Abstract
Understanding the functional connection that occurs for the three nuclear RNA polymerases to synthesize ribosome components during the ribosome biogenesis process has been the focal point of extensive research. To preserve correct homeostasis on the production of ribosomal components, cells might require the existence of proteins that target a common subunit of these RNA polymerases to impact their respective activities. This work describes how the yeast prefoldin-like Bud27 protein, which physically interacts with the Rpb5 common subunit of the three RNA polymerases, is able to modulate the transcription mediated by the RNA polymerase I, likely by influencing transcription elongation, the transcription of the RNA polymerase III, and the processing of ribosomal RNA. Bud27 also regulates both RNA polymerase II-dependent transcription of ribosomal proteins and ribosome biogenesis regulon genes, likely by occupying their DNA ORFs, and the processing of the corresponding mRNAs. With RNA polymerase II, this association occurs in a transcription rate-dependent manner. Our data also indicate that Bud27 inactivation alters the phosphorylation kinetics of ribosomal protein S6, a readout of TORC1 activity. We conclude that Bud27 impacts the homeostasis of the ribosome biogenesis process by regulating the activity of the three RNA polymerases and, in this way, the synthesis of ribosomal components. This quite likely occurs through a functional connection of Bud27 with the TOR signaling pathway.
Keywords: prefoldin-like, transcription, RNA polymerases, ribosome biogenesis, Saccharomyces cerevisiae
INTRODUCTION
In many eukaryotes, there are three nuclear RNA polymerases (RNA Pol I, II, III) responsible for synthesizing cellular RNAs. In plants, two other RNA pols, RNA Pol IV and V, have been described (Kwapisz et al. 2008; Haag and Pikaard 2011; Haag et al. 2014; Cuevas-Bermúdez et al. 2017). In yeast, 80% of transcriptional activity, and 50% in proliferating mammalian cells, is dedicated to the synthesis of ribosomal components (Xiao and Grove 2009); in yeast, this corresponds to the synthesis of the precursor of rRNA 35S by RNA Pol I, which generates the mature 18S, 25S, and 5.8S ribosomal RNAs (rRNAs), the synthesis of 5S rRNA by RNA Pol III, the expression of the mRNAs corresponding to all ribosomal proteins (RPs), and the synthesis of more than 250 proteins and 80 small nucleolar RNAs (snoRNAs) trans-acting factors mediated by RNA Pol II (Loewith and Hall 2011; Jakob et al. 2012; de la Cruz et al. 2018). This complex transcriptional activity, which results in yeast from total RNA Pol I transcription (60% total transcriptional activity), from ∼10% of all RNA Pol II transcription and 30% of all RNA Pol III activity (Warner 1999; de la Cruz et al. 2018), is properly regulated due to the tight coordination of the three RNA pols (Briand et al. 2001; Michels and Hernandez 2006).
Cells need to adjust their ribosome content to their extracellular environment to properly regulate their growth and maintain an adequate protein synthesis rate. In response to favorable conditions, cells up-regulate ribosome synthesis and grow faster, as for instance during cardiac hypertrophy, where cells increase their size and augment their ribosome content (Mayer and Grummt 2006; Iadevaia et al. 2014). In turn, growth inhibition triggers mechanisms to ensure lesser ribosome synthesis (Loewith and Hall 2011). This exquisite regulation is partly due to the fact that a significant proportion of global cellular energy and resources is spent on ribosome biogenesis. In exponentially growing yeast cells, about 200,000 ribosomes can be found, which implies the assembly of approximately 2000 ribosomes per minute (Warner 1999). In proliferating human HeLa cells, this number is about 3500 ribosomes per minute (Lewis and Tollervey 2000; Mayer and Grummt 2006).
In this complex scenario, it is obvious that mechanisms may exist to tightly coordinate the activity of the three RNA pols and other ribosome biogenesis steps to ensure a nonlimiting amount of all the ribosome components and factors devoted to their synthesis. Thus, different signaling pathways have been described as general regulators of ribosome biogenesis, which control various aspects of this multifaceted process, including transcription; basically, these pathways comprise the TOR signaling cascade via TORC1 and PKA kinases (Martin et al. 2004; Xiao and Grove 2009; Loewith and Hall 2011). The TOR pathway is active under favorable growth conditions where a high ribosome production rate is maintained. However, under stress conditions, this pathway is inhibited and ribosome biogenesis concomitantly ceases (Xiao and Grove 2009; González and Hall 2017). Some key regulators have been described to coordinate the synthesis of rRNAs, RP genes and the protein factors involved in ribosome biogenesis, such as those belonging to the rRNA and ribosome biogenesis (RRB) and the ribosome biogenesis (RiBi) regulons; among these factors, we find Sch9 and the S6K kinases, several transcription regulators like Rap1, Ifh1, Pbf1, Pbf2, Sfp1, Msn2/4, and Fhl1 and the components of the CURI complex which, in addition to Ifh1, includes the Ckb2, Utp22, and Rrp7 proteins (Martin et al. 2004; Rudra et al. 2005; Loewith and Hall 2011; Albert et al. 2016; Vizoso-Vázquez et al. 2018).
