Abstract
Background
Salt stress is one of the most damaging abiotic stresses in production of Upland cotton (Gossypium hirsutum). Upland cotton is defined as a medium salt-tolerant crop. Salinity hinders root development, shoots growth, and reduces the fiber quality.
Results
Our previous study verified a GhCIPK6a gene response to salt stress in G. hirsutum. The homologs of GhCIPK6a were analyzed in A2 (G. arboreum), D5 (G. raimondii), and AD1 (G. hirsutum) genomes. GhCIPK6a localized to the vacuole and cell membrane. The GhCBL1-GhCIPK6a and GhCBL8-GhCIPK6a complexes localized to the nucleus and cytomembrane. Overexpression of GhCIPK6a enhanced expression levels of co-expressed genes induced by salt stress, which scavenged ROS and involved in MAPK signaling pathways verified by RNA-seq analysis. Water absorption capacity and cell membrane stability of seeds from GhCIPK6a overexpressed lines was higher than that of wild-type seeds during imbibed germination stage. The seed germination rates and seedling field emergence percentages of GhCIPK6a overexpressed lines were higher than that of control line under salt stress. Moreover, overexpressing of GhCIPK6a in cotton increased lint percentage, and fiber length uniformity under salt stress.
Conclusions
We verified the function of GhCIPK6a by transformation and RNA-seq analysis. GhCIPK6a overexpressed lines exhibited higher tolerance to abiotic stresses, which functioned by involving in ROS scavenging and MAPK pathways. Therefore, GhCIPK6a has the potential for cotton breeding to improve stress-tolerance.
Keywords: CIPK, Salt stress, Co-expression, Upland cotton, Signaling pathway
Background
Soil salinity is one of the serious abiotic stresses that affect crop plant growth and development, and eventually reduce the yield and descend the quality [1, 2]. Plants initiate a series of adaptive mechanism and survival responses to abiotic stresses, and result in serial changes in gene expression [3–6]. These stress-response genes involve Ca2+-sensors [7, 8], transcription factors [9, 10], protein kinases [11–13], osmotic proteins [14–16], and hydroperoxidases [17, 18], in which protein kinases play a particularly important role in plant response. A group of serine-threonine kinases, designated as CBL-interacting protein kinases (CIPKs), which interact specifically with calcineurin B-like proteins (CBLs), has been characterized in plant genomes [19, 20].
CIPK proteins consist of a conserved N-terminal kinase catalytic domain, and a C-terminal regulatory domain separated from the kinase domain by a variable junction domain and the N-terminal kinase domain contains a putative activation loop phosphorylated by other protein kinases [21]. The highly conserved NAF/FISL domain in C-terminal of CIPK proteins is required and sufficient for interacting with CBL proteins, and functions as an auto-inhibitory domain [22]. Another motif, protein phosphatase interaction (PPI), is necessary for the interaction with abscisic acid-insensitive (ABI) protein [23]. The PPI motif controls the phosphorylation status of CIPK and PP2Cs (protein phosphatase 2C) [23–25]. CBLs generally interact with CIPKs to form CBL-CIPK complexes that regulate downstream target proteins. CIPK proteins and CBL-CIPK complexes were involved in various responsive processes in Arabidopsis and other plants, such as rice (Oryza sativa) [26–29], maize (Zea mays) [30–32], Populus euphratica [33, 34], canola (Brassica napus) [35, 36], eggplant (Solanum melongena) [37], tomato (Solanum lycopersicum) [38], foxtail millet (Setaria italica (L.) P. Beauv) [39], pineapple (Ananas comosus) [40] and cotton [41–44]. CIPKs and CBL-CIPK complex have been implicated in the plant’s response to abiotic stresses, biotic stresses, phytohormones, and nutrient deprivation [45–50].
In Arabidopsis, overexpression of AtCIPK6 gene increased the tolerance to salt stress [51, 52]. AtCBL4/SOS3 interacts with AtCIPK24/SOS2 in the roots to mediate Na+ extrusion via the action of the H+/Na+ antiporter SOS1 at the plasma membrane in SOS signaling pathway in Arabidopsis under salt stress [53, 54]. There were a novel SOS pathway, CBL10–CIPK8–SOS1, functions to transport accumulated Na+ out of cells to regulate salt tolerance [50]. The AtCBL1/9-AtCIPK23 complex activates the AKT1 channel at the plasma membrane to enhance K+ uptake under low K+ condition [24, 55, 56]. Meanwhile, AtCIPK23 and AtKC1 act synergistically to modulate the activity of AKT1 [57]. AtCBL9-AtCIPK23 complex was shown to interact with the nitrate transporter NRT1.1 during the primary nitrate response [49, 58, 59].
Upland cotton (G. hirsutum) is defined as a medium salt tolerant crop [60, 61]. Salinity hinders root development, shoots growth, and reduces the fiber quality [62]. Newly released Gossypium species genomes provide novel platform for functional genomics research [63–69]. GhCIPK homolog, GhCIPK1 is involved in the cotton fiber elongation process [70], and is shown to be co-expressed with and preferentially interact with GhCBL2/3 [71]. Heterologous expression of GhCIPK6 (KC465063) in Arabidopsis enhanced tolerance to salt treatment, and in transgenic cotton, this gene mediates the uptake of K+ under Ca2+ − deficient condition [42, 44].
In our previous study, we characterized a salt-responsive gene from cDNA libraries and Microarray results, GhCIPK6a (Accession number in GenBank: HM002633), homologous to AtCIPK6 [41]. GhCIPK6a also differently expressed in salt-tolerant and salt-sensitive Upland cotton varieties under salt stress by transcriptome analysis [72]. We isolated the full length of the GhCIPK6a gene and characterized its function in Upland cotton. Changes in physiological indexes showed that over-expression of GhCIPK6a in Upland cotton significantly strengthened the salt tolerance, which were verified that overexpressed GhCIPK6a enhanced co-expressed genes expression levels in stress signaling pathways by RNA-seq results. Meanwhile, the transgenic cotton grew better than wild type in saline field. Here we elucidate the function of GhCIPK6a and the potential use in transgenic cotton breeding.
Results
Isolation and basic analysis of GhCIPK6a
In present study, cDNA and genomic DNA sequences of GhCIPK6a were isolated from Upland cotton ‘Zhong G5’ [41]. A comparison of the genomic DNA and cDNA sequences using GSDS (http://gsds.cbi.pku.edu.cn/) [73] revealed that there was no intron in the genomic sequence of GhCIPK6a.
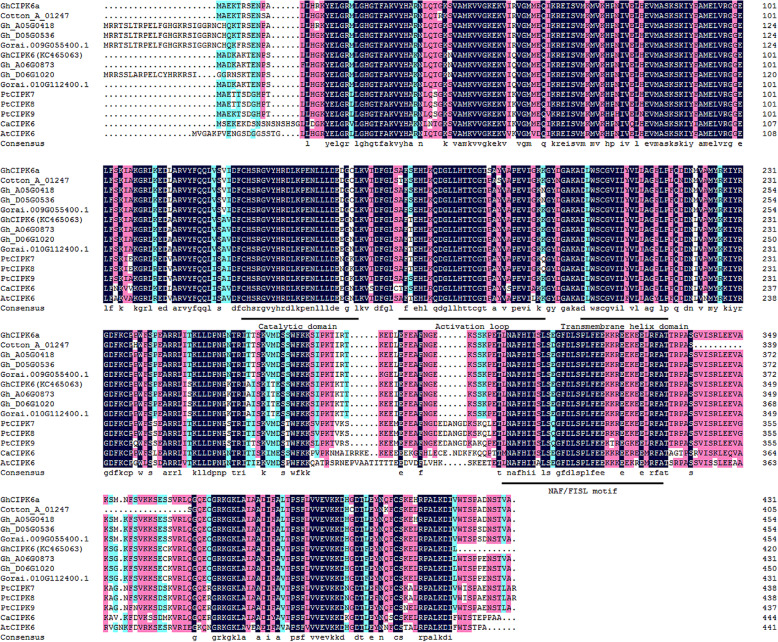
GhCIPK6a contained an open reading frame (ORF) of 1296 bp and encoded a protein of 431 amino acids and molecular weight 48.637 kD. Using ScanProsite online software (http://au.expasy.org/tools/scanprosite/), motif scan analysis showed that GhCIPK6a included a Serine/Threonine protein kinase catalytic domain at the N-terminal, which contained an ATP-binding region and an active site, and a CBL-interacting NAF/FISL module at the C-terminal. GhCIPK6a contains a transmembrane helix domain between amino acid residues 198 and 217 (Fig. 1).
Fig. 1.
Multiple sequence alignment of CIPK proteins using ClustalW implemented in DNAMAN. Conserved functional domains are underlined. At, Arabidopsis thaliana; Ca, Cicer arietinum; Pt, Populus trichocarpa; Gh, G. hirsutum; Cotton_A, G. arboreum; Gorai, G. raimondii. Numbers shown in the right indicate amino acid residue positions
To determine genomic location of GhCIPK6a, we compared GhCIPK6a to the genome sequences of D5 (G. raimondii) [65], A2 (G. arboreum) [63], and AD1 (G. hirsutum L. acc. TM-1) [66], and searched for homologs (Table 1). We identified four homologs in the D5 genome, including Gorai.001G032000.1, Gorai.009G055400.1, Gorai.010G112400.1, and Gorai.013G216700.1, in which, Gorai.009G055400.1 was the most identified to GhCIPK6a (Additional file 6 Fig. S1). In the A2 genome sequence, three homologs of GhCIPK6a were identified, including Cotton_A_01247, Cotton_A_09063, and Cotton_A_07248, and GhCIPK6a exhibited the highest similarity to Cotton_A_01247. The AD1 genome sequence harbored three homologs of GhCIPK6a, which genes ID was Gh_A05G0418, Gh_D05G0536, and Gh_D13G1983. Gh_A05G0418 and Gh_D05G0536 shared more than 99% amino acid sequence identity with GhCIPK6a. Phylogenetic analysis showed that all the CIPKs were classified as four groups (Group I to IV, Additional file 6 Fig. S1). GhCIPK6a was classified into Group III. The nucleotide sequence of GhCIPK6a shared 66.22% identity with AtCIPK6 (AT4G30960, http://www.arabidopsis.org/), and 70.72% with CaCIPK6 (EU492906.1, Cicer arietinum, http://www.ncbi.nlm.nih.gov/). Meanwhile, multiple sequence alignment of GhCIPK6 with related proteins were carried out (Fig. 1). These CIPK protein kinases included highly conserved functional domains, such as catalytic domain, activation loop, transmembrane helix domain, and NAF/FISL motif.
Table 1.
