Abstract
Few human pathogens have been the focus of as much concentrated worldwide attention as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of COVID-19. Its emergence into the human population and ensuing pandemic came on the heels of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), two other highly pathogenic coronavirus spillovers, which collectively have reshaped our view of a virus family previously associated primarily with the common cold. It has placed intense pressure on the collective scientific community to develop therapeutics and vaccines, whose engineering relies on a detailed understanding of coronavirus biology. Here, we present the molecular virology of coronavirus infection, including its entry into cells, its remarkably sophisticated gene expression and replication mechanisms, its extensive remodeling of the intracellular environment, and its multifaceted immune evasion strategies. We highlight aspects of the viral life cycle that may be amenable to antiviral targeting as well as key features of its biology that await discovery.
Keywords: plus-stranded RNA virus, virology, coronavirus, innate immunity, viral replication, virus entry, endoplasmic reticulum (ER), virus, pathogenesis, RNA polymerase, cellular immune response, endoplasmic reticulum, SARS-CoV-2
The Coronaviridae family of viruses are enveloped, single-stranded positive-sense RNA viruses grouped into four genera (alphacoronavirus, betacoronavirus, gammacoronavirus, and deltacoronavirus) that primarily infect birds and mammals, including humans and bats. The seven coronaviruses known to infect humans fall within the alpha- and betacoronavirus genera, whereas gamma- and deltacoronaviruses primarily infect birds. Coronaviruses have been studied for decades using the model betacoronavirus, murine hepatitis virus (MHV), and the human alphacoronavirus HCoV-229E. In humans, the circulating coronaviruses HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1 generally cause mild upper respiratory illness and collectively are associated with 10–30% of common cold cases (1). However, within the past two decades, three highly pathogenic coronaviruses have emerged into the human population as the result of spillover events from wildlife that can cause severe respiratory illness: severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002, Middle East respiratory syndrome coronavirus (MERS-CoV) emerged in 2011, and most recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in 2019. These outbreaks, together with estimates suggesting that hundreds to thousands of additional coronaviruses may reside in bats alone (2), highlight the potential for future coronavirus zoonotic transmission.
In this article, we provide an overview of the coronavirus life cycle with an eye toward its notable molecular features and potential targets for therapeutic interventions (Fig. 1). Much of the information presented is derived from studies of the betacoronaviruses MHV, SARS-CoV, and MERS-CoV, with a rapidly expanding number of reports on SARS-CoV-2. The first portion of the review focuses on the molecular basis of coronavirus entry and its replication cycle. We highlight several notable properties, such as the sophisticated viral gene expression and replication strategies that enable maintenance of a remarkably large, single-stranded, positive-sense (+) RNA genome and the extensive remodeling of cellular membranes to form specialized viral replication and assembly compartments. The second portion explores the mechanisms by which these viruses manipulate the host cell environment during infection including diverse alterations to host gene expression and immune response pathways. This article is intended as a more in-depth companion piece to our online “Coronavirus 101” lecture (https://youtu.be/8_bOhZd6ieM).
Figure 1.
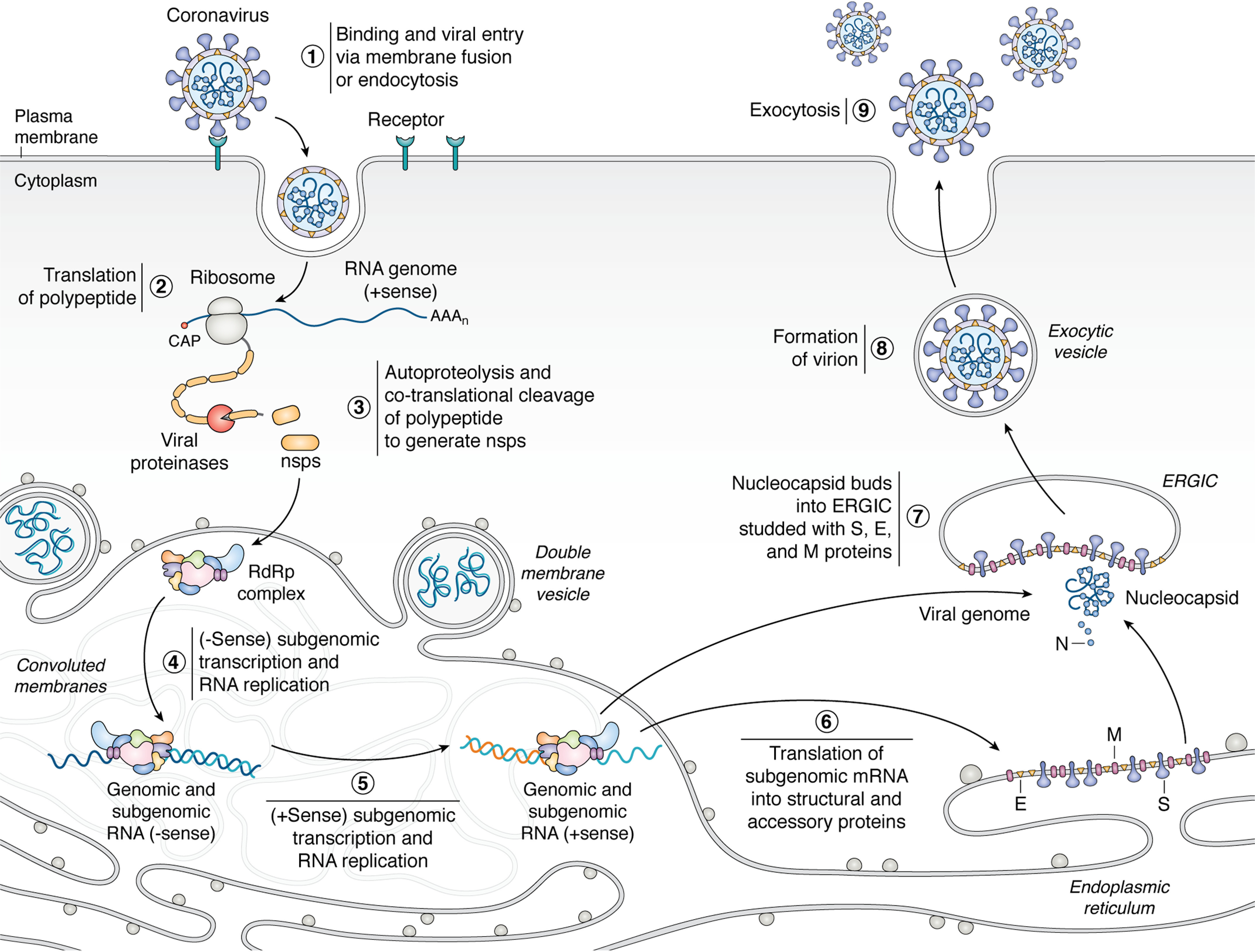
Coronaviruses engage with a host cell-surface receptor and deposit their RNA genomes into the host cytoplasm through endocytosis or directmembrane fusion (1). The positive-sense RNA genome is translated by the host translation machinery (2) to make polyproteins that are cotranslationally cleaved by proteases encoded in the polyprotein to generate components of RdRp complex (3). The RdRp complex uses the genome as a template to generate negative-sense subgenome and genome-length RNAs (4), which are in turn used as templates for synthesis of positive-sense full-length progeny genomes and subgenomic mRNAs (5). Transcription and replication occur in convoluted membranes (CM) adjacent to DMVs that are both derived from rough endoplasmic reticulum(see Fig. 6 for more details). The subgenomic mRNAs are translated into structural and accessory proteins (6). The positive sense genomic RNA is bound by nucleocapsid and buds into the ERGIC, which is decorated with structural proteins S, E, and M translated from positive-sense subgenomic RNAs (steps 6 and 7). The enveloped virion is then exported from the cell by exocytosis (steps 8 and 9).
Part I: The viral life cycle
Viral entry
Coronavirus particles consist of a ∼30-kb strand of positive-sense RNA that forms the genome; this genome is coated with nucleocapsid (N) protein and enclosed in a lipid bilayer containing three membrane proteins: spike (S), membrane (M), and envelope (E) (3). For all studied coronaviruses, the M protein is critical for incorporating essential viral components into new virions during morphogenesis, and N protein associates with the viral genome and M to direct genome packaging into new viral particles. The E protein forms an ion channel in the viral membrane and participates in viral assembly. The S protein is required for viral entry, as it binds to the target cell and initiates fusion with the host cell membrane (reviewed in Ref. 4). S is homotrimeric, with each subunit consisting of two domains, S1 and S2. S1 contains the receptor-binding domain (RBD) and engages with the host receptor, whereas S2 mediates subsequent membrane fusion to enable the virus to enter the host cytoplasm. Activation of the S protein fusion activity requires prior proteolytic cleavage at two sites. The first cleavage site is at the S1/S2 boundary, leading to structural changes in the S2 domain that place it in a prefusion conformation. This cleavage event also separates S2 from S1, although the two domains remain noncovalently associated. The second cleavage site is at S2′, which drives fusion of the viral and cellular membranes to enable release of the N-coated RNA genome into the cytoplasm.
Whereas coronaviruses use the above general strategy to enter target cells, the receptors and proteases used as well as subcellular sites of S cleavage differ depending on the virus (reviewed in Ref. 5). The S proteins of both SARS-CoV and SARS-CoV-2 use host ACE2 as their receptor (6–8) (Fig. 2). ACE2 is a cell-surface peptidase that hydrolyzes angiotensin II and is expressed in most organs, with particularly high expression in the epithelia of lung and small intestine (9). After ACE2 receptor binding, SARS-CoV and SARS-CoV-2 S proteins are subsequently cleaved and activated by the host cell-surface protease TMPRSS2 at the S1/S2 and S2′ sites, leading to membrane fusion (6, 10–12). Some coronavirus S proteins are precleaved at the S1/S2 site by the cellular protease furin during their biosynthesis in the producer cell, as has been shown for both MHV and MERS-CoV (13–15), priming them for entry upon receptor binding on the target cell. MERS-CoV S protein uses DPP4 as its receptor (16, 17), and multiple cellular proteases, including TMPRSS2, endosomal cathepsins, and furin, have been implicated in the subsequent cleavage at the S2′ site (16, 18, 19). The MHV S protein uses host CEACAM1a as its receptor and is subsequently cleaved at S2′ by lysosomal proteases (20, 21).
Figure 2.

Mechanism of SARS-CoV-2 viral entry. The SARS-CoV-2 S protein engages with the host ACE2 receptor and is subsequently cleaved at S1/S2 and S2′ sites by TMPRSS2 protease. This leads to activation of the S2 domain and drives fusion of the viral and host membranes. See section on 'viral entry' for details.
The extent to which specific coronaviruses fuse at the plasma membrane versus during endocytosis remains incompletely resolved. In the cases of SARS-CoV, MERS-CoV, and MHV, the involvement of endosomal and lysosomal proteases in cleavage of their S proteins suggests that entry can occur during endocytosis. MHV enters predominantly through clathrin-mediated endocytosis and fusion with lysosomal membranes, as lysosomal proteases activate the S protein (22, 23). For SARS-CoV and MERS-CoV, both the endocytic and direct membrane fusion pathways may be used for entry. Studies in which components of endocytosis and endosomal proteases have been blocked demonstrate that SARS-CoV and MERS-CoV can exploit the endocytic pathway to enter target cells (24–27). For these viruses, it is likely that the producer and target cell type influence which pathway they use for viral entry. For instance, when MERS-CoV S is precleaved in the producer cell, it gets activated by cell-surface proteases and enters the target cell by direct membrane fusion (28). In contrast, when MERS-CoV S is uncleaved in the producer cell, it enters the target cell through endocytosis and is instead activated by endosomal cathepsins. MERS-CoV with S that has not been precleaved during morphogenesis is incapable of infecting target cell types that have low expression of cathepsins. There are reports demonstrating that inhibition of endosomal cathepsins reduces the efficiency of SARS-CoV-2 entry, suggesting that this virus also exploits endocytosis as another route of entry in addition to direct membrane fusion (6, 29, 30).
There has already been considerable research on the SARS-CoV-2 S protein, given the crucial role it plays during viral entry (reviewed in Ref. 31). Comparing the SARS-CoV-2 S protein sequence with that of closely related SARS-CoV–like viruses revealed that almost all the residues important for ACE2 engagement are not conserved in SARS-CoV-2 (32), although the SARS-CoV-2 S RBD has a 10–20-fold higher binding affinity to ACE2 than SARS-CoV S RBD (33). The mechanistic basis for the enhanced binding affinity is not entirely clear, as ACE2 engagement is structurally similar between SARS-CoV S and SARS-CoV-2 S (34). However, there is a unique salt-bridge interaction present between SARS-CoV-2 S and ACE2, and this may contribute to the enhanced binding affinity. Furthermore, the S1/S2 site in SARS-CoV-2 S contains an insertion of polybasic residues (35, 36). The stretch of polybasic residues contains a furin recognition motif, and recent data suggest that furin can cleave at the S1/S2 site on SARS-CoV-2 S, but not SARS-CoV S, in producer cells (29, 37). This precleavage event is analogous to the processing of MERS-CoV S and MHV S, both of which also contain a furin cleavage site at S1/S2. A precleavage event at the S1/S2 site implies that SARS-CoV-2 S may only require cleavage at the S2′ site on the target cell surface, which would potentiate the membrane fusion process. Notably, acquisition of polybasic cleavage sites occurs during experimental selection for increased transmissibility and expanded tropism in other viruses, suggesting that it may have played a role in the bat-to-human spillover of SARS-CoV-2 (38–42). Further investigation into the properties of S protein from SARS-CoV-2 and other closely related viruses may provide insight into the origin of SARS-CoV-2 as well as the mechanism behind its high transmissibility.
Numerous therapeutic strategies are being explored to inhibit SARS-CoV-2 entry, including blocking ACE2 engagement, inactivating host proteases, and inhibiting S2-mediated membrane fusion. Neutralizing antibodies against SARS-CoV S display moderate efficacy in blocking SARS-CoV-2 infection due to significant differences in the epitope region (6, 35, 43, 44). A recent study isolated neutralizing antibodies capable of blocking the interaction between S and ACE2 from convalescent SARS-CoV-2 patients and demonstrated that they effectively reduce viral load in a mouse model, garnering optimism about the possible use of neutralizing antibodies for treatment (45). Other strategies include development of lipopeptides that block S2-mediated membrane fusion (46) and use of a clinically tested TMPRSS2 inhibitor (6). Not surprisingly, generating protective immunity against the S protein has been the major focus of SARS-CoV-2 vaccine efforts. S protein–directed vaccine platforms under development include production of recombinant S protein, use of nonpathogenic viral vectors to direct expression of S, and nucleic acid–based vaccines in which sequence encoding the S protein is delivered as an mRNA or on a DNA backbone (47). The viral vector and nucleic acid vaccine strategies rely on host ribosomes to translate the S sequence into protein, which would then be subsequently processed and presented to the immune system.
Genome organization, polyprotein synthesis, and proteolysis
Coronaviruses have one of the largest known genomes among RNA viruses, ranging from 27 to 32 kb in length, more than double the length of the average RNA virus genome, and encode for ∼22-29 proteins (48, 49). Given the constraints of eukaryotic translation, which generally allow one protein to be translated per mRNA with ribosome scanning beginning near the 5′ end, it is worth pausing to consider how this number of viral proteins can be synthesized from the genome with a single ribosome entry site. Coronaviruses achieve this feat through the use of large, multiprotein fusions (termed polyproteins, described below) that are subsequently processed into individual proteins (50), as well as through synthesis of sub-genome-length mRNAs using an unusual transcription mechanism (discussed in the subsequent section).
All of the viral nonstructural proteins (nsps) are encoded in two open reading frames (ORF1a and -b) that encompass roughly the first two-thirds of the viral genome (Fig. 3). ORF1a/b is translated from the 5′-capped RNA genome by cap-dependent translation to produce a shorter polyprotein (the ∼440–500-kDa pp1a, which includes nsps 1-11) or a longer polyprotein (the ∼740–810-kDa pp1ab, which includes nsp1 to -16), depending on whether the stop codon at the end of ORF1a is recognized or bypassed. Bypassing the ORF1a stop codon occurs through a −1 ribosomal frameshift in the overlapping region between ORF1a and -1b just upstream of the stop codon, enabling production of the larger pp1ab polyprotein. Frameshifting occurs with ∼20–50% efficiency (51) and is triggered by the presence of a slippery sequence, UUUAAAC, followed by an RNA pseudoknot structure (52), the disruption of which affects frameshifting efficiency (53). Whereas nsp1 to -11 from ORF1a are involved in a broad range of functions from blocking the initial immune response to functioning as cofactors for replication and transcription proteins, the core components of the replication and transcription machinery, such as the RNA-dependent RNA polymerase (RdRp), helicase, and other RNA-modifying enzymes, are present in the ORF1b portion of pp1ab. This frameshifting-based translational control strategy helps the virus maintain a stoichiometry of pp1a and pp1ab proteins that is optimal for infectivity and replication (54, 55). Due to this requirement of precise ratios of pp1a and pp1ab, frameshifting has been explored as a novel drug target (56, 57) similar to such efforts in HIV (58). These drugs typically prevent frameshifting by binding to RNA structures that are required for frameshifting (56, 57).
Figure 3.
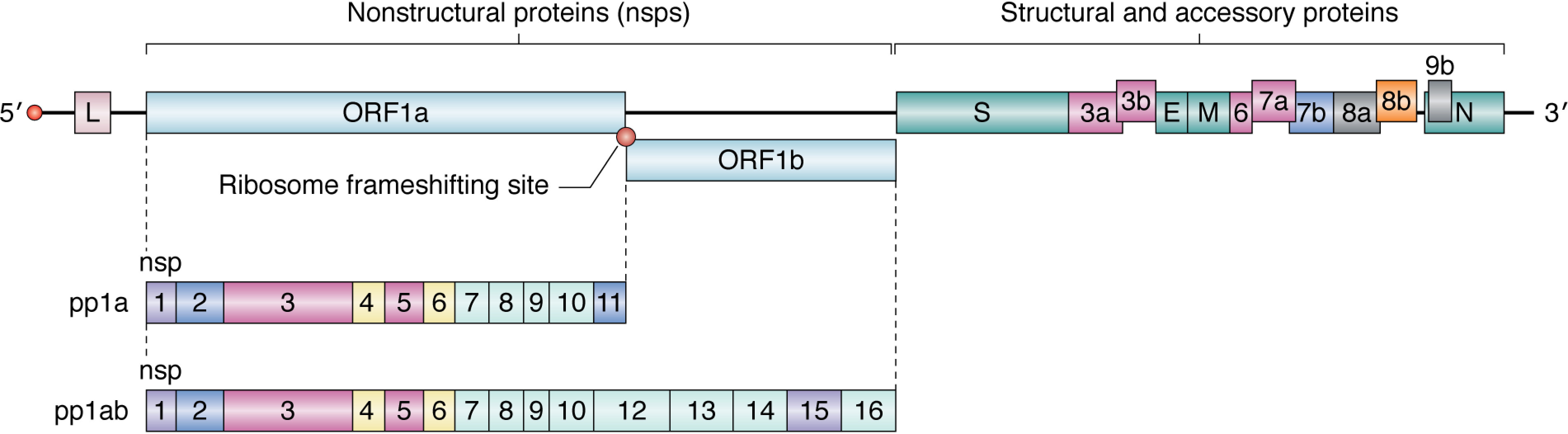
Genome organization of SARS-CoV. The RNA genome encodes two categories of proteins: nsps and structural and accessory proteins. The nonstructural proteins are encoded in ORF1a and ORF1b. Cap-dependent translation begins at ORF1a and produces pp1a, encompassing nsp1–11, or pp1ab, a longer polypeptide that includes nsp12–16. The production of either polypeptide depends on whether the stop codon at ORF1a is recognized by the ribosome or is bypassed through a change in the reading frame by the ribosome frameshifting site. The structural and accessory proteins are synthesized by translation of their respective subgenomic mRNAs (see Fig. 4). The proteins have been color-coded by functional categories for SARS-CoV (see Table 1).