As RNA pols I, II and III are multimeric protein complexes that contain five common subunits (Shpakovski et al. 1995; Cuevas-Bermúdez et al. 2017), it is tempting to speculate that factors may exist that influence the transcriptional activity of the three RNA pols via a common RNA pol subunit to regulate ribosome biogenesis, probably also in coordination with signaling pathways. One candidate for these regulatory factors could be yeast prefoldin-like Bud27 and its human ortholog URI (unconventional prefoldin RPB5-interactor). Both proteins bind the common Rpb5 subunit for the three RNA pols and have been proposed to be involved in TOR-dependent transcription programs (Gstaiger et al. 2003; Martínez-Fernández et al. 2015, 2018). Bud27 and URI are members of the prefoldin family of ATP-independent molecular chaperones, which are considered to function as scaffold proteins during the assembly of additional prefoldin family members (Mita et al. 2013; Martínez-Fernández et al. 2018). Both Bud27 and URI contain different domains: a Rpb5-binding domain, a PFD-binding domain and a URI-box (Martínez-Fernández et al. 2018). Bud27 also shows a nuclear localization signal (NLS) that is conserved in its human URI ortholog, and a leucine-rich nuclear export signal (NES) (Martínez-Fernández et al. 2018). Notably, the PFD-binding domain accounts for the association of Bud27 or URI with two members of the prefoldin complex, Pfd2 and Pfd6 (Mirón-García et al. 2013; Martínez-Fernández et al. 2018), but also appears necessary for the role of Bud27 during RNA Pol II transcription elongation (Mirón-García et al. 2014). Apart from this function, Bud27 and/or URI also participate in other relevant nuclear and cytoplasmic processes, some of of which are once again related to transcription: for example, the biogenesis and assembly of the three RNA pols in an Rpb5-dependent manner; the elongation of RNA Pol II-dependent transcription or RNA Pol III-dependent transcription via the association with chromatin remodeler complex RSC (Boulon et al. 2010; Cloutier and Coulombe 2010; Forget et al. 2010; Mirón-García et al. 2013, 2014; Martínez-Fernández et al. 2015, 2018; Vernekar and Bhargava 2015). Bud27 plays many other cellular functions. Thus, Bud27 has been proposed to coordinate translation initiation and cotranslational quality control (Deplazes et al. 2009). Bud27 also seems to act in DNA-repair processes (Muñoz-Galván et al. 2013). Both, Bud27 and URI have been considered to participate in TOR-dependent transcription programs (Gstaiger et al. 2003). Accordingly with their role in different fundamental cellular processes, human URI has been described to be related to cancer development (Theurillat et al. 2011; Tummala et al. 2014; Buren et al. 2016).
Notably for its relation with RNA Pol III, the decrease in RNA Pol III transcription observed in a bud27Δ null strain closely corresponds with the reduction in RNA Pol III occupancy, at least for genes types 2 (for tRNAs) and 3 (other genes that differ from those for tRNAs, and 5S rRNA) (Vernekar and Bhargava 2015). Similarly, the absence of Bud27 leads to defects in RNA Pol II transcription elongation, which is accompanied by reduced RNA Pol II occupancy, at least in some genes (Mirón-García et al. 2014). However, very little is known about the influence of Bud27 on both RNA Pol I activity and the transcription of the ribosomal components required for ribosome biogenesis.
In this work, we demonstrate that Bud27 influences the transcriptional activity of the three RNA pols by its absence impairing the synthesis of all ribosomal components and the ribosome biogenesis process. Our data indicate that Bud27 impacts the activity of RNA Pol I, probably through the transcription elongation step. Furthermore, the regulation of RNA Pol I activity may occur differently to mechanisms described to operate for RNA Pol II and III. We also show that Bud27 modulates 5S rRNA production by RNA Pol III, the maturation of rRNAs, mainly 7S pre-rRNAs processing that leads to mature 5.8S rRNAs and, notably, the production of the mRNAs of different RP and RRB/RiBi genes by RNA Pol II. Our results indicate that Bud27 associates with the ORF region of the RPs and RRB/RiBi genes, likely by its interaction with RNA Pol II in a transcription rate-dependent manner. Finally, we also suggest that Bud27 might modulate the activity of TORC1 kinase to influence the phosphorylation status of one of its targets, the S6 RP.
RESULTS
Bud27 modulates RNA Pol I transcription and rRNA processing
Rpb5 is a subunit shared by the three eukaryotic RNA pols. Several amino acid substitutions have been shown to affect the transcription mediated by any of the three transcription complexes (Zaros et al. 2007). We have previously demonstrated the influence of Bud27 on RNA Pol II-dependent transcription elongation, which is mediated by the physical and functional interaction of Bud27 with Rpb5 in coordination with the RSC chromatin remodeler complex (Mirón-García et al. 2014). Similarly, Bud27 modulates RNA Pol III-dependent transcription, probably also via its association with the RSC complex (Vernekar and Bhargava 2015). However, whether Bud27 influences RNA Pol I-dependent transcription, which represents ∼60% of total cellular transcriptional activity, and then, the rRNA synthesis for ribosome biogenesis, has not yet been explored.
