Abstract
Background
The genus Allium (Family: Amaryllidaceae) is an economically important group of crops cultivated worldwide for their use as a vegetable and spices. Alliums are also well known for their nutraceutical properties. Among alliums, onion, garlic, leek, and chives cultivated worldwide. Despite their substantial economic and medicinal importance, the genome sequence of any of the Allium is not available, probably due to their large genome sizes. Recently evolved omics technologies are highly efficient and robust in elucidating molecular mechanisms of several complex life processes in plants. Omics technologies, such as genomics, transcriptomics, proteomics, metabolomics, metagenomics, etc. have the potential to open new avenues in research and improvement of allium crops where genome sequence information is limited. A significant amount of data has been generated using these technologies for various Allium species; it will help in understanding the key traits in Allium crops such as flowering, bulb development, flavonoid biosynthesis, male sterility and stress tolerance at molecular and metabolite level. This information will ultimately assist us in speeding up the breeding in Allium crops.
Method
In the present review, major omics approaches, and their progress, as well as potential applications in Allium crops, could be discussed in detail.
Results
Here, we have discussed the recent progress made in Allium research using omics technologies such as genomics, transcriptomics, micro RNAs, proteomics, metabolomics, and metagenomics. These omics interventions have been used in alliums for marker discovery, the study of the biotic and abiotic stress response, male sterility, organ development, flavonoid and bulb color, micro RNA discovery, and microbiome associated with Allium crops. Further, we also emphasized the integrated use of these omics platforms for a better understanding of the complex molecular mechanisms to speed up the breeding programs for better cultivars.
Conclusion
All the information and literature provided in the present review throws light on the progress and potential of omics platforms in the research of Allium crops. We also mentioned a few research areas in Allium crops that need to be explored using omics technologies to get more insight. Overall, alliums are an under-studied group of plants, and thus, there is tremendous scope and need for research in Allium species.
Keywords: Allium, Genomics, Transcriptomics, Proteomics, Metabolomics, Metagenomics, Microrna
Introduction
Allium crops are cultivated globally due to their importance as a vegetable, condiment, and spice with medicinal properties. The genus Allium belongs to the family Amaryllidaceae of the order Asparagales. Telomerase sequence variation is responsible for the divergence of Allium from other genera belonging to Asparagales. The genus Allium comprises more than 920 species (Seregin, Anačkov & Friesen, 2015), and among them, onion, garlic, bunching onion, leek, and shallots are widely cultivated because of their economic importance. The total production value of Allium crops in 2014 was US$61348 million (http://www.fao.org/faostat/en/#data/QC), of which dry onion and garlic contributed a major share of 70% and 25%, respectively. China, India, and the United States of America are among the major Allium producing countries.
The biannual life cycle, high cross-pollination, and inbreeding depression in onion and vegetative propagation, and lack of flowering, especially in garlic, are the main barriers to the conventional breeding of these major Allium crops. Alliums are known to possess one of the largest genomes (10–30 pg/1C) among vegetable crops (Ricroch et al., 2005). Thus, sequencing and assembling of these genomes become difficult, leading to limited availability of markers (Chinnappareddy et al., 2013; Khosa et al., 2016a; Khosa et al., 2016b). Therefore, mapping and genomics-assisted breeding in Allium crops are lagging (Shigyo, Khar & Abdelrahman, 2018) compared with other crops, such as rice, wheat, and tomato. Presently, we are witnessing tremendous developments and data generation from various omics experiments such as genomics, transcriptomics, proteomics, metabolomics, and metagenomics. These approaches have the potential to increase the speed and accuracy of analysis ofthe complex molecular process, ultimately leading to the development of new strategies in breeding programs for Allium improvement.
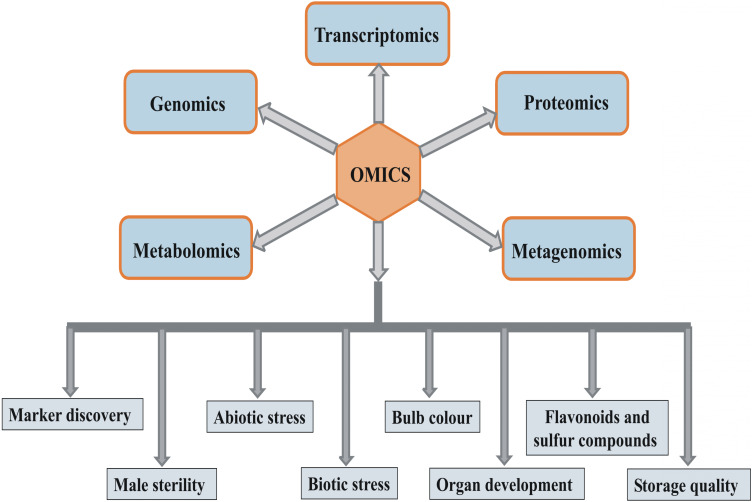
Next-generation sequencing (NGS) has recently been used for transcriptome analyses of hundreds of model as well as non-model plant species (Martin et al., 2013). RNA sequencing (RNA-seq) is a rapid and inexpensive technique that is independent of genome complexity, and thus, NGS has emerged as a method of choice for expression analyses in crops where genome sequence information is unavailable. In alliums, RNA-seq was used for studying telomeres, molecular mechanism underlying male sterility, flowering, abiotic stress, and bulb color, and for marker discovery (Duangjit et al., 2013; Abdelrahman et al., 2015; Kamenetsky et al., 2015; Fajkus et al., 2016; Baek, Kim & Kim, 2017; Yuan et al., 2018). The mRNA expression profile differs from the pattern of protein levels, and the correlation between them is often reported in the range of 0.3–0.5 (Voelckel, Gruenheit & Lockhart, 2017). It may be due to protein degradation, post-transcriptional and post-translational regulations (Voelckel et al., 2010). Protein levels are often closely associated with the trait compared to the transcript profile, and therefore, proteomics can be more reliable for studying plant development and responses to stress. Proteomics is an excellent research tool when genome sequence information is limited. A few proteomic studies have been conducted in Allium crops to determine abiotic stress tolerance (Chen et al., 2013; Qin et al., 2016) and garlic fertility (Shemesh-Mayer et al., 2015). Nowadays, metabolomics is used to gain insights into metabolites involved in specific cellular and developmental processes and stress responses in plants. Because of its rapid and accurate analyses, metabolomics can be used for selecting superior genotypes based on the quantity of metabolites of interest in the breeding program. Metabolomics has a vast potential for the characterization of flavonoids and sulfur (S) compounds in Allium crops. Garlic and onion metabolomes were studied for antimicrobial compounds (Farag et al., 2017), authentication of genotypes (Hrbek et al., 2018), and S-containing compounds (Nakabayashi & Saito, 2017). Thus, overall, omics approaches have a massive potential in Allium research (Fig. 1). Because Allium crops are yet unexplored in terms of omics compared with other crops, omics approaches can assist improvement in these crops.
Figure 1. Various omics approaches and their potential application in study of different areas of Allium crops improvement.
Survey Methodology
An extensive literature survey for published research papers in the area of omics approaches in Allium crops was conducted using different databases (e.g., Pubmed, Google Scholar, Science Direct, and Scopus). To collect all relevant information, we built our literature search by using the following keyword combinations: genomics and alliums, transcriptomics and alliums, proteomics and allium s, metabolomics and alliums, metagenomics, and alliums. These articles were critically studied and used as references for the present review article. Our literature survey was not limited to specific time, area, institute, and authors, and thus, a comprehensive and unbiased review was ensured.
Genomics
During the 19th century, the breeding approach, particularly in onions, was based only on mass selection. Further, the discovery of male sterility led to the development of F1 hybrids during the mid-20th century (McCallum et al., 2008). Recently, hybrids, as well as open-pollinated varieties, have considerably increased onion productivity. However, overall, limited improvement is observed in Allium crops due to some inherent reasons, such as high heterozygosity, inbreeding depression, sexual sterility in garlic, clonal propagation, sensitivity toward photoperiod, and temperature. The whole-genome sequence assembly, which can be a crucial resource for functional genomics in Allium crops, is not yet available for any of the Allium members.
Recently, NGS platforms have been routinely used for the discovery of markers such as SSRs and SNPs. Baldwin et al. (2012a) developed advanced genomic SSR markers using NGS for the double dhaploid population; 921 SSRs were recognized from the genomic sequence of 6.6 Mbp. Whole-genome scanning methods have developed numerous SNPs in onion. Advanced genotyping technologies and doubled haploid in onion allowed frequent identification of SNPs (Kim et al., 2014; Finkers et al., 2015; Shigyo, Fujito & Sato, 2019). The frequency of SNPs in onion is 1.7 SNPs/Kb, which is similar to that in other plant species. In massive onion germplasm, SNPs were validated, and the existing linkage map was mapped (Duangjit et al., 2013). A high-throughput genotyping approach, such as genotyping-by-sequencing (DArTseq), was utilized for the first time for diversity and structure analyses of a large garlic population with 417 accessions. These analyses have enabled in identifying unique and redundant accessions (Egea et al., 2017). Jo et al. (2017) developed an onion genetic map by using the genotyping-by-sequencing approach. High-fidelity SNPs that satisfied the segregation ratio criteria were used for mapping the F2 onion population (NW-001 X NW-002). The map generated was 1383 cM in length,with a marker interval of 8.08 cM. Markers developed from the genic region have greater transferability than those from noncoding areas. Recently, the double digest restriction site-associated DNA sequencing (ddRAD-seq) approach was used for SNP discovery in inbred lines of Korean short-day onion. A total of 1904 SNPs were discovered and used in population structure analyses and genetic relationship studies (Lee et al., 2018). Such SNPs or SSRs developed using NGS technologies can be helpful in developing precise linkage maps and managing genetic resources in onion as well as other related Allium species.
Nuclear genome
Onion possesses a genome of 1C=16.75 pg, which is several times larger than the rice genome (1C=0.6 pg) (Bennet & Smith, 2002). Other Allium species also exhibit larger genome sizes ranging from 7.4 Gb in Allium sibiricum to 72.9 Gb in A. validum (Shigyo, Khar & Abdelrahman, 2018). Among alliums, researchers have attempted sequencing the onion genome because of the economic importance of onions. Suzuki, Do & Mukai (2002) developed an onion BAC library consisting of 48,000 clones, which covered 0.39% of the genome. Kuhl et al. (2004) sequenced a normalized cDNA library yielding 18,388 sequences. The onion genome is low in GC content, and majority of the genome comprises repetitive sequences (75%–80%) (Do, Suzuki & Mukai, 2004). Among plants, the gene density of onion is the lowest (1 gene/168 Kb), as confirmed by BAC-mediated and whole-genome shotgun sequencing projects. Pilot onion genome sequencing of two BACs revealed AT-rich (64.8%) sequences and long transposable elements, as evidenced in other larger plant genomes. Utilizing the methyl filtration technique potentially reduces unspecified and transposon sequences and produces approximately 10% higher gene encoding sequences (Jakše et al., 2008). The SEQUON project was recently initiated by Netherland to sequence the onion genome; under this project, four TrueSeq genomic DNA libraries were sequenced using IlluminaHiSeq 2500 (Finkers et al., 2015). The resulted assembly was 10.8 Gb with 6.2 M contigs. Such a higher proportion of the genome coverage indicates the genome-wide distribution and sufficient divergence of ancient repeats, which can be assembled efficiently. In the present study, the authors propose that recently developed long-read sequencing platforms can further improve the assembly.
As mentioned earlier, majority of the Allium genome comprised repetitive sequences (Do, Suzuki & Mukai, 2004) that are composed of satellite DNA, tandem repeats, and transposable elements. A repeatome is the total quantity and types of repetitive DNA in the genome. The repeatome plays a crucial role in the evolution and organization of genomes (Maumus & Quesneville, 2014). Repeat DNA sequences in Allium crops were identified and mapped using cytogenetic techniques such as FISH (Kiseleva, Kirov & Khrustaleva, 2014; Mancia et al., 2015; Kirov et al., 2017). Hertweck (2013) performed comparative analyses of transposable elements from Asparagales and reported conservation as well as wide variation in the repeatomes of different taxa in Asparagales. Recently, repeatomes of few Allium species were studied using low-coverage genome sequencing by employing NGS and bioinformatic tools. More than 90% of the onion genome has been reported to be repetitive in nature (Fu et al., 2019). Repeatomes of A. cepa, A. sativum, and A. ursinum were dissected using NGS technology,and the most prevalent repeat was Ty3/gypsy elements. Comparative analyses revealed that these repeats are significantly diverged in these species; hence, the authors could not detect common clusters during the analyses (Peška et al., 2019; https://www.ibp.cz/local/data/allium/). Studies on repeatome scan help in understanding speciation and karyotype evolution processes in Allium species. Further, knowledge of repeated DNA can be exploited for marker discovery and breeding in alliums.
Telomere sequences play a key role in the phylogenetic divergence of Asparagales. The last switch is responsible for the divergence of Allium from other genera. The omics approach was used to study telomeres in Allium species. Fajkus et al. (2016) used RNA-seq data and other techniques to characterize unusual telomeric sequences in Allium species. Recently, Fajkus et al. (2019) characterized the telomerase RNA of different species in Asparagales by using transcriptome and CRISPR/Cas-mediated genome editing (https://www.ibp.cz/ local/data/telomeraserna/). Interesting findings of these studies can facilitate the understanding of the functions of telomerase and telomere in the divergence of various taxa.
