Abstract
Amyotrophic lateral sclerosis is a disease characterized by progressive paralysis and death. Most ALS-cases are sporadic (sALS) and patient heterogeneity poses challenges for effective therapies. Applying metabolite profiling on 77-sALS patient-derived-fibroblasts and 43-controls, we found ~25% of sALS cases (termed sALS-1) are characterized by transsulfuration pathway upregulation, where methionine-derived-homocysteine is channeled into cysteine for glutathione synthesis. sALS-1 fibroblasts selectively exhibited a growth defect under oxidative conditions, fully-rescued by N-acetylcysteine (NAC). [U–13C]-glucose tracing showed transsulfuration pathway activation with accelerated glucose flux into the Krebs cycle. We established a four-metabolite support vector machine model predicting sALS-1 metabotype with 97.5% accuracy. Both sALS-1 metabotype and growth phenotype were validated in an independent cohort of sALS cases. Importantly, plasma metabolite profiling identified a system-wide cysteine metabolism perturbation as a hallmark of sALS-1. Findings reveal that sALS patients can be stratified into distinct metabotypes with differential sensitivity to metabolic stress, providing novel insights for personalized therapy.
Keywords: Sporadic amyotrophic lateral sclerosis, Metabolomics, Stable isotope tracing, Transsulfuration, Cysteine, Methionine, Disease stratification
1. Background
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by progressive death of upper and lower motor neurons; symptoms include muscle weakness and atrophy, commonly leading to fatal paralysis within 5 years of disease onset (Kiernan et al., 2011). Approximately 90% of ALS patients have no familial history (a condition termed sporadic ALS; sALS), while the remaining 10% are due to recognized inherited gene mutations (Ghasemi and Brown Jr., 2017). Unfortunately, mechanisms leading to motor neuronal death in sALS are completely unknown and, as a consequence, there are neither viable disease biomarkers nor effective pharmacotherapies. Past ALS clinical trials have been largely unsuccessful (Miller et al., 2015) and currently there are only two approved drugs, Riluzole and Edaravone, both of which only prolong the lifetime of ALS cases by a few months. Conceivably, sALS can originate from multiple mechanistic triggers, and disease stratification (i.e., recognition of distinct disease-driver subtypes) holds potential to uncover new and effective targeted therapies.
As heterogeneity of the sALS patient population presents a major stumbling stone to rigorous mechanistic studies, stratifying sALS has become an area of considerable interest (Marin et al., 2016; Konrad et al., 2017). The worldwide ALS Stratification Prize - Using the Power of Big Data and Crowdsourcing for Catalyzing Breakthroughs in ALS was initiated in 2015, aiming to address the problem of ALS patient heterogeneity with consideration of key clinical distinctions, such as rankings on the ALS functional rating scale (Kuffner et al., 2015). Stratification efforts have identified several potential non-standard predictors of disease progression, including plasma uric acid, creatinine and surprisingly, blood pressure that may be related to sALS pathobiology (Kuffner et al., 2015). While past attempts at sALS case stratification have primarily relied on clinical parameters to anticipate disease progression, recognition of disease-associated metabolic types (i.e., metabotypes) is anticipated to be more telling, with potential to identify distinct molecular disease origins that inform rationally-targeted therapies.
Altered energy metabolism often precedes the clinical onset of ALS (Huisman et al., 2015). Moreover, alterations in energy metabolism have been inferred as a potential pathogenic mechanism underlying sALS (Valbuena et al., 2016; Kirk et al., 2014). It has recently been reported that hypermetabolism in ALS is associated with greater functional decline and accelerated mortality (Steyn et al., 2018). Notably, as the final products of genes, transcripts, and proteins, metabolites offer the most proximal insights into sALS disease mechanisms. Whereas specific metabolic perturbations that underlie bioenergetic alterations in sALS have remained elusive, understanding this phenomenon will likely inform on disease mechanisms and prove transformational for clinical care.
We sought to investigate whether the metabotype of dermal fibroblasts from sALS patients can enable patient subtyping and disease stratification that sheds light on underlying disease mechanisms. Notably, fibroblasts carry the same genetic composition as neurons, but unlike neural tissue, fibroblasts are readily accessible from patients for in vitro culture and analysis. Indeed, several studies have demonstrated the feasibility of fibroblasts as cellular models for ALS (Raman et al., 2015; Narayan et al., 2016; Allen et al., 2015; Codron et al., 2018) and prior studies of fibroblast metabolism have revealed bioenergetic alterations in sALS cases (Konrad et al., 2017; Kirk et al., 2014; Yang et al., 2015). Here, for the first time, we performed untargeted metabolite profiling on fibroblasts from sALS patients and healthy control subjects, in combination with assessments of cell viability and determination of plasma metabolites and gene expression profiles. Studies reveal a unique subset of sALS patients that display a distinct metabotype, typified by accelerated transsulfuration pathway-derived cysteine for support of GSH biosynthesis, along with hypermetabolism of glucose. Functionally, cells with this metabotype exhibit an increased susceptibility to metabolic stress which can be prevented by supplementation with the antioxidant NAC. The finding that sALS patients can be classified based on metabotype, associated with susceptibility to oxidative stress and protection by antioxidant treatment, offers a potentially critical first-step toward targeted personalized medicines that can provide effective therapy for this newly identified sALS patient metabotype.
2. Methods
2.1. Reagents
LC-MS grade acetonitrile (ACN), isopropanol (IPA) and methanol (MeOH) were purchased from Fischer Scientific. High purity deionized water (ddH2O) was filtered from Millipore (18 OMG). OmniTrace glacial acetic acid and ammonium hydroxide were obtained from EMD Chemicals. [U–13C] glucose, [2,3,3–2H]-serine were purchased from Cambridge Isotope Laboratory. Ammonium acetate and all other chemicals and standards were obtained from Sigma Aldrich in the best available grade.
2.2. Cell culture
A total of 77 sALS and 43 control fibroblast cell lines were obtained from cases and propagated as previously described (Konrad et al. Mol Neuorodeg 2017). 75,000 cells/well for each line were plated in duplicate 6-well plates and cultured in DMEM medium containing 5 mM glucose and 4 mM glutamine, 10% FBS,1% of 100× antibiotic/anti-mycotic (Ab+F; which contains sterile-filtered 10,000 units penicillin, 10 mg streptomycin and 25 μg amphotericin B per mL, and 2.5 μg/ml Plasmocin). Cells were harvested at 80% confluency and extracted for LC/MS metabolomic analysis. The mean passage number for all 77 studied sALS cell lines was 7.2 +/− 1.9, and the mean passage of all 43 control cell lines was 8.9 +/− 1.7.
2.3. Metabolite extraction
Each cell line was cultured and initially extracted as two independent biological replicates for LC/MS metabolomic data acquisition. Cells were washed twice with ice-cold PBS, followed by metabolite extraction in −70 °C 80:20 methanol:water (LC-MS grade methanol, Fisher Scientific). The tissue–methanol mixture was subjected to beadbeating for 45 s using a Tissuelyser cell disrupter (Qiagen). Extracts were centrifuged for 5 min at 5000 rpm to pellet insoluble material and supernatants were transferred to clean tubes. The extraction procedure was repeated two additional times and all three supernatants were pooled, dried in a Vacufuge (Eppendorf) and stored at −80 °C until analysis. The methanol-insoluble protein pellet was solubilized in 0.2 M NaOH at 95 °C for 20 min and protein was quantified using a BioRad DC assay. On the day of metabolite analysis, dried cell extracts were reconstituted in 70% acetonitrile at a relative protein concentration of 1 μg/ml, and 4 μl of this reconstituted extract was injected for LC/MS-based untargeted metabolite profiling.
A group of 240 patient-derived skin fibroblast cell extracts (120 samples with 2 biological replicates each) were analyzed by LC-QTOF metabolomics profiling in random sequence. Table S1 lists clinical characteristics, bioenergetics and metabotype information on all cell lines studied by untargeted metabolite profiling, microarray analysis and stable isotope tracing. To adjust for day-to-day and batch-to-batch LC/MS instrument drift, metabolite stability, and other experimental factors that may contribute to systematic error, metabolite measurements were normalized to one another using flanking quality control (QC) samples run at intervals of every 6 injections. These QC samples were prepared from a pool of all samples and this normalization procedure enabled the comparative analysis of data acquired over a 1-month collection period. Inter-fibroblast variability in metabolite abundances was assessed by the histogram distribution of coefficient of variance (CV) for 1172 detected metabolites in the control group, showing that > 50% of the observed features exhibit a CV value of 0.2–0.4 (Fig. S1).
Plasma metabolites were extracted by addition of 1 part plasma to 20 parts 70% acetonitrile in ddH2O (vol:vol). The mixture was briefly vortexed and then centrifuged for 5 min at 16,000 ×g to pellet precipitated proteins. An aliquot of the resulting extract (3 μl) was subjected to untargeted metabolite profiling, applying liquid chromatography coupled to both positive and negative ion monitoring mass spectrometry (LC/MS).
2.4. Untargeted metabolite profiling LC/MS
Cell extracts were analyzed by LC/MS as described previously (Chen et al., 2012; Ismailoglu et al., 2014), using a platform comprised of an Agilent Model 1290 Infinity II liquid chromatography system coupled to an Agilent 6550 iFunnel time-of-flight MS analyzer. Chromatography of metabolites utilized aqueous normal phase (ANP) chromatography on a Diamond Hydride column (Microsolv). Mobile phases consisted of: (A) 50% isopropanol, containing 0.025% acetic acid, and (B) 90% acetonitrile containing 5 mM ammonium acetate. To eliminate the interference of metal ions on chromatographic peak integrity and electrospray ionization, EDTA was added to the mobile phase at a final concentration of 6 μM. The mobile phase gradient used was: 0–1.0 min, 99% B; 1.0–15.0 min, to 20% B; 15.0 to 29.0, 0% B; 29.1 to 37 min, 99% B. Raw data were analyzed using MassHunter Profinder 8.0 and MassProfiler Professional (MPP) 14.9.1 software (Agilent technologies). Mann Whitney t-tests (p < 0.05) were performed to identify significant differences between groups.
2.5. Metabolite structure specification
To ascertain the identities of differentially expressed metabolites (P < 0.05), LC/MS data were searched against an in-house annotated personal metabolite database, created using MassHunter PCDL manager 7.0 (Agilent Technologies) based on monoisotopic neutral mass (< 5 ppm mass accuracy) and chromatographic retention times of pure standards. A molecular formula generator (MFG) algorithm in MPP was used to generate and score empirical molecular formulae, based on a weighted consideration of monoisotopic mass accuracy, isotope abundance ratios, and spacing between isotope peaks. A tentative compound ID was assigned when the PCDL database and MFG scores concurred for a given candidate molecule. Tentatively assigned molecules were confirmed based on a match of LC retention times and/or MS/MS fragmentation spectra for pure molecular standards.
2.6. Stable isotope tracing of [U-13C] glucose and [2,3,3-2H] serine
An in-house untargeted stable isotope tracing (USIT) workflow was employed using Agilent metabolite profiling software MassHunter Qualitative Analysis 7.0, MassProfinder 8.0 and MPP 14.9.1. Labelled metabolites were identified on the basis of differential abundance in cells cultured with supplemental heavy isotope-labelled metabolite vs. natural (light) isotope, as described previously (Leung et al., 2017; Chen et al., 2018). Isotope-enriched metabolites were identified on the basis of a previously curated database of isotopologues. Notably, the USIT workflow calculates and corrects for the natural abundance of 13C and 2H isotope in samples.
