Significance
We have compared gut microbiomes in the different stages of multiple sclerosis (MS) based on both microbial and functional analyses using fecal samples. Together with microbial composition data, metagenomic functional and metabolite data revealed a reduced level of microbial butyrate and propionate biosynthesis in the gut of relapsing remitting MS (RRMS). On the other hand, in the gut of secondary progressive MS (SPMS), we revealed an enhancement in microbial DNA mismatch repair, which was consistent with excessive fecal oxidation shown in sulfur metabolomic analysis. As elevated oxidative stress is closely associated with chronic neuroinflammation and neurodegeneration, the present result opens a way to microbiome data-assisted management of MS, useful for prevention of disease progression.
Keywords: secondary progressive multiple sclerosis, relapsing remitting multiple sclerosis, whole metagenomic sequencing, sulfur metabolomics, oxidative stress
Abstract
Multiple sclerosis (MS), an autoimmune disease of the central nervous system, generally starts as the relapsing remitting form (RRMS), but often shifts into secondary progressive MS (SPMS). SPMS represents a more advanced stage of MS, characterized by accumulating disabilities and refractoriness to medications. The aim of this study was to clarify the microbial and functional differences in gut microbiomes of the different stages of MS. Here, we compared gut microbiomes of patients with RRMS, SPMS, and two closely related disorders with healthy controls (HCs) by 16S rRNA gene and whole metagenomic sequencing data from fecal samples and by fecal metabolites. Each patient group had a number of species having significant changes in abundance in comparison with HCs, including short-chain fatty acid (SCFA)-producing bacteria reduced in MS. Changes in some species had close association with clinical severity of the patients. A marked reduction in butyrate and propionate biosynthesis and corresponding metabolic changes were confirmed in RRMS compared with HCs. Although bacterial composition analysis showed limited differences between the patient groups, metagenomic functional data disclosed an increase in microbial genes involved in DNA mismatch repair in SPMS as compared to RRMS. Together with an increased ratio of cysteine persulfide to cysteine in SPMS revealed by sulfur metabolomics, we postulate that excessive DNA oxidation could take place in the gut of SPMS. Thus, gut ecological and functional microenvironments were significantly altered in the different stages of MS. In particular, reduced SCFA biosynthesis in RRMS and elevated oxidative level in SPMS were characteristic.
Multiple sclerosis (MS) is an autoimmune demyelinating disease of the central nervous system (CNS), which is the major neurological disease with onset in young adulthood. Most patients experience signs of relapse with intervals of remission lasting for months to years during their early clinical stage. Patients who are staying in this stage are referred to as relapsing remitting MS (RRMS). Proportions of patients with RRMS may shift to the progressive disease form referred to as secondary progressive MS (SPMS) in years after onset of MS. Once transition to SPMS is established, neurological disability such as gait difficulty is accumulating gradually, as a result of progressive neuronal damage accompanied by neuroinflammation (1). Research in the last decades has led to the development of disease modifying drugs (DMDs) for RRMS, that are mainly targeting self-reactive T cells and B cells. The effect of the DMDs is very limited or unclear in SPMS, implying that pathogenesis of SPMS could be fundamentally different from that of RRMS. In fact, recent work, showing the involvement of cytotoxic T helper cells in SPMS, is supportive of this postulate (2, 3).
The number of patients with MS has been increasing for decades in developed countries (4), and this trend is probably most notable after World War II in Japan (4–6). Since genetic risk factors for MS are thought to be unchanged in the Japanese population during this period of time, nongenetic or environmental risk factors are thought to play significant roles. Cigarette smoking, lower exposure to sunlight, and early Epstein-Barr virus infection have been discovered as potential environmental risk factors for MS in Western countries (7); however, these factors are unlikely to explain the increase of MS in Asian countries. More recently, studies have indicated that the gut microbiome that appears to be influenced by modern lifestyle may have the crucial role in the pathogenesis of MS (8–12). Consistent with this, alterations of the gut microbiome were shown to affect functions of various T cell populations, thereby regulating the severity of neurological dysfunctions in mouse models of MS (13, 14). Moreover, dysbiosis or altered microbiome in the fecal samples of MS has been demonstrated in independent work from developed countries (10–12). To explore the functional implications of altered gut microbes in MS, researchers have transferred fecal microbiome from patients with MS or healthy controls into germ-free mice and observed development of an animal model of MS in those mice. The results showed that transfer of fecal samples from MS had a disease-enhancing effects, compared with transfer of control fecal samples (8, 9).
Although results of the aforementioned work are consistent with the idea linking the alteration of the gut microbiome with the pathogenesis of MS, it is obvious that reevaluation for a larger cohort is needed to obtain further insights. Moreover, despite the heterogeneity of MS regarding disease forms and stages, previous work did not touch on this issue and has not addressed whether there might exist differential patterns of gut microbiome changes between RRMS and SPMS. In the present study, we comparatively analyzed gut microbiomes from healthy controls, patients with RRMS, SPMS, and two related disorders—atypical MS and neuromyelitis optica spectrum disorder (NMOSD)—by using multiomics data of the 16S rRNA gene (16S), whole metagenomic sequences, and metabolites from fecal samples. Atypical MS is now considered a distinct disease condition, since spinal cord and optic nerves are mainly affected, but only few brain lesions are detected by magnetic resonance imaging (MRI) scans (6). NMOSD was previously regarded as a subtype of MS, but is now considered an independent disease, since the majority of the cases are characterized by the presence of anti-aquaporin 4 (AQP4) autoantibody and do not respond to DMDs used for MS (15).
The results have revealed structural and functional differences in the gut microbiome between the four MS-related disorders and identified rational candidates of gut species and functions, which could be involved in the development and progression of MS.
Results
Demographic Profiles of Patients and Controls.
Sixty-two patients with RRMS (mean age 39.0 y), 15 patients with SPMS (mean age 43.3 y), 21 patients with atypical MS (mean age 42.3 y), 20 patients with NMOSD (mean age 43.1 y), and 55 HCs (mean age 40.0 y) were recruited. Samples from 20 of the 62 RRMS patients were previously subjected to 16S analysis (12). The demographics of the subjects are shown in SI Appendix, Table S1. There were no significant differences in age and body mass index among all of the groups. On the other hand, female-to-male ratios were significantly different between the four patient and HC groups. However, the differences in gender ratios were not significant between the patient groups, and UniFrac analysis between female and male in the HC group did not reveal significant differences (SI Appendix, Table S2, see below), implying that differences in gender ratios do not substantially affect the gut microbiome structure. Furthermore, it appears that immunotherapy does not significantly affect the gut microbiome structure because UniFrac analysis did not reveal significant differences between patients with and without immunotherapy in the RRMS group (SI Appendix, Table S2, see below).
Comparison of Alpha- and Beta-Diversities in the Gut Microbiome between the Five Subject Groups.
