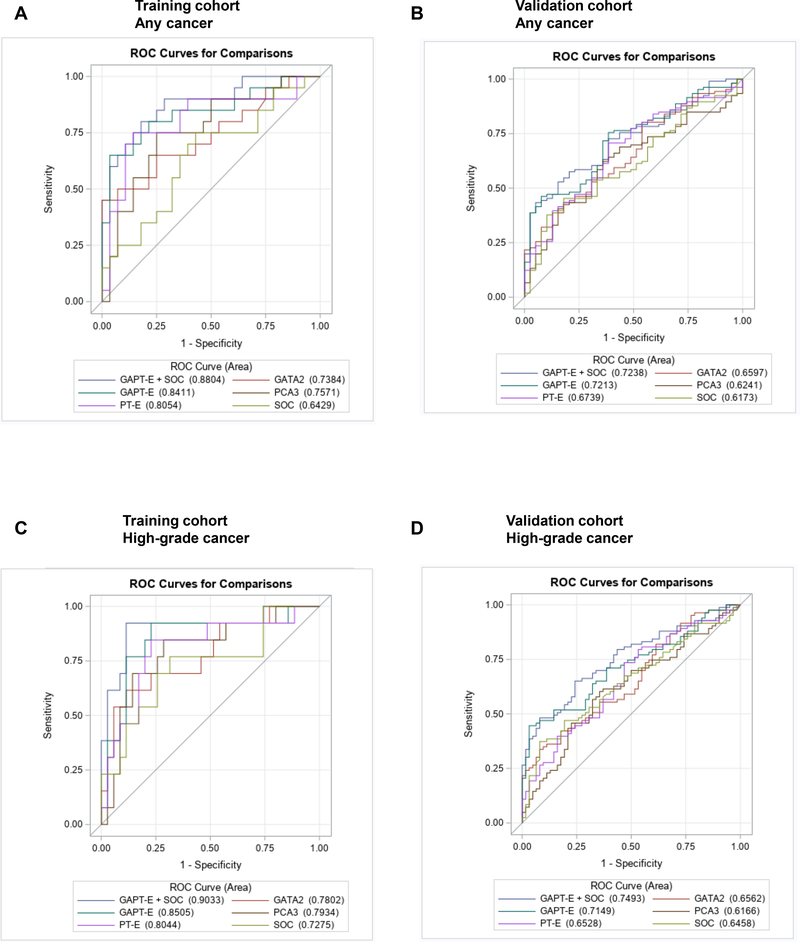
Next, we developed the GATA2, PCA3, and TMPRSS2-ERG (GAPT-E) and PCA3 plus TMPRSS2-ERG (PT-E) scores using logistic regression models in the training cohort (Table S2). ROC curve analysis of the training and validation cohort showed that tested biomarkers improved the detection of PCa (Fig. 3A and 3B) and high-risk disease (Fig. 3C and 3D), with the GAPT-E panel exhibiting the most robust results. Combining EV GATA2, PCA3, and TMPRSS2-ERG mRNA improved the prediction of any cancer and high-grade disease, when compared to individual biomarkers and PT-E and multivariable models of individual SOC variables (Table S3 and S4).
Fig. 3.
Receiver operating characteristic (ROC) curves show the performance of individual genes, SOC, PT-E, GAPT-E and GAPT-E plus SOC in discriminating any cancer (A and B) and high-grade disease (C and D) in the training (n=48) and validation (n=146) cohorts.
SOC = standard of care. PT-E= PCA3, and TMPRSS2-ERG. GAPT-E= GATA2, PCA3, and TMPRSS2-ERG.
