Summary
The musculoskeletal system is a striking example of how cell identity and position is coordinated across multiple tissues to ensure function. However, it is unclear upon tissue loss, such as complete loss of cells of a central musculoskeletal connecting tendon, whether neighboring tissues harbor progenitors capable of mediating regeneration. Here, using a zebrafish model, we genetically ablate all embryonic tendon cells and find complete regeneration of tendon structure and pattern. We identify two regenerative progenitor populations, sox10+ perichondrial cells surrounding cartilage and nkx2.5+ cells surrounding muscle. Surprisingly, laser ablation of sox10+ cells, but not nkx2.5+ cells, increases tendon progenitor number in the perichondrium, suggesting a mechanism to regulate attachment location. We find BMP signaling is active in regenerating progenitor cells and is necessary and sufficient for generating new scxa+ cells. Our work shows that muscle and cartilage connective tissues harbor progenitor cells capable of fully regenerating tendons and this process is regulated by BMP signaling.
Keywords: Zebrafish, tendon, ablation, regeneration, progenitor cells, BMP, scleraxis, Sox10, Nkx2.5
Introduction
Tendons provide crucial connections within the musculoskeletal system and are essential for movement. Human tendons are highly prone to injury and heal imperfectly through scar formation. These injuries can be challenging to treat clinically as the highly ordered tendon extracellular matrix organization is rarely restored, resulting in re-injury and limited mobility. Knowledge of tendon formation and repair has been advanced by the identification of the basic helix-loop-helix transcription factor Scleraxis (Scx) [1–3], which is the earliest marker of tendon fate. The expression of Tenomodulin (Tnmd), a type II transmembrane glycoprotein [4–6], and the assembly and growth of type I collagen fibrils signifies the beginning of tendon differentiation and functional maturation [7–10]. Throughout these processes, interactions between the forming muscles, tendons, and bones are essential for a properly formed musculoskeletal system [11–15], but their role in tendon healing has been less well defined.
Multiple mammalian models of tendon injury have improved our understanding of the tendon healing process. However, our ability to gain knowledge of regenerative mechanisms for tendon repair is restricted by the limited innate regenerative capabilities of adult mammalian tendons. Recent studies using mouse neonatal models of tendon injury have demonstrated regenerative healing that is dependent upon the involvement of Scx-lineage cells [16,17]. Adult studies have also identified multiple cell populations from the tendon as contributors to adult tendon repair [18–23], yet it is unknown if cells from adjacent tissues can participate in tendon regeneration upon complete loss or damage to the tissue. Zebrafish have robust regenerative potential, but their ability to regenerate tendons is unknown. Previous studies have shown that zebrafish tendons have molecular, structural, and mechanical similarities to mammalian tendons [24–27]. If zebrafish can regenerate their tendon tissue, this model would be ideal for identifying mechanisms underlying regenerative tendon healing, which would impact therapeutic approaches to treat tendon disease and injury.
Using a new genetic model that removes all scxa+ tendon cells and live imaging of the regenerative process, we show tendon cell regeneration is mediated by progenitors from neighboring tissues. Tendon cell ablation disrupts the morphology of the tendon matrix as well as the connecting cartilage and muscle. We find that zebrafish completely regenerate tendon attachment pattern and extracellular matrix structure and the morphology of the cartilage and muscle is restored. We determine that new tendon progenitor cells are recruited from sox10+ and nkx2.5+ cells surrounding the adjacent cartilage and muscle tissues. BMP signaling is active in the recruited progenitor cells and is sufficient and necessary for robust tendon cell regeneration. Collectively, our data show that after complete tendon cell loss, the surrounding tissues respond and serve as reservoirs of progenitor cells that mediate regeneration.
Results
Tendon cell ablation disrupts cartilage and muscle morphology
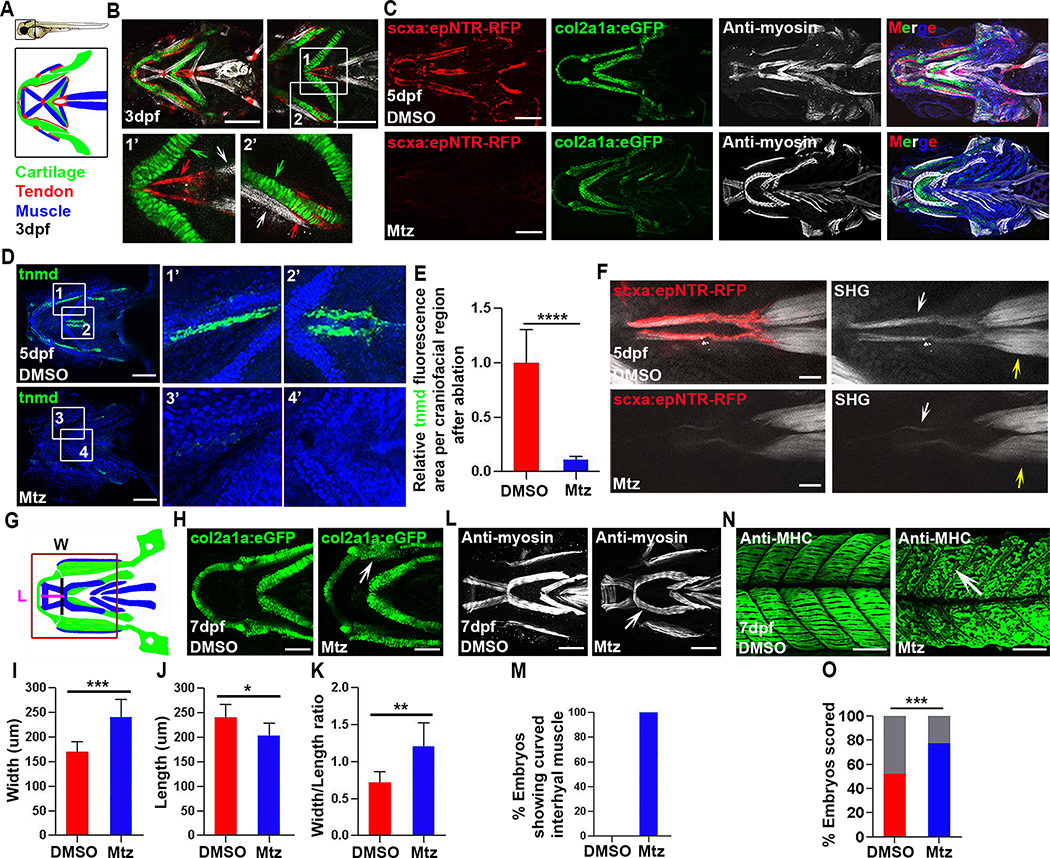
To investigate tendon regeneration in the context of a functioning musculoskeletal system, we have generated a genetic inducible tendon cell ablation model (Figures 1A and 1B) [28–32]. At 3 days post-fertilization (dpf), Tg(scxa:gal4-vp16; uas:epNTR-RFP), abbreviated as Tg(scxa:epNTR-RFP), is robustly expressed in all tendon populations in the craniofacial, pectoral fin, and myoseptal regions (Figures 1B and S1A). Expression of Tg(scxa:epNTR-RFP) resembles Tg(scxa:mcherry) and endogenous scxa expression (Figure S1A), and colocalizes with tenomodulin (tnmd) and collagen type 1a2 (col1a2), two mature tendon extracellular matrix (ECM) components [6,33], in the craniofacial and trunk myoseptal tendons (Figures S1B–S1E). We treated Tg(scxa:epNTR-RFP) embryos with Mtz from 3–5 or 5–7 dpf, stages at which the fish swim and move their jaws [34]. Following Mtz treatment, virtually all scxa+ tendon cells in Tg(scxa:epNTR-RFP) embryos were ablated, as demonstrated by the loss of RFP fluorescence (Figures 1C and S1I) and absence of tnmd transcripts (Figures 1D, 1E, and S1F). Terminal deoxynucleotidyl transferase-dUTP nick end labeling (TUNEL) assays confirmed apoptotic cell death following Mtz treatment (Figures S1G and S1H). To determine effects of tendon cell loss on the collagen fibrils of the tendon ECM, we examined collagen organization and density using second harmonic generation (SHG) imaging microscopy [35]. Due to its anatomical accessibility and functional contribution to craniofacial movements [36], we focused on the sternohyoideus (SH) tendon, which connects the SH muscles to the cartilages of the ventral pharynx [24,34]. We found that the SHG signal was disrupted in the SH tendon in ablated versus unablated controls (Figure 1F), demonstrating requirements for tendon cells in collagen matrix organization. Consistent with the tendon cell ablation, the embryos showed diminished swimming capacity, which may be due to disruption to the musculoskeletal system.
Figure 1. Tendon cell ablation disrupts the collagen matrix and cartilage and muscle morphology.
(A) Cartoons of a 3 dpf zebrafish embryo and the craniofacial muscle (Blue), tendon (Red), and cartilage (Green).
(B) Confocal images of craniofacial tendon (red), cartilage (green), and muscle (white) in Tg(scxa:gal4-vp16; uas:epNTR-RFP; col2a1a:eGFP) fish. Insets show higher magnification views of the sternohyoideus (SH) tendon region (white box 1 and 1’, red arrow, SH tendon. green arrow, ceratohyal cartilage. white arrow, SH muscle) and palatoquadrate region (white box 2 and 2’, red arrow, tendon. green arrow, palatoquadrate cartilage. white arrow, adductor mandibulae muscle).
(C) Tendon cell ablation in Tg(scxa:gal4-vp16; uas:epNTR-RFP; col2a1a: eGFP) fish. Top, unablated (DMSO). Bottom, ablated (Mtz). Blue in merged image is Hoechst staining.
(D) Fluorescent in situ for tnmd and Hoechst stained craniofacial regions in unablated (DMSO) and ablated (Mtz) embryos. White box 1, 3, 1’, 3’, palatoquadrate ligament region. White box 2, 4, 2’, 4’, SH tendon region.
(E) Quantification of tnmd fluorescence area in craniofacial region in unablated (DMSO, n=7) and ablated (Mtz, n=7) embryos. Two-tailed, unpaired Student’s t-test and data are mean ± s.d. ****, p<0.0001.
(F) Multiphoton images of SH tendons (red), fibrillar collagen (white arrow), and SH muscle (yellow arrow) in unablated (DMSO) and ablated (Mtz) embryos. SHG, second harmonic generation. Scale bar, 25 μm.
(G) Cartoon illustrating craniofacial region, ventral view with lines indicating areas used to measure width (W) and length (L) Red box shows the area analyzed in (H).
(H) Higher magnification views of lower jaw cartilage in control Tg(col2a1a: eGFP) (DMSO) and ablated (Mtz) embryos. White arrow, dysmorphic palatoquadrate cartilage.
(I-K) Quantification of width (I), length (J), and width/length ratio (K) for lower jaw cartilage in unablated (DMSO, n=8) and ablated (Mtz, n=8) embryos. Two-tailed, unpaired Student’s t-test and data are mean ± s.d. *, p<0.05. **, p<0.01. ***, p<0.001.
(L) Ventral views of craniofacial muscle in unablated (DMSO) and ablated (Mtz) embryos. White arrow, curved interhyal muscle.
(M) Percentage of embryos showing a curved interhyal muscle phenotype in unablated (DMSO, n=5, 0%) and ablated (Mtz, n=4, 100%) embryos.
(N) Confocal images of axial muscle in unablated (DMSO) and ablated (Mtz) embryos. White arrow, detached muscle.
(O) Percentage of embryos showing attachment and detachment phenotypes in unablated (DMSO, n=109) and ablated (Mtz, n=110) embryos. Grey bars show percentages of muscle fibers that remain attached in both conditions. Red and blue bars showed percentages of muscle fibers that detached in both conditions. Chi-squared test. ***, p<0.001.
dpf, days post fertilization. All images, anterior to the left. All scale bars are 100μm, except in (F) where noted. Tendon cell was ablated with Mtz at 3–5 dpf.
See also Figures S1 and S2
We next examined muscle and cartilage morphology in ablated Tg(scxa:epNTR-RFP; col2a1a:eGFP) fish. Analysis of col2a1a:eGFP+ chondrocytes revealed malformations in Meckel’s and ceratohyal cartilage of the lower jaw in tendon cell ablated animals compared with unablated controls at 7 dpf (Figures 1G and 1H). These included an increased width (distance between palatoquadrates)/length (distance between Meckel’s and ceratohyal) ratio of 1.21 ± 0.32 in ablated animals compared with 0.72 ± 0.14 in unablated controls (Figures 1I–1K) at all time points examined (Figures S2A and S2B). In contrast, the ethmoid plate, which also derives from cranial neural crest cells [37–39] and to which there are many fewer connected tendons (Figures S2C and S2D), was morphologically less affected by tendon cell ablation (Figures S2E and S2F). Craniofacial muscles also appeared abnormal, particularly the interhyal muscle, in ablated animals compared with unablated controls (Figures 1C, 1L, 1M, S2A, and S2B). Using a previously described muscle attachment assay [26], we found that the axial muscles of ablated animals had increased detachment frequency when challenged by electrical stimulation (Figures 1N and 1O). This indicates that tendon cells are required to maintain functional myotendinous junctions. Collectively, these analyses demonstrate a pivotal role for tendon cells in maintaining proper cartilage and muscle morphology. These results also suggest, in principle, that abnormalities observed in muscle or skeletal tissues could instead result from defects in tendons.