Characteristics of homologous genes in the A2, D5, and AD1 genomes
| Gene ID/Gene name | Genomic position | ORF length (bp) | DNA length (bp) | Protein length (aa) | Protein molecular mass (kd) | pI | Reference |
|---|---|---|---|---|---|---|---|
| GhCIPK6a (HM002633) | – | 1296 | 1296 | 431 | 48.71 | 9.00 | Zhang et al. 2011 [41] |
| Cotton_A_01247 | Ca03:32871827:32873122a | 1218 | 1296 | 405 | 45.80 | 9.04 | Li et al. 2014 [63] |
| Gorai.009G055400.1 | Chr09:4028418:4029980a | 1365 | 1563 | 454 | 51.29 | 9.35 | Paterson et al. 2012 [65] |
| Gh_A05G0418 | A05:4720527:4722095a | 1365 | 1596 | 454 | 51.41 | 9.31 | Zhang et al. 2015 [66] |
| Gh_D05G0536 | D05:4357435:4358998a | 1365 | 1564 | 454 | 51.30 | 9.26 | Zhang et al. 2015 [66] |
| GhCIPK6 (KC465063) | – | 1296 | – | 431 | 47.33 | 9.09 | He et al. 2013 [42] |
| Gorai.010G112400.1 | Chr10:21475391:21476686a | 1296 | 1296 | 431 | 48.49 | 9.04 | Paterson et al. 2012 [65] |
| Gh_A06G0873 | A06:33360018:33361313a | 1296 | 1296 | 431 | 48.55 | 9.10 | Zhang et al. 2015 [66] |
| Gh_D06G1020 | D06:21731900:21733431a | 1353 | 1532 | 450 | 50.79 | 9.31 | Zhang et al. 2015 [66] |
a, antisense strand; −-, no data
Subcellular localization of GhCIPK6a
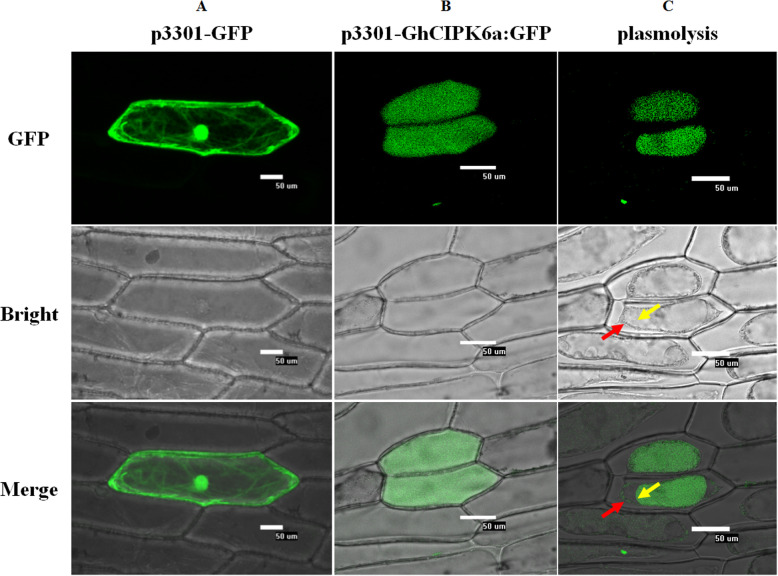
Subcellular localization analysis using SubLoc v1.0 (http://www.bioinfo.tsinghua.edu.cn/SubLoc/) indicated that GhCIPK6a was localized to the cytoplasm, which was confirmed by generating a construct by fusing GFP to C-terminal end of GhCIPK6a under the control of the CaMV 35S promoter, and transiently expressing the construct in onion epidermal cells. Indeed, fluorescence was specifically localized to the cytoplasm (Fig. 2b). To establish whether the GhCIPK6a: GFP fusion was present at the cell membrane; the onion epidermal cells were plasmolyzed in sucrose solution. The analysis demonstrated that the fusion protein was restricted to the vacuole and cell membrane (Fig. 2c).
Fig. 2.
Subcellular localization of the GhCIPK6a:GFP fusion protein in onion epidermal cells. a. Fluorescence microscopy images of cells expressing GFP protein. b. Fluorescence microscopy images of cells expressing the GhCIPK6a:GFP fusion. c. Fluorescence microscopy images of plasmolyzed cells in 30% sucrose solution. The cell membrane is marked with a red arrow, and the vacuole membrane is marked with a yellow arrow. Confocal images of epidermal cells were captured 22–24 h after transformation. Bar = 50 μm
Verification of the interaction between GhCIPK6a and GhCBLs in vivo
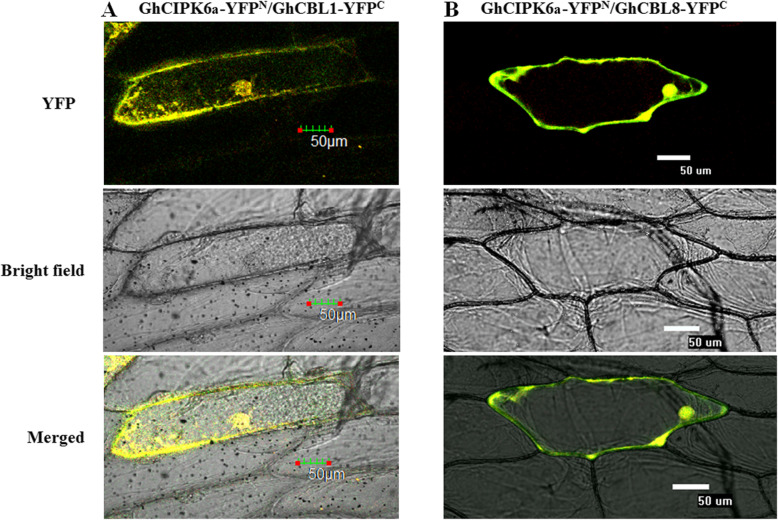
The activation of CIPKs was regulated by binding to one or more CBL proteins. It was previously reported that GhCIPK1 interacted with GhCBL2 and GhCBL3 [71]. So, we detected and investigated which GhCBL proteins interact with GhCIPK6a using the BiFC method.
Four GhCBL gene sequences obtained from NCBI (http://www.ncbi.nlm.nih.gov/): GhCBL1 (EU085038.1), GhCBL2 (EU085042.1), GhCBL3 (EU085040.1), and GhCBL8 (EU085041.1). When GhCIPK6a-YFPN and GhCBLs-YFPC fusion genes were co-expressed in onion epidermal cells using particle bombardment, yellow fluorescence signals were observed in the nucleus and cell membrane when GhCIPK6a co-expressed with GhCBL1 and GhCBL8 (Fig. 3). By contrast, no signal was observed when GhCIPK6a co-expressed with GhCBL2 or GhCBL3 in onion epidermal cells (data not shown).
Fig. 3.
BiFC assay demonstrated the interaction between GhCIPK6a and GhCBLs in onion epidermal cells. GhCIPK6a interacted with GhCBL1 (a) and GhCBL8 (b) in vivo. Confocal images of epidermal cells were captured 22–24 h after transformation. Bar = 50 μm
GhCIPK6a and salt-response genes significantly up regulated under salt treatment
In the salt-sensitive cultivar ‘Zhong G5’ [41], GhCIPK6a transcript accumulated to higher levels in the root, and expression level significantly increased by salt treatment for 1, 6, 12, and 24 h. In stem tissue, the GhCIPK6a expression was induced after a longer period of salt treatment than that in the root, and the fold-change was less than in the root (Additional file 7 Fig. S2a).
To investigate whether overexpression of GhCIPK6a in cotton enhanced salt tolerance, we transformed GhCIPK6a into Upland cotton cultivar ‘11–0516’. Eleven transgenic cotton individuals (T1 generation) were obtained (Additional file 8 Fig. S3a). Through kanamycin resistance assay, PCR analysis and Southern blotting assay, we obtained two T2 generation transgenic progeny plants that harbored two copies of insert fragment, which named by OE1 and OE2 (Additional file 8 Fig. S3b). We examined the expression level of GhCIPK6a in roots of transgenic and control plants at the three-leaf stage by qRT-PCR analysis. GhCIPK6a expression was significantly higher in OE2 than that in wild type line without treatment (Fig. 4). However, the expression level in OE1 line no significantly increased than wild-type cotton. Therefore, we further analyze the functions of GhCIPK6a gene using OE2 transgenic line.
Fig. 4.
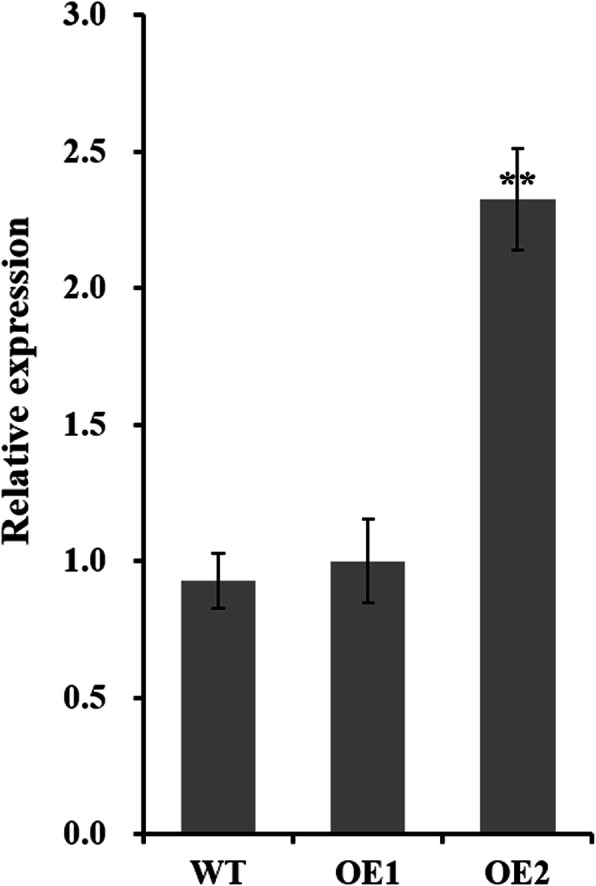
Relative expression of GhCIPK6a in the root tissues of transgenic lines (OE1 and OE2) and wild-type (WT) plants at the three-leaf stage under normal treatment. *, p-value < 0.05; **, p-value < 0.01
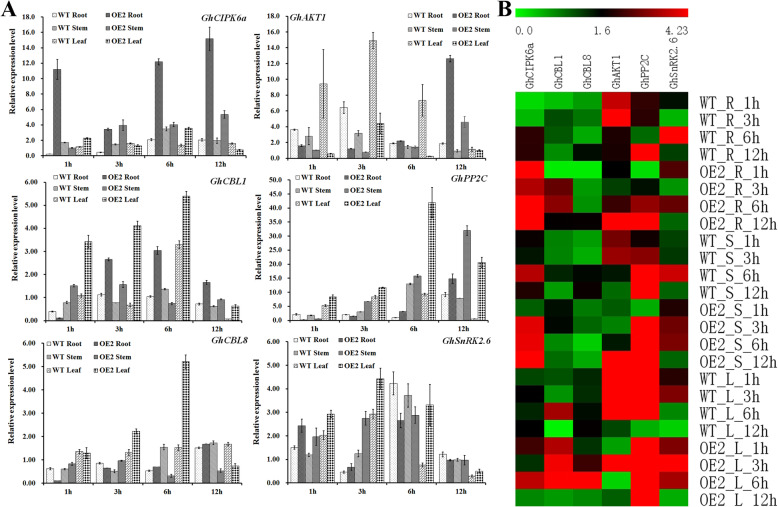
We also examined the expression level of GhCIPK6a in different tissues of transgenic cotton (OE2) under salt stress (Additional file 7 Fig. S2b). Overexpressed GhCIPK6a strongly induced by salt stress in roots of transgenic cotton, especially after 1, 3, 6, and 12 h salt treatments. In addition, the expression profiles of GhCIPK6a gene in other tissues were similar to that of wild type. Moreover, GhCIPK6a expression increased with increasing duration of exposure to salt stress (Fig. 5). To determine the function of GhCIPK6a under multiple abiotic stresses, we analyzed the cotton expression profiles of GhCIPK6a using public datasets from PLEXdb and GEO. GhCIPK6a gene was analyzed under multiple abiotic stresses, such as ABA, cold, drought, salinity, and alkalinity (pH) in G. hirsutum (Additional file 9 Fig. S4).
Fig. 5.
qRT-PCR analysis of GhCIPK6a expression and genes that were co-expressed with GhCIPK6a in different tissues under salt stress. a. Relative expression values of genes co-expressed with GhCIPK6a. b. Heatmap of GhCIPK6a expression and genes co-expressed with GhCIPK6a in different tissues under salt stress. WT, the wild type line, 11-0516; OE2, the transgenic cotton. R, root tissue; S, stem tissue; L, leaf tissue
CIPKs interact with CBLs and PP2Cs to form a complex that regulates the activity of K+ transporters [24, 74]. Therefore, we examined the expression level of GhAKT1 in transgenic cotton and wild type under salt stress and control (in hydroponic growth, Fig. 5) conditions. GhAKT1 expression only increased strongly in the root of the OE2 line at three-leaf stage after 12 h salt stress treatment, which exhibited similar tendency in stem. After salt treatment at three-leaf stage, GhAKT1 expression was strongly induced in all tissues of the wild-type line, especially the leaf. The increase in expression level might be associated with maintaining K+ homeostasis in root upon exposure to salt stress, which would enhance the salt tolerance of transgenic line in turn.
GhCIPK6a interacts with GhCBL1/GhCBL8, SnRK2.6 and PP2C proteins, respectively, to regulate the expression of downstream genes [24, 74]. Then, we analyzed the expression profiles of GhCBL1, GhCBL8, GhPP2C (DQ303437.1), and GhSnRK2.6 (JN872373) [12, 75] (Fig. 5). The transcript levels of GhCBL1 and GhCBL8 rose sharply soon after exposure to salt stress, and then decreased at 12 h after salt stress in leaves. During the same time, GhCBL1 expression was induced by salt stress in the stem of the transgenic line, OE2. GhPP2C was upregulated in all tissues of OE2 after salt treatment, especially the leaves, except in the roots. GhSnRK2.6 was strongly induced in all tissues of OE2 at one and three hours after treatment, but transcripts only accumulated in the leaves after 6 and 12 h stresses (Fig. 5).
