Abstract
A novel reverse-transcriptase loop mediated amplification (RT-LAMP) method targeting genes encoding the Spike (S) protein and RNA-dependent RNA polymerase (RdRP) of SARS-CoV-2 has been developed. The LAMP assay achieves a comparable limit of detection (25–50 copies per reaction) to commonly used RT-PCR protocols using clinical samples quantified by digital droplet PCR. Precision, cross-reactivity, inclusivity, and limit of detection studies were performed according to regulatory standards. Clinical validation of dual-target RT-LAMP (S and RdRP gene) achieved a PPA of 98.48 % (95 % CI 91.84%–99.96%) and NPA 100.00 % (95 % CI 93.84%–100.00%) based on the E gene and N2 gene reference RT-PCR methods. The method has implications for development of point of care technology using isothermal amplification.
Keywords: SARS-CoV-2, Covid, Diagnostics, Molecular, LAMP, PCR, Validation, Assay
1. Introduction
Over the last several decades, we have witnessed the rise of both known and novel viruses, including human immunodeficiency virus (HIV), Severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East Respiratory Syndrome (MERS-CoV), influenza H1N1, Ebola virus (EBOV), Dengue (DENV), Chikungunya (CHIK), Zika (ZIKV), and most recently 2019 novel coronavirus (SARS-CoV-2/COVID-19) (Reperant and Osterhaus, 2017; Li et al., 2020). Most of these emerging viral infections have been triggered by a direct zoonotic (animal-to-human) transmission event or enhancement, proliferation and spread of vectors such as the mosquito in new geographic areas. In December 2019 and early January 2020, a cluster of pneumonia cases from a novel coronavirus, SARS-CoV-2, was reported in Wuhan, China (Li et al., 2020; Zhou et al., 2020; Wu et al., 2020).
SARS-CoV-2 has now resulted in a global pandemic with the epicentre at the time of writing in Europe and North America (CDC COVID-19 Response Team, 2020). A common theme in the public health response to COVID19 and similar threats is the lack of rapidly deployable testing in the field to screen large numbers of individuals in exposed areas, international ports of entry, and testing in quarantine locations such as the home residences, as well as low-resourced areas (Cheng et al., 2020). This hampers case finding and increases the number of individuals at risk of exposure and infection. With the ease of travel across continents, delayed testing and lack of screening programs in the field, global human-to-human transmission will continue at high rates. These factors make a pandemic very difficult to contain. Early identification of the virus and rapid deployment of targeted point of care tests (POCT) can stem the spread through immediate quarantine of infected persons (Nguyen et al., 2020). We used existing viral genome sequences to develop a SARS-CoV-2 loop mediated amplification (LAMP) assay for clinical use (Forster et al., 2020; Lu et al., 2020a). LAMP relies on an alternate set of reagent chemistry that does not depend on or hinder critical elements of the RT-PCR supply chain which is now under duress (Ivanov, 2020). Our group has previously demonstrated the utility of LAMP for other infectious agents like malaria and dengue (Sigera et al., 2019; Mohon et al., 2019; Girma et al., 2018).
2. Materials and methods
2.1. Patient samples and ethics
Clinical samples used in this study were frozen (−80 °C) archived nasopharyngeal (NP) swabs in universal transport medium (UTM) at Alberta Precision Laboratories (Calgary, Canada), University of Washington, and University of California, San Francisco. Ethics approval for use of the archived samples was obtained from the Conjoint Health Research Ethics Board (CHREB) of the University of Calgary (REB20−0402). This study was approved by the institutional review board (IRB) at University of California, San Francisco (UCSF IRB #10−02598) as a no subject contact study with waiver of consent. The use of de-identified specimens were deemed non-human subject work by the University of Washington Institutional Review Board (IRB).
