Abstract
The cereal infecting fungus Fusarium graminearum is predicted to possess a single homologue of plant RALF (rapid alkalinisation factor) peptides. Fusarium mutant strains lacking FgRALF were generated and found to exhibit wildtype virulence on wheat and Arabidopsis floral tissue. Arabidopsis lines constitutively overexpressing FgRALF exhibited no obvious change in susceptibility to F. graminearum leaf infection. In contrast transient virus-mediated over-expression (VOX) of FgRALF in wheat prior to F. graminearum infection, slightly increased the rate of fungal colonisation of floral tissue. Ten putative Feronia (FER) receptors of RALF peptide were identified bioinformatically in hexaploid wheat (Triticum aestivum). Transient silencing of two wheat FER homoeologous genes prior to F. graminearum inoculation did not alter the subsequent interaction outcome. Collectively, our VOX results show that the fungal RALF peptide may be a minor contributor in F. graminearum virulence but results from fungal gene deletion experiments indicate potential functional redundancy within the F. graminearum genome. We demonstrate that virus-mediated over-expression is a useful tool to provide novel information about gene/protein function when results from gene deletion/disruption experimentation were uninformative.
Keywords: Virus-mediated over-expression, Triticum aestivum, Arabidopsis, Fusarium head blight, Fungal infection, Virus-induced gene silencing, Barley Stripe Mosaic Virus
1. Introduction
Fusarium Head Blight (FHB) is a disease that causes significant economic yield losses by reducing grain quality and safety in several cereal crops worldwide, such as wheat, barley, maize and oat. FHB disease is caused mainly by the Ascomycete fungus Fusarium graminearum (Backhouse, 2014). Like many highly successful plant pathogens during host plant infection and colonisation, F. graminearum is predicted to produce a diverse repertoire of secreted proteins, enzymes and secondary metabolites that modulate plant metabolism to suppress and/or re-programme plant defences (Brown et al., 2012, King et al., 2015, Rafiqi et al., 2012). Many interactions between a successful pathogen and its adapted plant host species rely upon the loss, acquisition or modification of effectors by the pathogen, as well as the presence of non-functional/weakly functional variant host proteins with roles in direct or indirect detection of these effectors (Jones et al., 2016). Thus, understanding the molecular functions of F. graminearum secreted proteins will help to elucidate the processes underlying wheat spike colonisation and fungal pathogenicity.
The gene designated FgRALF (FGRAMPH1_01G16205) codes for a protein that possesses the pfam domain RALF (Rapid alkalinisation factor; PF05498). RALF domain-containing proteins are predominately found in plants and play a role in plant development, such as regulating tissue expansion in sugarcane and negatively regulating pollen tube elongation in Arabidopsis (Li et al., 2016, Murphy and De Smet, 2014). An extensive database search revealed that different plant species possess multiple gene copies. For example, a total of 39, 43, 34 and 18 ralf genes were found in the reference genomes of Arabidopsis, rice, maize and soybean, respectively (Sharma et al., 2016). Although rapid-alkalinisation factor proteins are predominantly found in plants (both dicotyledonous and monocotyledonous species), this protein type has since been identified in a range of fungal species.
Thynne et al. (2017) analysed numerous fungal genomes searching for homologues of plant RALF proteins using RALF proteins sequences from Arabidopsis thalianaas the query sequence (do Canto et al., 2014). Twenty-six different species of fungi were found to possess RALF homologues from plants. Interestingly, all RALF domain containing species were plant pathogens, including three Basidiomycetes and 23 Ascomycetes species. However, ralfgenes are predicted to have been acquired independently in different fungal species. Phylogenetic analysis of peptide sequence similarity revealed that fungal RALF homologues are interspersed amongst the plant RALFs (Thynne et al., 2017). The same study proposed RALF homologues have diverged into four groups in Fusariumspecies; the F. graminearumsequence was placed within group III along with sequences from the cereal infecting species F. pseudograminearumand the tomato infecting species Fusarium oxysporumf. sp.radicis-lycopersiciCL57 (Thynne et al., 2017). Nevertheless, it is unknown if RALF domain containing proteins share similar functions in each group or if any correlated functions exists between the species groups.
For mutants of the tomato infecting species Fusarium oxysporum f.sp. lycopersici lacking the f-ralf gene (FOXG_21151) (group I) conflicting in planta phenotypes have been reported. Masachis et al., 2016 reported significant attenuation in virulence on plant roots, and increased expression of various defence genes in the host. In contrast, Thynne et al. (2017) demonstrated that the same F-RALF protein was not required for infection of F. oxysporum f.sp. lycopersici in roots of older tomato plants. Differences in how the pathogenicity tests were carried out may account for these contrasting results.
The Arabidopsis genome potentially encodes 39 RALF family proteins and although few components of the signalling pathway have been explored, the Feronia (FER) protein has been identified as a receptor for RALF1 (Campos et al., 2018, Haruta et al., 2014, Li et al., 2016). Arabidopsis AtRALF1 is a 120-amino acid peptide and contains the RALF domain between amino acids 58–119. FER is a receptor-like kinase (RLK) that contains an extracellular malectin-like protein, which is known to recognise and bind cell wall carbohydrates. AtRALF1 affects phosphorylation of FER and the key cell growth regulator H+-ATPase (Li et al., 2016). AtRALF1 was shown to initiate a downstream signalling cascade that led to apoplastic alkalinisation and inhibition of cell elongation of primary root (Haruta et al., 2014). Arabidopsis mutant plants lacking FER receptor were also more resistant to infection by F. oxysporum (Masachis et al., 2016).
In this study, we first analysed whether FgRALF, a group III RALF, is required for F. graminearum virulence on wheat floral tissue by generating and evaluated several independent single gene mutant strains lacking FgRALF. We then generated and tested the susceptibility of independent Arabidopsis lines constitutively overexpressing FgRALF for susceptibility to F. graminearum infections in leaves. In wheat, we explored whether Barley Stripe Mosaic virus mediated over-expression (BSMV-VOX) of FgRALF prior to fungal infection influenced the extend of disease development. For the final experimental approach, we investigated for the presence of predicted FER receptor encoding genes within the newly available hexaploid wheat genome (Appels et al., 2018). We used Barley Stripe Mosaic Virus Induced Gene silencing (BSMV-VIGS) to silence transiently all three homoeologous of the wheat FER gene prior to F. graminearum inoculation and the resulting interaction outcomes were explored in detail. Collectively, these data sets indicate that FgRALF potentially play a role during FHB infection.
2. Material and methods
2.1. Identification of putative Fusarium spp., wheat and Arabidopsis RALF genes
RALF proteins in Arabidopsis and wheat were retrieved from Plant Ensembl genome database release 44 (Bolser et al., 2016, Cunningham et al., 2019) and filtered using the BioMart tool for protein sequences that contain the Rapid Alkalinisation Factor domain (PF05498). Common wheat (Triticum aestivum) has a complex hexaploid genome (AABBDD) consisting of A, B and D homoeologous chromosome sets (International Wheat Genome Sequencing Consortium (IWGSC), 2018). For completeness, in cases where only one of the three wheat homoeologous contained the ralf domain, the other two homoeologues were also included in the bioinformatic analyses. RALF proteins from F. graminearum and some Fusarium species reported by Thynne et al. (2017) were used in this study.
Multiple protein sequences alignment of RALF proteins from Arabidopsis and Fusarium spp. was carried out using Geneious Alignment in Geneious 10 (Kearse et al., 2012). A tree was generated from protein alignment with Neighbour-Joining method using Jukes-Cantor distance model. Bootstrap analyses were based on 500 replicates.
2.2. F. graminearum gene deletion experiments
The FgRALF gene, FGRAMPH1_01G16205 (http://fungi.ensembl.org/Fusarium_graminearum/Info/Index) was deleted in F. graminearum wild-type strain PH-1 (NRRL 31084) for which the complete genome sequence is available (King et al., 2015). Gene deletion was done using the “split-marker” approach (Yu et al., 2004). The DNA flanks, 1030 bp 5′ and 1000 bp 3’ – sequence, and the hygromycin (hph) resistance gene split fragments (1246 bp 5′ and 858 bp 3’ – sequence) were amplified by polymerase chain reaction (PCR) with the primers listed in table S1. Hygromycin fragments were amplified from pHYG1.4 vector (Urban et al., 2003). PCRs were carried out in 25 μl volumes, containing 50 ng of DNA, 1 U Phusion High-Fidelity DNA polymerase (New England BioLabs Inc.), 10 pmol of each primer and 0.25 mM each deoxynucleoside triphosphate, in a standard buffer for 35 cycles with the following cycling parameters: denaturation at 98 °C for 10 s; annealing at 58 °C for 30 s; and DNA synthesis at 72 °C for 1 min. The resulting amplicon was gel purified using Qiagen gel extraction kit QIAstock. The fragments were inserted into the EcoRV restriction site of pGEM®-T Easy Vector (Promega) using Gibson assembly Master Mix kit (New England BioLabs Inc.) according to the manufacturer’s protocol to generate vectors pAMFg5.1 and pAMFg5.2.
The resulting vectors were used to transform protoplasts of F. graminearum strain PH-1, as described previously (Hohn and Desjardins, 1992). Briefly, both split-marker constructs contained in pAMFg5.1 and pAMFg5.2 were quantitatively amplified by PCR using HotStar TAQ polymerase (Qiagen) following the manufacturer’s instructions. PCR products were adjusted to a concentration of 1 μg μl−1 and 5 μl of each construct was mixed and transformed into 1 × 108 protoplasts of F. graminearum. Transformants were selected in REG medium (0.7% agarose, 0.2% Yeast Extract, 0.2% Casein-Hydrolysate (N-Z-Amine A), 0.8M sucrose) containing 75 μg/ml of hygromycin B. Hygromycin resistant transformants were then transferred to PDA agar plates containing hygromycin (10 μg/ml) for further analysis. For fungal genomic DNA extraction, transformants were grown in 10 ml potato dextrose medium in the presence of hygromycin (10 μg/ml) and DNA was extracted using DNeasy Plant Mini Kit (Qiagen).