GhCIPK6a enhanced salt tolerance during germination and seedling stages
Under salt stress, germination and emergence of cotton were the key stages. We determined the germination potential and seed germination rate of cotton seed using rolls of filter paper placed upright under salt treatment (150 mmol·L− 1 NaCl) and control (distilled water, CK) (Fig. 6a, Additional file 10 Fig. S5). The seed germination rate of the transgenic cotton lines (48.89 and 53.33%) was significantly higher than that of wild type (17.12%) under salt stress (Fig. 6a, Additional file 10 Fig. S5). But the differences between transgenic lines and wild type line in germination potential under salt stress significantly (data not shown). In addition, we also obtained the homozygotes of transgenic Arabidopsis, analyzed seed germination rate and growth under salt treatment. The transgenic Arabidopsis maintained higher seed germination rate and grew normal under NaCl concentration up to 200 mmol·L− 1 as well (data not shown).
Fig. 6.
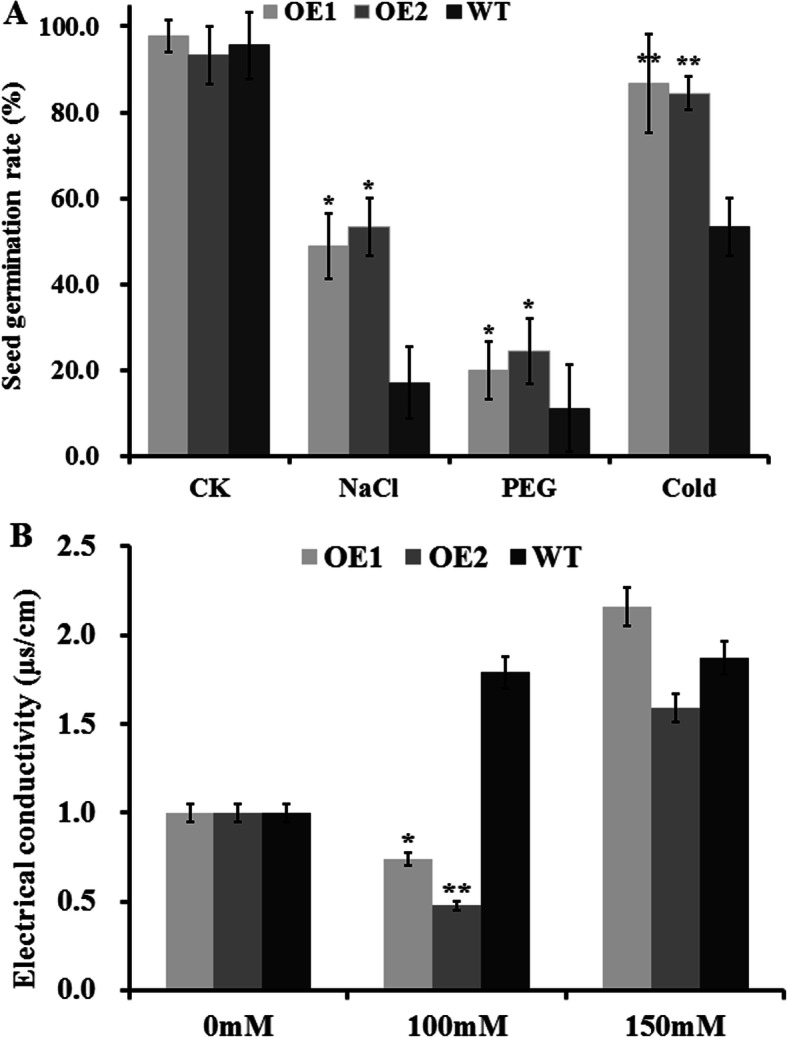
Analysis of seed germination rate and electrical conductivity of transgenic lines (OE1 and OE2) and wild-type (WT) line under abiotic stresses. a. Seed germination rate of transgenic lines (OE1 and OE2) and wild-type (WT) line under salt, PEG, and cold treatments. b. Electrical conductivity analysis of transgenic lines (OE1 and OE2) and wild-type (WT) line after soaking for 24 h in 0, 100, and 150 mmol·L− 1 NaCl solution. *, p-value < 0.05; **, p-value < 0.01
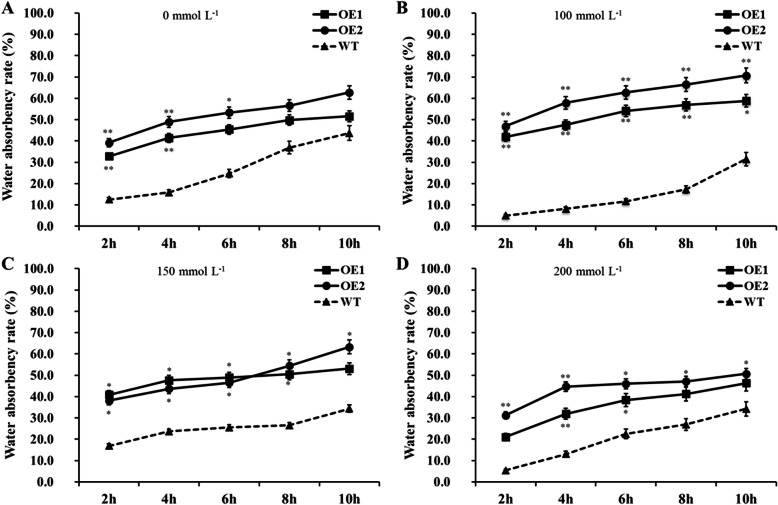
Meanwhile, we analyzed the changes in water absorbency rate of transgenic and wild type cotton seeds in imbibed germination stage (Fig. 7), and the effect of salt stress on cell membrane permeability by determining the electrical conductivity after soaking for 24 h in different concentrations of NaCl solution (Fig. 6b). The cell membrane of the transgenic lines (OE1 and OE2) was more stable than that of wild type, and water absorption capacity was higher than that of wild type during imbibed germination stage. Thus, transgenic cottonseeds maintain a higher seed germination rate under salt stress due to increased stability of cell membrane, which ensured that water was absorbed at normal rates under salt stress. Therefore, overexpression of GhCIPK6a in Upland cotton improved salt tolerance in seed germination stage through increasing the stability of cell membrane.
Fig. 7.
The water absorbency rate in imbibed germination stage of transgenic lines (OE1 and OE2) and wild-type (WT) line exposed to solution containing different NaCl concentrations (mmol·L− 1). a, control, represented the water absorbency rates of transgenic lines and wild type line exposed to distilled water (0 mmol·L− 1 NaCl concentration). B, C, and D represented the water absorbency rates of transgenic lines and wild type line exposed to solution with different NaCl concentrations, (b) 100 mmol·L− 1, (c) 150 mmol·L− 1, and (d) 200 mmol·L− 1, respectively. *, p-value < 0.05; **, p-value < 0.01
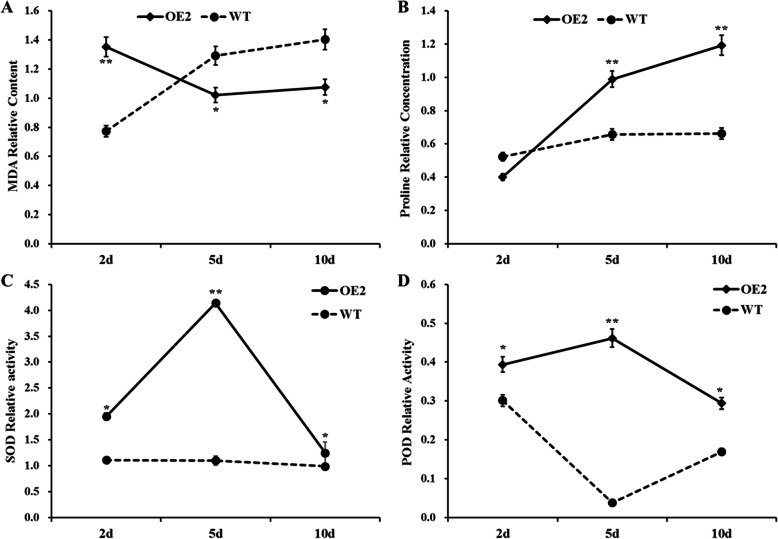
To confirm that overexpression of GhCIPK6a enhanced salt tolerance during seedling stage, we treated transgenic and wild-type seedlings at three-leaf stage by soaking the roots in hydroponic solution with 150 mmol·L− 1 NaCl. We determined the MDA and proline contents, and SOD and POD activities of seedlings exposed to salt stress. After 2, 5, and 10 d of salt treatment, the transgenic line maintained lower relative content of MDA than control, but relative content of proline and relative activities of POD and SOD were higher than those of wild-type (Fig. 8). Since MDA and proline promote membrane stability, and POD and SOD limit membrane lipid peroxidation by reducing the accumulation of H2O2. So the result suggests that over-expression of GhCIPK6a increases both the POD and SOD activities, and thereby reduces H2O2 accumulation and protects plant seedlings from membrane damage under salt stress.
Fig. 8.
Statistical analysis of physiological indexes in leaves of transgenic (OE2) line and wild-type (WT) line seedlings under 150 mmol·L− 1 NaCl treatment. a. The relative content of MDA in the leaves of transgenic line (OE2) and wild-type (WT) line after 2, 5, and 10 days of 150 mmol·L− 1 NaCl treatment. b. The relative concentration of proline in the leaves of transgenic line (OE2) and wild-type (WT) line after 2, 5, and 10 days of 150 mmol·L− 1 NaCl treatment. c and d. The relative activity of SOD (c) and POD (d) in the leaves of transgenic line (OE2) and wild-type (WT) line after 2, 5, and 10 days of 150 mmol·L− 1 NaCl treatment. *, p-value < 0.05; **, p-value < 0.01
Improved salt tolerance of GhCIPK6a transgenic cotton in field experiment
To evaluate the salt tolerance of the GhCIPK6a-overexpressed cotton plants, we planted the T6 and T7 generation transgenic lines and the control under two different conditions in the field experiment in Handan City, Hebei Province, China. The generation processes of transgenic lines were shown in Additional file 8 Fig. S3c and S3d. Table 2 showed the yield and fiber quality traits of transgenic lines and wild type control in 2016 and 2017.
Table 2.
Yield and fiber quality traits of GhCIPK6a transgenic lines and wild type under normal and saline conditions in 2016 and 2017
| Condition | Generation | Line ID | Boll number | Boll weight (g) | Lint percentage (%) | Fiber length (mm) | Fiber uniformity rate (%) | Fiber strength (cN/tex) | Fiber elongation (%) | Micronaire |
|---|---|---|---|---|---|---|---|---|---|---|
| Normal | T6 | OE1 | 14.7 | 5.3 | 38.0 | 30.4* | 85.9* | 29.9 | 6.9 | 5.2 |
| OE2 | 14.8 | 5.4 | 38.9* | 30.2 | 86.0** | 29.9 | 6.9 | 5.3* | ||
| WT | 14.7 | 5.5 | 37.0 | 29.9 | 85.1 | 29.6 | 6.9 | 5.2 | ||
| T7 | OE1 | 23.6* | 6.0 | 42.2 | 29.1 | 84.9 | 27.9 | 6.9 | 5.3 | |
| OE2 | 23.7** | 6.1 | 43.1 | 29.5 | 85.2 | 27.8 | 6.9 | 5.2 | ||
| WT | 22.1 | 6.2 | 42.1 | 29.6 | 85.4 | 26.8 | 6.9 | 5.2 | ||
| Saline | T6 | OE1 | 15.1* | 5.2 | 38.0* | 29.9 | 86.5** | 29.7 | 6.7 | 5.3 |
| OE2 | 15.9** | 5.3 | 38.3** | 30.2 | 86.0** | 29.9 | 6.7 | 5.3 | ||
| WT | 13.5 | 5.3 | 37.5 | 30.1 | 84.2 | 30.4 | 6.7 | 5.6 | ||
| T7 | OE1 | 22.4 | 5.5 | 43.0 | 30.8 | 85.0 | 28.8 | 6.8 | 5.5 | |
| OE2 | 22.1 | 5.7 | 43.3 | 30.7 | 85.3 | 28.5 | 6.7* | 5.5 | ||
| WT | 22.9 | 5.6 | 43.4 | 30.5 | 85.4 | 28.2 | 6.8 | 5.4 |
*, p-value < 0.05
**, p-value < 0.01
During seedling stage, the seedling field emergence percentage of GhCIPK6a overexpression lines and WT line were similar under normal condition. Under salt stress, the seedling field emergence percentage of OE lines was significantly higher than WT lines over 2 years (Fig. 9). In the flowering and boll periods, there was a hail disaster on June 28, 2016, which caused the boll number of OEs and WT lines less than that in 2017 (Table 2). Meanwhile, the transgenic lines recovered better than the WT line after hail disaster, especially under salt stress. The stronger resilience of OE lines was shown in more boll number and higher lint percentage than that in WT line, under salt condition in 2016 (Table 2). Otherwise, the fiber uniformity rate of GhCIPK6a overexpressed lines was significantly higher than that in WT line, which indicated that GhCIPK6a overexpressed lines showed stronger adaptability and resilience in extreme environments.
