Abstract
The complicated interplay of plant–pathogen interactions occurs on multiple levels as pathogens evolve to constantly evade the immune responses of their hosts. Many economically important crops fall victim to filamentous pathogens that produce small proteins called effectors to manipulate the host and aid infection/colonization. Understanding the effector repertoires of pathogens is facilitating an increased understanding of the molecular mechanisms underlying virulence as well as guiding the development of disease control strategies. The purpose of this review is to give a chronological perspective on the evolution of the methodologies used in effector discovery from physical isolation and in silico predictions, to functional characterization of the effectors of filamentous plant pathogens and identification of their host targets.
Keywords: bioinformatic effector predictions, effector host–target interactions, effectors, fungal phytopathogens, in planta methodologies, oomycete phytopathogens
The chronological progression of methodologies used in effector discovery; from physical isolation and in silico predictions, to functional characterization and host target identification.

1. INTRODUCTION
If people think Nature is their friend, then they sure don't need an enemy.
Kurt Vonnegut, Letter in Time magazine
1.1. The threats from filamentous phytopathogens
Our expanding global population forces us to intensify our crop production as we prepare to feed 2.2 billion more people by 2050. One of the main biotic challenges facing society to meeting these ever‐growing demands are filamentous plant pathogens. Oomycetes and fungi are the causal agents of some of the most notorious plant diseases and are a true threat to our global food security and community structures. Plant disease outbreaks have occurred throughout human history, some of the most infamous include the Irish potato famine caused by the oomycete Phytophthora infestans (Turner, 2005), Panama disease caused by Fusarium oxysporum f. sp. cubense (Gordon, 2017), and wheat stem rust caused by Puccinia graminis f. sp. tritici (Roelfs, 1985; Singh et al., 2011).
1.2. Effectors and the plant immune response
The elegantly described “zig‐zag” model by Jones and Dangl (2006) reveals a two‐tier immune response where pathogen‐associated molecular patterns (PAMPs) are first detected on host cell surfaces by pattern recognition receptors (PRRs), inducing pattern‐triggered immunity (PTI). To evade this response, pathogens secrete effector proteins that manipulate the host and aid colonization, yet in hosts that have the corresponding resistance (R) genes (Flor, 1971), these effectors are detected by receptors such as the intracellular Nod‐like receptors (NLRs) that induce effector‐triggered immunty (ETI), resulting in a hypersenstive response (HR) and programmed cell death (de Wit, 2016, Zhang et al., 2017).
Just as with all models, the story is more complicated and not all features of the plant–microbe interactions are accommodated. Effectors can be highly conserved, thus not under selective pressure to evade host detection, such as the members of the oomycete Crinkler (CRN) effector family or the core fungal effector NIS1 (Depotter and Doehlemann, 2019; Irieda et al., 2019) whilst other effectors are detected extracellularly (van der Burgh and Joosten, 2019).
Recent studies suggest that, rather than a two‐tier system of immunity, ETI and PTI activate different but interacting pathways leading to plant immunity. The activation of the paired Arabidopsis NLRs RRS1‐R and RPS4 by the bacterial effector AvrRps4 cannot induce HR without the presence of PAMPs (Ngou et al., 2020). Both co‐ and predelivery of AvrRps4 with PAMPs leads to an increased and prolonged expression of PTI‐associated defence genes such as BIK1, BAK1, and Rboh; the expression of these genes is not induced by effectors alone (Ngou et al., 2020). Similarly, ETI responses in Arabidopsis mutants lacking PRRs are greatly compromised, with the ETI‐induced reactive oxygen species (ROS) production being mediated by PRRs (Yuan et al., 2020). This suggests that PTI is a required component of ETI with mutual potentiation of immune mechanisms triggered by intracellular and cell‐surface receptors.
1.3. The importance of effector research
Hundreds of small proteins, predicted to be effectors, are secreted by filamentous phytopathogens during host colonization (Dean et al., 2005; Kämper et al., 2006; Yoshida et al., 2009; Duplessis et al., 2011). We have little understanding of the function of most of these putative effectors and each typically shares minimal or no sequence homology to proteins with previously defined functions. However, the effector repertoire of a pathogen is a major determinant of host specialization and can greatly impact whether the plant–pathogen interaction is successful or not based on the genotype of the host (Raffaele et al., 2010; Sánchez‐Vallet et al., 2018a)
Molecular studies have characterized over 60 fungal effectors across multiple species; however, this barely makes a dent in the candidate effector repertoire for each pathogenic species (Sperschneider et al., 2015). For example, the barley powdery mildew fungus Blumeria graminis f. sp. hordei alone is suspected to have roughly 7% of its genome encoding candidate secreted effector proteins (CSEPs) (Pedersen et al., 2012).
Identifying and characterizing the function of effector proteins will improve our understanding of their role in disease formation and influence our future strategies to combat pathogen infections. Fundamental effector research is a key part of devising new plant disease control strategies and this is detailed further in Sections 3.2 and 6 of this review. Effectors play an important role in crop breeding where, as well as being used to detect resistance genes in new cultivars, characterized effectors can be used to locate susceptibility loci in vulnerable crops (Vleeshouwers and Oliver, 2014). The development of mobile sequencing technology means that genes encoding effectors can also be used to detect the emergence of new strains of crop pathogens in the field and elude the severity of future disease outbreaks (Radhakrishnan et al., 2019). Effectors function in multiple ways, including inhibiting host enzymes, modulating plant immune responses, and targeting host gene‐silencing mechanisms. All features of effectors described in this article are summarized in Table 1, including their mode of action where known.
TABLE 1.
List of filamentous phytopathogen species and their effectors referred to in this review
| Effector | Size aa a | Uniprot ID | Biological function | Species | Disease name | Host | Reference |
|---|---|---|---|---|---|---|---|
| ATR13 | 187 | M4C367 | Secreted effector that acts as an elicitor of the HR specifically on plants carrying the defence protein RPP13 | Hyaloperonospora parasitica | Arabidopsis downy mildew | Arabidopsis thaliana | Sohn et al. (2007) |
| ATR1NDWsB | 311 | M4B6G6 | Secreted effector that acts as an elicitor of the HR specifically on plants carrying the cognate R defence protein RPP13 | H. parasitica | Arabidopsis downy mildew | A. thaliana | Sohn et al. (2007) |
| Avr1b‐1 | 204 | G5A9E5 | Uncharacterized | Phytophthora sojae | Stem and root rot of soybean | Soybean (Glycine max) | Shan et al. (2004); Dou et al. (2008) |
| Has been shown to reduce heterologously induced plant cell death | |||||||
| Avr1‐C039 | 89 | NA | Uncharacterized protein that is recognized in the host by direct binding of the NB‐LRR proteins RGA5, which together with RGA4 induces ETI | Magnaporthe oryzae | Rice blast | Rice (Oryza sativa) | Farman and Leong (1998) ; Cesari et al. (2013); Ribot et al. (2013) |
| Avr2 | 78 | Q8NID8 | Inhibits several apoplastic Cys proteases, including the tomato protease Rcr3, which is required for plant basal defence and induces HR in tomato races that carry the cognate R protein Cf‐2 | Cladosporium fulvum | Tomato leaf mould | Tomato (Solanum lycopersicum) | Rooney et al. (2005); van Esse et al. (2008); Song et al. (2009) |
| Avr3a | 147 | E2DWQ7 | Suppresses host BAK1/SERK3‐mediated immunity, by targeting and stabilizing host E3 ligase CMPG1 | Phytophthora infestans | Potato late blight | Potato (Solanum tuberosum) | Armstrong et al. (2005) |
| Avr4 | 135 | Q00363 | Chitin binding lectin, which inhibits plant chitinases to minimize chitin hydrolysis and also induces HR in tomato races that carry the cognate R protein Cf‐4 | C. fulvum | Tomato leaf mould | Tomato (S. lycopersicum) | Joosten et al. (1997); van den Burg et al. (2006) |
| Avr9 | 63 | P22287 | Induces necrosis by triggering HR in tomato containing the cognate R protein Cf‐9 | C. fulvum | Tomato leaf mould | Tomato (S. lycopersicum) | De Wit et al. (1985) |
| AVRa9 | 102 | N1J9M0 | Uncharacterized but is recognized by the intracellular MLA10 receptor in barley and results in HR | Blumeria graminis f. sp. hordei | Barley powdery mildew | Barley (Hordeum vulgare) | Saur et al. (2019a) |
| AvrL567‐A | 150 | Q6R661 | Triggers resistance responses in flax containing the cognate R proteins L5 and L6 | Melampsora lini | Flax rust | Flax (Linum usitatissimum) | Wang et al. (2007) |
| AvrL567‐D | 150 | Q1HBK6 | Triggers resistance responses in flax containing the cognate R protein L6 | M. lini | Flax rust | Flax (L. usitatissimum) | Wang et al. (2007) |
| AvrLm1 | 205 | Q258K5 | Interacts with the host protein (MAP) kinase 9 (BnMPK9), causing self‐increased protein accumulation and enhanced phosphorylation, resulting in the induction of cell death | Leptosphaeria maculans | Blackleg | Oilseed rape (Brassica napus) | Soyer et al. (2014); Fouché et al. (2018); Ma et al. (2018b) |
| Avr‐Pik and Avr‐PikD | 113 | C4B8C1 | Induces HR in rice races containing the corresponding cognate R protein Pik | M. oryzae | Rice blast | Rice (O. sativa) | Li et al. (2019); Yoshida et al. (2009) |
| AvrPikD is a novel allele of Avr‐Pik | |||||||
| AvrSr35 | 578 | A0A2I6B3G6 | Uncharacterized but interacts with the Sr35 immune receptor | Puccina graminis f. sp. tritici | Wheat stem rust | Wheat (Triticum sp.) | Salcedo et al. (2017) |
| AvrStb6 | 86 | A0A2K9YW36 | Uncharacterized, induces HR in wheat cultivars containing the cognate R protein Stb6 | Zymoseptoria tritici | Septoria leaf blotch | Wheat (Triticum sp.) | Zhong et al. (2017) |
| BAS1 | 115 | G5EHI7 | Induces an early, basal defence response such as ROS production and callose deposition in susceptible rice | M. oryzae | Rice blast | Rice (O. sativa) | Yang et al. (2017) |
| BEC1011 and BEC1054 | 118 | N1JJX4 | Noncatalytic homologue of fungal RNase that competitively binds host RNA to inhibit the degradation of the ribosomal RNA by RIPs, preventing host cell death | B. graminis f. sp. hordei | Barley powdery mildew | Barley (H. vulgare) | Pennington et al. (2019) |
| N1JJ94 | |||||||
| Capsicein | 98 | P15571 |
Induces incompatible HR Elicits leaf necrosis and causes the accumulation of pathogenesis‐related proteins |
Phytophthora capsici | Stem and fruit rot | Capsicum (Capsicum annuum) | Ricci et al. (1989) |
| Cce1 | 129 | A0A0D1C5E3 | Uncharacterized but may inhibit early PTI response in planta | Ustilago maydis | Corn smut | Maize (Zea mays) | Seitner et al. (2018) |
| Cinnamomin | 98 | P15569 | Induces incompatible HR | Phytophthora cinnamomi | Phytophthora root rot | >4,000 species including cinnamon (Cinnamomum verum) | Huet and Pernollet (1989) |
| Elicits leaf necrosis and causes the accumulation of pathogenesis‐related proteins | |||||||
| Cmu1 | 290 | A0A0D1DWQ2 | Interferes with the activity of host cytosolic chorismate mutase and inhibits the biosynthesis of salicylic acid required for plant defence signalling | U. maydis | Corn smut | Maize (Z. mays) | Djamei et al. (2011) |
| Cryptogein | 118 | P15570 | Induces incompatible HR | Phytophthora cryptogea | Tomato foot rot | Tomato (S. lycopersicum) | Ricci et al. (1989) |
| Elicits leaf necrosis and causes the accumulation of pathogenesis‐related proteins | |||||||
| Ecp1 | 96 | Q00364 | Extracellular protein that triggers Cf‐Ecp1 mediated resistance | C. fulvum | Tomato leaf mould | Tomato (S. lycopersicum) | Laugé et al. (1997) |
| Ecp2 | 165 | Q00365 | Extracellular protein that triggers Cf‐Ecp2 mediated resistance | C. fulvum | Tomato leaf mould | Tomato (S. lycopersicum) | Laugé et al. (1997) |
| Ecp6 | 222 | A0A1P8YXP5 | Ecp6 contains LysM domains, which bind to the fungal cell wall chitin with ultra‐high affinity, preventing detection by the host PRRs | C. fulvum | Tomato leaf mould | Tomato (S. lycopersicum) | De Jonge et al. (2010); Sánchez‐Vallet et al. (2013) |
| EPIC1 and EPIC2 | 126 | A1L015 | Inhibits several apoplastic Cys proteases, including the tomato protease Rcr3, which is required for plant basal defence | P. infestans | Potato late blight | Potato (S. tuberosum) | Song et al., 2009); Tian et al. (2007) |
| 125 | A1L017 | ||||||
| INF1 | 118 | Q01905 | A PAMP elicitor of plant cell death that targets the receptor kinase BAK1 | P. infestans | Potato late blight | Potato (S. tuberosum) | Kamoun et al. (1997); Chaparro‐Garcia et al. (2011) |
| MLP124266 and MLP1124499 | 69 | NA | Uncharacterized | Melampsora larici‐populina | Poplar rust | Poplar (Populus sp.) | de Guillen et al. (2019) |
| 50 | |||||||
| MoCDIP4 | 294 | G4MVX4 | Induces cell death in rice protoplasts | M. oryzae | Rice blast | Rice (O. sativa) | Chen et al. (2012) |
| NIS1 | 162 | N4VG36 | Targets the immune kinases BAK1 and BIK1 and disrupts downstream PTI responses | Colletotrichum orbiculare | Cucumber anthracnose | Cucumber (Cucumis sativus) | Yoshino et al. (2012); Irieda et al. (2019) |
| Para1 | 118 | P41801 | Induces incompatible HR in plants from the Solanaceae and Brassicaceae families | Phytophthora parasitica | Potato buckeye rot | Potato (S. tuberosum) | Kamoun et al. (1993) |
| Elicits leaf necrosis and causes the accumulation of pathogenesis‐related proteins | |||||||
| Pep1 | 178 | G0X7E8 | Inhibition of the plant oxidative burst by directly interacting with the peroxidase POX12, which generates ROS as a PTI response | U. maydis | Corn smut | Maize (Z. mays) | Doehlemann et al. (2009) |
| PiAvr2 | 116 | A0A2D1N523 | An RxLR effector that induces HR when interacting with the host NB‐LRR protein R2 | P. infestans | Potato late blight | Potato (S. tuberosum) | Saunders et al. (2012a) |
| Pit2 | 120 | A0A0D1EAR7 | Modulates host immunity by acting as a substrate mimic for apoplastic maize PLCPs, including CP1A, CP1B, XCP2, and CP2 processing of Pit2 releases the embedded inhibitor peptide PID14, which in turn blocks PLCP activity | U. maydis | Corn smut | Maize (Z. mays) | Mueller et al. (2013) |
| PsIsc1 | 210 | NA | An isochorismatase that supresses the precursor to the plant salicylate metabolism pathway and the subsequent salicylate‐mediated defences in planta | P. sojae | Stem and root rot of soybean | Soybean (G. max) | Liu et al. (2014) |
| PSTha5a23 | 108 | NA | Uncharacterized but is involved in PTI suppression | P. graminis f. sp. tritici | Wheat stem rust | Wheat (Triticum sp.) | Cheng et al. (2017) |
| PWL2 | 145 | A0A3G2LZW6 | Avirulence proteins in interactions involving weeping lovegrass and finger millet | M. oryzae | Rice blast | Rice (O. sativa) | Khang et al. (2010) |
| Rsp3 | 869 | A0A0D1DYI3 | Required for anthocyanin accumulation and blocks the antifungal activity of mannose‐binding maize proteins AFP1 and AFP2 | U. maydis | Corn smut | Maize (Z. mays) | Ma et al. (2018a); Seitner et al. (2018) |
| Six1/Avr3 | 154 | M1GN93 | Induces necrosis by triggering HR in tomato containing the cognate R protein I‐3 | Fusarium oxysporum f. sp. lycopersici | Tomato wilt | Tomato (S. lycopersicum) | Rep et al. (2004) |
| Six3/Avr2 | 163 | D0U2D2 | Avr2 suppresses PTI responses, such as growth inhibition, ROS production, MAPK activation, and callose deposition | F. oxysporum f. sp. lycopersici | Tomato wilt | Tomato (S. lycopersicum) | Houterman et al. (2009); Di et al. (2017) |
| Tin2 | 207 | NA | Masks a ubiquitin‐proteasome degradation motif in ZmTTK1 thereby stabilizing the anthocyanin biosynthesis pathway and decreases levels of metabolites available for plant defences | U. maydis | Corn smut | Maize (Z. mays) | Tanaka et al. (2014) |
| ToxA | 178 | P78737 | Proteinaceous toxin that causes necrotic lesions on infected leaves | Pyrenophora tritici‐repentis | Tan spot | Wheat (Triticum sp.) | Tomas et al. (1990); Ballance et al. (1996); Ciuffetti et al. (1997); Welti and Wang (2004) |
| Vd2LysM | 145 | G2X4U8 | Uncharacterized | Verticillium dahliae | Verticillium wilt | Tomato (S. lycopersicum) | de Jonge et al. (2013) |
| VdIsc1 | 190 | NA | An isochorismatase that supresses the precursor to the plant salicylate metabolism pathway and the subsequent salicylate‐mediated defences in planta | V. dahliae | Verticillium wilt | Multiple species | Liu et al. (2014) |
Abbreviations: HR, hypersensitive response; NB‐LRR, nucleotide‐binding domain (NB) and a leucine‐rich repeat (LRR); PAMP, pathogen‐associated molecular pattern; PLCP, papain‐like cysteine protease; PRR, pattern recognition receptor; PTI, pattern‐triggered immunity’ RIP, ribosome‐inactivating protein; ROS, reactive oxygen species.