In the two isolated gene replacement mutants selected - PH-1ΔFgRALF (1) and PH-1ΔFgRALF (8) - two diagnostic PCR fragments of 1.69kb and 1.32kb size were detectable using oligomer pairs AM117/GC1 (PCR 1) and CG2/AM118 (PCR 2), respectively. In both mutants PH-1ΔFgRALF (1) and PH-1ΔFgRALF (8), the FgRALF gene is absent using oligomer pairs MVOX1F/MVOX1R (PCR3).
2.3. Constitutive over-expression of FgRALF in Arabidopsis thaliana
Full-length FgRALF was PCR-amplified from cDNA extracted from F. graminearum-infected wheat cv. Bobwhite using Phusion High-Fidelity DNA polymerase (New England Biolabs). KpnI and MluI restriction sites were added to the 5′ and 3’ end of the FgRALF, respectively, using the primers listed in table S1.
Digestion by KpnI-HF and MluI-HF (New England Biolabs) enabled entry of FgRALF into the vector pMS37, downstream of a 35S promoter and upstream of an ocs terminator, using standard restriction enzyme cloning techniques (Sandkvist et al., 1995). The 35S:FgRALF:ocs cassette was then sub-cloned into the binary plant vector pMLBART by NotI (New England Biolabs) digestion (Gleave, 1992). Sequence-verified constructs were transformed into the Agrobacterium strain GV3101 for transformation of the Arabidopsis ecotype Columbia-erecta using the floral-dip method (Clough and Bent, 1998).
For selection of transgenic lines, seeds were surface-sterilised and grown on Murashige and Skoog (MS) medium supplemented with kanamycin (50μg ml−1) and carbenicillin (100μg ml−1). Independent transgenic T1 lines that segregated 3:1 were carried through and homozygous T3 lines were selected for Fusarium pathogenicity assays.
2.4. Identification of putative wheat Feronia receptors
Protein domain analysis of predicted FER and Feronia family genes in hexaploid wheat (Triticum aestivum) was carried out using the BioMart tool in Ensembl (Bolser et al., 2016). The wheat genome assembly used for this analysis was the IWGSCrefseq1 (International Wheat Genome Sequencing Consortium (IWGSC), 2018). Wheat genes coding for proteins contained both predicted kinase-like (PF07714) and malectin-like (PF12819) domains were filtered. These are the two protein-domains that are characteristic of Feronia protein family. Additionally, the previously identified seventeen members of Arabidopsis protein sequences belonging to the Feronia family were also extracted from Ensembl (Li et al., 2016). Multiple protein sequences alignment was carried out in ClustalW, linked to Geneious 10 (Kearse et al., 2012). A tree was generated from protein alignment with Neighbour-Joining method using Jukes-Cantor distance model. Bootstrap analyses were based on 1000 replicates.
2.5. RNA extraction and reverse-transcription (RT)-PCR
Total RNA was extracted from spring wheat cv. Bobwhite (water inoculated, infected with F. graminearum, or virus infected) tissue using TRIzol® reagent (Invitrogen, USA) according to the manufacturer’s protocol. For wheat cDNA, one microgram of total RNA was treated with RQ1 RNase-free DNase I (Promega, Madison, WI, U.S.A.) and was used for random primer-generated cDNA synthesis using High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, U.S.A.) according to the manufacturer’s instructions.
2.6. Barley Stripe Mosaic virus - Virus induced protein over-expression (BSMV-VOX) and virus induced gene silencing (BSMV-VIGS)
To explore in planta functions in wheat we used the BSMV-VIGS system described by Yuan et al. (2011), and the BSMV-VOX system described by Lee et al. (2012) which comprises of three T-DNA binary plasmids, pCaBS-α, pCaBS-β, and pCa-ɣbLICs.
The FgRALF and Fg08493 protein-overexpression constructs were generated by cloning each of the selected F. graminearum genes (FGSG_15123 and FGSG_08493) into pCassRZ-ɣb-2A-LIC using a ligation-independent cloning (LIC) strategy (Aslanidis and Dejong, 1990). Standard reverse transcription-polymerase chain reaction (RT-PCR) was used to generate cDNA clones of the selected genes using Phusion High Fidelity PCR Master with HF buffer (New England Biolabs, MA, USA). The starting template was total RNA extracted from wheat cv. Bobwhite spike tissue 5 days after inoculation with F. graminearum PH-1. Adaptors for LIC were incorporated at the 5′ and 3’ ends of the gene sequences for cloning into pCassRZ- ɣb-2A-LIC via PCR using primers described in table S1.
Gene-silencing constructs were generated by cloning fragments of the wheat TaFER1 and TaFER2 sequences into pCa-ɣbLIC in an antisense orientation, using also LIC strategy. In silico predictions by siFi21 software were used to select the most effective gene-specific fragments for silencing, ranging from 254 to 325 bp in size. This was done to ensure the selected fragments were not likely to direct off-target wheat gene silencing. The cDNA fragments were generated by standard reverse transcription-polymerase chain reaction (RT-PCR) from total RNA extracted from spring wheat cv. Bobwhite leaf tissue using Phusion High Fidelity PCR Master with HF buffer (New England Biolabs, MA, USA). The primers used are described in table S1. The BSMV:MCS4D construct containing just the multiple cloning site was used as a negative control construct for BSMV-VIGS.
For both BSMV-VOX and BSMV-VIGS, the BSMV pCaBS-α, pCaBS-β, and pCa-ɣbLICv derivatives were transformed separately into Agrobacterium tumefaciens GV3101 by electroporation.
Viral inoculation of N. benthamiana by agroinfiltration was carried out as previously described (Lee et al., 2012). The infiltrated N. benthamiana leaves were harvested at 5 days post-infiltration and ground using a mortar and pestle in 10 mM potassium phosphate buffer (pH 6.8) containing 1% cellite. For the BSMV-VIGS experiments the sap was mechanically inoculated into the fourth leaf of 38-days-old wheat plants. For BSMV-VOX experiments the sap was mechanically inoculated on both the first leaf below the flag leaf and the flag leaf on the main tiller of 42-days-old wheat plants. F. graminearum point inoculations took place when individual wheat tillers came into anthesis.
2.7. Quantitative RT-PCR
To test for the efficiency of silencing in the BSMV-VIGS experiments, RNA was extracted from silenced- and virus control-infected spikelets. RT-PCR was carried out and the resulting cDNA was diluted 1:20 with sterile deionised water and was analysed using SYBR Green Jumpstart ReadyMix (Sigma Aldrich). A 5-μL aliquot of diluted cDNA was used in a 20- μL PCR analysis, with an annealing temperature of 60 °C. Quantification of gene expression was carried out in an ABI 7500 Real-Time PCR system (Applied Biosystems). For normalisation of gene expression using quantitative RT-PCR in silenced-versus virus control-infected leaves, CDC48 was used as the reference gene (Lee et al., 2014). All primers were used at a final concentration of 400nM/reaction. The primers used for real-time PCR are described in table S1.
2.8. Agrobacterium-mediated expression of FgRALF in Nicotiana benthamiana and N. tabacum
Full-length FgRALF was PCR-amplified from cDNA generated as described above using primers with attB flanks. The AttB-flanked PCR product was cloned into the Gateway-compatible entry vector pDONR207 using BP clonase II enzyme mix (ThermoFisher) and transformed to the E. coli JM109 competent cells (Promega). Sequence-verified constructs were then recombined into the binary destination vector pEAQHTDEST3 (Sainsbury et al., 2009) using the LR clonase II enzyme mix (ThermoFisher) and used to transform JM109 cells. Sequence-verified constructs were subsequently transformed into A. tumefaciens GV3101 by electroporation for transient expression in the two Nicotiana species. Agrobacterium containing FgRALF was cultured and resuspended in agroinfiltration buffer to an OD600 of 1.0 for infiltration of 4-5-week-old Nicotiana plants. After 7 days, infiltrated leaves were assessed for cell death under white and UV light. Three experiments were carried out independently.
2.9. Plant growth conditions
Nicotiana benthamiana seeds were germinated in Levington F2+S compost (Everris Ltd.) and kept in a humid chamber in a controlled environment growth room at 23 °C:18 °C (day:night), 60% relative humidity with a 16-h photoperiod (180μmol m−2 per second of light). Plants 4-5-week-old were used for the preparation of BSMV-VIGS sap inoculum and for expression of recombinant FgRALF. The F. graminearum-susceptible wheat (Triticum aestivum) cultivar, Bobwhite, was grown in Rothamsted soil mix under the same conditions as described above. Arabidopsis thaliana seeds were grown in Levington F2+S compost (Everris Ltd.) in a controlled environment chamber at 20 °C:17 °C (day:night), 70% relative humidity with a 16-h photoperiod (200μmol m−2 per second of light). Seeds were stratified in the dark for 4 day at 5 °C before transfer to the growth chamber.
2.10. Fungal growth conditions and inoculations
The wild-type isolate of F. graminearum, PH-1, was grown on synthetic nutrient agar (SNA) plates for 8 days under constant illumination from one near-UV light and one white light (Leslie and Summerell, 2008). To induce fresh conidia formation, plates were washed with an overlay of TB3 (0.3% yeast extract, 0.3% Bacto Peptone, 20% sucrose) and two days later, spores were harvested and adjusted to a concentration of 1 × 105 spores ml−1. For wheat floral inoculations, when wheat spikes were at anthesis, 5μl of conidial suspension was pipetted into the wheat floral cavity between the palea and lemma of the first two florets of the 13th and 14th spikelets from the base of the wheat spike. For mock-inoculated plants, the florets were inoculated with 5μl of sterile distilled water. The inoculated plants were kept at high humidity for 48 h, of which for the first 24 h the humid chamber was covered to place the plants in darkness (Brown et al., 2010). Disease symptoms were scored by counting the number of symptomatic spikelets below the point of inoculation every three days until day 15 (Dilks et al., 2019).
