Abstract
Rationale
Increased protein synthesis of pro-fibrotic genes is a common feature in cardiac fibrosis and heart failure. Despite this observation, critical factors and molecular mechanisms for translational control of pro-fibrotic genes during cardiac fibrosis remain unclear.
Objective
To investigate the role of a bifunctional aminoacyl-tRNA synthetase (ARS), glutamyl-prolyl-tRNA synthetase (EPRS) in translational control of cardiac fibrosis.
Methods and Results
Results from re-analyses of multiple publicly available datasets of human and mouse heart failure, demonstrated that EPRS acted as an integrated node among the ARSs in various cardiac pathogenic processes. We confirmed that EPRS was induced at mRNA and protein levels (~1.5–2.5 fold increase) in failing hearts compared with non-failing hearts using our cohort of human and mouse heart samples. Genetic knockout of one allele of Eprs globally (Eprs+/–) using CRISPR-Cas9 technology or in a Postn-Cre-dependent manner (Eprsflox/+; PostnMCM/+) strongly reduces cardiac fibrosis (~50% reduction) in isoproterenol-, transverse aortic constriction-, and myocardial infarction-induced heart failure mouse models. Inhibition of EPRS using a prolyl-tRNA synthetase (PRS)-specific inhibitor, halofuginone (Halo), significantly decreases translation efficiency of proline-rich collagens in cardiac fibroblasts as well as TGF-β-activated myofibroblasts. Overexpression of EPRS increases collagen protein expression in primary cardiac fibroblasts under TGF-β stimulation. Using transcriptome-wide RNA-Seq and polysome profiling-Seq in Halo treated fibroblasts, we identified multiple novel Pro-rich genes in addition to collagens, such as Ltbp2 and Sulf1, which are translationally regulated by EPRS. SULF1 is highly enriched in human and mouse myofibroblasts. In the primary cardiac fibroblast culture system, siRNA-mediated knockdown of SULF1 attenuates cardiac myofibroblast activation and collagen deposition. Overexpression of SULF1 promotes TGF-β-induced myofibroblast activation and partially antagonizes anti-fibrotic effects of Halo treatment.
Conclusions
Our results indicate that EPRS preferentially controls translational activation of proline codon rich pro-fibrotic genes in cardiac fibroblasts and augments pathological cardiac remodeling.
Keywords: amino acyl-tRNA synthetases, fibrosis, halofuginone, heart failure, myofibroblast, proline, transcriptome
Subject Terms: Cardiomyopathy, Fibrosis, Heart Failure, Hypertrophy, Remodeling
Graphical Abstract

INTRODUCTION
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality worldwide. Heart failure (HF), a major manifestation of CVD, is often accompanied by cardiac fibrosis driven by increased pro-fibrotic protein synthesis following cardiac fibroblast (CF) activation1. According to the central dogma, protein synthesis requires gene transcription and mRNA translation, both of which can be regulated to control protein production. Transcriptional regulation during cardiac fibrosis has been extensively studied1, 2. However, the fact that protein expression often does not correlate with mRNA abundance3, 4 indicates translational control as another critical layer of regulation in promoting pro-fibrotic protein synthesis5, 6. Although increased pro-fibrotic protein synthesis has been observed in cardiac fibrosis4, 5, the underlying regulatory mechanism of pro-fibrotic mRNA translation in CFs has not been identified. This unaddressed question represents a critical gap in our understanding of the regulatory control of cardiac fibrosis in the pathologic remodeling process.
Human translation machinery is comprised of three major parts, ribosomes, translation factors (initiation/elongation/termination factors), and aminoacyl-transfer RNAs (aa-tRNAs)7, ligation products of amino acids and tRNAs that are catalyzed by aminoacyl-tRNA synthetases (ARSs)8, 9. Among all mammalian ARSs, glutamyl-prolyl-tRNA synthetase (EPRS) catalyzes the attachment of two amino acids, glutamic acid (E) and proline (P), to their cognate tRNAs for protein synthesis through two catalytic domains10. Since many pro-fibrotic proteins are proline-rich such as collagens, EPRS-mediated translational regulation probably plays a critical role in pro-fibrotic protein synthesis during cardiac fibrosis. As supporting evidence, in human genetics, hypoactive mutations in the PRS domain of EPRS leads to hypomyelinating leukodystrophy without causing any known cardiac dysfunction in patients11. These EPRS mutations imply that reduced PRS enzymatic activity may not have adverse effects on normal heart function while reducing proline-rich protein synthesis. However, the role of EPRS in cardiac disease is still unexplored. Further, an (E)PRS-specific inhibitor, halofuginone (Halo), blocks binding of (E)PRS to proline and tRNAPro and prevents their ligation12–14. Halo has been used to treat Duchenne Muscular Dystrophy (Akashi Therapeutics) by reducing fibrosis and increasing muscle strength (phase II clinical trials). Also, the Glaxo Smith Kline company confirmed its anti-fibrotic and cardiac protective activity in multiple mouse heart failure models15. However, the therapeutic mechanism of Halo has only been studied at the transcriptional level in triggering an amino acid starvation response (AASR) and a noncanonical TGF-β signaling pathway12, 15–18. The direct and preferential downstream transcripts regulated at the translational level by Halo-mediated inhibition of EPRS remains unclear. A previous study has revealed that decreased levels of the ribosome, a general translation machinery component, selectively regulated translation of a subset of transcripts in human hematopoiesis19. Therefore, as a general translation factor, EPRS may also have preferential translational targets during cardiac fibrosis, which has not been explored17, 18.
Here, we examined EPRS-mediated regulatory mechanisms of pro-fibrotic protein synthesis at the translatome-wide level in fibroblasts. We show that EPRS is induced in human and mouse failing hearts. EPRS is an integrated node downstream of various cardiac pathologic cues for translational control in cardiac fibrosis. Increased EPRS expression contributes to the elevated translation of proline (Pro)-rich (PRR) mRNAs via enhanced translation elongation in CFs. Genetic knockout of Eprs antagonized cardiac pathologic remodeling and fibrosis in different mouse heart failure models. Moreover, by using the EPRS inhibitor Halo, we found that EPRS inhibition selectively reduces PRR mRNA translation, such as collagens and other novel PRR genes, including LTBP2 and SULF1. Finally, we show that SULF1 is a novel biomarker for cardiac fibrosis, and that knockdown of SULF1 remarkably abolishes myofibroblast activation.
METHODS
A detailed description of materials and methods is provided in the online Supplemental Materials and the Major Resource Table. All data that support the findings of this study are available in the article and the online supplementary files. Additional technical information is available from the corresponding author upon request.
Human Specimens
All human samples of frozen cardiac tissues, including 17 samples from explanted failing hearts and 8 samples from non-failing donor hearts, as well as paraffin-embedded section slides from dilated cardiomyopathy (DCM), ischemic heart failure (IHF) or non-failing donor (NF) hearts, were acquired from the Cleveland Clinic. This study was approved by the Material Transfer Agreement between the University of Rochester Medical Center (URMC) and the Cleveland Clinic. All human samples were picked up randomly based on the presence or absence of heart failure by our collaborator, Dr. Wai Hong Wilson Tang at Cleveland Clinic. We are blinded from any clinical data. There may still be some sort of bias in the inclusion of human samples as exemplified as follows: 1) the sample size is limited and it may not fully reflect the outcome from a much larger population. 2) a sub-region of the left ventricle of the human hearts were collected for the experiments. Thus, it may not fully recapitulate the outcome in the whole heart. This is a limitation within human samples experiments in the current research.
Mice
The Eprs+/– heterozygous knockout (Eprs haploinsufficiency) chimera founder mice were produced in the University of Rochester Mouse Genome Editing Resource. We generated the Eprs targeted male chimera mouse on the C57BL/6J background and performed germline transmission. Using the CRISPR-Cas9 system, a gRNA (GCUAGAAUUGCAACUACGUCUGG) and a homology-directed recombination DNA template containing an insertion of tandem stop codons plus an adenosine residue (UAAUAAA) were used to introduce the early stop signal in exon 3 of the Eprs gene, resulting in a null allele. For experiments with Eprs+/– mice, control mice of the same age and gender from littermates or sibling mating were used. All animal procedures were performed following the National Institutes of Health (NIH) and University of Rochester Institutional guidelines. Three different HF models were used: (1) Isoproterenol (ISO) infusion model (20 mg/kg per day; neural hormonal model). (2) Transverse aortic constriction (TAC; surgical model). (3) Myocardial infarction (MI; surgical model). Eprs floxed mouse line Eprs_tm1c_B03 (C57BL6/N-Eprs<tm1c(EUBOMM)Hmgu>/Tcp) was purchased from The Center for Phenogenomics (Toronto, Canada) in the form of frozen sperm20. The Eprsflox/+ tm1c mouse line was rederived using In Vitro Fertilization performed by the Mouse Genome Editing Resource at URMC. The Eprsflox/+ tm1c mouse line was bred with PostnMCM/+ mice21 to obtain a tamoxifen-inducible Postn-Cre-driven Eprs conditional knockout (cKO) mouse line. We used one single initial dose of 30 μg/g of mouse body weight for tamoxifen (TMX) intraperitoneal injection followed by TMX food for the entire pathologic stimulus of TAC surgery. We performed power analysis for group size justification. Mice were randomized for experiments. Animal operations, including ISO infusion, TAC, and MI surgery, and echocardiography measurement, were performed blindly by the Microsurgical Core surgeons. Sections and histology analyses were performed blindly by the Histology Core.
Analysis of translational activity by polysome profiling
Polysome profiling for cytosolic translation was performed as previously described22. Cycloheximide (CHX, 100 μg/ml) was added to the cells for 15 mins before lysis to freeze ribosomes on mRNAs in the elongation phase. Approximately 107 cells were lysed in TMK lysis buffer (10 mM Tris-HCl pH 7.4, 100 mM KCl, 5 mM MgCl2, 1% Triton X-100, 0.5% Deoxycholate, 2 mM DTT) containing 100 μg/ml CHX, 4 U/ml RNase inhibitor (NEB) and proteinase inhibitor cocktail (Roche) on ice for 20 mins. Equal amounts of A260 absorbance from each sample were loaded onto a 10–50% sucrose gradient solution and centrifuged at 29,000 rpm for 4 hrs; 22 translation fractions were collected from each sample by Density Gradient Fractionation System (BRANDEL). Based on the UV absorbance curve, the 22 fractions were pooled into seven samples for total RNA extraction and RT-qPCR analysis, including free mRNP, 40S small ribosome subunit, 60S large ribosome subunit, 80S monosome, light polysomes (disome, trisome, etc.), and heavy polysomes (>5 ribosomes). For polysome-Seq, all the fractions were pooled into three samples, non-polysome (free mRNP, 40S, 60S subunit), light polysome (monosome, disome, trisome, tetrasome), and heavy polysome (≥5 ribosomes). Total RNA was extracted from the same volume of each pooled fraction with Trizol LS (ThermoFisher Scientific), and Renilla luciferase mRNA from in vitro transcription was used as RNA spike-in and loading control for RT-qPCR. The RNA-Seq and polysome-Seq data were analyzed by Genome Research Center of URMC23–27 and uploaded to the NCBI GEO database with ID of GSE136838.
Statistical analysis
All quantitative data were presented as mean ± SEM and analyzed using Prism 8.3.0 software (GraphPad). The statistical significance was analyzed by Prism 8.3.0 and R 3.5.1, and the non-parametric post hoc pairwise comparisons were performed using r package PMCMR27, 28. We used Kolmogorov-Smirnov test to assess if the data are normally distributed (N≥10), and parametric tests (unpaired student t test or ANOVA for normally distributed data) or non-parametric tests (unpaired Mann-Whitney test or Kruskal-Wallis test for not normally distributed data) were used accordingly. Since the normality cannot be assessed on the data with small sample size (N<10), we performed non-parametric tests across our data with small sample size (N<10). For comparison between 2 groups, non-parametric unpaired Mann-Whitney test was performed. For comparisons among ≥3 groups, non-parametric Kruskal-Wallis test was performed with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg method for type I error correction. For the counts of proline-rich genes, chi-square (ϰ2) test was performed. Two-sided P values <0.05 were considered to indicate statistical significance. Overall P values for comparisons among ≥3 groups were provided in Online Table XII. Specific statistical methods and post hoc tests were described in the figure legends.
