Abstract
Objective
The aim of this study was to examine differences in microRNA-497 (miR-497) expression during cartilage tissue formation and to test whether miR-497 directly interferes with Indian hedgehog (IHH) gene and inhibits IHH expression in human chondrocytes.
Design
At different cartilage development stages and different time points in bone matrix gelatin–induced endochondral ossification (BMG-ECO) rat models, the expression of miR-497 and the Ihh gene was monitored at the mRNA level. Bioinformatic analysis, gene mutation, dual luciferase reporter gene assays and gene expression assays at both the mRNA and protein levels in human chondrocytes were subsequently performed to validate the interaction between miR-497 and the IHH gene.
Results
The mRNA expression of miR-497 or the Ihh gene in BMG-ECO rats showed significant differences between the cartilage development stages and between different time points, and the trends in the expression of miR-497 and Ihh were reversed. Bioinformatic and dual luciferase reporter gene assays demonstrated a direct interaction between miR-497 and the IHH gene. Differential mRNA and protein expression profiles of the IHH gene in human chondrocytes after 48 hours of transfection with miR-497 mimics and a negative control indicated that miR-497 inhibited IHH expression.
Conclusion
Our study provided new clues for further functional and molecular mechanism studies of miR-497 in chondrogenesis and demonstrated a potential target for clinical therapy for cartilage degenerative disease.
Keywords: cartilage, miR-497, IHH, BMG-ECO, chondrogenesis
Introduction
MicroRNAs (miRNAs), a class of noncoding small RNAs of approximately 22 nucleotides, are endogenous, single-stranded RNAs.1 miRNAs are evolutionarily conserved and regulate more than one-third of all mammalian mRNAs, which are involved in almost all aspects of cells, such as proliferation, differentiation, communication, migration, and apoptosis.2,3 miRNAs are also implicated in the onset and development of diseases, and pathogenic or protective functions of tissue-specific miRNAs have also been demonstrated.4,5 In miRNAs, a seed sequence ranging from 6 to 8 nucleotides binds to a 3′ untranslated region (UTR) of target genes in a complementary fashion. This interaction represses the translation of miRNA target genes.6
In recent years, a number of miRNAs regulating chondrogenesis have been identified via interactions with their target genes. For example, the cartilage-specific miRNA, miR-140, is critical during chondrogenesis by targeting cartilage-related genes, such as HDAC4, CXCL12, SP1, DNPEP, BMP2, and SMAD3.7 During chondrogenic differentiation, the expression of miR-194, which mediates the differentiation process through repression of the expression of the transcription factor SOX5, is significantly downregulated.8 In our previous study, we constructed three small RNA libraries from the femoral head cartilage of Sprague-Dawley (SD) rats at postnatal days 0, 21, and 42, and identified dozens of miRNAs with significant differences during three developmental stages using a deep sequencing approach. The predicted targets of differentially expressed miRNAs were locally secreted factors and transcription factors associated with chondrocyte proliferation and differentiation.9 In our following study, miR-337 was also differently expressed in bone matrix gelatin–induced endochondral ossification (BMG-ECO) rat models and served as a repressor for TGFβ receptor type II expression.10
Based on our previous studies, we investigated the expression of miR-497 during 3 cartilage development stages of the femoral head. Using bioinformatic tools, we predicted that this miRNA targeted the IHH gene, which is associated with chondrogenesis and cartilage development. At different cartilage development stages and different time points of BMG-ECO rat models, we further examined the expression of miR-497 and the Ihh gene at the same time points and found that they exhibited opposite expression patterns and were significantly different in two rat models. Considering these findings, we combined computational analysis, gene mutation, dual luciferase reporter gene and gene expression assays to explore whether miR-497 directly targets IHH and inhibits IHH expression in human chondrocytes.
Methods
Bioinformatic Analyses
Two online bioinformatics tools, TargetScan (TargetScan6.2, http://www.targetscan.org/) and Miranda (http://www.microrna.org) were used to predict the target genes of miR-497 and their interactions according to the presence of binding sites in the seed region and the size of free energy between binding sites.
Rat Model Construction and Cartilage Collection
This study was approved by the Institutional Animal Ethics Committee of Xi’an Jiaotong University. SD rats were purchased from the Animal Centre of the Fourth Military Medical University, China. Rats were bred and kept in a climate-controlled environment and fed standard rodent chow and water ad libitum in the SPF animal house of the Department of Biochemistry and Molecular Biology, Xi’an Jiaotong University. The cartilage tissues of the femoral head were obtained from SD rats at postnatal days 0, 21, and 42 as previously described.9 In each group, 5 female and 5 male rats were sacrificed. The right sides were used for collecting cartilage and RNA extraction, and the left sides were used for paraffin sections and safranin O staining according to the routine histological treatment. Cartilage collection and RNA extraction were performed on the right leg, while paraffin sectioning and safranin O staining were conducted on the left side following a routine histological treatment.
