Abstract
Diagnosis of visceral leishmaniasis (VL) relies on invasive and risky aspirate procedures, and confirmation of cure after treatment is unreliable. Detection of Leishmania donovani antigens in urine has the potential to provide both a non-invasive diagnostic and a test of cure. We searched for L. donovani antigens in urine of VL patients from India and Sudan to contribute to the development of urine antigen capture immunoassays. VL urine samples were incubated with immobilised anti-L. donovani polyclonal antibodies and captured material was eluted. Sudanese eluted material and concentrated VL urine were analysed by western blot. Immunocaptured and immunoreactive material from Indian and Sudanese urine was submitted to mass spectrometry for protein identification. We identified six L. donovani proteins from VL urine. Named proteins were 40S ribosomal protein S9, kinases, and others were hypothetical. Thirty-three epitope regions were predicted with high specificity in the 6 proteins. Of these, 20 were highly specific to Leishmania spp. and are highly suitable for raising antibodies for the subsequent development of an antigen capture assay. We present all the identified proteins and analysed epitope regions in full so that they may contribute to the development of non-invasive immunoassays for this deadly disease.
Introduction
Visceral leishmaniasis (VL) is most commonly caused by Leishmania donovani in the Indian subcontinent and eastern Africa, whereas L. infantum is the agent in the Mediterranean, Middle East and South America. Both species are transmitted by female phlebotomine sand flies and symptomatic infection is considered fatal if untreated, therefore accurate diagnosis is crucial to patient outcome. India, Bangladesh and Nepal are aiming to eliminate VL as a public health problem and this relies on rapid case detection and confirmation of cure after treatment [1].
Routine diagnosis of VL is based on serology, commonly using the recombinant rK39 or rK28 antigens, followed by microscopic visualisation of the parasite in spleen, bone marrow or lymph node aspirate as confirmation. Conventional serology, which detects anti-Leishmania IgG, has several drawbacks: it is ineffective at confirming cure or relapse because it can remain positive for many years after successful treatment [2–6]; it is also less reliable in HIV co-infected cases where a negative result does not rule out leishmaniasis [7]. Molecular assays are sometimes applied, and may gain increased importance during VL elimination, however, non-invasive antigen detection would complement this as a diagnostic tool [8].
An ideal diagnostic for both primary VL cases and validating cure is the detection of parasite material in non-invasive samples such as urine or saliva, or a serological test that is specific for active infection [9]. As well, there is the need for low-cost, rapid and equipment-free diagnostics that can be used in low-resource settings at point-of-care with minimal training. Such assays may detect parasite DNA, for example by loop-mediated isothermal amplification (LAMP) [10, 11] or by recombinase polymerase amplification (RPA) [12], or may detect parasite antigens.
Several urine antigen capture immunoassays have been developed with the best established being the KAtex, a latex particle agglutination test that detects a carbohydrate antigen [13, 14]. The KAtex has a specificity of 84–100%, but poorer sensitivity of 47–87% [15–18] with the drawback that urine samples must be boiled before testing. However, the test is rapid, giving a result in less than 10 minutes and becoming negative for most patients 30 days post-treatment [18]. In addition, this urine antigen assay has shown utility in HIV/VL co-infection [19, 20]. Monoclonal and polyclonal antibodies against the undefined antigen in the KAtex test were later adapted to ELISA format [21].
Other assays have been reported that detect particular protein antigens of L. infantum in urine [22]. This approach required first identifying Leishmania proteins in VL urine by mass spectrometry, expressing them as recombinant antigens and raising antibodies that could be produced as highly specific and sensitive polyclonal or monoclonal antibodies [22].
An alternative approach is to raise antibodies to lysed whole parasite cells, containing a wide diversity of antigens and to use these to capture a range of undefined antigens from VL patient urine. Vallur et al. [23] reported the development of an ELISA using an affinity purified polyclonal rabbit antibody against L. donovani whole cell lysate. The assay was optimised by those authors and developed into an ELISA kit that performed well in detecting urine antigen in VL patients from both L. infantum and L. donovani endemic regions. Here we have undertaken a study using this antibody and other polyclonal anti-Leishmania antibodies, and by mass spectrometry, we have identified antigens in Indian and Sudanese VL urine, for the development of antigen capture assays.
Materials and methods
Ethics statement
Ethical permission was granted by the LSHTM Ethical Review Committee with approval number 11478, and as part of the EC-funded NIDIAG project. In India, the collection of samples was approved by the Ethics Committee of Banaras Hindu University, Varanasi. In Sudan, approval was by the Ethical Research Committee, Faculty of Medicine, University of Khartoum and the National Health Research Ethics Committee, Federal Ministry of Health, Sudan. Written informed consent was obtained from adult subjects included in the study or from the parents or guardians of individuals less than 18 years of age.
Two rabbits were used here to raise antiserum. Rabbit sera were produced in compliance with UK Home Office regulations and the Animals (Scientific Procedures) Act 1986, in authorised animal facilities, by licensed staff, and in accord with European regulations and the 3R policy of Refinement, Reduction and Replacement.
Processing and analysis of samples
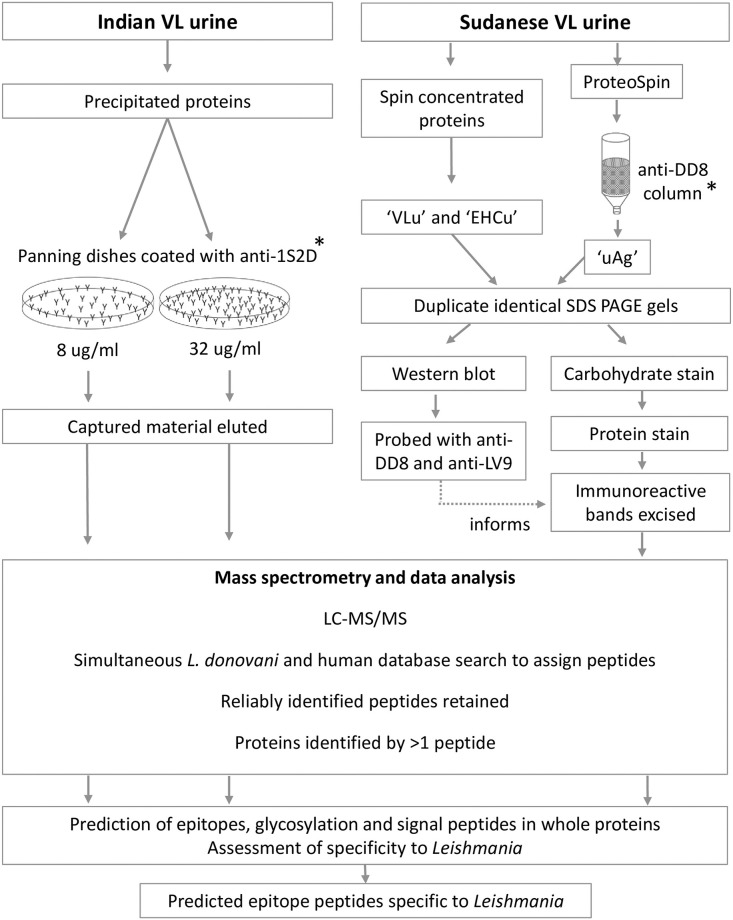
Fig 1 depicts the overall workflow of the urine samples in this study. We used two distinct methods in order to maximise the possibility of antigen capture.