Fig. 9.
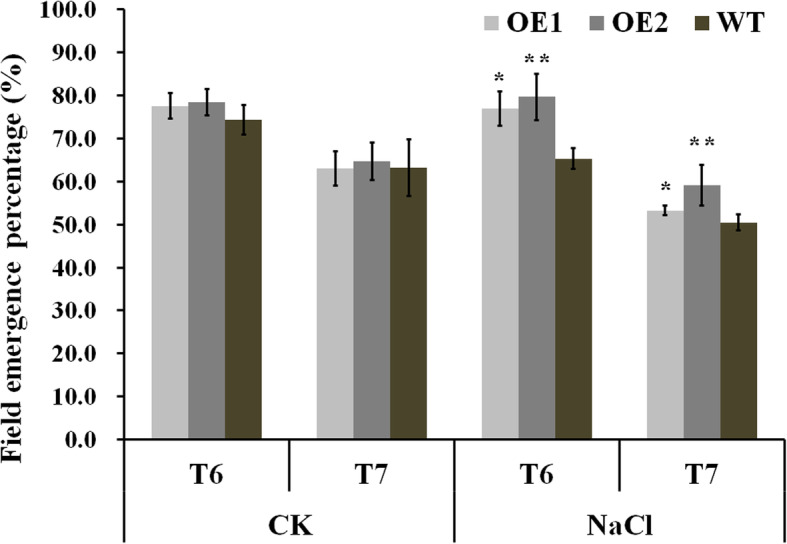
The seedling field emergence percentage of transgenic lines (OE1 and OE2) and wild-type (WT) line under normal and saline conditions in 2016 and 2017. *, p-value < 0.05; **, p-value < 0.01
In 2017, the boll number of transgenic lines was significantly higher than that in WT line under normal condition, which was no significant difference under salt stress. In addition, the boll weight and lint percentage were no significant differences between OE and WT lines. It can be speculated that GhCIPK6a overexpressed in Upland cotton could increase yield under normal condition. There was no influence in fiber quality trait. Under salt stress, there were also no differences in yield and fiber quality traits between overexpression and wild type lines (Table 2). Therefore, GhCIPK6a overexpressed in cotton increased the seedling field emergence percentage, seed cotton yield under salt condition, and maintained the stability of yield and fiber quality traits under extreme treatments.
We also evaluated the salt tolerance of transgenic and control cottons during flowering and boll setting stage in the field in Akesu City, Xinjiang Autonomous Region, China in 2013 (Additional file 11 Fig. S6). The salt content of the soil under the surface 5 to 10 cm was approximately 0.92%, which was significantly higher than the tolerance of cotton. Under severe salt stress, there were extensive necrosis in the leaves occurred in the wild type plants and ZG5, a salt sensitive cotton variety [41, 72]. However, the transgenic plants and Z9806 (a salt tolerant cotton variety [12]) could grow very well, and show no necrosis in leaves (Additional file 11 Fig. S6).
GhCIPK6a involved in MAPK signaling pathway and plant hormone signal transduction pathway to response to salt stress
In order to understand the mechanism of GhCIPK6a overexpressing to improve salt tolerance in transgenic cotton, we analyzed the transcriptome of GhCIPK6a-overespression (OE2) line and wild-type plants. A total of 252 genes were up- and 79 genes were down-regulated, respectively, in GhCIPK6a-overexpression compare to wild-type plants (Additional file 12 Fig. S7, Additional file 1 Table S1). In 252 up-regulated DEGs, GO-term analysis indicated 78 genes enriched in response to signaling, stress, ROS and stimulus progresses (Additional file 13 Fig. S8a, b). Among the 78 candidate genes, there were 33 genes response to salt, osmic, and drought stresses (Fig. 10a, b), eight genes involved in MAPK cascade (GO:0000165). The gene Gh_A11G1875 predicted in activation of MAPKK activity process (GO:0000186) and regulation of stomatal closure (GO:0090333). There was one gene, Gh_D02G0057, response to ABA stimulus (GO:0071215), and seven genes were response to gibberellin acid (GO:0009739). KEGG pathways analysis showed the 78 genes mainly enriched in to signal transduction pathways, especially in MAPK signaling pathway (ko04016) and plant hormone signal transduction pathway (ko04075) (Fig. 10a, b, Additional file 13 Fig. S8c).
Fig. 10.
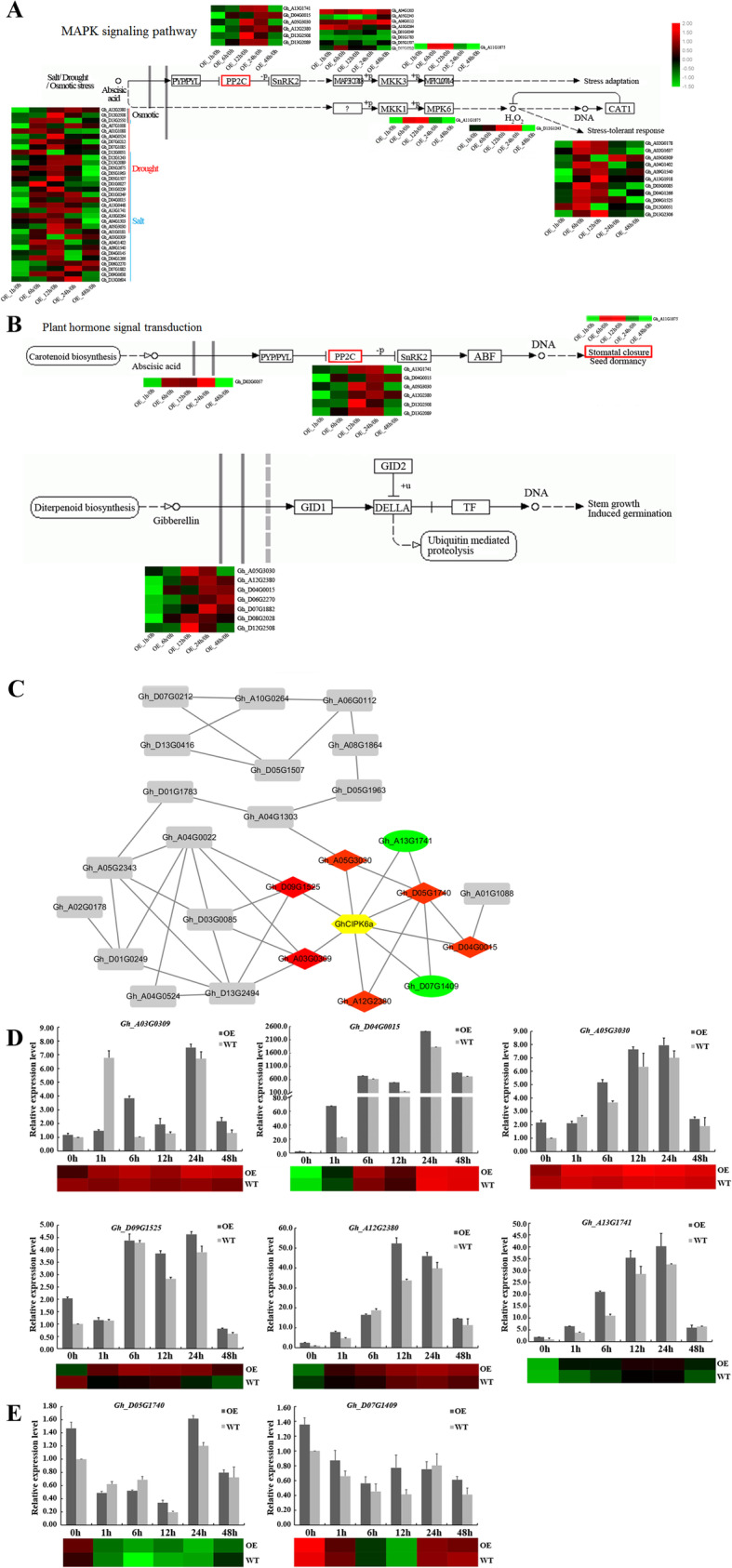
The specifically expressed DEGs in OE2 plants analysis. a and b. The DEGs enriched KEGG pathways and expression profile analysis. c. The PPI network predicted using STRING program. The genes with red rhombus background were co-expressed genes in up-regulated DEGs. The genes with green oval background were co-expressed genes from down-regulated DEGs. The genes with gray rectangular background were the ones predicted in the same PPI network with GhCIPK6a from up-regulated DEGs. d. Verification of expression profile of the up-regulated DEGs co-expressed with GhCIPK6a. e. Verification of expression profile of the down-regulated DEGs co-expressed with GhCIPK6a
Protein interactions among DEGs was detected using the online STRING program, there were 23 genes co-expressed with GhCIPK6a within 78 up-regulated genes, and two genes co-expressed with GhCIPK6a in 79 down-regulated DEGs (Additional file 2 Table S2). Combined the information of edges and nodes of up- and down-regulated genes, the PPI network was conducted using Cytoscape software (Fig. 10c).
In PPI network, there were eight genes predicted to co-express with GhCIPK6a directly, six up- and two down-regulated DEGs. Six up-regulated DEGs were Gh_A03G0309 (18.5 kD class I heat shock protein, HSP18.5-C), Gh_A05G3030 (Protein phosphatase 2C 37, PP2CA), Gh_A12G2380 (Probable protein phosphatase 2C 75, AHG1), Gh_A13G1741 (Protein phosphatase 2C 56, ABI1), Gh_D04G0015 (Probable protein phosphatase 2C 8, PP2C8), and Gh_D09G1525 (18.2 kD class I heat shock protein, HSP18.2), respectively. All PP2C genes were involved in MAPK signaling pathway (ko04016) and plant hormone signal transduction pathways (ko04075) (Fig. 10c). Two HSP genes were involved in Protein processing in endoplasmic reticulum (ko04141). Two down-regulated DEGs were Gh_D05G1740 (Serine/threonine-protein kinase STY46) and Gh_D07G1409 (Probable serine/threonine-protein kinase), which were no KEGG annotation, both enriched in the MAPKKK activity process (GO:0004709). All DEGs in the PPI network were verified using qRT-PCR (Fig. 10d, e, Additional file 14 Fig. S9) and were consistent with the RNA-seq data.
Overexpressed GhCIPK6a also improve the tolerance to osmotic and low-temperature stresses
To investigate the tolerance to abiotic stresses of transgenic cotton lines, we also determined seed germination potential and seed germination rate under drought (15% PEG 6000) and low-temperature stresses (distilled water, at 15 °C), respectively. Seed germination rate of transgenic cottons were significantly higher than wild type under drought (15% PEG 6000) and low-temperature stresses (Fig. 6a).
Discussion
To develop the gene sources for molecular breeding is a popular strategy used in genetic improvement to improve abiotic stresses. Here, GhCIPK6a (HM002633) has the potential to improve the salt tolerance in cotton. GhCIPK6a shared 66.22% nucleotide sequence identity with AtCIPK6. GhCIPK6a expression was significantly increased by salt treatment (Additional file 7 Fig. S2). Overexpression of GhCIPK6a enhanced the seed germination rate under various stress treatments.
The difference between GhCIPK6a (HM002633) and GhCIPK6 (KC465063)
Another GhCIPK6 gene (GhCIPK6(KC465063)) was previosly reported in Upland cotton [42, 44]. The GhCIPK6a (HM002633) identified here shared 78.56% nucleotide sequence identity with GhCIPK6 (KC465063), and 90.49% amino acid sequence identity. We further analyzed the GhCIPK6a (HM002633) and GhCIPK6 (KC465063) sequences in the A2, D5, and AD1 genomes (Table 1). The homolog IDs of GhCIPK6a (HM002633) in the A2, D5 and AD1 genomes are Cotton_A_01247, Gorai.009G055400.1, Gh_A05G0418 and Gh_D05G0536, respectively. There are three homologs of GhCIPK6 (KC465063), Gorai.010G112400.1 in the D5 genome, Gh_A06G0873 and Gh_D06G1020 in AD1 genome, but there was no homolog in the A2 genome. Each predicted chromosome locations of GhCIPK6a (HM002633) and GhCIPK6 (KC465063) in A2, D5, and AD1 genomes were shown in Table 1. Multiple sequence alignment and phylogenic analysis confirmed that the two GhCIPK6s were two different genes in sequence. The gene GhCIPK6 (KC465063) might be a paralog of GhCIPK6a (HM002633), created by cotton-specific duplication and evolution.
