Significance
The physical principles of membrane protein folding are not well understood. Because of the lack of water inside the cell membrane, the hydrophobic effect cannot drive the folding of membrane-embedded structural elements. Therefore, van der Waals packing interaction becomes a crucial driving force, which may imply that the membrane protein interior is tightly packed. Paradoxically, membrane proteins such as channels, transporters, receptors, and enzymes require cavities (i.e., voids, pockets, and pores) for function. Then, how do membrane proteins achieve the stability carrying out function? Using experiment and molecular dynamics simulation, we show that cavities in membrane proteins can be stabilized by favorable interaction with surrounding lipid molecules and play a pivotal role in balancing stability and flexibility for function.
Keywords: membrane protein stability, cavity, packing, GlpG, steric trapping
Abstract
Packing interaction is a critical driving force in the folding of helical membrane proteins. Despite the importance, packing defects (i.e., cavities including voids, pockets, and pores) are prevalent in membrane-integral enzymes, channels, transporters, and receptors, playing essential roles in function. Then, a question arises regarding how the two competing requirements, packing for stability vs. cavities for function, are reconciled in membrane protein structures. Here, using the intramembrane protease GlpG of Escherichia coli as a model and cavity-filling mutation as a probe, we tested the impacts of native cavities on the thermodynamic stability and function of a membrane protein. We find several stabilizing mutations which induce substantial activity reduction without distorting the active site. Notably, these mutations are all mapped onto the regions of conformational flexibility and functional importance, indicating that the cavities facilitate functional movement of GlpG while compromising the stability. Experiment and molecular dynamics simulation suggest that the stabilization is induced by the coupling between enhanced protein packing and weakly unfavorable lipid desolvation, or solely by favorable lipid solvation on the cavities. Our result suggests that, stabilized by the relatively weak interactions with lipids, cavities are accommodated in membrane proteins without severe energetic cost, which, in turn, serve as a platform to fine-tune the balance between stability and flexibility for optimal activity.
The van der Waals (vdW) packing interaction is one of the key molecular forces that stabilize proteins (1–5). In general, globular proteins are efficiently packed to minimize the size of cavities (i.e., voids, pockets, and pores) (6). The protein interior has a mean packing density (∼0.74) similar to the crystals of small organic molecules (0.70 to 0.78) and the close-packed sphere model (0.74 to 0.76) (6, 7). Proteins also have a liquid-like character. Protein structures are remarkably tolerant to amino acid substitutions, and the size distribution of cavities agrees with that predicted from the random-packed sphere model (7–9). Creating a cavity in the protein interior involves the loss of packing interaction, which incurs the free energy cost of 25 cal/mol to 30 cal/mol per unit cavity volume (Å3) (1, 2, 4). Nonetheless, in globular proteins, ∼15 cavities are found for every additional 100 residues with a broad size distribution from a few to ∼1,000 Å3 (7). Why are cavities so prevalent despite their unfavorable contribution to protein stability? Since the folding of globular proteins is majorly driven by the hydrophobic effect rather than by vdW packing, cavities may form randomly as a consequence of folding (7). Certain cavities are nevertheless strictly conserved playing a critical role in function such as catalysis, ligand binding, allostery, and transport (10–12).
Regarding the impact of cavities on protein stability, helical membrane proteins may serve as a counter model because packing interaction is a critical driving force in the folding (3–5, 13). The folding of helical membrane proteins can be divided into two thermodynamically distinct stages (13): In stage I, nonpolar segments in a polypeptide chain insert into the membrane as transmembrane (TM) helices, majorly driven by the hydrophobic effect and backbone hydrogen bonding (14, 15). In stage II, the inserted helices associate to form a compact native structure. In the latter stage, the hydrophobic effect cannot drive the compaction of the membrane-embedded structural elements because of the lack of water inside the lipid bilayer. Thus, to stabilize the native structure, attractive packing and polar interactions between TM helices should overcome favorable interactions between individual TM helices and solvating lipids (3–5, 13, 16–18).
With a similar energetic contribution of packing to the stability, globular proteins tend to accommodate large nonpolar and aromatic residues in the interior, while membrane proteins preferentially bury small residues such as Gly, Ala, and Ser (19–21). Hence, membrane proteins can pack closely, burying a larger fraction of residue areas than globular proteins (22). These studies suggest that membrane proteins extensively utilize packing to achieve their stability (4, 20). However, a number of studies suggest that membrane proteins are not tightly packed. In comparison to globular proteins, the average internal packing density of channels and transporters, which require pores and pockets for function, is low, and that of receptors and photosystems is similar (23). On average, membrane proteins have a larger fraction of the number of residues which contact cavities than globular proteins (19). A recent NMR relaxation study shows that the internal side chains of folded membrane proteins are highly dynamic compared to those of globular proteins (24). This implies that membrane proteins may not be as tightly packed as globular proteins, and the resulting low side-chain entropic cost can significantly compensate the lack of the hydrophobic effect as a driving force for membrane protein folding (24).
Here, we focus on elucidating how the two competing requirements, that is, packing for stability vs. cavities for function, are reconciled in the native structures of membrane proteins. Toward this goal, we test three hypotheses using the intramembrane protease GlpG of Escherichia coli as a model (Fig. 1A): 1) If cavities compromise the stability, improving packing by cavity-filling mutation will generally enhance the stability. 2) If cavities are critical to function, protein conformation can be locked into either inactive or active state by modifying the cavity size. 3) Lipid solvation can reduce the energetic cost of cavity formation such that cavities can be accommodated despite their unfavorable contribution to the stability.
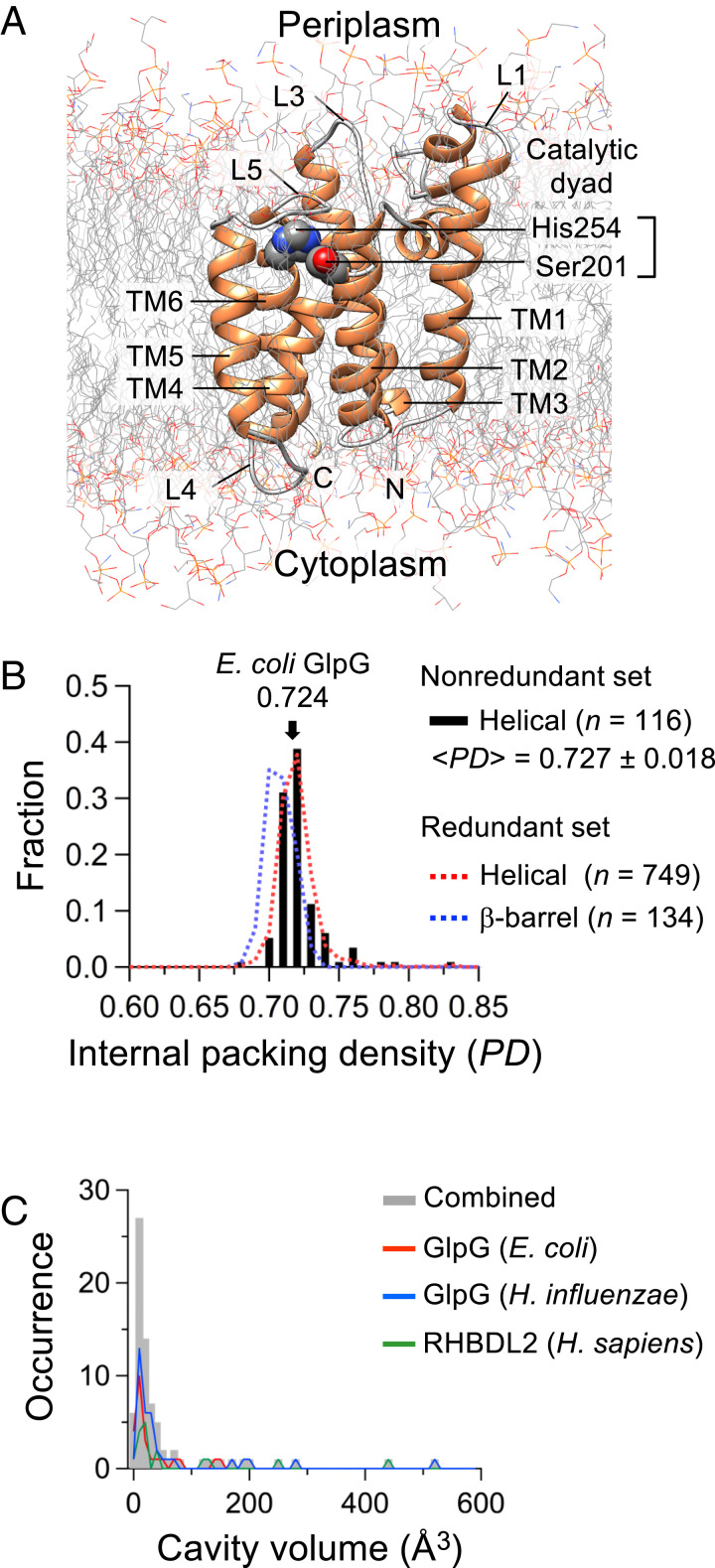
Fig. 1.