Fig 1. Sample processing and analysis workflow used in this study.
Asterisks indicate immunocapture methods. Anti-1S2D, -DD8 and -LV9 are rabbit antibodies against these respective strains of L. donovani. VLu, concentrated proteins from VL urine; EHCu, concentrated proteins from urine of Endemic Healthy Controls; uAg, material captured from VL urine by anti-L. donovani antibodies; LC-MS/MS, liquid chromatography tandem mass spectrometry.
Urine samples
Urine samples were from VL patients and healthy controls in two distinct VL endemic regions, namely the Indian state of Bihar and the Sudanese state of Gedaref. India: urine samples were collected from VL patients attending the Kala-Azar Medical Research Centre (KAMRC) clinic in Muzaffarpur, Bihar. Sudan: VL and endemic healthy control (EHC) urine samples were collected from field sites in Gedaref. In this work Leishmania-derived antigens were enriched from VL urine using antibodies, prior to mass spectrometry.
Generation of rabbit anti-L. donovani antibodies
Vallur et al. [23] described the generation of a rabbit anti-L. donovani polyclonal antibody against whole promastigote lysate antigen, which was affinity purified against soluble promastigote lysate antigen of L. donovani strain MHOM/SD/00/1S-2D. This antibody is hereafter referred to as anti-1S2D.
Here, we prepared soluble promastigote lysate antigens of L. donovani strains MHOM/IN/80/DD8 and MHOM/ET/67/LV9 (also known as HU3), as described [24]. Log phase promastigotes were washed 3 times in PBS, pelleted by centrifugation, flash frozen in liquid nitrogen and thawed 3 times, sonicated (Soniprep 150, MSE, UK) at 12 microns intensity for 3 x 30 seconds at 2-minute intervals on ice and finally centrifuged at 12,000 x g for 1 minute at 4°C. The supernatants containing soluble antigens were retained and used subsequently.
Soluble antigen (DD8 and LV9) was used to raise antiserum in two rabbits that were immunised percutaneously with two doses of the respective lysate. The first inoculation was with 200 μg—500 μg of lysate antigen (originating from approximately 107 parasites) with Freund’s complete adjuvant (F5881, Sigma-Aldrich). A second inoculation, four weeks later, was with the same amount of antigen, plus Freund’s incomplete adjuvant (F5506, Sigma-Aldrich). Five millilitres of blood was taken before inoculations and about 50 ml five months after the second inoculation.
The resulting polyclonal IgG was purified from both DD8 and LV9 antisera on protein A agarose (P3476, Sigma, UK), eluted with 0.1 M glycine, pH 2.7 and immediately neutralised with 60 μl of 1 M Tris pH 9.0. The purified IgG elutions are hereafter referred to as anti-DD8 and anti-LV9. An ELISA was performed to confirm the reactivity of the two antibodies with L. donovani antigen and that the pre-immune sera were non-reactive.
Indian VL urine
Precipitation of urine proteins
Total protein was precipitated from Indian VL patient urine: approximately 130 ml of urine (83 ml fresh and 52 ml previously frozen) was combined from 31 Indian VL patients and protease inhibitor cocktail (P8340, Sigma-Aldrich) was added at a dilution of 1:500. The urine was centrifuged at 2,500 x g for 3 minutes to pellet cellular debris, which was discarded. To the supernatant, we added 85 g solid ammonium sulphate to achieve 90% saturation then incubated the solution for one hour at ambient temperature (about 28°C) with mixing to allow proteins to precipitate. The urine/ammonium sulphate solution was centrifuged in aliquots at 3,900 x g at 20°C for 40 minutes and the supernatant was discarded. Protein pellets were then resuspended as two aliquots in a total of 4.25 ml PBS then desalted using PD10 columns (170851–01, GE Healthcare) by the centrifugation protocol and each replicate made up to 3 ml with PBS.
Immunocapture of proteins
To capture L. donovani antigens from the precipitated Indian VL urine protein, we performed immuno-panning (Fig 1, upper left). Anti-1S2D antibody was coated onto 5 cm diameter plastic dishes at 8 μg/ml (Dish 8) and, to increase antigen capture, at 35 μg/ml (Dish 35) in 1.5 ml coating buffer (0.1 M NaHCO3, pH 8.6) by incubation at 4°C overnight. The unbound antibody was removed, replaced with blocking buffer (coating buffer containing 5 mg/ml bovine serum albumin) and the dishes were incubated at 4°C for 1 hour. After blocking, both dishes were washed 5 times using PBS containing 0.05% v/v Tween 20 (PBST), with gentle agitation.
The first aliquot of urine protein solution was incubated in Dish 8 for 1 hour at room temperature with agitation before the unbound material was moved into Dish 35. Dish 8 was washed 3 times with PBST then another 3 times with PBS before bound material was eluted with a 15 minute incubation with 1 ml of 0.1 M glycine pH 2.7. Eluate was pipetted into a tube containing 32 μl neutralising buffer (1 M Tris pH 9.0) and sodium azide to a final concentration of 0.02% (w/v) then stored at -80°C.
Unbound material from Dish 35 was transferred back to Dish 8 for repetition of the whole process. From each of the two aliquots of urine protein solution, two elutions were made from each of the two dishes (thus n = 8 eluates), to maximise the chance of capturing Leishmania antigens. After each elution, the dishes were washed 10 times with PBS prior to re-use.