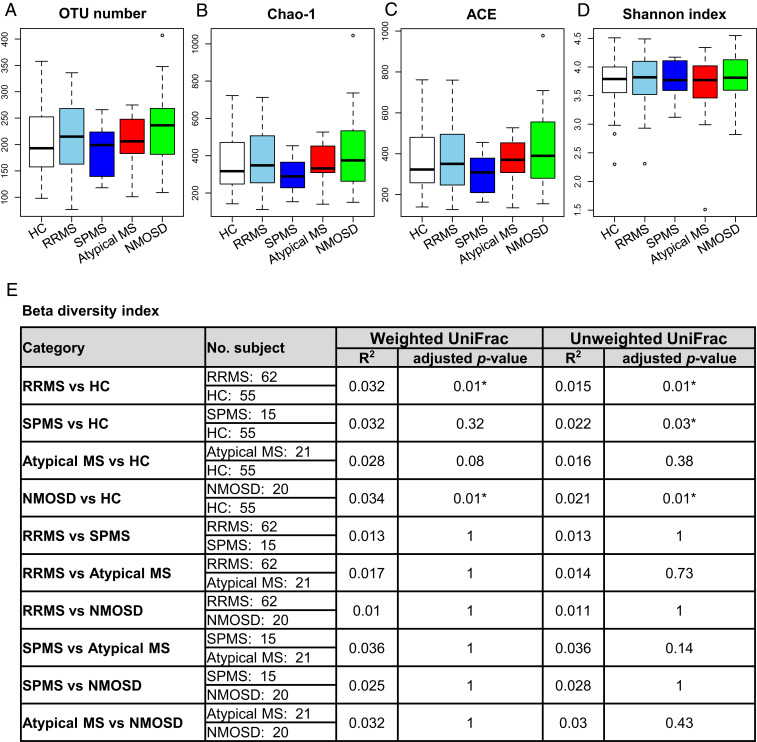
We obtained a total of 2,517,947 high-quality16S reads from the five subject groups by amplicon sequencing of the 16S rRNA gene V1–V2 region with MiSeq. We then randomly selected 3,000 reads per sample, accounting for a total of 354,000 reads from 118 samples. The four indexes: observed operational taxonomic unit (OTU) number, Chao-1, abundance-based coverage estimator (ACE), and the Shannon index did not show the significant differences in alpha-diversity between any pair of the five groups (Fig. 1 A–D).
Fig. 1.
Gut microbiome alpha- and beta-diversities in the five subject groups. (A–D) Alpha-diversity indexes. (A) The observed and (B) Chao 1-estimated OTUs, (C) the ACE, and (D) the Shannon index of fecal microbiome from the five groups based on 16S analysis. Each box plot represents median, interquartile range, minimum, and maximum values. (E) Beta-diversity indexes based on weighted and unweighted UniFrac analysis. R2 and adjusted P value in PERMANOVA between the five subject groups. UniFrac was based on the 16S data. *P < 0.05 based on the PERMANOVA with the Benjamini–Hochberg procedure for multiple group comparisons.
We next evaluated beta-diversity in the five groups by UniFrac distance using the 16S data. Permutational multivariate analysis of variance (PERMANOVA) showed significant differences in the overall gut microbiome structure (beta-diversity) between the RRMS and HC groups and between the NMOSD and HC groups based both on the weighted and unweighted UniFrac distance, and between the SPMS and HC groups based on the unweighted UniFrac distance (Fig. 1E). As described above, the observed differences in beta-diversity between the patient and HC groups were not significantly affected by differences in either gender or immunotherapy given to the patients (SI Appendix, Table S2).
Comparison of the Microbial Abundance between the Five Subject Groups.
We analyzed microbial abundances at various taxonomic levels. Taxonomic assignment was performed by mapping 16S reads to the microbial 16S and genome databases as described previously (12). The phylum level assignment identified six phyla with an average relative abundance of ≥0.1% in at least one of the five groups, and indicated significant changes in the abundance of Bacteroidetes between RRMS and HC (P = 0.0436) and that of Verrucomicrobia between RRMS and HC and between NMOSD and HC (P = 0.0275 and 0.0425, respectively; SI Appendix, Fig. S1A).
The genus level assignment identified 23 genera with an average relative abundance of ≥0.5% in at least one of the five groups, accounting for 93.5% of the total abundance (SI Appendix, Fig. S1B and Table S3). Among them, the abundance of Bifidobacterium was significantly higher in RRMS than in HC (P = 0.0475) and that of Streptococcus was also significantly higher in RRMS and SPMS than in HC (P = 0.0110 and 0.0110, respectively). The abundance of Megamonas was significantly lower in RRMS than in HC (P = 0.0102). Additionally, the abundance of Roseburia was significantly lower in SPMS and that of Alistipes was significantly higher in NMOSD than those in HC (P = 0.0420 and 0.0191, respectively).
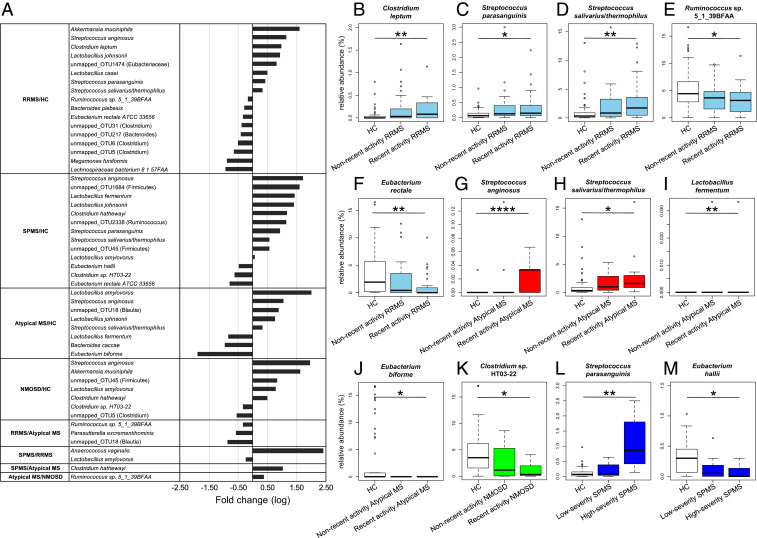
To explore possible association of microbial taxa with each patient group at the species level, we compared the abundance of 200 species-equivalent clusters with an average relative abundance of ≥0.1% in at least one of the five groups, consisting of 139 mapped_clusters (CLs) to known species with ≥97% identity and 61 unmapped_OTUs to known species with <97% identity. The comparison between all pairs of the five groups identified a total of 30 distinct species/OTUs having significant changes in abundance between any two groups (Fig. 2A and SI Appendix, Fig. S2 and Table S4). In the present analysis, we have identified 17 species/OTUs showing a significant difference in abundance between RRMS and HC including a greater abundance of Akkermansia muciniphila (mapped_CL208) in RRMS (P = 0.0259). Of these 17 species/OTUs, Eubacterium rectale (mapped_CL145), Streptococcus salivarius/thermophilus (mapped_CL267), Megamonas funiformis (mapped_CL428), unmapped_OTU5, unmapped_OTU6, and unmapped_OTU31 were also identified as those having significant changes between RRMS and HC in the previous study (12) (SI Appendix, Table S5).
Fig. 2.