RESULTS
EPRS is upregulated in human and mouse heart failure
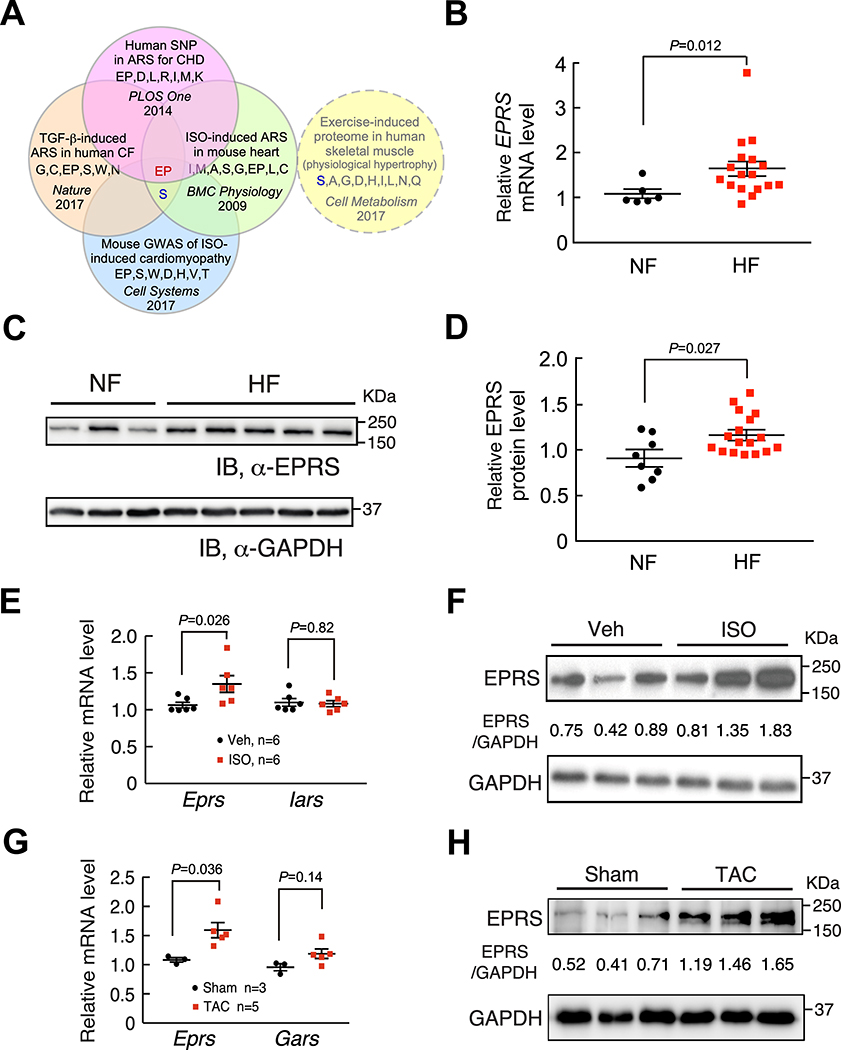
To screen for candidate translation factors involved in cardiac fibrosis, we re-analyzed RNA-Seq data of TGF-β (a typical pro-fibrotic cytokine29) treated human CFs from CVD patients4. We determined that cytoplasmic ARSs are the only significantly induced translation machinery components (Online Figure IA) among ribosome proteins and all translation factors (Online Figure IB, Online Table I)4. Among 20 ARSs, we identified EPRS as the key ARS involved in cardiac pathogenesis following screening of ARSs induced in TGF-β-activated human CFs4, human ARSs with genetic mutations in congenital heart disease30, mouse ARSs associated with isoproterenol (ISO)-induced cardiomyopathy by genome-wide association studies (GWAS)31, and ISO-induced ARSs in mouse failing hearts32 (Figure 1A). Multiple ARS proteins are induced in skeletal muscle of humans during exercise training, which is considered to be a mechanism to support protein synthesis and muscle growth33. EPRS is not among this group of ARSs required for physiologic muscle hypertrophy (Figure 1A, dotted circle). To validate this observation, we measured EPRS expression in our cohort of human and mouse heart samples. Our results showed that EPRS was induced at both mRNA and protein levels in failing hearts from heart failure patients compared to non-failure donor hearts (Figure 1B–D). Also, EPRS was induced at both mRNA and protein levels in the hearts of mice that underwent ISO infusion compared to those with vehicle treatment (Figure 1E and 1F). Consistently, EPRS was also induced at both mRNA and protein levels in the hearts of mice under TAC surgery compared to those with sham operations (Figure 1G and 1H). Taken together, these data indicate that EPRS may play a critical role in cardiac pathogenesis.
Figure 1. EPRS is upregulated in human and mouse heart failure.
(A) Glutamyl-prolyl-tRNA synthetase (EPRS) is a common hit of upregulated ARSs and ARSs with a genetic association in human and mouse heart disease, but not induced in human physiologic muscle hypertrophy.
(B) EPRS mRNA is increased in failing human hearts (n=17) compared to non-failing donor hearts (n=6). 18S rRNA is used as a loading control.
(C) EPRS protein is increased in failing human hearts compared to non-failing donor hearts. Representative Western blot results are shown.
(D) Quantification of Western blot results from all human heart samples indicates elevated expression of EPRS in failing human hearts (n=17) compared to non-failing donor hearts (n=8). GAPDH is used as a loading control for quantification.
(E-F) EPRS mRNA and protein expression are increased in the hearts from a 4-week ISO infusion induced mouse HF model compared to vehicle infused hearts while Iars (Isoleucyl-tRNA synthetase) mRNA remains unchanged. 18S rRNA and GAPDH are used as loading controls for mRNA and protein quantification, respectively.
(G-H) EPRS mRNA and protein expression are increased in the hearts from an 8-week TAC surgery-induced mouse HF model. Gars (glycyl-tRNA synthetase) is a negative control ARS in the TAC model. 18S rRNA and GAPDH are used as loading controls for mRNA and protein quantification, respectively.
Comparisons were performed by non-parametric unpaired Mann-Whitney test for B, D, E, G.
Single allele knockout of Eprs attenuates cardiac fibrosis under pathogenic stresses
To determine the role of EPRS during cardiac pathogenesis, we generated Eprs global KO mouse using CRISPR technology. The global Eprs KO mice were created by introducing two tandem premature stop codons plus a frame-shifting adenosine nucleotide in the third exon of the Eprs gene with protospacer adjacent motif (PAM) mutations (Online Figure IIA). TA-clone and DNA sequencing confirmed the successful insertion of premature stop codons into the Eprs genomic sequence (Online Figure IIB and IIC). Homozygous global Eprs KO mice are embryonic lethal. The heterozygous (het) appeared normal in weight (WT: 26.58±0.44g, n=8; Eprs+/–: 27.27±0.45g, n=6, male mice at the age of 3–4 months) and fertility (not shown). EPRS expression was reduced by almost half in the heart of Eprs+/– mice, based on quantitative measurements of EPRS expression using RT-qPCR and Western blot for heart tissue lysates (Online Figure IID) and immunostaining in the heart tissue sections (Online Figure IIE and IIF). These indicate that Eprs is an essential gene, and loss of one allele does not cause developmental defects in mice at baseline.
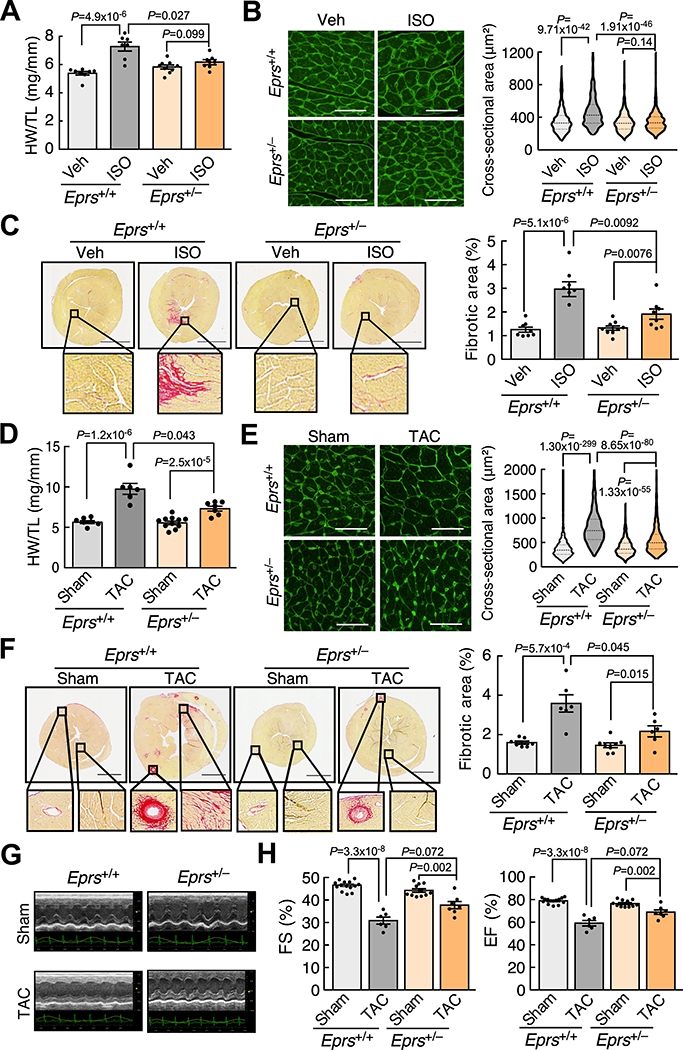
Given the induction of EPRS expression in human and mouse failing hearts, we next sought to determine the effects of reduced EPRS on the heart during cardiac remodeling in vivo. We first used a β-adrenergic receptor agonist ISO to treat mice via osmotic minipump implantation (20 mg/Kg/day) for four weeks. The hearts of WT and Eprs+/– mice were comparable in size at baseline, as indicated by heart weight/tibia length (HW/TL) ratio (Figure 2A) and wheat germ agglutinin (WGA) staining (Figure 2B). However, Eprs+/– mice exhibited a reduced cardiac hypertrophy phenotype compared to WT mice after ISO treatment (7.27±0.31 (Mean±SEM) in Eprs+/+ to 6.17±0.18 in Eprs+/– of HW/TL ratio and 474.8±7.59 μm2 in Eprs+/+ to 348.4±3.96 μm2 in Eprs+/– of left ventricle (LV) myocyte area) (Figure 2A and 2B). Picrosirius red staining showed that Eprs+/– mice had much less cardiac fibrosis (3.17±0.26% to 1.90±0.22%) after ISO treatment than WT mice (Figure 2C). Isolated adult CMs from WT and Eprs+/– mice responded to ISO and showed myocyte hypertrophy at similar levels (Online Figure IIG). These observations suggest that reduced myocyte hypertrophy in the ISO infusion mouse model is due to crosstalk between CFs and CMs as an indirect effect but not caused by reduced ability to translate proteins needed for the hypertrophic response in CMs.
Figure 2. Eprs haploinsufficiency attenuates cardiac hypertrophy and fibrosis in ISO infusion and TAC models.
(A) Eprs+/– mice show a reduced HW/TL (heart weight/tibia length) ratio in a 4-week ISO infusion mouse HF model compared to WT (Eprs+/+) mice. n=8/ 7/ 9/ 8 for Eprs+/+ Veh/ Eprs+/+ ISO/ Eprs+/– Veh/ Eprs+/– ISO, respectively.
(B) WGA staining of Eprs+/+ and Eprs+/– mice with ISO treatment. Cross-sectional area (CSA) of CMs was measured and quantified. n=4 hearts per group with 200–300 CMs measured per heart. Scale bar: 50 μm.
(C) Picrosirius red staining indicates decreased fibrotic area in the hearts from Eprs+/– mice after ISO infusion. Scale bar: 2 mm. n=8/ 7/ 9/ 8 for Eprs+/+ Veh/ Eprs+/+ ISO/ Eprs+/– Veh/ Eprs+/– ISO, respectively.
