Abstract
Why metalloenzymes often show dramatic changes in their catalytic activity when subjected to chemically similar but non-native metal substitutions is a long-standing puzzle. Here, we report on the catalytic roles of metal ions in a model metalloenzyme system, human carbonic anhydrase II (CA II). Through a comparative study on the intermediate states of the zinc-bound native CA II and non-native metal-substituted CA IIs, we demonstrate that the characteristic metal ion coordination geometries (tetrahedral for Zn2+, tetrahedral to octahedral conversion for Co2+, octahedral for Ni2+, and trigonal bipyramidal for Cu2+) directly modulate the catalytic efficacy. In addition, we reveal that the metal ions have a long-range (~10 Å) electrostatic effect on restructuring water network in the active site. Our study provides evidence that the metal ions in metalloenzymes have a crucial impact on the catalytic mechanism beyond their primary chemical properties.
Subject terms: Enzyme mechanisms, Metalloproteins, X-ray crystallography
Metalloenzymes often show different catalytic activities in the presence of non-native metal ions. Here, the authors report structures of carbonic anhydrase bound to zinc and several other metal ions and demonstrate that metal ion coordination geometries directly impact catalytic activity of the enzyme.
Introduction
Metalloproteins are ubiquitous in nature and play indispensable roles in key biological processes, such as DNA synthesis, chemical signaling, and cellular metabolism1,2. Due to their versatile chemical reactivity (acidity, electrophilicity, and/or nucleophilicity), incorporated metal ions add functionality to proteins and help catalyze some of the most intricate reactions in nature3,4. The issues of metal binding affinity and specificity of metal ions to proteins have been studied based on the metal coordination stereochemistry5,6 and semi-empirical and qualitative theories such as hard and soft acids and bases principle of Parr and Pearson7 and Irving–Williams series of divalent ion stability8,9. However, the role of metal ions in the functioning of proteins and the metal–protein relationships remain unclear at the atomic level. For example, metalloenzymes substituted by non-native metal ions often exhibit drastically different catalytic activities10,11, even when the substituted metal ions show chemical features broadly similar to the native one, such as ionic charge/size/mass, redox potential, electronic configuration, and allowed coordination geometry.
Among the various types of metalloenzymes, carbonic anhydrase (CA), the first enzyme recognized to contain zinc, is ubiquitous across all kingdoms of life and one of the most catalytically efficient enzymes ever known12–16. The enzyme catalyzes the reversible hydration of carbon dioxide (CO2) and thereby plays a critical role in respiration, particularly in the CO2 transport by way of blood-dissolved bicarbonate (HCO3−), and in intracellular pH homeostasis by maintaining CO2/HCO3− equilibrium. Within the wide classes of CA, CA II from human is well-suited to serve as a model system for investigating the role of metal ions because its overall structure is well-refined with atomic resolution (~1.0 Å). It possesses a well-defined active site containing a single metal-binding site (Fig. 1a, b), and the kinetic rates and fine details of the enzymatic mechanism have been studied extensively17–21 (Fig. 1c).
Fig. 1. Structure of native carbonic anhydrase II (Zn-CA II) and its catalytic mechanism.
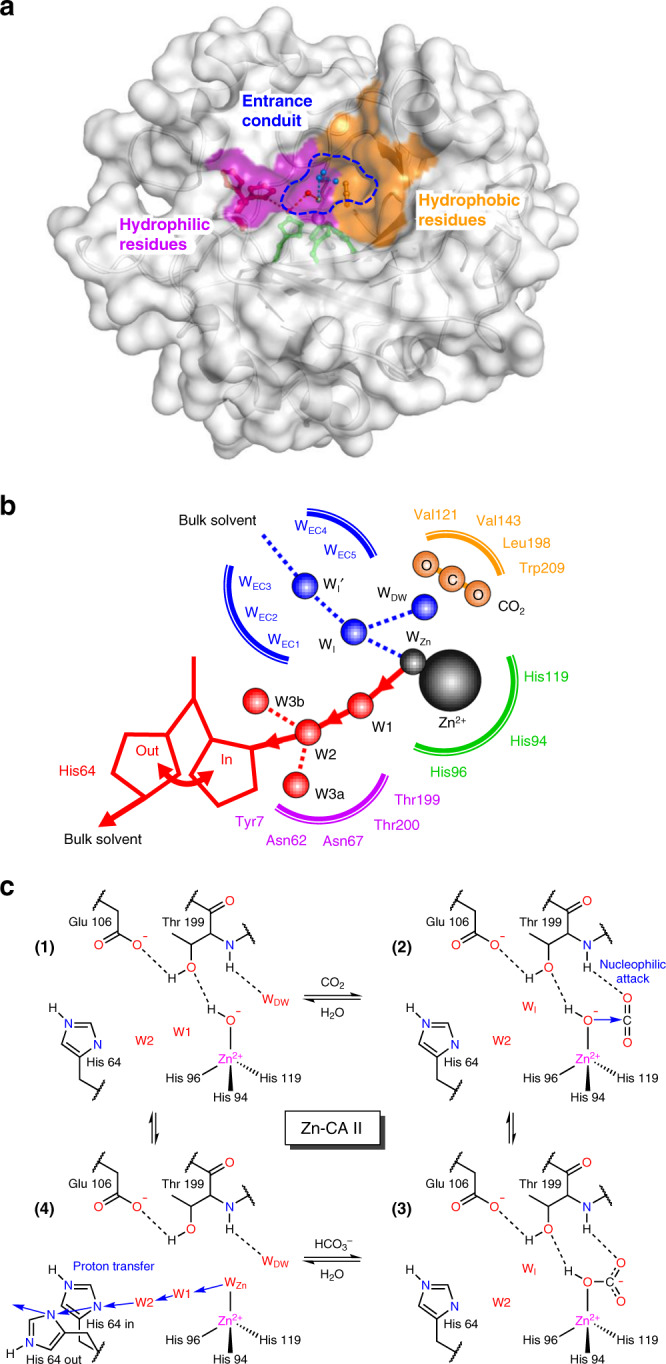
a The active site consists of zinc binding site, hydrophobic/hydrophilic regions, and entrance conduit (EC). b The water networks in the active site are responsible for the proton transfer (red) and substrate/product/water exchange (blue) during enzyme catalysis. c The CO2 hydration reaction mechanism of Zn-CA II. First, CO2 binds to the active site, leading to a nucleophilic attack by the zinc-bound hydroxyl ion onto CO2. HCO3− thus formed is subsequently displaced by the water molecule inflowing through EC. The HCO3− molecule likely binds to Zn2+ ion in a monodentate mode and its OH group is held at the Zn2+ ion due to the hydrogen bonding with Thr19952, 53. This product binding configuration leads to a weak interaction between the product and Zn2+ ion, thereby facilitating fast product dissociation54. Finally, proton transfer occurs via the network (WZn → W1 → W2 → His64) provided by the protein scaffold.