Structure and packing of GlpG. (A) Structural snapshot of the TM domain of E. coli GlpG (residues 91 to 272) from the MD simulation in the lipid bilayer (POPE:POPG, molar ratio = 3:1). (B) Distribution of the internal PD of membrane proteins of known structure (resolution <2.8 Å; n, the number of structures). The PD was calculated on the MP:PD server (http://proteinformatics.charite.de/mppd/links/). (C) Size distribution of the cavities in E. coli GlpG (PDB ID code 3B45) (55), H. influenzae GlpG (PDB ID code 2NR9) (32), and human RHBDL2 (modeled). The cavity volumes (Vms) were obtained on the CASTp server (http://sts.bioe.uic.edu/castp/index.html).
GlpG is a member of the widely conserved rhomboid protease family. Rhomboids are intramembrane serine proteases with a Ser−His catalytic dyad buried in the membrane (25). They regulate diverse biological processes such as epidermal growth factor signaling, mitochondrial quality control, cell adhesion, and cell-to-cell communication by cleaving a specific peptide bond in membrane-bound signaling proteins or enzymes (26). Extensive enzyme kinetic and structural studies have been carried out to elucidate the proteolytic mechanisms (27–36). Recently, GlpG has emerged as an important model for studying membrane protein folding (37–42).
So far, the contribution of packing to the stability and function of membrane proteins has been mainly studied using deletion (large-to-small) mutations (4, 37, 40). Here, using experiment and molecular dynamics (MD) simulation, we approach this problem in an opposite way; that is, the role of the structural cavities in GlpG is probed by cavity-filling (small-to-large) mutation. We find that, although compromising stability, cavities in membrane proteins are critical to facilitating conformational changes and balancing stability and flexibility for optimal activity. Cavities can be accommodated without severe energetic cost, through weak stabilization by lipid solvation, and this weak interaction fine-tunes the stability−flexibility balance.
Results
Stable Membrane Protein GlpG Has Packing Features Similar to Globular Proteins.
GlpG has moderate thermodynamic stability (ΔGoU = 5 kcal/mol to 6 kcal/mol) and resistance to thermal denaturation (Tm = ∼70 °C) in mild dodecylmaltoside (DDM) micelles (37, 38). The internal packing density (PD) of GlpG is 0.724, which falls within the typical range for globular (0.74 ± 0.03) and helical membrane proteins (0.73 ± 0.02; MP:PD, Membrane Protein Packing Database; Fig. 1B) of known structure (7, 43). Using a 1.4-Å-radius probe on the CASTp (Computed Atlas of Surface Topography of proteins) server (44), we found a total of 24 cavities with a broad size distribution (molecular surface volume, Vms = 2.0 Å3 to 141.7 Å3) in GlpG (Fig. 1C and SI Appendix, Table S1). This number corresponds to 13.4 cavities/100 residues, which is similar to the average frequency of cavities in globular proteins (∼15/100 residues) obtained with the same method (7). We also analyzed the packing of other rhomboid proteases. Since only one rhomboid structure is available from a distinct origin other than E. coli (Haemophilus influenzae GlpG, 40.1% sequence identity to E. coli GlpG) (32), we built a structural model of a distant homolog, human RHBDL2 (26.2% sequence identity to E. coli GlpG) (25), using homology modeling and structural refinement with MD simulation (SI Appendix, Fig. S1). The packing features of the three rhomboids, that is, the PD (0.702 to 0.737), and the frequency (8.6 to 19.1/100 residues) and size distribution (Vms = 3.9 Å3 to 523.6 Å3) of cavities, are comparable to those of globular proteins (SI Appendix, Fig. S2) (7, 43). Among the 24 cavities in E. coli GlpG, 13 overlapped with the cavities in the structurally equivalent regions of the other two rhomboids (SI Appendix, Fig. S3).
Analyzing the Impacts of Cavity-Filling Mutations on the Structure, Dynamics, and Packing of GlpG Using MD Simulation.
Among the 13 common cavities which are possibly conserved among rhomboid proteases, we selected five with Vms > 40 Å3 and designed 11 single cavity-filling mutations (Fig. 2). The cutoff volume corresponds to the approximate volume difference between Ala and Val, which allows room for small-to-large mutation (45). The fraction of electronegative atoms (N and O) surrounding each cavity ranges from 0.09 to 0.36 (SI Appendix, Table S2). Thus, the selected cavities have a largely nonpolar or moderately polar surface.
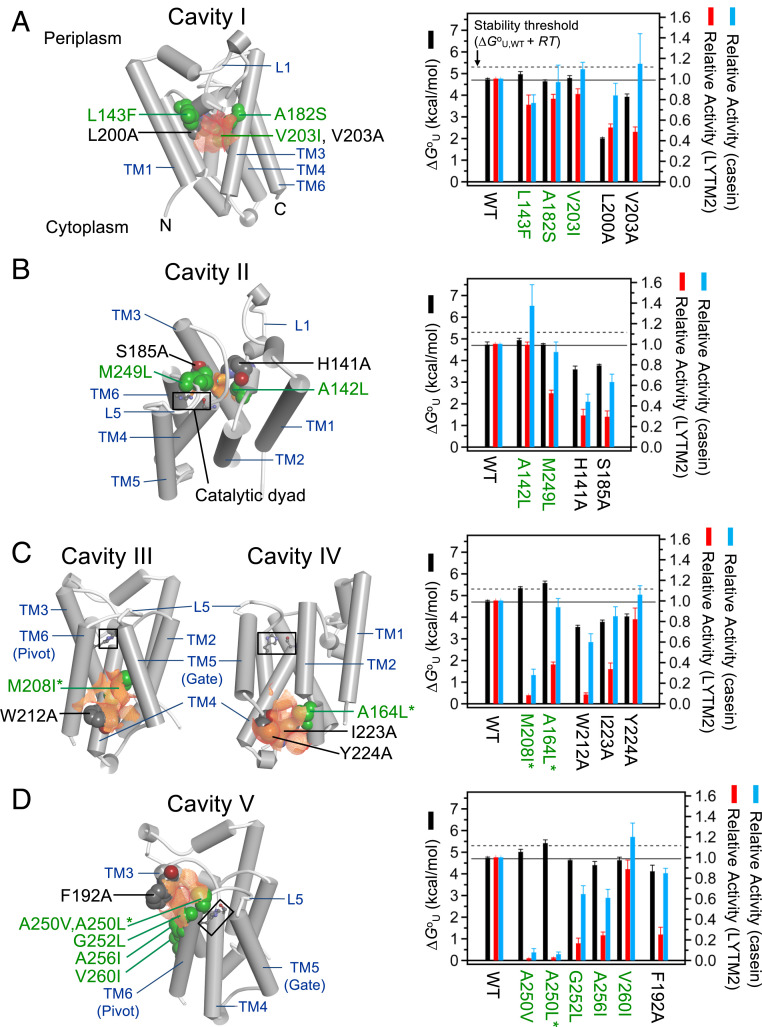
Fig. 2.
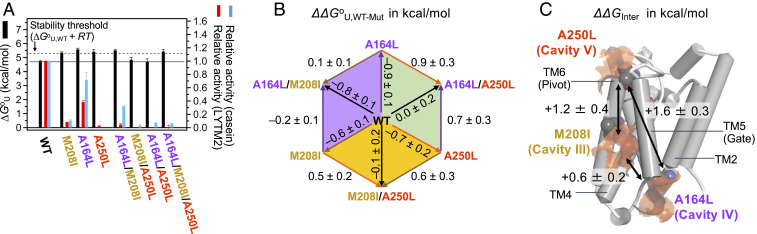
Impacts of mutations on the stability and activity of GlpG for cavities (A) I, (B) II, (C) III and IV, and (D) V. (Left) Locations of the cavities of interest (the surfaces in orange) and target residues for mutation (the spheres) in the WT structure. The catalytic dyad, Ser201(TM4)-His254 (TM6) (in the rectangular box), are shown in the ball-and-stick model. The text labels of the residues for small-to-large mutations are in green, and those for large-to-small mutations are in black. (Right) Thermodynamic stability (ΔGoU) of GlpG WT and variants and their activities relative to WT. The variants which are stabilized above the threshold level (ΔGoU,WT + RT: ΔGoU,WT, the stability of WT) are labeled with asterisks. Errors in ΔGoU denote ± SD from fitting. Errors in activity denote ± SEM (n = 3 to 4).
The effects of mutations on the structure and dynamics of GlpG were examined using all-atom MD simulations of wild-type (WT) and variants in a lipid bilayer. In the simulations up to 1.1 μs to 1.4 μs, the rmsds of the backbone heavy atoms relative to those of the reference WT structure reached a plateau within 200 ns to 600 ns (Fig. 3A), indicating the equilibration of protein conformation during simulation. The small plateau rmsd values at 1 Å to 2 Å imply the stability of the fold and the compatibility of the substituted residues with the WT structure. The residue rms fluctuations (rmsfs) of WT and variants show that the TM helices are rigid (<1 Å) and the interhelical loops are relatively flexible (<4 Å) (SI Appendix, Fig. S4A). The differences in rmsf between WT and a variant are small in both TM helical (<|∼0.2 Å|) and loop (<|∼2 Å|) regions (SI Appendix, Fig. S4B). Thus, the mutations did not noticeably alter the dynamics of WT.