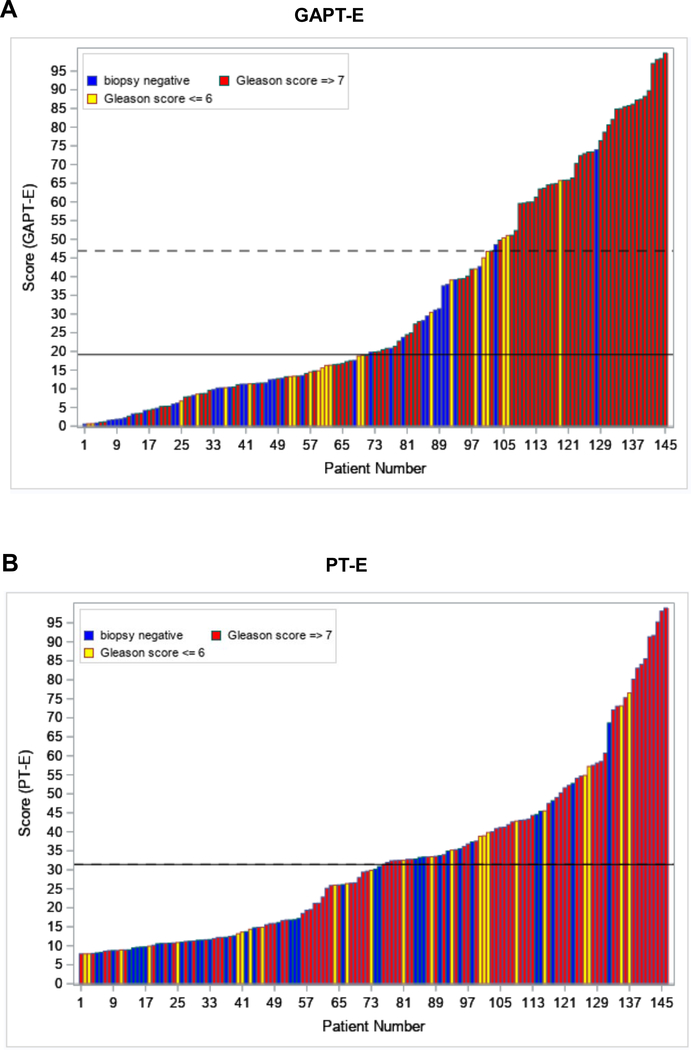
Finally, based in these results we tested the GAPT-E panel from each patient to derive a prediction score for initial biopsy result. Importantly, from this analysis we observed that adding GATA2 better discriminated high-risk prostate cancer in both the training (Table 3) and validation cohorts (Table 4). Of note, using the same GAPT-E cutoff score in the validation cohort as the one used in the training cohort, although the sensitivity decreased, the specificity improved by 74.6%, the NPV 11.1%, and the PPV 31.2% of high-grade disease when compared to a score based on SOC variables only. Indeed, adding GATA2 to PCA3 and TMPRSS2-ERG increased all these parameters, indicating that GATA2 substantially improves the prediction of high-risk disease prior to biopsy. Most importantly, applying a GAPT-E cut point of 46.9 for predicting high-grade disease would have avoided 69.8% of all biopsies (n=102) or 92.1% of unnecessary negative or low-grade biopsies (Fig. 4A). Otherwise, applying a PT-E (without GATA2) panel cut point of 31.4 for predicting high-grade disease would have only avoided 52.1% of all biopsies (n=76) or 61.9% of unnecessary negative biopsies (Fig. 4B).
Table 3.
Performance comparisons of GAPT-E to SOC at 90% fixed sensitivity – Cancer vs. No Cancer and High Grade Cancer (GS >=7) vs. GS<=6, No Cancer in the training cohort
| Cancer vs. No Cancer in the training cohort | ||||
| Sensitivity % (95% CI) | Specificity % (95% CI) | NPV % (95% CI) | PPV% (95% CI) | |
| *SOC | 90.0 (68.3–98.8) | 20.0 (7.71–38.6) | 75.0 (34.9–96.8) | 42.9 (27.7–59.0) |
| PT-E | 90.0 (68.3–98.8) | 63.3 (43.9–80.1) | 90.5 (69.6–98.8) | 62.1 (42.3–79.3) |
| GAPT-E | 90.0 (68.3–98.8) | 43.3 (25.5–62.6) | 86.7 (59.5–98.3) | 51.4 (34.0–68.6) |
| Improvement: GAPT-E vs. PT-E | −20.0 | 3.8 | −10.3 | |
| Improvement: GAPT-E vs. SOC | 23.3 | 11.7 | 8.5 | |
| High Grade Cancer (GS >=7) vs. GS<=6, No Cancer in the training cohort | ||||
| Sensitivity % (95% CI) | Specificity % (95% CI) | NPV % (95% CI) | PPV% (95% CI) | |
| *SOC | 92.3 (64.0–99.8) | 27.0 (13.8–44.1) | 90.9 (58.7–99.8) | 30.8 (17.0–47.6) |
| PT-E | 92.3 (64.0–99.8) | 54.1 (36.9–70.5) | 95.2 (76.2–99.2) | 41.4 (25.5–61.1) |
| GAPT-E | 92.3 (64.0–99.8) | 78.4 (61.8–90.2) | 96.7 (82.8–99.9) | 60.0 (36.1–80.1) |
| Improvement: GAPT-E vs. PT-E | 24.3 | 1.5 | 18.6 | |
| Improvement: GAPT-E vs. SOC | 51.4 | 5.8 | 29.2 | |
GAPT-E = urine EV GATA2, PCA3, and TMPRSS2-ERG; PT-E = urine EV PCA3, and TMPRSS2-ERG; SOC = standard of care; GS = Gleason score; NPV = negative predictive value; PPV = positive predictive value; CI = confidence interval
In the training data set SOC includes family history, PSA, and age.
