Abstract
Small molecules such as molecular oxygen, nitric oxide, and carbon monoxide play important roles in life, and many proteins require the transport of small molecules to and from the bulk solvent for their function. Ligand migration within a protein molecule is expected to be closely related to the overall structural changes of the protein, but the detailed and quantitative connection remains elusive. For example, despite numerous studies, how occluded ligand migration affects the kinetics and structural dynamics of the R-T transition remains unclear. To shed light on this issue, we chose homodimeric hemoglobin (HbI) with the I114F mutation (I114F), which is known to interfere with ligand migration between the primary and the secondary docking sites, and studied its kinetics and structural dynamics using time-resolved X-ray solution scattering. The kinetic analysis shows that I114F has three structurally distinct intermediates (I1, I2, and I3) as in the wild type (WT), but its geminate CO recombination occurs directly from I1 without the path via I2 observed in WT. Moreover, the structural transitions, which involve ligand migration (the transitions from I1 to I2 and from I3 to the initial state) are decelerated compared to WT. The structural analysis revealed that I114F involves generally smaller structural changes in all three intermediates compared to WT.
Keywords: picosecond time-resolved X-ray solution scattering, homodimeric hemoglobin, ligand migration, protein structural dynamics
Graphical Abstract
INTRODUCTION
Ligand binding to a protein is a fundamental process for the protein function in biology. The active site for ligand binding is usually located inside of the protein and surrounded by residues that stabilize the bound ligand or the intermediate structure of the substrate. Ligand binding to a protein requires the translocation of the ligand from the bulk solvent to the active site inside the protein, which is often facilitated by conformational changes of the protein.1–2 Therefore, the ligand migration pathway within the protein and its connection to protein conformational changes have been the topic of extensive studies.1–15 Heme proteins such as myoglobin (Mb), hemoglobin (Hb), and homodimeric hemoglobin (HbI), which play crucial roles in life by storing and delivering simple diatomic molecules such as molecular oxygen, nitric oxide, and carbon monoxide, have been targeted for such studies.16–17
HbI is an excellent model system to investigate the ligand-linked structural changes during allosteric regulation.18–20 Unlike Mb, which is a monomeric protein, the binding of the ligand in a multimeric protein is more complicated due to the additional effects of allosteric regulation, which generally proceeds by a series of structural changes.21 For example, the binding of the ligand (effector molecule) to one active site triggers subsequent structural changes that affect the reactivity of a distant active site. Hb is a textbook example, but its tetrameric structure naturally involves highly complex reaction dynamics.21–27 In this regard, HbI offers a simpler model system due to its homodimeric structure, and it has been studied to observe ligand migration during allosteric regulation with various site-directed mutations that can alter the ligand migration pathways.28–31
X-ray crystallography3–9, 29, 31 and optical and infrared spectroscopic tools10–15, 28, 30 have been used to investigate the ligand migration pathways in heme proteins. To gain better insight into ligand migration, a series of mutants, which can modify protein-ligand interactions and the ligand migration pathways by introducing a voluminous residue to block the putative transient ligand docking sites or to occlude possible migration pathways between these sites, have been made and investigated.15, 30–31 As a result of these comprehensive studies of the ligand migration pathways of heme proteins, for example, it is known that for Mb the ligand migrates between the cavities inside the protein, such as the primary docking site and Xe sites (secondary docking sites).6–8 Those cavities act as transient ligand docking sites, thereby reducing the possibility of the immediate rebinding to the heme group and consequently facilitating the easy escape of the ligand from the protein to the bulk solvent. It is also known that the migration of ligands requires structural changes in the protein that facilitate the movement of the ligand between internal cavities and the binding site.1–2 However, despite numerous studies, it remains elusive how occluded ligand migration affects the kinetics and structural dynamics of the R-T transition. To address this issue, information about both ligand migration and the global structural changes should be obtained, though this is not a trivial task. Here, we applied time-resolved X-ray solution scattering (TRXSS) to the I114F mutant of HbI (I114F) and compared the results with those from the wild type (WT) to investigate the relationship between ligand migration and the allosteric structural changes of HbI.
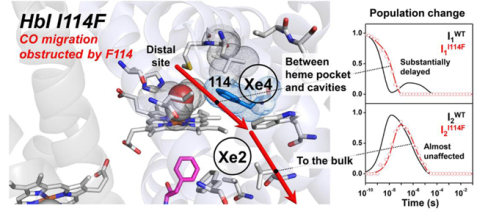
I114F is known to modify protein-ligand interaction by occluding possible ligand migration pathways between the heme pocket and the putative transient ligand docking sites.30, 32–33 As shown in Figure 1, the 114th residue, together with the 36th and 73rd residues, forms a channel for the migration of a photo-dissociated ligand between the primary docking site (distal pocket) and the secondary docking sites (Xe4 and Xe2 sites). Whereas this channel is relatively free from obstruction in WT, Phe114 in I114F makes this channel narrower.32 As a result, the geminate recombination would occur only from the primary docking site in I114F whereas it also occurs from the secondary docking sites in WT.30 In addition, since the steric hindrance imposed by Phe114 of the mutant restricts the ligand-linked heme movement deep into the subunit in the liganded state of I114F, as a result, the structure of the liganded state of I114F is mainly affected by the mutation. More specifically, the heme-heme distance and the subunit rotation angle in the liganded state of I114F are similar to those in the T state of WT, that is, the unliganded state of WT. Accordingly, the heme movement and the intersubunit rotation involved in the R-T transition are reduced in I114F.32
Figure 1.