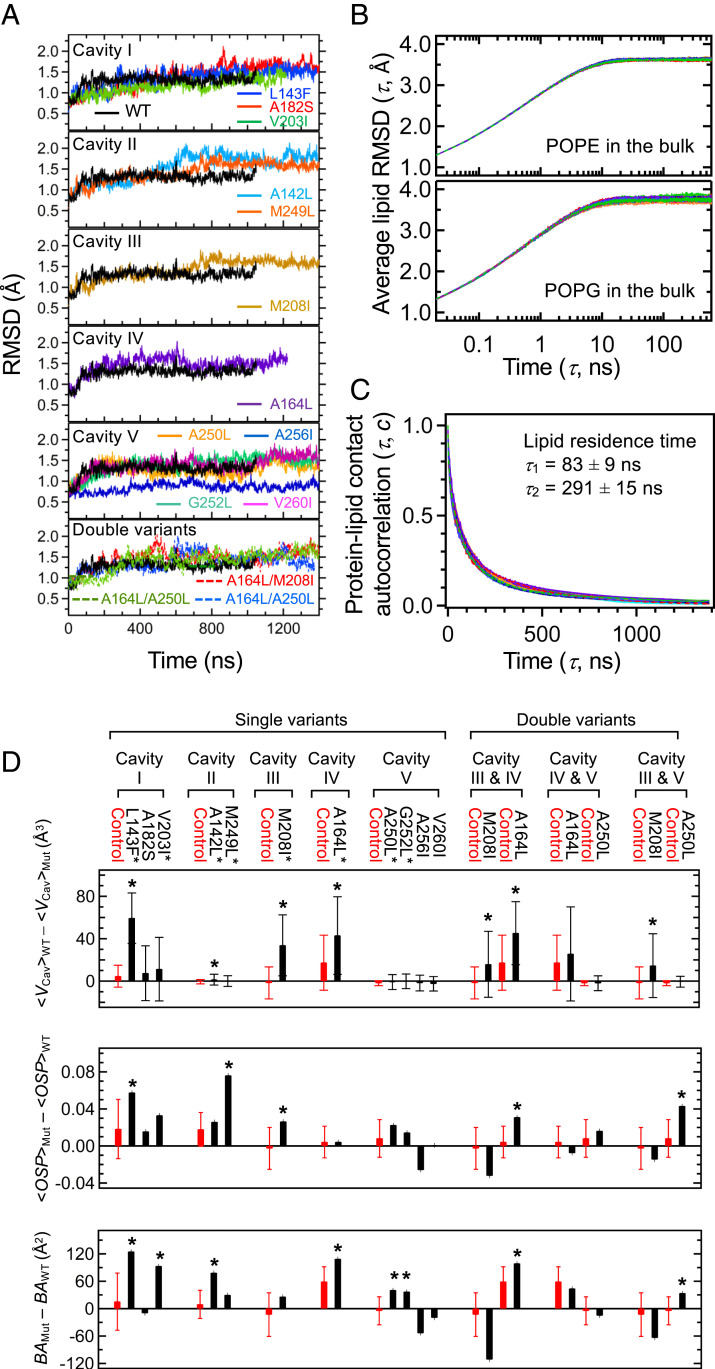
Fig. 3.
All-atom MD simulation of GlpG WT and variants in the lipid bilayer (POPE:POPG, molar ratio = 3:1). (A) The rmsds of the backbone heavy atoms relative to those in the crystal structure of WT (PDB ID code 2IC8) (35). (B) Equilibration of the lipid conformation in the bulk bilayer region measured by the average rmsd as a function of the time lag τ for (Top) POPE and (Bottom) POPG lipids. The color codes of the traces for WT and variants are the same as in A. (C) The lipid exchange at the protein surface measured by the lipid contact autocorrelation function on time. The color codes of the traces for WT and variants are the same as in A. (D) Impacts of the mutations on the packing in the cavities measured by the differences in cavity volume (<VCav>WT − <VCav>Mut), OSP (<OSPMut> ‒ <OSPWT>, averaged over the residues surrounding each cavity), and BA (BAMut ‒ BAWT, summed over the residues surrounding each cavity). “Control” (red bar) indicates the mean of each difference packing parameter calculated over WT and the variants which do not contain the mutation on the designated cavity. The error bar in “Control” corresponds to the SD. When a mutation induces an increase in packing exceeding the upper SD limit of the corresponding control, the mutation is regarded as “cavity filling” and marked with an asterisk.
All cavities of interest are lipid accessible, except for water-filled cavity II (Fig. 2). To ensure that lipid conformations and lipid−protein interactions reached an equilibrium during our simulation, we analyzed the rmsds of lipid molecules in the bulk bilayer and the resident times of lipid molecules at the protein surface. For the former, we selected lipid molecules located >40 Å away from the protein surface as the bulk lipids (45 ± 2 POPE, 1-palmitoyl-2-oleoyl-phosphatidylethanolamine, and 12 ± 1 POPG, 1-palmitoyl-2-oleoyl-phosphatidylglycerol). Then, we calculated the rmsd for each lipid molecule as a function of time lag, which was averaged over all selected lipid molecules (SI Appendix, Materials and Methods). For WT and variants, the average rmsds of POPE and POPG reached a plateau at 3.6 Å to 3.8 Å within ∼20 ns, indicating that our simulation can sample lipid conformations under equilibrium conditions (Fig. 3B). Next, to test the equilibration of protein−lipid interactions, we evaluated the autocorrelation function on time for all lipid heavy atoms in contact with the proteins (SI Appendix, Materials and Methods). The characteristic lipid residence times at the protein surface, τ1 and τ2 (the times when the correlation function decreased by 1/e and 1/e2 from the start, respectively), were 83 ± 9 ns and 291 ± 25 ns, respectively, which were 4 to 15 times longer than the relaxation of the bulk lipids (Fig. 3C). This result indicates that our simulation is long enough for lipid molecules to fully exchange at the protein surface as well as for protein conformations to equilibrate with the surrounding lipids.
Did the small-to-large mutations indeed improve packing in the targeted cavities? To answer this question, we took three approaches from MD simulation of WT and variants. Taking the structures from all time frames after the equilibration (>500 ns), we measured the volume of each cavity (VCav) using a grid-based method (46). With the equilibrated structures at 1 μs, we evaluated the occluded surface packing (OSP) (20) and buried surface area (BA) for each residue.
All cavities displayed a dynamic feature. The volume of each cavity fluctuated with the SD (σVol) of 20‒150% relative to the mean cavity volume (<VCav>; SI Appendix, Figs. S5 and S6). Despite the fluctuation, four mutations, L143F (cavity I), A142L (cavity II), M208I (cavity III), and A164L (cavity IV), effectively reduced the volume of the cavities targeted for mutation (i.e., <VCav>WT ‒ <VCav>Mut,targeted is larger than <<VCav>WT ‒ <VCav>Mut,not-targeted> + σ<VCav > WT‒ <VCav > Mut,not-targeted) (Fig. 3D and SI Appendix, Fig. S6). The difference in OSP and BA values between WT and a variant fluctuated as a function of residue number apparently in a random manner (SI Appendix, Figs. S7 and S8, respectively). Nevertheless, L143F (cavity I), M249L (cavity II), and M208I (cavity III) from the OSP analysis, and L143F (cavity I), V203I (cavity I), A142L (cavity II), A164L (cavity IV), A250L (cavity V), and G252L (cavity V) from the BA analysis, improved packing in the respective cavities (Fig. 3D). Overall, 8 out of 11 single small-to-large mutations improved packing in the targeted cavities by any measure, thus regarded as cavity-filling mutations.
A Benchmark Study to Validate Mutation-Induced Changes in Packing from MD Simulation.
How do the changes in packing induced by cavity-filling mutations influence the stability and function of GlpG? Before answering this question, we validated our approach for quantifying mutation-induced changes in packing by carrying out a benchmark MD simulation using bacteriorhodopsin (bR) as a model. Joh et al. (4) have measured the changes in cavity volume induced by a series of cavity-creating mutations from the crystal structures of bR WT and variants. From the relationship between the changes in stability and cavity volume, they have determined the contribution of vdW packing to the stability.
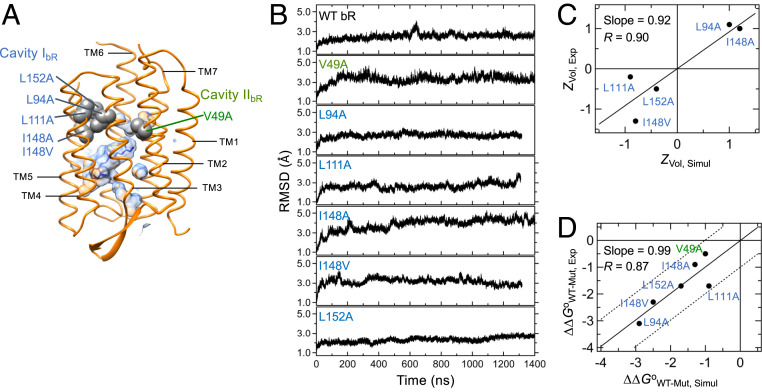
We constructed all-atomic models of WT and six variants of bR in DMPC (1,2-dimyristoyl- phosphatidylcholine) bilayers (Fig. 4A) and performed MD simulation up to ∼1.4 μs (Fig. 4B). In each system, the backbone rmsd relative to the corresponding crystal structure reached a plateau within 200 ns to 600 ns. On the basis of the spatial location of mutation, we targeted two cavities for volume analysis (Fig. 4A), that is, cavity IbR created by the mutations L94A, L111A, I148A, I148V, and L152A, and cavity IIbR created by V49A. Since we were interested in how well MD simulation can capture the local volume changes induced by mutation, the series of the volume changes of cavity IbR were compared between simulation (the volumes measured by the grid-based method) and experiment (the volumes measured using a 1.0-Å-radius probe) (SI Appendix, Table S3) (4, 46). We found a discrepancy which probably stemmed from the difference in the cavity detection method, or, more likely, the difference in the extent of protein flexibility allowed between the crystal lattices and the bilayers in silico. Nonetheless, the comparison of the normalized volume changes using Z scores (zi = (xi − μ)/s; xi is the volume of cavity IbR in each variant, and μ and s are the mean and SD of the cavity volumes in the variants, respectively) displayed a reasonable correlation between simulation and experiment (R = 0.90; Fig. 4C). Thus, our MD simulation can reliably capture the local volume changes induced by mutation.