Significantly increased or decreased microbial species/OTUs between the five subject groups and their correlation with clinical severity. (A) The list of 30 species/OTUs exhibiting significant changes in abundance between any two subject groups with fold-change (log10) between the related two groups. (B–F) Relative abundance of C. leptum (mapped_CL432; B), S. parasanguinis (mapped_CL5; C), S. salivarius/thermophilus (mapped_CL267; D), Ruminococcus sp. 5_1_39BFAA (mapped_CL372; E), E. rectale (mapped_CL145; F) in the HC, nonrecent activity RRMS, and recent activity RRMS groups. (G–J) Relative abundance of S. anginosus (mapped_CL121; G), S. salivarius/thermophilus (mapped_CL267; H), L. fermentum (mapped_CL129; I), E. biforme (mapped_CL429; J) in the HC, nonrecent activity atypical MS, and recent activity atypical MS groups. (K) Relative abundance of Clostridium sp. HT03-22 (mapped_CL169; K) in the HC, nonrecent activity NMOSD, and recent activity NMOSD groups. (L and M) Relative abundance of S. parasanguinis (mapped_CL5; L), E. hallii (mapped_CL78; M) in the HC, low-severity SPMS, and high-severity SPMS groups. Each box plot represents median, interquartile range, minimum, and maximum values. *P < 0.05, **P < 0.01, ****P < 0.0001 based on the Wilcoxon rank sum test with the Benjamini–Hochberg method for multiple group comparisons.
Association between the Microbial Abundance and Clinical Parameters.
We next examined the possible association between the microbial species and disease activity (annualized relapse rate) by comparing the abundance of the 30 species/OTUs between recent activity samples (those from patients who experienced at least one clinical relapse within a 1-y period before sample collection) and nonrecent activity samples (those from patients who had no experience of clinical relapse for more than 1 y before sample collection) in the RRMS, atypical MS, and NMOSD groups. The SPMS group was excluded because of its bias for recent activity samples (also see below). The results revealed that 5 of the 17 species/OTUs in the RRMS group, 4 of the 8 species/OTUs in the atypical MS groups, and 1 of the 7 species/OTUs in the NMOSD group had significant changes in abundance between the recent activity and HC samples but no significant change between the nonrecent activity and HC samples. In the RRMS group, the abundance of Streptococcus parasanguinis (mapped_CL5), S. salivarius/thermophilus (mapped_CL267), and Clostridium leptum (mapped_CL432) was significantly increased, and the abundance of E. rectale (mapped_CL145) and Ruminococcus sp. 5_1_39BFAA (mapped_CL372) was significantly decreased in the recent activity samples (P = 0.0301, 0.0013, 0.0047, 0.0013, and 0.0210, respectively; Fig. 2 B–F). In the atypical MS group, the abundance of Streptococcus anginosus (mapped_CL121) and S. salivarius/thermophilus (mapped_CL267) was significantly increased, and the abundance of Lactobacillus fermentum (mapped_CL129) and Eubacterium biforme (mapped_CL429) was significantly decreased in the recent activity samples (P = 0.00004, 0.0118, 0.0043, and 0.0485, respectively; Fig. 2 G–J). In the NMOSD group, the abundance of Clostridium sp. HT03-22 (mapped_CL169) was significantly decreased in the recent activity samples (P = 0.0128; Fig. 2K).
To evaluate the disease activity/severity of the SPMS group, we used disability progression rate (clinical severity score at sampling divided by disease duration) as an indicator of disease severity in SPMS. We adopted expanded disability status scale (EDSS; 0 represents no disabilities, 4 indicates sustained impairment of gait disturbance, and 10 means death by MS) as a clinical severity score (16), and classified the 15 SPMS patients into 7 high-severity and 8 low-severity SPMS patients based on the median disability progression rate of 0.35 for all of the 15 SPMS patients. The results revealed that the abundance of S. parasanguinis (mapped_CL5) was positively, and the abundance of Eubacterium hallii (mapped_CL78) was negatively associated with the high-severity SPMS group (P = 0.0194 and 0.0483, respectively; Fig. 2 L and M).
Functional Profiling of the Gut Microbiomes by Metagenomic Analysis.
We performed functional analysis using the metagenomic data obtained from sequencing of the same samples used for 16S analysis. Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (17), we identified a total of 6,163 KEGG orthologies (KOs) in the metagenomic data of the five subject groups. Of them, 582 had significant changes in abundance between all possible two groups. These KOs were involved in a total of 500 KEGG pathways (SI Appendix, Table S6). We then focused on the functional difference between HC and RRMS and between RRMS and SPMS, because development of RRMS from a healthy state and transition of RRMS into SPMS are key events in patients with MS.
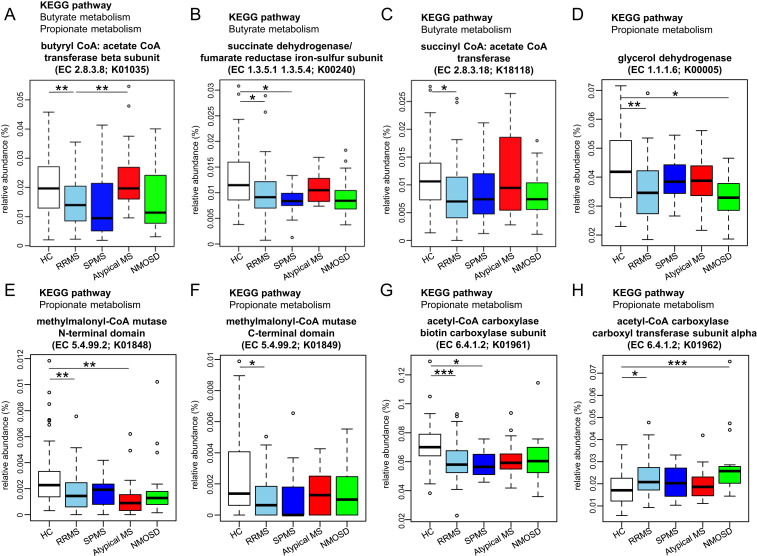
In comparison of HC and RRMS, we identified 97 KOs significantly enriched in RRMS (RRMS-enriched KOs), and 117 significantly depleted in RRMS (RRMS-depleted KOs) with P < 0.05 by Wilcoxon test (SI Appendix, Fig. S3A), and identified 83 pathways containing at least 1 of the 97 RRMS-enriched KOs or 1 of the 117 RRMS-depleted KOs. We ranked these individual pathways based on the ratio of RRMS-enriched KOs to RRMS-depleted KOs in principle. When no difference was revealed by this criterion, a pathway containing the KO with the lowest P value (Wilcoxon test) was ranked higher than another. In this ranking, we focused on the top seven pathways in all of which depleted KOs surpassed enriched KOs in RRMS (SI Appendix, Fig. S3B). Among the seven pathways, four pathways including citrate cycle (TCA cycle), carbon fixation pathway in prokaryotes, carbon metabolism, and oxidative phosphorylation were involved in energy acquisition and carbohydrate metabolism, two pathways of propanoate (propionate) metabolism and butanoate (butyrate) metabolism were closely associated with SCFA biosynthesis, and a pathway of porphyrin and chlorophyll metabolism was related to vitamin B12 biosynthesis. In the butyrate metabolism pathway, four enzymes (EC 2.8.3.8, EC 1.3.5.1, EC 1.3.5.4, and EC 2.8.3.18) represented the RRMS-depleted KOs (K01035, K00240, K00240, and K18118), respectively (Fig. 3 A–C). Among them, EC 2.8.3.8 (butyryl-CoA: acetate-CoA transferase beta-subunit; K01035) which is the main terminal enzyme for butyrate production (18–20) was significantly depleted in RRMS compared with that in HC or atypical MS (P = 0.0015 and 0.0064, respectively; Fig. 3A). In the propionate metabolism pathway, four enzymes (EC 2.8.3.8, EC 1.1.1.6, EC 5.4.99.2, and EC 6.4.1.2) represented the RRMS-depleted KOs (K01035, K00005, K01848-K01849, and K01961), respectively (Fig. 3 A and D–G) and one enzyme (EC 6.4.1.2) represented the RRMS-enriched KO (K01962; Fig. 3H).
