Abstract
A chemopreventive effect of aspirin (ASA) on lung cancer risk is supported by epidemiologic and preclinical studies. We conducted a randomized, double-blinded study in current heavy smokers to compare modulating effects of intermittent versus continuous low dose ASA on nasal epithelium gene expression and arachidonic acid (ARA) metabolism. Fifty-four participants were randomized to intermittent (ASA 81 mg daily for one week/placebo for one week) or continuous (ASA 81 mg daily) for 12 weeks. Low dose ASA suppressed urinary prostaglandin E2 metabolite (PGEM) (change of −4.55±11.52 from baseline 15.44 ±13.79 ng/mg creatinine for arms combined, p = 0.02), a surrogate of cyclooxygenase-mediated ARA metabolism, but had minimal effects on nasal gene expression of nasal or bronchial gene expression signatures associated with smoking, lung cancer and chronic obstructive pulmonary disease. Suppression of urinary PGEM correlated with favorable changes in a smoking associated gene signature (p<0.01). Gene set enrichment analysis (GESA) showed that ASA intervention led to 1079 enriched gene sets from the Canonical Pathways within the Molecular Signatures Database. In conclusion, low dose ASA had minimal effects on known carcinogenesis gene signatures in nasal epithelium of current smokers but results in wide-ranging genomic changes in the nasal epithelium, demonstrating utility of nasal brushings as a surrogate to measure gene expression responses to chemoprevention. PGEM may serve as a marker for smoking associated gene expression changes and systemic inflammation. Future studies should focus on NSAIDs or agent combinations with broader inhibition of pro-inflammatory ARA metabolism to shift gene signatures in an anti-carcinogenic direction.
Introduction
Lung cancer is the leading cause of cancer-related deaths both in men and women in the U.S. and worldwide. Over 234,000 new cases and 154,000 deaths of lung cancer are projected to occur in the US in 2018. Globally, more than 1.37 million lung cancer deaths occur every year [1]. While prevention of tobacco use and tobacco cessation efforts remain the most effective means to reduce the incidence of lung cancer, the risk of developing this disease is still elevated for years after successful quitting compared to never smokers. Effective and well-tolerated chemoprevention strategies for lung cancer are urgently needed.
Interest in aspirin (ASA) as a chemopreventive agent is based on epidemiologic cohort and case control studies showing reduction of precancerous lesions and cancers in lung, colon, breast, and prostate tissue [2–4]. An analysis of eight randomized trials of daily ASA for a mean duration of ≥ 4 years versus no aspirin reported a significant decrease in lung adenocarcinoma mortality with ASA use (HR=0.68, 95% CI 0.50–0.92) (5). In a large cohort study (VITamin And Lifestyle study), total NSAID use (>10 years) was associated with a borderline significant reduction in risk of lung cancer (HR 0.82; 95% C.I. 0.64–1.04), with the strongest association for adenocarcinoma (HR 0.59); this trend was limited to men (HR 0.66) and to long-term (≥ 10 years) former smokers (HR 0.65) [5]. The effects of ASA are also supported in chemically induced murine models of lung cancer where it inhibits tumorigenesis and lung cancer incidence [6, 7].
Exposure to cigarette smoke creates a field of injury throughout the entire respiratory tract by inducing a variety gene expression alterations associated with smoking [8, 9]; smoking cessation [10]; chronic obstructive pulmonary disease (COPD) [11, 12]; bronchial premalignant lesions [13]; and lung cancer [14]. There is significant overlap between bronchial and nasal smoking- and lung cancer-associated gene-expression changes [15], suggesting the ability to detect lung disease- related biology throughout the intra- and extra-thoracic airway.
Given the promise of ASA in reducing lung cancer risk, we sought to evaluate the effects of ASA on the airway field of injury via gene expression profiling of nasal brushings. Here, we present a study of current smokers randomized to a 12-week intervention of intermittent or continuous low dose (LD) ASA (NCT02123849) with gene expression from nasal brushings profiled at baseline, at end-of-intervention and at one week post intervention.
Materials and Methods
Study Design
The study was a single center randomized, double-blinded trial to determine the modulatory effects of intermittent ASA dosing (ASA 81 mg daily for one week alternating with placebo daily for one week) versus continuous ASA dosing (ASA 81 mg daily) for 12 weeks on nasal epithelium gene expression and arachidonic acid metabolism in current smokers. The intermittent schedule (1 week on/1 week off) was designed based on pre-clinical studies of potentially less toxic, alternative drug dosing schedules including studies of naproxen that showed equivalent efficacy of this weekly intermittent dosing schedule to daily continuous dosing in rodent models of urinary bladder and colon carcinogenesis [16,17]. The primary endpoint was treatment-associated modulation of a smoking-associated gene expression signature derived using bronchial and nasal brushings (n=119 genes) [18]. Secondary endpoints were assessment of the effects of treatment and discontinuation of treatment for one-week in continuous and intermittent dosing arms on 1) changes in cyclooxygenase (COX) and 5-lipoxygenase (LOX)-mediated arachidonic acid (ARA) metabolism; 2) persistence of the smoking gene expression signature in nasal epithelium one week off agent intervention; 3) modulation of additional gene expression signatures including: a nasal lung cancer signature (n=535 genes)[19], a bronchial smoking signature (n=81 rapidly reversible genes upon smoking cessation)[20], a bronchial lung cancer gene signature (n=23 genes)[21], a PI3K pathway activity signature (n=183 genes)[22], and a bronchial COPD signature (n=98 genes)[11]; 4) safety in current smokers of 12 week exposure to continuous versus intermittent ASA; 5) gender effects in the modulatory effects of intermittent and continuous ASA on a smoking-related gene expression signature; and 6) to explore in a discovery-driven fashion the effect of ASA intervention on whole-genome gene expression. The University of Arizona Human Subjects Protection Program approved the study and each participant provided written informed consent.
Study drug
ASA (81 mg) and matched placebo were provided by Bayer HealthCare to the National Cancer Institute, Division of Cancer Prevention (NCI DCP) and packaged and supplied to the study site by the NCI DCP Drug Repository, MRIGlobal, Kansas City, MO.
Study population
Current smokers at least 18 years of age with a ≥ 20 pack year tobacco exposure history and an average daily use of ≥10 cigarettes per day were recruited from the greater Tucson and Phoenix areas. Inclusion criteria included normal hematologic, biochemical and coagulation parameters, ability to participate in the trial and sign informed consent. Exclusion criteria included allergy to aspirin or NSAIDs; gastric intolerance to ASA or NSAIDs; history of gastric ulcer; ASA or NSAID use for more than 5 days per month within 3 months of enrollment; unwilling or unable to refrain from use of non-study ASA or NSAID; adult asthma; current, recent, or chronic use of leukotriene antagonists or glucocorticoids (systemic, topical and/or nasal sprays); requiring chronic anticoagulation or anti-platelet therapy; history of a bleeding disorder or hemorrhagic stroke; history of chronic sinusitis or recent nasal polyps; unwilling or unable to limit alcohol consumption; pregnant or lactating; inability to absorb an oral agent; current or history of lung cancer; other invasive cancer within the past five years except non-melanoma skin cancer.
