Abstract
Recent advances in 3D bioprinting have transformed the tissue engineering landscape by enabling the controlled placement of cells, biomaterials, and bioactive agents for the biofabrication of living tissues and organs. However, the application of 3D bioprinting is limited by the availability of cytocompatible and printable biomaterials that recapitulate properties of native tissues. Here, we developed an integrated 3D projection bioprinting and orthogonal photoconjugation platform for the precision tissue engineering of tailored microenvironments. By using a photoreactive thiol-ene gelatin bioink, soft hydrogels can be bioprinted into complex geometries and photopatterned with bioactive moieties in a rapid and scalable manner via digital light projection (DLP) technology. This enables localized modulation of biophysical properties such as stiffness and microarchitecture as well as precise control over spatial distribution and concentration of immobilized functional groups. As such, well-defined properties can be directly incorporated using a single platform to produce desired tissue-specific functions within bioprinted constructs. We demonstrated high viability of encapsulated endothelial cells and human cardiomyocytes using our dual process and fabricated tissue constructs functionalized with VEGF peptide mimics to induce guided endothelial cell growth for programmable vascularization. This work represents a pivotal step in engineering multifunctional constructs with unprecedented control, precision, and versatility for the rational design of biomimetic tissues.
Graphical Abstract
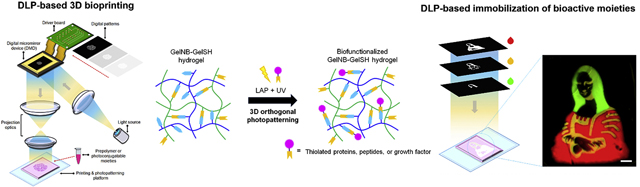
Introduction
Cells in vivo constantly experience dynamic changes to their surrounding microenvironment, thus an appropriate understanding of mechanisms that govern cellular processes including proliferation, differentiation, and maturation is critical to building functional tissue constructs [1]. Recent advances in 3D bioprinting technologies have emerged at the forefront amid biofabrication and tissue engineering by offering a multifaceted approach to enable control over mechanical properties, biological composition, and microarchitecture for investigating various cell-microenvironment interactions in response to well-defined biochemical and biophysical stimuli [2,3]. This has been accomplished by various bioprinting modalities including extrusion-based, inkjet-based, and light-based printers to produce heterogeneous cell-laden constructs including liver, cardiac, kidney, and cancer tissue mimics [4–7]. More recently the emergence of digital-light projection (DLP)-based 3D printing technologies have greatly enhanced the biofabrication of photopolymerizable hydrogel biomaterials such as gelatin methacrylate (GelMA), poly(ethylene glycol) diacrylate (PEGDA), glycidyl methacrylatehyaluronic acid (GM-HA), and thiol-ene gelatin for the production of complex tissue and organ substitutes [4,8–10]. In particular, DLP-based 3D printing systems enable higher spatial resolution at the micron scale (i.e. 3–5 μm) and more rapid printing times on the order of seconds due to the contactless plane-by-plane fabrication regime compared to traditional raster-like modalities [11]. Despite these advances in biofabrication, the limited selection of biocompatible and 3D printable biomaterials available consequently limits the functionality of 3D printed tissues as many still rely on utilizing conventional bioinks such as gelatin, collagen, alginate, hyaluronic acid, and polyethylene glycol (PEG) [12].
While numerous novel bioink biomaterials have been developed, it is challenging to satisfy the many interdependent conditions that will lead to ideal bioprinting as well as structural, compositional, and biological outcomes for specific tissue microenvironments [12]. One bottom-up approach to overcome these challenges is the concept of tailored post-print modifications through the conjugation of biologically active molecules to achieve desired functionality in printed constructs produced from common bioink derivatives [13]. For instance, using polydopamine chemistry, 3D printed scaffolds were immobilized with growth factors that have been shown to improve in vitro osteogenic differentiation by grafting human bone morphogenic protein-2 (rhBMP-2) on polycaprolactone (PCL) scaffolds [14]. In another study, 3D-printed poly(lactic acid) (PLA) scaffolds were chemically conjugated with calcium-deficient hydroxyapatite using polyethyleneimine to enhance mineral deposition and osteogenesis of human mesenchymal stem cells for bone tissue regeneration [15]. However, these chemistries are not compatible with bioprinted cellularized scaffolds and are restricted to only surface modification with little to no control over spatial distribution and concentration within the 3D constructs. Recently, the thiol-ene radical step growth reaction has gained attention for applications in tissue engineering and regenerative medicine as it combines the advantages of both photoinitiated systems and high selectivity under biocompatible reaction conditions [16,17]. Thiol-ene click chemistry involves the orthogonal coupling between macromers containing sulfur moieties and alkene groups to form a homogenous crosslinked network [16]. Due to the versatility and spatiotemporal control of this cytocompatible photoclick chemistry, its use as a polymerization and bioconjugation technique carries unique advantages to expand the scope of bioactive materials for creating functional biomimetic cell microenvironments. In particular, current biofunctionalization methods of thiol-ene hydrogels involving the use of traditional photolithography and multi-photon stereolithography with a nano to micron scale resolution for photoconjugation are limited in speed, flexibility, photopatterning volume, and throughput [18,19]. While several studies have demonstrated the feasibility of thiol-ene systems for both hydrogel biofabrication and photoconjugation [18,20–23], these methods are inadequate to implement as a scalable approach to build complex biomimetic multicomponent tissues.
Considering the orthogonality and photoreactive nature of thiol-ene click chemistry, we developed the first integrated 3D bioprinting and orthogonal bioconjugation platform that utilizes a rapid DLP-based approach to fabricate biomimetic tissues with programmable functionality at scale. Our system leverages the high resolution and spatiotemporal control offered by DLP-based 3D bioprinting technology coupled with a natural thiol-ene biopolymer system as a method to engineer tailorable 3D tissue environments. Importantly, the dual ability to rapidly print pure thiol-ene gelatin hydrogels and selectively photoconjugate thiolated proteins or cysteine-modified peptides enables a simple approach to directly modulate multidimensional properties within a single platform. Moreover, the architecture and mechanical properties of the hydrogel in addition to the spatial distribution as well as concentration of photoconjugated bioactive moieties can be locally tuned to form diverse soft functionalized matrices for precision tissue engineering.
2. Materials and Methods
Material syntheses
GelNB Synthesis
GelNB was synthesized using a previously described procedure [24]. Briefly, Type A gelatin from porcine skin (gel strength 300 g Bloom, Cat. #G2500, Sigma-Aldrich) was dissolved at 0.1 g/mL in 1X DPBS (Cat. #14190144, Gibco) in a round-bottom flask. Carbic anhydride (Cat. #247634–5g, Sigma-Aldrich) was added to the mixture to a concentration of 0.2 g/mL. After bringing the mixture to pH 8.0, the solution was reacted for 70 h at 50°C under constant stirring. An equal volume of 5X DPBS diluted from 10X DPBS (Cat. #SH302580, Thermo Fisher Scientific) heated to 37°C was then added to the mixture and stirred for 15 min to quench the reaction. The solution was then transferred to 12–14 kDa dialysis tubing (Cat. #132706, Spectrum Labs) and dialyzed against MilliQ water for one week with three dialysis changes per day. After dialysis, the solution was lyophilized for three days and then stored at −80°C until further use.
GelSH Synthesis
GelSH was synthesized by adapting previously established protocols [25]. Type A gelatin (Cat. #G2500, Sigma-Aldrich) was dissolved at 0.01 g/mL in 1X DPBS (Cat. #14190144, Gibco) in a round-bottom flask at 50°C prior to lowering the temperature to 30°C. Next, Traut’s reagent (Cat. #26101, Thermo Fisher Scientific) was added to the mixture to a final concentration of 0.4 mg/mL. The solution was then reacted for 15 h under constant stirring, then transferred to 12–14 kDa dialysis tubing (Cat. #132706, Spectrum Labs) and dialyzed for two days with three dialysis changes per day. The solution was dialyzed against 5 mM HCl on the first day and then against 1 mM HCl on the second day. After dialysis, the solution was lyophilized for three days and stored at −80°C until further use.
LAP Synthesis
LAP was synthesized using previously described procedures [26]. Briefly, 3.2 g of 2,4,6-trimethylbenzoyl chloride (Cat. #C965Y83, Thomas Scientific) was added dropwise to 3 g of dimethyl phenylphosphonite (Cat. #149470, Sigma-Aldrich) in a flask and allowed to react for 18 hours at room temperature under argon gas. The next day, the mixture was heated in a water bath to 50°C. 6.1 g lithium bromide (Cat. #213225, Sigma-Aldrich) dissolved in 100 mL of 2-butanone (Cat. #M209–4, Thermo Fisher Scientific) was added dropwise to the heated mixture via a syringe and reacted for 10 minutes, still at 50°C. During this time, the reaction formed a white precipitate within the flask, and the flask was then cooled for 4 hours at room temperature. The precipitate was then washed with 400 mL of 2-butanone in a filtration setup and allowed to dry overnight. Afterwards, the precipitate was collected and stored at room temperature under argon until further use.