Mitochondrial genome
Researchers have recently completed the sequencing of the mitochondrial genome of onion varieties (Table 1); the data obtained can help in understanding the male sterility mechanism in onion and its further use in breeding programs. The mitochondrial genome of onion (316 kb) containing the CMS-S male-sterile cytoplasm was sequenced using the Illumina NextSeq500 platform. They also identified cox1 as part of chimeric orf725, which is one of the candidate genes for CMS (Kim et al., 2016). The mitochondrial genome of the CMS-S-type onion variety “Momiji-3” was characterized which have a multi-chromosomal structure resulting from recombination events. Examination of transcript data revealed RNA editing at 635 positions, and a candidate gene for CMS in “Momiji-3” was also found to be orf725 (Tsujimura et al., 2019). Similarly, Kim, Yang & Kim (2019) compared the mitochondrial genome of two recently diverged cytoplasms—male-fertile and male-sterile CMS-T-like cytoplasms and obtained almost identical sequences. They also identified a chimeric gene, orf725, as a candidate gene for CMS. The mitochondrial genome of more Allium species with different cytoplasms should be sequenced for an improved understanding of the mitochondrial genome.
Table 1. List of mitochondrial and chloroplast genome sequence of Allium species.
| Species | Genome | Platform | Length (bp) | GC % | References |
|---|---|---|---|---|---|
| Allium cepa | Mitochondria | NextSeq500 | 316,363 | N.A. | Kim et al. (2016) |
| Allium cepa | Mitochondria | NextSeq 500 | 316,363 and 339,180 | N.A. | Kim, Yang & Kim (2019) |
| Allium cepa | Mitochondria | GS FLX | 316288 | 45.3 | Tsujimura et al. (2019) |
| Allium cepa | chloroplast | 454 FLX | 153 538 and 153 355 | 36.8 | Von Kohn, Kiełkowska & Havey (2013) |
| Allium cepa | Chloroplast | HiSeq2000 | 153,529, 153,440, and 153,568 | N.A. | Kim et al. (2015) |
|
Allium victorialis |
Chloroplast | HiSeq2000 | 154,074 | 36.48 | Lee et al. (2017) |
| Allium prattii | Chloroplast | Hiseq2000 | 154,482 | 37.02 | Jin et al. (2018) |
| Allium obliquum | Chloroplast | HiSeq 1500 | 152,387 | 36.8 | Filyushin et al. (2018) |
| Allium sativum | Chloroplast | HiSeq 1500 | 153,172 | N.A. | Filyushin et al. (2016) |
|
Allium fistulosum |
Chloroplast | HiSeq 2500 | 153,164 | 36.8 | Yusupov et al. (2019) |
|
Allium monanthum |
Chloroplast | Hiseq 2500 | 154,804 | 37 | Xie et al. (2019a); Xie et al. (2019b) |
| A. chrysanthum | Chloroplast | Hiseq 2500 | 153,621 | 36.8 | Xie et al. (2019a); Xie et al. (2019b) |
| A. rude | Chloroplast | Hiseq 2500 | 153,697 | 36.7 | Xie et al. (2019a); Xie et al. (2019b) |
| A. xichuanense | Chloroplast | Hiseq 2500 | 153,673 | 36.7 | Xie et al. (2019a); Xie et al. (2019b) |
| A. chrysocephalum, | Chloroplast | Hiseq 2500 | 153,710 | 36.8 | Xie et al. (2019a); Xie et al. (2019b) |
| A. maowenense, | Chloroplast | Hiseq 2500 | 153,608 | 36.8 | Xie et al. (2019a); Xie et al. (2019b) |
| A. herderianum | Chloroplast | Hiseq 2500 | 153,605 | 36.8 | Xie et al. (2019a); Xie et al. (2019b) |
| A. fistulosum | Chloroplast | HiSeq 4000 | 153,162 | 36.8 | Huo et al. (2019) |
| A. tuberosum | Chloroplast | HiSeq 4000 | 154,056 | 36.9 | Huo et al. (2019) |
| A. sativum | Chloroplast | HiSeq 4000 | 153,189 | 36.7 | Huo et al. (2019) |
| A. cepa | Chloroplast | HiSeq 4000 | 153,586 | 36.8 | Huo et al. (2019) |
|
Allium ovalifolium |
Chloroplast | HiSeq 4000 | 153,635 | 37 | Sun et al. (2019) |
Chloroplast genome
The chloroplast genome is known for its conserved nature, maternal inheritance, and collinear gene order. Therefore, the chloroplast genome is widely used for phylogeny and diversity analyses in plants. The chloroplast genome of several Allium species have been characterized using NGS platforms (Filyushin et al., 2018; Xie et al., 2019a; Yusupov et al., 2019) and used for marker discovery (Lee et al., 2017; Kim, Park & Yang, 2015), phylogenetic analyses (Xie et al., 2019b; Huo et al., 2019), and the study of male sterility (Von Kohn, Kiełkowska & Havey, 2013; Kim, Park & Yang, 2015; Table 1). The complete chloroplast genome is 152,387–154,482 bp in length,with a GC content of 36%–37% (Filyushin et al., 2016; Lee et al., 2017; Xie et al., 2019a; Xie et al., 2019b; Yusupov et al., 2019). Minor variations in the length of the chloroplast genome, among the Allium species, might be due to the occurrence of indels. The number of genes in this genome ranges from 114 to 141. Repeat analyses revealed that all chloroplast genomes contain similar repeat sequences (Huo et al., 2019). Information generated from chloroplast genome sequencing could be harnessed for studying and improving Allium crops.
Transcriptomics
Transcriptomes are being routinely studied using the RNA-seq approach because of the affordability of sequencing and availability of analyses pipelines for extracting meaningful and useful information. Transcriptomes facilitate transcript profiling of a tissue at a specific stage or time and enable validation and annotation of these putative differentially expressed transcripts. Sun et al. (2013) and Baldwin et al. (2012b) performed transcriptome characterization in garlic and doubled haploid onion respectively for the first time. Thereafter, several published articles have reported the use of RNA-seq for studying organ development, male sterility, marker discovery, abiotic stress response, and flavonoid synthesis in various Allium species (Table 2). Kim et al. (2015) attempted structural annotation of onion RNA-seq by using an integrated structural gene annotation pipeline. Similarly, Sohn et al. (2016) developed a draft reference transcript for onion by using long-read sequencing technology, suggesting that further progress in sequencing chemistry will help in whole-genome sequencing in alliums. Kazusa DNA Research Institute and Yamaguchi University, Japan, have developed a transcriptome database for Allium species—“AlliumTDB” (http://alliumtdb.kazusa.or.jp/). This database harbors transcriptome data of 12 different libraries of root, stem, bulb, and leaves of different species and their doubled haploids. It is a vital resource of genetic information in Allium crops. The desired information can be retrieved from the “KEYWORD” page (http://alliumtdb.kazusa.or.jp/keyword.html) by searching the keywords as genes, TFs etc. This key word search results into various available information on the searched terms from many databases like TAIR, NCBI NR, and RAP-DB. To discriminate sequences of transcription factors in AlliumTDB, we searched for the important transcription factors like WRKY (645), AP2/ERF (863), NAC (735), Myb (1970), DOF (293), HSF (256), ARF2 (123), ARF1 (95), LFY (6), C2H2-type zinc finger (1193), Basic-leucine zipper (759), GATA type Zinc finger (405) etc. The results have been summarized in to File S1 having the set name of unigene (DB), sequence name of unigene (AC), % identity (Iden) and unigene length (Annotation) etc. The sequences of WRKY4 TFs were filtered and aligned on Mega6 Software (Tamura et al., 2013) using Maximum Likelihood method (Nei & Kumar, 2000) under the JTT matrix-based model (Jones, Taylor & Thornton, 1992). The sequence diversity of all the sequences from A. cepa transcripts is presented in Fig. 2.
Table 2. List of the RNA seq analyses performed in Allium species.
| Species | Platform | Tissue | Purpose/observation | Reference |
|---|---|---|---|---|
| Allium sativum | Illumina HiSeq 2000 | Dormant and sprouting bud | Discovery of genes in sulfur assimilation | Sun et al. (2012) |
| Allium sativum | Illumina HiSeq 2500 | Whole plant of 10 and 45 days old | Developed SSR marker in garlic and studied their transferability to other Allium species | Liu et al. (2015) |
| Allium sativum | MiSeq v3 platform |
Leaves, basal plate, roots inflorescence, flowers, cloves | Organ specific transcriptome for identification key gene and mechanism of organ development | Kamenetsky et al. (2015) |
| Allium sativum | Roche 454-FLX | Leaves, pseudostems, and roots |
SNPs and indels discovery | Havey & Ahn (2016) |
| Allium sativum | Illumina HiSeq 2000 | Flower buds of male sterile and fertile lines | Energy deficiency of tapetum cells might be reason of male sterility in garlic | Shemesh-Mayer et al. (2015) |
| Allium sativum | Illumina HiSeq™ 2000 | Shoot apex | Genes differentially expressed in shoot apex were identified | Sun et al. (2013) |
| Allium cepa | Illumina HiSeq 2500 | Leaf | Discovery of NAC transcription factor in onion | Zheng et al. (2016) |
| Allium cepa | Illumina HiSeq 2000 | Outer, intermediate and inner scale | Programmed cell death in onion skin formation | Galsurker et al. (2017) |
| Allium cepa | Illumina HiSeq 2000 | Bulb | ISGAP was used for higher accuracy of transcript annotation | Kim et al. (2015) |
|
Allium cepa |
Roche 454 FLX platform | Bulbs, tissue from leaves, unopened umbels, bulbs, and roots |
SNP development based on saturated map | Duangjit et al. (2013) |
| Allium cepa | Illumina HiSeq 2000 | Leaf | Differentially expressed genes were identified at freezing temperature and development of SSR and SNPs markers | Han et al. (2016) |
| Allium cepa | HisSeq™ 2500 | Bulb | Dissected sucrose metabolism during bulb formation | Zhang et al. (2016) |
| Allium cepa | Illumina HiSeq™ 2000 | Anthers at the tetrad stage | Identification of differentially expressed gene in anthers of sterile and maintainer line | Yuan et al. (2018) |
| Allium cepa | Illumina HiSeq 2000 | Bulb | The AcPMS 1 involved in DNA mismatch Repair, is the best candidate for fertility restoration |
Kim et al. (2015) |
| Allium cepa | 454™ GS-FLX | Leaves and shoot | Developed toolkit for bulk PCR marker designing from transcriptome data | Baldwin et al. (2012b) |
| Allium cepa | Illumina HiSeq 2000 | Scales | Studied differential response of outer and inner scales to the heat treatment | Galsurker et al. (2018) |
| Allium cepa | Illumina HiSeq 4000 |
Bulb | Variation in bulb colour | Zhang et al. (2018) |
|
Allium cepa and interspecific hybrids |
Illumina HiSeq2000 | Leaves | Develoment of SNP markers from interspecific hybrids for introgression breeding | Scholten et al. (2016) |
| Allium cepa | PacBio_RSII platform, P4-C2 chemistry | Flowers, leaves, bulbs and roots | Long read sequencing was used to construct a draft reference transcripts | Sohn et al. (2016) |
| Allium porrum | Illumina HiSeq 2500 | Leaves | Identification of CONSTANS-like genes which play role in flowering | Liu et al. (2018) |
| Allium porrum | Illumina MiSeq | Leaves and cloves | Elucidation of organosulfur metabolic pathway | Mehra et al. (2019) |
| Allium fistulosum | Illumina HiSeq 2500 | Bulb | Study of saponin biosynthetic pathway and their possible role in fusarium resistance | Abdelrahman et al. (2017) |
| Allium fistulosum | Illumina HiSeq 2000 | Leaves, false stem, basal plate and root |
Gene for sulfur and selenium metabolism were identified, SSR marker were developed | Sun et al. (2016) |
| Allium fistulosum | GS-FLX and HiSeq 2000 | 2-week-old seedlings, leaf, roots, basal meristem, immature flower bract, mature bract, opened flowers, immature fruits and sliced pseudostem |
EST markers were developed for mapping and these marker will enable comparison with bulb onion | Tsukazaki et al. (2015) |
| Allium fistulosum | Illumina HiSeq 2000 | Leaves | Identified four genes for wax content and developed 1,558 SSR markers | Liu et al. (2014) |
| Allium fistulosum | Illumina HiSeq 2000 | Inflorescences | Differential gene expression in CMS and maintainer line | Liu et al. (2016) |
| A. fistulosum, shallots and 8 monosomic addition lines | HiSeq 2500 | Bulbs | Found hot spot for flavonoid synthesis on chromosome 5A | Abdelrahman et al. (2019) |
|
Allium tuberosum Rottler ex Spr |
Illumina HiSeq 2000 | Leaves, shoots and roots | Performed gene annotation and SSR identification in Chinese chive | Zhou et al. (2015) |
|
A. sativum, A. porrum, A. tuberosum, A. Fistulosum, A. ascalonicum, A. cepa, A. cepa var. agrogarum, A. chinense, A. macrostemon |
Illumina HiSeq 2500 | Leaf | PCD might involve in development of fistular leaves | Zhu et al. (2017) |
|
A. ursinum, A. cepa, A. angulosum, A. cernuum, A. ericetorum, A. fistulosum, A. nutans, A. sativum, A. scorodoprasum, A. senescens and A. vineale |
Illumina HiSeq 2000 | Leaves and roots | Reported that the unusual telomeric sequence present in Allium species, demonstrated the synthesis of telomere by telomerase | Fajkus et al. (2016) |
|
A. nutans, A. cepa, A. ursinum, A. angulosum, A. ericetorum, A. fistulosum, Tulbaghia violacea, Scilla peruviana, and Cestrum elegans |
Illumina NextSeq500 | Leave sand root | Studied telomeres in order Asparagales | Fajkus et al. (2019) |
Figure 2. Inferred Ancestral Sequences of WRKY4 like sequences from AlliumTDB.