(D) Single allele knockout of Eprs reduces HW/TL ratios in a TAC surgery-induced mouse HF model. n=7/ 6/ 11/ 7 for Eprs+/+ Sham/ Eprs+/+ TAC/ Eprs+/– Sham/ Eprs+/– TAC, respectively.
(E) WGA staining in Eprs+/+ and Eprs+/– mice with TAC surgery. n=4 hearts per group with 200–300 CMs measured per heart. Scale bar: 50 μm.
(F) Single allele knockout of Eprs reduces cardiac fibrosis in TAC surgery model. n=7/ 6/ 8/ 6 for quantification of picrosirius red staining for Eprs+/+ Sham/ Eprs+/+ TAC/ Eprs+/– Sham/ Eprs+/– TAC, respectively. Scale bar: 2 mm.
(G) Representative echocardiographic images suggest improved cardiac function in Eprs+/– mice compared to WT mice after TAC surgery.
(H) Fractional shortening (FS) and ejection fraction (EF) are partially recovered in Eprs+/– mice in TAC surgery-induced mouse HF model. n=14/ 6/ 11/ 7 for Eprs+/+ Sham/ Eprs+/+ TAC/ Eprs+/– Sham/ Eprs+/– TAC, respectively.
Comparisons were performed by non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction for A-F, H.
We then used TAC surgery, a pressure overload-induced cardiac hypertrophy model, to confirm the role of EPRS in HF. We found that Eprs+/– mice exhibited reduced cardiac hypertrophy (9.61±0.57 in Eprs+/+ to 7.31±0.30 in Eprs+/– of HW/TL ratio and 809.1±12.44 μm2 in Eprs+/+ to 555.2±9.12 μm2 in Eprs+/– of LV myocyte area) compared to WT mice after TAC surgery (Figure 2D and 2E). Cardiac fibrosis was reduced by ~40% (3.58±0.44% to 2.16±0.28%) in Eprs+/– mice compared to WT mice after TAC (Figure 2F). Moreover, echocardiography showed partially restored cardiac function in Eprs+/– mice compared to WT mice after TAC surgery (Figure 2G and 2H, Online Table II). To examine the anti-fibrotic effects of EPRS reduction in a more physiologically relevant model, we performed left anterior coronary artery ligation surgery to induce myocardial infarction in Eprs+/– versus WT mice. We found that cardiac fibrosis was significantly reduced from the apex (29.16±0.84% to 16.49±2.09%) to the base (8.24±0.73% to 1.94±0.26%) areas of the heart in Eprs+/– mice compared to WT mice after MI (Online Figure III). In summary, Eprs+/– mice exhibited reduced cardiac hypertrophy, fibrosis, and improved cardiac function in HF models under various stress conditions.
Postn-Cre-driven single allele knockout of Eprs attenuates cardiac fibrosis
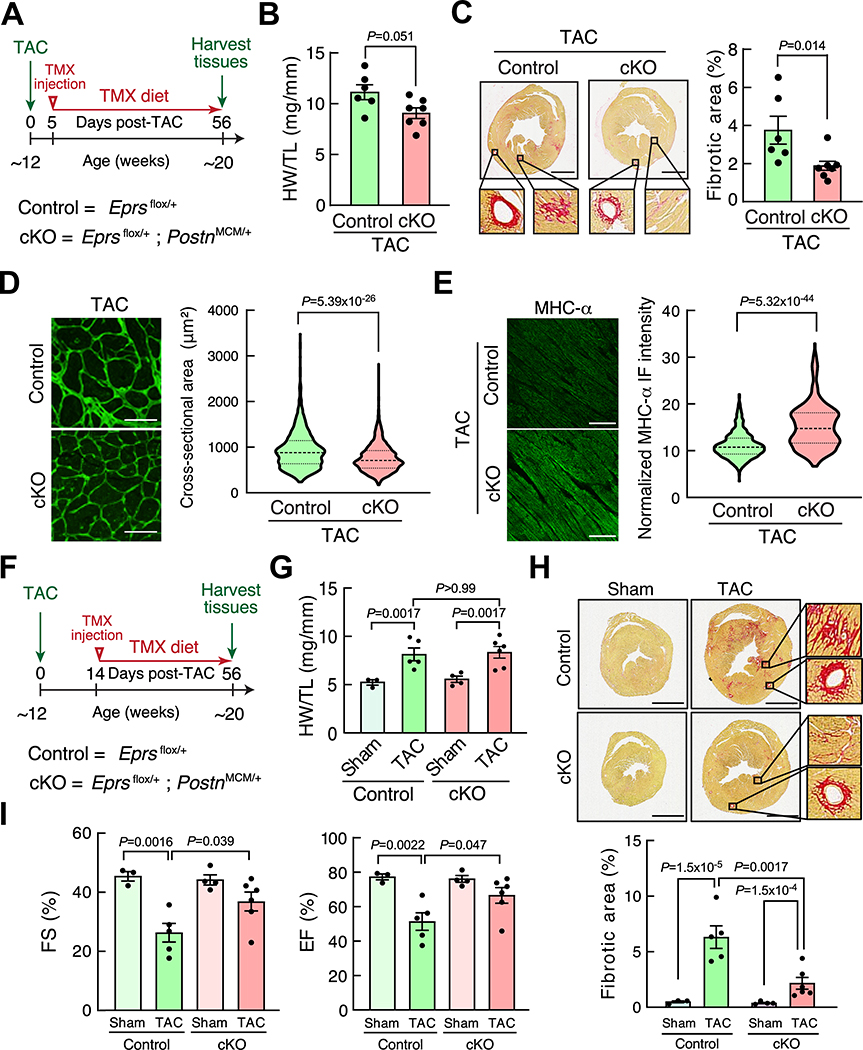
To test whether haploinsufficiency of EPRS in cardiac myofibroblasts reduces cardiac fibrosis, we obtained Eprs floxed mice from the International Mouse Phenotyping Consortium and bred with tamoxifen (TMX) inducible Postn-Cre mice (PostnMCM/+)21 to generate TMX-inducible Postn-Cre-driven single allele Eprs conditional knockout (cKO) mice (Online Figure IVA and IVB). We performed quantitative measurements of EPRS in cKO mice (Eprsflox/+; PostnMCM/+) and found that EPRS protein expression was reduced by ~45% in α-SMA positive CF cells under TAC surgical conditions (Online Figure IVC). The control (Eprsflox/+) and Eprs cKO (Eprsflox/+; PostnMCM/+) mice were subject to TAC surgery and fed with TMX food for two months (Figure 3A). We observed a reduced HW/TL ratio in Eprs cKO mice compared to control mice after TAC surgery (Figure 3B). More importantly, the fibrotic area was significantly reduced by ~49% in Eprs cKO mice (3.79±0.73% in control to 1.93±0.24% in cKO) compared to control mice as indicated by picrosirius red staining (Figure 3C). WGA staining for CMs and immunostaining for the sarcomeric protein, MHC-α, suggested reduced cardiac hypertrophy in Eprs cKO mice (Figure 3D and 3E), further confirming that the reduced hypertrophy phenotype is due to indirect effects from cellular communications between CMs and CFs. To examine the possible reversal effects of EPRS reduction after initiating cardiac fibrosis, we injected TMX in Eprs cKO mice 2 weeks post-TAC surgery followed by TMX food for the entire pathologic stimulus of TAC (Figure 3F). We found that Eprs genetic knockdown in Postn-positive CFs did not reverse cardiac hypertrophy (Figure 3G) but moderately reduced the established fibrosis (6.32±1.02% to 2.17±0.53%) (Figure 3H) and partially restored cardiac function (Figure 3I, Online Table III).
Figure 3. Eprs Postn-Cre-driven conditional knockout reduces cardiac hypertrophy and fibrosis in the TAC surgery model.
(A) Schematic of the experimental procedure for TAC surgery in tamoxifen (TMX)-induced Postn-Cre-driven Eprs conditional knockout (cKO, tm1d) and Eprsflox/+ mice (Control, tm1c) using a preventive model.
(B) HW/TL ratios suggest attenuated hypertrophy in Eprs cKO mice (n=7) after TAC surgery compared to Control mice (n=6).
(C) Picrosirius red staining indicates reduced cardiac fibrosis in Eprs cKO mice (n=7) compared to Control mice (n=6) after TAC surgery. Scale bar: 2 mm.
(D) WGA staining of heart tissue sections in Control and Eprs cKO mice with TAC surgery. n=4 hearts in Control group and n=5 in cKO group with >1000 CMs measured per heart. Scale bar: 50 μm.
(E) MHC-α protein expression in heart tissues of Control and Eprs cKO mice with TAC surgery. n=3 hearts per group with >400 CMs measured per heart. Scale bar: 50 μm.
(F) Schematic of the experimental procedure for TAC surgery in Eprs cKO and control mice using a reversal model.
(G) HW/TL ratios in Eprs cKO and Control mice after TAC surgery. n=3/ 5/ 4/ 6 for Control Sham/ Control TAC/ cKO Sham/ cKO TAC, respectively.
(H) Picrosirius red staining indicates reduced cardiac fibrosis in Eprs cKO mice compared to control mice after TAC surgery. n=3/ 5/ 4/ 6 for Control Sham/ Control TAC/ cKO Sham/ cKO TAC, respectively. Scale bar: 2 mm.
(I) Fractional shortening (FS) and ejection fraction (EF) are partially recovered in Eprs+/– mice in a TAC surgery-induced mouse HF model. n=3/ 5/ 4/ 6 for Control Sham/ Control TAC/ cKO Sham/ cKO TAC, respectively.
Comparisons were performed by non-parametric unpaired Mann-Whitney test for B-E, and non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction for G, H, I.
Taken together, these data suggest that global haploinsufficiency or Postn-Cre-driven single allele KO of Eprs attenuates cardiac remodeling under pathogenic stresses, and that sufficient expression of EPRS is essential for pathologic cardiac remodeling.