Determining Degree of Substitution
GelNB and GelSH substitution was characterized using 1H NMR (Supplementary Figure S1–3). For 1H NMR, 15–25 mg of sample was dissolved in 1 mL of D2O (Cat. #151882-10X1ML, Sigma-Aldrich) at 37°C. The sample was then analyzed with a 500 MHz Jeol NMR spectrometer against a gelatin standard. The degree of substitution was calculated to be approximately 44% and 22% for GelNB and GelSH, respectively, using previously outlined methods [27]. Briefly, the integration of the phenylalanine peaks at 7.5–6.9 ppm was used as an internal control, and the integration of the ε-amino groups of lysine/hydroxylysine at 2.8 ppm were compared between the standard and functionalized gelatins to determine the degree of substitution.
Prepolymer bioink preparation and 3D printing of GelNB-GelSH constructs
Stock solutions of 20% (w/v) GelNB, 15% (w/v) GelSH, and 4% (w/v) LAP were prepared by dissolving in 1X DPBS. To ensure proper dissolution, the GelNB and LAP solutions were warmed to 37°C, and the GelSH solution was warmed to 60°C. All solutions were stored at 4°C and heated to their respective temperatures prior to mixing. Based on the degree of substitution of thiol and norbornene groups for GelSH and GelNB, respectively, the solutions were mixed at a 1:1 molar ratio to maintain stoichiometric equivalence unless otherwise stated. All final prepolymer solution formulations were kept warm at 37°C prior to transferring to the 3D bioprinter.
A DLP-based 3D bioprinter was used to fabricate microscale acellular and cellularized GelNB-GelSH constructs as shown in Figure 1C. Briefly, the major components are as follows: (i) a 365 nm UV light LED source, (ii) a digital micromirror array device (DMD) composed of approximately 2 million micromirrors for modulating the projected optical patterns to control for selective photopolymerization; (iii) projection optics to focus the optical patterns from the DMD chip onto the fabrication plane; (iv) a motorized stage; and (v) a computer control unit. Digital patterns were designed in Adobe Photoshop. The printing chamber platform consisted of a polydimethylsiloxane (PDMS) base and methacrylated coverslip separated by two PDMS spacers, all mounted on a glass slide. Prepolymer solution was then carefully pipetted between the spacers, placed on the motorized stage, and subsequently exposed with DMD-projected light from above to produce the differently patterned constructs. See Supplementary Figure S4 for a brief schematic.
Figure 1.

Fabrication of biofunctionalized GelNB-GelSH hydrogels using an integrated DLP-based 3D projection bioprinting and orthogonal bioconjugation platform. (A) Schematic of photo-induced thiol-ene click chemistry reaction to produce GelNB-GelSH hydrogels and subsequent biofunctionalization. (B) Schematic of our DLP-based 3D bioprinter and orthogonal bioconjugation platform for the rapid fabrication of GelNB-GelSH hydrogels and photopatterning of bioactive moieties. (C) Rapid orthogonal photopatterning to immobilize multiple bioactive moieties within GelNB-GelSH hydrogels via digital light projection to form small to large volume complex designs. This is accomplished by sequential photopatterning onto the same hydrogel by using select digital patterns for each bioactive moiety. Scale bars = 250 μm. (D) Digital pattern of a retinal capillary network along with their corresponding brightfield images of printed GelNB-GelSH hydrogel showing high resolution features. Scale bars = 500 μm (i) and 250 μm (ii). (E) 3D printed GelNB-GelSH kidney-shaped constructs. The constructs were stained with dye post-printing for visualization. Scale bar = 5 mm.
To print larger structures such as the kidney-shaped constructs, a continuous 3D printer was used as previously described [28]. Briefly, the setup consists of a z-direction motorized glass build platform and a PDMS coated petri dish serving as the prepolymer reservoir where light is projected from below via the DMD chip. The 3D CAD model was sliced using a MATLAB script to generate a series of digital patterns used to sequentially photopolymerize the prepolymer solution in synchrony with the upwards movement of the build platform for continuous printing. Note that a high-concentration stoichiometric formulation of 3.33% (w/v) GelNB + 6.67% (w/v) GelSH + 1% (w/v) LAP was chosen to ensure structural integrity of the printed figurine.
Mechanical testing and swelling measurements
For physical characterization, GelNB-GelSH hydrogels composed of 1:1, 1:2, and 2:1 thiol:ene molar ratios were prepared to decouple the effects of macromer composition. Meanwhile, GelNB-GelSH hydrogels composed of 5% (w/v), 7.5% (w/v), and 15% (w/v) total gelatin content all with a stoichiometric 1:1 thiol:ene molar ratio were fabricated to determine the effects of overall macromer concentration. Optimized light exposure time was determined for each formulation to ensure complete coupling of norbornene to thiol groups by measuring the time required to reach a plateau of the average tissue deformation ratio (ATDR) which corresponds directly to the maximum stiffness (Supplementary Figure 6 and Supplementary Table S3). The tensile and compressive moduli of the GelNB-GelSH hydrogels were measured using the CellScale MicroSquisher (Waterloo, Canada) microscale mechanical testing apparatus. All samples were immersed in 1X DPBS at 37°C overnight prior to analysis. To measure the tensile modulus using the CellScale MicroSquisher, dog-bone-shaped specimens with overall dimensions of 6 mm (L) × 3 mm (W) with a 2 mm thickness and narrow section dimensions of 2 mm (L) × 1 mm (W) were used (see Supplementary Table S2 for printing parameters). Samples were first loaded and held in place by carefully puncturing it at 8 equidistant points (four at the top and four at the bottom) via two parallel-aligned 4-prong forks (Supplementary Figure S7 and Movie S1). Each sample was then pulled to 10% strain at a rate of 10 μm/s, held for 5 s, and the allowed to recover at a rate of 30 μm/s. The combination of these three steps constituted one repetition and three repetitions were performed per sample (n = 5). To measure the compressive modulus, a cantilever adhered to a platen was used to compress the sample to 10% strain at a rate of 3.33 μm/s, held for 2 s, followed by release at a rate of 10 μm/s. These steps were repeated three times to remove the effects of hysteresis and data from the third compression was used to calculate the modulus of the sample (n = 7). The resulting stress-strain curves for the tensile and compressive moduli measurements were then calculated by measuring the slope of each curve in its elastic region using a MATLAB code.
The stability of the GelNB-GelSH constructs were determined by measuring the swelling ratio (n = 4–6) (Supplementary Table S3). Briefly, samples were immersed in 1X DPBS at 37°C and imaged using a Leica DMI 6000-B microscope at 24 h, 72 h and 7 days post printing to obtain the hydrated cross-sectional area (Awet). Afterwards, samples were then washed three times with MilliQ water, dried in a 37°C oven for three days, and reimaged to measure the cross-sectional dry area (Adry). The swelling ratio for each time point was calculated by normalizing the average hydrated cross-sectional area to the average dry cross-sectional area (Awet/Adry). The average cross-sectional area of each construct was measured using ImageJ software.
Scanning electron microscopy (SEM)
GelNB-GelSH and GelMA hydrogel samples (printing parameters in Supplementary Table S4) were gradually dehydrated by first immersing in 20% ethanol overnight at 4°C. The next day, samples were subject to gradient dehydration in 30%, 40%, 50%, 60%, 70%, 80%, 90%, and 95% ethanol followed by 100% ethanol repeated three times total. Each soaking step was performed for 20 min at 4°C. The samples were then dried using a Tousimis AutoSamdri®−815A critical point dryer. Samples were then sputter coated with iridium for 7 s and imaged using a Zeiss Sigma 500 scanning electron microscope.
Cell culture maintenance and 3D bioprinting of cellularized constructs
Human umbilical vein endothelial cells (HUVECs) (Cat. #C2519A, Lonza) were maintained in Endothelial Cell Growth Medium-2 (EGM-2) (Cat. #CC-3162, Lonza) and passaged every 3–4 days at 80% confluence. C3H/10T1/2 (10T1/2) fibroblast cells (Cat. #CCL-226, ATCC) were maintained in Dulbecco Modified Eagle Medium (DMEM) (Cat. #11995–065, Gibco) supplemented with 10% (v/v) fetal bovine serum (Cat. #10082147, Gibco) and 1% (v/v) penicillin-streptomycin (Cat. #11548876, Gibco), and passaged every 3–4 days at 80–90% confluence. Human iPSCs (hiPSCs) were produced using previously established methods [4,29] and differentiated into iPSC-derived cardiomyocytes (iPSC-CMs) according to previously established protocols [30].
