Abstract
Objective
An adverse drug event (ADE) refers to an injury resulting from medical intervention related to a drug including harm caused by drugs or from the usage of drugs. Extracting ADEs from clinical records can help physicians associate adverse events to targeted drugs.
Materials and Methods
We proposed a cascading architecture to recognize medical concepts including ADEs, drug names, and entities related to drugs. The architecture includes a preprocessing method and an ensemble of conditional random fields (CRFs) and neural network–based models to respectively address the challenges of surrogate string and overlapping annotation boundaries observed in the employed ADEs and medication extraction (ADME) corpus. The effectiveness of applying different pretrained and postprocessed word embeddings for the ADME task was also studied.
Results
The empirical results showed that both CRFs and neural network–based models provide promising solution for the ADME task. The neural network–based models particularly outperformed CRFs in concept types involving narrative descriptions. Our best run achieved an overall micro F-score of 0.919 on the employed corpus. Our results also suggested that the Global Vectors for word representation embedding in general domain provides a very strong baseline, which can be further improved by applying the principal component analysis to generate more isotropic vectors.
Conclusions
We have demonstrated that the proposed cascading architecture can handle the problem of overlapped annotations and further improve the overall recall and F-scores because the architecture enables the developed models to exploit more context information and forms an ensemble for creating a stronger recognizer.
Keywords: adverse drug event, information extraction, named entity recognition, word embedding, electronic health record
INTRODUCTION
An adverse drug event (ADE) is “an injury resulting from medical intervention related to a drug” based on the definition of World Health Organization. An ADE includes the harm caused by the drug at normal dose (ie, adverse drug reaction) and the harm due to the use of a drug (eg, overdose or inappropriate dosage).1 An ADE is considered to be more comprehensive and clinically significant than adverse drug reaction, which also cause excess length of stay, extra costs, and mortality of patients.2,3 Extracting ADEs from clinical records can help physicians associate adverse events to the targeted drugs.4
Natural language processing has been applied by researchers to extract ADEs and other meaningful information from large amounts of unstructured records.5–8 Through the participation of the ADEs and medication extraction (ADME) in electronic health records (EHRs) track of the 2018 n2c2 shared task, we used the released ADME corpus to develop a cascading architecture that combined a conditional random field (CRF) model9 and 2 neural network models based on the bidirectional long short-term memory (LSTM)-CRF (BiLSTM-CRF). The training set of the ADME corpus includes 303 discharge summaries collected from the MIMIC-III (Medical Information Mart for Intensive Care III) clinical care database.10 The test data include exclusive 202 discharge summaries. The annotations contains 9 types of named entities including drugs, the strength, dosage, duration, frequency, form, route of administration, reason of a drug, and ADEs. The detail distributions of the number of the annotated entities are available in the Supplementary Appendix S1.
One major challenge of the released corpus is that the annotations among different entity types may have overlapping boundaries (the other observed challenges are described in Supplementary Appendix S1). Figure 1 displays examples in which we can observe 2 overlapping annotations. One is the phrase “[[narcotic]DRUG induced respiratory distress]ADE/REASON,” which was assigned to 2 medical concepts, reason and ADE, with one drug annotation being a substring of the above 2 annotations. Another is the word agitated, which belongs to both ADE and reason. The figure also demonstrates examples of arbitrary sentence breaking. Take the same phrase, narcotic induced respiratory distress, as an example. The phrase was segmented into 2 pieces: narcotic and induced respiratory distress.
Figure 1.
An example of the overlapping annotations.
Among all overlapping boundaries, the drug entity overlaps with the most types of entity including reason, frequency, ADE, form, strength, and route, while the ADE entity has the most overlapped instances. To handle the overlapping mention spans, the cascading architecture was proposed in which classifiers in the cascade were trained sequentially, and the output of one stage in the cascade affects the training instances given to the next.11 Similar approaches have been applied in several domains for improving the performance of individual classifiers. For instance, Zanoli et al,12 the winner of the Italian named entity recognition (NER) task at EVALITA 2009, developed a cascaded NER system combining the hidden Markov model and CRF to exploit contextual information from unlabeled data. Along the same line, Corbett and Copestake13 demonstrated that the F-measure of chemical NER can be improved by 0.06 by capitalizing on information generated by classifiers in the cascade. Esuli et al14 and Wang and Patrick15 also exhibited improved performances over traditional batch learning techniques for recognizing entities in the clinical domain. The idea of cascades is also leveraged in the field of computer vision. To illustrate, Heitz et al16 proposed a cascaded classification framework to combine off-the-shelf classifiers with the intention to improve the performance of each of them. The main advantage of the architecture of cascaded classifiers is that it enables cascaded classifiers to utilize more dependencies among similar or different subtasks of a target problem to enhance the overall classification performance. Unlike most previous works focused on the improvement of the final accuracy along with the improved computational efficiency, we adapted the idea of cascaded classifiers to not only improve the performance of the developed models, but also attack the problem of overlapped entities.
From our review of the methodologies employed by the official announced top 10 systems among the concepts subtask of the ADME track (our review of the top-performed methodologies are available in Supplementary Appendix S2), we realized that our method has several similar characteristics with other competing approaches. For instance, the main methodology employed by most top-performed teams, including ours, was based on a neural network architecture consisting of a core layer of the BiLSTM-CRF. Some systems also combined the neural network–based approaches with other machine learning algorithms to build an ensemble. The purpose of creating such an ensemble is to address the challenge of overlapping annotations by training individual models on subsets of nonoverlapping entity types and combining the results by using voting methods.