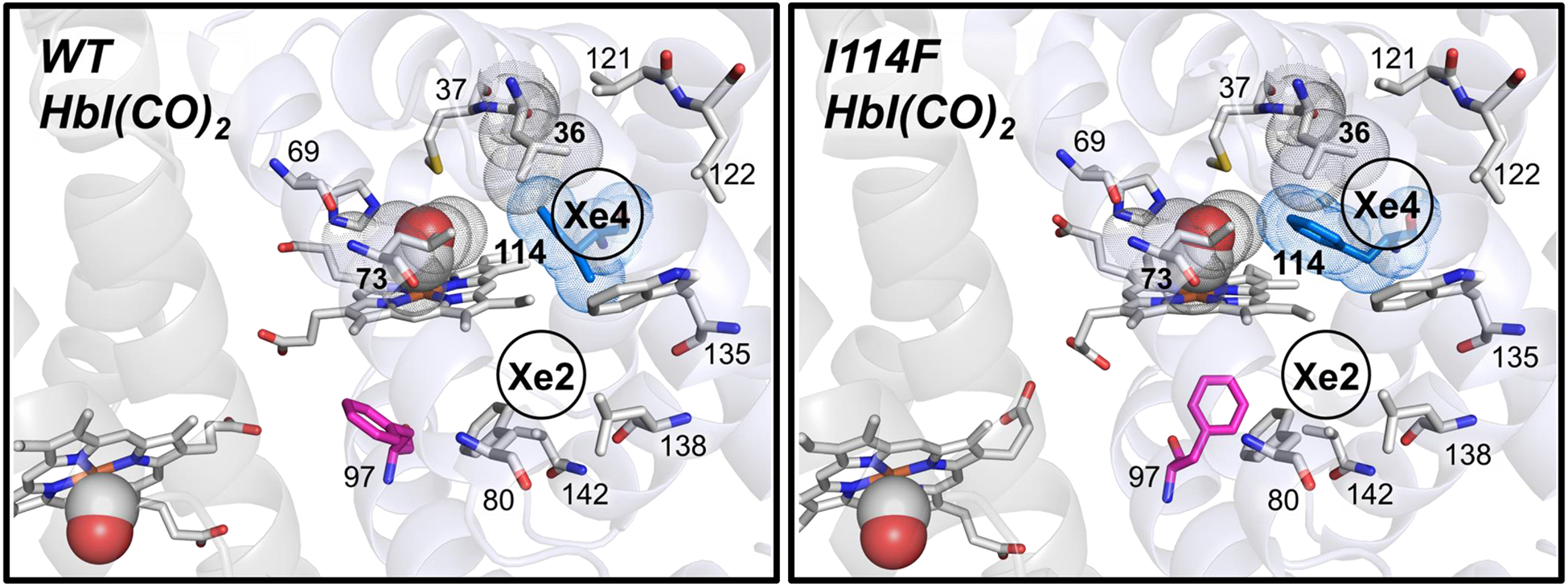
Close-up views of the heme environment of crystal structures of HbI(CO)2 for WT (left, pdb code: 3SDH34) and I114F (right, pdb code: 1JWN32). Heme and some residues forming secondary docking sites (Xe4 and Xe2 sites) are represented by the gray sticks, and heme-bounded CO molecules are shown as spheres. The 36th, 73rd, and 114th residues, which separate the primary and the secondary docking sites, are depicted with their van der Waals spheres (gray for the 36th and 73rd residues and blue for the 114th residue). Phe97 is shown as the magenta stick, which faces the subunit interface in WT but faces the proximal site in I114F. The positions of the Xe4 and Xe2 sites are labeled based on the crystal structure of Xe-exposed WT HbI(CO)2 (pdb code: 3G4631).
The static structures of I114F have been studied by X-ray crystallography32 and the ligand migration in I114F has been studied over wide time and temperature ranges using optical and infrared spectroscopy.30, 33 An X-ray crystallographic study has revealed detailed structural differences between the R and T states with the atomic resolutions,32 but could not access the intermediate structures and relevant kinetics. Fourier-transform infrared (FTIR) cryospectroscopy can discern the kinetics of CO migration,30 but it lacks information about global structural changes. TRXSS, also known as time-resolved X-ray liquidography (TRXL), is a relevant tool for probing the global structural changes linked to the CO migration and relevant kinetics.35–37 Due to its relatively low sensitivity to position of the CO molecule, the kinetics observed by TRXSS data reflect the kinetics associated with the overall conformation changes rather than the CO movement itself. However, since it is known that I114F interferes with the ligand migration pathway by obstructing the movement of the ligand migration between the primary and the secondary docking sites,30 it serves as an excellent model system to study the effect of an occluded ligand migration pathway on the kinetics of ligand migration and the structural dynamics of a protein. For example, ligand migration is expected to be decelerated in I114F compared to WT, but there has been no experimental evidence supporting this prediction.
EXPERIMENTAL METHODS
TRXSS data of I114F were collected at the 14-ID-B beamline of the Advanced Photon Source (APS). The solution sample of CO-bound I114F HbI(CO)2 sealed in a capillary with 3.5 mM concentration was excited by 35 ps long laser pulse at 532 nm. Subsequently, a ~100 ps long X-ray pulse probed the sample, and the scattered X-ray photons were recorded as a function of the time delay between the laser pump pulse and the X-ray probe pulse. Time-resolved scattering curves were collected at 36 pump-probe time delays ranging from 100 ps to 56.2 ms (four time points per decade). By taking the difference between the scattering curve measured at each time delay point and the reference scattering curve measured at −5 μs, we obtained time-resolved difference scattering curves, ΔS(q,t). The sample preparation, experiment, and the data processing are provided more in detail in the Supporting Information.
RESULTS AND DISCUSSION
The time-resolved difference X-ray solution scattering curves, ΔS (q, t), for I114F as shown in Figure 2 were measured after the photodissociation of CO ligands from the initial I114F HbI(CO)2 for time delays from 100 ps to 56.2 ms. To obtain the number of reaction intermediates and time constants of the reaction, we performed a singular value decomposition (SVD) analysis on the TRXSS data. The analysis revealed that the same kinetics framework involving three structurally distinct intermediates (termed I1, I2, and I3 according to their appearance time), which were previously identified for WT,35 can also be applied for I114F on the basis of the number of significant singular components (Figure S4). We fit the first three right singular vectors multiplied by their singular values with the sum of the exponentials, as shown in Figure S4d, and obtained six time constants of 640 (± 96) ps, 23 (± 1.5) ns, 990 (± 190) ns, 7.6 (± 1.0) μs, 3.2 (± 0.41) ms, and 29 (± 24) ms, whereas seven time constants were found for WT. In order to find the optimal kinetic model based on the observed time constants, the number of intermediates at each time delay should be determined. The number of intermediates in a certain time range can be obtained accurately through SVD analysis of reduced time ranges (see the Supporting Information for the detailed procedures). As a result, the optimal kinetic model for I114F (Figure 3a) is found and compared with that of WT (Figure 3b).35 Note that the latest two time constants are replaced by a bimolecular rate constant in the kinetic model. The main difference between I114F and WT is the fact that I114F has a different geminate CO recombination pathway, which is responsible for the lack of one time constant in I114F. As shown in Figure 2, the theoretical difference scattering curves (red curves) obtained from the kinetic model are in excellent agreement with the experimental difference scattering curves (black curves) at all time delays. From the kinetic analysis, we obtained the time-dependent population changes (Figure 3c) and the species-associated difference scattering (SADS) curves of each individual intermediate (Figure 3d). To distinguish the intermediates for WT and I114F, we labeled I1, I2, and I3 of WT as I1WT, I2WT, and I3WT, respectively, and likewise labeled those of I114F as I1I114F, I2I114F, and I3I114F, respectively.