2.2. LAMP primer design
Genomic sequences (cDNA) of the SARS-CoV-2 were retrieved from the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/) and multiple sequence alignment analysis (https://www.ebi.ac.uk/Tools/msa/clustalo/) was conducted with other related viruses. From the multiple sequence alignment, several regions unique to the SARS CoV-2 were identified. The primers were designed using the Primer Explorer V5 software (http://primerexplorer.jp/lampv5e/) by uploading the sequences of the S and RdRP gene. Initially, primers were designed with the default settings of the software and then screened based on the optimal self-dimer, 5’ end, and 3’end ΔG values. Additionally, the intervening sequence between F2/F1 and B1/B2 primer binding sites was manually set at a minimum of 18 nucleotides. Initial screening experiments were performed on several potential primer sets and two chosen based on amplification efficiency (data not shown). LAMP primer sets targeting unique regions of the Spike (S) protein gene and RNA-dependent RNA Polymerase gene (RdRP) were chosen (Table 1 ). For the external LAMP amplification control, primers were used against bacteriophage MS2 as previously described (Benzine et al., 2016).
Table 1.
Primer sets used in this study to perform RT-LAMP. Spike gene primer set is denoted S2 and RdRp primer set is denoted S3.
| Primer name | Sequence |
|---|---|
| S2-F3 | ATTCTAAGCACACGCCTAT |
| S2-B3 | GAAGATAACCCACATAATAAGCT |
| S2-F1P | ACCTATTGGCAAATCTACCAATGGTTTAGTGCGTGATCTCCCT |
| S2-B1P | ATCACTAGGTTTCAAACTTTACTTGCCTGTCCAACCTGAAGAAGA |
| S2-LPF | TTCTAAAGCCGAAAAACCCTG |
| S2-LPB | CATAGAAGTTATTTGACTCCTGGTG |
| S3-F3 | CACCTTATGGGTTGGGATT |
| S3-B3 | AACATATAGTGAACCGCCA |
| S3-F1P | GTTTGCGAGCAAGAACAAGTGAATGTGATAGAGCCATGCC |
| S3-B1P | ATACAACGTGTTGTAGCTTGTCACACATGACCATTTCACTCAA |
| S3-LPF | GGCCATAATTCTAAGCATGTTA |
| S3-LPB | ATTAGCTAATGAGTGTGCTCAAGTA |
2.3. In silico analysis of primer combinations to determine cross-reactivity and inclusivity
A Basic Local Alignment Search Tool (BLAST) search alignment (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for primers in set 2 (Spike gene) and set 3 (RdRP gene) were performed against a critical list of infectious agents that cause upper respiratory tract infections. A nucleotide local alignment using BLASTn with the default parameters was performed against the National Center of Biotechnology Information (NCBI) Nucleotide database (see Supplementary Data). The twelve RT-LAMP primers were aligned against 16,247 SARS-CoV-2 (taxid: 2697049) viral sequences in the NCBI nucleotide database available on August 20th, 2020. The output of the “discontiguous megablast” algorithm showed no divergence available between the 12 primers and the SARS-CoV-2 database and all of the query sequences showed 100 % identity against the expected sequences from the SARS-CoV-2 virus (92 genomes).
2.4. Design of artificial viral targets for LAMP assay verification
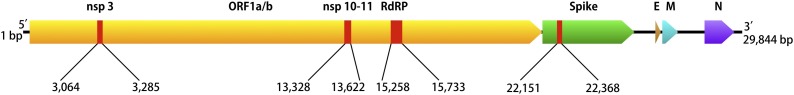
Four fragments of specific SARS-CoV-2 regions (ORF1ab (nsp3,10–11), RdRP (nsp 12), and spike (S)) were synthesized by SGI-DNA Inc. (San Diego, CA). Fragments were ligated to make one large concatenated DNA template using the BioXP3200 (SGI-DNA, San Diego, CA) automated Gibson assembly system. The final template was 1097 base pairs long containing a concatenated single artificial construct together with flanking plasmid sequence in that order (Fig. 1 ). To create a recombinant virus expressing the relevant RNA, the template containing the targeted sequences of interest was cloned into Sindbis Virus (SV) viral vector system (SINrep5) containing green fluorescent protein (EGFP) and then transfected into BHK21 cell lines (Bredenbeek et al., 1993; van Marle et al., 2003). The number of RNA genome copies was based on the number of fluorescent focus forming units generated by the recombinant SV vector.
Fig. 1.
Map of the gene fragments from SARS-CoV-2 (Genbank ID MT2078.1) that were used for synthesizing the genetic construct template. Four fragments of specific SARS-CoV-2 regions (ORF1ab (nsp3,10–11), RdRP (nsp 12), and spike (S)) were concatenated into a single artificial construct 1097 base pairs long.