To prepare the cells for 3D bioprinting, HUVECs and 10T1/2 cells were collected by treating with 0.05% trypsin-EDTA and 0.25% trypsin-EDTA, respectively, then aliquoted into 1.5 mL eppendorf tubes and centrifuged at 300xg for 5 min with the supernatant removed. Similarly, iPSC-CMs were collected by treating with 0.25% trypsin-EDTA for 5 min, aliquoted into 1.5 mL eppendorf tubes, and pelleted at 300 xg for 3 min with the supernatant removed. All pelleted cells were placed on ice and used within 2 h. For cell viability assessment, 3.33% (w/v) GelNB + 3.33% (w/v) GelSH + 0.4% (w/v) LAP prepolymer prewarmed to 37°C was added to each eppendorf to form a cell suspension of 5 million cells/mL comprised of mixture of HUVEC:10T1/2 cells at a ratio of 50:1. For iPSC-CMs, cells were encapsulated at a concentration of 40 million cells/mL. The prepolymer-cell bioink mixture was then carefully pipetted into the printing chamber of the DLP-based 3D printer and UV irradiated to create a 3 mm (L) × 3 mm (W) × 250 μm (H) cell-laden GelNB-GelSH construct for cell viability studies (see Supplementary Table S5). The HUVEC:10T1/2 and iPSC-CM cellularized GelNB-GelSH constructs were then immediately placed into a 24-well plate with 750 μL of their respective medias and media was exchanged every other day.
Cell viability and proliferation assessment
To assess HUVEC:10T1/2 and iPSC-CM cell viability, Live/Dead™ Viability/Cytotoxicity assay (Cat. #L3224, Thermo Fisher Scientific) and CellTiter Glo® 3D cell viability assay (Cat. #G9681, Promega) were performed at 24 h, 72 h, and 7 day time points. Briefly, triplicate samples of HUVEC:10T1/2 or iPSC-CM encapsulated GelNB-GelSH constructs were washed once with 1X DPBS and incubated in Live/Dead™ solution comprised of 2 uM calcein AM and 4 uM ethidium homodimer-1 in 1X DPBS for 30 min at 37°C. Following incubation, the stained samples were immediately imaged using a Leica DMI 6000-B microscope. Percentage viability was determined using ImageJ analysis and calculated by dividing the live cell count to the total cell count. Triplicate samples were also collected for each of the HUVEC:10T1/2 and iPSC-CM GelNB-GelSH constructs for quantification of the total ATP content using the CellTiter Glo® 3D cell viability assay. In general, samples were transferred to a 24-well plate and equal volumes of culture media and CellTiter-Glo® 3D Reagent was added. Samples were then shaken at 160 rpm at room temperature for 1 h to ensure full release of ATP content from each cell-laden construct. Next, 200 μL of supernatant was collected from each sample and transferred to a white opaque-walled 96-well plate and luminescence was measured with a Tecan Infinite® M200 PRO microplate reader. ATP content from each sample was calculated based on an ATP standard curved generated using ATP disodium salt (Cat. #P1132, Promega) according to the manufacturer’s instructions.
Preparation of fluorescently labeled ovalbumin and QK peptide conjugates
Ovalbumin Alexa Fluor 647 (Cat. #O34784, Thermo Fisher Scientific), ovalbumin Alex Fluor 555 (Cat# O34782, Thermo Fisher Scientific) and ovalbumin Alexa Fluor 488 (Cat. #O34781, Thermo Fisher Scientific) were used to demonstrate passive conjugation or the active photopatterning capability with our DLP-based 3D printer. NHS-PEG (3.4 kDa)-NHS (Cat. #PG2-NS-3k, Nanocs) and NHS-PEG (3.4 kDa)-thiol (Cat. #PG2-NSTH-3k, Nanocs) served as intermediate linkers for ovalbumin to the respective available ε-amino or norbornene groups in the GelNB-GelSH construct. Ovalbumin functionalization and photopatterning procedures were based on prior literature [31,32], and all solutions were prepared in 1X DPBS. Briefly, fluorescently labeled Alexa Fluor (AF) 488, 555 or 647 ovalbumin was first dissolved to create a 4 mg/mL stock solution. To conjugate the ovalbumin to the intermediate linker, we added 10 molar equivalents of the NHS-PEG linker, either NHS-PEG (3.4 kDa)-NHS or NHS-PEG (3.4 kDa)-thiol, to the ovalbumin stock solution and allowed to react at room temperature for 1 h on a rotator plate. The fluorescently labeled ovalbumin conjugates were stored at 4°C until further use.
The proangiogenic VEGF peptide mimic QK with a Cys-PEG6 modification on the N-terminus (Cys-PEG6-KLTWQELYQLKYKGI-NH2) was purchased from GenScript®. Briefly, the QK peptide was dissolved in 50 μL ultrapure water to yield a concentration of 1 mg/mL and mixed with an equal volume of 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (Cat. #BP300–100, Thermo Fisher Scientific) containing 800 mM NaCl and 20% (w/v) 1,2-propanediol. Next, 50 μL of 44 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) (Cat. #E7750–100MG, Sigma-Aldrich) and 48 mM 50 μL of N-hydroxysulfosuccinimide (sulfo-NHS) (Cat. #56485–250MG, Sigma-Aldrich) solutions were added to the QK peptide buffer solution and allowed to react under agitation for 15 min at room temperature. Meanwhile, 10 μL of 10 mg/mL Alexa Fluor™ 488 hydrazide sodium salt (Cat. #A10436, Thermo Fisher Scientific) dissolved in DMSO was mixed with 400 μL of 100 mM sodium acetate buffer pH 5.5 (Cat. # AM9740, Thermo Fisher Scientific) and added to the prepared QK peptide solution. The mixture was then allowed to react for 75 min at room temperature under constant agitation and the fluorescently labeled QK peptide was concentrated using a Pierce™ Protein Concentrator, 10 MWCO, 0.5 mL (Cat. # 88513, Thermo Fisher Scientific) by centrifuging the sample at 12,000 × g for 5 min. The supernatant containing the fluorescently labeled AF488-QK peptide was then adjusted to 1 mg/mL with 1X DPBS and stored at 4°C until further use.
Photopatterning of ovalbumin and QK peptide into GelNB-GelSH hydrogel
Active photopatterning of ovalbumin was performed by soaking printed GelNB-GelSH constructs in 1 mg/mL of AF488-ovalbumin-PEG-thiol solution containing 0.2% (w/v) LAP for 1 h. Digital patterns were then projected onto the GelNB-GelSH construct via UV irradiation using our DLP-based printer. Immediately after exposure the constructs were washed with 1X DPBS overnight. For passive conjugation of ovalbumin, GelNB-GelSH constructs were soaked in 1 g/mL of AF647-ovalbumin-PEG-NHS and allowed to react for 1 h at room temperature followed by washing in 1X DPBS overnight.
Similarly, QK peptide was actively photopatterned by soaking the GelNB-GelSH constructs in 1 mg/mL of QK peptide solution supplemented with 0.2% LAP (w/v) for 1 h at 37 °C, followed by selective UV irradiation with our DLP-based 3D printing platform and rinsed overnight in 1X PBS. Note, this same procedure was performed for both fluorescently labeled AF488-QK peptide for visualization and quantification of photoconjugation or non-fluorescently labeled QK peptide for the endothelial cell studies. Imaging was performed using a Leica DMI 6000-B fluorescent microscope and an Olympus FV1000 confocal microscope with 3D reconstructions produced using ImageJ software.
In our active photopatterning system, covalent conjugation can only be achieved upon UV irradiation in the presence of both LAP and the NHS-PEG-thiol linker (Supplementary Figure S8). All active photopatterning and passive conjugation experiments were conducted using 3.33% (w/v) GelNB + 3.33% (w/v) GelSH + 0.4% (w/v) LAP to ensure excess norbornene groups were available for conjugation. Meanwhile, the GelNB concentration was increased to 5% (w/v) and 6.66% (w/v) to demonstrate the effect of increased norbornene content on the amount of ovalbumin photoconjugation as shown in Figure 3C. See Supplementary Table S6, S7 and S8 for more printing information.
Figure 3.

Controlled presentation of immobilized proteins within GelNB-GelSH hydrogels using DLP-based photopatterning. (A) Dual mechanisms of protein conjugation showing active UV photopatterning with AF488-ovalbumin-PEG-thiol to excess norbornene groups, followed by passive spontaneous conjugation without UV irradiation of AF647-ovalbumin-PEG-NHS to available primary amine of GelNB or GelSH. Scale bars = 500 μm. (B) Changing photopatterning exposure time to vary the degree of ovalbumin conjugation to norbornene groups within the GelNB-GelSH hydrogel wherein: (i) shows AF488-ovalbumin-PEG-thiol at 15 s (top) and 30 s (bottom) exposure and (ii) shows a photopatterned overlap of AF647-ovalbumin-PEG-thiol into the central region. Scale bars = 500 μm. (C) Varying the degree of ovalbumin conjugation by changing the norbornene availability in the GelNB-GelSH hydrogel wherein greater conjugation is observed in regions with higher norbornene content. Scale bars = 500 μm. Dotted white lines depict border of the printed GelNB-GelSH hydrogel slab. (D) Plot of fluorescence intensity corresponding to regions (a-c) in the GelNB-GelSH hydrogel as indicated by the yellow line. (E) Confocal 3D reconstruction of AF488-ovalbumin-PEG-thiol conjugated to the GelNB-GelSH hydrogel displaying full depth of photopatterning throughout the 250 μm tall construct. Inset shows confocal top view image of the capillary photopattern. Scale bars = 250 μm and 200 um (inset). (F) Set of photopatterned designs used to form the Mona Lisa using (i) AF488-ovalbumin-PEG-thiol (green), (ii) AF555-ovalbumin-PEG-thiol (yellow), and (iii) AF647-ovalbumin-PEG-thiol (red). Scale bar = 250 μm.