Figure 2.
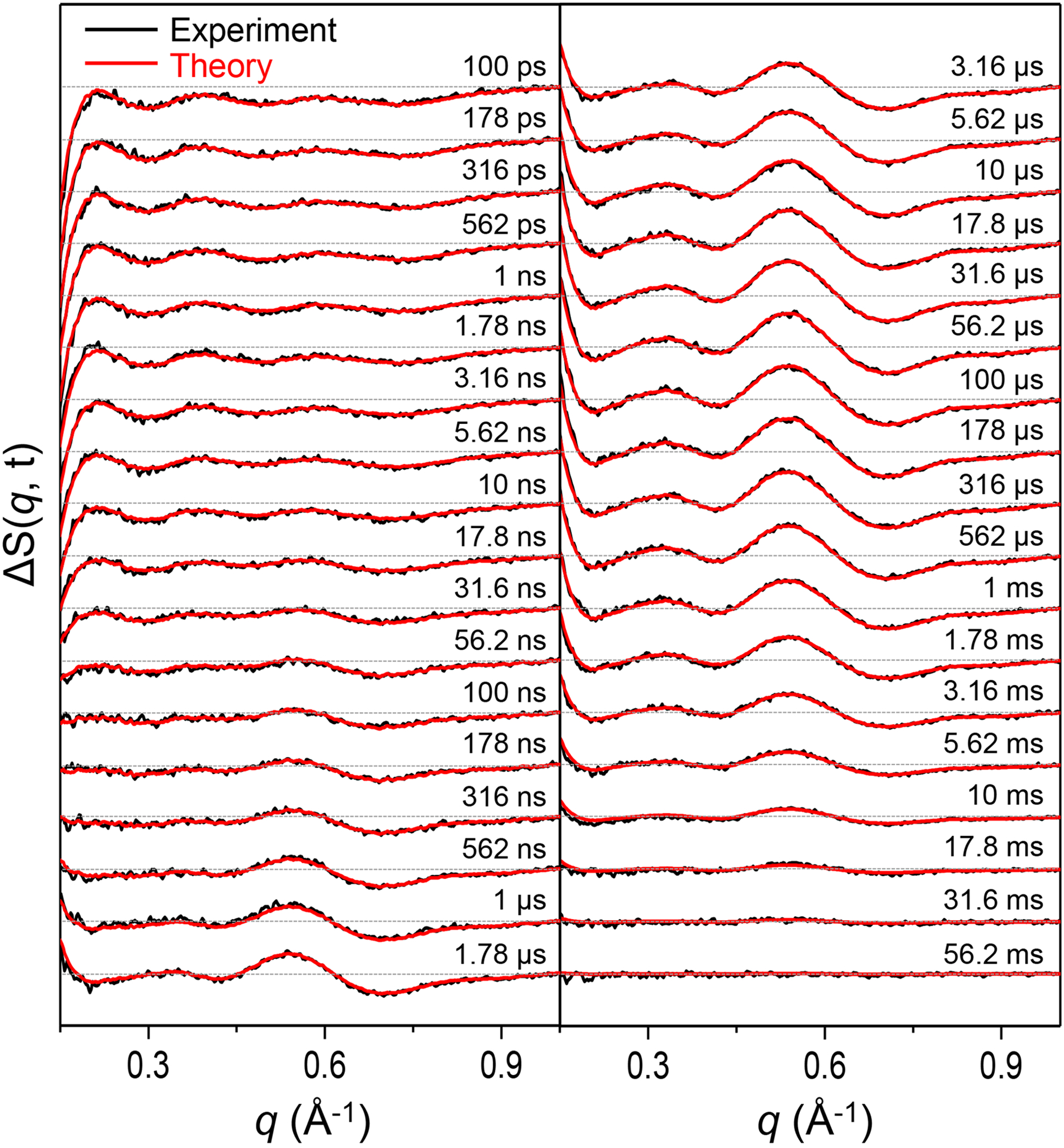
Time-resolved difference X-ray solution scattering curves, ΔS(q, t), measured for a solution sample of I114F. Experimental curves (black curves) are compared with theoretical curves (red curves) generated by linear combinations of left singular vectors based on the optimized kinetics model.
Figure 3.