2.5. LAMP assay conditions
The dual-target LAMP reaction was conducted using a combination of Warmstart® Rtx Reverse Transcriptase (New England Biolab, Whitby, ON) with Bst 2.0 Warmstart® DNA Polymerase (New England Biolab, Whitby, ON). In a 25 μL LAMP reaction mixture, 1.6 μM F1P and B1P, 0.8 μM LPF and LPB, 0.2 μM F3 and B3 primer concentrations, 8 mM MgSO4, 1.4 mM dNTPs, 8 units of Bst 2.0 WarmStart® DNA Polymerase and 15 units of Warmstart® RTx Reverse Transcriptase were used. The assay relied on 0.5 μL of 50X SYBR safe (Invitrogen) in the 25 μL reaction mixture for fluorescent detection. LAMP primers are reconstituted freshly each day and heated for 2 min at 95 °C prior to addition to the mastermix followed by immediate commencement of the LAMP reaction. The addition of 40 mM Guanidine hydrochloride (pH8.0) (Sigma, Oakville, ON) was used to optimize reaction conditions for RT-LAMP. For LAMP experiments, 12 μL of template was used in the 25 μL reaction mixture. Amplification was measured through increased relative fluorescence units (RFU) per minute in the CFX-96 Real-Time PCR detection system (Bio-Rad, Mississauga, ON). RFU values above 100 on the CFX-96 instrument were considered a qualitative positive when associated with a typical amplification curve before a 30-minute reaction time with concomitant amplification of the MS2 control. Amplification curves crossing the threshold after 30 min are considered negative. The 30-minute threshold was chosen based on pilot experiments in order to reduce false positives (data not shown).
2.6. Limit of detection and precision studies
Limit of detection of the LAMP assay was evaluated by using a nasopharyngeal (NP) swab sample infected with SARS-CoV-2 for which the viral load was quantified using digital droplet PCR. The NP sample (10 μL) was serially diluted to achieve the described copies of virus per LAMP reaction. Calculation of viral copy number using digital droplet PCR is described in detail in Supplementary Methods. Reproducibility of LOD (n = 24) studies at 25−50 copies per reaction were conducted in this way. Precision studies on separate days were performed using the artificial viral target described earlier. Precision studies were performed on two replicates, twice a day, for twenty days on a single CFX-96 instrument, and also 5 replicates daily for 5 days on 3 separate CFX-96 instruments.
2.7. Validation using clinical samples
Nasopharyngeal (NP) swabs (n = 100) and contrived samples (n = 24) were used in the analysis. Contrived samples were generated by using nasopharyngeal (NP) swabs infected with SARS-CoV-2 for which the viral load was quantified using digital droplet PCR. The contrived samples were at 100 copies of virus per reaction. Extraction of RNA from clinical NP swabs was performed using Promega SV Total RNA Isolation System (Madison, WI). The reference standard RT-PCR assays used in this study as a reference method were performed according to previous publications for the E gene (Corman et al. (2020)) and N2 gene (Centers for Disease Control and Prevention, 2020). The E gene RT-PCR was performed with modification according to the Alberta Public Health Laboratory reference method (Pabbaraju et al., 2020). The modifications included the addition of GC clamps at the 3′ end of the primers and the shortening and addition of a minor groove binding (MGB) moiety to the hydrolysis probe. Both E and N2 gene RT-PCR had to be positive for the sample to be considered a true positive. Four common human coronavirus samples (strain 0C43, NL63, 229E, and HKU1), inactivated MERS-CoV and SARS-CoV-1 (Zeptometrix Corp., Buffalo, NY), respiratory syncytial virus (RSV), and Influenza H1N1 clinical samples were directly tested by the S and RdRP LAMP primer sets to evaluate the specificity of the test but not included in the validation data set.