To liberate the individual nsps, pp1a and pp1ab are proteolytically processed in cis and in trans by two viral proteases encoded by nsp3 and nsp5. Nsp3 contains one or two papain-like proteases (PLpro1 and PLpro2), and nsp5 contains a chymotrypsin-like cysteine protease (3CLpro) (reviewed in Ref. 59). The 3CLpro catalyzes the proteolytic cleavage of all nsps downstream of nsp4 and is thus referred to as the main protease. Inhibitors of 3CLpro and PLpro have long been considered as potential drug targets, as their cleavage recognition sequences are distinct from other human proteases and they are essential to viral replication (60–62). Although PLpro is responsible for fewer cleavage events in pp1a, it additionally functions as a deubiquitinase and deISGylating (removal of conjugated interferon-stimulated gene 15 from cellular proteins) enzyme (63, 64), activities that contribute to evasion of the initial antiviral response (64). It is therefore possible that targeting PLpro would inhibit viral replication as well as prevent dysregulation of cellular signaling pathways that could lead to cell death in surrounding cells (65).
Replication and gene expression
A subset of nsps generated by proteolytic cleavage of the polyproteins come together to form the replication and transcription complexes (RTCs) that copy and transcribe the genome. RTCs reside in convoluted membrane structures (discussed in detail below) derived from rough endoplasmic reticulum (ER) and are anchored in place by viral transmembrane proteins nsp3, nsp4, and nsp6 (66–69). Similar to other positive-strand RNA viruses, replication of coronaviruses involves synthesis of the complementary full-length negative-strand RNA, which serves as a template for generation of positive-strand progeny genomes (70). The negative-strand templates get turned over via unknown mechanisms (71), and the positive-strand genomes are packaged into virions. Several cis-acting RNA elements in the 5′ and 3′ end of the genome are important for replication and transcription (reviewed in Refs. 72 and 73). These include conserved stem loop structures within ∼500 nucleotides of the 5′ end of the genome, structural elements in the 3′ UTR that are partially conserved across the different coronaviruses, and the 3′ poly(A) tail. Negative-strand synthesis is facilitated by the N protein interacting with both the poly(A) tail and the 5′ end of the genome to bring these termini in proximity (74).
In addition to genomic replication, the RTCs also carry out synthesis of subgenomic (sg RNA) mRNAs, which encode for the ORFs located in the 3′-proximal one-third of the genome. All sg mRNAs are co-terminal and contain a common 5′ leader sequence that is derived from the 5′ end of the viral genome (75). Placement of the common leader sequence at the 5′ end of all sg mRNAs involves an unusual and complex mechanism of discontinuous transcription (Fig. 4) (reviewed in Ref. 76). During negative-strand synthesis, the RdRp complex terminates or pauses at specific sites along the genome called transcription regulatory sequences (TRSs). The TRSs are present downstream of the common leader sequence at the 5′ end of the genome (TRS-L) and 5′ of every viral ORF along the body of the viral genome (TRS-B) except ORF1a and -1b. Complementarity between sequences in TRS-B on the newly synthesized negative sense RNA and TRS-L allows for the transcription complex to switch templates—effectively jumping from a given TRS-B to the TRS-L at the 5′ end of the genome. Transcription then continues, copying the leader sequence to complete the negative-strand sg RNA (77, 78). The negative-strand sg RNAs subsequently serve as templates to generate large numbers of sg mRNAs; the positive-strand RNAs far outnumber the negative-strand RNAs (79). Secondary structure analysis of the TRS-L region has shown that the context of the sequence and associated structures are important for ensuring that only the TRS-L, and not other TRS-B sequences, acts as the template for strand switching by the RdRp (80). The purpose of the 5′ leader sequence in all sg mRNAs, other than to potentially prime sg mRNA synthesis, is not completely understood. One study with SARS-CoV suggested that the 5′ leader sequence could be important for protection against cleavage by viral nsp1 (81), although the mechanism by which protection is rendered is unclear. The efficiency with which the template switch occurs is an important determinant of the levels of the different sg mRNAs and the ratio of sg mRNAs to genome-length RNA, as failed template switching leads to read-through at TRSs and increases the probability of producing genome-length RNA (reviewed in Ref. 80). Most of what is known about this regulation is from studies on arteriviruses, which belong to the same order (Nidovirales) as coronaviruses and synthesize sg mRNAs by a similar mechanism. The levels of several sg mRNAs are correlated with the stability (ΔG) of the duplex between TRS-L and TRS-B (77), and hence duplex stability was thought to be an important regulator of this process. However, a recent sequencing study with an arterivirus showed that some TRS-B sequences with 100% similarity to TRS-L core sequences were not used as switching points for the transcription complex, suggesting that whereas duplex stability is necessary, it is not sufficient to dictate template switching (82). Regulation of the levels of some sg mRNAs, such as the N protein sg mRNA in coronaviruses, was shown to be mediated by short- and long-range RNA-RNA interactions (83, 84).
Figure 4.
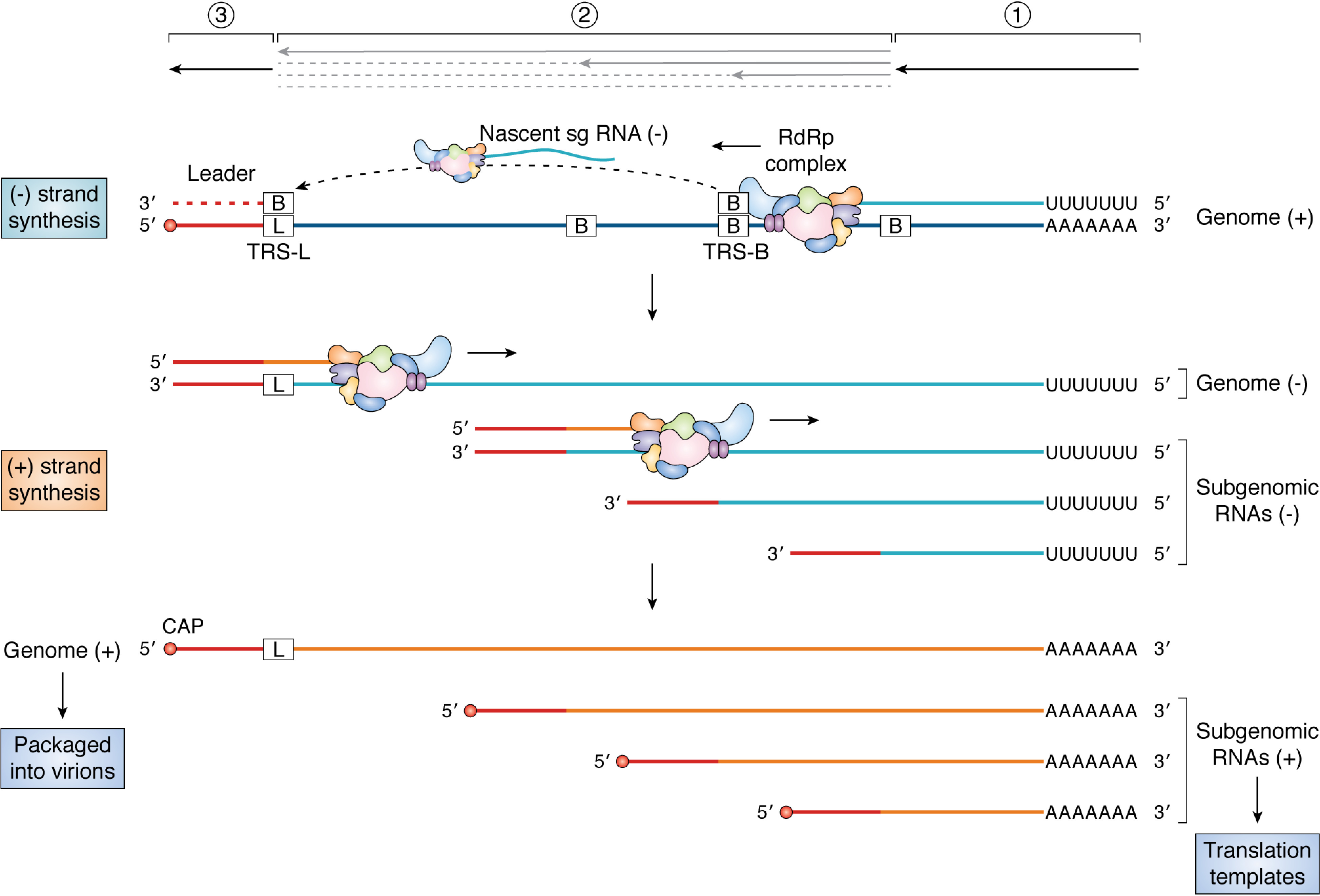
Discontinuous transcription. The RdRp complex initiates transcription at the 39 end of the positive-sense genome (1). Upon copying the TRS-B sequences present at specific sites along the genome body (2), the RdRp complex may “jump” to the TRS-L sequence (3) owing to complementarity between the TRS-B sequence on the nascent sg RNA and TRS-L sequence on the genome. Transcription is resumed on the new template, and the leader sequence (shown in red) is copied to complete the negative-strand sg RNA. The RdRp complex does not always switch templates at TRS-B sequences, resulting in the synthesis of genome-length negative-strand RNA. The negative-strand RNAs serve as templates for the synthesis of genome-length positive-strand RNAs or sg mRNAs.
Several proteins have also been implicated in regulating the levels of sg mRNAs and the switch between full-length negative-strand synthesis and sg RNA synthesis, although a clear picture of features that favor transcription or replication has not emerged. For example, the viral N protein (85) and the cellular kinase GSK-3 and helicase DDX1 (86) have been shown to be important for producing full-length negative-strand genomic RNA and long sg RNAs, suggesting a role in read-through of TRSs. However, the N protein also has helicase-like activity (87), promotes template switching, and appears dispensable for replication but required for efficient sg mRNA transcription (88). It is also possible that the transcription complex that carries out negative-strand synthesis is distinct from the version that carries out positive-strand synthesis (89).
Composition of the replication/transcription complex
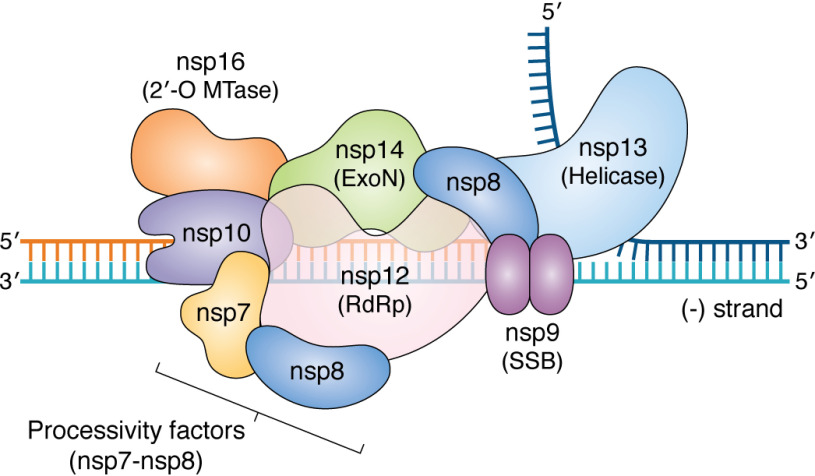
Coronavirus replication, discontinuous transcription, and RNA processing are orchestrated by a remarkably sophisticated replicase complex (Fig. 5). Unlike other RNA viruses, where replication is primarily dependent on the RdRp and a small number of cofactors, coronaviruses appear to use a multiprotein complex, including the RdRp (nsp12), processivity factors (nsp7-8), a helicase (nsp13), single-strand binding protein (nsp9), a proofreading exonuclease (nsp14), other cofactors (e.g. nsp10), and capping enzymes (e.g. nsp16). This is more reminiscent of replisomes from DNA-based organisms and is potentially a consequence of their unusually large genomes (90).
Figure 5.
Model of putative coronavirus replisome. Shown is a model of how the different proteins in the coronavirus replisome come together on the viral negative strand during synthesis of the positive-strand RNA. The core replicase is predicted to consist of the RdRp (nsp12), processivity factors (nsp7-8), and ExoN complex (nsp14, nsp10). The helicase is shown to be unwinding the dsRNA ahead of the replisome, and the SSB (nsp9) is shown as a dimer protecting single-stranded regions of the RNA. Additionally, the 2′-O-MTase (nsp16), which is predicted to be involved in RNA capping, is also indicated. The model is based on known structures and interactions between the proteins (see Refs. 92, 106, and 295–297 and references within) (298).
In vitro studies showed that whereas the SARS-CoV RdRp nsp12 has some minimal activity on its own, its activity and processivity are greatly stimulated in the presence of nsp7-nsp8 cofactors (91). Cryo-EM structures of the SARS-CoV and SARS-CoV-2 nsp12-nsp7-nsp8 tripartite complex revealed that nsp8 binds nsp12 as both a heterodimer (nsp7-nsp8) and by itself to stabilize the regions of nsp12 involved in RNA binding (92, 93). Whether the RdRp is capable of de novo initiation or requires a primer-template substrate remains heavily debated (94, 95). Coronavirus RdRps also have a conserved N-terminal domain that has nucleotidylation activity (NiRAN domain), which is essential for coronavirus replication (96). Structural homology analysis of the NiRAN domain suggests that it shares significant homology with the nucleotide-binding site of protein kinases (92), although how it might mediate nucleotidyltransferase or the function of this domain is not known.
In addition to its role as a processivity factor for the RdRp, nsp8 was first thought to function as a primase during replication (97, 98). However, whereas nsp8 has polyadenylation activity that is stimulated by the presence of a polyU stretch on the template strand, it is unable to incorporate other nucleotides on heteropolymeric templates (99), suggesting that it might not be a primase. Additionally, the cryo-EM structure with nsp7 and nsp12 does not suggest a mechanism for nucleotide incorporation by nsp8. It has been proposed that the presence of polyU sequences at the 5′ end of the negative-strand viral RNA could promote polyadenylation of the viral positive-strand RNAs by nsp8, but this remains to be experimentally validated. The poly(A) tail length also varies during infection (100), and it would be interesting to explore whether nsp8 has a role in this process.
One of the interacting partners of nsp8 in the RTC is nsp9, a single-strand (ss) nucleic acid–binding protein (101, 102) with no obvious sequence specificity or function. It binds ssDNA and ssRNA with equal affinity, although ssRNA is the presumed substrate during infection. Structural studies have shown that it dimerizes, and this is important for viral replication but dispensable for RNA binding (103). It is possible that nsp9 binds to single-stranded regions of the viral genome and protects them from nucleases, akin to the role played by ssDNA-binding proteins in DNA replication systems. Indeed, other ss nucleic acid–binding proteins are also known to play roles in recombination and homologous base pairing (104), processes that occur during discontinuous negative-strand synthesis in coronaviruses.
Another key component of the RTC is nsp13, a superfamily 1 (SF1) 5′ → 3′ helicase (105) that interacts with nsp12 (106) and several other components of the RTC. The functional role of helicases in replication of RNA viruses is largely unknown, although they are one of the most conserved proteins encoded by coronaviruses (reviewed in Ref. 107). Helicases use the energy from nucleotide hydrolysis to translocate on nucleic acids. In addition to its (d)NTPase activity, nsp13 also has a 5′-triphosphophatase activity, suggesting a role for it in RNA capping (108). The helicase domain of MERS and SARS-CoV nsp13 shows remarkable similarity to the cellular Upf1 helicase, a protein involved in the nonsense-mediated decay pathway. Based on this observation, it has been proposed that nsp13 could also play a role in quality control of RNAs (109).
One of the central outstanding questions about the role of helicases in RNA viruses is whether they function similarly to replicative helicases or if they are involved in unwinding local structures and removing obstacles for the polymerase. Replicative helicases typically work together with the polymerase to unwind the double-stranded nucleic acid ahead of the polymerase. The 5′ → 3′ directionality of the helicase is reminiscent of prokaryotic replisomes, where the helicase and polymerase translocate on different strands and the helicase helps in unwinding the duplex ahead of the polymerase. Thus, during the synthesis of full-length progeny genomes using the negative-strand RNA as a template, nsp13 could be bound to the positive-strand RNA and assist the RdRp as it copies the negative strand (Fig. 5). Cooperativity between the replicative helicase and polymerase is a conserved feature of DNA replisomes. The RdRp stimulates the activity of the helicase (106), but whether the helicase has a reciprocal effect on RdRp activity, similar to DNA replisomes, would be interesting to test. A non-mutually exclusive possibility is that the helicase facilitates RdRp template switching during discontinuous transcription by releasing subgenomic RNAs at TRS sites during negative-strand synthesis, similar to a role played by some other SF1 helicases in recombination (110).
Mechanisms underlying high-fidelity replication
RNA viruses typically have high mutation rates due to lack of RdRp proofreading activity, which promotes viral genetic diversity and increases their adaptive potential. However, the potential for accumulation of deleterious mutations leading to collapse of the viral population through error catastrophe caps the size of most RNA virus genomes to ∼15 kb (reviewed in Ref. 90). The ∼30-kb coronavirus genome far exceeds this threshold, indicating that they must have specialized mechanisms to counteract this mutational burden. In this regard, they are one of the few RNA viruses apart from toroviruses and roniviruses (which are also exceptionally large) that have an exonuclease activity and associated high-fidelity replication (111). The discovery of this exonuclease (nsp14-ExoN) in the coronavirus genome (112) showed for the first time the potential for proofreading activity in RNA viruses and explained how coronaviruses maintain their genome integrity. Indeed, the mutation rates of coronaviruses are an order of magnitude lower (10−6 to 10−7) than that of most RNA viruses, and mutating the SARS-CoV or MHV ExoN gene causes the error frequency to jump to that observed in many other RNA viruses (10−3 to 10−5) (113–115).
Active-site mutants that abolish the exonuclease activity of ExoN are lethal for HCoV-229E and transmissible gastroenteritis virus (TGEV) and cause impaired growth for MHV and SARS-CoV (112), suggesting that ExoN is important but may not be essential under all conditions. Why MHV and SARS-CoV but not HCoV-229E and TGEV can tolerate ExoN mutants is unclear, although it is possible that ExoN is essential only in alphacoronaviruses (HCoV and TGEV) and not in betacoronaviruses (MHV and SARS-CoV). It is also possible that the active-site mutation in SARS and MHV did not fully deactivate the enzyme or that other proteins in the replicase can compensate for the absence of an active ExoN. For example, nsp10 stimulates the catalytic activity of nsp14-ExoN to remove a mismatched nucleotide at the 3′ end of the RNA by >35-fold (116), and the high replication fidelity depends on the nsp10-nsp14 interaction (117). ExoN (nsp14) also interacts with the nsp12-nsp8-nsp7 tripartite complex (91), providing biochemical evidence for its role in proofreading during transcription/replication. Nsp10 also interacts with nsp16 (a potential RNA-modifying enzyme), and it has been proposed that all of these proteins could come together to form a larger complex during replication similar to DNA replisome complexes. In vitro biochemical studies comparing the activity of ExoN from MHV and SARS-CoV and HCoV-229E together with the accessory proteins could shed mechanistic light on these phenotypic differences between the ExoN mutants.