Eluates containing captured proteins were pooled into two volumes: one from Dish 8, the other from Dish 35. These were buffer exchanged into PBS and spin concentrated at 5°C in 3 kDa molecular weight cut-off (MWCO) Amicon Ultra filters (UFC500324, Millipore) at 14,000 x g in several 20 minute spins until the total volume of each was reduced to 100 μl.
Sudanese VL urine
Concentration of urine proteins
Sudanese VL patients’ urine from seven individuals was pooled, 0.4 ml from each to give 2.8 ml, and spin concentrated to 190 μl in Amicon Ultra 3 kDa MWCO tubes, with a final protein concentration of 12.5 mg/ml (hereafter ‘VLu’ for VL urine). Pooled EHC urine, 0.5 ml each from 14 individuals, was spin concentrated as above, down to about 100 μl (EHCu). Both urine concentrates were then washed to remove salts by making up to 0.5 ml with PBS and spinning down five times. These two concentrates, VLu and EHCu, were run in SDS-PAGE as described further below (Fig 1, upper right).
Affinity chromatography
Separately, another pool of Sudanese VL urine comprising 0.5 ml each of 57 individual urines with 1/100 protease inhibitor (P8340, Sigma-Aldrich) was concentrated with a ProteoSpin Urine Protein Concentration Micro Kit (17400, Norgen Biotek Corp) as per manufacturer’s instructions and in batches to avoid overloading the capacity of the kit to a final volume of 3.7 ml. To capture L. donovani antigens in urine, rabbit anti-DD8 was coupled to a cyanogen bromide-activated Sepharose matrix (C9142, Sigma-Aldrich) following manufacturer’s instructions and the 1.75 ml of gel was loaded into a disposable column. Due to limited amounts of anti-LV9 and anti-1S2D antibodies, anti DD8 was used for chromatography. The column was equilibrated using ten column volumes of PBS prior to use. Urine protein concentrate was incubated on the anti-DD8 column for 10 minutes, drained, washed 3 times with 10 ml PBS and bound material was eluted in 8 x 1 ml volumes of 0.1 M glycine pH 2.7, neutralised with 32 μl neutralising buffer per 1 ml eluate. Eluted fractions indicated to contain antigen by a dot blot were spin concentrated to give ‘VL urine antigen (uAg)’. Briefly, the dot blot consisted of 2 μl of each fraction dried onto nitrocellulose, blocked with PBS + 3% w/v non-fat milk powder (Premier International Foods, Spalding, UK) (PBSM), probed with anti-DD8 and detected with anti-rabbit IgG-horseradish peroxidase (HRP) (711-035-152, Jackson Immunoresearch) and DAB/CN substrate, with details as described below.
SDS-PAGE and western blotting
Both of the Sudanese VL urine products, namely the urine concentrate (VLu) and immune-selected urine antigen (uAg), were subjected to non-reducing SDS-PAGE in duplicate lanes of a 10% acrylamide Tricine gel as described by Schägger [25]. Half of the gel was stained firstly for carbohydrate using Pro Q Emerald 300 (P21857, ThermoFisher Scientific) visualised at 280 nm (Gel Doc, ThermoFisher Scientific) then the same gel was stained for protein using Sypro Ruby (S12000, ThermoFisher Scientific) and visualised with 488 nm excitation and 610 nm emission wavelengths (Typhoon Trio phosphorimager, Amersham Biosciences).
The other identical gel half was transferred onto 0.2 μm pore size nitrocellulose (10600015, GE Healthcare) at 160 mA for 120 minutes and air-dried. The membrane was then blocked using PBSM overnight at 4°C then washed in PBST, three times for 10 min each. Rabbit anti-DD8 and anti-LV9 were conjugated to HRP using a Lightning Link kit (701–0000, Expedeon) then diluted together at 1:400 and 1:500 respectively in PBST + 3% milk powder (PBSTM) and incubated on the blot for 2 hours with gentle agitation. The membrane was washed with PBST six times for 5 minutes each before addition of DAB/CN substrate solution (30 μg 4-chloro-1-naphthol dissolved in 5 ml methanol then added to 40 ml PBS, followed by the addition of 10 μg of 3,3’-diaminobenzidine dissolved in 5 ml methanol and finally 15 μl of 30% H2O2). The blot was incubated for 15 mins with gentle agitation in the dark before stopping the reaction by washing in deionised water several times.
Due to limited signal gained from these rabbit polyclonals conjugated directly to HRP, the blot was stripped using stripping buffer (0.2 M glycine, 0.1% SDS, 1% Tween 20, pH 2.2) for 10 min with agitation, followed by 2 x 10 min washes in PBS and 2 x 5 min washes in Tris buffered saline with Tween (20 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 7.6) (TBST). After stripping away the previous reaction, the blot was re-blocked and probed with the same two rabbit anti-L. donovani antibodies without HRP conjugation, followed by anti-rabbit IgG-HRP at 1:1000 and DAB/CN substrate as before. Bands in the carbohydrate/protein-stained gel half, corresponding to immunoreactive blot bands in the two VL urine sample types, were excised for mass spectrometry.
Mass spectrometry
The excised gel bands from Sudanese VLu and uAg, as well as the two Indian VL urine eluates from the panning dishes, were submitted to trypsin digest and liquid chromatography tandem mass spectrometry (LC-MS/MS) at the Advanced Proteomics Facility, Oxford University, UK.
For all samples, mass spectra were assigned to L. donovani peptides by a simultaneous search using Mascot (Matrix Science, UK) against both the L. donovani BPK282A1 reference [26] and UniProt Homo sapiens protein databases, which contained all the proteins of these organisms, as deduced from their genomes [27]. Criteria for identifying Leishmania peptides were being rank 1 (the peptide with the best match for a particular mass spectrum) and having a ppm of -10 to 10 (error on experimental peptide mass values, fraction expressed as parts per million). Proteins were identified when they contained 2 or more different peptides matching the above criteria and were taken forward to further analysis (Fig 1, lower panel). Once identified, peptides and proteins from VLu and uAg from Sudanese samples were considered together for subsequent analysis of their properties, as detailed below.