To confirm that the differences between GhCIPK6a (HM002633) and GhCIPK6 (KC465063) not only involved their nucleotide sequences, we analyzed the phosphorylated sites in amino acid sequences (Additional file 3 Table S3, Additional file 15 Fig. S10). Differences between amino acid sequences caused differences in the phosphorylated sites that might activate downstream genes.
Using public data, we analyzed the relative expression levels of GhCIPK6a (HM002633) and GhCIPK6 (KC465063) in different cotton tissues, at different stages of fiber development and subjected to various stresses (Additional file 9 Fig. S4). We found that the expression level of GhCIPK6a (HM002633) was strongly higher than GhCIPK6 (KC465063) in different tissues under different treatments. GhCIPK6a (HM002633) was more sensitive to salt stress than GhCIPK6 (KC465063). The transcripts of GhCIPK6a (HM002633) was up-regulated to a greater extent in roots of salt-tolerant and salt-sensitive cotton cultivars after different salt treatment times. Heterologous expression of GhCIPK6 (KC465063) significantly enhanced tolerance to salt, drought, and ABA treatments in transgenic Arabidopsis, but there was no evidence that this gene could increase abiotic tolerance when overexpressed in cotton. Therefore, abiotic stress had a greater effect on GhCIPK6a (HM002633) expression than that on GhCIPK6 (KC465063).
GhCIPK6a can be used in cotton stress-tolerance breeding
Seed germination stage is the most sensitive stage to salt stress for cotton development [76], and the seed germination rate of over-expressing GhCIPK6a lines was higher than that of wild type under abiotic stress. The effect of salt stress on cell membrane permeability was detected by determining the electrical conductivity after soaking the seeds for 24 h in different concentrations of NaCl solution since cotton is particularly sensitive to abiotic stresses at the germination stage. Compared with control, the transgenic line showed higher seed germination rate under varied stresses. In imbibed germination stage, the water absorbency rate of transgenic cotton also maintained a normal and stable level under salt stress. After 24 h of soaking in NaCl solution, the cell membrane of transgenic line (OE2) was more stable than control (Figs. 6, 7b).
Seed germination indexes include root length and hypocotyl length, which were measured at nine days after germination under salt and control treatments. There was no significant difference between the transgenic and wild-type cotton in seedlings growth under salt treatment and normal conditions (data not shown). In the normal and saline fields, the growth performance during the seedling and flowering stages were investigated, between GhCIPK6a overexpression lines and wild type line. The growth of transgenic and wild-type cottons was inhibited by salt stress without significant difference between them (data not shown). These results suggest that overexpression of GhCIPK6a does not relieve the inhibitory effect to seedlings growth of salt stress.
Under intense salt stress, overexpressed GhCIPK6a cotton plants could grow better than wild type cotton (Additional file 11 Fig. S6). The transgenic lines, and receptors, with salt sensitive and resistive varieties were planted in the natural salinity field in Akesu, Xinjiang Autonomous Region, China in 2013, which soil salt content below the surface 5 to 10 cm was approximately 0.92%. The OE2 line could grow better than other varieties in the salinity field. The damage degree of salt stress of OE2 cotton was lighter than controls (Additional file 11 Fig. S6). Meanwhile, seed germination, growth, yield and fiber quality traits of transgenic cotton lines were not decreased compare to wild type cotton, especially overexpressed GhCIPK6a plants showed higher adaptive capacity than receptor plants under extreme environments (Fig. 9, Table 2).
GhCIPK6a involved in multiple salt responsive pathways to response salt stress
In present study, we determined relative content of MDA in the shoots of transgenic and wild-type cotton under salt stress and normal conditions (Fig. 8). Under salt stress, relative MDA content in the OE2 line was maintained at around 1.00. However, in wild-type cotton, relative content of MDA increased with treatment time lasted. After 10 d exposure to salt stress, the MDA content of wild-type cotton was approximately 1.40-fold higher than control. The proline content in the OE2 and wild-type lines were both increased, but the increase was greater in OE2. Soil salt causes changes in physiological indexes while plants come across stress. Physiological indexes, including MDA, proline content, and the activity of antioxidant enzymes (POD and SOD), are typical parameters for evaluating abiotic stress tolerance. Plants with lower relative levels of MDA and higher levels of proline tend to maintain cell membrane stability under stresses [77]. Under salt treatment, the salt-tolerance cotton genotype displayed higher plant dry weight, photosynthesis and antioxidant enzymes activities [78]. Similarly, the activities of POD and SOD were higher in OE2 than wild type under salt stress, which implied that the transgenic cotton had a greater ability to scavenge the ROS in response to salt stress. Furthermore, the higher the activity of POD and SOD under stresses, the better the plant is able to scavenge over-produced ROS and protect its cells from ROS damage [16]. Among RNA-seq analysis results, there were some DEGs involved in the response to reactive oxygen species pathway (GO:0000302) in OE2 plants (Additional file 13 Fig. S8), which demonstrated GhCIPK6a overexpression could improve the express levels of peroxidase related genes to enhance ROS scavenging ability.
CIPKs are involved in a variety of stress responsive processes, function in multiple regulatory pathways, such as the SOS pathway, the Low-K+ response pathway [24, 74], and the ABA signaling pathway [79]. GhPP2C and GhSnRK2.6 were both strongly induced in OE2 plants under salt stress. Also, in RNA-seq analysis, there were four PP2C genes predicted to co-express with GhCIPK6a in transgenic plants under salt treatment (Fig. 10, Additional file 2 Table S2). It was speculated that GhCIPK6a is involved in the MAPK signaling pathway and plant hormone signal transduction pathway, by co-expressed with GhPP2C proteins.
The CIPK family of protein kinases regulate ion homeostasis by forming a complex with CBL proteins. The CBL4/CIPK6 complex in Arabidopsis modulates the AKT2 potassium channel in the plasma membrane [80]. In vitro CBL1, CIPK23, and AKT1 can act together to mediate the CBL-dependent enhancement of phosphorylation of target proteins by CIPKs [81]. CIPKs modulate the activity of the genes encoding K+ channels to mediate root K+ uptake, such as AtAKT1 and AtAKT2 in Arabidopsis [24, 55, 80, 82]. In wild halophyte Hordeum brevisubulatum, HbCIPK2, combined with HbCBL1, could activate HbVGKC1 to absorb K+, combined with HbCBL4/10 could modulate HbSOS1L to exclude Na+ [83]. Overexpression of GhCIPK6a in cotton might enhance tolerance to abiotic stress by interacting with GhCBLs, GhPP2C, and GhSnRK2.6, also regulates peroxidase activity, which were also verified using RNA-seq analysis between OE2 and WT cottons (Fig. 10, Additional file 13 Fig. S8). Thus, GhCIPK6a probably functions in various signal pathways that mediate response to abiotic stresses in cotton. In conclusion, GhCIPK6a could function in multiple stress responsive pathways, such as K+ uptake pathway (Fig. 5), ROS scavenging pathway (Fig. 8, Additional file 13 Fig. S8a), MAPK signaling pathway and plant hormone signal transduction pathway (Fig. 10a, b), to enhance salt tolerance in transgenic cotton. In one word, this work shows that GhCIPK6a overexpression can improve the salt tolerance in cotton.
Conclusions
We isolated the GhCIPK6a from Upland cotton under salt stress, and verified the function of GhCIPK6a by transformation and RNA-seq analysis. GhCIPK6a overexpressed lines exhibited higher seed germination rate than wild-type cotton under abiotic stresses, which functioned by involving in multiple stress responsive pathways, such as the K+ uptake pathway, the ROS scavenging pathway, MAPK signaling pathway and plant hormone signal transduction pathway. Moreover, GhCIPK6a overexpressed lines increased lint percentage, and fiber length uniformity under salt stress in the field. Therefore, GhCIPK6a has the potential for cotton breeding to improve stress-tolerance.
Methods
Plant materials and plant growth conditions
The Upland cotton cultivar ‘11–0516’ (a line bred from CCRI 12 in our laboratory) was used for genetic transformation. To amplify candidate gene sequences, cDNA and genomic DNA were isolated from the Upland cotton cultivar ‘Zhong G5’ [41, 72]. The ‘Zhong G5’seeds were provided by Chinese Academy of Agricultural Sciences primitively. The cultivars ‘11-0516’ and ‘Zhong G5’ are conserved by our laboratory now.
Upland cotton seedlings were grown in modified 1/2 Hoagland solution. Plants were grown at 28/20 °C, a light intensity of 600 mol·cm2∙s− 1, and a photoperiod of 14 h light/10 h dark [41]. At the three-leaf stage, half of the seedlings were transferred to plates containing 150 mmol·L− 1 NaCl solution in the growth medium; the remaining seedlings was grown in normal nutrient solution as control. Roots, stems, and leaves of the treated seedlings and control ones were sampled separately at 1, 2, 3, 6, and 12 h after exposure to salt stress, immediately frozen with liquid nitrogen and stored at − 80 °C for RNA extraction and qRT-PCR analysis.
Total RNA was extracted, qualified, and quantified, contaminating genomic DNA was digested with DNase I, and the RNA was reverse transcribed into cDNA following a previously described method [41].
Isolation and bioinformatics analysis of full-length GhCIPK6a
The primers which were designed using Primer Premier 5 software (PREMIER Biosoft International) and shown in Additional file 4 Table S4, were used to amplify the full-length cDNA of a previous reported CBL-interacting protein kinase (GW691274) from Upland cotton roots [41]. The fragment was obtained using a TaKaRa PCR Thermal Cycler Dice (TaKaRa Bio Inc.). The amplification conditions were as follows: 94 °C for 5 min, then 30 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 2 min, followed by 72 °C for 10 min. Then the fragment was purified and sequenced.
The amplified cDNA sequences were analyzed using Lasergene software (DNAStar, MD, USA). The gene structure was predicted using GSDS (Gene Structure Display Server, http://gsds.cbi.pku.edu.cn/) [73]. Protein conserved domains were predicted using ScanProsite online software (http://au.expasy.org/tools/scanprosite/).
Multiple sequence alignment of GhCIPK6a with CIPK proteins of other species was performed using the ClustalW program implemented in DNAMAN software, and the phylogenetic tree was constructed using MEGA5.2 software with the neighbor-joining method and the 1000 bootstrap test replicates.
Subcellular localization of GhCIPK6a
To make GFP-tagged GhCIPK6a, the coding region of GhCIPK6a was amplified with primers containing KpnI and BamHI restriction sites, and the PCR product was first cloned into pMD 18-T vector (TaKaRa), and then sequenced. After KpnI and BamHI digestion, the PCR fragment was cloned into the p3301-GFP plasmid, resulting in p3301-GhCIPK6a: GFP.
For subcellular localization in onion (Allium cepa) epidermal cells, the fusion construct p3301-GhCIPK6a: GFP was transformed into onion epidermal cells. The p3301-GFP vector was also introduced as control. The protocols of transformation and observation were referred as previously described [10]. The onion epidermal cells were plasmolyzed in 30% sucrose solution.
BiFC analysis of interaction between GhCIPK6a and GhCBLs
The fusion vectors pUC-GhCIPK6a-YFPN and pUC-GhCBLs-YFPC were constructed using gene-specific primers with restriction sites (Additional file 4 Table S4). The constructs were transiently co-expressed in onion epidermal cells [10]. After incubation for 22–24 h at 28 °C, YFP fluorescence in the transformed cells was observed by confocal laser scanning microscopy (Nikon, EZ C1).
Overexpression vector construction and cotton transformation
The coding sequence of GhCIPK6a was amplified by PCR using LA-Taq DNA polymerase and gene-specific primers containing restriction enzyme sites (Additional file 4 Table S4). GhCIPK6a was cloned and inserted into the XbaI-SacI site of the pBI121 vector, under the control of the CaMV 35S promoter. The recombinant plasmid was named pBI-GhCIPK6a. The construct was transferred into cotton cultivar (11–0516) using the pollen tube-pathway method [84]. Seedlings of independent lines of GhCIPK6a transgenic plants were identified using 5 g L− 1 kanamycin sulfate solution, and were further confirmed by amplification of NPTII, which primers were shown in Additional file 4 Table S4. The copy number of the inserted fragment was determined by Southern Blotting analysis.
