Key Points
Question
What is the association of having a very low number (“low-shot”) of training images with the performance of artificial intelligence algorithms for retinal diagnostics?
Findings
This cross-sectional study found that performance degradation occurred when using traditional algorithms with low numbers of training images. When using only 160 training images per class, traditional approaches had an area under the curve of 0.6585; low-shot methods using contrastive self-supervision outperformed this with an area under the curve of 0.7467.
Meaning
These findings suggest that low-shot deep learning methods show promise for use in artificial intelligence retinal diagnostics and may be beneficial for situations involving much less training data, such as rare retinal diseases or addressing artificial intelligence bias.
Abstract
Importance
Recent studies have demonstrated the successful application of artificial intelligence (AI) for automated retinal disease diagnostics but have not addressed a fundamental challenge for deep learning systems: the current need for large, criterion standard–annotated retinal data sets for training. Low-shot learning algorithms, aiming to learn from a relatively low number of training data, may be beneficial for clinical situations involving rare retinal diseases or when addressing potential bias resulting from data that may not adequately represent certain groups for training, such as individuals older than 85 years.
Objective
To evaluate whether low-shot deep learning methods are beneficial when using small training data sets for automated retinal diagnostics.
Design, Setting, and Participants
This cross-sectional study, conducted from July 1, 2019, to June 21, 2020, compared different diabetic retinopathy classification algorithms, traditional and low-shot, for 2-class designations (diabetic retinopathy warranting referral vs not warranting referral). The public domain EyePACS data set was used, which originally included 88 692 fundi from 44 346 individuals. Statistical analysis was performed from February 1 to June 21, 2020.
Main Outcomes and Measures
The performance (95% CIs) of the various AI algorithms was measured via receiver operating curves and their area under the curve (AUC), precision recall curves, accuracy, and F1 score, evaluated for different training data sizes, ranging from 5120 to 10 samples per class.
Results
Deep learning algorithms, when trained with sufficiently large data sets (5120 samples per class), yielded comparable performance, with an AUC of 0.8330 (95% CI, 0.8140-0.8520) for a traditional approach (eg, fined-tuned ResNet), compared with low-shot methods (AUC, 0.8348 [95% CI, 0.8159-0.8537]) (using self-supervised Deep InfoMax [our method denoted as DIM]). However, when far fewer training images were available (n = 160), the traditional deep learning approach had an AUC decreasing to 0.6585 (95% CI, 0.6332-0.6838) and was outperformed by a low-shot method using self-supervision with an AUC of 0.7467 (95% CI, 0.7239-0.7695). At very low shots (n = 10), the traditional approach had performance close to chance, with an AUC of 0.5178 (95% CI, 0.4909-0.5447) compared with the best low-shot method (AUC, 0.5778 [95% CI, 0.5512-0.6044]).
Conclusions and Relevance
These findings suggest the potential benefits of using low-shot methods for AI retinal diagnostics when a limited number of annotated training retinal images are available (eg, with rare ophthalmic diseases or when addressing potential AI bias).
This cross-sectional study evaluates whether deep learning methods using a relatively low number of training data sets are beneficial for automated retinal diagnostics.
Introduction
Artificial intelligence (AI) using deep learning (DL) systems (DLSs) has demonstrated promise for diagnosing retinal diseases.1,2 However, this research, to our knowledge, has not addressed a fundamental weakness of DL (ie, the need for prohibitively large, criterion standard–annotated retinal training data sets). When only a low number of training data are available (so-called low-shot learning situations),3 DL may perform worse. Deep learning systems may generalize poorly to other domains,3 which may entail differences in imaging systems, refractive status, photographic illumination, or patient population features, such as varying melanocytic content, that may affect retinal features.
There are several important practical settings in which this low-shot problem needs to be addressed when using DL for automated retinal image analysis. One setting is rare diseases (eg, serpiginous choroidopathy or angioid streaks in pseudoxanthoma elasticum) that are readily diagnosed by retinal specialists, but for which few images are available to train DL systems compared with the vast data sets available for training in diabetic retinopathy2 (DR) or age-related macular degeneration.4 Another setting is the paucity of quality images, for example, among individuals with retinal diseases and media opacities or among individuals from settings outside of epidemiologic studies or clinical trials where quality control of image acquisition might be limited. Another potentially important role of low-shot methods is to address AI bias due to the paucity or imbalance of data. For example, if training is from an epidemiologic study, such as the AREDS (Age-Related Eye Disease Study),5 in which few participants older than 85 years are present, the automated diagnoses might be biased against correct diagnoses for individuals older than 85 years when only a few training images for this age group were included in the training data set. When addressing bias, partitioning data into factorized sets of specific protected attributes (eg, age, sex, and race/ethnicity) may result in ever smaller data sets that may not provide a sufficient number of training images for a particular group (eg, women older than 85 years) and thereby may be biased against accurate diagnoses for that group.