Study procedures
Participants underwent a physical exam, clinical laboratory analysis, and assessments of medical history, concurrent medications, NSAIDs use, and tobacco use history at the eligibility evaluation. Participants who had taken NSAIDs within the preceding 2 weeks underwent a 4-week washout period before baseline specimen collection. Participants underwent baseline specimen collection of nasal brushing, urine, blood, and buccal cells. Participants were then randomized (1:1) to receive intermittent ASA dosing (ASA 81 mg daily for one week alternating with placebo daily for one week) or continuous ASA dosing (ASA 81 mg daily) for 12 weeks. The intermittent arm began with placebo for the first week so participants in this arm would be on ASA at end-of-intervention. For study visit scheduling conflicts, agent intervention was extended for 2 weeks until the rescheduled visit. A mid-study visit was conducted for adherence and adverse event (AE) checks and current tobacco use. An end-of-intervention visit was conducted to review AEs, clinical laboratory analysis, adherence check, and current tobacco use. Biospecimen collection of nasal brushing, urine, blood, and buccal cells was performed at the end of intervention and the 7–10 day post-intervention visits. Safety of agent intervention was assessed by self-reported AEs and clinical laboratory analysis. AEs were graded using the NCI Common Terminology Criteria for Adverse Events (CTCAE) v. 4.0. Upon study completion, participants were provided information on the Arizona Smokers’ Helpline to assist in smoking cessation.
Nasal brushing for gene expression analysis
Nasal epithelium brushings were collected using a nasal speculum to spread one nare while a standard cytology brush was inserted underneath the inferior nasal turbinate. The brush was rotated in place for 3 seconds and immediately placed in RNAProtect Cell solution. A second brushing from the same nare was similarly collected and processed. Samples were stored at −80°C prior to analysis.
Microarray data acquisition and data preprocessing
Total RNA was isolated from nasal brushings using Qiagen miRNeasy Mini Kit following manufacturer’s instruction. Integrity of the RNA samples was assessed by Agilent BioAnalyzer, and purity of the RNA was confirmed using a NanoDrop spectrophotometer. The total RNA was subsequently reverse-transcribed and the obtained cDNA was used as a template for in vitro transcription. The resulting antisense cRNA was purified using Nucleic Acid Binding Beads, and used as a template for reverse transcription to produce single-stranded DNA in the sense orientation. During this step dUTP was incorporated. The DNA was then fragmented using uracil DNA glycosylase and apurinic/apyrimidinic endonuclease 1 and labeled with DNA Labeling Reagent that is covalently linked to biotin using terminal deoxynucleotidyl transferase. In vitro transcription and cDNA fragmentation quality controls were carried out by running an mRNA Nano assay. The labeled fragmented DNA was hybridized to Human Gene Arrays 1.0ST for 16–18 hours at 45°C. The hybridized samples were washed and stained. The first staining with streptavidin-R-phycoerythrin was followed by signal amplification using a biotinylated goat anti-streptavidin antibody and another SAPE staining. Microarrays were immediately scanned. The resulting CEL files were summarized using Affymetrix Expression Console (version 1.1).
Gene expression values were generated for each Human Gene Arrays 1.0ST CEL file (n=120 total samples) using R statistical software (version 3.2.3) and the Robust Multiarray Average (RMA) algorithm (affy package) (23) with Entrez Gene-specific probeset mapping (version 20.0.0) from the Molecular and Behavioral Neuroscience Institute (Brainarray) at the University of Michigan [24]. The microarray array data is deposited in GEO under accession GSE124637. Standardized RMA quality metrics were assessed, including the normalized un-scaled standard error (NUSE, cutoff >1.05) and relative log expression (RLE, cutoff >0.1). Additionally, we conducted a principal component analysis (PCA) across all genes and samples and excluded samples that were greater than 2 standard deviations from mean of the first principal component. Samples with more than one failed quality metric were excluded from analysis (n=11). The PCA revealed a significant batch effect that was removed using ComBat [25]. The sex annotation of each sample was verified using expression levels of Y-chromosome specific genes.
Calculation of gene expression signature scores
For each previously published gene expression signatures, the corresponding processed gene expression data used to derive the signatures was downloaded from the Gene Expression Omnibus (GEO). Specifically, we downloaded the following datasets: GSE16008 for the smoking-associated gene expression signature derived from nasal and bronchial brushings, GSE80796 for the lung cancer-associated gene expression signature derived from nasal brushings, GSE7895 for the smoking-associated gene expression signature derived from bronchial brushings, GSE12815 for the PI3K activity signature, and GSE37147 for the COPD-associated gene expression signature derived from bronchial brushings. For each gene expression dataset (“signature data”), ComBat [25] was used to remove batch effects between the signature data and the ASA gene expression data. ComBat adjusted gene expression values were z-score normalized across the combined ASA and signature data. For each gene signature, principal component analysis was conducted across the signature data, and the first principal component was applied to the ASA data to generate gene signature scores (See Supplementary Figures S1 and S2 and Supplementary Methods). Additionally, we generated scores from a lung cancer-associated gene expression signature derived from bronchial brushings using the classifier described by Whitney et al. [21] to score the ASA data.
Analysis of urinary biomarkers of arachidonic acid metabolism
Prostaglandin E2 (PGE2) is a major COX-mediated ARA metabolism product. The major urinary metabolite of PGE2, 11α-hydroxy-9,15,-dioxo-2,3,4,5-tetranor-prostane-1,20-dioic acid (PGEM) was quantified by a sensitive and specific liquid chromatography tandem mass spectrometry assay [26]. Briefly, 1 ml urine was acidified to pH 3 with 1 M HCl, and PGEM was then converted to the O-methyloxime derivative by treatment with methyloxime HCl in sodium acetate buffer (pH 5). The methoximated PGEM was then extracted with C18 solid phase extraction columns. The eluate from solid phase extraction was mixed with the internal standard ([2H6]O-methyloxime PGEM), dried, reconstituted in mobile phase prior to injection onto the LC-MS system. The chromatographic separation was achieved by a C18 reverse phase column and a gradient mobile phase of ammonium acetate, acetonitrile, and acetic acid. The mass spectrometer was operated in negative ion mode utilizing electrospray ionization. Detection was through selected reaction monitoring (SRM), with the transition of m/z 385 to 336 monitored for PGE-M and the transition of m/z 391 to 339 for the internal standard. The assay was linear over the range of 0.3 – 125 ng/ml with an assay accuracy of > 90% and an inter-assay coefficient of variation of <10%.