Zebrafish completely regenerate tendon pattern and structure
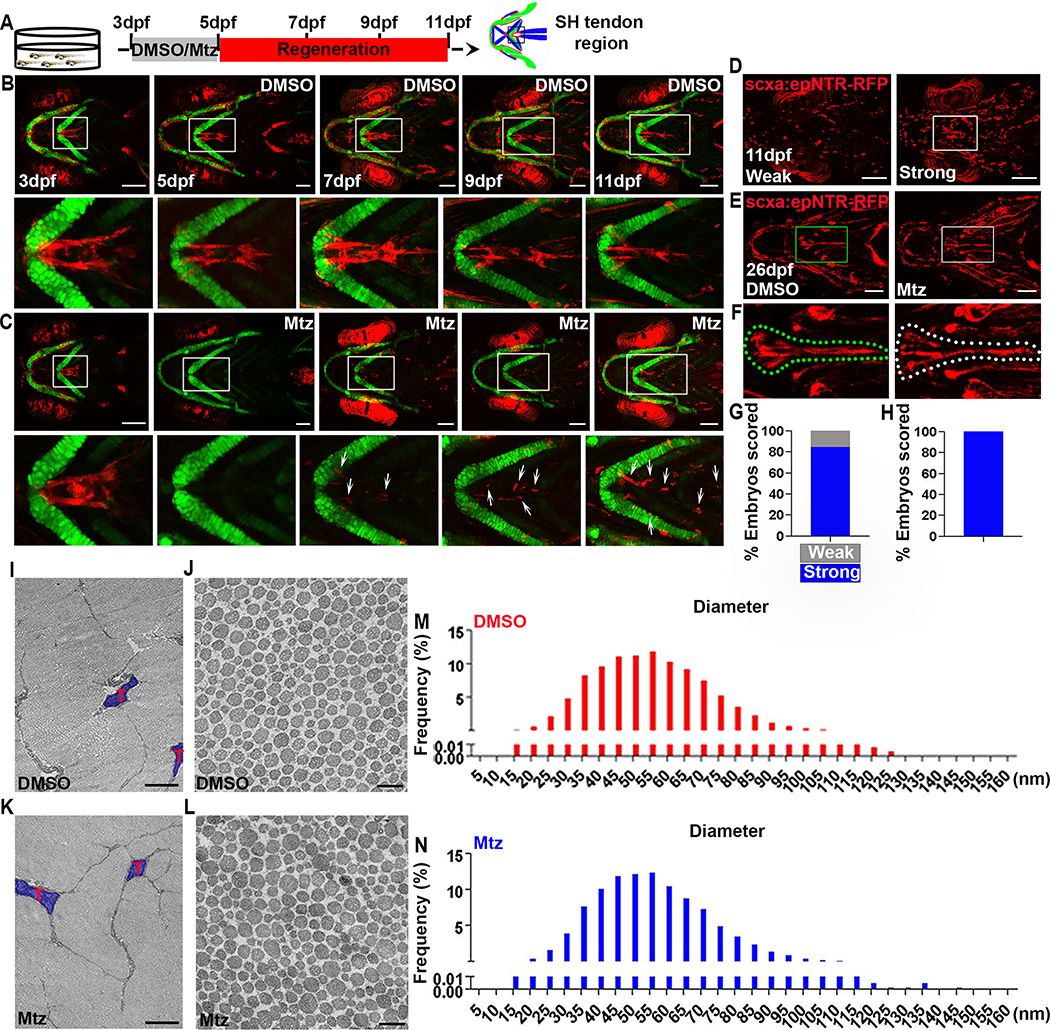
To determine if zebrafish can regenerate tendon cells, we monitored ablated animals at multiple time points. At 7 and 9 dpf, a few scattered scxa:epNTR-RFP+ cells were observed, and by 11 dpf, these cells became more prominent and organized (Figures 2A–2C). This regenerative response was very robust with 84.62% of the ablated animals showing a “strong scxa cell regeneration” response at 11 dpf, (Figures 2D and 2G), and 100% of the ablated animals resembling unablated controls by 26 dpf in terms of the distribution of scxa.epNTR-RFP+ tendon cells (Figures 2E, 2F, and 2H). We also observed tendon cell proliferation during regeneration (Figures S1J and S1K) and ablated animals were viable to adulthood (Figures S3D and S3E). Adults showed normal swimming, mating, and feeding behaviors. Tendon regeneration also restored cartilage and muscle defects, as analyzed using Alcian Blue staining and muscle-expressing Tg(mylz2:Amcyan) fish. At 26 dpf, there were no significant differences in cartilage and muscle morphology between ablated and control animals (Figures S2G–S2J), suggesting that regenerated tendon cells restored the attachment pattern of all components of the musculoskeletal system. Notably, at this stage, we observed recovered swimming capacity, with no difference with that of control animals. We also performed ablation at later stages, including 5–7 dpf, 30–33 dpf, and 90–93 dpf and observed the return of scxa:epNTR-RFP cells in all conditions (Figures S5J–S5L). The 5–7 dpf ablation showed similar regeneration to the 3–5 dpf ablation in terms of scxa:epNTR-RFP cells (Figure S5J). However, the later cell ablations at 30–33 dpf, and 90–93 dpf only resulted in qualitatively less scxa:epNTR-RFP cells returning after several days (Figures S5K and S5L). We also noticed variable expression of scxa:epNTR-RFP at adult stages in unablated animals, which is consistent with reports of silencing of Gal4/upstream activating sequence (UAS) systems in adult zebrafish [40]. Therefore, it is unclear whether the reduced scxa:epNTR-RFP cell number after adult ablation is due to alteration in regenerative capacity or reduced expression from the Gal4/UAS system.
Figure 2. Robust regeneration of zebrafish SH tendon pattern and composition.
(A) Experimental design of tendon cell ablation and regeneration. Tendon cells were ablated at 3–5dpf and regeneration was observed at multiple timepoints.
(B) Live images of craniofacial tendon cells at indicated time points in Tg(scxa:gal4-vp16; uas:epNTR-RFP; col2a1a:eGFP) fish. White boxes, SH tendon region in magnified view underneath. Scale bar, 100 μm.
(C) Live images of craniofacial tendon cell regeneration at indicated time points in ablated Tg(scxa:gal4-vp16; uas:epNTR-RFP; col2a1a:eGFP) fish. White boxes, SH tendon region in magnified view underneath. White arrows, regenerated tendon cells. Scale bar, 100 μm.
(D) Live images of tendon cell regeneration at 11 dpf. Left panel, weak regeneration. Right panel, strong regeneration. Weak (around 80 cells per head) versus strong (around 200 cells per head) was quantitatively defined. White box, SH tendon region. Scale bar, 100 μm.
(E) Live images of tendon cell regeneration at 26 dpf. Left panel, control. Right panel, regenerated. Green and white box, SH tendon region. Scale bar, 200 μm.
(F) Higher magnification views of SH tendon cell regeneration (from E) at 26 dpf. Left panel, control. Right panel, regenerated. Dashed green and white line demarcates the SH tendon.
(G) Quantification of tendon cell regeneration at 11 dpf (n=13). Strong regeneration (n=11, 84.62%). Weak regeneration (n=2, 15.38%).
(H) Percentage of fish showing a normal distribution of scxa:epNTR-RFP+ tendon cells at 26 dpf (n=11, 100%).
(I-L) Transmission electron microscopy analysis of SH tendon collagen fibril in control (DMSO) and regenerated (Mtz) at 180 dpf. T, tendon cell. Scale bar, 2 μm (I, K) and 0.2 μm (J, L).
(M, N) Distribution of collagen fibril diameter in control (M, DMSO) and regenerated (N, Mtz) at 180 dpf.
dpf, days post fertilization. nm, nanometer. All images (A-F), ventral view, anterior to the left. All images (I-L), transverse section view.
See also Figure S3
A critical part of successful regeneration is structural restoration of the tissue’s extracellular matrix (ECM). To compare the structural characteristics of regenerated tendons with that of controls, we employed two strategies. First, we visualized fibrillar collagen density and organization with multiphoton confocal microscopy. At 80 dpf, the intact SH tendon consists of two lobes, each of which is composed of scxa:epNTR-RFP+ tendon cells and collagen fibrils (Figure S3A). Although these tendons appeared smaller in size, the ablated animals regenerated two-lobed SH tendons containing scxa:epNTR-RFP+ tendon cells with densely organized collagen fibrils (Figure S3A). Second, we visualized collagen fibrils using transmission electron microscopy (TEM), and found that in 80 dpf regenerated SH tendons there were more small diameter fibrils at the expense of large diameter fibrils (Figures S3B and S3C). Interestingly, in both control and regenerated SH tendons at this stage, we observed cells with fibripositor morphology, which is a common feature of embryonic tendon [7,8] (Figure S3B). By 180 dpf, the ablated animals fully regenerated two-lobed SH tendons that had a size comparable to controls (Figures S3F and S3G). Moreover, TEM analysis revealed a similar distribution of collagen fibril diameter between regenerated and controls (Figures 2I–2L). At this stage, there was no increase in small sized diameter fibrils, which is characteristic of adult mammalian tendon healing [41] (Figures 2M, 2N, and S3H). Taken together, these findings indicate that tendon cell ablated zebrafish regenerate a functional SH tendon in terms of morphology and structure.
Tendon cell regeneration conserves developmental lineage relationships and identifies a unique contribution from two germ layers to the sternohyoideus tendon
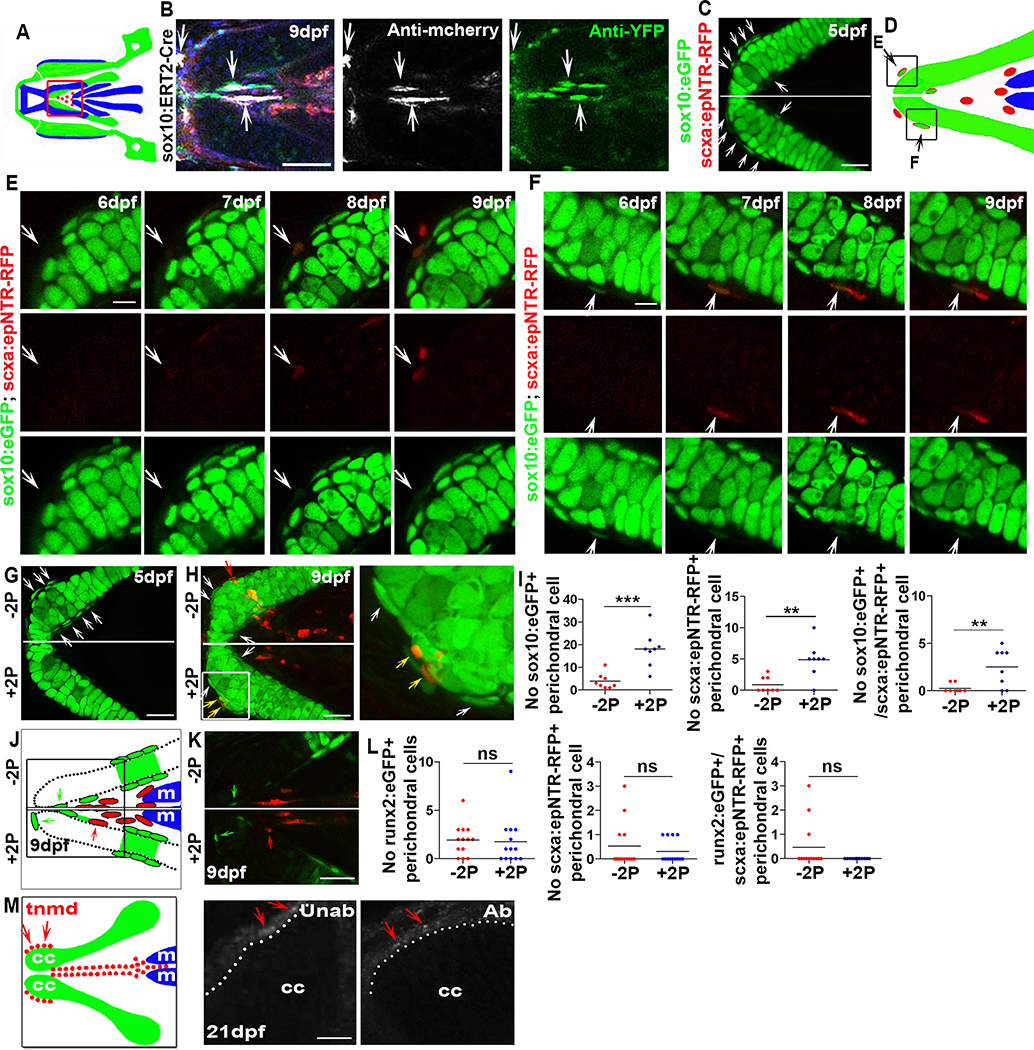
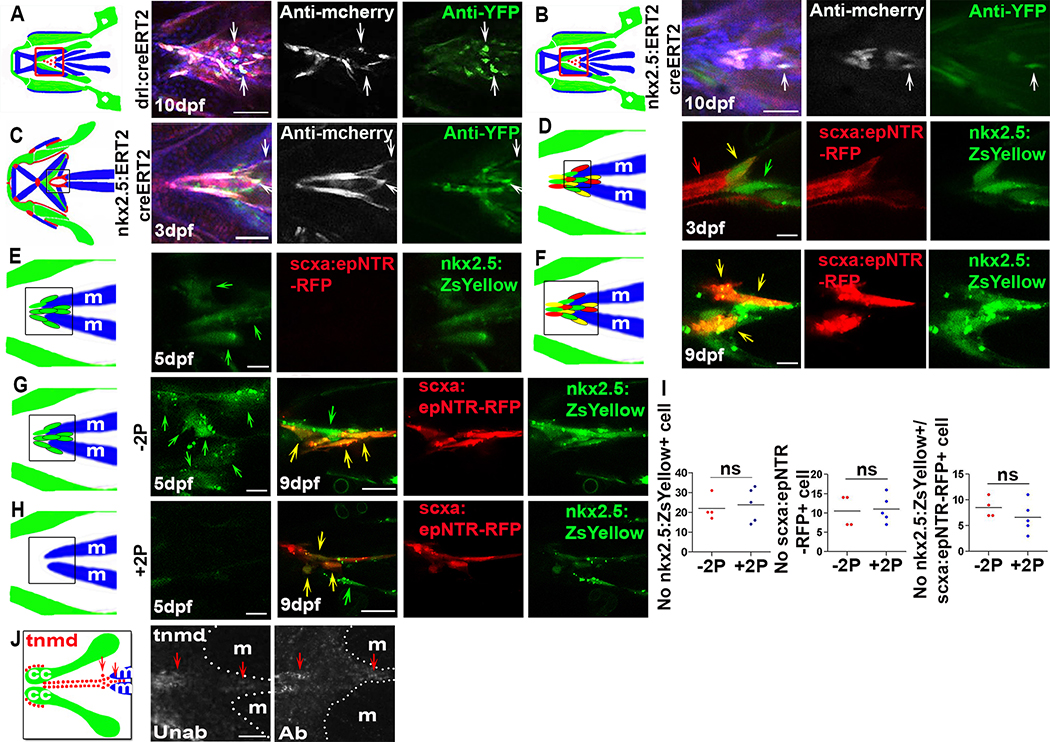
To identify the source of the regenerated tendon cells, we used an inducible Cre/Loxp recombination system to perform genetic fate-mapping experiments. We labelled neural crest or mesoderm cells and their progeny using Hydroxytamoxifen (4-HT)-inducible Cre drivers sox10 Tg(sox10:ERT2-Cre), draculin Tg(drl:creERT2) and nkx2.5 Tg(nkx2.5:ERT2CreERT2) with the Tg(ubi:Zebrabow) reporter, which efficiently labels cells with CFP or YFP after tissue-specific Cre-mediated recombination [42,43]. All the three driver lines have been previously shown to label specific cell lineages [44–47] and we could successfully label neural crest, mesoderm, and anterior lateral plate mesoderm, respectively (Figures S4A–S4G). We found that many cranial tendons and ligaments [25], as well as part of the regenerated SH tendon were derived from the sox10+ lineage (Figures 4A, 4B, and S4D). The mesoderm labeling drivers, draculin Tg(drl:creERT2) and nkx2.5 Tg(nkx2.5:ERT2CreERT2) [45,46,48] (Figures S4E–S4G), demonstrated that a subset of regenerated SH tendon cells originates from draculin+ and nkx2.5+-lineage mesodermal cells (Figures 3A and 3B). We also observed draculin-lineage mesodermal cells contributing to control unablated SH, pectoral fin, and axial tendons but not to other craniofacial tendons or ligaments (Figures S4H–S4K). Although both mesoderm-labeling lines were previously shown to mark lateral plate mesoderm, the Tg(drl:creERT2) line can also sporadically label somitic mesoderm [45], making it unclear which specific type of mesoderm contributes to the SH tendon. However, our results indicate that the SH tendon is unique to the cranial tendons in being derived from two germ layers, the neural crest and mesoderm. This result is consistent with studies showing mesodermal contributions to pharyngeal regions in other vertebrates [49,50]. Together, our work shows that the regenerated tendon cells of SH tendons derive from the same cellular lineages that form them during embryonic development.