Another potential low-shot situation arises when addressing the need to perform continuous disease phenotyping, as might be desirable when undertaking AI detection of like-kind disease presentations, with few evidentiary images to use for training (eg, images of polypoidal choroidal vasculopathy vs neovascular age-related macular degeneration or central serous retinopathy associated with choroidal neovascularization vs central serous retinopathy in the absence of choroidal neovascularization).
So-called low-shot methods that address cases in which few training images are available may be of help in these circumstances. To address these situations, this study evaluated (1) the decrease in performance of retinal diagnostic algorithms when the number of training data available was reduced substantially and (2) the different DLS strategies that may alleviate the challenges of small training data sets.
Methods
Classification Problem and Algorithms
This cross-sectional study was conducted from July 1, 2019, to June 21, 2020. An automated binary classification of DR using diagnostic DLSs was studied, involving nonreferable (stages 0 and 1) vs referable (stages 2, 3, and 4) classes.6 Because the research only included the use of publicly available data sets, the Johns Hopkins University School of Medicine deemed that this research was exempt from institutional review board review.
Baseline DLS
Several algorithms were evaluated. Traditional DL approaches for retinal diagnostics—used in many retinal AI studies1,2,4,7,8—have consisted of performing fine-tuning of a best-of-breed network (eg, ResNet, DenseNet, or Inception) or an ensemble thereof, pretrained using a fully supervised method.7,8 A more detailed background on clinical retinal applications of DL can be found in recent literature.1,2,4,7,8 To mirror this strategy, a ResNet509 network, pretrained on ImageNet (henceforth denoted as “RES_FT”) was used as a baseline to compare against other algorithms and applied to low-shot DR diagnostics. This ResNet50 network was unmodified from its original architecture.9 ResNet networks have an architecture that includes blocks with “bottleneck” and “skip” connections that make upstream layer activations available downstream, conferring important benefits for representation learning and backpropagation. These blocks are followed by 1 fully connected layer. A framework to train and test was implemented with a software stack using Keras, version 2.24 (Google LLC) with TensorFlow, version 1.14 (Google LLC) as a backend. Transfer learning was used by fine-tuning all layers of the network, pretrained on ImageNet (1000 classes). For optimization, stochastic gradient descent was used with a Nesterov momentum of 0.9, with a learning rate initially set to 1 × 10−3 and a learning rate decay with a factor of 0.1. Training was stopped after 5 epochs of no improvement evaluated on the validation set. Because ResNet expects that input size, images were preprocessed via resizing to 224 × 224 pixels, and then the intensity was normalized between −1 and 1.
Low-Shot DL Methods
For comparison with the baseline, additional methods using image encoding followed by classification were used. Images were represented (ie, encoded or embedded) via the same ResNet50 network, pretrained on only ImageNet. The embedded representation entailed taking the global pooling of the last convolutional layers of ResNet50. Subsequently, 1 of 3 classifiers (random forest, support vector machines, and K-nearest neighbors) was applied to the encoded images. The random forest method was denoted as RES_RF, the support vector machines method was denoted as RES_SVM, and the K-nearest neighbors method was denoted as RES_KNN.
For additional comparisons, this study used encoding via networks exploiting more recent contrastive self-supervision learning concepts, together with the transfer learning and classification strategies. Unlike full supervision, which trains networks by using input (ie, images)–output (ie, labels) pairs (like ResNet does), self-supervision is a style of unsupervised learning that does not require labels because it forces a network to learn by evaluating the data against itself. For instance, this can be done by asking a network to predict part of an image from another part of an image, to predict future frames of a video from past frames, or, in this case, to optimize representations by relating (using mutual information) local and global aspects of the data (images). In addition to requiring less-specific supervised information for training, this self-supervision approach forces the network to gain insight into generating representations of the data that are more applicable to the end tasks, which may be more amenable to generalization or for dealing with low-shot learning.
Examples of networks using such strategies include Deep InfoMax10,11 and, specifically, Augmented Multiscale Deep InfoMax (AMDIM)11 used here, which is trained by maximizing the similarity of a pair of local and global image representations, taken from different layers of the network, that were created from augmentations of the same source image, and by minimizing the similarity of pairs not created from the same source image (contrastive learning). Consequently, each image should be given as unique a representation as possible, while maintaining that the local and global representations are still heavily correlated under certain transformations.