Replication fidelity is inherently tied to viral fitness and, in most cases, changes to replication fidelity decrease fitness (reviewed in Ref. 90). This suggests that mutants with altered replication fidelity (such as the ExoN mutant) have potential therapeutic value as live attenuated vaccines (118). Indeed, the SARS-CoV ExoN mutant had decreased pathogenesis and did not revert to virulence even after persistent infection in vivo (118). The ExoN mutation did not revert to WT even over 250 viral passages, although it accumulated a variety of mutations that partially compensated for the replication defect and decreased the population sensitivity to mutagens (119). Several components of the replicase complex, including nsp8, nsp9, nsp12, and nsp13, had mutations in the coding region, underscoring the complexity and interdependence of the RTC and how that helps the virus circumvent the consequences of decreased fidelity. A better understanding of the mechanism of replication fidelity will also allow for the exploration of mutants that increase replication fidelity and thereby reduce diversity and potentially fitness of the population, as has been shown for polioviruses (120).
Recombination, which is generally high in RNA viruses and is linked to their virulence and pathogenicity (121), may also influence coronavirus diversity. In coronaviruses, recombination occurs as an inherent part of the replication cycle during the synthesis of sg RNAs and is tied to the ability of the RdRp to switch templates from the TRS-B sequence to the TRS-L sequence to copy the leader sequence from the 5′ end of the genome. Such recombination events can also occur between co-infecting coronaviruses with different genotypes (reviewed in Ref. 122). Recombination can lead to defective copies of RNA that can no longer be replicated (123) or recombinants with new properties, such as the ability to replicate in a new host (122), leading to new outbreaks. Mutational reversion and recombination-driven processes can pose significant challenges to the use of live attenuated vaccines (120), emphasizing the need to engineer recombination-resistant strains (124). A recent study suggests the involvement of nsp14-ExoN in mediating recombination frequency and junction site selection in several coronaviruses (125), opening up an exciting avenue of exploration for nsp14 in vaccine development.
Viral RNA processing
Capping the 5′ end of the viral mRNA is important for viral mRNA stability, translation initiation, and escape from the cellular innate immune system (126). Capping typically occurs co-transcriptionally in the nucleus, so RNA viruses that replicate in the cytoplasm encode their own enzymes or incorporate other strategies, such as cap snatching (as in bunyaviruses) (127), to protect the 5′ end of their RNAs. The coronavirus capping mechanism is not completely understood, although it appears to follow the canonical capping pathway. Capping begins with hydrolysis of the γ-phosphate of the 5′ end nucleotide; although not yet directly shown, this is thought to be mediated by the nucleotide triphosphatase activity of nsp13-helicase (128). This is followed by the addition of a guanosine monophosphate to the diphosphate RNA by a guanylyl transferase that has remained elusive in coronaviruses, although the NiRAN domain of nsp12 could be involved in this process (96). The guanosine is then methylated at the N7 position, likely by N7-methyltransferase (MTase) activity that resides in the C-terminal part of ExoN (nsp14) (129). Finally, nsp16 is thought to methylate the first and second nucleotides at the 2′-O position (130). This activity requires interaction with nsp10, which appears to improve substrate and RNA binding by nsp16 (131). The 2′-O-methylation is important for evasion of the type-I interferon (IFN) response (which is discussed below) (132). Of the enzymes involved in capping, the N7-MTase of nsp14 is an attractive antiviral target, as this domain exhibits a noncanonical MTase fold different from cellular MTases (129).
The 3′ end of coronavirus mRNAs are polyadenylated. The length of the polyadenylated tail regulates translation efficiency of the mRNAs (100) and is essential for negative-strand synthesis (133). Whereas polyadenylation-related elements, such as a AGUAAA hexamer and the poly(A) tail, work in concert to ensure polyadenylation of the genome (134), the precise mechanism by which this occurs is not known. It is also unclear whether the RdRp carries out the polyadenylation or if cellular poly(A) polymerases are recruited for this process.
Given that translation of coronavirus mRNAs relies on host cap–dependent translation machinery, a number of cellular cap-binding complex factors are candidates for therapeutic targeting (135, 136). Systematic mapping of the interaction between SARS-CoV-2 proteins and the host proteome has revealed interactions between viral proteins and host translation machinery, and an inhibitor of cap-dependent translation initiation reduced viral infectivity in cell culture (137). These data point to the possible effectiveness of a host-directed antiviral therapeutic strategy in treating COVID-19.
Replication/transcription complex proteins as drug targets
Whereas the complexity of the coronavirus replisome may have enabled the virus to expand its genome, it also presents numerous targets for the development of antivirals (138). Most prominent is the RdRp, as it is essential for the virus and lacks homologs in the host. Nucleoside analogs, which are nucleotide triphosphate (NTP) mimics, are commonly used RdRp inhibitors (139). However, designing nucleoside analogs as inhibitors is particularly challenging for coronaviruses due to the presence of the exonuclease, which can exise incorporated analogs and thus provide resistance. An exception to this has been the adenosine analog remedesivir, which is currently in phase 3 clinical trials for treating coronavirus infections (140). A recent in vitro study with purified RdRp-nsp8 complex from several coronaviruses showed that remedesivir incorporation blocks chain elongation 3 nucleotides downstream of its incorporation site, which potentially protects it from ExoN cleavage (141, 142). Additionally, remedesivir is selectively incorporated by the RdRp over the natural substrate ATP. Better in vitro reconstitution systems incorporating the other components of the RTC (nsp7-, nsp13-, and nsp14-exonuclease) will further help to elucidate the mechanism of inhibition.
It may also be of interest to develop nonnucleoside RdRp inhibitors, as have been developed for other RNA viruses, such as hepatitis C virus (143). Nonnucleoside inhibitors typically function allosterically and hence are potentially immune to the resistance conferred by the exonuclease activity of ExoN. Combining compounds that inhibit ExoN together with nucleoside analogs to inhibit the RdRp or using small molecules that increase the mutation load of the virus by other mechanisms that are not sensitive to the exonuclease are other viable options (144). Finally, other components of the RTC, such as the helicase (145), exonuclease (115), and capping machinery (131, 146), have also been considered as potential druggable targets.
Coronavirus replication occurs within heavily modified membranes
A defining feature of many positive-strand RNA viruses, including CoVs, is their ability to hijack and reform intracellular membranes to create a cellular niche for the replication of their RNA genome. Ultrastructural characterization of mainly MHV– and SARS-CoV–infected cells has revealed the membranes that anchor RTCs in CoV-infected cells to be quite striking, consisting of double membrane vesicles (DMVs) among other intricate convoluted membrane structures that isolate CoV RNA from the rest of the cellular environment (Fig. 6) (147–150). Conceptually, RTC formation leads to the concentration of viral replication machinery, spatially separating the sites of viral RNA replication from downstream virion assembly in the endoplasmic reticulum–Golgi intermediate compartment (ERGIC). Additionally, RTCs likely prevent detection of viral dsRNA replication products from innate immune sensors.
Figure 6.
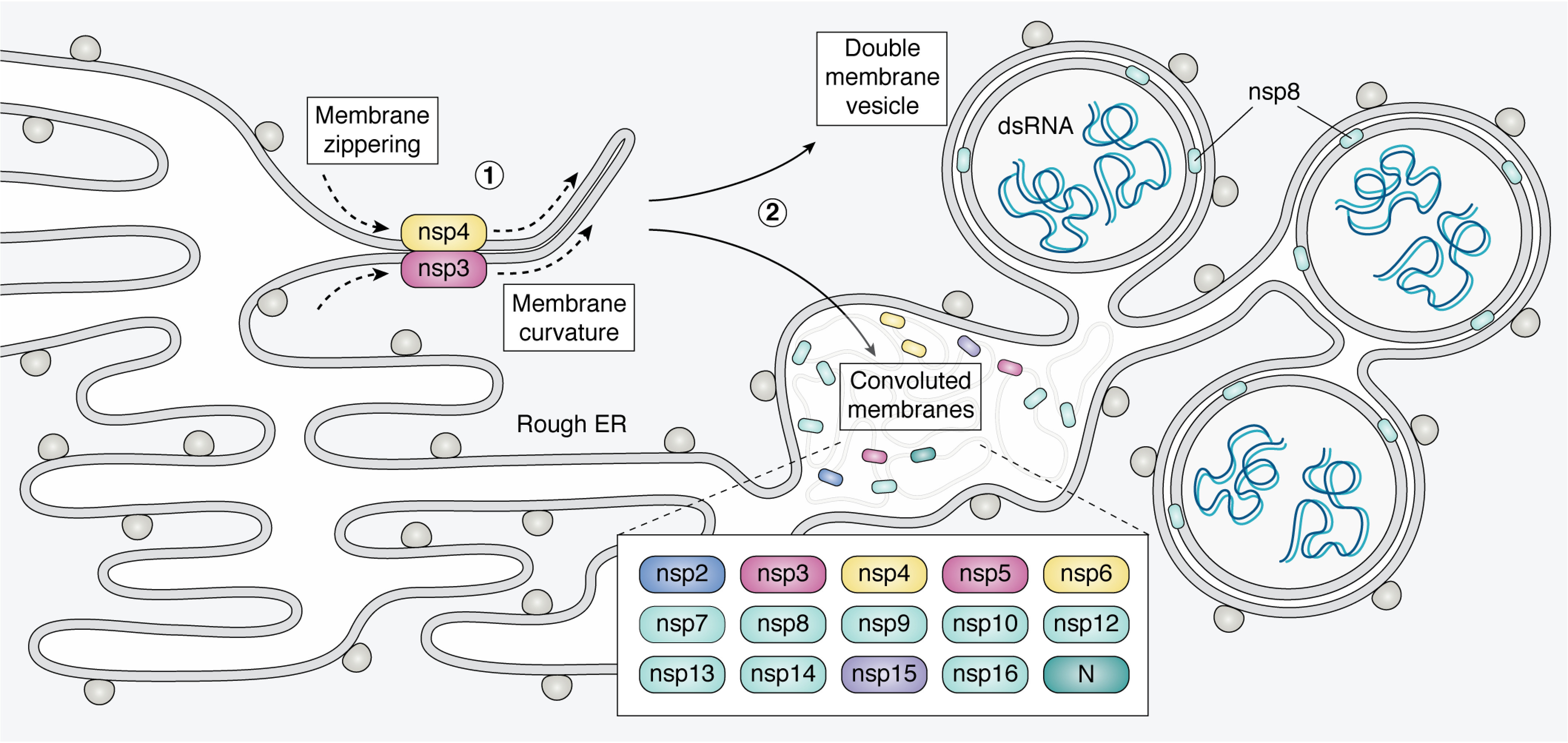
Diagram of convoluted membranes/double membrane vesicles. Coronavirus infection leads to ER membrane modification as RTCs are formed. Nsp3 and nsp4 are co-translationally embedded in the ER membrane and interact via their luminal loops. This leads to “zippering” of ER membranes and induced curvature (1). These interactions yield a complex array of convoluted membranes (CM) and DMVs that are contiguous with the rough ER (2). The protein components of RTCs are mainly localized to the convoluted membranes. The DMVs contain dsRNA, thought to be sequestered replication intermediates. The DMV inner membrane has no ribosomes, connections to the cytoplasm or connections to the rest of the network. The mechanism of DMV formation and the exact site of CoV RNA replication within this membrane network are currently unknown. See section, 'Coronavirus replication occurs within heavily modified membranes' for references.
The DMVs and convoluted membranes in CoV-infected cells form at the nuclear periphery and are derived from host ER membrane (66). The majority of the membrane manipulation is carried out by three nonstructural proteins with integral transmembrane domains: nsp3, nsp4, and nsp6 (67). Although biochemical characterization of these proteins is hindered by their hydrophobic nature, protein-protein interaction studies performed in cells suggest that nsp3, nsp4, and nsp6 can oligomerize and form complexes through their luminal loops (67–69). Expression of these RTC proteins individually in uninfected cells is sufficient to cause membrane proliferation and various perturbations of membrane morphology (69). Co-expression of nsp3 and nsp4 leads to their colocalization in perinuclear foci by fluorescence microscopy and the formation of membrane structures with increased curvature by EM (67). Because the specific interaction of nsp3 and nsp4 is required for these structures to form, it is hypothesized that nsp3 and nsp4 rearrange membranes and introduce curvature by a “zipper” mechanism, essentially bringing ER membranes together through nsp3/4 interactions (69) (Fig. 6). The nsp3/4 interaction also recruits other proteins, including nsp6, to anchor RTCs. Finally, triple transfection of nsp3, nsp4, and nsp6 together results in the formation of DMVs in uninfected cells (67). Due to the importance of membrane modification during viral replication, CoV transmembrane proteins may be attractive drug targets. In fact, a small molecule screen for antiviral activity yielded a compound that targets the transmembrane protein nsp6 and essentially blocks viral RNA replication and DMV formation (151).
During CoV infection, the inner membrane of the DMV is sealed while the outer membrane of the DMVs forms a contiguous network with the convoluted membranes and modified ER membranes (Fig. 6). When this network is isolated from cells, it is capable of producing both genomic and subgenomic RNAs in vitro even in the presence of RNases and proteases, but not detergent, thus implicating the membrane network in shielding viral RNA replication (152). The anchored RTC complexes consist of viral proteins nsp2–10, nsp12–16, and N protein, which have diverse enzymatic functions required for RNA replication as discussed above (150, 153, 154). The RTC microenvironment also includes numerous host proteins that participate in CoV biology, such as proteins involved in vesicular trafficking and translation initiation factors, the latter of which are suggestive of active translation near sites of viral RNA replication (154). The site of RNA replication inside this membrane network is currently unknown. Whereas viral RTC proteins labeled by immuno-EM primarily localize to convoluted membranes between DMVs, dsRNA (presumed to be of viral origin) labeled by the J2 antibody localizes inside the DMVs (Fig. 6). However, there is no experimental evidence demonstrating whether dsRNAs inside the DMV represent nascent viral transcripts, viral RNA replication byproducts, or even host dsRNAs. Recently, nascent viral RNA was visualized by metabolic labeling and quantitative EM autoradiography, revealing that viral transcription does in fact occur in association with the DMVs rather than convoluted membranes (155). The spatial resolution of this technique, while clearly demonstrating viral transcription within the vicinity of the DMVs, was not sufficient to pinpoint the localization of nascent viral RNA within DMVs and/or in association with DMV membranes. Because no visible pores or openings in the inner membrane of the DMV have been detected with conventional EM techniques, viral RNA synthesis regardless of locale would rely on a yet unidentified transport mechanism capable of moving viral proteins and/or RNA in and out of the DMV inner membrane (66, 155).
Viral packaging and egress
The assembly of an infectious CoV virion requires that its nucleocapsid, consisting of the viral RNA genome coated with N protein, and viral envelope coalesce into the same intracellular space. Viral glycoproteins that are incorporated into the envelope (M, E, and S proteins) are translated in the ER and retained at the site of budding in the ERGIC (Fig. 1). The ERGIC budding site is distinct from the site of viral genome synthesis in the RTC. The nucleocapsid core of the virion traffics from the RTC to ultimately bud into ERGIC membranes, which are decorated with M, E, and S protein and become the lipid envelope of the virion. The most abundant envelope component is the M protein, which plays a central role in viral egress. Outside of the context of infection, M protein expression alone is not sufficient to cause budding of virus-like particles, but co-expression with E (or N in the case of SARS-CoV) can result in virus-like particle formation in the absence of infection (156–158). During infection, the M protein nucleates virion components within the ERGIC budding compartment, as M directly interacts with the virion proteins E, N, and S and the CoV genomic RNA (159–162). The E protein, while not highly abundant in the envelope, is critical for viral envelope curvature and maturation and can form membrane ion channels, although the significance of this latter activity is not yet appreciated (163, 164). S protein assembly into virions is enhanced by C-terminal dilysine, dibasic, or tyrosine-based endoplasmic reticulum retention signals (165–167). Although the retention signals are quite divergent among CoVs, all serve to maintain S near the ERGIC-localized M protein, ensuring M-S interaction at the site of virion assembly. Following budding of the nucleocapsid core into the M-, E-, and S-containing ERGIC membranes, the newly enveloped virion then leaves the cell through the exocytic pathway.
Although CoV replication produces an abundance of unique viral RNAs in the cell (positive-strand genomic RNAs, positive-strand sg mRNAs, and negative-strand RNAs), purified CoV virions house mainly full genome-length RNA (159, 168, 169). Conceptually, this specificity is thought to be driven by a packaging signal unique to the genome-length RNA. In MHV, a packaging signal has been mapped to ORF1b within the nsp15 gene (a region absent in sg RNAs) and is predicted to form a bulged stem-loop structure with repeating AGC/GUAAU motifs (170, 171). This packaging signal specifically binds both the N and M proteins, but the order in which these interactions occur is not clear (reviewed in Ref. 172). N protein must have broad RNA-binding activity, as it ultimately coats the length of the viral genome to form the nucleocapsid component of the virion and additionally forms complexes with sg RNAs (160, 173, 174). Thus, an additional role of M in recognizing the packaging signal and selecting full-length genomic RNA is an attractive model for genome packaging specificity, at least in the context of MHV infection (175). In contrast, the packaging signal identified in MHV is absent from other lineages of β-coronaviruses, including SARS-CoV and MERS (reviewed in Ref. 172), leaving us with little understanding of how other CoVs selectively package genome-length RNAs.
Part II: Viral manipulation of the host
Viruses depend on host processes to complete their life cycle. In addition to employing cellular machines like the ribosome to translate their proteins and manipulating cellular membranes during RNA synthesis and viral morphogenesis, several coronavirus proteins modify the cellular environment in ways that may influence viral pathogenesis and replication in vivo. In this section, we discuss the roles of coronavirus proteins in altering the cellular signaling landscape as well as the ability of the virus to modulate host gene expression and its interactions with and counteraction of the host immune response.
Accessory proteins and viral pathogenicity
Coronavirus genomes contain a number of genes concentrated in the 3′ region of the genome that encode for accessory proteins that are largely dispensable for viral replication and growth in vitro (176–182). The SARS-CoV genome encodes for eight accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b), which are the best-studied set of accessory proteins among β-coronaviruses (183, 184). Accessory proteins are specific to each CoV genus and exhibit little homology across the family; as such, this set of eight proteins are specific to human and animal isolates of SARS-CoV (185). Additionally, no significant amino acid sequence similarity is shared between SARS-CoV accessory proteins and other known viral or cellular proteins, providing little insight to predict functional roles (186). Despite being nonessential for viral replication in cultured cells, the accessory proteins presumably modulate virus-host interactions that are important during in vivo infection, including cell proliferation, programmed cell death, pro-inflammatory cytokine production, and IFN signaling (see Table 1) (186, 187). Many SARS-CoV accessory proteins can also be incorporated into virions or virus-like particles during infection, potentially suggesting minor structural roles (187).