Marker discovery
The limitation of DNA markers is one of the hurdles in the breeding ofAllium crops and their improvement (Chinnappareddy et al., 2013; Khosa et al., 2016a; Khosa et al., 2016b). The RNA-seq approach has been widely used in the discovery of microsatellites SSRs and SNPs in several species. In alliums, few attempts have been made to develop markers from transcriptome data and linkage maps. Baldwin et al. (2012a) and Baldwin et al. (2012b) performed RNA-seq of the doubled haploid bulb onion “CUDH2150” and the genetically distant “Nasik Red” by using 454™ sequencings, and the mapping of reads revealed 16836 indels and SNPs. These markers have been further used for developing a linkage map of more than 800 cM covering all marker linkage groups. Transcriptomes of two inbred lines of onion were sequenced on the Roche-454 platform, and 3364 SNPs were identified on 1716 cDNA contigs. These SNPs were further used in the genetic mapping of different mapping populations (Duangjit et al., 2013). RNA-seq was used for identifying SNPs from male-fertile and male-sterile onion genotypes. From 141 contigs, 430 homozygous SNPs were identified and further used for identifying candidate genes for male sterility (Kim et al., 2015). Similarly, Shigyo, Fujito & Sato (2019) highlighted the use of RNA-seq-derived SNP markers in the development of high-density maps. A garlic transcriptome was sequenced using the Illumina platform that yielded 135360 unigenes. The EST data were used for developing 1506 SSR markers (Liu et al., 2015).
Similarly, microsatellite, SNP, and indel markers were discovered from the transcriptome data of garlic (Havey & Ahn, 2016; Chand et al., 2015). Tsukazaki et al. (2015) used the transcriptome shotgun assembly and Illumina HiSeq 2000 for RNA-seq analyses in A. fistulosum. Analyses of 54904 unigenes led to the discovery of 2396 SSRs, 9002 SNPs, and 4335 indels. These markers were used in the genetic mapping of A. fistulosum in comparison with bulb onion due to the availability of common markers. Similarly, EST-SSR markers were developed for Welsh onion (Yang et al., 2015) and Chinese chive (Zhou et al., 2015) from their respective transcriptome data.
Male sterility
Cytoplasmic male sterility is one of the most crucial traits of Allium crops and is routinely exploited for hybridization. However, the mechanism of male sterility is not well characterized at the molecular level in alliums. In onion, several molecular markers have been developed for selecting CMS genotypes. Kim et al. (2015) used BSA and RNA-seq analyses to identify candidate genes for restoring fertility. Several SNPs that differentiate male-sterile and male-fertile bulks were identified, along with 14 contigs that showed their perfect association withthe Ms locus. Among these contigs, AcPMS1 involved in the DNA repair mechanism is the best candidate gene and the most reliable marker for selecting the restorer line. Further, the transcriptome of the tetrad stage anther from male-sterile and maintainer lines was analyzed. Five genes that are associated with male sterility in onion were identified, of which two were cytoplasmic (atp9 and cox1) and three were coded by the nucleus (SERK1, AG, and AMS) (Yuan et al., 2018). Differential gene expression in male-sterile and maintainer lines of Welsh onion was investigated using the transcriptome approach, and more than 4000 unigenes were found to be differentially expressed, including genes known for their role in male sterility, such as F-type ATPase , NADH dehydrogenase , and cytochrome c oxidase (Liu et al., 2016). These studies have revealed that CMS in alliums is governed by both mitochondrial as well as nuclear genes and the interaction between them. The male-sterile garlic genotype showed tapetal hypertrophy, which is a cause of sterility. Integrated transcriptome and proteome analyses revealed several differentially expressed genes and proteins in male-sterile and fertile genotypes. These candidate genes (AP3, ms2, mmd1, gpat2, nad7, ccmC, cox2, 18S rRNA, flavanol synthase , and sod) were involved in male sterility in garlic. Physiological and molecular analyses indicated that respiratory restriction or non-regulated programed cell death (PCD) might result in energy deficiency, and in turn, sterile pollens (Shemesh-Mayer et al., 2015).
Abiotic stress
Plants are consistently subjected to various types of environmental stress because of their sessile nature and due to weather uncertainty in the field. This stress is responsible for significant yield reduction and economic loss of up to 50% (Atkinson & Urwin, 2012). Because climate change affects crops, we need to develop stress-tolerant and climate-smart varieties of crops. Roots of Allium crops are shallow, with a maximum root depth of 0.18 meter, and thus, a slight deepening of the water level renders onion prone to drought stress (Drinkwater & Janes, 1955). Flooding, waterlogging, and salinity severely affect the yield of Allium crops (Yiu et al., 2009; Sta-Baba et al., 2010).
The abiotic stress response in Allium crops has not been studied in detail; RNA-seq has enormous potential in elucidating these stress response mechanisms. The shallot is well adapted to abiotic stress compared with onion (Currah, 2002). Differential transcriptomic and metabolomics analyses were performed in the doubled haploids of onion and shallots. Genes involved in amino acid, osmoprotectant, and flavonoid biosynthesis were significantly upregulated in the doubled haploids of shallot (Abdelrahman et al., 2015). These genes might play a role in improved adaptability of shallot to abiotic stress, and thus, they may be used as candidate genes in onion breeding for stress tolerance. The response of cold-tolerant and -susceptible onion genotypes was studied using a transcriptomic approach at cold and freezing temperatures. A total of 491 genes were differentially expressed at freezing temperatures among cold-tolerant and susceptible onions (Han et al., 2016). These genes need to be explored further for better adaptation of onion crops growing at high altitudes to low temperature. Dufoo-Hurtado et al. (2013) reported the upregulation of phenolic metabolism-related genes in garlic after low-temperature conditioning of “seed” cloves. NAC (NAM, ATAF, and CUC) transcription factors play an essential role in plant stress response as well as development. Zheng et al. (2016) identified 39 NAC genes from onion leaves by using RNA-seq and classified them into five groups of NAC transcription factors.
Biotic stress
Majority of alliums constitute economically important cash crops, and significant losses occur due to diseases and pests in these crops. Agrochemicals applied to manage biotic stress lead to an increase in the cost of production, ultimately reducing the income of farmers. There are minimal resistance genes available in Allium crops. RNA-seq can prove to be an excellent tool for discovering novel genes for pest and disease resistance in crops such as Allium, where the draft genome sequence is not available. Till date, no single report has published on transcriptome analyses in alliums concerning biotic stress responses. In onions, limited resistance sources are available, and therefore, related or wild species need to be explored for disease resistance . Scholten et al. (2016) performed transcriptome analyses of interspecific hybrids of A. roylei and A. fistulosum and onion varieties. SNP markers developed from these data were used for linkage and QTL mapping. QTL for resistance to Botrytis squamosa was identified on the 6th chromosome of A. roylei. Steroidal saponins are involved in plant defense against different biotic stress. Abdelrahman et al. (2017) isolated the saponin compound called alliospiroside A from the monosomic addition lines of A. fistulosum and reported its antagonistic activity against Fusarium. RNA-seq analyses of these monosomic lines and normal lines led to the identification of 50 unigenes for saponin biosynthesis and found the upregulation of these genes in the monosomic addition line of A. fistulosum. Cuticlar and epicuticular wax depositions on onion leaves are known to involve in biotic stress tolerance, especially against onion thrips (Damon, Groves & Havey, 2014; Silva et al., 2015). RNA-seq analyses of a waxy and non-waxy mutant of Welsh onion revealed differential expression of 798 genes. Four of these genes were validated using qPCR, and COG annotation revealed that they were involved in lipid biosynthesis and defense response in plants (Liu et al., 2014). Such studies might help in understanding wax deposition in other Allium species and thus in developing pest- and disease-resistant varieties. Another report on garlic reported the identification of the presence of four known garlic allexiviruses; A, C, E, and X from transcriptome data (Kamenetsky et al., 2015). Thus, transcriptome analysis is a highly sensitive method for identifying known and unknown viruses infecting Allium crops. It can be used for studying plant–virus interaction in a tissue-specific manner. Mohapatra & Nanda (2018) identified chitinase genes from transcriptome data by using an in silico approach. Similarly, RNA-seq data deposited in databases could be mined for the discovery of genes involved in plant defense response.
Organ development
Organ development in plants is a highly organized and precisely regulated process in response to indigenous signals as well as environmental cues. Flowering and bulb development is a critical process in Allium crops. Gene regulation plays a vital role in a smooth transition of growth phases in the life cycle of the plant; however, little information is available about gene interaction and regulation. Expression analyses using transcriptome has enabled extraction of this information for a large number of genes at the global level in less time and at reduced costs. Therefore, in non-model crops, such as alliums, NGS-based expression analyses helped in understanding the mechanism of plant development.
Transcriptome analyses of dormant and sprouting garlic shoot apices were performed to elucidate differential gene expression and reveal the role of genes in dormancy as well as sprouting in garlic. More than 20000 unigenes were found to be upregulated in the sprouting shoot apex. Several candidate genes (ENHYDROUS, DAG1, DAM, DTH8, etc.) involved in dormancy and sprouting of the shoot apex were identified (Sun et al., 2013). Kamenetsky et al. (2015) developed an organ-specific transcriptome catalog for fertile garlic. Different organs exhibited a variation in gene expression profiles, and the highest number of organ-specific reads were reported from the flower. Genes in organosulfur metabolism exhibited an organ-specific profile, whereas several genes involved in flowering and bulb formation were also expressed in leaves, indicating the role of signal transduction. Flowering is a major challenge in garlic breeding, and thus, such studies in fertile garlic will open avenues for understanding the genetic mechanism underlying the reproductive process in garlic as well as other alliums. The CONSTANS and CONSTANS -like (COL) genes are known for their prime role in flowering. Transcriptome analyses of leek led to the identification of 17 putative ApCOL genes, some of which exhibited high similarity with COL genes from other species involved in the modulation of flowering and heading date (Liu et al., 2018). Different leaf morphologies are observed in alliums, such as flat, solid, and fistular. Cellular studies have revealed that fistular leaves arise from a solid precursor by PCD (Ni et al., 2015). A molecular evidence for this finding was provided using transcriptome analyses of nine important species of Allium with different leaf morphologies (Zhu et al., 2017). Phylogenetic analyses of transcriptome revealed that genes associated with PCD presented rapid diversification in fistular leaf species or showed conserved nature in solid leaf species in evolutionary history. Several potential genes involved in plant PCD were subjected to positive selection or evolved in species having fistula-type leaves. This varying selection pattern of PCD-related genes might play an essential role in the development of cavities in fistular leaves. FLOWERING LOCUS T (FT) is the major component of the floral signal molecule as well as other plant development processes. Transcriptome analyses of the doubled haploid onion led to the discovery of six FT-like genes that play a role in flowering and bulb formation. AcFT2 promotes flowering, whereas AcFT1 favors bulb formation, and AcFT4 is antagonistic to AcFT1 (Lee et al., 2013). These functions of AcFT genes were further confirmed through transgenic studies in Arabidopsis. Thus, FT-like gene regulation should be studied in non-flowering and non-bulb-forming alliums and their wild relatives for a further in-depth understanding of their role in alliums. Khosa et al. (2016a) and Khosa et al. (2016b) performed RNA-seq analyses for the doubled haploid bulb onion “CUDH2107” from leaves, various stages of flowering, and roots. They identified several genes involved in flowering and male fertility in onion, which are the orthologs of rice genes. Sucrose metabolism during onion bulb development is a crucial phenomenon (Mallor et al., 2011). Zhang et al. (2018) identified key genes expressed during sucrose metabolism through transcriptome analysis. They performed RNA-seq analyses in three bulb stages (15, 30, and 40 days after bulb swelling), and KEGG analyses revealed that “sucrose and starch metabolism”-related genes were dominant. Expression of the sucrose transporter was the highest during early stages. Sucrose synthase and invertase seem to be involved in sucrose breakdown. Fructose and glucose contents gradually increased from early to later stages, whereas sucrose content decreased. Sucrose metabolism and bulb growth suggested that 30–40 DAS is a period of rapid expansion of bulb (Zhang et al., 2018). . Desiccation and senescence processes lead to the formation of the onion bulb skin. The bulb skin plays a crucial role in the storage and shelf life of the onion bulb and influence its consumer acceptance (Chope et al., 2012). Scanning electron microscopy revealed that the desiccation of the outer scale occurs from inside out, and DNA degradation indicated the critical role of PCD in skin formation. Gene ontology enrichment of transcripts from outer, intermediate, and inner scales revealed an increase in processes related to defense response, PCD processes, carbohydrate metabolism, and flavonoid biosynthesis in the outer scale, whereas the inner scale exhibited increased metabolism and developmental growth processes (Galsurker et al., 2017). These results indicated that the metabolism for bulb skin formation occurs only in the outer scales of onion.
Flavonoids and bulb color
Bulb color is one of the important quality traits in onion. Various bulb colors exist, such as white, yellow, red, pink, chartreuse, and gold. Variation in this trait is governed by mutations in structural and regulatory genes of the flavonoid biosynthesis pathway (Khandagale & Gawande, 2019). Baek, Kim & Kim (2017) performed RNA-seq and bulk segregant analyses of yellow and F2 white plants to develop a marker linked to the C locus that determines the white bulb color in onion. Ninety-seven genes, including previously identified genes of the flavonoid pathway, exhibited more than five-fold expression in the yellow bulk.