Here, the specific type of network11 used different image augmentations, such as random resized, cropped, and color-jittered versions of the same image, to compute the similarity between the global and local representations. With the use of a network called AMDIM-large11 (implemented as a variant of ResNet) pretrained on ImageNet, each image was encoded into its 2 representations, the local representation taken from the layer that outputs a 7 × 7 spatial map with 2560 channels and the global representation taken from the final layer with dimension 2560. Both representations were used during the pretrained phase. This representation can be thought of as functionally equivalent (being global) to the ResNet50 representation. Both representations were used for downstream classification.
This study developed a custom algorithm using the original AMDIM’s local representation as input, to train a randomly initialized ResNet34 network (we refer to this extension henceforth as DIM). This strategy was found to have good success in a related study.12 This network took input images downsampled to 128 × 128 pixels and normalized using ImageNet settings. For training this network, Adam13 was used, with the learning rate set to 1 × 10−3, β1 = 0.9, and β2 = 0.99. The global encoding of the image output by the pretrained AMDIM-Large network was also used in a similar manner as ResNet50 above (ie, it was used as input to 3 classifiers [random forest, support vector machines, and K-nearest neighbors]; the random forest method was denoted as DIM_RF, the support vector machines method was denoted as DIM_SVM, and the K-nearest neighbors method was denoted as DIM_KNN).
Data Set
A data set consisting originally of 88 692 public domain fundi from 44 346 participants6 from EyePACS was used (2 fundi, 1 each eye, per patient). Preprocessing included the following steps: each image was cropped to the retina circumscribed square, padded when necessary, and resized to the resolutions already described. Nongradable fundi were not removed, to make AI diagnostic tasks more challenging.
Data Partitions
Fundi were partitioned, as is done traditionally,1,4,6,7,8 into training, validation, and testing sets. The number of training exemplars were limited to N-shots, where N was the number of fundi per class, for a total of 2N training samples for each experiment. In the rest of this article, the number N referring to the cardinality of the training set will be understood to also include validation samples. Experiments were conducted by varying N logarithmically—between N = 10 and N = 5120 shots (see eTables 1, 2, and 3 in the Supplement for the values of N used). Training samples were chosen randomly from the complete data set. The selection of training, validation, and testing followed strict patient partitioning, meaning that all samples for a given patient were restricted to lie within the same partition. The RES_FT method further split the 2N training samples randomly into an 80% training and 20% validation split, whereas DIM was trained for 20 epochs directly because its feature encoder was fixed. Cardinality of the test data set was kept constant at 1766 samples, and the same testing samples were used across all experiments, algorithms, and N values, for fair comparison. In addition, testing was class balanced with 883 samples per class, allowing for more transparent performance analysis.
Performance Metrics
Statistical analysis was performed from February 1 to June 21, 2020. The performance and 95% CIs of all algorithms were evaluated using accuracy, receiver operating characteristic curve, area under the receiver operating characteristic curve (AUC), precision recall curve, and an F1 score computed for all algorithms and across all experiments for all N values.
Results
Table 1,11 Table 2,11 Table 3,11 and the Figure report the accuracy, AUC, and F1 score for the baseline (RES_FT) and alternate low-shot DL (LSDL) methods under investigation, including RES_RF, RES_SVM, RES_KNN, DIM_RF, DIM_SVM, and DIM_KNN, shown in this article for 4 experiments associated with decreasing values of training shots of 5120, 160, 40, and 10, to capture the main behavior of these algorithms. The outcomes of all experiments are reported in tabular (eTables 1, 2, and 3 in the Supplement) and plot format. Plots also include 95% CIs, receiver operating characteristic curves, and precision recall curves (eFigures 1-13 in the Supplement).
Table 1. Accuracy of Algorithms for Retinal Diagnosis That Address Bias in Artificial Intelligence.