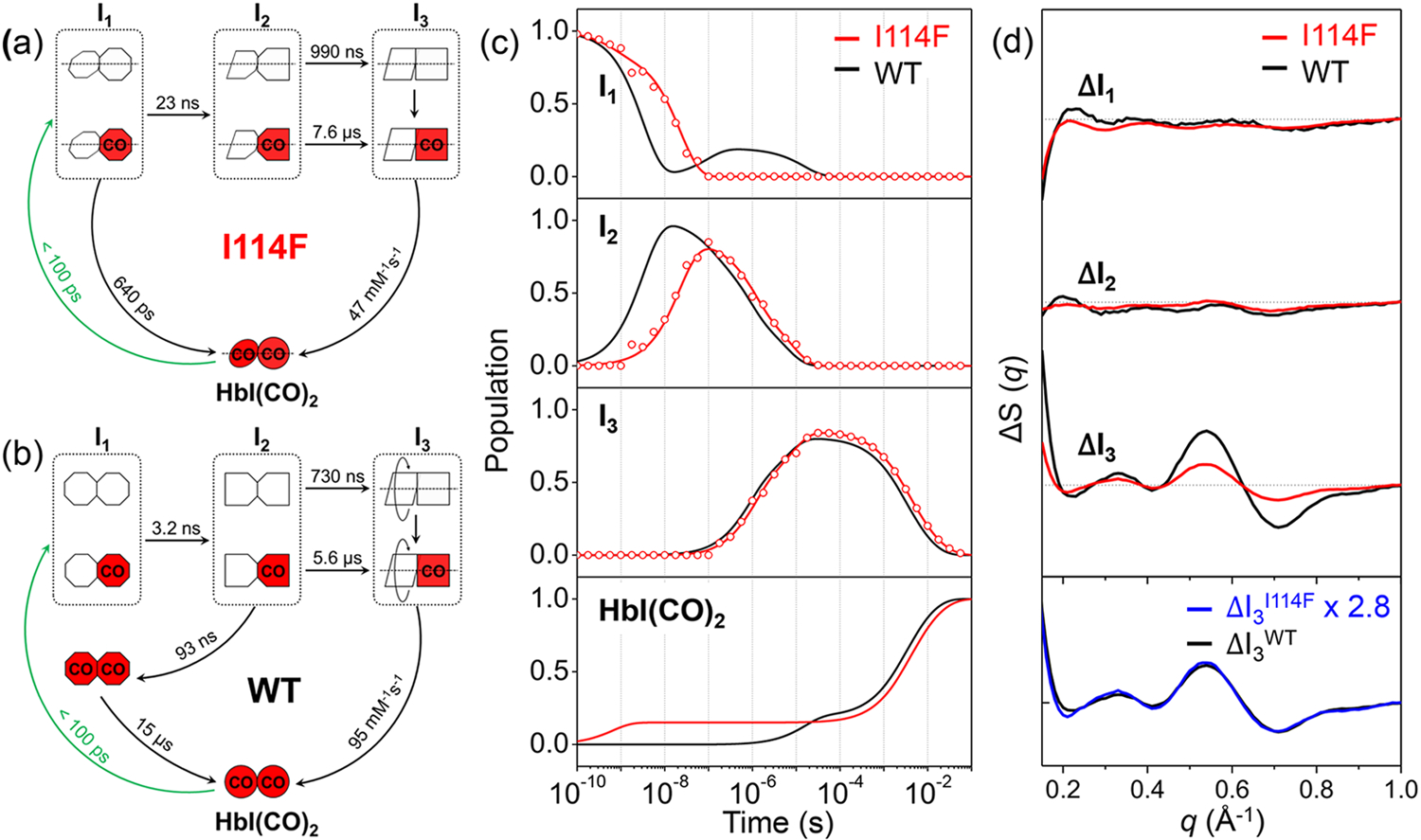
(a, b) Kinetic models for I114F and WT. The red (with “CO”) and white symbols represent ligated and photolyzed subunits, respectively. To indicate the three structurally distinguishable intermediates with different tertiary structures, the subunit of each intermediate is presented with a different shape. For all three intermediates and HbI(CO)2 of I114F, two subunits are described as rotating with respect to the other, reflecting the quaternary structural difference with respect to the R state of WT, that is, the liganded state of WT. For WT, only I3 is represented in such a way, indicating that the quaternary transition involving subunit rotation occurs only in I3. In the kinetic model for WT, two connected red octagons represent a ligated form of I1, which is formed by the geminate recombination of CO with I2 and structurally indistinguishable from the photolyzed forms (two connected white octagons and the white-red mixed ones) of I1. (c) Population changes of the three intermediates and initial HbI(CO)2 as a function of time for I114F (red) and WT (black). The lines correspond to the populations obtained from the kinetics analysis. The open circles correspond to the optimized populations obtained by fitting the experimental curve at each time point with a linear combination of SADS curves for the three intermediates. (d) SADS curves of the three intermediates for I114F (red) and WT (black). The scaled curve of I3I114F (blue) has a shape nearly identical to that of I3WT, but with a smaller magnitude by 2.8 times.
The kinetic models for I114F and WT are shown in Figures 3a and 3b,35 respectively. Both have three structurally distinct intermediates. The earliest intermediate, I1, is formed by laser excitation from the initial state within our experimental time resolution of ~100 ps. In I114F, 16% of I1 goes back to the initial I114F HbI(CO)2 by the geminate recombination of CO with a time constant of 640 ps, and the rest transforms into the second intermediate, I2, with a time constant of 23 ns, which is substantially (about 7 times) slower than that of WT (3.2 ns). The geminate recombination pathway has the smaller fraction (16 %) in spite of its faster time constant, compared to the I1-to-I2 transition pathway, indicating that the geminate recombination and the I1-to-I2 transition processes are not kinetically controlled (see the Supporting Information for details). The subsequent biphasic I2-to-I3 transitions, which are interpreted as the R-T transitions of two substates of fully and partially photolyzed forms, occur with time constants of 990 ns and 7.6 μs in I114F, similar to those of WT (730 ns and 5.6 μs). The last intermediate, I3, goes back to the initial I114F HbI(CO)2 state via the bimolecular CO recombination with a bimolecular rate constant of 47 mM−1s−1, which is two times slower than that of WT (95 mM−1s−1).
The unveiled kinetics shows that there are two significantly decelerated transitions in I114F. Because the occluded ligand migration pathway in I114F30, 32 is expected to decelerate the ligand migration between the primary and the secondary docking sites and TRXSS measures global structural changes rather than the ligand migration process itself, we can infer that the decelerated structural transitions manifested in the TRXSS data of I114F would involve CO migration between the primary and the secondary docking sites. The first decelerated process is the I1-to-I2 transition. Since the I1 intermediate is formed directly after photodissociation within 100 ps, CO should still likely be in the primary docking site. Thus, the delayed I1-to-I2 transition by the mutation means that the structural transition from I1 to I2 involves the migration of CO from the primary to the secondary docking sites. The significant deceleration of the I1-to-I2 transition observed in I114F was not observed in TRXSS studies on other mutants such as F97Y35 and T72V37 where their ligand migration pathways are not expected to be affected by the mutations, supporting that the I1-to-I2 transition is related to ligand migration from the primary to the secondary docking sites. The second decelerated process is the bimolecular CO rebinding of I3. As Phe114 makes the access of CO to the active site difficult, the bimolecular CO recombination can be slowed as well, which is consistent with the results of flash photolysis and oxygen equilibrium binding studies.30, 32–33 On the other hand, other major processes, the biphasic structural transitions from I2 to I3, in I114F are not delayed and have rates similar to those of WT. This means that the I2-to-I3 transition is not likely to involve the movement of CO between the primary and the secondary docking sites. According to a previous structural analysis of WT, it is known that I1WT and I2WT have structures close to the R state of WT, whereas I3WT has a structure close to the T state of WT.35 Therefore, we expect that the CO migrates from the inside of the protein to the bulk during the I2-to-I3 transition.