Figure 4. Newly-induced SH tendon cells have conserved neural crest developmental origins and arise from cartilage attachment sites.
(A) A cartoon showing the craniofacial region at 9dpf. Red box indicates SH tendon region.
(B) sox10:ERT2-Cre based neural crest cell lineage tracing of regenerated SH tendon cells (white arrow) in ablated Tg(scxa:gal4-vp16; uas:epNTR-RFP; uas-E1b:NfsB-mcherry; sox10:ERT2-Cre; ubi:zebrabow) fish. Blue color in merged image was Hoechst staining. Scale bar, 50 μm.
(C) Confocal images of tendon cell ablated Tg(scxa:gal4-vp16; uas:epNTR-RFP; sox10:eGFP) fish showing sox10:eGFP+ perichondral cells (white arrow) at the ceratohyal cartilage attachment site. Scale bar, 25 μm.
(D) A cartoon showing ceratohyal cartilage attachment site with black boxes indicate the regions analyzed in (E) and (F). Black arrow, scxa:epNTR-RFP+/sox10:eGFP+ cell.
(E) Live images of new cell induction in Tg(scxa:gal4-vp16; uas:epNTR-RFP; sox10:eGFP) fish at indicated time points. White arrow, a perichondral cell that turned on sox10:eGFP and scxa:epNTR-RFP during regeneration. Scale bar, 10 μm.
(F) Live images of new cell induction in Tg(scxa:gal4-vp16; uas:epNTR-RFP; sox10:eGFP) fish at indicated time points. White arrow, a sox10:eGFP + perichondral cell turned on scxa:epNTR-RFP and decreased sox10:eGFP during regeneration. Scale bar, 10 μm.
(G) Two photon images of ceratohyal cartilage attachment site in tendon cell ablated Tg(scxa:gal4-vp16; uas:epNTR-RFP; sox10:eGFP) fish showing sox10:eGFP+ perichondral cells (white arrow) at one side that was not two photon laser ablated (−2P). Two photon laser ablation of sox10:eGFP+ perichondral cells was performed on the contralateral side (+2P) at 5dpf and on average 10 cells were ablated (with a range of 8 to 14 from fish to fish). Scale bar, 25 μm.
(H) Confocal images of ceratohyal cartilage attachment site in tendon cell ablated Tg(scxa:gal4-vp16; uas:epNTR-RFP; sox10:eGFP) fish after two photon laser ablation (+2P) of one ceratohyal region as in (G). White arrow, sox10:eGFP+ perichondral cells. Yellow arrow, sox10:eGFP+/scxa:epNTR-RFP+ tendon cells. Red arrow, scxa:epNTR-RFP+ tendon cells. White box area was magnified to the right. Scale bar, 25 μm.
(I) Quantification of sox10:eGFP+, scxa:epNTR-RFP+, and sox10:eGFP+/scxa:epNTR-RFP+ perichondral cells at 9dpf without two photon (−2P) and the contralateral with two photon (+2P) ablation. Two-tailed, unpaired Student’s t-test and data are mean. **, p<0.01. ***, p<0.001.
(J) A cartoon showing ceratohyal cartilage region with black box indicates attachment site. Green arrow, runx2:eGFP+ perichondral cells. Red arrow, scxa:epNTR-RFP+ tendon cell.
(K) Confocal images of ceratohyal cartilage attachment site in Tg(scxa:gal4-vp16; uas:epNTR-RFP; runx2:eGFP) fish. Two photon ablation of runx2:eGFP+ perichondral cells was performed at 5dpf. Green arrow, runx2:eGFP+ perichondral cells. Red arrow, scxa:epNTR-RFP+ tendon cell. Scale bar, 25 μm.
(L) Quantification of runx2:eGFP+, scxa:epNTR-RFP+, and runx2:eGFP+/scxa:epNTR-RFP+ perichondral cells at 9dpf without two photon (−2P) and the contralateral with two photon (+2P) ablation. Two-tailed, unpaired Student’s t-test and data are mean. ns, no significance.
(M) tnmd expression in ceratohyal cartilage attachment site (red arrow) in unablated and ablated fish at 21dpf.
dpf, days post fertilization. Unab, unablated. Ab, ablated. cc, ceratohyal cartilage. m, SH muscle. 2P, two photon. Tendon cell was ablated at 3–5dpf. All images, ventral view, anterior to the left.
See also Figures S4 and S5
Figure 3. Newly-induced SH tendon cells have conserved mesodermal developmental origins and arise from muscle attachment sites.
(A) drl:creERT2 based mesodermal cell lineage tracing for regenerated SH tendon cells (white arrows) in ablated Tg(scxa:gal4-vp16; uas:epNTR-RFP; uas-E1b:NfsB-mcherry; drl:creERT2; ubi:zebrabow) fish. Red box in cartoon shows region analyzed. Blue color in merged image was Hoechst staining. Scale bar, 50 μm.
(B) nkx2.5:ERT2creERT2 based anterior lateral plate mesoderm cell lineage tracing for regenerated SH tendon cells (white arrows) in ablated Tg(scxa:gal4-vp16; uas:epNTR-RFP; uas-E1b:NfsB-mcherry; nkx2.5:ERT2creERT2; ubi:zebrabow) fish. Blue color in merged image was Hoechst staining. Scale bar, 50 μm.
(C) nkx2.5:ERT2creERT2 based cell lineage tracing of ontogenetic SH tendon cells (white arrow) in Tg(scxa:gal4-vp16; uas:epNTR-RFP; uas-E1b:NfsB-mCherry; nkx2.5:ERT2creERT2; ubi:zebrabow) fish. Black box in cartoon shows the area analyzed. Scale bar, 25 μm.
(D) Confocal images of SH muscle attachment site in Tg(scxa:gal4-vp16; uas:epNTR-RFP; nkx2.5:ZsYellow) fish. Red arrow, scxa:epNTR-RFP+ cells. Green arrow, nkx2.5:ZsYellow+ cells. Yellow arrow, nkx2.5:ZsYellow+/scxa:epNTR-RFP+ cells. Black box in cartoon shows the area analyzed. Scale bar, 5 μm.
(E) Confocal images of SH muscle attachment site in Tg(scxa:gal4-vp16; uas:epNTR-RFP; nkx2.5:ZsYellow) fish. Green arrow, nkx2.5:ZsYellow+ cells. Black box in cartoon shows the area analyzed. Scale bar, 5 μm.
(F) Confocal images of SH muscle attachment site in Tg(scxa:gal4-vp16; uas:epNTR-RFP; nkx2.5:ZsYellow) fish. Yellow arrow, nkx2.5:ZsYellow+/scxa:epNTR-RFP+ cells. Black box in cartoon shows the area analyzed. Scale bar, 5 μm.
(G, H) Two photon (5dpf) and confocal (9dpf) images of SH muscle attachment site in Tg(scxa:gal4-vp16; uas:epNTR-RFP; nkx2.5:ZsYellow) fish with (+2P, H) and without (−2P, G) two photon ablation. Two photon ablation of nkx2.5:ZsYellow+ cells was performed at 5dpf. Yellow arrow, nkx2.5:ZsYellow+/scxa:epNTR-RFP+ cell. Green arrow, nkx2.5:ZsYellow+ cell. Black box in cartoon shows region analyzed. Scale bar, 25 μm.
(I) Quantification of nkx2.5:ZsYellow+, scxa:epNTR-RFP+, and nkx2.5:ZsYellow+/scxa:epNTR-RFP+ cells in SH muscle attachment site at 9dpf with (+2P) and without (−2P) two photon ablation. Two-tailed, unpaired Student’s t-test and data are mean. ns, no significance.
(J) tnmd expression in SH muscle attachment site (red arrow) in unablated and ablated fish at 21 dpf.
dpf, days post fertilization. Unab, unablated. Ab, ablated. m, SH muscle. 2P, two photon. Tendon cell was ablated at 3–5dpf for all, except (C, D). All images, ventral view, anterior to the left.
See also Figures S4 and S5
To determine if regenerating SH tendon cells derive from the scxa+-lineage, we generated a Tg(uas:creERT2) transgenic line and combined it with Tg(scxa:gal4-vp16) to mark tendon cells. We first assessed the specificity of expression and recombination efficiency of this line. Upon crossing to Tg(ubi:Zebrabow) and hydroxytamoxifen treatment, we observed reporter expression in tendon regions (Figures S4L and S4M). When focused on SH tendon, we found a subset of cells were labelled by YFP after recombination (Figure S4L). The inability to label all tendon cells could be due to our detection methods, level of transgene expression, and/or recombination rate of the reporter in our tissue. Nevertheless, we found that this transgene was specific in only labeling scxa+ tendon cells as we never observed recombination in other tissues. Upon complete ablation of the tendon cells with two-day treatments of Mtz from 5–7 dpf, we never observed scxa+/YFP+ tendon cells in the regenerated SH tendon region (Data not shown). To determine if scxa+ cells have the potential to contribute to regeneration, we performed a partial ablation with one-day Mtz treatments from 5–6 dpf. Here, pre-existing scxa+ cells partially contributed to SH tendon cell regeneration (Figure S4N). Together, this indicates that scxa+ cells have the potential to contribute to SH tendon cell regeneration, but under conditions of complete ablation, it appears that the regenerated SH tendon derives from non-scxa+-lineage cells.
Tendon cell regeneration is mediated by progenitor cells that surround muscle and cartilage at musculoskeletal attachment sites
To visualize cells being recruited to tendon fates during regeneration, we performed live-imaging and focused on two components of the SH tendon, the SH muscle and ceratohyal cartilage attachment sites. In the SH muscle attachment site, live imaging of the ablated Tg(scxa:epNTR-RFP; col2a1a:eGFP) fish demonstrated cells turning on scxa:epNTR-RFP (Figure S5A). We thought these cells might be from connective tissue surrounding muscle based on the observations that nkx2.5+ cells contribute to the SH tendon in unablated and ablated animals (Figures 3B and 3C) and that the Tg(nkx2.5:ZsYellow) reporter line labels cells in the SH tendon and surrounding the muscle in unablated fish (Figure 3D). Therefore, we live imaged Tg(scxa:epNTR-RFP; nkx2.5:ZsYellow) fish. Immediately after ablation we only observed nkx2.5:ZsYellow+ cells surrounding the muscle near the attachment site (Figure 3E). At 4 days post ablation, we observed nkx2.5:ZsYellow+ cells co-expressing scxa:epNTR-RFP (Figure 3F), indicating the nkx2.5:ZsYellow+ cells surrounding the muscle turn on scxa.epNTR-RFP during the regenerative process. Laser ablation of nkx2.5:ZsYellow+ cells after scxa:epNTR-RFP+ cell ablation did not alter the numbers of newly generated nkx2.5:ZsYellow+ cells or nkx2.5:ZsYellow+/scxa:epNTR-RFP+ cells compared with controls (Figures 3G–3I). It is possible that this result is due to our inability to completely eliminate all cells surrounding the muscle capable of forming tendon or that cells originate from locations outside the ablated region.