2.7. Microarray analysis
Total skin fibroblast RNA was extracted from 27 sALS patients and 27 controls using Trizol reagent (Invitrogen). RNA was further processed using the Agilent Absolutely RNA miRNA Kit. The total RNA concentration and RNA sample integrity were verified using a Bio-Analyzer 2100 (Agilent Technologies, Waldbronn, Germany). The quality of isolated RNA was determined using an Agilent 2200 TapeStation system and Bioanalyzer with mRNA labeling and microarray processing that were performed according to the manufacturers recommendations. miRNA labeling was done using an Agilent miRNA Complete Labeling and Hyb Kit with gene expression and miRNA data extracted using Agilent Feature Extraction Software.
Extracted mRNA and miRNA data were analyzed using the differential expression workflow in GeneSpring GX 14.9.1 (Agilent Technologies, CA) described as follows: signal intensities for each mRNA probe was normalized to 75th percentile values, with baseline transformation for gene expression analysis. Separately, miRNA data was normalized to 90th percentile values and baseline transformed for miRNA analysis. The differential miRNA list (P < 0.05, FC > 1.2) was used to identify the gene targets based on a target prediction database incorporated in GeneSpring GX (e.g. TargetScan, PicTar, microRNA.org). The differentially expressed mRNA was combined with validated miRNA targets and metabolites for integrated multi-Omics pathway analysis using databases linked to the Kyoto Encyclopedia of Genes and Genomes (KEGG), Wiki pathways and Biocyc.
2.8. Fibroblast cell viability measurements and validation
Cells were seeded in 96 well imaging plates (cellvis, P96–1.5H-N) and cultured in phenol red- and glucose-free DMEM, supplemented with 5 mM galactose, 1 mM pyruvate and 2 mM glutamine. At 24, 48 and 72 h after seeding, 3 technical replicate wells for each patient cell line were stained with 1 μg/ml Hoechst 33342, 1 μg/ml propidium Iodide and 1 uM Calcein AM (thermo) for 20 min at 37 °C. Plates were subsequently imaged using a high content imager (Imagexpress pico, Molecular Devices). Nuclei were segmented using ImageJ software based on the Hoechst channel, and viable cells were counted after setting thresholds for minimum Calcein and maximum propidium Iodide fluorescence intensities, indicative of true viable cells. Rates of change in viable cell counts were calculated by fitting a line to cell counts over the three times of measurement.
2.9. Statistics
All values are averages of at least three independent measurements. Error bars indicate standard deviation (S.D.) or standard error of the mean (S.E.M.). Statistically significant differences between two groups were estimated by unpaired two-tailed Student’s t-test with significance set at p < 0.05.
3. Results
3.1. Untargeted metabolite profiling identifies a distinct sALS metabotype in case-derived fibroblasts
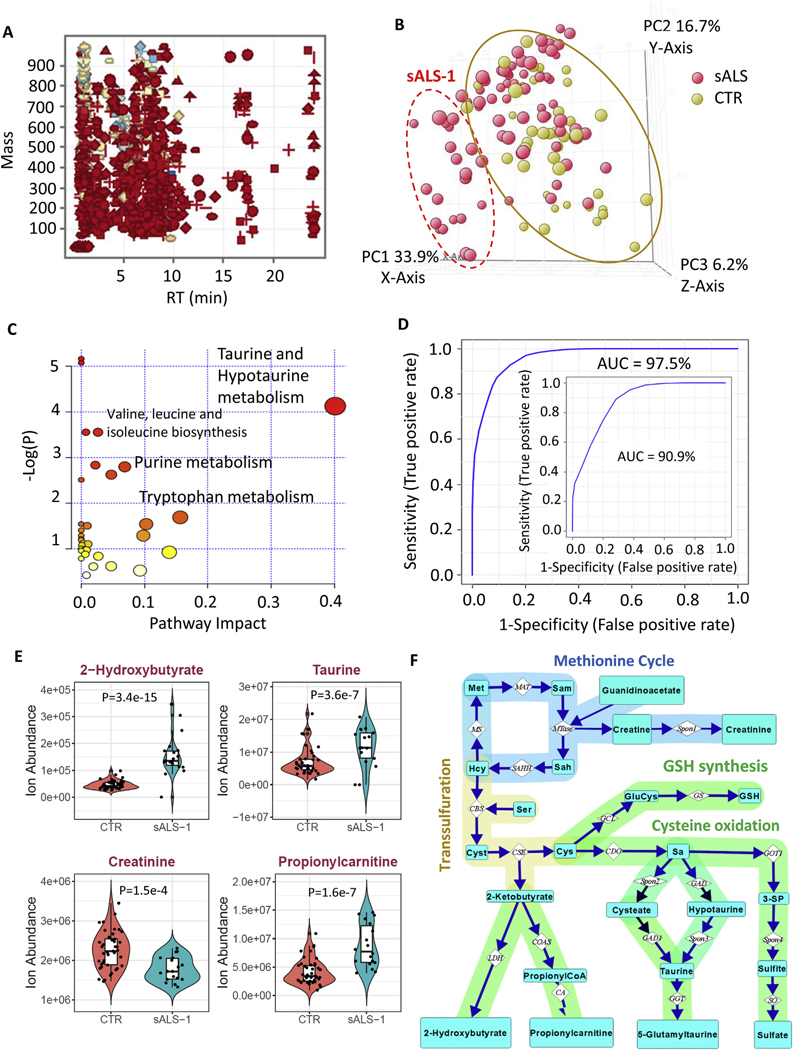
Table 1 summarizes the clinical characteristics of 77 sALS cases and 43 healthy controls studied herein. Patient-derived fibroblast cell lines had been previously genotyped and found to have no known familial ALS-associated mutations and were typically studied within 7–9 cell passages (Table 1). Dermal fibroblast cell lines were established from all cases and 1172 quality control-normalized metabolite features were detected by LC/MS in greater than 70% of cell extracts from either sALS or control fibroblast groups (Fig. 1A). Metabolite profiling and nonparametric Mann-Whitney statistical analysis (P < 0.05) revealed that 507 of the 1172 recognized features are differentially expressed in sALS fibroblasts (Table S2). Principal component analysis (PCA) did not completely separate sALS as a whole from the control group (Fig. 1B), but a distinct subgroup of 18/77 sALS patient-derived cell lines with PC1 score greater than 13 and PC2 score less than 8 clustered together, separating from controls based on principal component 1 (PC1) which accounted for 33.9% of total variance.
Table 1.
Clinical characteristics of the sALS and control cohorts.
| Subject | sALS-Male | sALS-Female | sALS-Total | CTR-Male | CTR-Female | CTR-Total |
|---|---|---|---|---|---|---|
| N | 43 | 34 | 77 | 23 | 20 | 43 |
| BMI | 26.54 ± 4.55 | 26.04 ± 6.41 | 26.32 ± 5.41 | NA | NA | NA |
| Age at Symptom Onset | 56.28 ± 9.92 | 62.44 ± 8.45 | 59.00 ± 9.74 | NA | NA | NA |
| Age at Skin BX | 57.65 ± 9.76 | 63.858 ± 0.44 | 60.39 ± 9.66 | 64.83 ± 8.95 | 56.95 ± 5.19 | 61.16 ± 8.36 |
| *ALSFRS-R at Biopsy | 32.81 ± 8.61 | 32.38 ± 7.80 | 32.62 ± 8.21 | NA | NA | NA |
| % Rate of Decline | 1.20 ± 0.84 | 1.08 ± 0.56 | 1.141 ± 0.72 | NA | NA | NA |
| #FVC (%) | 74.78 ± 25.61 | 75.84 ± 29.05 | 75.25 ± 27.00 | NA | NA | NA |
| Lumbosacral onset | 11 | 12 | 23 | NA | NA | NA |
| Bulbar onset | 8 | 9 | 17 | NA | NA | NA |
| Cervical onset | 21 | 12 | 33 | NA | NA | NA |
| Other onset | 2 | 2 | 4 | NA | NA | NA |
Fig. 1.
Untargeted metabolite profiling of fibroblast lines identifies an sALS subgroup with enhanced transsulfuration pathway and glucose hypermetabolism. A: mass and retention time scatter plot of 1172 quality control-normalized metabolite features detected in greater than 70% of samples in either 77 sALS or 43 control groups. B: PCA score plot showing incomplete separation of 77 sALS (red circles) from 43 controls (yellow circles), but a subgroup of 18 sALS patients (identified in the dashed red oval as sALS-1) was readily separated from controls. C: integrated enrichment and pathway topology analysis of differential metabolites in sALS-1 by Metaboanalyst 4.0, recognizing taurine and hypotaurine in the transsulfuration pathway as the most significant pathway impact and lowest matching P-value. Notably, the circle size is proportional to the pathway impact value and the colour of the circle represents the associated pathway P-value scaled as a heat map from yellow (high P-value) to red (low-P value). D: ROC curve derived from SVM model predicting 18 sALS-1 subgroup from 43 randomly selected controls. The AUC of the prediction is 97.5%. The inset showed the ROC curve and AUC for a repeat experiment comprising the same 18 sALS-1 subset vs. 18 randomly selected controls. E: Ion abundance of 4 sALS-1 predictor metabolites (2-hydroxybutyrate, taurine, propionylcarnitine and creatinine). Data was plotted as violin plots to display data frequency, with jitter points showing individual fibroblast lines and an inset boxplot presenting median value, interquartile range and the 95% confidence interval. P value was calculated by 2-tailed Student’s t-test, sALS-1, n = 18; CTR, n = 43. F: Schematic representation of metabolites and enzymes involved in the methionine cycle, methylation and transsulfuration pathways. Abbreviation of metabolites and enzymes are as follows: Met, methionine; Hcy: homocysteine; Sam: S-adenosylmethionine; Sah: S-adenosylhomocysteine; Cyst: cystathionine; Sa: sulfoalanine; 3-SP: 3-sulfopyruvate. The enzyme abbreviations were adapted from the KEGG pathway. MAT: methionine adenosyltransferase; MS: methionine synthase; SAHH: S-adenosylhomocysteine hydrolase; MTase: methyltransferase; CBS: cystathionine β-synthase; CSE: cystathionine γ-lyase; GCL: glutamate cysteine ligase; GS: GSH synthase; CDO: cysteine dioxygenase; LDH: lactate dehydrogenase; COAS: propionylCoA synthetase; CA: carnitine acyltransferase; GAD1: glutamate decarboxylase 1; GGT: Gamma-glutamyltransferase; GOT1: glutamic-oxaloacetic transaminase 1; SO: sulfite oxidase; Spon1, Spon2 and Spon3 refer to non-enzymatic spontaneous degradation process.
To confirm that the sALS subgroup (denoted sALS-1) could be reproducibly distinguished from the control group, we repeated the metabolic profiling in an independent replication experiment, comparing the same 18 sALS-1 cases vs. a group of 18 randomly selected control cell lines. Results confirmed 42 metabolites to be differentially-expressed in both the original study and the replication study (Fig. S2, Table S3). Among these metabolites, significant increases were observed for 2-hydroxybutyrate, taurine, and C3-C5 carnitines, along with significant decreases in creatinine and several amino acids. Integrated enrichment and pathway topology analysis of these differentially-expressed metabolites (applying Metaboanalyst 4.0) revealed the taurine/hypotaurine pathway to be the most significantly impacted (Fig. 1C).