Number of amino acids including signal peptide.
2. THE CHRONOLOGICAL PERSPECTIVE OF FINDING EFFECTORS
There is nothing like looking, if you want to find something.
J. R. R. Tolkien, The Hobbit, or There and Back Again
2.1. The proteomics approach
Some of the best‐characterized effector proteins come from the biotrophic fungal pathogen Cladosporium fulvum, the causal agent of tomato leaf mould and an early model system for fungal effector discovery. C. fulvum avirulence (Avr) effectors are a classic example of the gene‐for‐gene model. The detection of the Avr effector by the host carrying the cognate R gene can induce a strong immune response in the plant and inhibit C. fulvum colonization (Flor, 1971; De Wit et al., 1986).
Early in planta studies took advantage of the fact that C. fulvum only colonizes the tomato leaf apoplast. Secreted proteins could be isolated by collecting apoplastic wash fluid from C. fulvum‐infected tomato leaves and studying the effects of this fluid on a range of tomato varieties (De Wit et al., 1985). When fluid collected from plants infected with C. fulvum races harbouring the avr9 gene was infiltrated into the near‐isogenic tomato leaves carrying the Cf‐9 gene a strong HR was triggered. Treating this fluid with proteases confirmed the Cf‐9‐mediated HR was triggered by proteinaceous entities (De Wit et al., 1986). The subsequent purification of the small Avr9 (Figure 1) then led to the first fungal Avr gene to be cloned, whilst its low expression profile in vitro suggested for the first time that the host plant plays an important role in inducing Avr expression (Schottens‐Toma and de Wit, 1988; van Kan et al., 1991; Van den Ackerveken et al., 1992, 1994). The mature Avr9 is a 28 amino acid protein with a high percentage of cysteines (n = 6), features that become important in many subsequent effector identification stories (van Kan et al., 1991).
FIGURE 1.
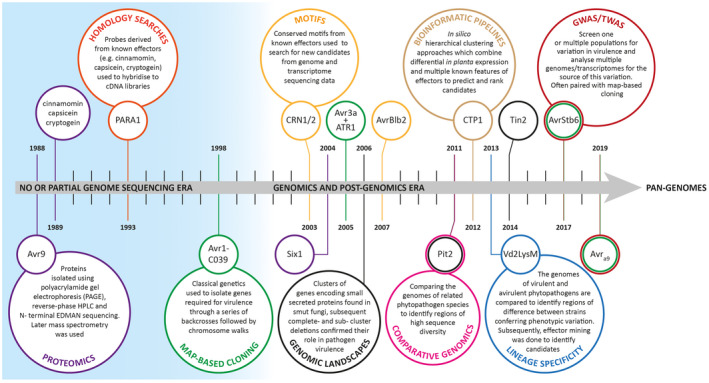
A timeline showing the progression of filamentous plant pathogen effector prediction and identification from the pregenomic era to the present day. The first effectors identified using these methods are included as well as the elicitins used for homology‐based searches. Increasingly, pangenome data are used to predict core and novel candidates but as yet none have been characterized using this technique. For a recent review of pangenomics see Golicz et al. (2019). Details on individual effectors named are given in Table 1.
This apoplastic proteomics approach was successfully used to identify additional small cysteine‐rich C. fulvum effectors such as Avr4 (Schottens‐Toma and de Wit, 1988; van den Burg et al., 2006) and was employed to identify Six1 (Avr3) and Six3 (Avr2) in Fusarium oxysporum f. sp. lycopersici (Fol) (Rep et al., 2004; Houterman et al., 2007; Houterman et al., 2009).
2.2. Homology searches
Once an effector has been cloned, the sequence can be used to identify homologous candidates in closely related species. Three elicitins were isolated from Phytophthora spp. using proteomics techniques: cryptogein (P. cryptogea), cinnamomin (P. cinnamomi), and capsicein (P. capsici) (Huet and Pernollet, 1989, Ricci et al., 1989). Primers were deigned based on conserved regions of the elicitin amino acid sequences and used to probe cDNA libraries from P. parasitica, leading to the discovery of the host‐specific elicitor protein PARA1 (Kamoun et al., 1993).
2.3. Genetic mapping
Prior to the genomics era, the isolation of Avr proteins from intracellular colonizing fungal pathogens such as Magnaporthe oryzae and haustoria‐producing pathogens was unsuccessful using the proteomics approach. Instead, in the case of the rice blast fungus M. oryzae, map‐based cloning techniques were used to clone Avrs such as Avr1‐CO39 (Farman and Leong, 1998). Avr1‐CO39 was mapped to a region on chromosome 1 by a series of backcrosses of the progeny of the virulent isolate Guy11 and the avirulent isolate 2539 (Smith and Leong, 1994). Later, a chromosome‐walking strategy led to the physical mapping and identification of Avr1‐CO39. The identity of the Avr1‐CO39 locus was confirmed by transforming the virulent Guy11 strain with cosmids from the Avr1‐CO39 genetic interval. This resulted in a loss of pathogenicity on rice cultivars containing the corresponding functional CO39 resistance gene (Farman and Leong, 1998).
2.4. Always lagging behind
By the end of the 20th century, over 30 bacterial Avr genes had been cloned and characterized by screening cosmid libraries, with almost all of these coming from two host‐specific species of Pseudomonas and Xanthomonas (Leach and White, 1996; De Wit, 1997). In comparison, using proteomics and genetic mapping, only eight fungal phytopathogen Avr genes had been successfully identified and confirmed to be effectors (Laugé and De Wit, 1998). But all this was about to change.
2.5. Sanger and next‐generation sequencing of pathogen genomes
In the early 2000s, the Fungal Genome Initiative (FGI) was established following the publication of a white paper (Birren et al., 2003) to promote the sequencing in the public domain of fungal genomes belonging to species important to human health, agriculture, and industry. By 2017 a total of 191 genomes of fungal plant pathogens had been sequenced, including the economically important M. oryzae, Fusarium graminearum, and Botrytis cinerea (Dean et al., 2005, 2012; Cuomo et al., 2007; Amselem et al., 2011; Aylward et al., 2017). This, together with the publication of numerous oomycete genomes, including the late potato blight pathogen Phytophthora infestans (Haas et al., 2009), as well as extensive in planta and in vitro transcriptome data sets, has led to an explosion in effector discovery. These techniques for effector discovery are summarized in Table 2.
TABLE 2.
Approaches and techniques deployed for effector discovery and the initial proteins/genes successfully isolated
| Technique | Effector | Species | Reference |
|---|---|---|---|
| Proteomics | Avr9 | Cladosporium fulvum | Schottens‐Toma and de Wit (1988); van Kan et al. (1991) |
| Six1 | Fusarium oxysporum f. sp. lycopersici | Rep et al. (2004) | |
| Map‐based cloning | Avr1‐CO39 | Magnaporthe grisea | Farman and Leong (1998) |
| Avr3a | Phytophthora infestans | Armstrong et al. (2005) | |
| ATR1 | Hyaloperonospora parasitica | Rehmany et al. (2005) | |
| Homology searches | PARA1 | Phytophthora parasitica | Kamoun et al. (1993) |
| INF1 | P. infestans | Kamoun et al. (1997) | |
| Motifs/secretion peptides | Crn1 and Crn2 | P. infestans | Torto et al. (2003) |
| AvrBlb2 | P. infestans | Win et al. (2007); Oh et al. (2009) | |
| Genomic landscapes | Tin2 | Ustilago maydis | Kämper et al. (2006); Brefort et al. (2014) |
| Comparative genomics | Pit2 | U. maydis | Doehlemann et al. (2011) |
| Bespoke bioinformatic pipelines | CTP1 | Melampsora larici‐populina | Saunders et al. (2012b); Petre et al. (2015) |
| Lineage‐specific | Vd2LysM | Verticillium dahliae | de Jonge et al. (2013); Kombrink et al. (2017) |
| GWAS/TWAS | AvrStb6 | Zymoseptoria tritici | Zhong et al. (2017) |
| Avra9 | Blumeria graminis f. sp. hordei | Saur et al. (2019a) |
3. REFINING EFFECTOR PREDICTION
Truth, like gold, is to be obtained not by its growth, but by washing away from it all that is not gold.
Leo Tolstoy, Diaries
3.1. Secretion
As the de Wit et al. studies demonstrated, a key feature of effectors is secretion by the pathogen into the host (De Wit et al., 1985; Asai and Shirasu, 2015). Therefore, early studies in effector discovery using sequencing data focused on the predicted secretome.
In a bid to identify extracellular effector proteins, Torto et al. (2003) used their PEX‐finder algorithm to mine transcript datasets of the potato pathogen P. infestans. The algorithm searched for a specific amino acid sequence known as a signal peptide followed by a cleavage site commonly found at the N‐terminus of secreted proteins (Nielsen and Krogh, 1998; Torto et al., 2003). Of the 261 cDNAs predicted to code for secreted proteins, 78 had no matches to those found in the public databases, a feature common to candidate effectors. Using high‐throughput functional expression assays this study led to the discovery of a large complex family of effectors called crinklers (CRNs), which are found throughout the pathogenic oomycetes (Schornack et al., 2010; Amaro et al., 2017).
However, some characterized secreted effectors lack a signal peptide. For example, the effectors, PsIsc1 and VdIsc1, produced by Phytophthora sojae and Verticillium dahliae, respectively, have been shown to be unconventionally secreted into the respective host to suppress salicylate (SA)‐mediated defences in planta (Liu et al., 2014).
Another difficulty is that such broad criteria leaves a large pool of possible effector candidates that are demanding in both time and resources to functionally characterize, with studies often having low discovery rates. The Magnaporthe grisea effector MC69, essential for appressoria formation (Motaung et al., 2017), was the only candidate from 1,306 putative secreted proteins that was found to be required for pathogenicity following large‐scale gene disruptions (Yoshida et al., 2009; Saitoh et al., 2012).
3.2. Domains
The C. fulvum effector Ecp6 sequesters the fungal cell wall protein chitin, preventing chitin fragment detection by the host PRRs, and thereby evades a host immune response (De Jonge et al., 2010). Ecp6 contains LysM domains that bind to chitin with ultrahigh affinity, therefore outcompeting host immune receptors (Sánchez‐Vallet et al., 2013). The LysM domain found in Ecp6 has now been identified in over 302 putative effectors from 62 published fungal genomes, and is conserved among effectors targeting the chitin detection aspect of plant immunity (De Jonge and Thomma, 2009; Lee et al., 2014).
On the other hand, the Avr2 effector from C. fulvum and the EPIC1 and EPIC2 effectors from P. infestans both target the tomato defence protease Rcr3 (Song et al., 2009) yet are unrelated and share no sequence similarity, thus relying on the presence of conserved domains could cause many possible candidates to be overlooked.
3.3. Motifs
The first four oomycete Avr effectors cloned, ATR13 and ATR1NDWsB from the downy mildew Hyaloperonospora parasitica (Allen et al., 2004; Rehmany et al., 2005), Avr3a from P. infestans (Armstrong et al., 2005), and Avr1b‐1 from P. sojae (Shan et al., 2004), showed no sequence similarity except for two conserved motifs at the N‐terminus. These RxLR and DEER motifs have since been identified as N‐terminal host targeting domains and, in P. infestans, the RxLR motif in the Avr3a effector is required for translocation into potato cells (Whisson et al., 2007; Bos et al., 2010).
RxLR effectors have been identified in multiple Phytophthora, Albugo, and Hyaloperonospora species, with 568 RxLR genes being found in P. infestans alone, making this the largest oomycete effector family to date (Anderson et al., 2015). Rapid variation and host specialization are attributed to the general lack of sequence similarity in filamentous pathogen effectors, yet this mostly contributes to the variation in the C‐terminus of oomycete effector sequences, leaving the N‐terminal motifs largely conserved (Win et al., 2007). Conserved motifs such as RxLR and the more downstream DEER are used as powerful bioinformatic tools to isolate putative effector repertoires from genomic sequences (Jiang et al., 2008; Raffaele and Kamoun, 2012).
Within pathogenic fungi there is limited evidence for conserved translocation motifs. One possible exception is the [YFC]xC motif found in Blumeria graminis f. sp. hordei and Puccinia spp., members of the phyla Ascomycota and Basidiomycota, respectively (Godfrey et al., 2010; Duplessis et al., 2011). The evolutionary distance between these two fungi suggests a deep homology in the conservation of this motif, linked to a biotrophic lifestyle that uses haustoria‐based feeding.
However, the general lack of sequence similarity or conserved domains means that bioinformatic approaches to effector prediction need to go beyond sequence homology.
3.4. Structure
The structural properties of proteins are more highly conserved than amino acid sequences (Illergård et al., 2009) and therefore could be used as a tool for effector prediction. The structural similarities between the two sequenced M. oryzae effectors Avr1‐CO39 and Avr‐Pia were found using two‐ and three‐dimensional nuclear magnetic resonance (NMR) experiments (de Guillen et al., 2015) and led to the discovery of the Magnaporthe Avr and ToxB‐like effector family (MAX), which contains half of all cloned M. oryzae Avrs despite sharing less than 25% sequence identity (de Guillen et al., 2015).
The structural analysis of four RxLR oomycete effectors showed the presence of a conserved C‐terminus 3‐α‐helix fold (Boutemy et al., 2011; Yaeno et al., 2011). This WY domain, named after the interacting tryptophan and tyrosine residues, hints to a core, stable protein scaffold as a source of protein function (Wirthmueller et al., 2013).