Physiological index determination
To investigate the tolerance of transgenic cotton lines to abiotic stress, germination assays were performed using filter paper roll upright method. Thirty seeds of each transgenic or control cotton line were placed in a roll of filter paper, supplemented with distilled water (Control), NaCl solution (150 mmol·L− 1) and PEG6000 solution (15%, w/w), respectively, and maintained at 28 °C for 9 days, three replicates. The seed germination rate was investigated on the ninth day. The seed germination rate = the number of germinated seeds at the ninth day / the total number of seeds × 100%. Root length and hypocotyl length of seedlings were measured on the ninth day. In addition, to evaluate the chilling tolerance of the transgenic lines, seeds were placed at 15 °C for 15 d and the seed germination rate was obtained, three replicates. To determine the effect of salt stress on germination stage, water absorptivity rates were measured during the imbibition stage. Twenty seeds of each transgenic or control line exposed to solution containing different NaCl concentrations (0, 100, 150, and 200 mmol·L− 1), and maintained at 28 °C, four replicates. The weight of seeds was determined each 2 h. The water absorbency rate = (the weight of seeds after water absorption – the initial weight of seeds before soaking) / the initial weight of seeds before soaking × 100%.
For seedling growth assay, transgenic seedlings and control were grown in modified 1/2 Hoagland solution. At the three-leaf stage, transgenic and control cotton seedlings were transferred to 1/2 Hoagland solution or solution supplemented with 150 mmol·L− 1 NaCl. Roots of salt-stressed and control seedlings were sampled at 1, 2, 3, 6, and 12 h after treatment, immediately frozen in liquid nitrogen, and then stored at − 80 °C until used for RNA extraction and qRT-PCR analysis. After growing for 2, 5, and 10 days under salt stress, leaves were sampled for determination of physiological indexes, including the MDA and proline contents, and antioxidant enzyme (SOD and POD) activity, followed previous methods [85–87]. All assays include three biological replications with six plants each replication and error bars indicate the standard deviation (SD).
Field phenotype assay
To evaluate the salt tolerance of transgenic cotton lines during the whole growth and development period, field trials were carried out in Quzhou Experimental Station of China Agricultural University at Handan City (36°78′N, 114°92′E), Hebei Province, China, in summer season of 2016 to 2017. Cotton plants were planted in four row plots with 13 holes each row. Plots were 4 m in length with 80 cm row spacing for the experiment. Plants were spaced 33 cm in rows. Each hole planted eight seeds. The field design followed a randomized complete block design with four replications, respectively. Field management followed the local conventional standard field practices. The saline field was irrigated with 0.4% saline water before sowing, and the control field was irrigated with fresh water [12]. The receptor cultivar ‘11–0516’ was used as control in normal and salinity conditions.
The seedling field emergence percentage was investigated after sowing 30 days, which were used to reflect the status of the seedling emergency and survival. The seedling emergency percentage = the number of survival seedling / the number of total sowing seeds (eight seeds per hole× 13 holes per row × four rows) × 100%. Boll number was surveyed from 44 individuals in each plot on Sept 15th. Yield related traits consisted of boll weight (g) and lint percent (%). Fiber length (mm), fiber length uniformity (%), fiber strength (cN·tex− 1), fiber elongation (%), and Micronaire were determined with HVI 1000 (Uster® HVISPECTRUM, Spinlab, USA) by the Cotton Quality Supervision, Inspection and Testing Center, Ministry of Agriculture, Anyang, Henan Province, China.
Expression analysis based on public expression data of cotton
Public cotton expression data (Additional file 5 Table S5) were obtained from PLEXdb (http://www.plexdb.org/index.php) and Gene expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). Transcriptome differences between the root tissues of two cotton cultivars (salt-tolerant ‘Zhong 07’ and salt-sensitive ‘Zhong G5’) were evaluated after 3, 12, and 48 h of exposure to salt stress [72], and expression profiles of candidate genes were also analyzed. All microarray data was normalized using robust multichip analysis (RMA). The expression profiles of several candidate genes were examined using quantitative RT-PCR in transgenic cotton plants exposed to salt stress. The heatmap was constructed by Mev software (http://mev.tm4.org/).
RNA sequencing and analysis
Total RNA was extracted from the root samples of OE2 transgenic line and the receptor cultivar ‘11–0516’ seedlings, which were treated for 0, 1, 6, 12, 24 and 48 h, respectively, and three biological replicates. RNA-seq libraries were prepared and sequenced on an Illumina Hiseq2500 platform (Biomarker Technology Corporation, Beijing, China). All raw data were submitted in the NCBI sequence read archive (http://www.ncbi.nlm.nih.gov/sra) with project ID PRJNA644135 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA644135).
Significant differential expression analysis was using the DEGSeq method [88], defined as the fold change (salt treated cv 0 h) > 2 and FDR < 0.01, that analysis was performed using BMKCloud (www.biocloud.net). Gene Ontology (GO) analysis was conducted by WEGO online software. The DEGs enriched pathways analysis was used Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Protein–protein interaction (PPI) networks was established using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) v11 (http://string-db.org/) and Cytoscape app (http://apps.cytoscape.org/apps/stringapp) [89, 90]. A combined score ≥ 0.4 was chosen for PPI network construction.
Quantitative RT-PCR analysis
Quantitative RT-PCR analyzed the expression profiles of candidate genes in transgenic cotton plants and wild type subjected to salt stress. Quantitative RT-PCR was performed in triplicate with an ABI PRISM® 7500 Real-Time PCR System using the SYBR® Premix Ex Taq™ Kit (TaKaRa, DRR041A), according to the manufacturer’s instruction. The gene-specific primers were shown in Additional file 4 Table S4. The GhUBQ7 transcript was used as a normalization control to quantify relative levels. Relative expression levels were calculated using the 2-∆∆t method [91, 92]. Standard deviation was calculated from three biological replicates.
Statistical analysis
Statistical analyses were performed using SPSS software (13.0 for Windows Evaluation Version, SPSS Inc., LEAD Technologies, USA). All significant differences were identified using a t-test and marked as *, p-value < 0.05; **, p-value < 0.01.
Supplementary information
Additional file 1: Table S1. Up- and down-regulated DEGs in OE2 cotton treated by salt stress screening from RNA-seq analysis.
Additional file 2: Table S2. The edges and nodes information of Co-expression genes with GhCIPK6a in PPI network predicted using STRING online program.
Additional file 3: Table S3. Prediction of phosphorylated sites of GhCIPK6a (HM002633) and GhCIPK6 (KC465063) by KinasePhos (http://kinasephos.mbc.nctu.edu.tw/).
Additional file 4: Table S4. Primers used in vectors constructed and qRT-PCR. The underlined sequences were the restriction enzymes sequences.
Additional file 5: Table S5. Public cotton expression data from PLEXdb (http://www.plexdb.org/index.php) and the Gene expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/).
Additional file 6: Figure S1. Phylogenic analysis of GhCIPK6a (HM002633) and CIPK homolog proteins from other species. Full-length amino acid sequences were aligned using the integrated ClustalW and phylogenetic tree was constructed using the neighbor-joining method implemented in MEGA5.2 (1000 bootstrap test replicates). Black triangles indicated GhCIPK6a (HM002633) and homologs from the A2, D5, and AD1 genomes, respectively. Black circles indicated GhCIPK6 (KC465063), and homologs from the D5 and AD1 genomes, respectively. The percentage of replicate trees was shown at the branches. Different colored branches represented different groups. Proteins, located in the branch marked by a blue triangle, were chosen for multiple alignments using DNAMAN software.
Additional file 7: Figure S2. Expression analysis of GhCIPK6a in different tissues after salt treatment. A. Expression analysis in Upland cotton cultivar ‘Zhong G5’; B. Expression profile in OE2 and wild-type lines.
Additional file 8: Figure S3. PCR analysis and Southern blotting assay of positive transgenic plants. A. Identification of positive T1 transgenic plants by amplifying the resistant gene NPTII. The samples marked in the red box were the positive T1 GhCIPK6a transgenic plants. M, D2000 DNA ladder; 1–11, positive individuals: 11 J100–1, 11 J100–2, 11 J100–7, 11 J100–8, 11 J100–12, 11 J100–15, 11 J100–21, 11 J100–24, 11 J100–27, 11 J100–34, 11 J100–35, respectively. CK+: positive control. The samples out of the red box were not involved here. B. Southern blotting assay of transgenic cotton plants using the resistance gene NPTII as the probe. In the red box: M, D2000 DNA ladder; 1, OE1 (12D44); 2, OE2 (12D47), which were the sample used in present study. 3–8, represents different individuals, which were 12D48-12D53, respectively. C and D. The genealogical diagram of OE1 (12D44) and OE2 (12D47) lines and their offspring.
Additional file 9: Figure S4. Expression profiles of GhCIPK6s under abiotic stress using public datasets. I, public data from PLEXdb. II, GSE50770, analyzed the crosstalk between genes that are responsive to multiple abiotic stresses, including ABA, cold, drought, salinity, and alkalinity (pH) in G. hirsutum. III, Transcriptome analysis between roots of the salt-tolerant cultivar ‘Zhong 07’ and salt-sensitive cultivar ‘Zhong G5’ after 3, 12, and 48 h of 150 mmol·L− 1 NaCl treatment.
Additional file 10: Figure S5. The germination performance of transgenic lines (OE1 and OE2) and wild type (WT) line under salt treatment (NaCl) and control (CK) conditions. Bar = 1 cm.
Additional file 11: Figure S6. Salt tolerance assay of transgenic and control cottons during flowering and boll setting stage in the field in Akesu, Xinjiang Autonomous Region, China in 2013. The salt content of the soil under the surface 5 to 10 cm was approximately 0.92%.
Additional file 12: Figure S7. Venn diagrams of up- and down-regulated DEGs among different salt stress time points in between OE2 and WT plants, identified from RNA-seq analysis.
Additional file 13: Figure S8. GO-term and KEGG analysis of up- and down-regulated DEGs. A. GO-term analysis of specific up- and down-regulated DEGs in OE plants following salt treatment. B. KEGG pathway analysis of 78 candidate DEGs.
Additional file 14: Figure S9. Validation of express profiles of candidate DEGs by qRT-PCR. A. RNA-seq analysis and qRT-PCR analysis of the co-expressed DEGs in PPI network. B. Correlation of the expression profiles between RNA-seq and qRT-PCR analysis.
Additional file 15: Figure S10. Analysis of differences in amino acid sequences and phosphorylation sites between GhCIPK6a (HM002633) and GhCIPK6 (KC465063). A. Sequence alignment of GhCIPK6a (HM002633) and GhCIPK6 (KC465063). B. Schematic of predicted phosphorylation sites of GhCIPK6s by KinasePhos (http://kinasephos.mbc.nctu.edu.tw/).
Acknowledgements
We thank Dr. Lida Zhang (Shanghai Jiaotong University) for helpful discussion and Mr. Junbo Zhen (Hebei Academy of Agriculture and Forestry Sciences) for attending part work in gene cloning. We thank Dr. Heng Jian and Dr. Xiaoli Tian (China Agricultural University) for providing vectors used in the BiFC assay and subcellular localization assay, respectively. We thank Dr. Kunbo Wang (Chinese Academy of Agricultural Sciences) for kindly providing ‘Zhong G5’ seeds.
Abbreviations
- ABA
Abscisic acid
- ABI
Abscisic acid-insensitive
- BiFC
Bimolecular fluorescence complementation
- CaMV
Cauliflower mosaic virus
- CBL
Calcineurin B-like proteins
- CIPK
CBL-interacting protein kinases
- DEG
Differentially expressed genes
- GEO
Gene expression Omnibus
- GFP
Green fluorescent protein
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
Mitogen-activated protein kinases
- MDA
Dicarboxylic aldehyde
- OE
Overexpression
- PLEXdb
Plant Expression Database
- POD
Peroxidase
- PP2C
Protein phosphatase 2C
- PPI
Protein phosphatase interaction
- qRT-PCR
Quantitative real-time PCR
- ROS
Reactive oxygen species
- SD
Standard deviation
- SnRK
Sucrose-nonfermenting-related kinases
- SOD
Superoxide dismutase
- SOS
Salt overly sensitive
- WT
Wild type
Authors’ contributions
YS performed all experiments, data analysis and prepared the manuscript. AG attended to prepare the RNA samples for RNA-seq. YH attended discussion and revised the manuscript. YW maintained experimental platform and performed bench work. JH conceived the experiments, provided experimental platform, and revised the manuscript. All authors approved of the final manuscript.