In addition to the techniques applied in the preprocessing and the final ensemble steps, one significant difference of our implemented method and the others is the choice of the pretrained word embedding. Most of the top-ranked teams utilized the entire MIMIC-III dataset to create pretrained word embeddings with the word2vec package while we used a pretrained embedding released by Moen and Ananiadou.17 A variety of approaches have been proposed to learn the word representations, along with several publicly available pretrained word representations, such as word2vec (GN) trained with Google News,18 Global Vectors for word representation (GloVe),19 and fastText20 trained with Wikipedia and the pretrained model released by Moen and Ananiadou17 in the biomedical domain. Recently, there has been an emphasis on further improving the pretrained vectors through combinations21,22 or postprocessing algorithms.23,24 The selection of the pretrained word embeddings is known to have a larger impact on the performance of sequence labeling task25 than many other hyperparameters. In light of this, in addition to present our cascaded architecture for the ADME track, we study the effectiveness of different word embedding methods applied for the problem of ADME in EHRs.
MATERIALS AND METHODS
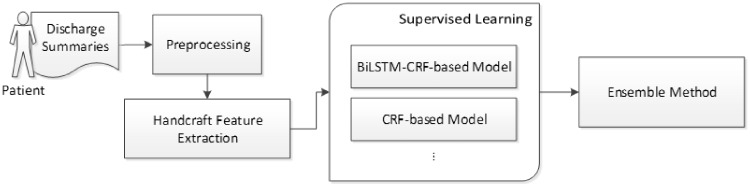
Figure 2 demonstrates the general workflow applied by all top-ranked systems. We elaborate on all the steps in the following subsections.
Figure 2.
The general workflow applied by top-ranked systems.
Preprocessing step
The typical preprocessing applied by all teams including tokenization, sentence segmentation, and the extraction of part-of-speech (PoS) information. Some teams further exploited off-the-shelf natural language processing tools such as cTAKES (clinical Text Analysis Knowledge Extraction System) and MetaMap to extract medication information. In our implementation, a discharge summary was preprocessed by our clinical toolkit26 to segment sentences and generate the tokens and corresponding PoS information based on MedPost.27 The numerical normalization method proposed in our previous work28 was employed to normalize variations in the numerical parts of each token.
However, the detailing of preprocessing steps of the ADME track turns out to be quite complicated due to the following challenges. For the situation of overlapping boundaries illustrated in Figure 1, we generated 3 training sets, each of which contains nonoverlapping annotations. The rationale to compile the 3 sets is to enable the development of a cascading architecture to train our supervised learning models, each of which uses information collected from the output of the previous classifier as additional information in the cascade. Owing to the reasons that the drug entity is the most frequent overlapping type and all other entities considered in the ADME track are related to drugs, we decided to include annotations of the drug entity and other entities that do not overlap with drugs in the first subtraining set. On the other hand, because recognizing ADE requires wide context information, we reserved the annotations of the ADE entity and excluded entities overlapping with ADEs (ie, drugs and reasons) in the last subtraining set so that the recognizer developed based on that can have more context information to perform a finer learning. The second subtraining set finally contained annotations for entities overlapped with drugs or ADEs.
Furthermore, some sections, such as “DISCHARGE MEDICATIONS” and “DISCHARGE INSTRUCTIONS,” may contain ordered or unordered lists of items. The descriptions in those sections may also contain a variety of arbitrary line breaks which cause sentence breaking errors. We therefore exploited our section recognizer,29 along with 31 keywords collected from the training set of the ADME corpus, to identify sections and classify them into 4 categories. For each category, we developed regular patterns to fix the line-breaking errors after sentence breaking.
Handcrafted feature extraction
Table 1 summarizes the features extracted for a target word. Those features were commonly used in the NER tasks, and most of them were also adopted by the other top-ranked teams. Note that the last feature shown in Table 1 was extracted only for the CRF model.
Table 1.
Handcraft features extracted for the ADME task.
| Feature name | Description |
|---|---|
| PoSa | PoS information generated in the preprocessing step was extracted for the current word. |
| Context wordsa | A context window of 7 was set to extract the current word and its surrounding words. |
| Chunk | The chunk information of the current word was extracted. |
| Morphological featuresa | The morphological features such as prefixes and suffixes defined in our previous work28 were extracted, which were empirically shown to provide clues for classifying the type of concepts. |
| Orthographic featuresa | The orthographic features defined in our previous work28 were extracted, which were empirically shown to be able to detect patterns of named entities. |
| Common medical abbreviations | Whether the current word matched with the common medical abbreviations defined in the annotation guideline.30 The following list of names and the corresponding entity types was used:
|
| ADE, drug and disease dictionary featuresa | The dictionaries used in our previous work31 were encoded based on the occurrence encoding presented in our previous work.29 The encoded information of the current word was extracted. |
| Word cluster featuresa | The cluster number where the current word belongs to was extracted as a feature. The cluster was generated by using the k-means algorithm from the word embedding vectors. |
ADE: adverse drug event; ADME: adverse drug events and medication extraction; PoS: part of speech.
aFeature also used by other top-ranked teams.