Resolving the structure of known effector proteins provides a useful tool for supporting the candidacy of putative effectors. One of the early effectors to be structurally resolved was ToxA produced by the tan spot fungus, Pyrenophora tritici‐repentis. The ToxA crystal structure was resolved using X‐ray crystallography (1.65 Å) and revealed a novel β‐sandwich fold (Sarma et al., 2005). Later, the resolution of the flax rust, Melampsora lini, effectors AvrL567‐A and ‐D showed a similar β‐sandwich fold hinting at the structural homology of unrelated effector proteins (Wang et al., 2007).
Recently the structures of two candidate effectors in the poplar rust fungus, Melampsora larici‐populina, were resolved using NMR. One, MLP124266, is the first fungal protein to present a knottin‐like structure (Postic et al., 2017) whilst the other, MLP1124499, shares structural similarity with members of the Nuclear Transport Factor‐2 (NTF2) superfamily. In both cases these candidate effectors show no sequence homology with structurally similar proteins and are the first examples of effectors with these structures (de Guillen et al., 2019).
3.5. Rich in cysteines but not in size
The additional criteria for candidate effector selection often require secreted proteins to be small and cysteine‐rich (Sperschneider et al., 2015). The presence of multiple cysteines enables the formation of stabilizing disulphide bridges (De Wit et al., 1986; Doehlemann et al., 2009).
Relying on such broad criteria can be problematic as, despite many known effectors sharing these features, these are not universal requirements. NIS1, first described in the cucumber anthracnose fungus Colletotrichum orbiculare (Yoshino et al., 2012), is conserved across both Basidiomycota and Ascomycota (Irieda et al., 2019), but contains no cysteines.
Relying on the size of mature peptides as a parameter for effector identification can also be problematic. The maximum size of a small protein in effector discovery can be anything from 150 to 400 amino acids (Bowen et al., 2009; Saunders et al., 2012b). However, even the larger size limits would exclude the P. graminis f. sp. tritici effector AvrSr35 with a mature length of 578 amino acids (Salcedo et al., 2017).
With these issues in mind, bioinformatic pipelines have been developed to encompass multiple criteria to refine effector prediction.
3.6. Bespoke bioinformatic pipelines
Saunders et al. developed an in silico analysis pipeline that moved away from reliance on sequence similarity‐based methods for effector identification and included physiological functions such as expression profiles, taxonomic information, and genomic features of potential candidates (Saunders et al., 2012b). To identify the repertoire of potential effectors within two rust fungus genomes, a clustering algorithm grouped candidates into families and ranked their likelihood of being effectors based on the knowledge that filamentous pathogen effectors have a least one of eight specific properties. These properties included the absence of recognized Pfam domains, similarities to haustorial proteins, and the presence of internal repeats. The number of candidates continued to functional analysis using this pipeline was greatly reduced (Saunders et al., 2012b). This approach has limitations as it is dependent on the thresholds based on a priori assumptions about effector properties; the number of missed effectors remains to be seen.
At each step of the general pipeline for effector prediction and subsequent characterization, in silico tools, whether bioinformatical software or web‐based servers, have been developed to aid effector refinement. The presence of signal peptides, transmembrane motifs, or GPI anchors can all be predicted using tools such as SignalP (www.cbs.dtu.dk/services/SignalP/), TMHMM (www.cbs.dtu.dk/services/TMHMM/), and PredGPI (gpcr.biocomp.unibo.it/predgpi/pred.htm), which use neural networks or hidden Markov modelling to recognize motifs within protein sequences associated with these features (Pierleoni et al., 2008; Armenteros et al., 2019). The subcellular localization of candidate effectors can also be predicted by searching for chloroplast or mitochondrial transit peptides or nuclear localization signals using tools such as WoLF‐PSORT (wolfpsort.hgc.jp/) or LOCALIZER (localizer.csiro.au/) (Horton et al., 2007; Sperschneider et al., 2017). Machine learning has also resulted in the development of web‐based tools that can predict with 89% accuracy whether proteins in the predicted secretome are effectors or not. EffectorP2.0 (effectorp.csiro.au/) takes into account the net charge and serine/cysteine content of proteins to prioritize candidate effectors for further functional validation (Sperschneider et al., 2018).
3.7. Genomic landscape and transposable elements
Many fungal plant pathogens exhibit a two‐speed genome, with distinct genomic compartments evolving at different rates. Alongside core stable regions, which are slow to evolve and often contain genes involved in metabolism, are hypervariable areas with high recombination and richness in repetitive sequences, including transposable elements (TEs). This genomic landscape and the presence of TEs serve to drive adaptive evolution (Faino et al., 2016) and these hypervariable regions often are the location of genes associated with pathogenicity, including effectors (Fouché et al., 2018; Jones et al., 2018).
In M. oryzae and Zymoseptoria tritici, TEs are associated with pathogenicity clusters and are seen to flank the first characterized Z. tritici effector, AvrStb6 (Bao et al., 2017; Zhong et al., 2017). TEs have also been shown to interfere with effector gene expression via epigenetic control. For example, AvrLm1 in Leptosphaeria maculans, located in a TE‐rich genomic region, showed distinct histone methylation that acts to temporarily suppress expression during colonization to evade host recognition (Soyer et al., 2014; Fouché et al., 2018). This suggests that the variability of the genomic region or the proximity to TEs maybe useful factors in refining the search for candidate effectors.
Following the sequencing, genome assembly and annotation of the tumour‐forming maize smut fungus Ustilago maydis, c.18% of genes encoding secreted proteins were found to be arranged into 12 discrete clusters within the genome (Kämper et al., 2006). These clusters were co‐regulated by a central pathogen‐development regulator and expression induced in tumour tissue. Deletions of five clusters caused clear changes in virulence, including the largest cluster, 19A, which caused a strong attenuation in virulence and reduced tumour formation upon deletion (Kämper et al., 2006; Brefort et al., 2014). Subsequent subdeletions of 19A members led to the identification of the effector Tin2, required for anthocyanin production (Brefort et al., 2014; Tanaka et al., 2014).
3.8. Comparative genomics
By comparing the genomes of U. maydis and Sporisorium reilianum, Schirawski et al. (2010) found that effector clusters and pathogenicity‐related regions were more highly diverged between the close relatives than the rest of the genome. This comparison led to the identification of the pit gene cluster involved in tumour formation in U. maydis (Doehlemann et al., 2011). Within this cluster the secreted effector Pit2, involved in plant defence suppression and cysteine protease inhibition, was found (Doehlemann et al., 2011; Mueller et al., 2013). This same comparison was used to locate gene clusters and candidate effectors in S. reilianum, and whilst genes that have a partial impact on disease severity have been identified, as yet no candidates strongly attenuate virulence (Ghareeb et al., 2019).
3.9. Lineage‐specific elements
Novel effectors were identified in the asexual fungus V. dahliae, where chromosome reshuffling has led to the formation of lineage‐specific (LS) regions of plasticity in the genome (de Jonge et al., 2013). These LS regions are enriched with retrotransposon and repetitive sequence elements, as well as being the location of many candidate effectors. Contrary to the two‐speed genome hypothesis, these LS regions show strong levels of conservation with little to no single nucleotide polymorphisms (SNPs) being identified, even within the intergenic regions (Depotter et al., 2019). In one such LS region, four putative effectors were identified, including the LysM domain containing effector Vd2LysM, which was only found in the VdLs17 strain (de Jonge et al., 2013).
3.10. Sequence divergence
Molecular variation in filamentous phytopathogen genes is known to be essential for altering pathogen–host interaction outcome and can provide insight into the evolution of virulence (Allen et al., 2008). Polymorphisms in effector sequences among isolates can impact on virulence and are involved in host adaptation; this makes them promising targets for disease control strategies.
The genomes of four isolates of the wheat yellow stripe rust fungus Puccinia striiformis f. sp. tritici were resequenced and assessed for SNPs. Proteins that displayed nonsynonymous substitutions between isolates that differed in virulence on specific wheat cultivars were identified (Cantu et al., 2013). This led to five secreted polymorphic candidate effectors being nominated for further characterization from a predicted secretome of 2,999 proteins.
This sequence divergence has also proved useful in identifying pathogens in the field. Using the Oxford Nanopore MinION sequencer, 242 highly variable genes were used to collect real‐time population dynamics data of P. st riiformis f. sp. tritici isolates in Ethiopia (Radhakrishnan et al., 2019). This Mobile And Real‐time PLant disEase (MARPLE) diagnostic system can be used to monitor the emergence of plant pathogen strains, but can also be adapted to include newly characterized effectors within the panel of genes. Going forward, MARPLE will allow for the monitoring of mutations and the detection of effector evolution that may be linked to gain of virulence of phytopathogens, all within the confines of the field.
3.11. Association mapping in the sequencing era
In silico predictions of effectors, whilst allowing us to rapidly screen whole genomes for candidates, lack discriminatory power and often result in candidate effectors having no clear impact on pathogen virulence. Genome‐wide association studies (GWAS) and quantitative trait locus (QTL) mapping can identify loci associated with heritable phenotypic variation, such as virulence, thereby complementing techniques to identify and clone Avr effectors recognized by known host resistance proteins (Plissonneau et al., 2017). The Zymoseptoria tritici effector AvrStb6 was isolated in this way (Zhong et al., 2017). Using crosses between two Swiss strains of Z. tritici, QTL mapping found a confidence interval containing nine candidates for AvrStb6. Combining this with a GWAS study from over 100 different natural isolates led to one candidate, a small cysteine‐rich secreted protein that was not present in the original Z. tritici genome annotation (Zhong et al., 2017).
An additional benefit of using GWAS in effector discovery is that the natural variation in SNP calling identified in wild populations can be used to quantify how each SNP contributes to pathogen virulence (Sánchez‐Vallet et al., 2018b). Integrating GWAS with transcriptome dataset, referred to as transcriptome‐wide association studies (TWAS) (Wainberg et al., 2019), identified the link between genes and traits across populations and has been used to discover Blumeria graminis f. sp. hordei Avra effectors, including Avra9 (Saur et al., 2019a).
4. FUNCTIONAL CHARACTERIZATION
Make your work to be in keeping with your purpose.
Leonardo da Vinci, The Practice of Painting
4.1. Knock out or knock down: let's be disruptive
One of the simplest ways to determine the pathogenicity of a candidate effector is to disrupt the encoding gene and determine whether the virulence on a susceptible host or the Avr phenotype on a resistance genotype is compromised. Early transformation studies of the C. fulvum effectors relied on double homologous recombination to insert a selectable marker into the target gene encoding a known effector such as ecp1 and ecp2, thus disrupting them (Laugé et al., 1997). Later sequencing technology allowed transformations without the need for cloning. Mutants of the corn smut fungus Ustilago maydis were made using PCR‐based protocols combined with protoplast transformation to generate candidate effector knockout mutants (Schulz et al., 1990; Kämper, 2004). This method is widely used and has successfully facilitated the functional characterization of U. maydis effectors, including Rsp3 and Cce1 (Ma et al., 2018a; Seitner et al., 2018).
Agrobacterium tumefaciens‐mediated transformation (ATMT) is another method to disrupt genes and is widely used in plant transformations. ATMT was first used in fungi in budding yeast in 1995 and then the technique was adapted for use in filamentous fungi, including M. oryzae (Bundock et al., 1995; Rho et al., 2001). This method relies on the targeted insertion of a selectable marker into the fungal genome from a disarmed Ti plasmid of transformed Agrobacterium to disrupt the gene of interest. The selectable marker is incorporated into the fungal genome via homologous recombination, a process that occurs easily in yeast. This mechanism, however, is highly variable in filamentous fungi, where nonhomologous end‐joining (NHEJ) appears to be the dominant DNA repair pathway over homologous recombination (Meyer et al., 2007; Villalba et al., 2008). The Ku70 protein is part of a complex that regulates the NHEJ pathway (Ninomiya et al., 2004), and its deletion has led to the increase of homologous recombination in M. oryzae from <25% to 80% (Kershaw and Talbot, 2009). Combining ATMT with the generation of ∆Ku70 mutants led to the characterization of the Z. tritici Avr effector AvrStb6 (Zhong et al., 2017).
Another, more recent, method of gene disruption is using the genome‐editing system CRISPR‐Cas9. Originally identified as an immune mechanism in bacteria and archaea, the CRISPR‐Cas9 system is used as a genome‐editing tool in plants and animals, and was adapted by Nødvig et al. (2015) for use in filamentous fungi (Mali et al., 2013; Fauser et al., 2014; Nødvig et al., 2015). This technique has led to targeted gene disruption and consequent characterization of effectors in the oomycete P. sojae and the fungal pathogen U. maydis (Fang and Tyler, 2016; Schuster et al., 2018).
There are, however, difficulties in producing stable transformants in phytopathogens that are obligate biotrophs (Thomas et al., 2001; Lorrain et al., 2019). In these cases, knockdown technologies such as host‐induced gene silencing (HIGS) are more successful. The HIGS assay detailed in Figure 2 has led to the identification of many effectors, including the barley powdery mildew Blumeria graminis f. sp. hordei ribonuclease‐like effectors BEC1054 and BEC1011 (Nowara et al., 2010; Pliego et al., 2013; Pennington et al., 2019).
FIGURE 2.
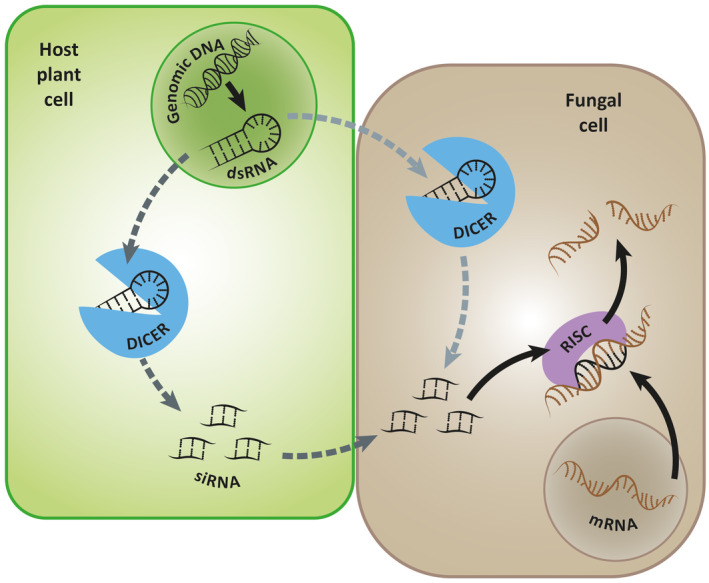
The host‐induced gene silencing (HIGS) construct encodes an inverted sequence that forms a hairpin double‐stranded (ds) RNA following transcription and is introduced into the host plant either by transient or stable transformation. The dsRNA is processed to form small interfering RNA (siRNA), either before or after delivery to the pathogen cell using the plants innate RNAi machinery. Once inside the fungal cells the siRNA silences the target effector genes by interfering with the target mRNA transcripts (Koch et al., 2018). The movement of small RNA between host and pathogen is detailed by Wang and Dean (2020).
Gene disruption assays do have their limitations even when successful transformants are produced. Many effector mutants display no associated phenotype. Genetic redundancies, where multiple effectors have the same function, or buffering, where the host compensates or interfers in signalling using alternative pathways, may result in false‐negative results (Hillmer et al., 2017; Tyler, 2017).
4.2. In planta expression
When a candidate effector is heterologously expressed in planta various functional assays can be used to determine the virulence activities of the protein.
Necrosis assays monitor for the induction of HR‐like cell death, which can be a result of Avr/R protein/guardee protein interactions or be directly induced by the candidate effector. These assays were first carried out using the model plant Nicotiana tabacum (tobacco), which is infiltrated with transformed Agrobacterium that delivers the effector gene expressed from an inducible promoter into the plant cell for transient protein production (Kamoun et al., 1999; Qutob et al., 2002; Ma et al., 2012).