Funding
This research was supported in part by National Natural Science Foundation of China (Project No. 31530053) and National Key R & D Program for Crop Breeding (Project No. 2016YFD0100305), which provided the fund for our research, but the funding organizations were not involved in the design of the study, data collection, analysis of the data, or the writing of the manuscript.
Availability of data and materials
All transcriptome raw data are available at NCBI project ID PRJNA644135 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA644135).
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ying Su, Email: suying0425@cau.edu.cn.
Anhui Guo, Email: guoah@cau.edu.cn.
Yi Huang, Email: huangyi@oilcrops.cn.
Yumei Wang, Email: yumeiwang001@126.com.
Jinping Hua, Email: jinping_hua@cau.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02548-4.
References
- 1.Mai WX, Tian CY, Li L. Localized salt accumulation: the main reason for cotton root length decrease during advanced growth stages under drip irrigation with mulch film in a saline soil. J Arid Land. 2014;6(3):361–370. [Google Scholar]
- 2.Murshed R, Lopez-Lauri F, Sallanon H. Effect of salt stress on tomato fruit antioxidant systems depends on fruit development stage. Physiol Mol Biol Plants. 2014;20(1):15–29. doi: 10.1007/s12298-013-0209-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krasensky J, Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot. 2012;63(4):1593–1608. doi: 10.1093/jxb/err460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018;217(2):523–539. doi: 10.1111/nph.14920. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, Guo Y. Unraveling salt stress signaling in plants. J Integr Plant Biol. 2018;60(9):796–804. doi: 10.1111/jipb.12689. [DOI] [PubMed] [Google Scholar]
- 6.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134(1):43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinhorst L, Kudla J. Calcium and reactive oxygen species rule the waves of signaling. Plant Physiol. 2013;163(2):471–485. doi: 10.1104/pp.113.222950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golldack D, Luking I, Yang O. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011;30(8):1383–1391. doi: 10.1007/s00299-011-1068-0. [DOI] [PubMed] [Google Scholar]
- 10.Su Y, Liang W, Liu Z, Wang Y, Zhao Y, Ijaz B, Hua J. Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. J Plant Physiol. 2017;218:222–234. doi: 10.1016/j.jplph.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Fujii H, Zhu JK. Osmotic stress signaling via protein kinases. Cell Mol Life Sci. 2012;69(19):3165–3173. doi: 10.1007/s00018-012-1087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Y, Wang YM, Zhen JB, Zhang X, Chen ZW, Li L, Huang Y, Hua JP. SnRK2 homologs in Gossypium and GhSnRK2.6 improved salt tolerance in transgenic upland cotton and Arabidopsis. Plant Mol Biol Report. 2017;35(4):442–456. [Google Scholar]
- 13.Shinozawa A, Otake R, Takezawa D, Umezawa T, Komatsu K, Tanaka K, Amagai A, Ishikawa S, Hara Y, Kamisugi Y, et al. SnRK2 protein kinases represent an ancient system in plants for adaptation to a terrestrial environment. Commun Biol. 2019;2(1):30. doi: 10.1038/s42003-019-0281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang X, Zhang L, Natarajan SK, Becker DF. Proline mechanisms of stress survival. Antioxid Redox Signal. 2013;19(9):998–1011. doi: 10.1089/ars.2012.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saadia M, Jamil A, Akram NA, Ashraf M. A study of proline metabolism in canola (Brassica napus L.) seedlings under salt stress. Molecules. 2012;17(5):5803–5815. doi: 10.3390/molecules17055803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki N, Koussevitzky S, Mittler R, Miller G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012;35(2):259–270. doi: 10.1111/j.1365-3040.2011.02336.x. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal PK, Shukla PS, Gupta K, Jha B. Bioengineering for salinity tolerance in plants: state of the art. Mol Biotechnol. 2013;54(1):102–123. doi: 10.1007/s12033-012-9538-3. [DOI] [PubMed] [Google Scholar]
- 18.Hayat S, Hayat Q, Alyemeni MN, Ahmad A. Proline enhances antioxidative enzyme activity, photosynthesis and yield of Cicer arietinum L. exposed to cadmium stress. Acta Bot Croat. 2013;72(2):323–335. [Google Scholar]
- 19.Mao J, Manik SM, Shi S, Chao J, Jin Y, Wang Q, Liu H. Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes. 2016;7(9):62. doi: 10.3390/genes7090062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanyal SK, Rao S, Mishra LK, Sharma M, Pandey GK. Plant stress responses mediated by CBL-CIPK phosphorylation network. Enzymes. 2016;40:31–64. doi: 10.1016/bs.enz.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Batistic O, Kudla J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta. 2004;219(6):915–924. doi: 10.1007/s00425-004-1333-3. [DOI] [PubMed] [Google Scholar]
- 22.Albrecht V, Ritz O, Linder S, Harter K, Kudla J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001;20(5):1051–1063. doi: 10.1093/emboj/20.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta M, Guo Y, Halfter U, Zhu JK. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci U S A. 2003;100(20):11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan WZ, Lee SC, Che YF, Jiang YQ, Luan S. Mechanistic analysis of AKT1 regulation by the CBL-CIPK-PP2CA interactions. Mol Plant. 2011;4(3):527–536. doi: 10.1093/mp/ssr031. [DOI] [PubMed] [Google Scholar]
- 25.Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S. A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci U S A. 2007;104(40):15959–15964. doi: 10.1073/pnas.0707912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KN, Lee JS, Han H, Choi SA, Go SJ, Yoon IS. Isolation and characterization of a novel rice Ca2+−regulated protein kinase gene involved in responses to diverse signals including cold, light, cytokinins, sugars and salts. Plant Mol Biol. 2003;52(6):1191–1202. doi: 10.1023/b:plan.0000004330.62660.a2. [DOI] [PubMed] [Google Scholar]
- 27.Kanwar P, Sanyal SK, Tokas I, Yadav AK, Pandey A, Kapoor S, Pandey GK. Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium. 2014;56(2):81–95. doi: 10.1016/j.ceca.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Xiang Y, Huang Y, Xiong L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007;144(3):1416–1428. doi: 10.1104/pp.107.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong G, Liu X, Qiu P, Wu X, Du Z, Zhang J, Yang L, Wu Z. Rice grassy stunt virus p5 interacts with two protein components of the plant-specific CBL-CIPK Ca(+2) signaling network of rice. Virus Genes. 2017;53(3):446–453. doi: 10.1007/s11262-017-1437-z. [DOI] [PubMed] [Google Scholar]
- 30.Lin XE, Ji HQ, Niu JH, Hu YM, Fu ZJ, Liu ZH, Tang JH. Cloning one CIPK gene from a thermo-sensitive genic self-incompatible line in maize expressing under different temperatures. Agric Sci China. 2011;10(6):813–819. [Google Scholar]
- 31.Tai FJ, Wang Q, Yuan ZL, Yuan ZH, Li HY, Wang W. Characterization of five CIPK genes expressions in maize under water stress. Acta Physiol Plant. 2013;35(5):1555–1564. [Google Scholar]
- 32.Zhang F, Li L, Jiao Z, Chen Y, Liu H, Chen X, Fu J, Wang G, Zheng J. Characterization of the calcineurin B-like (CBL) gene family in maize and functional analysis of ZmCBL9 under abscisic acid and abiotic stress treatments. Plant Sci. 2016;253:118–129. doi: 10.1016/j.plantsci.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 33.Lv FL, Zhang HC, Xia XL, Yin WL. Expression profiling and functional characterization of a CBL-interacting protein kinase gene from Populus euphratica. Plant Cell Rep. 2014;33(5):807–818. doi: 10.1007/s00299-013-1557-4. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Lv F, Han X, Xia X, Yin W. The calcium sensor PeCBL1, interacting with PeCIPK24/25 and PeCIPK26, regulates Na+/K+ homeostasis in Populus euphratica. Plant Cell Rep. 2013;32(5):611–621. doi: 10.1007/s00299-013-1394-5. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Ren F, Zhou L, Wang QQ, Zhong H, Li XB. The Brassica napus Calcineurin B-like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signaling. J Exp Bot. 2012;63(17):6211–6222. doi: 10.1093/jxb/ers273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang HF, Yang B, Liu WZ, Li HW, Wang L, Wang BY, Deng M, Liang WW, Deyholos MK, Jiang YQ. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.) BMC Plant Biol. 2014;14:8. doi: 10.1186/1471-2229-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Jiang MM, Ren L, Liu Y, Chen HY. Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.) Mol Gen Genomics. 2016;291(4):1769–1781. doi: 10.1007/s00438-016-1218-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Zhou X, Liu S, Yu A, Yang C, Chen X, Liu J, Wang A. Identification and functional analysis of tomato CIPK gene family. Int J Mol Sci. 2019;21(1):110. doi: 10.3390/ijms21010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J, Yu A, Du Y, Wang G, Li Y, Zhao G, Wang X, Zhang W, Cheng K, Liu X, et al. Foxtail millet (Setaria italica (L.) P. Beauv) CIPKs are responsive to ABA and abiotic stresses. PLoS One. 2019;14(11):e0225091. doi: 10.1371/journal.pone.0225091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aslam M, Fakher B, Jakada BH, Zhao L, Cao S, Cheng Y, Qin Y. Genome-wide identification and expression profiling of CBL-CIPK gene family in pineapple (Ananas comosus) and the role of AcCBL1 in abiotic and biotic stress response. Biomolecules. 2019;9(7):293. doi: 10.3390/biom9070293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Zhen JB, Li ZH, Kang DM, Yang YM, Kong J, Hua JP. Expression profile of early responsive genes under salt stress in upland cotton (Gossypium hirsutum L.) Plant Mol Biol Report. 2011;29(3):626–637. [Google Scholar]
- 42.He L, Yang X, Wang L, Zhu L, Zhou T, Deng J, Zhang X. Molecular cloning and functional characterization of a novel cotton CBL-interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem Biophys Res Commun. 2013;435(2):209–215. doi: 10.1016/j.bbrc.2013.04.080. [DOI] [PubMed] [Google Scholar]
- 43.Wang JJ, Lu XK, Yin ZJ, Mu M, Zhao XJ, Wang DL, Wang S, Fan WL, Guo LX, Ye WW, et al. Genome-wide identification and expression analysis of CIPK genes in diploid cottons. Genet Mol Res. 2016;15(4):15048852. doi: 10.4238/gmr15048852. [DOI] [PubMed] [Google Scholar]
- 44.Guo K, Tu LL, He YH, Deng JW, Wang MJ, Huang H, Li ZH, Zhang XL. Interaction between calcium and potassium modulates elongation rate in cotton fiber cells. J Exp Bot. 2017;68(18):5161–5175. doi: 10.1093/jxb/erx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Batistic O, Kudla J. Analysis of calcium signaling pathways in plants. Biochim Biophys Acta. 2012;1820(8):1283–1293. doi: 10.1016/j.bbagen.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 46.Tang RJ, Zhao FG, Garcia VJ, Kleist TJ, Yang L, Zhang HX, Luan S. Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc Natl Acad Sci U S A. 2015;112(10):3134–3139. doi: 10.1073/pnas.1420944112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu QY, An LJ, Li WL. The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014;33(2):203–214. doi: 10.1007/s00299-013-1507-1. [DOI] [PubMed] [Google Scholar]
- 48.Liu P, Duan Y, Liu C, Xue Q, Guo J, Qi T, Kang Z, Guo J. The calcium sensor TaCBL4 and its interacting protein TaCIPK5 are required for wheat resistance to stripe rust fungus. J Exp Bot. 2018;69(18):4443–4457. doi: 10.1093/jxb/ery227. [DOI] [PubMed] [Google Scholar]
- 49.Yasuda S, Aoyama S, Hasegawa Y, Sato T, Yamaguchi J. Arabidopsis CBL-interacting protein kinases regulate carbon/nitrogen-nutrient response by phosphorylating ubiquitin ligase ATL31. Mol Plant. 2017;10(4):605–618. doi: 10.1016/j.molp.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Yin X, Xia Y, Xie Q, Cao Y, Wang Z, Hao G, Song J, Zhou Y, Jiang X: CBL10-CIPK8-SOS1, a novel SOS pathway, functions in Arabidopsis to regulate salt tolerance. J Exp Bot 2019, http://doi: 10.1093/jxb/erz549. [DOI] [PMC free article] [PubMed]
- 51.Chen L, Wang QQ, Zhou L, Ren F, Li DD, Li XB. Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol Biol Rep. 2013;40(8):4759–4767. doi: 10.1007/s11033-013-2572-9. [DOI] [PubMed] [Google Scholar]