Here we investigated whether Bud27 could impact RNA Pol I activity. To explore the influence of the absence of Bud27 on the production of rRNAs, the steady-state levels of the pre- and mature rRNA species were analyzed (See Supplemental Fig. S2 for a scheme of pre-rRNA processing in yeast). Since Bud27 is not essential, a bud27Δ null strain and its otherwise isogenic wild-type counterpart were exponentially grown in liquid YPD at 30°C before being shifted at 37°C and maintained exponentially growing for up to 12 h; at 37°C, the bud27Δ strain clearly grew more slowly than its otherwise isogenic wild-type counterpart (a doubling time of 3.7 h vs. 1.7 h, respectively. See Supplemental Fig. S3 for growth curves at 37°C). Total RNA was extracted from each strain and analyzed by northern blot hybridization. As shown in Figure 1A, no major differences were detected for most of the pre-rRNAs intermediates and mature rRNAs analyzed from both strains at any temperature. However, a slight, but significant, increase in the levels of 7S pre-rRNAs was detected in the bud27Δ null strain compared to its wild-type counterpart (Fig. 1A). Despite the accumulation of 7S pre-rRNAs, the steady-state levels of the mature 5.8S rRNAs remained practically unaffected following the temperature shift likely because these mature RNAs are very stable and their levels normally change as the result of their dilution by cellular duplication (Kressler et al. 1998).
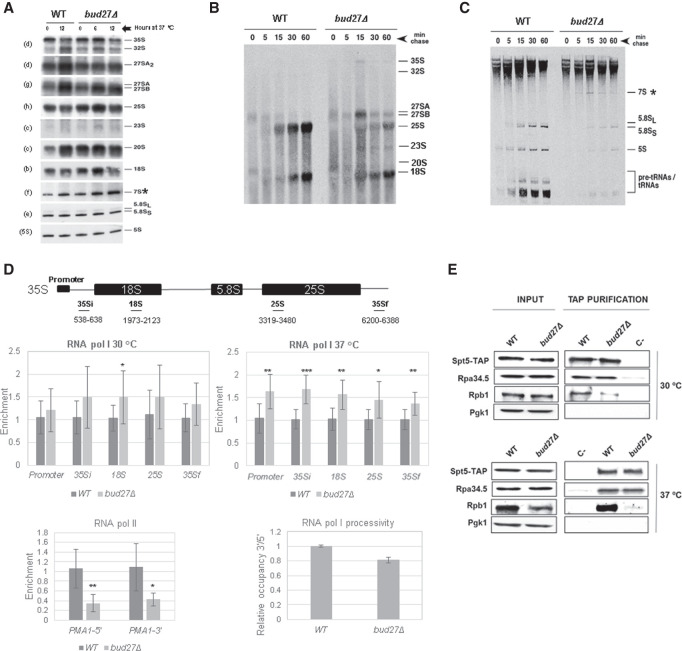
FIGURE 1.
Bud27 is required for the optimal synthesis and maturation of the rRNAs transcribed by RNA polymerase I. (A) Northern blot analysis of the pre- and mature rRNAs of the wild-type and bud27Δ strains. Cells were grown in YPD medium at 30°C and shifted to 37°C for 6 or 12 h. Total RNA was extracted and equal amounts (5 µg) were subjected to northern hybridization. Probes, in brackets, are described in Supplemental Table S2 and Supplemental Figure S2. 7S pre-rRNA is indicated by an asterisk. (B,C) The wild-type and bud27Δ strains were transformed with YCplac33 (CEN, URA3), grown at 30°C in SD-Ura to the mid-log phase and shifted to 37°C for 12 h. Cells were pulse-labeled with [5,6-3H]uracil for 2 min, followed by a chase with unlabeled uracil largely in excess for the indicated times. Total RNA was extracted from each sample and 20,000 c.p.m. were loaded and separated on 1.2% agarose-6% formaldehyde gel (B) or on 7% polyacrylamide-8 M urea gel (C) before being transferred to nylon membranes and visualized by fluorography. The positions of the different pre- and mature rRNAs are indicated. We assume that 7S pre-rRNAs correspond to the species labeled by an asterisk, which were only evident in bud27Δ cells. (D) RNA Pol I and Pol II occupancy (Rpb8-TAP occupancy) was analyzed by chromatin immunoprecipitation (ChIP) in the wild-type and bud27Δ cells containing a functional TAP-tagged version of Rpb8, which is a common subunit to the three RNA pols. Cells were grown in YPD at 30°C or shifted to 37°C for 12 h. RNA pols were precipitated using Dynabeads Pan Mouse IgG as described in the Materials and Methods. The occupancy on the rDNA unit was analyzed for the indicated amplicons. The values found for the immunoprecipitated PCR products were compared to those of the total input, and the ratio of each PCR product of transcribed genes to a nontranscribed region of chromosome V was calculated. The average and standard deviations of three biological replicates are shown. RNA Pol II occupancy on PMA1 gene (5′ and 3′ regions) was analyzed at 30°C. Statistical significance, by t-Student. (*) P < 0.05, (**) P < 0.005, (***) P < 001. The lowest right panel shows the processivity analysis of the data from RNA Pol I occupancy, at 37°C. (E) Wild-type and bud27Δ cells containing a functional TAP-tagged version of Spt5 were grown in YPD medium at 30°C or shifted to 37°C for 12 h. Whole-cell extracts were performed and subjected to pull-down with an anti-TAP resin. The obtained immunoprecipitates were analyzed by western blot with antibodies against TAP (Spt5), Rpa34.5 (RNA Pol I), Rpb1 (RNA Pol II), and Pgk1 as a control.