In 1999 the P. infestans and C. fulvum effectors Inf1 and Avr9, respectively, were transformed into either wild‐type or Cf‐9 transgenic N. tabacum using this method. The assay showed that INF1 was capable of inducing necrosis in wild‐type tobacco whilst Avr9 could only do so in transgenic tobacco expressing the corresponding R gene Cf‐9 (Kamoun et al., 1999). Later Avr9 and Cf‐9 were transiently coexpressed in N. tabacum using agroinfiltration to confirm the induction of HR in the nonhost plant following expression of the Avr/R gene pairs (Van der Hoorn et al., 2000).
Effector characterization in nonhost dicotyledonous model plants maybe more suited to high‐throughput screening than in cereal hosts. However, these highly artificial scenarios do have several limitations. A negative screen with no visible phenotype upon recombinant expression may indicate either the candidate is not an effector or the effector target/receptor is lacking in the model species. On the otherhand, HR‐induced necrosis in an effector screen may not be caused by a specific effector/target interaction but by nonhost resistance (NHR) triggered by detection of the candidate (Kettles et al., 2017). Although of interest, by definition the latter scenario would not occur in native host interactions. Therefore, expression assays in the native host maybe the more useful for functional characterization.
Candidate effectors can be transiently expressed in protoplast cells and cell death monitored via the reduction in expression of a co‐transfected reporter gene such as β‐glucuronidase (GUS) or luciferase (Chen et al., 2006; Lu et al., 2016). This approach was used to identify the cell death‐inducing properties of five M. oryzae effectors, including MoCDIP4 (M. oryzae cell death inducing protein 4), in rice protoplasts (Chen et al., 2012) and the NLR‐mediated recognition of four newly identified barley powdery mildew avirulence effectors, including AVRa9, in barley (Saur et al., 2019a).
Cell‐death suppression assays are used to detect the alteration of the plant immune response induced by a known cell death elicitor. The overexpression of the stem rust candidate effector PSTha5a23 in Nicotiana benthamiana suppresses P. infestans INF1‐triggered cell death, indicating that PSTha5a23 plays a role in controlling plant defence responses (Cheng et al., 2017).
An alternative method of expressing effectors in plant cells uses the bacterial type III secretion system (T3SS) derived from the tomato bacterial speck pathogen Pseudomonas syringe pv. tomato DC3000 (He et al., 2004). This system was first adapted for filamentous plant pathogens by Sohn et al. (2007) to deliver oomycete effector proteins into Arabidopsis. Sohn et al. showed that, by fusing the downy mildew (H. parasitica) effectors ATR1 and ATR13 to the N‐terminal secretion‐translocation signals of the P. syringae effectors AvrRpm1 and AvrRps4, the effectors could be secreted into Arabidopsis plant cells and contribute to pathogen virulence. Since then, the T3SS has been used to functionally characterize candidate effectors from multiple oomycetes, including P. infestans and Hyaloperonospora arabidopsidis (Whisson et al., 2007; Fabro et al., 2011). Despite T3SS being used to screen effector candidates of stem rust (P. graminis f. sp. tritici) and bean rust (Uromyces appendiculatus), this system is rarely used for fungal effector characterization and has limited success on cereals (Upadhyaya et al., 2014; Qi et al., 2019; Saur et al., 2019b). These problems are linked to the required unfolding and refolding of effectors prior to insertion, especially those rich in cysteine–cysteine bridges.
As well as monitoring for necrosis, or lack thereof, the in planta growth of another pathogenic species can be used as a proxy to determine the role in virulence effectors play. Stable transformants of the nonhost Arabidopsis that expressed candidate poplar rust fungus (M. larici‐populina) effectors were inoculated with the oomycete pathogen H. arabidopsidis. Eleven of 16 effectors tested supported greater sporulation of this native Arabidopsis pathogen, suggesting that the effectors had the capacity to interfere with processes in a nonhost plant to favour pathogenesis (Germain et al., 2018).
4.3. The viral overexpression system
Due to the limited effectiveness of both T3SS and Agrobacterium‐mediated transient expression in most cereal species, viruses have been developed as efficient vectors for heterologous protein expression (viral overexpression, VOX) (Lee et al., 2012).
The barley stripe mosaic virus (BSMV) was first verified as a tool for protein expression when used to overexpress the luciferase reporter gene in protoplast cells and later to express green fluorescent protein (GFP) in planta (Joshi et al., 1990; Haupt et al., 2001; Lawrence and Jackson, 2001). The BSMV vector was adapted for use in the VOX system and used to characterize the function of the fungal effector ToxA (Manning et al., 2010) (Figure 3). However, the compact nature of the virus results in a negative correlation between fragment size and stability of the viral vector (Avesani et al., 2007; Bruun‐Rasmussen et al., 2007). BSMV‐VOX has been widely used for heterologous expression of proteins up to 150 amino acids; however, as previously stated there is no agreed size limit for an effector (Figure 3a; Bouton et al., 2018).
FIGURE 3.
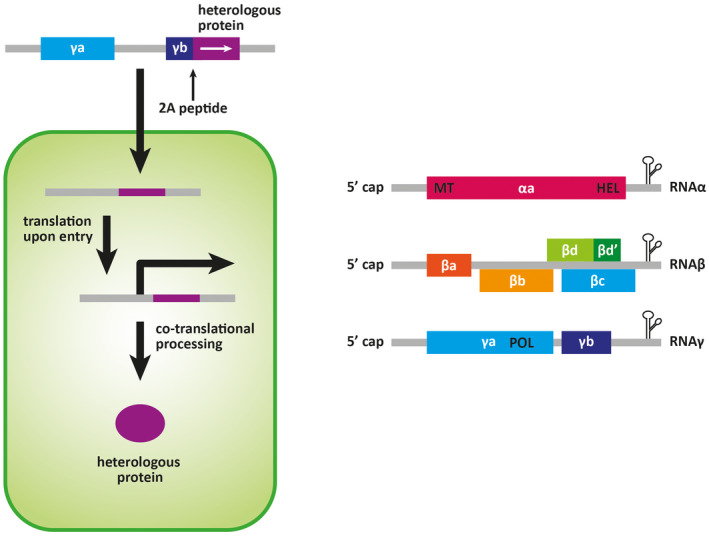
The BSMV‐VOX technology adapted from Lee et al. (2012). (a) Virus‐mediated overexpression (VOX) system. The heterologous protein coding sequence is inserted in the γ genome of barley stripe mosaic virus (BSMV), upstream of the in‐frame stop codon in the γb open reading frame (ORF). A gene for the autoproteolytic peptide 2A is also inserted between the 3′ terminus of the γb ORF and the gene of interest for processing the fusion protein during translation, thus releasing the heterologous protein of interest. (b) The BSMV genome is composed of three RNAs that are capped at the 5′ end and form a tRNA‐like hairpin secondary structure at the 3′ terminus. RNAα encodes the αa replicase protein containing methyltransferase and helicase domains. RNAβ encodes coat and movement proteins whilst RNAγ encodes the polymerase (POL) component of replicase, and the cysteine‐rich γb protein involved in viral pathogenicity.
Another limitation of BSMV for use in effector discovery is that this virus has a tripartite RNA genome (Figure 3b). The heterologous protein is inserted into the γ genome yet all three subgenomes are required to combine for successful expression in planta making BSMV‐VOX unsuitable for high‐throughput screening assays.
The foxtail mosaic virus (FoMV) has been adapted for use in VOX systems in cereals (Bouton et al., 2018). Vectors derived from FoMV such as PV101 avoid many of the caveats of those from BSMV. FoMV has a monopartite RNA genome and the PV101 vector can be used to successfully express proteins up to 600 amino acids in size. In addition, unlike BSMV vectors, PV101 allows for heterologous expression of proteins in their native form, including possible signal peptides, without the need for processing from proteases that may only be 90% efficient (Bouton et al., 2018). In situations where the effector expressed from the VOX vector rapidly triggers R protein‐mediated defences, virus spread is halted and therefore the phenotypic readout in the bioassay is the lack of systemic spread of the recombinant virus (Saintenac et al., 2018).
4.4. Where do they go?
Knowing the localization of candidate effectors within host tissues not only demonstrates that the protein can be translocated from the pathogen to its host, but also suggests where the effector target(s) may be found. Traditionally in situ hybridization assays were done where antibodies were raised against the effector or an added epitope tag and detected using transmission electron microscopy (TEM). Translocation of fungal effectors into the host cell was first shown using an immunocytochemical approach in rusts. The gold‐ and fluorescence‐labelling of four independently raised antibodies to the RTP1p protein in Uromyces fabae and its homolog in Uromyces striatus showed that in later stages of infection RTP1p translocated from the extrahaustorial matrix to inside the plant cell itself (Kemen et al., 2005).
For apoplastic effectors, localization was often determined by means of their isolation. The C. fulvum effectors Avr2, Avr4, Avr9, and Ecp6 were directly isolated from the apoplastic fluid, whereas the P. infestans protease inhibitor EPIC1 was isolated from the apoplast after antibodies were raised (Joosten et al., 1997; Rooney et al., 2005; Tian et al., 2007; Bolton et al., 2008). Whilst successful, these approaches are laborious, expensive, and not suited to high‐throughput screening of either apoplastic or cytoplasmic effector candidates (Dalio et al., 2017).
The nuclear localization of the P. infestans CRN effectors was determined using N‐terminal GFP tagging and confocal microscopy. By overexpression five GFP‐CRN (without the signal peptide) fusion proteins in planta the effectors were shown to accumulate within plant cell nuclei (Schornack et al., 2010). High‐throughput screening of 61 candidate effectors (ChECs) from the anthracnose fungus Colletotrichum higginsianum using this method found that whilst nine of the ChECs were imported into the nucleus, others localized to the Golgi bodies, microtubules, and peroxisomes, all novel targets for fungal effectors (Robin et al., 2018).
The U. maydis effectors Cmu1 and Tin2 have been shown to localize to the maize cytoplasm; however, this could not be demonstrated when fluorescently tagged (Djamei et al., 2011; Tanaka et al., 2014; Tanaka et al., 2015). This may be due to the tags inhibiting the partial unfolding of the effectors, thereby preventing their translocation, or the incorrect refolding of the tags themselves upon entering the cytoplasm (Lo Presti et al., 2015).
Whilst investigating the translocation of M. oryzae effectors into rice cells, fluorescent‐tagged cytoplasmic effectors were seen to first accumulate in the plant‐membrane derived infection structure, the biotrophic interfacial complex (BIC), prior to delivery into the cytoplasm, whereas tagged apoplastic effectors localized to the invasion hyphae (Mosquera et al., 2009; Khang et al., 2010). The BIC’s role in effector translocation could only be confirmed by the addition of a nuclear localization signal (NLS) to cytoplasmic effectors, causing artificial accumulation in the nucleus of the neighbouring rice cells. This approach concentrated the fluorescent signal into discrete foci observable using live cell imaging (Khang et al., 2010).
For apoplastic effectors it is difficult to distinguish between apoplastic or cytoplasmic localization when the fluorescently tagged candidates appear to localize to the plasma membrane or cell wall. Enlarging the apoplastic space by the stepwise addition of hypertonic solutions, a process known as plasmolysis, revealed that the U. maydis host‐peroxidase inhibitor Pep1 was indeed apoplastic and was evenly distributed throughout the enlarged space (Oparka, 1994; Doehlemann et al., 2009).
Alternatively, the BirA assay does not require the use of large fluorescent tags that may interfere with effector function or localization. BirA, developed by Lo Presti et al., is based on the bacterial enzyme biotin ligase that biotinylates any protein that has a short (15 amino acids) peptide Avitag (Lo Presti et al., 2017). Maize lines that expressed the biotin ligase in the cytoplasm were infected with transformed U. maydis strains that had either the Cmu1 or the Tin2 effectors tagged with the Avitag. Biotinylation was detected via immunoprecipitation of extracted proteins using streptavidin‐coated magnetic beads, thus confirming the tagged effectors had met the biotin ligase in the host cytoplasm (Lo Presti et al., 2017).
5. EFFECTOR INTERACTIONS
…to manage a system effectively, you might focus on the interactions of the parts rather than their behavior taken separately.
Russell L. Ackoff and Fred Emery, On purposeful systems
Arguably the Holy Grail of effector characterization is to identify the exact molecular targets of each effector and/or the molecules used by the plant to bind to them. This can lead to defining the precise sequences and molecular interactions occurring at the point(s) of direct contact. The former is very challenging because the effector sequences do not give many clues as to their function(s).
5.1. A shot in the dark: unbiased screening
Unbiased “forward” screening to find protein–protein interactions (PPI) is a common technique used in many aspects of molecular biology. The yeast two‐hybrid system (Y2H), first developed 30 years ago, allows for the large‐scale screening of cDNA libraries derived from pathogen‐infected plants for effector target identification (Fields and Song, 1989; Mukhtar et al., 2011). Interactions detected by Y2H screens must be validated by additional PPI assays as this approach is prone to false positives.
The most common Y2H validation technique is co‐immunoprecipitation (Co‐IP). Co‐immunoprecipitation is used to screen effector interactors in heterologous systems. When 20 candidate poplar rust fungus (M. larici‐populina) effectors were tagged with GFP and expressed in N. benthamiana, five were found to specifically interact with plant proteins by pull‐down assays using anti‐GFP followed by protein purification (Figure 4a) (Petre et al., 2015).
FIGURE 4.
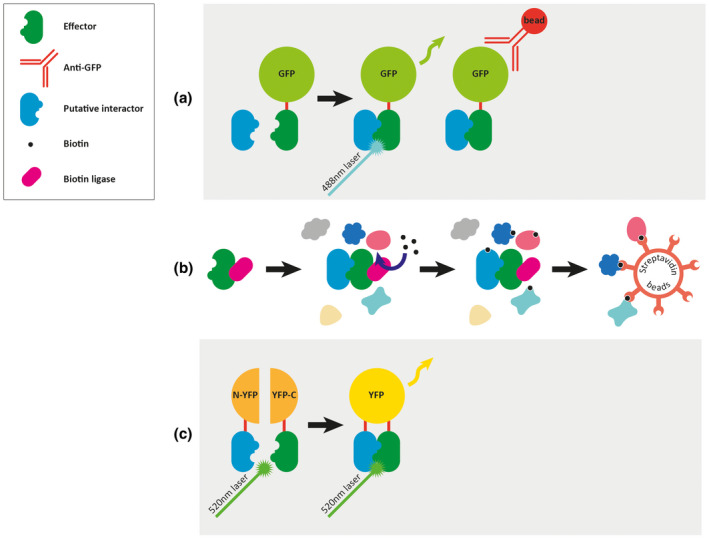
Protein–protein interaction techniques. (a) Co‐immunoprecipitation, effectors are tagged with a peptide sequence such as green fluorescent protein (GFP) and expressed in planta. Antibodies are used to pull down the protein complexes that can then be analysed using liquid chromatography and mass spectrometry (LC‐MS/MS) (Petre et al., 2017). (b) Biotinylation, effectors are fused to mutant biotin ligase enzymes and expressed in vivo. The fusion protein catalyses the biotinylation of interacting and proximal proteins in the presence of biotin. The biotinylated proteins are captured using streptavidin beads (Roux et al., 2012). (c) Bimolecular fluorescence complementation, the effector and putative interactors are tagged with nonfluorescent fragments of yellow fluorescent protein (YFP). Direct interaction of the tagged effectors results in YFP reassembly visualized in vivo or quantified using flow cytometry (Kerppola, 2008; Graciet and Wellmer, 2010; Miller et al., 2015).
Biotinylation is also used for proximity labelling based on tools such as BioID (Li et al., 2017). A benefit of proximity labelling over co‐immunoprecipitation is the possibility of identifying proteins that only weakly or transiently interact with the target (Figure 4b). Recently a new proximity labelling tool, TurboID, has been shown to provide more efficient labelling in planta compared to BioID and can also reduce the biotin incubation time from 16 hr to 10 min (Branon et al., 2018; Zhang et al., 2019). These new advances in PPI technology pave the way for higher‐throughput effector interaction screening in planta.