Arabidopsis inoculations were done using a detached leaf assay method with some modifications (Chen et al., 2006). Briefly, 4-week-old plants were placed on square plates (10 cm × 10 cm) containing 1% water agar with the adaxial surface facing upwards. Rosette leaves were wounded by puncture of the adaxial surface over the mid-rib with a glass Pasteur pipette and 5 μL of inoculum (5 × 105 spores ml−1) supplemented with 20 μM deoxynivalenol (DON) was deposited on the fresh wound on the adaxial surface. The mock inoculation was similarly applied using sterile distilled water (5 μL) amended with the same DON concentration. Plates were kept in the dark for three days and then exposed to low light (40 μmol m−2 per second of light) for an additional fourdays until assessment. At seven days post inoculation (dpi), the inoculated leaves were photographed and disease levels were quantified by measuring the proportion of lesioned/necrotic area compared to the total leaf area. These analyses were done using the LemnaGrid software module (LemnaTec GmbH, Aachen, Germany).
For Arabidopsis floral inoculation (Urban et al., 2002), Arabidopsis plants ecotype Landsberg erecta (Ler-0) with 2–3 open flowers but no siliques (Growth stage 6) (Boyes et al., 2001) were spray inoculated with F. graminearum conidia suspension (1 × 106 spores.ml−1) using 15ml spray bottles. Each plant received approximately 0.5ml of suspension. Control plants received a similar volume of sterile distilled water. Inoculated plants were kept in Perspex boxes (50 x 50 × 100 cm) at 100% humidity for 7 days, with the first 24h in the dark. At 7 dpi, visible infection symptoms on the flowers and developing siliques were assessed using the Fusarium – Arabidopsis Disease (FAD) scoring system described in Urban et al. (2002).
F. graminearum infection experiments were done at least two or three times for negative and positive results, respectively.
2.11. Fusarium graminearum growth rate on PDA media
F. graminearum strains were grown on potato dextrose agar (PDA) plates in the dark at 25 °C. After three days, the diameter of fungal colonies was measured. The diameter of 15 plates for each strain was measured and compared (Leslie and Summerell, 2008).
2.12. Statistical analyses
For infection of wheat spikes inoculated with F. graminearum wild-type and mutants, the area under disease progress curve (AUDPC) was calculated based on the number of infected spikelets over time until 15 dpi. The values were analysed using a linear mixed effect model (LMM) test, while adjusting for multiple comparisons using Tukey’s method. Data was analysed in R using packages and code described in Schandry (2017).
For the following experiments, Genstat for Windows 19th Edition was used (VSN International, 2017). Proportion data from VOX and VIGS experiments were analysed using a generalised linear mixed model (GLMM) assuming a binomial distribution with logit link function. Significance of difference between calculated means was determined using least significant difference (LSD) at the 5% level of significance.
The statistical design for Arabidopsis leaf inoculation assays consisted of randomised blocks. Forty-eight plants were used in total for each experiment (eight plants/independent line). Disease was quantified by expressing the diseased leaf area relative to the total leaf area. Mean disease levels for each genotype were compared using a multi-stratum analysis of variance (ANOVA). Independent lines were compared with the wild type using a Dunnett’s test.
Arabidopsis–Fusarium disease susceptibility data generated using the FAD scoring system (Urban et al., 2002) following flower spray inoculation were analysed using a proportional-odds model fitted as a GLM assuming a multinomial distribution (with three classes and logit link function) for the counts of plants (12 in total per genotype) classified by disease score.
For fungal growth tests, the experimental design was randomised with 15 plates/strain. Diameter measurements were subject to ANOVA followed by two-sided Student’s t-test to compare the two independents mutant strains (PH-1ΔFgRALF (1) and (8)) with FgPH-1 WT.
Graphic visualisations were done using the ggplot2 (Wickham, 2009) package in R.
3. Results
3.1. FgRALF is closely related to four putative RALF peptides from Arabidopsis thaliana
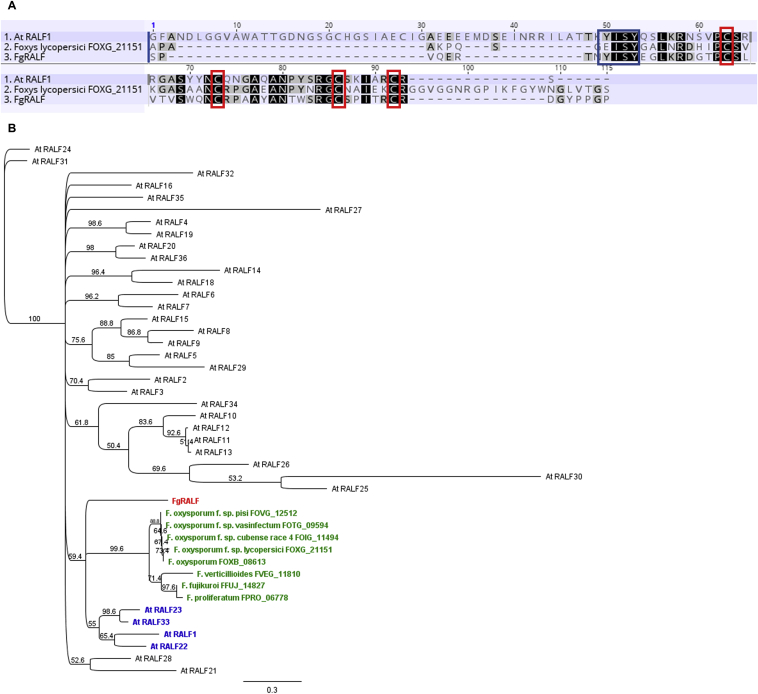
Analysis of the F. graminearum genome detected a predicted protein (FGRAMPH1_01G16205, FGSG_15123, UniProtKB A0A0E0SJI5) that possesses the pfam domain RALF (Rapid alkalinisation factor; PF05498). Previous studies demonstrated that this gene is a homologue within the plant family of secreted regulatory peptides, termed rapid alkalinising factors (RALF) (Thynne et al., 2017). Plant RALFs are secreted as pre-propeptides and proteolytically processed to a bioactive peptide (Srivastava et al., 2009). However, the predicted F. graminearum RALF (FgRALF) lacks a propeptide sequence and consists only of the mature peptide preceded by an N-terminal secretion signal (Fig. 1A). The mature FgRALF protein is predicted to be 59 aa in length and to contain four conserved cysteine residues and the ‘YISY’ motif, both of which have been shown to be required for the alkalinising activity of plant RALFs (Pearce et al., 2010).
Fig. 1.
Alignment and analysis of the rapid alkalinisation factor (RALF) domain predicted in the F. graminearum sequence. A) Amino-acid sequence alignment of the predicted mature F. graminearum FgRALF (FGRAMPH1_01G16205), Arabidopsis thaliana AtRALF1 (AT1G02900) and F. oxysporum f.sp. lycopersici (FOXG_21151) proteins. Conserved residues are indicated in black. Cysteine residues predicted to form disulfide bonds are in red boxes. The conserved isoleucine residue essential for biological activity of AtRALF1 is in the blue box. B) Neighbour-Joining consensus tree of RALF proteins alignment from Arabidopsis thaliana and selected Fusarium species. The designated FgRALF, is shown in red text; RALF genes from other Fusarium species are shown in green text and the closely related RALF genes from Arabidopsis are shown in blue text.
In order to identify which of the Arabidopsis RALF genes were more closely related to FgRALF in F. graminearum, a neighbour-joining phylogenetic tree was built. Fig. 1B shows that FgRALF and other putative RALF proteins from different Fusarium species are most closely related to the clade containing the well-studied AtRALF23, AtRALF33, AtRALF22 and AtRALF1 sequences. These four AtRALF have been shown to be involved in plant immunity (Stegmann et al., 2017).
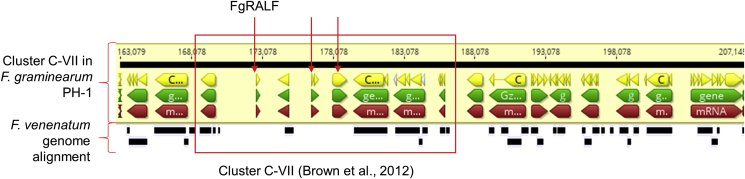
Of the fungal genera harbouring RALF homologues, the genus Fusarium has the most diverse array of RALF homologues. Interestingly, two sequenced F. oxysporum isolates that are not reported to be plant pathogens (a human pathogen and non-pathogenic isolate) carry mutations in the conserved cysteine residues suggesting loss of function (Thynne et al., 2017). Similar mutations were checked for in the non-pathogenic Fusarium species, Fusarium venenatum which is closely related to F. graminearum. In the fully completed and assembled F. venenatum genome, no FgRALF homologue was found, suggesting this gene is probably absent in this species (King et al., 2018). In F. graminearum, FgRALF is in the sub-telomere region of chromosome 3 within the small gene cluster C-VII. This cluster was previously predicted to be enriched for small secreted proteins (Brown et al., 2012). Blast analysis of this cluster within the F. venenatum genome (King et al., 2018) identified that not only FgRALF, but at least two more genes in the cluster are absent in F. venenatum (Fig. 2).
Fig. 2.