We explored in more detail whether the loss-of-function of Bud27 was associated with defects in rRNA transcription dependent on RNA Pol I activity and/or pre-rRNA processing. To do so, we analyzed the kinetics of rRNA maturation by pulse-chase labeling with [5,6-3H]uracil. Both the bud27Δ mutant and the otherwise isogenic wild-type strain were transformed with YCplac33 plasmid (CEN, URA3, see the “Materials and Methods” section) to make them prototrophic for uracil. Then, cells were exponentially grown in minimal medium lacking uracil (SD-Ura) at 30°C before being shifted to 37°C and maintained exponentially growing for up to 12 h. Under these conditions, the bud27Δ mutant cells clearly grew more slowly than the wild-type cells (a doubling time of about 4.0 h versus 2.7 h, respectively. See Supplemental Fig. S3). Then, cultures were pulse-labeled for 1 min with [5,6-3H]uracil and chased for 5, 15, 30, and 60 min with nonradioactive uracil considerably in excess. Total RNA was extracted from each sample. Uracil incorporation was measured by scintillation counting and the same counts per RNA extract were analyzed by gel electrophoresis, followed by transfer to nylon membranes and fluorography. As a result, uracil incorporation into pre- and mature rRNAs reduced by approximately twofold in the bud27Δ mutant compared to the wild-type counterpart, as estimated from the counts detected in the RNA samples (data not shown) and from the relative exposure required to provide equal signals in the nylon membranes after fluorography (Fig. 1B,C). These results suggest less RNA Pol I activity in the bud27Δ mutant cells, and also evidence that deletion of BUD27 modestly affects rRNA processing. An analysis of high-molecular-weight RNAs revealed that the maturation of 35S and 27S pre-rRNAs was slightly delayed in the bud27Δ mutant, as shown by their persistence in the chase at later time points (Fig. 1B). A delay in 35S pre-rRNA processing was further reflected by small amounts of the well-defined 23S pre-rRNA intermediate appearing. Consistently with some delay during the processing of 27S pre-rRNAs, the synthesis of the mature 25S rRNA was also retarded, unlike that of 18S rRNA, which was apparently unaffected (Fig. 1B). The analysis of low-molecular-weight pre- and mature RNAs showed a marked accumulation of species with the gel mobility of 7S pre-rRNAs and delayed mature 5.8S rRNAs production (Fig. 1C). Taken together, these data indicate that Bud27 directly contributes to both the transcription by RNA Pol I and subsequent co- or posttranscriptional events by influencing further 35S pre-rRNA processing, which is its sole primary transcript. In particular, the absence of Bud27 generally delayed the maturation of 25S and 5.8S rRNAs, and led to mild 7S pre-rRNA accumulation, while no effects were detected in 18S rRNA maturation.
To further address the role of Bud27 in RNA Pol I transcription, we performed RNA Pol I ChIP experiments (by qPCR ChIP assays) to analyze the occupancy of this polymerase along the 35S rDNA unit. As Bud27 has been shown to be involved in TOR-dependent transcription programs and lack of Bud27 partially overlaps TOR inhibition responses (Gstaiger et al. 2003), and in line with what occurs when TOR is inhibited by rapamycin addition (Claypool et al. 2004) or under nutrient starvation (Torreira et al. 2017), we addressed whether reduced RNA Pol I occupancy along the 35S rDNA unit could account for the diminished RNA Pol I activity observed in the bud27Δ mutant. For this purpose, we used exponentially growing cells in YPD at 30°C of the bud27Δ mutant and the otherwise isogenic wild-type strain containing a functional TAP-tagged version of Rpb8, which is a common subunit to the three RNA pols. Surprisingly, our results (Fig. 1D, left upper panel) showed no significant differences of RNA Pol I occupancy throughout the 35S rDNA region or its promoter in the bud27Δ cells compared to wild-type cells, except for a minor, but statistically significant, increase in RNA Pol I occupancy at the 18S rDNA unit. These data suggest that transcription initiation by RNA Pol I is not altered in bud27Δ mutant cells, rather the RNA Pol I complex may transiently stall at the 5′ end of the 35S rDNA unit once transcription advances. These results point to a specific role of Bud27 in RNA Pol I transcription, which seems to differ from that occurring upon TOR inhibition by rapamycin or nutrient starvation and results in clearly diminished RNA Pol I occupancy (Claypool et al. 2004; Torreira et al. 2017) that could account for Bud27 modulating RNA Pol I-dependent transcription elongation. As a control, we also performed RNA Pol II ChIP analyses by taking the advantage of purifying Rpb8-TAP to also precipitate RNA Pol II. As expected, and in accordance with previously reported data (Mirón-García et al. 2014), the results of the ChIP analyses, which were done with cells grown at 30°C, revealed a significantly decreased RNA Pol II occupancy in the PMA1 gene (Fig. 1D). We validated the RNA Pol I occupancy data by analyzing the amount of global RNA Pol I associated with chromatin from the chromatin-enriched fractions obtained by the yChEFs procedure (Cuevas-Bermúdez et al. 2019) using western blot experiments run with anti-Rpa34.5 antibodies against its Rpa34.5 subunit. As shown in Supplemental Figure S4, no apparent differences in the association with the chromatin of RNA Pol I (Rpa34.5) from bud27Δ or wild-type cells were detected. In contrast, and in agreement with previously reported data (Vernekar and Bhargava 2015), RNA Pol III associated less efficiently with chromatin in bud27Δ cells, as evidenced by a different enrichment in the Rpc160 largest subunit of RNA Pol III (Supplemental Fig. S4).