Similarly, regulatory genes for the MBW complex were upregulatedin the yellow bulk. SNPs in the gene coding for glutathione S-transferase revealed linkage with the C locus. Transcriptome analyses of white and dark red onions led to the identification of 16 DEGs that play a critical role in the flavonoid biosynthesis pathway. Flavonoid 3′, 5′-hydroxylase (F3′5′H) and dihydroflavonol 4-reductase (DFR) genes play crucial roles in the biosynthesis of dark red bulbs. Further, the ratio F3′5′H: F3′H plays an essential role in the diversity of bulb color (Zhang et al., 2018).
MicroRNAs
MicroRNAs (miRNAs) are a group of small, endogenous noncoding RNAs (ncRNAs) comprising approximately 18–25 nucleotides that regulate gene expression in animals, plants, and protozoans. miRNAs control approximately 60% protein-coding gene activities and regulate numerous cellular processes (Bartel, 2009). miRNAs regulate gene expression through translational repression or target mRNA degradation (Chekulaeva & Filipowicz, 2009). Among Allium crops, two reports have reported discovery of miRNA in garlic and onion by using the in silico approach (Panda et al., 2014; Kohnehrouz, Bastami & Nayeri, 2018). Fusarium oxysporum f. sp. cepae (FOC) is the most devastating pathogen that infects roots and basal plates of garlic. (Chand et al., 2017) performed small RNA sequencing of a FOC-resistant line after infection and discovered 45 miRNAs responsive to FOC. Overexpression of miR164a, miR168a, and miR393 resulted in decreased fungal growth as well as upregulation of defense genes. Similarly, in an earlier study, Chand, Nanda & Joshi (2016) reported that miR394 functions as a negative modulator of FOC resistance in garlic. miRNA thus plays a huge role in the regulation of genes in Allium crops in response to stress as well as development, and thus, for its application in crop improvement, more miRNAs and their targets need to be discovered. Recently, we performed genome-wide identification of miRNA in Thrips tabaci, along with their expression profiling and target prediction (Balan et al., 2018). These miRNAs and their targets can be used for the management of onion thrips by using RNA interference technology.
Proteomics
Proteomics is the high-throughput study of proteins expressed in specific tissues in an individual at a specific time or developmental stage. It plays a key role in interactive or meta-omics analyses, as it complements gene expression and metabolomic studies and hence is more helpful in crops, such as alliums, where limited genomic information is available (Table 3). Proteome alteration in sprouts after low-temperature conditioning of garlic cloves was studied (Dufoo-Hurtado et al., 2015). Differentially synthesized proteins were analyzed using 2-DE and LC-ESI-MS/MS and were found to be involved in various biological processes. This study complemented other studies reporting changes in physiology, biochemistry, and molecular biology of cloves after low-temperature treatment (Dufoo-Hurtado et al., 2013; Guevara-Figueroa et al., 2015). Proteomic changes during freeze–thaw injury and recovery in onion scales were studied. Majority of injury-related proteins (IRPs) were found to be antioxidants, stress proteins, molecular chaperones, and proteins of energy metabolism; these IRPs were induced as a first response to mitigate injury. Recovery-related proteins involved in injury repair facilitates cellular homeostasis, cell wall remodeling, reactive oxygen species scavenging, defense against possible post-thaw infection, and regulating the energy budget to sustain these processes (Chen et al., 2013). Plant’s ability to recover from freeze–thaw injury is a critical factor contributing to tolerance to this stress. Similarly, proteome level changes in response to toxic metals, such as copper and sodium selenite, were also reported in onion (Qui et al., 2015; Karasinski et al., 2017). 2-DE and MS approaches identified 47 differential abundant protein spots between lower and upper epidermis of scales in red and yellow onions; among these, 31 were reported to be unique proteins. These differential proteins were involved in flavonoid synthesis, response to stress, and cell division (Wu et al., 2016). While studying gametogenesis and sterility in garlic, significant differences were observed in the 2-DE profile between sterile and fertile genotypes. A variation was observed in protein maps of different stages of microsporogenesis (Shemesh-Mayer et al., 2013). These differential proteins need to be investigated using MS to elucidate their identity and potential role in fertility.
Table 3. List of the proteomics analyses performed in Allium species.
| Species | Organ/tissue | Platform | Trait/aim | Reference |
|---|---|---|---|---|
| Allium sativum L | Garlic sprouts | 2-DE, LC-ESI-MS/MS | Low temperature conditioning alters the proteome | Dufoo-Hurtado et al. (2015) |
| Allium cepa var. agrogarum | Roots | 2-DE, AutoFlex TOF/TOF II | Proteomic response of roots to Cu stress | Qin et al. (2016) |
| Allium sativum L | Bulbs | 2-DE, XCT mass spectrometer | Characterizatio of Copper–Zinc Superoxide dismutase | Sfaxi et al. (2012) |
| Allium cepa L | Scale epidermis | 2-DE, MALDI-TOF/TOF | Studied differential protein abundance in upper and lower epidermis | Wu et al. (2016) |
| Allium cepa L | Scales | 2-DE, 4700 MALDITOF/TOF | Studied the proteomic response during and after recovery of freeze-thaw injury | Chen et al. (2013) |
| Allium sativum L | Anthers | 2-DE | Study revealed proteomic differences between fertile and sterile genotypes, developmental stages | Shemesh-Mayer et al. (2013) |
| Allium cepa L | Roots | nanoLC-ESI-QOrbitrap-MS | Studied the effect of sodium selenite at proteome level | Karasinski et al. (2017) |
Metabolomics
Plants produce large numbers of metabolites of diversified structures and physical properties in varying abundance. These metabolites vary with the genotype, plant development, environmental conditions, storage, and processing. Metabolite profiling has now been implemented on a broad range of plant species, including but not limited to, tomato, potato, rice, wheat, strawberry, Medicago, Arabidopsis and Allium species. Approximately, 13000 onion accessions are held in gene banks worldwide (Böttcher et al., 2017), but only a small portion of this diverse Allium resource is exploited due to the paucity of information (Soininen et al., 2014a; Soininen et al., 2014b; Soininen et al., 2012). Allium crops are extensively consumed for their broad array of health-promoting effects, which are assigned to the presence of different profiles of organosulfur compounds (OSCs), such asalk(en)yl cysteine sulfoxides, S-allyl cysteine, thiosulfinates (mainly allicin), diallyl sulfides, vinyldithiins, and (E)- and (Z)-ajoene (Block et al., 2010; Ramirez et al., 2017). Metabolites present in Allium species may represent their adaptation to different environmental cues during the domestication process. They also govern important characters in alliums, such as color, pungency, taste, stress tolerance, and medicinal properties. A wide range of modern techniques, such as Liquid Chromatography-Mass Spectrometry (LC–MS), Gas Chromatography–MS (GC-MS), Nuclear Magnetic Resonance (NMR), LC-Fourier Transform Ion Cyclotron Resonance (FTICR)-MS, LC-tandem quadrupole (QqQ)-MS, LC-Quadrupole Time-of-Flight (QTOF)-MS/MS, and Ultrahigh Performance Liquid Chromatography-coupled Electrospray Ionization Quadrupole Time-of-Flight Mass Spectrometry (UHPLC/ESI-QTOFMS), have been used to explore, determine, and characterize the metabolic profile of Allium species (Soininen et al., 2014a; Soininen et al., 2014b; Soininen et al., 2012; Nakabayashi et al., 2016; Böttcher et al., 2017; Molina-Calle et al., 2017; Hrbek et al., 2018; Table 4).
Table 4. List of the metabolomic analyses performed in Allium species.
| Species | Organ/tissue | Platform | Trait/aim | Reference |
|---|---|---|---|---|
| Allium sativum | Clove | LC-MS/MS | Low temperature conditioning led to high phenolic and anthocyanin content | Dufoo-Hurtado et al. (2013) |
| Allium cepa L. | Bulb | UHPLC/ESI-QTOFMS | Variation in metabolite profiles of bulbs due to genetic and environmental factors | Böttcher et al. (2018) |
| Finnish onions,, German long shallot, French leek and Chinese garlic | Bulb | NMR and HPLCMS | Quantification of metabolite in Allium species | Soininen et al. (2014a),Soininen et al. (2014b) |
| Allium sativum, Allium cepa L. | Dried bulbs | GC/MS, UPLC/MS | Effect of drying methods on chemical composition was investigated | Farag et al. (2017) |
| Allium sativum L. | Cloves | DART-HR-OrbitrapMS, HPLC-ESI-HR-TOFMS | Authenticity of garlic from different geographical locations was assessed | Hrbek et al. (2018) |
| Allium sativum L | GC/MS, HPLC-MS/MS | Developed rapid, simple and efficient method for sensory evaluation of garlic | Liu et al. (2019) | |
| Allium cepa L. | Fresh and stored bulbs | NMR | Highlighted the use of Metabolomics for food authentication | Saviano et al. (2019) |
| Allium cepa L. | Farag et al. (2019) | |||
| Allium cepa | Bulb | LC/ESI-QTOFMS | Metabolic profiling of onion cultivars | Böttcher et al. (2017) |
| 35 Allium species | Bulb and leaves | HPLC-ESI-HRMS and NMR | Identification and quantification of metabolite with α-glucosidase inhibitory activity |
Schmidt, Nyberg & Staerk (2014) |
| Allium sativum | Cloves | LC–QTOF MS/MS | Analysed the differential composition of fresh and black garlic | Molina-Calle et al. (2017) |
| Allium cepa, Allium sativum, Allium fistulosum | Bulbs | LC-FTICR-MS | Performed chemical assignment of structural isomers of S-containing metabolites | Nakabayashi et al. (2016) |
| Allium sativum | Bulbs | HPLC and LC-FTICR-MS |
Sulphur containing metabolites were analysed | Yoshimoto et al. (2015) |
| Allium cepa | Bulbs | 1H NMR and LC–MS/MS | Bioactive compounds were identified | Soininen et al. (2012) |
| Allium porrum L | bulbs | 1H NMR and HPLC–MS | Demonstrated metabolome profile of different varieties | Soininen et al. (2014a),Soininen et al. (2014b) |
| Allium roylei | Leaves, bulbs, and roots-basal stems | HPLC | Bioactive compounds from different organs were analysed | Abdelrahman et al. (2014) |
| A. fistulosum, shallots and 8 monosomic addition lines | Bulbs | LC-QqQ-MS | Metabolites in flavonoid pathway were studied | Abdelrahman et al. (2019) |
Among alliums, metabolic profiling is so far performed in onion bulbs, considering cultivars and cultivation year (Böttcher et al., 2018); change in organosulfur metabolites after heat treatment in onion (Kim et al., 2016); differentiation profiling of black and fresh garlics (Molina-Calle et al., 2017); low-temperature conditioning of “seed” cloves (Dufoo-Hurtado et al., 2013); volatile profiling of onion and garlic after subjecting to different drying methods (Farag et al., 2017); metabolic profiling of leaves, bulbs, roots, and basal stems in A. roylei and A. porrum L. (Abdelrahman et al., 2014; Soininen et al., 2014a; Soininen et al., 2012); authentication of the geographic origin of garlic (Hrbek et al., 2018); bulb color and flavonoids (Zhang et al., 2018; Abdelrahman et al., 2019); and metabolic profiling of fresh and stored onion bulbs (Saviano et al., 2019). Metabolomics, along with the artificial neural network, was used for sensory evaluation of garlic and garlic products by using 89 quality indicators (Liu et al., 2019). Farag et al. (2019) studied the effect of the pickling process on onion metabolome and reported a decrease in secondary metabolites; thus, pickling using brine negatively affects the health benefits of onion. Functions and application of some metabolites are presented in Table S2. Onions produce discrete metabolites, including fructo-oligosaccharides (Galdón et al., 2009), OSCs (Block et al., 2010), and flavonoids (Olsson, Gustavsson & Vagen, 2010). In plants, flavonoids are present as flavonoid aglycones, flavonoid O-glycosides, flavonoid C-glycosides, and/or flavonoid O-, C-glycosides (Niessen, 2006). A total of seven different flavonols were identified and characterized from red onions; however, concentrations of flavonoids vary among genotypes and species (Bonaccorsi et al., 2005). These flavonoids determine the color of the onion bulb and tolerance to biotic and abiotic stresses in alliums (Khandagale & Gawande, 2019). The metabolomic approach in Allium is focused toward exploring compositional difference, characterization of enzyme activity, and detection and chemical assignment of metabolites in different genotypes and cultivars (Schmidt, Nyberg & Staerk, 2014; Yoshimoto et al., 2015; Nakabayashi et al., 2016; Molina-Calle et al., 2017). Little work has been performed to explore the metabolic response of Allium with respect to abiotic and biotic stresses, which affect plant growth and yield. Recently, Abdelrahman et al. (2016) attempted the dissection of Trichoderma l ongibrachiatum-induced defense response of onion against Fusarium through metabolite profiling. More than 25 metabolites were accumulated in a significant amount in the plant when onion seeds were primed with T. longibrachiatum. These metabolites are produced in abiotic and biotic stresses and increase onion resistance to F. oxysporum. Therefore, there is a wide scope to explore these areas, which might help to decipher resistance and/or tolerance mechanisms in alliums.