Fig. 3.
Significantly increased or decreased KEGG orthologies in RRMS associated with butyrate or propionate metabolism. (A–G) Relative abundance of the KEGG orthologies (KOs) involved in the pathway of butyrate metabolism (A–C) or propionate metabolism (A and D–G) that were significantly decreased in RRMS compared with HC. (H) Relative abundance of the KO involved in the pathway of propionate metabolism that was significantly increased in RRMS compared with HC. Each box plot represents median, interquartile range, minimum, and maximum values. *P < 0.05, **P < 0.01, ***P < 0.001 based on Wilcoxon rank sum test with the Benjamini–Hochberg method for multiple group comparisons.
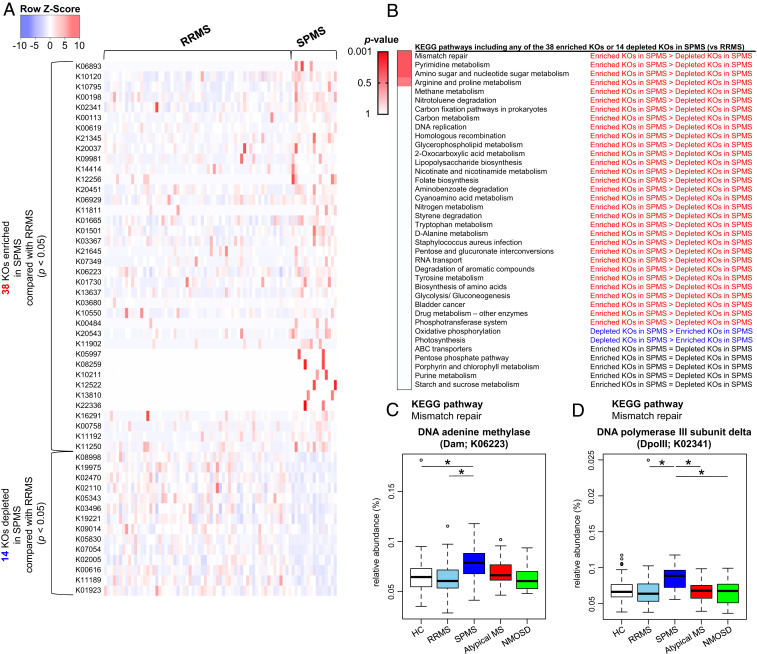
We next compared metagenomic genes between RRMS and SPMS as described in comparison between HC and RRMS. We identified 38 KOs significantly enriched in SPMS (SPMS-enriched KOs), and 14 significantly depleted in SPMS (SPMS-depleted KOs) with P < 0.05 by the Wilcoxon test (Fig. 4A). We have subsequently identified 38 pathways containing at least 1 of the SPMS-enriched KOs or 1 of the SPMS-depleted KOs. We ranked these individual pathways based on the ratio of SPMS-enriched KOs to SPMS-depleted KOs. In this ranking for the pathway, the top was a mismatch repair pathway associated with microbial DNA damage, in which enriched KOs surpassed depleted KOs in SPMS (Fig. 4B). In this pathway, two enzymes, DNA adenine methylase (Dam) and DNA polymerase III subunit delta (DpoIII) corresponded to the SPMS-enriched KOs (K06223 and K02341), respectively. The abundance of Dam (K06223) was significantly increased in SPMS compared to that in HC or RRMS (P = 0.0309 and 0.0442, respectively; Fig. 4C) and the abundance of DpoIII (K02341) was significantly increased in SPMS compared with RRMS, atypical MS, or NMOSD (P = 0.0114, 0.0440, and 0.0440, respectively; Fig. 4D).
Fig. 4.
Differences in functional profile of gut microbiome between RRMS and SPMS. (A) The list of 38 significantly enriched and 14 significantly depleted KEGG orthologies (KOs) in the SPMS group compared to those in the RRMS group (Wilcoxon test, P < 0.05). The Z-score based on the abundance of each KO is depicted from lowest (blue) to highest (red) according to the scale shown at the Top. (B) List of KEGG pathways including at least 1 of the 38 significantly enriched KOs (SPMS-enriched KOs) or 1 of the 14 significantly depleted KOs (SPMS-depleted KOs) in the comparisons between the RRMS and SPMS groups. These individual pathways are ranked based on the ratios of SPMS-enriched KOs to SPMS-depleted KOs. P value is based on the Fisher’s exact test. (C and D) Relative abundance of the two significantly enriched KOs in SPMS compared with RRMS which are included in the mismatch repair pathway. The relative abundance of DNA adenine methylase (Dam; C) and DNA polymerase III subunit delta (DpoIII; D) between the five subject groups. Each box plot represents median, interquartile range, minimum, and maximum values. *P < 0.05 based on Wilcoxon rank sum test with the Benjamini–Hochberg method for multiple group comparisons.
Comparison of Functions of Three Major Nutrient Metabolism.
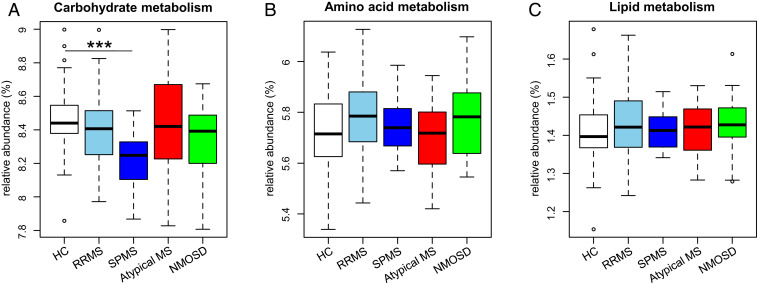
We also examined metabolism pathways related to three major nutrients (carbohydrates, proteins, and lipids) at the KEGG subcategory level to evaluate the possible effects of diet on the gut microbiome, by comparing the abundance of genes in these metabolism pathways among the five groups. The results revealed that genes for the carbohydrate metabolism were significantly decreased in SPMS compared to those in HC (P = 0.0009; Fig. 5), while genes related with amino acid and lipid metabolisms were not significantly different among the HC and patient groups.
Fig. 5.
Metagenomic functional profile of three major nutrient metabolisms in the five subject groups. (A–C) Relative abundance of carbohydrate metabolism (A), amino acid metabolism (B), and lipid metabolism (C) at the KEGG subcategory level in the five groups. Each box plot represents median, interquartile range, minimum, and maximum values. *P < 0.05, **P < 0.01, ***P < 0.001 based on Wilcoxon rank sum test with the Benjamini–Hochberg method for multiple group comparisons.
Levels of Short-Chain Fatty Acid and Oxidative Stress Marker in the Feces of Patients with RRMS and SPMS.