- 52.Tripathi V, Parasuraman B, Laxmi A, Chattopadhyay D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009;58(5):778–790. doi: 10.1111/j.1365-313X.2009.03812.x. [DOI] [PubMed] [Google Scholar]
- 53.Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci U S A. 2002;99(12):8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008;56(3):505–516. doi: 10.1111/j.1365-313X.2008.03612.x. [DOI] [PubMed] [Google Scholar]
- 55.Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007;52(2):223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- 56.Li L, Kim BG, Cheong YH, Pandey GK, Luan S. A Ca (2)+ signaling pathway regulates a K(+) channel for low-K response in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103(33):12625–12630. doi: 10.1073/pnas.0605129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang XP, Chen LM, Liu WX, Shen LK, Wang FL, Zhou Y, Zhang Z, Wu WH, Wang Y. AtKC1 and CIPK23 synergistically modulate AKT1-mediated low-potassium stress responses in Arabidopsis. Plant Physiol. 2016;170(4):2264–2277. doi: 10.1104/pp.15.01493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vert G, Chory J. A toggle switch in plant nitrate uptake. Cell. 2009;138(6):1064–1066. doi: 10.1016/j.cell.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138(6):1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Chinnusamy V, Jagendorf A, Zhu JK. Understanding and improving salt tolerance in plants. Crop Sci. 2005;45(2):437–448. [Google Scholar]
- 61.Iqbal K, Azhar FM, Khan IA. Ehsan-Ullah: variability for drought tolerance in cotton (Gossypium hirsutum) and its genetic basis. Int J Agric Biol. 2011;13(1):61–66. [Google Scholar]
- 62.Brugnoli E, Bjorkman O. Growth of cotton under continuous salinity stress: influence on allocation pattern, stomatal and non-stomatal components of photosynthesis and dissipation of excess light energy. Planta. 1992;187(3):335–347. doi: 10.1007/BF00195657. [DOI] [PubMed] [Google Scholar]
- 63.Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 64.Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33(5):524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 65.Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin DC, Llewellyn D, Showmaker KC, Shu SQ, Udall J, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492(7429):423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 66.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 67.Fang L, Wang Q, Hu Y, Jia Y, Chen J, Liu B, Zhang Z, Guan X, Chen S, Zhou B, et al. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat Genet. 2017;49(7):1089–1098. doi: 10.1038/ng.3887. [DOI] [PubMed] [Google Scholar]
- 68.Wang M, Tu L, Yuan D, Zhu SC, Li J, Liu F, Pei L, Wang P, Zhao G, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet. 2019;51(2):224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- 69.Hu Y, Chen JD, Fang L, Zhang ZY, Ma W, Niu YC, Ju LZ, Deng JQ, Zhao T, Lian JM, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–748. doi: 10.1038/s41588-019-0371-5. [DOI] [PubMed] [Google Scholar]
- 70.Gao P, Zhao PM, Wang J, Wang HY, Wu XM, Xia GX. Identification of genes preferentially expressed in cotton fibers: a possible role of calcium signaling in cotton fiber elongation. Plant Sci. 2007;173(1):61–69. [Google Scholar]
- 71.Gao P, Zhao PM, Wang J, Wang HY, Du XM, Wang GL, Xia GX. Co-expression and preferential interaction between two calcineurin B-like proteins and a CBL-interacting protein kinase from cotton. Plant Physiol Biochem. 2008;46(10):935–940. doi: 10.1016/j.plaphy.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 72.Guo J, Shi G, Guo X, Zhang L, Xu W, Wang Y, Su Z, Hua J. Transcriptome analysis reveals that distinct metabolic pathways operate in salt-tolerant and salt-sensitive upland cotton varieties subjected to salinity stress. Plant Sci. 2015;238:33–45. doi: 10.1016/j.plantsci.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 73.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanchez-Barrena MJ, Martinez-Ripoll M, Albert A. Structural biology of a major signaling network that regulates plant abiotic stress: the CBL-CIPK mediated pathway. Int J Mol Sci. 2013;14(3):5734–5749. doi: 10.3390/ijms14035734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Z, Ge X, Yang Z, Zhang C, Zhao G, Chen E, Liu J, Zhang X, Li F. Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.) BMC Genet. 2017;18(1):54. doi: 10.1186/s12863-017-0517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sattar S, Hussnain T, Javaid A. Effect of NaCl salinity on cotton (Gossypium arboreum L.) grown on MS medium and in hydroponic cultures. J Anim Plant Sci. 2010;20(2):87–89. [Google Scholar]
- 77.Ben Rejeb K, Abdelly C, Savoure A. How reactive oxygen species and proline face stress together. Plant Physiol Biochem. 2014;80:278–284. doi: 10.1016/j.plaphy.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 78.Ibrahim W, Qiu CW, Zhang C, Cao F, Shuijin Z, Wu F. Comparative physiological analysis in the tolerance to salinity and drought individual and combination in two cotton genotypes with contrasting salt tolerance. Physiol Plant. 2019;165(2):155–168. doi: 10.1111/ppl.12791. [DOI] [PubMed] [Google Scholar]
- 79.Danquah A, de Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv. 2014;32(1):40–52. doi: 10.1016/j.biotechadv.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 80.Held K, Pascaud F, Eckert C, Gajdanowicz P, Hashimoto K, Corratge-Faillie C, Offenborn JN, Lacombe B, Dreyer I, Thibaud JB, et al. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 2011;21(7):1116–1130. doi: 10.1038/cr.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hashimoto K, Eckert C, Anschutz U, Scholz M, Held K, Waadt R, Reyer A, Hippler M, Becker D, Kudla J. Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J Biol Chem. 2012;287(11):7956–7968. doi: 10.1074/jbc.M111.279331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu LL, Ren HM, Chen LQ, Wang Y, Wu WH. A protein kinase, calcineurin B-like protein-interacting protein kinase 9, interacts with calcium sensor calcineurin B-like protein 3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiol. 2013;161(1):266–277. doi: 10.1104/pp.112.206896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang H, Feng H, Zhang J, Ge R, Zhang L, Wang Y, Li L, Wei J, Li R: Emerging crosstalk between two signaling pathways coordinates K+ and Na+ homeostasis in the halophyte Hordeum brevisubulatum. J Exp Bot 2020, doi: 10.1093/jxb/eraa191. [DOI] [PubMed]
- 84.Zhou G, Weng J, Zheng Y, Huang J, Qian S, Liu G. Introduction of exogenous DNA into cotton embryos. Methods Enzymol. 1983;101:433–481. doi: 10.1016/0076-6879(83)01032-0. [DOI] [PubMed] [Google Scholar]
- 85.Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39(1):205–207. [Google Scholar]
- 86.Misra HP, Fridovich I. The generation of superoxide radical during the autoxidation of hemoglobin. J Biol Chem. 1972;247(21):6960–6962. [PubMed] [Google Scholar]
- 87.Peever TL, Higgins VJ. Electrolyte leakage, lipoxygenase, and lipid-peroxidation induced in tomato leaf tissue by specific and nonspecific elicitors from Cladosporium fulvum. Plant Physiol. 1989;90(3):867–875. doi: 10.1104/pp.90.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 89.Demchak B, Hull T, Reich M, Liefeld T, Smoot M, Ideker T, Mesirov JP. Cytoscape: the network visualization tool for GenomeSpace workflows. F1000Research. 2014;3:151. doi: 10.12688/f1000research.4492.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 92.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Up- and down-regulated DEGs in OE2 cotton treated by salt stress screening from RNA-seq analysis.
Additional file 2: Table S2. The edges and nodes information of Co-expression genes with GhCIPK6a in PPI network predicted using STRING online program.
Additional file 3: Table S3. Prediction of phosphorylated sites of GhCIPK6a (HM002633) and GhCIPK6 (KC465063) by KinasePhos (http://kinasephos.mbc.nctu.edu.tw/).
Additional file 4: Table S4. Primers used in vectors constructed and qRT-PCR. The underlined sequences were the restriction enzymes sequences.
Additional file 5: Table S5. Public cotton expression data from PLEXdb (http://www.plexdb.org/index.php) and the Gene expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/).
Additional file 6: Figure S1. Phylogenic analysis of GhCIPK6a (HM002633) and CIPK homolog proteins from other species. Full-length amino acid sequences were aligned using the integrated ClustalW and phylogenetic tree was constructed using the neighbor-joining method implemented in MEGA5.2 (1000 bootstrap test replicates). Black triangles indicated GhCIPK6a (HM002633) and homologs from the A2, D5, and AD1 genomes, respectively. Black circles indicated GhCIPK6 (KC465063), and homologs from the D5 and AD1 genomes, respectively. The percentage of replicate trees was shown at the branches. Different colored branches represented different groups. Proteins, located in the branch marked by a blue triangle, were chosen for multiple alignments using DNAMAN software.
Additional file 7: Figure S2. Expression analysis of GhCIPK6a in different tissues after salt treatment. A. Expression analysis in Upland cotton cultivar ‘Zhong G5’; B. Expression profile in OE2 and wild-type lines.
Additional file 8: Figure S3. PCR analysis and Southern blotting assay of positive transgenic plants. A. Identification of positive T1 transgenic plants by amplifying the resistant gene NPTII. The samples marked in the red box were the positive T1 GhCIPK6a transgenic plants. M, D2000 DNA ladder; 1–11, positive individuals: 11 J100–1, 11 J100–2, 11 J100–7, 11 J100–8, 11 J100–12, 11 J100–15, 11 J100–21, 11 J100–24, 11 J100–27, 11 J100–34, 11 J100–35, respectively. CK+: positive control. The samples out of the red box were not involved here. B. Southern blotting assay of transgenic cotton plants using the resistance gene NPTII as the probe. In the red box: M, D2000 DNA ladder; 1, OE1 (12D44); 2, OE2 (12D47), which were the sample used in present study. 3–8, represents different individuals, which were 12D48-12D53, respectively. C and D. The genealogical diagram of OE1 (12D44) and OE2 (12D47) lines and their offspring.
Additional file 9: Figure S4. Expression profiles of GhCIPK6s under abiotic stress using public datasets. I, public data from PLEXdb. II, GSE50770, analyzed the crosstalk between genes that are responsive to multiple abiotic stresses, including ABA, cold, drought, salinity, and alkalinity (pH) in G. hirsutum. III, Transcriptome analysis between roots of the salt-tolerant cultivar ‘Zhong 07’ and salt-sensitive cultivar ‘Zhong G5’ after 3, 12, and 48 h of 150 mmol·L− 1 NaCl treatment.
Additional file 10: Figure S5. The germination performance of transgenic lines (OE1 and OE2) and wild type (WT) line under salt treatment (NaCl) and control (CK) conditions. Bar = 1 cm.
Additional file 11: Figure S6. Salt tolerance assay of transgenic and control cottons during flowering and boll setting stage in the field in Akesu, Xinjiang Autonomous Region, China in 2013. The salt content of the soil under the surface 5 to 10 cm was approximately 0.92%.
Additional file 12: Figure S7. Venn diagrams of up- and down-regulated DEGs among different salt stress time points in between OE2 and WT plants, identified from RNA-seq analysis.
Additional file 13: Figure S8. GO-term and KEGG analysis of up- and down-regulated DEGs. A. GO-term analysis of specific up- and down-regulated DEGs in OE plants following salt treatment. B. KEGG pathway analysis of 78 candidate DEGs.
Additional file 14: Figure S9. Validation of express profiles of candidate DEGs by qRT-PCR. A. RNA-seq analysis and qRT-PCR analysis of the co-expressed DEGs in PPI network. B. Correlation of the expression profiles between RNA-seq and qRT-PCR analysis.
Additional file 15: Figure S10. Analysis of differences in amino acid sequences and phosphorylation sites between GhCIPK6a (HM002633) and GhCIPK6 (KC465063). A. Sequence alignment of GhCIPK6a (HM002633) and GhCIPK6 (KC465063). B. Schematic of predicted phosphorylation sites of GhCIPK6s by KinasePhos (http://kinasephos.mbc.nctu.edu.tw/).
Data Availability Statement
All transcriptome raw data are available at NCBI project ID PRJNA644135 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA644135).