The active site of CA II is located at the base cavity of a 15 Å depth from the surface and is further subdivided into three regions comprised of hydrophobic and hydrophilic regions, with an entrance conduit (EC) in-between22–25 (Fig. 1a). These regions are responsible for substrate binding, proton transfer, and substrate/product/water exchange during catalysis, respectively (Fig. 1b). The active site zinc ion is tetrahedrally coordinated to the protein by the imidazole groups of three histidine residues, with the remaining tetrahedral site occupied by a solvent molecule (water or hydroxide ion, depending upon pH). The catalytic zinc ion in CA II serves as a Lewis acid; its primary role is to lower the pKa of the Zn-bound water from 10 to 7, allowing the formation of a zinc-bound hydroxide ion at physiological pH26. The zinc ion can be substituted by other physiologically relevant transition metal ions such as Co2+, Ni2+, Cu2+, Cd2+, and Mn2+ which results in drastic changes in the catalytic activity of CA II (~50% active to completely inactive)21. It has been also reported that the metal substitutions may induce alternative catalytic activities of CA II other than CO2/HCO3− conversion27, for instance, reduction of nitrite to nitric oxide in presence of copper28.
Previous structural studies had suggested that different metal coordination geometries in the non-native CA II may play an important role in their catalysis29,30, but no clear evidence was presented to support such a claim. Our present study focuses on investigating the detailed structural changes in CA II during the CO2/HCO3− conversion catalysis and correlating these variations to the relevant catalytic mechanisms. These experimental insights offer us a fresh peek into the origin of the activity alterations caused by non-native metal substitutions.
To study the role of metal ions in CA II, we selected four divalent transition-metal ions (Zn2+, Co2+, Ni2+, and Cu2+) that induce drastic changes in CA II activity (100%, ~50%, ~2%, and 0%, respectively)31,32. The catalytic intermediate states of the metal-free (apo, as a control) and the four metal-bound CA IIs were prepared by cryocooling protein crystals under CO2 pressures from 0 (no CO2 pressurization) to 20 atm33,34.
We show that the characteristic metal ion coordination geometries directly modulate the catalytic processes, including substrate binding, its conversion to product, and product binding. In addition, we reveal that the metal ions have a long-range (~10 Å) electrostatic effect on restructuring the water network at the active site, affecting the product displacement and the proton transfer process. The cumulative effect of such alterations provides mechanistic insights into the overall reduction of the enzymatic activity in the non-native metal-substituted CA IIs.
Results
The role of metal ion coordination geometries
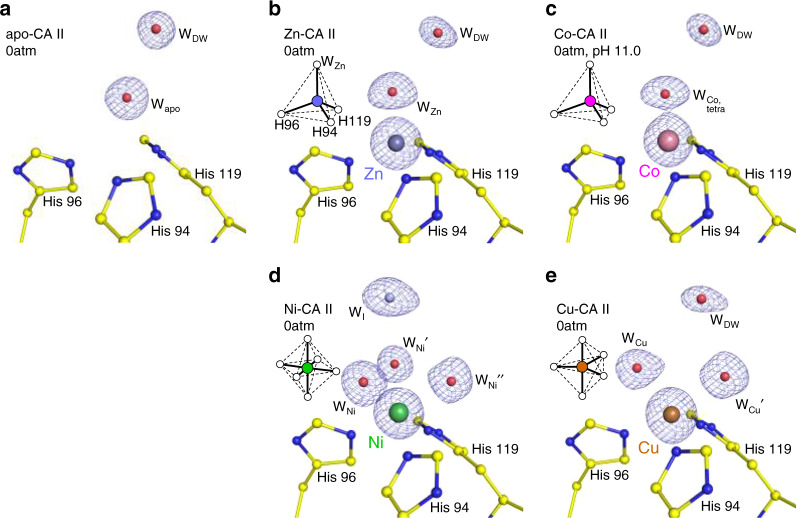
The coordination geometry around the metal binding site in CA II, when no CO2 pressure is applied, is shown in Fig. 2. The metal-free apo-CA II shows an electron density map reflecting the presence of a water molecule in the metal binding site (Fig. 2a). In Zn-CA II and Co-CA II (pH 11.0), the metal ions display tetrahedral coordination with three histidine residues (His94, His96, and His119) and a water molecule (Fig. 2b, c). In contrast, Ni-CA II contains three bound water molecules, completing an octahedral (hexa-coordinate) geometry (Fig. 2d). Finally, Cu-CA II possesses two bound water molecules, arranged in trigonal bipyramidal (penta-coordinate) geometry (Fig. 2e).
Fig. 2. Metal coordination geometry in CA II without CO2 pressurization.
a In apo-CA II, the metal binding site is vacant. b, c Zn- and Co-CA II show tetrahedral, d Ni-CA II octahedral, and e Cu-CA II trigonal bipyramidal coordination geometry. The electron density (2Fo–Fc, blue) is contoured at 2.2σ. All structures were obtained at pH 7.8 except for (c) which is obtained at pH 11.0. The intermediate water (WI) in (d) is colored in steel blue for clarity.
Next, we investigated the effect of metal coordination geometry on the efficacy of substrate (CO2) and product (HCO3−) binding. The apo- and Zn-CA II structures cryocooled at 20 atm CO2 pressure are shown in Fig. 3. The apo-CA II shows a clear binding of CO2 molecule, replacing deep water, WDW present within the active site, suggesting that the CO2 binding at the active site is mostly dictated by the metal-free protein scaffold (Fig. 3a). The native holoenzyme Zn-CA II shows CO2 binding almost identical to that in apo-CA II (Fig. 3b, c). The CO2 molecule is located 2.9 Å away from the Zn-bound water (WZn), in a configuration conducive for the nucleophilic attack (Fig. 3d).
Fig. 3. Substrate/product binding in apo- and Zn-CA II.
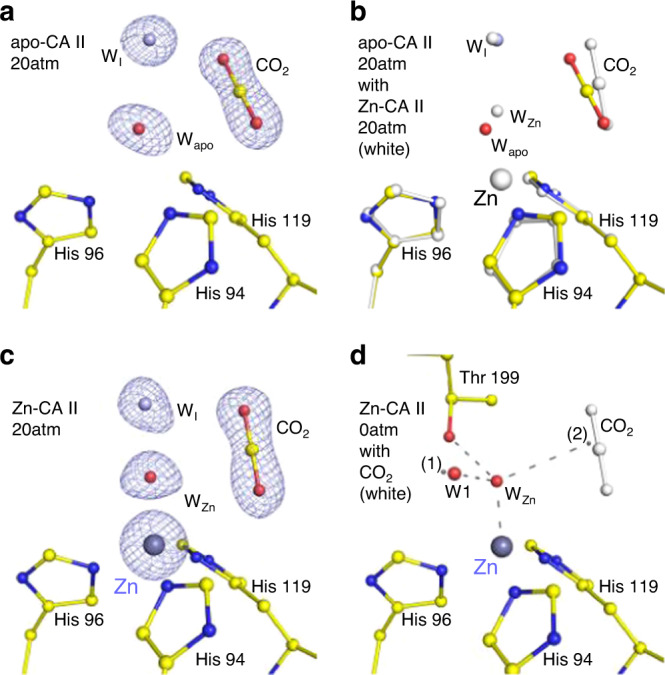
The intermediate water (WI) is colored in steel blue for clarity. The electron density (2Fo–Fc, blue) is contoured at 2.2σ. a, b At 20 atm of CO2 pressure, apo-CA II shows clear binding of CO2 without the need of Zn2+ ion. c Zn-CA II shows similar binding of CO2 as in apo-CA II while maintaining tetrahedral metal coordination. d Upon CO2 binding (white) in Zn-CA II, WZn is located at the center of the hypothetical tetrahedral arrangement made up of Zn2+ ion, Thr199-Oγ1, position (1) (close to W1), and position (2) (close to the carbon atom in CO2). In this configuration, a hybridized lone pair in WZn directly faces CO2 molecule at a distance, appropriate for efficient nucleophilic attack. Distance between the position (2) and C atom of CO2 is merely 0.36 Å.