Fig. 4.
Benchmark MD simulation of WT and variants of bR in a DMPC bilayer. (A) The structure of WT bR (PDB ID code 1PY6) (68). The major internal cavities (the surfaces), the mutated residues for cavity-creating mutations (the spheres), and bound retinal (the sticks). (B) The rmsds of the backbone heavy atoms of WT and variants during MD simulation. (C) Correlation between the normalized cavity volumes (Z scores) determined from experiment (4) and simulation. (D) Correlation between the protein stabilities determined from experiment (4) and simulation. The dashed lines indicate the deviation by ±1.0 kcal/mol from the fitted linear line.
Next, we further attempted to predict the mutation-induced stability changes using the volume changes measured from MD simulation. Since a given mutation influenced not only the volume of the cavity targeted by the mutation but also that not targeted, it was difficult to reliably predict the stability changes only using the volume changes of a single cavity targeted for mutation, which was the approach with the static crystal structures (4). Therefore, at our benchmarking level, we constructed a simple linear response model to predict the stability changes (ΔΔGoU,WT-Mut,Simul) using the volume changes of both cavities,
| [1] |
where ΔVI,WT-Mut and ΔVII,WT-Mut denote the mutation-induced volume changes of cavities IbR and IIbR, respectively, obtained from MD simulation, the coefficients cI and cII represent the packing contributions to the stability in the respective cavities, and b is related to the contribution of lipid or water solvation to the stability changes upon mutation. These coefficients were fitted to the model. Intriguingly, the stability changes can be modeled within the errors of ±1.0 kcal/mol with a reasonable correlation (R = 0.87) (Fig. 4D and SI Appendix, Table S4). Thus, our MD simulation approach can serve not only to evaluate the mutation-induced changes in cavity volume but also to build a model for reliable prediction of the free energy changes of membrane proteins (see Lipid Solvation on Cavity Fine-tunes Stability).
Impacts of Native Cavities in the N Subdomain on Stability and Activity.
Next, we investigated the impacts of cavity-filling mutations on the thermodynamic stability (ΔGoU) and activity of GlpG. To measure the stability in DDM micelles, we employed steric trapping, which couples spontaneous unfolding of a doubly biotinylated protein to competitive binding of bulky monovalent streptavidin (mSA) (SI Appendix, Fig. S9A) (38). Mutations were made in the background of the double-biotin variant, 172/267-BtnPyr2 (172/267: the residues for cysteine substitution; BtnPyr: a thiol-reactive biotin label with a pyrene fluorophore) (SI Appendix, Fig. S9B). The 172/267-BtnPyr2 possesses the same global stability as WT without biotin labels (38). Steric trapping allows for precise determination of protein stability (an SE in ΔGoU ≤ ±0.2 kcal/mol) directly under native conditions (SI Appendix, Fig. S10).
Proteolytic activity of GlpG was measured using the second TM segment of E. coli lactose permease (LYTM2) (47) as a substrate in DDM micelles and large DMPC:CHAPS(3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate) bicelles (SI Appendix, Figs. S11 and S12). Bicelles were used to test GlpG activity in a bilayer environment. For LYTM2, the activities of variants relative to WT in micelles were highly correlated with those in bicelles (SI Appendix, Fig. S13). We also measured the activity for the generic water-soluble substrate, casein (SI Appendix, Fig. S14). Casein, which would access the catalytic dyad directly from the bulk water, served as a probe to test the intactness of the active site upon mutation.
The impact of mutation on the stability and activity of GlpG exhibited a unique dependence on the targeted cavity. Cavity I is surrounded by the residues mainly from the N-terminal half of GlpG (TM1–L1–TM2–TM3–L3: N subdomain) and penetrates the protein interior near the bilayer center. Three small-to-large mutations on cavity I, including the cavity-filling mutations L143F and V203I, did not significantly alter the stability and activity relative to WT (Fig. 2A).
Next, we targeted the water-filled cavity near the catalytic dyad (cavity II) also surrounded by the residues mainly from the N subdomain (Fig. 2B). This cavity is known as a water retention site, providing water molecules required for proteolysis (48). Despite the improved packing in cavity II by the mutations A142L and M249L, the WT stability level was retained (Fig. 2B). Our MD simulation indicates that the water molecules remained bound in the cavity upon mutation (SI Appendix, Fig. S15). Probably due to the unperturbed water retention, A142L did not affect the activity for both TM and water-soluble substrates. In comparison, M249L at the junction between L5 and TM6 selectively reduced the activity for LYTM2 to 52 ± 3%, while retaining the activity for casein at 92 ± 10%. It has been suggested that the opening of the L5 loop enables the access of the scissile bond in a substrate to the active site (27, 49) (see the next section). Thus, it is likely that the improved packing by M249L at the L5−TM6 junction partially inhibited the opening of L5, leading to the activity reduction preferentially for the TM substrate.
A number of structural and folding studies indicate that the N subdomain of GlpG serves as a rigid structural template, while the C subdomain possesses conformational plasticity undergoing subglobal unfolding (28, 31, 38, 39, 49). Cavity-filling mutations in the already stable N subdomain did not increase the stability, probably because the improved packing induced a strain without a gain of stability. It is also possible that, for lipid-accessible cavity I, the cavity-filling mutations modified the lipid−cavity interactions to counteract the stabilization by the improved packing (see Lipid Solvation on Cavity Fine-tunes Stability).
Impacts of Native Cavities in the C Subdomain on Stability and Activity.
We targeted three cavities (cavities III to V; Fig. 2 C and D) in the C subdomain of GlpG (TM4–L4–TM5–L5–TM6) harboring the catalytic dyad Ser201/His254. Crystal structures show that the C-terminal segment TM5–L5–TM6 is subject to conformational changes. In a majority of apo structures, TM5 is packed against TM2, and the L5 loop caps the catalytic dyad (Fig. 2C) (35). In comparison, TM5 is tilted away from TM2, or L5 forms an “open cap” or disordered conformation in the structures bound with several mechanism-based inhibitors and two apo structures (SI Appendix, Fig. S16) (27, 28, 31, 49, 50). On the basis of the dramatic activation induced by deletion mutations at the TM2–TM5 interface and the plasticity of TM5 and L5, it has been suggested that TM substrates bind to the TM2−TM5 interface and that TM5 serves as a “gate” controlling the access of TM substrates to the catalytic dyad along with the L5 cap (27, 49, 51). However, chemical cross-linking between TM2 and TM5 retains activity, and a modeling study predicts that substrate binding may not require opening of TM5 (34, 50). Thus, the gating role of TM5 has been debated.
Interestingly, the mutations targeting the cavities near TM5 (M208I on cavity III and A164L on cavity IV) effectively reduced the volume of the two cavities by 30 to 60% (SI Appendix, Fig. S6) and enhanced the stability (ΔΔGoU,WT-Mut > RT, thermal energy = ∼0.6 kcal/mol; R, gas constant; T, absolute temperature, 298 K). Cavity III deeply intrudes into the TM4−TM5 interface (Fig. 2C). M208I on this cavity stabilized GlpG by +0.6 ± 0.2 kcal/mol, substantially decreasing the activity to 8 ± 1% for the TM substrate LYTM2. This inactivation is unexpected because the mutated site is not only distant from the catalytic dyad but also does not directly interfere with substrate binding at the TM2−TM5 interface. Cavity IV is located at the substrate binding site, but distant from the catalytic dyad (Fig. 2C). A164L on this cavity induced the largest stabilization (+0.9 ± 0.2 kcal/mol) among tested mutations, decreasing the activity to 38 ± 3% for LYTM2. Notably, both M208I and A164L displayed larger relative activity for casein (28 ± 6% and 94 ± 9%, respectively) than that for LYTM2. Thus, the activity loss for the TM substrate was not caused by the disruption of the active site. From MD simulation, all tested mutations did not perturb the hydrogen bond between Ser201 and His254 (SI Appendix, Table S5), which activates Ser201 for the nucleophilic attack on the scissile peptide bond (35).
The gain of interaction (i.e., the enhanced stability) and loss of function by the cavity-filling mutations near TM5 implies the stabilization of the gate-closed conformation. Supporting this, MD simulation shows that M208I shrank the substrate binding site by tilting of TM5 (SI Appendix, Fig. S17), slightly decreasing the Cα−Cα distance between Phe153 (TM2) and Trp236 (TM5) by up to ∼0.8 Å (SI Appendix, Fig. S18). These results suggest a critical role of the cavities near TM5 in mediating the gating motion for substrate binding.