| Algorithma | Accuracy, % (95% CI) | |||
|---|---|---|---|---|
| 10 Samples | 40 Samples | 160 Samples | 5120 Samples | |
| RES_FT | 51.47 (49.14-53.80) | 54.93 (52.61-57.25) | 59.85 (57.56-62.14) | 74.29 (72.25-76.33) |
| RES_KNN | 50.23 (47.90-52.56) | 51.47 (49.14-53.80) | 58.61 (56.31-60.91) | 61.04 (58.77-63.31) |
| RES_SVM | 50.45 (48.12-52.78) | 55.04 (52.72-57.36) | 63.19 (60.94-65.44) | 73.16 (71.09-75.23) |
| RES_RF | 50.91 (48.58-53.24) | 57.36 (55.05-59.67) | 63.08 (60.83-65.33) | 69.20 (67.05-71.35) |
| DIM | 55.95 (53.63-58.27)b | 62.74 (60.48-65.00)b | 65.23 (63.01-67.45) | 75.71 (73.71-77.71) |
| DIM_KNN | 52.10 (49.77-54.43) | 54.59 (52.27-56.91) | 59.34 (57.05-61.63) | 64.33 (62.10-66.56) |
| DIM_SVM | 54.53 (52.21-56.85) | 62.06 (59.80-64.32) | 67.04 (64.85-69.23)b | 76.39 (74.41-78.37)b |
| DIM_RF | 55.72 (53.40-58.04) | 59.12 (56.83-61.41) | 63.14 (60.89-65.39) | 71.06 (68.94-73.18) |
Abbreviations: DIM, Deep InfoMax; KNN, K-nearest neighbors; RES_FT, traditional fine-tuned ResNet algorithm; RF, random forest; SVM, support vector machine.
Algorithms include: a traditional fine-tuned ResNet algorithm (denoted as RES_FT), RES_RF and RES_SVM (ResNet encoding fed into an RF or SVM classifier), Augmented Multiscale Deep InfoMax encoding11 yielding local and global features, fed to a classifier, consisting of either ResNet (using only local features, and denoted as DIM), and 3 other classifiers using the global features of DIM and either DIM_SVM, or DIM_RF.
Best-performing algorithm for number of samples used for training.
Table 2. ROC Area Under the Curve of Algorithms for Retinal Diagnosis That Address Bias in Artificial Intelligence.
| Algorithma | ROC area under the curve (95% CI) | |||
|---|---|---|---|---|
| 10 Samples | 40 Samples | 160 Samples | 5120 Samples | |
| RES_FT | 0.5178 (0.4909-0.5447) | 0.5671 (0.5404-0.5938) | 0.6585 (0.6332-0.6838) | 0.8330 (0.8140-0.8520) |
| RES_KNN | 0.5076 (0.4807-0.5345) | 0.5221 (0.4952-0.5490) | 0.6148 (0.5887-0.6409) | 0.6549 (0.6296-0.6802) |
| RES_SVM | 0.4992 (0.4722-0.5262) | 0.5912 (0.5648-0.6176) | 0.6787 (0.6539-0.7035) | 0.8078 (0.7875-0.8281) |
| RES_RF | 0.5055 (0.4786-0.5324) | 0.5940 (0.5676-0.6204) | 0.6742 (0.6493-0.6991) | 0.7564 (0.7340-0.7788) |
| DIM | 0.5778 (0.5512-0.6044)b | 0.6760 (0.6511-0.7009)b | 0.7467 (0.7239-0.7695)b | 0.8348 (0.8159-0.8537) |
| DIM_KNN | 0.5248 (0.4979-0.5517) | 0.5625 (0.5358-0.5892) | 0.6134 (0.5873-0.6395) | 0.6884 (0.6638-0.7130) |
| DIM_SVM | 0.5440 (0.5172-0.5708) | 0.6525 (0.6271-0.6779) | 0.7455 (0.7227-0.7683) | 0.8581 (0.8405-0.8757)b |
| DIM_RF | 0.5706 (0.5440-0.5972) | 0.6234 (0.5975-0.6493) | 0.7039 (0.6798-0.7280) | 0.7985 (0.7778-0.8192) |
Abbreviations: DIM, Deep InfoMax; KNN, K-nearest neighbors; RES_FT, traditional fine-tuned ResNet algorithm; RF, random forest; ROC, receiver operating curve; SVM, support vector machine.
Algorithms include a traditional fine-tuned ResNet algorithm (denoted as RES_FT), RES_RF and RES_SVM (ResNet encoding fed into an RF or SVM classifier), Augmented Multiscale Deep InfoMax encoding11 yielding local and global features, fed to a classifier, consisting of either ResNet (using only local features, and denoted as DIM), and 3 other classifiers using the global features of DIM and either DIM_SVM, or DIM_RF.
Best-performing algorithm for number of samples used for training.