The urinary leukotriene E4 (LTE4), the terminal product of 5-LOX mediated ARA metabolism, was quantified by a sensitive and specific liquid chromatography tandem mass spectrometry assay [27]. Briefly, 5 ml urine was acidified to pH 3 with 1M HCl and mixed with the internal standard ([2H3]LTE4 (1 ng)) and extracted with C18 solid phase extraction columns. The eluate from solid phase extraction was dried and reconstituted in an aliquot of methanol and filtered using a 0.2 μm Spin-X filter. The filtrate was dried and reconstituted in an aliquot of methanol/water (50/50) prior to injection onto the LC-MS system. The chromatographic separation was achieved by a C18 reverse phase column and a gradient mobile phase of ammonium acetate, acetic acid, and acetonitrile. The mass spectrometer was operated in negative ion mode utilizing electrospray ionization. Detection was through SRM, with the transition of m/z 438 to 333 monitored for LTE4 and the transition of m/z 441 to 336 for the internal standard. The assay was linear over the range of 6.25 – 2,500 pg/ml with an assay accuracy of >90% and an inter-assay coefficient of variation of <12%.
Urinary biomarker levels were normalized by urinary creatinine concentrations measured using a creatinine assay kit (Diazyme Laboratories).
Statistical methods
The study planned to randomize 56 eligible participants to have at least 40 participants (20 per arm) with the complete set of evaluable specimens for biomarker analysis, based on an estimated attrition rate of 25%. A one-sided two-sample t-test at a significance level of 5% was performed to evaluate whether intermittent ASA is non-inferior to continuous ASA in changes of the gene signature scores. Additional regression analysis was performed to adjust for baseline levels and/or potential confounders, e.g. gender or BMI. A two-sided two-sample t-test was used to compare the baseline values of gene signature scores between the intervention arms and the baseline values of PGEM and LTE4 and changes in PGEM and LTE4 levels between the intervention arms. A two-sided paired t test was performed to evaluate the changes in gene signature scores, PGEM and LTE4 overall, by intervention arm, and by gender. All of the secondary analyses are considered exploratory so no correction for multiple comparisons were used. The Fisher’s exact test was used to compare the frequency of adverse events between the intervention arms.
Bioinformatics analysis to identify ASA-associated gene expression alterations
Differential gene expression was calculated between sample collection time points: 1) baseline to end-of-intervention, 2) baseline to one-week post intervention, and 3) end-of-intervention to one-week post intervention in samples that passed quality control metrics (n=109). These analyses reflect the genomic effects of ASA exposure, ASA exposure with persistence beyond one week, and ASA withdrawal, respectively. Differential gene expression was calculated for each comparison using linear modeling via the R package limma [28], adjusting for ASA dosing arm and blocking by subject. Adjusted p-values (FDR) < 0.25 were considered significant. The EnrichR tool [29] was used to explore the functional role of significant genes. Additional pathway analyses were conducted using gene set enrichment analysis (GSEA) [30] and genes rank ordered by t-statistic for each ASA effect from the linear modeling results. GSEA was used to assess enrichment of biology presumed to be modulated by aspirin exposure, and gene sets related to repair and wound healing from the Molecular Signatures Database (MSigDB) [31]. GSEA was also used to screen the curated Canonical Pathways (C2) within MSigDB and adjusted p-values (FDR) < 0.05 were considered significant. The most highly enriched genes from the significant gene sets were compiled using the Leading Edge Analysis tool.
We also compared ASA-associated gene expression alterations between the two dosing arms. Paired t-tests were computed between baseline and end-of-intervention within each arm. The results of this analysis were used to construct dosing arm specific ranked lists of genes by t-statistic and gene sets (the top 100 most up- and down-regulated genes). GSEA was used to establish ASA-associated gene expression changes between the two arms using these ranked lists and gene sets.
Results
Participant demographics
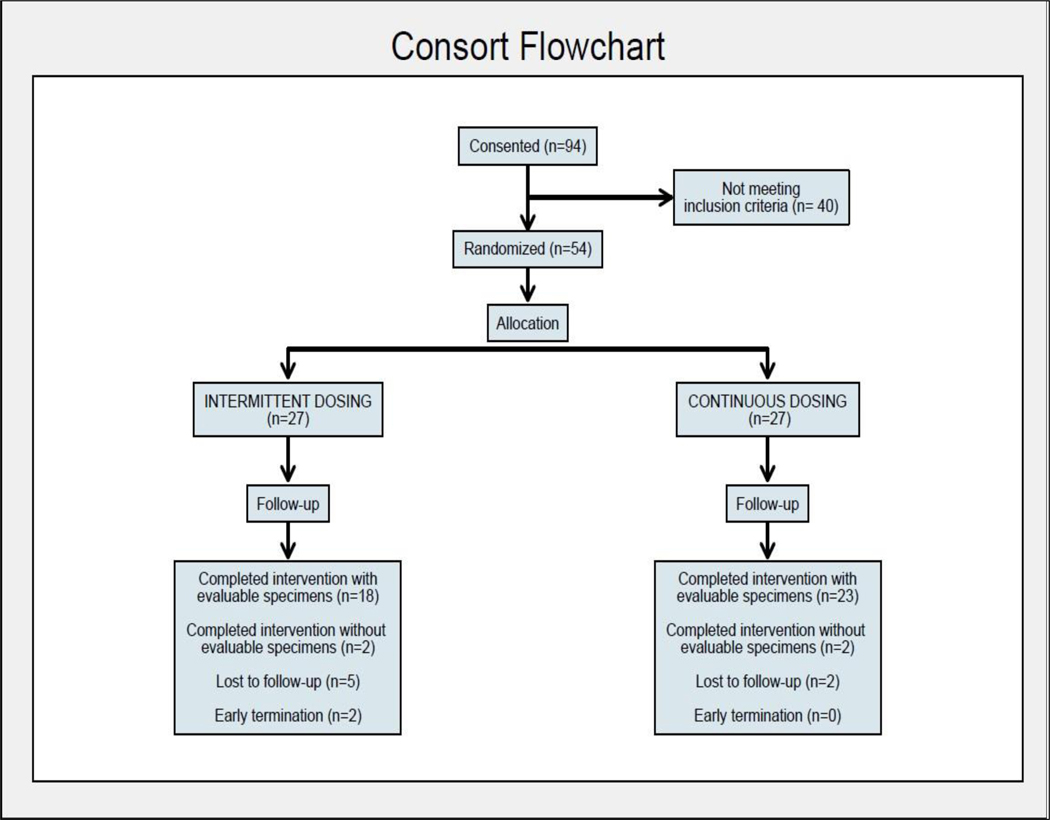
From June 2014 through May 2016, 54 participants (30 male/24 female) were randomized to the intermittent ASA arm (N=27; 15 male/12 female) or continuous ASA arm (N=27; 15 male/12 female) (See Consort Diagram, Fig. 1). Eighteen participants in the intermittent arm and 23 in the continuous arm completed study intervention with complete sets of evaluable biospecimens. Participant demographics and characteristics are summarized in Table 1. The majority of participants were White Non-Hispanic; Hispanic/Latino represented 15% and 22%, respectively, for intermittent and continuous ASA arms. Mean age of participants was 52 years in both arms. There were no significant differences by intervention arm for age, race/ethnicity, height/weight, pack year tobacco exposure and daily alcohol use except higher BMI in the continuous ASA arm. All participants had high urinary cotinine levels consistent with self-reported current heavy tobacco use, which were similar between arms and remained high during the study (data not shown).