Sufficient EPRS is required for efficient translation of Pro-rich collagen proteins
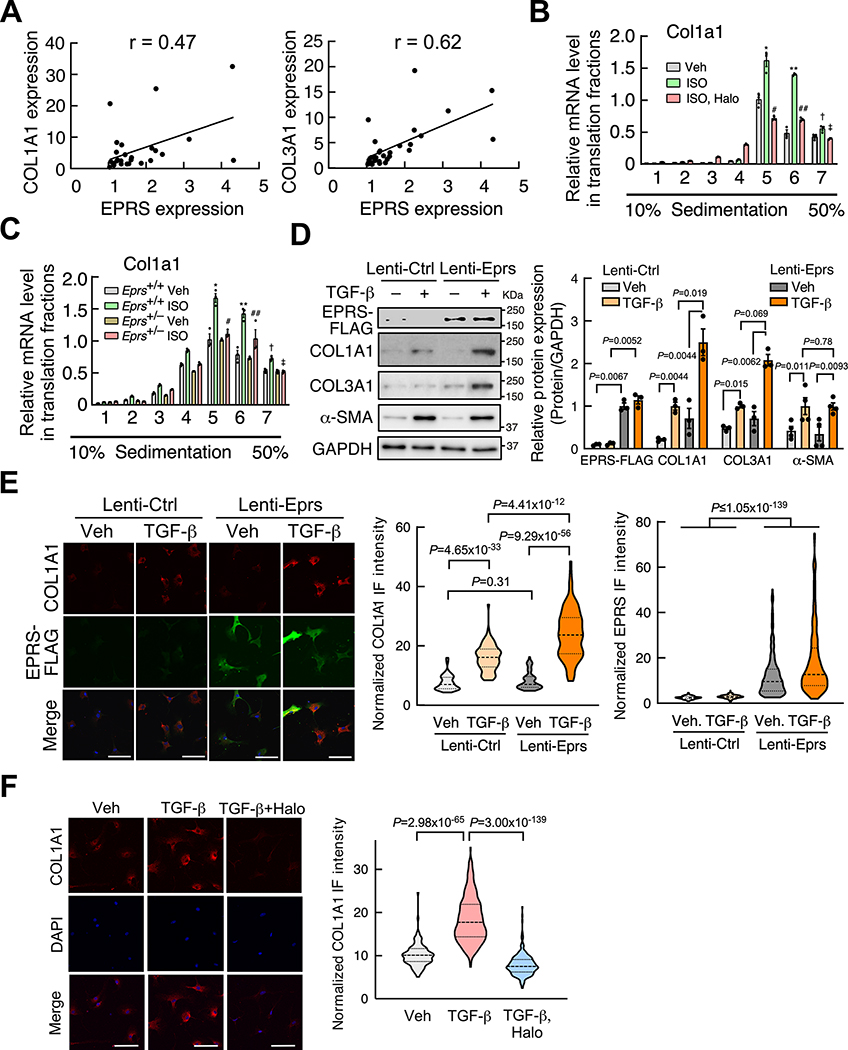
Pearson correlation analyses in human HF samples indicate that EPRS expression correlates with collagen expression (Pearson correlation coefficient r = 0.47, 0.62 for COL1A1 and COL3A1, respectively), but not ANF and BNP expression (Figure 4A and Online Figure VA). This observation suggests that EPRS may directly regulate the expression of collagens in CFs. In primary cultured adult mouse CFs, we found that pro-hypertrophic (ISO and Ang II), as well as pro-fibrotic agonists (TGF-β and IL-11), significantly increased Eprs expression by 1.3–2.7 fold (Online Figure VB). We confirmed that EPRS expression was increased by ISO at the mRNA and protein levels in mouse CFs due to increased transcription (Online Figure VB and VC). Expression of Pro-rich (PRR) collagen genes, including COL1A1 and COL3A1, was not markedly altered at the mRNA level (<1.5 folds) (Online Figure VD and VE). However, COL1A1 and COL3A1 protein levels were significantly induced by ISO (3–5 folds), suggesting preferential translational regulation of collagen genes by ISO (Online Figure VE). In contrast, ISO stimulus did not significantly alter the mRNA expression of Acta2 and Postn, while TGF-β remarkably increased their expression (Online Figure VD). Therefore, we first chose ISO treatment for examining the translational regulatory effects of EPRS in primary adult mouse CFs. The polysome profiling-RT-qPCR assay showed significantly increased association of Col1a1 and Col3a1 mRNAs with heavy polysomes after ISO treatment and reduced polysome association following Halo exposure, suggesting that ISO-enhanced translation efficiency of collagen transcripts is reversed by Halo (Figure 4B and Online Figure VF upper panel). Furthermore, the haploinsufficiency of EPRS resulted in a similar translational reduction of collagen mRNAs in the isolated mouse CFs (Figure 4C and Online Figure VF lower panel). To provide evidence for EPRS function in promoting pro-fibrotic PRR protein synthesis, we overexpressed EPRS using lentivirus infection in primary mouse CFs as a gain-of-function model. We found that the protein expression of Pro-rich COL1A1 and COL3A1 was significantly induced when CFs were stimulated by TGF-β compared to the cells infected with control lentivirus (Figure 4D, 4E, and Online Figure VG). To examine whether EPRS inhibition reduces collagen synthesis in activated myofibroblasts, we treated primary mouse CFs with vehicle versus TGF-β for 24 hrs to activate CF-to-myofibroblast conversion and then treated the cells with Halo and measured collagen expression in myofibroblasts using immunofluorescence. We observed that collagen protein expression was significantly reduced by Halo treatment (Figure 4F). These results indicate that a sufficient amount of EPRS and its activity are critical to maintenance of efficient translation of proline-rich collagens upon ISO stimulation in cardiac fibroblasts.
Figure 4. EPRS is required for efficient Pro-rich collagen translation in cardiac fibroblasts.
(A) EPRS expression is correlated with the expression of collagens in human heart samples (n=23). Pearson correlation coefficient was presented.
(B) Polysome-RT-qPCR assay indicates that EPRS inhibition by Halo reverses ISO-induced polysome association with COL1A1 mRNA in primary mouse CFs. * P=0.0013, ** P=3.0×10−8, † P=0.0013, Veh. vs. ISO; # P=7.2×10−6, ## P=9.2×10−5, ‡ P=1.4×10−4, ISO vs. ISO, Halo by non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction.
(C) Polysome-associated COL1A1 mRNA is reduced in primary Eprs+/– CFs compared to WT (Eprs+/+) CFs after ISO treatment. * P=7.7×10−4, ** P=6.3×10−6, † P=0.021, Eprs+/+ Veh. vs. Eprs+/+ ISO; # P=0.0083, ## P=0.0047, ‡ P=0.0019, Eprs+/+ ISO vs. Eprs+/– ISO by non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction.
(D) EPRS overexpression induces Pro-rich collagen protein expression but not non-Pro-rich α-SMA protein. Lentiviral EPRS and control lentivirus were used to infect primary mouse CFs with or without TGF-β (10 ng/ml) treatment. Left panel: representative images of immunoblot. Right panel: quantitative analysis of IB images from n=3–4 replicated experiments.
(E) EPRS overexpression induces COL1A1 protein expression in primary mouse CFs indicated by IF. Scale bar: 100 μM. n>120 cells for COL1A1 and n>150 cells for EPRS protein expression from 3 biologically replicated experiments.
(F) Halo inhibits COL1A1 protein expression in TGF-β-activated myofibroblasts. 10 ng/ml TGF-β was used to treat primary mouse CFs. Halo: 100 nM. Scale bar: 100 μm. n>200 cells from 3 biologically replicated experiments.
Comparisons were performed by non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction for D-F.
Global identification of novel Pro-rich proteins as preferential EPRS targets
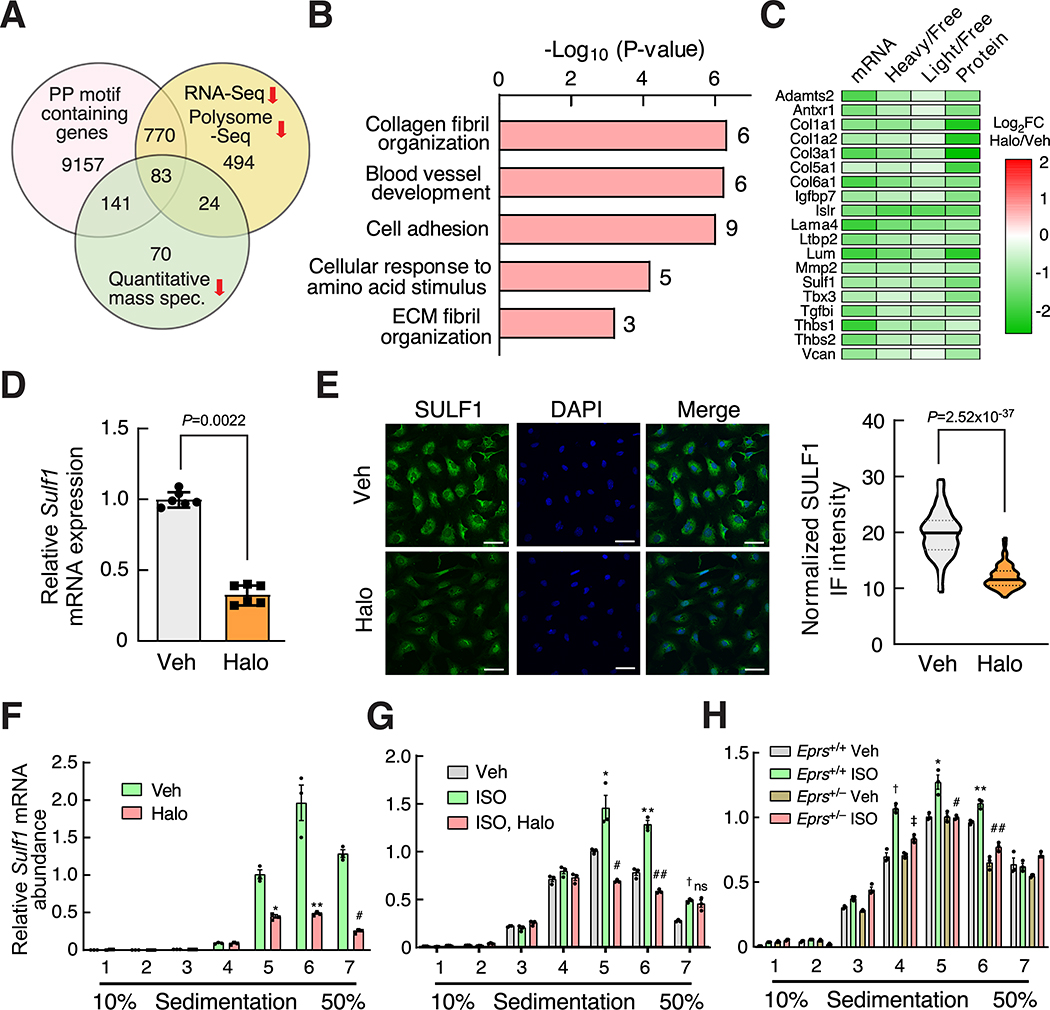
As we have shown that collagens, as exemplary PRR genes, are translationally regulated by EPRS, we sought to determine whether EPRS regulates preferential mRNA targets as opposed to all the transcripts in fibroblasts. To address this question, we chose NIH/3T3 mouse fibroblasts as a cell model system to perform high throughput screening based on high reproducibility and its ability to recapitulate gene regulatory events from primary CFs. We treated fibroblasts with vehicle or low-dose Halo (100 nM) and performed RNA-Seq (transcriptome profiling) and polysome profiling-Seq (translatome profiling). We identified novel PRR gene pathways which are preferentially regulated by EPRS via enhanced translation elongation at Pro-rich codons and antagonized by Halo (Figure 5A). We categorized differentially regulated genes into four groups (Area 1–4) based on their change at the steady-state RNA level and the ratio between a heavy (or light) polysome fraction and a non-polysome fraction (translation efficiency, TE) (Figure 5B and Online Figure VIA, Online Table IV and V).
Figure 5. Pro-rich genes are preferential translational targets of EPRS.
(A) Schematic for RNA-Seq and polysome profiling-Seq (polysome-Seq) procedure in Halo treated NIH/3T3 mouse fibroblasts. Polysome-free fractions include free mRNP, 40S ribosome subunit, 60S ribosome subunit. Light polysome fractions include 80S monosome, disome, trisome, and tetrasome. Heavy polysome fractions include polysome fractions with ≥5 ribosomes.
(B) Differentially expressed genes identified by RNA-Seq and polysome-Seq are indicated by dot plot. Translation efficiency (TE) is indicated by the ratio of heavy polysome and polysome-free fraction. The colored dots indicate statistically significantly changed genes identified by either RNA-Seq or polysome-Seq. The genes with Padj<0.05 in either of three different groups (ratio of Halo vs. Veh. treated samples for total RNA, non-polysome, and heavy polysome) were considered as significantly changed genes. All significantly changed genes were divided into four areas based on log2FC of total mRNAs and heavy polysome mRNAs. Data were submitted to GEO database (GSE136838).
(C) Translationally dysregulated genes are indicated by overlapping changed genes in the same area of heavy polysome (B) and light polysome (Online Figure VIA). Genes decreased (Area 1) or increased (Area 3) at translation and steady-state mRNA levels are shown.
(D) A majority of genes show a synergistic change at the mRNA and translational levels after EPRS inhibition by Halo. The number of genes is shown with changes at both translation efficiency (the ratio of heavy/light polysome to polysome-free fraction) and steady-state mRNA levels in all four areas.
(E) KEGG signaling pathway analyses indicate that ECM-receptor interaction and ribosome biogenesis are the top enriched pathways in Area 1 and Area 3, respectively. DAVID Bioinformatics Resources 6.8 was used.
(F) PP motif analyses suggest that there are more PP motif-bearing genes in the translationally decreased gene cluster (Area 1) compared to the translationally increased gene cluster (Area 3).
(G) Distribution of genes according to the number of PP motifs (0, 1, 2, 3, ≥4) indicate that the genes in Area 1 contain more PP motifs compared to genes in Area 3.
(H) Frequency of PP motif normalized by protein length or gene number is significantly higher in Area 1 compared to Area 3.
Comparisons were performed by Chi-Square test for gene number counts for F, G and non-parametric unpaired Mann-Whitney test for quantification of PP motif frequency for H.