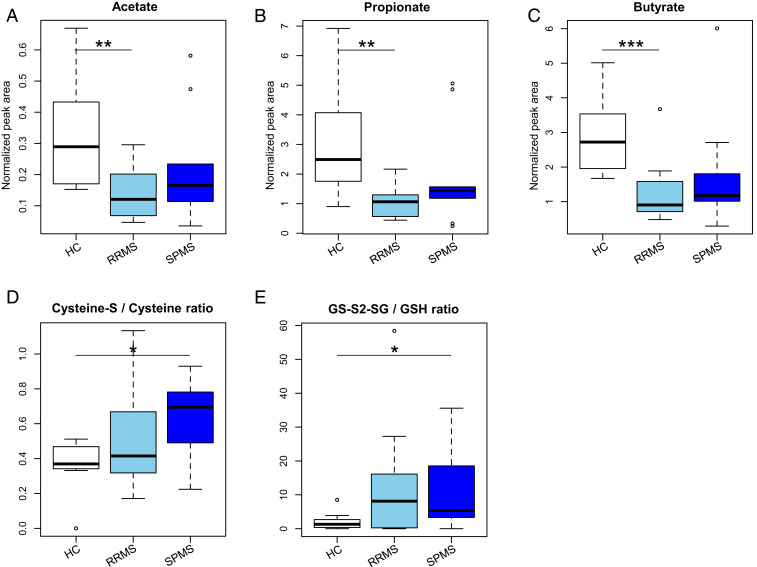
We also analyzed the levels of SCFAs in fecal samples from 12 patients with RRMS, 9 patients with SPMS, and 8 HCs (SI Appendix, Table S7). The results showed that the levels of fecal acetate, propionate, and butyrate in RRMS were significantly lower than those in HC (P = 0.0055, 0.0030, and 0.0007, respectively). Samples from SPMS also showed tendencies for reduction of the SCFAs as compared to those from HCs (Fig. 6 A–C).
Fig. 6.
Levels of short chain fatty acids and an index of oxidative stress in the feces of the patients with RRMS and SPMS. (A–C) The concentration of acetate (A), propionate (B), and butyrate (C) in the feces of the patients with RRMS, SPMS, and HC. (D and E) The ratio of cysteine persulfide to cysteine (D) and glutathione persulfide to glutathione (E) in the feces of the patients with RRMS, SPMS, and HC. Each box plot represents median, interquartile range, minimum, and maximum values. *P < 0.05, **P < 0.01, ***P < 0.001, based on Wilcoxon rank sum test.
Since an increased abundance of two genes involved in the mismatch repair pathway implied a high level of oxidative state in the SPMS samples, we additionally measured levels of sulfur compounds in the same fecal samples used for the SCFA analysis (SI Appendix, Table S7). We detected 19 sulfur compounds, of which cysteine and the oxidized form (cysteine-S), and glutathione (GSH) and the oxidized form (GS-S2-SG) were pairs of the oxidative and reductive forms useful for evaluation of the oxidative state, respectively (21). The ratio of cysteine-S (oxidative form) to cysteine (reductive form) and that of GS-S2-SG (oxidative form) to GSH (reductive form) were significantly higher in SPMS than in HC (P = 0.0152 and P = 0.0432), respectively (Fig. 6 D and E). We did not detect any other pairs of persulfide (oxidative form) and nonpersulfide (reductive form) in the present study. These findings further support the notion that an enhanced level of DNA mismatch repair is possibly caused by excessive oxidative stress in the gut of patients with SPMS. These results suggested a close association between excessive oxidative stress and an enhanced level of DNA mismatch repair in the gut of patients with SPMS.
Discussion
The aim of this study is to gain insights into the pathogenesis of the CNS autoimmune diseases. We first compared the fecal microbiome structure from four disease forms (RRMS, SPMS, atypical MS, and NMOSD) with healthy controls. The number of patients with RRMS and SPMS is much smaller in Japan than that of Western countries, but has been rapidly increasing (6), reminiscent of the increase of inflammatory bowel diseases (IBDs) in Asian countries (22, 23). This may be linked with westernization in lifestyle and subsequent alterations of gut microbiomes. Although the other two forms (atypical MS and NMOSD) are known to be relatively common in Japan, it appears that these forms may also be increasing.
All alpha-diversity indexes were similar among the gut microbiomes of the five groups. However, beta-diversity based on UniFrac metrics revealed significant differences in the overall composition of gut microbiomes between the RRMS and HC, between the SPMS and HC, and between the NMOSD and HC groups, indicating that the gut microenvironment in RRMS, SPMS, and NMOSD significantly differed from that in the HC group. Similar results were also obtained in previous work on the comparison between RRMS and HCs (12).
Several previous studies reported significant changes in gut microbiomes in RRMS compared with HCs, and identified the specific bacteria that might be associated with this disease (8–12). In a previous study in Japan, we identified 19 species that were reduced in RRMS. Of note, 14 of these species belonged to Clostridia XIVa and IV clusters (12). These clusters were highlighted as they contain a number of species capable of producing butyrate, which is potentially protective for MS, owing to its ability to suppress pathogenic T cells and enhance remyelination (24–26). Consistently, oral intake of butyrate significantly ameliorated the clinical course of an animal model of MS (27, 28). In the present study, we identified 9 species significantly reduced in RRMS as compared with HC. Notably, 8 of them belonged to Clostridia XIVa and IV clusters. The corresponding bacteria included E. rectale (mapped_CL145), M. funiformis (mapped_CL428), unmapped_OTU5, unmapped_OTU6, and unmapped_OTU31, whose reduction in RRMS was shown in the previous study (12). Notably, E. rectale (mapped_CL145), which was closely associated with recent activity in RRMS patients has the potential to most robustly produce butyrate among the gut bacteria (19, 20) and is the dominant species in the normal gut microbiomes (29). Moreover, we confirmed a reduced butyrate production in the gut of RRMS at metagenomic functional and metabolite levels, and we also revealed a reduced propionate biosynthesis in the gut of RRMS. A recent study showed that longitudinal supplementation of propionate significantly improved various clinical parameters in patients with MS accompanied by induction of colonic regulatory T cells (30). These results are in harmony with reduced butyrate and propionate production in the gut as characteristic of RRMS.
An increase of A. muciniphila (mapped_CL208) in RRMS compared with HC was highlighted in Western populations (8, 9, 11). In the present study, consistent results were obtained in Japanese populations. Published data indicate that A. muciniphila could play an important role in the pathogenesis of MS through the induction of proinflammatory T helper 1 (Th1) cells (9). The Th1-inducing ability of A. muciniphila was also highlighted in the field of cancer immunotherapy (31). However, as the abundance of this bacterium in Japanese populations is relatively small, biological implications in Japanese populations remain unclear at present.