Using the Find Minimal Entities-Forward Selection Algorithm - provided in the Agilent MassProfilerProfessional software package (version 14.9.1), along with a support vector machine (SVM) learning algorithm and receiver operative characteristics (ROC) curve-based biomarker analysis using metaboanalyst 4.0 (https://www.metaboanalyst.ca/MetaboAnalyst/upload/RocUploadView.xhtml), we generated a 4-metabolite linear SVM model (comprising 2-hydroxybutyrate, taurine, propionylcarnitine and creatinine) that recognized the sALS-1 subgroup with an overall accuracy of 97% in the original group (with 1/45 false-positives and 1/18 false-negatives; Fig. 1D) and 90.9% accuracy in the replication analysis. This SVM model was generated using a leave-oneout cross validation approach. Independent targeted quantitative analysis confirmed the observed decreased level of creatinine, and increased levels of 2-hydroxybutyrate, taurine, and propionylcarnitine (Fig. 1E).
Remarkably, this group of 4 metabolites share interconnectivity in the methionine cycle, methylation and transsulfuration pathways (Fig. 1F). Under metabolic stress, supplies of L-cysteine for glutathione synthesis can become rate-limiting, triggering the diversion of homocysteine into the transsulfuration pathway for production of cystathionine as the first step in cysteine biosynthesis, at the expense of use for methionine regeneration. In the transsulfuration pathway, cystathionine is cleaved to cysteine and 2-ketobutyrate, with the latter species undergoing enzymatic reduction to 2-hydroxybutyrate. An additional fate for 2-ketobutyrate is entry into TCA cycle via addition to propionylcarnitine for fatty acid β-oxidation in mitochondria (Fig. 1F). Creatine is synthesized from guanidinoacetate in an S-adenosylmethionine (SAM) dependent reaction (Wyss and Kaddurah-Daouk, 2000) and can spontaneously degrade to creatinine (Fig. 1F). Collectively, these untargeted metabolite profiling findings indicate selective transsulfuration pathway activation in the sALS-1 subset of ALS cases.
3.2. Serine incorporation into glutathione (GSH) is increased in sALS-1
We next asked whether the observed increase in transsulfuration may be linked to increased glutathione (GSH) synthesis in cells of the sALS-1 metabotype. To test this possibility, sALS-1 and control fibroblasts were grown in 2 mM [2,3,3-2H]-serine supplemented medium, followed by a determination of the extent of deuterium enrichment in GSH. Notably, we observed that the accuracy of the ALS-1 SVM prediction model decreased most markedly when 2-hydroxybutyrate was excluded from model building - overall accuracies of excluding 2-hydroxybutyrate, taurine, creatinine and propionyl-carnitine from SVM model generation were 0.81, 0.95, 0.94 and 0.94, respectively. Because performing stable isotope tracing on a larger number of cell lines would be costly and impractical to perform, stable isotope tracing experiments were performed to compare the three sALS fibroblast cell lines with highest levels of intracellular 2-hydroxybutyrate, compared with three randomly selected control cell lines. Power analysis, considering the variability of all 18 sALS-1 and 43 control cell lines and an effect size of 3.78, indicated that 3 cell lines per group would be sufficient to detect significant between group differences (p < 0.05; two-tailed t-test).
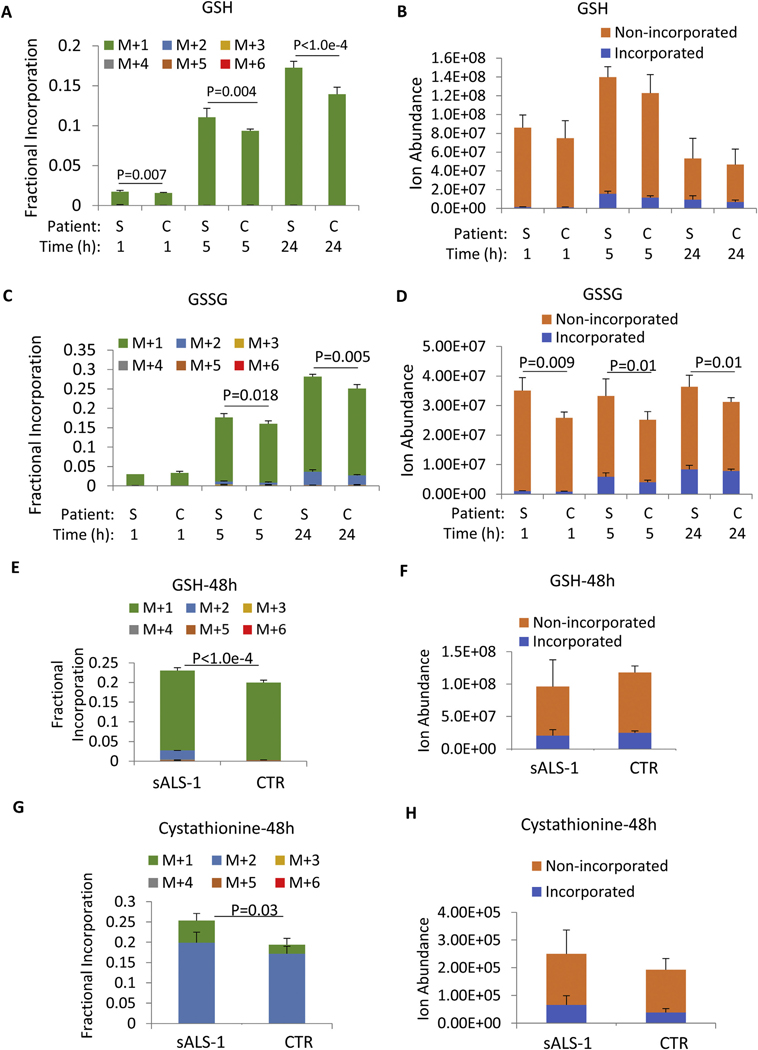
Notably, serine metabolism to glycine would predictably result in a singly-deuterated glycine when incorporated into de novo synthesized GSH. Indeed, sALS-1 fibroblasts in medium supplemented with [2,3,3-2H]-serine demonstrated a modest, but significant, increase in isotopic incorporation into GSH relative to controls after growth for 1 h, 5 h, and 24 h, (Fig. 2A, B). Similarly, glutathione disulfide (GSSG) in sALS-1 fibroblasts showed a significantly elevated serine-derived glycine incorporation, and significantly increase absolute levels of GSSG relative to control fibroblasts after growth for 5 h and 24 h (Fig. 2C, D).
Fig. 2.
[2,3,3-2H]-serine incorporations into GSH, GSSG and cystathionine syntheses are increased in sALS-1. A, C: Fractional isotopic enrichment of GSH and GSSG after 1 h, 5 h, and 24 h cell culture in [2,3,3-2H]-serine medium. B, D: Incorporated and non-incorporated ion abundance of GSH and GSSG after 1 h, 5 h, and 24 h of cell culture in [2,3,3-2H]-serine medium. E, G: Fractional isotopic enrichment of GSSG and cystathionine after 48 h cell culture in [2,3,3-2H]-serine medium. F, H: Incorporated and non-incorporated ion abundance of GSH and cystathionine after 48 h of cell culture in [2,3,3-2H]-serine medium. “S” denotes sALS-1 patients and “C” denotes controls. M + n (n = 1 to 6) refers to the number of incorporated deuterium. All data represent mean ± S.D., n = 2 from 3 cell lines per group. P value was calculated from 2-tailed Student’s t-test.
The biosynthesis of cytosolic GSH is tightly regulated by cysteine availability and the activity of γ-glutamylcysteinyl ligase (GCL) (Lu, 2013; Liu et al., 2009; van Klaveren et al., 1997). As shown in Fig. 1F, cystathionine is synthesized by cystathionine β-synthase (CBS) via condensation of homocysteine with serine. Hence, the extent of [2,3,3-2H]-serine incorporation into cystathionine can also serve to assess activity of the transsulfuration pathway. Surprisingly, we did not detect any serine incorporation into cystathionine at 1 h, 5 h and 24 h after [2,3,3-2H]-serine supplementation, indicating that transsulfuration from homocysteine to de novo synthesized cysteine was insignificant during this period. One potential explanation for this apparent lack of de novo cysteine production is that cystine in the culture medium was ample to support GSH production over the 24 h period of study.
To assess whether cysteine/cystine entry from the growth medium is responsible for delayed activation of de novo cysteine biosynthesis, we cultured cells with [2,3,3-2H]-serine for 48 h to allow for depletion of cystine in the medium. At this time, increased incorporation of serine-derived cysteine for GSH synthesis was observed in the sALS-1 group vs. control (as demonstrated by M + 2 isotopologues, in addition to a predominant serine-derived M + 1 glycine incorporation), while the total GSH pool was not significantly altered (Fig. 2E, F). In accord with utilization of serine for de novo cysteine synthesis after 48 h, we also observed increased incorporation of deuterium from serine into cystathionine in the sALS-1 vs. control fibroblast groups (Fig. 2G–H). Therefore, increased cysteine demand in the sALS-1 cell group, at least in part for GSH synthesis, resulted in accelerated transsulfuration of homocysteine to cysteine, concomitant with increased NADH dependent reduction of alpha-ketobutyrate to 2-hydroxybutyrate (Fig. 1F). Notably, the cystathionine γ-lyase/hydrogen sulfide system was reported to be essential for maintaining cellular GSH (Lee et al., 2014). Thus, upregulation of the transsulfuration pathway in sALS-1 can serve to maintain GSH levels as a potential means for protection against oxidative cell stress.
3.3. [U–13C]-glucose incorporation into GSH synthesis is increased in sALS-1
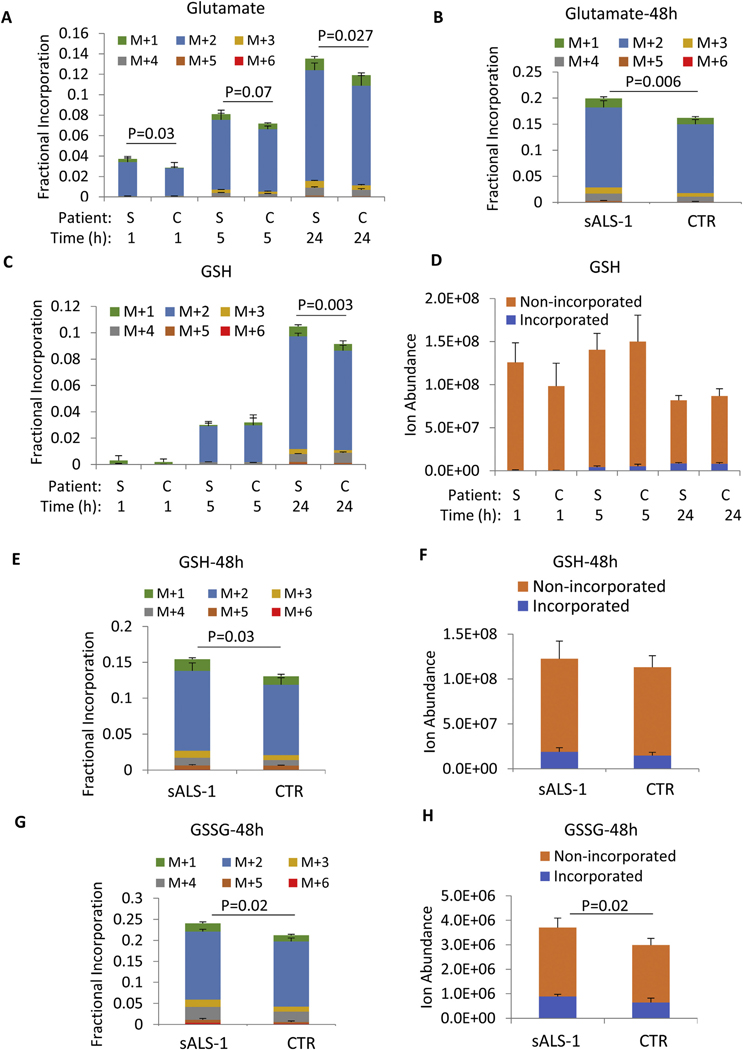
We next explored the relative utilization of glutamate for GSH synthesis. Glucose enters the TCA cycle as acetyl-CoA and contributes carbon atoms to produce α-ketoglutarate and its transamination product, glutamate. We compared the indirect contribution of [U-13C]-glucose-derived glutamate for GSH synthesis in sALS-1 vs. control fibroblast cell lines. To this end, 5 mM [U–13C]-glucose was added to glucose-free DMEM medium and 13C incorporation into glutamate was quantified at 1 h, 5 h, 24 h and 48 h. At 1 h, 13C enrichment of intracellular glucose approximated 100%, and incorporation into glutamate was observed at all time points predominantly as the M + 2 isotopologue (Fig. 3A, B). sALS-1 fibroblasts showed significantly increased GSH incorporation from glucose-derived glutamate (M + 2) at 24 h and 48 h (Fig. 3C, E). Notably, [U–13C]-glucose can also incorporate into serine, and serine-derived glycine can be further incorporated into GSH. However, under the cell culture conditions used, incorporation of glucose into glycine (M + 1) was much less than into glutamate M + 2 (Fig. 3C, E). Despite increases in GSH incorporation from glutamate at 24 h and 48 h, the total GSH pool was similar in sALS-1 and control fibroblasts (Fig. 3D, F), indicating that both GSH synthesis and consumption are increased in sALS-1 patient-derived fibroblasts. This interpretation is further strengthened by the accelerated incorporation of 13C into GSSG and the increased total GSSG pool in sALS-1 fibroblasts (Fig. 3G–H). Together, these findings indicate an accelerated rate of GSH oxidation and enhanced NADPH consumption to provide antioxidant defense in fibroblasts from patients with the sALS-1 metabotype.