Figure 1.
Consort Flow Diagram
Table 1.
Baseline characteristics for randomized participants.
| Overall (N=54) | Intermittent ASA (N=27) | Continuous ASA (N=27) | p-value1 | |
|---|---|---|---|---|
| Age, yr | 52±8 | 52±8 | 52±8 | 0.91 |
| Male, N | 30 | 15 | 15 | 1.00 |
| Race, N | ||||
| White/Black or African Am/Asian/Am Indian or Alaskan Native/More than one race |
45/5/1/1/2 | 21/3/0/1/2 | 24/2/1/0/0 |
0.46 |
| Hispanic or Latino, N | 10 | 4 | 6 | 0.73 |
| Weight, kg | 83.5±21.1 | 81.0±20.2 | 85.9±22.1 | 0.40 |
| Height, cm | 171. 9±11.0 | 173.5±12.3 | 170.2±9.6 | 0.27 |
| BMI, kg/m2 | 28.0±5.2 | 26.5±4.6 | 29.4±5.6 | 0.04 |
| Pack-years | 35.7±12.9 | 33.7±9.7 | 37.7±15.3 | 0.25 |
| Urinary cotinine levels (ng/mg Cr) |
5793.21±4519.80 | 5538.03±2585.74 | 6048.39±5900.98 | 0.68 |
| Current Drinker (1–2 serving/day) |
40 (74.07%) | 19 (70.37%) | 21 (77.78%) | 0.76 |
derived from two-sample t test for continuous variables and Fisher’s exact test for categorical variables
Adherence and Safety
Participants took 98% of the assigned pills on average by pill count. Both the intermittent and continuous ASA interventions were well tolerated. Adverse events deemed possibly or probably related to study agent included dyspepsia, dry mouth, decreased hematocrit, bruising, ALT increase and platelet decrease with no significant difference in adverse events between the study intervention arms (Table 2). All AEs were Grade 1 or 2 events treated with supportive care intervention and were self-limited. While these AEs were not unexpected given the safety profile of ASA, the two AEs of decreased hematocrit may have been related to ASA-induced GI bleeding which is a significant toxicity. For these AEs, hematocrit normalized in follow-up without need for medical treatment.
Table 2.
Summary of study intervention related adverse events.
| Adverse Events | Intermittent ASA Arm (N = 27) | Continuous ASA Arm (N = 27) |
|---|---|---|
| Gastrointestinal disorders | ||
| Dyspepsia | 1 (3.7%) | 2 (7.4%) |
| Dry mouth | 1 (3.7%) | 0 |
| Blood and lymphatic system | ||
| Hematocrit decreased | 1 (3.7%) | 1 (3.7%) |
| Injury, poisoning and procedural complications |
||
| Bruising | 1 (3.7%) | 0 |
| Investigations | ||
| Alanine aminotransferase increased |
0 | 1 (3.7%) |
| Platelet count decreased | 0 | 1 (3.7%) |
LD ASA intervention has minimal effects on smoking, lung cancer and COPD associated nasal and bronchial gene expression signatures:
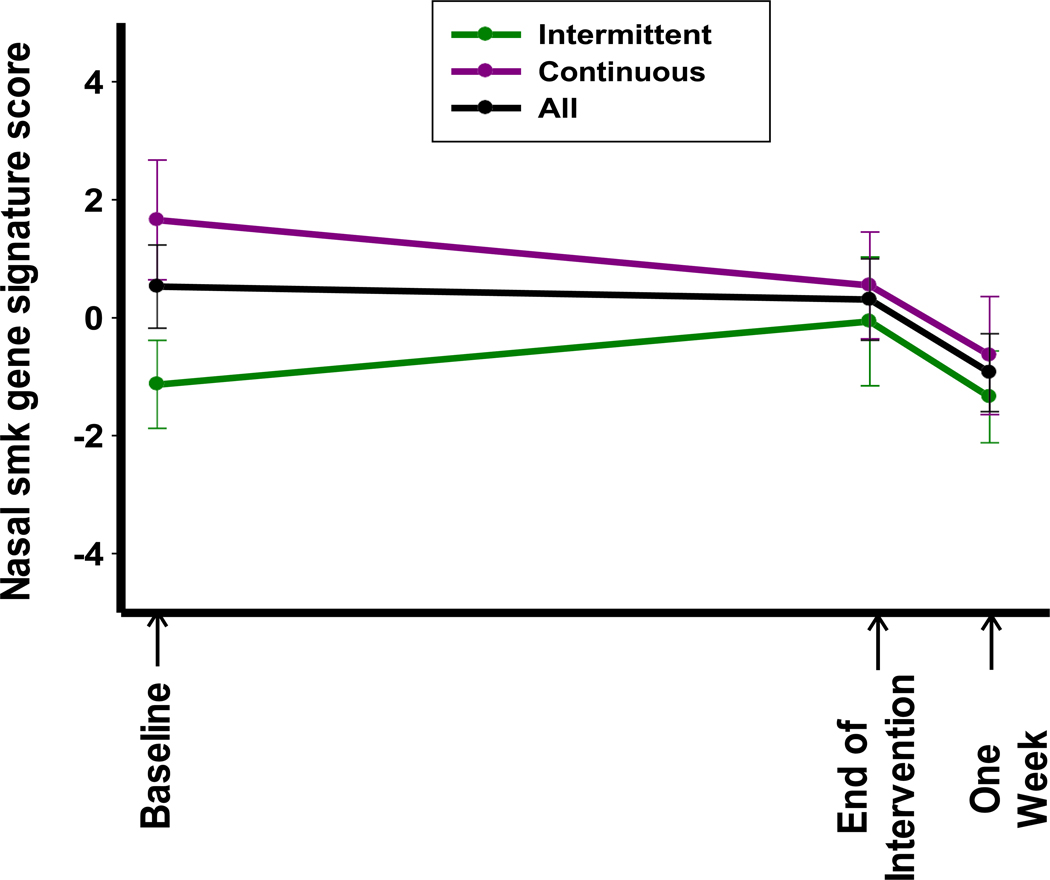
High quality microarray data was generated for 37 of the 40 participant samples at baseline and 36 of 40 samples at end-of-intervention and one-week post intervention. These samples were scored based on several previously published smoking, lung cancer, and COPD associated gene expression signatures based on nasal and bronchial brushings (See Supplementary Methods, Figures S1 and S2, Tables S1–5, Supplementary Data 1). Data for the primary endpoint analysis of modulation of the smoking associated gene expression signature score derived from nasal and bronchial brushings (18) are summarized in Table 3. There was no significant change in this gene signature score at end of intervention compared to baseline for the individual intervention arms (change of −1.11 ± 4.05 from 1.66 ± 4.76, p = 0.21, in the continuous arm; change of 1.39 ± 4.41 from −1.13 ± 2.89, p = 0.26, in the intermittent arm). There was a significant difference in the baseline gene signature score between arms, which limited the analysis for non-inferiority for the intermittent arm effect. There was a significant change in the score at one-week post agent from baseline in the continuous arm (change of −1.95 ± 3.42 from 1.66 ± 4.76, p = 0.02). The smoking associated gene expression signature derived from bronchial brushings also demonstrated a significant change in the score at one-week post agent from baseline in the continuous arm (Supplementary Table S2, change of 1.32 ± 2.67 from −0.99 ± 3.91, p = 0.04). In both cases, however, the direction of change was unfavorable (toward current smoker signature scores in the signature data, Supplementary Figure S2). Finally, there was a significant increase in the bronchial PI3K activity score at end of intervention in the continuous arm (Supplementary Table S4, change of 0.77 ± 1.69 from −0.60 ± 1.92, p < 0.05) that was in an unfavorable direction as higher scores are associated with activated PI3K in the signature data (Supplementary Figure S2).