As defects in rRNA synthesis and processing were observed in the bud27Δ mutant cells upon the shift to 37°C for 12 h (Fig. 1B,C), we also performed RNA Pol I ChIP experiments in bud27Δ and the otherwise isogenic wild-type cells under these growth conditions. As also shown in Figure 1D, the qPCR analysis showed that the amount of RNA Pol I occupancy increased along the entire 35S rDNA and its promoter in the mutant cells, which was statistically significant in this case. Yet despite this general increase, a reduction in RNA Pol I occupancy from the beginning to the end of the rDNA unit was observed in the bud27Δ cells (Fig. 1D, right upper panel). These data suggest and reinforce the idea of a defect in RNA Pol I transcription elongation in bud27Δ cells, but also point out a role of functional Bud27 in controlling the binding of RNA Pol I to the 35S rDNA region. To know if lack of Bud27 could influence RNA Pol I transcription elongation, we measured the processivity of RNA Pol I by using the ChIP assay results, as previously reported (Schneider et al. 2007). We compared the RNA Pol I association near the 5′ and 3′ ends of the 35S rRNA gene and calculated the ratio between the ChIP signals at the 3′ end relative to the 5′ end, which can be then used to estimate the enzyme's relative processivity. As shown in Figure 1D (right lower panel), we observed a reproducible decrease of ∼20% in the distribution of RNA Pol I at the 3′ of the rDNA gene versus the 5′ region, at 37°C. These results suggest that deleting BUD27 may lead to an altered efficiency in RNA Pol I elongation along the rDNA gene.
RNA Pol I transcription elongation and rRNA processing depend on the Spt4/5 transcription factor, which physically interacts with RNA Pol I. Consequently, spt4Δ and spt5 mutants affect rRNA processing and rRNA synthesis (Schneider et al. 2006; Anderson et al. 2011; Viktorovskaya et al. 2011; Scull and Schneider 2019). To investigate whether BUD27 deletion could impair the association between RNA Pol I and the Spt5/4 complex, we analyzed the physical interaction between Spt5 and RNA Pol I by TAP purification of TAP-tagged Spt5 in both the bud27Δ mutant and wild-type cells grown in YPD at 30°C or upon a shift to 37°C for 12 h. As shown in Figure 1E, no significant alteration to the amount of the RNA Pol I Rpa34.5 subunit copurified with Spt5 resulted from the bud27Δ mutation either at 30°C or upon the shift to 37°C. Strikingly, BUD27 deletion affected the physical interaction between Spt5 and RNA Pol II, which was analyzed with antibodies against its Rpb1 largest subunit (Fig. 1E), in consonance with the previous reporter role of Bud27 in RNA Pol II-mediated transcription elongation (Mirón-García et al. 2014).
Taken together, these data evidence that lack of Bud27 affects RNA Pol I activity and rRNA processing, probably by altering transcription elongation in an Bud27-specific manner that seems to differ from this occurring upon TOR-inhibition by rapamycin addition or nutrient starvation (Claypool et al. 2004; Torreira et al. 2017).
Bud27 modulates transcription by RNA Pol III
We also analyzed the functional relation between Bud27 and RNA Pol III and its contribution to ribosome biogenesis. Our data revealed that the steady-state levels of 5S rRNA, a transcript synthesized by RNA Pol III, were apparently not affected in the bud27Δ null strain at either of the tested temperatures (Fig. 1A). In line with this, previous data have reported no impact on the transcription of the type 1 (5S rRNA) gene in a bud27Δ mutant (Vernekar and Bhargava 2015). To gain more insight into the role of Bud27 in RNA Pol III-dependent transcription and into its contribution to rRNA synthesis, we further examined whether Bud27 was required for the transcription and/or processing of 5S rRNA. To do so, the kinetics of the processing and production of 5S rRNA was also analyzed by pulse-chase labeling with [5,6-3H]uracil after a shift to 37°C for 12 h. As shown in Figure 1C, 5S rRNA production was clearly delayed upon BUD27 deletion. These results evidence a defect in RNA Pol III activity for the synthesis of 5S rRNA, contrarily to what has been previously proposed (Vernekar and Bhargava 2015). However, these data did not contradict those of the steady-state levels of 5S rRNA when considering the high cellular stability of this rRNA component. To investigate whether a reduction in 5S rRNA synthesis could result from a deficiency in the RNA Pol III occupancy to the 5S rRNA gene (type 1) (Shukla and Bhargava 2018), we performed RNA Pol III ChIP assays using as bait a TAP-tagged Rpb8 expressed in bud27Δ or wild-type cells grown at 30°C in YPD medium. Notably as shown in Figure 2B, the RNA Pol III occupancy in the 5S rRNA gene clearly decreased in the bud27Δ mutant compared to its otherwise isogenic wild-type counterpart. Similar results were obtained when the ChIP analyses were performed upon a shift to 37°C for 12 h (Fig. 2B, bottom panel).
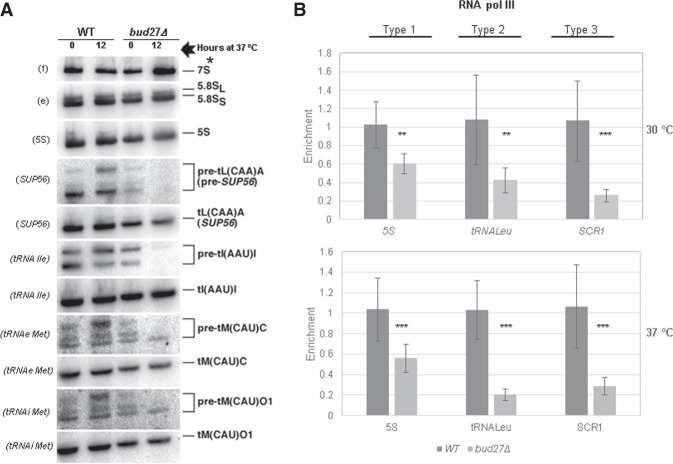
FIGURE 2.