In Co-CA II (pH 11.0) cryocooled at 20 atm CO2 pressure, dual binding of CO2 and HCO3− is observed (Fig. 4a). Upon CO2 binding, the tetrahedral coordination is maintained, but an unusual expansion to octahedral coordination is observed upon HCO3− binding (Fig. 4b). In the transformed octahedral geometry, the HCO3− molecule is bound in a bidentate mode to the Co2+ ion along with an additional water molecule. Compared to the monodentate binding mode in Zn-CA II, the negative charge on the bidentate HCO3− can be distributed among the two oxygen atoms bound to Co2+ ion, allowing stronger product binding to the metal ion (Supplementary Fig. 1a–f). Unlike Zn-CA II (Supplementary Fig. 2), the Co-CA II intermediates obtained at different pH values (7.8 and 11.0) reveal that the HCO3− molecule is firmly bound to Co2+ ion with full occupancy at lower pH (Fig. 4c, d), but this binding affinity weakens as pH increases (Fig. 4a). The result suggests that, during the catalytic cycle, deprotonation of the Co2+-bound water may lead to dissociation of the HCO3− molecule from the Co2+ ion, due to the charge–charge repulsion between the formed hydroxide ion and the HCO3− molecule. Following the HCO3− dissociation, the tetrahedral coordination is restored (Fig. 2c).
Fig. 4. Substrate/product binding in Co-CA II.
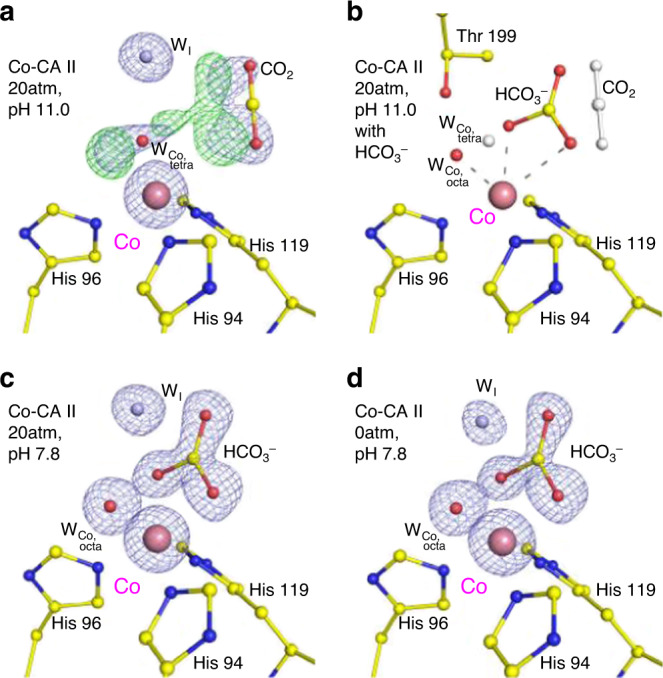
The intermediate water (WI) is colored in steel blue for clarity. The electron density (2Fo–Fc, blue) and the difference map (Fo–Fc, green) are contoured at 2.2σ and 7.0σ, respectively. a, b At 20 atm of CO2 pressure, Co-CA II at pH 11.0 shows superposition of CO2 binding (~50% occupancy, white) with tetrahedral coordination and HCO3− binding (~50% occupancy) with octahedral coordination. c, d Co-CA II at pH 7.8 shows complete binding of HCO3−, showing octahedral coordination even in absence of added CO2. It is likely that the captured HCO3− is converted from the CO2 absorbed in the crystal from ambient air.
On the other hand, at 20 atm CO2 pressure, Ni-CA II shows octahedral coordination comprising the bidentate HCO3− and a water molecule in a similar manner to that of Co-CA II (Fig. 5a, Supplementary Fig. 1g–i). It is noted that one of the three bound water molecules experiences steric hindrance with the CO2-binding configuration in Zn-CA II (Fig. 5b). Thus, it is likely that the CO2 molecule entering the active site pushes away one of the Ni-bound water molecules, and then a nucleophilic attack occurs from one of the two remaining water molecules, forming HCO3−. Unlike Co-CA II (Fig. 4a, c, d), the Ni-CA II intermediates obtained at different pH values (7.8 and 11.0) indicate that the HCO3− binding affinity is almost unresponsive to pH variation (Supplementary Fig. 3). The result suggests that the deprotonation of the Ni2+-bound water is insufficient to facilitate HCO3− dissociation in the stable octahedral coordination, and that the bound HCO3− is directly displaced by two incoming water molecules in Ni-CA II. Finally, in Cu-CA II, no clear electron density of CO2 or HCO3− is visible (Fig. 5c). The faint and diffused electron density suggests that a CO2 molecule encounters a severe steric hindrance from one of Cu2+-bound water molecules. Even if the CO2 molecule adopts proper orientation as in Zn-CA II, the bound CO2 position remains far too distant (3.9 Å) from the spare Cu2+-bound water molecule for any effective interaction. Moreover, the significantly distorted geometry negates the scope of any nucleophilic attack (Fig. 5d). The inefficient substrate binding and the unfavorable distorted geometry explain the complete enzymatic inactivity of Cu-CA II.
Fig. 5. Substrate/product binding in Ni- and Cu-CA II.
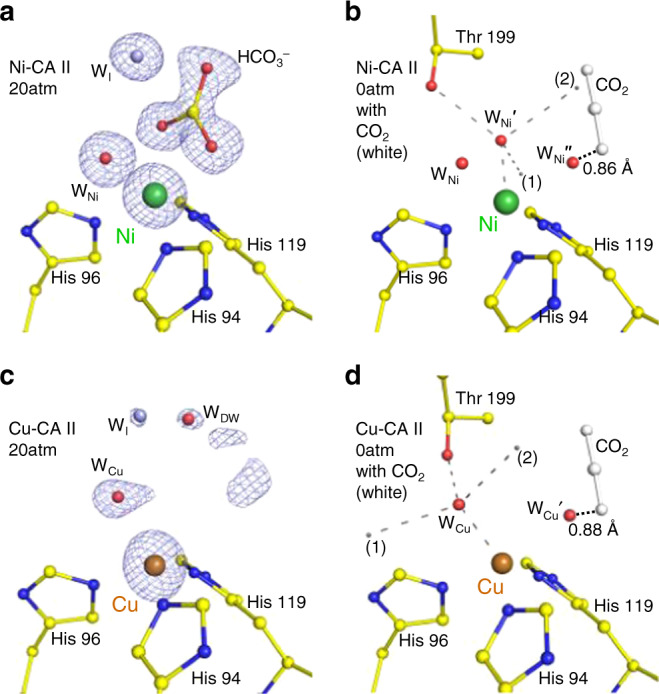
The intermediate water (WI) is colored in steel blue for clarity. The electron density (2Fo–Fc, blue) is contoured at 2.2σ. a At 20 atm of CO2 pressure, Ni-CA II maintains octahedral coordination with HCO3− binding. b Compared to the WZn geometry in Zn-CA II (Fig. 3d), the nucleophilic attack geometry around WNi′ has steric hindrance on CO2 molecule (adapted from Zn-CA II, 20 atm, white) and is distorted away. Distance between the position (2) and C atom of CO2 is 1.55 Å. c Cu-CA II shows only disordered electron density in the CO2/HCO3− binding site. d The nucleophilic attack geometry around WCu has steric hindrance on CO2 molecule (adapted from Zn-CA II, 20 atm) and is significantly distorted away. Distance between the position (2) and C atom of CO2 is 2.93 Å.