Table 4.
Performance comparisons of GAPT-E to SOC at 90% fixed sensitivity in the training cohort – Cancer vs. No Cancer and High Grade Cancer (GS >=7) vs. GS<=6, No Cancer in the validation cohort
| Cancer vs. No Cancer in the validation cohort | ||||
| Sensitivity % (95% CI) | Specificity % (95% CI) | NPV % (95% CI) | PPV% (95% CI) | |
| *SOC | 86.9 (79.0–92.7) | 15.4 (5.86–30.5) | 30.0 (11.9–54.3) | 73.8 (65.2–81.2) |
| PT-E | 54.2 (44.3–63.9) | 69.2 (52.4–83.0) | 35.5 (24.8–47.3) | 82.9 (72.0–90.8) |
| GAPT-E | 57.0 (47.1–66.5) | 66.7 (44.6–76.6) | 36.1 (25.1–48.3) | 82.4 (71.8–90.3) |
| Improvement: GAPT-E vs. PT-E | 2.5 | 0.6 | −0.5 | |
| Improvement: GAPT-E vs. SOC | 51.3 | 6.1 | 8.6 | |
| High Grade Cancer (GS >=7) vs. GS<=6, No Cancer in the validation cohort | ||||
| Sensitivity % (95% CI) | Specificity % (95% CI) | NPV % (95% CI) | PPV% (95% CI) | |
| *SOC | 84.3 (74.7–91.4) | 17.5 (9.1–29.1) | 45.8 (25.6–67.2) | 57.4 (48.1–66.3) |
| PT-E | 55.4 (44.1–66.3) | 61.9 (48.8–73.9) | 51.3 (39.6–63.0) | 65.7 (53.4–76.7) |
| GAPT-E | 47.0 (35.9–58.3) | 92.1 (82.4–97.4) | 56.9 (46.7–66.6) | 88.6 (75.4–96.2) |
| Improvement: GAPT-E vs. PT-E | 30.2 | 5.6 | 22.9 | |
| Improvement: GAPT-E vs. SOC | 74.6 | 11.1 | 31.2 | |
GAPT-E = urine EV GATA2, PCA3, and TMPRSS2-ERG; PT-E = urine EV PCA3, and TMPRSS2-ERG; SOC = standard of care; GS = Gleason score; NPV = negative predictive value; PPV = positive predictive value; CI = confidence interval
In the validation set SOC includes race, family history, PSA, and age
Fig. 4.
Waterfall plot of A) GAPT-E and B) PT-E scores in relation to biopsy outcomes across the validation cohort (n=146). Dashed line is high-grade cutoff and solid line is cutoff for any cancer. For the PT-E test the high-grade and any cancer cutoffs are the same, hence the lines are coincident. Each colored bar represents an individual patient’s score, increasing from left to right. Blue, biopsy negative; Yellow, GS≤6; and Red, GS≥7.
GAPT-E = GATA2, PCA3, and TMPRSS2-ERG; PT-E= PCA3, and TMPRSS2-ERG.
Collectively, these results demonstrate that urine EV GATA2 mRNA levels improves the detection of clinically significant PCa and may help tailor the management of this disease.
DISCUSSION
Overdiagnosis of indolent PCa is an important issue worldwide 26. Although the high sensitivity of PSA has significantly improved the detection of PCa, its low specificity has raised the number of unnecessary invasive diagnostic procedures. Consequently, there is an unmet clinical need to identify novel biomarkers to detect clinically significant PCa and avoid unnecessary biopsies.