Bud27 is required for the optimal synthesis and maturation of 5S rRNA and tRNAs transcribed by RNA polymerase III. (A) Northern analyses of the selected pre- and mature tRNAs and 5S rRNA of wild-type and bud27Δ strains. Cells were grown in YPD medium at 30°C and shifted to 37°C for 12 h. Total RNA was extracted and equal amounts (5 µg) were subjected to northern hybridization. Probes, in brackets, are described in Supplemental Table S2. 7S pre-rRNAs are indicated by an asterisk. Note that the doublets shown in the pre-tRNA panels are different precursor forms of the corresponding tRNAs. (B) RNA Pol III occupancy (Rpb8-TAP occupancy) was analyzed by chromatin immunoprecipitation (ChIP) in the wild-type and bud27Δ cells containing a functional Rpb8-TAP tagged, which is a common subunit to the three RNA pols. Cells were grown in YPD at 30°C or shifted to 37°C for 12 h. RNA pols were precipitated using Dynabeads Pan Mouse IgG as described in Materials and Methods. Occupancy in the type 1 (5S rDNA), type 2 (tRNALeu, SUP56), and type 3 (SCR1) genes was analyzed. The values found for the immunoprecipitated PCR products were compared to those of the total input, and the ratio of each polymerase chain reaction (PCR) product of transcribed genes to a nontranscribed region of chromosome V was calculated. The average and standard deviations of three biological replicates are shown. Statistical significance, by t-Student. (**) P < 0.01, (***) P < 0.05.
The defect in RNA Pol III transcription observed in the bud27Δ mutant was not specific of 5S rRNA, but also extended to type 2 (tRNAs) genes. In fact the kinetics of the processing and production of tRNAs analyzed by pulse-chase labeling with [5,6-3H]uracil after a shift to 37°C for 12 h demonstrated that the bulk of pre-tRNAs was clearly delayed in the bud27Δ strain compared to the wild-type control (Fig. 1C). Furthermore, the steady-state levels of the distinct pre- and mature tRNA species analyzed by northern blot hybridization from the total RNA isolated from wild-type and bud27Δ cells showed that pre-tRNA levels clearly lowered in the bud27Δ strain upon the shift to 37°C (Fig. 2A). Notably, our results also evidenced slight differences between the wild-type and bud27Δ strains at 30°C, such as those in the pre-tRNA levels for leucine (Sup56) and for the initiator methionine tRNAs, unlike what has been previously reported (Vernekar and Bhargava 2015), and also in the band intensities for leucine (SUP56) and isoleucine pre-tRNA, which suggests mild defects in pre-tRNA processing. However, the levels of the analyzed mature tRNAs did not significantly change, which could likely be due to their cellular stability. In agreement with the diminished RNA Pol III activity, an equivalent decrease in RNA Pol III occupancy was evidenced for the leucine tRNA (tL(CAA)A) gene. Furthermore, a major reduction in RNA Pol III occupancy was observed for the SCR1 gene (type 3). These results were consistent with the lesser association of Rpc160, which is the largest subunit of RNA Pol III, with chromatin in bud27Δ cells (Supplemental Fig. S4). Similar results were obtained when the ChIP analyses were performed upon a shift to 37°C for 12 h, although the reduction in RNA Pol III occupancy in the leucyl-tRNA was clearly exacerbated (Fig. 2B, bottom panel).
Notably, these defects were apparently not the mere consequence of a defective Bud27-dependent assembly of RNA Pol III. In fact, correcting the assembly of the three RNA pols in the bud27Δ mutant strain by RPB5 overexpression at either 30°C (Mirón-García et al. 2013) or 37°C, as the suppression of the cytoplasmic mislocalization of subunits of the RNA pols revealed (Supplemental Fig. S5), did not modify the results in Figures 1A–C and 2A (data not shown). Moreover, under the RPB5 overexpression conditions, the bud27Δ mutant but not the wild-type strain was hypersensitive to mycophenolic acid (Supplemental Fig. S6), a nucleotide triphosphate (NTP)-depleting drug used to detect S. cerevisiae strains defective in transcription elongation (Shaw et al. 2001).
Altogether, our data indicated that Bud27 contributed to the efficient RNA Pol III-dependent transcription of 5S rRNA, but also to the transcription and processing of other types of RNA Pol III-driven genes. The reduced transcription activity was mainly dependent on RNA Pol III occupancy for 5S rRNA, but also for the other RNA Pol III genes, which agrees with previous data for type 2 and type 3 genes (Vernekar and Bhargava 2015). Notably, the defects caused by loss of Bud27 did not seem dependent mainly on defective RNA Pol III assembly, unlike what has been previously proposed (Vernekar and Bhargava 2015). Similarly, RPB5 overexpression did not suppress the RNA Pol I transcriptional defects observed in a bud27Δ strain (data not shown). Moreover, these defects were not caused by significant differences in the rDNA gene copy number, estimated by qPCR, between the wild-type and the bud27Δ mutant strain (Supplemental Fig. S7).