Electrostatic effects of metal ions on active-site water network
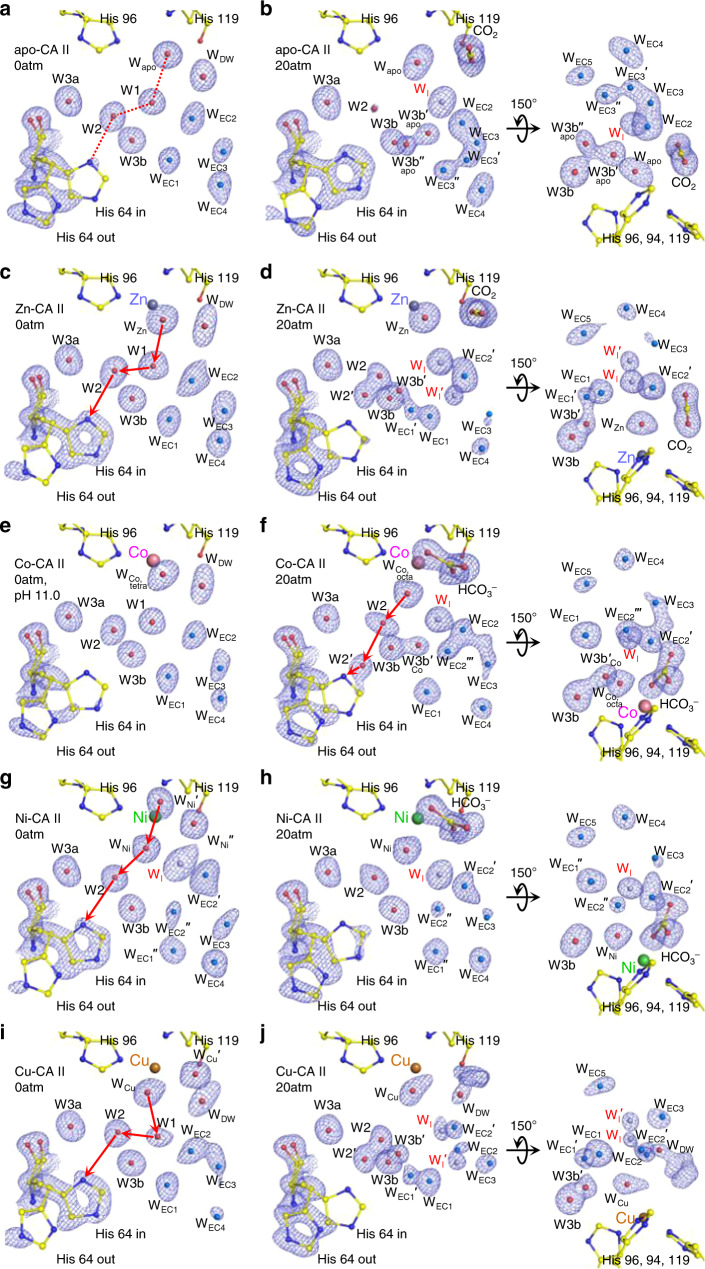
Figure 6 shows the proton transfer pathway and the water network in the EC of CA II. The metal-free apo-CA II shows the well-defined pathway (WApo → W1 → W2 → His64, Fig. 6a) in the absence of CO2. Upon CO2 binding, the pathway is disrupted in such a way that W1 disappears and an intermediate water WI emerges (Figs. 3a and 6b). Zn-CA II largely resembles the apoenzyme in terms of both the same well-defined pathway (WZn → W1 → W2 → His64) utilized for proton transfer (Fig. 6c) and the dynamics of W1/WI upon CO2 binding (Figs. 3c and 6d). This observation clearly suggests that the primary water network necessary for the proton transfer is organized by the protein scaffold without the need for metal ions. However, in comparison to apo-CA II (Fig. 6b), Zn-CA II shows significantly modified dynamics of W2 and His64, which are believed to be critical for efficient proton transfer (Fig. 6d). Additionally, in Zn-CA II, the significantly modified dynamics of the EC waters stabilizes another intermediate water WI′, that in turn bridges WI with the bulk solvent outside the protein, thereby facilitating the replenishment of WZn and W1 during the catalytic cycle (Figs. 1b and 6d). These observations suggest that the Zn2+ ion produces a long-range (~10 Å) electrostatic field in which water structure and dynamics in the active site are fine-tuned to facilitate the proton transfer and the water/substrate/product exchange.
Fig. 6. Active site in CA II showing proton transfer pathway and EC water network (WEC1 ~ WEC5).
The electron density (2Fo–Fc) is contoured at 1.7σ except for EC waters at 1.5σ. The EC waters are colored in aqua marine and the intermediate waters (WI and WI′) in steel blue for clarity. W2′ is an alternative position of W2. The possible proton transfer pathways in the metal-CA IIs are depicted as red arrows. All structures were obtained at pH 7.8 except for e at pH 11.0. a, b Apo-CA II shows well-ordered water arrangement (dotted red line) with His64 favored in outward conformation at 0 atm CO2 pressure. Upon CO2 binding, His64 moves inward and water molecules show highly dynamical motions, stabilizing WI. c, d Zn-CA II shows His64 favored in inward conformation at 0 atm CO2 pressure. Upon CO2 binding, W2, W3b, and WEC waters show significantly different dynamics with His64 moving outward, and an additional intermediate water (WI′) is stabilized with the WEC molecules. The motions of W3b and WEC1 turn on the dynamic interplay between the proton transfer and EC water networks. e, f Co-CA II shows similar arrangement initially as in Zn-CA II. However, upon full HCO3− binding, the dynamical motions of EC waters are different and the intermediate water WI′ is less stabilized. Note that, in Co-CA II, proton transfer seems to occur while the product is still bound. g, h Ni-CA II initially shows altered water arrangements due to octahedral coordination. Upon HCO3− binding, significantly reduced water dynamical motions are recognized. i, j Cu-CA II shows unexpectedly similar dynamical motions of active site waters and His64 as in Zn-CA II.
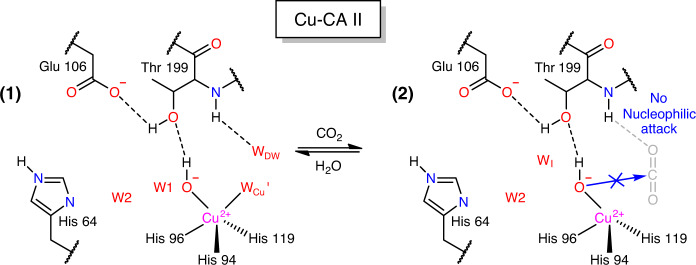
In the absence of CO2, Co-CA II forms the same proton transfer network as in Zn-CA II (Fig. 6e). Once CO2 or HCO3− binds, W1 disappears and WI appears like what happens in Zn-CA II (Figs. 4a, c, and 6f). However, as the deprotonation of Co2+-bound water should occur prior to the HCO3− dissociation, it is likely that the proton transfer occurs via the altered network (WCo,octa → W2 → His64) while the product is still bound to the Co2+ ion (Fig. 6f). In addition, Co-CA II shows modified dynamics of W2, His64, and EC waters as compared to Zn-CA II (Fig. 6f). Meanwhile, in Ni-CA II, octahedral coordination is stabilized throughout the entire catalytic cycle, and consequently, W1 is absent due to its steric hindrance with one of the Ni-bound water molecules (Figs. 2d, 5a, and 6g). Based on the CO2 binding configuration in Zn-CA II and the bidentate HCO3− binding observed in Ni-CA II, it is most likely that the substrate-to-product conversion occurs via the nucleophilic attack from WNi′ to CO2 (Fig. 5b). This result suggests that the proton transfer occurs possibly via the modified pathway WNi′ → WNi → W2 → His64 (Fig. 6g). Also, unlike Zn-CA II and Co-CA II, W2 in Ni-CA II shows significantly different dynamics and WI′ is destabilized, a plausible reflection of the altered electrostatic environment (Fig. 6h). Finally, Cu-CA II reveals that the possible proton transfer pathway (WCu → W1 → W2 → His64) is well-defined (Fig. 6i) and W2 and His64 dynamics is surprisingly similar to that in Zn-CA II (Fig. 6j). This result corroborates well with our conjecture that it is the lack of efficient substrate binding and unfavorable distorted geometry for the nucleophilic attack that are responsible for the complete inactivity of Cu-CA II.