Microbial metagenomics
A large number of microorganisms inhabit around, on, or in plants in a natural ecosystem. The interaction of the microbial community with plants leads to plant growth promotion in different ways, such as nutrient mobilization and secretion of plant growth hormone, enzymes, antibiotics, and other beneficial compounds (Verma et al., 2018). The exploration of microbial diversity interacting with crop-specific plants is limited because only a few percentage of microbes can be cultured and characterized; to explore a large number of microbes, the metagenomics approach can be applied as it is independent of culture (Krishna et al., 2019). Metagenomics approaches are being applied to explore the significance of uncultivated microbes interacting with different Allium species (Yurgel et al., 2018; Knerr et al., 2018; Qiu et al., 2018; Huang, 2018; Matthews, Pierce & Raymond, 2019). A recent study conducted by Chen et al. (2018) revealed that the crop rotation of Chinese cabbage (Brassica rapa subsp. pekinensis) with potato onion (A. cepa var. aggregatum Don.) significantly reduces the incidence and disease index of clubroot disease caused by Plasmodiophora (Plasmodiophora brassicae) in Chinese cabbage. A reduction in disease was observed due to the reduction in secondary plasmodia of Plasmodiophora. A similar study conducted by Nishioka et al. (2019) revealed that cucumber crop rotation and mixed cropping with Allium species (A. cepa and A. fistulosum) suppress the Fusarium wilt. 16S rRNA gene sequencing by using Illumina MiSeq revealed the dominant presence of the Flavobacterium genus. The predominance of Flavobacterium inhibits the multiplication of pathogens in soil (Nishioka et al., 2019). The metagenomics study of Chinese leek (A. tuberosum) by using high-throughput 16S rRNA gene Illumina sequencing revealed the predominance of Proteobacteria, Acidobacteria, Bacteroidetes, Cyanobacteria , and Planctomycetes microbial communities responsible for potential antifungal and nematicidal activities (Huang, 2018). The metagenomics of onion bulb was conducted to study bulb-rotting bacterial and fungal pathogens during storage; as under storage environment, a significant amount of onion bulbs are affected by diseases. The metagenomics for potential pathogen analysis, which was performed for both diseased and healthy bulbs under storage conditions, revealed the abundance of the following bacterial and fungal taxa: Acinetobacter, Burkholderia, Citrobacter, Enterobacteriaceae, Gluconobacter, Pseudomonas, Botrytis, Nectriaceae, Penicillium, Wickerhamomyces and Candida. These taxa are also abundant in the disease-free bulb, but the presence of various fermenters in diseased bulbs suggests that fermenters play a crucial role in onion bulb rotting, along with abiotic factors (Yurgel et al., 2018). These studies have highlighted the significance of alliums metagenomics; in the future, alliums metagenomics should be further explored to identify bacterial and fungal strains exhibiting bactericidal, fungicidal, nematicidal, and growth-promoting activities as well asgenes and proteins for crop protection and production. Till date, the rhizosphere of Allium crops is largely unexplored, and the metagenomics approach can help in discovering the diversity as well as an abundance of microbes associated with Allium roots in varying soil type and climatic conditions.
Meta-omics approach
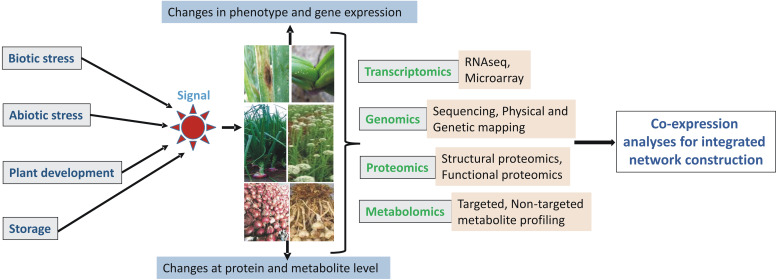
The information generated from these individual omics experiments needs to be correlated with each other to draw meaningful and precise conclusions for further knowledge-based breeding for elite crop development. The omics era has revolutionized research in complex biological processes and traits in many model organisms. Thus, we need to perform integrated omics studies in Allium crops for a quick and improved understanding of molecular mechanisms of various traits (Fig. 3). Integrated transcriptome and proteome have been performed in garlic for studying flower and pollen development (Shemesh-Mayer et al., 2015).
Figure 3. Schematic representation of integrated ‘omics’ analyses in Allium crops.
Similarly, RNA-seq and metabolomics have been integrated into the study of the abiotic stress response (Abdelrahman et al., 2015), onion bulb color (Zhang et al., 2018), and flavonoids (Abdelrahman et al., 2019). From these studies, authors have suggested that separate omics analyses by using any single approach can be insufficient for the precise elucidation of various traits and their molecular mechanisms. These integrated omics interventions further enable researchers to precisely associate genetic variation with a metabolite or protein and thus ultimately help in selecting candidate genes with greater confidence for future metabolic and protein engineering in Allium crops for the development of noble varieties.
Conclusion and future perspective
Although Allium crops are widely cultivated and consumed, research in these crops is challenging due to their biennial nature, cross-pollination, inbreeding depression, photoperiod sensitivity, and lack of flowering. However, with the development of advanced omics technologies and their affordability, research in these crops is gaining attention and pace by overcoming these challenges.
Due to the large size of the genome in Allium species, the availability of genome sequences can be a major breakthrough in Allium research. Rapid progress in sequencing technologies can definitely enable researchers to complete Allium sequencing projects. Increased number of doubled haploid lines need to be developed for alliums growing in different photoperiod conditions and used for genome sequencing. Genome sequencing projects for onion and shallots are under progress at Wageningen University and Yamaguchi University, respectively. To complete these daunting tasks, a coordinated effort is needed. The plastid genome of several Allium species has been characterized and needs to be used for male sterility and phylogenetic analyses. With the advancement in sequencing technologies and reduction in cost, recently, the use of RNA-seq has significantly increased in diverse alliums. These studies have attempted the elucidation of complex molecular mechanisms involved in male sterility, bulb development, bulb color, flowering, and stress responses in alliums. Several putative unigenes conferring the aforementioned traits have been identified from RNA-seq datasets. Biotic and abiotic stresses are the key factors that affect both the quality and yield of Allium crops; yet, limited information is available regarding molecular response to stress in these crops. Thus, transcriptomic analyses under stress will help in understanding the stress physiology and molecular biology of alliums. miRNAs play a crucial role in gene regulation in plant development and stress response in plants; however, information on small RNAs is also lacking in alliums. Generally, wild relatives harbor several economically vital traits, such as abiotic stress tolerance, disease resistance, and secondary metabolites. Thus, wild alliums need to be investigated using transcriptomics to identify genes responsible for these traits as well as those for domestication. Further, these domestication-associated genes can be modified using transgenic or genome editing tools to cultivate these wild relatives. The level of proteins and metabolites often differ with the mRNA expression profile of corresponding genes. Therefore, proteomic and metabolomic analyses are crucial because they analyze the end product of the central dogma, and thus, these omics tools should be used in an integrated manner for the proper unraveling of the trait. A few studies have reported the use of proteomics and metabolomics in understanding biological processes, such as stress response, flowering, and metabolic profiling for geographic origin authentication and analyses of OSCs. However, these efforts are not sufficient, and a few areas, such as bolting in onion; flowering in short-day garlic; storability of onion and garlic; high TSS onion for processing, pungency; and aroma, disease and pest resistance need to be addressed using these omics technologies. These studies need to be conducted comparatively on short-day and long-day onions to obtain some clues on the domestication of onion and garlic. Recently developed breeding techniques, such as CRISPR, may be used in alliums for targeted genome editing of key genes involved in haploid induction, male sterility, and virus resistance. However, to validate specificity of mutations, a reference genome is required; thus,the research fraternity working on Allium crops is eagerly waiting for the release of the draft sequence of onion. Availability of sequence information can open new avenues and accelerate Allium research, such as molecular breeding, functional genomics, and gene editing.
Supplemental Information
Funding Statement
This work was supported by HORTDOGRSIL201800400084, Indian Council of Agricultural Research. Kiran Khandagale was supported by a Savitribai Phule Pune University Post-doctorate fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Kiran Khandagale, Ram Krishna, Praveen Roylawar, Avinash B. Ade, Ashwini Benke, Bharat Shinde, Major Singh and Suresh Gawande conceived and designed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Ashutosh Rai analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
This is a literature review and thus does not have any raw data.
References
- Abdelrahman et al. (2016).Abdelrahman M, Abdel-Motaal F, El-Sayed M, Jogaiah S, Shigyo M, Ito SI, Tran LSP. Dissection of Trichoderma longibrachiatum-induced defense in onion (Allium cepa L.) against Fusarium oxysporum f. sp. cepa by target metabolite profiling. Plant Science. 2016;246:128–138. doi: 10.1016/j.plantsci.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Abdelrahman et al. (2017).Abdelrahman M, El-Sayed M, Sato S, Hirakawa H, Ito SI, Tanaka K, et al. Shigyo M. RNA-sequencing-based transcriptome and biochemical analyses of steroidal saponin pathway in a complete set of Allium fistulosum—A. cepa monosomic addition lines. PLOS ONE. 2017;12(8):e0181784. doi: 10.1371/journal.pone.0181784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelrahman et al. (2014).Abdelrahman M, Hirata S, Ito SI, Yamauchi N, Shigyo M. Compartmentation and localization of bioactive metabolites in different organs of Allium roylei. Bioscience, Biotechnology, and Biochemistry. 2014;78(7):1112–1122. doi: 10.1080/09168451.2014.915722. [DOI] [PubMed] [Google Scholar]
- Abdelrahman et al. (2019).Abdelrahman M, Hirata S, Sawada Y, Hirai MY, Sato S, Hirakawa H, Mine Y, Tanaka K, Shigyo M. Widely targeted metabolome and transcriptome landscapes of Allium fistulosum–A. cepa chromosome addition lines revealed a flavonoid hot spot on chromosome 5A. Scientific Reports. 2019;9(1):3541. doi: 10.1038/s41598-019-39856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelrahman et al. (2015).Abdelrahman M, Sawada Y, Nakabayashi R, Sato S, Hirakawa H, El-Sayed M, Hirai MY, Saito K, Yamauchi N, Shigyo M. Integrating transcriptome and target metabolome variability in doubled haploids of Allium cepa for abiotic stress protection. Molecular Breeding. 2015;35(10):195. doi: 10.1007/s11032-015-0378-2. [DOI] [Google Scholar]
- Atkinson & Urwin (2012).Atkinson NJ, Urwin PE. The interaction of plant biotic and abiotic stresses: from genes to the field. Journal of Experimental Botany. 2012;63(10):3523–3543. doi: 10.1093/jxb/ers100. [DOI] [PubMed] [Google Scholar]
- Baek, Kim & Kim (2017).Baek G, Kim CW, Kim S. Development of a molecular marker tightly linked to the C locus conferring a white bulb color in onion (Allium cepa L.) using bulked segregant analysis and RNA-Seq. Molecular Breeding. 2017;37(8):94. doi: 10.1007/s11032-017-0697-6. [DOI] [Google Scholar]
- Balan et al. (2018).Balan RK, Ramasamy A, Hande RH, Gawande SJ, Krishna Kumar NK. Genome-wide identification, expression profiling, and target gene analysis of micro RNA s in the Onion thrips, Thrips tabaci Lindeman (Thysanoptera: Thripidae), vectors of tospoviruses (Bunyaviridae) Ecology and Evolution. 2018;8(13):6399–6419. doi: 10.1002/ece3.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin et al. (2012a).Baldwin S, Pither-Joyce M, Wright K, Chen L, McCallum J. Development of robust genomic simple sequence repeat markers for estimation of genetic diversity within and among bulb onion (Allium cepa L.) populations. Molecular Breeding. 2012a;30(3):1401–1411. doi: 10.1007/s11032-012-9727-6. [DOI] [Google Scholar]