Dark Agouti (DA) rats were obtained from the Section of Medical Inflammation Research, Lund University, Sweden. DA rats are an inbred strain and are more sensitive to various joint diseases. DA rats have been used to construct BMG-ECO and Kashin-Beck disease models to study cartilage formation and cartilage pathology, respectively.10,11 Following a previously described protocol, femur and tibia shafts from adult DA rats were collected and cut into chips of approximately 1 cm3 in size. After freezing, sequential extraction was performed to remove lipids from the bones, demineralize the matrix, and extract soluble proteins and protein polysaccharides. Next, the bone chips were subjected to vacuum freeze-drying and stored at −20°C for implantation.10,12 Thirty adult DA rats, including 15 females and 15 males, received syngeneic BMG implantation in the supraspinatus and rhomboid muscles. Rats were randomly divided into 3 groups, with each comprising 5 female and 5 male rats. At 1, 2, and 3 weeks after implantation, the rats were sacrificed to harvest the generated cartilaginous tissues. Parts of tissues were used for paraffin sections and performing toluidine blue staining following a routine histological treatment. Collected cartilage and harvested cartilaginous tissues were immediately used for RNA extraction or stored at −80°C for further RNA extraction.
RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction
According to the manufacturer’s instructions and a modified method, total RNA was extracted from cartilage or generated cartilaginous tissues with TRIzol (Invitrogen, Waltham, MA, USA).13 Briefly, the cartilage or tissue was collected as quickly as possible, homogenized by grinding in liquid nitrogen, and then transferred into DEPC-treated 1.5 mL tubes filled with 1 mL TRIzol reagent. Next, RNA was dissolved in RNase-free water and quantified by NanoDrop2000 (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA from human chondrocytes was directly extracted by TRIzol reagent.
First-strand cDNA was synthesized using a Revertaid First Strand cDNA Synthesis kit (Fermentas, Burlington, Ontario, Canada) according to the manufacturer’s instructions. A specific stem-loop primer was used for miR-497 reverse transcription. Real-time quantitative polymerase chain reaction (RT-qPCR) was performed with FastStart Universal SYBR Green Master (Roche, Indianapolis, IN, USA) for quantification. The relative expression of miRNA was normalized by rno-let-7a or U6 small nuclear RNA, and relative expression of mRNA was normalized by Gapdh or GAPDH. Each sample was tested in triplicate. The primer information for miRNAs and genes is shown in Table 1.
Table 1.
Primers of miRNAs and Genes for RT-qPCR and Primers for 3′-UTR Fragments.
| Name | Accession Number | Sequence (From 5′ to 3′) |
|---|---|---|
| rno-miR-497 Stem-loop | MIMAT0003383 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTACAAA |
| rno-miR-497 Forward | CGCCAGCAGCACACUGUGG | |
| rno-miR-497 Reverse | GTGCAGGGTCCGAGGT | |
| rno-let-7a Stem-loop | MIMAT0000774 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACTAT |
| rno-let-7a Forward | CGCTGAGGTAGTAGGTTGT | |
| rno-let-7a Reverse | GTGCAGGGTCCGAGGT | |
| Rat Ihh Forward | CCCACGTTCATTGCTCTGTC | |
| Rat Ihh Reverse | ACACGCTCCCCAGTTTCTAG | |
| Rat Gapdh Forward | NM_017008 | AAGATGGTGAAGGTCGGTGT |
| Rat Gapdh Reverse | TGACTGTGCCGTTGAACTTG | |
| hsa-miR-497 Stem-loop | MIMAT0002820 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAAAC |
| hsa-miR-497 Forward | CGCCAGCAGCACACUGUGG | |
| hsa-miR-497 Reverse | GTGCAGGGTCCGAGGT | |
| Human U6 Forward | NR_004394 | CTCGCTTCGGCAGCACA |
| Human U6 Reverse | AACGCTTCACGAATTTGCGT | |
| Human IHH Forward | NM_002181 | ATCATCTTCAAGGACGAGGAGA |
| Human IHH Reverse | GGGCCTTTGACTCGTAATACAC | |
| Human GAPDH Forward | NM_002046 | CCAAGGTCATCCATGACAACT |
| Human GAPDH Reverse | CAGGGATGATGTTCTGGAGAG | |
| 3′-UTR Forward for WT | NM_002181 | CGAGCTCAGCTGGCCCTGGAAGGGACCT |
| 3′-UTR Reverse for WT | CGAGCTCACAGAACTCTGCCCACAGACA | |
| 3′-UTR Forward for MUT | GTTGAACGACGAAAATTCCCTGGGAGCCAGCA | |
| 3′-UTR Reverse for MUT | GAATTTTCGTCGTTCAACTGAGGCGCAAGCCCA |
Recombinant Construction and Sequence Mutagenesis
Primers for the 3′-UTR fragment of the IHH gene containing putative miR-497 binding sites is shown in Table 1. The fragment was cloned into the pmirGLO luciferase vector (Promega, Fitchburg, WI, USA) downstream of the firefly luciferase gene between the SacI and XbaI enzyme incision sites. The PCR product was identified by restriction enzyme digestion and sequencing. The DNA sequences of amplified fragments were consistent with the RCh38/hg38 reference sequence from UCSC. This recombinant was referred to as wild-type vector (WT).
Mutated IHH 3′-UTR sequences, containing all six nucleotides in the putative seed-pair region, were synthesized using PCR-directed mutation. In a similar manner, the mutated fragment was cloned into the pmirGLO vector and validated by restriction enzyme digestion and sequencing assays. Recombinants harboring A-T and C-G substitution were referred to as the mutant (MUT). The primer information for mutated fragments is shown in Table 1.