Discussion
Our results provide advanced insights into the role of metal ions and the metal-protein relationship for the CA II catalytic mechanism. In the absence of metal ions, the protein scaffold provides a fundamental structural template necessary for the catalytic activity. The protein scaffold helps usher a substrate molecule from the outside bulk solvent into the active site through desolvating and positioning it at a configuration conducive for nucleophilic attack. The protein scaffold also provides well-ordered water networks in the vicinity of the active site, which can be utilized for proton transfer and substrate/product/water exchange. Metal ions then bring a key property for the CA II catalytic activity in generating hydroxyl ion at neutral pH and retaining it at the active site. Beyond their primary Lewis acid property, metal ions are directly involved in the catalytic mechanism via their coordination geometry and long-range electrostatic effects. The most efficient native Zn-CA II preserves a tetrahedral coordination and fine-tunes the water network embedded within the protein scaffold (Fig. 1c). The tetrahedral coordination allows efficient conversion of substrate into product, and the long-range electrostatic field orchestrates the structure and dynamics of water network in the active site, imperative for the rapid product displacement and fast proton transfer. In comparison, semi-efficient Co-CA II shows similar catalytic behavior up to the product formation stage as in Zn-CA II, but the expansion of the metal coordination geometry from tetrahedron to octahedron during the catalytic cycle alters the product displacement and proton transfer process (Fig. 7). The significantly less efficient Ni-CA II maintains octahedral coordination and shows altered electrostatic effects, hampering efficient conversion from substrate to product, product displacement, and proton transfer (Fig. 8). Finally, completely inactive Cu-CA II suggests substantial steric hindrance encountered by the substrate in the active site and poor geometry for product conversion due to the trigonal bipyramidal coordination (Fig. 9).
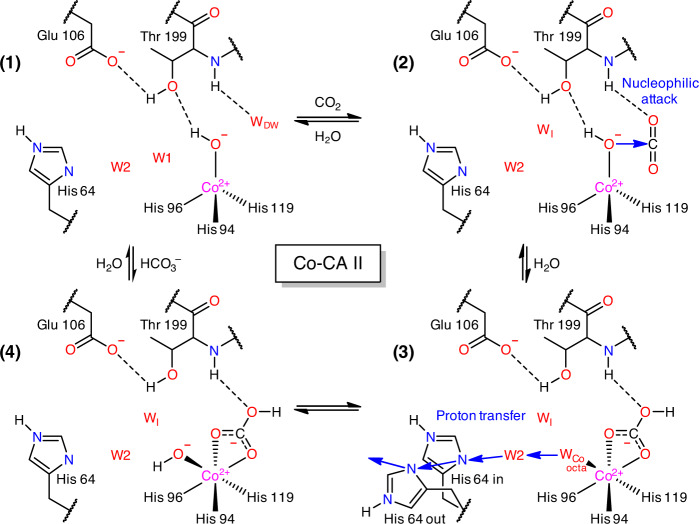
Fig. 7. Proposed catalytic mechanism of Co-CA II.
In Co-CA II, CO2 binding and the catalytic conversion of CO2 to HCO3− occur in the same way as in the Zn-CA II with tetrahedral geometry. However, the HCO3− displacement and proton transfer process are significantly altered due to the coordination expansion to octahedral geometry during catalysis. This octahedral coordination allows bidentate binding mode of HCO3− and reorganization of negative charge of HCO3− toward Co2+ ion, allowing stronger HCO3− binding to metal ion. To dissociate the product, proton transfer first occurs via an altered pathway (possibly, WCo,octa → W2 → His64) and WCo,octa is converted into the hydroxyl ion. This negatively charged hydroxyl ion then pushes away the bound product, and the tetrahedral coordination is restored for the next catalytic cycle.
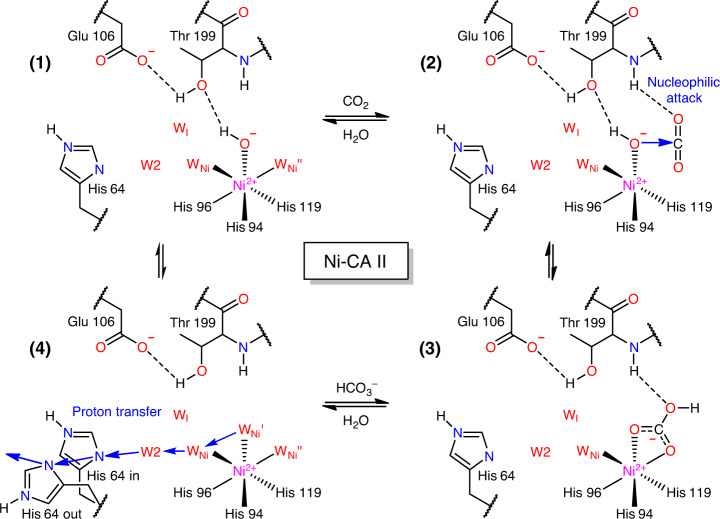
Fig. 8. Proposed catalytic mechanism of Ni-CA II.
In Ni-CA II, octahedral coordination is maintained throughout the whole catalytic cycle. The significant consequence is that one of the three bound water molecules experiences steric hindrance with the CO2 binding. In addition, the nucleophilic attack geometry is distorted (Fig. 5b), suggesting less efficient conversion into HCO3−. The formed HCO3− is strongly bound to Ni2+ ion in a bidentate mode as in the Co-CA II but is directly displaced by two inflowing water molecules. Finally, proton transfer occurs via an altered network (possibly, WNi′ → WNi → W2 → His64) to restore the catalytic cycle.
Fig. 9. Proposed catalytic mechanism of Cu-CA II.
In Cu-CA II, one of the two bound water molecules in the trigonal bipyramid coordination experiences significant steric hindrance from CO2 molecule, hindering adoption of proper configuration for nucleophilic attack. In addition, even if CO2 binds to the active site temporarily, the nucleophilic attack geometry is too distant (3.9 Å) and significantly distorted (Fig. 5d).