- Baldwin et al. (2012b).Baldwin S, Revanna R, Thomson S, Pither-Joyce M, Wright K, Crowhurst R, Fiers M, Chen L, Macknight R, McCallum JA. A toolkit for bulk PCR-based marker design from next-generation sequence data: application for development of a framework linkage map in bulb onion (Allium cepa L.) BMC Genomics. 2012b;13(1):637. doi: 10.1186/1471-2164-13-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel (2009).Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block et al. (2010).Block E, Dane AJ, Thomas S, Cody RB. Applications of direct analysis in real time mass spectrometry (DART-MS) in Allium chemistry, 2-Propenesulfenic and 2-propenesulfinic acids, diallyltrisulfane S-oxide, and other reactive sulfur compounds from crushed garlic and other Alliums. Journal of Agricultural and Food Chemistry. 2010;58(8):4617–4625. doi: 10.1021/jf1000106. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi et al. (2005).Bonaccorsi P, Caristi C, Gargiulli C, Leuzzi U. Flavonolglucoside profile of southern Italian red onion (Allium cepa L.) Journal of Agricultural and Food Chemistry. 2005;53(7):2733–2740. doi: 10.1021/jf048152r. [DOI] [PubMed] [Google Scholar]
- Böttcher et al. (2017).Böttcher C, Krähmer A, Stürtz M, Widder S, Schulz H. Comprehensive metabolite profiling of onion bulbs (Allium cepa) using liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry. Metabolomics. 2017;13(4):35. doi: 10.1007/s11306-017-1175-1. [DOI] [Google Scholar]
- Böttcher et al. (2018).Böttcher C, Krähmer A, Stürtz M, Widder S, Schulz H. Effect of cultivar and cultivation year on the metabolite profile of onion bulbs (Allium cepa L.) Journal of Agricultural and Food Chemistry. 2018;66(12):3229–3238. doi: 10.1021/acs.jafc.8b00254. [DOI] [PubMed] [Google Scholar]
- Chand, Nanda & Joshi (2016).Chand SK, Nanda S, Joshi RK. Regulation of miR394 in response to fusarium oxysporum f, sp. cepae (FOC) Infection in Garlic (Allium sativum L) Frontiers in Plant Science. 2016;7:258. doi: 10.3389/fpls.2016.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand et al. (2017).Chand SK, Nanda S, Mishra R, Joshi RK. Multiple garlic (Allium sativum L.) microRNAs regulate the immunity against the basal rot fungus Fusarium oxysporum f, sp. Cepae. Plant Science. 2017;257:9–21. doi: 10.1016/j.plantsci.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Chand et al. (2015).Chand SK, Nanda S, Rout E, Joshi RK. Mining, characterization and validation of EST derived microsatellites from the transcriptome database of Allium sativum L. Bioinformation. 2015;11(3):145–150. doi: 10.6026/97320630011145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva & Filipowicz (2009).Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Current Opinion in Cell Biology. 2009;21(3):452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2013).Chen K, Renaut J, Sergeant K, Wei H, Arora R. Proteomic changes associated with freeze-thaw injury and post-thaw recovery in onion (Allium cepa L.) scales. Plant, Cell & Environment. 2013;36(4):892–905. doi: 10.1111/pce.12027. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2018).Chen S, Zhou X, Yu H, Wu F. Root exudates of potato onion are involved in the suppression of clubroot in a Chinese cabbage-potato onion-Chinese cabbage crop rotation. European Journal of Plant Pathology. 2018;150(3):765–777. doi: 10.1007/s10658-017-1307-5. [DOI] [Google Scholar]
- Chinnappareddy et al. (2013).Chinnappareddy LRD, Khandagale K, Chennareddy A, Ramappa VG. Molecular markers in the improvement of Allium crops. Czech Journal of Genetics and Plant Breeding. 2013;49(4):131–139. doi: 10.17221/111/2013-CJGPB. [DOI] [Google Scholar]
- Chope et al. (2012).Chope GA, Cools K, Hammond JP, Thompson AJ, Terry LA. Physiological, biochemical and transcriptional analysis of onion bulbs during storage. Annals of Botany. 2012;109(4):819–831. doi: 10.1093/aob/mcr318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currah (2002).Currah L. Onions in the tropics: cultivars and country reports. Allium Crop Science: Recent Advances. 2002;2(17.66):379–407. [Google Scholar]
- Damon, Groves & Havey (2014).Damon SJ, Groves RL, Havey MJ. Variation for epicuticular waxes on onion foliage and impacts on numbers of onion thrips. Journal of the American Society for Horticultural Science. 2014;139(4):495–501. doi: 10.21273/JASHS.139.4.495. [DOI] [Google Scholar]
- Do, Suzuki & Mukai (2004).Do GS, Suzuki G, Mukai Y. Genomic organization of a novel root alliinase gene, ALL1, in onion. Gene. 2004;325:17–24. doi: 10.1016/j.gene.2003.09.033. [DOI] [PubMed] [Google Scholar]
- Drinkwater & Janes (1955).Drinkwater WO, Janes BE. Effects of irrigation and soil moisture on maturity, yield and storage of two onion hybrids. Proceedings of the American Society for Horticultural Science. 1955;66:267–278. [Google Scholar]
- Duangjit et al. (2013).Duangjit J, Bohanec B, Chan AP, Town CD, Havey MJ. Transcriptome sequencing to produce SNP-based genetic maps of onion. Theoretical and Applied Genetics. 2013;126(8):2093–2101. doi: 10.1007/s00122-013-2121-x. [DOI] [PubMed] [Google Scholar]
- Dufoo-Hurtado et al. (2015).Dufoo-Hurtado MD, Huerta-Ocampo J. Á, Barrera-Pacheco A, Barba de la Rosa AP, Mercado-Silva EM. Low temperature conditioning of garlic (Allium sativum L.)seed cloves induces alterations in sprouts proteome. Frontiers in Plant Science. 2015;6:332. doi: 10.3389/fpls.2015.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufoo-Hurtado et al. (2013).Dufoo-Hurtado MD, Zavala-Gutiérrez KG, Cao CM, Cisneros-Zevallos L, Guevara-González RG, Torres-Pacheco I, Vázquez-Barrios ME, Rivera-Pastrana DM, Mercado-Silva EM. Low-temperature conditioning of seed cloves enhances the expression of phenolic metabolism related genes and anthocyanin content in ‘coreano’garlic (Allium sativum) during plant development. Journal of Agricultural and Food Chemistry. 2013;61(44):10439–10446. doi: 10.1021/jf403019t. [DOI] [PubMed] [Google Scholar]
- Egea et al. (2017).Egea LA, Mérida-García R, Kilian A, Hernandez P, Dorado G. Assessment of genetic diversity and structure of large garlic (Allium sativum) germplasm bank, by diversity arrays technology genotyping-by-sequencing platform (DArTseq) Frontiers in Genetics. 2017;8:98. doi: 10.3389/fgene.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajkus et al. (2016).Fajkus P, Peška V, Sitová Z, Fulnečková J, Dvořáčková M, Gogela R, Sýkorová E, Hapala J, Fajkus J. Allium telomeres unmasked: the unusual telomeric sequence (CTCGGTTATGGG) n is synthesized by telomerase. The Plant Journal. 2016;85(3):337–347. doi: 10.1111/tpj.13115. [DOI] [PubMed] [Google Scholar]
- Fajkus et al. (2019).Fajkus P, Peška V, Závodník M, Fojtová M, Fulnečková J, Dobias Š, Kilar A, Dvořáčková M, Zachová D, Nečasová I, Sims J. Telomerase RNAs in land plant. Nucleic Acids Research. 2019;47(18):9842–9856. doi: 10.1093/nar/gkz695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag et al. (2017).Farag M, Ali S, Hodaya R, El-Seedi H, Sultani H, Laub A, Eissa TF, Abou-Zaid F, Wessjohann L. Phytochemical profiles and antimicrobial activities of Allium cepa red cv. and A. sativum subjected to different drying methods: a comparative MS-based metabolomics. Molecules. 2017;22(5):761. doi: 10.3390/molecules22050761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag et al. (2019).Farag MA, Tawfike AF, Donia MS, Ehrlich A, Wessjohann LA. Influence of pickling process on Allium cepa and citrus limon metabolome as determined via mass spectrometry-based metabolomics. Molecules. 2019;24(5):928. doi: 10.3390/molecules24050928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filyushin et al. (2016).Filyushin MA, Beletsky AV, Mazur AM, Kochieva EZ. The complete plastid genome sequence of garlic Allium sativum L. Mitochondrial DNA Part B. 2016;1(1):831–832. doi: 10.1080/23802359.2016.1247669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filyushin et al. (2018).Filyushin MA, Beletsky AV, Mazur AM, Kochieva EZ. Characterization of the complete plastid genome of lop-sided onion Allium obliquum L. (Amaryllidaceae) Mitochondrial DNA Part B. 2018;3(1):393–394. doi: 10.1080/23802359.2018.1456369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkers et al. (2015).Finkers R, van Workum W, van Kaauwen M, Huits H, Jungerius A, Vosman B, Scholten OE. Wageningen University and Research; Wageningen: 2015. SEQUON-sequencing the onion genome. [DOI] [Google Scholar]
- Fu et al. (2019).Fu J, Zhang H, Guo F, Ma L, Wu J, Yue M, Zheng X, Qiu Z, Li L. Identification and characterization of abundant repetitive sequences in Allium cepa. Scientific Reports. 2019;9(1):1–7. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdón et al. (2009).Galdón BR, Rodríguez CT, Rodríguez ER, Romero CD. Fructans and major compounds in onion cultivars (Allium cepa) Journal of Food Composition and Analysis. 2009;22(1):25–32. doi: 10.1016/j.jfca.2008.07.007. [DOI] [Google Scholar]
- Galsurker et al. (2017).Galsurker O, Doron-Faigenboim A, Teper-Bamnolker P, Daus A, Fridman Y, Lers A, Eshel D. Cellular and molecular changes associated with onion skin formation suggest involvement of programmed cell death. Frontiers in Plant Science. 2017;7:2031. doi: 10.3389/fpls.2016.02031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galsurker et al. (2018).Galsurker O, Doron-Faigenboim A, Teper-Bamnolker P, Daus A, Lers A, Eshel D. Differential response to heat stress in outer and inner onion bulb scales. Journal of Experimental Botany. 2018;69(16):4047–4064. doi: 10.1093/jxb/ery189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Figueroa et al. (2015).Guevara-Figueroa T, López-Hernández L, Lopez MG, Hurtado MD, Vázquez-Barrios ME, Guevara-Olvera L, Guevara González RG, Rivera-Pastrana DM, Torres-Robles H, Mercado-Silva EM. Conditioning garlic seed cloves at low temperature modifies plant growth, sugar, fructan content, and sucrose sucrosefructosyltransferase (1-SST) expression. Scientia Horticulturae. 2015;189:150–158. doi: 10.1016/j.scienta.2015.03.030. [DOI] [Google Scholar]
- Han et al. (2016).Han J, Thamilarasan SK, Natarajan S, Park JI, Chung MY, Nou IS. De novo assembly and transcriptome analysis of bulb onion (Allium cepa L.) during cold acclimation using contrasting genotypes. PLOS ONE. 2016;11(9):e0161987. doi: 10.1371/journal.pone.0161987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havey & Ahn (2016).Havey MJ, Ahn YK. Single nucleotide polymorphisms and indel markers from the transcriptome of garlic. Journal of the American Society for Horticultural Science. 2016;141(1):62–65. doi: 10.21273/JASHS.141.1.62. [DOI] [Google Scholar]
- Hertweck (2013).Hertweck KL. Assembly and comparative analysis of transposable elements from low coverage genomic sequence data in Asparagales. Genome. 2013;56(9):487–494. doi: 10.1139/gen-2013-0042. [DOI] [PubMed] [Google Scholar]
- Hrbek et al. (2018).Hrbek V, Rektorisova M, Chmelarova H, Ovesna J, Hajslova J. Authenticity assessment of garlic using a metabolomic approach based on high resolution mass spectrometry. Journal of Food Composition and Analysis. 2018;67:19–28. doi: 10.1016/j.jfca.2017.12.020. [DOI] [Google Scholar]
- Huang (2018).Huang YH. Comparison of rhizosphere and endophytic microbial communities of Chinese leek through high-throughput 16S rRNA gene Illumina sequencing. Journal of Integrative Agriculture. 2018;17(2):359–367. doi: 10.1016/S2095-3119(17)61731-3. [DOI] [Google Scholar]
- Huo et al. (2019).Huo Y, Gao L, Liu B, Yang Y, Kong S, Sun Y, Yang Y, Wu X. Complete chloroplast genome sequences of four Allium species: comparative and phylogenetic analyses. Scientific Reports. 2019;9(1):1–14. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakše et al. (2008).Jakše J, Meyer JD, Suzuki G, McCallum J, Cheung F, Town CD, Havey MJ. Pilot sequencing of onion genomic DNA reveals fragments of transposable elements, low gene densities, and significant gene enrichment after methyl filtration. Molecular Genetics and Genomics. 2008;280(4):287–292. doi: 10.1007/s00438-008-0364-z. [DOI] [PubMed] [Google Scholar]
