Abstract
Epidemiological studies show that SARS-CoV-2 infection leads to severe symptoms only in a fraction of patients, but the determinants of individual susceptibility to the virus are still unknown. The major histocompatibility complex (MHC) class I exposes viral peptides in all nucleated cells and is involved in the susceptibility to many human diseases. Here, we use artificial neural networks to analyze the binding of SARS-CoV-2 peptides with polymorphic human MHC class I molecules. In this way, we identify two sets of haplotypes present in specific human populations: the first displays weak binding with SARS-CoV-2 peptides, while the second shows strong binding and T cell propensity. Our work offers a useful support to identify the individual susceptibility to COVID-19 and illustrates a mechanism underlying variations in the immune response to SARS-CoV-2. A record of this paper’s transparent peer review process is included in the Supplemental Information.
Keywords: SARS-CoV-2, HLA, haplotypes, artificial neural networks, T cell propensity, peptides
Highlights
-
•
Binding of SARS-CoV-2 peptides to HLA molecules is computed
-
•
Weakly or strongly binding haplotypes are identified in human populations
-
•
Results explain variations in the individual immune response to SARS-CoV-2
The response to SARS-CoV-2 infection differs from person to person, with some patients developing more severe symptoms than others. In this paper, Caterina La Porta and Stefano Zapperi show that the immune recognition of SARS-CoV-2 viral peptides differs widely among individuals and could thus explain why they may respond differently to the virus.
Introduction
SARS-CoV-2, the coronavirus causing the COVID-19 pandemic, is the seventh coronavirus known to infect humans. SARS-CoV, MERS-CoV, and SARS-CoV-2 can cause severe disease, whereas HCoV-HKU1, HCoV-NL63, HCoV-OC43, and HCoV-229E are associated with mild symptoms (Corman et al., 2018). For a successful infection, multiple elements of the host immune response must be overcome, including both innate and adaptive immunities (Mandl et al., 2015). The human leukocyte antigen (HLA) system or the major histocompatibility complex (MHC) is a very polymorphic region of the human genome, which plays an important role in the individual genetic susceptibility to human diseases (Dendrou et al., 2018). For example, in infectious diseases, HIV infection was shown to be highly correlated with HLA-A∗29, HLA-B∗35, and HLA-B∗57 (Hill, 1998; Mallal et al., 2002; Carrington et al., 1999; Goulder and Watkins, 2008; Mekue et al., 2019; Valenzuela-Ponce et al., 2018), and H1N1 flu was shown to be associated with several HLAs (Falfán-Valencia et al., 2018; Luckey et al., 2019). Association between disease severity and HLA was also reported in SARS-CoV patients (Lin et al., 2003; Ng et al., 2004; Chen et al., 2006; Keicho et al., 2009; Spìnola, 2016).
According to the structure and function of its genes, the human MHC has been classified into three main regions: class I, class II, and class III. The class I regions are located on the most telomeric part of the human MHC and include three highly polymorphic HLA genes, known as classical (class Ia: HLA-A, HLA-B, and HLA-C), and three lowly polymorphic HLA genes, known as non-classical (class Ib: HLA-E, HLA-F, and HLA-G) (Shiina et al., 2009). These molecules display small protein fragments at the cell surface, mostly originated in the cytosol. Thus, when a cell expresses foreign proteins, due, for example, to a viral infection, peptides created in the cytosol through proteasome-dependent processes could bind MHC class I. This MHC-peptide complex can then be exposed on the cellular membrane, triggering an immune response if it is recognized by CD8+ T cells (Maffei et al., 1997; Goldberg and Rizzo, 2015). Among HLA class I, HLA-B is the most polymorphic classical class I gene, with 4,077 alleles identified to date in different human populations, followed by HLA-A (3,285 alleles) and HLA-C (2,801 alleles) (González-Galarza et al., 2015).
Due to the rapid spread of the SARS-CoV-2 virus, a crucial question is to understand why there is individual susceptibility to the virus in the population. Epidemiological studies show that only a fraction of the infected individuals experience severe respiratory symptoms due to SARS-CoV-2 (Wu et al., 2020), with an infection fatality ratio estimated for China at around 0.6% and increasing with age (Verity et al., 2020). Another recent work estimated that in France, on an average, 2.6% of the infected individuals were hospitalized, while 0.53% died, again with dependence on the age of the patient (Salje et al., 2020). In light of these results, it would be extremely important to identify possible susceptible subjects in advance to protect them with adequate prevention strategies (Lipsitch et al., 2020).