LASTZ alignment between F. graminearum and F. venenatum genomes. Alignment between the previously predicted eight members gene cluster C-VII (Brown et al., 2012) within the F. graminearum PH-1 genome and the comparable gene cluster in the non-pathogenic species F. venenatum. The alignment was carried out in Geneious 10. The top black line indicates the genome sequence of F. graminearum PH-1 chromosome 3. The yellow, green and red arrows indicate the coding sequence, genomic sequence and mRNA sequence, respectively. The position of the FgRALF and the other missing genes in F. venenatum are indicated with the red arrows. At the bottom of the figure, the black lines indicate the alignment of the F. venenatum genome sequence.
We also checked for the presence of RALF proteins in hexaploid wheat by filtering proteins containing RALF domain (PF05498) on the predicted wheat proteome. We found 33 wheat protein sequences containing a predicted RALF domain. In cases where only one of the three wheat homoeologous contained the RALF domain, the other two homoeologues were also included in the analysis, resulting in a total of 36 sequences (Table S2). The wheat RALF mature peptides shared between 17.4% and 46.9% identity with AtRALF1. Most of the sequences (19) contain the ‘YISY’ motif and four cysteines residues. Three sequences contain the “ISY”, which is also suggested to be enough for the alkalinising activity in plant RALFs (Masachis et al., 2016), but carry mutations in the first two cysteine residues. The remaining 14 sequences have between one and three mutations in the ‘YISY’ motif (Table S2).
3.2. Effect of FgRALF gene deletion in F. graminearum virulence
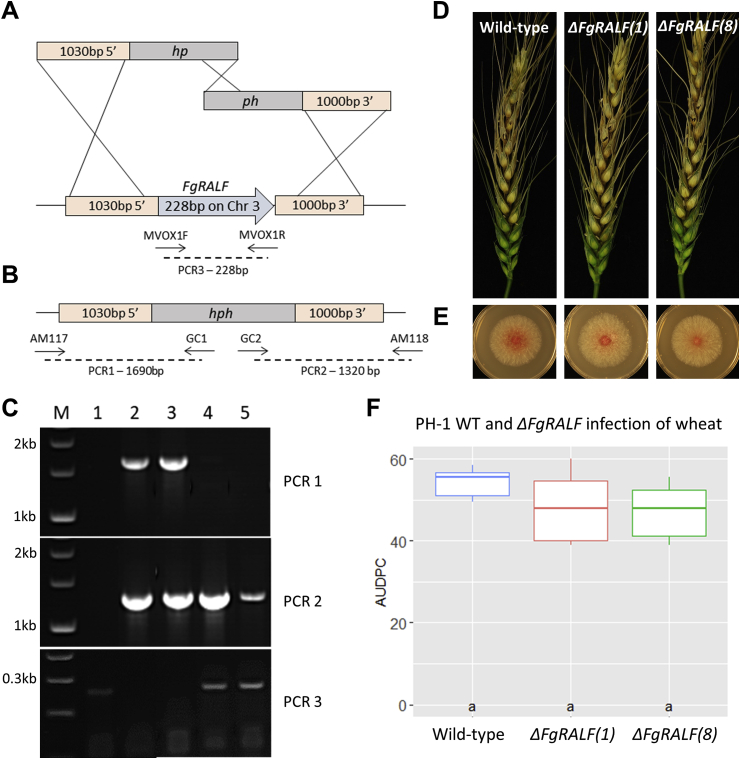
To test the role of RALF in fungal virulence, a population of transformants was generated, and two independent PH-1ΔFgRALF gene deletion mutants (1 and 8) were selected for further analysis. Molecular characterisation of the selected mutants is shown in Fig. 3A–C. No statistically significant differences were observed in in vitro fungal growth when grown on rich media (PDA) between wild type and the two PH-1ΔFgRALF (1) and (8) strains (Fig. 3E). Colony colour, conidia spore morphology and spore germination rates were indistinguishable between wild-type and mutant strains.
Fig. 3.
Deletion and functional characterisation of FgRALF. (A) Genomic 5′ flank (1030bp) and 3′ flank (1000bp) (bars) were amplified with primers and fused to parts of the hph hygromycin resistance gene. Fused PCR fragments were used in a split-marker strategy to replace FgRALF. (B) Anticipated diagnostic PCR for successful gene replacement of FgRALF. (C) Results of diagnostic PCR and expected sizes indicated in (A) and (B). Loadings are: M—λ DNA-BstEII digest, 1–5 WT and transformants FgRALF (1), (8), (5) and (6). FgRALF (1) and (8) have the ΔFgRALF null allele and lost the 228bp ralf fragment. (D) Wheat spikes inoculated with wild-type, FgRALF null mutants (1) and (8) strains 12 days post-inoculation. Spore-droplet inoculated spikelets are marked with black dots. (E) FgPH-1 WT and FgRALF null mutants (1) and (8) strains growth on PDA for 3 days. No differences in fungal growth was observed (n = 15) (p > 0.5 – t-test). (F) Analysis of area under the disease progression curve (AUDPC). AUDPC values, represented in the box-plot, as estimated by a linear mixed effects model coloured by strain. Letters above the strain names indicate the significance group (p < 0.05).
The wild type and the two FgRALF-deleted strains were tested for infectivity and disease formation in wheat floral tissue using the fully susceptible cv. Bobwhite via point inoculation. In two independent experiments, using six spikes per strain, no reduction in initial infection or FHB symptoms development were observed compared to the wild type (Fig. 3C and E). In two further pathogenicity test, F. graminearum strain PH-1wt and PH-1ΔFgRALF (1) and (8) were spray-inoculated into Arabidopsis (Ler-0) floral tissue (Urban et al., 2002). No significant differences in disease phenotype were observed (Fig. S1).
3.3. Agrobacterium-mediated expression of FgRALF in Nicotiana benthamiana and N. tabacum
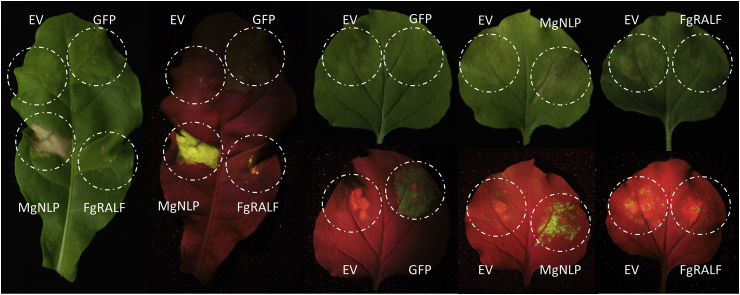
Transient expression of fungal proteins in Nicotiana benthamiana is widely used to screen for effector functionality in regard to cell-death inducing ability. To test whether FgRALF is recognised in a non-natural host species and activates plant defences or plant cell death, we used an Agrobacterium-mediated transient effector expression system in Nicotiana benthamiana and N. tabacum. This system utilises the Cowpea mosaic virus (CPMV)-derived pEAQ-HT expression vector which facilitates high-level and long-lasting recombinant protein expression (Sainsbury et al., 2009). Full length FgRALF with a signal peptide was cloned into pEAQ-HT for in planta expression. As a positive control for cell death induction, the Z. tritici protein MgNLP was used, and the negative controls were GFP-expressing vector and empty vector (Kettles et al., 2017, Motteram et al., 2009). Consistent with previous results, MgNLP expressed using pEAQ-HT induced a strong cell death phenotype as indicated by macroscopic symptoms visible under white light and accumulation of cell death-related auto-fluorescent compounds visible under UV light (Fig. 4). No cell death was detected in GFP-expressing control leaves or empty vector (Fig. 4). FgRALF-expressing leaves also did not show any cell-death visible symptoms both under white light and UV light. These results suggest that FgRALF is not recognised in this non-host plant species.
Fig. 4.
Agrobacterium-mediated expression of FgRALF in Nicotiana tabacum and Nicotiana benthamiana does not induce a defence response. Leaves photographed at 7 days post agroinfiltration. The same leaves are photographed under white light and UV light. Empty vector (EV) was used as a negative control and MgNLP was included as a positive control for necrosis. GFP was included as a control for heterologous protein expression. The N. tabacum leaves are on the left.
3.4. Overexpression of FgRALF in wheat using BSMV-VOX
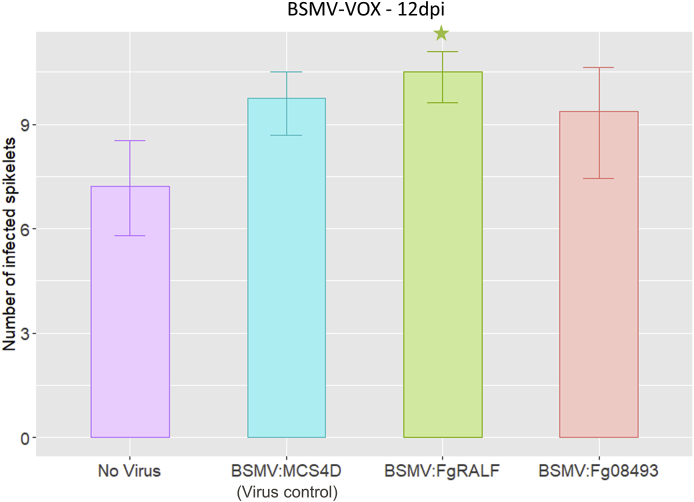
To explore whether pre-expression of FgRALF in wheat floral tissue prior to inoculation with F. graminearum would alter the interaction outcome we deployed the transient BSMV-VOX expression system which is very suitable for small proteins (Lee et al., 2012). In these experiments, all the statistical analyses were carried out comparing the treatments with the BSMV:MCS4D control, where the only addition to the viral genome is a multiple cloning site (MCS) from pBluescript K. The virus infection itself appears to have an effect on FHB disease outcome and therefore the virus inoculated plants are the more suitable control for comparison (Wing-Sham Lee, personal communication). The combined results from four BSMV-VOX experiments are presented in Fig. 5, where typically between 10 and 13 spikes per construct were tested in each experiment. For these experiments the gene Fg08493 (FGRAMPH1_01G10097, UniProtKB ID I1RW39) which is predicted to encode a small secreted protein (54 amino acids), containing two cysteine residues that is upregulated during the symptomless phase of F. graminearum infection (Dilks et al., 2019) was chosen as an additional control. The Fg08493 treatment did not induce more or less FHB disease and showed similar infection progress to the MCS virus control. Whereas transient overexpression of FgRALF resulted in a small but statistically significant increase in the number of visibly diseased spikelets below the F. graminearum inoculation points in wheat spikes compared to BSMV:MCS4D (p < 0.05) (Fig. 5).