5.2. Split‐marker complementation
The effector Pep1 is essential for the pathogenicity of the corn smut fungus U. maydis (Doehlemann et al., 2009). The direct interaction between Pep1 and the plant peroxidase POX12 was validated using the bimolecular fluorescence complementation (BiFC) assay (Figure 4c), which involves two parts of a fluorescent marker being fused to candidate interactors. Only when the interactors meet can the full‐length fluorescent marker assemble and be detected. Alternatively, the firefly‐derived enzyme luciferase can be used for split‐marker complementation. This has the advantage over BiFC for in planta studies because luciferase does not require excitation by light for detection, thereby eliminating autofluorescence interference (Li et al., 2011). However, using split‐marker complementation for PPI validation is not infallible as heterologous overexpression of proteins in N. benthamiana can affect protein localization and therefore interactors.
5.3. Structural interactions: pinpointing the surface contacts and their strengths
Knowledge of effector structures whilst in complex with their targets gives us a greater insight into the molecular basis of these cross‐kingdom interactions.
The C. fulvum effector Avr4 was one of the first to be characterized from a family of effectors that bind to and protect fungal cell‐wall chitin from host chitinase (Joosten et al., 1997; van den Burg et al., 2006). Recently the crystalline structure of Avr4 in complex with its chitin ligand (resolved to 1.95Å) has highlighted the residues required for this function (Hurlburt et al., 2018). Structural mutant studies have also shown that recognition of the Avr4 by the cognate Cf‐4 immune receptor does not depend on the same ligand binding as previously thought (Hurlburt et al., 2018).
The crystal structure of the rice intracellular NLR immune receptor Pik in complex with the M. oryzae effector Avr‐Pik (1.6Å resolution) reveals molecular details of the recognition event that leads to HR‐induced cell death (Maqbool et al., 2015). The effector surface involved in this interaction was also identified as being involved in the surface interactions between Avr‐Pia and the NLR‐RATX1 in M. oryzae (Ortiz et al., 2017).
In the past decade protein structures are increasingly being resolved without the need to form crystals or use damaging X‐rays but by using cryo‐electron microscopy. This technique is widely used to resolved proteins in complexes and has been used to show both inactive Arabidopsis NLR complex ZAR1‐RKS1 and the intermediate form when the complex interacts with a protein modified by the bacterial effector AvrAC (Xanthomonas campestris pv. campestris) (Wang et al., 2019). Cryo‐e, despite gaining popularity in structural biology, is unable to resolve proteins smaller than 65 kDa, a size exclusion that would include many fungal and oomycete effectors (Muench et al., 2019).
The strength of effector–target interactions can be determined using isothermal titration calorimetry whereby direct measurement of the heat that is either released or absorbed during the molecular binding event gives a complete thermodynamic picture of the reaction, including affinity, enthalpy, and stoichiometry (Duff et al., 2011). For the conserved M. oryzae MAX effector Avr1‐CO39, isothermal titration calorimetry was used to confirm that direct interaction with the heavy‐metal associated (HMA) domain of the rice NLR RGA5 was required for effector binding (Guo et al., 2018).
A greater understanding of how structural interactions aid the specificity of Avr recognition is vital for future work in developing sustainable disease resistance in important food crops.
6. EXPLOITING EFFECTOR DISCOVERIES TO CONTROL CROP PLANT DISEASES
Knowing is not enough; we must apply. Willing is not enough; we must do.
Johann Wolfgang von Goethe, Wilhelm Meister's Journeyman Years
The ultimate goal of effector discovery, from identification to characterization to target interactions, is to apply this knowledge to the control of multiple pathogens that threaten our food security.
6.1. “Effectoromics”
For over 100 years disease resistance loci have been introduced into crops and subsequently shuffled through traditional breeding techniques, whether that be as individual genes or stacked to achieve often only short‐lived resistance to pathogens (Vleeshouwers et al., 2011; Langner et al., 2018). Despite this, the search for novel R genes with durable or broad‐spectrum resistance remains ongoing.
The term “effectoromics” is used to describe the use of effectors in high‐throughput screening for R protein function in either the germplasm of crop cultivars or a sexually compatible species. Avr effectors can be harnessed to screen rapidly for HR phenotypes, a hallmark of an ETI response (Vleeshouwers and Oliver, 2014). Well‐established techniques of transient overexpression of Avrs using viral vectors such as potato virus X (PVX) in conjunction with agroinfiltration have been widely used for the identification and cloning of R genes in solanaceous species such as potato, tomato, and wild Solanum species (Takken et al., 2000; Du et al., 2014).
The search for broad‐spectrum or more robust R genes for breeding purposes maybe more nuanced than previously thought as multiple unrelated R genes can recognize the same pathogen effector (Aguilera‐Galvez et al., 2018).
6.2. Screening with necrosis‐inducing effectors to remove host susceptibility loci
The necrosis‐inducing effector ToxA was isolated from the wheat tan spot fungus P. tritici‐repentis in 1996. Infiltration of purified ToxA into the apoplastic space of a susceptible wheat cultivar containing the Tsn1 susceptibility (S) gene is itself sufficient to induce tan spot symptoms (Tomas et al., 1990; Ballance et al., 1996; Ciuffetti et al., 1997; Welti and Wang, 2004). Wheat breeders routinely use the purified toxin to screen all new wheat germplasm to eliminate susceptible lines from their breeding programmes. This method is preferred over screening for molecular markers linked to the corresponding Tsn1 locus due to the ease of application and speed of results (Vleeshouwers and Oliver, 2014). Tsn1 removal from all newly commercially released wheat varieties has improved resistance to tan spot disease and Australia has seen a 26% reduction in ToxA‐sensitive wheat grown in the 10 years prior to 2016 (See et al., 2018).
7. KEEPING TRACK OF EFFECTOR DISCOVERIES IN MULTIPLE SPECIES IN AN INCREASINGLY DATA‐RICH WORLD
A place for everything, and everything in its place.
Idiom from 17th century
In the past two decades effector discovery and characterization have exploded with regard to crop pests and pathogens. This key information is found in multiple original research publications, review articles, UniProt, individual pathogen genome browsers, and species‐specific websites. However, to aid future research and guide the direction of work the genotype and fine phenotyping data surrounding these discoveries and new insights needs to be FAIR (Findable, Accessible, Interoperable, and Reusable) to molecular plant pathologists as well as the wider life sciences communities.
Publicly available repositories of curated data regarding proteins with confirmed roles in pathogenicity and virulence are a fundamental tool for effector study. The Pathogen–Host Interactions database (PHI‐base, www.phi‐base.org) is a manually curated database comprising over 6,780 genes from 268 pathogens of over 210 hosts (September 2019), of which 60% are plants (Urban et al., 2020). Within the PHI‐base (version 4.8), 799 interaction entries involve 731 distinct functionally characterized fungal or oomycete effectors from over 40 species. Collectively, these effector entries and their considerable metadata can be used for comparative studies, genome landscape explorations, the enrichment of transcriptome/proteome data sets, PPI network predictions, as well as the starting point for potentially novel artificial intelligence approaches.
8. CONCLUSIONS AND OUTLOOK
“Would you tell me, please, which way I ought to go from here?”
“That depends a good deal on where you want to get to,” said the Cat.
Lewis Carroll, Alice in Wonderland
Effectors are the mysterious molecular tools evolved and used by plant pathogens in multiple ways. Effector studies are of vital importance in addressing the global food security challenge, yet the explosion in research efforts aimed at understanding effector biology over the last few decades has left us with a dichotomy in our knowledge. Due to early focus on a small number of pathosystems, whether due to experimental convenience or the economic impact of the disease, for some pathogens, such as M. oryzae, we have resolved three‐dimensional protein structures and know interacting surfaces of multiple effectors and their interactors. In other cases, important crop pathogens such as F. graminearum and the newly emerging pathogens Ramularia collo‐cygni and Corynespora cassiicola, although several hundred candidate effectors have been predicted, each lacks functional characterization (McGrann et al., 2016; Lopez et al., 2018).
The arrival of full genome sequencing almost two decades ago has been a double‐edged sword. Bioinformatic pipelines and the development of prediction software has sped up the refinement of putative effectors whilst simultaneously highlighting the vastness of the gene repertoires to be investigated. For effector characterization, the future efficiency not only depends on the development of ultrahigh‐throughput functional assays but also their use in combination with lower‐throughput novel and well‐established techniques such as QTL mapping and GWAS (Plissonneau et al., 2017).
Whilst multiple developments in effector discovery have increased our understanding of these enigmatic proteins, arguably the explosion in effector research can be attributed to the development of three approaches: genome sequencing, bespoke bioinformatic pipelines, and Agrobacterium‐mediated transient expression in planta. Armed with only an annotated genome, even understudied conifer‐infecting fungal pathogens can be screened for the presence of putative effector proteins (Raffaello and Asiegbu, 2017). With this in mind, genome reannotations and improvements to prediction algorithms continuously widen the pool of effector candidates available, especially in well‐studied crop pathogens (Zhong et al., 2017; Frantzeskakis et al., 2018). Therefore, perhaps the greatest roadblock to effector discovery is the accuracy of genome assembly and annotation, an issue that will take at least 5–10 years to resolve with the inclusion of pangenomes (Cissé and Stajich, 2019).
The genome annotation of multiple isolates through the construction of pathogen pangenomes allows for intraspecific genome analysis and will provide insight into the links between high polymorphisms and host specificity. The use of pangenome analyses has already led to the differentiation between core candidate effectors and novel candidate effectors in Z. tritici and M. oryzae (Singh et al., 2019; Badet et al., 2019). Machine‐learning‐based prediction tools as well as the robotic implementation of practical molecular techniques should help to fast track the progress from effector prediction to characterization. This anticipated progress will undoubtedly erode some of the disparity in our interspecies knowledge and lift the veil on the enigmatic filamentous phytopathogen effector repertoire. Many novel functions, locations, interactions, and generic underlying themes remain to be discovered.
ACKNOWLEDGMENTS
We would like to acknowledge the invaluable input given by friends and colleagues during the drafting of this manuscript, including Drs Catherine Walker and Ana Machado Wood. Special thanks go to Dr Jason Rudd for encouragement following the reading of the first draft and Amy Dodd for creating the figures. C.K. is funded by the UK Biotechnology and Biological Sciences Research Council (BBSRC) and the Lawes Trust, Doctoral Training Programme grant (BB/M008770/1). K.H.K. receives BBSRC grant‐aided support as part of the Institute Strategic Programme Designing Future Wheat Grant (BB/P016855/1). The PHI‐base is funded from two UK BBSRC grants BB/K020056/1 and BB/S020020/1.
Kanja C, Hammond‐Kosack KE. Proteinaceous effector discovery and characterization in filamentous plant pathogens. Molecular Plant Pathology. 2020;21:1353–1376. 10.1111/mpp.12980
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study.
REFERENCES
- Aguilera‐Galvez, C. , Champouret, N. , Rietman, H. , Lin, X. , Wouters, D. , Chu, Z. et al (2018) Two different R gene loci co‐evolved with Avr2 of Phytophthora infestans and confer distinct resistance specificities in potato. Studies in Mycology, 89, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, R.L. , Bittner‐Eddy, P.D. , Grenville‐Briggs, L.J. , Meitz, J.C. , Rehmany, A.P. , Rose, L.E. et al (2004) Host–parasite coevolutionary conflict between Arabidopsis and downy mildew. Science, 306, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Allen, R.L. , Meitz, J.C. , Baumber, R.E. , Hall, S.A. , Lee, S.C. , Rose, L.E. et al (2008) Natural variation reveals key amino acids in a downy mildew effector that alters recognition specificity by an Arabidopsis resistance gene. Molecular Plant Pathology, 9, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro, T.M. , Thilliez, G.J. , Motion, G.B. and Huitema, E. (2017) A perspective on CRN proteins in the genomics age: evolution, classification, delivery and function revisited. Frontiers in Plant Science, 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem, J. , Cuomo, C.A. , van Kan, J.A. , Viaud, M. , Benito, E.P. , Couloux, A. et al (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genetics, 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, R.G. , Deb, D. , Fedkenheuer, K. and McDowell, J.M. (2015) Recent progress in RXLR effector research. Molecular Plant‐Microbe Interactions, 28, 1063–1072. [DOI] [PubMed] [Google Scholar]
- Armenteros, J.J.A. , Tsirigos, K.D. , Sønderby, C.K. , Petersen, T.N. , Winther, O. , Brunak, S. et al (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature Biotechnology, 37, 420–423. [DOI] [PubMed] [Google Scholar]
- Armstrong, M.R. , Whisson, S.C. , Pritchard, L. , Bos, J.I. , Venter, E. , Avrova, A.O. et al (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proceedings of the National Academy of Sciences of the United States of America, 102, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, S. and Shirasu, K. (2015) Plant cells under siege: plant immune system versus pathogen effectors. Current Opinion in Plant Biology, 28, 1–8. [DOI] [PubMed] [Google Scholar]
- Avesani, L. , Marconi, G. , Morandini, F. , Albertini, E. , Bruschetta, M. , Bortesi, L. et al (2007) Stability of Potato virus X expression vectors is related to insert size: implications for replication models and risk assessment. Transgenic Research, 16, 587–597. [DOI] [PubMed] [Google Scholar]
- Aylward, J. , Steenkamp, E.T. , Dreyer, L.L. , Roets, F. , Wingfield, B.D. and Wingfield, M.J. (2017) A plant pathology perspective of fungal genome sequencing. IMA Fungus, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badet, T. , Oggenfuss, U. , Abraham, L. , McDonald, B.A. and Croll, D. (2020) A 19‐isolate reference‐quality global pangenome for the fungal wheat pathogen Zymoseptoria tritici. BMC Biology, 18, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballance, G. , Lamari, L. , Kowatsch, R. and Bernier, C. (1996) Cloning, expression and occurrence of the gene encoding the Ptr necrosis toxin from Pyrenophora tritici‐repentis . Molecular Plant Pathology On‐Line. [http://www.bspp.org.uk/mppol/]1996/1209ballance [Google Scholar]
- Bao, J. , Chen, M. , Zhong, Z. , Tang, W. , Lin, L. , Zhang, X. et al (2017) PacBio sequencing reveals transposable elements as a key contributor to genomic plasticity and virulence variation in Magnaporthe oryzae . Molecular Plant, 10, 1465–1468. [DOI] [PubMed] [Google Scholar]
- Birren, B. , Fink, G. and Lander, E. (2003) A white paper for fungal comparative genomics. Available at: https://www.broadinstitute.org/files/shared/fungi/fgi/FGI_02_whitepaper_2003.pdf [Accessed March 2020].