Quantification of photoconjugation in GelNB-GelSH constructs
To qualitatively measure the relative amount of photoconjugation of ovalbumin or QK peptide within the GelNB-GelSH constructs, printed samples were photopatterned using AF488-labeled ovalbumin or QK peptide according to the above described methods. Upon rinsing the samples in 1X PBS overnight at 37 °C, images were taken with a Leica DMI 6000-B fluorescent microscope. Measurements of the relative fluorescent intensity were determined using ImageJ software (n=5). The plot profile function in ImageJ was also used to measure the change in fluorescent intensity across the capillary photopatterned slab shown in Figure 3C.
To quantify the concentration of photoconjugated ovalbumin or QK peptide within the GelNB-GelSH constructs, triplicate samples of 2 mm (L) × 2 mm (W) × 250 μm (H) slabs were all photopatterned with AF488-labeled ovalbumin or QK peptide at 20 s exposure and placed in a 48-well plate. Using adapted protocols [33], 200 μL of prewarmed 0.3 mg/mL collagenase (Cat. # C9891–25MG, Sigma-Aldrich) in 1X PBS was added to each well and digested at 37°C for 1 h. Non-photopatterned GelNB-GelSH slabs were also digested in the same manner and served as background controls. Meanwhile, a 7-point standard curve was prepared using serially diluted AF488-labeled ovalbumin or QK peptide 1 mg/mL stock solution that was also exposed for 20 s to account for minor photobleaching of the samples due to brief UV exposure. Finally, 200 μL of the digested sample supernatants and standards were loaded into a flat bottom black 96-well plate and read with a Tecan Infinite® M200 PRO microplate reader at an excitation of 490 nm and emission of 525 nm. Concentration was calculated based on the slope of the standard curve (n=5).
Assessment of endothelial cell behavior to QK peptide
To assess the chemotactic response of photopatterned QK peptide on endothelial cell migration within GelNB-GelSH hydrogels, a 3D printed daisy flower construct with HUVEC:10T1/2 cells (50:1) encapsulated at 20 million cells/mL in the central floral disc was fabricated using our integrated DLP-based printer. To visualize endothelial cell migration, HUVECs and 10T1/2 cells were stained with CellTracker™ Green CMFDA Dye (Cat. #C2925, Invitrogen). Cells were rinsed once with 1XPBS followed by incubation for 45 min at 37°C in serum-free M199 basal media (Cat. #11150059, Thermo Fisher Scientific) for HUVECs and DMEM basal media (Cat. # 11995–065, Gibco) for 10T1/2 cells each supplemented with 5 μM of CellTracker™ Green CMFDA Dye. After incubation, the cells were rinsed with 1X PBS to remove residual dye and their respective culture medium was replaced and allowed to recover overnight prior to encapsulation. The following four groups were tested: 1) full petal photopatterned +QK peptide, 2) single petal photopatterned +QK peptide, 3) full petal photopatterned −QK peptide control, and 4) non-photopatterned −QK peptide control. Note that the QK peptide photopatterned daisy constructs were fabricated as illustrated in Supplementary Figure S9. Briefly, full daisy or single petal regions were first printed and then incubated for 1 h at 37°C in 1 mg/mL QK peptide in ultrapure water prior to photopatterning with our DLP-based platform and subsequent endothelial cell encapsulation in the central region. The samples were cultured in EGM-2 media with media changes every other day and at time points of 0, 24, 48, and 72 h post-printing the daisy constructs were imaged using a Leica DMI 6000-B fluorescent microscope to visualize endothelial cell migration from the central region to the surrounding petals. ImageJ cell counter was used to quantify the average cell counts per petal for each group over time (n = 6). All QK peptide photopatterning cell experiments were conducted using 3.33% (w/v) GelNB + 3.33% (w/v) GelSH + 0.4% (w/v) LAP to ensure excess norbornene groups were available for conjugation as well as having appropriate mechanical properties to support cell viability based on the prior the cell viability experiments. See Supplementary Table S7 for further printing information.
Guided vascularization of a developing cardiac tissue mimic
To examine directed endothelial cell migration within a cellularized tissue construct, a developing cardiac tissue mimic was fabricated as a proof-of-concept to demonstrate the efficacy of photoconjugated QK-peptide to guide vascular growth as shown in Figure 4G. Briefly, as illustrated in Supplementary Figure S10, 50 million cells/mL of iPSC-CMs were first 3D printed to create the cardiac tissue portion followed by incubation for 1 h at 37°C in 1 mg/mL QK peptide in M199 basal media, and subsequently photopatterned with a vascular digital pattern. After rinsing the photopatterned cardiac tissue construct overnight in 1XPBS, CellTracker Green™ stained HUVEC:10T1/2 cells (50:1, 20 million cells/mL) representing the endothelial cell population of the sinus venosus was printed into the complementary region. The samples were cultured in EGM-2 media with media changes every other day and at time points of 0, 24, 48, and 72 h post-printing the daisy constructs were imaged using a Leica DMI 6000-B fluorescent microscope to visualize endothelial cell migration from the sinus venosus to the cardiac tissue region (n = 3). See Supplementary Table S8 for further printing information.
Figure. 4.
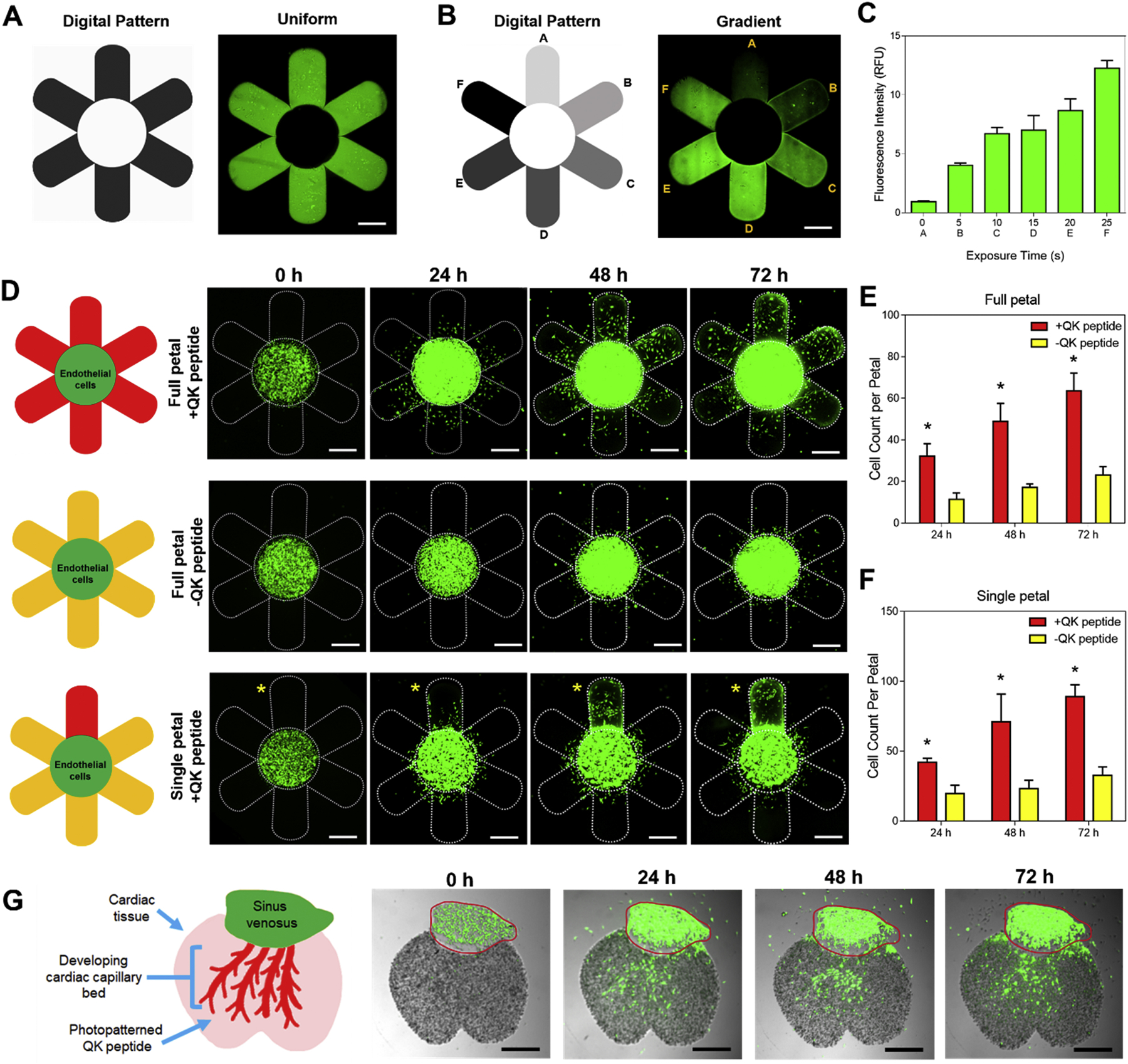
Photopatterning of the VEGF mimic QK peptide for guidance of endothelial migration, proliferation, and directionality. Representative fluorescent images of a printed GelNB-GelSH daisy construct showing (A) uniform and (B) gradient (i.e. petals: A = 0 s, B = 5 s, C = 15 s, D = 20 s, E = 25 s, F = 30 s exposure) photopatterning of AF488-QK peptide. Scale bars = 500 μm. (C) Plot of relative fluorescence intensity corresponding to each petal in the gradient photopatterned daisy flower (n = 5). (D) Representative fluorescent images of 3D bioprinted GelNB-GelSH daisy constructs showing the migratory response of encapsulated CellTracker Green™ stained HUVEC:10T1/2 (50:1) cells from the central floral disc to the petal regions over 72 h. Yellow asterisk denotes the single photopatterned +QK peptide petal. Scale bars = 500 μm. (E) Average cell counts per petal in the full +QK peptide petal design compared to the full non-photopatterned -QK peptide petal control. (F) Average cell counts per petal in the single photopatterned +QK peptide petal compared to the surrounding non-photopatterned -QK peptide petal controls. * = significance between groups (p<0.05, n = 6). (G) 3D printed developing cardiac tissue mimic composed of iPSC-CMs showing CellTracker Green™ stained HUVEC:10T1/2 (50:1) cells growing from the sinus venosus to the capillary bed region in response to photopatterned +QK peptide over 72 h. Red outline denotes printed region of the sinus venosus (n = 3). Scale bar = 500 μm.