Epitope prediction
Epitope prediction was carried out to indicate short, Leishmania-specific peptides within the identified proteins that may be more easily synthesised for proceeding with antibody generation and test development. The complete amino acid sequences of proteins identified by 2 or more peptides were obtained through UniProtKB [27] and submitted to B-cell epitope prediction by BepiPred 2.0 [28]. The epitope score threshold was set at 0.65 (on a scale of 0 to 1), which gave a specificity of 99% and sensitivity of 2%, with an alternative threshold of 0.55 for higher sensitivity (specificity 81.6%, sensitivity 29%). Minimum length was 8 residues, with no maximum.
Specificity of proteins and epitopes to L. donovani
Specificity of the mass spectrometry-identified peptides to Leishmania was ensured by a simultaneous search of the data against both human and Leishmania proteomes, with only those matching to Leishmania being taken forward to ensure that they were not chance matches to human peptides. This was followed by sequence analysis to confirm genus specificity. Proteins, and predicted epitope peptides from these proteins, were assessed for sequence similarity to other species using a BLASTP search against the NCBI non-redundant (nr) database with no species restriction and using the default settings. For epitope sequences, the BLASTP search was optimised for short sequences. BLAST output was assessed for the sequence identity between the query and biologically-relevant species, i.e., human pathogens and commensals. Sequence similarity to other Leishmania species was tolerated if L. donovani was a complete match.
Signal peptide prediction
Proteins were submitted to SignalP 4.1 [29] to predict the presence of a signal peptide, which provided evidence that the protein may be secreted from the cell by this method, although Leishmania also has other secretory pathways. SignalP 4.1 is optimised to distinguish between transmembrane domains and signal peptides [29].
Glycosylation prediction
N-linked glycosylation of proteins was predicted using NetNGlyc [30], which identified potential glycosylation sites via the R-group nitrogen atom of asparagine in the Asn-X-Ser/Thr motif, where X could be any amino acid residue except Pro. Glycosylation could indicate that the protein sequence underlying the glycan is inaccessible to capture antibodies, or that the glycan itself could be investigated as a diagnostic antigen.
Protein properties
The possible identities of hypothetical proteins identified by mass spectrometry were sought using online tools that identify domains and protein features based on sequence similarity with known proteins: NCBI domain enhanced lookup time (DELTA) BLAST tool [31]; InterPro [32]; BlastKOALA (KEGG Orthology and Links Annotation) via KEGG [33]. Additional information on protein features was from Expasy Prosite [34] and published literature.
Results
Using mass spectrometry, we identified four L. donovani proteins in Indian VL urine and two in Sudanese VL urine. Epitope prediction returned 33 B-cell epitopes of 8 or more amino acids, within the complete protein sequences of the 6 proteins. Of the 33, 20 had complete identity only with Leishmania spp. in a BLAST search of the NCBI nr database of all organisms. Details of peptides and proteins from each urine source are described further below. Full protein sequences, annotated with the detected peptides and predicted epitopes, are shown in S1 Fig.
Indian VL urine antigen capture
In total from Indian VL urine, 19 different peptides were identified as those of L. donovani after a search of the mass spectra against L. donovani and human protein databases simultaneously; 11 and 12 from each panning dish respectively, with four present in both dish eluates (S1 Table).
Four proteins, one of which occurred on both panning dishes, were confidently identified by more than one peptide (Table 1). In addition to proteins, 10 solo peptides (i.e., the only representatives of their parent protein) were identified with a highly reliable ID by mass spectrometry and high specificity to L. donovani (S2 Table).
Table 1. Leishmania donovani proteins identified by more than one peptide in urine of Indian and Sudanese visceral leishmaniasis patients.
| Sample | Origin of peptide | Leishmania peptides identified by MS | Parent protein (UniProtKB/GenBank) | Predicted epitope sequences with high specificity to Leishmania (aa length) |
|---|---|---|---|---|
| Indian VL urine | Dish 8 | EYEELR; ALAEGQER; AKAEAEAAR | Hypothetical protein (LdBPK_191140 / XP_003860289.1) | VDDRTHREA [9] |
| QRQRQHAHA [9] | ||||
| RRQRHTSP [8] | ||||
| RNRPESSH [8] | ||||
| Dishes 8 & 35 | LSRSMEVR; LSSVQAGEVR | 40S ribosomal protein S9 (LdBPK_070760 / XP_003865205.1) | SSRRASTTKPGPPPRAS [17] | |
| GMQLVGELNDSLD [13] | ||||
| LDQQPSVGTTT [11] | ||||
| Dish 35 | FLDKLR; RSSQSSTSATYR | Hypothetical protein (LdBPK_323250 / XP_003863736.1) | SDNGASPGSRSPRSSRRSSQSSTS [24] | |
| SPAHQRSRAGASRSASRQG [19] | ||||
| STKRPRQSAVYG [12] | ||||
| Dish 35 | ALISPSVLR; LSDAPRVCR | Protein kinase (LdBPK_262110 / XP_003861796.1) | NSSSYSGSLGSPAS [14] | |
| VSPVRRNSSSTAL [13] | ||||
| ANGGNSSSNSYT [12] | ||||
| QQQQQSSNRPS [11] | ||||
| AGTARLGSSS [10] | ||||
| RSTPRAGMP [9] | ||||
| Sudanese VL urine | VLu | EFVVSGAALR; ITDMQREIR | Hypothetical protein (LdBPK_160110 / XP_003859699.1) | VRFRPNASLADGDAKSSAHGTVTQYGSPA‡ [29] |
| VLu | ITSDEVLR; TVNEDLSR | Protein kinase (LdBPK_351070 / XP_003864692.1) | ANDDSESATRVEGLQVMSDINSIPL [25] | |
| DGQQIKVSSSGGGSSSKGSSNSTGS [25] | ||||
| KEERQRMHA [9] |
VLu: concentrated proteins from VL patient urine; aa: amino acid.
‡ This peptide was identified by the lower epitope score threshold of 0.55 because the protein did not contain epitopes >8 residues at a threshold of 0.65.