Considering the active translation state in both light and heavy polysome fractions, we redefined the four groups by overlapping genes from heavy and light polysome-associated transcripts (Figure 5C). In the overlapped region of the Venn diagram, 1371 genes were downregulated at translation and steady-state mRNA levels (Area 1), and 775 genes were upregulated at both levels (Area 3). This finding supports the hypothesis that only a small cluster of genes were preferential translational targets of EPRS. Furthermore, most of the significantly changed genes were located in Area 1 and 3 (Figure 5D and Online Figure VIB). This observation indicates a coordinated regulatory effect between steady-state mRNA level and translation efficiency under EPRS inhibitory conditions. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the genes in Area 1 (reduced at RNA and TE levels) using DAVID software34 revealed multiple pro-fibrotic pathways, including ECM-receptor interaction (11 collagen genes such as Col1a1/1a2/3a1/4a6/5a1/6a1, four integrin genes Itga7/a8/b3/b4, etc.) (Online Figure VIC) and proteoglycans (Fgfr1, Pdgfra/rb, Tgfb2/b3, Tgfbr1/r2, Thbs1/3, etc.) (Figure 5E and Online Table VI). In contrast, the genes in Area 3 (induced at RNA and TE levels) are enriched in general translation factors, including ribosome biogenesis, ribosome (17 cytosolic and 6 mitochondrial ribosome proteins), aminoacyl-tRNA biosynthesis (16 ARSs including Eprs) (Online Figure VID), and amino acid biosynthesis (Figure 5E), which suggests activation of a compensatory amino acid starvation response upon EPRS inhibition12, 15.
Previous studies demonstrated that the formation of a peptide bond inside iterated proline or proline with other amino acids is much slower than that of the non-proline-bearing peptide bond, and thus Pro-Pro (PP) motifs function as ribosome pausing sites during translation elongation35, 36. This is an evolutionarily conserved mechanism from bacteria to humans and is also unique to the Pro codon among all genetic codons 37, 38. Therefore, we hypothesize that the translation of genes bearing more PP motifs is more sensitive to mild EPRS inhibition than that of non-PP-bearing genes. Consistent with this hypothesis, the Area 1 gene cluster contains a larger number of PP motif-bearing genes compared to Area 3 (Figure 5F, Online Table VII). The proportion of genes bearing more than four PP motifs in Area 1 is much higher than that in Area 3 (Figure 5G, Online Table VII). The overall frequency of PP motifs in Area 1 is also significantly higher than that in Area 3, as indicated by the number of PP motifs per 10,000 amino acids or per transcript (Figure 5H, Online Table VII). Major cardiac fibrosis marker genes in the Area 1 gene cluster, such as Col3a1 and Col1a1 (and other collagen genes), contain highly enriched Pro and Gly genetic codons due to abundant Pro-Pro-Gly (PPG) motifs (Supplemental sequence information) compared to overall codon composition in the mouse genome (Online Figure VIIA and VIIB). Polysome profiling followed by RT-qPCR confirmed that collagen transcripts were reduced in light polysome (fraction 5) and heavy polysome fractions (fraction 6 and 7), suggesting inhibition of EPRS by Halo causes ribosome stalling at PRR codons and decreased translation elongation of collagen mRNAs (Online Figure VIIC and VIID). In addition, gene regulation changed in the opposite direction at the mRNA and TE levels in Area 2 and 4 in which a small number of genes are located (Online Figure VIIE and VIIF, Online Table V). Taken together, these results suggest that PP motif-bearing genes are preferential targets of EPRS, and mild inhibition of EPRS by Halo leads to inefficient translation of these Pro codon rich genes.
SULF1 and LTBP2 are novel EPRS downstream effectors
To screen for specific novel downstream effectors of EPRS as potential target genes for anti-fibrotic drug treatment, we performed high throughput bioinformatic screening by integrating genomic, transcriptomic, translatomic, and proteomic analyses (Figure 6A). We first performed in silico bioinformatic analyses to uncover conserved PRR genes that bear PP genetic codon motifs across the reviewed human and mouse proteins from Uniprot proteomic sequences39 at the genome-wide scale (Online Table VIII), including PPG-rich ECM genes such as collagens among many Pro-rich proteins. Next, we overlapped PP motif bearing genes (Online Table VIII) with downregulated genes in our transcriptomic and translatomic analyses (Figure 5B) and proteins that were reduced at the steady-state level in human CFs upon low-dose Halo treatment in the published quantitative proteomic data15 (Online Figure VIIIA, Online Table IX). As expected, eight collagen genes (i.e., COL1A1/1A2/3A1/5A1/6A1/6A2/6A3/12A1) were translationally reduced in mouse fibroblasts, and their steady-state protein level was also decreased in human CFs after Halo treatment. Another six collagen genes (COL4A6/5A3/11A1/16A1/27A1/28A1) were reduced at the translation efficiency level in mouse fibroblasts, and five collagen genes (COL4A1/5A2/7A1/14A1/18A1) were reduced at the protein level in human CFs. Intriguingly, two recent new cardiac fibrosis marker genes cytoskeleton associated protein 4 (CKAP4)40 and interleukin 11 (IL-11)4 were found as PRR genes (Supplemental sequence information) downregulated in our polysome-Seq analysis (Online Table IV and VIII). We also observed that GTPBP2 (a ribosome rescue factor for dissembling stalled ribosomes)41 (Online Figure VIIIA) and EIF5A (an elongation factor)35, 36 are both induced by Halo treatment (Online Table V). The induction of GTPBP2 indicates enhanced recycling of stalled ribosomes during Halo exposure. EIF5A is required for ribosome readthrough of consecutive Pro codons in PRR genes, and thus its increase serves as another compensatory response upon Halo-driven inhibition of Pro codon decoding35, 36. These results suggest a previously unappreciated cellular adaptive response to promote the recycling of stalled ribosomes caused by EPRS inhibition and facilitates the readthrough of PP motifs. In contrast, most housekeeping genes are not decreased at translational efficiency or steady-state protein levels, including all histone genes, a majority of DNA polymerase complex genes, RNA polymerase complex genes, 38 ARSs, all heterogeneous nuclear ribonucleoproteins, GAPDH, vimentin, β-actin among many others (Online Table IV). These observations further support the idea of EPRS-mediated selective regulation of PRR mRNA translation.
Figure 6. SULF1 is a novel EPRS downstream target.
(A) Integrated genomic, transcriptomic, translatomic, and proteomic15 analyses identify 83 PP motif-containing genes as preferential downstream targets of EPRS.
(B) Gene Ontology (GO) analyses performed for 83 Halo-downregulated PRR genes from (A). Top five enriched pathways from the GO analysis (Biological Process) are shown. The number on the right of bars indicates the number of genes in each GO term. DAVID Bioinformatics Resources 6.8 was used.
(C) The log2FC change of mRNA, translation, and steady-state protein levels of all the genes from (B) are shown by heatmap. Steady-state protein level changes were obtained from a published database15.
(D-E) Sulf1 mRNA (D) and protein (E) expression is reduced by Halo-mediated EPRS inhibition in primary mouse CFs. 18S rRNA is used as a loading control. Scale bar: 50 μM. Comparisons were performed by non-parametric unpaired Mann-Whitney test for D, E.
(F) Polysome-associated Sulf1 mRNA is reduced in Halo treated primary mouse CFs. * P=2.6×10−4, ** P=3.1×10−6, # P=4.9×10−7, Halo vs. Veh. by non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction.
(G) EPRS inhibition reverses ISO-induced polysome association of Sulf1 mRNA in primary mouse CFs. * P=4.6×10−4, ** P=4.7×10−5, † P=7.9×10−4 Veh. vs. ISO; # P=6.0×10−8, ## P=1.3×10−8, ns: P=0.42 ISO vs. ISO, Halo by non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction.
(H) Polysome-associated Sulf1 mRNA is reduced in primary Eprs+/– CFs compared to WT (Eprs+/+) CFs after ISO treatment. † P=2.2×10−9, * P=6.6×10−4, ** P=9.8×10−5 Eprs+/+ Veh. vs. Eprs+/+ ISO; ‡ P=3.4×10−5, # P=4.4×10−4, ## P=1.9×10−7, Eprs+/+ ISO vs. Eprs+/– ISO by non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction.
By overlapping the three datasets, a group of 83 PP motif-containing genes was shown to be downregulated at translational and steady-state protein levels (Figure 6A, Online Table IX). Gene Ontology (GO) analyses revealed the top five pathways, including collagen fibril organization (COL1A1, COL1A2, COL3A1, COL5A1, ADAMTS2, LUM), cell adhesion (SULF1, TGFBI, VCAN, THBS1, THBS2, LAMA4, IGFBP7, LSLR), and extracellular fibril organization (COL3A1, COL5A1, LTBP2) among others, many of which are involved in cardiac pathologic remodeling (Figure 6B and Online Table X). Steady-state mRNA expression, translation efficiency, and steady-state protein expression are reduced for the major EPRS preferential target genes from the top five GO pathways (Figure 6C). Among these EPRS targets, LTBP2 (latent TGF-β-binding protein 2) has recently been discovered as an ISO-induced fibrosis marker protein42 and shown in our screen as downregulated at steady-state mRNA and polysome associated mRNA levels in Halo-treated mouse fibroblasts (Online Figure VIIIB). Mouse LTBP2 protein bears 17 PRR motifs (human LTBP2 has 20 PRR motifs) such as PPP and PPG (Supplemental sequence information). The polysome-associated Ltbp2 mRNA was significantly reduced by Halo treatment and Eprs haploinsufficiency in primary mouse CFs (Online Figure VIIIC–E).
We have a particular interest in SULF1, which is an endosulfatase that selectively removes 6-O-sulfate from heparin sulfate proteoglycans (HSPGs) and regulates signaling transduction of multiple growth factors43, 44; it could be considered as a pharmacologic target for anti-fibrosis drug screening based on its enzymatic activity. SULF1 contains PRR codons such as the PPG motif in both human and mouse (plus PPD and PPT motifs in the human gene and PPR motif in the mouse gene). Multi-omics data suggest that SULF1 was downregulated at mRNA, translation, and steady-state protein levels by Halo (Figure 6C). We confirmed that SULF1 mRNA and protein expression were downregulated by Halo in mouse CFs by RT-qPCR and IF (Figure 6D and 6E). Polysome profiling-RT-qPCR was performed to confirm downregulation of SULF1 at the translational level. Halo treatment reduced the polysome association of Sulf1 mRNA (Figure 6F) and attenuated ISO-induced Sulf1 mRNA translation (Figure 6G). Moreover, the translation efficiency of Sulf1 mRNA was dramatically reduced in isolated primary adult mouse CFs from Eprs+/– mice compared to WT mice under ISO treatment (Figure 6H). In summary, we validated LTBP2 and SULF1 as novel authentic EPRS-regulated, Halo-responsive Pro codon rich genes.