Fig. 3.
[U–13C]-glucose incorporation into GSH synthesis is increased in sALS-1. A, B, C, E, G: Glutamate, GSH and GSSG fractional incorporation after 1 h, 5 h, 24 h and 48 h of [U-13C] glucose isotope enrichment. D, F, H: incorporated and non-incorporated ion abundance of GSH and GSSG after 1 h, 5 h, 24 h and 48 h of [U-13C] glucose isotope enrichment. “S” denotes sALS-1 patients and “C” denotes controls. M + n (n = 1 to 6) refers to the number of incorporated 13C. All data represent mean ± S.D., n = 2 from 3 cell lines per group, cell lines studied were the same as those described in Fig. 2. P value was calculated from 2-tailed Student’s t-test.
3.4. [U–13C]-glucose incorporation into TCA cycle intermediates and nucleotides is accelerated in sALS-1
Stable isotope tracing studies also revealed that increased GSH synthesis in sALS-1 fibroblasts was associated with an accelerated overall rate of glycolysis, TCA cycle, and additional linked metabolic pathways. We analyzed targeted and untargeted 13C tracing results from cells in [U-13C]-glucose medium at 1 h, 5 h and 24 h. sALS-1 fibroblasts exhibited increased fractional incorporation of [U-13C]-glucose into lactate, TCA cycle intermediates, amino acids (aspartate and alanine), and C2 and C4 acyl-carnitines (Fig. 4A–K). Interestingly, regardless of changes in total metabolite abundance (Fig. S3–4), the rate of glucose incorporation into each of the aforementioned species was increased at 24 h. We further confirmed this observed increase of glucose flux into TCA cycle intermediates and cataplerotic products in an independent experiment, where cells were cultured for 48 h in [U-13C]-glucose-supplemented medium (Fig. S5). Notably, most TCA cycle intermediates exhibited < 15% enrichment from 13C in glucose, except for citrate, which exhibited a 40% isotopic enrichment after 24 h. This is consistent with the fact that rapidly proliferating fibroblasts operate a truncated TCA cycle, where citrate is diverted to the cytosol for oxidation to oxoglutarate by isocitrate dehydrogenase 1, concomitant with NADPH production and generation of acetyl-CoA for fatty acid synthesis (Ghesquiere et al., 2014). Our data indicate that sALS-1 fibroblasts exhibit an accelerated TCA cycle flux, both oxidative and reductive, supportive of increased energy metabolism. This hypermetabolic phenotype is consistent with an increase in mitochondrial bioenergetics that was previously demonstrated generically for sALS vs. control fibroblasts (Konrad et al., 2017; Kirk et al., 2014).
Fig. 4.
[U–13C]-glucose incorporation into lactate, TCA cycle intermediates and related metabolites is increased in sALS-1. A-K: Fractional incorporation after 1 h, 5 h, and 24 h of [U-13C] glucose isotope enrichment. “S” denotes sALS-1 patients and “C” denotes controls. M + n (n = 1 to 6) refers to the number of incorporated 13C. All data represent mean ± S.D., n = 2 from 3 cell lines per group, cell lines studied were the same as those described. P value was calculated from 2-tailed Student’s t-test. Note: Due to the overlapping of succinate M + 2 with the reference ion (m/z 119.03) used for mass accuracy correction, the calculated M + 2 incorporation in succinate was much smaller than the actual incorporated value. However, the relative incorporation of [U-13C] glucose to succinate is significantly increased in sALS-1 compared to the control under the same condition.
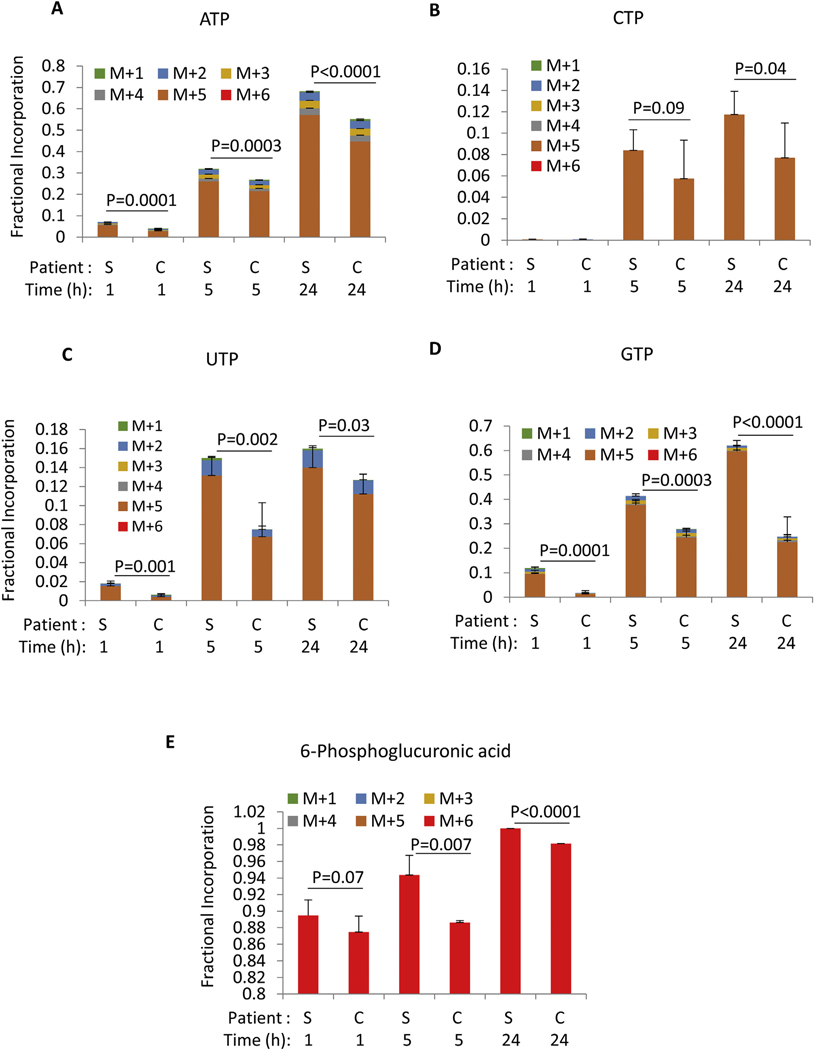
In accord with an accelerated TCA cycle, sALS-1 fibroblasts exhibited a significant increase in glucose incorporation into nucleotide triphosphate pools after 24 h (mostly via the ribose M + 5 isotopologue), whereas total levels of nucleotide triphosphate remained unchanged (Fig. 5 A–D, Fig. S6 A–D). This can be explained by increased rates of both de novo nucleotide synthesis and nucleotide phosphate consumption. Notably, in sALS-1 cells, 6-phosphogluconate, a precursor of NADPH, ribulose-5-phosphate, and ribose 5-phosphate production via the pentose phosphate pathway showed a rapid increase in fractional enrichment that reached 85% by 1 h (Fig. 5E). Increased fractional enrichment and decreased total intracellular levels (Fig. S6E) indicate an accelerated rate of 6-phosphogluconate consumption in sALS-1 patient derived fibroblasts.
Fig. 5.
[U–13C] glucose incorporation into nucleotide triphosphates and pentose phosphate intermediate is increased in sALS-1. A-K: fractional incorporation after 1 h, 5 h and 24 h of isotope enrichment. “S” denotes sALS-1 patients and “C” denotes controls. M + n (n = 1 to 6) refers to the number of incorporated 13C. All data represent mean ± S.D., n = 2 from 3 cell lines per group, cell lines studied were those described in Fig. 2. P value was calculated from 2-tailed Student’s t-test.
Taken together, these findings indicate that sALS fibroblasts exhibit increased glucose influx into the TCA cycle, cataplerotic amino acid synthesis, accelerated GSH synthesis, and pentose phosphate pathway activity, providing antioxidant NADPH and ribose for support of nucleotide biosynthesis.
3.5. Mitochondrial membrane potential is increased in sALS-1 fibroblasts
We previously demonstrated that mitochondrial bioenergetics is perturbed in skin fibroblasts from a group of 171 sALS patients, compared with 98 control fibroblast cell lines (Konrad et al., 2017). Specifically, sALS fibroblasts showed a significant increase in the ratio of mitochondrial membrane potential (MMP) to total mitochondrial mass (MMP:MM ratio). Since the sALS patient- and control-fibroblasts utilized in the present study were all included in the previously studied cohort, we performed meta-analysis of the prior bioenergetics data obtained from the sALS-1 study cells, relative to non-stratified sALS cell lines (designated sALS-2) and control fibroblasts. Based on this analysis, we found that sALS-1 cell lines (i.e., those with an accelerated transsulfuration pathway), exhibited a greater increase in MMP:MM ratio than both ALS-2 and control cell lines (Fig. S7). Interestingly, metabolite profiling and glucose tracing of ribose incorporation into ATP synthesis revealed that the total intracellular ATP pools did not differ between these groups, despite a significantly increased ribose incorporation into ATP of ALS-1 fibroblasts (Fig. 5A, S6A). This finding is in agreement with the published reports using a chemiluminescent approach to quantify ATP levels (Kawamata et al., 2017). Together, these data support the hypothesis that sALS-1 patient-derived fibroblasts have increased bioenergetic demands (Konrad et al., 2017).
3.6. Integrated transcriptomic and metabolomic pathway analysis reveals super-transsulfuration activation in sALS-1 fibroblasts
We performed RNA microarray and microRNA analysis to investigate whether glucose and transsulfuration hypermetabolism can be explained by selective transcriptional changes in sALS-1 fibroblasts. Toward this end, transcripts and microRNA targets from 27 sALS fibroblasts were compared to those from 27 randomly selected control subjects. Of the 27 sALS patient cell lines studied, 11 belonged to the sALS-1 metabotype and 16 were sALS-2. Table S4 summarizes our finding of differentially expressed mRNA and miRNA targets comparing sALS-1 to sALS-2 and control fibroblast cell lines (ANOVA, uncorrected P < 0.05). Due to the comprehensive nature of gene annotation databases (mostly cancer and disease related), we first curated a searchable database to include candidate gene sets from previously published sALS studies, then identified targeted genes based on differentially-expressed transcripts and microRNA targets in the sALS subgroups.