Sudanese VL urine antigen capture
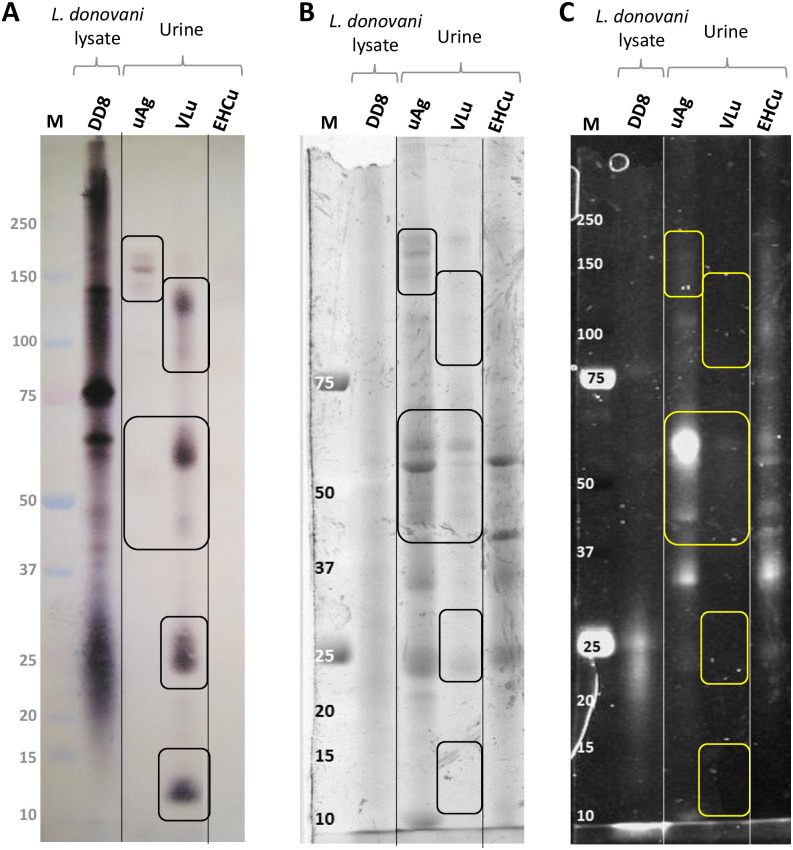
L. donovani peptides and proteins were identified by mass spectrometry of excised gel bands corresponding to immunocaptured urine antigen (uAg) and immunoreactive VL urine components (VLu) (Fig 2). Protein and carbohydrate staining of the SDS PAGE gel revealed that very few of these antigens were glycoproteins (Fig 2B and 2C).
Fig 2. Sudanese VL patients’ urine (VLu) and immunocaptured urine antigen (uAg) detected by rabbit anti-L. donovani DD8 and -LV9 by western blot.
Regions submitted to mass spectrometry are broadly indicated by boxes. A) western blot, B) corresponding gel stained for proteins, C) the same gel as B, stained for carbohydrates. Molecular mass marker (M) sizes in kiloDaltons. Lanes are: lysate antigen of L. donovani strain DD8; uAg, VL urine material eluted from an anti-L. donovani DD8 immunochromatography column; VLu, concentrated VL urine; EHCu, concentrated urine from endemic healthy controls. Vertical lines indicate where the photographs were spliced to remove non-relevant lanes from the original images, and realigned based on the molecular weight marker.
Mass spectrometry of the Sudanese VL urine material (uAg and VLu) excised from the SDS PAGE gel confidently (>1 peptide) identified two proteins and nine additional solo peptides (Table 1 and S2 Table). The peptides with a confident mass spectrometry ID had complete identity only to proteins from Leishmania genus according to a BLAST search, but part of some sequences occurred in other human pathogens or commensals. Specificity to Leishmania was improved when two peptides from the same protein were considered together.
Specificity of proteins to L. donovani
In total, we identified six proteins with confidence (>1 peptide) from Indian (4 proteins) and Sudanese (2 proteins) VL urine that showed high specificity to Leishmania spp., although not necessarily to L. donovani (Table 1). From Indian VL urine, protein kinase (LdBPK_262110) and hypothetical proteins (LdBPK_191140, LdBPK_323250) showed very high sequence similarity to several Leishmania species including L. infantum and species causing cutaneous leishmaniasis, and moderate identity to Trypanosoma cruzi proteins. The 40S ribosomal protein S9 (LdBPK_070760) also had high sequence identity to that of other Leishmania species, but little to any other genera.
The two L. donovani proteins identified in Sudanese VL urine, hypothetical protein (LdBPK_160110) and protein kinase (LdBPK_351070), also had very high homology to L. infantum with minor sequence differences between these and other Leishmania species. Moderate homology was found between the hypothetical protein and those of T. cruzi, whereas the protein kinase had high homology with T. cruzi proteins.
In addition to the proteins, 19 solo peptides were identified in total by all methods, 10 from Indian and 9 from Sudanese VL urine (S2 Table). Seven of these were from named proteins and all others were from hypothetical proteins. Interestingly, there was no overlap between L. donovani peptides from Indian and Sudanese VL urine.
Epitope prediction
All 6 Leishmania proteins identified in the Indian and Sudanese VL urine were submitted to epitope prediction using BepiPred 2.0 and together contained 33 peptides at a score threshold of 0.65 which optimised specificity and 119 epitopes at a threshold score of 0.55, which provided higher sensitivity (S3 Table). None of the 6 proteins contained signal peptides and 4 contained potential N-linked glycosylation sites (S3 Table).
Potential VL urine antigens
The 33 epitope peptides predicted with high specificity within the identified Leishmania proteins, and one additional with lower specificity, were assessed for specificity to L. donovani. Twenty of the 34 epitope sequences were selected as they had complete sequence identity to L. donovani and little or no identity to sequences from other relevant species i.e., human pathogens or commensals. Specific peptides or possible epitope regions indicated for production of antibodies for use in antigen capture assays are detailed in Table 1.
Discussion
Validated tests for urine antigen in infectious diseases include those for schistosomiasis [35], tuberculosis [36], Legionella pneumophila and Streptococcus pneumoniae [37], among others. Here, we captured L. donovani antigens from VL patient urine by two methods using immobilised anti-L. donovani antibodies in panning and chromatography. Both methods yielded parasite peptides which we identified by mass spectrometry. These led to identification of six L. donovani proteins from urine of VL patients from India and Sudan. The proteins were predicted to contain highly species-specific epitopes that therefore make good candidates for targets of a non-invasive urine antigen capture immunoassay for VL. We additionally identified 19 single peptides with very high identity to L. donovani, which provide evidence of additional parasite proteins in VL urine.