Fig. 5.
BSMV-VOX of FgRALF in wheat. Graph representing number of visibly diseased spikelets by F. graminearum below the inoculation points in wheat spikes. A minimum of 10 spikes per virus treatment in each experiment were analysed. Data shown were collected at 12 days’ post F. graminearum-inoculation. The green star denotes the treatment where statistically significant differences in number of diseased spikelets, relative to BSMV:MCS4D control (blue bar), were observed (p < 0.05 from GLMM analysis). This graph represents a total of four combined experiment for all treatments except BSMV:Fg08493, which was included in one experiment. “No Virus” represents wheat plants with no virus inoculation prior to F. graminearum infection. “BSMV:MCS4D” represents control virus treatment where the only addition to the viral genome is a multiple cloning site (MCS). “BSMV:FgRALF” represents BSMV expressing the FgRALF gene. “BSMV:Fg08493” represents BSMV expressing the Fg08493 gene.
3.5. Effect of FgRALF overexpression on Arabidopsis F. graminearum susceptibility
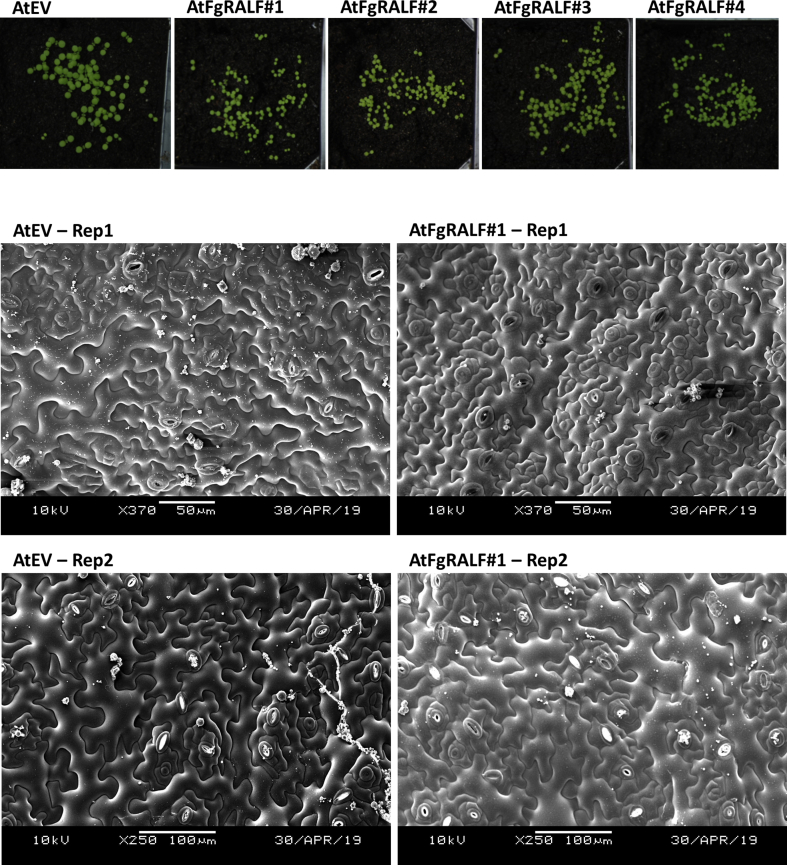
To test whether expression of FgRALF in Arabidopsis prior to F. graminearum inoculation would alter the interaction outcome, we generated via stable transformation and then selected through molecular analyses the required Arabidopsis lines in subsequent generations. In total, four homozygous T3 Arabidopsis transgenic lines (ecotype Columbia erecta) expressed FgRALF under the control of the constitutive CaMV 35S promoter (hereafter called AtFgRALF) were produced. We observed that AtFgRALF seedlings were initially smaller than EV controls up to the 2-week old stage (Fig. S2). However, after this age, no visible morphological differences were observed among control and overexpressing lines. Similar results were observed by Matos et al. (2008) when stably overexpressed AtRALF1 in Arabidopsis. Scanning electron microscopy analyses revealed no differences in number of stomata, open stomata and epidermal cells in the young, smaller AtFgRALF seedlings compared to EV seedlings (Fig. S2).
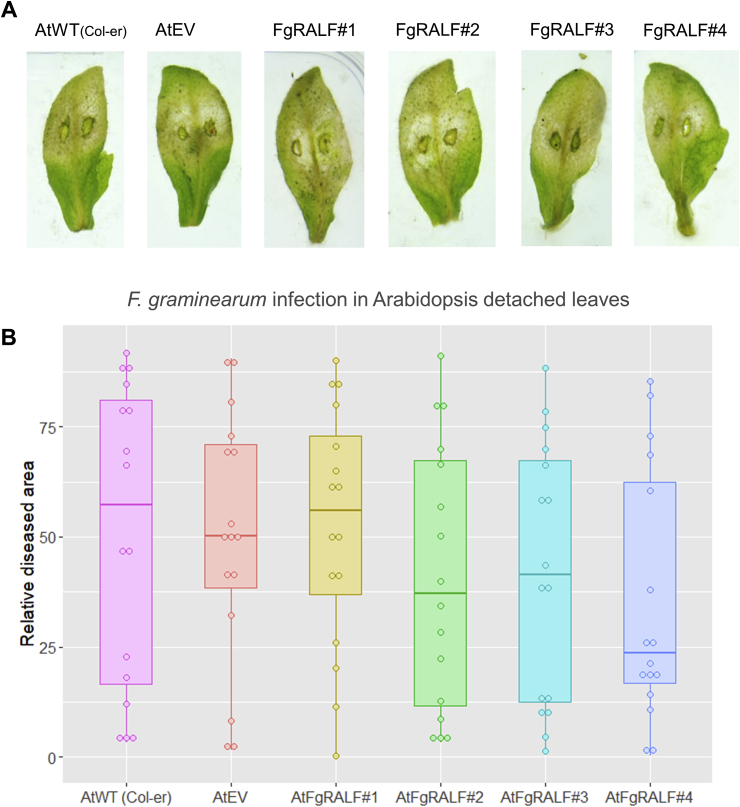
The effect of FgRALF over-expression on susceptibility to F. graminearum was tested in 4-week old Arabidopsis plants using detached leaf inoculation (Chen et al., 2006). We decided to use F. graminearum leaf inoculation instead of floral inoculation for this assay because the latter usually leads to very strong infection in wild-type Landsberg erecta plants (Urban et al., 2002). Therefore, it would be more difficult to assess disease enhancement. F. graminearum spores were placed onto Arabidopsis detached leaves supplemented with 20μM DON. The levels of disease on the AtFgRALF overexpressing lines were similar to that observed on the control plants (p > 0.05) (Fig. 6A–B).
Fig. 6.
Infection symptoms on Arabidopsis leaves following inoculation with F. graminearum. A) The appearance of representative detached leaves 6 days after spore droplet inoculation. B) Box-plot and dot-plot of 16 infected leaves from 8 different plants of each transgenic line at 7dpi; typical infection symptoms were recorded as the percentage (from 0 to 100) of necrotic area in each leaf. There were no visible differences between infection symptoms on FgRALF-expressing leaves compared with the wild-type (p > 0.05). AtWT (Col-er)- wild-type (Col-er), AtEV- Col-er harbouring an empty vector (EV), AtRALF#1 to #4 are four independent Col-er expressing FgRALF.
3.6. The wheat genome encodes two predicted paralogous loci coding for the receptor like kinase Feronia
The identification of 36 putative RALF proteins in the predicted wheat proteome, prompted us to explore the wheat genome for sequences with a high level of similarity to AtRALF1 receptor Feronia (AtFER). In Arabidopsis, AtFER forms a distinct clade with 16 closely related proteins within the superfamily of receptor-like kinases (RLKs), termed “The Feronia family”. All members of the Feronia family are distinguished from other RLKs by having an extracellular malectin-like (PF12819) domain (Li et al., 2016).