Table 3.
Baseline and changes in the gene expression signature score calculated based on a smoking-associated gene expression signature derived from nasal and bronchial brushings by Zhang (18).
| Overall | Intermittent ASA | Continuous ASA | P1 | |
|---|---|---|---|---|
| All participants | ||||
| Baseline | 0.53±4.29; N=37 | −1.13±2.89; N=15 | 1.66±4.76; N=22 | 0.05 |
| Change at End of Intervention from Baseline | −0.14±4.31; N=36 (p=0.85) |
1.39±4.41; N=14 (p=0.26) |
−1.11±4.05 (p=0.21) |
0.04 |
| Change at one-week post agent from Baseline |
−1.21±3.48; N=35 (p<0.05) |
−0.10±3.38; N=14 (p=0.91) |
−1.95±3.42; N=21 (p=0.02) |
0.06 |
| Change at one-week post agent from End of Intervention | −1.33±4.42; N=34 (p=0.09) |
−1.42±4.80; N=13 (p=0.31) |
−1.28±4.28; N=21 (p=0.19) |
0.53 |
| Male | N=21 | N=10 | N=11 | |
| Baseline | −0.12±4.26; N=19 | −1.37±3.79; N=8 | 0.80±4.53 | 0.29 |
| Change at End-of-Intervention from Baseline | 0.45±5.04; N=18 (p=0.71) |
2.05±6.22; N=7 (p=0.42) |
−0.57±4.13 (p=0.66) |
0.15 |
| Change at one-week post agent from Baseline | −0.58±4.08; N=17 (p=0.57) |
0.61±4.35; N=7 (p=0.72) |
−1.41±3.89; N=10 (p=0.28) |
0.17 |
| Change at one-week post agent from End-of-Intervention | −1.60±4.92; N=16 (p=0.21) |
−1.26±6.76; N=6 (p=0.67) |
−1.80±3.85; N=10 (p=0.17) |
0.42 |
| Female | N=19 | N=8 | N=11 | |
| Baseline | 1.20±4.33; N=18 | −0.85±1.61; N=7 | 2.51±5.05 | 0.06 |
| Change at End-of-Intervention from Baseline | −0.72±3.48; N=18 (p=0.39) |
0.74±1.54; N=7 (p=0.25) |
−1.65±4.08 (p=0.21) |
0.05 |
| Change at one-week post agent from Baseline |
−1.81±2.78; N=18 (p=0.01) |
−0.81±2.17; N=7 (p=0.36) |
−2.45±3.03 (p=0.02) |
0.12 |
| Change at one-week post agent from End-of-Intervention | −1.09±4.04; N=18 (p=0.27) |
−1.55±2.82; N=7 (p=0.20) |
−0.80±4.77 (p=0.59) |
0.64 |
a two-sided two-sample t test was used to compare baseline values between treatment arms; a one- sided two-sample t test (non-inferiority test) was used to determine whether the changes in the intermittent arm were not inferior to the continuous arm.
derived from two-sided paired t test
ASA decreases COX-mediated ARA metabolite PGEM but does not alter 5-LOX-mediated ARA metabolite LTE4:
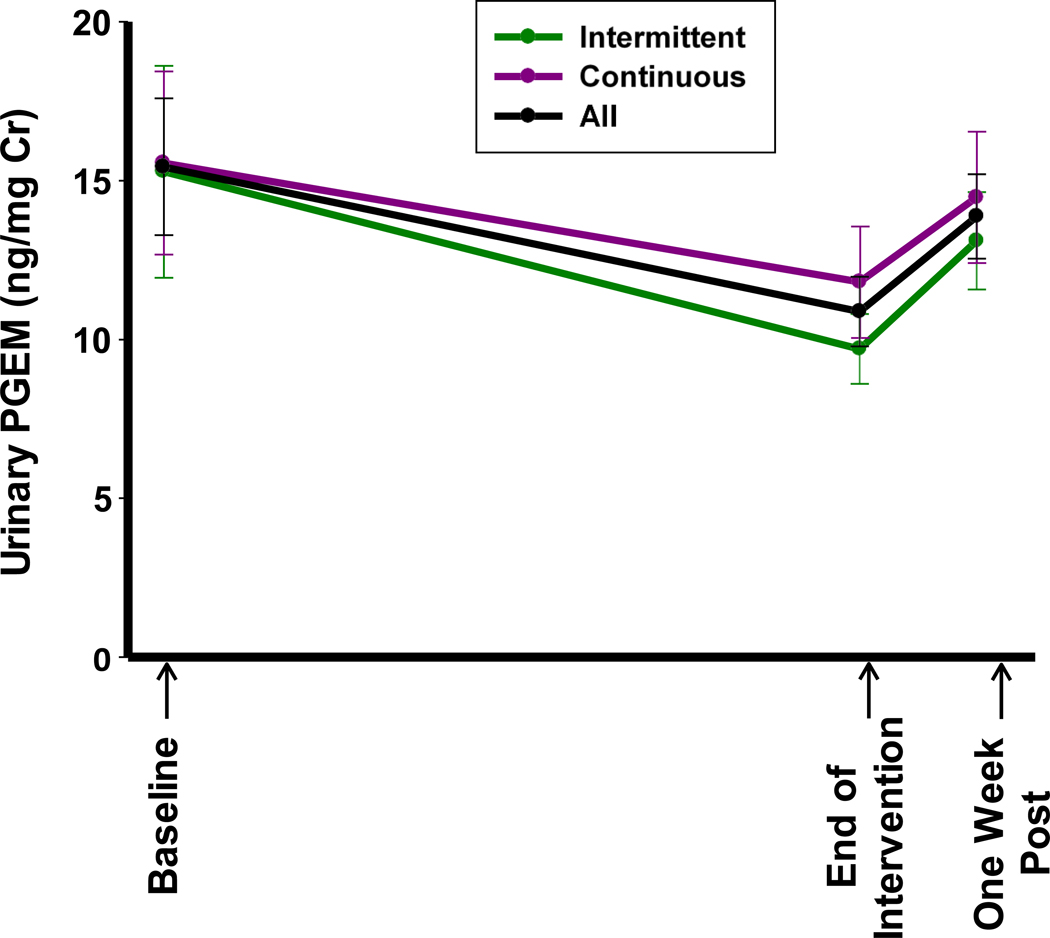
Urinary PGEM and LTE4 levels were analyzed for 41 participants. For the intervention arms combined, PGEM significantly decreased from baseline to end-of-intervention (Supplementary Table S6, change of −4.55±11.52 from baseline levels of 15.44 ±13.79 ng/mg creatinine, p=0.02), consistent with an expected biochemical effect of ASA. When PGEM was analyzed by intervention arm no significant changes from baseline to end-of-intervention were noted. From end-of-intervention to one-week post agent, there was a significant increase in PGEM towards baseline values for combined intervention arms (p=0.01) (Supplementary Table S6, Fig. 3). The observed changes in PGEM were driven mostly by the changes in males but not in females. Conversely, there were no significant changes in urinary LTE4 levels overall, within the study arm or stratified by gender at the end-of-intervention and one-week post agent intervention (Supplementary Table S7).