The dietary habits have changed dramatically over the last century in developed countries, characterized by reduced dietary fiber and increased high-fat consumption (32). The tendency is most conspicuous in Asian countries, such as Japan. Although the deep-rooted Japanese traditional foods are enriched in dietary fiber derived from grains, beans, and vegetables, the prevailing “westernization” in Japan has led to the decreased intake of dietary fiber (33). In the research field of IBDs, which is also increasing like MS, epidemiological data suggest that the increase of IBDs in Japan can be linked to a reduced intake of dietary fiber, associated with a lower production of butyrate in the gut (34). Accordingly, we may speculate that a reduction of butyrate production in MS may result from “westernization.” Published data in healthy volunteers and gnotobiotic mice have revealed that dietary changes may rapidly affect the gut microbiome structure, and survival of E. rectale is highly dependent on uptake of dietary fiber (35, 36). The available data allow us to hypothesize that dietary changes led to the altered microbiome, such as a reduction of butyrate-producing species which lowers the threshold for developing MS in Japanese populations.
In the process of analyzing the gut microbiome structures in the subgroups of MS, the multiomics data disclosed the intriguing differences between SPMS and RRMS, which is in line with our hypothesis that transition from RRMS to SPMS may be driven by alterations of the gut microbiome (37). Metagenomic functional data disclosed an increase in microbial genes involved in DNA mismatch repair in SPMS as compared to RRMS, which we presumed to be caused by excessive oxidative stress. Supportive of this, sulfur metabolomics has revealed that a ratio of cysteine persulfide to cysteine, which is an indicator of oxidation, is significantly increased in the feces of patients with SPMS, allowing us to postulate that excessive DNA oxidation could take place in the gut of SPMS patients. Regarding the mechanism for the enhanced oxidative stress in the gut of SPMS patients, attention could be directed to the relative abundance of the genus Streptococcus (6.40%), which was the highest in SPMS among the five subject groups and significantly increased in SPMS compared to that in the HC group. Additionally, the abundance of S. parasanguinis (mapped_CL5) was strongly correlated with high-severity SPMS. Interestingly, most of the streptococcal species detected in the present study were oral streptococci (38), which produce hydrogen peroxide as a byproduct of aerobic metabolism (39). As such, it is possible to speculate on a partial causality between the increase in Streptococcus and the enhanced oxidative stress in the gut of SPMS patients.
We showed the significant reduction of the microbial carbohydrate metabolism in the gut of SPMS patients. As many of anaerobic species metabolize carbohydrates to produce SCFAs, carbon dioxide, and hydrogen (40), such an alteration in the gut of SPMS patients may lead to the depletion of gut-derived hydrogen, which is a ubiquitous molecule with an antioxidant property (41). As such, the reduced carbohydrate metabolism could be a mechanism for the enhanced oxidative states in the gut of SPMS patients. On the other hand, experimental studies have revealed that gut-derived hydrogen is rapidly transported into systemic circulation and ameliorates brain disease models by reducing oxidative stress (41, 42). Furthermore, innate immune-mediated oxidative injury in the CNS has been proposed as an important process underlying the progression of MS (43–45). Taken together, the reduction in microbial carbohydrate metabolism could be linked to SPMS pathogenesis through the depletion of gut-derived reductive hydrogen and subsequently enhanced oxidative stress.
It is a limitation of our study that our patient cohort was relatively small and derived from a single facility in Japan, although the strategy to focus on a population of the same nationality has a significant merit, considering the differences of the gut microbiome between different nations (29). Another limitation is that the proportion of treatment-naïve patients is very low in the cohort, although we showed the status of immunotherapy did not significantly affect the gut microbiome structure. Notably, early introduction of DMDs is being recommended in the clinical guidelines, and the difficulty of collecting treatment-naïve samples is widely appreciated. Finally, this is neither a prospective nor interventional study, and most data are not functionally validated, although the functional implications of reduced SCFAs and increased oxidative stress are supported by other studies.
Based on the multiomics data from four distinct MS-related disorders, we conclude that the ecological and functional microenvironment of the gut is differentially altered in the different stages of MS. Collective data indicate that the alterations of the gut microbiome in Japanese patients with MS may be at least partly explained by changes in dietary habits. Among the new information obtained, oxidative stress linked with SPMS may provide a target of therapy.
Materials and Methods
This study was approved by the National Center of Neurology and Psychiatry Ethics Committee (A2016-132) and the Research Ethics Committee of Waseda University and RIKEN Center for Integrative Medical Sciences (H30-4) (6). Signed informed consent was obtained from all subjects who provided specimens.
Participants.
At first, we recruited 98 patients with MS and 31 patients with NMOSD from ages 20 to 80. To avoid the effect of age on the gut microbiome, we subsequently excluded 11 patients with NMOSD whose onset age was over 50. This exclusion was based on a previous report that patients with late onset-NMOSD (onset age, over 50) have distinct clinical features compared to those with early onset (46). All 98 patients with MS included in this study fulfilled the McDonald criteria for diagnosis (47). In this study, we divided the total patients with MS into three clinical phenotypes based on a clinical and MRI-based method. The patients with MS-specific brain MRI lesions (48) were categorized as having typical MS, whereas the patients without these MS-specific brain MRI lesions were categorized as having atypical MS. Among the 77 patients with typical MS, RRMS was defined based on a relapsing remitting clinical course, whereas SPMS was diagnosed retrospectively by an attending physician based on an establishment of a sustained period of worsening neurological impairments (49). All patients with NMOSD included in this study were positive for serum anti-AQP4 antibody and fulfilled the international consensus diagnosis criteria for NMOSD (15). No patients had an active relapse at the time of study enrollment. Annualized relapse rate was defined as the number of relapses 1 y prior to sample collection. Clinical severity of the patients was evaluated by EDSS score which is a measure of neurological impairment based on clinical assessment of MS (16). Patients suffering from infectious diseases and treated with antibiotics during the collection of fecal samples were excluded from the study. Twelve patients with RRMS, two patients with SPMS, and two patients with atypical MS were not receiving any immunotherapies within at least 2 mo prior to sampling. We obtained 16S and metagenomic sequences from fecal samples of 55 age-adjusted healthy individuals from 104 healthy Japanese individuals (29, 50). Moreover, we obtained fecal samples for metabolite analysis from 12 patients with RRMS, 9 patients with SPMS, and 8 newly recruited healthy individuals. Detailed clinical characteristics of the enrolled patients and HCs are shown in SI Appendix, Tables S1 and S7.
DNA Preparation.
In accordance with a previously described method (51), freshly collected fecal samples were transported at 4 °C to the laboratory in a plastic bag containing a disposable oxygen-absorbing and carbon dioxide-generating agent in which anaerobes sensitive to oxygen can survive. In the laboratory, the fecal samples were suspended in phosphate-buffered saline containing 20% glycerol, immediately frozen using liquid nitrogen, and stored at −80 °C until use. Bacterial DNA was isolated and purified from the fecal samples according to enzymatic lysis methods.
Amplicon Sequencing of the 16S rRNA Gene V1–V2 Region.
The 16S V1–V2 region was amplified by PCR with barcoded 27Fmod (5′-agrgtttga- tymtggctcag-3′) and the reverse primer 338R (5′-tgctgcctcccgtaggagt-3′) (51). PCR was performed using 50 μL of 1× Ex Taq PCR buffer composed of 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, and 1.5 mM MgCl2 in the presence of 250 μM dNTPs, 1 unit Ex Taq polymerase (Takara Bio), forward and reverse primers (0.2 μM), and ∼20 ng of template DNA. Thermal cycling was performed in a 9700 PCR System (Life Technologies Japan) and the cycling conditions were as follows: initial denaturation at 96 °C for 2 min, followed by 20 cycles of denaturation at 96 °C for 30 s, annealing at 55 °C for 45 s, and extension at 72 °C for 1 min, with a final extension at 72 °C. PCR amplicons were purified using AMPure XP magnetic purification beads (Beckman Coulter Inc.) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies Japan). An equal amount of each PCR amplicon was mixed and subjected to multiplexed amplicon sequencing with MiSEq (2 × 300 paired-end run), according to the manufacturer’s instructions.