- Bolton, M.D. , van Esse, H.P. , Vossen, J.H. , de Jonge, R. , Stergiopoulos, I. , Stulemeijer, I.J. et al (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Molecular Microbiology, 69, 119–136. [DOI] [PubMed] [Google Scholar]
- Bos, J.I. , Armstrong, M.R. , Gilroy, E.M. , Boevink, P.C. , Hein, I. , Taylor, R.M. et al (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proceedings of the National Academy of Sciences of the United States of America, 107, 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutemy, L.S. , King, S.R. , Win, J. , Hughes, R.K. , Clarke, T.A. , Blumenschein, T.M. et al (2011) Structures of Phytophthora RXLR effector proteins a conserved but adaptable fold underpins functional diversity. Journal of Biological Chemistry, 286, 35834–35842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton, C. , King, R.C. , Chen, H. , Azhakanandam, K. , Bieri, S. , Hammond‐Kosack, K.E. et al (2018) Foxtail mosaic virus: A viral vector for protein expression in cereals. Plant Physiology, 177, 1352–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, J.K. , Mesarich, C.H. , Rees‐George, J. , Cui, W. , Fitzgerald, A. , Win, J. et al (2009) Candidate effector gene identification in the ascomycete fungal phytopathogen Venturia inaequalis by expressed sequence tag analysis. Molecular Plant Pathology, 10, 431–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon, T.C. , Bosch, J.A. , Sanchez, A.D. , Udeshi, N.D. , Svinkina, T. , Carr, S.A. et al (2018) Efficient proximity labeling in living cells and organisms with TurboID. Nature Biotechnology, 36, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brefort, T. , Tanaka, S. , Neidig, N. , Doehlemann, G. , Vincon, V. and Kahmann, R. (2014) Characterization of the largest effector gene cluster of Ustilago maydis . PLoS Pathogens, 10, e1003866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun‐Rasmussen, M. , Madsen, C.T. , Jessing, S. and Albrechtsen, M. (2007) Stability of barley stripe mosaic virus–induced gene silencing in barley. Molecular Plant‐Microbe Interactions, 20, 1323–1331. [DOI] [PubMed] [Google Scholar]
- Bundock, P. , Den Dulk‐Ras, A. , Beijersbergen, A. and Hooykaas, P. (1995) Trans‐kingdom T‐DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae . The EMBO Journal, 14, 3206–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg, H.A. , Harrison, S.J. , Joosten, M.H. , Vervoort, J. and de Wit, P.J. (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Molecular Plant‐Microbe Interactions, 19, 1420–1430. [DOI] [PubMed] [Google Scholar]
- van der Burgh, A.M. and Joosten, M.H. (2019) Plant immunity: thinking outside and inside the box. Trends in Plant Science, 24, 587–601. [DOI] [PubMed] [Google Scholar]
- Cantu, D. , Segovia, V. , Maclean, D. , Bayles, R. , Chen, X. , Kamoun, S. et al (2013) Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics, 14, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari, S. , Thilliez, G. , Ribot, C. , Chalvon, V. , Michel, C. , Jauneau, A. et al (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR‐Pia and AVR1‐CO39 by direct binding. The Plant Cell, 25, 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro‐Garcia, A. , Wilkinson, R.C. , Gimenez‐Ibanez, S. , Findlay, K. , Coffey, M.D. , Zipfel, C. et al (2011) The receptor‐like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana . PLoS ONE, 6, e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Tao, L. , Zeng, L. , Vega‐Sanchez, M.E. , Umemura, K. and Wang, G.‐L. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein–protein interactions in rice. Molecular Plant Pathology, 7, 417–427. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Songkumarn, P. , Venu, R.C. , Gowda, M. , Bellizzi, M. , Hu, J. et al (2012) Identification and characterization of in planta‐expressed secreted effector proteins from Magnaporthe oryzae that induce cell death in rice. Molecular Plant‐Microbe Interactions, 26, 191–202. [DOI] [PubMed] [Google Scholar]
- Cheng, Y. , Wu, K. , Yao, J. , Li, S. , Wang, X. , Huang, L. et al (2017) PST ha5a23, a candidate effector from the obligate biotrophic pathogen Puccinia striiformis f. sp. tritici, is involved in plant defense suppression and rust pathogenicity. Environmental Microbiology, 19, 1717–1729. [DOI] [PubMed] [Google Scholar]
- Ciuffetti, L.M. , Tuori, R.P. and Gaventa, J.M. (1997) A single gene encodes a selective toxin causal to the development of tan spot of wheat. The Plant Cell, 9, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cissé, O.H. and Stajich, J.E. (2019) FGMP: assessing fungal genome completeness. BMC Bioinformatics, 20, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo, C.A. , Güldener, U. , Xu, J.‐R. , Trail, F. , Turgeon, B.G. , di Pietro, A. et al (2007) The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science, 317, 1400–1402. [DOI] [PubMed] [Google Scholar]
- Dalio, R.J. , Herlihy, J. , Oliveira, T.S. , McDowell, J.M. and Machado, M. (2017) Effector biology in focus: a primer for computational prediction and functional characterization. Molecular Plant‐Microbe Interactions, 31, 22–33. [DOI] [PubMed] [Google Scholar]
- Dean, R.A. , Talbot, N.J. , Ebbole, D.J. , Farman, M.L. , Mitchell, T.K. , Orbach, M.J. et al (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature, 434, 980–986. [DOI] [PubMed] [Google Scholar]
- Dean, R. , van Kan, J.A. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , di Pietro, A. , Spanu, P.D. et al (2012) The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depotter, J.R. and Doehlemann, G. (2019) Target the core: durable plant resistance against filamentous plant pathogens through effector recognition. Pest Management Science, 76, 426–431. [DOI] [PubMed] [Google Scholar]
- Depotter, J.R. , Shi‐Kunne, X. , Missonnier, H. , Liu, T. , Faino, L. , van den Berg, G.C. et al (2019) Dynamic virulence‐related regions of the plant pathogenic fungus Verticillium dahliae display enhanced sequence conservation. Molecular Ecology, 28, 3482–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di, X. , Cao, L. , Hughes, R.K. , Tintor, N. , Banfield, M.J. and Takken, F.L. (2017) Structure–function analysis of the Fusarium oxysporum Avr2 effector allows uncoupling of its immune‐suppressing activity from recognition. New Phytologist, 216, 897–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei, A. , Schipper, K. , Rabe, F. , Ghosh, A. , Vincon, V. , Kahnt, J. et al (2011) Metabolic priming by a secreted fungal effector. Nature, 478, 395. [DOI] [PubMed] [Google Scholar]
- Doehlemann, G. , Linde, K.V.D. , Aßmann, D. , Schwammbach, D. , Hof, A. , Mohanty, A. et al (2009) Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathogens, 5, e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann, G. , Reissmann, S. , Aßmann, D. , Fleckenstein, M. and Kahmann, R. (2011) Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis‐induced tumour formation. Molecular Microbiology, 81, 751–766. [DOI] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Chen, Y. , Wang, Q. , Wang, X. et al (2008) Conserved C‐terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. The Plant Cell, 20, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J. , Rietman, H. and Vleeshouwers, V.G. (2014) Agroinfiltration and PVX agroinfection in potato and Nicotiana benthamiana . Journal of Visualized Experiments, 83, e50971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff, M.R. Jr., Grubbs, J. and Howell, E.E. (2011) Isothermal titration calorimetry for measuring macromolecule‐ligand affinity. Journal of Visualized Experiments, 55, e2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis, S. , Cuomo, C.A. , Lin, Y.‐C. , Aerts, A. , Tisserant, E. , Veneault‐Fourrey, C. et al (2011) Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proceedings of the National Academy of Sciences of the United States of America, 108, 9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse, H.P. , van't Klooster, J.W. , Bolton, M.D. , Yadeta, K.A. , van Baarlen, P. , Boeren, S. et al (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. The Plant Cell, 20, 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabro, G. , Steinbrenner, J. , Coates, M. , Ishaque, N. , Baxter, L. , Studholme, D.J. et al (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathogens, 7, e1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faino, L. , Seidl, M.F. , Shi‐Kunne, X. , Pauper, M. , van den Berg, G.C. , Wittenberg, A.H. et al (2016) Transposons passively and actively contribute to evolution of the two‐speed genome of a fungal pathogen. Genome Research, 26, 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y. and Tyler, B.M. (2016) Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Molecular Plant Pathology, 17, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farman, M.L. and Leong, S.A. (1998) Chromosome walking to the AVR1‐CO39 avirulence gene of Magnaporthe grisea: discrepancy between the physical and genetic maps. Genetics, 150, 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser, F. , Schiml, S. and Puchta, H. (2014) Both CRISPR/Cas‐based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana . The Plant Journal, 79, 348–359. [DOI] [PubMed] [Google Scholar]
- Fields, S. and Song, O.‐K. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annual Review of Phytopathology, 9, 275–296. [Google Scholar]
- Fouché, S. , Plissonneau, C. and Croll, D. (2018) The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Current Opinion in Microbiology, 46, 34–42. [DOI] [PubMed] [Google Scholar]
- Frantzeskakis, L. , Kracher, B. , Kusch, S. , Yoshikawa‐Maekawa, M. , Bauer, S. , Pedersen, C. et al (2018) Signatures of host specialization and a recent transposable element burst in the dynamic one‐speed genome of the fungal barley powdery mildew pathogen. BMC Genomics, 19, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain, H. , Joly, D.L. , Mireault, C. , Plourde, M.B. , Letanneur, C. , Stewart, D. et al (2018) Infection assays in Arabidopsis reveal candidate effectors from the poplar rust fungus that promote susceptibility to bacteria and oomycete pathogens. Molecular Plant Pathology, 19, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghareeb, H. , Zhao, Y. and Schirawski, J. (2019) Sporisorium reilianum possesses a pool of effector proteins that modulate virulence on maize. Molecular Plant Pathology, 20, 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey, D. , Böhlenius, H. , Pedersen, C. , Zhang, Z. , Emmersen, J. and Thordal‐Christensen, H. (2010) Powdery mildew fungal effector candidates share N‐terminal Y/F/WxC‐motif. BMC Genomics, 11, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golicz, A.A. , Bayer, P.E. , Bhalla, P.L. , Batley, J. and Edwards, D. (2019) Pangenomics comes of age: from bacteria to plant and animal applications. Trends in Genetics, 36, 132–145. [DOI] [PubMed] [Google Scholar]
- Gordon, T.R. (2017) Fusarium oxysporum and the fusarium wilt syndrome. Annual Review of Phytopathology, 55, 23–39. [DOI] [PubMed] [Google Scholar]
- Graciet, E. and Wellmer, F. (2010) The plant N‐end rule pathway: structure and functions. Trends in Plant Science, 15, 447–453. [DOI] [PubMed] [Google Scholar]
- de Guillen, K. , Ortiz‐Vallejo, D. , Gracy, J. , Fournier, E. , Kroj, T. and Padilla, A. (2015) Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathogens, 11, e1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guillen K., Lorrain C., Tsan P., Barthe P., Petre B., Saveleva N., Rouhier N., Duplessis S., Padilla A., Hecker A. (2019) Structural genomics applied to the rust fungus Melampsora larici‐populina reveals two candidate effector proteins adopting cystine knot and NTF2‐like protein folds. Scientific Reports, 9, 18084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L. , Cesari, S. , de Guillen, K. , Chalvon, V. , Mammri, L. , Ma, M. et al (2018) Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces. Proceedings of the National Academy of Sciences of the United States of America, 115, 11637–11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H. , Handsaker, R.E. , Cano, L.M. et al (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393. [DOI] [PubMed] [Google Scholar]
- Haupt, S. , Duncan, G.H. , Holzberg, S. and Oparka, K.J. (2001) Evidence for symplastic phloem unloading in sink leaves of barley. Plant Physiology, 125, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, S.Y. , Nomura, K. and Whittam, T.S. (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochimica et Biophysica Acta, 1694, 181–206. [DOI] [PubMed] [Google Scholar]
- Hillmer, R.A. , Tsuda, K. , Rallapalli, G. , Asai, S. , Truman, W. , Papke, M.D. et al (2017) The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genetics, 13, e1006639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, P. , Park, K.J. , Obayashi, T. , Fujita, N. , Harada, H. , Adams‐Collier, C.J. et al (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Research, 35, W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman, P.M. , Speijer, D. , Dekker, H.L. , de Koster, C.G. , Cornelissen, B.J. and Rep, M. (2007) The mixed xylem sap proteome of Fusarium oxysporum‐infected tomato plants. Molecular Plant Pathology, 8, 215–221. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Ma, L. , van Ooijen, G. , de Vroomen, M.J. , Cornelissen, B.J. , Takken, F.L. et al (2009) The effector protein Avr2 of the xylem‐colonizing fungus Fusarium oxysporum activates the tomato resistance protein I‐2 intracellularly. The Plant Journal, 58, 970–978. [DOI] [PubMed] [Google Scholar]
- Huet, J.‐C. and Pernollet, J.‐C. (1989) Amino acid sequence of cinnamomin, a new member of the elicitin family, and its comparison to cryptogein and capsicein. FEBS Letters, 257, 302–306. [DOI] [PubMed] [Google Scholar]
- Hurlburt, N.K. , Chen, L.‐H. , Stergiopoulos, I. and Fisher, A.J. (2018) Structure of the Cladosporium fulvum Avr4 effector in complex with (GlcNAc) 6 reveals the ligand‐binding mechanism and uncouples its intrinsic function from recognition by the Cf‐4 resistance protein. PLoS Pathogens, 14, e1007263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illergård, K. , Ardell, D.H. and Elofsson, A. (2009) Structure is three to ten times more conserved than sequence—a study of structural response in protein cores. Proteins: Structure Function, and Bioinformatics, 77, 499–508. [DOI] [PubMed] [Google Scholar]
- Irieda, H. , Inoue, Y. , Mori, M. , Yamada, K. , Oshikawa, Y. , Saitoh, H. et al (2019) Conserved fungal effector suppresses PAMP‐triggered immunity by targeting plant immune kinases. Proceedings of the National Academy of Sciences of the United States of America, 116, 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, R.H. , Tripathy, S. , Govers, F. and Tyler, B.M. (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proceedings of the National Academy of Sciences of the United States of America, 105, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323. [DOI] [PubMed] [Google Scholar]
- Jones, D.A. , Bertazzoni, S. , Turo, C.J. , Syme, R.A. and Hane, J.K. (2018) Bioinformatic prediction of plant–pathogenicity effector proteins of fungi. Current Opinion in Microbiology, 46, 43–49. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. and Thomma, B.P. (2009) Fungal LysM effectors: extinguishers of host immunity? Trends in Microbiology, 17, 151–157. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Kombrink, A. , Shinya, T. , Desaki, Y. , Bours, R. et al (2010) Conserved fungal LysM effector Ecp6 prevents chitin‐triggered immunity in plants. Science, 329, 953–955. [DOI] [PubMed] [Google Scholar]
- de Jonge, R. , Bolton, M.D. , Kombrink, A. , van den Berg, G.C. , Yadeta, K.A. and Thomma, B.P. (2013) Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Research, 23, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, M. , Vogelsang, R. , Cozijnsen, T.J. , Verberne, M.C. and de Wit, P. (1997) The biotrophic fungus Cladosporium fulvum circumvents Cf‐4‐mediated resistance by producing unstable AVR4 elicitors. The Plant Cell, 9, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, R.L. , Joshi, V. and Ow, D. (1990) BSMV genome mediated expression of a foreign gene in dicot and monocot plant cells. The EMBO Journal, 9, 2663–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. , Klucher, K.M. , Coffey, M.D. and Tyler, B.M. (1993) A gene encoding a host‐specific elicitor protein of Phytophthora parasitica . Molecular Plant‐Microbe Interactions, 6, 573–573. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , van West, P. , de Jong, A.J. , de Groot, K.E. , Vleeshouwers, V.G. and Govers, F. (1997) A gene encoding a protein elicitor of Phytophthora infestans is down‐regulated during infection of potato. Molecular Plant‐Microbe Interactions, 10, 13–20. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Honée, G. , Weide, R. , Laugé, R. , Kooman‐Gersmann, M. , de Groot, K. et al (1999) The fungal gene Avr9 and the oomycete gene inf1 confer avirulence to potato virus X on tobacco. Molecular Plant‐Microbe Interactions, 12, 459–462. [Google Scholar]