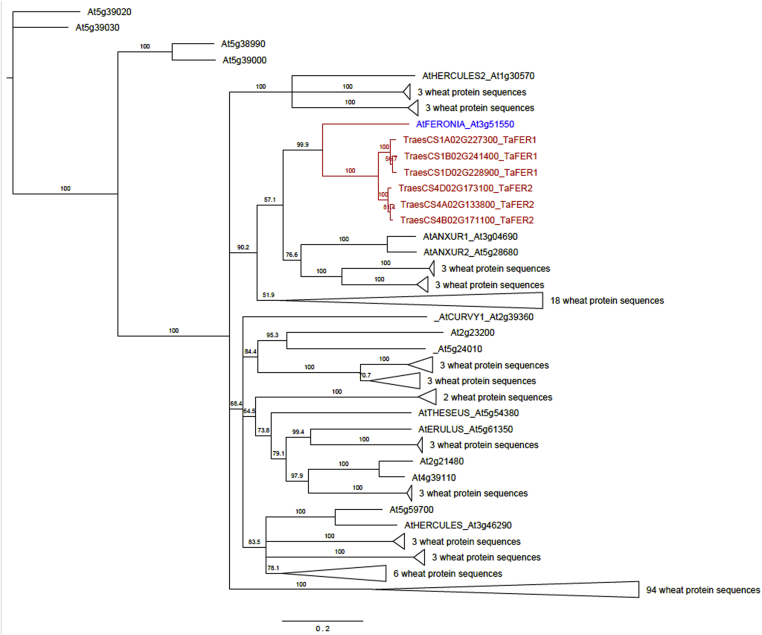
The BioMart tool in Ensembl Plants (Bolser et al., 2016) was used to find wheat proteins containing both the kinase-like domain (PF07714) and the malectin-like domain (PF12819). A total of 156 sequences encoding Feronia-like proteins were identified in the reference wheat (Triticum aestivum) genome (variety Chinese Spring). Protein alignments identified wheat sequences corresponding to all 17 members of the Arabidopsis Feronia family. A phylogenetic analysis was then used to identify the wheat protein sequences most closely related to AtFER (Fig. 7 and Fig. S3).
Fig. 7.
Neighbour-joining (NJ) tree based on the wheat (T. aestivum) and Arabidopsis protein sequences containing both predicted kinase-like (PF07714) and malectin-like (PF12819) domains. The numbers indicate the NJ bootstrap values for 1000 replicates. Due to the large number of protein sequences the tree was collapsed. The full expanded tree is represented in Fig. S3. The protein name in blue indicates the Arabidopsis Feronia (AtFER). The names in red indicate the wheat sequences closely related to AtFER (TaFER1 and TaFER2). The names in black are the other members of Arabidopsis and wheat Feronia families.
Two wheat genes, consisting of three homoeologues each, named TaFER1 and TaFER2 were selected as the AtFER orthologues (Table 1, Fig. 7). TaFER1 is located on chromosome 1 and TaFER2 is located on chromosome 4. For both loci, the A, B and D genomes homoeologues were identified and these exhibited a high level of sequence conservation (80.8–81.5% sequence identify) (Table 1).
Table 1.
Gene ID and similarity to Feronia from Arabidopsis of putative Feronia protein sequences in wheat.
| Gene name | Gene IDa | Chrom. Locationb | % ID with AtFERc |
|---|---|---|---|
| TaFER1 | TraesCS1D01G228900 | 1D | 80.8 |
| TaFER1 | TraesCS1B01G241400 | 1B | 80.9 |
| TaFER1 | TraesCS1A01G227300 | 1A | 81 |
| TaFER2 | TraesCS4D01G173100 | 4D | 81.3 |
| TaFER2 | TraesCS4B01G171100 | 4B | 81 |
| TaFER2 | TraesCS4A01G133800 | 4A | 81.5 |
Gene ID according IWGSC v1.0 (International Wheat Genome Sequencing Consortium).
Chromosome locations according IWGSC.
Percentage of identity with Arabidopsis thaliana Feronia protein sequence.
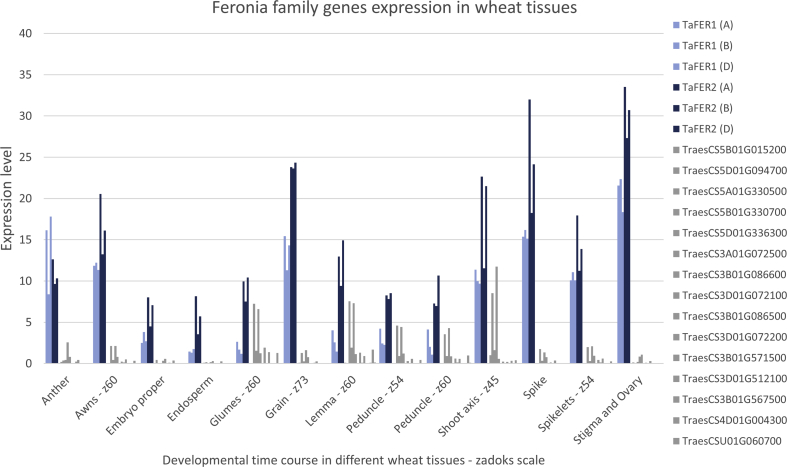
Analysis of the wheat gene expression data available within the Wheat eFP Browser (Ramirez-Gonzalez et al., 2018, Winter et al., 2007) demonstrated that all homoeologues of TaFER1 and TaFER2 had the highest expression levels present in different floral tissues as well as during the early and late stages of grain development (Fig. S4). Whereas the expression of other wheat genes predicted to be part of the Feronia family were either absent or at very low levels in these tissue types (Fig. S4). The identification of two putative FER genes in wheat both with high expression identified in floral tissues throughout development suggests these homoeologues may have one or more so far uncharacterised functions in wheat floral tissue.
3.7. Barley Stripe Mosaic virus – Virus inducted gene silencing (BSMV- VIGS) of Feronia genes in wheat
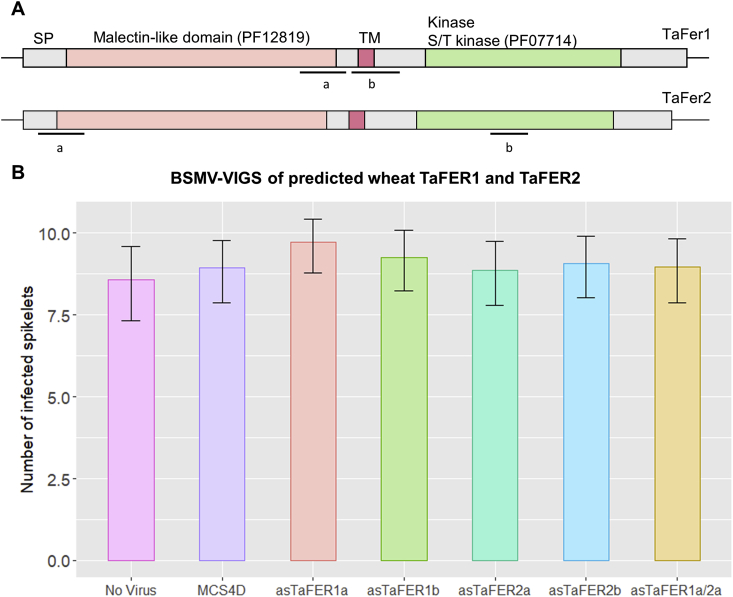
In a BSMV-VIGS experiment, a short fragment of a transcribed sequence of a plant gene is inversely inserted into a cloned virus genome and the recombinant virus is then inoculated onto test plants, triggering Post-Transcriptional Gene Silencing (PTGS) (Lee et al., 2012). Five BSMV-VIGS constructs were generated: two constructs each for TaFER1 and TaFER2 which target different regions (designated ‘a’ and ‘b’) in the transcripts. These constructs were named TaFER1a, TaFER1b, TaFER2a and TaFER2b (Fig. 8A), and the fifth construct contained concatenated ‘a’ fragments of TaFER1 and TaFER2 that target both sets of gene transcripts simultaneously in the same plant, named TaFER1a/2a (Table S3). Off-target silencing was not predicted to arise from any of the gene fragments (Luck et al., 2019). BSMV:MCS4D was used as a virus control. Fifteen days after virus inoculation, wheat spikes at anthesis were point inoculated with F. graminearum spores or sterile distilled water. Although Feronia is suggested to be involved in plant development, no visible effect on plant development were observed after silencing with any of the TaFER constructs.
Fig. 8.
BSMV-VIGS of Feronia genes in wheat. A) Diagrammatical representation of wheat TaFer1 and TaFer2 protein sequence showing both predicted kinase-like (PF07714) and malectin-like (PF12819) protein domains. Bars extending from the termini of the predicted protein structures indicate nontranslated cDNA regions up- and downstream of the coding regions. Bars below each gene model indicate the a and b regions amplified to generate different Barley stripe mosaic virus-mediated virus-induced gene silencing (BSMV-VIGS) constructs targeting these genes. SP = signal peptide, TM = transmembrane region. B) Graph representing number of visibly diseased spikelets below the F. graminearum inoculated points in wheat spikes at 15 dpi. “No Virus” represents wheat plants with no virus inoculation prior F. graminearum infection. “MCS4D″ represents control virus treatment where the only addition to the viral genome is a multiple cloning site (MCS). The BSMV-VIGS silencing constructs include asTaFER1a, asTaFER1b, asTaFER1a/2a, asTaFER2a and asTaFER2b. This graph represents a total of three combined experiment. Treatments did not present statistically significant differences in number of diseased spikelets, relative to BSMV:MCS4D control (p > 0.05 from GLMM analysis).