In conclusion, we examined the role of various metal ions in carbonic anhydrase catalysis beyond their primary chemical property as a Lewis acid. We demonstrated that metal ions are directly involved in the enzymatic mechanism via their coordination geometry and long-range electrostatics to orchestrate intricate water dynamics. Our experimental results can be used as direct input for theoretical and computational studies on the role of metal ions, which we anticipate could open a new window to the study of metal–protein relationships, drug discovery targeting metalloenzymes, engineering of natural metalloenzymes, rational design of de novo metalloenzymes, and synthesis of supramolecular analogues to metalloenzymes.
Methods
Protein expression and purification
The native Zn-CA II was expressed in a recombinant strain of Escherichia coli [BL21 (DE3) pLysS] containing a plasmid encoding the CA II gene35. Purification was carried out using affinity chromatography36. Briefly, bacterial cells were enzymatically lysed with hen egg white lysozyme, and the lysate was placed onto an agarose resin coupled with p-(aminomethyl)-benzene-sulfonamide which binds CA II. The protein on the resin was eluted with 0.4 M sodium azide, in 100 mM Tris-HCl pH 7.0. The azide was removed by extensive buffer exchange against 10 mM Tris-HCl pH 8.0.
Apo-CA II (zinc free) was then prepared by incubating Zn-CA II in a zinc chelation buffer (100 mM pyridine 2,6-dicarboxylic acid, 25 mM MOPS pH 7.0) at 20 °C for 18 h. The resulting protein was then run through an affinity column with benzylsulfonamide resin to remove residual Zn-CA II. The chelating agent was then removed by buffer-exchange against 50 mM Tris-HCl pH 7.818. The loss of zinc ion was examined using the esterase kinetic assay and further confirmed in the crystallographic structure. The enzyme activity was revived by an addition of 1 mM ZnCl2.
Esterase kinetic assay
The CO2/HCO3− conversion catalytic activity of CA II can be measured directly by stopped flow assays, monitoring labeled CO2/HCO3− conversion using mass spectroscopy, or indirectly by monitoring the innate esterase activity spectroscopically37,38. In this study, the esterase activity assays were performed as a control to ensure zinc was fully chelated from recombinant CA II. The 4-nitrophenyl acetate (4-NPA) molecule is cleavable by CA II and thus used here as a colorimetric substrate. CA II cleaves the ester bond of 4-NPA generating 4-nitrophenol, which is spectroscopically absorbent at 348 nm in the ultraviolet–visible spectrum. Thus, the reaction can be monitored spectroscopically at 348 nm39.
In a 96 deep-well plate, aliquots of 50 μL of 0.1 mg mL−1 CA II in storage buffer were added to each well. To initiate the reaction, 200 μL of 0.8 mM 4-NPA dissolved in 3% acetone in water was added to the sample well. The well plate was then immediately inserted into the plate reader (Synergy HTX, BioTek, Winooski, WI, USA). Absorbance at 348 nm was recorded every 8 s for 10 min. The absorbance data of Apo- and Zn-CA II are plotted in Supplementary Fig. 4.
Crystallization and non-native metal substitution
Crystals of CA II were obtained using the hanging drop vapor diffusion method40. A 10 μl drop of equal volumes of protein (5 μl) and the well-solution (5 μl) was equilibrated against 500 μl of the well-solution (1.3 M sodium citrate, 50 mM Tris-HCl pH 7.8) at RT (~20 °C)41. Crystals grew to an approximate ~30 × 100 × 200 μm3 in size in a few days. To prepare non-native metal substituted CA II, the apo-CA II crystals were transferred into soaking solutions of cobalt, nickel and copper salt (100 mM CoCl2, 100 mM NiCl2, 10 mM CuCl2 along with 1.3 M sodium citrate, 50 mM Tris-HCl with pH 7.8). The crystals were incubated for 2–3 days to let the Co2+, Ni2+ and Cu2+ ions infuse into the active site42. The CA II crystals at pH 11.0 were obtained with 3-(cyclohexylamino)propanesulfonic acid buffer instead of using Tris-HCl.
Cryocooling under CO2 pressure
Cryo-trapping the intermediate states of Zn-CA II was previously achieved by cryocooling CA II crystals under CO2 pressure33,34, leading to the capture of CO2 in the active site of CA II43. More recently, series of intermediate states have been tracked in CA II by controlling the internal CO2 pressure levels25,44. In this study, the CO2 entrapment was carried out using a high-pressure cryo-cooler for X-ray crystallography (HPC-201, Advanced Design Consulting, USA). The apo-, Zn-, Co-, Ni-, and Cu-CA II crystals were first soaked in a cryo-solution containing 35% (v/v) glycerol supplemented to the soaking solution. The crystals were then coated with mineral oil to prevent dehydration, and loaded into the base of high-pressure tubes33. The coated mineral oil worked as a CO2 buffering medium as well, aiding in the absorption of CO2 into the crystals45. The crystals were pressurized at room temperature in the pressure tubes with CO2 gas at 0 atm (no pressurization) and 20 atm. After a wait of about 5 min, the crystals were cryocooled in liquid nitrogen (77 K). Once the CO2 bound crystals were fully cryocooled, the CO2 gas pressure was withdrawn, and the crystal samples were stored in a liquid nitrogen dewar for subsequent X-ray data collection.
X-ray diffraction and data collection
Diffraction data were collected at Pohang Light Source II (wavelength of 0.9793 Å, beam size of 100 μm) under nitrogen cold stream (100 K). Data were collected using the oscillation method in intervals of 1° step on an ADSC Quantum 270 CCD detector (Area Detector Systems Corporation, USA) with a crystal-to-detector distance of 120 mm. A total of 360 images were collected on each of the CA II crystal data sets.
For each data set, a new fresh pressure-cryocooled crystal was used. The absorbed X-ray dose for a single data set was less than 5 × 105 Gy, which is much less than the Henderson dose limit of 1.2 × 107 Gy46. Moreover, we have checked that X-ray radiation dose at least up to 107 Gy does not induce apparent changes in the active site. The result confirms that the active site structures described in our study are unaffected by the X-ray radiation. Indexing, integration, and scaling were performed by using HKL200047. The data processing statistics are given in Supplementary Table 1.
Structure determination and model refinement
The CA II structures were determined using the CCP4 program suite48. Prior to refinement, a random 5% of the data were flagged for Rfree analysis. The previously reported crystal structures (PDB codes of 5DSR and 5YUK for apo- and metal substituted CA II) were used as the initial phasing models25,49. The maximum likelihood refinement (MLH) was carried out using REFMAC550. The refined structures were manually checked using the molecular graphics program COOT51. Reiterations of MLH were carried out with anisotropic B factor.
On completion of the structural refinements as described above, systematic refinements were further carried out to accurately determine the partial occupancies of the His 64 in and the His 64 out configurations. A total of 99 structures were prepared for each of the CA II structures, in which the occupancies of the His 64 in and the His 64 out configurations were changed in incremental steps of 1% (i.e., the first structure with 1% in and 99% out, the second structure with 2% in and 98% out, …, the 99th structure with 99% in and 1% out). MLH refinements were carried out in parallel for all the 99 structures. After MLH refinements, the overall R-factor as a function of partial occupancy of the His 64 in configuration was obtained, and it was fitted into a quadratic function (Supplementary Fig. 5). The partial occupancy values of the His 64 configurations were determined where the overall R-factor is minimized. Details on the final refinement statistics are given in Supplementary Table 1. All structural figures were rendered with PyMol (Schrödinger, LLC).