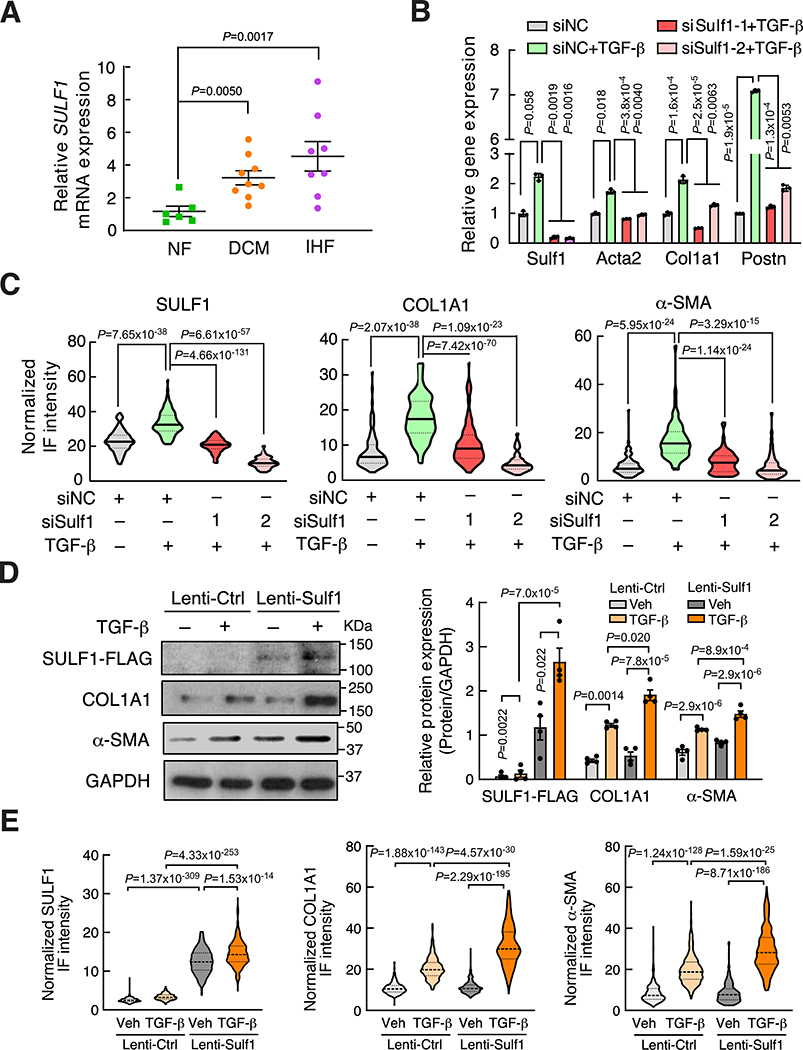
SULF1 is a myofibroblast marker and required for cardiac fibroblast activation
As we have shown that SULF1 is a preferential downstream target of EPRS, we sought to examine the expression of SULF1 during cardiac fibrosis and heart failure. Sulf1 mRNA was dominantly expressed in primary mouse CFs rather than CMs (Online Figure IXA). By screening a high throughput microarray-based gene expression database45 of human heart failure, we discovered that SULF1 mRNA expression was significantly induced in dilated cardiomyopathy (DCM) and ischemic heart failure (IHF) patients compared to non-failing donor hearts (Online Figure IXB). We confirmed this phenomenon using our validation cohort of human HF samples (Figure 7A). In our human heart samples, SULF1 expression is positively correlated with the expression of COL1A1 (Pearson correlation coefficient r = 0.60) and COL3A1 (Pearson correlation coefficient r = 0.67) (Online Figure IXC and IXD), indicating a potential functional correlation of SULF1 with CF activation. Indeed, SULF1 is highly expressed in α-SMA positive myofibroblasts in failing human hearts (Online Figure IXE–G) and in the heart of the TAC-induced mouse HF model (Online Figure IXH and IXI). These data indicate SULF1 as a myofibroblast marker gene during cardiac pathologic remodeling and heart failure.
Figure 7. SULF1 is required for cardiac fibroblast activation and collagen deposition.
(A) RT-qPCR confirms increased SULF1 expression in the hearts of DCM (n=9) and IHF (n=8) patients compared to non-failing donor heart tissues (n=6) in a validation cohort of human samples. 18S rRNA is used as a loading control for mRNA measurement.
(B) Knockdown of SULF1 by siRNA attenuates TGF-β-induced cardiac fibroblast activation as indicated by reduced expression of fibroblast activation marker genes in primary mouse CFs. 18S rRNA is used as a loading control.
(C) Knockdown of SULF1 reduces COL1A1 and α-SMA protein expression in TGF-β-treated primary mouse CFs indicated by quantification of normalized immunofluorescence (IF) intensity. n>120 cells from three biological replicates were analyzed.
(D) SULF1 overexpression induces myofibroblast marker protein expression. Sulf1-overexpressing and control lentiviruses were used to infect primary mouse CFs with or without TGF-β (10 ng/ml) treatment. Left panel: representative images of immunoblot. Right panel: quantitative analysis of IB images from n=4 replicated experiments.
(E) SULF1 overexpression induces COL1A1 and α-SMA protein expression in primary mouse CFs indicated by quantification of normalized immunofluorescence (IF) intensity. n>200 cells for COL1A1 and n>230 cells for α-SMA quantification from 3 biologically replicated experiments. Quantification of SULF1, COL1A1, and α-SMA protein expression is shown based on IF staining in Online Figure XC and XD.
Comparisons were performed by non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction for A-E.
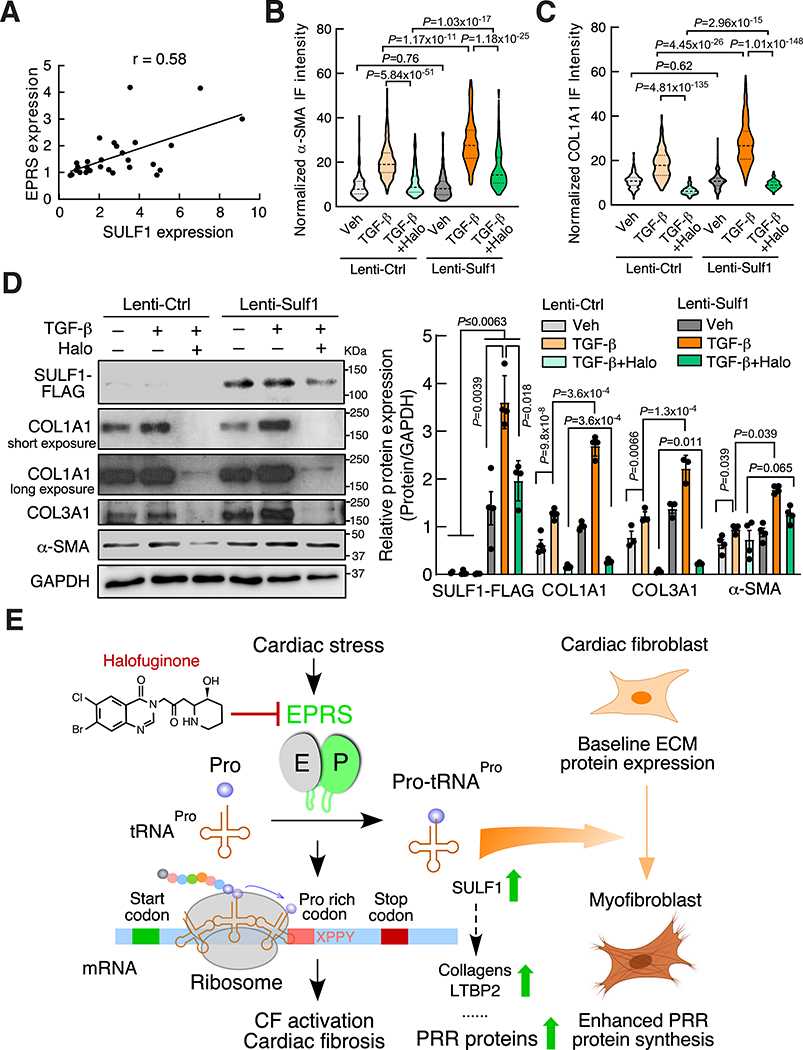
Given the dominant expression of SULF1 in CFs and the positive correlation between SULF1 and collagens, we next examined the effects of knockdown of SULF1 on myofibroblast activation. We transfected mouse CFs with two SULF1-specific siRNAs, and both siRNAs significantly reduced Sulf1 mRNA expression. TGF-β induced the expression of the myofibroblast activation marker gene while knockdown of Sulf1 dampened marker gene expression (Figure 7B). Immunofluorescence imaging results indicate that reduction of Sulf1 remarkably inhibited TGF-β induced CF activation and collagen deposition (Figure 7C, Online Figure XA and XB). We overexpressed SULF1 in primary mouse CFs under vehicle and TGF-β treatment and measured expression of the myofibroblast activation marker proteins. We found that SULF1 overexpression induced α-SMA and collagen expression at the protein levels (Figure 7D, 7E, Online Figure XC and XD). In our human heart samples, SULF1 expression is positively correlated (Pearson correlation coefficient r = 0.58) with the expression of EPRS (Figure 8A), providing a piece of evidence that SULF1 acts as a downstream effector of EPRS. We overexpressed SULF1 in primary mouse CFs followed by treatment with Halo and found that SULF1 partially rescued expression of α-SMA and collagens (Figure 8B–D), suggesting that combined influence of SULF1 and other EPRS downstream effector genes contribute to cardiac fibroblast-to-myofibroblast activation. Taken together, our results suggest SULF1 is required for cardiac fibroblast activation and can be used as a myofibroblast marker during cardiac fibrosis and heart failure.
Figure 8. SULF1 overexpression partially antagonizes the anti-fibrotic effect of Halo-driven EPRS inhibition.
(A) SULF1 expression is correlated with EPRS in the validation cohort of human subjects. Pearson correlation coefficient was presented.
(B-C) SULF1 overexpression induces α-SMA and COL1A1 protein expression and rescues their expression from Halo suppression in primary mouse CFs indicated by IF. n>150 cells from 3 biologically replicated experiments were used for quantification.
(D) SULF1 overexpression partially rescues TGF-β-activated myofibroblast marker protein expression from Halo suppression in primary mouse CFs. Left panel: representative images of immunoblot. Right panel: quantitative analysis of IB images from n=3–4 replicated experiments.
(E) Schematic model of EPRS-mediated translational activation of PRR proteins during cardiac fibrosis. XPPY: PP represents ProPro; X and Y represent any amino acid.
Comparisons were performed by non-parametric Kruskal-Wallis test with Conover-Iman method for post hoc pairwise comparisons and Benjamini-Hochberg correction for B-D.
DISCUSSION
In these studies, we investigated translational control mechanisms of pro-fibrotic gene expression in cardiac fibrosis (Figure 8E). We found that alteration in EPRS expression in CFs is a conserved aspect of pathologic cardiac remodeling in human HF patients as well as in HF mouse models. Using both global and Postn-Cre-driven single allele Eprs conditional knockout mouse models, we demonstrated that reducing EPRS antagonizes pathologic cardiac fibrosis in vivo. Mechanistically, we identified a subset of novel preferential translational target genes of EPRS and discovered that EPRS regulates translation of PRR transcripts, which construct most ECM and secretory signaling molecules. Among those targets, we identified several novel PRR genes that were not previously well studied in the heart (e.g., LTBP2, SULF1). These genes may serve as novel anti-fibrotic targets for development of drug therapies to treat HF. This study demonstrates the in vivo function of EPRS in cardiac remodeling and provides a strong rationale for using translation factors (e.g., EPRS) or their downstream effectors (e.g., SULF1) as novel therapeutic targets to treat cardiac fibrosis.
We demonstrate that genetic knockout of one allele of the Eprs gene can effectively reduce cardiac remodeling and fibrosis (Figure 2). Postn-Cre-driven conditional knockout of Eprs further confirms that fibroblast-derived EPRS contributes significantly to cardiac fibrosis under cardiac stress conditions (Figure 3). These genetic manipulations mimic mild translational inhibition using the (E)PRS-specific inhibitor Halo in multiple heart failure disease mouse models15. This new therapeutic approach targeting an evolutionarily conserved, ubiquitous housekeeping translation factor can be used to treat cardiac fibrosis of multiple etiologies and is generalizable to antagonize fibrosis in various organs46. Recent studies have shown that EPRS directly forms a complex with TGF-β1 receptor, Janus kinases, and STAT6 and regulates ECM expression via activation of TGF-β1 signaling in lung and liver17, 18. This suggests that EPRS may synergistically regulate the expression of Pro-rich ECM at both transcriptional and translational levels during cardiac fibrosis.