Table 1.
SARS-CoV proteins and their functions with SARS-CoV-2 variations
aa, amino acid(s); ↑, up-regulation/activation. Shown is amino acid divergence from Ref. 189 relative to the consensus sequence of SARS-CoV and SARS-like CoVs. Color coding of table rows denotes general functional categories for SARS-CoV proteins:  viral entry and structural protein;
viral entry and structural protein;  host shutoff;
host shutoff;  unknown;
unknown;  immune antagonist;
immune antagonist;  DMV formation;
DMV formation;  replication and transcription;
replication and transcription;  apoptosis induction;
apoptosis induction;  other accessory protein function.
other accessory protein function.
| Name | Functions | SARS-CoV-2 variations | References |
|---|---|---|---|
| I. Structural proteins | |||
| S | Spike protein, host cell receptor binding for viral entry, JNK ↑, ERK ↑, CCL2 ↑ | 27 aa substitutions (6 in RBD, 6 in subdomain, and 4 in known peptide antigen for SARS-CoV); no aa substitutions in CoV-2 polybasic cleavage site compared with CoV consensus sequence; cleavage site is absent in SARS-CoV | 256, 299, 300 |
| E | Envelope protein, viral assembly and release, p38 MAPK ↑ | 164, 253, 301–303 | |
| M | Membrane protein, virion shape, membrane curvature | 304, 305 | |
| N | Nucleocapsid protein, binding to RNA genome, genome tethering to RTCs, type I IFN production and signaling inhibition, viral suppressor of RNA silencing, AP-1 ↑, JNK ↑, p38 MAPK ↑ | 5 aa substitutions | 252, 254, 306–308 |
| II. Nonstructural proteins | |||
| nsp1 | Cellular mRNA degradation and translation inhibition, type I IFN inhibition | 7 aa substitutions | 81, 195, 196, 201 |
| nsp2 | Unknown | 61 aa substitutions | |
| nsp3 | Papain-like protease, polypeptide cleaving, type I IFN production and signaling inhibition, IL-6 ↑ | 102 aa substitutions | 309, 310 |
| nsp4 | DMV formation | 36 aa substitutions | 311, 312 |
| nsp5 | 3CLpro, polypeptides cleaving, type I IFN signaling inhibition | 5 aa substitutions | 313, 314 |
| nsp6 | Restricting autophagosome expansion, DMV formation | 21 aa substitutions | 67, 315 |
| nsp7 | Processivity factor for RdRp, cofactor with nsp8 and nsp12 | 91–93 | |
| nsp8 | Processivity factor for RdRp, cofactor with nsp7 and nsp12 | 4 aa substitutions | 91–93, 97, 98 |
| nsp9 | Single-strand nucleic acid–binding protein | 1 aa substitution | 101, 102 |
| nsp10 | Catalytic activity of Nsp14-ExoN ↑, scaffold protein for nsp14 and nsp16 | 2 aa substitutions | 91, 116, 117 |
| nsp11 | Unknown | ||
| nsp12 | RdRp | 17 aa substitutions | 92, 94, 95 |
| nsp13 | SF1 5′ to 3′ RNA helicase, 5′-triphosphophatase activity | 105, 108 | |
| nsp14 | Proofreading exonuclease, N7- methyltransferase | 15 aa substitutions | 112, 114, 116, 129 |
| nsp15 | Endoribonuclease, evasion of dsRNA sensors | 17 aa substitutions | 316–318 |
| nsp16 | 2′-O-Methyltransferase, down-regulates the activities of RIG-I and MDA5 (MHV) | 12 aa substitutions | 319, 320 |
| III. Accessory proteins | |||
| 3a | NF-κB↑, JNK ↑, IL-8 ↑, RANTES ↑, ion-channel activity, necrosis, pyroptosis, apoptosis induction, and cell cycle arrest | 16 aa substitutions | 250, 258 |
| 3b | Type I IFN production and signaling inhibition, JNK ↑, ERK ↑, apoptosis and necrosis induction, and cell cycle arrest | Truncated to 22 aa, with 6 aa substitutions (154 aa in SARS-CoV) | 249, 259, 321 |
| 6 | Type I IFN production and signaling inhibition and cellular DNA synthesis ↑ | No aa substitutions compared with CoV consensus sequence but only 69% aa identity with SARS-CoV | 190, 234, 322–324 |
| 7a | NF-κB ↑, JNK ↑, p38 MAPK ↑, host translation inhibition, apoptosis induction, and cell cycle arrest | 5 aa substitutions | 205, 250 |
| 7b | Unknown | 4 aa substitutions | |
| 8a | Caspase-dependent apoptosis induction | Encoded by single ORF8 gene (121 aa) | 275 |
| 8b | Cellular DNA synthesis ↑, ATF6 branch of UPR ↑ | 325, 326 | |
| 9b | Caspase-dependent apoptosis induction | 1 aa substitution | 276 |
Given the variability of accessory genes between coronaviruses, they may be linked to virus-specific pathogenicity. That said, annotation of the SARS-CoV-2 genome has identified a similar set of accessory genes as SARS-CoV (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, and ORF9b), albeit with some notable differences among the putative type-I IFN signaling antagonists (188, 189). These include a premature stop codon in ORF3b resulting in a truncated and likely nonfunctional 20-amino acid protein, and relatively lower (69%) amino acid similarity of the ORF6 protein. This may suggest differences in the susceptibility of SARS-CoV and SARS-CoV-2 to host IFN responses (190).
The ORF8 region of the SARS-CoV genome, encoding for ORF8a and ORF8b proteins, displays major variation among human and animal isolates of SARS-CoV (191). Animal isolates contain a single ORF8 gene, whereas this region forms two separate genes in human isolates. However, SARS-CoV and SARS-CoV-2 isolated from patients during early phases of outbreaks closely resemble animal isolates with a single ORF8 gene, likely representing the first zoonotic transmission (192, 193). It remains unclear whether these variances arise from genomic instability or if there is adaptive evolutionary pressure for these changes that may be related to the functional role of ORF8 proteins.
Functional roles have yet to be established for the majority of accessory proteins of other alpha- or gammacoronaviruses. Moreover, much of our understanding of these proteins in the betacoronaviruses derives from transfection or overexpression systems rather than during infection of cultured cells or in vivo. Further development of animal models is paramount to advancing our mechanistic understanding in this area. Nonetheless, the propensity for these genes to be maintained in coronavirus genomes suggests underlying functional importance.
Host shutoff
Numerous RNA and DNA viruses inhibit cellular gene expression by directly targeting mRNAs in order to redirect resources toward viral gene expression and dampen innate immune responses (194). In coronaviruses, this “host shutoff” activity is best characterized for nsp1, which uses an unusual two-part mechanism to restrict translation of mRNA that involves translational repression by 40S binding as well as mRNA cleavage (195, 196). Nsp1 itself does not have detectable RNase activity, but its expression causes cleavage of mRNAs such as IFN-β near the 5′ end of transcripts, perhaps by recruitment of a host endonuclease (195). Viral RNAs as well as some highly structured 5′ UTRs, including certain internal ribosome entry site sequences, are resistant to cleavage, although still susceptible to translational repression (81). The 5′ common leader sequence is necessary and sufficient to confer protection to viral and reporter RNAs from nsp1-induced endonucleolytic cleavage (81, 197). Whereas nsp1 specifically targets RNA polymerase II–transcribed RNAs in cells (195), the spectrum of mRNAs that are cleaved in response to nsp1 is unclear. However, if it functions analogously to mRNA-targeting host shutoff factors in other viruses, host transcripts may be broadly down-regulated (198–200).
Mutations in SARS-CoV nsp1 that block its interaction with the 40S ribosome inhibit both the translational repression and RNA cleavage functions, but an RNA cleavage–deficient mutant retains the translational repression activity (201–204). Thus, nsp1-induced RNA cleavage may occur subsequent to translational repression. Unlike SARS-CoV and SARS-CoV-2 nsp1, MERS nsp1 does not contain the region of the protein that mediates interaction with the 40S, although it still represses host translation. Instead, it targets translationally competent transcripts of nuclear origin and spares virus-like reporter RNAs that are introduced directly into the cytoplasm (203).
The accessory protein ORF7a has also been shown to participate in SARS-CoV host shutoff by reducing total protein synthesis (205). Additional studies are needed to clarify the relative contribution of ORF7a and nsp1 to the translational repression seen during infection as well as to decipher the mechanisms underlying translational repression and mRNA cleavage.
Immune antagonism
Coronavirus-induced dampening of host antiviral responses and an overexuberant pro-inflammatory host response (e.g. cytokine storm) have been linked to the disease pathology associated with infection (206–210). Infection with the circulating human coronaviruses (HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1) rarely causes severe disease, and that which occurs is largely associated with comorbidities (211). However, the highly pathogenic human coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 can cause significant acute respiratory disease syndrome (212). Samples from SARS, MERS, and COVID-19 patients show limited induction of antiviral IFN cell signaling pathways (206, 213–216). Additionally, SARS patients exhibiting high initial virus titers and increased inflammatory monocyte-macrophages and neutrophil accumulation in the lungs were associated with marked elevation of pro-inflammatory cytokines and chemokines (217, 218). Pro-inflammatory cytokines and chemokines recruit inflammatory cells to the sites of infection. Subsequently, neutrophils and cytotoxic T cells, along with these cytokines, induce severe lung tissue damage, including vascular leakage, and stimulate pulmonary fibrosis (219). Recent work analyzing pro-inflammatory profiles among COVID-19 patients identified a similar subset of cytokines and chemokines to be markedly up-regulated (220, 221).
Coronaviruses engage and counteract the immune system in a variety of ways (Fig. 7), which collectively are hypothesized to underlie the disease pathology. SARS-CoV in particular encodes multiple factors that directly antagonize pattern recognition receptors (PRRs) and simultaneously target the expression of IFN-signaling molecules induced by viral recognition. Many of these factors also further stimulate pro-inflammatory cellular responses. These multifaceted interactions with the immune system presumably contribute to the highly restricted induction of type I IFNs during coronavirus infection, while stimulating production of pro-inflammatory molecules. Below we summarize how individual coronavirus proteins modulate the host innate, inflammatory, and adaptive immune responses.
Figure 7.
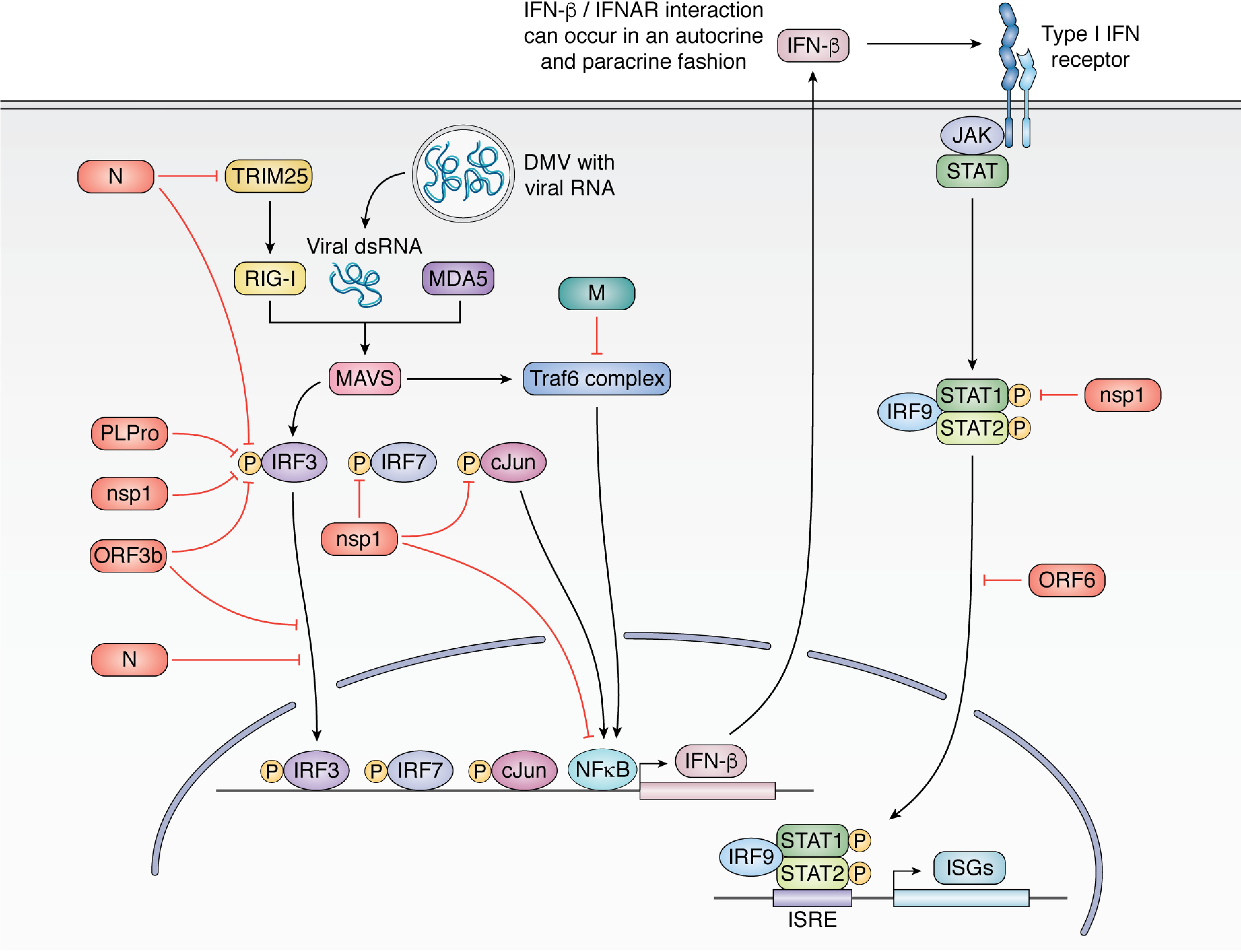
Innate immune antagonism by SARS-CoV. SARS-CoV inhibits multiple arms of the type I IFN response, resulting in strongly dampened IFN-β production during infection. The N protein inhibits recognition of the foreign viral RNA by inhibiting TRIM25 activation of RIG-I and also inhibiting IRF3 phosphorylation. PlPro, nsp1, and ORF3b also inhibit IRF3 phosphorylation, and ORF3b and N further inhibit IRF3 translocation to the nucleus. Nsp1 additionally targets IRF7 and c-Jun phosphorylation. M inhibits assembly of the Traf6 complex, thereby reducing NF-κB import into the nucleus. Together, these activities result in reduced type I IFN production (IFN-β). IFN-β signals in an autocrine and paracrine fashion to activate ISGs through JAK/STAT signaling. Nsp1 inhibits STAT1 phosphorylation, and ORF6 inhibits STAT1 translocation to the nucleus, further dampening ISG production.
Modulation of the host innate immune response
In contrast to viruses like Sendai virus and influenza A, all assayed coronaviruses have shown limited induction of IFN-β and other type I IFNs in tissue culture models, in mice, and in patient samples, including MERS-CoV, HCoV-229E, SARS-CoV, and SARS-CoV-2 (206, 214–216, 222). Type I IFNs remain suppressed even during co-infection of Sendai virus and SARS-CoV, highlighting the ability of coronaviruses to actively silence immune effector expression (201). Suppression by MERS-CoV is particularly robust, as it down-regulates IFN-β ∼60-fold more than SARS-CoV and 300-fold more than HCoV 229E, indicating that differences in viral gene sequences between the coronaviruses influence this response (184).
The strong down-regulation of type I IFN during CoV infection suggests that these viruses are highly sensitive to the presence of IFN, and administration of IFNs has been proposed as a therapeutic for SARS-CoV and SARS-CoV-2. IFN-β dramatically (5 × 104–fold) reduces SARS-CoV RNA copies in cell culture, and IFN-α reduced viral titer in macaques 1 × 104–fold. In cell culture, pretreatment with either IFN-α or IFN-β followed by SARS-CoV infection or post-treatment with IFN-β decreased viral replication (223, 224). Similar results showing antiviral effects of type I IFN treatment were recently described in tissue culture models with SARS-CoV-2 (190, 225), highlighting the potential for the rational design of a live attenuated vaccine with mutations in key immune agonist genes, discussed below.
Replication intermediates produced during RNA virus infection can be recognized by two PRRs: RIG-I and MDA5. RIG-I preferentially recognizes short dsRNA with 5′ di- and triphosphates, whereas MDA5 preferentially recognizes long dsRNA, which is formed as an intermediate during RNA copying (226, 227). MHV is primarily recognized by MDA5, as MDA5 but not RIG-I knockout cells show strong IFN-β induction following infection (228). Interestingly, no coronavirus inhibitor of MDA5 has yet to be identified, which is notable, given CoV targeting of many other arms of the innate immune response.
Multiple SARS-CoV proteins antagonize the host innate immune response, including ORF3b, ORF6, nsp1, N, M, and PLpro (Fig. 7). For example, N protein inhibits TRIM25 ubiquitylation, thereby limiting activation of the RIG-I PRR that recognizes viral dsRNA with a 5′ di- or triphosphate (229). Downstream of PRR activation, IFN-β, and other type I IFNs are transcriptionally induced by the phosphorylation and dimerization of IRF3 and -7, which then traffic to the nucleus to initiate transcription. PLpro, nsp1, ORF3b, and N all inhibit IRF3 phosphorylation, blocking its nuclear entry and type I IFN transcription (63, 201, 230–232). Nsp1 further inhibits IRF7 activation and reduces c-Jun expression and phosphorylation (201). Type I IFNs can also be turned on by NF-κB, but NF-κB–responsive promoter activation is inhibited by both the viral M and nsp1 proteins (233). Once type I IFNs are produced, they then signal through the JAK/STAT pathway to induce interferon-stimulated genes (ISGs) in an autocrine and paracrine fashion. Coronaviruses target this pathway as well; nsp1 induces degradation of IFN-β RNA during host shutoff, ORF6 inhibits STAT1 translocation to the nucleus, and nsp1 inhibits STAT1 phosphorylation, inhibiting downstream induction of ISGs (195, 234).
Experiments with viral deletion mutants using reverse genetics have begun to parse out the overlapping contributions of each of these viral proteins. Consistent with its role as an essential virulence factor, deletion of nsp1 severely attenuates infection in in vivo mouse models with MHV and renders mice immune to a subsequent challenge with WT virus (235). Although viruses lacking ORF3b or ORF6 do not exhibit reduced viral replication in tissue culture or in mouse models of SARS-CoV, this may be due to functional redundancy with other IFN antagonists, which could contribute to the pathogenicity of SARS-CoV (178). In this regard, expression of SARS-CoV ORF6 (but not other SARS-CoV accessory proteins) during infection with an attenuated version of MHV leads to increased viral replication in cell culture and increased virulence in mice (236).
dsRNA produced during RNA virus replication can also trigger host translation shutoff through induction of the antiviral 2′-5′ oligoadenylate synthetase and RNase L. 2′-5′ oligoadenylate synthetase synthesizes RNAs with unique 2′-5′ linkages that activate RNase L, which broadly antagonizes translation by cleaving host and viral RNAs, including ribosomal RNAs, restricting viral replication. MERS NS4b and MHV ns2 are 2′,5′-phosphodiesterases that directly cleave the 2′-5′ RNAs that activate RNase L, thereby inhibiting cellular detection of viral replication intermediates. Deletion of the MHV ns2 protein or mutation of its catalytic residues results in increased IFN-γ in the liver of infected mice (237–240). MHV nsp15 (EndoU) also targets the production of dsRNA by endonucleolytically degrading stretches of polyU RNA made during copying of the viral poly(A) tail, and mutation of nsp15 results in a 200-fold increase in IFN-β induction over WT (241). In addition to the above mechanisms of immune antagonism, coronaviruses appear to also reduce the immunogenicity of the dsRNA they produce by sequestering them in the DMVs (147, 242).