Figure 3.
Baseline and changes in urinary PGEM levels
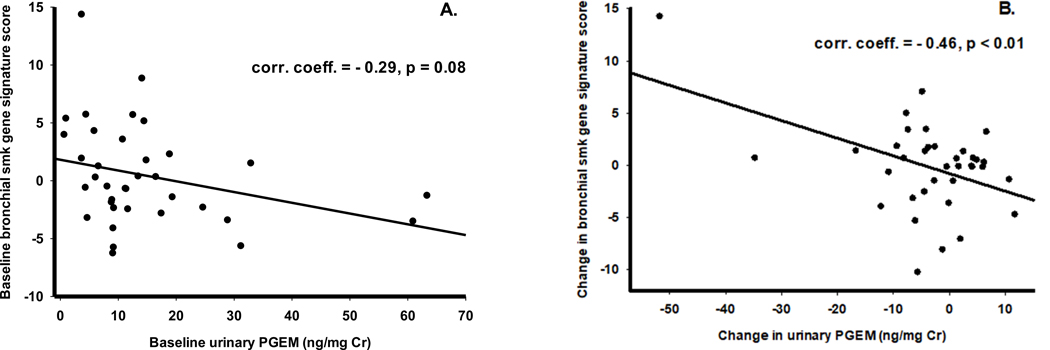
Participants with higher baseline levels of PGEM and greater post-intervention PGEM suppression had more favorable modulation of the nasal and bronchial epithelium derived smoking associated gene expression signature scores (Supplementary Tables S8 A, B). Higher scores for the nasal and bronchial epithelium derived 119-gene smoking signature (closer to scores for never smokers) were associated with lower urinary PGEM levels (less COX2 activation) (r = −0.29, P=0.08) (Fig. 4A). The correlation between change in nasal smoking signature score from baseline to end of intervention with PGEM was significant, indicating those with more suppressed PGEM levels have greater (more favorable) increases in smoking scores (r = −0.46; P=<0.01) (Fig. 4B).
Figure 4.
Correlations between smoking gene signature (16) scores and urinary PGEM levels. A: Correlation between baseline scores and PGEM levels; B: Correlation between changes (change at end-of study from baseline) in scores and PGEM levels.
ASA induces wide-ranging genomic changes by gene set enrichment analysis (GSEA):
Differential expression analysis did not yield differentially expressed genes between baseline and end-of-intervention time points. There were 720 and 161 genes (FDR<0.25) found to be differentially expressed between baseline and one-week post intervention and end-of-intervention and one-week post intervention, respectively.
GSEA showed enrichment of gene sets reflecting prostanoid and arachidonic acid metabolism (Supplementary Table S9). The Gene Ontology arachidonic acid mono-oxygenase activity and metabolic process pathways were enriched among genes up-regulated at baseline compared to end-of-intervention (GSEA, FDR<0.05). Gene Ontology prostanoid metabolic process and biosynthetic process pathways were enriched among genes up-regulated at end-of-intervention compared to baseline (GSEA, FDR<0.05). Furthermore, a screen of the MSigDB curated Canonical Pathways, a collection of gene sets that represents diverse biological processes, identified 1079, 787, and 112 gene sets significantly enriched between baseline and end-of-intervention, baseline and one-week post intervention, and end-of-intervention and one-week post intervention, respectively (GSEA, FDR<0.05). Sixty-six percent of gene sets significantly enriched (n=521) from the baseline to one-week post intervention analysis were present in the baseline to end-of-intervention analysis, presumably reflecting persistent ASA-associated genomic changes (Supplementary Data 2). Fifty-two percent of the significant gene sets (n=577) from the baseline to end-of-intervention analysis were not present in the baseline to one-week post intervention analysis, presumably reflecting rapidly reversible ASA-associated genomic changes (Supplementary Data 3). The Leading Edge Analysis Tool identified genes in the persistently altered pathways to be related to the biology of ribosomes, histones, proteasomes, chemokines, and the mitochondrial electron transport chain, whereas genes in the rapidly reversible pathways are associated with cellular signaling and immune function, and include numerous kinases and second messengers. We also found the two dosing arms to be significantly and concordantly enriched (GSEA, FDR<0.05), however, the continuous dosing arm yielded a higher number of significantly enriched gene sets from the curated Canonical Pathways than the intermittent dosing arm (1022 versus 470 gene sets in the baseline to end-of-intervention analysis), supporting a stronger genomic effect of continuous dosing.
Additionally, two repair and wound healing-related gene sets were enriched among genes up-regulated one-week post intervention compared to end-of intervention and two additional pathways were enriched among genes up-regulated at one-week post interventions compared to baseline (GSEA, FDR<0.05) (Supplementary Table S10). The results suggest that a subset of the gene expression changes identified are associated with regeneration of the epithelium after repeat brushing collected one-week post intervention.
Discussion
We investigated the effect of 12 weeks of LD ASA 81 mg/d on an intermittent versus a continuous daily schedule in current heavy smokers on a comprehensive set of nasal and bronchial epithelium-derived gene expression signatures associated with smoking, lung cancer and COPD. Overall, and for each dosing arm, we did not observe a statistically significant modulation of the smoking, lung cancer, and COPD gene expression signatures. We showed that LD ASA was effective in suppressing COX-mediated ARA metabolism in high risk smokers. Through the GSEA, we found that LD ASA led to wide-ranging genomic changes in the nasal epithelium, with the continuous dosing resulting in a higher number of enriched gene sets than the intermittent dosing.