Metagenomic Sequencing.
To obtain metagenomic sequences, we performed whole-genome shotgun sequence analysis with IonProton (200-base reads with the Proton I chip) platforms according to the manufacturer’s protocol.
Analysis of the 16S rRNA Gene V1–V2 Region.
Two paired-end reads were merged using the fastq-join program based on overlapping sequences. Reads with an average quality value of <25 and inexact matches to both universal primers were filtered out. Filter-passed reads were used after trimming off both primer sequences. For chimera checking and taxonomy assignment of the 16S rRNA data, we constructed our own databases from three publicly available databases: Ribosomal Database Project (RDP) v. 10.27, CORE (microbiome.osu.edu/), and a reference genome sequence database obtained from the National Center for Biotechnology Information (NCBI) FTP site (ftp://ftp.ncbi.nih.gov/genbank/, December 2011). Reads having BLAST match lengths <90% with the representative sequence in the three databases were considered as chimeras and removed. Filtered reads were obtained from quality-control processes based on the previously described method (12). Specifically, 3,000 high-quality 16S reads, with an average quality value of >25, were randomly chosen from all filter-passed reads for each sample, and both primer sequences were trimmed. To evaluate alpha-diversity, the number of OTUs for each sample was obtained by clustering the 3,000 16S reads with a 97% identity threshold. To analyze beta-diversity, UniFrac distance analysis which is a phylogenic tree-based metric was used (52). To reveal the species level taxonomic assignment, the 16S reads were then mapped to 87,558 full-length (FL)-16S sequences as described in a previous article (12), based on a similarity search against 87,558 nonredundant FL-16S sequences using BLAST with ≥97% identity and ≥90% coverage in sequence alignment. The FL-16S sequences mapped by 16S reads were further clustered using USEARCH5 with a 97% identity cutoff to generate clusters of FL-16S equivalent to OTUs at the species level, defined as “mapped_CLs” in this study. Taxonomic assignment of 16S reads was performed based on the 97% FL-16S clusters to which they were mapped. Unmapped 16S reads were subjected to conventional clustering using USEARCH5 with a 97% identity cutoff to obtain the OTUs defined as “unmapped_OTUs” and were assigned to a higher-level taxon based on a similarity search against the 16S database. The number of 16S reads that mapped to each cluster and formed each unmapped_OTU was used to estimate the bacterial composition at the species, genus, and phylum levels.
Analysis of the Metagenomic Sequencing.
In Ion Proton reads, low-quality reads with an average quality value of <20 or <75 base pairs (bp) were removed and exact duplicates, 5′/3′ duplicates, reverse complement exact duplicates, and reverse complement 5′/3′ duplicates were removed with PRINSEQ. Human-derived reads were removed by mapping them to the human genome (HG19) with Bowtie2. The high-quality reads were then assembled into contigs using MEGAHIT (v.1.2.4). Contigs with a length of <500 bp were removed. Gene prediction was performed using Prodigal (53). The nucleotide sequences of predicted genes were clustered using CD-HIT-est v.4.6 with a ≥95% sequence identity and ≥90% coverage. The clustered gene sequences were then translated to proteins. Functional annotation of the predicted proteins was performed using BLASTP v.2.2.26 against the KEGG database (release 63) with an E-value of 1e−5. We mapped 1 million high-quality reads onto the nonredundant gene set using bowtie2 to obtain the quantified functional composition in each sample.
Analysis of Sulfur Metabolomics.
Freshly collected fecal samples were transported at 4 °C to the laboratory in a plastic bag containing a disposable oxygen-absorbing and carbon dioxide-generating agent. In the laboratory, the fecal samples were frozen and stored at −80 °C until use. Sulfur metabolomic analysis was performed by Sulfur Index service in Japan with liquid chromatography coupled to a tandem mass spectrometry (LC-MS/MS) system as described previously (54, 55). Briefly, the sulfur-containing compounds in the samples were extracted by adding methanol and converted to fluorescent derivatives with monobromobimane. The target metabolite levels were determined from the peak area by mass chromatography and represented as relative amounts after normalization with the peak area of the internal standard (D-camphor-10-sulfonic acid). In the process of data analysis, we excluded one HC subject in the comparison of the ratio of GS-S2-SG to GSH because GSH compounds were not detected in the sample from this subject.
Quantification of Fecal SCFAs.
For SCFA analysis, the prepared solution for the analysis of sulfur index was partially utilized, and 10 μL of the solution was added into 40 μL of Milli-Q water. Then, 50 μL of 50 mM 3-nitrophenylhydrazine hydrochloride (75% methanol), 50 μL of 50 mM 1-ethyl-3-(3-dimethylamino-propyl) carbodiimide hydrochloride (75% methanol), 7.5% pyridine (75% methanol), and internal standards (75% methanol) were added into the solution. After mixing for 30 min at room temperature, 5 μL of formic acid was added. The resulting solution was applied to the LC-MS/MS analysis, utilizing the “LC/MS/MS Method Package for Short Chain Fatty Acids” obtained from the Shimadzu Corporation according to the recommended procedures of the manufacturer.
Statistical Analysis.
All statistical analyses were conducted with R version 4.0.0 or Prism. A Wilcoxon rank sum test was used to compare the microbiome data between subject groups using the Benjamini–Hochberg procedure to compare data from more than two groups. A Kruskal–Wallis test and χ2 test were used for the comparison of patient demographic data. A Fisher’s exact test was used for the ranking of enriched or depleted metabolic pathways. We conducted a PERMANOVA to compare the overall microbiome structure between subject groups using the Benjamini–Hochberg procedure to compare data from more than two groups, as appropriate. The significance level was set at P value <0.05.
Supplementary Material
Acknowledgments
The authors thank H. Morita for technical support. We thank C. Shindo, E. Iioka, H. Yamaguchi, N. Yamashita, R. Kurokawa, and Y. Hattori for technical support. We thank A. Tomita, D. Noto, M. Nakamura, N. Chihara, T. Matsuoka, and R. Ikeguchi for obtaining informed patient consent and gathering samples. We thank I. Ohtsu and T. Nitta at Euglena Co., Ltd. for analyzing sulfur metabolomics and SCFAs in the fecal samples. This study was supported by the Practical Research Project for Rare/Intractable Diseases from Japan Agency for Medical Research and Development, AMED (16ek0109189h0001, 17ek0109189h0002, 18ek0109189h0003, and 19ek0109417h0001) and AMED-Core Research for Evolutional Science and Technology (18gm1010011h0001 and 19gm1010011h0002).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011703117/-/DCSupplemental.
Data Availability.
The 16S V1–V2 region sequences from 118 patients with MS and NMOSD that were analyzed in the present study were deposited in DDBJ/GenBank/EMBL with the accession number DRA007703 for the 16S sequences and DRA007704 for the metagenomic sequences.