Work in animal models using a mouse-adapted strain of SARS-CoV, which resembles human disease, has further bolstered the connection between a dysregulated innate immune response and disease pathology (209). Genetic knockout of the IFN-α/β receptor or inflammatory monocyte-macrophage depletion during infection protected SARS-CoV–infected mice, demonstrating the role of a vigorous pro-inflammatory response in lethal SARS-CoV infection and identifying these pathways as potential therapeutic targets in patients infected with a highly pathogenic coronavirus.
Modulation of host pro-inflammatory response and programmed cell death pathways
A number of SARS-CoV proteins have been implicated in modulating pro-inflammatory immune responses, likely contributing to the cytokine storm detected in infected patients. Mitogen-activated protein kinase (MAPK) pathways are critical in relaying environmental stress to host cellular stress responses to elicit appropriate physiological responses, such as cellular proliferation, differentiation, development, inflammatory responses, and apoptosis (243). There are three major MAPK pathways in mammals: the extracellular signal–regulated kinase (ERK), p38 MAPK, and the stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK), which are all targeted during SARS-CoV infection (244). Phosphorylation of ERK and JNK has been observed in SARS-CoV–infected cells, and increased levels of phosphorylated p38 MAPK have been found in both cell culture studies and in the leukocytes of SARS patients and have been linked to abnormal interleukin-6 (IL-6) and interleukin-8 (IL-8) cytokine profiles in these patients (244, 245). COVID-19 patients also exhibit a similar pattern of immune dysregulation, with elevated levels of IL-6 in particular, which correlates with severe disease pathology and mortality (221, 246). Work in murine models has shown that elevated IL-6 levels play a major role in driving acute lung injury akin to that observed in both SARS and COVID-19 patients, and loss of IL-6 alleviates the severity of acute lung injury (247).
In cell culture experiments, overexpression of SARS-CoV structural (S, M, N, and E) and various accessory proteins (ORF3a, ORF3b, and ORF7a) has been associated with the activation or interference with MAPK and NF-κB signaling pathways, correlating with dramatic expression changes at cytokine and chemokine promoters, such as CCL2, IL-8, and RANTES (summarized in Table 1) (248–254). However, interference with host NF-κB activity occurs with some cell-type variability and varying degrees of effects on cytokine induction (250–252, 255). Nonetheless, this likely contributes to the up-regulation of cytokines and chemokines associated with acute respiratory disease syndrome, asthma, and pulmonary fibrosis, which is consistent with pro-inflammatory profiles observed during SARS-CoV infection and accounts for patient deaths (256).
Another trigger of pro-inflammatory responses during infection with highly pathogenic coronaviruses is activation of host programmed cell death pathways. Necroptosis and pyropotosis are forms of highly inflammatory cell death that are observed during infection with cytopathic viruses and likely contribute to the molecular mechanisms underlying the severe lung pathology associated with SARS, MERS, and COVID-19 (257). Cell death through these mechanisms leads to a wave of local inflammation involving increased secretion of pro-inflammatory cytokines and chemokines leading to further tissue damage. SARS-CoV-2–infected patients exhibit elevated levels of the cytokine IL-1β, which is associated with pyroptosis (220). In particular, expression of the SARS-CoV ORF3a protein induces caspase-independent necrotic cell death and also initiates an inflammatory cascade through activation of the NLRP3 inflammasome contributing to pyroptosis (258). The SARS-CoV ORF3b protein has also been shown to induce necrosis (259). Notably, the ORF3b protein is truncated to 20 aa in SARS-CoV-2 and is likely nonfunctional, suggesting differences in the underlying mechanisms driving virally induced necrotic cell death in SARS and COVID-19 patients (188).
Whereas noninflammatory apoptosis often serves as a host antiviral response during infection (260), infection-induced activation or modulation of host apoptotic machinery may also induce death of particular cell types that enhance viral egress and pathogenesis (261). During human coronavirus infection, virally induced apoptosis can occur in a variety of cell types beyond those of the respiratory tract, including immune cells such as macrophages, monocytes, T lymphocytes, and dendritic cells (262–266). A molecular understanding of the pro-apoptotic roles for coronavirus proteins largely comes from studies of SARS-CoV or homologous MHV proteins that investigate the pro-apoptotic roles of their structural proteins and unique accessory proteins (summarized in Table 1). Expression of S, N, E, M, ORF3a, ORF3b, ORF7a, OR8a, or ORF9b proteins in various cells lines have all been shown to trigger apoptosis mediated through various pathways, including the PERK pathway through the unfolded protein response, cytochrome c release, and caspase-dependent apoptosis pathways (259, 264, 267–276). At present, it remains unclear whether SARS-CoV–induced cell death functions as an immune evasion tactic, an exit strategy to enhance viral spread, or an indirect consequence of viral replication.
Neutralizing antibodies and memory B-cell response
Protective immunity requires preexisting antibodies, memory B cells, and memory T-cell responses. B- and T-cell responses can be detected within 1 week following the onset of symptoms in both SARS and COVID-19 patients (277, 278). Following infection with SARS-CoV in particular, neutralizing antibodies develop within 2–3 weeks, likely against the S protein (279, 280). In contrast, COVID-19 patients may develop an antibody response earlier due to viral titers peaking earlier (281–284). The primary target of neutralizing antibodies in SARS-CoV is the RBD of the S protein, a region of the protein that is significantly different in SARS-CoV-2 (285, 286). As such, only a small number of previously identified monoclonal antibodies to SAR-CoV bind and neutralize SARS-CoV-2 (287, 288). A number of strategies are being employed to develop therapeutic monoclonal antibodies against SARS-CoV-2, including mouse immunization and hybridoma isolation and cloning of B-cell sequences from convalescent human patients, which has previously been successful in treating SARS patients (44, 289–291).
Importantly, neutralizing antibody titers and the memory B-cell responses, while robust against SARS-CoV (and likely for SARS-CoV-2), are relatively short-lived in recovered patients. Neutralizing antibody titers consistently decline over time and cannot be detected in most SARS-recovered patients 6 years following the onset of symptoms, and memory B-cell responses cannot be detected as early as 3.5 years post-infection (292). These responses may also be short-lived for at least a subset of COVID-19 patients (44). In contrast, memory T-cell responses persist up to 6 years post-infection in a large subset of SARS-recovered patients (292). Whereas T-cell responses are critical for controlling infection and memory T cells are present in higher numbers and often elicit faster responses post-infection than memory B cells, memory T cells alone likely cannot provide adequate long-term protective immunity (293). Importantly, vaccinated animal models have also shown increased immunopathology associated with detrimental T-cell responses; thus, further investigation is critical in understanding coronavirus specific T-cell responses, particularly in the context of vaccine development (294).
Waning protective immunity among previously infected individuals opens up questions regarding the susceptibility of reinfection. Future studies defining immune correlates of protection following SARS-CoV-2 infection are critical and will inform both vaccine strategies and disease management.
Conclusions
Coronavirus spillovers have provoked three epidemics in the last 20 years, and our ability to counteract future emergent viruses will be influenced by how deeply we understand the mechanistic details of coronavirus replication and the virus-host interaction. Despite immense progress, significant questions still exist. For example, much remains to be learned about the mechanisms by which the multicomponent replicase complex executes its sophisticated genome replication, transcription, and RNA-processing functions. The membrane reorganization necessary to form viral replication and transcription compartments is also a central facet of coronavirus biology, yet how the various stages of the viral replication cycle are coordinated and organized within these vesicles and the mechanism of dsRNA sequestration in the DMVs are largely unknown. Virus-host interactions that influence the innate and adaptive immune response are of obvious importance, as they presumably underlie aspects of coronavirus pathogenesis that can differ markedly between viral strains and are central to vaccine development. Many viral accessory proteins appear to antagonize the innate immune system, yet their relative contributions and roles during infection are not clear. Despite these significant unknowns, the current pace of SARS-CoV-2 research is truly remarkable and a testament to the power of cooperative and collaborative science. As these new discoveries are rapidly layered onto the existing foundational work in the coronavirus field, our understanding of this fascinating group of viruses will surely be refined and reshaped more rapidly than for any pathogen in human history.
Acknowledgments
We apologize to those who conducted the significant amount of research we were unable to cite herein and acknowledge that the current pace of the coronavirus field means that many new findings will have emerged by the time this manuscript is published. All figures were designed in collaboration with Biorender and are available as editable templates at BioRender.com.
Funding and additional information—This work was supported by National Institutes of Health Grant CA136367 and a COVID-19 Excellence in Research Award from the Laboratory for Genomics Research (to B. A. G.). B. A. G. is also an investigator of the Howard Hughes Medical Institute. J. M. T. was supported by American Cancer Society Postdoctoral Award 131370-PF-17-245-01-MPC, and M. L. was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Predoctoral Award PGSD3-516787– 2018. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- MHV
- murine hepatitis virus
- SARS
- severe acute respiratory syndrome
- CoV
- coronavirus
- MERS
- Middle East respiratory syndrome
- N
- nucleocapsid
- S
- spike
- M
- membrane
- E
- envelope
- RBD
- receptor-binding domain
- nsp
- nonstructural protein
- RdRp
- RNA-dependent RNA polymerase
- RTC
- replication and transcription complex
- ER
- endoplasmic reticulum
- sg RNA
- subgenomic RNA
- TRS
- transcription regulatory sequence
- ss
- single-strand
- TGEV
- transmissible gastroenteritis virus
- MTase
- N7-methyltransferase
- IFN
- interferon
- DMV
- double membrane vesicle
- ERGIC
- endoplasmic reticulum–Golgi intermediate compartment
- PRR
- pattern recognition receptor
- ISG
- interferon-stimulated gene
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal–regulated kinase
- SAPK
- stress-activated protein kinase
- JNK
- c-Jun N-terminal kinase
- IL
- interleukin
- ExoN
- exonuclease
- 3CLpro
- chymotrypsin-like cysteine protease
- PLpro
- papain-like protease
- SF
- superfamily 1
- RANTES
- regulated on activation normal T cell expressed and secreted.
References
- 1. Paules C. I., Marston H. D., and Jama A. F. (2020) Coronavirus infections—more than just the common cold. JAMA 10.1001/jama.2020.1097 [DOI] [PubMed] [Google Scholar]
- 2. He B., Zhang Y., Xu L., Yang W., Yang F., Feng Y., Xia L., Zhou J., Zhen W., Feng Y., Guo H., Zhang H., and Tu C. (2014) Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J. Virol. 88, 7070–7082 10.1128/JVI.00631-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siu Y. L., Teoh K. T., Lo J., Chan C. M., Kien F., Escriou N., Tsao S. W., Nicholls J. M., Altmeyer R., Peiris J. S. M., Bruzzone R., and Nal B. (2008) The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 82, 11318–11330 10.1128/JVI.01052-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li F. (2016) Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3, 237–261 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Millet J. K., and Whittaker G. R. (2015) Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 202, 120–134 10.1016/j.virusres.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., and Pöhlmann S. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li W. H., Moore M. J., Vasilieva N., Sui J. H., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., Choe H., and Farzan M. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hamming I., Timens W., Bulthuis M. L. C., Lely A. T., Navis G. J., and van Goor H. (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus: a first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637 10.1002/path.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glowacka I., Bertram S., Müller M. A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T. S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., and Pöhlmann S. (2011) Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85, 4122–4134 10.1128/JVI.02232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., and Taguchi F. (2010) Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 84, 12658–12664 10.1128/JVI.01542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., and Gallagher T. (2011) A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 85, 873–882 10.1128/JVI.02062-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Millet J. K., and Whittaker G. R. (2014) Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 111, 15214–15219 10.1073/pnas.1407087111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frana M. F., Behnke J. N., Sturman L. S., and Holmes K. V. (1985) Proteolytic cleavage of the E2-glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell-fusion. J. Virol. 56, 912–920 10.1128/JVI.56.3.912-920.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Haan C. A. M., Stadler K., Godeke G.-J., Bosch B. J., and Rottier P. J. M. (2004) Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 78, 6048–6054 10.1128/JVI.78.11.6048-6054.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qian Z., Dominguez S. R., and Holmes K. V. (2013) Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS ONE 8, e76469 10.1371/journal.pone.0076469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K. C., Chen Y.-H., Zhang L., and Wang X. (2013) Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 23, 986–993 10.1038/cr.2013.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Krämer-Kühl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., Hahn V,. T., Simmons G., Hofmann H., and Pöhlmann S. (2013) The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 87, 5502–5511 10.1128/JVI.00128-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shirato K., Kawase M., and Matsuyama S. (2013) Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 87, 12552–12561 10.1128/JVI.01890-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blau D. M., Turbide C., Tremblay M., Olson M., Létourneau S., Michaliszyn E., Jothy S., Holmes K. V., and Beauchemin N. (2001) Targeted disruption of the Ceacam1 (MHVR) gene leads to reduced susceptibility of mice to mouse hepatitis virus infection. J. Virol. 75, 8173–8186 10.1128/jvi.75.17.8173-8186.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan K., Zelus B. D., Meijers R., Liu J.-H., Bergelson J. M., Duke N., Zhang R., Joachimiak A., Holmes K. V., and Wang J.-H. (2002) Crystal structure of murine sCEACAM1a[1,4]: a coronavirus receptor in the CEA family. EMBO J. 21, 2076–2086 10.1093/emboj/21.9.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burkard C., Verheije M. H., Wicht O., van Kasteren S. I., van Kuppeveld F. J., Haagmans B. L., Pelkmans L., Rottier P. J. M., Bosch B. J., and de Haan C. A. M. (2014) Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 10, e1004502 10.1371/journal.ppat.1004502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eifart P., Ludwig K., Böttcher C., de Haan C. A. M., Rottier P. J. M., Korte T., and Herrmann A. (2007) Role of endocytosis and low pH in murine hepatitis virus strain A59 cell entry. J. Virol. 81, 10758–10768 10.1128/JVI.00725-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simmons G., Gosalia D. N., Rennekamp A. J., Reeves J. D., Diamond S. L., and Bates P. (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 102, 11876–11881 10.1073/pnas.0505577102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., and Jiang C. (2008) SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 18, 290–301 10.1038/cr.2008.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., and Sugamura K. (2007) Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 81, 8722–8729 10.1128/JVI.00253-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou N., Pan T., Zhang J., Li Q., Zhang X., Bai C., Huang F., Peng T., Zhang J., Liu C., Tao L., and Zhang H. (2016) Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV). J. Biol. Chem. 291, 9218–9232 10.1074/jbc.M116.716100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park J.-E., Li K., Barlan A., Fehr A. R., Perlman S., McCray P. B., and Gallagher T. (2016) Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U.S.A. 113, 12262–12267 10.1073/pnas.1608147113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., and Li F. (2020) Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 117, 11727–11734 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., and Xiao G. (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., and Garry R. F. (2020) The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wan Y., Shang J., Graham R., Baric R. S., and Li F. (2020) Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94, 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., and McLellan J. S. (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., and Wang X. (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- 35. Walls A. C., Park Y.-J., Tortorici M. A., Wall A., McGuire A. T., and Veesler D. (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N. G., and Decroly E. (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 176, 104742 10.1016/j.antiviral.2020.104742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoffmann M., Kleine-Weber H., and Pöhlmann S. (2020) A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784.e5 10.1016/j.molcel.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheng J., Zhao Y., Xu G., Zhang K., Jia W., Sun Y., Zhao J., Xue J., Hu Y., and Zhang G. (2019) The S2 subunit of QX-type infectious bronchitis coronavirus spike protein is an essential determinant of neurotropism. Viruses 11, 972 10.3390/v11100972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Follis K. E., York J., and Nunberg J. H. (2006) Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology 350, 358–369 10.1016/j.virol.2006.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Menachery V. D., Dinnon K. H., Yount B. L., McAnarney E. T., Gralinski L. E., Hale A., Graham R. L., Scobey T., Anthony S. J., Wang L., Graham B., Randell S. H., Lipkin W. I., and Baric R. S. (2020) Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 94, e01774–19 10.1128/jvi.01774-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alexander D. J., and Brown I. H. (2009) History of highly pathogenic avian influenza. Rev. Sci. Tech. 28, 19–38 10.20506/rst.28.1.1856 [DOI] [PubMed] [Google Scholar]
- 42. Ito T., Goto H., Yamamoto E., Tanaka H., Takeuchi M., Kuwayama M., Kawaoka Y., and Otsuki K. (2001) Generation of a highly pathogenic avian influenza A virus from an avirulent field isolate by passaging in chickens. J. Virol. 75, 4439–4443 10.1128/JVI.75.9.4439-4443.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., et al. (2020) Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tay M. Z., Poh C. M., Rénia L., MacAry P. A., and Ng L. F. P. (2020) The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374 10.1038/s41577-020-0311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., et al. (2020) A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368, 1274–1278 10.1126/science.abc2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., et al. (2020) Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 30, 343–355 10.1038/s41422-020-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Corey B. L., Mascola J. R., Fauci A. S., and Collins F. S. (2020) A strategic approach to COVID-19 vaccine R&D. Science 368, 948–950 10.1126/science.abc5312 [DOI] [PubMed] [Google Scholar]
- 48. Kim D., Lee J.-Y., Yang J.-S., Kim J. W., Kim V. N., and Chang H. (2020) The architecture of SARS-CoV-2 transcriptome. Cell 38, 661–623 10.1016/j.cell.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gorbalenya A. E., Enjuanes L., Ziebuhr J., and Snijder E. J. (2006) Nidovirales: evolving the largest RNA virus genome. Virus Res. 117, 17–37 10.1016/j.virusres.2006.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brian D. A., and Baric R. S. (2005) Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287, 1–30 10.1007/3-540-26765-4_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Irigoyen N., Firth A. E., Jones J. D., Chung B. Y. W., Siddell S. G., and Brierley I. (2016) High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 12, e1005473 10.1371/journal.ppat.1005473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bredenbeek P. J., Pachuk C. J., Noten A. F., Charité J., Luytjes W., Weiss S. R., and Spaan W. J. (1990) The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 18, 1825–1832 10.1093/nar/18.7.1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Plant E. P., Sims A. C., Baric R. S., Dinman J. D., and Taylor D. R. (2013) Altering SARS coronavirus frameshift efficiency affects genomic and subgenomic RNA production. Viruses 5, 279–294 10.3390/v5010279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Plant E. P., Rakauskaite R., Taylor D. R., and Dinman J. D. (2010) Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 84, 4330–4340 10.1128/JVI.02480-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Plant E. P., and Dinman J. D. (2008) The role of programmed-1 ribosomal frameshifting in coronavirus propagation. Front. Biosci. 13, 4873–4881 10.2741/3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Park S.-J., Kim Y.-G., and Park H.-J. (2011) Identification of RNA pseudoknot-binding ligand that inhibits the −1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J. Am. Chem. Soc. 133, 10094–10100 10.1021/ja1098325 [DOI] [PubMed] [Google Scholar]