In this paper, we propose a method to identify the dependence on HLA class I polymorphic alleles of the individual immune response to SARS-CoV-2. We focus on this class of the MHC, since it is expressed by all nucleated cells, including antigen-presenting cells. In practice, we estimate the aggressiveness of COVID-19 based on the compatibility between the specific HLA I polymorphism and SARS-CoV-2 peptides. Because experimental characterization of neoantigens is costly and time consuming, there is a growing effort in the development of computational methods that are able to predict peptide-MHC binding and the subsequent immune response. Supervised neural network machine learning approaches are currently showing an increasing performance and are widely used as in silico epitope prediction tools (Paul et al., 2020; Jurtz et al., 2017; O’Donnell et al., 2018). Here, we use two of these epitope prediction algorithms to compute binding affinities between SARS-CoV-2 peptides and 79 HLA class I. Similar calculations are performed to identify peptides for vaccine development (Campbell et al., 2020).
We compare our predictions for SARS-CoV-2 with analogous predictions for SARS-CoV and HCoV-OC43, a coronavirus responsible for the common cold. We also assess the stability and T cell propensity of these peptides for a smaller number of HLA alleles (Trolle and Nielsen, 2014). Using this method, we identify a set of weakly binding haplotypes and assess their prevalence in specific human subpopulations, as well as a set of strongly binding haplotypes for which we also compute peptide stability and T cell propensity (Trolle and Nielsen, 2014). All together, our strategy paves the way to the development of a general screening method to assess individual COVID-19 susceptibility in the population.
Results
To compute the binding affinities of coronavirus peptides, we combine the predictions of two state-of-the-art methods (Paul et al., 2020): netMHCpan (Jurtz et al., 2017) and MHCflurry (O’Donnell et al., 2018), both based on artificial neural networks. The combination of the two methods allows us to have a more robust result that is independent of the artificial neural networks used. We consider 79 common polymorphic HLA class I alleles supported by both methods and combine their predictions for the binding affinities for peptides of lengths 8–11. These 79 HLA alleles are present in a considerable fraction of the human population as illustrated in Figure S1. We scan peptides that are produced by proteasome degradation (Nielsen et al., 2005) considering only the structural proteins of SARS-CoV-2, which are the most abundant proteins in coronaviruses (Bar-On et al., 2020). We then compared the results obtained from the structural proteins of SARS-CoV and HCOV-OC43.
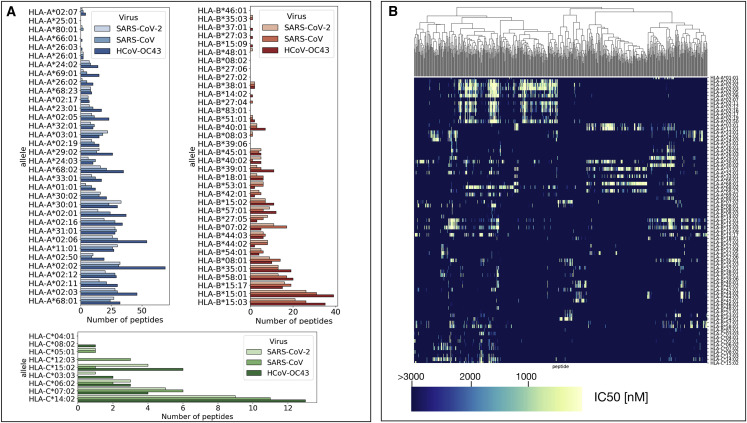
As shown in Figures S2–S4 for HLA-A, HLA-B, and HLA-C alleles and SARS-CoV-2 peptides, binding affinities are broadly distributed with a peak at high affinities so that the majority of peptides display weak binding to the HLA. Furthermore, the distributions differ between the various alleles (Figures S2–S4), confirming the presence of a heterogeneous binding pattern. To encapsulate the binding affinity distributions into a simple parameter, we counted all the peptides displaying a strong binding affinity () for each of the 79 alleles. We carried out the same analysis for all the three coronaviruses. Figure 1 A displays the number of strongly binding peptides for each allele showing that there is a close similarity between SARS-CoV-2 and SARS-CoV. In particular, alleles with a few strongly binding peptides in SARS-CoV-2 also display small numbers in SARS-CoV, while alleles with many strongly binding peptides in SARS-CoV-2 also show many strong peptides in SARS-CoV (Figure 1A). We can also observe similarities between SARS-CoV-2 and HCoV-OC43, but typically, HCoV-OC43 displays more strongly binding peptides than SARS-CoV-2 or SARS-CoV (see Figure 1A).