Finally, we tested mutations on cavity V at the TM3−TM6 interface in the periplasmic side (Fig. 2D). TM6 harboring the catalytic His254 is also subject to conformational changes (SI Appendix, Fig. S16). Mechanism-based inhibitors covalently bound to Ser201 induce a slight outward (52) or inward (28, 53) pivot motion at the periplasmic end of TM6. However, the involvement of this small-amplitude motion in the catalytic mechanism has been elusive.
The stepwise increase of the side-chain volume on cavity V by A250V and A250L gradually increased the stability by +0.4 ± 0.2 and +0.7 ± 0.2 kcal/mol, respectively, dramatically decreasing the activity for both LYTM2 and casein to <5% (Fig. 2D). G252L and A256I on the same cavity also substantially decreased the activity for LYTM2 to 15 to 25%, retaining the activity for casein at 60 to 65%. Thus, the activity loss for the TM substrate by these mutations was not necessarily due to the disruption of the active site. V260I at the lipid-exposed position outside of cavity V fully restored the activity for both LYTM2 and casein (>85%) without altering the stability. In the crystal structures, the pivot motion of TM6 involves the rotation of small Ala250 and Gly252 on TM6 against bulky Val188, Gln189, and Tyr192 on TM3 (SI Appendix, Fig. S19). Therefore, it is likely that the increases in the side-chain volume on cavity V inhibit the pivot motion of TM6 and that this motion has a large impact on the proteolysis mechanism.
As a control, we tested the impacts of cavity-creating mutations on all cavities (Fig. 2). As expected, these mutations decreased the stability to various extents (ΔΔGoU,WT-Mut = –0.6 kcal/mol to ‒2.7 kcal/mol). H141A (cavity II), which is known to disrupt the water retention and conduction toward the active site (31), similarly reduced the activity for LYTM2 and casein to 30 ± 6% and 44 ± 8%, respectively (Fig. 2B). Except for this mutation and Y224A (cavity IV), the latter of which had a minor impact on the stability and activity (Fig. 2C), all other cavity-creating mutations substantially reduced the activity for LYTM2 to 10 to 50% regardless of the degree of destabilization, retaining the activity for casein at 60 to 120%. Thus, the activity loss for the TM substrate induced by the cavity expansion did not stem from either the disruption of the active site or the conformational destabilization (see Are All Cavities Critical to Function?).
Additivity, Cooperativity, and Propagation of Stabilizing Mutations.
Our finding that all stabilizing mutations are mapped onto the more flexible C subdomain and substantially reduce the activity suggests that these mutations stabilize inactive conformations of GlpG by inhibiting the functionally important “gating” (A164L and M208I) or “pivot” (A250L) motion. Next, we tested whether these stabilization effects were additive by measuring the stability and activity of the pairwise double and triple variants (Fig. 5A). All of these variants were almost inactive for both substrates. Although some of the variants were stabilized, the individual stabilization effects were not additive.
Fig. 5.
Additivity and cooperativity of stabilizing cavity-filling mutations. (A) Impacts of the double and triple stabilizing mutations on the stability and activity. (B) Thermodynamic cycle analysis describing the stability changes induced by the stabilizing single and combined double mutations. (C) Cooperativity between the stabilizing mutations. Pairwise interaction energies (ΔΔGInter) were calculated using Fig. 5B (SI Appendix, Materials and Methods). The mutated residues (the spheres) and the residues in the catalytic dyad (the sticks) are shown. Errors denote ± SD from fitting.
To track down the molecular origin of the nonadditivity, we carried out a thermodynamic cycle analysis to determine the free energy of interaction (ΔΔGInter) between the substituted residues (SI Appendix, Materials and Methods and Fig. 5B) (54). Slight negative cooperativity (ΔΔGInter = +0.6 ± 0.2 kcal/mol) occurred between M208I (cavity III) and A164L (cavity IV), which are spatially close (Fig. 5C). Interestingly, larger negative cooperativity was observed between M208I (cavity III) and A250L (cavity V) (ΔΔGInter = +1.2 ± 0.4 kcal/mol) and between A164L (cavity IV) and A250L (cavity V) (ΔΔGInter = +1.6 ± 0.3 kcal/mol), which are farther separated. This result indicates that the flexible C subdomain of GlpG is an ensemble of multiple disjoint conformations, and stabilizing one destabilizes another. Thus, the conformation stabilized by A250L (the TM6 pivot locked) is not likely to simultaneously populate with that stabilized by M208I or A164L (the TM5 gate locked). The negative cooperativity is further supported by MD simulation showing that the improved packing by the single cavity-filling mutations was suppressed in either cavity when these mutations were combined in the double variants (Fig. 3D).
By design, steric trapping captures transient opening of the native tertiary contacts at a specific biotin pair, allowing measurement of the local stability of the region encompassing the biotin pair (38). We compared the mutation-induced stability changes measured at the biotin pair in the C subdomain (172/267-BtnPyr2) to those measured at the N subdomain (95/172-BtnPyr2). The same mutations that had stabilized the C subdomain (ΔΔGoU,WT-Mut = +0.6 kcal/mol to +1.0 kcal/mol) did not stabilize the N subdomain as much (ΔΔGoU,WT-Mut = −0.1 kcal/mol to +0.3 kcal/mol) (SI Appendix, Fig. S20). Thus, the stabilization by the cavity-filling mutations was only locally effective in the C subdomain, not globally propagated to the N subdomain.
Lipid Solvation on Cavity Fine-tunes Stability.
The crystal structures and our MD simulation indicate that all of the cavities of interest except for water-filled cavity II interact with detergent or lipid molecules. For WT, the patterns of the cavity−detergent interaction in the crystal structure are remarkably similar to the cavity−lipid interaction in the simulation (Fig. 6, Left and Middle) (55, 56). For these cavities, we found an interesting correlation between the changes in stability upon mutation and the associated changes in lipid interaction. To quantitatively describe the cavity−lipid interaction, for the time frames from MD simulation (>500 ns), we measured the number of lipid atoms within each cavity (SI Appendix, Fig. S21) as well as the distance between a specific residue in each cavity and its closest approaching lipid atom (SI Appendix, Fig. S22).
Fig. 6.
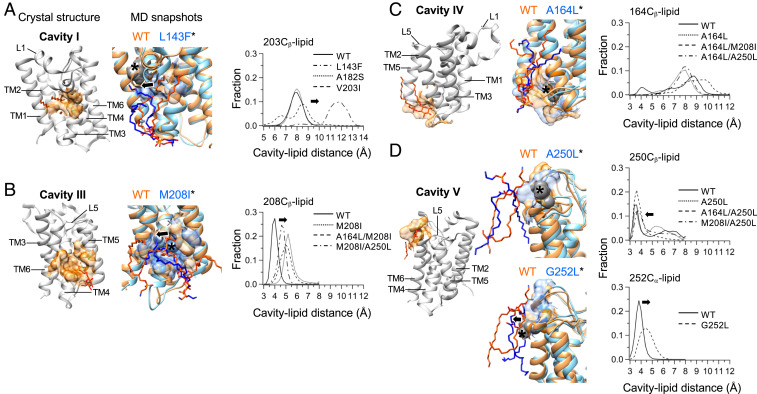
Impacts of cavity-filling mutations on lipid−protein interaction for cavities (A) I, (B) III, (C) IV, and (D) V. (Left) Crystallographic detergent molecules (the sticks in red) bound to the cavities in WT GlpG (PDB ID code 3B45). (Middle) The structural snapshots of WT (orange) and variant (cyan) at 1 μs from MD simulation are superimposed with the closest approaching lipid molecules in the stick model. The residues surrounding each cavity are shown in the surfaces. The site of each mutation is marked with an asterisk. The movement of lipid chain segments induced by mutation is indicated with black arrows. (Right) The distance distributions between a specific residue atom and the closest-approaching lipid heavy atoms in each designated cavity. In D, the structural snapshots (Middle) and cavity–lipid distances (Right) of WT and A250L are shown at the top, and those of WT and G252L are at the bottom.
For the cavity-filling mutations, L143F (cavity I), M208I (cavity III), and G252L (cavity V), the increase in packing was coupled to noticeable displacement of lipid atoms from the cavities targeted for mutation (Fig. 6, Right), while, to less extent, for A164L (cavity IV) and A250L (cavity V). Under the assumption that the mutational impacts on the lipid−protein interactions are only local near the cavities targeted for mutation, the packing−desolvation coupling offers an opportunity to quantify the strengths of lipid−protein interactions relative to those of protein packing, which yields various outcomes in the stability (SI Appendix, Fig. S23). This assumption is supported by the fact that the cavity-filling mutations distinctively improve the packing in the cavity targeted for mutation (Fig. 3D). Toward this goal, we employed the formulation suggested by Fleming and Engelman (3) to study TM helix−helix interactions,
| [2] |
ΔΔGpacking denotes the free energy change induced by the change in protein packing upon mutation (i.e., Δ<VCav>WT-Mut = <VCav>WT – <VCav>Mut determined from MD simulation). σpacking is the vdW packing contribution to the protein stability per unit cavity volume (σpacking = –29 cal/mol/Å3) (2–4). ΔΔGPL and ΔΔGLL represent the free energy changes in lipid solvation (or desolvation) upon mutation and the associated lipid reorganization, respectively. We define their sum (ΔΔGPL + ΔΔGLL) as the free energy change in “lipid exchange” (ΔΔGEx,PL), which can be interpreted as the preference (or disfavor) of partitioning a lipid segment in a cavity relative to the surrounding lipids.