Bud27 is required for the normal accumulation of the mRNAs of ribosomal protein genes
As stated above, in addition to transcription by RNA Pol II (Mirón-García et al. 2014), Bud27 contributes to transcription by RNA Pol I and III and, therefore, to the transcription of ribosomal components. As previously mentioned, lack of Bud27 lowers the mRNA levels of certain genes transcribed by RNA Pol II at 37°C, such as PYK1 (Mirón-García et al. 2014). We wondered if Bud27 could significantly influence the transcription of RP genes, and if inactivating Bud27 could also impact ribosome biogenesis via RP production. Thus, we explored whether the loss-of-function of Bud27 would also affect the steady-state levels of the mRNAs of selected genes corresponding to RP proteins of the large and small r-subunit. Otherwise isogenic wild-type and bud27Δ cells were grown at 30°C and then shifted to 37°C for up to 12 h. Then, total RNA was isolated and the mRNA levels of selected RP genes were assessed by RT-qPCR. As shown in Figure 3A, the bud27Δ strain led to a substantial decrease in mRNA accumulation for all tested RP genes, especially when shifting cells from 30°C to 37°C.
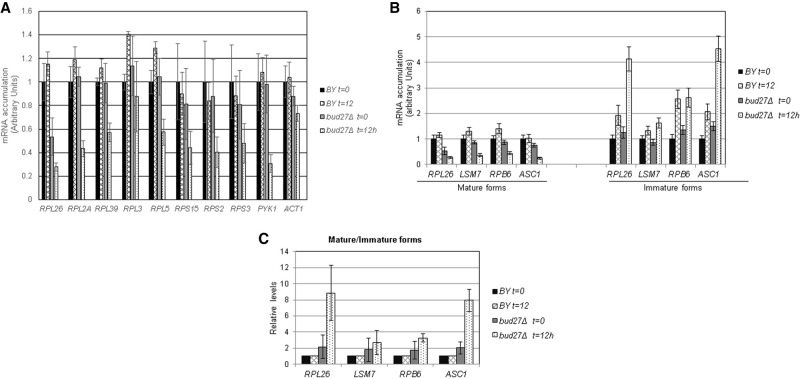
FIGURE 3.
Lesser accumulation of mRNAs of the RP genes in the absence of Bud27. (A) Analysis of the mRNA accumulation of several RP genes by RT-qPCR in the wild-type and bud27Δ strains. Cells were grown in YPD medium at 30°C and shifted to 37°C for 12 h. The mRNA levels of PYK1 were used as a control of a gene whose expression decreases and those of ACT1 as the control of a gene with no significant alteration. The used oligonucleotides are listed in Supplemental Table S1. (B) Analysis by RT-qPCR of the mRNA accumulation of the immature and mature forms of some intron-containing genes in the wild-type and bud27Δ strains. Cells were grown in YPD medium at 30°C and shifted to 37°C for 12 h. (C) Ratios for the immature versus mature forms of the genes analyzed in B.
Some RP genes contain introns, which have been suggested to modulate the expression of the corresponding RP genes in response to different stresses (Petibon et al. 2016). To investigate whether intron maturation is impaired in the absence of Bud27, and how it influences the drop observed in the accumulation of mature RP mRNAs in a bud27Δ strain, the levels of the mature and immature forms of RPL26 mRNA were also analyzed by RT-qPCR. As shown in Figure 3B, the immature form of this mRNA accumulated when bud27Δ cells were shifted to 37°C for 12 h, while the mature form decreased and, thus, led to a higher immature/mature mRNA RPL26 ratio than that observed in the corresponding wild-type strain (Fig. 3C). Notably, this alteration was not exclusive of intron-containing RP genes, but was also observed for the mRNAs of other intron-containing genes, such as those of LSM7, RPB6, or ASC1, which codes for the Asc1 protein that interacts with ribosomes and regulates translation (Baum et al. 2004). Therefore, lack of Bud27 generally alters mRNA splicing.
Altogether, our results suggest that Bud27 seems to be required for the proper accumulation of mRNAs of RP genes, likely due to a positive role played during their transcription, but also by the contribution of Bud27 to the general mRNA splicing process.
Bud27 globally regulates the expression of RP and ribosome biogenesis regulon genes
By bearing in mind the impact of Bud27 inactivation on the accumulation of the mRNA levels for selected RP genes, we studied the mRNA levels for all the RP and RiBi genes in a genome-wide analysis by RNA-seq in a bud27Δ mutant and its otherwise isogenic wild-type strain at the permissive temperature of 30°C. Two biological replicates were used with high technical reproducibility (see Supplemental Fig. S8). As shown in Figure 4, the RP genes expression was down-regulated in the bud27Δ mutant compared to its wild-type counterpart. Although this decrease was weak when globally analyzed, the results in Figure 3 reflect that this effect was exacerbated when shifting cells to 37°C, at least for the analyzed genes. A less marked, but still significant decrease, was also observed for the global expression of the RiBi genes (Fig. 4). The RiBi genes required for 60S r-subunit biogenesis were similarly affected to those required for 40S r-subunit biogenesis (data not shown). Notably, the drop in mRNA accumulation for the RP genes in the bud27Δ mutant versus its otherwise isogenic wild-type control became more evident when all the RP genes were represented on a scatterplot (as shown in Fig. 7, panel C). This also occurred for the RiBi genes, albeit to a lesser extent.
FIGURE 4.