Figure 1.
Characterization of the Binding Heterogeneity of SARS-CoV-2 and SARS-CoV Peptides Are Similar and Differs for HCoV-OC43
(A) The number of strongly binding peptides () for SARS-Cov-2, SARS-Cov, and HCOV-OC43 estimated for 79 class I HLA alleles by combining predictions from netMHCpan and MHCflurry.
(B) The binding affinities () of SARS-Cov-2 peptides are shown in the clustered colormap for 79 class I HLA alleles. Only peptides with at least one binding affinity smaller than 1,000 nM are included.
To confirm that our results do not depend on the particular cutoff chosen, we also repeated the analysis with a smaller cutoff for strong binding (). In Figure S5, we compare the number of strongly binding peptides in SARS-CoV-2 for the two different cutoffs. The outcome is very similar, apart from small quantitative differences. To obtain a cutoff-independent assessment of the binding of viral peptides to each HLA molecule, we computed a total binding affinity by weighting the binding affinity of all the peptides, as described in the STAR Methods section. The values of for the binding among SARS-CoV-2, SARS-CoV, and HCoV-OC43 peptides to all the considered HLA molecules are reported in Figure S6. Comparing the patterns in Figures 1A and S6, we can see that the HLA molecules with few strongly binding peptides are also those with higher values of , confirming the robustness of our results.
A visual representation of how much strongly binding peptides are shared among different HLA alleles is provided in Figure 1B for the case of SARS-CoV-2. The figure shows that some peptides display strong affinity (in yellow) for a number of HLA molecules, but in general, peptides only bind strongly to relatively small numbers of HLA molecules, highlighting the heterogeneity of MHC-peptide interactions across human haplotypes.
To understand the similarities between SARS-CoV-2 and SARS-CoV in more depth, we report in Figure S7A peptides that are common to both viruses and that bind strongly to more than one HLA molecule. We observe that some peptides bind strongly to up to 8 HLA molecules. Conversely, for each HLA molecule, there are strongly binding peptides that are unique to SARS-CoV-2 or SARS-CoV. The numbers of these peptides are summarized in Figure S7B for each HLA molecule.
We then analyzed the stability between peptides and HLA class I as well as T cell propensity. To this end, we used NetTepi (Trolle and Nielsen, 2014) to find the putative T cell epitopes for SARS-CoV-2 and SARS-CoV considering all 13 alleles available for this method. These alleles are widely frequent in human populations: the HLA-A alleles are present in around 60% of the populations, while the HLA-B alleles are present in around 30% of the populations. As shown in Figure S8A, the number of highly ranked peptides are very similar in the case of SARS-CoV-2 and SARS-CoV. Highly ranked peptides that are common to the two coronaviruses are reported in Figure S8B, together with their rank. In Figure S9, we display distributions of binding stability, T cell propensity, and T cell epitope score for the 13 supported HLA alleles and all the strongly binding peptides previously identified in SARS-CoV-2. By ranking the HLA molecules as based on the T cell epitope score, we can identify the list of HLA molecules that are most likely to bind SARS-CoV-2 peptides that are recognized by T cells.