References
- 1.Compston A., Coles A., Multiple sclerosis. Lancet 372, 1502–1517 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Raveney B. J. et al., Eomesodermin-expressing T-helper cells are essential for chronic neuroinflammation. Nat. Commun. 6, 8437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C. et al., Extrapituitary prolactin promotes generation of Eomes-positive helper T cells mediating neuroinflammation. Proc. Natl. Acad. Sci. U.S.A. 116, 21131–21139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallin M. T. et al.; US Multiple Sclerosis Prevalence Workgroup , The prevalence of MS in the United States: A population-based estimate using health claims data. Neurology 92, e1029–e1040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyake S., Yamamura T., Gut environmental factors and multiple sclerosis. J. Neuroimmunol. 329, 20–23 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Osoegawa M. et al.; Research Committee of Neuroimmunological Diseases , Temporal changes and geographical differences in multiple sclerosis phenotypes in Japanese: Nationwide survey results over 30 years. Mult. Scler. 15, 159–173 (2009). [DOI] [PubMed] [Google Scholar]
- 7.McKay K. A., Jahanfar S., Duggan T., Tkachuk S., Tremlett H., Factors associated with onset, relapses or progression in multiple sclerosis: A systematic review. Neurotoxicology 61, 189–212 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Berer K. et al., Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. U.S.A. 114, 10719–10724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cekanaviciute E. et al., Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U.S.A. 114, 10713–10718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J. et al., Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 6, 28484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jangi S. et al., Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7, 12015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyake S. et al., Dysbiosis in the Gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS One 10, e0137429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadowaki A. et al., Gut environment-induced intraepithelial autoreactive CD4(+) T cells suppress central nervous system autoimmunity via LAG-3. Nat. Commun. 7, 11639 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokote H. et al., NKT cell-dependent amelioration of a mouse model of multiple sclerosis by altering gut flora. Am. J. Pathol. 173, 1714–1723 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wingerchuk D. M. et al.; International Panel for NMO Diagnosis , International consensus diagnostic criteria for neuromyelitis optica spectrum disorders. Neurology 85, 177–189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurtzke J. F., Rating neurologic impairment in multiple sclerosis: An expanded disability status scale (EDSS). Neurology 33, 1444–1452 (1983). [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M., New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis P., Young P., Holtrop G., Flint H. J., Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 12, 304–314 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Vital M., Karch A., Pieper D. H., Colonic butyrate-producing communities in humans: An overview using omics data. mSystems 2, e00130-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parada Venegas D. et al., Short chain fatty acids (SCFAs)-Mediated Gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10, 277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada K. et al., Characterization of sulfur-compound metabolism underlying wax-ester fermentation in Euglena gracilis. Sci. Rep. 9, 853 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prideaux L., Kamm M. A., De Cruz P. P., Chan F. K., Ng S. C., Inflammatory bowel disease in Asia: A systematic review. J. Gastroenterol. Hepatol. 27, 1266–1280 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Ng S. C., Epidemiology of inflammatory bowel disease: Focus on Asia. Best Pract. Res. Clin. Gastroenterol. 28, 363–372 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Atarashi K. et al., Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Furusawa Y. et al., Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Chen T., Noto D., Hoshino Y., Mizuno M., Miyake S., Butyrate suppresses demyelination and enhances remyelination. J. Neuroinflammation 16, 165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haghikia A. et al., Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity 43, 817–829 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Mizuno M., Noto D., Kaga N., Chiba A., Miyake S., The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PLoS One 12, e0173032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishijima S. et al., The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 23, 125–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duscha A. et al., Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell 180, 1067–1080.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Routy B. et al., Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Maslowski K. M., Mackay C. R., Diet, gut microbiota and immune responses. Nat. Immunol. 12, 5–9 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Nakaji S. et al., Trends in dietary fiber intake in Japan over the last century. Eur. J. Nutr. 41, 222–227 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Andoh A. et al., Multicenter analysis of fecal microbiota profiles in Japanese patients with Crohn’s disease. J. Gastroenterol. 47, 1298–1307 (2012). [DOI] [PubMed] [Google Scholar]
- 35.David L. A. et al., Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desai M. S. et al., A dietary fiber-deprived Gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadowaki A., Saga R., Lin Y., Sato W., Yamamura T., Gut microbiota-dependent CCR9+CD4+ T cells are altered in secondary progressive multiple sclerosis. Brain 142, 916–931 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreth J., Merritt J., Qi F., Bacterial and host interactions of oral streptococci. DNA Cell Biol. 28, 397–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L., Kreth J., The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid. Med. Cell. Longev. 2012, 717843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki Y. et al., Are the effects of alpha-glucosidase inhibitors on cardiovascular events related to elevated levels of hydrogen gas in the gastrointestinal tract? FEBS Lett. 583, 2157–2159 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Ohsawa I. et al., Hydrogen acts as a therapeutic antioxidant by selectively reducing cytotoxic oxygen radicals. Nat. Med. 13, 688–694 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Fujita K. et al., Hydrogen in drinking water reduces dopaminergic neuronal loss in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson’s disease. PLoS One 4, e7247 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendiola A. S. et al., Transcriptional profiling and therapeutic targeting of oxidative stress in neuroinflammation. Nat. Immunol. 21, 513–524 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiner H. L., A shift from adaptive to innate immunity: A potential mechanism of disease progression in multiple sclerosis. J. Neurol. 255 (suppl. 1), 3–11 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Lassmann H., van Horssen J., Mahad D., Progressive multiple sclerosis: Pathology and pathogenesis. Nat. Rev. Neurol. 8, 647–656 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Collongues N. et al., Characterization of neuromyelitis optica and neuromyelitis optica spectrum disorder patients with a late onset. Mult. Scler. 20, 1086–1094 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Thompson A. J. et al., Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 17, 162–173 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Horowitz A. L., Kaplan R. D., Grewe G., White R. T., Salberg L. M., The ovoid lesion: A new MR observation in patients with multiple sclerosis. AJNR Am. J. Neuroradiol. 10, 303–305 (1989). [PMC free article] [PubMed] [Google Scholar]
- 49.Rovaris M. et al., Secondary progressive multiple sclerosis: Current knowledge and future challenges. Lancet Neurol. 5, 343–354 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Takayasu L. et al., A 3-dimensional mathematical model of microbial proliferation that generates the characteristic cumulative relative abundance distributions in gut microbiomes. PLoS One 12, e0180863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S. W. et al., Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 20, 241–253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lozupone C., Lladser M. E., Knights D., Stombaugh J., Knight R., UniFrac: An effective distance metric for microbial community comparison. ISME J. 5, 169–172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyatt D. et al., Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawano Y. et al., Involvement of the yciW gene in l-cysteine and l-methionine metabolism in Escherichia coli. J. Biosci. Bioeng. 119, 310–313 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Tanaka N., Kawano Y., Satoh Y., Dairi T., Ohtsu I., Gram-scale fermentative production of ergothioneine driven by overproduction of cysteine in Escherichia coli. Sci. Rep. 9, 1895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S V1–V2 region sequences from 118 patients with MS and NMOSD that were analyzed in the present study were deposited in DDBJ/GenBank/EMBL with the accession number DRA007703 for the 16S sequences and DRA007704 for the metagenomic sequences.