Using the (E)PRS inhibitor Halo, we discovered that the EPRS-PRR axis acts as a novel translational control pathway to regulate pro-fibrotic Pro-rich mRNA translation and cardiac fibrosis (Figure 8E). Pro is unique among the 20 amino acids because it forms a cyclic structure between the α-amine group and the side chain. This specific structure dramatically slows ribosome decoding of two consecutive Pro codons37. This unique feature of Pro probably explains why Halo specifically inhibits PRR mRNA translation, while preserving global mRNA translation by inducing expression of housekeeping genes such as ARSs and ribosome proteins (Figure 5E). Myc, a positive transcriptional regulator of ribosome biogenesis47, is also upregulated by Halo (Figure 5B, Online Table V). This may partially explain the increased expression of ribosomal genes. In evolution, Pro-rich protein genes are largely introduced in the genome of multicellular organisms in comparison to single cellular species38. EF-P (Elongation factor P in prokaryotes)/eIF5A (Elongation factor 5 in eukaryotes) protein is introduced to facilitate efficient peptide bond formation for Pro dipeptidyl motifs35, 36, 38; its upregulation by Halo treatment represents an adaptive response to facilitate ribosome readthrough of PP motifs. The majority of Pro-rich proteins are transmembrane or secretory proteins since the Pro-rich peptides usually play important roles in penetrating the plasma membrane for transmembrane localization or secretion48, 49. Many of these proteins are either ECM or ligands/receptors for cellular signaling, which promote cardiac fibrosis. siRNA-mediated knockdown of cysteinyl-tRNA synthetase (CysRS) by ~50% in mouse fibroblast cells did not reduce collagen protein expression (Online Figure XIA). Compared to Halo, Leucinol (leucyl-tRNA synthetase inhibitor) did not inhibit collagen protein expression at a range of low to high doses (Online Figure XIB). These data further support the specific action of (E)PRS inhibition in reducing collagen expression and fibrogenesis. In another aspect, Postn (containing no PRR motif) was significantly downregulated at the polysome-associated mRNA level (Online Table V). We also measured POSTN protein expression by IB and IF in TGF-β-activated WT and Eprs+/– CFs treated with vehicle or Halo. We found that POSTN protein expression was slightly reduced in Eprs+/– CFs and dramatically inhibited by Halo treatment (Online Figure XIIA–C). These results suggest that POSTN protein expression may be affected by Halo through downregulation of one or more putative Pro-rich regulatory proteins indirectly. We cannot exclude the possibility of some Pro-rich proteins acting as cardioprotective factors, and thus beneficial effects from EPRS inhibition may be from the combined net outcome. In addition to EPRS canonical aminoacylation function, previous studies have shown that EPRS exerts a noncanonical effect in translational silencing of inflammation-related mRNAs such as VEGFA in monocyte/Mφ as an innate immune response8, 50–53. It remains unknown as to whether EPRS from immune cells plays a role in regulating the inflammatory response via either the canonical or noncanonical mechanisms.
During the PRR gene screen, we identified 83 PRR genes in addition to collagens (Online Table IX and X), which are downregulated by Halo at the posttranscriptional level and may play critical roles in cardiac fibrosis. Induced EPRS can increase Pro-tRNAPro levels, a building block for the synthesis of Pro-rich proteins (i.e., collagens, LTBP2, etc.). We also observed that translationally reduced mRNAs by Halo tend to have reduced steady-state mRNA levels (Figure 5D and Online Figure VIB). Previous studies have shown that optimal genetic codon composition and translation capacity of mRNAs in human cells significantly influence baseline mRNA stability54, 55. We assume that translational repression of PRR mRNA is coupled with enhanced mRNA decay due to reduced translation elongation rates at PP motifs and diminished ribosome-mediated protection of the transcripts during EPRS inhibition. Based on our data, we conclude that cardiac stress-induced EPRS promotes cardiac fibrosis via increased Pro-tRNAPro and enhanced stabilization and translation of PRR pro-fibrotic mRNAs in cardiac fibroblasts.
Among EPRS target PRR genes, we discovered that SULF1 could serve as a biomarker for cardiac fibrosis. Genetic knockdown of SULF1 by siRNA inhibits myofibroblast activation stimulated by TGF-β. SULF1 and SULF2 are two redundant homolog enzymes of heparan sulfate 6-O-endosulfatases56. SULF1 catalyzes the removal of 6-O-sulfate from heparan sulfate chains and modulates the function of multiple growth factors44, 57, 58. Genetic knockout of Sulf1 in mouse genome did not cause any developmental, behavioral, histological, and aging-related phenotypes, but knockout of Sulf2 led to reduced body weight56, 57. A previous study reported that SULF1 was induced by TGF-β in both cultured human lung fibroblasts in vitro and murine lungs in vivo. Further, siRNA-mediated knockdown of SULF1 promotes α-SMA expression and TGF-β signaling59, which is inconsistent with our results here. This discrepancy may suggest that the function of SULF1 in cardiac fibroblasts is different from that of lung fibroblasts. A recent report showed that SULF1 is predominately expressed in endothelial cells and cardiac fibroblasts in the murine heart, and that overexpressed SULF1 induces a pro-fibrotic gene expression signature in mouse embryonic fibroblasts60, which supports our findings to some extent. However, global knockout of SULF1 attenuates angiogenesis and cardiac repair after ischemic injury without affecting interstitial fibrosis in non-infarcted regions60. Notably, SULF2, most highly expressed in monocytes and macrophages, could be secreted by these myeloid cells and compensate for the loss of SULF1 function in vivo. In our study, we showed the necessity for fibroblast-derived SULF1 in myofibroblast activation in primary CF culture. Therefore, it is worthwhile to further elucidate the pathophysiologic function of endothelial cell and cardiac fibroblast-derived SULF1 in cell type-specific KO mouse models in vivo. These studies will provide insight into whether cell type-specific loss of SULF1 is beneficial or detrimental during cardiac remodeling, especially in non-ischemic heart disease, in which there is less activation of the inflammation and immune response, and lower expression of SULF2. SULF1 has been reported to attenuate intracellular signaling downstream of multiple heparan sulfate-binding growth factors, including VEGFA, FGF2, HB-EGF, and HGF61. It remains a question of how the separate and combined sulfate modifications and downstream responses (e.g., binding affinity with cardiac fibroblast surface heparan sulfate and interactions with their cognate receptors) of these growth factors are changed upon knockdown or inhibition of SULF1. On the other hand, overexpression of SULF1 partially antagonizes the inhibitory effects from Halo treatment in CFs, indicating other downstream targets (e.g., collagens, LTBP2, etc.) of EPRS may contribute to the pro-fibrotic process. Our studies suggest that a better understanding of the mechanisms of translational control in cardiac fibrosis has broad implications for developing novel treatment strategies to combat CVD by either targeting a specific translation factor or its downstream effectors.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
TGF-β and IL-11 increase the synthesis of pro-fibrotic proteins during cardiac fibrosis.
Many pro-fibrotic genes contain Pro genetic codon rich motifs such as collagens.
Glutamyl-prolyl-tRNA synthetase (EPRS) is an essential housekeeping enzyme required for ligating Pro to tRNAPro for the synthesis of Pro-containing proteins.
Halofuginone is an (E)PRS-specific enzymatic inhibitor that is cardioprotective in multiple heart failure mouse models.
What New Information Does This Article Contribute?
This study reveals translational control mechanisms of pro-fibrotic gene expression in cardiac fibrosis.
EPRS mRNA and protein expression are induced in failing human hearts and mouse hearts undergoing pathologic cardiac remodeling.
Demonstration of the in vivo function of EPRS in cardiac remodeling — Eprs+/– mice and Postn-Cre-driven tamoxifen-inducible Eprs conditional knockout mice show reduced pathologic cardiac fibrosis under stress, suggesting that mildly reducing EPRS expression is cardioprotective.
Identification of novel preferential translational target genes of EPRS — We observed that EPRS regulates translation of Pro-rich (PRR) transcripts, which comprises most extracellular matrix and secretory signaling molecules.
We validated one of these targets, Sulfatase 1 (SULF1) as a myofibroblast marker protein in human and mouse heart failure and a potential target gene for anti-fibrosis treatment.
In cardiac fibroblasts, the synthesis of pro-fibrotic proteins is upregulated by cardiac stressors to activate extracellular matrix deposition and impair cardiac function. In this study, we discovered an EPRS-PRR gene axis that influences translational homeostasis of pro-fibrotic proteins and promotes pathologic cardiac remodeling and fibrosis. We identified EPRS as a common node downstream of multiple cardiac stressors and as a novel regulatory factor which facilitates pro-fibrotic mRNA translation in cardiac fibrosis. Global and Postn-Cre-driven genetic ablation of EPRS can effectively reduce cardiac fibrosis. This study reveals a novel translational control mechanism that modulates cardiac fibrosis and heart function. Mild inhibition of PRR mRNA translation may represent a general therapeutic strategy for the treatment of heart disease. These findings provide novel insights into translational control mechanisms of cardiac fibrosis and will promote the development of novel therapeutics using inhibition of pro-fibrotic translation factors or their downstream effectors.
Acknowledgments
We are grateful to Omar Hedaya, Qiuqing Wang (Biostatistician from Cleveland Clinic), and Xueya Cai (Research Associate Professor of Biostatistics and Computational Biology at the University of Rochester School of Medicine and Dentistry) for critical reading of the manuscript and biostatistical consulting, respectively. We thank for Drs. Chen Yan, Craig Morrell, and Bradford Berk for their helpful discussions. We appreciate the technical assistance from Orazio Slivano and Erika Flores Medina (Aab CVRI), and Mohan Amy (Aab CVRI) for histology and animal surgical operations, respectively. NIH/3T3 cells were provided by Dr. Eric Small from Aab CVRI at URMC. PostnMerCreMer mice were transferred from Eric Small’s lab and originally generated by Dr. Jeffery Molkentin at Cincinnati Children’s Hospital. Jordan Rhen contributed to mouse strain breeding and maintenance. Rommel Morales provided efforts for detection of SULF1 protein level in human blood samples. The Eprs+/– mouse line was produced by Dr. Lin Gan from the Mouse Genome Editing Core at URMC. None of the authors have any financial conflict of interest related to the research described in this manuscript.
SOURCES OF FUNDING
This work was supported in part by National Institutes of Health grants R01 HL132899 and R01 HL147954 (to P.Y.), University of Rochester CTSA award number UL1 TR002001 from the National Center for Advancing Translational Sciences of the National Institutes of Health (content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health), start-up funds from Aab Cardiovascular Research Institute of University of Rochester Medical Center (to P.Y.), and American Heart Association Postdoctoral Fellowship 19POST34400013 (to J.W.).
Nonstandard Abbreviations and Acronyms
- ARS
aminoacyl-tRNA synthetase
- CF
cardiac fibroblast
- CM
cardiomyocyte
- CVD
cardiovascular disease
- ECM
extracellular matrix
- EPRS
glutamyl-prolyl-tRNA synthetase
- Halo
halofuginone
- HF
heart failure
- ISO
isoproterenol
- Pro-tRNAPro
prolyl tRNAPro, proline ligated to tRNAPro
- PRR
proline-rich (XPPY codon motifs)
- TAC
transverse aortic constriction
- TMX
tamoxifen
Footnotes
DISCLOSURES
None.