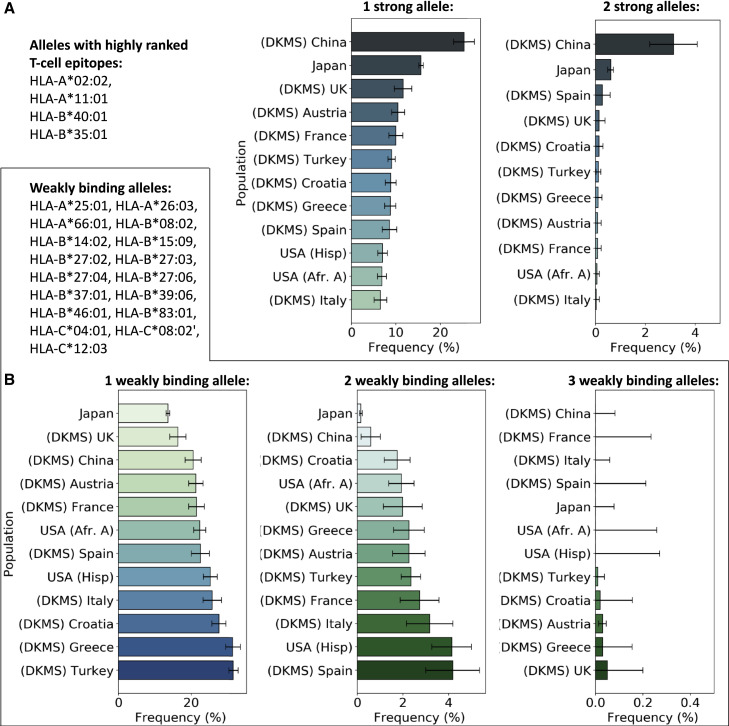
Having characterized the binding propensity of each molecule to SARS-CoV-2 peptides, we thus investigated how individuals from different human populations are likely to respond to SARS-CoV-2 infection in terms of peptide presentation by HLA class I. To this end, we collected haplotype frequencies from different human populations (i.e., Europeans, Chinese, Japanese, Hispanic, and African Americans) and inspected the prevalence of weakly binding molecules, defined as those who display no strongly binding peptide from SARS-CoV-2 structural proteins. This list included: HLA-A∗25:01, HLA-A∗26:03, HLA-A∗66:01, HLA-B∗08:02, HLA-B∗14:02, HLA-B∗15:09, HLA-B∗27:02, HLA-B∗27:03, HLA-B∗27:04, HLA-B∗27:06, HLA-B∗37:01, HLA-B∗39:06, HLA-B∗46:01, HLA-B∗83:01, HLA-C∗04:01, HLA-C∗08:02, and HLA-C∗12:03. Since HLA is codominant and all the alleles (A, B, and C) were expressed by each individual, we determined the haplotype that contains three, two, or one of the alleles contained in the list. We then plotted their prevalence in the different human populations (see Figure 2 B). The results showed that haplotypes with three weakly binding alleles are generally quite rare, amounting to up to two individuals over 1,000. The frequency of haplotypes with two weakly binding alleles is around 1%–4% depending on the populations, with the exception of Japan, where those haplotypes amount only to 0.16%, and Germans of Chinese origin, where this frequency is 0.59%. Finally, haplotypes with only one weakly binding peptide were more common, showing frequencies of around 20% with small variations among different populations.
Figure 2.
Response to SARS-CoV-2 across Human Populations
(A) Frequencies of haplotypes containing one or two strong alleles, defined as those with highly ranked T cell epitopes.
(B) Frequencies of haplotypes containing one, two, and three weakly binding alleles for SARS-CoV-2. Error bars are estimated 95% confidence intervals.
We finally report the frequency of haplotypes containing either one or two of the HLA alleles that are most likely to bind SARS-CoV2 peptides and be recognized by T cells. Figure 2A shows variations of the frequency in the populations, with Chinese and Japanese displaying the highest frequency of these haplotypes.
Discussion
The possibility to screen the population and predict a score of aggressiveness for each specific individual is a critical issue to develop personalized therapeutic strategies and to mitigate the effects of the infection in the shortest time. To reach this final goal, we focused on HLA class I, which is involved in presenting viral peptides to CD8+ T cells, mounting the immune response. A more complete picture of the immune response could be obtained by studying HLA class II molecules, but the performance of peptide-HLA class II binding prediction algorithms is still inferior to that of HLA class I predictors (Andreatta et al., 2018).