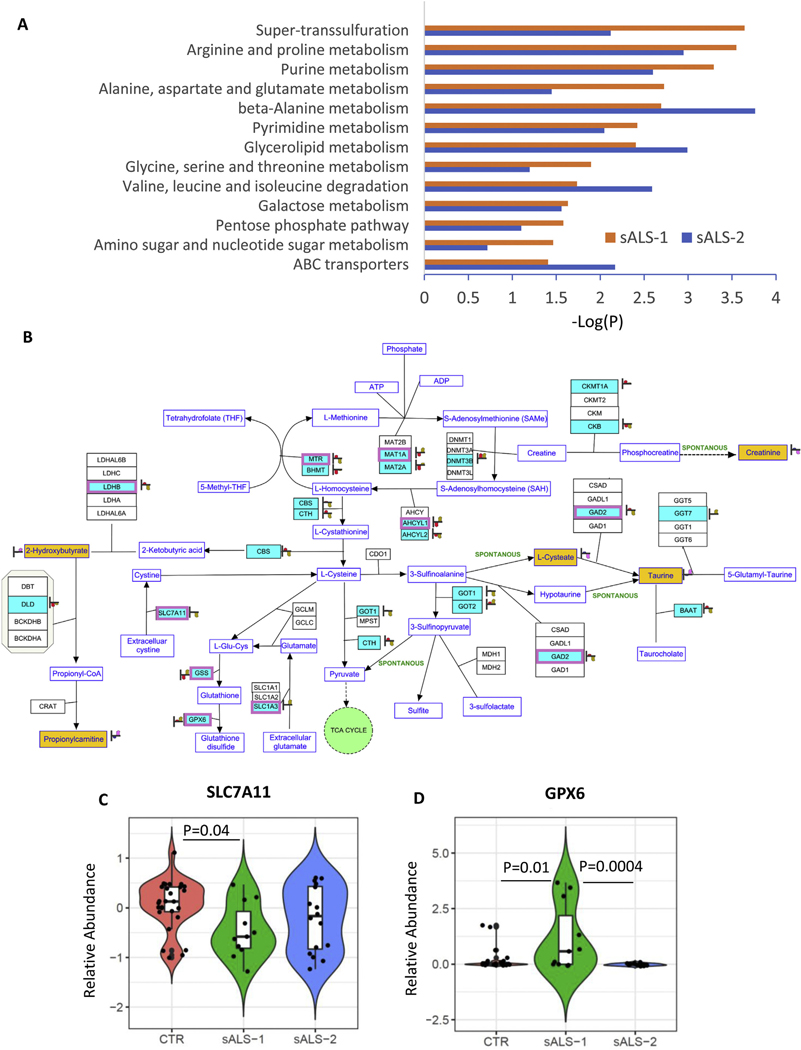
Using the curated database, we integrated transcriptome, microRNAome and metabolome data into metabolic pathways queried by KEGG, Wiki-pathways and BioCyc. The results identified the super-transsulfuration pathway to be among the top significantly enriched pathways in sALS-1 fibroblasts (p = 2.3 x e−4 by Fischer’s exact test, Fig. 6A). Note that sALS-1 fibroblasts showed a much smaller enrichment p-value than sALS-2 cells, in accord with the transsulfuration pathway being selectively accelerated in sALS-1 vs. sALS-2 and control fibroblasts.
Fig. 6.
Pathway Integration of transcript and metabolite changes in sALS-1 and sALS-2 compared to controls. A: Significant enrichments of integrated multiomic data into metabolic pathways in sALS-1 and sALS-2 compared to controls (P = 2.3e-4, Fisher’s exact-test). Differentially expressed messenger RNA (mRNA) and microRNA (miRNA) targets were obtained from 11 sALS-1, 16 sALS-2 and 27 age and gender matched controls. Differentially expressed intracellular metabolites were obtained from 18 sALS-1 relative to 18 controls. The metabolic pathways used for mapping were queried by KEGG, Wiki-pathways and Biocyc and imported into Agilent GeneSpring 14.9.1 for pathway analysis. B: Visualization of significant mapping of the differentially expressed mRNA, miRNA targets and metabolites involved in the super-transsulfuration pathway in sALS-1 compared to control group. Light blue entities with purple boxes: differentially expressed mRNA (P < 0.05, 2-tailed Student’s t-test); light blue entities without purple box: differentially expressed miRNA targets (P < 0.05, 2-tailed Student’s t-test); yellow entities: intracellular metabolites. Heat strips next to entities represent normalized abundance in control (left heat strip, red) and sALS-1 (right heat strip, brown). C: Distinctive expression of cystine-glutamate antiporter (SLC7A11) mRNA in sALS-1 compared to sALS-2 and control group. D: Distinctive expression glutathione peroxidase 6 (GPX6) mRNA in sALS-1 compared to sALS-2 and control group. P value was calculated from 2-tailed Student’s t-test.
We next mapped the multiomic data (i.e. differentially expressed mRNA, microRNA targets and metabolites in sALS-1, compared to control cell lines) representing the transsulfuration pathway to concomitantly visualize differentially-expressed transcripts along with metabolites in sALS-1 fibroblasts. Differentially-expressed mRNA transcripts (entities depicted in Fig. 6B in light blue with purple boxes; P < 0.05) and differentially expressed microRNA targets (entities depicted in Fig. 6B in light blue without purple boxes; P < 0.05) were not only detected in the methionine cycle and homocysteine to cysteine transsulfuration pathway, but also in the GSH and cysteine metabolic pathways (both synthesis and oxidation/degradation).
The cystine/glutamate antiporter (system xc-/SLC7A11) contributes importantly to GSH synthesis by transporting cystine into the cell. With cystine entry, SLC7A11 concomitantly anti-ports glutamate, potentially leading to excitotoxicity. This dual action of SLC7A11 may be highly relevant to ALS because of its key roles in both oxidative stress and excitotoxicity. SLC7A11 was previously reported to be dysregulated in ALS patients and in animal models of familial ALS disease (Mesci et al., 2015; Albano et al., 2013; Figuera-Losada et al., 2017). Notably, significant downregulation of SLC7A11 was found in sALS-1, but not sALS-2, compared to controls (Fig. 6C). Conversely, GPX6, an isoform of glutathione peroxidase involved in the oxidation of GSH to GSSG, was found to increase in sALS-1, but not sALS-2, relative to controls (Fig. 6D). Decreased SLC7A11 and increased GPX6 expression could be important contributors to transsulfuration pathway activation in sALS-1 case-derived fibroblasts.
Taken together, sALS-1 case-derived fibroblasts show distinct transcriptional changes involved in cystine and glutamate transport, as well as GSH synthesis/oxidation, compared to sALS-2 and control fibroblasts.
3.7. NAC selectively rescues metabolically-stressed sALS-1 fibroblasts
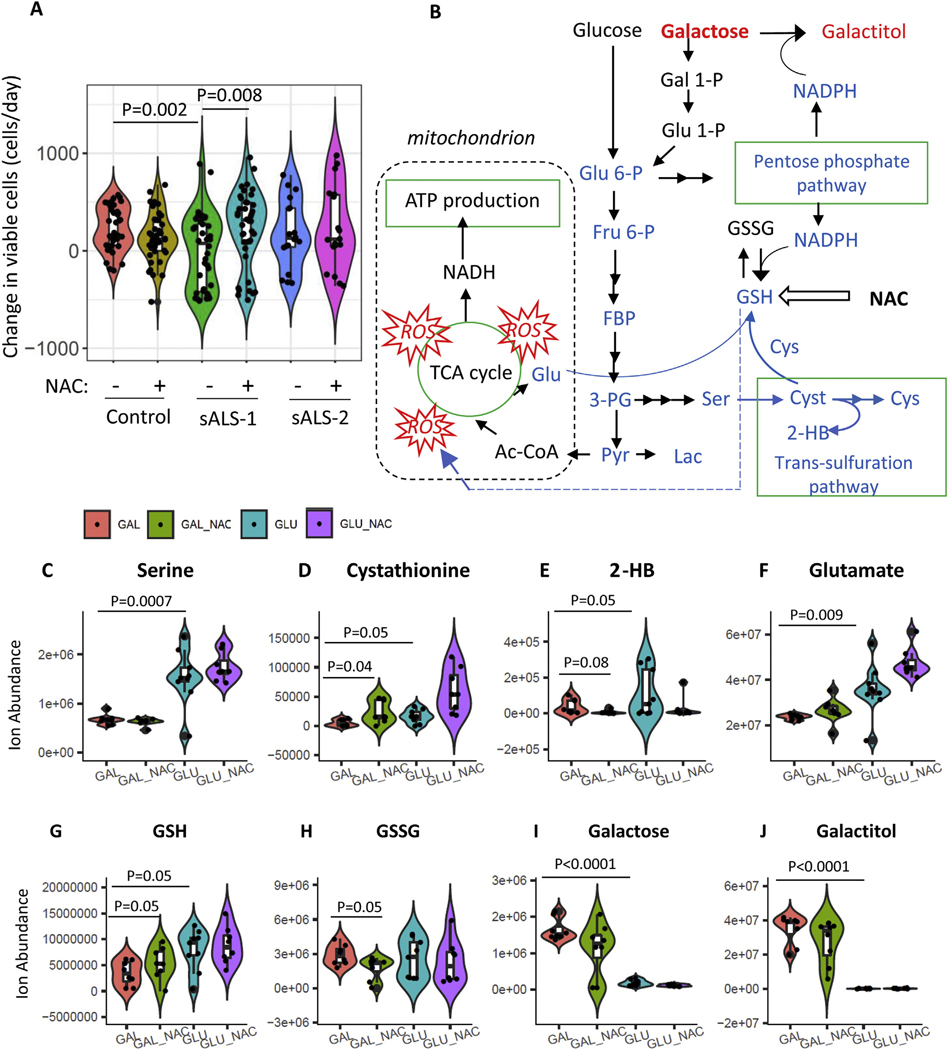
To further investigate whether cysteine availability and transsulfuration pathway activation are biologically relevant for sALS-1 fibroblast cell viability, we compared the effect of NAC supplementation on cell proliferation under metabolic stress conditions, where galactose was substituted for glucose as a bioenergetic fuel in the fibroblast culture media. Whereas the production of pyruvate via glycolytic metabolism of glucose yields 2 net molecules of ATP, the production of pyruvate via glycolytic metabolism of galactose yields no net ATP. Thus, galactose medium forces cells to rely on mitochondrial oxidative phosphorylation (OXPHOS) and has been used to test fibroblasts for oxidative defects (Robinson et al., 1992).
Compared to sALS-2 and control fibroblasts, sALS-1 metabotype fibroblasts exhibit upregulated oxidative phosphorylation in glucose-containing medium (Fig. 4–5 and Fig. S5–6), and predictably cannot upregulate OXPHOS to a further extent in galactose-containing medium. As shown in Fig. 7A, sALS-1 fibroblasts displayed significantly impaired viability when grown in galactose medium for 24 h, and this growth defect was completely rescued by NAC supplementation. In contrast, sALS-1 cell viability in glucose-containing medium was not different from control and sALS-2 fibroblasts, and NAC supplementation had no effect on cell viability in any of the cell groups when grown in glucose medium (data not shown).
Fig. 7.