In addition, I114F has a different geminate recombination pathway compared to WT, which supports our interpretation of the CO migration during the transition between I1 and I2. In WT, the geminate recombination occurs from the I2 intermediate. A part of I2 transforms into a ligated form of I1 (the two red octagons in Figure 3b), and then subsequently returns to the initial liganded state. In contrast, in I114F, the geminate recombination occurs from I1 not I2. According to a FTIR cryospectroscopic study which is sensitive to the CO position, the geminate recombination occurs only from the primary docking site in I114F, in contrast in WT, it occurs from both of the primary and the secondary docking sites.30 By comparing the previous FTIR study and our TRXSS results, we can infer the location of the ligand (CO molecule) in the I1 and I2 intermediates. The geminate recombination in I114F is likely to occur directly from rather the primary docking site than the secondary docking sites, since Phe114 residue disturbs the movement of the ligand from the secondary to the primary docking sites, thereby blocking the I2-to-I1 transition. These differences in the geminate recombination kinetics between the I114F and WT suggest that the ligand in the I1 and I2 intermediates resides in the primary and the secondary docking sites, respectively.
The SADS curves shown in Figure 3d, extracted from the kinetic analysis, contain the structural information of each intermediate. The difference in the amplitude between the SADS curves of I114F and WT is attributed to the difference in the magnitude between the structural change occurring in I114F and WT (see the Supporting Information for details). As shown in the upper panel of Figure 3d, the SADS curves of I114F have smaller amplitudes than those of the WT for all intermediates, suggesting that the degree of structural change is smaller in I114F. Notably, in the case of I3, which was identified as the intermediate arising from the R-T transition, the SADS curves of I114F and WT have nearly identical shapes, whereas the magnitude of the SADS curve of I114F is 2.8 times smaller than that of WT (the lower panel of Figure 3d). The identical shapes with different magnitudes suggest that the trend of structural changes between the liganded state and I3 in WT is maintained in I114F similarly, while the amount of the structural change is less pronounced in I114F (see the Supporting Information for details). This observation is consistent with the result expected from the previous crystallographic study.32 Due to the restricted heme movement, the liganded state of I114F has closer structure to rather the T state of WT than the R state of WT.
We conducted a structure refinement analysis on the SADS curves to extract key structural parameters of each intermediate structure according to the protocol established in our previous study on WT.35 The detailed procedures are given in the SI. The refined candidate structures are obtained from the analysis and their theoretical scattering patterns show excellent agreement with the experimental SADS curves (Figure S9). The three key structural parameters, the displacement of the residues, the heme-heme distance, and the subunit rotation angle, are extracted from the sets of the refined candidate structures (Figure 4). The first and the other two are relevant to the tertiary and quaternary structural changes, respectively.
Figure 4.
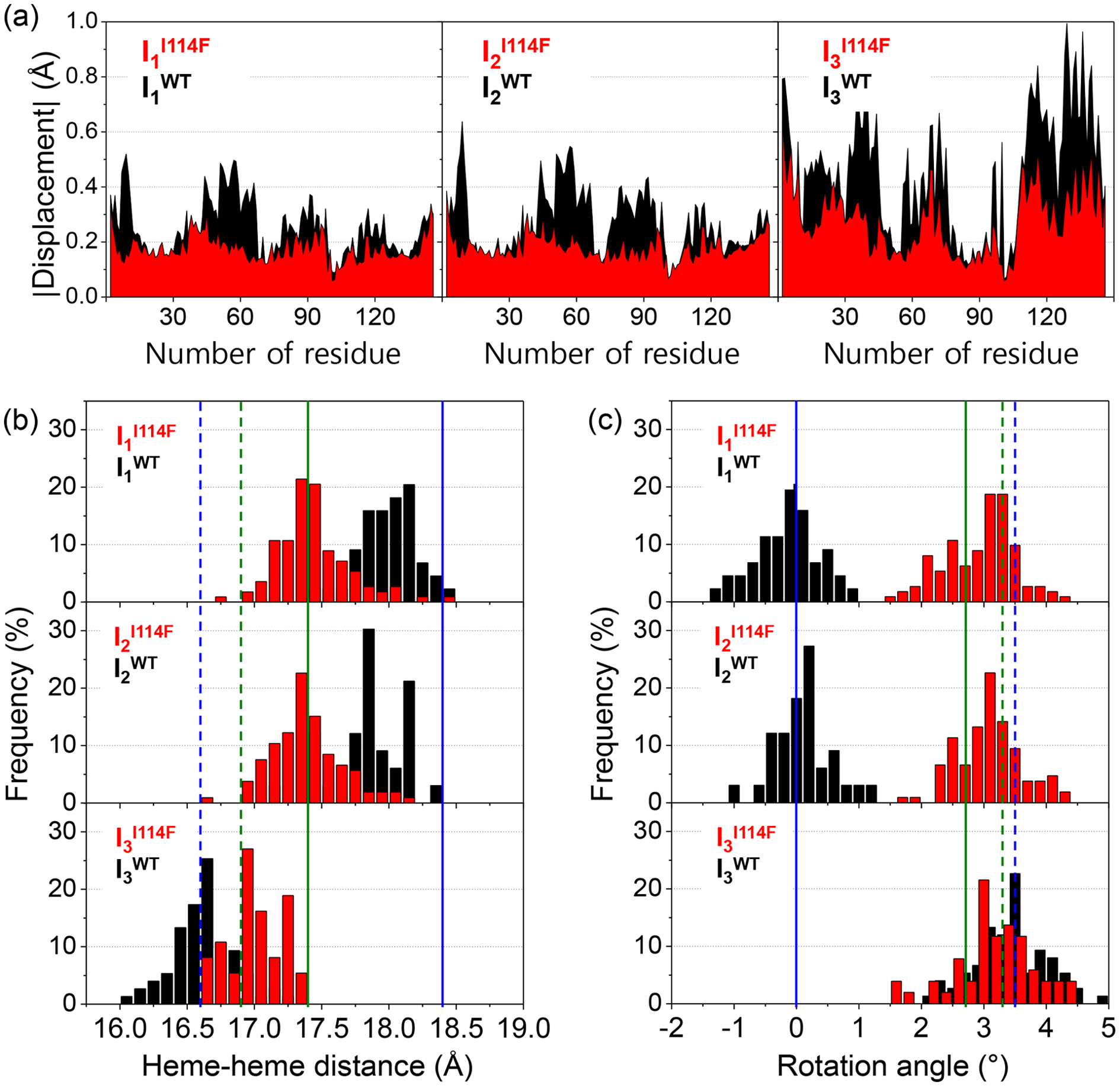
(a) Absolute displacement as a function of the residue for the candidate structures of three intermediates for I114F (red) and WT (black). For the calculation of the displacement, the reference structure for I114F is its liganded crystal structure, whereas for WT it is its liganded crystal structure. (b, c) The occurrence distribution of (b) the heme-heme distance and (c) the subunit rotation angle for the candidate structures of I114F (red) and WT (black) with respect to the R state of WT. The values from the crystal structures for the liganded state of WT and I114F are indicated by the blue and green vertical solid lines, respectively, and those for the unliganded state of WT and I114F are likewise indicated by the blue and green vertical dotted lines, respectively. The pdb codes for liganded WT, unliganded WT, liganded I114F, and unliganded I114F are 3SDH,34 4SDH,34 1JWN,32 and 1JZK,32 respectively.