SARS-CoV-2 and SARS-CoV share 80% of the genome, while the similarity between SARS-CoV-2 and HCOV-OC43 is only 50% (Bar-On et al., 2020). We have thus compared the variations in binding affinities between coronavirus-derived peptides and a large number of HLA class I molecules for SARS-CoV-2, SARS-CoV, and HCoV-OC43. Our results show that the binding patterns are similar for SARS-CoV-2 and SARS-CoV, while they differ more for HCoV-OC43. We then identified a list of HLA class I alleles, where the binding with SARS-CoV-2 peptides is particularly weak. This means that these HLA alleles have a smaller probability to immediately start an adaptive immune response. In contrast, we also identified the list of HLA alleles whose binding with SARS-CoV-2 peptides is particularly strong and are most likely to activate a T cell response.
Our results clearly show the heterogeneity of the human population in responding to SARS-CoV-2 infection. To quantify this heterogeneity, we computed the frequency of haplotypes that are predicted to display a weak SARS-CoV-2 peptide-HLA class I binding in different human populations. In particular, we measured the prevalence of haplotypes that contain one, two, or three weakly binding alleles. Individuals with these haplotypes are likely to display a weaker immune response to SARS-Cov-2 infection. In this way, we developed a clear parameter that can be useful to screen the population.
Earlier studies in SARS-CoV revealed association between the presence of HLA-B∗46:01, one of the weakly binding alleles in our list, and the observation of severe symptoms in a cohort of Taiwanese patients (Lin et al., 2003). Similar associations were found with HLA-B∗07:01 (Keicho et al., 2009) and HLA-C∗08-01 (Chen et al., 2006) that are, however, not among the alleles we were able to study with our method. Since SARS-CoV-2 and SARS-CoV share most of the genome and we show that they display similar HLA binding profiles, it is expected that the weakly binding HLA molecules are similar for both viruses. While these early studies confirm the relevance of our approach, in light of our results it would be more appropriate to investigate the correlations between disease severity and the complete haplotype, instead of focusing on individual alleles.
Furthermore, we also identified the HLA haplotypes associated with a strong combined peptide affinity, stability, and T cell propensity and measured their prevalence in different human populations. We found that these haplotypes are more present in Asian populations. This might be a relevant parameter to study the diffusion of the disease across the world. Simulations of diffusion of the virus might take this effect into account. Altogether, our strategy could be the basis to develop individualized tests to assess the immune susceptibility to COVID-19 in the population. To reach this goal, it would be important to extend our analysis also to HLA class II molecules.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and Algorithms | ||
| netMHCpan 4.0 | Jurtz et al., 2017 | https://services.healthtech.dtu.dk/service.php?NetMHCpan-4 |
| MHCflurry | O'Donnell et al., 2018 | https://github.com/openvax/mhcflurry |
| NetChop 3.1 | Nielsen et al., 2005 | http://www.cbs.dtu.dk/services/NetChop/ |
| NetTepi | Trolle and Nielsen, 2014 | http://www.cbs.dtu.dk/services/NetTepi/ |
| Code for analysis pipeline | This paper | https://github.com/ComplexityBiosystems/hla-covid |
| Other | ||
| Sequence data, statistical analyses, and raw data | This paper | https://github.com/ComplexityBiosystems/hla-covid |
Resource Availability
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Caterina A. M. La Porta (caterina.laporta@unimi.it).
Materials Availability
This study did not generate new unique reagents or materials.
Data and Code Availability
The source codes generated to obtain the results presented in this paper are available at https://github.com/ComplexityBiosystems/hla-covid. Binding affinities for SARS-CoV-2, SARS-CoV and HCoV-OC43 are reported in Data S1. The protein sequences used in this paper are also available at https://github.com/ComplexityBiosystems/hla-covid/. Haplotype frequencies for different population are retrieved from the Allele Frequency Net Database (http://www.allelefrequencies.net/) (González-Galarza et al., 2015) and available at https://github.com/ComplexityBiosystems/hla-covid/.