NAC selectively rescues sALS-1 fibroblasts exposed to metabolic stress. A: sALS-1 fibroblasts show a distinct loss of viability phenotype in galactose medium, rescued by 24 h 2 mM NAC supplementation, compared to controls and sALS-2 group. n = 18–40 from 3 cell lines per group, Data was plotted as violin plots to display data frequency, with jitter points showing individual fibroblast lines and an inset boxplot presenting median value, interquartile range and the 95% confidence interval. P value was calculated from 2-tailed Student’s t-test. B: Schematic depicting key metabolites in galactose glycolysis, TCA cycle, pentose phosphate pathway and transsulfuration pathway that contribute to galactose induced oxidative stress and subsequent NAC rescue in sALS-1. Glu 6-P, glucose 6-phosphate; Gal 1-P, galactose 1-phosphate; Glu 1-P, glucose 1-phosphate; Fru 6-P, fructose 6-phosphate; FBP, Fructose 1,6-bisphosphate; 3PG, 3-phosphoglycerate; Pyr, pyruvate; F1P, fructose-1-phosphate; Lac, lactate; Ac-CoA, Acetyl-CoA; Glu; Glutamate; Ser, serine; Cyst, cystathionine; 2-HB, 2-hydroxybutyrate; Cys, cysteine. Blue denotes decreased and red denotes increased metabolites and associated pathways in galactose compared to glucose medium. C-J: key metabolite changes sALS-1 comparing galactose and glucose media with and without NAC treatment. n = 8–9 from 3 cell lines per group. P value was calculated from 2-tailed Student’s t-test.
Besides forcing OXPHOS, galactose is known to induce oxidative stress (Du et al., 2019; Aydin et al., 2018; Budni et al., 2017). Consistently, we found that key metabolites related to antioxidant defense are altered in galactose-relative to glucose-containing sALS-1 fibroblast growth media (Fig. 7, B–J). Serine, cystathionine and 2-hydroxybutyrate, the precursor and products of the transsulfuration pathway, were all significantly decreased upon growth in galactose medium compared to glucose medium (Fig. 7C–E), indicating the capacity of sALS-1 cells to compensate for heightened oxidative stress in galactose-containing medium by upregulating transsulfuration was compromised. Diminished synthesis of serine from glycolysis and synthesis of glutamate from the TCA cycle (Fig. 7F) may underlie the observed decrease in GSH synthesis (Fig. 7G), while relative GSSG levels remained unchanged (Fig. 7H). Additionally, a decreased rate of glucose-6-phoshate production in galactose-containing medium results in decreased pentose phosphate flux and NADPH production. A slowed rate of glycolysis also leads to galactose build-up, and subsequent activation of galactose reduction to galactitol, which consumes NADPH (Fig. 7B, I–J). Notably, the combination of increased galactose reduction to galactitol and decreased NADPH production from pentose phosphate pathway would predictably impair the recycling of GSH from GSSG. NAC supplemention effectively prevented galactose-induced oxidative stress, manifest by increased GSH and decreased GSSG along with enhanced cell viability (Fig. 7A, G–H). These results support the view that that sALS-1 fibroblasts are operating OXPHOS and transsulfuration at near-maximal capacity in glucose-containing medium and have a limited capacity for further upregulation when challenged with an oxidative stressor. Our findings indicate that NAC supplementation provides sALS-1 fibroblasts with sufficient antioxidant capacity to enable metabolic diversion from transsulfuration to energy generation.
3.8. sALS-1 patient plasma displays altered transsulfuration pathway metabolites
To identify novel disease biomarkers, we explored the possibility that sALS-1 patient plasma can reveal indications of altered metabolism, in accord with that found from studies of patient-derived fibroblasts. Toward this end, plasma samples from the same 18 sALS-1 cases that yielded fibroblasts with the accelerated transsulfuration and glucose hypermetabolism metabotype, were compared to plasmas from 20 age/sex-matched control patients. Targeted and untargeted metabolite profiling detected > 1000 metabolite features, of which 85 were found to be differentially-abundant in sALS-1 cases, compared to controls (Fig. 8A). PCA analysis revealed that plasma metabolite abundances from sALS-1 cases cluster and separate from controls based on levels of these 85 metabolites (Fig. 8B). Notably, the metabolite differences included decreased levels of some amino acids (glutamine, glutamate, methionine, tyrosine) and creatinine, along with increased levels of taurine-related species (i.e., taurine and glutamyltaurine, bile acids and 2-hydroxybutyrate), purine metabolites, phospholipids, ceramides and fatty acid amides. Two species that were initially unknown and found to be present in the majority of sALS-1 cases were subsequently identified as Riluzole and its primary metabolite, hydroxyriluzole glucuronide. Mapping all identified differentially abundant sALS-1 plasma metabolites to KEGG pathways, revealed taurine/hypotaurine metabolism as the pathway with highest impact score and most significant p-value (Fig. 8C), in accord with findings from sALS-1 fibroblast studies. Notably, increased plasma taurine and 2-hydroxybutyrate (both transsulfuration pathway intermediates), along with decreased creatinine (methionine cycle-related methylation product; Fig. 1F), exhibited similar alterations in plasma to that found in fibroblasts, suggesting a systemic metabolic perturbation in sALS-1 patients, and the potential to effectively use plasma profiling for facile clinical recognition of patients with the sALS-1 metabotype.
Fig. 8.
sALS-1 showed distinct plasma metabolite profiles from controls and indications of altered taurine and hypotaurine metabolic pathway. A: heatmap of differential plasma metabolites in sALS-1 compared to controls. Metabolites related to altered taurine and transsulfuration pathway, amino acids, lipid metabolism and purine nucleosides were highlighted in red (upregulated) and blue (downregulated) boxes. ALS drug Riluzole and its drug metabolite hydroxyriluzole glucuronide were also highlighted. B: PCA score plot showing the separation of sALS-1 from controls based on the 85 differentially expressed plasma metabolites (sALS-1, n = 18; controls, n = 20). C: Mapping the differentially expressed plasma metabolites into KEGG pathway using Pathway analysis module of MetaboAnalyst 3.0 recognizes taurine and hypotaurine in the transsulfuration pathway as the most significant pathway impact and lowest matching P-value.
We then asked whether sALS-1 could be distinguished from sALS-2 patients by the clinical characteristics described in Table 1. Based on the available clinical data for 18 sALS-1 and 55 sALS-2 cases, sALS-1 patients did not show a significant difference from ALS-2 cases in terms of age at the onset of symptoms, ALS severity at the time of biopsy (based on ALSFRS-R, an ALS functional rating scale–revised), rate of functional decline, FVC (Forced vital capacity, %), and BMI(body mass index). However, sALS-1 fibroblasts did exhibit a frequency distribution difference from ALS-2 cells regarding the site of initial disease onset (Fig. S8). Notably, sALS-1 cases presented with a higher frequency of lumbosacral onset (50%), while sALS-2 had > 50% cervical onset. These data raise an interesting question as to whether patient metabotype plays may be impacted by the site of disease initiation, which in some studies has been shown to play a role in disease prognosis (Calvo et al., 2017).
3.9. Independent validation of growth defect of sALS-1 fibroblasts under oxidative culture conditions
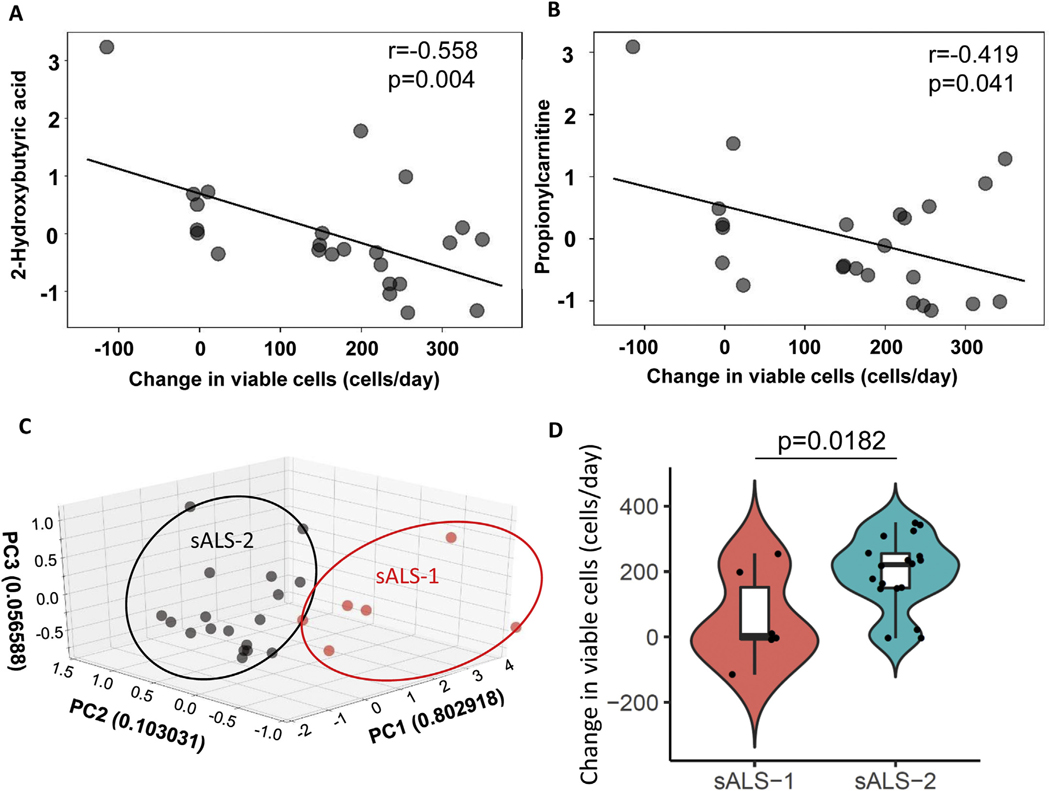
In an independent cohort of 24 sALS patient fibroblasts, we performed a targeted analysis of the 4 sALS-1 distinguishing metabolites (2-hydroxybutyrate, taurine, propionylcarnitine and creatinine) and simultaneously assessed cell proliferation in galactose medium. We found a significant inverse correlation for two metabolites (2-hydroxybutyrate and propionylcarnitine, Fig. 9A–B) with an attenuated rate of cell proliferation when grown in glucose-free galactose-containing (oxidative) medium. Our four-metabolite SVM model, which was trained on the original sALS dataset (Fig. 1D), identified 6 of the 24 sALS patients as sALS-1 and the remainder as sALS-2 (Fig. 9C). As anticipated for this validation cohort, the average proliferation rate of sALS-1 patient-derived fibroblasts in galactose medium was significantly lower than cells from sALS-2 cases (Fig. 9D). Together, these findings affirm that the sALS-1 metabotype is characterized by a growth disadvantage under oxidative conditions.
Fig. 9.
An independent validation set of sALS-1 fibroblasts show attenuated growth under oxidative conditions. Inverse correlation between cell growth rate and levels of (A) 2-hydroxybutyrate (Pearson’s = −0.558, p = 0.004) and (B) propionylcarnitine (Pearson’s = −0.419, p = 0.041) in medium containing galactose as primary energy source (n = 24). C: PCA score plot based on 4 sALS-1-distinguishing metabolites that classify sALS-1 vs sALS-2 cases (sALS-1, n = 6; sALS-2, n = 18). D: Violin plot of cell viability over time in galactose medium, comparing sALS-1 and sALS-2 cases (p = 0.0182).