Structural analysis of the bound water molecules
To compare the bound water molecules in the active site and the EC, we carefully refined water molecules based on the PDB and COOT validation checks and the electron density maps (cutoff level of 1σ in 2Fo–Fc electron density map). We have tested the consistency and reproducibility of the bound water molecules in the active site and the EC carefully. There were several closely positioned water molecules in the active site and the EC of the CA II structures. Since most of these waters exist transiently, it was allowed that they can be located closer than the normal stably bound water molecules. In this regard, water molecules closely located near the active site and EC regions were not excluded in the final coordinates. The important bound water molecules addressed in the main paper are listed in Supplementary Table 2. The distance information between CO2, HCO3−, Thr199, and important water molecules is listed in Supplementary Table 3.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors would like to thank the staff at Pohang Light Source II for their support in data collection. This work was initiated by the support of Samsung Science and Technology Foundation (SSTF-BA1702-04) and further supported by the National Research Foundation of Korea (NRF) grant (NRF-2019R1A2C1004274) funded by the Korea government (MSIT).
Source data
Author contributions
C.U.K. conceived the research, J.K.K., C.L., S.W.L., J.T.A. ran the experiments, J.K.K. and C.U.K. analysed the data. J.K.K., A.A., R.M., C.-M.G., and C.U.K. wrote the paper. All authors contributed to the overall scientific interpretation and edited the paper.
Data availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://wwpdb.org/) as [PDB code 6LUU [10.2210/pdb6luu/pdb] (0 atm CO2 pressure, pH 7.8), 6LUV [10.2210/pdb6luv/pdb] (20 atm, pH 7.8)] for apo-CA II, [6LUW [10.2210/pdb6luw/pdb] (0 atm, pH 7.8), 6LUX [10.2210/pdb6lux/pdb] (20 atm, pH 7.8), 6LUY [10.2210/pdb6luy/pdb] (0 atm, pH 11.0), 6LUZ [10.2210/pdb6luz/pdb] (20 atm, pH 11.0)] for Zn-CA II, [6LV1 [10.2210/pdb6lv1/pdb] (0 atm, pH 7.8), 6LV2 [10.2210/pdb6lv2/pdb] (20 atm, pH 7.8), 6LV3 [10.2210/pdb6lv3/pdb] (0 atm, pH 11.0), 6LV4 [10.2210/pdb6lv4/pdb] (20 atm, pH 11.0)] for Co-CA II, [6LV5 [10.2210/pdb6lv5/pdb] (0 atm, pH 7.8), 6LV6 [10.2210/pdb6lv6/pdb] (20 atm, pH 7.8), 6LV7 [10.2210/pdb6lv7/pdb] (0 atm, pH 11.0), 6LV8 [10.2210/pdb6lv8/pdb] (20 atm, pH 11.0)] for Ni-CA II, and [6LV9 [10.2210/pdb6lv9/pdb] (0 atm, pH 7.8), 6LVA [10.2210/pdb6lva/pdb] (20 atm, pH 7.8)] for Cu-CA II. Two earlier structures [5DSR [10.2210/pdb5dsr/pdb] and 5YUK [10.2210/pdb5yuk/pdb]] were used for structure determination. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Ian Davis and the other, anonymous reviewer(s) for their contribution to the peer review of this week. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-18425-5.
References
- 1.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. CA: University Science Books Mill Valley; 1994. [Google Scholar]
- 2.Holm RH, Kennepohl P, Solomon EI. Structural and functional aspects of metal sites in biology. Chem. Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 3.Ragsdale SW. Metals and their scaffolds to promote difficult enzymatic reactions. Chem. Rev. 2006;106:3317–3337. doi: 10.1021/cr0503153. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, et al. Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem. Rev. 2014;114:4366–4469. doi: 10.1021/cr400479b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rulisek L, Vondrasek J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J. Inorg. Biochem. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 6.Dudev T, Lim C. Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem. Rev. 2003;103:773–788. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- 7.Parr RG, Pearson RG. Absolute hardness—companion parameter to absolute electronegativity. J. Am. Chem. Soc. 1983;105:7512–7516. [Google Scholar]
- 8.Irving H, Williams RJP. Order of stability of metal complexes. Nature. 1948;162:746–747. [Google Scholar]
- 9.Sigel H, Mccormic DB. On discriminating behavior of metal ions and ligands with regard to their biological significance. Acc. Chem. Res. 1970;3:201. [Google Scholar]
- 10.Parkin G. Synthetic analogues relevant to the structure and function of zinc enzymes. Chem. Rev. 2004;104:699–767. doi: 10.1021/cr0206263. [DOI] [PubMed] [Google Scholar]
- 11.Valdez CE, Smith QA, Nechay MR, Alexandrova AN. Mysteries of metals in metalloenzymes. Acc. Chem. Res. 2014;47:3110–3117. doi: 10.1021/ar500227u. [DOI] [PubMed] [Google Scholar]
- 12.Spiro, T. G. Zinc Enzymes. (J. Wiley New York, 1983).
- 13.Davenport HW. The early days of research on carbonic anhydrase. Ann. N. Y. Acad. Sci. 1984;429:4–9. doi: 10.1111/j.1749-6632.1984.tb12310.x. [DOI] [PubMed] [Google Scholar]
- 14.Chegwidden, W. R., Carter, N. D. & Edwards, Y. H. The Carbonic Anhydrases: New Horizons. Vol. 90 (Birkhäuser, 2013).
- 15.Frost, S. C. & McKenna, R. Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications. Vol. 75 (Springer Science & Business Media, 2013).
- 16.Supuran, C. T. & De Simone, G. Carbonic anhydrases as biocatalysts: from theory to medical and industrial applications. (Elsevier, 2015).