Cell Culture and Transfection
HEK293T cells were cultured in DMEM high glucose medium (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA) and incubated at 37°C in a humidified incubator with 5% CO2. The mature miR-497 sequence (miR-497 mimics) and negative control (mimic NC) were ordered from GenePharm Corporation (Shanghai, China). Sequences from 5′ to 3′ are listed as follows: NC mimic sense UUCUCCGAACGUGUCACGUTT, antisense ACGUGACACGUUCGGAGAATT; miR-497 mimic sense CAGCAGCACACUGUGGUUUGU, antisense AAACCACAGUGUGCUGCUGUU. HEK293T cells were seeded into 48-well plates at 2.4 × 104 cells/well. After 24 hours, 50 nM miR-497 mimics were cotransfected with 10 ng WT or 10 ng MUT into HEK293T cells by Lipofectamine 2000 (Invitrogen, Waltham, MA, USA). Simultaneously, 50 nM mimic NC was cotransfected with 10 ng WT and 10 ng MUT.
C28/I2 chondrocytes, a human cell line, were kindly provided by Dr. Mary B. Goldring from the Harvard Institutes of Medicine, Boston, MA, USA. Chondrocytes were cultured in DMEM/F12 (HyClone, Logan, UT, USA) containing 10% FBS as previously described.14,15 Cells were seeded into 6-well plates at 2 × 105 cells/well. After 48 hours, 50 nM miR-497 mimic or mimic NC were transfected into C28/I2 cells by Lipofectamine 2000. Moreover, a blank group was used as a control and a reagent group as a mock.
Dual Luciferase Reporter Gene Assay
Forty-eight hours after cotransfection into HEK293T cells, the activity of the luciferase reporter gene was detected using a Dual-Luciferase Reporter Assay System (Promega, Fitchburg, WI, USA) on a plate-reading luminometer (Thermo, Waltham, MA, USA). Each sample was tested in quadruplicate. The relative luciferase activity was normalized against a blank control and calculated as the ratio of firefly to renilla luciferase activity.
Protein Extraction and Western Blotting
Forty-eight hours after transfection, C28/I2 cells were lysed in RIPA lysis buffer (Beyotime, Beijing, China). Protein concentrations were determined using a bicinchoninic acid (BCA) assay kit (Pierce, Waltham, MA, USA). Briefly, total protein was separated on a 10% SDS/PAGE gel and transferred to a PVDF membrane (Millipore, Burlington, MA, USA) according to standard procedures in a Bio-Rad system. GAPDH was used as a loading control on the same membrane. The primary antibodies were anti-IHH (ab52919, Abcam, Cambridge, UK) or anti-GAPDH (TA-08, ZSGB-BIO, Beijing, China). The secondary antibodies used to detect the signal were anti-mouse (31430, Thermo) or anti-rabbit IgG (31460, Thermo). The signal intensity of target proteins was determined by an enhanced chemiluminescence system (Millipore).
Statistical Analyses
Data are presented as the mean ± standard error of the mean (SEM) from 3 independent experiments. All data were first tested for a normal distribution using the Shapiro-Wilk test before differences were analyzed. One-way analysis of variance was performed among the groups, followed by post hoc tests using the Student-Newman-Keuls (SNK) test to analyze significant differences between the groups. P < 0.05 was considered statistically significant.
Results
Bioinformatic Prediction of IHH as a miR-497 Target Gene
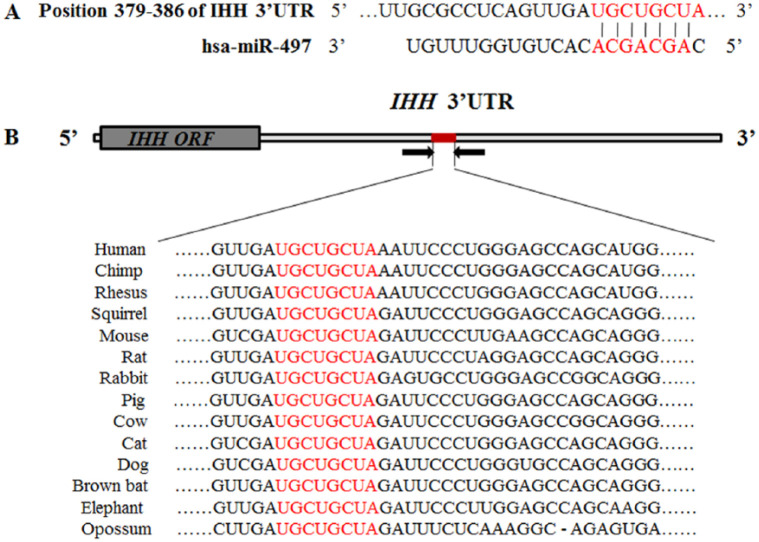
TargetScan and Miranda online tools predicted that miR-497 was a candidate miRNA for targeting the IHH gene at nucleotides 379-386 of its 3′-UTR. The complementary sequences in the IHH 3′-UTR and miR-497 seed sequence are shown in red (Fig. 1A). The sequences in the binding site of the IHH 3′-UTR were highly conserved among 14 species in vertebrates, including human, chimp, rhesus, squirrel, mouse, rat, and rabbit, suggesting its potential function. The binding sites of the IHH gene 3′-UTR are shown in red (Fig. 1B).