At the cartilage attachment site, we observed many sox10:eGFP+ cells in the ceratohyal cartilage perichondrium attachment site compared with other regions of the ceratohyal after ablation (Figure 4C). We next observed sox10:eGFP− perichondrial cells become sox10:eGFP+/scxa:epNTR-RFP+ cells (Figures 4D and 4E), and sox10:eGFP+ perichondrial cells turn on scxa:epNTR-RFP and appear to reduce sox10:eGFP expression (Figures 4D and 4F), indicating the sox10:eGFP+ perichondrial cells may represent a progenitor cell source for regeneration and upon becoming tendon cells they decrease sox10:eGFP expression. Interestingly, there were also some sox10:eGFP+/scxa:epNTR-RFP+ cells in unablated animals, putative perichondrial cells transitioning to tendon cells (Figures S5B and S5C). To test whether ablating sox10:eGFP+ perichondrial cells affected scxa+ cell induction, we used a two photon laser ablation system [51] to remove only sox10:eGFP+ perichondrial cells from one ceratohyal cartilage, leaving the contralateral side unablated (Figure 4G). Unexpectedly, 4 days post ablation, we observed increased numbers of sox10:eGFP+ and sox10:eGFP+/scxa:epNTR-RFP+ cells compared with the contralateral control (Figures 4H and 4I). Although we did not allow these fish to develop beyond 9 dpf, it would be interesting to test whether laser ablation of these and/or the nkx2.5:ZsYellow+ cells affects tendon maturation compared to the control unablated side. We next tested whether the increase in scxa:epNTR-RFP+ cells was specific to ablating sox10:eGFP+ perichondrial cells or occurred after deleting any perichondrial cell population. Because runx2+ marks perichondrial cells becoming osteoblasts [52,53] (Figures S5D–S5G), we used laser ablation to remove runx2:eGFP+ perichondrial cells from one ceratohyal after scxa:epNTR-RFP+ cell ablation. We did not see significant changes in the number of runx2:eGFP+, scxa:epNTR-RFP+, or runx2:eGFP+/ scxa:epNTR-RFP+ cells (Figures 4J–4L), suggesting the runx2+ cells are not required for regeneration. Moreover, an osc.eGFP reporter line visualizing osteoblast formation demonstrated these scxa:epNTR-RFP+ cells were not osteoblasts (Figures S5H and S5I). This indicates that the increase in scxa:epNTR-RFP+ cells is a unique response to the laser ablation of the sox10:eGFP+ perichondrial cells. It also suggests the possible presence of a mechanism that limits the induction of additional cells in the peri chondrium to restrict the location of the regenerating attachment site. Finally, to confirm that the new cells in the attachment sites differentiated into tendon, we examined tnmd expression and detected transcripts in both attachment regions (Figures 3J and 4M). Taken together, the induction of new tendon cells in areas surrounding cartilage and muscle would suggest that multiple anatomic locations contain progenitor cells capable of mediating tendon regeneration.
BMP signaling is required and sufficient for tendon cell regeneration
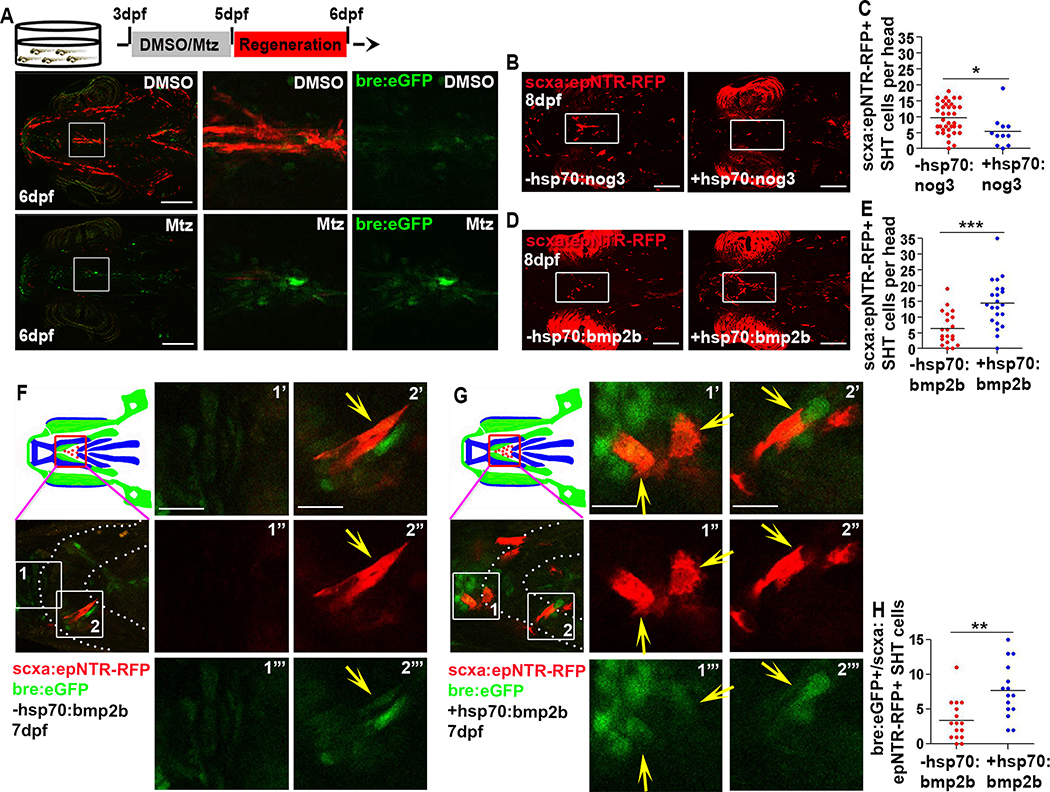
To identify key molecular mechanisms regulating this regenerative process, we performed a candidate screen of signaling pathways with roles in musculoskeletal development. We analyzed expression between 5–6 dpf of transgenic reporters for Notch [54], Wnt [55], and BMP [56] signaling and Sonic hedgehog [57] expression, following ablation. No discernible changes were observed between ablated and control fish transgenic for Notch, Sonic hedgehog, and Wnt reporters (Figures S6A–S6C), suggesting these pathways may not be involved in early tendon cell regeneration or that these transgenic lines were unable to detect changes in signaling. In contrast, ablated animals showed an increase in the activation of the BMP-responsive element (bre.eGFP) reporter at 6 dpf (Figures 5A, S6D, S6J, and S6K). Further investigation found that bre.eGFP was co-expressed with scxa:epNTR-RFP+ tendon cells in the SH tendon (Figure S6E) and staining for phospho-Smad1/5 confirmed active BMP signaling in the SH tendon and surrounding cells at 3 dpf in unablated fish (Figure S6F). bre.eGFP signal was also active in tissues surrounding the developing mouth, Meckel’s cartilage, and in the mandibulohyoid junction, which connects the intermandibularis posterior muscles to interhyal muscles (Figure S6G). Treatment with Dorsomorphin, a potent inhibitor of BMP signaling [58], compromised SH tendon development (Figures S6H and S6I). Together, these results suggest BMP signaling is involved in SH tendon development and that the pathway reinitiates during tendon cell regeneration.
Figure 5. BMP signaling is necessary and sufficient for SH tendon cell regeneration.
(A) Live images of tendon cells (red) and cells responding to BMP signaling (green) in unablated (DMSO) and ablated (Mtz) Tg(scxa:gal4-vp16; uas:epNTR-RFP; bre:eGFP) fish. Scale bar, 200μm.
(B) Live images of SHT cell regeneration in non hsp70:nog3 and hsp70:nog3 fish. Insets show the SHT region analyzed. Scale bar, 100 μm. Ablation was performed at 3–5 dpf and for all heat shock experiments (B-E), ablated embryos were heat shocked at 5, 6, and 7 dpf and each time for one hour.
(C) Quantification of SHT cell number in non hsp70:nog3 (n=37, average = 9 cells) and hsp70:nog3 (n=11, average = 5 cells) fish at 8 dpf. Two-tailed, unpaired Student’s t-test and data are mean. *, p<0.05.
(D) Live images of SHT cell regeneration in non hsp70:bmp2b and hsp70: bmp2b fish. Insets show SHT region analyzed. Scale bar, 100 μm.
(E) Quantification of SHT cell number in non hsp70:bmp2b (n=20, average=6 cells) and hsp70: bmp2b (n=22, average =14 cells) fish at 8 dpf. Two-tailed, unpaired Student’s t-test and data are mean. ***, p<0.001. (B-E) heat shock and SHT cell number quantification were performed blinded to the genotype of the fish, after quantification, fish were genotyped to determine the presence of the transgene.
(F, G) Live images of SHT cell regeneration in non-hsp70:bmp2b (F) and hsp70: bmp2b (G) fish. Yellow arrow, bre:eGFP+/scxa:epNTR-RFP+ cell. Red box in cartoon showed SHT region analyzed. White dashed line demarcated ceratohyal cartilage. Boxes 1 and 2 are magnified to the right (1’-2’”) Scale bar, 20 μm.
(H) Quantification of bre:eGFP+/scxa:epNTR-RFP+ SHT cell number in non-hsp70:bmp2b (n=16, average = 3 cells) and hsp70:bmp2b (n=15, average = 8 cells) treatment at 7 dpf. Each dot represented the cell number from individual fish. Ablation was performed at 3–5 dpf and ablated embryos were heat shocked at 5, and 6 dpf and each time for one hour. Two-tailed, unpaired Student’s t-test and data were mean. **, p<0.01.
dpf, days post fertilization. SHT, sternohyoideus tendon. All images, anterior to the left. Tendon cell was ablated at 3–5dpf.
To test the function of BMP signaling in tendon cell regeneration, we used chemical and genetic approaches to inhibit the BMP pathway. First, we used LDN-193189, an inhibitor of BMP signaling [59], to treat the ablated animals at 6–8dpf. In contrast to the DMSO control, LDN-193189-treated animals had compromised SH tendon cell regeneration, with a 54.72% reduction in the number of regenerating cells (Figure S6L). This phenotype could be reproduced by Dorsomorphin treatment, which reduced the number of regenerating tendon cells by 34.52% (Figure S6M). Interestingly, treating the unablated fish with LDN-193189 at the same stage did not affect SH tendon cell development (Figure S7A). Next, we used genetic mis-expression of noggin through the heat shock promoter Tg(scxa:epNTR-RFP; hsp70:nog3) to antagonize the BMP pathway. Consistent with the pharmacological inhibition, embryos found to carry the hsp70:nog3 allele by subsequent genotyping had a diminished regenerative response, with a 43.47% decrease in the number of regenerating cells (Figures 5B and 5C). To test if activation of the BMP pathway could promote the regenerative response, we used the Tg(hsp70:bmp2b) reporter line and found a significant increase of 133.33% in the number of scxa:epNTR-RFP+ cells (Figures 5D and 5E). Notably, BMP activation also increased the number of bre:eGFP+/scxa:epNTR-RFP+ cells (Figures 5F–5H). Our finding that activating BMP signaling promotes SH tendon cell appearance is surprising given previous studies suggesting BMP signaling negatively regulates tendon formation and is pro-osteogenic [2,60]. However, our work is consistent with other studies that have shown BMP and, in particular, GDF family members can promote tendon formation and enhance tendon repair in a chicken injury model [61–64]. Collectively, our analyses show that the BMP pathway is both necessary and sufficient for early tendon cell regeneration.
BMP inhibition blocks the induction of new tendon cells at musculoskeletal attachment sites
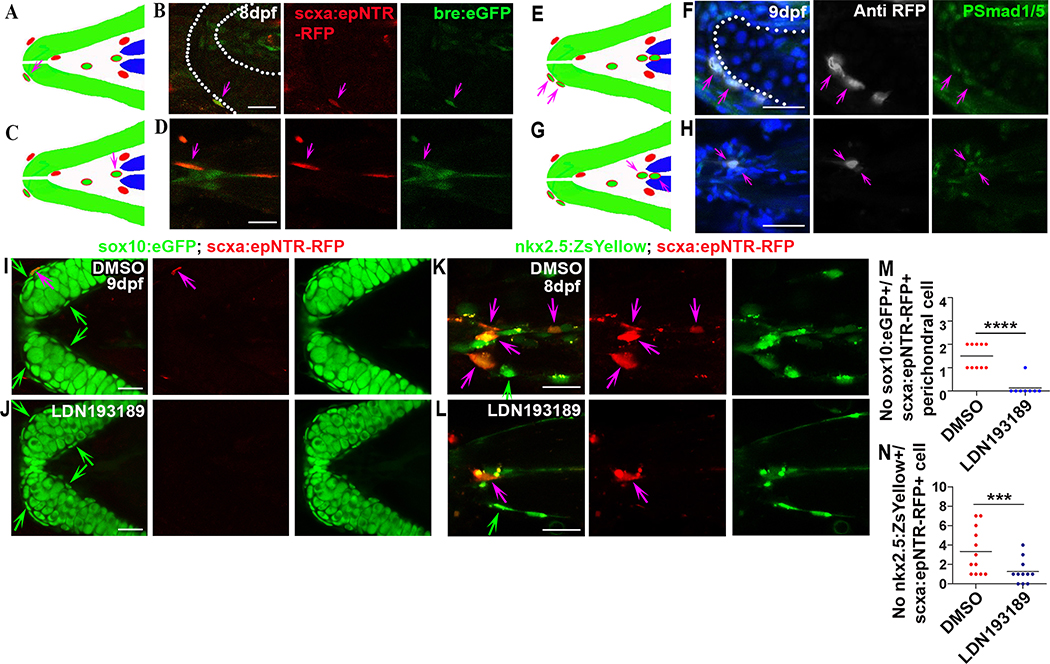
Next, to better understand how BMP signaling is affecting the regenerative process, we focused on both attachment sites. The reasons for focusing on the attachment sites are based on fluorescent in situ hybridization results and live imaging data. We screened the expression of BMP ligands and receptors, including bmp2b, bmp4, bmp7b, acvr1l, bmpr1aa, bmpr1ab, bmpr1ba, and bmpr1bb, in the ablated animals. Among these, bmp2b, bmp4, and bmpr1ba were found to be highly expressed in the attachment sites (Figures S7B–S7K), and bmp2b and bmpr1ba were slightly upregulated in SH muscle attachment sites in the ablated animals compared to controls (Figures S7H–S7K). qPCR also revealed significant upregulation of bmp2b and bmpr1ba in the craniofacial region (Figure S7L), suggesting a potential involvement of bmp2b and bmpr1ba in regeneration. To confirm localized BMP signaling at the attachments, we examined Tg(scxa:epNTR-RFP; bre:eGFP) fish and found bre:eGFP+/scxa:epNTR-RFP+ cells in attachment sites surrounding cartilage and muscle (Figures 6A–6D). Similarly, scxa.epNTR-RFP+ cells surrounding muscle and cartilage were also phospho-Smad1/5 staining positive (Figures 6E–6H), indicating these cells are directly responding to BMP signaling. To ask whether inhibiting BMP signal might block new cell induction in these sites, we treated the ablated animals with either DMSO or LDN193189 and quantified the number of sox10:eGFP+/scxa:epNTR-RFP+ and nkx2.5:ZsYellow+/scxa:epNTR-RFP+ cells. As a result, we found LDN193189 treatment impeded new scxa:epNTR-RFP+ cell induction in both sites (Figures 6I–6N), indicating BMP signaling is required for robust induction of scxa.epNTR-RFP+ cells at attachment regions.