- Jin et al. (2018).Jin FY, Xie DF, Zhou SD, He XJ. Characterization of the complete chloroplast genome of Allium prattii. Mitochondrial DNA Part B. 2018;3(1):153–154. doi: 10.1080/23802359.2018.1436994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo et al. (2017).Jo J, Purushotham PM, Han K, Lee HR, Nah G, Kang BC. Development of a genetic map for onion (Allium cepa L.) using reference-free genotyping-by-sequencing and SNP assays. Frontiers in Plant Science. 2017;8:1606. doi: 10.3389/fpls.2017.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, Taylor & Thornton (1992).Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kamenetsky et al. (2015).Kamenetsky R, Faigenboim A, Mayer ES, Michael TB, Gershberg C, Kimhi S, Esquira I, Shalom SR, Eshel D, Rabinowitch HD, Sherman A. Integrated transcriptome catalogue and organ-specific profiling of gene expression in fertile garlic (Allium sativum L.) BMC Genomics. 2015;16(1):12. doi: 10.1186/s12864-015-1212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasinski et al. (2017).Karasinski J, Wrobel K, Corrales Escobosa AR, Konopka A, Bulska E, Wrobel K. Allium cepa L. response to sodium selenite (Se (IV)) studied in plant roots by a lc-ms-based proteomic approach. Journal of Agricultural and Food Chemistry. 2017;65(19):3995–4004. doi: 10.1021/acs.jafc.7b01085. [DOI] [PubMed] [Google Scholar]
- Khandagale & Gawande (2019).Khandagale K, Gawande S. Genetics of bulb colour variation and flavonoids in onion. The Journal of Horticultural Science and Biotechnology. 2019;94(4):522–532. doi: 10.1080/14620316.2018.1543558. [DOI] [Google Scholar]
- Khosa et al. (2016a).Khosa JS, Lee R, Bräuning S, Lord J, Pither-Joyce M, McCallum J, Macknight RC. Doubled haploid ‘CUDH2107” as a reference for bulb onion (Allium cepa L.) research: development of a transcriptome catalogue and identification of transcripts associated with male fertility. PLOS ONE. 2016a;11(11):e0166568. doi: 10.1371/journal.pone.0166568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosa et al. (2016b).Khosa JS, McCallum J, Dhatt AS, Macknight RC. Enhancing onion breeding using molecular tools. Plant Breeding. 2016b;135(1):9–20. doi: 10.1111/pbr.12330. [DOI] [Google Scholar]
- Kim et al. (2016).Kim B, Kim K, Yang TJ, Kim S. Completion of the mitochondrial genome sequence of onion (Allium cepa L.) containing the CMS-S male-sterile cytoplasm and identification of an independent event of the ccmF N gene split. Current Genetics. 2016;62(4):873–885. doi: 10.1007/s00294-016-0595-1. [DOI] [PubMed] [Google Scholar]
- Kim, Yang & Kim (2019).Kim B, Yang TJ, Kim S. Identification of a gene responsible for cytoplasmic male-sterility in onions (Allium cepa L.) using comparative analysis of mitochondrial genome sequences of two recently diverged cytoplasms. Theoretical and Applied Genetics. 2019;132(2):313–322. doi: 10.1007/s00122-018-3218-z. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2015).Kim S, Kim CW, Park M, Choi D. Identification of candidate genes associated with fertility restoration of cytoplasmic male-sterility in onion (Allium cepa L.) using a combination of bulked segregant analysis and RNA-seq. Theoretical and Applied Genetics. 2015;128(11):2289–2299. doi: 10.1007/s00122-015-2584-z. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2014).Kim S, Kim MS, Kim YM, Yeom SI, Cheong K, Kim KT, Jeon J, Kim S, Kim DS, Sohn SH, Lee YH, Lee YH. Integrative structural annotation of de novo RNA-Seq provides an accurate reference gene set of the enormous genome of the onion (Allium cepa L.) DNA Research. 2014;22(1):19–27. doi: 10.1093/dnares/dsu035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Park & Yang (2015).Kim S, Park JY, Yang T. Comparative analysis of the complete chloroplast genome sequences of a normal male-fertile cytoplasm and two different cytoplasms conferring cytoplasmic male sterility in onion (Allium cepa L.) The Journal of Horticultural Science and Biotechnology. 2015;90(4):459–468. doi: 10.1080/14620316.2015.11513210. [DOI] [Google Scholar]
- Kirov et al. (2017).Kirov IV, Kiseleva AV, Van Laere K, Van Roy N, Khrustaleva LI. Tandem repeats of Allium fistulosum associated with major chromosomal landmarks. Molecular Genetics and Genomics. 2017;292(2):453–464. doi: 10.1007/s00438-016-1286-9. [DOI] [PubMed] [Google Scholar]
- Kiseleva, Kirov & Khrustaleva (2014).Kiseleva AV, Kirov IV, Khrustaleva LI. Chromosomal organization of centromeric Ty3/gypsy retrotransposons in Allium cepa L. and Allium fistulosum L. Russian Journal of Genetics. 2014;50(6):586–592. doi: 10.1134/S102279541404005X. [DOI] [PubMed] [Google Scholar]
- Knerr et al. (2018).Knerr AJN, Wheeler D, Schlatter D, Sharma-Poudyal D, du Toit LJ, Paulitz TC. Arbuscularmycorrhizal fungal communities in organic and conventional onion crops in the Columbia Basin of the Pacific Northwest United States. Phytobiomes Journal. 2018;2(4):194–207. doi: 10.1094/PBIOMES-05-18-0022-R. [DOI] [Google Scholar]
- Kohnehrouz, Bastami & Nayeri (2018).Kohnehrouz BB, Bastami M, Nayeri S. In silico identification of novel microRNAs and targets using EST analysis in Allium cepa L. Interdisciplinary Sciences: Computational Life Sciences. 2018;10(4):771–780. doi: 10.1007/s12539-017-0240-9. [DOI] [PubMed] [Google Scholar]
- Krishna et al. (2019).Krishna R, Ansari WA, Verma JP, Singh M. Role of plant growth promoting microorganisms in sustainable agriculture and nanotechnology. Cambridge: Woodhead Publishing; 2019. Modern molecular and omics tools for understanding the plant growth-promoting rhizobacteria; pp. 39–53. [Google Scholar]
- Kuhl et al. (2004).Kuhl JC, Cheung F, Yuan Q, Martin W, Zewdie Y, McCallum J, Catanach A, Rutherford P, Sink KC, Jenderek M, Prince JP, Town CD, Prince JP. A unique set of 11, 008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders Asparagales and Poales. The Plant Cell. 2004;16(1):114–125. doi: 10.1105/tpc.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2017).Lee J, Chon J, Lim J, Kim EK, Nah G. Characterization of complete chloroplast genome of Allium victorialis and its application for barcode markers. Plant Breeding and Biotechnology. 2017;5(3):221–227. doi: 10.9787/PBB.2017.5.3.221. [DOI] [Google Scholar]
- Lee et al. (2018).Lee JH, Natarajan S, Biswas MK, Shirasawa K, Isobe S, Kim HT, Nou IS, Park JI, Seong CN, Nou IS. SNP discovery of Korean short day onion inbred lines using double digest restriction site-associated DNA sequencing. PLOS ONE. 2018;13(8):e0201229. doi: 10.1371/journal.pone.0201229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2013).Lee R, Baldwin S, Kenel F, McCallum J, Macknight R. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nature Communications. 2013;4:2884. doi: 10.1038/ncomms3884. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2018).Liu C, Tang Q, Cheng C, Xu Y, Yang Z, Dai Z, Su J. Identification of putative CONSTANS-like genes from the de novo assembled transcriptome of leek. Biologiaplantarum. 2018;62(2):269–276. [Google Scholar]
- Liu et al. (2019).Liu J, Liu L, Guo W, Fu M, Yang M, Huang S, Zhang F, Liu Y. A new methodology for sensory quality assessment of garlic based on metabolomics and an artificial neural network. RSC Advances. 2019;9(31):17754–17765. doi: 10.1039/C9RA01978B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2016).Liu Q, Lan Y, Wen C, Zhao H, Wang J, Wang Y. Transcriptome sequencing analyses between the cytoplasmic male sterile line and its maintainer line in welsh onion (Allium fistulosum L.) International Journal of Molecular Sciences. 2016;17(7):1058. doi: 10.3390/ijms17071058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2014).Liu Q, Wen C, Zhao H, Zhang L, Wang J, Wang Y. RNA-Seq reveals leaf cuticular wax-related genes in Welsh onion. PLOS ONE. 2014;9(11):e113290. doi: 10.1371/journal.pone.0113290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2015).Liu T, Zeng L, Zhu S, Chen X, Tang Q, Mei S, Tang S. Large-scale development of expressed sequence tag-derived simple sequence repeat markers by deep transcriptome sequencing in garlic (Allium sativum L.) Molecular Breeding. 2015;35(11):204. doi: 10.1007/s11032-015-0399-x. [DOI] [Google Scholar]
- Mallor et al. (2011).Mallor C, Balcells M, Mallor F, Sales E. Genetic variation for bulb size, soluble solids content and pungency in the Spanish sweet onion variety Fuentes de Ebro. Response to selection for low pungency. Plant Breeding. 2011;130(1):55–59. doi: 10.1111/j.1439-0523.2009.01737.x. [DOI] [Google Scholar]
- Mancia et al. (2015).Mancia FH, Sohn SH, Ahn YK, Kim DS, Kim JS, Kwon YS, Kim CW, Lee TH, Hwang YJ. Distribution of various types of repetitive DNAs in Allium cepa L. based on dual color FISH. Horticulture, Environment, and Biotechnology. 2015;56(6):793–799. doi: 10.1007/s13580-015-1100-3. [DOI] [Google Scholar]
- Martin et al. (2013).Martin L, Fei Z, Giovannoni J, Rose JKC. Catalyzing plant science research with RNA-seq. Frontiers in Plant Science. 2013;4:66. doi: 10.3389/fpls.2013.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, Pierce & Raymond (2019).Matthews A, Pierce S, Raymond B. Rhizobacterial community assembly patterns vary between crop species. Frontiers in Microbiology. 2019;10:581. doi: 10.3389/fmicb.2019.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maumus & Quesneville (2014).Maumus F, Quesneville H. Deep investigation of Arabidopsis thaliana junk DNA reveals a continuum between repetitive elements and genomic dark matter. PLOS ONE. 2014;9(4):e94101. doi: 10.1371/journal.pone.0094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum et al. (2008).McCallum J, Thomson S, Pither-Joyce M, Kenel F, Clarke A, Havey MJ. Genetic diversity analysis and single-nucleotide polymorphism marker development in cultivated bulb onion based on expressed sequence tag–simple sequence repeat markers. Journal of the American Society for Horticultural Science. 2008;133(6):810–818. doi: 10.21273/JASHS.133.6.810. [DOI] [Google Scholar]
- Mehra et al. (2019).Mehra R, Jasrotia RS, Mahajan A, Sharma D, Iquebal MA, Kaul S, Dhar MK. Transcriptome analysis of Snow Mountain Garlic for unraveling the organosulfur metabolic pathway. Genomics. 2019 doi: 10.1016/j.ygeno.2019.07.014. [DOI] [PubMed] [Google Scholar]
- Mohapatra & Nanda (2018).Mohapatra RK, Nanda S. In silico analysis of onion chitinases using transcriptome data. Bioinformation. 2018;14(8):440. doi: 10.6026/97320630014440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Calle et al. (2017).Molina-Calle M, de Medina VS, Priego-Capote F, de Castro MDL. Establishing compositional differences between fresh and black garlic by a metabolomics approach based on LC–QTOF MS/MS analysis. Journal of Food Composition and Analysis. 2017;62:155–163. doi: 10.1016/j.jfca.2017.05.004. [DOI] [Google Scholar]
- Nakabayashi & Saito (2017).Nakabayashi R, Saito K. Ultrahigh resolution metabolomics for S-containing metabolites. Current Opinion in Biotechnology. 2017;43:8–16. doi: 10.1016/j.copbio.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Nakabayashi et al. (2016).Nakabayashi R, Sawada Y, Aoyagi M, Yamada Y, Hirai MY, Sakurai T, Kamoi T, Rowan D, Saito K. Chemical assignment of structural isomers of sulfur-containing metabolites in garlic by liquid chromatography- fourier transform ion cyclotron resonance- mass spectrometry. The Journal of nutrition. 2016;146(2):397S–402S. doi: 10.3945/jn.114.202317. [DOI] [PubMed] [Google Scholar]
- Nei & Kumar (2000).Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Ni et al. (2015).Ni XL, Su H, Zhou YF, Wang FH, Liu WZ. Leaf-shape remodeling: programmed cell death in fistular leaves of Allium fistulosum. Physiologiaplantarum. 2015;153(3):419–431. doi: 10.1111/ppl.12255. [DOI] [PubMed] [Google Scholar]
- Nishioka et al. (2019).Nishioka T, Marian M, Kobayashi I, Kobayashi Y, Yamamoto K, Tamaki H, Suga H, Shimizu M. Microbial basis of Fusarium wilt suppression by Allium cultivation. Scientific Reports. 2019;9(1):1715. doi: 10.1038/s41598-018-37559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, Gustavsson & Vagen (2010).Olsson ME, Gustavsson KE, Vagen IM. Quercetin and isorhamnetin in sweet and red cultivars of onion (Allium cepa L.) at harvest, after field curing, heat treatment, and storage. Journal of Agricultural and Food Chemistry. 2010;58(4):2323–2330. doi: 10.1021/jf9027014. [DOI] [PubMed] [Google Scholar]
- Panda et al. (2014).Panda D, Dehury B, Sahu J, Barooah M, Sen P, Modi MK. Computational identification and characterization of conserved miRNAs and their target genes in garlic (Allium sativum L.) expressed sequence tags. Gene. 2014;537(2):333–342. doi: 10.1016/j.gene.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Peška et al. (2019).Peška V, Mandáková T, Ihradská V, Fajkus J. Comparative dissection of three giant genomes: Allium cepa, Allium sativum, and Allium ursinum. International Journal of Molecular Sciences. 2019;20(3):733. doi: 10.3390/ijms20030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin et al. (2016).Qin R, Ning C, Björn LO, Li S. Proteomic analysis of Allium cepa var, agrogarum L. roots under copper stress. Plant and Soil. 2016;401(1–2):197–212. doi: 10.1007/s11104-015-2741-9. [DOI] [Google Scholar]