Figure 1.
Schematic diagrams of the IHH gene targeted by miR-497, and high conservation was observed in the binding site of the IHH 3′-UTR among 14 species in vertebrates. (A) Schematic diagram of the pairing relationship between miR-497 and the IHH 3′-UTR predicted by TargetScan and Miranda tools. The putative seed-pair region is shown in red. (B) High conservation of the target site in the IHH 3′-UTR among 14 species in vertebrates. The binding site of the IHH gene 3′-UTR is shown in red.
Downregulation of miR-497 and Reversed Expression of Ihh during Cartilage Development
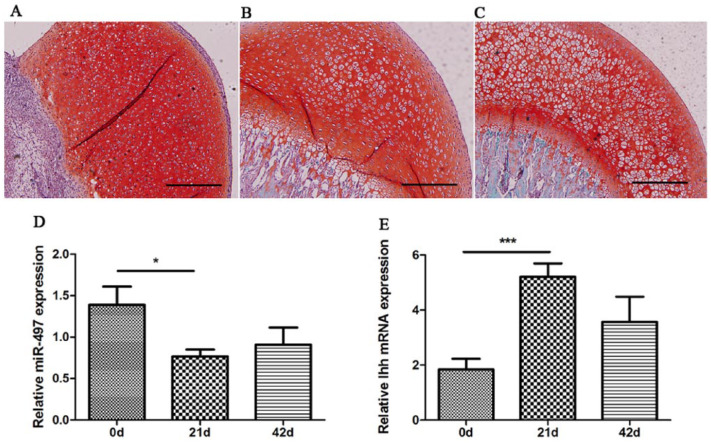
The femoral heads at postnatal days 0, 21, and 42 from SD rats were equivalent to newborn (0 years), ablactation (5 years), and juvenile (11 years) human developmental stages. Day 0, 21, and 42 represent only cartilage tissue without bone structure, the epiphyseal plate and primary ossification center formation, and the mature articular structure and secondary ossification center formation, respectively.16 Different cartilage developmental stages were identified by staining with hematoxylin and eosin and safranin O according to our previous studies.9,10 The process during the first two stages represents chondrogenesis during which cartilage-specific genes, such as aggrecan and Col2a1, are significantly upregulated. The third stage mainly involves articular-epiphyseal growth cartilage, which remains under the permanent articular cartilage and is responsible for the growth and shaping of the epiphysis.9,17 In the current study, safranin O staining was used to show this process in Figure 2A-C. The relative expression of miR-497 was significantly downregulated at postnatal days 0 and 21, suggesting its involvement in cartilage development (Fig. 2D). In contrast, the chondrogenesis-related gene Ihh showed reversed expression during the process (Fig. 2E).
Figure 2.
Relative expression of miR-497 and Ihh detected by RT-qPCR at different stages of rat cartilage development. (A-C) Representative histological images of safranin O staining at postnatal days 0, 21, and 42, respectively. The scale bar is 200 µm. (D) The downregulated expression of miR-497 in the femoral head and knee articular cartilage from Sprague-Dawley rats at postnatal day 0, 21 and 42, respectively. (E) The reversed expression of Ihh at postnatal days 0, 21, and 42. *P < 0.05, ***P < 0.001.
Downregulation of miR-497 and Reversed Expression of Ihh during Cartilage Formation
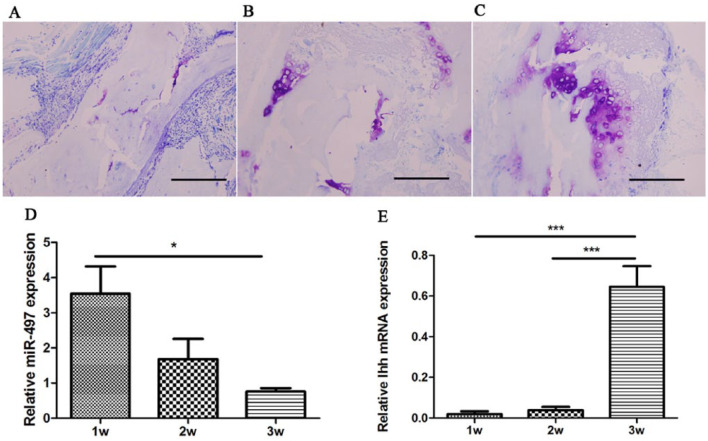
BMG planted into allogeneic rats induced endochondral ossification. The implanted bone gelatin provides the milieu for the bone morphogenetic potential of migratory mesenchymal cells from muscles. The transfer of insoluble bone morphogenetic protein (BMP) from the matrix to migratory mesenchyma1 cells accounts for chondrogenesis and endochondral ossification.12,18 The histological results of hematoxylin and eosin examination revealed small portions of cartilage 1 week after surgery; within 2 to 3 weeks, an abundant chondrocyte mass was produced. The number of chondrocytes peaked at 3 weeks, and chondrocytes gradually decreased and were replaced by bone tissue at 4 weeks.10 Therefore, the first 3 weeks reflected chondrogenesis. In the current study, chondrogenesis was displayed via toluidine blue staining as shown in Figure 3A-C. The number of generated chondrocytes was increased during chondrogenesis. Using RT-qPCR, we tested the expression of miR-497 and Ihh at 3 time points after surgery. The level of Ihh expression was significantly increased during the first 3 weeks and peaked at 3 weeks, consistent with the function of stimulating chondrocyte proliferation (Fig. 3E). The expression of miR-497 was significantly decreased during chondrocyte proliferation, suggesting its involvement in chondrogenesis (Fig. 3D).