Studies using similar methods to identify pathogen antigens in urine include those of Abeijon et al. [22, 38] who identified L. infantum proteins tryparedoxin peroxidase, superoxide dismutase and nuclear transport factor 2, by mass spectrometry of concentrated Brazilian VL urine excised from gel bands, as well as L. donovani proteins encoded by genes Ld-mao1, Ld-ppi1, Ld-clp1 and Ld-mad1 from Indian and Kenyan VL urine. We did not identify any of these proteins, and in addition, none of the peptides and proteins that we found occurred in both Indian and Sudanese samples. This suggests that there could be many proteins or protein fragments of parasite origin in the urine of VL patients. However, differences between the studies were that we used an antibody (1S2D) to capture antigens while other studies did not. We searched mass results against a single genome database to identify peptides, which may have limited our findings, whereas Abeijon et al used a wide range of genomes [38]. However, we performed a rigorous quality control in order to exclude false peptide matches to Leishmania of peptides that were actually of human origin.
The proteins identified here are likely to be intracellular, based on their identities and features. However, Leishmania has various secretion pathways, therefore the proteins may be secreted by non-classical mechanisms [39]. Four proteins did contain potential glycosylation sites, a feature more common to surface proteins involved directly in host-parasite interactions [40, 41]. The carbohydrate and protein staining of the SDS-PAGE gel also indicated that very few proteins were glycosylated, and those that were, were not detected by antibodies on the corresponding blot. This may be expected as the rabbit antibodies were raised against a soluble lysate antigen from a preparation method that favours intracellular contents rather than cell membranes. By comparison, the KAtex assay that detects a carbohydrate antigen uses an antibody raised against whole intact parasites [13].
Three of the 6 proteins identified in VL urine were hypothetical proteins, defined by the presence of start and stop codons in their genomic sequences but without experimental evidence for the protein itself. These proteins were submitted to several protein domain identification tools to reveal possible similarities to well characterised proteins and elucidate their features and possible functions in Leishmania (Table 2).
Table 2. Functions of Leishmania donovani proteins identified in urine of Indian and Sudanese patients with visceral leishmaniasis.
| VL urine origin | Protein accession number | Features | Functions |
|---|---|---|---|
| India | LdBPK_191140a | Predicted zinc finger RING-type domain; predicted coil regions | Ubiquitination pathway and other intracellular protein processing pathways. |
| India | LdBPK_323250a | DENN domain | Involved in GTP/GDP exchange and occur in other proteins that regulate membrane traffic in eukaryotes. |
| India | LdBPK_070760 | 40S ribosomal S9 protein | Protein subunit of the 40S ribosomal subunit. |
| India | LdBPK_262110 | Protein kinase | Add phosphate groups in cell signalling pathways. |
| Sudan | LdBPK_351070 | Protein kinase | Add phosphate groups in cell signalling pathways. |
| Sudan | LdBPK_160110a | MORN repeat motif | Unknown. |
a hypothetical protein.
The great difference in western blot and SDS-PAGE gel profiles between the concentrated Sudanese VL urine (VLu) and urine antigens (uAg) purified from the same sample type was unexpected. However, the sensitivity of antibodies used may detect proteins that are not readily visible on the gel. In addition, the process of stripping and re-probing the blot could have somewhat altered the composition of the blot. However, the EHCu remained negative, indicating very little non-specific reaction. Heating the urine, which we did not do, could have led to more specific reactions [42], however, this favours carbohydrate antigens as it denatures proteins and we sought to retain as many of the conformational protein epitopes as possible by running non-reduced samples on gels.
Coiled protein regions are often made by repeats of a few of the same amino acids, as identified here in hypothetical protein LdBPK_191140, and this repetition can lead to high antigenicity, making them good targets for immunodiagnostics [43–45]. The geographic overlap of VL with Chagas disease, caused by T. cruzi, particularly in Brazil, makes it imperative to identify a Leishmania antigen that will not cross-react with this other trypanosomatid. Although the proteins identified here had very high sequence identity with L. donovani or L. infantum, some had moderate and one had high homology with T. cruzi proteins. Therefore, a polyclonal antibody against a complete L. donovani protein could also react with T. cruzi proteins. For this reason, as done here, selecting shorter and more species-specific peptide sequences is preferable because these contain fewer epitopes and generate a more specific antibody response, although it is unclear whether these epitopes would be linear or conformational.
Based on the species specificity of the epitope regions, we suggest that combining several of these, either for raising a mixed polyclonal antiserum, or later combining individual antibodies in the assay, may improve specificity and sensitivity of the prospective assay. This was found by Abeijon et al. [22, 46] where combined assays had improved performance over individual analyte assays.
A strength of our study was that the criteria for identifying parasite proteins in VL urine took into account the large number and amount of human proteins in the sample [47]. Initial steps to enrich for Leishmania antigens using specific antibodies assisted in achieving peptide identification by mass spectrometry. Further, by searching mass data simultaneously against both human and L. donovani protein databases, we were able to exclude mass spectra that had a better match to human peptide sequences, thus excluding potential false positive matches to L. donovani. The dual searching did not exclude possible matches to bacterial peptides, therefore we searched both the peptides and the proteins against the NCBI nr database to exclude this possibility.
A potential limitation to our technique is that while we have presented L. donovani protein identities here, there is no certainty that these proteins exist as whole proteins in VL urine as we can only identify short peptides by mass spectrometry. However, the presence of the peptides suggests that at least fragments of the proteins do occur in urine, along with many host-derived proteins [47, 48]. We used a large volume of urine from multiple VL patients in order to capture the peptides that were identified. However, with more abundant samples, it would have been beneficial to conduct replicates to investigate the frequency of these peptides in VL urine. Progression from identifying VL urinary antigens to having a prototype rapid diagnostic test (RDT) relies on follow-up to synthesise antigens and raise antibodies. An alternative method to develop monoclonal antibodies directly against precipitated urine proteins could also be attempted, followed by screening for sensitivity and specificity for VL, as used for malaria [49].
Conclusions
We used various methods to capture L. donovani antigens from VL urine from India and Sudan, including panning of urine proteins with immobilised anti-L. donovani antibodies and visualisation of immunoreactive bands in VL urine on a western blot. All methods yielded L. donovani proteins by mass spectrometry of captured or immunoreactive material and 5 of 6 proteins were highly specific to Leishmania. In addition, epitope prediction revealed 20 Leishmania-specific B-cell epitopes that we present here, that make ideal candidates for synthesis and to generate antiserum for antigen capture assay development.
Supporting information
(PDF)
Bold, underlined text indicates peptides identified in urine by mass spectrometry. Boxed text indicates predicted epitopes within each protein.