Core cascading architecture for supervised learning
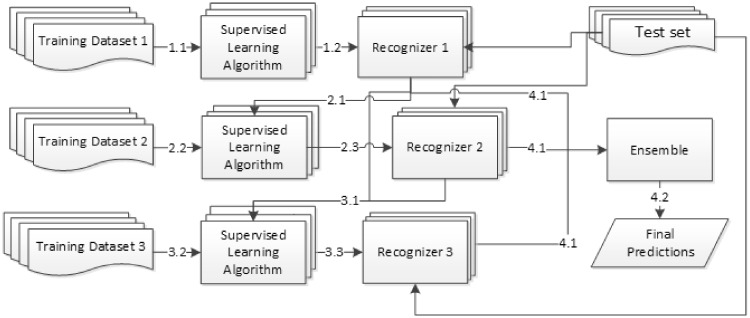
As described in the preprocessing section, we compiled 3 nonoverlapping subtraining sets. To learn from the compiled datasets at the same time making use of the outcome from the preceding models in order, we applied a cascading architecture to develop our recognizers. The workflow is shown in Figure 3. In the first stage (steps 1.1-1.3), the first subtraining set was used to train the first set of supervised learning models including 1 CRF model and 2 BiLSTM-CRF models. The developed models are similar to most of the stand-alone NER systems, which just produced a sequence of predicted labels learned from the training set. In the second stage, the output from the trained recognizer (step 2.1), along with the features extracted from the second subtraining set, which composed of a different set of nonoverlapping labels (step 2.2), were merged to train the second set of NER models (step 2.3). By collecting the outputs of the first and second sets of recognizers (step 3.1) and combining them with the features extracted from the third training set (step 3.2), the last set of NER models was built (step 3.3). In the prediction time, the outputs of all models (9 models in our implementation; step 4.1) were combined by an ensemble algorithm to produce the final predictions (step 4.2).
Figure 3.
A cascading architecture developed for the ADME task.
For each stage in the cascade, we used CRFs, the LSTM-BiLSTM-CRF network,32 and a convolutional neural network (CNN)-BiLSTM-CRF network33 to develop our NER models. In the first stage, the handcraft features described in the previous section were extracted for all models. In the second and third stages, the handcraft features along with the output from the preceding stage(s) were extracted for building the models. The annotations were represented in the BIO notation, where B denotes the beginning of an entity, I denotes inside but not at beginning of an entity, and O indicates outside of an entity. All sentences, including those that did not contain any annotations, were included in our subtraining sets.
For CRF, the linear chain architecture was used.9 For BiLSTM-CRFs, we used the structure developed in our previous work for the task of family history information extraction.34 The structure consists of 3 layers: the character sequence representation layer, the word sequence representation layer based on LSTM, and the CRF inference layer. Herein, we created 2 BiLSTM-CRFs, whose difference is their character representation layers. The first BiLSTM-CRF used CNN with max-pooling to capture the morphological information, whereas the second one used LSTM. The generated character embedding was then concatenated with the pretrained word embedding vectors and the aforementioned handcrafted features, which were represented by a randomly initialized 20 dimensional vectors to form the input vector of the BiLSTM sequence layer. Hereafter, we refer to them as C-BiLSTM-CRF and L-BiLSTM-CRF, respectively.
In the last stage of the cascading architecture is an ensemble algorithm. The algorithm uses a voting method to judge the final predictions and the corresponding boundaries. The input of the algorithm includes all cascaded models’ predictions. The algorithm can be configured into 3 modes: strict, the lenient, and optimal (the detail of the 3 modes is described at Supplementary Appendix S3). In the present study, we applied the optimal model, which first assigns the tokens with labels predicted by all models. For each token, the algorithm examines whether the assigned entity type label received votes exceeding the optimal voting count estimated by using a holdout development set. If the number of votes of an entity type label does not receive the required counts, the label is discarded. If there are more than 1 label met the required vote counts, the label with higher number of votes was output. If the vote ties and the assigned labels are different but allowed be overlapped (estimated based on the training set), the labels are assigned as such, or the actual labels are determined based on the label distribution estimated on the training set.
Pretrained word embedding methods
In the architecture of our C- and L-BiLSTM-CRF models, a pretrained word embedding was used for the word sequence representation layer. Table 2 shows the pretrained word representations considered in the study.
Table 2.
The pretrained word embeddings used in the study
| Name | Corpus | Dimension | Vocab. size |
|---|---|---|---|
| GloVea | Wikipedia and English Gigaword | 200 | 400 000 |
| fastTextb | Wikipedia | 300 | 2 519 370 |
| nlplabc | PubMed and PMC | 200 | 2 231 684 |
| word2vecGN | Google News | 300 | 3 000 000 |
| Numberbatchd | Hybrid of ConceptNet, word2vecGN and GloVe | 300 | 417 194 |
| BioWordVece | PubMed and MIMIC-III | 200 | 16 545 451 |
| word2vecMIMIC | MIMIC-III | 300 | 320 313 |
| ConcatenatedVec | Hybrid of GloVe, fastText, and word2vecMIMIC | 700 | 228 763 |
| AddedVec | Hybrid of fastText and word2vecMIMIC | 300 | 46 404 |
| PurifiedVec | Postprocessed GloVe vectors | 200 | 400 000 |
MIMIC-III: Medical Information Mart for Intensive Care III; PMC: PubMed Central.