Figure 6. BMP signaling is necessary for induction of SH tendon cells at musculoskeletal attachment sites.
Cartoon (A) and confocal images (B) of ceratohyal cartilage attachment site showing bre:eGFP+/scxa:epNTR-RFP+ perichondral cell (purple arrow, 53% of scxa:epNTR-RFP+ cells). White dashed line in (B) demarcated ceratohyal cartilage. Scale bar, 25 μm.
Cartoon (C) and confocal images (D) of SH muscle attachment site showing bre.eGFP+/scxa:epNTR-RFP+ cell (purple arrow). Scale bar, 25 μm.
Cartoon (E) and confocal images (F) of ceratohyal cartilage attachment site showing P-Smad1/5+/scxa:epNTR-RFP+ perichondral cells (purple arrows). Blue color in merged image was Hoechst staining. White dashed line in (F) demarcated ceratohyal cartilage. Scale bar, 25 μm.
Cartoon (G) and confocal images (H) of SH muscle attachment site showing P-Smad1/5+/scxa:epNTR-RFP+ cells (purple arrows). Blue color in merged image was Hoechst staining. Scale bar, 25 μm.
(I, J) Confocal images of ceratohyal cartilage attachment site in ablated Tg(scxa:gal4-vp16; uas:epNTR-RFP; sox10:eGFP) fish upon DMSO (I) or LDN193189 (J) treatment. Chemical treatments were performed 6–8dpf. Green arrow, sox10:eGFP+ perichondral cell. Purple arrow, sox10:eGFP+/scxa:epNTR-RFP+ perichondral cell. Scale bar, 25 μm.
(K, L) Confocal images of SH muscle attachment site in ablated Tg(scxa:gal4-vp16; uas:epNTR-RFP; nkx2.5:ZsYellow) fish upon DMSO (K) or LDN193189 (L) treatment. Chemical treatments were performed 6–8dpf. Green arrow, nkx2.5:ZsYellow+ cell. Purple arrow, nkx2.5:ZsYellow+/scxa:epNTR-RFP+ cell. Scale bar, 20 μm.
(M) Quantification of sox10:eGFP+/scxa:epNTR-RFP+ perichondral cells in ceratohyal cartilage attachment site upon DMSO (46% of scxa:epNTR-RFP+ cells) or LDN193189 (25% of scxa:epNTR-RFP+ cells) treatment. Two-tailed, unpaired Student’s t-test and data were mean. ****, p<0.0001.
(N) Quantification of nkx2.5:ZsYellow+/scxa:epNTR-RFP+ cells in SH muscle attachment site upon DMSO (75% of scxa:epNTR-RFP+ cells) or LDN193189 (73% of scxa:epNTR-RFP+ cells) treatment. Two-tailed, unpaired Student’s t-test and data were mean. ***p<0.001.
dpf, days post fertilization. All images, anterior to the left. Tendon cell was ablated at 3–5dpf.
Discussion
Tendon injuries present many clinical treatment challenges, mainly due to their poor regenerative potential in humans and a limited understanding of their biology. Most studies investigating tendon repair have used adult animals such as the mouse, rat, and rabbit, where tendon injuries were induced through overuse by treadmill running [65], through induction of inflammation and degeneration of the matrix by collagenase injection [66], or acutely through partial or complete physical injury [21,22,67]. Although these studies have advanced our understanding of how mammalian tendons respond to injury and have identified endogenous cues for repair [21,22], rarely do these animal models achieve complete regeneration which limits our understanding of the processes guiding regenerative healing.
Unlike adult mammals, zebrafish can fully regenerate almost all injured organs [42,68–74]. However, their ability to regenerate tendon and the underlying cellular mechanisms have not been studied. In this study, we have established a new model of tendon regeneration to reveal what cell types and molecular pathways could replace the missing tendon cells (Figure 7). Using this system, we find that after selectively ablating all tendon cells at 3–5 dpf, tendon matrix organization and the morphology of its connecting muscle and cartilage tissues are subsequently disrupted. These defects are later restored upon the re-articulation of new tendon cells to the muscle-skeletal interfaces and the regeneration of the tendon matrix. We find that, at late embryonic and larval stages, progenitors located in neighboring regions surrounding cartilage and muscle are activated to replace the missing tendon cells. Laser ablation of the perichondrial sox10:eGFF+ cells results in increased scxa:epNTR-RFP+ cells. In addition, BMP signaling is required and sufficient for the early recruitment of the attachment progenitor cells to become scxa+/tnmd+ tendon cells. Together, this work identifies new sources of progenitor cells capable of becoming tendon and highlights the plasticity of connective tissues surrounding the musculoskeletal system.
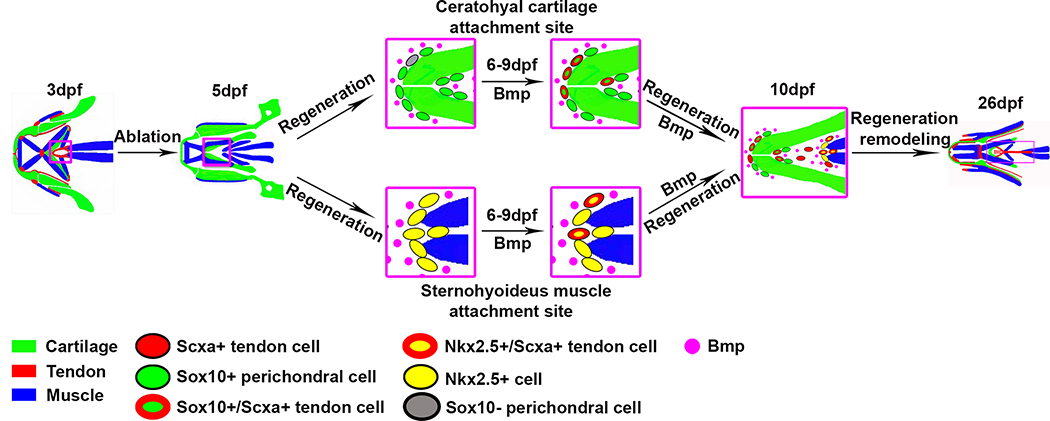
Figure 7. A model for SH tendon cell regeneration in zebrafish.
During the initial stage of regeneration, the cells located in the ceratohyal perichondrium expressing sox10:eGFP and cells surrounding the SH muscle expressing nkx2.5:ZsYellow turn on scxa:epNTR-RFP. There are also sox10:eGFP- perichondrial cells that turn on sox10:eGFP and scxa:epNTR-RFP. These cells arise from the neural crest/sox10-lineage and the mesoderm/nkx2.5-lineage, respectively. During regeneration, these cells in the attachment regions express BMP ligands and receptors, and the scxa:epNTR-RFP+ cells in the peri-ochondrium and surrounding the muscle co-express bre:eGFP+ or P-Smad1/5+, indicating that they are targets of BMP signaling. Loss of BMP signaling depletes the number of cells turning on scxa:epNTR-RFP expression, suggesting BMP signaling is necessary to convert sox10:eGFP perichondrium and nkx2.5:ZsYellow cells to tendon fates. By 26 dpf, ablated fish regenerate the distribution of scxa:epNTR-RFP+ tendon cells, and have restored cartilage and muscle morphology.
See also Figure S7
Additionally, there are many critical questions that would be interesting to investigate on how the dynamic processes of regeneration are achieved. For example, how do the new cells communicate with each other to establish a functional tendon and re-establish the proper attachment pattern? Are the cells surrounding cartilage and muscle heterogeneous with differing abilities to regenerate tendon and are there other cell sources that are able to contribute to tendon regeneration? It is also unknown how BMP signaling regulates these specific cell populations at their distinct locations. Although our data suggests BMP signaling is required for the specification of scxa+ cells from nkx2.5+ or sox10+ cells (Figures 6I–6N), we cannot rule out a role for BMP signaling in scxa+ cell proliferation. It is also unclear whether prolonged BMP inhibition or activation affect the long term regenerative response and if BMP activation generally promotes scxa+ cell number and tendon formation in non-regenerative conditions. Certainly, transforming growth factor β (TGFβ) family members, such as GDF5 can promote tendon formation in other assays [61,64]. In our loss of function experiments, BMP inhibition had no effect on non-ablated animals at later stages (6–8 dpf), suggesting BMP is required for the appearance of scxa+ cells specifically during regeneration. It also should be noted that although the matrix is disrupted following scxa+ cell loss, some matrix remains, and it would be interesting to test whether this remnant matrix contains signals that direct cell recruitment at the proper locations.
Another question that remains is whether this plasticity is an inherent property of their developmental lineage relationships in forming the connections between the tendon and cartilage or muscle. Notably, the presence of doubly positive perichondrial sox10:eGFP+; scxa:epNTR-RFP+ cells in non-ablated animals could suggest that cells in the perichondrium normally contribute to the attachment and tendon during zebrafish development. Moreover, the existence of sox10:eGFP-cells in the perichondrium that turn on sox10:eGFP and scxa:epNTR-RFP implies a complex cellular composition at the tendon and cartilage interface that is capable of contributing to tendon regeneration. In developing mouse limb regions, Sox9+/Scx+ cells form the tendon-bone attachment structure, termed enthesis [75,76], but the identity of the equivalent cells in the zebrafish is unclear. In addition, the role of Sox9+/Scx+ cells in murine tendon healing is unknown. Multipotent stem or progenitor cells have been identified in the periosteum and these cells have been shown to differentiate into chondrocytes, osteoblasts, and adipocytes, and contribute to bone fracture healing in mice [77], or participate in zebrafish jawbone regeneration and axolotl skeletal regeneration [74,78]. However, these studies did not examine the ability of these cells to contribute to tendon. Lineage tracing in mice using an alpha smooth muscle actin (αSMA) inducible Cre, revealed an expansion of αSMA+ cells in the periosteum towards the bone insertion and patellar tendon injury [20]. This suggests that the mammalian periosteum may contribute to tendon healing and would be consistent with the perichondrium/periosteum containing progenitors with multilineage differentiation potential.
For the muscle-associated connective tissue, there is a developmental association between the forming muscle connective tissue and tendon. Co-expression of Scx and the muscle connective tissue marker, Tcf4, is observed in embryonic limb and craniofacial regions [79,80]. However, the lineage relationship between cells in these expression domains and whether Tcf4+ cells can contribute to tendon in injury models is unknown. Recent single cell analysis has identified two connective tissue cell populations in the adult muscle interstitial compartment: fibroadipogenic progenitors that are marked by Pdgfra and Scx+ cells capable of tenogenic differentiation [81]. In this single cell analysis study, it was hypothesized that Scx+ cells in the muscle may serve as potential reservoirs for repair after injury, but this was not tested. The relationship of these cells to the nkx2.5+/scxa+ cells in our work is unknown as we do not observe scxa+ cells in muscle tissue. Altogether, little is known regarding zebrafish muscle connective tissue cells, and direct analysis of muscle connective tissue markers such as pdgfra and tcf4 is confounded by their expression in other cell types, including neural crest [82], tendon cells themselves [83], and the branchial arches [84].
It is becoming clear that cell fate transitions or plasticity has emerged as a potential mechanism for tissue repair following injury. Depending on the type of injury, there may be differences in the cell types recruited and whether fate-transition is observed. In most cases of partial damage or chronic inflammation, cells remaining in or surrounding the damaged tissue respond, undergo transitions in fate and contribute to new cell formation. Examples of this include dedifferentiation of specialized cell types in epithelia tissues [85], heart regeneration in fish [68,69], and transdifferentiation in liver regeneration [71,86]. In the cases where part of or entire tissues are removed such as fin regeneration in fish [87] and limb regeneration in axolotl [88,89], a blastema forms and drives regeneration. However, many studies have shown that the blastema contains multiple progenitor cells that are lineage restricted [72,90], which, together, points to limited inter-tissue cellular plasticity during regeneration. Interestingly, connective tissue cells appear to be the exception in that they can form dermal and muscle connective tissues as well as skeletal tissues in the regenerating axolotl limb [78]. Our findings provide new molecular insights into cellular plasticity during tissue repair outside the context of a blastema and suggest that progenitor cells from adjacent and distinct connective tissues can serve as reservoirs to replace the lost tendon cells. In the future, it would be interesting to evaluate if mouse and human adult bone and muscle connective tissues have cells that could function in tendon repair and may provide a new cell source for the development of treatments for tendon injuries.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact Jenna Galloway (jenna_galloway@hms.harvard.edu). Plasmids and zebrafish lines used in this study are available upon request.