The absolute displacements of the residues for the three intermediates, related to the tertiary structural changes, are shown in Figure 4a. This figure visualizes the degree of movement of the residues with respect to the reference structure. Here, we defined (i) the position of a residue as the distance between the Cɑ atom and the iron atom of the heme in the same subunit, and (ii) the displacement of a residue as the difference in the positions between the candidate and reference structures. The reference structures of WT and I114F are their own static crystal structures of the liganded state (pdb codes WT: 3SDH,34 and I114F: 1JWN32). For all three intermediates, the area under the absolute displacement curve of I114F is smaller than that of WT, meaning that I114F undergoes less structural changes during the transition to each intermediate. In the case of I3, the magnitudes of the displacements are much smaller in I114F than in WT, whereas the directions of the displacements in I114F are similar to those in WT. Hence, the magnitude of the structural changes involved in the transition to I3 is greatly reduced in I114F, whereas the direction of the structural changes remains the same in WT and I114F. This is fully consistent with the fact that the shape of the SADS curve of I3I114F is identical to that of I3WT but with a smaller magnitude by 2.8 times.
The occurrence distribution of the heme-heme distance and the subunit rotation angle of the intermediates for I114F and WT are also obtained and compared in Figures 4b and 4c, respectively. They are the key structural parameters about the quaternary structural transition as the cooperative ligand binding of HbI is modulated by the hydrogen-bonding network between two hemes32 and the subunit rotation.34, 38–41 In WT, the transitions from the liganded state (heme-heme distance = 18.4 Å, subunit rotation angle = 0˚) to I1WT (heme-heme distance = 18.0 ± 0.2 Å, subunit rotation angle = −0.1 ± 0.5˚) and to I2WT (17.9 ± 0.3 Å and 0.1 ± 0.5˚) involve very small changes in the heme-heme distance and subunit rotation angle. The major structural changes during the photoreaction of WT occur in I3WT (16.6 ± 0.2 Å and 3.4 ± 0.6˚), whose structure is close to the T state of WT (16.6 Å and 3.5˚). In I114F, the heme-heme distances of both I1I114F (17.4 ± 0.3 Å) and I2I114F (17.4 ± 0.3 Å) are identical to that of the liganded state of I114F (17.4 Å) but much smaller than that of the R state of WT (18.4 Å). The heme-heme distance of I3I114F (17.0 ± 0.3 Å) is similar to that of the unliganded state of I114F (16.9 Å) and the T state of WT (16.6 Å). The subunit rotation angles of I1I114F (2.9 ± 0.6˚), I2I114F (3.1 ± 0.6˚), and I3I114F (3.3 ± 0.6˚) are also close to that of the T state of WT (3.5˚). As can be seen in Figures 4b and 4c, the extent of quaternary structural changes such as the heme-heme distance change and intersubunit rotation are smaller in I114F, compared to WT. The smaller quaternary structural changes in I114F are attributed to the quaternary structure of the liganded state of I114F. Specifically, the starting structure of the R-T transition (the initial I114F HbI(CO)2) in I114F is already shifted toward the T state of WT, whereas the end structure (I3I114F) is still similar to the T state of WT. Accordingly, the amount of structural changes involved in the R-T transition of I114F is smaller than that of WT (Figure S10). The restricted heme movement by Phe114 reduces the degree of quaternary structural change, which is consistent with conclusions drawn from the crystal structures.32
CONCLUSIONS
In this work, we harnessed the power of TRXSS to investigate the detailed reaction kinetics and intermediate structures.The effects of the occluded ligand migration pathway on the kinetics and structural dynamics were elucidated by comparing the TRXSS data from WT and I114F, which is known to occlude ligand migration between the primary and the secondary docking sites. The kinetic analysis of the TRXSS data revealed that I114F has a kinetic framework with three structurally distinct intermediates (I1, I2, and I3), as in WT. From a comparative study with WT, we found that two structural transitions (the I1-to-I2 transition and the bimolecular recombination of I3) are significantly delayed in I114F. Therefore, we suggest that they involve ligand migration between the primary and the secondary docking sites. We also found that the geminate CO recombination occurs from the I1 intermediate instead of the I2 intermediate in I114F. A structural analysis using the SADS curves of the three intermediates revealed that the restriction of heme movement by Phe114 reduces the degree of allosteric structural change, which is consistent with results from the crystal structures. The delayed and different structural transitions caused by the obstructed ligand migration indicate that the mutation causes both the deceleration of ligand migration and the alteration of the conformation change during the protein dynamics.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Institute for Basic Science (IBS-R004). We acknowledge BioCARS staff for support and helpful discussions. Use of the BioCARS Sector 14 at the APS was supported by the National Institutes of Health (NIH) National Institute of General Medical Sciences Grant P41GM103543. The time-resolved setup at Sector 14 was funded in part through collaboration with P. Anfinrud (NIH/NIDDK) through the Intramural Research Program of the NIDDK. Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. DE-AC02-06CH11357.