3. Results
3.1. Analytical studies using dual-target SARS-CoV-2 RT-LAMP
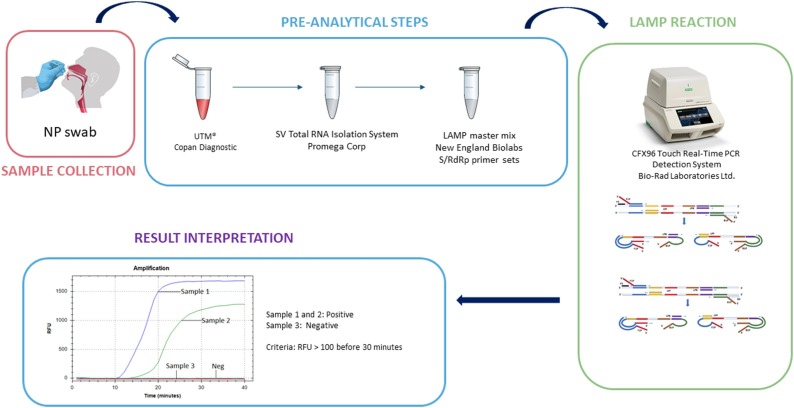
The workflow used to conduct RT-LAMP is depicted in Fig. 2 . The primer sequences used to target the Spike gene (set S2) and RdRP gene (Set 3) are listed in Table 1. The limit of detection was evaluated using a patient sample (NP swab in UTM viral load confirmed by digital droplet PCR). The quantified sample was serially diluted to achieve a range from 100 to 12.5 copies per reaction. The LOD was confirmed at 25 copies per reaction when using 40 mM Guanidine hydrochloride (pH8.0) in the reaction mix (Table 2 ). Twenty four replicates from a serial dilution containing 25−50 copies of SARS-CoV-2 which equates to 1-2X LOD (patient sample NP swab in UTM viral load confirmed by digital droplet PCR) per reaction were tested using dual-target RT-LAMP (Table 3 ). Twenty-three of 24 samples (95.8 %) were positive. In silico analysis confirmed that no significant cross-reactivity that affects LAMP reactions that rely on six primers per reaction were present (Supplementary Table 1). Finally, further studies were performed for precision on a daily basis (2 replicates for 20 days twice day and 5 replicates on 3 instruments daily for 5 days) that demonstrated 100 % concordance (Supplementary Tables 2 and 3).
Fig. 2.
Workflow used to analyze samples in this study. Images were obtained from the Centers for Disease Control (www.cdc.gov) and Bio-Rad Laboratories (www.bio-rad.com).
Table 2.
The limit of detection was evaluated using a patient sample (NP swab in UTM viral copies confirmed by digital droplet PCR) serially diluted to achieve a range from 100 to 12.5 copies per reaction for RT-LAMP. A single representative experiment is shown (minutes for a positive call). R – replicate; Neg – negative; UTM – universal transport medium; NTC – no template control.
| Patient sample NP UTM | R1 | R2 | R3 |
|---|---|---|---|
| 100 copies/reaction | 16.38 | 16.38 | 16.38 |
| 50 copies/reaction | 15.11 | 12.04 | 15.07 |
| 25 copies/reaction | 13.27 | 18.77 | 12.76 |
| 12.5 copies/reaction | 16.51 | 15.42 | Neg |
| NTC | Neg | Neg | Neg |
Table 3.
Replicates (n = 24) from a serial dilution containing approximately 25-50 copies of SARS-CoV-2 (patient sample NP swab in UTM viral copies confirmed by digital droplet PCR) per reaction at approximately 1-2X LOD was tested using dual target RT-LAMP.