DATA AND CODE AVAILABILITY
The published article includes all datasets generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish husbandry and transgenic lines
Zebrafish were raised and maintained under standard laboratory conditions at 28.5°C. The transgenic lines used in this study are listed in key resources table. All animal work was approved by the MGH IACUC (protocol #2012N000167).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti RFP | Rockland | 600-401-379 |
| Mouse anti mcherry | Living Colors | 632543 |
| Mouse anti MF 20 | Developmental Studies Hybridoma Bank | RRID; AB_2147781 |
| Mouse anti Collagen II | Developmental Studies Hybridoma Bank | II-II6B3 |
| Mouse anti myosin heavy chain (MHC) | Developmental Studies Hybridoma Bank | A4.1025 |
| Rabbit anti GFP | ThermoFisher Scientific | A-21311 |
| Phospho-Smad1/5 | Cell Signaling Technology | 9516T |
| Goat anti-Rabbit Alexa Fluor 488 | ThermoFisher Scientific | A-11008 |
| Goat anti-Mouse Alexa Fluor 488 | ThermoFisher Scientific | A-21121 |
| Goat anti-Rabbit Alexa Fluor 594 | ThermoFisher Scientific | A-11012 |
| Goat anti-Mouse Alexa Fluor 647 | ThermoFisher Scientific | A-21241 |
| Donkey anti-Rabbit Alexa Fluor 647 | abcam | ab150067 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| N-Phenylthiourea | Sigma | P7629 |
| DMSO | Sigma | 276855 |
| Metronidazole | Sigma | M3761 |
| (Z)-4-Hydroxytamoxifen | Sigma | H7904 |
| Dorsomorphin | Cayman Chemical | 866405-64-3 |
| LDN-193189 | Cayman Chemical | 1062368-62-0 |
| Experimental Models: Organisms/Strains | ||
| scxa:gal4-vp16 | This study | N/A |
| uas:creERT2 | This study | N/A |
| uas:epNTR-RFP | [28] | y268Tg |
| uas-E1b:Kaede | [93] | s1999tTg |
| ubi:Zebrabow | [43] | a131Tg |
| uas:Eco.Nsfb-mcherry | [94] | rw0144Tg |
| sox10:ERT2-Cre | [44] | zf760Tg |
| col2a1a:eGFP | [95] | nu13Tg |
| scxa:mcherry | Galloway lab, MGH, USA | fb301Tg |
| mylz2:Amcyan | [96] | N/A |
| mylz2:mcherry | [96] | N/A |
| tp1:eGFP | [54] | um14Tg |
| shha:eGFP | [57] | sb15Tg |
| otm:d2eGFP | [55] | kyu1Tg |
| bre:eGFP | [56] | pt510Tg |
| hsp70:nog3 | [97] | fr14Tg |
| hsp70:bmp2b | [97] | fr13Tg |
| drl:creERT2 | [45] | N/A |
| runx2:eGFP | [72] | zf259Tg |
| osc:eGFP | [72] | hu4008Tg |
| nkx2.5:ZsYellow | [98] | fb7Tg |
| nxk2.5:ERT2CreERT2 | [46] | fb8Tg |
| Tübingen | Galloway lab, MGH, USA | N/A |
| Oligonucleotides | ||
| Primers, see Table S1–S3 | This paper | N/A |
| Recombinant DNA | ||
| 10xuas-creERT2-polyA | This paper | N/A |
| scxa-gal4-vp16 | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | Galloway lab, MGH, USA | N/A |
| GraphPad Prism 5 | Galloway lab, MGH, USA | N/A |
| Adobe Photoshop CS6 | Galloway lab, MGH, USA | N/A |
| Microsoft Excel | Galloway lab, MGH, USA | N/A |
METHOD DETAILS
Generation of transgenic lines for tendon cell ablation and fate mapping
To generate the Tg(scxa:gal4-vp16) transgenic line, a BAC harboring the cis-regulatory elements of scxa (CH211 251g8; Accession #BX255894) was modified to contain Tol2 sites and gal4-vp16 in place of the first exon, and this construct was co-injected into one-cell stage Tg(uas-E1b:Kaede) embryos with Tol2 transposase mRNA. The positive F0 founders demonstrated Kaede expression in the craniofacial region and myosepta and were raised to adulthood. Upon sexual maturity, the Tg(scxa:gal4-vp16; uas-E1b:Kaede) fish were out-crossed to Tg(uas:epNTR-RFP) to generate the ablation line Tg(scxa:gal4-vp16; uas:epNTR-RFP). We generated multiple ablation lines (line 1, 2, 3, 4, 5) with similar expression patterns and line 5 was chosen for this study based on the most faithful and specific expression in the tendons.
To generate the Tg(10xUAS:creERT2) strain, abbreviated as Tg(uas:creERT2), gateway cloning technology (Life Technologies) was used to perform the LR recombination reaction with the pENTR5’−10xUAS, pME-CreERT2 and p3E-polyA. The entire uas:creERT2 construct containing a gamma-crystallin GFP cassette [99] was co-injected with Tol2 transposase mRNA into one-cell stage wild-type AB embryos. The successful F0 founders were identified three days later based on the green lens and stable F1 transgenic lines were established by out-crossing to wild-type AB fish and raised to adulthood.
Tamoxifen treatment and genetic fate mapping
The following transgenic fishes: Tg(scxa:gal4-vp16; uas:epNTR-RFP; uas:creERT2; ubi.Zebrabow), Tg(scxa:gal4-vp16; uas:epNTR-RFP; uas.nfsb-mcherry; sox10:ERT2-Cre; ubi.Zebrabow), Tg(scxa:gal4-vp16; uas:epNTR-RFP; uas.nfsb-mcherry; drl:creERT2; ubi.Zebrabow), and Tg(scxa:gal4-vp16; uas:epNTR-RFP; uas.nfsb-mcherry; nkx2.5:ERT2creERT2; ubi.Zebrabow) at indicated time points were treated with 20uM (Z)-4-Hydroxytamoxifen (4-HT) solution (Sigma, H7904) for 34h (14hpf-2dpf for sox10:ERT2-Cre), 48h (1dpf-3dpf for uas:creERT2), 42h (6hpf-2dpf for drl:creERT2), and 38hpf (10hpf-2dpf for nkx2.5:ERT2creERT2) at 28.5°C. The Cre/loxP-mediated recombination efficacy was confirmed by examining the fluorescence change following treatment. After 4-HT treatment, ablation was performed for 1 or 2 days at indicated time points and ablated animals were then transferred to housing system for regeneration.
Chemical treatments
For tendon cell ablation, animals at indicated time points were treated with 5mM Mtz (Sigma, M3761) in embryo medium in the dark for 1 or 2 days at 28.5°C. The efficacy of ablation was confirmed by checking the RFP fluorescence. At the end of ablation, the treated animals were fixed for staining and analysis or transferred to petri dishes or the zebrafish housing system to raise for regeneration assays. For Dorsomorphin (Cayman, 866405–64-3) treatments, 6 dpf ablated embryos (1 day post ablation) were incubated in 10uM Dorsomorphin for 3 days. For LDN-193189 (Cayman, 1062368–62-0) treatment, 6 dpf ablated embryos were bathed in 10uM LDN-193189 for 2 days. For Dorsomorphin treatment at 1–3 dpf, 1 dpf embryos were bathed in 20uM Dorsomorphin for 2 days.
EdU labeling and TUNEL
For EdU labeling assays, embryos at 9 dpf were soaked in 500uM EdU solution in DMSO for 20 minutes on ice or were injected with EdU into the heart or intestine, followed by 4 hours at 28.5°C and then fixed 1 hour at room temperature. EdU positive cells were visualized after staining using the Click-iT® EdU Alexa Fluor® 488 Imaging Kit (Thermo Fisher Scientific, C10337). For TUNEL assay, embryos at the indicated stages were stained using the Click-iT® Plus TUNEL Assay (Life Technologies, C10617) with properly extended treatment of Proteinase K.
Whole-mount in situ hybridization (WISH)
WISH was performed as previously described [25] using DIG (Roche, 11277073910) labeled anti-sense RNA probes against scxa, tnmd, col1a2. Probes were synthesized using Sp6 or T7 RNA polymerase (Roche, 10810274001 or 10881767001). Signals were visualized using BCIP/NBT (Fisher, OB020501/FERR0841) at a concentration of 175/225ug/ml or using a TSA Plus Fluorescein Evaluation Kit (PerkinElmer, NEL741E001KT). After staining, embryos were positioned with the ventral or lateral side facing the objective and images were taken from single plane using a ZEISS Axio Imager.D2 microscope.
Fluorescent in situ hybridization (FISH) on isolated lower jaw
FISH was performed as previously described [25] using DIG (Roche, 11277073910) labeled anti-sense RNA probes against bmp2b, bmp4, bmp7b, bmpr1aa, bmpr1ab, bmpr1ba, bmpr1bb, acvr1l. Probes were synthesized using Sp6 or T7 RNA polymerase (Roche, 10810274001 or 10881767001). Signals were visualized using a TSA Plus Cyanine 3 System (PerkinElmer, NEL744001KT). After staining, lower jaws were washed 1–4 hours in 2% H2O2 and positioned with the ventral side facing the objective and images were taken using a Leica SP8 inverted confocal scanning microscope. The primer sequences used for making BMP probes are listed in Table S2.
Heat-shock
Tg(scxa:gal4-vp16; uas:epNTR-RFP) line was crossed to Tg(hsp70:nog3) or Tg(hsp70:bmp2b) lines and tendon cells were ablated at 3–5dpf and ablated animals were heat shocked by transferring them into a pre-warmed incubator and heat shock was performed for 1 hour at 37°C at indicated time points (See figure legend). After heat shock, the animals were transferred into housing system for regeneration. For testing the responsiveness of Tg(bre:eGFP) embryo to BMP activation and inhibition, Tg(bre:eGFP) was crossed to Tg(hsp70:nog3) or Tg(hsp70:bmp2b) lines and embryos were heat shocked for 1 hour at 37°C at indicated time points (See figure legend). The primer sequences used for geno- typing Tg(hsp70:nog3) and Tg(hsp70:bmp2b) fish are listed in Table S1.
Whole-mount antibody staining
Whole-mount antibody staining was performed at 4°C unless otherwise stated. Embryos were fixed in 4% PFA for 1 hour at room temperature (RT) or overnight at 4°C, followed by PBST (PBS+0.6% Triton X-100) washes and dehydration to methanol series, and stored in 100% methanol for at least 30 minutes at −20°C. Embryos were then rehydrated to methanol series and washed with PBST and subjected to Proteinase K treatment. Following PBST washes and re-fixation with 4% PFA for 20 minutes at RT, embryos were washed with PBST and blocked with PBST/10% normal goat serum/1% DMSO for 1 hour at RT. Embryos were incubated with primary antibodies diluted in blocking solution overnight at 4°C (4 days for 26 day fish). Embryos were washed with PBST at least 2 hours with changes every 30 minutes and then incubated with secondary antibodies overnight at 4°C (2 days for 26 day fish at RT). Hoechst (Thermo Scientific, H3570) was added to the secondary antibody solution if nuclear staining was needed. Embryos were washed with PBST for 2X 30 minutes before imaging. The primary antibodies used were: Rabbit anti RFP (Rockland, 600–401-379), Mouse anti RFP (abcam, ab109809), Rabbit anti Phospho-Smad1/5 (Cell Signaling, 9516T), Mouse anti mcherry (Living Colors, 632543), Mouse anti MF 20 (DSHB, University of Iowa), Mouse anti Collagen II (DSHB, II-II6B3), Mouse anti myosin heavy chain (MHC) (A1025, DSHB), Rabbit anti GFP (Thermo Scientific, A21311). Secondary antibodies used were: Goat anti-Rabbit Alexa Fluor 488 (Thermo Scientific, A-11008), Goat anti-Mouse Alexa Fluor 488 (Thermo Scientific, A21121), Goat anti-Rabbit Alexa Fluor 594 (Thermo Scientific, A11012), Goat anti-Mouse Alexa Fluor 647 (Thermo Scientific, A21241), Donkey anti-Rabbit Alexa Fluor 647 (Abcam, ab150067).
Phospho-Smad1/5 staining
Staining was performed at 4°C unless otherwise stated. Embryos were fixed in 4% PFA for 1 hour at RT or overnight at 4°C, followed by PBST (PBS+0.1% Tween 20) washes and dehydration to methanol series, and stored in 100% methanol for at least 30 minutes at −20°C. Embryos were rehydrated to methanol series and washed with PBST. Lower jaws were collected for PBST washes and re-fixation with 4% PFA for 20 minutes at RT, Lower jaws were washed with PBST and blocked with 4% BSA (Fisher Scientific, BP1605–100) for 2 hours at RT. Lower jaws were incubated with Rabbit anti Phospho-Smad1/5 overnight at 4°C. Lower jaws were washed with PBST at least 2 hours with changes every 30 minutes and then incubated with secondary antibodies overnight at 4°C. Hoechst was added to the secondary antibody solution to stain the nucleus. Lower jaws were washed with PBST for 2 hours at RT before imaging. For imaging, lower jaw was embedded in 4% methylcellulose and imaging was taken using a Leica SP8 inverted confocal scanning microscope.
Total RNA isolation, RT-PCR and qPCR
Zebrafish total RNA was extracted from embryo heads of indicated time points using Trizol reagent (Life Technologies, 15596026) as described previously [100]. Reverse transcription was performed using a reverse transcriptase kit (Thermo Scientific, 18091200) according to the manufacturer’s protocol. Transcribed cDNA and primers were added to SYBR green PCR master mix (Life Technologies, 4367659). The StepOnePlus real-time PCR system (Applied Biosystems) was used to obtain the cycle threshold value. The relative expression of each gene was determined after being normalized to beta actin. The primer sequences used for qPCR are listed in Table S3.
Muscle stimulation
Tg(scxa:gal4-vp16; uas:epNTR-RFP) embryos were incubated in Mtz for tendon cell ablation at 3–5dpf and were then subjected to electrical stimulation at 7dpf as previously described [26].
Transmission Electron Microscopy
SH tendons were collected and prepared for electron microscopy by immersion in 1.5% formaldehyde/1.5% glutaraldehyde (Tousimis Research Corporation, #1010A) in DMEM (Gibco, 21063029) containing 0.05% tannic acid (Fisher Scientific, MK-1674–125) overnight followed by an extensive rinse in Dulbecco’s serum-free media (SFM), then post fixation in 1% OsO4 for 1 hour at room temperature. The samples were washed in SFM and then dehydrated in a graded series of ethanol from 30% to 100%, rinsed in propylene oxide and infiltrated in Spurrs epoxy over a total time of 2 hours, accelerated via microwave energy. Samples were polymerized at 70°C over 18 hours. 40nm ultra-thin sections were cut on a Leica EM UC7 ultramicrotome using a Diatome diamond knife and sections were mounted on formvar-coated copper palladium grids and post-stained in 8% ethanolic uranyl acetate followed by lead citrate. Sections were imaged in FEI G20 TEM at 120kV and photographed using a side-entry model XR41 AMT 2K x 2K camera. Montaging was accomplished using proprietary AMT software with 20% overlap and reconstructed with FIJI. Measurement of collagen fibril diameters was performed manually on montage images using ImageJ. Specifically, four to eight montage images were analyzed for each sample. For analysis at 80 dpf, a total of 122531 and 184242 fibrils were measured for control (n=2) and regenerated (n=3) SH tendons. For analysis at 180dpf, a total of 147007 and 155856 fibrils were measured for control (n=2) and regenerated (n=2) SH tendons, respectively. For area calculation for each collagen fibril, the formula A=π(D/2)22 (A, Area; D, Diameter) was used.