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We express our profound regard and gratitude to the late Professor Marleen Boelaert of the Institute of Tropical Medicine, Antwerp, Belgium, for initiating and so generously and effectively leading the NIDIAG project, of which this work was part. We also thank Osman Ahmed, Osama Eisa and Alfarazdeg Saad for sample collection in Sudan.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
TM was funded by the Sir Halley Stewart Trust (http://www.sirhalleystewart.org.uk/). The views expressed within this report are those of the authors and not necessarily those of the Trust. TM was additionally supported by the John Henry Memorial Fund (UK Charity number: 1118007). This work was part of the NIDIAG network research partnership supported by the European Commission under the Health Cooperation Work Programme of the 7th Framework Programme (Grant agreement no. 260260, https://cordis.europa.eu/project/id/260260). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Regional Strategic Framework for elimination of kala-azar from the South East Asia Region (2011–2015). 2012.
- 2.De Almeida Silva L, Romero HD, Prata A, Costa RT, Nascimento E, Carvalho SF, et al. Immunologic tests in patients after clinical cure of visceral leishmaniasis. Am J Trop Med Hyg. 2006;75(4):739–43. 10.4269/ajtmh.2006.75.739 [DOI] [PubMed] [Google Scholar]
- 3.Gidwani K, Picado A, Ostyn B, Singh SP, Kumar R, Khanal B, et al. Persistence of Leishmania donovani antibodies in past visceral leishmaniasis cases in India. Clin Vaccine Immunol. 2011;18(2):346–8. 10.1128/cvi.00473-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh DP, Sundar S, Mohapatra TM. The rK39 strip test is non-predictor of clinical status for kala-azar. BMC Res Notes. 2009;2:187 10.1186/1756-0500-2-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zijlstra EE, Nur Y, Desjeux P, Khalil EA, El-Hassan AM, Groen J. Diagnosing visceral leishmaniasis with the recombinant K39 strip test: experience from the Sudan. Trop Med Int Health. 2001;6(2):108–13. 10.1046/j.1365-3156.2001.00680.x [DOI] [PubMed] [Google Scholar]
- 6.Reiter-Owona I, Rehkaemper-Schaefer C, Arriens S, Rosenstock P, Pfarr K, Hoerauf A. Specific K39 antibody response and its persistence after treatment in patients with imported leishmaniasis. Parasitol Res. 2015:1–9. 10.1007/s00436-015-4801-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cota GF, de Sousa MR, Demarqui FN, Rabello A. The diagnostic accuracy of serologic and molecular methods for detecting visceral leishmaniasis in HIV infected patients: meta-analysis. PLoS Negl Trop Dis. 2012;6(5):e1665 10.1371/journal.pntd.0001665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundar S, Singh OP. Molecular diagnosis of visceral leishmaniasis. Mol Diagn Ther. 2018;22(4):443–57. 10.1007/s40291-018-0343-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Technical Report Series. 975. Research priorities for Chagas disease, human African trypanosomiasis and leishmaniasis. Geneva, Switzerland: World Health Organization, 2012. [PubMed]
- 10.Verma S, Singh R, Sharma V, Bumb RA, Negi NS, Ramesh V, et al. Development of a rapid loop-mediated isothermal amplification assay for diagnosis and assessment of cure of Leishmania infection. BMC Infect Dis. 2017;17(1):223 10.1186/s12879-017-2318-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukhtar M, Ali SS, Boshara SA, Albertini A, Monnerat S, Bessell P, et al. Sensitive and less invasive confirmatory diagnosis of visceral leishmaniasis in Sudan using loop-mediated isothermal amplification (LAMP). PLoS Negl Trop Dis. 2018;12(2):e0006264 10.1371/journal.pntd.0006264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mondal D, Ghosh P, Khan MA, Hossain F, Bohlken-Fascher S, Matlashewski G, et al. Mobile suitcase laboratory for rapid detection of Leishmania donovani using recombinase polymerase amplification assay. Parasit Vectors. 2016;9(1):281 10.1186/s13071-016-1572-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attar ZJ, Chance ML, el-Safi S, Carney J, Azazy A, El-Hadi M, et al. Latex agglutination test for the detection of urinary antigens in visceral leishmaniasis. Acta Trop. 2001;78(1):11–6. [DOI] [PubMed] [Google Scholar]
- 14.Sarkari B, Chance M, Hommel M. Antigenuria in visceral leishmaniasis: detection and partial characterisation of a carbohydrate antigen. Acta Trop. 2002;82(3):339–48. [DOI] [PubMed] [Google Scholar]
- 15.Sundar S, Agrawal S, Pai K, Chance M, Hommel M. Detection of leishmanial antigen in the urine of patients with visceral leishmaniasis by a latex agglutination test. Am J Trop Med Hyg. 2005;73(2):269–71. [PubMed] [Google Scholar]
- 16.Diro E, Techane Y, Tefera T, Assefa Y, Kebede T, Genetu A, et al. Field evaluation of FD-DAT, rK39 dipstick and KATEX (urine latex agglutination) for diagnosis of visceral leishmaniasis in northwest Ethiopia. Trans R Soc Trop Med Hyg. 2007;101(9):908–14. 10.1016/j.trstmh.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 17.Rijal S, Boelaert M, Regmi S, Karki BMS, Jacquet D, Singh R, et al. Evaluation of a urinary antigen-based latex agglutination test in the diagnosis of kala-azar in eastern Nepal. Trop Med Int Health. 2004;9(6):724–9. 10.1111/j.1365-3156.2004.01251.x [DOI] [PubMed] [Google Scholar]
- 18.Salam MA, Khan MG, Mondal D. Urine antigen detection by latex agglutination test for diagnosis and assessment of initial cure of visceral leishmaniasis. Trans R Soc Trop Med Hyg. 2011;105(5):269–72. 10.1016/j.trstmh.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 19.Riera C, Fisa R, Lopez P, Ribera E, Carrio J, Falco V, et al. Evaluation of a latex agglutination test (KAtex) for detection of Leishmania antigen in urine of patients with HIV-Leishmania coinfection: value in diagnosis and post-treatment follow-up. Eur J Clin Microbiol Infect Dis. 2004;23(12):899–904. 10.1007/s10096-004-1249-7 [DOI] [PubMed] [Google Scholar]
- 20.Vogt F, Mengesha B, Asmamaw H, Mekonnen T, Fikre H, Takele Y, et al. Antigen detection in urine for noninvasive diagnosis and treatment monitoring of visceral leishmaniasis in human immunodeficiency virus coinfected patients: an exploratory analysis from Ethiopia. Am J Trop Med Hyg. 2018. 10.4269/ajtmh.18-0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarkari B, Chance M, Hommel M. A capture ELISA for the diagnosis of visceral leishmaniasis using a monoclonal antibody against a leishmanial. Iran Biomed J. 2005;9(3):117–22. [Google Scholar]