This is the embedding we used during the n2c2 ADME track, which is available at http://evexdb.org/pmresources/vec-space-models/.
The first to sixth embeddings listed in Table 2 are publicly available pretrained vectors. In our implementation, if a word was listed in the pretrained embedding, the corresponding vector was assigned. Otherwise, the corresponding embedding was randomly initialized. In both cases, the embeddings were updated during training by the backpropagation step. During the participation of the ADME track, the nlplab embedding was used.
In addition to the pretrained vectors, we generated the following 4 word representations. The first was word2vecMIMIC: a self-trained word representation trained by using the skip-gram algorithm18 on the MIMIC-III corpus.10 The same preprocessing procedure was applied on the MIMIC-III corpus before generating the representation. The second was ConcatedVec, in which we directly concatenated the embeddings of GloVe, fastText, and the self-trained word2vecMIMIC. Third was AddedVec, in which the embeddings of fastText and the self-trained word2vecMIMIC were added by using the vector addition. The last to be generated was PurifiedVec, a postprocessed vector, by applying the principal component analysis on the GloVe embedding to generate a more isotropic vectors. Here, we followed the suggestion by Mu et al23 to postprocessed the first dominating D/100 dimensions, where D is equals to 200 for GloVe.
RESULTS
The official evaluation metrics in terms of precision (P), recall (R), and micro F-measure (F) were used to report the performance of the proposed methods. The evaluation includes 2 boundary matching methods: (1) the lenient mode, in which an overlapped boundary between the gold annotation and the system’s prediction is allowed; and (2) the strict mode, in which the boundary of the system’s prediction must exact match with the gold annotation. The lenient evaluation mode was the primary evaluation metric used by the organizers of the ADME track therefore we only listed the PRF-scores under the lenient mode.
Official evaluation results
We used a holdout development set to study the performance of the developed models along with the proposed cascading architecture. The hyper-parameters used for the C- and L-BiLSTM-CRF models as well as the parameters of the ensemble algorithm were also tuned on the developed set (the organizers of the ADME track released their training data by sets, and we used the last set released by the organizers as the development set [38 summaries] and the other data as the training set [265 summaries]). The final parameters are listed in Supplementary Appendix S3. The 3 best performed configurations on the development set were chosen as the configurations for the 3 submitted runs for the ADME track. The first run was based on the cascading architecture with the C-BiLSTM-CRF model. The second run is the full cascading system including CRF, C-BiLSTM-CRF, and L-BiLSTM-CRF models, which was ranked second on the development set. The last is a cascading system, including the C- and L-BiLSTM-CRF models, which was the best-performing model on the development set. The official results on the test set are shown in Table 3. Note that the word embedding used by all runs was nlplab, which listed in Table 2.
Table 3.
Official overall evaluation results on the test set under the lenient evaluation mode
| Submitted run | P | R | F |
|---|---|---|---|
| Run 1: C-BiLSTM-CRF | 0.935 | 0.892 | 0.913 |
| Run 2: all models | 0.944 | 0.892 | 0.917 |
| Run 3: all BiLSTM-CRF models | 0.939 | 0.900 | 0.919 |
BiLSTM-CRF: bidirectional long short-term memory conditional random field; C-BiLSTM-CRF: convolutional neural network bidirectional long short-term memory conditional random field; F: micro F-score; P: precision; R: recall.
Overall, we can see that the 2 ensemble runs have better F-scores than do those of the individual cascading run (run 1). Specifically run 3, with the 2 BiLSTM-CRF models, achieved the highest RF-scores, whereas run 2, with all 3 models, had the best P. Similar to our observation on the development set, the inclusion of the CRF model’s predictions did not improve the overall F-score on the test set.
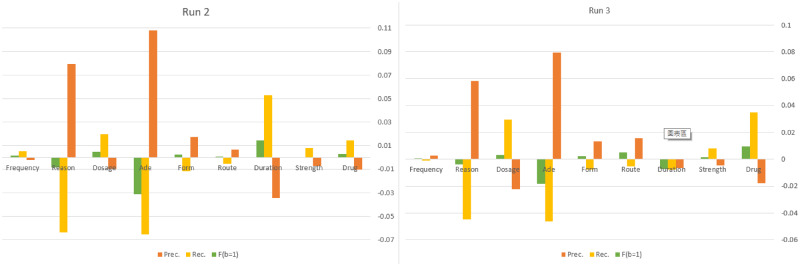
Figure 4 further shows the detailed PRF-scores for each entity type, in which the F-scores of run 1 were used as the baseline, with the PRF-scores are shown in Table 4. Overall, we can observe that the individual cascading run performed best in recognizing the strength entity type, while it also had the lowest F-score for the ADE type. On the other hand, except for ADEs, reasons, and durations, both ensemble systems achieved better F-scores in entity types, including drugs, strengths, routes, forms, dosages, and frequencies. Both systems had better recalls in entity types, including drugs, strengths, and dosages, and better P-scores in types including reason, ADEs, forms, and routes. In particular, the P-scores of the ADE and reason entity types were significantly improved with the cost of reduced R-scores. This may be due to the strict threshold settings for the 2 entities, which require that they have more than half of the votes. For example, for the ADE type, our setting (the optimal mode) requires all involved models to have committed the annotated boundaries, or the annotations may be ignored (in case there is no overlapping) or sliced into a smaller overlapped piece.