Imaging
All live or fixed embryos from WISH or antibody staining were mounted in 4% methyl cellulose (Sigma, M0387) on a thin glass slide (Gold Seal, 3323), or in 1% low melt agarose (RPI, 9012–36-6) in an imaging dish, or mounted in glycerol (Sigma, G7757) on a glass slide, or put into a slot made from 4% agarose (Bio-Excell, 61133056), depending on the experiment. Embryos were positioned with the ventral or lateral side facing the objective and images were taken either from single plane or Z-Stack using a ZEISS Axio Zoom.V16 Stereo microscope, Nikon ECLIPSE Ti-E inverted confocal microscope, and Leica SP8 inverted confocal laser scanning microscope, depending on the experiment.
Multiphoton imaging
0.2% tricaine-anesthetized 5dpf embryos were put into a slot made from 4% agarose and embryos were positioned with the ventral side of the head facing the objective. 1% low melt agarose was added to the slot to immobilize the embryos and a thin coverslip was put on the low melt agarose to facilitate the imaging. For imaging the SH tendons at 80 and 180dpf, fishes were tricaine-anesthetized and fixed in 4% PFA for 1 hour at room temperature and SH tendons were either incompletely or completely dissected and collected using two forceps (Fine Science Tools, 11252–00). Dissected SH tendons were put on a flat agarose surface and covered with a coverslip. Images were taken from a Z-Stack acquisition using an Olympus FVMPE-RS multiphoton microscopy system.
Multiphoton laser cell ablation
0.2% tricaine-anesthetized 5dpf embryos were embedded in 1% low melting agarose and embryos were positioned with the ventral side facing the objective. To perform the cell ablation, the individual target cell either from perichondrium or muscle junction area was selected and focused in a single plane and irradiated for 3s by a multiphoton laser at 800nm (35%). The ablation was confirmed by the loss of transgene expression and activation of second harmonic generation signal. After ablation, embryos were transferred into system for regeneration.
Alcian blue staining
26 dpf juveniles were fixed with 4% PFA at room temperature (RT) for 2hours. Fish were washed with PBS for 5 minutes at RT, treated with 50% ethanol for 10 minutes at RT on a rotating platform, and stained with staining solution (66.5% ethanol, 0.19M MgCl2, 0.02% alcian blue stock) overnight at RT on rotating platform. Fish were washed with ddH2O for 15 minutes at RT, and bleached (0.8% W/V KOH, 0.1% Tween20, 0.9% H2O2) until pigmentation was removed. Fish were washed with PBS for 5 minutes at RT and fixed with 4% PFA for 1 hour at RT. Images were taken using a Nikon Eclipse 80i microscope with a Nikon DS Ri 1 camera.
QUANTIFICATION AND STATISTICAL ANALYSIS
Embryos at each time point were randomly grouped into different treatments for each experiment. No treated embryos were excluded from the analysis unless they died during the process. No statistical method was used to predetermine sample size and sample sizes were indicated in each figure legend. For meas urements of tnmd fluorescence area, the whole craniofacial region of each embryo was selected and properly thresholded in ImageJ and the whole area was used for analysis. For quantifying the SH tendon cell number in heat shock experiments, samples and analysis were blinded to treatments prior to genotyping. For analysis of two conditions, two-tailed, unpaired Student’s t-tests were used and for multiple conditions comparison, one-way ANOVA with tukey’s post-hoc analysis was used. For analyzing muscle detachment phenotype, statistical analysis was performed using Chi test. P values were indicated in each figure legend and data were presented as mean or mean ± s.d.
Supplementary Material
Generation of a zebrafish tendon cell ablation line
Robust tendon cell regeneration in zebrafish
sox10+ and nkx2.5+ cells from attachment sites contribute to tendon cell regeneration
BMP signaling is sufficient and required for tendon cell regeneration
Using a new tendon cell ablation model, Niu et al. show that zebrafish can fully regenerate properly patterned and structurally correct tendons. Cellular regeneration of the tendon is driven by BMP signaling and newly recruited progenitors from connective tissue surrounding cartilage and muscle, specifically at musculoskeletal attachment sites.
Acknowledgments
We thank L. Zon (Boston Children’s Hospital) for providing the Tg(drl:creERT2) line; K. Poss (Department of Cell Biology, Duke University) for providing the Tg(shha:eGFP) line; H. Burgess (Division of Developmental Biology, NICHD, NIH) for providing the Tg(uas:epNTR-RFP) and Tg(uas-E1b:NfsB-mcherry) lines; M. Mullins (Department of Cell and Developmental Biology, University of Pennsylvania) for providing the Tg(hsp70l:bmp2b) line; B. Roman (Department of Biological Sciences, University of Pittsburgh) for providing the Tg(bre:eGFP) line; D. Langenau (Cancer Center, MGH) for providing the Tg(mylz2:Amcyan) and Tg(mylz2:mcherry) lines; C. Burns (Cardiovascular Research Center, MGH) for providing Tg(nkx2.5:ZsYellow) and Tg(nkx:ERT2CreERT2) lines; S. Fisher (School of Medicine, Boston University) for providing Tg(runx2:eGFP) line. We also thank Y (Zoey). Li (Wuxi Biologics, Shanghai, China) for providing the cartoons for zebrafish embryos and adults; D. Keene and S. Tufa (Micro Imaging Center, Shriners Hospital for Children, Portland, Oregon) for providing TEM service; S. Schulte-Merker (Institute for Cardiovascular Organogenesis and Regeneration, University of Münster, Münster, Germany) and K. Kawakami (Division of Molecular and Developmental Biology, National Institute of Genetics, Mishima, Japan) for providing the constructs and protocols for making scxa:gal4-vp16 construct [91,92]. We are grateful to C. Tabin (Harvard Medical School), L. Zon (Boston Children’s Hospital), J. Rajagopal (Center for Regenerative Medicine, MGH) for their critical reading and suggestions of the manuscript. J.L.G. and X.N. were supported by NIH/NICHD HD069533, NIH/NIDCR DE024771, NIH/NIAMS AR071554, and awards from the Charles H. Hood Foundation and the Harvard Stem Cell Institute. A.S. and T.F.S. were supported by NIH/NIAMS AR067797, NIH/NIDCR DE013828, NIH/NIDCR DE023050.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cserjesi P, Brown D, Ligon KL, Lyons GE, Copeland NG, Gilbert DJ, Jenkins N. a, and Olson EN (1995). Scleraxis: a basic helix-loophelix protein that prefigures skeletal formation during mouse embryogenesis. Development 121, 1099–1110. [DOI] [PubMed] [Google Scholar]
- 2.Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, and Tabin CJ (2001). Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development (Cambridge, England) 128, 3855–3866. [DOI] [PubMed] [Google Scholar]
- 3.Brent AE, Schweitzer R, and Tabin CJ (2003). A somitic compartment of tendon progenitors. Cell 113, 235–248. [DOI] [PubMed] [Google Scholar]
- 4.Shukunami C, Oshima Y, and Hiraki Y (2001). Molecular cloning of tenomodulin, a novel Chondromodulin-I related gene. Biochemical and Biophysical Research Communications 280, 1323–1327. [DOI] [PubMed] [Google Scholar]
- 5.Docheva D, Hunziker EB, Fassler R, and Brandau O (2005). Tenomodulin Is Necessary for Tenocyte Proliferation and Tendon Maturation. Molecular and Cellular Biology 25, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shukunami C, Takimoto A, Oro M, and Hiraki Y (2006). Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Developmental Biology 298, 234–247. [DOI] [PubMed] [Google Scholar]
- 7.Canty EG, Lu Y, Meadows RS, Shaw MK, Holmes DF, and Kadler KE (2004). Coalignment of plasma membrane channels and protrusions (fibropositors) specifies the parallelism of tendon. Journal of Cell Biology 165, 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canty EG, Starborg T, Lu Y, Humphries SM, Holmes DF, Meadows RS, Huffman A, O’Toole ET, and Kadler KE (2006). Actin filaments are required for fibripositor-mediated collagen fibril alignment in tendon. Journal of Biological Chemistry 281, 38592–38598. [DOI] [PubMed] [Google Scholar]
- 9.Kalson NS, Starborg T, Lu Y, Mironov A, Humphries SM, Holmes DF, and Kadler KE (2013). Nonmuscle myosin II powered transport of newly formed collagen fibrils at the plasma membrane. Proceedings of the National Academy of Sciences 110, E4743–E4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalson NS, Lu Y, Taylor SH, Holmes DF, and Kadler KE (2015). A structure-based extracellular matrix expansion mechanism of fibrous tissue growth. eLife 4, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edom-Vovard F, Schuler B, Bonnin MA, Teillet MA, and Duprez D (2002). Fgf4 positively regulates scleraxis and tenascin expression in chick limb tendons. Developmental Biology 247, 351–366. [DOI] [PubMed] [Google Scholar]
- 12.Brent AE (2005). Genetic analysis of interactions between the somitic muscle, cartilage and tendon cell lineages during mouse development. Development 132, 515–528. [DOI] [PubMed] [Google Scholar]
- 13.Huang AH, Riordan TJ, Pryce B, Weibel JL, Watson SS, Long F, Lefebvre V, Harfe BD, Stadler HS, Akiyama H, et al. (2015). Musculoskeletal integration at the wrist underlies the modular development of limb tendons. Development 142, 2431–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blitz E, Viukov S, Sharir A, Shwartz Y, Galloway JL, Pryce BA, Johnson RL, Tabin CJ, Schweitzer R, and Zelzer E (2009). Bone Ridge Patterning during Musculoskeletal Assembly Is Mediated through SCX Regulation of Bmp4 at the Tendon-Skeleton Junction. Developmental Cell 17, 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasson P, DeLaurier A, Bennett M, Grigorieva E, Naiche LA, Papaioannou VE, Mohun TJ, and Logan MPO (2010). Tbx4 and Tbx5 Acting in Connective Tissue Are Required for Limb Muscle and Tendon Patterning. Developmental Cell 18, 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ansorge HL, Hsu JE, Edelstein L, Adams S, Birk DE, and Soslowsky LJ (2012). Recapitulation of the Achilles tendon mechanical properties during neonatal development: a study of differential healing during two stages of development in a mouse model. Journal of orthopaedic research : official publication of the Orthopaedic Research Society 30, 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell K, Chien C, Bell R, Laudier D, Tufa SF, Keene DR, Anda- rawis-Puri N, and Huang AH (2017). Novel Model of Tendon Regeneration Reveals Distinct Cell Mechanisms Underlying Regenerative and Fi- brotic Tendon Healing. Scientific Reports 7, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi Y, Ehirchiou D, Kilts TM, Inkson CA, Embree MC, Sonoyama W, Li L, Leet AI, Seo BM, Zhang L, et al. (2007). Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nature Medicine 13, 1219–1227. [DOI] [PubMed] [Google Scholar]
- 19.Nichols AEC, Best KT, and Loiselle AE (2019). The cellular basis of fibrotic tendon healing: challenges and opportunities. Translational Research 209, 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyment NA, Hagiwara Y, Matthews BG, Li Y, Kalajzic I, and Rowe DW (2014). Lineage tracing of resident tendon progenitor cells during growth and natural healing. PLoS One 9, e96113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvey T, Flamenco S, and Fan C-M (2019). A Tppp3+Pdgfra+ tendon stem cell population contributes to regeneration and reveals a shared role for PDGF signalling in regeneration and fibrosis. Nature Cell Biology 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Zhang X, Huang H, Xia Y, Yao YF, Mak AFT, Yung PSH, Chan KM, Wang L, Zhang C, et al. (2017). Osteocalcin expressing cells from tendon sheaths in mice contribute to tendon repair by activating hedgehog signaling. eLife 6, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Best KT, and Loiselle AE (2019). Scleraxis lineage cells contribute to organized bridging tissue during tendon healing and identify a subpopulation of resident tendon cells. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 33, 8578–8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGurk PD, Swartz ME, Chen JW, Galloway JL, and Eberhart JK (2017). In vivo zebrafish morphogenesis shows Cyp26b1 promotes tendon condensation and musculoskeletal patterning in the embryonic jaw. PLoS Genetics 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JW, and Galloway JL (2014). The development of zebrafish tendon and ligament progenitors. Development 141, 2035–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, and Schilling TF (2014). Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions. eLife 2014, e02372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah RR, Nerurkar NL, Wang CC, and Galloway JL (2015). Tensile Properties of Craniofacial Tendons in the Mature and Aged Zebrafish. 867–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabor KM, Bergeron SA, Horstick EJ, Jordan DC, Aho V, Porkka-Heiskanen T, Haspel G, and Burgess HA (2014). Direct activation of the Mauthner cell by electric field pulses drives ultrarapid escape responses. Journal of Neurophysiology 112, 834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pisharath H, Rhee JM, Swanson MA, Leach SD, and Parsons MJ (2007). Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mechanisms of Development 124, 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, and Stainier DYR (2007). Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Developmental Dynamics 236, 1025–1035. [DOI] [PubMed] [Google Scholar]
- 31.Zhang R, Han P, Yang H, Ouyang K, Lee D, Lin YF, Ocorr K, Kang G, Chen J, Stainier DYR, et al. (2013). In vivo cardiac reprogramming contributes to zebrafish hearTregeneration. Nature 498, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Cao J, Dickson AL, and Poss KD (2015). Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 522, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Léjard V, Brideau G, Blais F, Salingcarnboriboon R, Wagner G, Roehrl, Noda M, Duprez D, Houillier P, and Rossert J (2007). Scleraxis and NFATc regulate the expression of the pro-a1(I) collagen gene in tendon fibroblasts. Journal of Biological Chemistry 282, 17665–17675. [DOI] [PubMed] [Google Scholar]
- 34.Schilling TF, and Kimmel CB (1997). Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development 124, 2945–2960. [DOI] [PubMed] [Google Scholar]
- 35.Williams RM, Zipfel WR, and Webb WW (2005). Interpreting second-harmonic generation images of collagen I fibrils. Biophysical Journal 88, 1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernández LP, Barresi MJF, and Devoto SH (2002). Functional morphology and developmental biology of zebrafish: reciprocal illumination from an unlikely couple. Integrative and comparative biology 42, 222–231. [DOI] [PubMed] [Google Scholar]