The selection of LD ASA, the dosing schedules and duration of intervention may have led to the minimal effects on the pre-selected gene expression signatures. While a body of data supports the development of LD ASA in lung cancer chemoprevention [32, 33], it is possible that regular-strength ASA might have yielded more robust modulation of the gene signatures in the anti-carcinogenic direction. An additional arm of regular-strength ASA would have strengthened the study by addressing dose-response in modulating risk of tobacco exposure and lung cancer. While a short duration intervention of regular-strength ASA is feasible for proof-of-concept chemoprevention studies, only LD ASA has a safety profile amenable to long-term use as a chemoprevention agent. Additionally, the use of fixed low dose ASA may have influenced modulation of the gene expression signatures, as a recent analysis of ASA primary prevention trials of cardiovascular events and secondary prevention of colorectal cancer showed an interaction between LD ASA effects and body size, with LD ASA effect seen only in participants <70 kg [34]. We tested an intermittent schedule of ASA based on preclinical intermittent dosing models versus a continuous daily dosing schedule, with a greater effect on broad genomic changes noted with continuous daily dosing. While the small number of participants per dosing arm limits interpretation of these data, the relatively short duration of effect of ASA on cyclooxygenase (96 hours) may have translated to recovery back to baseline of gene expression modulatory effects over the intermittent arm’s week-long dosing holiday.
The optimal duration of LD ASA to potentially modulate tobacco- and lung cancer-related gene expression in the nasal epithelium as a surrogate for the respiratory epithelium is unknown. The study’s relatively short (12 week) intervention is broadly consistent with a number of preclinical biomarker studies of NSAIDs and other classes of chemoprevention agents in murine models of cigarette smoke exposure [35, 36]. However, murine models may not adequately represent the pharmacologic modulation of human chronically smoke exposed respiratory epithelium as murine models require high doses of both carcinogen and chemopreventive agent to detect an effect in a short period of time.
LD ASA intervention in this smokers cohort led to a significant decrease in PGEM, a surrogate marker of COX-mediated ARA metabolism, for the intervention arms combined but not when analyzed separately, possibly owing to the small sample size of the individual arms. Our study cohort had elevated baseline levels of urinary PGEM as well as LTE4, a surrogate marker of the 5-LOX mediated ARA metabolism, comparable to those previously reported for current smokers [37] and indicating increased systemic inflammation. PGEM decrease was driven mostly in males, who had higher baseline PGEM levels compared to females and which may reflect higher COX-2 activity in the males. Therefore, males with high levels of baseline PGEM may have had greater prostaglandin inhibition with LD ASA related to greater COX-2 inhibition. An important finding of this study is that greater suppression of PGEM levels by ASA intervention was associated with greater modulation of smoking scores in a favorable direction. Urinary PGEM may therefore serve as a useful biomarker to select at risk heavy smokers who may preferentially benefit from ASA or other inhibitors of ARA metabolism. We did not find significant changes in urinary LTE4 levels, over the intervention, an expected outcome given that ASA’s affinity for the lipoxygenase binding sites is low or not at all. That LTE4 levels did not increase even in participants in the highest tertile of baseline PGEM (data not shown) suggests that LD ASA did not shunt ARA metabolism through the pro-inflammatory 5-LOX metabolic pathway. This contrasts with a celecoxib intervention in current smokers in which shunting through 5-LOX occurred in participants with high baseline PGEM [27].
We identified ASA-associated gene expression alterations in the nasal epithelium. We did not detect statistically significant differential gene expression between baseline and end-of-intervention; however, using GSEA, we identified 1079 pathways enriched with ASA treatment, including prostanoid metabolism pathways. Biological pathways enriched with ASA treatment and subsequently reversed after one week post intervention included pathways associated with cellular signaling and immune function, and numerous kinases and second messengers interferon and inflammation response. In contrast, biological pathways enriched with ASA treatment and persisted after one week post intervention were associated pathways related to the biology of ribosomes, histones, proteasomes, chemokines, and the mitochondrial electron transport chain. We also found concordant enrichment between the two dosing arms. However, continuous dosing yielded a higher number of significantly enriched gene sets than intermittent dosing, suggesting a stronger genomic effect of continuous dosing.
Two repair and wound healing associated pathways were enriched among genes up-regulated one-week post intervention compared to end-of-intervention, possibly related to injury to the nasal epithelium induced by repeat nasal brushing. The effect of repeat brushing may have also contributed to the significant change we observed in the bronchial smoking score in an unfavorable direction between one week post intervention and end-of-intervention. Future studies may need to increase the interval or alternate the site on which the brushing is obtained.
In conclusion, our study showed that LD ASA had minimal effects on known carcinogenesis gene signatures in nasal epithelium and suggests that interventions with single agents or rational combinations of agents that more broadly inhibit pro-inflammatory ARA metabolism may be needed to shift the signature scores in the anti-carcinogenic direction. GSEA showed that ASA treatment is associated with wide-ranging genomic changes in the nasal epithelium, demonstrating the potential utility of using nasal brushing as a surrogate to measure gene expression responses to chemoprevention. Additionally, PGEM may serve as a marker for smoking associated gene expression changes and systemic inflammation to select at risk individuals for chemoprevention studies.
Supplementary Material
Figure 2.
Baseline and changes in nasal smoking gene signature score (16)
Acknowledgments:
This work was supported by a contract (HHSN261201200031I) from the National Cancer Institute and the Arizona Cancer Center Support Grant (CA023074). The authors wish to thank Bayer Healthcare for supplying aspirin and placebo. The authors also wish to acknowledge Frances Minter, Ann De Jong, Valerie Butler, Bonita Weible, Jerilyn San Jose, Catherine Cordova, and Wade Chew for their excellent assistance in the performance of the clinical study and endpoint assays. We thank Diane Ruedas for assistance in preparation of the manuscript.