Table 3. F1 Score of Algorithms for Retinal Diagnosis That Address Bias in Artificial Intelligence.
| Algorithma | 10 Samples | 40 Samples | 160 Samples | 5120 Samples |
|---|---|---|---|---|
| RES_FT | 0.5648 | 0.4865 | 0.4925 | 0.7291 |
| RES_KNN | 0.5536 | 0.5263 | 0.5863 | 0.5870 |
| RES_SVM | 0.6067 | 0.5340 | 0.5812 | 0.7011 |
| RES_RF | 0.5893 | 0.5962 | 0.6414 | 0.6739 |
| DIM | 0.6381 | 0.5846 | 0.7022b | 0.7360 |
| DIM_KNN | 0.5447 | 0.5674 | 0.5864 | 0.6082 |
| DIM_SVM | 0.6513b | 0.6086 | 0.6600 | 0.7446b |
| DIM_RF | 0.5660 | 0.6316b | 0.6498 | 0.6985 |
Abbreviations: DIM, Deep InfoMax; KNN, K-nearest neighbors; RES_FT, traditional fine-tuned ResNet algorithm; RF, random forest; SVM, support vector machine.
Algorithms include a traditional fine-tuned ResNet algorithm (denoted as RES_FT), RES_RF and RES_SVM (ResNet encoding fed into an RF or SVM classifier), Augmented Multiscale Deep InfoMax encoding11 yielding local and global features, fed to a classifier, consisting of either ResNet (using only local features, and denoted as DIM), and 3 other classifiers using the global features of DIM and either DIM_SVM, or DIM_RF.
Best algorithm for number of samples used for training.
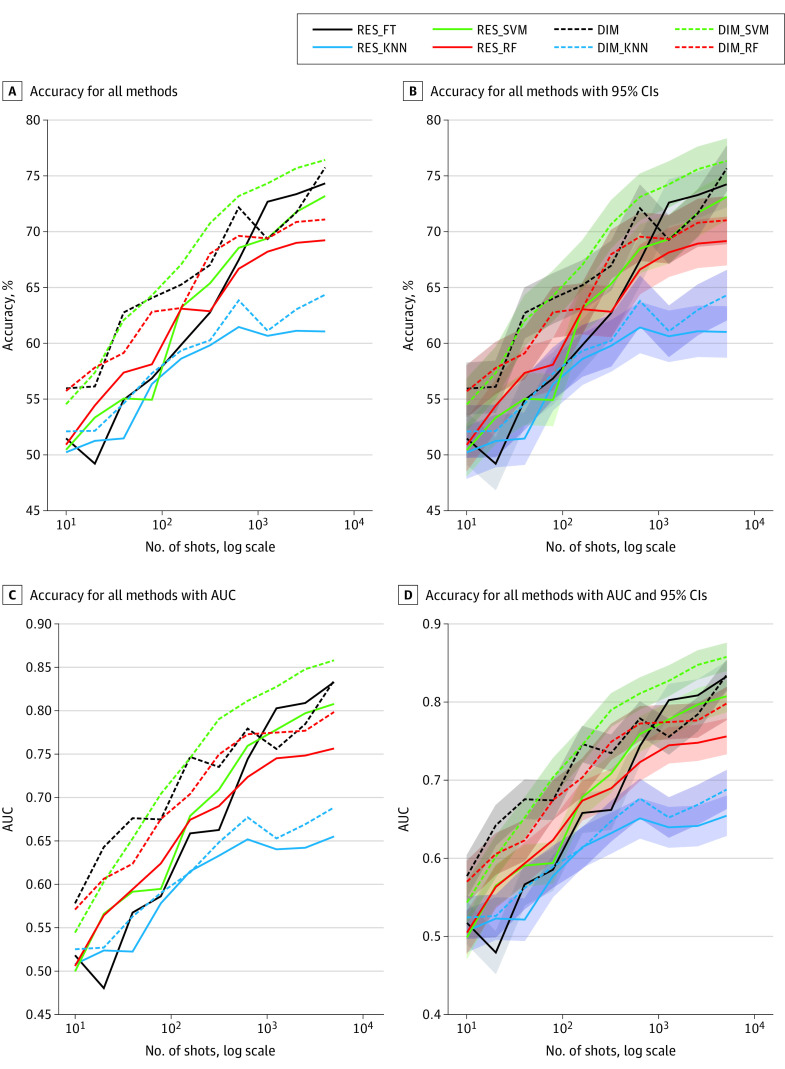
Figure. Comparison of Accuracy.
A, Accuracy for all methods across all low-shot experiments. B, Accuracy with 95% CIs. C, Accuracy with area under the curve (AUC). D, Accuracy with AUC and 95% CIs. DIM indicates AMDIM concatenated with ResNet34; DIM_KNN, AMDIM with K-nearest neighbors; DIM_RF, AMDIM with random forest; DIM_SVM, AMDIM with support vector machines; RES_FT, ResNet50 network; RES_KNN, ResNet50 network with K-nearest neighbors; RES_RF, ResNet50 network with random forest; and RES_SVM, ResNet50 network with support vector machines.