- Kämper, J. (2004) A PCR‐based system for highly efficient generation of gene replacement mutants in Ustilago maydis . Molecular Genetics and Genomics, 271, 103–110. [DOI] [PubMed] [Google Scholar]
- Kämper, J. , Kahmann, R. , Bölker, M. , Ma, L.‐J. , Brefort, T. , Saville, B.J. et al (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis . Nature, 444, 97. [DOI] [PubMed] [Google Scholar]
- van Kan, J.A. , van den Ackerveken, G. and de Wit, P. (1991) Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Molecular Plant‐Microbe Interactions, 4, 52–59. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , Kemen, A.C. , Rafiqi, M. , Hempel, U. , Mendgen, K. , Hahn, M. et al (2005) Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Molecular Plant‐Microbe Interactions, 18, 1130–1139. [DOI] [PubMed] [Google Scholar]
- Kerppola, T.K. (2008) Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annual Review of Biophysics, 37, 465–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw, M.J. and Talbot, N.J. (2009) Genome‐wide functional analysis reveals that infection‐associated fungal autophagy is necessary for rice blast disease. Proceedings of the National Academy of Sciences of the United States of America, 106, 15967–15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettles, G.J. , Bayon, C. , Canning, G. , Rudd, J.J. and Kanyuka, K. (2017) Apoplastic recognition of multiple candidate effectors from the wheat pathogen Zymoseptoria tritici in the nonhost plant Nicotiana benthamiana . New Phytologist, 213, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang, C.H. , Berruyer, R. , Giraldo, M.C. , Kankanala, P. , Park, S.Y. , Czymmek, K. et al (2010) Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell‐to‐cell movement. The Plant Cell, 22, 1388–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A. , Stein, E. and Kogel, K.‐H. (2018) RNA‐based disease control as a complementary measure to fight Fusarium fungi through silencing of the azole target cytochrome P450 lanosterol C‐14 α‐demethylase. European Journal of Plant Pathology, 152, 1003–1010. [Google Scholar]
- Kombrink, A. , Rovenich, H. , Shi‐Kunne, X. , Rojas‐Padilla, E. , van den Berg, G.C. , Domazakis, E. et al (2017) Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Molecular Plant Pathology, 18, 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner, T. , Kamoun, S. and Belhaj, K. (2018) CRISPR crops: plant genome editing toward disease resistance. Annual Review of Phytopathology, 56, 479–512. [DOI] [PubMed] [Google Scholar]
- Laugé, R. and de Wit, P.J. (1998) Fungal avirulence genes: structure and possible functions. Fungal Genetics and Biology, 24, 285–297. [DOI] [PubMed] [Google Scholar]
- Laugé, R. , Joosten, M.H. , van den Ackerveken, G.F. , van den Broek, H.W. and de Wit, P.J. (1997) The in planta‐produced extracellular proteins ECP1 and ECP2 of Cladosporium fulvum are virulence factors. Molecular Plant‐Microbe interactions, 10, 725–734. [Google Scholar]
- Lawrence, D.M. and Jackson, A. (2001) Requirements for cell‐to‐cell movement of Barley stripe mosaic virus in monocot and dicot hosts. Molecular Plant Pathology, 2, 65–75. [DOI] [PubMed] [Google Scholar]
- Leach, J.E. and White, F.F. (1996) Bacterial avirulence genes. Annual Review of Phytopathology, 34, 153–179. [DOI] [PubMed] [Google Scholar]
- Lee, W.S. , Hammond‐Kosack, K.E. and Kanyuka, K. (2012) Barley stripe mosaic virus‐mediated tools for investigating gene function in cereal plants and their pathogens: virus‐induced gene silencing, host‐mediated gene silencing, and virus‐mediated overexpression of heterologous protein. Plant Physiology, 160, 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.‐S. , Rudd, J.J. , Hammond‐Kosack, K.E. and Kanyuka, K. (2014) Mycosphaerella graminicola LysM effector‐mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Molecular Plant‐Microbe Interactions, 27, 236–243. [DOI] [PubMed] [Google Scholar]
- Li, J.‐F. , Bush, J. , Xiong, Y. , Li, L. and McCormack, M. (2011) Large‐scale protein–protein interaction analysis in Arabidopsis mesophyll protoplasts by split firefly luciferase complementation. PLoS ONE, 6, e27364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Li, J. , Wang, L. and Di, L.J. (2017) Proximity labeling of interacting proteins: application of BioID as a discovery tool. Proteomics, 17, 1700002. [DOI] [PubMed] [Google Scholar]
- Li, J. , Wang, Q. , Li, C. , Bi, Y. , Fu, X. and Wang, R. (2019) Novel haplotypes and networks of AVR‐Pik alleles in Magnaporthe oryzae . BMC Plant Biology, 19, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Song, T. , Zhang, X. , Yuan, H. , Su, L. , Li, W. et al (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nature Communications, 5, 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti, L. , Lanver, D. , Schweizer, G. , Tanaka, S. , Liang, L. Tollot, M. et al (2015) Fungal effectors and plant susceptibility. Annual Review of Plant Biology, 66, 513–545. [DOI] [PubMed] [Google Scholar]
- Lo Presti, L. , Zechmann, B. , Kumlehn, J. , Liang, L. , Lanver, D. Tanaka, S. et al (2017) An assay for entry of secreted fungal effectors into plant cells. New Phytologist, 213, 956–964. [DOI] [PubMed] [Google Scholar]
- Lopez, D. , Ribeiro, S. , Label, P. , Fumanal, B. , Venisse, J.‐S. , Kohler, A. et al (2018) Genome‐wide analysis of Corynespora cassiicola leaf fall disease putative effectors. Frontiers in Microbiology, 9, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, C. , Gonçalves Dos Santos, K.C. , Germain, H. , Hecker, A. and Duplessis, S. (2019) Advances in understanding obligate biotrophy in rust fungi. New Phytologist, 222, 1190–1206. [DOI] [PubMed] [Google Scholar]
- Lu, X. , Kracher, B. , Saur, I.M. , Bauer, S. , Ellwood, S.R. , Wise, R. et al (2016) Allelic barley MLA immune receptors recognize sequence‐unrelated avirulence effectors of the powdery mildew pathogen. Proceedings of the National Academy of Sciences of the United States of America, 113, E6486–E6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Lukasik, E. , Gawehns, F. and Takken, F.L. (2012) The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves In Bolton M. D. & Thomma B. P. H. J. (Eds.), Methods in Molecular Biology (Vol. 835, pp. 61–74). New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- Ma, L.‐S. , Wang, L. , Trippel, C. , Mendoza‐Mendoza, A. , Ullmann, S. , Moretti, M. et al (2018a) The Ustilago maydis repetitive effector Rsp3 blocks the antifungal activity of mannose‐binding maize proteins. Nature Communications, 9, 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Djavaheri, M. , Wang, H. , Larkan, N.J. , Haddadi, P. , Beynon, E. et al (2018b) Leptosphaeria maculans effector protein AvrLm1 modulates plant immunity by enhancing MAP kinase 9 phosphorylation. iScience, 3, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P. , Yang, L. , Esvelt, K.M. , Aach, J. , Guell, M. , Dicarlo, J.E. et al (2013) RNA‐guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, V.A. , Chu, A.L. , Scofield, S.R. and Ciuffetti, L.M. (2010) Intracellular expression of a host‐selective toxin, ToxA, in diverse plants phenocopies silencing of a ToxA‐interacting protein, ToxABP1. New Phytologist, 187, 1034–1047. [DOI] [PubMed] [Google Scholar]
- Maqbool, A. , Saitoh, H. , Franceschetti, M. , Stevenson, C. , Uemura, A. , Kanzaki, H. et al (2015) Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife, 4, e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrann, G.R. , Andongabo, A. , Sjökvist, E. , Trivedi, U. , Dussart, F. Kaczmarek, M. et al (2016) The genome of the emerging barley pathogen Ramularia collo‐cygni . BMC Genomics, 17, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, V. , Arentshorst, M. , El‐Ghezal, A. , Drews, A.‐C. , Kooistra, R. , van den Hondel, C.A. et al (2007) Highly efficient gene targeting in the Aspergillus niger kusA mutant. Journal of Biotechnology, 128, 770–775. [DOI] [PubMed] [Google Scholar]
- Miller, K.E. , Kim, Y. , Huh, W.‐K. and Park, H.‐O. (2015) Bimolecular fluorescence complementation (BiFC) analysis: advances and recent applications for genome‐wide interaction studies. Journal of Molecular Biology, 427, 2039–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera, G. , Giraldo, M.C. , Khang, C.H. , Coughlan, S. and Valent, B. (2009) Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1‐4 as biotrophy‐associated secreted proteins in rice blast disease. The Plant Cell, 21, 1273–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaung, T.E. , Saitoh, H. and Tsilo, T.J. (2017) Large‐scale molecular genetic analysis in plant‐pathogenic fungi: a decade of genome‐wide functional analysis. Molecular Plant Pathology, 18, 754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, A.N. , Ziemann, S. , Treitschke, S. , Aßmann, D. and Doehlemann, G. (2013) Compatibility in the Ustilago maydis–maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathogens, 9, e1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench, S.P. , Antonyuk, S.V. and Hasnain, S.S. (2019) The expanding toolkit for structural biology: synchrotrons, X‐ray lasers and cryoEM. IUCrJ, 6, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.‐R. , Dreze, M. , Epple, P. , Steinbrenner, J. , Moore, J. et al (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou, B.P.M. , Ahn, H.‐K. , Ding, P. , Redkar, A. , Brown, H. , Ma, Y. et al (2020) Estradiol‐inducible AvrRps4 expression reveals distinct properties of TIR‐NLR‐mediated effector‐triggered immunity. Journal of Experimental Botany, 71, 2186–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H. and Krogh, A. (1998) Prediction of signal peptides and signal anchors by a hidden Markov model. Proceedings of International Conference on Intelligent Systems for Molecular Biology, 6, 122–130. [PubMed] [Google Scholar]
- Ninomiya, Y. , Suzuki, K. , Ishii, C. and Inoue, H. (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end‐joining. Proceedings of the National Academy of Sciences of the United States of America, 101, 12248–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nødvig, C.S. , Nielsen, J.B. , Kogle, M.E. and Mortensen, U.H. (2015) A CRISPR‐Cas9 system for genetic engineering of filamentous fungi. PLoS ONE, 10, e0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowara, D. , Gay, A. , Lacomme, C. , Shaw, J. , Ridout, C. , Douchkov, D. et al (2010) HIGS: host‐induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . The Plant Cell, 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.‐K. , Young, C. , Lee, M. , Oliva, R. , Bozkurt, T.O. , Cano, L.M. et al (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi‐blb2. The Plant Cell, 21, 2928–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J. (1994) Plasmolysis: new insights into an old process. New Phytologist, 126, 571–591. [Google Scholar]
- Ortiz, D. , de Guillen, K. , Cesari, S. , Chalvon, V. , Gracy, J. , Padilla, A. et al (2017) Recognition of the Magnaporthe oryzae effector AVR‐Pia by the decoy domain of the rice NLR immune receptor RGA5. The Plant Cell, 29, 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, C. , van Themaat, E.V.L. , McGuffin, L.J. , Abbott, J.C. , Burgis, T.A. , Barton, G. et al (2012) Structure and evolution of barley powdery mildew effector candidates. BMC Genomics, 13, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington, H.G. , Jones, R. , Kwon, S. , Bonciani, G. , Thieron, H. , Chandler, T. et al (2019) The fungal ribonuclease‐like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathogens, 15, e1007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre, B. , Saunders, D.G. , Sklenar, J. , Lorrain, C. , Win, J. , Duplessis, S. et al (2015) Candidate effector proteins of the rust pathogen Melampsora larici‐populina target diverse plant cell compartments. Molecular Plant‐Microbe Interactions, 28, 689–700. [DOI] [PubMed] [Google Scholar]
- Petre, B. , Win, J. , Menke, F.L. and Kamoun, S. (2017) Protein–protein interaction assays with effector–GFP fusions in Nicotiana benthamiana In Periyannan S. (Eds.), Wheat Rust Diseases. Methods in Molecular Biology (Vol. 1659, pp. 85–98). New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- Pierleoni, A. , Martelli, P.L. and Casadio, R. (2008) PredGPI: a GPI‐anchor predictor. BMC Bioinformatics, 9, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliego, C. , Nowara, D. , Bonciani, G. , Gheorghe, D.M. , Xu, R. , Surana, P. et al (2013) Host‐induced gene silencing in barley powdery mildew reveals a class of ribonuclease‐like effectors. Molecular Plant‐Microbe Interactions, 26, 633–642. [DOI] [PubMed] [Google Scholar]
- Plissonneau, C. , Benevenuto, J. , Mohd‐Assaad, N. , Fouché, S. , Hartmann, F.E. and Croll, D. (2017) Using population and comparative genomics to understand the genetic basis of effector‐driven fungal pathogen evolution. Frontiers in Plant Science, 8, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic, G. , Gracy, J. , Périn, C. , Chiche, L. and Gelly, J.‐C. (2017) KNOTTIN: the database of inhibitor cystine knot scaffold after 10 years, toward a systematic structure modeling. Nucleic Acids Research, 46, D454–D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, M. , Yu, M. , Grayczyk, J. , Darben, L.M. , Rieker, M.E.G. , Seitz, J. et al (2019) Candidate effectors from Uromyces appendiculatus, the causal agent of rust on common bean, can be discriminated based on suppression of immune responses. Frontiers in Plant Science, 10, 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob, D. , Kamoun, S. and Gijzen, M. (2002) Expression of a Phytophthora sojae necrosis‐inducing protein occurs during transition from biotrophy to necrotrophy. The Plant Journal, 32, 361–373. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan, G.V. , Cook, N.M. , Bueno‐Sancho, V. , Lewis, C.M. , Persoons, A. , Mitiku, A.D. et al (2019) MARPLE, a point‐of‐care, strain‐level disease diagnostics and surveillance tool for complex fungal pathogens. BMC Biology, 17, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaello, T. and Asiegbu, F.O. (2017) Small secreted proteins from the necrotrophic conifer pathogen Heterobasidion annosum sl. (HaSSPs) induce cell death in Nicotiana benthamiana . Scientific Reports, 7, 8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele, S. and Kamoun, S. (2012) Genome evolution in filamentous plant pathogens: why bigger can be better. Nature Reviews Microbiology, 10, 417–430. [DOI] [PubMed] [Google Scholar]
- Raffaele, S. , Farrer, R.A. , Cano, L.M. , Studholme, D.J. , Maclean, D. , Thines, M. et al (2010) Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science, 330, 1540–1543. [DOI] [PubMed] [Google Scholar]
- Rehmany, A.P. , Gordon, A. , Rose, L.E. , Allen, R.L. , Armstrong, M.R. , Whisson, S.C. et al (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. The Plant Cell, 17, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep, M. , van der Does, H.C. , Meijer, M. , van Wijk, R. , Houterman, P.M. , Dekker, H.L. et al (2004) A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Molecular Microbiology, 53, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Rho, H.‐S. , Kang, S. and Lee, Y.‐H. (2001) Agrobacterium tumefaciens‐mediated transformation of the plant pathogenic fungus, Magnaporthe grisea . Molecules and Cells (Springer Science and Business Media BV), 12, 407–411. [PubMed] [Google Scholar]
- Ribot, C. , Cesari, S. , Abidi, I. , Chalvon, V. , Bournaud, C. , Vallet, J. et al (2013) The Magnaporthe oryzae effector AVR 1–CO39 is translocated into rice cells independently of a fungal‐derived machinery. The Plant Journal, 74, 1–12. [DOI] [PubMed] [Google Scholar]
- Ricci, P. , Bonnet, P. , Huet, J.C. , Sallantin, M. , Beavouis‐Cante, F. , Bruneteau, M. et al (1989) Structure and activity of proteins from pathogenic fungi Phytophthora eliciting necrosis and acquired resistance in tobacco. European Journal of Biochemistry, 183, 555–563. [DOI] [PubMed] [Google Scholar]