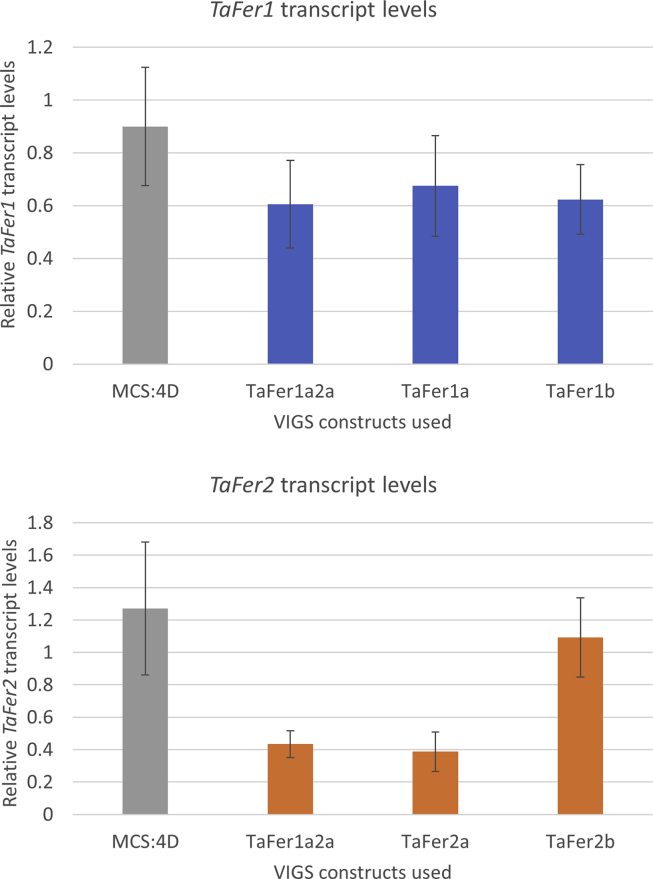
Efficient knock-down of TaFer1 transcript levels was achieved with both of the BSMV-VIGS constructs designed (TaFer1a and TaFer1b), with a reduction in TaFer1 transcript levels of around 30% in wheat spikes of plants subjected to BSMV-VIGS when sampled at 3 dpi (Fig. S5). Only one of the two BSMV-VIGS constructs designed to target TaFer2 was effective. Construct ‘a’ induced around 65% of wheat TaFer2 transcript levels in wheat spikes of plants subjected to BSMV-VIGS asTaFer2a when sampled at 3 dpi (Fig. S5). The silencing efficiency for the double construct was similar to the best construct targeting a single gene.
A generalised linear model (GLM) statistical analysis revealed no significant difference in F. graminearum infections between any of the asFER constructs and the MCS4D virus control treatment (Fig. 8B and C).
4. Discussion
The aim of this study was to explore whether a homologue of plant RALF protein identified in the highly problematic cereal infecting ascomycete fungus F. graminearum (group III) contributes to fungal virulence. This was done using four complementary approaches which assessed RALF protein function in the pathogen, in a non-cereal and a cereal plant host species. Previously only a RALF gene from F. oxysporum f.sp. lycopersici (group I) had been assessed for a role in fungal virulence (Masachis et al., 2016, Thynne et al., 2017).
We first tested the effect of FgRALF gene deletion on F. graminearum virulence. Pathogenicity tests with two independently generated FgRALF gene deletion mutants were found not to alter the Fusarium disease phenotype when compared to F. graminearum wild-type strain on the floral tissue of both wheat and Arabidopsis. However, the use of reverse genetics to assess gene function in fungi can often be complicated by genetic redundancy (Aguileta et al., 2012). In addition, by comparing the two data sets available on RALF protein analyses in F. oxysporum f. sp. lycopersici, the effect of f-ralf gene deletion mutants in fungal virulence appears to be influenced by other components, for example plant growth conditions, plant fitness and methods of disease assessment (Masachis et al., 2016, Thynne et al., 2017).
When FgRALF was transiently over-expressed in N. benthamiana and N. tabacum no obvious host responses were induced. Therefore, recognition of FgRALF by a non-natural host species appears unlikely. A previous study has shown that Nicotiana is a non-natural host species for F. graminearum (Urban et al., 2002). It is plausible that FgRALF could function as a suppressor of plant defences, but this was not formally tested in this study.
The enhanced levels of Fusarium infection obtained from the BSMV-VOX experiment are interesting. Heterologous proteins are generally expressed at low to moderate levels from the BSMV vector (Bouton et al., 2018). Possibly the predominant vascular colonisation by the virus preferentially targets the FgRALF and therefore any induced physiological and cellular changes may occur at vascular locations. Although not an exclusively vascular colonising species, a proportion of the F. graminearum hyphae are vascular associated during all phases of the colonisation (Brown et al., 2012, Dilks et al., 2019). Alternatively, the FgRALF could be operating in combination with the known suppressors produced by the BSMV to provide a beneficial effect on fungal colonisation. In BSMV, the γb protein is suggested to have a role in suppressing plant defence mechanisms (Jackson et al., 2009). The results presented in Fig. 5 indicate that Fusarium infections are considerably enhanced in a BSMV-VOX experiment, because the virus control (BSMV:MCS4D) treatment was more susceptible to FHB than the no virus treated control plants. Further studies involving in planta transcriptome analyses might be able to resolve whether there are either major or more subtle spatial and temporal differences in gene expression during the F. graminearum – wheat floral interaction in the presence and absence of BSMV colonisation and FgRALF expression. In contrast, overexpression of FgRALF in stably transformed Arabidopsis plants did not achieve the same outcome, possibly because the colonisation pattern of F. graminearum in Arabidopsis plants is different to that occurring in its natural host wheat. Additionally, if there is a virus effect contributing to FgRALF activity, this component is absent in stably transformed plants.
While some aspects of the role of F-RALF in F. oxysporum f.sp. lycopersici – host interaction remain undetermined, both studies suggest that F-RALF-triggered plant responses are mediated by the plant’s receptor-like kinase (RLK) Feronia (FER) (Masachis et al., 2016, Thynne et al., 2017). To explore the host contribution, we identified genes belonging to the Feronia family in hexaploid wheat. A total of 156 sequences predicted to contain both malectin-like and kinase-like domains were identified. At two wheat loci, three homoeologues closest related to AtFER were present (Fig. 7). These homoeologous gene sets were also found to be highly expressed in wheat spikes.
By transiently silencing the two most highly expressed wheat FER genes, namely TaFER1 and TaFER2, either individually or simultaneously, no evidence for a role for FER receptor in the F. graminearum - wheat floral interaction was obtained. We hypothesise that this could be due mainly three reasons: TaFER is not a receptor of FgRALF and therefore does not play a role in F. graminearum infection on wheat. TaFer is a receptor of FgRALF, but this recognition does not play a role in F. graminearum wheat interaction. TaFer is a receptor of FgRALF with a role in pathogenicity, but other TaFER homologues in wheat may function also as a receptor for FgRALF when other TaFERs are reduced in expression. A survey of the sequenced wheat genome revealed that there were potentially 36 genes that code for RALF proteins.
Group III related fungal RALF sequences have not previously been tested for function. The earlier phylogenetic studies suggested that fungal RALF genes were acquired through horizontal gene transfer (HGT) (Thynne et al., 2017). The additional analyses done in the study using recently fully assembled and annotated fungal genomes, reinforce this view. The very closely related non-pathogenic species Fusarium venenatum lacks this sequence whilst other closely related phytopathogenic Fusarium species, such as F. pseudograminearum, contains a ralf homologue. Sequence analysis of a few ralf homologues identified in the genomes of non-phytopathogen fungal species demonstrated that non-functional proteins are encoded (Thynne et al., 2017). Interestingly, more distally related plant pathogenic species for example Leptosphaeria maculans which infects a range of Brassicas also contain a predicted ralf homologue. RALF homologues have been found mostly in plant pathogens and have a sporadic distribution in the fungal kingdom, probably suggesting acquisition of this gene through HGT (Thynne et al., 2017). The phylogenetics analyses have also revealed that amongst the fungal genera harbouring RALF homologues, the genus Fusarium has the most diverse array of RALF homologues. Therefore, this gene could still have a role during Fusarium colonisation in the plant, but the latest results show its function is not still clear (Masachis et al., 2016, Thynne et al., 2017).
In this study using various highly complementary experimental techniques and inoculation methods, FgRALF has been shown not be required for fungal virulence, however, based on our VOX results, this gene may still be important during FHB disease formation in wheat.
Declaration of competing interest
The authors declare no competing financial interest.
Acknowledgements
WSL, MU and KHK received UK Biotechnology and Biological Sciences Research Council (BBSRC) grant-aided support as part of the Institute Strategic Programmes 20:20 Wheat (BB/J00426X/) and Designing Future Wheat (BB/P016855/1). The CAPES Foundation of Brazil is thanked for AKMW’s PhD scholarship (BEX 1266-13-6). CW was supported by a BBSRC University of Nottingham Doctoral Training Partnership (DTP). We thank Dr Michael Hammond-Kosack for helping with Feronia homoeologues identification, Dr Tom Ashfield for providing access to the LemnaGrid software module and Prof George Lomonossoff for supplying the pEAQ vector.