Figure 3.
Relative expression of miR-497 and Ihh detected by RT-qPCR in endochondral ossification models induced by bone morphogenetic gelatin. (A-C) Representative histological images of toluidine blue staining at 1, 2, and 3 weeks, respectively. The scale bar is 200 µm. (D) The downregulated expression of miR-497 at 1, 2, and 3 weeks. (E) The reversed expression of Ihh at 1, 2, and 3 weeks (For interpretation of the references to colours in this figure legend, refer to the online version of this article). *P < 0.05, ***P < 0.001.
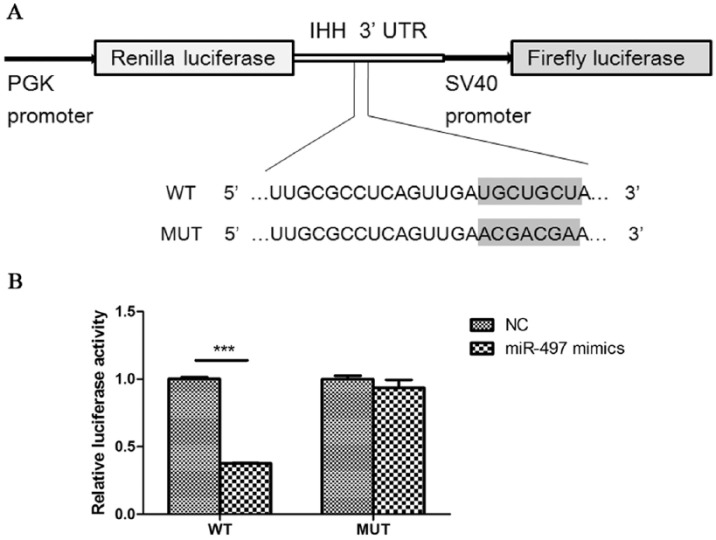
Successful Wild-Type Recombinant and Mutant Construction and Direct Regulation of IHH by miR-497
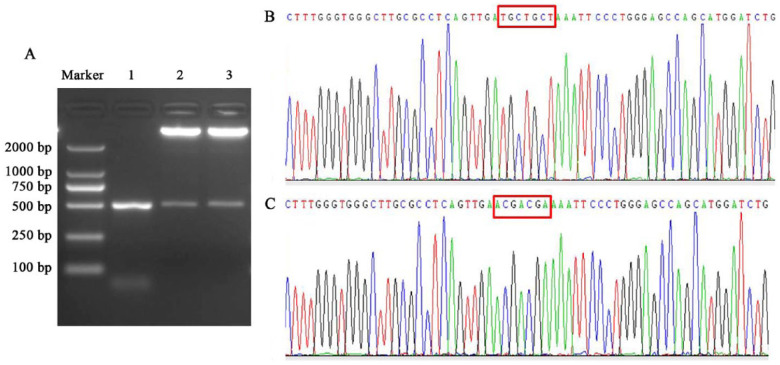
The WT and MUT recombinants were validated by restriction enzyme digestion and sequencing methods. The products of restriction enzyme digestion included two bands. One band represented the pmirGLO vector, which was 7350 bp, and the second band represented PCR products, which were 500 bp, the same size as the products before cloning into the vector (Fig. 4A). The sequencing result of the amplified fragments in the WT recombinant was consistent with the RCh38/hg38 reference sequences from the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgTracks) and NCBI database (http://www.ncbi.nlm.nih.gov/gquery/) (Fig. 4B). The mutated sequence in the binding sites of the MUT was according to the A-T and C-G substitution criterion (Fig. 2B).
Figure 4.
The results of restriction enzyme digestion and sequencing. (A) The results of restriction enzyme digestion. The DL2000 marker shows the size of DAN fragments, lane 1 shows the PCR product of the IHH 3′-UTR, and lanes 2 and 3 show the restriction enzyme digestion results of the wild-type recombinant and mutant. (B) Partial sequencing result of the wild-type recombinant. Nucleotides in the red boxes are located in the binding site in the IHH 3′-UTR. (C) Partial sequencing result of the mutant. Nucleotides in the red boxes are located in the binding site in the IHH 3′-UTR.
The luciferase activity in HEK293T cells cotransfected with miR-497 mimics and WT recombinant was significantly decreased, resulting in a 62.7% reduction compared with that in cells transfected with the negative control (Fig. 5A). Nevertheless, the luciferase activity of miR-497 mimics cotransfected with MUT showed no significant difference compared with that of the negative control, suggesting that miR-497 directly binds to the IHH 3′-UTR and represses its expression (Fig. 5B).