- Robin, G.P. , Kleemann, J. , Neumann, U. , Cabre, L. , Dallery, J.‐F. , Lapalu, N. et al (2018) Subcellular localization screening of Colletotrichum higginsianum effector candidates identifies fungal proteins targeted to plant peroxisomes, Golgi bodies, and microtubules. Frontiers in Plant Science, 9, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfs, A. P. (1985) Wheat and rye stem rust In Roelfs A. P. & Bushnell W. R. (Eds.), Diseases, Distribution, Epidemiology, and Control (pp. 3–37). San Diego: Harcourt Brace Jovanovich. [Google Scholar]
- Rooney, H.C.E. , Klooster, J.W.V.T. , van der Hoorn, R.A.L. , Joosten, M.H.A.J. , Jones, J.D.G. and de Wit, P.J.G.M. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf‐2‐dependent disease resistance. Science, 308, 1783–1786. [DOI] [PubMed] [Google Scholar]
- Roux, K.J. , Kim, D.I. , Raida, M. and Burke, B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. Journal of Cell Biology, 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac, C. , Lee W.S., Cambon F., Rudd J. J., King R.C., Marande W., et al (2018) Wheat receptor‐kinase‐like protein Stb6 controls gene‐for‐gene resistance to fungal pathogen Zymoseptoria tritici . Nature Genetics, 50, 368–374. [DOI] [PubMed] [Google Scholar]
- Saitoh, H. , Fujisawa, S. , Mitsuoka, C. , Ito, A. , Hirabuchi, A. , Ikeda, K. et al (2012) Large‐scale gene disruption in Magnaporthe oryzae identifies MC69, a secreted protein required for infection by monocot and dicot fungal pathogens. PLoS Pathogens, 8, e1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo, A. , Rutter, W. , Wang, S. , Akhunova, A. , Bolus, S. , Chao, S. et al (2017) Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science, 358, 1604–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Vallet, A. , Saleem‐Batcha, R. , Kombrink, A. , Hansen, G. , Valkenburg, D.‐J. , Thomma, B.P. et al (2013) Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLife, 2, e00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Vallet, A. , Fouché, S. , Fudal, I. , Hartmann, F.E. , Soyer, J.L. , Tellier, A. et al (2018a) The genome biology of effector gene evolution in filamentous plant pathogens. Annual Review of Phytopathology, 56, 21–40. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Vallet, A. , Hartmann, F.E. , Marcel, T.C. and Croll, D. (2018b) Nature's genetic screens: using genome‐wide association studies for effector discovery. Molecular Plant Pathology, 19, 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma, G.N. , Manning, V.A. , Ciuffetti, L.M. and Karplus, P.A. (2005) Structure of Ptr ToxA: an RGD‐containing host‐selective toxin from Pyrenophora tritici‐repentis . The Plant Cell, 17, 3190–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, D.G. , Breen, S. , Win, J. , Schornack, S. , Hein, I. , Bozkurt, T.O. et al (2012a) Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. The Plant Cell, 24, 3420–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, D.G. , Win, J. , Cano, L.M. , Szabo, L.J. , Kamoun, S. and Raffaele, S. (2012b) Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PLoS ONE, 7, e29847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur, I.M. , Bauer, S. , Kracher, B. , Lu, X. , Franzeskakis, L. , Müller, M.C. et al (2019a) Multiple pairs of allelic MLA immune receptor‐powdery mildew AVRA effectors argue for a direct recognition mechanism. eLife, 8, e44471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur, I.M. , Bauer, S. , Lu, X. and Schulze‐Lefert, P. (2019b) A cell death assay in barley and wheat protoplasts for identification and validation of matching pathogen AVR effector and plant NLR immune receptors. Plant Methods, 15, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski, J. , Mannhaupt, G. , Münch, K. , Brefort, T. , Schipper, K. , Doehlemann, G. et al (2010) Pathogenicity determinants in smut fungi revealed by genome comparison. Science, 330, 1546–1548. [DOI] [PubMed] [Google Scholar]
- Schornack, S. , van Damme, M. , Bozkurt, T.O. , Cano, L.M. , Smoker, M. , Thines, M. et al (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proceedings of the National Academy of Sciences of the United States of America, 107, 17421–17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottens‐Toma, I.M. and de Wit, P.J. (1988) Purification and primary structure of a necrosis‐inducing peptide from the apoplastic fluids of tomato infected with Cladosporium fulvum (syn. Fulvia fulva). Physiological and Molecular Plant Pathology, 33, 59–67. [Google Scholar]
- Schulz, B. , Banuett, F. , Dahl, M. , Schlesinger, R. , Schäfer, W. , Martin, T. et al (1990) The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain‐related motif. Cell, 60, 295–306. [DOI] [PubMed] [Google Scholar]
- Schuster, M. , Schweizer, G. and Kahmann, R. (2018) Comparative analyses of secreted proteins in plant pathogenic smut fungi and related basidiomycetes. Fungal Genetics and Biology, 112, 21–30. [DOI] [PubMed] [Google Scholar]
- See, P.T. , Marathamuthu, K. , Iagallo, E. , Oliver, R. and Moffat, C. (2018) Evaluating the importance of the tan spot ToxA–Tsn1 interaction in Australian wheat varieties. Plant Pathology, 67, 1066–1075. [Google Scholar]
- Seitner, D. , Uhse, S. , Gallei, M. and Djamei, A. (2018) The core effector Cce1 is required for early infection of maize by Ustilago maydis . Molecular Plant Pathology, 19, 2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, W. , Cao, M. , Leung, D. and Tyler, B.M. (2004) The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps 1b . Molecular Plant‐Microbe Interactions, 17, 394–403. [DOI] [PubMed] [Google Scholar]
- Singh, R.P. , Hodson, D.P. , Huerta‐Espino, J. , Jin, Y. , Bhavani, S. , Njau, P. et al (2011) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annual Review of Phytopathology, 49, 465–481. [DOI] [PubMed] [Google Scholar]
- Singh, P.K. , Mahato, A.K. , Jain, P. , Rathour, R. , Sharma, V. and Sharma, T.R. (2019) Comparative genomics reveals the high copy number variation of a retro transposon in different Magnaporthe isolates. Frontiers in Microbiology, 10, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. and Leong, S. (1994) Mapping of a Magnaporthe grisea locus affecting rice (Oryza sativa) cultivar specificity. Theoretical and Applied Genetics, 88, 901–908. [DOI] [PubMed] [Google Scholar]
- Sohn, K.H. , Lei, R. , Nemri, A. and Jones, J.D. (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana . The Plant Cell, 19, 4077–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. , Win, J. , Tian, M. , Schornack, S. , Kaschani, F. , Ilyas, M. et al (2009). Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proceedings of the National Academy of Sciences of the United States of America, 106, 1654–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer, J.L. , el Ghalid, M. , Glaser, N. , Ollivier, B. , Linglin, J. , Grandaubert, J. et al (2014) Epigenetic control of effector gene expression in the plant pathogenic fungus Leptosphaeria maculans . PLoS Genetics, 10, e1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Dodds, P.N. , Gardiner, D.M. , Manners, J.M. , Singh, K.B. and Taylor, J.M. (2015) Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathogens, 11, e1004806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Catanzariti, A.‐M. , Deboer, K. , Petre, B. , Gardiner, D.M. , Singh, K.B. et al (2017) LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Scientific Reports, 7, 44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Dodds, P.N. , Gardiner, D.M. , Singh, K.B. and Taylor, J.M. (2018) Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Molecular Plant Pathology, 19, 2094–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken, F.L. , Luderer, R. , Gabriëls, S.H. , Westerink, N. , Lu, R. , de Wit, P.J. et al (2000) A functional cloning strategy, based on a binary PVX‐expression vector, to isolate HR‐inducing cDNAs of plant pathogens. The Plant Journal, 24, 275–283. [DOI] [PubMed] [Google Scholar]
- Tanaka, S. , Brefort, T. , Neidig, N. , Djamei, A. , Kahnt, J. , Vermerris, W. et al (2014) A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. eLife, 3, e01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, S. , Djamei, A. , Presti, L.L. , Schipper, K. , Winterberg, S. , Amati, S. et al (2015) Experimental approaches to investigate effector translocation into host cells in the Ustilago maydis/maize pathosystem. European Journal of Cell Biology, 94, 349–358. [DOI] [PubMed] [Google Scholar]
- Thomas, S.W. , Rasmussen, S.W. , Glaring, M.A. , Rouster, J.A. , Christiansen, S.K. and Oliver, R.P. (2001) Gene identification in the obligate fungal pathogen Blumeria graminis by expressed sequence tag analysis. Fungal Genetics and Biology, 33, 195–211. [DOI] [PubMed] [Google Scholar]
- Tian, M. , Win, J. , Song, J. , van der Hoorn, R. , van der Knaap, E. and Kamoun, S. (2007) A Phytophthora infestans cystatin‐like protein targets a novel tomato papain‐like apoplastic protease. Plant Physiology, 143, 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas, A. , Feng, G. , Reeck, G. , Bockus, W. and Leach, J. (1990) Purification of a cultivar‐specific toxin from Pyrenophora tritici‐repentis, causal agent of tan spot of wheat. Molecular Plant‐Microbe Interactions, 3, 221–224. [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A. et al (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Research, 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, R.S. (2005) After the famine: plant pathology, Phytophthora infestans, and the late blight of potatoes, 1845–1960. Historical Studies in the Physical and Biological Sciences, 35, 341–370. [Google Scholar]
- Tyler, B.M. (2017) The fog of war: how network buffering protects plants’ defense secrets from pathogens. PLoS Genetics, 13, e1006713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya, N.M. , Mago, R. , Staskawicz, B.J. , Ayliffe, M.A. , Ellis, J.G. and Dodds, P.N. (2014) A bacterial type III secretion assay for delivery of fungal effector proteins into wheat. Molecular Plant‐Microbe Interactions, 27, 255–264. [DOI] [PubMed] [Google Scholar]
- Urban, M. , Cuzick, A. , Seager, J. , Wood, V. , Rutherford, K. , Venkatesh, S.Y. et al (2020) PHI‐base: the pathogen–host interactions database. Nucleic Acids Research, 48, D613–D620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ackerveken, G. , Dunn, R. , Cozijnsen, A. , Vossen, J. , van den Broek, H. and de Wit, P. (1994) Nitrogen limitation induces expression of the avirulence gene avr9 in the tomato pathogen Cladosporium fulvum . Molecular and General Genetics, 243, 277–285. [DOI] [PubMed] [Google Scholar]
- Van den Ackerveken, G.F. , van Kan, J.A. and de Wit, P.J. (1992) Molecular analysis of the avirulence gene avr9 of the fungal tomato pathogen Cladosporium fulvum fully supports the gene‐for‐gene hypothesis. The Plant Journal, 2, 359–366. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A. , Laurent, F. , Roth, R. and de Wit, P.J. (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr 9/Cf‐9‐induced and Avr 4/Cf‐4‐induced necrosis. Molecular Plant‐Microbe Interactions, 13, 439–446. [DOI] [PubMed] [Google Scholar]
- Villalba, F. , Collemare, J. , Landraud, P. , Lambou, K. , Brozek, V. , Cirer, B. et al (2008) Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non‐homologous end joining. Fungal Genetics and Biology, 45, 68–75. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G. and Oliver, R.P. (2014) Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Molecular Plant‐Microbe Interactions, 27, 196–206. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G. , Raffaele, S. , Vossen, J.H. , Champouret, N. , Oliva, R. , Segretin, M.E. et al (2011) Understanding and exploiting late blight resistance in the age of effectors. Annual Review of Phytopathology, 49, 507–531. [DOI] [PubMed] [Google Scholar]
- Wainberg, M. , Sinnott‐Armstrong, N. , Mancuso, N. , Barbeira, A.N. , Knowles, D.A. , Golan, D. et al (2019) Opportunities and challenges for transcriptome‐wide association studies. Nature Genetics, 51, 592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. and Dean, R.A. (2020) Movement of small RNAs in and between plants and fungi. Molecular Plant Pathology, 21, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.‐I.‐A. , Gunčar, G. , Forwood, J.K. , Teh, T. , Catanzariti, A.‐M. , Lawrence, G.J. et al (2007) Crystal structures of flax rust avirulence proteins AvrL567‐A and‐D reveal details of the structural basis for flax disease resistance specificity. The Plant Cell, 19, 2898–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Wang, J. , Hu, M. , Wu, S. , Qi, J. , Wang, G. et al (2019) Ligand‐triggered allosteric ADP release primes a plant NLR complex. Science, 364, eaav5868. [DOI] [PubMed] [Google Scholar]
- Welti, R. and Wang, X. (2004) Lipid species profiling: a high‐throughput approach to identify lipid compositional changes and determine the function of genes involved in lipid metabolism and signaling. Current Opinions in Plant Biology, 7, 337–344. [DOI] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. et al (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Win, J. , Morgan, W. , Bos, J. , Krasileva, K.V. , Cano, L.M. , Chaparro‐Garcia, A. et al (2007) Adaptive evolution has targeted the C‐terminal domain of the RXLR effectors of plant pathogenic oomycetes. The Plant Cell, 19, 2349–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller, L. , Maqbool, A. and Banfield, M.J. (2013) On the front line: structural insights into plant–pathogen interactions. Nature Reviews Microbiology, 11, 761–776. [DOI] [PubMed] [Google Scholar]
- de Wit, P.J. (1997) Pathogen avirulence and plant resistance: a key role for recognition. Trends in Plant Science, 2, 452–458. [Google Scholar]
- de Wit, P.J. (2016) Cladosporium fulvum effectors: weapons in the arms race with tomato. Annual Review of Phytopathology, 54, 1–23. [DOI] [PubMed] [Google Scholar]
- de Wit, P.J. , Hofman, A.E. , Velthuis, G.C. and Kuć, J.A. (1985) Isolation and characterization of an elicitor of necrosis isolated from intercellular fluids of compatible interactions of Cladosporium fulvum (syn. Fulvia fulva) and tomato. Plant Physiology, 77, 642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit, P.J. , Buurlage, M.B. and Hammond, K.E. (1986) The occurrence of host‐, pathogen‐ and interaction‐specific proteins in the apoplast of Cladosporium fulvum (syn. Fulvia fulva) infected tomato leaves. Physiological and Molecular Plant Pathology, 29, 159–172. [Google Scholar]
- Yaeno, T. , Li, H. , Chaparro‐Garcia, A. , Schornack, S. , Koshiba, S. , Watanabe, S. et al (2011) Phosphatidylinositol monophosphate‐binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proceedings of the National Academy of Sciences of the United States of America, 108, 14682–14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Liu, L. , Wang, Y. , Wang, C. , Yan, J. , Liu, Y. et al (2017) Overexpression of BAS1 in rice blast fungus can promote blast fungus growth, sporulation and virulence in planta. Saudi Journal of Biological Sciences, 24, 1884–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K. , Saitoh, H. , Fujisawa, S. , Kanzaki, H. , Matsumura, H. , Yoshida, K. et al (2009) Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae . The Plant Cell, 21, 1573–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino, K. , Irieda, H. , Sugimoto, F. , Yoshioka, H. , Okuno, T. and Takano, Y. (2012) Cell death of Nicotiana benthamiana is induced by secreted protein NIS1 of Colletotrichum orbiculare and is suppressed by a homologue of CgDN3. Molecular Plant‐Microbe Interactions, 25, 625–636. [DOI] [PubMed] [Google Scholar]
- Yuan, M. , Jiang, Z. , Bi, G. , Nomura, K. , Liu, M. , He, S.Y. et al (2020) Pattern‐recognition receptors are required for NLR‐mediated plant immunity. bioRxiv. [preprint]. 10.1101/2020.04.10.031294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Dodds, P.N. and Bernoux, M. (2017) What do we know about NOD‐like receptors in plant immunity? Annual Review of Phytopathology, 55, 205–229. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Song, G. , Lal, N.K. , Nagalakshmi, U. , Li, Y. , Zheng, W. et al (2019) TurboID‐based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor‐mediated immunity. Nature Communications, 10, 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Z. , Marcel, T.C. , Hartmann, F.E. , Ma, X. , Plissonneau, C. , Zala, M. et al (2017) A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytologist, 214, 619–631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.