Figure 4.
Entity type performance comparison for submitted runs using Run 1 as the baseline.
Table 4.
The performance of the C-BiLSTM-CRF model (run 1) for each entity type on the test set under the lenient evaluation mode
| Entity Type | P | R | F |
|---|---|---|---|
| Drug | 0.9306 | 0.8742 | 0.931 |
| Strength | 0.9558 | 0.9452 | 0.9741 |
| Duration | 0.7767 | 0.6534 | 0.8161 |
| Route | 0.9284 | 0.9337 | 0.947 |
| Form | 0.9249 | 0.9153 | 0.9494 |
| ADE | 0.4328 | 0.2784 | 0.3875 |
| Dosage | 0.8969 | 0.8829 | 0.9267 |
| Reason | 0.5849 | 0.521 | 0.6267 |
| Frequency | 0.8479 | 0.828 | 0.9695 |
ADE: adverse drug event; C-BiLSTM-CRF: convolutional neural network bidirectional long short-term memory conditional random field; F: micro F-score; P: precision; R: recall.
Performance comparison with different word embedding methods
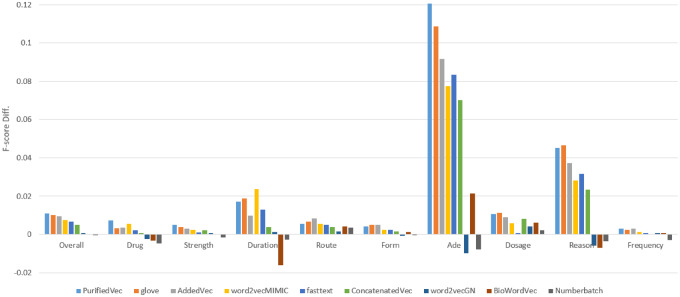
In this section, we further study the effectiveness of different word embeddings listed in Table 2. Here, we only applied the listed embeddings with the C-BiLSTM-CRF model because it was the best-performing single model on the development set. Each embedding was used for training 3 models on the 3 subtraining sets. The proposed cascading architecture configured with the same setting for run 1 was used to generate the final predictions. In all developed models, the embeddings were updated during training by the backpropagation step. Figure 5 depicts the performance comparison in which the F-score of the C-BiLSTM-CRF model with the nlplab embedding was used as the baseline (detailed results are available at Supplementary Appendix S4).
Figure 5.
Performance comparison for models with different word embeddings. Here the y-axis is the difference between the F-scores of each model for different entity types and that of the model with the nlplab embedding.
From the results shown in Figure 5, we can see that replacing nlplab with other pretrained embeddings resulted in improved F-scores, in addition to the Numberbatch embedding. The most suitable pretrained embedding for the ADME track is GloVe because it demonstrates more improvement on the F-score than others do. Further, if we take the combinations or postprocessing techniques into consideration, the model with the PurifiedVec achieved the best overall F-score, and performed better than the others in terms of the drug, strength, ADEs, and frequency concepts.
DISCUSSION
Performance comparison with different machine learning algorithms
Under the proposed cascading architecture, we employed 3 machine learning algorithms: CRF, C-BiLSTM-CRF, and L-BiLSTM-CRF. The overall PRF-scores of the 3 individual models on the development and test sets are listed in Table 5.
Table 5.
The microevaluation results on the development and test sets under the optimal mode for the 3 machine learning algorithms
| Dataset | Algorithm | P | R | F |
|---|---|---|---|---|
| Development | CRF | 0.937 | 0.892 | 0.914 |
| C-BiLSTM-CRF | 0.929 | 0.913 | 0.921 | |
| L-BiLSTM-CRF | 0.934 | 0.907 | 0.920 | |
| Test set | CRF | 0.946 | 0.871 | 0.907 |
| C-BiLSTM-CRF | 0.935 | 0.892 | 0.913 | |
| L-BiLSTM-CRF | 0.937 | 0.890 | 0.913 |
C-BiLSTM-CRF: convolutional neural network bidirectional long short-term memory conditional random field; CRF: conditional random field; F: micro F-score; L-BiLSTM-CRF: long short-term memory bidirectional long short-term memory conditional random field P: precision; R: recall.
We can see that the 2 BiLSTM-CRF models consistently achieved better recall and F-scores on both the development and test sets, indicating that the architecture can learn generalizable morphological and lexical patterns of the target concepts. The characteristics of the CNN and LSTM character sequence representation layers of the 2 models are different: the CNN approach takes only n-grams into account, without considering the position information, while the LSTM approach considers all characters and concerns their positions. Intuitively, the L-BiLSTM-CRF model should be superior to the C-BiLSTM-CRF model because the former one exploits more information. However, our results demonstrated that the C-BiLSTM-CRF model achieved a slightly better overall F-score. In our experiments, the training time of the L-BiLSTM-CRF model increased 11.7% relative to that of the C-BiLSTM-CRF model, so the C-BiLSTM-CRF model should be preferred because of its higher computational efficiency.
On the other hand, the CRF models have better P-scores, but underperformed in comparison with neural network–based models in terms of RF-scores. The CRF models only relied on the handcrafted features and seem to have lower generalizability in particular for concepts such as ADEs and reasons. Compared with other entity types, the annotations for these 2 types tend to have more narrative descriptions, such as “lip and tongue swelling” for ADE and “prevent more clots from forming” for reason, which can be successfully recognized by neural network–based models but not by CRFs.