Inspection of the results, as shown in Table 1,11 Table 2,11 and Table 3,11 reveals that the AI algorithms tested demonstrated that the LSDL methods reported performed on par with traditional DLS method for a high number of shots (N = 5120) but did comparatively much better for lower numbers of shots (N = 10, 40, or 160 samples per class) and underwent a more graceful performance degradation as the number of training samples decreased. Specifically, when trained with sufficiently large data sets (N = 5120), the LSDL methods had slight improvements over the baseline DLS method (AUC, 0.8330 [95% CI, 0.8140-0.8520]) for the widely used fine-tuned method (RES_FT) compared with the LSDL using DIM_SVM (AUC, 0.8581 [95% CI, 0.8405-0.8757]), as well as using DIM (AUC, 0.8348 [95% CI, 0.8159-0.8537]).
However, when fewer training images were available (N = 160), the baseline fine-tuned ResNet50 (RES_FT) degraded more, with an AUC of 0.6585 (95% CI, 0.6332-0.6838), compared with the LSDL, which performed well considering the reduction in the size of the data set (eg, DIM: AUC, 0.7467 [95% CI, 0.7239-0.7695]) (Table 211). At a very low number of shots (N = 10), the fine-tuned RES_FT approach had poor performance, close to chance (AUC, 0.5178 [95% CI, 0.4909-0.5447]), while the LSDL still appeared to perform relatively better (eg, DIM: AUC, 0.5778 [95% CI, 0.5512-0.6044]) and underwent a more graceful and less catastrophic overall degradation.
Discussion
This evaluation of DL for automated DR diagnostics with small training data sets suggests that the performance of the baseline DLS using ResNet50 and fine-tuning, exemplifying one of the popular DL methods applied to retinal diagnostics, deteriorated substantially when the training size was relatively low (≤160 samples per class), when compared with other proposed methods that had better characteristics with a low number of images, principally methods that exploited AMDIM for representation learning. Results for the LSDL methods suggested that the performance was acceptable when training with relatively small sizes (n = 160) and also points to a more graceful degradation in performance when using a very low number of shots, with training sizes as small as 10.
To interpret the performance of each method in the aggregate, we believe that the level of performance decreases for the fine-tuned ResNet method—as the number of training exemplars decreases—because it may be hard for the network to do fine-tuning of network weights and learn to do an effective transfer from the original domain to the target domain with a lower number of exemplars. This may be less of a problem for methods that use image encodings with support vector machines because they will successfully train with fewer exemplars compared with methods using fully connected layers. More important, networks that use novel contrastive self-supervision strategies that depart from full supervision and use information theoretic approaches, such as the AMDIM-based methods, may have an edge in low-shot situations. Support vector machines, by trying to perform linear or nonlinear discrimination in high-dimensional spaces, act as a proxy for mutual information.10 Other methods (not used in this study) that perform self-supervision and contrastive predictive coding12 like AMDIM, or rely on mutual information between local and global representation of images, should also be studied in the future. The success of these methods is due to the properties that result from self-supervision; the original self-supervised objective that AMDIM was trained under tries to make each image representation as unique as possible while being invariant to different views of the data. It also aims to make representations that contain locally consistent information across structural locations in the image.10 Consequently, the global representation should act as a summary of the image already, which is amenable to methods such as support vector machines. On the other hand, as each image is given as unique a representation as possible, overfitting becomes more likely owing to the presentation of few data points, resulting in possible downsides and slightly lower accuracy in some situations, which may explain how DIM_SVM overtakes DIM at higher shots.
These findings should support additional pursuits and investigation of LSDL methods for endeavors in which the clinical AI diagnosis must rely on smaller and possibly unbalanced data sets. Application to rare retinal diseases is one such use, in which only much smaller training data sets are typically available. For instance, one may never have 88 692 images of serpiginous choroidopathy, but if a measurable performance can be detected with only 10 images, then it may be possible to assemble and grade enough images to design a DLS that is able to identify serpiginous choroidopathy.