In an attempt to improve the detection of clinically significant PCa a wide variety of biomarkers have been studied. In this context, based on their high prostate organ specificity, the long non-coding RNA PCA3 27 and the oncogenic gene fusion rearrangement TMPRSS2-ERG 16 have been analyzed in EVs and used to develop several clinical diagnostic and prognostic tests.
A major strength of the present study is that it is the first to describe the utility of GATA2 as a urine biomarker that reflects both prostate specificity and PCa aggressiveness. Our results showed that urine GATA2 EV mRNA levels correlate with PCa tissue expression and significantly decrease after prostatectomy, supporting its use as a urine biomarker for the detection of PCa. Importantly, these results laid the foundation to uncover that GATA2 mRNA levels in urine EVs may discriminate PCa and high-grade disease. Furthermore, in this study, we confirm the results of previous reports 13, 14 describing the utility of non-DRE urine EV PCA3 and TMPRSS2-ERG mRNA analysis for the detection of PCa. Most notably, when adding GATA2 to PCA3 and TMPRSS2-ERG the detection of high-risk PCa was substantially improved.
A limitation of our study is that GATA2 is not exclusively expressed in prostate cells. In this context, it is worth noting that urine EV GATA2 mRNA expression did not completely disappear after prostatectomy suggesting that other cells within the urinary track may secret EVs containing GATA2 mRNA. Indeed public available transcriptomic datasets show that GATA2 mRNA is also expressed at low levels in the bladder28, 29. Moreover, although EV GATA2 mRNA levels discriminate between Gleason score 6 and Gleason score ≥7, it did not differentiate between Gleason score 3+4 vs 4+3 and >7. These results may be a consequence of several factors which comprise: i) limited number of patients with high Gleason score included in the study, ii) lack of correlation studies between PCa volume and EV GATA2 mRNA expression, and iii) pooled analysis of EVs. Ideally, new technologies that allow the quantification and analysis of single EV molecular content, when complemented with PCa volume, may improve the accuracy of EV biomarkers in discriminating high-grade PCa. Other limitations include the lack of MRN imaging studies and multiple serial biopsies in most of the patients analyzed, as well as the inability to study free-PSA as part of the SOC variables which can be a source of potential error.
Importantly, to translate urine EV GATA2 mRNA detection into the clinic several careful considerations will be required. For example, the selection of the best commercial kit that isolates EVs in a timelier manner instead of serial ultracentrifugation30, as well as the identification of clinical variables that might influence urine EV GATA2 mRNA expression such as infection, digital rectal examination, sexual activity, exercise and diet, among others, will need to be considered. Finally, the GAPT-E scores and cutoff points will require further validation in independent larger prospective studies. Ideally, these studies should be designed to evaluate how GAPT-E may complement existing tests, such as Prostate Health Index 12 and OPKO-4K15, which already have shown to better discriminate high-risk PCa.
CONCLUSIONS
In men with suspicion of PCa, urine EV GATA2 mRNA can discriminate high-grade disease and be used to tailor prostate biopsy.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Shannon Fields, Kerith Wang, Marlon Suarez, and Rachel Weil for their help in sample collection and the Biostatistics Shared Resource for their support in the statistical analysis.
Financial support
This work was supported by the National Cancer Institute at the National Institutes of Health (R01 CA207311) and NCI Cancer Center Support Grant 5P30CA056036-17.
Footnotes
The authors declare no conflicts of interest.