Figure 5.
miR-497 directly targets the IHH gene. (A) Schematic diagram of the wild-type recombinant and mutant. The shaded section of the wild-type recombinant represents the binding site with the seed sequences of miR-497. The mutation sequence in the mutant results in no binding with miR-497. (B) Dual luciferase reporter assays for miR-497 mimics/negative control cotransfected with the wild-type recombinant and mutant, respectively. WT, wild-type recombinant; MUT, mutant; NC, negative control. ***P < 0.001.
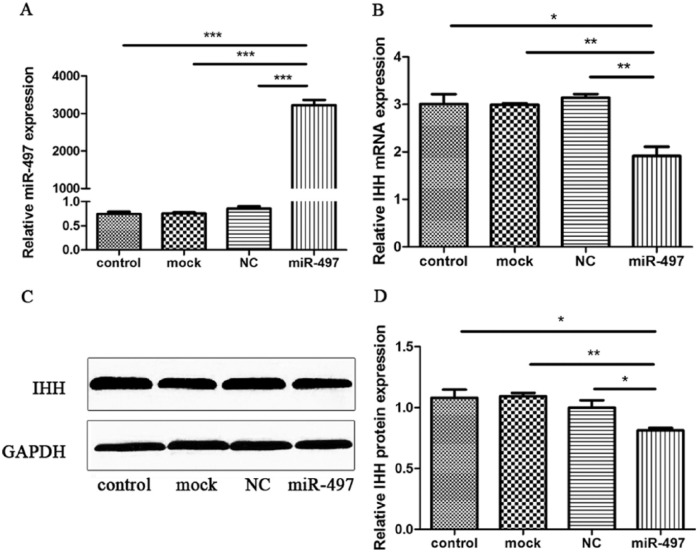
miR-497 Regulates the Endogenous Expression of IHH in Human Chondrocytes
C28/I2 cells were transfected with miR-497 mimics or negative control to determine whether endogenously expressed IHH is regulated by miR-497. The RT-qPCR results of miR-497 expression 48 hours after transfection indicated that compared with the negative control, mock, or control, miR-497 mimics caused mature miR-497 levels to increase by more than 3000-fold, suggesting high transfection efficiency (Fig. 6A). The posttransfection expression of IHH showed that compared with the negative control, mock, or control, miR-497 mimics significantly decreased the level of IHH (Fig. 6B). Western blotting results also indicated that compared with the negative control, mock, or control, miR-497 mimics could decrease the IHH protein level (Fig. 6C). Three independent Western blotting experiments were performed, and GAPDH was used as a reference gene. The relative expression of IHH was significantly decreased (Fig. 6D). Collectively, these data demonstrated that miR-497 represses endogenous IHH expression at the mRNA and protein levels in human chondrocytes.
Figure 6.
miR-497 represses the endogenous expression of IHH in human chondrocytes. (A) The RT-qPCR results of miR-497 relative expression in C28/I2 cells 48 hours after transfection. (B) The RT-qPCR results of IHH relative expression in C28/I2 cells 48 hours after transfection. (C) Western blotting results of IHH with GAPDH as an internal control. (D) Statistical analysis of Western blotting results from 3 independent Western blotting experiments. NC, negative control. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In the current study, the expression of miR-497 was significantly different at 3 time points in a postnatal rat model and BMG-ECO rat model. Although the developmental stages in different experiments were not exactly equal, the different time points reflected the cartilage developmental stages and chondrogenesis. Hence, miR-497 is associated with cartilage formation. According to bioinformatic analyses and reversed expression of miR-497 and Ihh during chondrogenesis, miR-497 probably targeted the IHH gene to play a role in chondrogenesis.
To validate the direct interaction between miR-497 and IHH, we mutated the complementary bases in the 3′-UTR of IHH, which binds to the seed sequence of miR-497. Then, dual luciferase activity assays were performed with cotransfection of WT recombinant with miR-497 mimics and NC mimics. The significantly decreased activity in the WT recombinant with miR-497 mimics compared with that with NC mimics demonstrated that miR-497 inhibits the expression of IHH. Nevertheless, the dual luciferase activity following cotransfection of the MUT with mimics compared with that following cotransfection of NC mimics showed no significant difference, indicating a direct interaction between miR-497 and the IHH gene. Using C28/I2 cells, we further validated that miR-497 inhibits endogenous IHH gene expression at both the mRNA and protein levels in human chondrocytes.