4. Discussion
In an effort to identify metabolic biomarkers for stratification of sALS patients, we applied untargeted metabolite profiling to patient-derived dermal fibroblasts. This effort confidently identified a subgroup of sALS fibroblasts (i.e., sALS-1) with a distinct transsulfuration pathway upregulation. Importantly, we identify this metabotype also in cognate plasma from sALS-1 cases and show that this subgroup exhibits perturbed mRNAs that encode proteins relavent to transsulfuration/cysteine metabolism. We further demonstrate that the altered metabolic pathways in sALS-1 patient-derived fibroblasts are linked to increased GSH synthesis and accelerated glucose metabolism. Notably, these metabolic alterations are associated with impaired cell viability under oxidative stress, which is highly responsive to supplementation with the antioxidant, NAC. This interplay between glucose hypermetabolism and increased antioxidant demand raises the possibility that upregulated antioxidant defenses serve to meet hypermetabolic demands in sALS-1 patient cases and define this disease subtype as candidates for future targeted therapies.
It is known that a subset of ALS patients expend approximately 10% more resting brain energy than healthy individuals (Cistaro et al., 2012; Weijs, 2011; Desport et al., 2001). Overall, hypermetabolism is a feature of more than half of the ALS population and was recently shown to be a deleterious prognostic factor. Indeed, patients with hypermetabolism have a 20% worse prognosis than those that do not exhibit the hypermetabolism phenotype (Vucic, n.d.; Jesus et al., 2017). Until now, hypermetabolism in ALS has mostly been considered in relation to glucose metabolism. Indeed, 18F-FDG-PET imaging is widely applied clinically to assess and discriminate CNS differences in glucose uptake that distinguish hypermetabolism in both familial and sALS patients (Cistaro et al., 2012; Canosa et al., 2016; Turner and Leigh, 2000). Enhanced 18F-FDG-PET uptake correlates with cognitive impairment in ALS patients and provides a convenient means to identify altered metabolic activity (Canosa et al., 2016).
Using stable isotope tracing, we demonstrate a selective enhancement of transsulfuration pathway flux in fibroblasts derived from the sALS-1 patient subtype, associated with accelerated rates of GSH synthesis and glucose metabolism. Notably, the sALS-1 subgroup exhibits increased intracellular and plasma levels of transsulfuration pathway metabolites 2-hydroxybutyrate and taurine (correlating with one another) along with accelerated de novo synthesis of GSH. Attenuated cystine entry into ALS-1 fibroblasts may act to accelerate the conversion of homocysteine to cysteine via the transsulfuration pathway, fueling an increased demand for GSH that we surmise arises in sALS-1 patients due to greater antioxidant demand. We propose that increased glucose utilization and GSH synthesis are respectively a cause and consequence of oxidant stress in the sALS-1 fibroblast metabotype, found in approximately 25% (18/77) of sALS cases in our test cohort. In accord with this finding, a recent report correlated in vivo ALS staging with glucose metabolism patterns and demonstrated that four neuropathological stages of ALS correlate with discriminative regional brain glucose metabolism patterns that associate with both failing respiratory function (measured as forced vital capacity) and disease duration (van Weehaeghe et al., 2017). Therefore, sALS-1 cases are likely to comprise a spectrum of glucose hypermetabolism phenotypes sharing a distinct metabotype tuned by cysteine availability for GSH synthesis and defense against oxidative stress.
The finding that similar changes in transsulfuration metabolites occur in both fibroblasts and plasma from sALS-1 cases is intriguing, as it may provide an opportunity for facile stratification of sALS cases based on plasma biomarkers. Remarkably, changes in plasma levels of 2-hydroxybutyrate, taurine, amino acids and creatinine have been previously reported in ALS patients (Wuolikainen et al., 2016; Lawton et al., 2014; Lawton et al., 2012; Patel et al., 2013). As mentioned above, 2-hydroxybutyrate derives from 2-ketobutyrate, produced from cystathionine in the transsulfuration pathway via its NADH-dependent reduction, catalyzed by lactate dehydrogenase B (Fig. 1F). Elevated levels of plasma 2-hydroxybutyrate and 2-ketobutyrate have previously been shown in several studies involving different cohorts of ALS cases and ascribed to disease-associated oxidative stress (Wuolikainen et al., 2016; Lawton et al., 2014; Lawton et al., 2012). Moreover, decreased plasma creatinine has previously been suggested to be an ALS risk factor (van Eijk et al., n.d.; Chio et al., 2014). Besides taurine, bile acids (e.g., cholic acid) and bile acid conjugates (e.g., taurocholic acid) were found to increase in sALS-1 plasma (Fig. 8A). Bile acids play a neuroprotective role in a diverse spectrum of age-related neurodegenerative disorders (Ackerman and Gerhard, 2016). There are two primary bile acids produced by the liver in humans: cholic acid and chenodeoxycholic acid. These primary bile acids can undergo conjugation with glycine or taurine prior to secretion in the bile as glycocholic acid, taurocholic acid, glycochenodeoxycholic acid, and taurochenodeoxycholic acid. Therefore, upregulated plasma levels of taurine and taurocholic acid could represent an additional adaptive defense mechanism that opposes heightened oxidative stress in ALS-1 cases.
5. Conclusion
We describe the first stratification of a subgroup of sALS skin fibroblasts based on metaboytpe and identify a distinct patient cohort characterized by increased transsulfuration and glucose hypermetabolism. We show that the observed metabolite changes are similarly detected in sALS-1 plasma and find that sALS-1 fibroblasts are uniquely sensitive to metabolic/oxidative stress and selectively protected by supplementation with the antioxidant NAC. Therefore, we infer that facile plasma profiling could be employed to confidently stratify sALS cases, recognizing the sASL-1 subtype for targeted benefit using antioxidant therapies. We hypothesize that the metabolic rewiring that results in upregulation of the transsulfuration pathway in these patients initially arises as an adaptation to limiting cysteine and hence GSH availability, triggered by hypermetabolism that accelerates the rate of GSH oxidation. Knowledge obtained from metabotype stratification of skin-derived fibroblasts from sALS-1 cases offers a much-needed first step toward targeted precision medicines for therapy of sALS patients.
Supplementary Material
Acknowledgements
We thank Dr. Hiroshi Mitsumoto (Columbia University) and the COSMOS initiative for providing the fibroblast and plasma samples utilized in this work. Fibroblast samples and clinical data were obtained from the NINDS Repository.
Funding
Work on this manuscript was supported by NIHR01NS093872 to GM, SG, and LS, NIH R01NS062055 to GM, NIH P01HD06724 to SG, and an ALS Association grant 15-IIP-195 to GM.
Abbreviations:
- ALS
Amyotrophic lateral sclerosis
- sALS
sporadic ALS
- sALS-1
sALS subgroup with enhanced transsulfuration metabotype
- sALS-2
non-stratified sALS
- TCA
tricarboxylic acid cycle
- DMEM
Dulbecco modified Eagle medium
- PBS
Phosphate buffered saline
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC/MS
Liquid chromatography/mass spectrometry
- QC
Quality control
- PCA
Principal component analysis
- USIT
untargeted stable isotope tracing
- ROC
Receiver operating characteristic
- AUC
Area under the curve
- SVM
Support vector machine
- NAC
N-acetylcysteine
- NADPH
Nicotinamide adenine dinucleotide phosphate
- ATP
Adenosine triphosphate
- GSH
Glutathioine
- GSSG
GSH disulfide
- SLC7A11
glutamate cystine antiporter
- ALSFRS
ALS Functional Rating Scale
- BMI
Body mass index
- FVC
Forced vital capacity
- MMP
Mitochondrial membrane potential
- MM
Mitochondrial mass. M + n: Isotopologue having n number of incorporated heavy isotope(s)
Footnotes
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. Raw data are available on-line at the following site: https://wcm.box.com/s/za0e3gz79y3lh443gmqv4efyhu6pnbta
Declaration of Competing Interest
Nothing to report.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nbd.2020.105025.
References
- Ackerman HD, Gerhard GS, 2016. Bile Acids in Neurodegenerative Disorders. Front. Aging Neurosci. 8, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano R, Liu X, Lobner D, 2013. Regulation of system x(c)- in the SOD1-G93A mouse model of ALS. Exp. Neurol. 250, 69–73. [DOI] [PubMed] [Google Scholar]
- Allen SP, Duffy LM, Shaw PJ, Grierson AJ, 2015. Altered age-related changes in bioenergetic properties and mitochondrial morphology in fibroblasts from sporadic amyotrophic lateral sclerosis patients. Neurobiol. Aging 36, 2893–2903. [DOI] [PubMed] [Google Scholar]
- Aydin S, Yanar K, Simsek B, Cebe T, Sitar ME, Belce A, Cakatay U, 2018. Galactose-induced aging model in rat testicular tissue. J. Coll. Physicians Surg. Pak. 28, 501–504. [DOI] [PubMed] [Google Scholar]
- Budni J, Garcez ML, Mina F, Bellettini-Santos T, da Silva S, Luz APD, Schiavo GL, Batista-Silva H, Scaini G, Streck EL, Quevedo J, 2017. The oral administration of D-galactose induces abnormalities within the mitochondrial respiratory chain in the brain of rats. Metab. Brain Dis. 32, 811–817. [DOI] [PubMed] [Google Scholar]
- Calvo A, Moglia C, Lunetta C, Marinou K, Ticozzi N, Ferrante GD, Scialo C, Soraru G, Trojsi F, Conte A, Falzone YM, Tortelli R, Russo M, Chio A, Sansone VA, Mora G, Silani V, Volanti P, Caponnetto C, Querin G, Monsurro MR, Sabatelli M, Riva N, Logroscino G, Messina S, Fini N, Mandrioli J, 2017. Factors predicting survival in ALS: a multicenter Italian study. J. Neurol. 264, 54–63. [DOI] [PubMed] [Google Scholar]
- Canosa A, Pagani M, Cistaro A, Montuschi A, Iazzolino B, Fania P, Cammarosano S, Ilardi A, Moglia C, Calvo A, Chio A, 2016. 18F-FDG-PET correlates of cognitive impairment in ALS. Neurology 86, 44–49. [DOI] [PubMed] [Google Scholar]
- Chen Q, Park HC, Goligorsky MS, Chander P, Fischer SM, Gross SS, 2012. Untargeted plasma metabolite profiling reveals the broad systemic consequences of xanthine oxidoreductase inactivation in mice. PLoS One 7, e37149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Kirk K, Shurubor YI, Zhao D, Arreguin AJ, Shahi I, Valsecchi F, Primiano G, Calder EL, Carelli V, Denton TT, Beal MF, Gross SS, Manfredi G, D’Aurelio M, 2018. Rewiring of glutamine metabolism is a bioenergetic adaptation of human cells with mitochondrial DNA mutations. Cell Metab. 10.1016/j.cmet.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Calvo A, Bovio G, Canosa A, Bertuzzo D, Galmozzi F, Cugnasco P, Clerico M, De Mercanti S, Bersano E, Cammarosano S, Ilardi A, Manera U, Moglia C, Sideri R, Marinou K, Bottacchi E, Pisano F, Cantello R, Mazzini L, Mora G, 2014. Piemonte, S. Valle d’Aosta Register for Amyotrophic Lateral, Amyotrophic lateral sclerosis outcome measures and the role of albumin and creatinine: a population-based study. JAMA Neurol 71, 1134–1142. [DOI] [PubMed] [Google Scholar]