Effectiveness of the proposed cascading architecture
We have proposed a cascading architecture combining cascading information and the ensemble method to deal with the challenge of overlapping boundaries. Although combining the results of noncascaded recognizers (the information collected from the output of the previous classifier was ignored in the cascade) via an ensemble can also address this challenge, we observed that the proposed cascaded architecture can exploit more context information to improve the performance of recognizing drug-related entities. Overall, by comparing the recognition performance of the cascaded recognizer and noncascaded recognizer both trained on the third subtraining set, we observed that the cascaded recognizer had higher recall on the test set under both the strict and lenient evaluation modes, resulting in improved F-scores, around 0.025. In particular, for the recognition of ADEs, which turned out to be the most difficult among the 9 types in the ADME track, we observed that the cascaded recognizer can recognize more ADEs co-occurring with drugs described in the same sentences, such as “[cefepime]DRUG was discontinued secondary to drug [rash]ADE,” “An [ACEi]DRUG was held due to [intolerance]ADE in the past,” and “… he reports always getting [diahrrea]ADE with his [chemotherapy]DRUG.” On the other hand, the noncascaded classifier could recognize more individually mentioned ADEs such as “excessively somnolent” in the sentence, “This was subsequently discontinued when he was found to be [excessively somnolent]ADE,” whose induced drugs were mentioned far from the mentions’ resident sentences. However, the cases are occurred less in the ADME corpus, resulting in a reduced F-score by 0.011. Furthermore, although we did not find overlapping ADEs and drugs in the test set, such cases do appear in the training set. We believe that the proposed cascaded recognizer can recognize more overlapping ADE-drug mentions at the prediction time due to the preceding observations.
The previously mentioned slight improvement also revealed the limitation of the current implementation in identifying ADEs. One way to improve the performance of the ADE recognition based on the proposed cascading architecture is to improve the recognition performance of the classifiers developed in the preceding stages. We conducted an experiment on the test set to study the performance ceiling, by replacing the system-predicted named entities corresponding to each preceding stage with the gold annotations, and observed that the recall can be significantly improved by 0.440, resulting in an improved F-score of 0.472 in the case of the CRF model. However, even with the improvement, the recall of ADEs is still far from satisfaction. Studying the false negative cases revealed that most of the ADE mentions that appeared in the test set were not described in the training set or even annotated as the “reason” type. One possible solution could be to incorporate external knowledge sources like SNOMED CT (Systematized Nomenclature of Medicine Clinical Terms) and RxNorm, or other standard terminologies described in Goss et al.35 Another considerable solution is to adopt an attention-based neural network architecture, which could leverage more contextual information to alleviate the problem.
Effectiveness of different word embeddings
Similar to the observation of Reimers and Gurevych25 and Ma and Hovy,33 the use of GloVe embedding turns out to give the best overall F-score in the ADME task. Reimers and Gurevych25 described that the coverage of the employed embedding have a high impact on the achieved performance. Among the employed embeddings, the Numberbatch embedding has the lowest coverage (16.5%) on the test set, resulting in the lowest overall F-score. However, it is surprising to see that the models with the 2 high coverage embeddings trained from the MIMIC-III corpus (BioWordVec: 68.2%; word2vecMIMIC: 55.3%) did not outperform the model with GloVe embedding, which was trained on the general domain. Our in-house word2vecMIMIC has lower coverage but a higher F-score than BioWordVec does, which may be because we applied the same preprocessing methods before generating the embedding. As shown in Figure 4, the inclusion of domain-specific embeddings like word2vecMIMIC can improve the effectiveness in recognizing domain-specific concepts like drugs, but the general domain embeddings such as fastText and GloVe are preferred in cases involving narrative descriptions like ADEs and reasons. The results coincide with the observation of Wang et al36 in which they stated that the word embeddings trained on biomedical domain corpora do not necessarily have better performance than those trained on general domain corpora do for any downstream biomedical natural language processing task.
Finally, the best performed embedding in our experiments is the postprocessed GloVe vectors. Unlike other embedding, both the PR-scores of the model with the PurifiedVec can be improved as shown in Figure 5. We have further applied the same postprocessing on the word2vecMIMIC embedding, but did not see any improvement on the overall F-score. The results demonstrated that the principal component analysis–based postprocessing operation could be a candidate technique to refine the original word representations. In the future, we would like to consider other advanced word embedding techniques such as ELMo37 or the postprocessing method for dimensionality reduction,38 or variance normalization and dynamic embedding.39
Effectiveness of the preprocessing step and handcrafted features
As described in the Preprocessing Step section, we developed methods to process descriptions containing lists of items and arbitrary line breaks to resolve sentence breaking errors. To investigate the sensitivity of the developed neural network–based models on such errors, we conducted an experiment to compare the performance of the C-BiLSTM-CRF models on the corresponding test sets with the GloVe embedding trained on the preprocessed and nonpreprocessed training sets, respectively. We observed that the model trained on the nonpreprocessed dataset had a better P-score (+0.035) but a lower R-score (–0.055), which resulted in a reduction of 0.01 of the overall F-score. Further examination revealed that the model obtained lower recall and F-scores for all medical concepts except the drug concept, and is particularly deficient in recognizing the duration and form concepts, whose F-scores were reduced by more than 0.02. These concepts usually appear in narratives described in ordered or unordered lists that often contain arbitrary line breaks. The narratives may be incorrectly segmented, leading to noisy contextual information or extremely lengthy sentences (based on our estimation, the longest sentences in the nonpreprocessed and preprocessed datasets contain 1253 words and 480 words, respectively. On average, the sentences in the nonpreprocessed dataset are longer than those in the preprocessed dataset [15.57 words vs. 12.95 words]), which may hinder the training of a reliable model.