Statistical analysis
All data are expressed as a mean ± standard deviation (SD). Statistical analyses were performed with GraphPad Prism version 6.0 Software by one-way ANOVA with Tukey’s post-hoc comparison. Significant differences were considered when p<0.05.
3. Results and Discussion
3.1. Photoclickable thiol-ene gelatin hydrogel synthesis and printability
In this study, we synthesized a pure gelatin thiol-ene prepolymer composed of two precursors: norbornene-functionalized gelatin (GelNB) and thiol-functionalized gelatin (GelSH) (Figure 1A). Norbornene was chosen as the alkene functional group due to its low homopolymerization and strained bicyclic structure, which allows for a more rapid reaction than electron‐deficient enes [16]. The photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) was added to the GelNB-GelSH prepolymer to enable photopolymerization using our DLP-based 3D projection bioprinter (Figure 1B) along with subsequent orthogonal photopatterning to form diverse biofunctionalized tissue matrices at scale (Figure 1C). Due to the combined significant ring strain relief by the addition of a thiyl radical to the double bond in norbornene and rapid abstraction of hydrogen from the thiol, the overall step-growth polymerization kinetics proceed very quickly [17]. As such, we demonstrated rapid 3D printing, on the order of seconds, of complex GelNB-GelSH hydrogels in a retinal capillary design (Figure 1D). With this printing system we were able to achieve constructs possessing fine micron scale resolution features as small as 10–30 μm with visible pixel detail on the edges. Given the inherently soft mechanical properties of pure gelatin-based biomaterials, the ability to fabricate larger 3D printed structures can be challenging and typically requires the addition of synthetic biomaterials to provide structural integrity. Since our GelNB-GelSH hydrogels rapidly form a homogenous gel network under photopolymerization, we were able to readily print centimeter scale structures by employing a continuous, DLP-based 3D printing approach. In this case, a pair of kidney-shaped constructs (Figure 1E, Supplementary Figure S11) each measuring approximately 1 cm (L) × 0.7 cm (W) × 0.4 cm (H) was fabricated without the need for additional photo-absorbers typically required to control for excess free radical initiation or propagation that negatively effects the resolution, consistent with other works 3D printing thiol-ene gelatin [8]. General features on the kidney shape were well-resolved with smooth contours, no physical collapse, and the structure robustly remained intact upon manual manipulation.
3.2. Modulation of physical properties of GelNB-GelSH hydrogels
Given the printability of our GelNB-GelSH material, we next investigated the impact of different molar ratios of norbornene to thiol groups to modulate the physical properties of the printed hydrogels. Unlike chain-growth polymerization systems, such as gelatin methacrylate (GelMA), in which increased gel network density is often associated with higher macromer concentration [34,35], the orthogonal nature of our thiol-ene GelNB-GelSH prepolymer allows us to decouple the effects of available norbornene and thiol groups to alter the mechanical properties. Based on the measured degree of substitution for GelNB (~44%) and GelSH (~22%) (Supplementary Figures S1–S3), a set of GelNB-GelSH hydrogels of 1:1, 1:2, and 2:1 molar ratio of norbornene to thiol functional groups was fabricated. Relative to hydrogels prepared with 1:1 GelNB:GelSH, a change to 1:2 GelNB:GelSH resulted in a significant increase in tensile modulus as well as compressive modulus from 0.5 ± 0.3 kPa to 1.3 ± 0.4 kPa and 1.1 ± 0.3 kPa to 4.9 ± 1.3 kPa, respectively (Figure 2A,B). This can be attributed to the excess thiol groups present in the 1:2 GelNB:GelSH formulation resulting in a denser network due to both the thiol-ene orthogonal coupling as well as the formation of disulfide bonds from the excess thiol moieties [36]. Notably, these mechanical properties are much softer than other thiol-ene gels, such as those developed by Bertlein et al. [8], which uses a very short cross linker (DTT) for provide its thiol groups. This means our material is more applicable towards soft tissues, as opposed to stiffer models, such as those required by cancer models. While the swelling ratio (i.e. ~15% and ~17% for 1:1 and 1:2 GelNB:GelSH, respectively) was not significantly different between the hydrogels (Figure 2C), closer inspection of the ultrastructure via scanning electron microscopy (SEM) revealed a denser crosslinked network in the 1:2 GelNB:GelSH formulation (Figure 2D). In contrast, hydrogels prepared using a 2:1 GelNB:GelSH formulation with an excess of available norbornene groups showed a significantly lower tensile and compressive modulus of 0.8 ± 0.4 kPa and 1.7 ± 0.4 kPa, respectively, compared to the 1:2 GelNB:GelSH case of excess thiol groups (Figure 2A,B). Since fewer crosslinks are formed in the 2:1 GelNB:GelSH hydrogel, a looser gel network is created which is reflected upon the significantly higher swelling ratio observed (Figure 2C) along with a more porous ultrastructure (Figure 2D). We further demonstrated that by fixing the molar ratio to 1:1 GelNB:GelSH, varying the total macromer concentration from 5% (w/v) to 15% (w/v) was directly proportional to an increase in tensile (i.e. ~0.5 to ~5 kPa) and compressive moduli (i.e. ~1 to ~11 kPa) and decreased swelling ratio (i.e. ~15% to ~9%) as expected due to greater crosslinking density (Figure 2A–C). In general, the compressive moduli of the printed GelNB-GelSH hydrogels were higher than the tensile moduli. Though it can be difficult in practice for soft materials like hydrogels, characterizing the tensile modulus in addition to the compressive modulus provides valuable insight for tissue engineering applications in which a graft or a scaffold is required to sustain both stretching and compressive forces [37,38]. Furthermore, all GelNB-GelSH hydrogels remained stable with no observable dimensional changes over 7 days upon reaching swelling equilibrium and possessed a more homogeneous gel network ultrastructure compared to chain growth polymerized GelMA (Supplementary Figure S5). Overall, these results showcase the possibility to modularly tune the physical characteristics of GelNB-GelSH hydrogels by altering the macromer components to control the degree of crosslinking independently of total gelatin content.
Figure 2.

Physical characterization of printed GelNB-GelSH hydrogels and cytocompatibility of encapsulated cells. Plots of (A) tensile modulus, (B) compressive modulus, and (C) swelling ratio over time. * = significance between time points (p<0.05, n = 4–7). (D) Representative SEM images showing the ultrastructure and porosity of the GelNB-GelSH hydrogels with varied molar ratios of norbornene to thiol functional groups. Scale bar = 1 μm. (E) Representative fluorescent images showing Live/Dead™ (green = live, red = dead) stained iPSC-CMs and HUVEC:10T1/2 cells (50:1) over 7 days in culture. Scale bars = 250 μm. CellTiter Glo® 3D cell viability assay quantification of ATP content for (F) iPSC-CMs and (G) HUVEC:10T1/2 cells encapsulated in GelNB-GelSH constructs over 7 days in culture. * = significance between time points (p<0.05, n = 3). For cell viability assays in (E), (F), and (G), GelNB:GelSH (2:1) (i.e. 3.33% (w/v) GelNB + 3.33% (w/v) GelSH) constructs were used.