- 22.Abeijon C, Alves F, Monnerat S, Wasunna M, Mbui J, Viana AG, et al. Development of a multiplexed assay for detection of Leishmania donovani and Leishmania infantum protein biomarkers in urine samples of patients with visceral leishmaniasis. J Clin Microbiol. 2019;57(5):e02076–18. 10.1128/jcm.02076-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallur AC, Tutterrow YL, Mohamath R, Pattabhi S, Hailu A, Abdoun AO, et al. Development and comparative evaluation of two antigen detection tests for Visceral Leishmaniasis. BMC Infect Dis. 2015;15(1):384 10.1186/s12879-015-1125-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharyya T, Ayandeh A, Falconar AK, Sundar S, El-Safi S, Gripenberg MA, et al. IgG1 as a potential biomarker of post-chemotherapeutic relapse in visceral leishmaniasis, and adaptation to a rapid diagnostic test. PLoS Negl Trop Dis. 2014;8(10):e3273 10.1371/journal.pntd.0003273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1(1):16–22. 10.1038/nprot.2006.4 [DOI] [PubMed] [Google Scholar]
- 26.Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, et al. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21(12):2143–56. 10.1101/gr.123430.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D69. 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45(W1):W24–W9. 10.1093/nar/gkx346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–6. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 30.Gupta R, Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 2002:310–22. [PubMed] [Google Scholar]
- 31.Boratyn GM, Schaffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. Domain enhanced lookup time accelerated BLAST. Biol Direct. 2012;7:12 10.1186/1745-6150-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–d9. 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41(Database issue):D344–7. 10.1093/nar/gks1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corstjens PL, De Dood CJ, Kornelis D, Fat EM, Wilson RA, Kariuki TM, et al. Tools for diagnosis, monitoring and screening of Schistosoma infections utilizing lateral-flow based assays and upconverting phosphor labels. Parasitology. 2014;141(14):1841–55. 10.1017/s0031182014000626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suwanpimolkul G, Kawkitinarong K, Manosuthi W, Sophonphan J, Gatechompol S, Ohata PJ, et al. Utility of urine lipoarabinomannan (LAM) in diagnosing tuberculosis and predicting mortality with and without HIV: prospective TB cohort from the Thailand Big City TB Research Network. Int J Infect Dis. 2017;59:96–102. 10.1016/j.ijid.2017.04.017 [DOI] [PubMed] [Google Scholar]
- 37.Athlin S, Iversen A, Ozenci V. Comparison of the ImmuView and the BinaxNOW antigen tests in detection of Streptococcus pneumoniae and Legionella pneumophila in urine. Eur J Clin Microbiol Infect Dis. 2017;36(10):1933–8. 10.1007/s10096-017-3016-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abeijon C, Kashino SS, Silva FO, Costa DL, Fujiwara RT, Costa CH, et al. Identification and diagnostic utility of Leishmania infantum proteins found in urine samples from patients with visceral leishmaniasis. Clin Vaccine Immunol. 2012;19(6):935–43. 10.1128/cvi.00125-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverman JM, Chan SK, Robinson DP, Dwyer DM, Nandan D, Foster LJ, et al. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 2008;9(2):R35 10.1186/gb-2008-9-2-r35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anish C, Martin CE, Wahlbrink A, Bogdan C, Ntais P, Antoniou M, et al. Immunogenicity and diagnostic potential of synthetic antigenic cell surface glycans of Leishmania. ACS Chem Biol. 2013;8(11):2412–22. 10.1021/cb400602k [DOI] [PubMed] [Google Scholar]
- 41.Rodrigues JA, Acosta-Serrano A, Aebi M, Ferguson MA, Routier FH, Schiller I, et al. Parasite glycobiology: a bittersweet symphony. PLoS Pathog. 2015;11(11):e1005169 10.1371/journal.ppat.1005169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brito-Santos F, Ferreira MdF, Trilles L, Muniz MdM, Veloso dos Santos VG, Carvalho-Costa FA, et al. Preheating of urine improves the specificity of urinary cryptococcal antigen testing using the lateral flow assay. PLoS Negl Trop Dis. 2017;11(5):e0005304 10.1371/journal.pntd.0005304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burns JM, Shreffler WG, Benson DR, Ghalib HW, Badaro R, Reed SG. Molecular characterization of a kinesin-related antigen of Leishmania chagasi that detects specific antibody in African and American visceral leishmaniasis. Proc Natl Acad Sci U S A. 1993;90(2):775–9. 10.1073/pnas.90.2.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goto Y, Carter D, Reed SG. Immunological dominance of Trypanosoma cruzi tandem repeat proteins. Infect Immun. 2008;76(9):3967–74. 10.1128/iai.00604-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goto Y, Coler RN, Reed SG. Bioinformatic identification of tandem repeat antigens of the Leishmania donovani complex. Infect Immun. 2007;75(2):846–51. 10.1128/iai.01205-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abeijon C, Campos-Neto A. Potential non-invasive urine-based antigen (protein) detection assay to diagnose active visceral leishmaniasis. PLoS Negl Trop Dis. 2013;7(5):e2161 10.1371/journal.pntd.0002161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santucci L, Candiano G, Petretto A, Bruschi M, Lavarello C, Inglese E, et al. From hundreds to thousands: widening the normal human urinome. J Proteom. 2014;(0). 10.1016/j.jprot.2014.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adachi J, Kumar C, Zhang Y, Olsen J, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7(9):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Markakpo US, Bosompem KM, Dzodzomenyo M, Danso-Appiah A, Essuman EE, Anyan WK, et al. Minimising invasiveness in diagnostics: developing a rapid urine-based monoclonal antibody dipstick test for malaria. Trop Med Int Health. 2016;21(10):1263–71. 10.1111/tmi.12744 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Bold, underlined text indicates peptides identified in urine by mass spectrometry. Boxed text indicates predicted epitopes within each protein.
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.