- 57. Ahn D.-G., Lee W., Choi J.-K., Kim S.-J., Plant E. P., Almazán F., Taylor D. R., Enjuanes L., and Oh J.-W. (2011) Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antiviral Res. 91, 1–10 10.1016/j.antiviral.2011.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brakier-Gingras L., Charbonneau J., and Butcher S. E. (2012) Targeting frameshifting in the human immunodeficiency virus. Expert Opin. Ther. Targets 16, 249–258 10.1517/14728222.2012.665879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ziebuhr J., Snijder E. J., and Gorbalenya A. E. (2000) Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81, 853–879 10.1099/0022-1317-81-4-853 [DOI] [PubMed] [Google Scholar]
- 60. Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., and Hilgenfeld R. (2020) Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368, 409–412 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kumar V., Shin J. S., Shie J.-J., Ku K. B., Kim C., Go Y. Y., Huang K.-F., Kim M., and Liang P.-H. (2017) Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLPro inhibitors. Antiviral Res. 141, 101–106 10.1016/j.antiviral.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anand K., Ziebuhr J., Wadhwani P., Mesters J. R., and Hilgenfeld R. (2003) Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300, 1763–1767 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- 63. Clementz M. A., Chen Z., Banach B. S., Wang Y., Sun L., Ratia K., Baez-Santos Y. M., Wang J., Takayama J., Ghosh A. K., Li K., Mesecar A. D., and Baker S. C. (2010) Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84, 4619–4629 10.1128/JVI.02406-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mielech A. M., Kilianski A., Baez-Santos Y. M., Mesecar A. D., and Baker S. C. (2014) MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 450–451, 64–70 10.1016/j.virol.2013.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Báez-Santos Y. M., St John S. E., and Mesecar A. D. (2015) The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 115, 21–38 10.1016/j.antiviral.2014.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Knoops K., Kikkert M., Worm S. H. E. V. D., Zevenhoven-Dobbe J. C., van der Meer Y., Koster A. J., Mommaas A. M., and Snijder E. J. (2008) SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6, e226 10.1371/journal.pbio.0060226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Angelini M. M., Akhlaghpour M., Neuman B. W., and Buchmeier M. J. (2013) Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio 4, e00524–13 10.1128/mBio.00524-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hagemeijer M. C., Ulasli M., Vonk A. M., Reggiori F., Rottier P. J. M., and de Haan C. A. M. (2011) Mobility and interactions of coronavirus nonstructural protein 4. J. Virol. 85, 4572–4577 10.1128/JVI.00042-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hagemeijer M. C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen En Henegouwen P. M., Vonk A. M., Rottier P. J. M., Reggiori F., and de Haan C. A. M. (2014) Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology 458-459, 125–135 10.1016/j.virol.2014.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sawicki S. G., and Sawicki D. L. (2005) Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 287, 31–55 10.1007/3-540-26765-4_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang T., and Sawicki S. G. (2001) Mouse hepatitis virus minus-strand templates are unstable and turnover during viral replication. Adv. Exp. Med. Biol. 494, 491–497 10.1007/978-1-4615-1325-4_71 [DOI] [PubMed] [Google Scholar]
- 72. Sola I., Mateos-Gómez P. A., Almazán F., Zúñiga S., and Enjuanes L. (2011) RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 8, 237–248 10.4161/rna.8.2.14991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Madhugiri R., Fricke M., Marz M., and Ziebuhr J. (2016) Coronavirus cis-acting RNA elements. Adv. Virus Res. 96, 127–163 10.1016/bs.aivir.2016.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lo C. Y., Tsai T. L., Lin C. N., Lin C. H., and Wu H. Y. (2019) Interaction of coronavirus nucleocapsid protein with the 5′- and 3′- ends of the coronavirus genome is involved in genome circularization and negative-strand RNA synthesis. FEBS J. 286, 3222–3239 10.1111/febs.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lai M. M., Patton C. D., and Stohlman S. A. (1982) Further characterization of mRNA's of mouse hepatitis virus: presence of common 5-”end nucleotides. J. Virol. 41, 557–565 10.1128/JVI.41.2.557-565.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sola I., Almazán F., Zúñiga S., and Enjuanes L. (2015) Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2, 265–288 10.1146/annurev-virology-100114-055218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sola I., Moreno J. L., Zúñiga S., Alonso S., and Enjuanes L. (2005) Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 79, 2506–2516 10.1128/JVI.79.4.2506-2516.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zúñiga S., Sola I., Alonso S., and Enjuanes L. (2004) Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78, 980–994 10.1128/jvi.78.2.980-994.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sawicki S. G., Sawicki D. L., and Siddell S. G. (2007) A contemporary view of coronavirus transcription. J. Virol. 81, 20–29 10.1128/JVI.01358-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Di H., McIntyre A. A., and Brinton M. A. (2018) New insights about the regulation of Nidovirus subgenomic mRNA synthesis. Virology 517, 38–43 10.1016/j.virol.2018.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Huang C., Lokugamage K. G., Rozovics J. M., Narayanan K., Semler B. L., and Makino S. (2011) SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 7, e1002433 10.1371/journal.ppat.1002433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Di H., Madden J. C., Morantz E. K., Tang H.-Y., Graham R. L., Baric R. S., and Brinton M. A. (2017) Expanded subgenomic mRNA transcriptome and coding capacity of a nidovirus. Proc. Natl. Acad. Sci. U.S.A. 114, E8895–E8904 10.1073/pnas.1706696114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Moreno J. L., Zúñiga S., Enjuanes L., and Sola I. (2008) Identification of a coronavirus transcription enhancer. J. Virol. 82, 3882–3893 10.1128/JVI.02622-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mateos-Gómez P. A., Morales L., Zúñiga S., Enjuanes L., and Sola I. (2013) Long-distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J. Virol. 87, 177–186 10.1128/JVI.01782-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schelle B., Karl N., Ludewig B., Siddell S. G., and Thiel V. (2005) Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 79, 6620–6630 10.1128/JVI.79.11.6620-6630.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wu C.-H., Chen P.-J., and Yeh S.-H. (2014) Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe 16, 462–472 10.1016/j.chom.2014.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Grossoehme N. E., Li L., Keane S. C., Liu P., Dann C. E., Leibowitz J. L., and Giedroc D. P. (2009) Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 394, 544–557 10.1016/j.jmb.2009.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zúñiga S., Cruz J. L. G., Sola I., Mateos-Gómez P. A., Palacio L., and Enjuanes L. (2010) Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 84, 2169–2175 10.1128/JVI.02011-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sawicki S. G., and Sawicki D. L. (1986) Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57, 328–334 10.1128/JVI.57.1.328-334.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Smith E. C., Sexton N. R., and Denison M. R. (2014) Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu. Rev. Virol. 1, 111–132 10.1146/annurev-virology-031413-085507 [DOI] [PubMed] [Google Scholar]
- 91. Subissi L., Posthuma C. C., Collet A., Zevenhoven-Dobbe J. C., Gorbalenya A. E., Decroly E., Snijder E. J., Canard B., and Imbert I. (2014) One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 111, E3900–E3909 10.1073/pnas.1323705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kirchdoerfer R. N., and Ward A. B. (2019) Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 10, 2342 10.1038/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., et al. (2020) Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368, 779–782 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Velthuis, Te A. J. W., Arnold J. J., Cameron C. E., van den Worm S. H. E., and Snijder E. J. (2010) The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 38, 203–214 10.1093/nar/gkp904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ahn D.-G., Choi J.-K., Taylor D. R., and Oh J.-W. (2012) Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 157, 2095–2104 10.1007/s00705-012-1404-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lehmann K. C., Gulyaeva A., Zevenhoven-Dobbe J. C., Janssen G. M. C., Ruben M., Overkleeft H. S., van Veelen P. A., Samborskiy D. V., Kravchenko A. A., Leontovich A. M., Sidorov I. A., Snijder E. J., Posthuma C. C., and Gorbalenya A. E. (2015) Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 43, 8416–8434 10.1093/nar/gkv838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Imbert I., Guillemot J.-C., Bourhis J.-M., Bussetta C., Coutard B., Egloff M.-P., Ferron F., Gorbalenya A. E., and Canard B. (2006) A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25, 4933–4942 10.1038/sj.emboj.7601368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Velthuis, Te A. J. W., van den Worm S. H. E., and Snijder E. J. (2012) The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 40, 1737–1747 10.1093/nar/gkr893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tvarogová J., Madhugiri R., Bylapudi G., Ferguson L. J., Karl N., and Ziebuhr J. (2019) Identification and characterization of a human coronavirus 229E nonstructural protein 8-associated RNA 3′-terminal adenylyltransferase activity. J. Virol. 93, e00291–19 10.1128/JVI.00291-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wu H. Y., Ke T.-Y., Liao W.-Y., and Chang N.-Y. (2013) Regulation of coronaviral poly(A) tail length during infection. PLoS ONE. 8, e70548 10.1371/journal.pone.0070548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Egloff M.-P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E. J., Gorbalenya A. E., Cambillau C., and Canard B. (2004) The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U.S.A. 101, 3792–3796 10.1073/pnas.0307877101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L. L. M., Diprose J., Alderton D., Walsh M., et al. (2004) The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12, 341–353 10.1016/j.str.2004.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Miknis Z. J., Donaldson E. F., Umland T. C., Rimmer R. A., Baric R. S., and Schultz L. W. (2009) Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J. Virol. 83, 3007–3018 10.1128/JVI.01505-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Marceau A. H. (2012) Functions of single-strand DNA-binding proteins in DNA replication, recombination, and repair. Methods Mol. Biol. 922, 1–21 10.1007/978-1-62703-032-8_1 [DOI] [PubMed] [Google Scholar]
- 105. Adedeji A. O., Marchand B., Velthuis, Te A. J. W., Snijder E. J., Weiss S., Eoff R. L., Singh K., and Sarafianos S. G. (2012) Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE 7, e36521 10.1371/journal.pone.0036521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z., and Rao Z. (2019) Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 47, 6538–6550 10.1093/nar/gkz409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lehmann K. C., Snijder E. J., Posthuma C. C., and Gorbalenya A. E. (2015) What we know but do not understand about nidovirus helicases. Virus Res. 202, 12–32 10.1016/j.virusres.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ivanov K. A., Thiel V., Dobbe J. C., van der Meer Y., Snijder E. J., and Ziebuhr J. (2004) Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78, 5619–5632 10.1128/JVI.78.11.5619-5632.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hao W., Wojdyla J. A., Zhao R., Han R., Das R., Zlatev I., Manoharan M., Wang M., and Cui S. (2017) Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 13, e1006474 10.1371/journal.ppat.1006474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Raney K. D., Byrd A. K., and Aarattuthodiyil S. (2013) Structure and mechanisms of SF1 DNA helicases. Adv. Exp. Med. Biol. 767, 17–46 10.1007/978-1-4614-5037-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Smith E. C., and Denison M. R. (2013) Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 9, e1003760 10.1371/journal.ppat.1003760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Minskaia E., Hertzig T., Gorbalenya A. E., Campanacci V., Cambillau C., Canard B., and Ziebuhr J. (2006) Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 5108–5113 10.1073/pnas.0508200103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Eckerle L. D., Lu X., Sperry S. M., Choi L., and Denison M. R. (2007) High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 81, 12135–12144 10.1128/JVI.01296-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Eckerle L. D., Becker M. M., Halpin R. A., Li K., Venter E., Lu X., Scherbakova S., Graham R. L., Baric R. S., Stockwell T. B., Spiro D. J., and Denison M. R. (2010) Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6, e1000896 10.1371/journal.ppat.1000896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Smith E. C., Blanc H., Surdel M. C., Vignuzzi M., and Denison M. R. (2013) Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 9, e1003565 10.1371/journal.ppat.1003565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., and Decroly E. (2012) RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U.S.A. 109, 9372–9377 10.1073/pnas.1201130109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Smith E. C., Case J. B., Blanc H., Isakov O., Shomron N., Vignuzzi M., and Denison M. R. (2015) Mutations in coronavirus nonstructural protein 10 decrease virus replication fidelity. J. Virol. 89, 6418–6426 10.1128/JVI.00110-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Graham R. L., Becker M. M., Eckerle L. D., Bolles M., Denison M. R., and Baric R. S. (2012) A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 18, 1820–1826 10.1038/nm.2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Graepel K. W., Lu X., Case J. B., Sexton N. R., Smith E. C., and Denison M. R. (2017) Proofreading-deficient coronaviruses adapt for increased fitness over long-term passage without reversion of exoribonuclease-inactivating mutations. mBio 8, e01503–17 10.1128/mBio.01503-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Vignuzzi M., Wendt E., and Andino R. (2008) Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 14, 154–161 10.1038/nm1726 [DOI] [PubMed] [Google Scholar]
- 121. Xiao Y., Rouzine I. M., Bianco S., Acevedo A., Goldstein E. F., Farkov M., Brodsky L., and Andino R. (2017) RNA recombination enhances adaptability and is required for virus spread and virulence. Cell Host Microbe 22, 420 10.1016/j.chom.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Su S., Wong G., Shi W., Liu J., Lai A. C. K., Zhou J., Liu W., Bi Y., and Gao G. F. (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 24, 490–502 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Nagy P. D., and Simon A. E. (1997) New insights into the mechanisms of RNA recombination. Virology 235, 1–9 10.1006/viro.1997.8681 [DOI] [PubMed] [Google Scholar]
- 124. Graham R. L., Deming D. J., Deming M. E., Yount B. L., and Baric R. S. (2018) Evaluation of a recombination-resistant coronavirus as a broadly applicable, rapidly implementable vaccine platform. Commun. Biol. 1, 179–110 10.1038/s42003-018-0175-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Gribble J., Pruijssers A. J., Agostini M. L., Anderson-Daniels J., Chappell J. D., Lu X., Stevens L. J., Routh A. L., and Denison M. R. (2020) The coronavirus proofreading exoribonuclease mediates extensive viral recombination. bioRxiv 10.1101/2020.04.23.057786 [DOI] [PMC free article] [PubMed]
- 126. Hyde J. L., and Diamond M. S. (2015) Innate immune restriction and antagonism of viral RNA lacking 2′-O methylation. Virology 479–480, 66–74 10.1016/j.virol.2015.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Cheng E., and Mir M. A. (2012) Signatures of host mRNA 5′ terminus for efficient hantavirus cap snatching. J. Virol. 86, 10173–10185 10.1128/JVI.05560-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ivanov K. A., and Ziebuhr J. (2004) Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 78, 7833–7838 10.1128/JVI.78.14.7833-7838.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., and Guo D. (2009) Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 106, 3484–3489 10.1073/pnas.0808790106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E. J., Canard B., and Decroly E. (2010) In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 6, e1000863 10.1371/journal.ppat.1000863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., and Canard B. (2011) Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 7, e1002059 10.1371/journal.ppat.1002059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B. W., Ziebuhr J., Szretter K. J., Baker S. C., Barchet W., Diamond M. S., Siddell S. G., Ludewig B., and Thiel V. (2011) Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12, 137–143 10.1038/ni.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Lin Y. J., Liao C. L., and Lai M. M. (1994) Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 68, 8131–8140 10.1128/JVI.68.12.8131-8140.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Peng Y.-H., Lin C.-H., Lin C. N., Lo C. Y., Tsai T. L., and Wu H. Y. (2016) Characterization of the role of hexamer AGUAAA and poly(A) tail in coronavirus polyadenylation. PLoS ONE 11, e0165077 10.1371/journal.pone.0165077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Müller C., Schulte F. W., Lange-Grünweller K., Obermann W., Madhugiri R., Pleschka S., Ziebuhr J., Hartmann R. K., and Grünweller A. (2018) Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res. 150, 123–129 10.1016/j.antiviral.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Cencic R., Desforges M., Hall D. R., Kozakov D., Du Y., Min J., Dingledine R., Fu H., Vajda S., Talbot P. J., and Pelletier J. (2011) Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J. Virol. 85, 6381–6389 10.1128/JVI.00078-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., O'Meara M. J., Rezelj V. V., Guo J. Z., Swaney D. L., Tummino T. A., Hüttenhain R., Kaake R. M., Richards A. L., Tutuncuoglu B., et al. (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., and Canard B. (2014) SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antiviral Res. 101, 122–130 10.1016/j.antiviral.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Pruijssers A. J., and Denison M. R. (2019) Nucleoside analogues for the treatment of coronavirus infections. Curr. Opin. Virol. 35, 57–62 10.1016/j.coviro.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Agostini M. L., Andres E. L., Sims A. C., Graham R. L., Sheahan T. P., Lu X., Smith E. C., Case J. B., Feng J. Y., Jordan R., Ray A. S., Cihlar T., Siegel D., Mackman R. L., Clarke M. O., et al. (2018) Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 9, e00221–18 10.1128/mBio.00221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Gordon C. J., Tchesnokov E. P., Feng J. Y., Porter D. P., and Götte M. (2020) The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 295, 4773–4779 10.1074/jbc.AC120.013056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gordon C. J., Tchesnokov E. P., Woolner E., Perry J. K., Feng J. Y., Porter D. P., and Gotte M. (2020) Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 295, 6785–6797 10.1074/jbc.ra120.013679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Cannalire R., Tarantino D., Astolfi A., Barreca M. L., Sabatini S., Massari S., Tabarrini O., Milani M., Querat G., Mastrangelo E., Manfroni G., and Cecchetti V. (2018) Functionalized 2,1-benzothiazine 2,2-dioxides as new inhibitors of Dengue NS5 RNA-dependent RNA polymerase. Eur. J. Med. Chem. 143, 1667–1676 10.1016/j.ejmech.2017.10.064 [DOI] [PubMed] [Google Scholar]