3.3. Soft GelNB-GelSH hydrogels as highly cytocompatible matrices
In the context of tissue engineering, gelatin is a naturally occurring protein derivative of collagen and has been highly regarded as a matrix scaffolding biomaterial given its intrinsic biological properties, biocompatibility, and biodegradability [39]. Specifically, several studies have demonstrated the encapsulation of various cell types within gelatin-based thiol-ene hydrogels including porcine chondrocytes, human hepatocellular carcinoma cells, human fibroblasts, and human keratinocytes [8,22,40]. However, with the advent of induced-pluripotent stem cell (iPSC) technology and their growing use in regenerative medicine, disease modeling, and drug discovery, few studies have investigated the biocompatibility of iPSC-derived cell types in gelatin thiol-ene hydrogel systems. To evaluate the potential of our GelNB-GelSH hydrogel as a supportive matrix for iPSCs, we encapsulated iPSC-derived cardiomyocytes (iPSC-CMs) in a soft hydrogel formulation chosen to fall within the stiffness range of the developing heart (i.e. ~5 kPa) to ensure optimal cell survival [41]. Live/Dead™ staining images showed good viability with very few detectable dead cells within the GelNB-GelSH hydrogels over 7 days (Figure 2E), which correlates to the consistent metabolic activity measured using CellTiter-Glo® 3D cell viability assay (Figure 2F). Moreover, the high cellular density maintained that is necessary for cell-cell interactions led to strong synchronized contraction being observed uniformly throughout the entire tissue construct starting at 72 h post-encapsulation (Supplementary Movie S2). Since vascularization plays an important role in providing vital nutrients to cells as well as promoting the maturation of tissues [4,42], we also assessed the viability of encapsulated human umbilical cord endothelial cells (HUVECs) co-cultured with C3H/10T1/2 (10T1/2) fibroblast cells to act as the supportive perivascular cells and mimic the native vascular population [43]. From the Live/Dead™ fluorescent images (Figure 2E), our GelNB-GelSH hydrogels supported high cell viability, spreading, and proliferation as measured by the increase in metabolic activity over 7 days in culture (Figure 2G). Confluent regions of the printed construct also showed the cells adopting a cobblestone-like endothelial cell morphology near the surface of the hydrogel. Overall, our GelNB-GelSH hydrogels were able to support greater than 86% and 94% viability across all time points for the encapsulated iPSC-CMs and HUVEC:10T1/2 cells, respectively (Supplementary Table S1). Together, these findings confirm the potential of our 3D bioprinted GelNB-GelSH hydrogels for fabricating functional cell-laden tissue constructs for cardiac and vascular tissue engineering applications.
3.4. Spatiotemporally controlled presentation of DLP-based photopatterned proteins within GelNB-GelSH hydrogels
A key advantage of thiol-ene hydrogel systems is the capability to control the presentation of bioactive moieties to introduce additional biofunctional properties such as chemotactic guidance and differentiation factors to dictate cell fate. Using the same chemistry for GelNB-GelSH hydrogel assembly, thiolated proteins or peptide growth factors can be directly conjugated to available norbornene moieties within the hydrogel upon selective UV irradiation with our DLP-based 3D projection printing platform. Here, we used fluorescently labeled ovalbumin to explore multiple techniques to tune the degree of conjugation for the generation of patterned hydrogel substrates. In the first case, we demonstrated active UV photopatterning of AF488-ovalbumin-PEG-thiol into a capillary design followed by passive conjugation of AF647-ovalbumin-PEG-NHS in the complementary regions (Figure 3A). The latter conjugation proceeds spontaneously via N-hydroxysuccinimide (NHS)-ester chemistry whereby an amide bond is formed with an available primary amine of GelNB or GelSH. Due to patterned conjugation of AF488-ovalbumin-PEG-thiol within the hydrogel, secondary crosslinking within the same capillary region is greatly reduced as shown by the absence of AF647-ovalbumin-PEGNHS (Figure 3Aii). Next, we examined the degree of photoconjugation as a function of the UV irradiation exposure time (Figure 3B). Increasing the exposure time resulted in stronger fluorescence intensity which is directly correlated to greater AF488-ovalbumin-PEG-thiol conjugation within the GelNB-GelSH hydrogel as shown in the 15 s and 30 s exposed capillary patterns in Figure 3Bi. Varying the exposure time also enables us to change the quantity of immobilized protein within a selected area and control the amount of subsequent photoconjugation available for different proteins within the same region. This was demonstrated by photopatterning AF647-ovalbumin-PEG-thiol over partial regions of the 15 s and 30 s exposed AF488-ovalbumin-PEG-thiol patterns (Figure 3B). Merged images displaying an overlap of the patterns within the 15 s region showed both AF488-ovalbumin-PEG-thiol and AF647-ovalbumin-PEG-thiol visibly present compared to the 30 s region where the AF488-ovalbumin-PEG-thiol predominates. These findings illustrate a simple and direct approach to deterministically pattern multiple bioactive conjugates of varying amounts using exposure time within a specified area. Not only can this technique produce gradient designs, but it could also be useful in applications where preparing combinations of different factors is necessary to decouple synergistic effects in complex tissue systems. As a final variable, we assessed the effect of norbornene content within the hydrogels to modify the degree of photoconjugation. Here, photopatterning exposure time was kept constant while three different hydrogels of increasing excess norbornene groups were tested (Figure 3C). As expected, hydrogels possessing a greater excess of norbornene resulted in a greater degree of AF488-ovalbumin-PEG-thiol photoconjugation as measured by the increasing fluorescence intensity of the capillary designs observed between the 2:1 to 3:1 to 4:1 GelNB:GelSH portions of the hydrogel (Figure 3D). Confocal 3D reconstruction of our photopatterned capillary design further demonstrated the ability of our platform to attain full-depth photoconjugation throughout a 250 μm tall construct while maintaining clearly defined features of at least 50 μm resolution (Figure 3E). At present, rapidly photopatterning large volumes on the millimeter scale as well as real-time interchange between complex geometric designs is challenging using multi-photon techniques which function in a raster-like fashion or traditional photolithography that involves multiple processing steps [18,19]. Our platform takes advantage of the benefits of DLP-based projection patterning used in the 3D bioprinting of hydrogels to rapidly photopattern the entire plane without compromising resolution, while also allowing for quick exchange of digital patterns and bioconjugates to form intricate designs. We confirmed this capability by generating a multi-colored Mona Lisa portrait measuring approximately 2 mm by 3 mm in area by successively photopatterning three distinct digital patterns designed for each color of fluorescently labeled ovalbumin-PEG-thiols (i.e. AF488, AF555, and AF647). The fluorescent images in Figure 3F show well-defined shapes for each of the photopatterned regions which overlay to form the complete portrait. This is particularly useful in the biofabrication of tissues wherein rapid construction is critical to maintain cell viability in addition to having the flexibility to incorporate small to large scale hierarchical components. Altogether, the inherent tunability of our platform and GelNB-GelSH hydrogel system to adjust for different parameters to produce complex photoconjugated designs opens unlimited possibilities to engineer customized biofunctional matrices in a reproducible and robust manner.