On the basis of our validated MD simulation approach for predicting the free energy changes of membrane proteins (Fig. 4), we evaluated the contribution of protein packing vs. lipid solvation to the stability of GlpG using Eq. 2. The weak stabilization by the mutation L143F on cavity I (–ΔΔGoU,WT-Mut = –0.2 ± 0.2 kcal/mol) stems from the stabilization by the improved packing (ΔΔGpacking = –1.7 ± 0.6 kcal/mol), which is canceled out by the unfavorable lipid desolvation (ΔΔGEx,PL = +1.5 ± 0.6 kcal/mol) (Fig. 6A). The unfavorable lipid desolvation indicates that cavity I in WT has been stabilized by interacting lipids. The stabilization by M208I on cavity III (–ΔΔGoU,WT-Mut = –0.6 ± 0.1 kcal/mol) is the outcome of the coupling between the improved packing (ΔΔGpacking = –1.0 ± 0.4 kcal/mol) and the weakly unfavorable lipid desolvation (ΔΔGEx,PL = +0.4 ± 0.4 kcal/mol) (Fig. 6 B, Right). In contrast, A164L (–ΔΔGoU,WT-Mut = –0.9 ± 0.1 kcal/mol) is mainly stabilized by the improved packing (ΔΔGpacking = –0.8 ± 0.8 kcal/mol; Fig. 6 C, Right) on cavity IV with a negligible contribution from lipid solvation (ΔΔGEx,PL = –0.1 ± 0.8 kcal/mol). A250L does not improve packing on cavity V (ΔΔGpacking = ∼0 kcal/mol) but induces favorable lipid solvation (ΔΔGEx,PL = –0.7 ± 0.2 kcal/mol), which stabilizes the protein (–ΔΔGoU,WT-Mut = –0.7 ± 0.2 kcal/mol) (Fig. 6D). Although A250L induces a subtle change in lipid solvation (Fig. 6 D, Right Top), the attraction of lipid atoms to cavity V is observed for the double variants, A164L/A250L and M208I/A250L, indicating a possible creation of a higher-affinity lipid interaction site by substituted Leu250. Despite the displacement of lipids by G252L from cavity V (Fig. 6 D, Right Bottom), the energetic contribution is negligible (ΔΔGEx,PL = +0.2 ± 0.1 kcal/mol) without improving either packing or stability (ΔΔGpacking and –ΔΔGoU,WT-Mut of ∼0 kcal/mol).
Our perturbation approach using mutation suggests that lipid-accessible cavities in GlpG fine-tune the stability through changes in protein−lipid interactions in various modes. Importantly, lipid interaction on a cavity can moderately stabilize the protein. We note that the stability was measured in DDM micelles. Thus, the weakly favorable lipid−protein interactions (ΔΔG = ‒1.5 to –0.1 kcal/mol) quantified here are actually between detergents and the protein.
Discussion
The impacts of structural cavities on thermodynamic stability and activity of membrane proteins have not been systematically investigated. Instead, extensive mutagenesis, thermal denaturation and computational studies have been carried out to obtain thermostable variants or lock protein conformation into a specific functional state suitable for structure determination (57–60). Using the membrane-integrated enzyme GlpG as a tractable model and cavity-filling mutation as a probe, we demonstrated the pivotal role of structural cavities in modulating stability, activity, and lipid interactions of membrane proteins.
Do Native Cavities Compromise Stability?
We tested our first hypothesis that improving protein packing by cavity-filling mutation generally stabilizes a membrane protein. Among 11 small-to-large single mutations, only 3 were significantly stabilizing, the effects of which were nonetheless modest (ΔΔGoU,WT-Mut = ∼1 kcal/mol). Studies on globular proteins indicate that the stability gain by single cavity-filling mutations is ∼1 kcal/mol at best, while stabilization up to ∼5 kcal/mol has been reported using multiple mutations (61, 62). Thus, the modest stabilization observed for GlpG seems to be a maximal level that can be reached by single mutations.
All stabilizing cavity-filling mutations were mapped onto the more flexible C subdomain (i.e., the region with room for further stabilization) of functional importance. Therefore, native cavities indeed compromise stability, but stabilization by improved packing strongly depends on the structural, dynamic, and functional context of a protein.
Are All Cavities Critical to Function?
Despite the apparently random spatial distribution, certain cavities seem to be more important to function. GlpG can be inactivated via the selective stabilization of the flexible C subdomain by cavity-filling mutations on the same subdomain. This result is consistent with our second hypothesis that a membrane protein can be strategically locked into a specific functional state by modifying the cavity size. This also suggests that cavities play a critical role in facilitating functional motions in a membrane protein by balancing stability and flexibility. We expected that cavity-creating mutations might enhance GlpG activity by providing even more flexibility. However, most of the cavity-creating mutations on the selected cavities reduced the activity for the TM substrate without perturbing the active site. Probably, the cavity expansion increased the probability of nonproductive motions of the structural elements near the cavities. This result suggests that the conformational changes involved in the proteolysis of TM substrates are highly coordinated motions controlled by the size and shape of the native cavities.
Our result exemplifies a stability−activity trade-off by selective stabilization of a conformational subset of membrane proteins, which is in line with the studies of the M2 proton channel of influenza A virus, G protein coupled receptors (GPCRs) and transporters (57–60, 63–66). Interestingly, stabilizing mutations are found at a surprisingly high frequency for the M2 channel (∼60% of tested mutations) (66). The frequency of finding stabilizing mutations in membrane proteins is known to be higher (∼10% of tested mutations) than in globular proteins (57, 59, 64, 67, 68). In the extreme case of the M2 channel, a homotetrameric pore formed by single-spanning TM helices, the functional and stabilizing traits spatially overlap running through the tetrameric pore (66). Thus, the natural sequences of the channel seem optimized for functional conformational changes with a potential for further stabilization (65, 66). In contrast, only six stabilizing mutations (either ΔΔGU,WT-Mut < –RT or ΔTm,WT-Mut < –3 °C) have been found from ∼200 mutations on E. coli GlpG (<5% of tested mutations) (37, 38, 40, 48). This discrepancy can be explained by the modular architecture of GlpG (38, 39, 42), which endows an optimal balance between the functional efficiency sustained by the flexible C subdomain and the structural stability assured by the rigid N subdomain. Here, when mutations are made on the C subdomain with conformational flexibility and functional importance, we indeed find stabilizing mutations at a high frequency (∼27%, 3 out of 11 cavity-filling or cavity-creating mutations).
Using the strategy of cavity-filling mutation, we were able to not only reliably map the functionally important conformational change (gating by TM5) which had been previously suggested (51) but also newly identify another (pivoting by TM6). Crystallographic studies indicate that the pivot motions of TM6 induced by mechanism-based inhibitors involve a series of side-chain rotations from the active site along the way to the substrate binding site (34, 52). This may imply that the continuous TM5–L5–TM6 segment undergoes a concerted motion controlling substrate binding. However, the negative cooperativity between cavities III/IV and cavity V suggests that the pivot motion of TM6 is not necessarily coupled to the gating motion of TM5, playing a distinct functional role. The pivot motion may be critical to the opening of the L5 cap to expose the catalytic dyad to a scissile bond, explaining why inhibiting the pivot motion by A250L almost abrogated the activity for water-soluble casein (Fig. 2D). A recent intriguing crystallographic study capturing the structures of the catalytic intermediates of GlpG shows that the opening of the L5 cap precedes the opening of the TM5 gate (27). Our study suggests that the negative cooperativity between the two motions is the physical origin that controls the early catalytic mechanisms.
How Strong Is Membrane Protein−Lipid Interaction?
Interaction with lipids is critical to the stability and function of membrane proteins. Structural studies of membrane proteins have found lipid molecules at the clefts formed between packed TM helices (“annular lipids”) or in the tertiary or quaternary contacts with a high specificity (69–72). Despite the importance, the energetics of protein−lipid interaction is poorly understood. A recent study using nanoelectrospray ionization mass spectrometry shows that the free energy of association between lipids and a membrane protein can be up to ∼‒7 kcal/mol (73). The free energy calculations of protein−lipid interaction using MD simulation yield –0.5 kcal/mol to –1 kcal/mol for nonspecific or weak interaction, and –1 kcal/mol to –10 kcal/mol for specific or strong interaction (74). Although the detergent−protein interactions that we quantified may not be the same as those with lipids, the cavity solvation by detergents (ΔΔG = –1.5 kcal/mol to –0.1 kcal/mol) falls within the weak interaction regime.
The weak lipid interaction may have implications in the stability and function of membrane proteins. It can reduce the energetic cost of cavity formation caused by the loss of packing. Thus, cavities can be accommodated in membrane proteins without severe stability loss. The weak solvation would also facilitate conformational changes of membrane proteins, assuring the structural flexibility critical to the function of enzymes, transporters, and GPCRs. Other MD simulation studies on GlpG suggest that bound lipids near the active site may compete with substrate binding (75). Intriguingly, highly deformed lipids surrounding GlpG boost its translational diffusion within the membrane, enhancing the activity (76).