| NP swab sample (25−50 copies per reaction) | S and RdRP RT-LAMP | Minutes for positive call | MS2 LAMP | Minutes for positive call | E-gene PCR | Ct value-E gene |
|---|---|---|---|---|---|---|
| Replicate 1 | Pos | 11.78 | Pos | 11.05 | Pos | 31.79 |
| Replicate 2 | Pos | 12.47 | Pos | 10.87 | Pos | 31.13 |
| Replicate 3 | Pos | 18.2 | Pos | 10.69 | Pos | 31.33 |
| Replicate 4 | Neg | N/A | Pos | 10.9 | Pos | 31.58 |
| Replicate 5 | Pos | 11.28 | Pos | 10.76 | Pos | 31.17 |
| Replicate 6 | Pos | 10.18 | Pos | 11.05 | Pos | 31.86 |
| Replicate 7 | Pos | 11.28 | Pos | 11.05 | Pos | 31.71 |
| Replicate 8 | Pos | 11.71 | Pos | 10.95 | Pos | 30.61 |
| Replicate 9 | Pos | 9.32 | Pos | 11.04 | Pos | 30.76 |
| Replicate 10 | Pos | 11.04 | Pos | 10.57 | Pos | 31.02 |
| Replicate 11 | Pos | 10.88 | Pos | 10.86 | Pos | 30.82 |
| Replicate 12 | Pos | 10.52 | Pos | 10.96 | Pos | 30.86 |
| Replicate 13 | Pos | 21.98 | Pos | 11.33 | Pos | 30.94 |
| Replicate 14 | Pos | 11.15 | Pos | 11.01 | Pos | 30.86 |
| Replicate 15 | Pos | 21.09 | Pos | 10.9 | Pos | 32.3 |
| Replicate 16 | Pos | 12.67 | Pos | 10.34 | Pos | 31.12 |
| Replicate 17 | Pos | 10.98 | Pos | 11.2 | Pos | 31.08 |
| Replicate 18 | Pos | 16.13 | Pos | 10.79 | Pos | 31.65 |
| Replicate 19 | Pos | 9.94 | Pos | 10.7 | Pos | 30.39 |
| Replicate 20 | Pos | 11.02 | Pos | 10.62 | Pos | 31.84 |
| Replicate 21 | Pos | 25.28 | Pos | 10.64 | Pos | 31.53 |
| Replicate 22 | Pos | 10.32 | Pos | 10.97 | Pos | 31.04 |
| Replicate 23 | Pos | 10.94 | Pos | 10.34 | Pos | 31.62 |
| Replicate 24 | Pos | 10.71 | Pos | 10.57 | Pos | 31.62 |
| Positive Control | Pos | 6.78 | Pos | 9.02 | Pos | 20.23 |
| Negative Control 1 | Neg | NA | Neg | NA | Neg | NA |
| Negative Control 2 | Neg | NA | Neg | NA | Neg | NA |
| Negative Control 3 | Neg | NA | Neg | NA | Neg | NA |
3.2. Clinical validation of SARS-CoV-2 LAMP using clinical and contrived samples
A clinical validation sample set of nasopharyngeal swabs were used in the analysis. Given no gold standard exists, percent positive agreement (PPA) and negative percent agreement (NPA) were calculated. Reference methods included two RT-PCR (E gene and N2 gene) methods employed by reference laboratories. Dual-target RT-LAMP (S and RdRP gene) achieved a PPA of 98.48 % (95 % CI 91.84%–99.96%) and NPA 100.00 % (95 % CI 93.84%–100.00%) based on the E gene and N2 gene reference RT-PCR methods (Table 4 ). One false negative sample by RT-LAMP was positive by both E gene (Ct 33.5) and N2 gene (Ct 35.1). One sample out of 124 was strongly positive by the N2 gene (Ct 17.4) and negative by E gene and considered a true positive. No cross-reactivity was observed with known circulating respiratory viruses, namely (HCoV) OC43, 229E, NL63, HKU1; RSV; and influenza virus A (H1N1) pdm09 (data not shown).
Table 4.
Validation of dual-target RT-LAMP (S and RdRP genes) using a validation set of clinical samples. PPA - positive percent agreement; NPA - negative percent agreement.
| RT-LAMP (S and RdRP gene) | RT-PCR (E and N2 gene) |
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 65 | 0 | 65 |
| Negative | 1 | 58 | 59 |
| Total | 66 | 58 | 124 |
| PPA | 98.48 % (95 % CI 91.84%–99.96%) | ||
| NPA | 100.00 % (95 % CI 93.84%–100.00%) | ||
4. Discussion
The global pandemic with SARS-CoV-2 has resulted in the need for diagnostic test development at a scale never seen before. Rapid deployment of validated laboratory-developed diagnostic tests or commercial tests is essential to the containment of the virus as it allows for self-quarantine measures to be imposed in a strategic fashion before widespread community transmission occurs (Cheng et al., 2020; Nguyen et al., 2020). Diagnostic tests have to be analytically sensitive in order to not to miss any cases in the acute phase of viremia (Sigera et al., 2019). As such, nucleic acid amplification tests serve this purpose. In particular, RT-PCR has been employed as the primary diagnostic counter-measure (Nalla et al., 2020). However, reagent supply chains for key items are under immense pressure. Local solutions to reagent sources have become paramount because barriers to trade of these selected items have been a concern.