- 17.Liang JY, Lipscomb WN. Theoretical-study of carbonic anhydrase-catalyzed hydration of Co2—a brief review. Int. J. Quantum Chem. 1989;36:299–312. [Google Scholar]
- 18.Håkansson K, Carlsson M, Svensson LA, Liljas A. Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes. J. Mol. Biol. 1992;227:1192–1204. doi: 10.1016/0022-2836(92)90531-n. [DOI] [PubMed] [Google Scholar]
- 19.Lindskog, S. & Silverman, D. N. in The Carbonic Anhydrases 175–195 (Springer, 2000). [DOI] [PubMed]
- 20.Silverman DN, McKenna R. Solvent-mediated proton transfer in catalysis by carbonic anhydrase. Acc. Chem. Res. 2007;40:669–675. doi: 10.1021/ar7000588. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamurthy VM, et al. Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein-ligand binding. Chem. Rev. 2008;108:946–1051. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liljas A, et al. Crystal structure of human carbonic anhydrase C. Nat. N. Biol. 1972;235:131–137. doi: 10.1038/newbio235131a0. [DOI] [PubMed] [Google Scholar]
- 23.Eriksson A, Kylsten P, Jones T, Liljas A. Proteins: structures and functions. Genetics. 1988;4:283–293. doi: 10.1002/prot.340040407. [DOI] [PubMed] [Google Scholar]
- 24.Christianson DW, Fierke CA. Carbonic anhydrase: evolution of the zinc binding site by nature and by design. Acc. Chem. Res. 1996;29:331–339. [Google Scholar]
- 25.Kim JK, et al. Active-site solvent replenishment observed during human carbonic anhydrase II catalysis. IUCrJ. 2018;5:93–102. doi: 10.1107/S2052252517017626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCall KA, Huang C, Fierke CA. Function and mechanism of zinc metalloenzymes. J. Nutr. 2000;130:1437S–1446S. doi: 10.1093/jn/130.5.1437S. [DOI] [PubMed] [Google Scholar]
- 27.Piazzetta P, Marino T, Russo N, Salahub DR. The role of metal substitution in the promiscuity of natural and artificial carbonic anhydrases. Coord. Chem. Rev. 2017;345:73–85. [Google Scholar]
- 28.Andring JT, Kim CU, McKenna R. Structure and mechanism of copper-carbonic anhydrase II: a nitrite reductase. IUCrJ. 2020;7:287–293. doi: 10.1107/S2052252520000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hakansson K, Wehnert A. Structure of cobalt carbonic anhydrase complexed with bicarbonate. J. Mol. Biol. 1992;228:1212–1218. doi: 10.1016/0022-2836(92)90327-g. [DOI] [PubMed] [Google Scholar]
- 30.Hakansson K, Wehnert A, Liljas A. X-ray analysis of metal-substituted human carbonic anhydrase II derivatives. Acta Crystallogr. D Biol. Crystallogr. 1994;50:93–100. doi: 10.1107/S0907444993008790. [DOI] [PubMed] [Google Scholar]
- 31.Lindskog S, Nyman PO. Metal-binding properties of human erythrocyte carbonic anhydrases. Biochim. Biophys. Acta. 1964;85:462–474. doi: 10.1016/0926-6569(64)90310-4. [DOI] [PubMed] [Google Scholar]
- 32.Coleman JE. Metal ion dependent binding of sulphonamide to carbonic anhydrase. Nature. 1967;214:193–194. doi: 10.1038/214193a0. [DOI] [PubMed] [Google Scholar]
- 33.Kim CU, Kapfer R, Gruner SM. High-pressure cooling of protein crystals without cryoprotectants. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005;61:881–890. doi: 10.1107/S090744490500836X. [DOI] [PubMed] [Google Scholar]
- 34.Kim CU, Wierman JL, Gillilan R, Lima E, Gruner SM. A high-pressure cryocooling method for protein crystals and biological samples with reduced background X-ray scatter. J. Appl. Crystallogr. 2013;46:234–241. doi: 10.1107/S0021889812045013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forsman C, Behravan G, Osterman A, Jonsson B-H. Production of active human carbonic anhydrase ll in E. coli. Acta Chem. Scand. 1988;42:314–318. doi: 10.3891/acta.chem.scand.42b-0314. [DOI] [PubMed] [Google Scholar]
- 36.Khalifah RG, Strader DJ, Bryant SH, Gibson SM. Carbon-13 nuclear magnetic resonance probe of active-site ionizations in human carbonic anhydrase B. Biochemistry. 1977;16:2241–2247. doi: 10.1021/bi00629a031. [DOI] [PubMed] [Google Scholar]
- 37.Coleman JE. Mechanism of action of carbonic anhydrase—substrate sulfonamide and anion binding. J. Biol. Chem. 1967;242:5212–5219. [PubMed] [Google Scholar]
- 38.Tu C, Silverman D. Catalysis of cobalt (II)-substituted carbonic anhydrase II of the exchange of oxygen-18 between carbon dioxide and water. Biochemistry. 1985;24:5881–5887. doi: 10.1021/bi00342a029. [DOI] [PubMed] [Google Scholar]
- 39.Tashian RE, Douglas DP, Yu YS. Esterase and hydrase activity of carbonic anhydrase. I. From primate erythrocytes. Biochem. Biophys. Res. Commun. 1964;14:256–261. doi: 10.1016/0006-291x(64)90445-0. [DOI] [PubMed] [Google Scholar]
- 40.McPherson, A. Preparation and Analysis of Protein Crystals. (John Wiley & Sons, 1982).
- 41.Fisher SZ, et al. Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: insights into the proton transfer mechanism. Biochemistry. 2007;46:2930–2937. doi: 10.1021/bi062066y. [DOI] [PubMed] [Google Scholar]
- 42.Avvaru BS, et al. Comparison of solution and crystal properties of Co(II)-substituted human carbonic anhydrase II. Arch. Biochem. Biophys. 2010;502:53–59. doi: 10.1016/j.abb.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Domsic JF, et al. Entrapment of carbon dioxide in the active site of carbonic anhydrase II. J. Biol. Chem. 2008;283:30766–30771. doi: 10.1074/jbc.M805353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim CU, et al. Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals. Proc. Natl Acad. Sci. USA. 2016;113:5257–5262. doi: 10.1073/pnas.1520786113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim CU, Hao Q, Gruner SM. Solution of protein crystallographic structures by high-pressure cryocooling and noble-gas phasing. Acta Crystallogr. D Biol. Crystallogr. 2006;62:687–694. doi: 10.1107/S0907444906014727. [DOI] [PubMed] [Google Scholar]
- 46.Henderson R. Cryo-protection of protein crystals against radiation damage in electron and X-ray diffraction. Proc. R. Soc. B. 1990;241:6–8. [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Collaborative CP. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.West D, et al. Structural and kinetic effects on changes in the CO(2) binding pocket of human carbonic anhydrase II. Biochemistry. 2012;51:9156–9163. doi: 10.1021/bi301155z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Krebs JF, Ippolito JA, Christianson DW, Fierke CA. Structural and functional importance of a conserved hydrogen bond network in human carbonic anhydrase II. J. Biol. Chem. 1993;268:27458–27466. [PubMed] [Google Scholar]
- 53.Xue Y, Liljas A, Jonsson BH, Lindskog S. Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II. Proteins. 1993;17:93–106. doi: 10.1002/prot.340170112. [DOI] [PubMed] [Google Scholar]
- 54.Liljas A. Carbonic anhydrase under pressure. IUCrJ. 2018;5:4–5. doi: 10.1107/S2052252517018012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://wwpdb.org/) as [PDB code 6LUU [10.2210/pdb6luu/pdb] (0 atm CO2 pressure, pH 7.8), 6LUV [10.2210/pdb6luv/pdb] (20 atm, pH 7.8)] for apo-CA II, [6LUW [10.2210/pdb6luw/pdb] (0 atm, pH 7.8), 6LUX [10.2210/pdb6lux/pdb] (20 atm, pH 7.8), 6LUY [10.2210/pdb6luy/pdb] (0 atm, pH 11.0), 6LUZ [10.2210/pdb6luz/pdb] (20 atm, pH 11.0)] for Zn-CA II, [6LV1 [10.2210/pdb6lv1/pdb] (0 atm, pH 7.8), 6LV2 [10.2210/pdb6lv2/pdb] (20 atm, pH 7.8), 6LV3 [10.2210/pdb6lv3/pdb] (0 atm, pH 11.0), 6LV4 [10.2210/pdb6lv4/pdb] (20 atm, pH 11.0)] for Co-CA II, [6LV5 [10.2210/pdb6lv5/pdb] (0 atm, pH 7.8), 6LV6 [10.2210/pdb6lv6/pdb] (20 atm, pH 7.8), 6LV7 [10.2210/pdb6lv7/pdb] (0 atm, pH 11.0), 6LV8 [10.2210/pdb6lv8/pdb] (20 atm, pH 11.0)] for Ni-CA II, and [6LV9 [10.2210/pdb6lv9/pdb] (0 atm, pH 7.8), 6LVA [10.2210/pdb6lva/pdb] (20 atm, pH 7.8)] for Cu-CA II. Two earlier structures [5DSR [10.2210/pdb5dsr/pdb] and 5YUK [10.2210/pdb5yuk/pdb]] were used for structure determination. Source data are provided with this paper.