REFERENCES
- 1.Davis J and Molkentin JD. Myofibroblasts: trust your heart and let fate decide. J Mol Cell Cardiol. 2014;70:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Travers JG, Kamal FA, Robbins J, Yutzey KE and Blaxall BC. Cardiac Fibrosis: The Fibroblast Awakens. Circ Res. 2016;118:1021–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Beyer A and Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165:535–50. [DOI] [PubMed] [Google Scholar]
- 4.Schafer S, Viswanathan S, Widjaja AA, Lim WW, Moreno-Moral A, DeLaughter DM, Ng B, Patone G, Chow K, Khin E, Tan J, Chothani SP, Ye L, Rackham OJL, Ko NSJ, Sahib NE, Pua CJ, Zhen NTG, Xie C, Wang M, Maatz H, Lim S, Saar K, Blachut S, Petretto E, Schmidt S, Putoczki T, Guimaraes-Camboa N, Wakimoto H, van Heesch S, Sigmundsson K, Lim SL, Soon JL, Chao VTT, Chua YL, Tan TE, Evans SM, Loh YJ, Jamal MH, Ong KK, Chua KC, Ong BH, Chakaramakkil MJ, Seidman JG, Seidman CE, Hubner N, Sin KYK and Cook SA. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 2017;552:110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chothani S, Schafer S, Adami E, Viswanathan S, Widjaja AA, Langley SR, Tan J, Wang M, Quaife NM, Pua CJ, D’Agostino G, Shekeran SG, George BL, Lim S, Cao EY, van Heesch S, Witte F, Felkin LE, Christodoulou EG, Dong J, Blachut S, Patone G, Barton PJR, Hubner N, Cook SA and Rackham OJL. Widespread Translational Control of Fibrosis in the Human Heart by RNA-Binding Proteins. Circulation. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann CL, Kanda M, Worth CL, Schafer S, Calviello L, Merriott R, Patone G, Hummel O, Wyler E, Obermayer B, Mucke MB, Lindberg EL, Trnka F, Memczak S, Schilling M, Felkin LE, Barton PJR, Quaife NM, Vanezis K, Diecke S, Mukai M, Mah N, Oh SJ, Kurtz A, Schramm C, Schwinge D, Sebode M, Harakalova M, Asselbergs FW, Vink A, de Weger RA, Viswanathan S, Widjaja AA, Gartner-Rommel A, Milting H, Dos Remedios C, Knosalla C, Mertins P, Landthaler M, Vingron M, Linke WA, Seidman JG, Seidman CE, Rajewsky N, Ohler U, Cook SA and Hubner N. The Translational Landscape of the Human Heart. Cell. 2019;178:242–260 e29. [DOI] [PubMed] [Google Scholar]
- 7.Kim S and Coulombe PA. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat Rev Mol Cell Biol. 2010;11:75–81. [DOI] [PubMed] [Google Scholar]
- 8.Yao P and Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med. 2013;5:332–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao P, Poruri K, Martinis SA and Fox PL. Non-catalytic Regulation of Gene Expression by Aminoacyl-tRNA Synthetases. Top Curr Chem. 2013. [DOI] [PubMed] [Google Scholar]
- 10.Arif A, Yao P, Terenzi F, Jia J, Ray PS and Fox PL. The GAIT translational control system. Wiley Interdiscip Rev RNA. 2018;9:e1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendes MI, Gutierrez Salazar M, Guerrero K, Thiffault I, Salomons GS, Gauquelin L, Tran LT, Forget D, Gauthier MS, Waisfisz Q, Smith DEC, Simons C, van der Knaap MS, Marquardt I, Lemes A, Mierzewska H, Weschke B, Koehler W, Coulombe B, Wolf NI and Bernard G. Bi-allelic Mutations in EPRS, Encoding the Glutamyl-Prolyl-Aminoacyl-tRNA Synthetase, Cause a Hypomyelinating Leukodystrophy. Am J Hum Genet. 2018;102:676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller TL, Zocco D, Sundrud MS, Hendrick M, Edenius M, Yum J, Kim YJ, Lee HK, Cortese JF, Wirth DF, Dignam JD, Rao A, Yeo CY, Mazitschek R and Whitman M. Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat Chem Biol. 2012;8:311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Sun L, Yang XL and Schimmel P. ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase. Nature. 2013;494:121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon NH, Fox PL and Kim S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat Rev Drug Discov. 2019. [DOI] [PubMed] [Google Scholar]
- 15.Qin P, Arabacilar P, Bernard RE, Bao W, Olzinski AR, Guo Y, Lal H, Eisennagel SH, Platchek MC, Xie W, Del Rosario J, Nayal M, Lu Q, Roethke T, Schnackenberg CG, Wright F, Quaile MP, Halsey WS, Hughes AM, Sathe GM, Livi GP, Kirkpatrick RB, Qu XA, Rajpal DK, Faelth Savitski M, Bantscheff M, Joberty G, Bergamini G, Force TL, Gatto GJ Jr., Hu E and Willette RN. Activation of the Amino Acid Response Pathway Blunts the Effects of Cardiac Stress. J Am Heart Assoc. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundrud MS, Koralov SB, Feuerer M, Calado DP, Kozhaya AE, Rhule-Smith A, Lefebvre RE, Unutmaz D, Mazitschek R, Waldner H, Whitman M, Keller T and Rao A. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 2009;324:1334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song DG, Kim D, Jung JW, Nam SH, Kim JE, Kim HJ, Kim JH, Pan CH, Kim S and Lee JW. Glutamyl-Prolyl-tRNA Synthetase Regulates Epithelial Expression of Mesenchymal Markers and Extracellular Matrix Proteins: Implications for Idiopathic Pulmonary Fibrosis. Front Pharmacol. 2018;9:1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song DG, Kim D, Jung JW, Nam SH, Kim JE, Kim HJ, Kim JH, Lee SJ, Pan CH, Kim S and Lee JW. Glutamyl-prolyl-tRNA synthetase induces fibrotic extracellular matrix via both transcriptional and translational mechanisms. FASEB J. 2019;33:4341–4354. [DOI] [PubMed] [Google Scholar]
- 19.Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, Jovanovic M, Ellis SR, Fulco CP, Engreitz JM, Schutz S, Lian J, Gripp KW, Weinberg OK, Pinkus GS, Gehrke L, Regev A, Lander ES, Gazda HT, Lee WY, Panse VG, Carr SA and Sankaran VG. Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell. 2018;173:90–103 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF and Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, SC JL, Aronow BJ, Tallquist MD and Molkentin JD. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun. 2016;7:12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao P, Wu J, Lindner D and Fox PL. Interplay between miR-574–3p and hnRNP L regulates VEGFA mRNA translation and tumorigenesis. Nucleic Acids Res. 2017;45:7950–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M and Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y, Smyth GK and Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 26.Love MI, Huber W and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: URL https://www.R-project.org/ 2016. [Google Scholar]
- 28.Pohlert T The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). R package. http://CRAN.R-project.org/package=PMCMR 2014.
- 29.Teekakirikul P, Eminaga S, Toka O, Alcalai R, Wang L, Wakimoto H, Nayor M, Konno T, Gorham JM, Wolf CM, Kim JB, Schmitt JP, Molkentin JD, Norris RA, Tager AM, Hoffman SR, Markwald RR, Seidman CE and Seidman JG. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-beta. J Clin Invest. 2010;120:3520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Da M, Feng Y, Xu J, Hu Y, Lin Y, Ni B, Qian B, Hu Z and Mo X. Association of aminoacyl-tRNA synthetases gene polymorphisms with the risk of congenital heart disease in the Chinese Han population. PLoS One. 2014;9:e110072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rau CD, Romay MC, Tuteryan M, Wang JJ, Santolini M, Ren S, Karma A, Weiss JN, Wang Y and Lusis AJ. Systems Genetics Approach Identifies Gene Pathways and Adamts2 as Drivers of Isoproterenol-Induced Cardiac Hypertrophy and Cardiomyopathy in Mice. Cell Syst. 2017;4:121–128 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galindo CL, Skinner MA, Errami M, Olson LD, Watson DA, Li J, McCormick JF, McIver LJ, Kumar NM, Pham TQ and Garner HR. Transcriptional profile of isoproterenol-induced cardiomyopathy and comparison to exercise-induced cardiac hypertrophy and human cardiac failure. BMC Physiol. 2009;9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MM, Dasari S, Konopka AR, Johnson ML, Manjunatha S, Esponda RR, Carter RE, Lanza IR and Nair KS. Enhanced Protein Translation Underlies Improved Metabolic and Physical Adaptations to Different Exercise Training Modes in Young and Old Humans. Cell Metab. 2017;25:581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT and Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 35.Schuller AP, Wu CC, Dever TE, Buskirk AR and Green R. eIF5A Functions Globally in Translation Elongation and Termination. Mol Cell. 2017;66:194–205 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H and Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–8. [DOI] [PubMed] [Google Scholar]
- 37.Buskirk AR and Green R. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2017;372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandal A, Mandal S and Park MH. Genome-wide analyses and functional classification of proline repeat-rich proteins: potential role of eIF5A in eukaryotic evolution. PLoS One. 2014;9:e111800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.UniProt C UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A and van Rooij E. Single-Cell Sequencing of the Healthy and Diseased Heart Reveals Cytoskeleton-Associated Protein 4 as a New Modulator of Fibroblasts Activation. Circulation. 2018;138:166–180. [DOI] [PubMed] [Google Scholar]
- 41.Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH and Ackerman SL. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park S, Ranjbarvaziri S, Lay FD, Zhao P, Miller MJ, Dhaliwal JS, Huertas-Vazquez A, Wu X, Qiao R, Soffer JM, Rau C, Wang Y, Mikkola HKA, Lusis AJ and Ardehali R. Genetic Regulation of Fibroblast Activation and Proliferation in Cardiac Fibrosis. Circulation. 2018;138:1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sala-Newby GB, George SJ, Bond M, Dhoot GK and Newby AC. Regulation of vascular smooth muscle cell proliferation, migration and death by heparan sulfate 6-O-endosulfatase1. FEBS Lett. 2005;579:6493–8. [DOI] [PubMed] [Google Scholar]
- 44.Gorsi B, Liu F, Ma X, Chico TJ, v A, Kramer KL, Bridges E, Monteiro R, Harris AL, Patient R and Stringer SE. The heparan sulfate editing enzyme Sulf1 plays a novel role in zebrafish VegfA mediated arterial venous identity. Angiogenesis. 2014;17:77–91. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Morley M, Brandimarto J, Hannenhalli S, Hu Y, Ashley EA, Tang WH, Moravec CS, Margulies KB, Cappola TP, Li M and consortium MA. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105:83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rockey DC, Bell PD and Hill JA. Fibrosis--a common pathway to organ injury and failure. N Engl J Med. 2015;372:1138–49. [DOI] [PubMed] [Google Scholar]
- 47.van Riggelen J, Yetil A and Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–9. [DOI] [PubMed] [Google Scholar]
- 48.Qi F, Motz M, Jung K, Lassak J and Frishman D. Evolutionary analysis of polyproline motifs in Escherichia coli reveals their regulatory role in translation. PLoS Comput Biol. 2018;14:e1005987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guidotti G, Brambilla L and Rossi D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol Sci. 2017;38:406–424. [DOI] [PubMed] [Google Scholar]
- 50.Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM and Fox PL. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell. 2012;149:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M and Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao P, Potdar AA, Ray PS, Eswarappa SM, Flagg AC, Willard B and Fox PL. The HILDA complex coordinates a conditional switch in the 3’-untranslated region of the VEGFA mRNA. PLoS Biol. 2013;11:e1001635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venkata Subbaiah KC, Wu J, Potdar A and Yao P. hnRNP L-mediated RNA switches function as a hypoxia-induced translational regulon. Biochem Biophys Res Commun. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, Hand JM, Wright M and Bazzini AA. Translation affects mRNA stability in a codon-dependent manner in human cells. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narula A, Ellis J, Taliaferro JM and Rissland OS. Coding regions affect mRNA stability in human cells. RNA. 2019;25:1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holst CR, Bou-Reslan H, Gore BB, Wong K, Grant D, Chalasani S, Carano RA, Frantz GD, Tessier-Lavigne M, Bolon B, French DM and Ashkenazi A. Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS One. 2007;2:e575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA and Emerson CP Jr. SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development. 2007;134:3327–38. [DOI] [PubMed] [Google Scholar]
- 58.Ai X, Do AT, Kusche-Gullberg M, Lindahl U, Lu K and Emerson CP Jr. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J Biol Chem. 2006;281:4969–76. [DOI] [PubMed] [Google Scholar]
- 59.Yue X, Li X, Nguyen HT, Chin DR, Sullivan DE and Lasky JA. Transforming growth factor-beta1 induces heparan sulfate 6-O-endosulfatase 1 expression in vitro and in vivo. J Biol Chem. 2008;283:20397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korf-Klingebiel M, Reboll MR, Grote K, Schleiner H, Wang Y, Wu X, Klede S, Mikhed Y, Bauersachs J, Klintschar M, Rudat C, Kispert A, Niessen HW, Lubke T, Dierks T and Wollert KC. Heparan Sulfate-Editing Extracellular Sulfatases Enhance Vascular Endothelial Growth Factor Bioavailability for Ischemic Heart Repair. Circ Res. 2019. [DOI] [PubMed] [Google Scholar]
- 61.Hammond E, Khurana A, Shridhar V and Dredge K. The Role of Heparanase and Sulfatases in the Modification of Heparan Sulfate Proteoglycans within the Tumor Microenvironment and Opportunities for Novel Cancer Therapeutics. Front Oncol. 2014;4:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.