- Qiu et al. (2018).Qiu Z, Li N, Lu X, Zheng Z, Zhang M, Qiao X. Characterization of microbial community structure and metabolic potential using IlluminaMiSeq platform during the black garlic processing. Food Research International. 2018;106:428–438. doi: 10.1016/j.foodres.2017.12.081. [DOI] [PubMed] [Google Scholar]
- Ramirez et al. (2017).Ramirez DA, Locatelli DA, Gonzalez RE, Cavagnaro PF, Camargo AB. Analytical methods for bioactive sulfur compounds in Allium: an integrated review and future directions. Journal of Food Composition and Analysis. 2017;61:4–19. doi: 10.1016/j.jfca.2016.09.012. [DOI] [Google Scholar]
- Ricroch et al. (2005).Ricroch A, Yockteng R, Brown SC, Nadot S. Evolution of genome size across some cultivated Allium species. Genome. 2005;48(3):511–520. doi: 10.1139/g05-017. [DOI] [PubMed] [Google Scholar]
- Saviano et al. (2019).Saviano G, Paris D, Melck D, Fantasma F, Motta A, Iorizzi M. Metabolite variation in three edible Italian Allium cepa L. by NMR-based metabolomics: a comparative study in fresh and stored bulbs. Metabolomics. 2019;15(8):105. doi: 10.1007/s11306-019-1566-6. [DOI] [PubMed] [Google Scholar]
- Schmidt, Nyberg & Staerk (2014).Schmidt JS, Nyberg NT, Staerk D. Assessment of constituents in Allium by multivariate data analysis, high-resolution α-glucosidase inhibition assay and HPLC-SPE-NMR. Food chemistry. 2014;161:192–198. doi: 10.1016/j.foodchem.2014.03.062. [DOI] [PubMed] [Google Scholar]
- Scholten et al. (2016).Scholten OE, van Kaauwen MP, Shahin A, Hendrickx PM, Keizer LP, Burger K, van Heusden AW, van der Linden CG, Vosman B. SNP-markers in Allium species to facilitate introgression breeding in onion. BMC Plant Biology. 2016;16(1):187. doi: 10.1186/s12870-016-0879-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seregin, Anačkov & Friesen (2015).Seregin AP, Anačkov G, Friesen N. Molecular and morphological revision of the Allium saxatile group (Amaryllidaceae): geographical isolation as the driving force of underestimated speciation. Botanical Journal of the Linnean Society. 2015;178(1):67–101. doi: 10.1111/boj.12269. [DOI] [Google Scholar]
- Sfaxi et al. (2012).Sfaxi IH, Ezzine A, Coquet L, Cosette P, Jouenne T, Marzouki MN. Combined proteomic and molecular approaches for cloning and characterization of copper–zinc superoxide dismutase (Cu, Zn-SOD2) from garlic (Allium sativum) Molecular Biotechnology. 2012;52(1):49–58. doi: 10.1007/s12033-011-9473-8. [DOI] [PubMed] [Google Scholar]
- Shemesh-Mayer et al. (2015).Shemesh-Mayer E, Ben-Michael T, Rotem N, Rabinowitch HD, Doron-Faigenboim A, Kosmala A, Perlikowski D, Sherman A, Kamenetsky R. Garlic (Allium sativum L.) fertility: transcriptome and proteome analyses provide insight into flower and pollen development. Frontiers in Plant Science. 2015;6:271. doi: 10.3389/fpls.2015.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh-Mayer et al. (2013).Shemesh-Mayer E, Winiarczyk K, Błaszczyk L, Kosmala A, Rabinowitch HD, Kamenetsky R. Male gametogenesis and sterility in garlic (Allium sativum L.): barriers on the way to fertilization and seed production. Planta. 2013;237(1):103–120. doi: 10.1007/s00425-012-1748-1. [DOI] [PubMed] [Google Scholar]
- Shigyo, Fujito & Sato (2019).Shigyo M, Fujito S, Sato S. Toward repositioning Allium vegetable crops as global commodities: opportunities and challenges for future application of omics and phytomedomics platforms to crop improvement. ActaHortic. 2019;1251:1–8. [Google Scholar]
- Shigyo, Khar & Abdelrahman (2018).Shigyo M, Khar A, Abdelrahman M., editors. The allium genomes. Springer; 2018. [Google Scholar]
- Silva et al. (2015).Silva VCPD, Bettoni MM, Bona C, Foerster LA. Morphological and chemical characteristics of onion plants (Allium cepa L.) associated with resistance to onion thrips. ActaScientiarum. Agronomy. 2015;37(1):85–92. [Google Scholar]
- Sohn et al. (2016).Sohn SH, Ahn YK, Lee TH, Lee JE, Jeong MH, Seo CH, Chandra R, Kwon YS, Kim CW, Kim DS, Won SY, Kim JS, Choi D. Construction of a draft reference transcripts of onion (Allium cepa) using long-read sequencing. Plant Biotechnology Reports. 2016;10(6):383–390. doi: 10.1007/s11816-016-0409-4. [DOI] [Google Scholar]
- Soininen et al. (2014a).Soininen TH, Jukarainen N, Auriola SO, Julkunen-Tiitto R, Karjalainen R, Vepsäläinen JJ. Quantitative metabolite profiling of edible onion species by NMR and HPLC–MS. Food Chemistry. 2014a;165:499–505. doi: 10.1016/j.foodchem.2014.05.132. [DOI] [PubMed] [Google Scholar]
- Soininen et al. (2012).Soininen TH, Jukarainen N, Julkunen-Tiitto R, Karjalainen R, Vepsäläinen JJ. The combined use of constrained total-line-shape 1H NMR and LC–MS/MS for quantitative analysis of bioactive components in yellow onion. Journal of Food composition and Analysis. 2012;25(2):208–214. doi: 10.1016/j.jfca.2011.09.001. [DOI] [Google Scholar]
- Soininen et al. (2014b).Soininen TH, Jukarainen N, Soininen P, Auriola SO, Julkunen-Tiitto R, Oleszek W, Stochmal A, Karjalainen RO, Vepsäläinen J. Metabolite profiling of leek (Allium porrum L) cultivars by 1H NMR and HPLC–MS. Phytochemical Analysis. 2014b;25(3):220–228. doi: 10.1002/pca.2495. [DOI] [PubMed] [Google Scholar]
- Sta-Baba et al. (2010).Sta-Baba R, Hachicha M, Mansour M, Nahdi H, Ben Kheder M. Response of onion to salinity. African Journal of Plant Science and Biotechnology. 2010;4:7–12. [Google Scholar]
- Sun et al. (2019).Sun K, He J, Xiang Q, Wang X, Guan WB, Zhao L. The complete chloroplast genome of Allium ovalifolium var, leuconeurum, an endemic plant in Southwest China. Mitochondrial DNA Part B. 2019;4(1):1681–1682. doi: 10.1080/23802359.2019.1602009. [DOI] [Google Scholar]
- Sun et al. (2013).Sun XD, Ma GQ, Cheng B, Li H, Liu SQ. Identification of differentially expressed genes in shoot apex of garlic (Allium sativum L.) using Illumina sequencing. Journal of Plant Studies. 2013;2(2):136–148. doi: 10.5539/jps.v2n2p136. [DOI] [Google Scholar]
- Sun et al. (2016).Sun XD, Yu XH, Zhou SM, Liu SQ. De novo assembly and characterization of the Welsh onion (Allium fistulosum L.) transcriptome using Illumina technology. Molecular Genetics and Genomics. 2016;291(2):647–659. doi: 10.1007/s00438-015-1131-6. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2012).Sun X, Zhou S, Meng F, Liu S. De novo assembly and characterization of the garlic (Allium sativum) bud transcriptome by Illumina sequencing. Plant Cell Reports. 2012;31(10):1823–1828. doi: 10.1007/s00299-012-1295-z. [DOI] [PubMed] [Google Scholar]
- Suzuki, Do & Mukai (2002).Suzuki G, Do GS, Mukai Y. Efficient storage and screening system for onion BAC clones. Breeding Science. 2002;52(2):157–159. doi: 10.1270/jsbbs.52.157. [DOI] [Google Scholar]
- Tamura et al. (2013).Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura et al. (2019).Tsujimura M, Kaneko T, Sakamoto T, Kimura S, Shigyo M, Yamagishi H, Terachi T. Multichromosomal structure of the onion mitochondrial genome and a transcript analysis. Mitochondrion. 2019;46:179–186. doi: 10.1016/j.mito.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Tsukazaki et al. (2015).Tsukazaki H, Yaguchi S, Sato S, Hirakawa H, Katayose Y, Kanamori H, et al. Hamada M. Development of transcriptome shotgun assembly-derived markers in bunching onion (Allium fistulosum) Molecular Breeding. 2015;35(1):55. doi: 10.1007/s11032-015-0265-x. [DOI] [Google Scholar]
- Verma et al. (2018).Verma JP, Jaiswal DK, Krishna R, Prakash S, Yadav J, Singh V. Characterization and screening of thermophilic Bacillus strains for developing plant growth promoting consortium from hot spring of Leh and Ladakh Region of India. Frontiers in Microbiology. 2018;9:1293. doi: 10.3389/fmicb.2018.01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelckel, Gruenheit & Lockhart (2017).Voelckel C, Gruenheit N, Lockhart P. Evolutionary transcriptomics and proteomics: insight into plant adaptation. Trends in Plant Science. 2017;22(6):462–471. doi: 10.1016/j.tplants.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Voelckel et al. (2010).Voelckel C, Mirzaei M, Reichelt M, Luo Z, Pascovici D, Heenan PB, Schmidt S, Janssen B, Haynes PA, Lockhart PJ. Transcript and protein profiling identify candidate gene sets of potential adaptive significance in New Zealand Pachycladon. BMC Evolutionary Biology. 2010;10(1):151. doi: 10.1186/1471-2148-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Kohn, Kiełkowska & Havey (2013).Von Kohn C, Kiełkowska A, Havey MJ. Sequencing and annotation of the chloroplast DNAs and identification of polymorphisms distinguishing normal male-fertile and male-sterile cytoplasms of onion. Genome. 2013;56(12):737–742. doi: 10.1139/gen-2013-0182. [DOI] [PubMed] [Google Scholar]
- Wu et al. (2016).Wu S, Ning F, Wu X, et al. Wang W. Proteomic characterization of differential abundant proteins accumulated between lower and upper epidermises of fleshy scales in onion (Allium cepa L.) Bulbs. PLOS ONE. 2016;11(12):e0168959. doi: 10.1371/journal.pone.0168959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie et al. (2019a).Xie DF, Jin FY, Yang X, Li H, Xie FM, He XJ. The complete chloroplast genome of a wild onion species Allium monanthum (Alliaceae) Mitochondrial DNA Part B. 2019a;4(1):854–855. doi: 10.1080/23802359.2019.1572462. [DOI] [Google Scholar]
- Xie et al. (2019b).Xie DF, Yu HX, Xie C, Deng YQ, Chen J, Yu Y, Zhou SD, He X. Phylogeny of Chinese Allium species in Section Daghestanica and adaptive evolution of Allium (Amaryllidaceae, Allioideae) species revealed by the chloroplast complete genome. Frontiers in Plant Science. 2019b;10:460. doi: 10.3389/fpls.2019.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2015).Yang L, Wen C, Zhao H, Liu Q, Yang J, Liu L, Wang Y. Development of polymorphic genic SSR markers by transcriptome sequencing in the welsh onion (Allium fistulosum L.) Applied Sciences. 2015;5(4):1050–1063. doi: 10.3390/app5041050. [DOI] [Google Scholar]
- Yiu et al. (2009).Yiu JC, Juang LD, Fang DYT, Liu CW, Wu SJ. Exogenous putrescine reduces flooding-induced oxidative damage by increasing the antioxidant properties of Welsh onion. Scientiahorticulturae. 2009;120(3):306–314. [Google Scholar]
- Yoshimoto et al. (2015).Yoshimoto N, Onuma M, Mizuno S, Sugino Y, Nakabayashi R, Imai S, Tsuneyoshi T, Sumi S, Saito K. Identification of a flavin-containing S-oxygenating monooxygenase involved in alliin biosynthesis in garlic. The Plant Journal. 2015;83(6):941–951. doi: 10.1111/tpj.12954. [DOI] [PubMed] [Google Scholar]
- Yuan et al. (2018).Yuan Q, Song C, Gao L, Zhang H, Yang C, Sheng J, Ren J, Chen D, Wang Y. Transcriptome de novo assembly and analysis of differentially expressed genes related to cytoplasmic male sterility in onion. Plant Physiology and Biochemistry. 2018;125:35–44. doi: 10.1016/j.plaphy.2018.01.015. [DOI] [PubMed] [Google Scholar]
- Yurgel et al. (2018).Yurgel SN, Abbey L, Loomer N, Gillis-Madden R, Mammoliti M. Microbial communities associated with storage onion. Phytobiomes. 2018;2(1):35–41. doi: 10.1094/PBIOMES-12-17-0052-R. [DOI] [Google Scholar]
- Yusupov et al. (2019).Yusupov Z, Deng T, Liu C, Lin N, Tojibaev K, Sun H. The complete chloroplast genome of Allium fistulosum. Mitochondrial DNA Part B. 2019;4(1):489–490. doi: 10.1080/23802359.2018.1545532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2018).Zhang C, Li X, Zhan Z, Cao L, Zeng A, Chang G, Liang Y. Transcriptome sequencing and metabolism analysis reveals the role of cyanidin metabolism in dark-red onion (Allium cepa L.) Bulbs. Scientific Reports. 2018;8(1):14109. doi: 10.1038/s41598-018-32472-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2016).Zhang C, Zhang H, Zhan Z, Liu B, Chen Z, Liang Y. Transcriptome analysis of sucrose metabolism during bulb swelling and development in onion (Allium cepa L.) Frontiers in Plant Science. 2016;7:1425. doi: 10.3389/fpls.2016.01425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng et al. (2016).Zheng X, Tang S, Zhu S, Dai Q, Liu T. Identification of an NAC transcription factor family by deep transcriptome sequencing in onion (Allium cepa L.) PLOS ONE. 2016;11(6):e0157871. doi: 10.1371/journal.pone.0157871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou et al. (2015).Zhou SM, Chen LM, Liu SQ, Wang XF, Sun XD. De novo assembly and annotation of the Chinese chive (Allium tuberosum Rottler ex Spr.) transcriptome using the Illumina platform. PLOS ONE. 2015;10(7):e0133312. doi: 10.1371/journal.pone.0133312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu et al. (2017).Zhu S, Tang S, Tan Z, Yu Y, Dai Q, Liu T. Comparative transcriptomics provide insight into the morphogenesis and evolution of fistular leaves in Allium. BMC Genomics. 2017;18(1):60. doi: 10.1186/s12864-016-3474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
This is a literature review and thus does not have any raw data.