Method Details
Calculation of Binding Affinities for Individual Peptides
We downloaded the fasta sequences for SARS-CoV-2 (GenBank: MN908947.3), SARS-CoV (NCBI Reference Sequence: NC_004718.3) and HCoV-OC43 (NCBI Reference Sequence: NC_006213.1). We restrict our analysis to the most abundant structural proteins (Bar-On et al., 2020): S,N,E,M for SARS-Cov and SARS-Cov-2 and S,N,E,M,HE for HCOV-OC43. In order to estimate binding affinities for peptides, we combine two recent algorithms based on artificial neural networks (ANN): netMHCpan 4.0 (Jurtz et al., 2017) and MHCflurry (O’Donnell et al., 2018). NetMHCpan uses a pan-allele approach to provide predictions for binding affinities of peptides to any MHC molecule by an ANN trained on a combination of more than 180000 quantitative binding data (Jurtz et al., 2017). MHCflurry uses instead an allele specific algorithm where each MHC allele is associated with 8-16 neural networks trained on affinity measurements (O’Donnell et al., 2018). Here, we run netMHCpan 4.0 and MHCflurry on a set of 79 HLA-A, HLA-B and HLA-C alleles supported by both algorithm. We run netMHCpan 4.0 predictions on the DTU server (https://services.healthtech.dtu.dk/service.php?NetMHCpan-4.0) while MHCflurry predictions are obtained using the epitopepredict python code (https://github.com/dmnfarrell/epitopepredict). In both cases, we scan all the peptides of lengths 8-11 for the proteins of interest. We only consider peptides that are likely to be produced by proteasome degradation. To this end, we employ NetChop 3.1 (Nielsen et al., 2005) a neural network based algorithm that scans proteins for probable cleavage sites of the human proteasome. We next compare the predictions for the binding affinities obtained by netMHCpan 4.0 and MHCflurry for each peptide and MHC allele. As shown in Figure S10, there is a strong correlation between the two predictions but in some cases the two predictions sometimes display large differences. We discard these values considering only peptides for which , where and are the predictions for binding affinity () obtained by the two algorithms. The binding affinity is then taken to be the average of and . Finally, peptides for which both and are smaller than 1000 nM are defined as strongly binding. We thus count the number of strongly binding peptides for each allele.
Calculation of the Total Binding Affinity to HLA Molecules
Consider a set of n peptides with concentrations with that can bind with HLA molecules with dissociation constants . We denote by the concentration of free HLA molecules and by the concentration of HLA molecules bound to a peptide i. According to the law of mass action, we have that
| (Equation 1) |
The probability for a HLA molecule to be bound by any peptide i can be written as
| (Equation 2) |
For peptides with uniform concentration , we can write
| (Equation 3) |
where is a measure of the total binding affinity of all the peptides to a given HLA molecule. To estimate , we use the predicted binding affinities as a proxy for the dissociation constants . The binding affinity is strictly equal to the dissociation constant only for non-competitive binding, while the two quantities are just proportional in competitive binding assays (Cheng and Prusoff, 1973; Lazareno and Birdsall, 1993).
Identification of T Cell Epitopes
To identify potential T cell epitopes, we use NetTepi 1.0 server (https://services.healthtech.dtu.dk/service.php?NetTepi-1.0) which combines estimates for peptide-MHC binding affinity, peptide-MHC stability and T cell propensity (Trolle and Nielsen, 2014). Peptides are then ranked against a set of 200000 natural peptides to obtain a global rank score. Here we scan all SARS-Cov-2 and SARS-Cov peptides with lengths 8–11 from the 4 structural viral proteins and retain the peptides with rank score lower than 2%. We perform the calculations for all the available class I MHC allele using the default values for the relative weight on stability prediction and the relative weight on T cell propensity prediction. The alleles suppored by NetTepi are well represented in human populations. In particular, the supported HLA-A alleles are present in around 60% of the populations, while the HLA-B are present in around 30% of the populations.
Quantification of Haplotype Frequencies and Statistical Analysis
We consider populations with a sample size larger than 1000 individuals and containing data for all the three classical polymorphic HLA genes. We include data from the German Bone Marrow Donor File (Deutsche KnochenMarkSpenderdate, DKMS) which provides thousands of haplotypes for Germans with different origins. We also include a large dataset from Japan, sample over more than 18000 individuals, and two large datasets from the United States of America (African-Americans and Hispanics). Confidence intervals for haplotype frequencies f are estimated assuming binomial statistics (i.e. , with for a 95% confidence interval, where N is the sample size). When we use instead the rule of three: . Statistical analysis is implemented in python and available within the released code https://github.com/ComplexityBiosystems/hla-covid.