- 144. Agostini M. L., Pruijssers A. J., Chappell J. D., Gribble J., Lu X., Andres E. L., Bluemling G. R., Lockwood M. A., Sheahan T. P., Sims A. C., Natchus M. G., Saindane M., Kolykhalov A. A., Painter G. R., Baric R. S., et al. (2019) Small-molecule antiviral β-d-N4-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 93, e01348–19 10.1128/JVI.01348-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Adedeji A. O., Singh K., Calcaterra N. E., DeDiego M. L., Enjuanes L., Weiss S., and Sarafianos S. G. (2012) Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob. Agents Chemother. 56, 4718–4728 10.1128/AAC.00957-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ferron F., Decroly E., Selisko B., and Canard B. (2012) The viral RNA capping machinery as a target for antiviral drugs. Antiviral Res. 96, 21–31 10.1016/j.antiviral.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Gosert R., Kanjanahaluethai A., Egger D., Bienz K., and Baker S. C. (2002) RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76, 3697–3708 10.1128/jvi.76.8.3697-3708.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Goldsmith C. S., Tatti K. M., Ksiazek T. G., Rollin P. E., Comer J. A., Lee W. W., Rota P. A., Bankamp B., Bellini W. J., and Zaki S. R. (2004) Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10, 320–326 10.3201/eid1002.030913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Snijder E. J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J. J. M., van der Meulen J., Koerten H. K., and Mommaas A. M. (2006) Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80, 5927–5940 10.1128/JVI.02501-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Ulasli M., Verheije M. H., de Haan C. A. M., and Reggiori F. (2010) Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 12, 844–861 10.1111/j.1462-5822.2010.01437.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Lundin A., Dijkman R., Bergström T., Kann N., Adamiak B., Hannoun C., Kindler E., Jónsdóttir H. R., Muth D., Kint J., Forlenza M., Müller M. A., Drosten C., Thiel V., and Trybala E. (2014) Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 10, e1004166 10.1371/journal.ppat.1004166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. van Hemert M. J., van den Worm S. H. E., Knoops K., Mommaas A. M., Gorbalenya A. E., and Snijder E. J. (2008) SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4, e1000054 10.1371/journal.ppat.1000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Denison M. R., Spaan W. J., van der Meer Y., Gibson C. A., Sims A. C., Prentice E., and Lu X. T. (1999) The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73, 6862–6871 10.1128/JVI.73.8.6862-6871.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. V'kovski P., Gerber M., Kelly J., Pfaender S., Ebert N., Braga Lagache S., Simillion C., Portmann J., Stalder H., Gaschen V., Bruggmann R., Stoffel M. H., Heller M., Dijkman R., and Thiel V. (2019) Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. Elife 8, 25 10.7554/eLife.42037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Snijder E. J., Limpens R. W. A. L., de Wilde A. H., de Jong A. W. M., Zevenhoven-Dobbe J. C., Maier H. J., Faas F. F. G. A., Koster A. J., and Bárcena M. (2020) A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 18, e3000715 10.1371/journal.pbio.3000715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Bos E. C., Luytjes W., van der Meulen H. V., Koerten H. K., and Spaan W. J. (1996) The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218, 52–60 10.1006/viro.1996.0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Vennema H., Godeke G. J., Rossen J. W., Voorhout W. F., Horzinek M. C., Opstelten D. J., and Rottier P. J. (1996) Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15, 2020–2028 10.1002/j.1460-2075.1996.tb00553.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Huang Y., Yang Z.-Y., Kong W.-P., and Nabel G. J. (2004) Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 78, 12557–12565 10.1128/JVI.78.22.12557-12565.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kuo L., and Masters P. S. (2013) Functional analysis of the murine coronavirus genomic RNA packaging signal. J. Virol. 87, 5182–5192 10.1128/JVI.00100-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Narayanan K., Maeda A., Maeda J., and Makino S. (2000) Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74, 8127–8134 10.1128/jvi.74.17.8127-8134.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Nguyen V. P., and Hogue B. G. (1997) Protein interactions during coronavirus assembly. J. Virol. 71, 9278–9284 10.1128/JVI.71.12.9278-9284.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Opstelten D. J., Raamsman M. J., Wolfs K., Horzinek M. C., and Rottier P. J. (1995) Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131, 339–349 10.1083/jcb.131.2.339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Wilson L., Mckinlay C., Gage P., and Ewart G. (2004) SARS coronavirus E protein forms cation-selective ion channels. Virology 330, 322–331 10.1016/j.virol.2004.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Venkatagopalan P., Daskalova S. M., Lopez L. A., Dolezal K. A., and Hogue B. G. (2015) Coronavirus envelope (E) protein remains at the site of assembly. Virology 478, 75–85 10.1016/j.virol.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Lontok E., Corse E., and Machamer C. E. (2004) Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 78, 5913–5922 10.1128/JVI.78.11.5913-5922.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Schwegmann-Wessels C., Al-Falah M., Escors D., Wang Z., Zimmer G., Deng H., Enjuanes L., Naim H. Y., and Herrler G. (2004) A novel sorting signal for intracellular localization is present in the S protein of a porcine coronavirus but absent from severe acute respiratory syndrome-associated coronavirus. J. Biol. Chem. 279, 43661–43666 10.1074/jbc.M407233200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. McBride C. E., Li J., and Machamer C. E. (2007) The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 81, 2418–2428 10.1128/JVI.02146-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Makino S., Yokomori K., and Lai M. M. (1990) Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J. Virol. 64, 6045–6053 10.1128/JVI.64.12.6045-6053.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Escors D., Izeta A., Capiscol C., and Enjuanes L. (2003) Transmissible gastroenteritis coronavirus packaging signal is located at the 5′ end of the virus genome. J. Virol. 77, 7890–7902 10.1128/jvi.77.14.7890-7902.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Fosmire J. A., Hwang K., and Makino S. (1992) Identification and characterization of a coronavirus packaging signal. J. Virol. 66, 3522–3530 10.1128/JVI.66.6.3522-3530.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Chen S.-C., van den Born E., van den Worm S. H. E., Pleij C. W. A., Snijder E. J., and Olsthoorn R. C. L. (2007) New structure model for the packaging signal in the genome of group IIa coronaviruses. J. Virol. 81, 6771–6774 10.1128/JVI.02231-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Masters P. S. (2019) Coronavirus genomic RNA packaging. Virology 537, 198–207 10.1016/j.virol.2019.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Baric R. S., Nelson G. W., Fleming J. O., Deans R. J., Keck J. G., Casteel N., and Stohlman S. A. (1988) Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 62, 4280–4287 10.1128/JVI.62.11.4280-4287.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Cologna R., Spagnolo J. F., and Hogue B. G. (2000) Identification of nucleocapsid binding sites within coronavirus-defective genomes. Virology 277, 235–249 10.1006/viro.2000.0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Narayanan K., Chen C.-J., Maeda J., and Makino S. (2003) Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 77, 2922–2927 10.1128/jvi.77.5.2922-2927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. de Haan C. A. M., Masters P. S., Shen X., Weiss S., and Rottier P. J. M. (2002) The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296, 177–189 10.1006/viro.2002.1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Haijema B. J., Volders H., and Rottier P. J. M. (2004) Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 78, 3863–3871 10.1128/jvi.78.8.3863-3871.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Yount B., Roberts R. S., Sims A. C., Deming D., Frieman M. B., Sparks J., Denison M. R., Davis N., and Baric R. S. (2005) Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79, 14909–14922 10.1128/JVI.79.23.14909-14922.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Ontiveros E., Kuo L., Masters P., and Perlman S. (2001) Analysis of nonessential gene function in recombinant MHV-JHM. Gene 4 knockout recombinant virus. Adv. Exp. Med. Biol. 494, 83–89 10.1007/978-1-4615-1325-4_13 [DOI] [PubMed] [Google Scholar]
- 180. Shen S., Wen Z. L., and Liu D. X. (2003) Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication. Virology 311, 16–27 10.1016/S0042-6822(03)00117-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Hodgson T., Britton P., and Cavanagh D. (2006) Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 80, 296–305 10.1128/JVI.80.1.296-305.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Casais R., Davies M., Cavanagh D., and Britton P. (2005) Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 79, 8065–8078 10.1128/JVI.79.13.8065-8078.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Marra M. A., Jones S. J. M., Astell C. R., Holt R. A., Brooks-Wilson A., Butterfield Y. S. N., Khattra J., Asano J. K., Barber S. A., Chan S. Y., Cloutier A., Coughlin S. M., Freeman D., Girn N., Griffith O. L., et al. (2003) The genome sequence of the SARS-associated coronavirus. Science 300, 1399–1404 10.1126/science.1085953 [DOI] [PubMed] [Google Scholar]
- 184. Rota P. A., Oberste M. S., Monroe S. S., Nix W. A., Campagnoli R., Icenogle J. P., Peñaranda S., Bankamp B., Maher K., Chen M.-H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J. L., et al. (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300, 1394–1399 10.1126/science.1085952 [DOI] [PubMed] [Google Scholar]
- 185. Lai M. M., and Cavanagh D. (1997) The molecular biology of coronaviruses. Adv. Virus Res. 48, 1–100 10.1016/S0168-1702(96)01421-9, 10.1016/S0065-3527(08)60286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Liu D. X., Fung T. S., Chong K. K.-L., Shukla A., and Hilgenfeld R. (2014) Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 109, 97–109 10.1016/j.antiviral.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Narayanan K., Huang C., and Makino S. (2008) SARS coronavirus accessory proteins. Virus Res. 133, 113–121 10.1016/j.virusres.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., et al. (2020) Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Lokugamage K. G., Hage A., Schindewolf C., Rajsbaum R., and Menachery V. D. (2020) SARS-CoV-2 is sensitive to type I interferon pretreatment. bioRxiv 10.1101/2020.03.07.982264 [DOI] [PMC free article] [PubMed]
- 191. Satija N., and Lal S. K. (2007) The molecular biology of SARS coronavirus. Ann. N. Y. Acad. Sci. 1102, 26–38 10.1196/annals.1408.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Chinese SARS Molecular Epidemiology Consortium (2004) Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303, 1666–1669 10.1126/science.1092002 [DOI] [PubMed] [Google Scholar]
- 193. Su Y. C., Anderson D. E., Young B. E., Linster M., Zhu F., Jayakumar J., Zhuang Y., Kalimuddin S., Low J. G. H., Tan C. W., Chia W. N., Mak T. M., Octavia S., Chavatte J. -M., Lee R. T. C., et al. (2020) Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. bioRxiv 11, e01610–e01620 10.1128/mBio.01610-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Abernathy E., and Glaunsinger B. (2015) Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 479–480, 600–608 10.1016/j.virol.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., and Makino S. (2006) Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U.S.A. 103, 12885–12890 10.1073/pnas.0603144103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.-T. K., and Makino S. (2008) Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 82, 4471–4479 10.1128/JVI.02472-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Tanaka T., Kamitani W., DeDiego M. L., Enjuanes L., and Matsuura Y. (2012) Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 86, 11128–11137 10.1128/JVI.01700-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Clyde K., and Glaunsinger B. A. (2011) Deep sequencing reveals direct targets of gammaherpesvirus-induced mRNA decay and suggests that multiple mechanisms govern cellular transcript escape. PLoS ONE 6, e19655–12 10.1371/journal.pone.0019655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Rodriguez W., Srivastav K., and Muller M. (2019) C19ORF66 broadly escapes virus-induced endonuclease cleavage and restricts Kaposi's sarcoma-associated herpesvirus. J. Virol. 93, e00373–19 10.1128/JVI.00373-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Gaucherand L., Porter B. K., Levene R. E., Price E. L., Schmaling S. K., Rycroft C. H., Kevorkian Y., McCormick C., Khaperskyy D. A., and Gaglia M. M. (2019) The influenza A virus endoribonuclease PA-X usurps host mRNA processing machinery to limit host gene expression. Cell Rep. 27, 776–791.e7 10.1016/j.celrep.2019.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Wathelet M. G., Orr M., Frieman M. B., and Baric R. S. (2007) Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 81, 11620–11633 10.1128/JVI.00702-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Lokugamage K. G., Narayanan K., Huang C., and Makino S. (2012) Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 86, 13598–13608 10.1128/JVI.01958-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Lokugamage K. G., Narayanan K., Nakagawa K., Terasaki K., Ramirez S. I., Tseng C.-T. K., and Makino S. (2015) Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 89, 10970–10981 10.1128/JVI.01352-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Kamitani W., Huang C., Narayanan K., Lokugamage K. G., and Makino S. (2009) A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 16, 1134–1140 10.1038/nsmb.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Kopecky-Bromberg S. A., Martínez-Sobrido L., and Palese P. (2006) 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 80, 785–793 10.1128/JVI.80.2.785-793.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Law H. K. W., Cheung C. Y., Ng H. Y., Sia S. F., Chan Y. O., Luk W., Nicholls J. M., Peiris J. S. M., and Lau Y. L. (2005) Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 106, 2366–2374 10.1182/blood-2004-10-4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Spiegel M., Schneider K., Weber F., Weidmann M., and Hufert F. T. (2006) Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J. Gen. Virol. 87, 1953–1960 10.1099/vir.0.81624-0 [DOI] [PubMed] [Google Scholar]
- 208. Yen Y.-T., Liao F., Hsiao C.-H., Kao C.-L., Chen Y.-C., and Wu-Hsieh B. A. (2006) Modeling the early events of severe acute respiratory syndrome coronavirus infection in vitro. J. Virol. 80, 2684–2693 10.1128/JVI.80.6.2684-2693.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Channappanavar R., Fehr A. R., Vijay R., Mack M., Zhao J., Meyerholz D. K., and Perlman S. (2016) Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19, 181–193 10.1016/j.chom.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Menachery V. D., Eisfeld A. J., Schäfer A., Josset L., Sims A. C., Proll S., Fan S., Li C., Neumann G., Tilton S. C., Chang J., Gralinski L. E., Long C., Green R., Williams C. M., et al. (2014) Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 5, e01174–14 10.1128/mBio.01174-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Cabeça T. K., Granato C., and Bellei N. (2013) Epidemiological and clinical features of human coronavirus infections among different subsets of patients. Influenza Other Respir. Viruses 7, 1040–1047 10.1111/irv.12101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Gralinski L. E., and Baric R. S. (2015) Molecular pathology of emerging coronavirus infections. J. Pathol. 235, 185–195 10.1002/path.4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. McCray P. B., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H. P., Halabi C., Sigmund C. D., Meyerholz D. K., Kirby P., Look D. C., and Perlman S. (2007) Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81, 813–821 10.1128/JVI.02012-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Spiegel M., Pichlmair A., Martínez-Sobrido L., Cros J., García-Sastre A., Haller O., and Weber F. (2005) Inhibition of β interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79, 2079–2086 10.1128/JVI.79.4.2079-2086.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Ireland D. D. C., Stohlman S. A., Hinton D. R., Atkinson R., and Bergmann C. C. (2008) Type I interferons are essential in controlling neurotropic coronavirus infection irrespective of functional CD8 T cells. J. Virol. 82, 300–310 10.1128/JVI.01794-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Funk C. J., Wang J., Ito Y., Travanty E. A., Voelker D. R., Holmes K. V., and Mason R. J. (2012) Infection of human alveolar macrophages by human coronavirus strain 229E. J. Gen. Virol. 93, 494–503 10.1099/vir.0.038414-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Franks T. J., Chong P. Y., Chui P., Galvin J. R., Lourens R. M., Reid A. H., Selbs E., Mcevoy C. P. L., Hayden C. D. L., Fukuoka J., Taubenberger J. K., and Travis W. D. (2003) Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 34, 743–748 10.1016/S0046-8177(03)00367-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Nicholls J. M., Poon L. L. M., Lee K. C., Ng W. F., Lai S. T., Leung C. Y., Chu C. M., Hui P. K., Mak K. L., Lim W., Yan K. W., Chan K. H., Tsang N. C., Guan Y., Yuen K. Y., et al. (2003) Lung pathology of fatal severe acute respiratory syndrome. Lancet 361, 1773–1778 10.1016/S0140-6736(03)13413-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Ludwicka-Bradley A., Silver R. M., and Bogatkevich G. S. (2011) Coagulation and autoimmunity in scleroderma interstitial lung disease. Semin. Arthritis Rheum. 41, 212–222 10.1016/j.semarthrit.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., et al. (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395, 1054–1062 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Blanco-Melo D., Nilsson-Payant B. E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T. X., Oishi K., Panis M., Sachs D., Wang T. T., Schwartz R. E., Lim J. K., Albrecht R. A., and tenOever B. R. (2020) Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e9 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., and Doerr H. W. (2003) Treatment of SARS with human interferons. Lancet 362, 293–294 10.1016/S0140-6736(03)13973-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Haagmans B. L., Kuiken T., Martina B. E., Fouchier R. A. M., Rimmelzwaan G. F., van Amerongen G., van Riel D., de Jong T., Itamura S., Chan K.-H., Tashiro M., and Osterhaus A. D. M. E. (2004) Pegylated interferon-α protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 10, 290–293 10.1038/nm1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Mantlo E., Bukreyeva N., Maruyama J., Paessler S., and Huang C. (2020) Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 179, 104811 10.1016/j.antiviral.2020.104811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Schmidt A., Schwerd T., Hamm W., Hellmuth J. C., Cui S., Wenzel M., Hoffmann F. S., Michallet M.-C., Besch R., Hopfner K.-P., Endres S., and Rothenfusser S. (2009) 5′-Triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U.S.A. 106, 12067–12072 10.1073/pnas.0900971106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T. S., Fujita T., and Akira S. (2008) Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene-I and melanoma differentiation–associated gene 5. J. Exp. Med. 205, 1601–1610 10.1084/jem.20080091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Roth-Cross J. K., Bender S. J., and Weiss S. R. (2008) Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 82, 9829–9838 10.1128/JVI.01199-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., and Cao C. (2017) The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 91, e02143–16 10.1128/JVI.02143-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Devaraj S. G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C. J., Tseng C.-T. K., Baker S. C., and Li K. (2007) Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 282, 32208–32221 10.1074/jbc.M704870200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Zhou P., Li H., Wang H., Wang L.-F., and Shi Z. (2012) Bat severe acute respiratory syndrome-like coronavirus ORF3b homologues display different interferon antagonist activities. J. Gen. Virol. 93, 275–281 10.1099/vir.0.033589-0 [DOI] [PubMed] [Google Scholar]