Funding Sources
This work was supported by the Institute for Basic Science (IBS-R004).
ABBREVIATIONS
- HbI
homodimeric hemoglobin
- I114F
I114F mutant of HbI
- WT
wild type of HbI
- Mb
myoglobin
- Hb
hemoglobin
- TRXSS
time-resolved X-ray solution scattering
- FTIR
Fourier-transform infrared
- TRXL
time-resolved liquidography
- SVD
singular value decomposition
- SADS
species-associated difference scattering.
Footnotes
The authors declare no competing financial interest.
Supporting Information.
The Supporting Information is available free of charge on the ACS Publication website at DOI: https://doi.org/10.1021/acs.jpcb.9b11749
Sample preparation; Data collection; Data processing; SVD analysis; Kinetic model; kinetic analysis; Origin of the small amplitude in the SADS curve of I114F; Relation between the amplitude of the SADS curve and the magnitude of the structural change; Structure refinement; Photoconversion yield; and Table and Figures (PDF)
REFERENCES
- 1.Brunori M, Structural dynamics of myoglobin. Biophys Chem 2000, 86 (2–3), 221–230. [DOI] [PubMed] [Google Scholar]
- 2.Brunori M; Gibson QH, Cavities and packing defects in the structural dynamics of myoglobin. EMBO Rep 2001, 2 (8), 674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlichting I; Berendzen J; Phillips GN Jr.; Sweet RM, Crystal structure of photolysed carbonmonoxy-myoglobin. Nature 1994, 371 (6500), 808–812. [DOI] [PubMed] [Google Scholar]
- 4.Teng TY; Srajer V; Moffat K, Photolysis-induced structural changes in single crystals of carbonmonoxy myoglobin at 40 K. Nat. Struct. Biol 1994, 1 (10), 701–705. [DOI] [PubMed] [Google Scholar]
- 5.Hartmann H; Zinser S; Komninos P; Schneider RT; Nienhaus GU; Parak F, X-ray structure determination of a metastable state of carbonmonoxy myoglobin after photodissociation. Proc. Natl. Acad. Sci. U.S.A 1996, 93 (14), 7013–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunori M; Vallone B; Cutruzzola F; Travaglini-Allocatelli C; Berendzen J; Chu K; Sweet RM; Schlichting I, The role of cavities in protein dynamics: crystal structure of a photolytic intermediate of a mutant myoglobin. Proc. Natl. Acad. Sci. U.S.A 2000, 97 (5), 2058–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu K; Vojtchovsky J; McMahon BH; Sweet RM; Berendzen J; Schlichting I, Structure of a ligand-binding intermediate in wild-type carbonmonoxy myoglobin. Nature 2000, 403 (6772), 921–923. [DOI] [PubMed] [Google Scholar]
- 8.Ostermann A; Waschipky R; Parak FG; Nienhaus GU, Ligand binding and conformational motions in myoglobin. Nature 2000, 404 (6774), 205–208. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt M; Nienhaus K; Pahl R; Krasselt A; Anderson S; Parak F; Nienhaus GU; Srajer V, Ligand migration pathway and protein dynamics in myoglobin: a time-resolved crystallographic study on L29W MbCO. Proc. Natl. Acad. Sci. U.S.A 2005, 102 (33), 11704–11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alben JO; Beece D; Bowne SF; Doster W; Eisenstein L; Frauenfelder H; Good D; McDonald JD; Marden MC; Moh PP; Reinisch L; Reynolds AH; Shyamsunder E; Yue KT, Infrared spectroscopy of photodissociated carboxymyoglobin at low temperatures. Proc. Natl. Acad. Sci. U.S.A 1982, 79 (12), 3744–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry ER; Sommer JH; Hofrichter J; Eaton WA, Geminate recombination of carbon monoxide to myoglobin. J. Mol. Biol 1983, 166 (3), 443–451. [DOI] [PubMed] [Google Scholar]
- 12.Carver TE; Rohlfs RJ; Olson JS; Gibson QH; Blackmore RS; Springer BA; Sligar SG, Analysis of the kinetic barriers for ligand binding to sperm whale myoglobin using site-directed mutagenesis and laser photolysis techniques. J. Biol. Chem 1990, 265 (32), 20007–20020. [PubMed] [Google Scholar]
- 13.Balasubramanian S; Lambright DG; Boxer SG, Perturbations of the distal heme pocket in human myoglobin mutants probed by infrared spectroscopy of bound CO: correlation with ligand binding kinetics. Proc. Natl. Acad. Sci. U.S.A 1993, 90 (10), 4718–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim M; Jackson TA; Anfinrud PA, Ultrafast rotation and trapping of carbon monoxide dissociated from myoglobin. Nat Struct Biol 1997, 4 (3), 209–14. [DOI] [PubMed] [Google Scholar]
- 15.Nienhaus K; Deng P; Kriegl JM; Nienhaus GU, Structural dynamics of myoglobin: effect of internal cavities on ligand migration and binding. Biochemistry 2003, 42 (32), 9647–9658. [DOI] [PubMed] [Google Scholar]
- 16.Chapman SK; Daff S; Munro AW, Heme: The most versatile redox centre in biology? Struct Bond 1997, 88, 39–70. [Google Scholar]
- 17.Anderson JL; Chapman SK, Ligand probes for heme proteins. Dalton Trans 2005, (1), 13–24. [DOI] [PubMed] [Google Scholar]
- 18.Chiancone E; Vecchini P; Verzili D; Ascoli F; Antonini E, Dimeric and tetrameric hemoglobins from the mollusc Scapharca inaequivalvis. Structural and functional properties. J. Mol. Biol 1981, 152 (3), 577–592. [DOI] [PubMed] [Google Scholar]