Acknowledgments
We thank Ludwig-Maximilian University Munich and the Internationales Begegnungszentrum München, where this work was completed, for their hospitality. S.Z. also acknowledges support from the Alexander von Humboldt Foundation through the Humboldt Research Award and thanks Friedrich-Alexander-Universität Erlangen-Nürnberg for their hospitality.
Author Contributions
C.A.M.L.P. and S.Z. designed the project, performed the research, analyzed the data, and wrote the paper.
Declaration of Interests
The authors declare no competing interests.
Published: September 10, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cels.2020.08.011.
Supplemental Information
References
- Andreatta M., Trolle T., Yan Z., Greenbaum J.A., Peters B., Nielsen M. An automated benchmarking platform for MHC class II binding prediction methods. Bioinformatics. 2018;34:1522–1528. doi: 10.1093/bioinformatics/btx820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Ponce H., Alva-Hernández S., Garrido-Rodríguez D., Soto-Nava M., García-Téllez T., Escamilla-Gómez T., García-Morales C., Quiroz-Morales V.S., Tapia-Trejo D., del Arenal-Sánchez S. Novel HLA class I associations with HIV-1 control in a unique genetically admixed population. Sci. Rep. 2018;8:6111. doi: 10.1038/s41598-018-23849-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y.M., Flamholz A., Phillips R., Milo R. Sars-CoV-2 (COVID-19) by the numbers. eLife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K.M., Steiner G., Wells D.K., Ribas A., Kalbasi A. Prediction of Sars-CoV-2 epitopes across 9360 HLA class I alleles. bioRxiv. 2020 biorxiv.org/content/10.1101/2020.03.30 [Google Scholar]
- Carrington M., Nelson G.W., Martin M.P., Kissner T., Vlahov D., Goedert J.J., Kaslow R., Buchbinder S., Hoots K., O’Brien S.J. HLA and HIV-1: heterozygote advantage and B∗ 35-Cw∗ 04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- Chen Y.-M.A., Liang S.-Y., Shih Y.-P., Chen C.-Y., Lee Y.-M., Chang L., Jung S.-Y., Ho M.-S., Liang K.-Y., Chen H.-Y. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J. Clin. Microbiol. 2006;44:359–365. doi: 10.1128/JCM.44.2.359-365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Prusoff W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendrou C.A., Petersen J., Rossjohn J., Fugger L. HLA variation and disease. Nat. Rev. Immunol. 2018;18:325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- Falfán-Valencia R., Narayanankutty A., Reséndiz-Hernández J.M., Pérez-Rubio G., Ramírez-Venegas A., Nava-Quiroz K.J., Bautista-Félix N.E., Vargas-Alarcón G., Castillejos-López M.D.J., Hernández A. An increased frequency in HLA Class I alleles and haplotypes suggests genetic susceptibility to influenza A (H1N1) 2009 pandemic: a case-control study. J. Immunol. Res. 2018;2018:1–12. doi: 10.1155/2018/3174868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A.C., Rizzo L.V. MHC structure and function - antigen presentation. Part 2. Einstein (Sao Paulo) 2015;13:157–162. doi: 10.1590/S1679-45082015RB3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Galarza F.F., Takeshita L.Y., Santos E.J., Kempson F., Maia M.H., da Silva A.L., Teles e Silva A.L., Ghattaoraya G.S., Alfirevic A., Jones A.R., Middleton D. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43:D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Watkins D.I. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A.V. The immunogenetics of human infectious diseases. Annu. Rev. Immunol. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keicho N., Itoyama S., Kashiwase K., Phi N.C., Long H.T., Ha L.D., Ban V.V., Hoa B.K., Hang N.T.L., Hijikata M. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum. Immunol. 2009;70:527–531. doi: 10.1016/j.humimm.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazareno S., Birdsall N.J. Estimation of competitive antagonist affinity from functional inhibition curves using the gaddum, schild and Cheng-Prusoff equations. Br. J. Pharmacol. 1993;109:1110–1119. doi: 10.1111/j.1476-5381.1993.tb13737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Tseng H.