3.5. Biofunctionalized GelNB-GelSH hydrogels enable programmable endothelial cell growth
3D bioprinting technology has emerged as an excellent method for the placement of biomaterials and cells with high spatiotemporal precision to build biomimetic tissue and organ substitutes. Consequently, an equally important factor to demonstrate the potential clinical translation of bioprinted tissues is the ability to control cell fate over time to ensure predictable biological responses such as migration, proliferation, and differentiation in vitro or in vivo [44]. Building on this concept, we applied our integrated DLP-based 3D bioprinting and orthogonal bioconjugation platform to fabricate a 3D chemotactic guidance model to produce programmable tissue vascularization. We hypothesized that photopatterning the VEGF peptide mimic, QK with a cysteine-PEG6 modification on the N-terminus (i.e. Cys-PEG6-KLTWQELYQLKYKGINH2), within our GelNB-GelSH hydrogels would permit control over encapsulated endothelial cell migration, proliferation, and directionality as it has been demonstrated previously to modulate angiogenic response [45,46]. To enable visualization of photopatterned QK peptide, we used fluorescently labeled AF488-QK peptide and photopatterned it into a daisy flower to demonstrate control over both uniform and gradient designs as shown in Figure 4A and Figure 4B, respectively, wherein longer exposure times resulted in greater fluorescence intensity (Figure 4C). In this case, an exposure time of 20 s was chosen for all subsequent cell studies which corresponds to an approximate concentration of 0.36 ± 0.03 μg/mL QK peptide photoconjugated within the GelNB-GelSH hydrogel. To visually track endothelial cell response, a 3D printed daisy flower construct was created comprising of acellular petals and a cellularized central floral disc composed of encapsulated HUVEC:10T1/2 cells (50:1 ratio) stained with CellTracker™ Green (Figure 4D and Supplementary Figure S8A). Bioactivity of the QK peptide was maintained post-photopatterning as confirmed by the significantly greater number of cells observed in the petal regions of the full photopatterned +QK peptide group compared to the non-photopatterned -QK peptide control (Figure 4E). In particular, the effect of QK peptide on cell migratory response was immediate at 24 h with cells reaching more than 500 μm in distance to the petal periphery by 72 h. In addition, cell proliferation was also observed as shown by the visible enlargement of the central floral disc in the full photopatterned +QK peptide group. While photopatterning may have induced small changes in mechanical properties of the petal regions, these effects were found to be negligible in influencing endothelial cell migration in the full petal photopatterned controls absent of QK peptide (Supplementary Figure S9C). Directionality of endothelial cells due to the presence of photopatterned QK peptide was also assessed using a single petal photopatterned +QK peptide design (Figure 4D). Analogous to the full photopatterned +QK peptide group, significant preferential migration of cells was observed towards the single photopatterned +QK peptide petal compared to the remaining five petals across 72 h in culture (Figure 4F). In addition, redistribution of cell density within the central flower disc occurred showing visibly higher cell density located near the border of the single photopatterned +QK peptide petal (Figure 4D). Based on the bioactive potency of photopatterned QK peptide, as a proof-of-concept, we investigated a practical application by recapitulating the developmental event of recruiting vascular cells from the sinus venous to form the cardiac capillary bed (Figure 4G) [47]. We first 3D printed the cardiac portion composed of iPSC-CMs at tissue scale density (i.e. 50 million cells/mL) and subsequently photopatterned QK peptide into a vascular digital pattern followed by bioprinting of the endothelial sinus encapsulated with HUVEC:10T1/2 cells (50:1 ratio) stained with CellTracker™ Green (Supplementary Figure S10A). Over the course of 72 h, visible migration of the endothelial cell population was observed that was guided by the photopatterned QK peptide within the cardiac tissue region. Meanwhile, there was no observable endothelial cell migration into the cardiac region in the photopatterned controls absent of QK peptide as well as non-photopatterned -QK peptide controls (Supplementary Figure S10B). In general, this work showcases the relevant application of our DLP-based orthogonal bioconjugation system to study endothelial cell guidance and lays the groundwork for future studies examining dynamic processes for the controlled de novo formation of vascular networks.
4. Conclusions
Our findings highlight the potential application of our integrated DLP-based 3D bioprinting and orthogonal bioconjugation platform as an advanced biomanufacturing method for precision tissue engineering. In particular, our platform enables us to produce programmable cell niches through biofunctionalized 3D printed hydrogels with predetermined environmental cues to control for different cellular behavioral outcomes. Namely, the versatility offered in our thiol-ene GelNB-GelSH hydrogel system enables higher-ordered modulation of desired mechanical properties as well as gel network density through varying the molar ratio of norbornene to thiol moieties. The pure gelatin composition within our soft GelNB-GelSH hydrogels also served as a conducive environment to support high cell viability of iPSC-CMs and endothelial cells to enable rapid 3D bioprinting of complex tissue geometries with micron scale resolution. By using our DLP-based orthogonal photoconjugation technique to immobilize proteins and peptide growth factors, additional biochemical properties could be directly designed into 3D bioprinted cell matrices with high spatiotemporal precision in a scalable manner. In addition to sequential 3D bioprinting and photopatterning as demonstrated in this study to form heterogenous photoconjugated regions, it is also possible to perform photopatterning during 3D printing to achieve homogenous photoconjugated constructs in a one-step process to improve the workflow of future applications. The methods established in this study for combining bioprinting and orthogonal photoconjugation can be later adopted to fully 3D models, and we aim to expand peptide immobilization into 3D geometries, allowing us to explore long term vasculature and tissue development. Additionally, in the realm of regenerative medicine, this technology would open new directions to build transformative 3D bioprinted cell matrices that can predictably guide desired cell fate by utilizing the expansive library of bioactive peptides, proteins, or growth factors available in literature for broad applications such as tissue repair, disease modeling, and stem cell engineering.
Supplementary Material
Acknowledgements
The authors would like to thank Leilani Kwe, Jacob Stupin, Xinyue (Cinny) Ma and Henry Hwang for their assistance with this project. This work was supported in part by grants from the National Institutes of Health (R01EB021857, R21AR074763, R21HD100132, R21 EY031122, R33HD090662, NS047101) and National Science Foundation (1542148, 1644967, 1903933). Scholarship funding for Dr. Claire Yu was provided by the Natural Sciences and Engineering Research Council (NSERC) Postdoctoral Fellowship Scholarship of Canada under Grant No. PDF-487240-2016. Furthermore, this material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program for Jacob Schimelman under Grant No. DGE-1650112.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
The raw/processed data required to reproduce these findings can be shared by the authors upon request.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Barthes J, Özçelik H, Hindié M, Ndreu-Halili A, Hasan A, Vrana NE, Cell Microenvironment Engineering and Monitoring for Tissue Engineering and Regenerative Medicine: The Recent Advances, Biomed Res. Int 2014 (2014) 1–18. doi: 10.1155/2014/921905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li J, Chen M, Fan X, Zhou H, Recent advances in bioprinting techniques: approaches, applications and future prospects, J. Transl. Med 14 (2016) 271. doi: 10.1186/s12967-016-1028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ng WL, Lee JM, Zhou M, Chen Y-W, Lee K-XA, Yeong WY, Shen Y-F, Vat polymerization-based bioprinting—process, materials, applications and regulatory challenges, Biofabrication. 12 (2020) 022001. doi: 10.1088/1758-5090/ab6034. [DOI] [PubMed] [Google Scholar]
- [4].Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng G-S, Sheikh F, Chien S, Chen S, Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting, Proc. Natl. Acad. Sci 113 (2016) 2206–2211. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Noor N, Shapira A, Edri R, Gal I, Wertheim L, Dvir T, 3D Printing of Personalized Thick and Perfusable Cardiac Patches and Hearts, Adv. Sci 6 (2019) 1900344. doi: 10.1002/advs.201900344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, Lewis JA, Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips, Sci. Rep 6 (2016) 34845. doi: 10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Heinrich MA, Bansal R, Lammers T, Zhang YS, Michel Schiffelers R, Prakash J, 3D‐Bioprinted Mini‐Brain: A Glioblastoma Model to Study Cellular Interactions and Therapeutics, Adv. Mater 31 (2019) 1806590. doi: 10.1002/adma.201806590. [DOI] [PubMed] [Google Scholar]
- [8].Bertlein S, Brown G, Lim KS, Jungst T, Boeck T, Blunk T, Tessmar J, Hooper GJ, Woodfield TBF, Groll J, Thiol-Ene Clickable Gelatin: A Platform Bioink for Multiple 3D Biofabrication Technologies, Adv. Mater 29 (2017) 1703404. doi: 10.1002/adma.201703404. [DOI] [PubMed] [Google Scholar]