An ablation study was conducted to gain a better insight into the impact of the extracted handcrafted features on the neural network–based models. To this end, we used the same aforementioned C-BiLSTM-CRF architecture trained on the preprocessed dataset as the baseline. Results indicated that the handcrafted features did not contribute equally on the recognition of all medical concepts. For example, the inclusion of the dictionary features improved the P-scores for recognizing all medical concepts, while the R-scores for ADEs, duration, and route concepts remain the same or were slightly increased to contribute to better F-scores. However, concepts such as drug, strength, and frequency had lower R- and F-scores, which may be due to the insufficient coverage of the employed lexicons, the noise introduced in the dictionary matching process, and the possibly overlapping annotations among these concepts. For the overall F-scores, a performance degradation was observed when eliminating the PoS, morphological, or orthographic features, respectively. This reveals that the combined contribution of these features with the overall recognition performance exceeded that of learning these features through neural networks alone. On the other hand, the inclusion of chunk and dictionary features turned out to reduce the overall F-scores. An additional study will be required to understand whether the inclusion of these features is redundant or not for the deep learning models. Detailed results of the previous 2 studies are available in Supplementary Appendix S4.
CONCLUSION
We have proposed a cascading architecture combining 2 well-known sequence labeling models including CRFs and BiLSTM-CRF along with the study of the effectiveness of a variety of word representation techniques to deal with the task of ADME. The developed system can recognize medical concepts including ADEs, drug names, and entities related to drugs, which was officially ranked seventh of 28 teams in the concepts subtask of the ADME track. From our experimental results, we can come to the following conclusions. First, with the proposed cascading architecture, we can deal with the problem of overlapped annotations and improve the overall RF-scores because the architecture renders cascaded recognizers more context information. Second, the 2 popular sequence labeling techniques provide promising solution for the ADME task. CRFs with well-defined features come very close to the performance of the stronger BiLSTM-CRF models, except in the concept type involving narrative descriptions. The GloVe embedding provides a very strong baseline, which can be further improved by applying postprocessing to eliminate the common mean vector and a few top dominating directions from the word vectors. Last, in accordance with the observation of other previous works, the empirical results show that BiLSTM-CRF models using either CNN- or LSTM-char layers achieved similar overall F-scores, but the C-BiLSTM-CRF model should be preferred due to its higher computational efficiency.
FUNDING
This work was supported by Ministry of Science and Technology, R.O.C. grant number MOST-106-2221-E-143-007-MY3.
AUTHOR CONTRIBUTIONS
H-JD conceived the presented idea, planned the experiments and supervised the project. H-JD and C-HS carried out the experiments. H-JD and C-HS verified the analytical methods and contributed to the interpretation of the results. H-JD and C-SW supervised the findings of this work. H-JD wrote the manuscript with support from C-HS and C-SW. All authors discussed the results and contributed to the final manuscript.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1. Nebeker JR, Barach P, Samore MH.. Clarifying adverse drug events: a clinician's guide to terminology, documentation, and reporting. Ann Intern Med 2004; 140 (10): 795–801. [DOI] [PubMed] [Google Scholar]
- 2. Bates DW, Cullen DJ, Laird N, et al. Incidence of adverse drug events and potential adverse drug events: implications for prevention. JAMA 1995; 274 (1): 29–34. [PubMed] [Google Scholar]
- 3. Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP.. Adverse drug events in hospitalized patients: excess length of stay, extra costs, and attributable mortality. JAMA 1997; 277 (4): 301–6. [PubMed] [Google Scholar]
- 4. Leape LL, Bates DW, Cullen DJ, et al. Systems analysis of adverse drug events. JAMA 1995; 274 (1): 35–43. [PubMed] [Google Scholar]
- 5. Harpaz R, Callahan A, Tamang S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Saf 2014; 37 (10): 777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aramaki E, Miura Y, Tonoike M, et al. Extraction of adverse drug effects from clinical records. Stud Health Technol Inform 2010; 160 (Pt 1): 739–43. [PubMed] [Google Scholar]
- 7. Gurulingappa H, Mateen‐Rajpu A, Toldo L.. Extraction of potential adverse drug events from medical case reports. J Biomed Semantics 2012; 3 (1): 15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kang N, Singh B, Bui C, Afzal Z, van Mulligen EM, Kors JA.. Knowledge-based extraction of adverse drug events from biomedical text. BMC Bioinformatics 2014; 15 (1): 64.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labeling sequence data. In: Proceedings of the 18th International Conference on Machine Learning (ICML); 2001: 282–9.
- 10. Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Viola P, Jones M.. Robust real-time object detection. Int J Comput Vis 2001; 57 (2): 137–54. [Google Scholar]
- 12. Zanoli R, Pianta E, Giuliano C.. Named entity recognition through redundancy driven classifiers. In: Proceedings of EVALITA; 2009: 9.