Corresponding Editor: Prof. G.M. Gadd
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.funbio.2020.05.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
References
- Aguileta G., Lengelle J., Chiapello H., Giraud T., Viaud M., Fournier E., Rodolphe F., Marthey S., Ducasse A., Gendrault A., Poulain J., Wincker P., Gout L. Genes under positive selection in a model plant pathogenic fungus, Botrytis. Infect. Genet. Evol. 2012;12:987–996. doi: 10.1016/j.meegid.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Appels R., Eversole K., Feuillet C., Keller B., Rogers J., Stein N., Pozniak C.J., Choulet F., Distelfeld A., Poland J. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361 doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- Aslanidis C., Dejong P.J. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhouse D. Global distribution of Fusarium graminearum, F. asiaticum and F. boothii from wheat in relation to climate. Eur. J. Plant Pathol. 2014;139:161–173. [Google Scholar]
- Bolser D., Staines D.M., Pritchard E., Kersey P. Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 2016;1374:115–140. doi: 10.1007/978-1-4939-3167-5_6. [DOI] [PubMed] [Google Scholar]
- Bouton C., King R.C., Chen H., Azhakanandam K., Bieri S., Hammond-Kosack K.E., Kanyuka K. Foxtail mosaic virus: a viral vector for protein expression in cereals. Plant Physiol. 2018;177:1352–1367. doi: 10.1104/pp.17.01679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.A., Antoniw J., Hammond-Kosack K.E. The predicted secretome of the plant pathogenic fungus Fusarium graminearum: a refined comparative analysis. PloS One. 2012;7 doi: 10.1371/journal.pone.0033731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.A., Urban M., Van De Meene A.M.L., Hammond-Kosack K.E. The infection biology of Fusarium graminearum: defining the pathways of spikelet to spikelet colonisation in wheat ears. Fungal Biol. 2010;114:555–571. doi: 10.1016/j.funbio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Campos W.F., Dressano K., Ceciliato P.H.O., Guerrero-Abad J.C., Silva A.L., Fiori C.S., Morato do Canto A., Bergonci T., Claus L.A.N., Silva-Filho M.C., Moura D.S. Arabidopsis thaliana rapid alkalinization factor 1-mediated root growth inhibition is dependent on calmodulin-like protein 38. J. Biol. Chem. 2018;293:2159–2171. doi: 10.1074/jbc.M117.808881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.W., Steed A., Harden C., Nicholson P. Characterization of Arabidopsis thaliana-Fusarium graminearum interactions and identification of variation in resistance among ecotypes. Mol. Plant Pathol. 2006;7:391–403. doi: 10.1111/j.1364-3703.2006.00349.x. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S., Cummins C., Davidson C., Dodiya K.J., Gall A., Giron C.G., Gil L., Grego T., Haggerty L., Haskell E., Hourlier T., Izuogu O.G., Janacek S.H., Juettemann T., Kay M., Laird M.R., Lavidas I., Liu Z., Loveland J.E., Marugan J.C., Maurel T., McMahon A.C., Moore B., Morales J., Mudge J.M., Nuhn M., Ogeh D., Parker A., Parton A., Patricio M., Abdul Salam A.I., Schmitt B.M., Schuilenburg H., Sheppard D., Sparrow H., Stapleton E., Szuba M., Taylor K., Threadgold G., Thormann A., Vullo A., Walts B., Winterbottom A., Zadissa A., Chakiachvili M., Frankish A., Hunt S.E., Kostadima M., Langridge N., Martin F.J., Muffato M., Perry E., Ruffier M., Staines D.M., Trevanion S.J., Aken B.L., Yates A.D., Zerbino D.R., Flicek P. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilks T., Halsey K., De Vos R.P., Hammond-Kosack K.E., Brown N.A. Non-canonical fungal G-protein coupled receptors promote Fusarium head blight on wheat. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Canto A.M., Ceciliato P.H.O., Ribeiro B., Morea F.A.O., Garcia A.A.F., Silva-Filho M.C., Moura D.S. Biological activity of nine recombinant AtRALF peptides: implications for their perception and function in Arabidopsis. Plant Physiol. Biochem. 2014;75:45–54. doi: 10.1016/j.plaphy.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Gleave A.P. A versatile binary vector system with a T-DNA organizational-structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Haruta M., Sabat G., Stecker K., Minkoff B.B., Sussman M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science. 2014;343:408–411. doi: 10.1126/science.1244454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.O., Lim H.S., Bragg J., Ganesan U., Lee M.Y. Hordeivirus replication, movement, and pathogenesis. Annu. Rev. Phytopathol. 2009;47:385–422. doi: 10.1146/annurev-phyto-080508-081733. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Vance R.E., Dangl J.L. Intracellular innate immune surveillance devices in plants and animals. Science. 2016;354 doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettles G.J., Bayon C., Canning G., Rudd J.J., Kanyuka K. Apoplastic recognition of multiple candidate effectors from the wheat pathogen Zymoseptoria tritici in the nonhost plant Nicotiana benthamiana. New Phytol. 2017;213:338–350. doi: 10.1111/nph.14215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R., Brown N.A., Urban M., Hammond-Kosack K.E. Inter-genome comparison of the Quorn fungus Fusarium venenatum and the closely related plant infecting pathogen Fusarium graminearum. BMC Genom. 2018;19:269. doi: 10.1186/s12864-018-4612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R., Urban M., Hammond-Kosack M.C.U., Hassani-Pak K., Hammond-Kosack K.E. The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum. BMC Genom. 2015;16:544. doi: 10.1186/s12864-015-1756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Hammond-Kosack K.E., Kanyuka K. Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 2012;160:582–590. doi: 10.1104/pp.112.203489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.S., Rudd J.J., Hammond-Kosack K.E., Kanyuka K. Mycosphaerella graminicola LysM effector-mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol. Plant Microbe Interact. 2014;27:236–243. doi: 10.1094/MPMI-07-13-0201-R. [DOI] [PubMed] [Google Scholar]
- Leslie J.F., Summerell B.A. John Wiley & Sons; 2008. The Fusarium Laboratory Manual. [Google Scholar]
- Li C., Wu H.M., Cheung A.Y. FERONIA and her pals: functions and mechanisms. Plant Physiol. 2016;171:2379–2392. doi: 10.1104/pp.16.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck S., Kreszies T., Strickert M., Schweizer P., Kuhlmann M., Douchkov D. siRNA-finder (si-Fi) software for RNAi-target design and off-target prediction. Front. Plant Sci. 2019;10 doi: 10.3389/fpls.2019.01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S., Segorbe D., Turra D., Leon-Ruiz M., Furst U., El Ghalid M., Leonard G., Lopez-Berges M.S., Richards T.A., Felix G., Di Pietro A. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.43. [DOI] [PubMed] [Google Scholar]
- Matos J.L., Fiori C.S., Silva-Filho M.C., Moura D.S. A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett. 2008;582:3343–3347. doi: 10.1016/j.febslet.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Motteram J., Kufner I., Deller S., Brunner F., Hammond-Kosack K.E., Nurnberger T., Rudd J.J. Molecular characterization and functional analysis of MgNLP, the sole NPP1 domain-containing protein, from the fungal wheat leaf pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 2009;22:790–799. doi: 10.1094/MPMI-22-7-0790. [DOI] [PubMed] [Google Scholar]
- Murphy E., De Smet I. Understanding the RALF family: a tale of many species. Trends Plant Sci. 2014;19:664–671. doi: 10.1016/j.tplants.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Pearce G., Yamaguchi Y., Munske G., Ryan C.A. Structure-activity studies of RALF, Rapid Alkalinization Factor, reveal an essential - YISY - motif. Peptides. 2010;31:1973–1977. doi: 10.1016/j.peptides.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Rafiqi M., Ellis J.G., Ludowici V.A., Hardham A.R., Dodds P.N. Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr. Opin. Plant Biol. 2012;15:477–482. doi: 10.1016/j.pbi.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Ramirez-Gonzalez R.H., Borrill P., Lang D., Harrington S.A., Brinton J., Venturini L., Davey M., Jacobs J., van Ex F., Pasha A., Khedikar Y., Robinson S.J., Cory A.T., Florio T., Concia L., Juery C., Schoonbeek H., Steuernagel B., Xiang D., Ridout C.J., Chalhoub B., Mayer K.F.X., Benhamed M., Latrasse D., Bendahmane A., International Wheat Genome Sequencing C., Wulff B.B.H., Appels R., Tiwari V., Datla R., Choulet F., Pozniak C.J., Provart N.J., Sharpe A.G., Paux E., Spannagl M., Brautigam A., Uauy C. The transcriptional landscape of polyploid wheat. Science. 2018;361 doi: 10.1126/science.aar6089. [DOI] [PubMed] [Google Scholar]
- Sainsbury F., Thuenemann E.C., Lomonossoff G.P. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 2009;7:682–693. doi: 10.1111/j.1467-7652.2009.00434.x. [DOI] [PubMed] [Google Scholar]
- Sandkvist M., Bagdasarian M., Howard S.P., Dirita V.J. Interaction between the autokinase epse and epsl in the cytoplasmic membrane is required for extracellular secretion in vibrio-cholerae. EMBO J. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schandry N. A practical guide to visualization and statistical analysis of R. solanacearum infection data using R. Front. Plant Sci. 2017;8:623. doi: 10.3389/fpls.2017.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Hussain A., Mun B.G., Imran Q.M., Falak N., Lee S.U., Kim J.Y., Hong J.K., Loake G.J., Ali A., Yun B.W. Comprehensive analysis of plant rapid alkalization factor (RALF) genes. Plant Physiol. Biochem. 2016;106:82–90. doi: 10.1016/j.plaphy.2016.03.037. [DOI] [PubMed] [Google Scholar]
- Srivastava R., Liu J.X., Guo H.Q., Yin Y.H., Howell S.H. Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 2009;59:930–939. doi: 10.1111/j.1365-313X.2009.03926.x. [DOI] [PubMed] [Google Scholar]
- Stegmann M., Monaghan J., Smakowska-Luzan E., Rovenich H., Lehner A., Holton N., Belkhadir Y., Zipfel C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science. 2017;355:287–289. doi: 10.1126/science.aal2541. [DOI] [PubMed] [Google Scholar]
- Thynne E., Saur I.M.L., Simbaqueba J., Ogilvie H.A., Gonzalez-Cendales Y., Mead O., Taranto A., Catanzariti A.M., McDonald M.C., Schwessinger B., Jones D.A., Rathjen J.P., Solomon P.S. Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol. Plant Pathol. 2017;18:811–824. doi: 10.1111/mpp.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban M., Daniels S., Mott E., Hammond-Kosack K. Arabidopsis is susceptible to the cereal ear blight fungal pathogens Fusarium graminearum and Fusarium culmorum. Plant J. 2002;32:961–973. doi: 10.1046/j.1365-313x.2002.01480.x. [DOI] [PubMed] [Google Scholar]
- Urban M., Mott E., Farley T., Hammond-Kosack K. The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 2003;4:347–359. doi: 10.1046/j.1364-3703.2003.00183.x. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PloS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.H., Hamari Z., Han K.H., Seo J.A., Reyes-Dominguez Y., Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Yuan C., Li C., Yan L.J., Jackson A.O., Liu Z.Y., Han C.G., Yu J.L., Li D.W. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PloS One. 2011;6 doi: 10.1371/journal.pone.0026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.