The dual-target RT-LAMP test for SARS-CoV-2 developed in this study has comparable analytical sensitivity and specificity, limit of detection, precision, and achieved excellent agreement compared to the reference RT-PCR methods used internationally. The addition of guanidine hydrochloride (pH8.0) at 40 mM in the LAMP reaction improves the limit of detection (25–50 copies per reaction) to a level comparable to RT-PCR methods. LAMP does not rely on the same reagents as RT-PCR and thus alleviates pressure on key supply chain items. The LAMP method is amenable to high throughput testing in either 96-well or 384-well. Other groups have presented RT-LAMP solutions in the literature (Huang et al., 2020; Lu et al., 2020b; Park et al., 2020; Yan et al., 2020; Lamb et al., 2020; Fowler et al., 2020). The LAMP solutions differ in several ways: first, the target genes of choice vary between studies as do the specific primer sequences chosen; second, the limits of detection reported vary in terms of SARS-CoV-2 copies detected per reaction; third, the detection systems vary from thermocycler-based detection for laboratory developed test (LDT) solutions to near-patient solutions based on visual detection of dyes or fluorophores; fourth, the extent to which data reflect requirements for clinical validation. The RT-LAMP assay described here is unique in the SARS-CoV-2 LAMP literature to date in that it offers validation data meeting current regulatory standards which include precision studies on several instruments, reproducibility studies over 20 days, a robust clinical validation sample set, and a limit of detection equal or superior to other LAMP studies. These data should enable a clinical laboratory to perform this assay as a LDT.
Additionally, the LAMP assay chemistry presented in this work is able to detect SARS-CoV-2 in UTM without the need for a kit-based RNA extraction method using lyophilized reagents and visual detection (manuscript in preparation). This format may be of particular interest to resource-limited settings. Limitations of the study include not testing other sample types such as alternate swabs, nasal washes, saliva, sputum, or stool. This work is ongoing with a special emphasis on swab-free testing and direct visualization. LAMP presents a much needed alternative approach to SARS-CoV-2 diagnostic testing that is available for deployment immediately in a laboratory-developed test format as it relies on other key reagents that do not cannibalize RT-PCR reagents. In the future, the LAMP chemistry has potential to be adapted to a microfluidic device POCT to be deployed in the community, either at ports of entry, homes, pharmacies, or workplaces.
Funding statement
Funding for this study was obtained from the Canadian Institutes for Health Research (NFRFR-2019-00015, DRP,CC), Genome Canada (DRP), and the M.J. Murdock Charitable Trust (KRJ).
CRediT authorship contribution statement
Abu Naser Mohon: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Writing - original draft, Writing - review & editing. Lisa Oberding: Data curation, Methodology. Jana Hundt: Methodology, Writing - review & editing. Guido van Marle: Resources, Writing - review & editing. Kanti Pabbaraju: Data curation, Resources, Writing - review & editing. Byron M. Berenger: Data curation, Resources, Writing - review & editing. Luiz Lisboa: Writing - review & editing. Thomas Griener: Methodology, Writing - review & editing. Markus Czub: Methodology, Writing - review & editing. Cody Doolan: Methodology, Writing - review & editing. Venice Servellita: Methodology. Charles Y. Chiu: Funding acquisition, Investigation, Supervision, Writing - review & editing. Alexander L. Greninger: Funding acquisition, Investigation, Supervision, Writing - review & editing. Keith R. Jerome: Funding acquisition, Investigation, Supervision, Writing - review & editing. Dylan R. Pillai: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors report no declarations of interest. CD is an employee of Illucidx Inc. ANM and DRP have patents filed related to LAMP technology.