The cavity stabilization by solvation is not limited to membrane proteins. For globular proteins, theoretical studies predict that the free energy of transfer of water from the bulk to internal cavities (ΔGhydration) widely varies from 5 kcal/mol to ‒12 kcal/mol, highly depending on the polarity of the cavity surface (77). An experimental study has shown that cavity hydration can stabilize a globular protein by 1.5 kcal/mol to 2.0 kcal/mol (78). Therefore, for both globular and membrane proteins, cavity solvation by either water or lipids can counterbalance the energetic cost caused by the loss of protein packing (24).
Concluding Remark.
Here, we have elucidated a versatile role of cavities in balancing the stability and flexibility for activity of membrane proteins. Our strategy employing cavity-filling mutation in combination with experiment and simulation may serve as a tool for mapping conformational changes critical to function and provide guidance for membrane protein design and engineering.
Materials and Methods
Detailed information on materials, modeling of RHBDL2, MD simulation, expression and purification of GlpG, biotin labeling, stability and activity determination, and thermodynamic cycle analysis can be found in SI Appendix.
Structural Modeling of Human Rhomboid Protease RHBDL2.
The sequence alignment of RHBDL2 and E. coli GlpG in the predicted TM helices was obtained from the previous literature and used for the comparative modeling of RHBDL2 (the core TM helices, TM1 to TM6) on the Rosetta software with three structural templates of E. coli GlpG (2IC8, 2XOV, and 3B45) (25, 35 55, 79, 80). The modeled structure with the lowest energy was chosen as the starting point of MD simulation for further relaxation in a POPC explicit membrane and water using the CHARMM (Chemistry at Harvard Macromolecular Mechanics)-27 force field (81).
MD Simulation of GlpG.
We first simulated for GlpG WT, and then moved on for each variant based on the equilibrated WT conformation. The WT system was built with the crystal structure of E. coli GlpG (Protein Data Bank [PDB] ID code 2IC8; ref. 35) embedded in a lipid bilayer (POPE:POPG = 231:77) constructed using the web-based CHARMM-GUI (graphical user interface) membrane builder (82). The composite system was immersed in the TIP3P water solvent (83), followed by a charge neuralization and ionization with 150 mM NaCl in a box of 100 × 100 × 81 Å3. Intermolecular and intramolecular potential energies were enumerated based on the CHARMM36 force field (84). All simulations were massively parallelized in the GPU (graphics processing unit)-accelerated IBM Power8 machine with a 2-fs time step in the semiisotropic isobaric and isothermal ensemble of 1 atm and 310 K.
Preparation and Biotin Labeling of GlpG.
The TM domain of GlpG (residues 87 to 276) encoded in pET15b vector was expressed in E. coli BL21(DE3)RP strain with an N-terminal His6-tag (38). Purified double cysteine variants (172C/267C or 95C/172C) at 0.5 mM Tris-(2-carboxyethyl) phosphine hydrochloride (TCEP, Pierce) was incubated with a 20-fold molar excess of the thiol-reactive biotin derivative with a pyrene fluorophore (BtnPyr-IA) in the base buffer, 50 mM Tris-(hydroxymethyl) aminomethane hydrochloride (TrisHCl), 200 mM NaCl, pH 8.0, and 0.2% DDM overnight in the dark at 25 °C.
Preparation of mSA.
WT mSA, and mSA-S45A (Kd,biotin = 9.0 nM) and mSA-S27A (Kd,biotin = 1.4 nM) variants with reduced biotin affinities, were prepared as described previously (38). Each variant contained a single cysteine mutation S83C in the active subunit to conjugate the thiol-reactive dabcyl quencher (Dabcyl Plus C2 maleimide, Anaspec).
Determining Thermodynamic Stability (ΔGoU) of GlpG.
ΔGoUs of GlpG variants were determined using steric trapping, that is, by measuring the attenuated second binding of mSA coupled to unfolding. Binding was detected by quenching of pyrene monomer fluorescence (λEx. = 345 nm and λEm. = 390 nm) from BtnPyr labels conjugated to the double-biotin variants of GlpG (172/267-BtnPyr2 or 95/172-BtnPyr2) upon binding of mSA labeled with dabcyl quencher (38); 1 μM GlpG was titrated with an mSA variant, mSA-S45A or mSA-S27A, in 5 mM DDM, 0.25 mM TCEP, 20 mM sodium phosphate, and 200 mM NaCl (pH 7.5) at 25 °C. The second binding phase in the binding isotherm was fitted to the equation to obtain ΔGoU (SI Appendix, Materials and Methods).
Activity Assays of GlpG.
Proteolytic activity of GlpG was measured using the cleavage rates of the second TM segment of E. coli lactose permease fused to staphylococcal nuclease (SN-LYTM2) in DDM micellar or DMPC/CHAPS bicellar solution, or Bodipy-FL labeled casein (Thermo Fisher) in DDM solution. For SN-LYTM2, the position at the five residues upstream from the scissile bond was mutated to Cys for labeling with thiol-reactive environment-sensitive fluorophore iodoacetyl-7-nitrobenz-2-oxa-1,3-diazol (IA-NBD, Setareh Biotech). Time-dependent change of NBD or Bodipy fluorescence, the initial slope of which represented the activity, was monitored with λEx. = 485 nm and λEm. = 535 nm.
Supplementary Material
Acknowledgments
We thank the H.H. laboratory members for critical comments, and Lin Song for helping with the OSP analysis. Special thanks go to anonymous reviewers for constructive comments. This work was supported by NIH Grants R01GM118685 (to H.H.) and R01GM126189 (to G.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917770117/-/DCSupplemental.
Data Availability.
All data supporting the findings of this study are available within this article and SI Appendix.
References
- 1.Kellis J. T. Jr., Nyberg K., Fersht A. R., Energetics of complementary side-chain packing in a protein hydrophobic core. Biochemistry 28, 4914–4922 (1989). [DOI] [PubMed] [Google Scholar]
- 2.Eriksson A. E. et al., Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 255, 178–183 (1992). [DOI] [PubMed] [Google Scholar]
- 3.Fleming K. G., Engelman D. M., Specificity in transmembrane helix−helix interactions can define a hierarchy of stability for sequence variants. Proc. Natl. Acad. Sci. U.S.A. 98, 14340–14344 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joh N. H., Oberai A., Yang D., Whitelegge J. P., Bowie J. U., Similar energetic contributions of packing in the core of membrane and water-soluble proteins. J. Am. Chem. Soc. 131, 10846–10847 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mravic M. et al., Packing of apolar side chains enables accurate design of highly stable membrane proteins. Science 363, 1418–1423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards F. M., Areas, volumes, packing and protein structure. Annu. Rev. Biophys. Bioeng. 6, 151–176 (1977). [DOI] [PubMed] [Google Scholar]
- 7.Liang J., Dill K. A., Are proteins well-packed? Biophys. J. 81, 751–766 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim W. A., Sauer R. T., Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature 339, 31–36 (1989). [DOI] [PubMed] [Google Scholar]
- 9.Wen J., Chen X., Bowie J. U., Exploring the allowed sequence space of a membrane protein. Nat. Struct. Biol. 3, 141–148 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Kadirvelraj R., Sennett N. C., Polizzi S. J., Weitzel S., Wood Z. A., Role of packing defects in the evolution of allostery and induced fit in human UDP-glucose dehydrogenase. Biochemistry 50, 5780–5789 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Kuang Q., Purhonen P., Hebert H., Structure of potassium channels. Cell. Mol. Life Sci. 72, 3677–3693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tseng Y. Y., Liang J., Estimation of amino acid residue substitution rates at local spatial regions and application in protein function inference: A Bayesian Monte Carlo approach. Mol. Biol. Evol. 23, 421–436 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Popot J. L., Engelman D. M., Membrane protein folding and oligomerization: The two-stage model. Biochemistry 29, 4031–4037 (1990). [DOI] [PubMed] [Google Scholar]
- 14.Hessa T. et al., Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Cao Z., Hutchison J. M., Sanders C. R., Bowie J. U., Backbone hydrogen bond strengths can vary widely in transmembrane helices. J. Am. Chem. Soc. 139, 10742–10749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gratkowski H., Lear J. D., DeGrado W. F., Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. U.S.A. 98, 880–885 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou F. X., Merianos H. J., Brunger A. T., Engelman D. M., Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. U.S.A. 98, 2250–2255 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowie J. U., Membrane protein folding: How important are hydrogen bonds? Curr. Opin. Struct. Biol. 21, 42–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamian L., Liang J., Helix-helix packing and interfacial pairwise interactions of residues in membrane proteins. J. Mol. Biol. 311, 891–907 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Eilers M., Shekar S. C., Shieh T., Smith S. O., Fleming P. J., Internal packing of helical membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 5796–5801 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adamian L., Nanda V., DeGrado W. F., Liang J., Empirical lipid propensities of amino acid residues in multispan alpha helical membrane proteins. Proteins 59, 496–509 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Oberai A., Joh N. H., Pettit F. K., Bowie J. U., Structural imperatives impose diverse evolutionary constraints on helical membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 17747–17750 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hildebrand P. W., Rother K., Goede A., Preissner R., Frömmel C., Molecular packing and packing defects in helical membrane proteins. Biophys. J. 88, 1970–1977 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Brien E. S. et al., Membrane proteins have distinct fast internal motion and residual conformational entropy. Angew. Chem. Int. Ed. Engl. 59, 11108–11114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemberg M. K., Freeman M., Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 17, 1634–1646 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman M., The rhomboid-like superfamily: Molecular mechanisms and biological roles. Annu. Rev. Cell Dev. Biol. 30, 235–254 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Cho S., Baker R. P., Ji M., Urban S., Ten catalytic snapshots of rhomboid intramembrane proteolysis from gate opening to peptide release. Nat. Struct. Mol. Biol. 26, 910–918 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho S., Dickey S. W., Urban S., Crystal structures and inhibition kinetics reveal a two-stage catalytic mechanism with drug design implications for rhomboid proteolysis. Mol. Cell 61, 329–340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickey S. W., Baker R. P., Cho S., Urban S., Proteolysis inside the membrane is a rate-governed reaction not driven by substrate affinity. Cell 155, 1270–1281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urban S., Lee J. R., Freeman M., Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–182 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Wu Z. et al., Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat. Struct. Mol. Biol. 13, 1084–1091 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Lemieux M. J., Fischer S. J., Cherney M. M., Bateman K. S., James M. N., The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc. Natl. Acad. Sci. U.S.A. 104, 750–754 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strisovsky K., Sharpe H. J., Freeman M., Sequence-specific intramembrane proteolysis: Identification of a recognition motif in rhomboid substrates. Mol. Cell 36, 1048–1059 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Y., Ha Y., Large lateral movement of transmembrane helix S5 is not required for substrate access to the active site of rhomboid intramembrane protease. J. Biol. Chem. 288, 16645–16654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Zhang Y., Ha Y., Crystal structure of a rhomboid family intramembrane protease. Nature 444, 179–180 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Ben-Shem A., Fass D., Bibi E., Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl. Acad. Sci. U.S.A. 104, 462–466 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker R. P., Urban S., Architectural and thermodynamic principles underlying intramembrane protease function. Nat. Chem. Biol. 8, 759–768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo R. et al., Steric trapping reveals a cooperativity network in the intramembrane protease GlpG. Nat. Chem. Biol. 12, 353–360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min D., Jefferson R. E., Bowie J. U., Yoon T. Y., Mapping the energy landscape for second-stage folding of a single membrane protein. Nat. Chem. Biol. 11, 981–987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paslawski W. et al., Cooperative folding of a polytopic α-helical membrane protein involves a compact N-terminal nucleus and nonnative loops. Proc. Natl. Acad. Sci. U.S.A. 112, 7978–7983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z., Jumper J. M., Freed K. F., Sosnick T. R., On the interpretation of force-induced unfolding studies of membrane proteins using fast simulations. Biophys. J. 117, 1429–1441 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schafer N. P., Truong H. H., Otzen D. E., Lindorff-Larsen K., Wolynes P. G., Topological constraints and modular structure in the folding and functional motions of GlpG, an intramembrane protease. Proc. Natl. Acad. Sci. U.S.A. 113, 2098–2103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose A., Theune D., Goede A., Hildebrand P. W., MP:PD—A data base of internal packing densities, internal packing defects and internal waters of helical membrane proteins. Nucleic Acids Res. 42, D347–D351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dundas J. et al., CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116-8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Counterman A. E., Clemmer D. E., Volumes of individual amino acid residues in gas-phase peptide ions. J. Am. Chem. Soc. 121, 4031–4039 (1999). [Google Scholar]
- 46.Paramo T., East A., Garzón D., Ulmschneider M. B., Bond P. J., Efficient characterization of protein cavities within molecular simulation trajectories: trj_cavity. J. Chem. Theory Comput. 10, 2151–2164 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Maegawa S., Ito K., Akiyama Y., Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry 44, 13543–13552 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Zhou Y., Moin S. M., Urban S., Zhang Y., An internal water-retention site in the rhomboid intramembrane protease GlpG ensures catalytic efficiency. Structure 20, 1255–1263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., Ha Y., Open-cap conformation of intramembrane protease GlpG. Proc. Natl. Acad. Sci. U.S.A. 104, 2098–2102 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoll S. et al., Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures. EMBO J. 33, 2408–2421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker R. P., Young K., Feng L., Shi Y., Urban S., Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc. Natl. Acad. Sci. U.S.A. 104, 8257–8262 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue Y., Ha Y., Catalytic mechanism of rhomboid protease GlpG probed by 3,4-dichloroisocoumarin and diisopropyl fluorophosphonate. J. Biol. Chem. 287, 3099–3107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ticha A. et al., General and modular strategy for designing potent, selective, and pharmacologically compliant inhibitors of rhomboid proteases. Cell. Chem. Biol. 24, 1523–1536 e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horovitz A., Double-mutant cycles: A powerful tool for analyzing protein structure and function. Fold. Des. 1, R121–R126 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., Maegawa S., Akiyama Y., Ha Y., The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG. J. Mol. Biol. 374, 1104–1113 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vinothkumar K. R., Structure of rhomboid protease in a lipid environment. J. Mol. Biol. 407, 232–247 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serrano-Vega M. J., Magnani F., Shibata Y., Tate C. G., Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc. Natl. Acad. Sci. U.S.A. 105, 877–882 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bozzi A. T. et al., Crystal structure and conformational change mechanism of a bacterial Nramp-family divalent metal transporter. Structure 24, 2102–2114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen K. Y., Zhou F., Fryszczyn B. G., Barth P., Naturally evolved G protein-coupled receptors adopt metastable conformations. Proc. Natl. Acad. Sci. U.S.A. 109, 13284–13289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smirnova I., Kasho V., Sugihara J., Kaback H. R., Trp replacements for tightly interacting Gly-Gly pairs in LacY stabilize an outward-facing conformation. Proc. Natl. Acad. Sci. U.S.A. 110, 8876–8881 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borgo B., Havranek J. J., Automated selection of stabilizing mutations in designed and natural proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 1494–1499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karpusas M., Baase W. A., Matsumura M., Matthews B. W., Hydrophobic packing in T4 lysozyme probed by cavity-filling mutants. Proc. Natl. Acad. Sci. U.S.A. 86, 8237–8241 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen K. M., Keri D., Barth P., Computational design of G protein-coupled receptor allosteric signal transductions. Nat. Chem. Biol. 16, 77–86 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Scott D. J., Kummer L., Tremmel D., Plückthun A., Stabilizing membrane proteins through protein engineering. Curr. Opin. Chem. Biol. 17, 427–435 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Stouffer A. L. et al., The interplay of functional tuning, drug resistance, and thermodynamic stability in the evolution of the M2 proton channel from the influenza A virus. Structure 16, 1067–1076 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stouffer A. L., Nanda V., Lear J. D., DeGrado W. F., Sequence determinants of a transmembrane proton channel: An inverse relationship between stability and function. J. Mol. Biol. 347, 169–179 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Bowie J. U., Stabilizing membrane proteins. Curr. Opin. Struct. Biol. 11, 397–402 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Faham S. et al., Side-chain contributions to membrane protein structure and stability. J. Mol. Biol. 335, 297–305 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Gonen T. et al., Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438, 633–638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knowles P. F., Watts A., Marsh D., Spin-label studies of lipid immobilization in dimyristoylphosphatidylcholine-substituted cytochrome oxidase. Biochemistry 18, 4480–4487 (1979). [DOI] [PubMed] [Google Scholar]
- 71.Laganowsky A. et al., Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanders C. R., Mittendorf K. F., Tolerance to changes in membrane lipid composition as a selected trait of membrane proteins. Biochemistry 50, 7858–7867 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cong X. et al., Determining membrane protein-lipid binding thermodynamics using native mass spectrometry. J. Am. Chem. Soc. 138, 4346–4349 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Hedger G., Shorthouse D., Koldsø H., Sansom M. S. P., Free energy landscape of lipid interactions with regulatory binding sites on the transmembrane domain of the EGF receptor. J. Phys. Chem. B 120, 8154–8163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White S. H., Bondar A. N., Lipid-mediated helix gating in the GlpG rhomboid protease from Escherichia coli. Biophys. J. 100, 358 (2011). [Google Scholar]
- 76.Kreutzberger A. J. B., Ji M., Aaron J., Mihaljević L., Urban S., Rhomboid distorts lipids to break the viscosity-imposed speed limit of membrane diffusion. Science 363, 497–504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olano L. R., Rick S. W., Hydration free energies and entropies for water in protein interiors. J. Am. Chem. Soc. 126, 7991–8000 (2004). [DOI] [PubMed] [Google Scholar]
- 78.Takano K., Funahashi J., Yamagata Y., Fujii S., Yutani K., Contribution of water molecules in the interior of a protein to the conformational stability. J. Mol. Biol. 274, 132–142 (1997). [DOI] [PubMed] [Google Scholar]
- 79.Vinothkumar K. R. et al., The structural basis for catalysis and substrate specificity of a rhomboid protease. EMBO J. 29, 3797–3809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leaver-Fay A. et al., ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brooks B. R. et al., CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid wate. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 84.Best R. B. et al., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within this article and SI Appendix.