- 37.Kague E, Gallagher M, Burke S, Parsons M, Franz-Odendaal T, and Fisher S (2012). Skeletogenic Fate of Zebrafish Cranial and Trunk Neural Crest. PLoS One 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, and Schilling TF (2005). Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development 132, 3977–3988. [DOI] [PubMed] [Google Scholar]
- 39.Mongera A, Singh AP, Levesque MP, Chen Y-Y, Konstantinidis P, and Nusslein-Volhard C (2013). Genetic lineage labeling in zebrafish un-covers novel neural crest contributions to the head, including gill pillar cells. Development 140, 916–925. [DOI] [PubMed] [Google Scholar]
- 40.Pang SC, Wang HP, Zhu ZY, and Sun YH (2015). Transcriptional Activity and DNA Methylation Dynamics of the Gal4/UAS System in Zebrafish. Marine Biotechnology 17, 593–603. [DOI] [PubMed] [Google Scholar]
- 41.Dunkman AA, Buckley MR, Mienaltowski MJ, Adams SM, Thomas SJ, Kumar A, Beason DP, lozzo RV, Birk DE, and Soslowsky LJ (2014). The injury response of aged tendons in the absence of biglycan and decorin. Matrix Biology 35, 232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurevich DB, Nguyen PD, Siegel AL, Ehrlich OV, Sonntag C, Phan JMN, Berger S, Ratnayake D, Hersey L, Berger J, et al. (2016). Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science 353, aad9969. [DOI] [PubMed] [Google Scholar]
- 43.Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, Ciruna B, Sanes JR, Lichtman JW, and Schier AF (2013). Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140, 2835–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rochard LJ, Ling ITC, Kong Y, and Liao EC (2015). Visualization of chondrocyte intercalation and directional proliferation via zebrabow clonal cell analysis in the embryonic Meckel’s cartilage. Journal of Visualized Experiments 2015, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosimann C, Panáková D, Werdich AA, Musso G, Burger A, Lawson KL, Carr LA, Nevis KR, Sabeh MK, Zhou Y, et al. (2015). Chamber identity programs drive early functional partitioning of the heart. Nature Communications 6, 8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guner-Ataman B, Paffett-Lugassy N, Adams MS, Nevis KR, Jahangiri L, Obregon P, Kikuchi K, Poss KD, Burns CE, and Burns CG (2013). Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development (Cambridge) 140, 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paffett-Lugassy N, Novikov N, Jeffrey S, Abrial M, Guner-Ataman B, Sakthivel S, Burns CE, and Burns CG (2017). Unique developmental trajectories and genetic regulation of ventricular and outflow tract progenitors in the zebrafish second heart field. Development (Cambridge) 144, 4616–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felker A, Nieuwenhuize S, Dolbois A, Blazkova K, Hess C, Low LWL, Burger S, Samson N, Carney TJ, Bartunek P, et al. (2016). In vivo performance and properties of Tamoxifen metabolites for CreERT2 control. PLoS One 11, e0152989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noden DM (1983). The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. The American Journal of Anatomy 168, 257–276. [DOI] [PubMed] [Google Scholar]
- 50.Sefton EM, Piekarski N, and Hanken J (2015). Dual embryonic origin and patterning of the pharyngeal skeleton in the axolotl ( Ambystoma mexicanum ). Evolution & Development 17, 175–184. [DOI] [PubMed] [Google Scholar]
- 51.Liu C, Wu C, Yang Q, Gao J, Li L, Yang D, and Luo L (2016). Macrophages Mediate the Repair of Brain Vascular Rupture through Direct Physical Adhesion and Mechanical Traction. Immunity 44, 1162–1176. [DOI] [PubMed] [Google Scholar]
- 52.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R, Gao Y, Inada M, et al. (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to mat- urational arrest of osteoblasts. Cell 89, 755–764. [DOI] [PubMed] [Google Scholar]
- 53.St-Jacques B, Hammerschmidt M, and McMahon A (1999). Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Development 13, 2072–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, and Leach SD (2009). Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mechanisms of Development 126, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimizu N, Kawakami K, and Ishitani T (2012). Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/B-catenin signaling-reporter transgenic zebrafish. Developmental Biology 370, 71–85. [DOI] [PubMed] [Google Scholar]
- 56.Laux DW, Febbo JA, and Roman BL (2011). Dynamic analysis of BMP-responsive smad activity in live zebrafish embryos. Developmental Dynamics 240, 682–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shkumatava A (2004). Sonic hedgehog, secreted by amacrine cells, acts as a short-range signal to direct differentiation and lamination in the zebrafish retina. Development 131, 3849–3858. [DOI] [PubMed] [Google Scholar]
- 58.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, and Peterson RT (2008). Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nature Chemical Biology 4, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, and Peterson RT (2008). Structureactivity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorganic and Medicinal Chemistry Letters 18, 4388–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto-Shiraishi YI, and Kuroiwa A (2013). Wnt and BMP signaling cooperate with Hox in the control of Six2 expression in limb tendon precursor. Developmental Biology 377, 363–374. [DOI] [PubMed] [Google Scholar]
- 61.Wolfman NM, Hattersley G, Cox K, Celeste AJ, Nelson R, Yamaji N, Dube JL, DiBlasio-Smith E, Nove J, Song JJ, et al. (1997). Ectopic induction of tendon and ligament in rats by growth and differentiation factors 5, 6, and 7, members of the TGF-b gene family. J. Clin. Invest. 100, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lou J, Tu Y, Ludwig FJ, Zhang J, and Manske PR (1999). Effect of bone morphogenetic protein-12 gene transfer on mesenchymal progenitor cells. Clinical orthopaedics and related research, 333–339. [DOI] [PubMed] [Google Scholar]
- 63.Lou J, Tu Y, Burns M, Silva MJ, and Manske P (2001). BMP-12 gene transfer augmentation of lacerated tendon repair. Journal of Orthopaedic Research 19, 1199–1202. [DOI] [PubMed] [Google Scholar]
- 64.Lorda-Diez CI, Montero JA, Choe S, Garcia-Porrero JA, and Hurle JM (2014). Ligand- and stage-dependent divergent functions of BMP signaling in the differentiation of embryonic skeletogenic progenitors in vitro. Journal of Bone and Mineral Research 29, 735–748. [DOI] [PubMed] [Google Scholar]
- 65.Pingel J, Fredberg U, Mikkelsen LR, Schjerling P, Heinemeier KM, Kjaer M, Harisson A, and Langberg H (2013). No inflammatory gene-expression response to acute exercise in human Achilles tendinopathy. European Journal of Applied Physiology 113, 2101–2109. [DOI] [PubMed] [Google Scholar]
- 66.Solchaga LA, Bendele A, Shah V, Snel LB, Kestler HK, Dines JS, and Hee CK (2014). Comparison of the effect of intra-tendon applications of recombinant human platelet-derived growth factor-BB, plateletrich plasma, steroids in a rat achilles tendon collagenase model. Journal of Orthopaedic Research 32, 145–150. [DOI] [PubMed] [Google Scholar]
- 67.O’Brien EJO, Frank CB, Shrive NG, Hallgrimsson B, and Hart DA (2012). Heterotopic mineralization (ossification or calcification) in tendinopathy or following surgical tendon trauma. International Journal of Experimental Pathology 93, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jopling C, Sleep E, Raya M, Marti M, Raya A, and Belmonte JCI (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, MacRae CA, Stainier DYR, and Poss KD (2010). Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 464, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goldman D (2014). Müller glial cell reprogramming and retina regeneration. Nature Reviews Neuroscience 15, 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He J, Lu H, Zou Q, and Luo L (2014). Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 146, 789–800.e8. [DOI] [PubMed] [Google Scholar]
- 72.Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-merker S, et al. (2011). Bone Regenerates via Dedifferentiation of Osteoblasts in the Zebrafish Fin. Developmental Cell 20, 713–724. [DOI] [PubMed] [Google Scholar]
- 73.Mokalled MH, Patra C, Dickson AL, Endo T, Stainier DYR, and Poss KD (2016). Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 354, 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paul S, Schindler S, Giovannone D, de Millo Terrazzani A, Mariani FV, and Crump JG (2016). Ihha induces hybrid cartilage-bone cells during zebrafish jawbone regeneration. Development (Cambridge) 143, 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blitz E, Sharir A, Akiyama H, and Zelzer E (2013). Tendon-bone attachment unit is formed modularly by a distinct pool of Scx-and Sox9-positive progenitors. Development (Cambridge) 140, 2680–2690. [DOI] [PubMed] [Google Scholar]
- 76.Sugimoto Y, Takimoto A, Akiyama H, Kist R, Scherer G, Nakamura T, Hiraki Y, and Shukunami C (2012). Scx+/Scx9+ progenitors contribute to the establishment of the junction between cartilage and ten- don/ligament. Development (Cambridge) 140, 2280–2288. [DOI] [PubMed] [Google Scholar]
- 77.Debnath S, Yallowitz AR, McCormick J, Lalani S, Zhang T, Xu R, Li N, Liu Y, Yang YS, Eiseman M, et al. (2018). Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature 562, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCusker CD, Diaz-Castillo C, Sosnik J, Phan Q, A., and Gardiner DM (2016). Cartilage and bone cells do not participate in skeletal regeneration in Ambystoma mexicanum limbs. Developmental Biology 416, 26–33. [DOI] [PubMed] [Google Scholar]
- 79.Kardon G, Harfe BD, and Tabin CJ (2003). A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Developmental Cell 5, 937–944. [DOI] [PubMed] [Google Scholar]
- 80.Tokita M, and Schneider RA (2009). Developmental origins of species- specific muscle pattern. Developmental Biology 331, 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giordani L, He GJ, Negroni E, Sakai H, Law JYC, Siu MM, Wan R, Corneau A, Tajbakhsh S, Cheung TH, et al. (2019). High-Dimensional Single-Cell Cartography Reveals Novel Skeletal Muscle-Resident Cell Populations. Molecular Cell 74, 609–621.e6. [DOI] [PubMed] [Google Scholar]
- 82.Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, and Postlethwait JH (2008). MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nature Genetics 40, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sugg KB, Markworth JF, Disser NP, Rizzi AM, Talarek JR, Sarver DC, Brooks SV, and Mendias CL (2018). Postnatal tendon growth and remodeling require platelet-derived growth factor receptor signaling. American Journal of Physiology - Cell Physiology 314, C389–C403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brockschmidt A, Todt U, Ryu S, Hoischen A, Landwehr C, Birnbaum S, Frenck W, Radlwimmer B, Lichter P, Engels H, et al. (2007). Severe mental retardation with breathing abnormalities (Pitt - Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Human Molecular Genetics 16, 1488–1494. [DOI] [PubMed] [Google Scholar]
- 85.Lin B, Srikanth P, Castle AC, Nigwekar S, Malhotra R, Galloway JL, Sykes DB, and Rajagopal J (2018). Modulating Cell Fate as a Therapeutic Strategy. Cell Stem Cell 23, 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raven A, Lu WY, Man TY, Ferreira-Gonzalez S, O’Duibhir E, Dwyer BJ, Thomson JP, Meehan RR, Bogorad R, Koteliansky V, et al. (2017). Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pfefferli C, and Jazwinska A (2015). The art of fin regeneration in zebrafish. Regeneration 2, 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McCusker CD, Athippozhy A, Diaz-Castillo C, Fowlkes C, Gardiner DM, and Voss SR (2015). Positional plasticity in regenerating Amybstoma mexicanum limbs is associated with cell proliferation and pathways of cellular differentiation Regeneration and repair. BMC Developmental Biology 15, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haas BJ, and Whited JL (2017). Advances in Decoding Axolotl Limb Regeneration. Trends in Genetics 33, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, and Tanaka EM (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65. [DOI] [PubMed] [Google Scholar]
- 91.Bussmann J, and Schulte-Merker S (2011). Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development 138, 4327–4332. [DOI] [PubMed] [Google Scholar]
- 92.Suster ML, Abe G, Schouw A, and Kawakami K (2011). Transposon-mediated BAC transgenesis in zebrafish. Nature Protocols 6, 1998–2021. [DOI] [PubMed] [Google Scholar]
- 93.Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, and Parsons MJ (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Developmental Biology 304, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Agetsuma M, Aizawa H, Aoki T, Nakayama R, Takahoko M, Goto M, Sassa T, Amo R, Shiraki T, Kawakami K, et al. (2010). The habenula is crucial for experience-dependent modification of fear responses in zebrafish. Nature Neuroscience 13, 1354–1356. [DOI] [PubMed] [Google Scholar]
- 95.Dale RM, and Topczewski J (2011). Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Developmental Biology 357, 518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith ACH, Raimondi AR, Salthouse CD, Ignatius MS, Blackburn JS, Mizgirev IV, Storer NY, Jong J.L.O. De, Chen AT, Zhou Y, et al. (2018). High-throughput cell transplantation establishes that tumor- initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. 115, 3296–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chocron S, Verhoeven MC, Rentzsch F, Hammerschmidt M, and Bakkers J (2007). Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Developmental Biology 305, 577–588. [DOI] [PubMed] [Google Scholar]
- 98.Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, Gu A, Mosimann C, Sondalle S, Peterson RE, et al. (2011). Latent TGF-pbinding protein 3 identifies a second heart field in zebrafish. Nature 474, 645–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davidson A (2003). Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Developmental Biology 263, 191–202. [DOI] [PubMed] [Google Scholar]
- 100.Niu X, Hong J, Zheng X, Melville DB, Knapik EW, Meng A, and Peng J (2014). The nuclear pore complex function of sec13 protein is required for cell survival during retinal development. Journal of Biological Chemistry 289, 11971–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.