Acknowledgments
Dr. Ranmalee Amarasekara, Dr. Tara Winstone, and Barbara Chow for expert technical assistance, Daniel Castaneda Mogollon for performing bioinformatics analysis, and Omar Abdullah and Noah Toppings for research analytical support.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2020.113972.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Benzine J.W., Brown K.M., Agans K.N., Godiska R., Mire C.E., Gowda K., Converse B., Geisbert T.W., Mead D.A., Chander Y. Molecular diagnostic field test for point-of-care detection of ebola virus directly from blood. J. Infect. Dis. 2016;214:S234–S242. doi: 10.1093/infdis/jiw330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P.J., Frolov I., Rice C.M., Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC COVID-19 Response Team Severe outcomes among patients with coronavirus disease 2019 (COVID-19) - United States, February 12-March 16, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. Centers for Disease Control and Prevention.
- Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., Dittrich S., Yansouni C.P. Diagnostic testing for severe acute respiratory syndrome-related Coronavirus-2: a narrative review. Ann. Intern. Med. 2020;172(11):726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. Bull. Eur. Sur Mal Transm. Eur. Commun Dis. Bull. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U. S. A. 2020;117(17):9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler V.L., Armson B., Gonzales J.L., Wise E.L., Howson E.L.A., Vincent-Mistiaen Z., Fouch S., Maltby C.J., Grippon S., Munro S., Jones L., Holmes T., Tillyer C., Elwell J., Sowood A., Santos H., de Peyer O., Dixon S., Hatcher T., Sivanesan S., Knight H., Laxman S., Walsh C., Andreou M., Morant N., Clark D., Houghton R., Moore N., Cortes N., Kidd S.P. A reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 within nasopharyngeal and oropharyngeal swabs at Hampshire Hospitals NHS Foundation Trust. Preprint. Infect. Dis. (except HIV/AIDS) 2020 [Google Scholar]
- Girma S., Cheaveau J., Mohon A.N., Marasinghe D., Legese R., Balasingam N., Abera A., Feleke S.M., Golassa L., Pillai D.R. Prevalence and epidemiological characteristics of asymptomatic malaria based on ultrasensitive diagnostics: a cross-sectional study. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018;69(6):1003–1010. doi: 10.1093/cid/ciy1005. [DOI] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D. Predicting the impacts of epidemic outbreaks on global supply chains: a simulation-based analysis on the coronavirus outbreak (COVID-19/SARS-CoV-2) case. Transp. Res. Part E Logist Transp. Rev. 2020;136 doi: 10.1016/j.tre.2020.101922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet Lond. Engl. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int. J. Mol. Sci. 2020;21(8):2826. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohon A.N., Getie S., Jahan N., Alam M.S., Pillai D.R. Ultrasensitive loop mediated isothermal amplification (US-LAMP) to detect malaria for elimination. Malar. J. 2019;18:350. doi: 10.1186/s12936-019-2979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer/probe sets and one assay kit. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Duong Bang D., Wolff A. 2019 novel coronavirus disease (COVID-19): paving the road for rapid detection and point-of-Care diagnostics. Micromachines. 2020:11. doi: 10.3390/mi11030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabbaraju, K., Wong, A.A., Douesnard, M., Ma, R., Gill, K., Dieu, P., Fonseca, K., Zelyas, N., Tipples, G.A. Development and validation of reverse transcriptase-PCR assays for the testing of SARS-CoV-2. J. Assoc. Med. Microbiol. Infect. Dis. (in press). [DOI] [PMC free article] [PubMed]
- Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.-J., Kim S.-J., Maeng J.-S. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. J. Mol. Diagn. 2020;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reperant L.A., Osterhaus A.D.M.E. AIDS, Avian flu, SARS, MERS, Ebola, Zika… what next? Vaccine. 2017;35:4470–4474. doi: 10.1016/j.vaccine.2017.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigera P.C., Amarasekara R., Rodrigo C., Rajapakse S., Weeratunga P., De Silva N.L., Huang C.H., Sahoo M.K., Pinsky B.A., Pillai D.R., Tissera H.A., Jayasinghe S., Handunnetti S., Fernando S.D. Risk prediction for severe disease and better diagnostic accuracy in early dengue infection; the Colombo dengue study. BMC Infect. Dis. 2019;19:680. doi: 10.1186/s12879-019-4304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., Ethier J., Silva C., Mac Vicar B.A., Power C. Human immunodeficiency virus type 1 envelope-mediated neuropathogenesis: targeted gene delivery by a Sindbis virus expression vector. Virology. 2003;309:61–74. doi: 10.1016/s0042-6822(02)00139-3. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.