- 232. Kopecky-Bromberg S. A., Martínez-Sobrido L., Frieman M., Baric R. A., and Palese P. (2007) Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81, 548–557 10.1128/JVI.01782-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Siu K.-L., Kok K.-H., Ng M.-H. J., Poon V. K. M., Yuen K.-Y., Zheng B.-J., and Jin D.-Y. (2009) Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3·TANK·TBK1/IKKε complex. J. Biol. Chem. 284, 16202–16209 10.1074/jbc.M109.008227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Frieman M., Yount B., Heise M., Kopecky-Bromberg S. A., Palese P., and Baric R. S. (2007) Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81, 9812–9824 10.1128/JVI.01012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Züst R., Cervantes-Barragán L., Kuri T., Blakqori G., Weber F., Ludewig B., and Thiel V. (2007) Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3, e109 10.1371/journal.ppat.0030109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Pewe L., Zhou H., Netland J., Tangudu C., Olivares H., Shi L., Look D., Gallagher T., and Perlman S. (2005) A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79, 11335–11342 10.1128/JVI.79.17.11335-11342.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Thornbrough J. M., Jha B. K., Yount B., Goldstein S. A., Li Y., Elliott R., Sims A. C., Baric R. S., Silverman R. H., and Weiss S. R. (2016) Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio 7, e00258–12 10.1128/mBio.00258-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Roth-Cross J. K., Stokes H., Chang G., Chua M. M., Thiel V., Weiss S. R., Gorbalenya A. E., and Siddell S. G. (2009) Organ-specific attenuation of murine hepatitis virus strain A59 by replacement of catalytic residues in the putative viral cyclic phosphodiesterase ns2. J. Virol. 83, 3743–3753 10.1128/JVI.02203-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Zhao L., Jha B. K., Wu A., Elliott R., Ziebuhr J., Gorbalenya A. E., Silverman R. H., and Weiss S. R. (2012) Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11, 607–616 10.1016/j.chom.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Comar C. E., Goldstein S. A., Li Y., Yount B., Baric R. S., and Weiss S. R. (2019) Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. mBio 10, 1814–1816 10.1128/mBio.00319-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Hackbart M., Deng X., and Baker S. C. (2020) Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. U.S.A. 117, 8094–8103 10.1073/pnas.1921485117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Versteeg G. A., Bredenbeek P. J., van den Worm S. H. E., and Spaan W. J. M. (2007) Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 361, 18–26 10.1016/j.virol.2007.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Morrison D. K. (2012) MAP kinase pathways. Cold Spring Harb. Perspect. Biol. 4, a011254 10.1101/cshperspect.a011254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Lim Y. X., Ng Y. L., Tam J. P., and Liu D. X. (2016) Human coronaviruses: a review of virus-host interactions. Diseases 4, 26 10.3390/diseases4030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Wang W.-K., Chen S.-Y., Liu I.-J., Kao C.-L., Chen H.-L., Chiang B.-L., Wang J.-T., Sheng W.-H., Hsueh P.-R., Yang C.-F., Yang P.-C., and Chang S.-C, and Severe Acute Respiratory Syndrome Research Group of the National Taiwan University College of Medicine/NTU Hospital (2004) Temporal relationship of viral load, ribavirin, interleukin (IL)-6, IL-8, and clinical progression in patients with severe acute respiratory syndrome. Clin. Infect. Dis. 39, 1071–1075 10.1086/423808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Giamarellos-Bourboulis E. J., Netea M. G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., et al. (2020) Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 27, 992–1000.e3 10.1016/j.chom.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Goldman J. L., Sammani S., Kempf C., Saadat L., Letsiou E., Wang T., Moreno-Vinasco L., Rizzo A. N., Fortman J. D., and Garcia J. G. N. (2014) Pleiotropic effects of interleukin-6 in a “two-hit” murine model of acute respiratory distress syndrome. Pulm. Circ. 4, 280–288 10.1086/675991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Liu M., Yang Y., Gu C., Yue Y., Wu K. K., Wu J., and Zhu Y. (2007) Spike protein of SARS-CoV stimulates cyclooxygenase-2 expression via both calcium-dependent and calcium-independent protein kinase C pathways. FASEB J. 21, 1586–1596 10.1096/fj.06-6589com [DOI] [PubMed] [Google Scholar]
- 249. Varshney B., and Lal S. K. (2011) SARS-CoV accessory protein 3b induces AP-1 transcriptional activity through activation of JNK and ERK pathways. Biochemistry 50, 5419–5425 10.1021/bi200303r [DOI] [PubMed] [Google Scholar]
- 250. Kanzawa N., Nishigaki K., Hayashi T., Ishii Y., Furukawa S., Niiro A., Yasui F., Kohara M., Morita K., Matsushima K., Le M. Q., Masuda T., and Kannagi M. (2006) Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-κB activation. FEBS Lett. 580, 6807–6812 10.1016/j.febslet.2006.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Dosch S. F., Mahajan S. D., and Collins A. R. (2009) SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-κB pathway in human monocyte macrophages in vitro. Virus Res. 142, 19–27 10.1016/j.virusres.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. He R., Leeson A., Andonov A., Li Y., Bastien N., Cao J., Osiowy C., Dobie F., Cutts T., Ballantine M., and Li X. (2003) Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 311, 870–876 10.1016/j.bbrc.2003.10.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Jiménez-Guardeño J. M., Nieto-Torres J. L., DeDiego M. L., Regla-Nava J. A., Fernandez-Delgado R., Castaño-Rodriguez C., and Enjuanes L. (2014) The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 10, e1004320 10.1371/journal.ppat.1004320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Surjit M., Liu B., Jameel S., Chow V. T. K., and Lal S. K. (2004) The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 383, 13–18 10.1042/BJ20040984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. DeDiego M. L., Nieto-Torres J. L., Regla-Nava J. A., Jiménez-Guardeño J. M., Fernandez-Delgado R., Fett C., Castaño-Rodriguez C., Perlman S., and Enjuanes L. (2014) Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 88, 913–924 10.1128/JVI.02576-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Chen I.-Y., Chang S. C., Wu H. Y., Yu T.-C., Wei W.-C., Lin S., Chien C.-L., and Chang M.-F. (2010) Upregulation of the chemokine (C-C motif) ligand 2 via a severe acute respiratory syndrome coronavirus spike-ACE2 signaling pathway. J. Virol. 84, 7703–7712 10.1128/JVI.02560-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Fink S. L., and Cookson B. T. (2005) Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 73, 1907–1916 10.1128/IAI.73.4.1907-1916.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Yue Y., Nabar N. R., Shi C.-S., Kamenyeva O., Xiao X., Hwang I.-Y., Wang M., and Kehrl J. H. (2018) SARS-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 9, 904 10.1038/s41419-018-0917-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Khan S., Fielding B. C., Tan T. H. P., Chou C.-F., Shen S., Lim S. G., Hong W., and Tan Y.-J. (2006) Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 122, 20–27 10.1016/j.virusres.2006.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Benedict C. A., Norris P. S., and Ware C. F. (2002) To kill or be killed: viral evasion of apoptosis. Nat. Immunol. 3, 1013–1018 10.1038/ni1102-1013 [DOI] [PubMed] [Google Scholar]
- 261. Amara A., and Mercer J. (2015) Viral apoptotic mimicry. Nat. Rev. Microbiol. 13, 461–469 10.1038/nrmicro3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Gu J., and Korteweg C. (2007) Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 170, 1136–1147 10.2353/ajpath.2007.061088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Collins A. R. (2002) In vitro detection of apoptosis in monocytes/macrophages infected with human coronavirus. Clin. Diagn. Lab. Immunol. 9, 1392–1395 10.1128/cdli.9.6.1392-1395.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Yang Y., Xiong Z., Zhang S., Yan Y., Nguyen J., Ng B., Lu H., Brendese J., Yang F., Wang H., and Yang X.-F. (2005) Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 392, 135–143 10.1042/BJ20050698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Ying T., Li W., and Dimitrov D. S. (2016) Discovery of T-cell infection and apoptosis by Middle East respiratory syndrome coronavirus. J. Infect. Dis. 213, 877–879 10.1093/infdis/jiv381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Chu H., Zhou J., Wong B. H.-Y., Li C., Chan J. F.-W., Cheng Z.-S., Yang D., Wang D., Lee A. C.-Y., Li C., Yeung M.-L., Cai J.-P., Chan I. H.-Y., Ho W.-K., To K. K.-W., et al. (2016) Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 213, 904–914 10.1093/infdis/jiv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Chow K. Y. C., Yeung Y. S., Hon C. C., Zeng F., Law K. M., and Leung F. C. C. (2005) Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells. FEBS Lett. 579, 6699–6704 10.1016/j.febslet.2005.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Tsoi H., Li L., Chen Z. S., Lau K.-F., Tsui S. K. W., and Chan H. Y. E. (2014) The SARS-coronavirus membrane protein induces apoptosis via interfering with PDK1-PKB/Akt signalling. Biochem. J. 464, 439–447 10.1042/BJ20131461 [DOI] [PubMed] [Google Scholar]
- 269. DeDiego M. L., Nieto-Torres J. L., Jiménez-Guardeño J. M., Regla-Nava J. A., Álvarez E., Oliveros J. C., Zhao J., Fett C., Perlman S., and Enjuanes L. (2011) Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 7, e1002315 10.1371/journal.ppat.1002315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Liao Y., Fung T. S., Huang M., Fang S. G., Zhong Y., and Liu D. X. (2013) Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 87, 8124–8134 10.1128/JVI.00626-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Padhan K., Minakshi R., Towheed M. A. B., and Jameel S. (2008) Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 89, 1960–1969 10.1099/vir.0.83665-0 [DOI] [PubMed] [Google Scholar]
- 272. Tan Y.-J., Teng E., Shen S., Tan T. H. P., Goh P.-Y., Fielding B. C., Ooi E. E., Tan H. C., Lim S. G., and Hong W. (2004) A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 78, 6723–6734 10.1128/JVI.78.13.6723-6734.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Tan Y.-X., Tan T. H. P., Lee M. J. R., Tham P.-Y., Gunalan V., Druce J., Birch C., Catton M., Fu N. Y., Yu V. C., and Tan Y.-J. (2007) Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 81, 6346–6355 10.1128/JVI.00090-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Schaecher S. R., Touchette E., Schriewer J., Buller R. M., and Pekosz A. (2007) Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J. Virol. 81, 11054–11068 10.1128/JVI.01266-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Chen C.-Y., Ping Y.-H., Lee H.-C., Chen K.-H., Lee Y.-M., Chan Y.-J., Lien T.-C., Jap T.-S., Lin C.-H., Kao L.-S., and Chen Y.-M. A. (2007) Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 196, 405–415 10.1086/519166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Sharma K., Åkerström S., Sharma A. K., Chow V. T. K., Teow S., Abrenica B., Booth S. A., Booth T. F., Mirazimi A., and Lal S. K. (2011) SARS-CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLoS ONE 6, e19436 10.1371/journal.pone.0019436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Tan Y.-J., Goh P.-Y., Fielding B. C., Shen S., Chou C.-F., Fu J.-L., Leong H. N., Leo Y.-S., Ooi E. E., Ling A. E., Lim S. G., and Hong W. (2004) Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 11, 362–371 10.1128/cdli.11.2.362-371.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Thevarajan I., Nguyen T. H. O., Koutsakos M., Druce J., Caly L., van de Sandt C. E., Jia X., Nicholson S., Catton M., Cowie B., Tong S. Y. C., Lewin S. R., and Kedzierska K. (2020) Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 26, 453–455 10.1038/s41591-020-0819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Nie Y., Wang G., Shi X., Zhang H., Qiu Y., He Z., Wang W., Lian G., Yin X., Du L., Ren L., Wang J., He X., Li T., Deng H., et al. (2004) Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J. Infect. Dis. 190, 1119–1126 10.1086/423286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Temperton N. J., Chan P. K., Simmons G., Zambon M. C., Tedder R. S., Takeuchi Y., and Weiss R. A. (2005) Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg. Infect. Dis. 11, 411–416 10.3201/eid1103.040906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Pan Y., Zhang D., Yang P., Poon L. L. M., and Wang Q. (2020) Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 20, 411–412 10.1016/S1473-3099(20)30113-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Kim J. Y., Ko J.-H., Kim Y., Kim Y.-J., Kim J.-M., Chung Y.-S., Kim H. M., Han M.-G., Kim S. Y., and Chin B. S. (2020) Viral load kinetics of SARS-CoV-2 infection in first two patients in Korea. J. Korean Med. Sci.. 35, e86 10.3346/jkms.2020.35.e86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., et al. (2020) SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 382, 1177–1179 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Peiris J. S. M., Chu C. M., Cheng V. C. C., Chan K. S., Hung I. F. N., Poon L. L. M., Law K. I., Tang B. S. F., Hon T. Y. W., Chan C. S., Chan K. H., Ng J. S. C., Zheng B. J., Ng W. L., Lai R. W. M., et al. HKU/UCH SARS Study Group (2003) Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361, 1767–1772 10.1016/S0140-6736(03)13412-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X., Hensley L. E., Prabakaran P., Rockx B., Sidorov I. A., Corti D., Vogel L., Feng Y., Kim J.-O., Wang L.-F., et al. (2007) Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. U.S.A. 104, 12123–12128 10.1073/pnas.0701000104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Li F., Li W., Farzan M., and Harrison S. C. (2005) Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864–1868 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- 287. Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., and Ying T. (2020) Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 9, 382–385 10.1080/22221751.2020.1729069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Wang C., Li W., Drabek D., Okba N. M. A., van Haperen R., Osterhaus A. D. M. E., van Kuppeveld F. J. M., Haagmans B. L., Grosveld F., and Bosch B. J. (2020) A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 11, 2251–2256 10.1038/s41467-020-16256-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Cheng Y., Wong R., Soo Y. O. Y., Wong W. S., Lee C. K., Ng M. H. L., Chan P., Wong K. C., Leung C. B., and Cheng G. (2005) Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 24, 44–46 10.1007/s10096-004-1271-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Soo Y. O. Y., Cheng Y., Wong R., Hui D. S., Lee C. K., Tsang K. K. S., Ng M. H. L., Chan P., Cheng G., and Sung J. J. Y. (2004) Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin. Microbiol. Infect. 10, 676–678 10.1111/j.1469-0691.2004.00956.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Yeh K.-M., Chiueh T.-S., Siu L. K., Lin J.-C., Chan P. K. S., Peng M.-Y., Wan H.-L., Chen J.-H., Hu B.-S., Perng C.-L., Lu J.-J., and Chang F.-Y. (2005) Experience of using convalescent plasma for severe acute respiratory syndrome among healthcare workers in a Taiwan hospital. J. Antimicrob. Chemother. 56, 919–922 10.1093/jac/dki346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Tang F., Quan Y., Xin Z.-T., Wrammert J., Ma M.-J., Lv H., Wang T.-B., Yang H., Richardus J. H., Liu W., and Cao W.-C. (2011) Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 186, 7264–7268 10.4049/jimmunol.0903490 [DOI] [PubMed] [Google Scholar]
- 293. Kalia V., Sarkar S., Gourley T. S., Rouse B. T., and Ahmed R. (2006) Differentiation of memory B and T cells. Curr. Opin. Immunol. 18, 255–264 10.1016/j.coi.2006.03.020 [DOI] [PubMed] [Google Scholar]
- 294. Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., and Baric R. S. (2011) A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 85, 12201–12215 10.1128/JVI.06048-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Brunn, von A., Teepe C., Simpson J. C., Pepperkok R., Friedel C. C., Zimmer R., Roberts R., Baric R., and Haas J. (2007) Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE 2, e459 10.1371/journal.pone.0000459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., DeDiego M. L., Enjuanes L., and Guo D. (2008) Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS ONE 3, e3299 10.1371/journal.pone.0003299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Imbert I., Snijder E. J., Dimitrova M., Guillemot J.-C., Lécine P., and Canard B. (2008) The SARS-coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 133, 136–148 10.1016/j.virusres.2007.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Snijder E. J., Decroly E., and Ziebuhr J. (2016) The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus. Res. 96, 59–126 10.1016/bs.aivir.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Delmas B., and Laude H. (1990) Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 64, 5367–5375 10.1128/JVI.64.11.5367-5375.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Beniac D. R., Andonov A., Grudeski E., and Booth T. F. (2006) Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 13, 751–752 10.1038/nsmb1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Nieto-Torres J. L., DeDiego M. L., Álvarez E., Jiménez-Guardeño J. M., Regla-Nava J. A., Llorente M., Kremer L., Shuo S., and Enjuanes L. (2011) Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology 415, 69–82 10.1016/j.virol.2011.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. DeDiego M. L., Álvarez E., Almazán F., Rejas M. T., Lamirande E., Roberts A., Shieh W.-J., Zaki S. R., Subbarao K., and Enjuanes L. (2007) A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 81, 1701–1713 10.1128/JVI.01467-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Kuo L., and Masters P. S. (2003) The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77, 4597–4608 10.1128/jvi.77.8.4597-4608.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Neuman B. W., Kiss G., Kunding A. H., Bhella D., Baksh M. F., Connelly S., Droese B., Klaus J. P., Makino S., Sawicki S. G., Siddell S. G., Stamou D. G., Wilson I. A., Kuhn P., and Buchmeier M. J. (2011) A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 174, 11–22 10.1016/j.jsb.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.-Y., and Altmeyer R. (2005) Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 86, 1423–1434 10.1099/vir.0.80671-0 [DOI] [PubMed] [Google Scholar]
- 306. Chang C.-K., Sue S.-C., Yu T.-H., Hsieh C.-M., Tsai C.-K., Chiang Y.-C., Lee S.-J., Hsiao H.-H., Wu W.-J., Chang W.-L., Lin C.-H., and Huang T.-H. (2006) Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 13, 59–72 10.1007/s11373-005-9035-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Cui L., Wang H., Ji Y., Yang J., Xu S., Huang X., Wang Z., Qin L., Tien P., Zhou X., Guo D., and Chen Y. (2015) The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. Virol. 89, 9029–9043 10.1128/JVI.01331-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Hurst K. R., Koetzner C. A., and Masters P. S. (2009) Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 83, 7221–7234 10.1128/JVI.00440-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Lei J., Kusov Y., and Hilgenfeld R. (2018) Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 149, 58–74 10.1016/j.antiviral.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Serrano P., Johnson M. A., Chatterjee A., Neuman B. W., Joseph J. S., Buchmeier M. J., Kuhn P., and Wüthrich K. (2009) Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3. J. Virol. 83, 12998–13008 10.1128/JVI.01253-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Gadlage M. J., Sparks J. S., Beachboard D. C., Cox R. G., Doyle J. D., Stobart C. C., and Denison M. R. (2010) Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J. Virol. 84, 280–290 10.1128/JVI.01772-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Beachboard D. C., Anderson-Daniels J. M., and Denison M. R. (2015) Mutations across murine hepatitis virus nsp4 alter virus fitness and membrane modifications. J. Virol. 89, 2080–2089 10.1128/JVI.02776-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Stobart C. C., Sexton N. R., Munjal H., Lu X., Molland K. L., Tomar S., Mesecar A. D., and Denison M. R. (2013) Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J. Virol. 87, 12611–12618 10.1128/JVI.02050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Zhu X., Wang D., Zhou J., Pan T., Chen J., Yang Y., Lv M., Ye X., Peng G., Fang L., and Xiao S. (2017) Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J. Virol. 91, 51–14 10.1128/JVI.00003-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Cottam E. M., Whelband M. C., and Wileman T. (2014) Coronavirus NSP6 restricts autophagosome expansion. Autophagy 10, 1426–1441 10.4161/auto.29309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Bhardwaj K., Sun J., Holzenburg A., Guarino L. A., and Kao C. C. (2006) RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 361, 243–256 10.1016/j.jmb.2006.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Deng X., Hackbart M., Mettelman R. C., O'Brien A., Mielech A. M., Yi G., Kao C. C., and Baker S. C. (2017) Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U.S.A. 114, E4251–E4260 10.1073/pnas.1618310114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Zhang L., Li L., Yan L., Ming Z., Jia Z., Lou Z., and Rao Z. (2018) Structural and biochemical characterization of endoribonuclease Nsp15 encoded by Middle East respiratory syndrome coronavirus. J. Virol. 92, 286–216 10.1128/JVI.00893-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., and Guo D. (2011) Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 7, e1002294 10.1371/journal.ppat.1002294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Shi P., Su Y., Li R., Liang Z., Dong S., and Huang J. (2019) PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. 265, 57–66 10.1016/j.virusres.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Freundt E. C., Yu L., Park E., Lenardo M. J., and Xu X.-N. (2009) Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J. Virol. 83, 6631–6640 10.1128/JVI.00367-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Hussain S., and Gallagher T. (2010) SARS-coronavirus protein 6 conformations required to impede protein import into the nucleus. Virus Res. 153, 299–304 10.1016/j.virusres.2010.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Zhao J., Falcón A., Zhou H., Netland J., Enjuanes L., Pérez Breña P., and Perlman S. (2009) Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J. Virol. 83, 2368–2373 10.1128/JVI.02371-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Hussain S., Perlman S., and Gallagher T. M. (2008) Severe acute respiratory syndrome coronavirus protein 6 accelerates murine hepatitis virus infections by more than one mechanism. J. Virol. 82, 7212–7222 10.1128/JVI.02406-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Sung S.-C., Chao C.-Y., Jeng K.-S., Yang J.-Y., and Lai M. M. C. (2009) The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology 387, 402–413 10.1016/j.virol.2009.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Law P. Y. P., Liu Y.-M., Geng H., Kwan K. H., Waye M. M.-Y., and Ho Y.-Y. (2006) Expression and functional characterization of the putative protein 8b of the severe acute respiratory syndrome-associated coronavirus. FEBS Lett. 580, 3643–3648 10.1016/j.febslet.2006.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]