Publisher's Disclaimer: DISCLAIMER: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our subscribers we are providing this early version of the article. The paper will be copy edited and typeset, and proof will be reviewed before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to The Journal pertain.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I et al. : Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 68: 394, 2018 [DOI] [PubMed] [Google Scholar]
- 2.Potosky AL, Miller BA, Albertsen PC et al. : The role of increasing detection in the rising incidence of prostate cancer. JAMA, 273: 548, 1995 [PubMed] [Google Scholar]
- 3.Lavallee LT, Binette A, Witiuk K et al. : Reducing the Harm of Prostate Cancer Screening: Repeated Prostate-Specific Antigen Testing. Mayo Clin Proc, 91: 17, 2016 [DOI] [PubMed] [Google Scholar]
- 4.van Vugt HA, Roobol MJ, Kranse R et al. : Prediction of prostate cancer in unscreened men: external validation of a risk calculator. Eur J Cancer, 47: 903, 2011 [DOI] [PubMed] [Google Scholar]
- 5.Loeb S: Guideline of guidelines: prostate cancer screening. BJU Int, 114: 323, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Siravegna G, Marsoni S, Siena S et al. : Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol, 14: 531, 2017 [DOI] [PubMed] [Google Scholar]
- 7.Diaz LA Jr., Bardelli A: Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol, 32: 579, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan JCM, Massie C, Garcia-Corbacho J et al. : Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer, 17: 223, 2017 [DOI] [PubMed] [Google Scholar]
- 9.Tkach M, Thery C: Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell, 164: 1226, 2016 [DOI] [PubMed] [Google Scholar]
- 10.Chevli KK, Duff M, Walter P et al. : Urinary PCA3 as a predictor of prostate cancer in a cohort of 3,073 men undergoing initial prostate biopsy. J Urol, 191: 1743, 2014 [DOI] [PubMed] [Google Scholar]
- 11.Govers TM, Caba L, Resnick MJ: Cost-Effectiveness of Urinary Biomarker Panel in Prostate Cancer Risk Assessment. J Urol, 200: 1221, 2018 [DOI] [PubMed] [Google Scholar]
- 12.Catalona WJ, Bartsch G, Rittenhouse HG et al. : Serum pro-prostate specific antigen preferentially detects aggressive prostate cancers in men with 2 to 4 ng/ml prostate specific antigen. J Urol, 171: 2239, 2004 [DOI] [PubMed] [Google Scholar]
- 13.McKiernan J, Donovan MJ, Margolis E et al. : A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer in Patients with Prostate-specific Antigen 2–10ng/ml at Initial Biopsy. Eur Urol, 74: 731, 2018 [DOI] [PubMed] [Google Scholar]
- 14.McKiernan J, Donovan MJ, O’Neill V et al. : A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol, 2: 882, 2016 [DOI] [PubMed] [Google Scholar]
- 15.Vickers AJ, Cronin AM, Aus G et al. : A panel of kallikrein markers can reduce unnecessary biopsy for prostate cancer: data from the European Randomized Study of Prostate Cancer Screening in Goteborg, Sweden. BMC Med, 6: 19, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlins SA, Day JR, Lonigro RJ et al. : Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur Urol, 70: 45, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Neste L, Hendriks RJ, Dijkstra S et al. : Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur Urol, 70: 740, 2016 [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Bravo V, Carceles-Cordon M, Hoshida Y et al. : The role of GATA2 in lethal prostate cancer aggressiveness. Nat Rev Urol, 14: 38, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang YT, Wang K, Fazli L et al. : GATA2 as a potential metastasis-driving gene in prostate cancer. Oncotarget, 5: 451, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He B, Lanz RB, Fiskus W et al. : GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proc Natl Acad Sci U S A, 111: 18261, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendriksen PJ, Dits NF, Kokame K et al. : Evolution of the androgen receptor pathway during progression of prostate cancer. Cancer Res, 66: 5012, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Robinson JL, Tzou KS, Parker AS et al. : GATA2 expression and biochemical recurrence following salvage radiation therapy for relapsing prostate cancer. Br J Radiol, 90: 20170174, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vidal SJ, Rodriguez-Bravo V, Quinn SA et al. : A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell, 27: 223, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Bravo V, Pippa R, Song WM et al. : Nuclear Pores Promote Lethal Prostate Cancer by Increasing POM121-Driven E2F1, MYC, and AR Nuclear Import. Cell, 174: 1200, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohm M, Locke WJ, Sutherland RL et al. : A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene, 28: 3847, 2009 [DOI] [PubMed] [Google Scholar]
- 26.Sandhu GS, Andriole GL: Overdiagnosis of prostate cancer. J Natl Cancer Inst Monogr, 2012: 146, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bussemakers MJ, van Bokhoven A, Verhaegh GW et al. : DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res, 59: 5975, 1999 [PubMed] [Google Scholar]
- 28.Consortium GT: Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348: 648, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhlen M, Fagerberg L, Hallstrom BM et al. : Proteomics. Tissue-based map of the human proteome. Science, 347: 1260419, 2015 [DOI] [PubMed] [Google Scholar]
- 30.Ma C, Jiang F, Ma Y et al. : Isolation and Detection Technologies of Extracellular Vesicles and Application on Cancer Diagnostic. Dose Response, 17: 1559325819891004, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.