Most bones develop through the process of bone formation known as endochondral ossification. During this process, cartilage models are formed through condensation of mesenchymal cells, followed by differentiation into chondrocytes and secretion of cartilage extracellular matrix components.19,20 Proliferating chondrocytes in the center region of cartilage anlagen differentiate into prehypertrophic and hypertrophic cells and produce mineralized extracellular matrix with subsequent replacement by bone.21 Ihh is expressed by prehypertrophic and early hypertrophic chondrocytes; as a member of the hedgehog family of secreted ligands, Ihh stimulates chondrocyte proliferation and inhibits chondrocyte hypertrophy during chondrogenesis. Additionally, Ihh regulates osteoblast differentiation in the perichondrium.19 Ihh-deficient mice reportedly display markedly reduced chondrocyte proliferation, premature chondrocyte hypertrophy, and failure of osteoblast development in endochondral bones.22
In the current study, the expression of Ihh in cartilage tissue from the femoral head peaked at postnatal day 21 in rats. At this time point, the specific genes involved in cartilage development, such as Sox9, Aggrecan, ColIIα1, were upregulated.9 In BME-ECO rat models, the expression of Ihh in newly generated cartilage tissue peaked at 3 weeks when an abundant chondrocyte mass was produced.10 Ihh and parathyroid hormone–related protein (PTHrP), which is expressed in the periarticular perichondrium and early proliferating chondrocytes, form a negative feedback loop that plays a significant role in controlling chondrocyte maturation.23 In addition, Ihh promotes the expression of PTHrP, which could upregulate chondrocyte proliferation, and gives rise to delayed chondrocyte hypertrophy. Hence, PTHrP indirectly negatively regulates Ihh expression by preventing proliferative chondrocytes from converting into hypertrophic cells.24 Moreover, Ihh has a PTHrP-independent pathway that positively regulates chondrocyte proliferation mainly through the transcription factors of the Gli family.25,26 Hedgehog signals can upregulate CCND1 and CCND2 for cell cycle acceleration.27 Moreover, the BMP and IHH pathways participate in a positive feedback loop. BMP signaling induces IHH expression, but the effect of BMP signaling on proliferation and hypertrophy are IHH-independent. In contrast, IHH induces expression of various BMPs, but the effects of IHH on proliferation are independent of BMP signaling, indicating the existence of a positive feedback loop between BMPs and IHH.28,29
In the present work, IHH was directly targeted by miR-497 and showed reversed expression with miR-497 during the process of cartilage formation. Human C28/I2 chondrocytes retain chondrocytic morphology, maintain continuous proliferation in monolayer culture and express chondrocyte-specific genes, particularly anabolic and catabolic extracellular matrix genes.30,31 The gain of miR-497 function in human C28/I2 chondrocytes further demonstrated that miR-497 likely affects chondrogenesis via the IHH gene and associated pathways. miR-497 belongs to the miR-15/107 family, which is implicated in human cancers, cardiovascular disease and neurodegenerative disease.32,33 These miRNAs regulate the expression of genes involved in cell division, metabolism, stress response, and angiogenesis in vertebrate species.32 A recent study also demonstrated that miR-497 could induce cell cycle arrest by targeting Cdc25 and Ccnd in skeletal muscle stem cells.34 Therefore, miR-497 might regulate the cell cycle in chondrocytes through the IHH pathway in chondrogenesis. Considering that manipulation of some cartilage development-associated miRNAs, such as miR-101 and miR-140, has a marked therapeutic effect on osteoarthritis,35,36 a cartilage degenerative disease, miR-497 is a potential target miRNA for the therapy for diseases involving cartilage degradation. However, further functional studies on miR-497 in vitro and in vivo are needed in the future.
In conclusion, the current study revealed that miR-497 is implicated in chondrogenesis and directly targets the IHH gene and inhibits its endogenous expression in human chondrocytes at both the mRNA and protein levels. Our study provided new clues for further functional and molecular mechanism studies of miR-497 in chondrogenesis and the development of novel clinical therapies for cartilage degenerative disease.
Footnotes
Acknowledgments and Funding: The authors would like to acknowledge all the individuals who have participated in or helped with our research. This study was supported by the Natural Science Foundation of Shaanxi Province (No. 2018JM7081; 2018JM7039), the National Natural Science Foundation of China (No. 31371298; 81301151), and the Key Project of Social Development & Science and Technology of Shaanxi Province (No. 2016SF-130).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Ethical approval for this study was obtained from the Medical Ethics Committee of Xi’an Jiaotong University.
Animal Welfare: The present study followed international, national, and/or institutional guidelines for humane animal treatment and complied with relevant legislation.
ORCID iD: Jianbing Ma  https://orcid.org/0000-0001-6513-5713
https://orcid.org/0000-0001-6513-5713
References
- 1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281-97. [DOI] [PubMed] [Google Scholar]
- 2. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15-20. [DOI] [PubMed] [Google Scholar]
- 3. Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817-21. [DOI] [PubMed] [Google Scholar]
- 4. Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19(R2):R169-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143(5):703-9. [DOI] [PubMed] [Google Scholar]