Artificial intelligence bias is also a concern for both AI in general and medical and retinal diagnostics applications, or for predicting the need for health care,14,15 where partitioning of the data into specific factors, such as race/ethnicity or origin, sex, age, or other protected attributes, would likely yield sometimes balanced but often very small training data sets for specific partitions (eg, individuals of African descent with the neovascular form of age-related macular degeneration or individuals from large epidemiologic studies or large randomized clinical trials who are 85 years or older). In this case, the lack of large amounts of training data could be associated with degraded and unequal performance across protected attributes. Low-shot deep learning approaches using better representation learning in techniques such as AMDIM, paired with cascaded classifiers, could help address the problem of AI bias, to enable diagnostic systems that exhibit similar accuracy16 regardless of factors such as race/ethnicity or age. Low-shot deep learning can offer an option to address both balance and the need to train with much smaller data sets, which can complement other approaches to AI bias formulated on the synthesis of additional data via generative methods (P. M. Burlina, PhD, N. Joshi, BS, P. William, BS, K. D. Pacheco, MD, N. M. Bressler, MD, written communication, 2020),17 disentanglement,18 or “blindness” to factors of variation.19,20,21 Other approaches of interest that should be investigated and that are associated with these challenges include the use of anomaly detection techniques.19,20
Limitations
This study has some limitations. It examined some promising avenues for encoding data by using contrastive unsupervised techniques to address the challenge of low-shot learning. Other LSDL21 and debiasing (P. M. Burlina, PhD, N. Joshi, BS, P. William, BS, K. D. Pacheco, MD, N. M. Bressler, MD, written communication, 2020)15,16,21 techniques exist and continue to be developed by the research community and should also be compared and investigated for clinical retinal diagnostics. Only 1 retinal disease, DR, was evaluated in this analysis. There may be other situations with potential low-shot training of AI for diagnostics that may not be well addressed by this approach and that may need other solutions, including situations in which additional samples are needed for additional training of the AI retinal diagnostics. Also, while the end point in this study was performance, as measured by metrics such as accuracy, F1 score, or AUC, which relate to the “equal odds” and ”equal accuracy” criteria of AI bias,16 there may be other fairness requirements that are not well captured by those metrics but that merit use.
Other limitations include the need to identify and then include the biological variability that may exist with each disease. For example, there may be different amounts of melanocytes affecting retinal pigmentation in individuals of Asian vs European vs African descent, different refractive errors affecting optic nerve appearances in the setting of pathologic myopia, or different media opacities across individuals of various ages. In these situations, LSDL may degrade when insufficient data are present, as was seen for stringent situations (eg, N = 10). These limitations may help guide future studies.
Conclusions
This investigation evaluated the change in performance in the automated diagnosis of DR from retinal images using much fewer training data, termed low-shot learning. The results show that the performance of widely used DLS methods relying on full supervision that typically require large training data sets degrades substantially when used with limited data sets. The DL approaches in this investigation that used representation learning via self-supervised models suggest that there is promise in using these approaches for low-shot learning; such methods appear to perform better and have more graceful performance degradation as the number of training images is decreased. These low-shot methods could address AI approaches to retinal diagnostics when a limited number of retina images are available for training, as may be the case for rare ophthalmic diseases or to address potential AI bias due to training sets that may have few examples for certain groups with specific protected factors (eg, race/ethnicity, sex, or age) or combinations.
eTable 1. Performance of Algorithms: Accuracy
eTable 2. Performance of Algorithms: ROC AUC
eTable 3. Performance of Algorithms: F1 Score
eFigure 1. Accuracy for All Methods and Number of Shots
eFigure 2. Accuracy and Confidence Intervals for All Shots
eFigure 3. ROC AUC for All Shots
eFigure 4. ROC AUC for All Shots with 95% Confidence Intervals
eFigure 5. F1 Score for All Shots
eFigure 6. N=10 Shots Results: ROCs and Confidence Intervals, All, Two-Curve Comparisons of Methods
eFigure 7. N=40 Shots Results: ROCs and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 8. N=160 Shots Results: ROCs and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 9. N=5120 Shots Results: ROCs and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 10. N=10 Shots Results: PR Curves and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 11. N=40 Shots Results: PR Curves and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 12. N=160 Shots Results: PR Curves and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 13. N=5120 Shots Results: PR Curves and Confidence Intervals, All Methods, Two by Two Comparison of Methods