- 19.Antonini E; Ascoli F; Brunori M; Chiancone E; Verzili D; Morris RJ; Gibson QH, Kinetics of ligand binding and quaternary conformational change in the homodimeric hemoglobin from Scapharca inaequivalvis. J. Biol. Chem 1984, 259 (11), 6730–6738. [PubMed] [Google Scholar]
- 20.Mozzarelli A; Bettati S; Rivetti C; Rossi GL; Colotti G; Chiancone E, Cooperative oxygen binding to scapharca inaequivalvis hemoglobin in the crystal. J. Biol. Chem 1996, 271 (7), 3627–3632. [DOI] [PubMed] [Google Scholar]
- 21.Perutz MF, Mechanisms of cooperativity and allosteric regulation in proteins. Q. Rev. Biophys 1989, 22 (2), 139–237. [DOI] [PubMed] [Google Scholar]
- 22.Perutz MF, Stereochemistry of cooperative effects in haemoglobin. Nature 1970, 228 (5273), 726–739. [DOI] [PubMed] [Google Scholar]
- 23.Baldwin J; Chothia C, Haemoglobin: the structural changes related to ligand binding and its allosteric mechanism. J. Mol. Biol 1979, 129 (2), 175–220. [DOI] [PubMed] [Google Scholar]
- 24.Shaanan B, Structure of human oxyhaemoglobin at 2.1 A resolution. J. Mol. Biol 1983, 171 (1), 31–59. [DOI] [PubMed] [Google Scholar]
- 25.Fermi G; Perutz MF; Shaanan B; Fourme R, The crystal structure of human deoxyhaemoglobin at 1.74 A resolution. J. Mol. Biol 1984, 175 (2), 159–174. [DOI] [PubMed] [Google Scholar]
- 26.Eaton WA; Henry ER; Hofrichter J; Mozzarelli A, Is cooperative oxygen binding by hemoglobin really understood? Nat. Struct. Biol 1999, 6 (4), 351–358. [DOI] [PubMed] [Google Scholar]
- 27.Adachi S; Park SY; Tame JR; Shiro Y; Shibayama N, Direct observation of photolysis-induced tertiary structural changes in hemoglobin. Proc. Natl. Acad. Sci. U.S.A 2003, 100 (12), 7039–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiancone E; Elber R; Royer WE Jr.; Regan R; Gibson QH, Ligand binding and conformation change in the dimeric hemoglobin of the clam Scapharca inaequivalvis. J. Biol. Chem 1993, 268 (8), 5711–5718. [PubMed] [Google Scholar]
- 29.Knapp JE; Pahl R; Srajer V; Royer WE Jr., Allosteric action in real time: Time-resolved crystallographic studies of a cooperative dimeric hemoglobin. Proc. Natl. Acad. Sci. U.S.A 2006, 103 (20), 7649–7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nienhaus K; Knapp JE; Palladino P; Royer WE Jr.; Nienhaus GU, Ligand migration and binding in the dimeric hemoglobin of Scapharca inaequivalvis. Biochemistry 2007, 46 (49), 14018–14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knapp JE; Pahl R; Cohen J; Nichols JC; Schulten K; Gibson QH; Srajer V; Royer WE Jr., Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis. Structure 2009, 17 (11), 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knapp JE; Gibson QH; Cushing L; Royer WE Jr., Restricting the ligand-linked heme movement in Scapharca dimeric hemoglobin reveals tight coupling between distal and proximal contributions to cooperativity. Biochemistry 2001, 40 (49), 14795–14805. [DOI] [PubMed] [Google Scholar]
- 33.Nichols JC; Royer WE Jr.; Gibson QH, An optical signal correlated with the allosteric transition in Scapharca inaequivalvis HbI. Biochemistry 2006, 45 (51), 15748–15755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Royer WE Jr.; Hendrickson WA; Chiancone E, Structural transitions upon ligand binding in a cooperative dimeric hemoglobin. Science 1990, 249 (4968), 518–521. [DOI] [PubMed] [Google Scholar]
- 35.Kim KH; Muniyappan S; Oang KY; Kim JG; Nozawa S; Sato T; Koshihara SY; Henning R; Kosheleva I; Ki H; Kim Y; Kim TW; Kim J; Adachi S; Ihee H, Direct Observation of Cooperative Protein Structural Dynamics of Homodimeric Hemoglobin from 100 ps to 10 ms with Pump-Probe X-ray Solution Scattering. J Am Chem Soc 2012, 134 (16), 7001–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JG; Kim TW; Kim J; Ihee H, Protein Structural Dynamics Revealed by Time-Resolved X-ray Solution Scattering. Accounts Chem Res 2015, 48 (8), 2200–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JG; Muniyappan S; Oang KY; Kim TW; Yang C; Kim KH; Kim J; Ihee H, Cooperative protein structural dynamics of homodimeric hemoglobin linked to water cluster at subunit interface revealed by time-resolved X-ray solution scattering. Struct. Dyn 2016, 3 (2), 023610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Royer WE Jr.; Hendrickson WA; Chiancone E, The 2.4-A crystal structure of Scapharca dimeric hemoglobin. Cooperativity based on directly communicating hemes at a novel subunit interface. J. Biol. Chem 1989, 264 (35), 21052–21061. [PubMed] [Google Scholar]
- 39.Condon PJ; Royer WE Jr., Crystal structure of oxygenated Scapharca dimeric hemoglobin at 1.7-A resolution. J. Biol. Chem 1994, 269 (41), 25259–25267. [DOI] [PubMed] [Google Scholar]
- 40.Royer WE Jr., High-resolution crystallographic analysis of a co-operative dimeric hemoglobin. J. Mol. Biol 1994, 235 (2), 657–681. [DOI] [PubMed] [Google Scholar]
- 41.Royer WE Jr.; Heard KS; Harrington DJ; Chiancone E, The 2.0 A crystal structure of Scapharca tetrameric hemoglobin: cooperative dimers within an allosteric tetramer. J. Mol. Biol 1995, 253 (1), 168–186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