- [9].Lim KS, Levato R, Costa PF, Castilho MD, Alcala-Orozco CR, Van Dorenmalen KMA, Melchels FPW, Gawlitta D, Hooper GJ, Malda J, Woodfield TBF, Bio-resin for high resolution lithography-based biofabrication of complex cell-laden constructs, Biofabrication. 10 (2018) 034101. doi: 10.1088/1758-5090/aac00c. [DOI] [PubMed] [Google Scholar]
- [10].Lin H, Zhang D, Alexander PG, Yang G, Tan J, Cheng AWM, Tuan RS, Application of visible light-based projection stereolithography for live cell-scaffold fabrication with designed architecture, Biomaterials. 34 (2013) 331–339. doi: 10.1016/j.biomaterials.2012.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yu C, Schimelman J, Wang P, Miller KL, Ma X, You S, Guan J, Sun B, Zhu W, Chen S, Photopolymerizable Biomaterials and Light-Based 3D Printing Strategies for Biomedical Applications, Chem. Rev (2020). doi: 10.1021/acs.chemrev.9b00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jakus AE, Rutz AL, Shah RN, Advancing the field of 3D biomaterial printing, Biomed. Mater 11 (2016) 014102. doi: 10.1088/1748-6041/11/1/014102. [DOI] [PubMed] [Google Scholar]
- [13].Zhang Y, Post-3D printing modification for improved biomedical applications, Int. J. Bioprinting 3 (2017). doi: 10.18063/IJB.2017.02.001. [DOI] [Google Scholar]
- [14].Lee SJ, Lee D, Yoon TR, Kim HK, Jo HH, Park JS, Lee JH, Kim WD, Kwon IK, Park SA, Surface modification of 3D-printed porous scaffolds via mussel-inspired polydopamine and effective immobilization of rhBMP-2 to promote osteogenic differentiation for bone tissue engineering, Acta Biomater. 40 (2016) 182–191. doi: 10.1016/J.ACTBIO.2016.02.006. [DOI] [PubMed] [Google Scholar]
- [15].Jaidev LR, Chatterjee K, Surface functionalization of 3D printed polymer scaffolds to augment stem cell response, Mater. Des 161 (2019) 44–54. doi: 10.1016/J.MATDES.2018.11.018. [DOI] [Google Scholar]
- [16].Hoyle CE, Bowman CN, Thiol-Ene Click Chemistry, Angew. Chemie Int. Ed 49 (2010) 1540–1573. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- [17].Hoyle CE, Lee TY, Roper T, Thiol-enes: Chemistry of the past with promise for the future, J. Polym. Sci. Part A Polym. Chem 42 (2004) 5301–5338. doi: 10.1002/pola.20366. [DOI] [Google Scholar]
- [18].DeForest CA, Polizzotti BD, Anseth KS, Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments., Nat. Mater 8 (2009) 659–64. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shadish JA, Benuska GM, DeForest CA, Bioactive site-specifically modified proteins for 4D patterning of gel biomaterials, Nat. Mater (2019) 1. doi: 10.1038/s41563-019-0367-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS, A Versatile Synthetic Extracellular Matrix Mimic via Thiol-Norbornene Photopolymerization, Adv. Mater 21 (2009) 5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Azagarsamy MA, Anseth KS, Bioorthogonal Click Chemistry: An Indispensable Tool to Create Multifaceted Cell Culture Scaffolds, (2012). doi: 10.1021/mz300585q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Greene T, Lin C-C, Modular Cross-Linking of Gelatin-Based Thiol–Norbornene Hydrogels for in Vitro 3D Culture of Hepatocellular Carcinoma Cells, (2015). doi: 10.1021/acsbiomaterials.5b00436. [DOI] [PubMed] [Google Scholar]
- [23].Grim JC, Brown TE, Aguado BA, Chapnick DA, Viert AL, Liu X, Anseth KS, A Reversible and Repeatable Thiol-Ene Bioconjugation for Dynamic Patterning of Signaling Proteins in Hydrogels, ACS Cent. Sci 4 (2018) 909–916. doi: 10.1021/acscentsci.8b00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Munoz Z, Shih H, Lin C-C, Gelatin hydrogels formed by orthogonal thiol-norbornene photochemistry for cell encapsulation, Biomater. Sci 2 (2014) 1063–1072. doi: 10.1039/c4bm00070f. [DOI] [PubMed] [Google Scholar]
- [25].Kommareddy S, Amiji M, Preparation and evaluation of thiol-modified gelatin nanoparticles for intracellular DNA delivery in response to glutathione, Bioconjug. Chem 16 (2005) 1423–1432. doi: 10.1021/bc050146t. [DOI] [PubMed] [Google Scholar]
- [26].Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS, Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility, Biomaterials. 30 (2009) 6702–6707. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hoch E, Hirth T, Tovar GEM, Borchers K, Chemical tailoring of gelatin to adjust its chemical and physical properties for functional bioprinting, J. Mater. Chem. B 1 (2013) 5675–5685. doi: 10.1039/c3tb20745e. [DOI] [PubMed] [Google Scholar]
- [28].Zhu W, Tringale KR, Woller SA, You S, Johnson S, Shen H, Schimelman J, Whitney M, Steinauer J, Xu W, Yaksh TL, Nguyen QT, Chen S, Rapid continuous 3D printing of customizable peripheral nerve guidance conduits, Mater. Today 21 (2018) 951–959. doi: 10.1016/J.MATTOD.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Volarevic V, Markovic BS, Gazdic M, Volarevic A, Jovicic N, Arsenijevic N, Armstrong L, Djonov V, Lako M, Stojkovic M, Ethical and Safety Issues of Stem Cell-Based Therapy, Int. J. Med. Sci 15 (2018) 36–45. doi: 10.7150/ijms.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yu C, Ma X, Zhu W, Wang P, Miller KL, Stupin J, Koroleva-Maharajh A, Hairabedian A, Chen S, Scanningless and continuous 3D bioprinting of human tissues with decellularized extracellular matrix, Biomaterials. 194 (2018) 1–13. doi: 10.1016/J.BIOMATERIALS.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Grim JC, Brown TE, Aguado BA, Chapnick DA, Viert AL, Liu X, Anseth KS, A Reversible and Repeatable Thiol-Ene Bioconjugation for Dynamic Patterning of Signaling Proteins in Hydrogels, ACS Cent. Sci 4 (2018) 909–916. doi: 10.1021/acscentsci.8b00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jivan F, Fabela N, Davis Z, Alge DL, Orthogonal click reactions enable the synthesis of ECM-mimetic PEG hydrogels without multi-arm precursors, J. Mater. Chem. B 6 (2018) 4929–4936. doi: 10.1039/C8TB01399C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Odedra D, Chiu LLY, Shoichet M, Radisic M, Endothelial cells guided by immobilized gradients of vascular endothelial growth factor on porous collagen scaffolds, Acta Biomater. 7 (2011) 3027–3035. doi: 10.1016/j.actbio.2011.05.002. [DOI] [PubMed] [Google Scholar]
- [34].Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A, Cell-laden microengineered gelatin methacrylate hydrogels, Biomaterials. 31 (2010) 5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Benton JA, DeForest CA, Vivekanandan V, Anseth KS, Photocrosslinking of gelatin macromers to synthesize porous hydrogels that promote valvular interstitial cell function., Tissue Eng. Part A 15 (2009) 3221–30. doi: 10.1089/ten.TEA.2008.0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].B.-S. A, M.D. H, C.E. K, N. L, B.-S. A, M.D. H, C.E. K, N. L, Thiolated polymers: Stability of thiol moieties under different storage conditions, Sci. Pharm 70 (2002) 331–339. doi: 10.3797/scipharm.aut-02-32. [DOI] [Google Scholar]
- [37].Butler DL, Goldstein SA, Guilak F, Functional Tissue Engineering: The Role of Biomechanics, J. Biomech. Eng 122 (2000) 570. doi: 10.1115/1.1318906. [DOI] [PubMed] [Google Scholar]
- [38].Xiao Y, Friis EA, Gehrke SH, Detamore MS, Mechanical testing of hydrogels in cartilage tissue engineering: beyond the compressive modulus., Tissue Eng. Part B. Rev 19 (2013) 403–12. doi: 10.1089/ten.TEB.2012.0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Echave MC, Burgo LS, Pedraz JL, Orive G, Gelatin as Biomaterial for Tissue Engineering, Curr. Pharm. Des 23 (2017) 3567–3584. doi: 10.2174/0929867324666170511123101. [DOI] [PubMed] [Google Scholar]
- [40].Xu K, Fu Y, Chung W, Zheng X, Cui Y, Hsu IC, Kao WJ, Thiol-ene-based biological/synthetic hybrid biomatrix for 3-D living cell culture., Acta Biomater. 8 (2012) 2504–16. doi: 10.1016/j.actbio.2012.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Young JL, Engler AJ, Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro, Biomaterials. 32 (2011) 1002–1009. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Moon JJ, West JL, Vascularization of engineered tissues: approaches to promote angiogenesis in biomaterials., Curr. Top. Med. Chem 8 (2008) 300–10. http://www.ncbi.nlm.nih.gov/pubmed/18393893 (accessed June 6, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhu W, Qu X, Zhu J, Ma X, Patel S, Liu J, Wang P, Lai CSE, Gou M, Xu Y, Zhang K, Chen S, Direct 3D bioprinting of prevascularized tissue constructs with complex microarchitecture, Biomaterials. 124 (2017) 106–115. doi: 10.1016/j.biomaterials.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chan G, Mooney DJ, New materials for tissue engineering: towards greater control over the biological response, Trends Biotechnol. 26 (2008) 382–392. doi: 10.1016/J.TIBTECH.2008.03.011. [DOI] [PubMed] [Google Scholar]
- [45].D’Andrea LD, Iaccarino G, Fattorusso R, Sorriento D, Carannante C, Capasso D, Trimarco B, Pedone C, Targeting angiogenesis: Structural characterization and biological properties of a de novo engineered VEGF mimicking peptide, Proc. Natl. Acad. Sci 102 (2005) 14215–14220. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang L, Zhao M, Li S, Erasquin UJ, Wang H, Ren L, Chen C, Wang Y, Cai C, “Click” immobilization of a VEGF-mimetic peptide on decellularized endothelial extracellular matrix to enhance angiogenesis., ACS Appl. Mater. Interfaces 6 (2014) 8401–6. doi: 10.1021/am501309d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bernanke DH, Velkey JM, Development of the Coronary Blood Supply: Changing Concepts and Current Ideas, Anat Rec (New Anat). 269 (2002) 198–208. doi: 10.1002/ar.10139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