-K., Trejaut J.A., Lee H.-L., Loo J.-H., Chu C.-C., Chen P.-J., Su Y.-W., Lim K.H., Tsai Z.-U. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Swerdlow D.L., Finelli L. Defining the epidemiology of Covid-19—studies needed. N. Engl. J. Med. 2020;382:1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- Luckey D., Weaver E.A., Osborne D.G., Billadeau D.D., Taneja V. Immunity to influenza is dependent on MHC II polymorphism: study with 2 HLA transgenic strains. Sci. Rep. 2019;9:19061. doi: 10.1038/s41598-019-55503-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei A., Papadopoulos K., Harris P.E. MHC class I antigen processing pathways. Hum. Immunol. 1997;54:91–103. doi: 10.1016/s0198-8859(97)00084-0. [DOI] [PubMed] [Google Scholar]
- Mallal S., Nolan D., Witt C., Masel G., Martin A.M., Moore C., Sayer D., Castley A., Mamotte C., Maxwell D. Association between presence of HLA-B∗ 5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- Mandl J.N., Ahmed R., Barreiro L.B., Daszak P., Epstein J.H., Virgin H.W., Feinberg M.B. Reservoir host immune responses to emerging zoonotic viruses. Cell. 2015;160:20–35. doi: 10.1016/j.cell.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekue L.M., Nkenfou C.N., Ndukong E., Yatchou L., Dambaya B., Ngoufack M.N., Kameni J.K., Kuiaté J.R., Ndjolo A. HLA A∗ 32 is associated to HIV acquisition while B∗ 44 and B∗ 53 are associated with protection against HIV acquisition in perinatally exposed infants. BMC Pediatr. 2019;19:249. doi: 10.1186/s12887-019-1620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.H., Lau K.M., Li L., Cheng S.H., Chan W.Y., Hui P.K., Zee B., Leung C.B., Sung J.J. Association of human-leukocyte-antigen class I (B∗ 0703) and class II (DRB1∗ 0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J. Infect. Dis. 2004;190:515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Lund O., Keşmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- O’Donnell T.J., Rubinsteyn A., Bonsack M., Riemer A.B., Laserson U., Hammerbacher J. MHCflurry: open-source class I MHC binding affinity prediction. Cell Syst. 2018;7:129–132.e4. doi: 10.1016/j.cels.2018.05.014. [DOI] [PubMed] [Google Scholar]
- Paul S., Croft N.P., Purcell A.W., Tscharke D.C., Sette A., Nielsen M., Peters B. Benchmarking predictions of mhc class i restricted T cell epitopes in a comprehensively studied model system. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje H., Tran Kiem C.T., Lefrancq N., Courtejoie N., Bosetti P., Paireau J., Andronico A., Hozé N., Richet J., Dubost C.-L. Estimating the burden of Sars-CoV-2 in France. Science. 2020;369:208–211. doi: 10.1126/science.abc3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T., Hosomichi K., Inoko H., Kulski J.K., Jan The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- Spìnola H. HLA loci and respiratory infectious diseases. Respir. Res. 2016;2:56–66. [Google Scholar]
- Trolle T., Nielsen M. NetTepi: an integrated method for the prediction of T cell epitopes. Immunogenetics. 2014;66:449–456. doi: 10.1007/s00251-014-0779-0. [DOI] [PubMed] [Google Scholar]
- Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G.T., Fu H. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Bushman M., Kishore N., Niehus R., de Salazar P.M., Cowling B.J., Lipsitch M., Leung G.M. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat. Med. 2020;26:506–510. doi: 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source codes generated to obtain the results presented in this paper are available at https://github.com/ComplexityBiosystems/hla-covid. Binding affinities for SARS-CoV-2, SARS-CoV and HCoV-OC43 are reported in Data S1. The protein sequences used in this paper are also available at https://github.com/ComplexityBiosystems/hla-covid/. Haplotype frequencies for different population are retrieved from the Allele Frequency Net Database (http://www.allelefrequencies.net/) (González-Galarza et al., 2015) and available at https://github.com/ComplexityBiosystems/hla-covid/.