- 7. Zhang R, Ma J, Yao J. Molecular mechanisms of the cartilage-specific microRNA-140 in osteoarthritis. Inflamm Res. 2013;62(10):871-7. [DOI] [PubMed] [Google Scholar]
- 8. Xu J, Kang Y, Liao WM, Yu L. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS One. 2012;7(3):e31861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun J, Zhong N, Li Q, Min Z, Zhao W, Sun Q, et al. MicroRNAs of rat articular cartilage at different developmental stages identified by Solexa sequencing. Osteoarthritis Cartilage. 2011;19(10):1237-45. [DOI] [PubMed] [Google Scholar]
- 10. Zhong N, Sun J, Min Z, Zhao W, Zhang R, Wang W, et al. MicroRNA-337 is associated with chondrogenesis through regulating TGFBR2 expression. Osteoarthritis Cartilage. 2012;20(6):593-602. [DOI] [PubMed] [Google Scholar]
- 11. Min Z, Zhao W, Zhong N, Guo Y, Sun M, Wang Q, et al. Abnormality of epiphyseal plate induced by selenium deficiency diet in two generation DA rats. APMIS. 2015;123(8):697-705. [DOI] [PubMed] [Google Scholar]
- 12. Urist MR, Iwata H, Ceccotti PL, Dorfman RL, Boyd SD, McDowell RM, et al. Bone morphogenesis in implants of insoluble bone gelatin. Proc Natl Acad Sci U S A. 1973;70(12):3511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li D, Ren W, Wang X, Wang F, Gao Y, Ning Q, et al. A modified method using TRIzol reagent and liquid nitrogen produces high-quality RNA from rat pancreas. Appl Biochem Biotechnol. 2009;158(2):253-61. [DOI] [PubMed] [Google Scholar]
- 14. Tan L, Peng H, Osaki M, Choy BK, Auron PE, Sandell LJ, et al. Egr-1 mediates transcriptional repression of COL2A1 promoter activity by interleukin-1β. J Biol Chem. 2003;278(20):17688-700. [DOI] [PubMed] [Google Scholar]
- 15. Wang W, Zhong B, Sun J, Cao J, Tian J, Zhong N, et al. Down-regulated HS6ST2 in osteoarthritis and Kashin-Beck disease inhibits cell viability and influences expression of the genes relevant to aggrecan metabolism of human chondrocytes. Rheumatology (Oxford). 2011;50(12):2176-86. [DOI] [PubMed] [Google Scholar]
- 16. Fox JG. Laboratory animal medicine. Changes and challenges. Cornell Vet. 1985;75(1):159-70. [PubMed] [Google Scholar]
- 17. Drissi H, Zuscik M, Rosier R, O’Keefe R. Transcriptional regulation of chondrocyte maturation: potential involvement of transcription factors in OA pathogenesis. Mol Aspects Med. 2005;26(3):169-79. [DOI] [PubMed] [Google Scholar]
- 18. Terashima Y, Urist MR. Chondrogenesis in outgrowths of muscle tissue onto modified bone matrix in tissue culture. Clin Orthop Relat Res. 1977;(127):248-56. [PubMed] [Google Scholar]
- 19. Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332-6. [DOI] [PubMed] [Google Scholar]
- 20. Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40(1):46-62. [DOI] [PubMed] [Google Scholar]
- 21. Cooper KL, Oh S, Sung Y, Dasari RR, Kirschner MW, Tabin CJ. Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions. Nature. 2013;495(7441):375-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13(16):2072-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273(5275):613-22. [DOI] [PubMed] [Google Scholar]
- 24. Lai LP, Mitchell J. Indian hedgehog: its roles and regulation in endochondral bone development. J Cell Biochem. 2005;96(6):1163-73. [DOI] [PubMed] [Google Scholar]
- 25. Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128(24):5099-108. [DOI] [PubMed] [Google Scholar]
- 26. Pan Y, Wang C, Wang B. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the sonic hedgehog-regulated mouse development. Dev Biol. 2009;326(1):177-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katoh Y, Katoh M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review). Int J Mol Med. 2008;22(3):271-5. [PubMed] [Google Scholar]
- 28. Seki K, Hata A. Indian hedgehog gene is a target of the bone morphogenetic protein signaling pathway. J Biol Chem. 2004;279(18):18544-9. [DOI] [PubMed] [Google Scholar]
- 29. Minina E, Wenzel HM, Kreschel C, Karp S, Gaffield W, McMahon AP, et al. BMP and Ihh/PTHrP signaling interact to coordinate chondrocyte proliferation and differentiation. Development. 2001;128(22):4523-34. [DOI] [PubMed] [Google Scholar]
- 30. Toegel S, Wu SQ, Piana C, Unger FM, Wirth M, Goldring MB, et al. Comparison between chondroprotective effects of glucosamine, curcumin, and diacerein in IL-1beta-stimulated C-28/I2 chondrocytes. Osteoarthritis Cartilage. 2008;16(10):1205-12. [DOI] [PubMed] [Google Scholar]
- 31. Claassen H, Schicht M, Brandt J, Reuse K, Schadlich R, Goldring MB, et al. C-28/I2 and T/C-28a2 chondrocytes as well as human primary articular chondrocytes express sex hormone and insulin receptors—useful cells in study of cartilage metabolism. Ann Anat. 2011;193(1):23-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Finnerty JR, Wang WX, Hébert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402(3):491-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang G, Xiong G, Cao Z, Zheng S, You L, Zhang T, et al. miR-497 expression, function and clinical application in cancer. Oncotarget. 2016;7(34):55900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sato T, Yamamoto T, Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat Commun. 2014;5:4597. [DOI] [PubMed] [Google Scholar]
- 35. Dai L, Zhang X, Hu X, Liu Q, Man Z, Huang H, et al. Silencing of miR-101 prevents cartilage degradation by regulating extracellular matrix–related genes in a rat model of osteoarthritis. Mol Ther. 2015;23(8):1331-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Si HB, Zeng Y, Liu SY, Zhou ZK, Chen YN, Cheng JQ, et al. Intra-articular injection of microRNA-140 (miRNA-140) alleviates osteoarthritis (OA) progression by modulating extracellular matrix (ECM) homeostasis in rats. Osteoarthritis Cartilage. 2017;25(10):1698-707. [DOI] [PubMed] [Google Scholar]