References
- 1.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402-2410. doi: 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 2.Ting DSW, Liu Y, Burlina P, Xu X, Bressler NM, Wong TY. AI for medical imaging goes deep. Nat Med. 2018;24(5):539-540. doi: 10.1038/s41591-018-0029-3 [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Yao Q, Kwok J, Ni LM. Generalizing from a few examples: a survey on few-shot learning. Preprint. Posted April 10, 2019. Last revised March 29, 2020. arXiv 1904.05046. [Google Scholar]
- 4.Burlina PM, Joshi N, Pekala M, Pacheco KD, Freund DE, Bressler NM. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135(11):1170-1176. doi: 10.1001/jamaophthalmol.2017.3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Age-Related Eye Disease Study Research Group A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119(10):1417-1436. doi: 10.1001/archopht.119.10.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuadros J, Bresnick G. EyePACS: an adaptable telemedicine system for diabetic retinopathy screening. J Diabetes Sci Technol. 2009;3(3):509-516. doi: 10.1177/193229680900300315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faes L, Liu X, Wagner SK, et al. A clinician’s guide to artificial intelligence: how to critically appraise machine learning studies. Transl Vis Sci Technol. 2020;9(2):7. doi: 10.1167/tvst.9.2.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi RY, Coyner AS, Kalpathy-Cramer J, Chiang MF, Campbell JP. Introduction to machine learning, neural networks, and deep learning. Transl Vis Sci Technol. 2020;9(2):14. doi: 10.1167/tvst.9.2.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He K, Zhang X, Ren S, Sun J Deep residual learning for image recognition. Paper presented at: Proceedings of IEEE/CT Conference on Computer Vision and Pattern Recognition. 2016:770-778. [Google Scholar]
- 10.Hjelm RD, Fedorov A, Lavoie-Marchildon S, et al. Learning deep representations by mutual information estimation and maximization. Preprint. Posted August 20, 2018. Last revised February 22, 2019. arXiv 1808.06670. [Google Scholar]
- 11.Bachman P, Hjelm RD, Buchwalter W. Learning representations by maximizing mutual information across views. Preprint. Posted June 3, 2019. Last revised July 8, 2019. arXiv 1906.00910. [Google Scholar]
- 12.Hénaff OJ, Srinivas A, De Fauw J, et al. Data-efficient image recognition with contrastive predictive coding. Prepint. Posted May 22, 2019. Last revised July 1, 2020. arXiv 1905.09272. [Google Scholar]
- 13.Kingma DP, Ba J. Adam: a method for stochastic optimization. Preprint. Posted December 22, 2014. Last revised January 30, 2017. arXiv 1412.6980. [Google Scholar]
- 14.Parikh RB, Teeple S, Navathe AS. Addressing bias in artificial intelligence in health care. JAMA. 2019;22(24):2377-2378. doi: 10.1001/jama.2019.18058 [DOI] [PubMed] [Google Scholar]
- 15.Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366(6464):447-453. doi: 10.1126/science.aax2342 [DOI] [PubMed] [Google Scholar]
- 16.Mehrabi N, Morstatter F, Saxena N, Lerman K, Galstyan A. A survey on bias and fairness in machine learning. Preprint. Posted August 23, 2019. Last revised September 17, 2019. arXiv 1908.09635. [Google Scholar]
- 17.Burlina PM, Joshi N, Pacheco KD, Liu TYA, Bressler NM. Assessment of deep generative models for high-resolution synthetic retinal image generation of age-related macular degeneration. JAMA Ophthalmol. 2019;137(3):258-264. doi: 10.1001/jamaophthalmol.2018.6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul W, Wang I-J, Alajaji F, Burlina PM. Unsupervised semantic attribute discovery and control in generative models. Preprint. arXiv 11169. [Google Scholar]
- 19.Burlina PM, Joshi N, Wang I-J Where’s Wally now? deep generative and discriminative embeddings for novelty detection. Paper presented at: Proceedings of IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2019:11507-11516. [Google Scholar]
- 20.Chalapathy R, Chawla S. Deep learning for anomaly detection: a survey. Preprint. Posted January 10, 2019. Last revised January 23, 2019. arXiv 1901.03407. [Google Scholar]
- 21.Zhang BH, Lemoine B, Mitchell M Mitigating unwanted biases with adversarial learning. Paper presented at: Proceedings of the AAAI/ACM Conference on AI, Ethics, and Society. February 2018; New Orleans, LA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Performance of Algorithms: Accuracy
eTable 2. Performance of Algorithms: ROC AUC
eTable 3. Performance of Algorithms: F1 Score
eFigure 1. Accuracy for All Methods and Number of Shots
eFigure 2. Accuracy and Confidence Intervals for All Shots
eFigure 3. ROC AUC for All Shots
eFigure 4. ROC AUC for All Shots with 95% Confidence Intervals
eFigure 5. F1 Score for All Shots
eFigure 6. N=10 Shots Results: ROCs and Confidence Intervals, All, Two-Curve Comparisons of Methods
eFigure 7. N=40 Shots Results: ROCs and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 8. N=160 Shots Results: ROCs and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 9. N=5120 Shots Results: ROCs and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 10. N=10 Shots Results: PR Curves and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 11. N=40 Shots Results: PR Curves and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 12. N=160 Shots Results: PR Curves and Confidence Intervals, All Methods, Two by Two Comparison of Methods
eFigure 13. N=5120 Shots Results: PR Curves and Confidence Intervals, All Methods, Two by Two Comparison of Methods