- 13. Corbett P, Copestake A.. Cascaded classifiers for confidence-based chemical named entity recognition. BMC Bioinformatics 2008; 9 (S11): S4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esuli A, Marcheggiani D, Sebastiani F.. An enhanced CRFs-based system for information extraction from radiology reports. J Biomed Inform 2013; 46 (3): 425–35. [DOI] [PubMed] [Google Scholar]
- 15. Wang Y, Patrick J. Cascading classifiers for named entity recognition in clinical notes. In: Proceedings of the Workshop on Biomedical Information Extraction; 2009: 42–9.
- 16. Heitz G, Gould S, Saxena A, Koller D. Cascaded classification models: combining models for holistic scene understanding. In: Advances in Neural Information Processing Systems 21 (NIPS 2008); 2009: 1–8.
- 17. Moen S, Ananiadou TSS. Distributional semantics resources for biomedical text processing. In: Proceedings of the Fifth International Symposium on Languages in Biology and Medicine (LBM); 2013.
- 18. Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. arXiv2013. Sep 7 [E-pub ahead of print].
- 19. Pennington J, Socher R, Manning CD. GloVe: global vectors for word representation. In: Proceedings of the Empirical Methods in Natural Language Processing (EMNLP 2014); 2014; 12: 1532–43.
- 20. Bojanowski P, Grave E, Joulin A, Mikolov T.. Enriching word vectors with subword information. Trans Assoc Comput Linguist 2017; 5: 135–46. [Google Scholar]
- 21. Garten J, Sagae K, Ustun V, Dehghani M. Combining distributed vector representations for words. In: Proceedings of the NAACL-HLT; 2015: 95–101.
- 22. Roberts K. Assessing the corpus size vs. similarity trade-off for word embeddings in clinical NLP. In: Proceedings of the Clinical Natural Language Processing Workshop (ClinicalNLP); 2016: 54–63.
- 23. Mu J, Bhat S, Viswanath P. All-but-the-top: Simple and effective postprocessing for word representations. arXiv2018 Mar 19. [E-pub ahead of print].
- 24. Wu Y, Xu J, Jiang M, Zhang Y, Xu H.. A study of neural word embeddings for named entity recognition in clinical text. AMIA Annu Symp Proc 2015; 2015: 1326–33. [PMC free article] [PubMed] [Google Scholar]
- 25. Reimers N, Gurevych I. Optimal hyperparameters for deep lstm-networks for sequence labeling tasks. arXiv2017. Aug 16 [E-pub ahead of print].
- 26. Chang N-W, Dai H-J, Jonnagaddala J, Chen C-W, Tsai R-H, Hsu W-L.. A context-aware approach for progression tracking of medical concepts in electronic medical records. J Biomed Inform 2015; 58 (S): S150–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith L, Rindflesch T, Wilbur WJ.. MedPost: a part of speech tagger for BioMedical text. Bioinformatics 2004; 20 (14): 2320–1. [DOI] [PubMed] [Google Scholar]
- 28. Tsai R-H, Sung C-L, Dai H-J, Hung H-C, Sung T-Y, Hsu W-L.. NERBio: using selected word conjunctions, term normalization, and global patterns to improve biomedical named entity recognition. BMC Bioinformatics 2006; 7 (Suppl 5): S11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dai H-J, Syed-Abdul S, Chen C-W, Wu C-C.. Recognition and evaluation of clinical section headings in clinical documents using token-based formulation with conditional random fields. Biomed Res Int 2015; 2015: 873012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buchan K, Bari MD, Stubbs A. Annotation guidelines for the adverse drug event (ADE) and medication extraction challenge. 2018. https://n2c2.dbmi.hms.harvard.edu/files/ADE_Annotation_Guideline_final.pdf. Accessed December 9, 2018.
- 31. Dai H-J, Touray M, Wang C-K, Jonnagaddala J, Syed-Abdul S.. Feature engineering for recognizing adverse drug reactions from Twitter posts. Information 2016; 7: 27. [Google Scholar]
- 32. Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural architectures for named entity recognition. In: Proceedings of NAACL-HLT 2016; 2016: 260–70.
- 33. Ma X, Hovy E. End-to-end sequence labeling via bi-directional LSTM-CNNs-CRF. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics; 2016: 1064–74.
- 34. Wang F-D, Wang C-K, Dai H-J. Family history information extraction with neural sequence labeling model. In: Proceedings of BioCreative/OHNLP Challenge 2018; 2018.
- 35. Goss FR, Zhou L, Plasek JM, et al. Evaluating standard terminologies for encoding allergy information. J Am Med Inform Assoc 2013; 20 (5): 969–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y, Liu S, Afzal N, et al. A comparison of word embeddings for the biomedical natural language processing. J Biomed Inform 2018; 87: 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peters ME, Neumann M, Iyyer M, et al. Deep contextualized word representations. In: Proceedings of NAACL-HLT 2018; 2018: 2227–37.
- 38. Raunak V. Effective dimensionality reduction for word embeddings. In: Proceedings of the Learning from Limited Labeled Data (LLD) Workshop; 2017.
- 39. Wang B, Chen F, Wang A, Kuo C-C. Post-processing of word representations via variance normalization and dynamic embedding. arXiv2019. Feb 4 [E-pub ahead of print].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.