- Cistaro A, Valentini MC, Chio A, Nobili F, Calvo A, Moglia C, Montuschi A, Morbelli S, Salmaso D, Fania P, Carrara G, Pagani M, 2012. Brain hypermetabolism in amyotrophic lateral sclerosis: a FDG PET study in ALS of spinal and bulbar onset. Eur. J. Nucl. Med. Mol. Imaging 39, 251–259. [DOI] [PubMed] [Google Scholar]
- Codron P, Cassereau J, Vourc’h P, Veyrat-Durebex C, Blasco H, Kane S, Procaccio V, Letournel F, Verny C, Lenaers G, Reynier P, Chevrollier A, 2018. Primary fibroblasts derived from sporadic amyotrophic lateral sclerosis patients do not show ALS cytological lesions. Amyotroph Lateral Scler Frontotemporal Degener 19, 446–456. [DOI] [PubMed] [Google Scholar]
- Desport JC, Preux PM, Magy L, Boirie Y, Vallat JM, Beaufrere B, Couratier P, 2001. Factors correlated with hypermetabolism in patients with amyotrophic lateral sclerosis. Am. J. Clin. Nutr. 74, 328–334. [DOI] [PubMed] [Google Scholar]
- Du ZD, Yu S, Qi Y, Qu TF, He L, Wei W, Liu K, Gong SS, 2019. NADPH oxidase inhibitor apocynin decreases mitochondrial dysfunction and apoptosis in the ventral cochlear nucleus of D-galactose-induced aging model in rats. Neurochem. Int. 124, 31–40. [DOI] [PubMed] [Google Scholar]
- Figuera-Losada M, Thomas AG, Stathis M, Stockwell BR, Rojas C, Slusher BS, 2017. Development of a primary microglia screening assay and its use to characterize inhibition of system xc(−) by erastin and its analogs. Biochem Biophys Rep 9, 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemi M, Brown RH Jr., 2017. Genetics of amyotrophic lateral sclerosis. Cold Spring Harb Perspect Med. 10.1101/cshperspect.a024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghesquiere B, Wong BW, Kuchnio A, Carmeliet P, 2014. Metabolism of stromal and immune cells in health and disease. Nature 511, 167–176. [DOI] [PubMed] [Google Scholar]
- Huisman MH, Seelen M, van Doormaal PT, de Jong SW, de Vries JH, van der Kooi AJ, de Visser M, Schelhaas HJ, van den Berg LH, Veldink JH, 2015. Effect of Presymptomatic body mass index and consumption of fat and alcohol on amyotrophic lateral sclerosis. JAMA Neurol 72, 1155–1162. [DOI] [PubMed] [Google Scholar]
- Ismailoglu I, Chen Q, Popowski M, Yang L, Gross SS, Brivanlou AH, 2014. Huntingtin protein is essential for mitochondrial metabolism, bioenergetics and structure in murine embryonic stem cells. Dev. Biol. 391, 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesus P, Fayemendy P, Nicol M, Lautrette G, Sourisseau H, Preux PM, Desport JC, Marin B, Couratier P, 2017. Hypermetabolism is a deleterious prognostic factor in patients with amyotrophic lateral sclerosis. Eur. J. Neurol. 10.1111/ene.13468. [DOI] [PubMed] [Google Scholar]
- Kawamata H, Peixoto P, Konrad C, Palomo G, Bredvik K, Gerges M, Valsecchi F, Petrucelli L, Ravits JM, Starkov A, Manfredi G, 2017. Mutant TDP-43 does not impair mitochondrial bioenergetics in vitro and in vivo. Mol. Neurodegener. 12, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, Zoing MC, 2011. Amyotrophic lateral sclerosis. Lancet 377, 942–955. [DOI] [PubMed] [Google Scholar]
- Kirk K, Gennings C, Hupf JC, Tadesse S, D’Aurelio M, Kawamata H, Valsecchi F, Mitsumoto H, Groups APCS, Manfredi G, 2014. Bioenergetic markers in skin fibroblasts of sporadic amyotrophic lateral sclerosis and progressive lateral sclerosis patients. Ann. Neurol. 76, 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad C, Kawamata H, Bredvik KG, Arreguin AJ, Cajamarca SA, Hupf JC, Ravits JM, Miller TM, Maragakis NJ, Hales CM, Glass JD, Gross S, Mitsumoto H, Manfredi G, 2017. Fibroblast bioenergetics to classify amyotrophic lateral sclerosis patients. Mol. Neurodegener. 12, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffner R, Zach N, Norel R, Hawe J, Schoenfeld D, Wang L, Li G, Fang L, Mackey L, Hardiman O, Cudkowicz M, Sherman A, Ertaylan G, Grosse-Wentrup M, Hothorn T, van Ligtenberg J, Macke JH, Meyer T, Scholkopf B, Tran L, Vaughan R, Stolovitzky G, Leitner ML, 2015. Crowdsourced analysis of clinical trial data to predict amyotrophic lateral sclerosis progression. Nat. Biotechnol. 33, 51–57. [DOI] [PubMed] [Google Scholar]
- Lawton KA, Cudkowicz ME, Brown MV, Alexander D, Caffrey R, Wulff JE, Bowser R, Lawson R, Jaffa M, Milburn MV, Ryals JA, Berry JD, 2012. Biochemical alterations associated with ALS. Amyotroph. Lateral Scler. 13, 110–118. [DOI] [PubMed] [Google Scholar]
- Lawton KA, Brown MV, Alexander D, Li Z, Wulff JE, Lawson R, Jaffa M, Milburn MV, Ryals JA, Bowser R, Cudkowicz ME, Berry JD, Northeast ALSC, 2014. Plasma metabolomic biomarker panel to distinguish patients with amyotrophic lateral sclerosis from disease mimics. Amyotroph Lateral Scler Frontotemporal Degener 15, 362–370. [DOI] [PubMed] [Google Scholar]
- Lee ZW, Low YL, Huang S, Wang T, Deng LW, 2014. The cystathionine gammalyase/hydrogen sulfide system maintains cellular glutathione status. Biochem. J. 460, 425–435. [DOI] [PubMed] [Google Scholar]
- Leung KY, Pai YJ, Chen Q, Santos C, Calvani E, Sudiwala S, Savery D, Ralser M, Gross SS, Copp AJ, Greene NDE, 2017. Partitioning of one-carbon units in folate and methionine metabolism is essential for neural tube closure. Cell Rep. 21, 1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Rush T, Zapata J, Lobner D, 2009. Beta-N-methylamino-l-alanine induces oxidative stress and glutamate release through action on system xc(−). Exp. Neurol. 217, 429–433. [DOI] [PubMed] [Google Scholar]
- Lu SC, 2013. Glutathione synthesis. Biochim. Biophys. Acta 1830, 3143–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin B, Couratier P, Arcuti S, Copetti M, Fontana A, Nicol M, Raymondeau M, Logroscino G, Preux PM, 2016. Stratification of ALS patients’ survival: a population-based study. J. Neurol. 263, 100–111. [DOI] [PubMed] [Google Scholar]
- Mesci P, Zaidi S, Lobsiger CS, Millecamps S, Escartin C, Seilhean D, Sato H, Mallat M, Boillee S, 2015. System xC- is a mediator of microglial function and its deletion slows symptoms in amyotrophic lateral sclerosis mice. Brain 138, 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RG, Block G, Katz JS, Barohn RJ, Gopalakrishnan V, Cudkowicz M, Zhang JR, McGrath MS, Ludington E, Appel SH, Azhir A, 2015. N.P.I. Phase 2 Trial, Randomized phase 2 trial of NP001-a novel immune regulator: Safety and early efficacy in ALS. Neurol Neuroimmunol Neuroinflamm 2 e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan M, Seeley KW, Jinwal UK, 2016. Identification of Apo B48 and other novel biomarkers in amyotrophic lateral sclerosis patient fibroblasts. Biomark. Med 10, 453–462. [DOI] [PubMed] [Google Scholar]
- Patel SS, Molnar MZ, Tayek JA, Ix JH, Noori N, Benner D, Heymsfield S, Kopple JD, Kovesdy CP, Kalantar-Zadeh K, 2013. Serum creatinine as a marker of muscle mass in chronic kidney disease: results of a cross-sectional study and review of literature. J. Cachexia. Sarcopenia Muscle 4, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman R, Allen SP, Goodall EF, Kramer S, Ponger LL, Heath PR, Milo M, Hollinger HC, Walsh T, Highley JR, Olpin S, McDermott CJ, Shaw PJ, Kirby J, 2015. Gene expression signatures in motor neurone disease fibroblasts reveal dysregulation of metabolism, hypoxia-response and RNA processing functions. Neuropathol. Appl. Neurobiol. 41, 201–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BH, Petrova-Benedict R, Buncic JR, Wallace DC, 1992. Nonviability of cells with oxidative defects in galactose medium: a screening test for affected patient fibroblasts. Biochem Med Metab Biol 48, 122–126. [DOI] [PubMed] [Google Scholar]
- Steyn FJ, Ioannides ZA, van Eijk RPA, Heggie S, Thorpe KA, Ceslis A, Heshmat S, Henders AK, Wray NR, van den Berg LH, Henderson RD, McCombe PA, Ngo ST, 2018. Hypermetabolism in ALS is associated with greater functional decline and shorter survival. J. Neurol. Neurosurg. Psychiatry 89, 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MR, Leigh PN, 2000. Positron emission tomography (PET)–its potential to provide surrogate markers in ALS. Amyotroph Lateral Scler Other Motor Neuron Disord 1 (Suppl. 2), S17–S22. [DOI] [PubMed] [Google Scholar]
- Valbuena GN, Rizzardini M, Cimini S, Siskos AP, Bendotti C, Cantoni L, Keun HC, 2016. Metabolomic analysis reveals increased aerobic glycolysis and amino acid deficit in a cellular model of amyotrophic lateral sclerosis. Mol. Neurobiol. 53, 2222–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk RPA, Eijkemans MJC, Ferguson TA, Nikolakopoulos S, Veldink JH, van den Berg LH, 2017. Monitoring disease progression with plasma creatinine in amyotrophic lateral sclerosis clinical trials. J. Neurol. Neurosurg. Psychiatry. 10.1136/jnnp-2017-317077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Klaveren RJ, Demedts M, Nemery B, 1997. Cellular glutathione turnover in vitro, with emphasis on type II pneumocytes. Eur. Respir. J. 10, 1392–1400. [DOI] [PubMed] [Google Scholar]
- van Weehaeghe D, Ceccarini J, Willekens SM, de Vocht J, van Damme P, van Laere K, 2017. Is there a glucose metabolic signature of spreading TDP-43 pathology in Amyotrophic Lateral Sclerosis? Q J Nucl Med Mol Imaging. 10.23736/S1824-4785.17.03009-6. [DOI] [PubMed] [Google Scholar]
- Vucic S, 2017. Hypermetabolism appears to be an adverse prognostic biomarker in amyotrophic lateral sclerosis: a potential for therapeutic intervention? Eur. J. Neurol. 10.1111/ene.13470. [DOI] [PubMed] [Google Scholar]
- Weijs PJ, 2011. Hypermetabolism, is it real? The example of amyotrophic lateral sclerosis. J. Am. Diet. Assoc. 111, 1670–1673. [DOI] [PubMed] [Google Scholar]
- Wuolikainen A, Jonsson P, Ahnlund M, Antti H, Marklund SL, Moritz T, Forsgren L, Andersen PM, Trupp M, 2016. Multi-platform mass spectrometry analysis of the CSF and plasma metabolomes of rigorously matched amyotrophic lateral sclerosis. Parkinson’s disease and control subjects, Mol Biosyst 12, 1287–1298. [DOI] [PubMed] [Google Scholar]
- Wyss M, Kaddurah-Daouk R, 2000. Creatine and creatinine metabolism. Physiol. Rev. 80, 1107–1213. [DOI] [PubMed] [Google Scholar]
- Yang S, Zhang KY, Kariawasam R, Bax M, Fifita JA, Ooi L, Yerbury JJ, Nicholson GA, Blair IP, 2015. Evaluation of skin fibroblasts from amyotrophic lateral sclerosis patients for the rapid study of pathological features. Neurotox. Res. 28, 138–146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.