Abbreviations
- AE
Adverse event
- ASA
Aspirin
- ARA
Arachidonic Acid
- BMI
body mass index
- CTCAE
Common Terminology Criteria for Adverse Events
- COPD
chronic obstructive pulmonary disease
- cRNA
Circular RNA
- COX
Cyclooxygenase
- dUTP
2’-Deoxyuridine, 5’-Triphosphate
- FDR
False Discovery Rate
- GEO
Gene Expression Omnibus
- GSEA
Gene Set Enrichment Analysis
- LTE4
leukotriene E4
- LOX
lipoxygenase
- MSigDB
Molecular Signatures Database
- NSAID
nonsteroidal anti-inflammatory drug
- PGEM
prostaglandin E metabolite
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Mills EJ, Wu P, Alberton M, Kanters S, Lanas A, Lester R. Low-dose Aspirin and Cancer Mortality: A Meta-analysis of Randomized Trials. American Journal of Medicine 2012;125(6):560–567. [DOI] [PubMed] [Google Scholar]
- 3.Algra AM, Rothwell PM. Effects of regular aspirin on long-term cancer incidence and metastasis: a systematic comparison of evidence from observational studies versus randomised trials. Lancet Oncology 2012;13(5):518–527. [DOI] [PubMed] [Google Scholar]
- 4.Bosetti C, Rosato V, Gallus S, Cuzick J, La Vecchia C. Aspirin and cancer risk: a quantitative review to 2011. Annals of Oncology 2012;23(6):1403–1415. [DOI] [PubMed] [Google Scholar]
- 5.Slatore CG, Au DH, Littman AJ, Satia JA, White E. Associaton of Nonsteroidal Anti- Inflammatory Drugs with Lung Cancer: Results from a Large Cohort Study. Cancer Epidemiology Biomarkers & Prevention 2009;18(4):1203–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castonguay A, Rioux N, Duperron C, Jalbert G. Inhibition of lung tumorigenesis by NSAIDS: A working hypothesis. Experimental Lung Research 1998;24(4):605–615. [DOI] [PubMed] [Google Scholar]
- 7.Saini RK, Sanyal SN. Chemopreventive Effect of Nonsteroidal Anti-inflammatory Drugs on 9,10- Dimethylbenz[a]anthracene-Induced Lung Carcinogenesis in Mice. Oncology Research 2009;17(11–12):505–518. [DOI] [PubMed] [Google Scholar]
- 8.Mao L, Lee JS, Kurie JM, et al. Clonal genetic alterations in the lungs of current and former smokers. J Natl Cancer Inst. 1997;89(12):857–862 [DOI] [PubMed] [Google Scholar]
- 9.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med (Berl). 200 Jan:85(1): 39–53. Epub 2006 Nov 8. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Lee JJ, Tang H, Fan YH, Xiao L, Ren H, et al. Impact of smoking cessation on global gene expression in the bronchial epithelium of chronic smokers. Cancer Prev Res (Phila) 2008. July;1(2):112–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steiling K, van den Berge M, Hijazi K, Florido R, Campbell J, Liu G, Xiao J, Zhang X, Duclos G, Drizik E, Si H, Perdomo C, Dumont C, Coxson HO, Alekseyev YO, Sin D, Pare P, Hogg JC, McWilliams A, Hiemstra PS, Sterk PJ, Timens W, Chang JT, Sebastiani P, O’Connor GT, Bild AH, Postma DS, Lam S, Spira A, Lenburg ME. A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am J Respir Crit Care Med. 2013. May 1;187(9):933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boudewijn IM, Faiz A, Steiling K, van der Wiel E, Telenga ED, Hoonhorst SJM, et al. Nasal gene expression differentiates COPD from controls and overlaps bronchial gene expression. Respir Res 2017;18:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beane J, Mazzilli SA, Tassinari AM, Liu G, Zhang X, Liu H, et al. Detecting the presence and progression of premalignant lung lesions via airway gene expression. Clin Cancer Res 2017;23:5091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev. Res. 2011;4:803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sridhar S, Schembri F, Zeskind J, Shah V, Gustafson AM, Steiling K, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genom. 2008;9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lubet RA, Scheiman JM, Bode A, White J, Minasian L, Juliana MM, Boring DL, Steele VE, Grubbs CJ. 2015. Prevention of chemically induced urinary bladder cancers by naproxen: Protocols to reduce gastric toxicity in humans do not alter preventive efficacy. Cancer prevention research (Philadelphia, Pa). 8(4):296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammed A, Janakiram NB, Madka V, Zhang Y, Singh A, Biddick L, Li Q, Lightfoot S, Steele VE, Lubet R et al. 2018a. Abstract 4983: Intermittent dosing regimens of naproxen and aspirin inhibit azoxymethane-induced rat colon adenoma progression to adenocarcinoma and carcinoma invasion. Cancer research. 78(13 Supplement):4983–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Sebastiani P, Liu G, Schembri F, Zhang X, Dumas YM, et al. Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiological Genomics 2010;41(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Rogers JF, Gerrein J, Anderlind C, Liu G, Zhang S, Alekseyev Y, et al. Shared Gene Expression Alterations in Nasal and Bronchial Epithelium for Lung Cancer Detection. J Natl Cancer Inst. 2017. July 1;109(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beane J, Sebastinani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biology 2007;8:R201.1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitney DH, Elashoff MR, Porta-Smith K, Gower AC, Vachani A, Ferguson JS, et al. Derivation of a bronchial genomic classifier for lung cancer in a prospective study of patients undergoing diagnostic bronchoscopy. BMC Med Genomics. 2015. May 6;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gustafson AM, Soldi R, Anderlind C et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Science Translational Medicine 2010;2(26). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCall MN, Jaffee HA, Irizarry RA. fRMA ST: Frozen robust multiarray analysis for Affymetrix Exon and Gene ST arrays. Bioinformatics 2012; 28:3153–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp.
- 25.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, Liu C. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS One 2011:, 6, e17238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphey LJ, Williams MK, Sanchez SC, Byrne LM, Csiki I, Oates JA, et al. Quantification of the major urinary metabolite of PGE(2) by a liquid chromatographic/mass spectrometric assay: determination of cyclo oxygenase-specific PGE(2) synthesis in healthy humans and those with lung cancer. Analytical Biochemistry 2004;334(2):266–275. [DOI] [PubMed] [Google Scholar]
- 27.Duffield-Lillico AJ, Boyle JO, Zhou XK et al. Levels of Prostaglandin E Metabolite and Leukotriene E-4 Are Increased in the Urine of Smokers: Evidence that Celecoxib Shunts Arachidonic Acid into the 5-Lipoxygenase Pathway. Cancer Prevention Research 2009;2(4):322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W and Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 201543(7), pp. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;128(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences Oct 2005, 102 (43) 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberzon A, Subramanian A, Pinchback R, Thorvalds Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics, Volume 27, Issue 12, 15 June 2011, Pages 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsoi KKF, Ho JMW, Chan FCH, Sung JJY. Long-term use of low-dose aspirin for cancer prevention: A 10-year population cohort study in Hong Kong Int J Cancer 2018. December 21. doi: 10.1002/ijc.32083. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 33.Ye S, Lee M, Lee D, Ha EH, Chun EM. Association of Long-term Use of Low-Dose Aspirin as Chemoprevention With Risk of Lung Cancer. JAMA Netw Open. 2019. March 1;2(3):e190185. doi: 10.1001/jamanetworkopen.2019.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothwell PM, Cook NR, Gaziano JM, Prioce JF, Belch JFF, Roncaglioni MC, Morimoto T, Mehta Z. Effects of aspirin on risks of vascular events and cancer according to bodyweight and dose: analysis of individual patient data from randomized trials. The Lancet 2018;392:387–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Maestra S, D’Agostini F, Izzotti A, Micale RT, Mastracci L, Camoirano A, Balansky R, Trosko JE, Steele VE, De Flora S. Modulation by aspirin and naproxen of nucleotide alterations and tumors in the lung of mice exposed to environmental cigarette smoke since birth. Carcinogenesis. 2015. December;36(12):1531–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Izzott A, Balansky R, Ganchev G, Iltcheva M, Longobardi M, Pulliero A, Camozzirano A, D’Agostini F, Geretto M, Micale RT, La Maestra S, Miller MS, Steele VE, De Flora S. Early and late effects of aspirin and naproxen on microRNAs in the lung and blood of mice, either unexposed or exposed to cigarette smoke.Oncotarget. 2017. August 24;8(49):85716–85748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gross ND, Boyle JO, Morrow JD, et al. Levels of prostaglandin E metabolite, the major urinary metabolite of prostaglandin E2, are increased in smokers. Clin Cancer Res. 2005;11(16):6087–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.