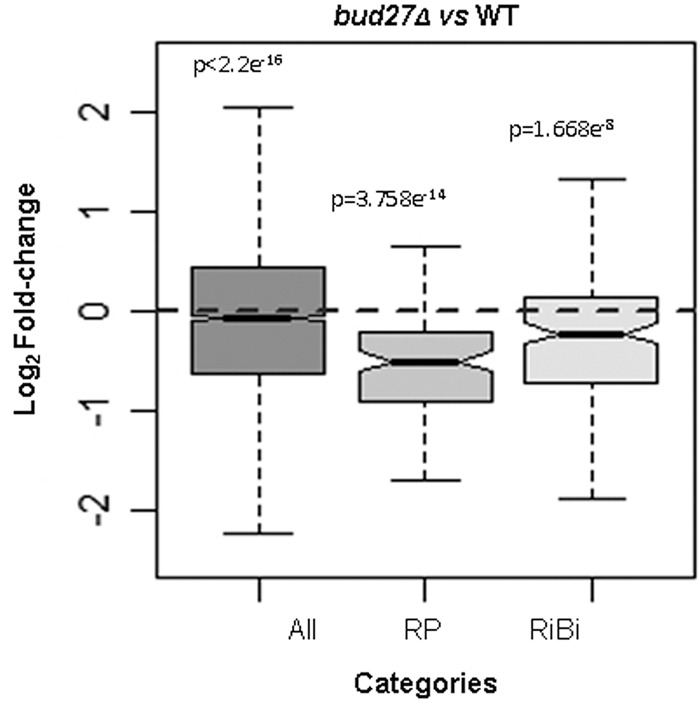
Comparison of the transcriptomic expression profile of the RP and RiBi genes from bud27Δ and wild-type yeast cells. Box plots showing the effects of BUD27 deletion on the expression levels of the indicated classes of genes. Statistical testing was done by the Wilcoxon test. Data represent the differentially expressed genes (more than twofold). Cells were grown in YPD at 30°C.
FIGURE 7.
Global transcriptomic expression analysis of a bud27Δ mutant and rapamycin-treated wild-type cells. (A) Venn diagrams representing the differentially expressed genes (more than twofold) in the bud27Δ versus wild-type cells and the rapamycin-treated versus untreated wild-type cells. Cells were grown in YPD at 30°C. Rapamycin was added at the 400 ng/mL concentration. P-values are represented for each analyzed data set. (B) A correlation scatterplot showing the relation between the RNA-seq data of the bud27Δ versus wild-type cells and the rapamycin-treated versus untreated wild-type cells. The Spearman correlation coefficient is indicated. The color scale indicates the density of overlapping genes (from colder to warmer colors as density increases). (C) A similar correlation analysis as in B depicting the correlation coefficients of the RP, RiBi, and the other protein-coding genes, separately. Tendency lines are adjusted to the specific gene groups indicated by the color scheme.
Taken together, we conclude that Bud27 is required for optimal expression of RP genes and, to a lesser extent, of most RiBi genes.
Genome-wide analysis of Bud27 binding is seen to be associated with RP genes
In light of the results described so far, we were interested in understanding if Bud27 would act locally to modulate RP and also RiBi gene transcription. It has been reported that Bud27 can be detected in isolated chromatin fractions (Mirón-García et al. 2014). Thus, we analyzed the actual binding locations of Bud27 in the genome. To do so, we performed a ChIP-seq analysis using a TAP-tagged Bud27 strain grown at 30°C on YPD medium, and found that it was bound to the chromatin of many protein-coding genes (Fig. 5A). The metagene analysis showed that, on average, the genes belonging to the RP and RiBi regulons displayed greater Bud27 occupancy than the average protein-coding genes (Fig. 5B). Interestingly, Bud27 showed a tendency to accumulate toward the 3′ end of the RP gene bodies, whereas the RiBi gene bodies were more evenly occupied (Fig. 5B).
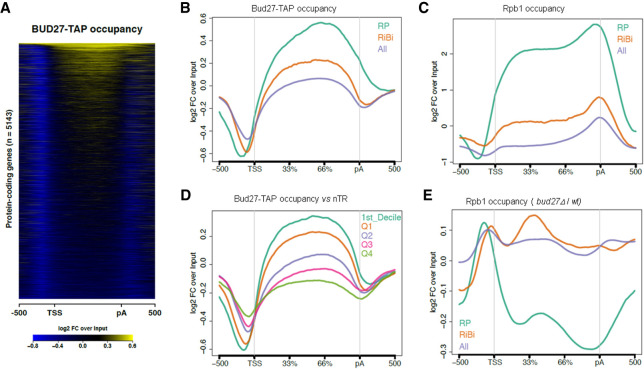
FIGURE 5.
ChIP-seq analysis of Bud27 occupancy. (A) Heatmap of the input-normalized Bud27-TAP ChIP-seq signals over a region that includes the gene body 500 bp upstream of the transcription start site (TSS), and 500 bp downstream from the cleavage and polyadenylation sites (pA). The heatmap includes all the protein-coding genes with TSS and pA annotations. (B) The input-normalized average metagene profile of the occupancy of Bud27-TAP over gene bodies and flanking regions of all protein-coding genes from Figure 5A and the subsets of the RP (n = 129) and RiBi (n = 236) genes. (C) The same as in panel B for Rpb1 ChIP-seq. (D) The average metagene profile of the occupancy of Bud27-TAP over the gene bodies and flanking regions of all the protein-coding genes, separated by the nascent transcription rate level (nTR). Q1–Q4 indicates the nTR quartiles. D1 refers to the first nTR decile. (E) The average Rpb1 ChIP-seq occupancy change profile (bud27Δ /wt ratio) over the three groups of the analyzed genes.
The relative Bud27 occupancy levels observed among the three groups of analyzed genes, in which RPs were more occupied than RiBi, and RiBi were, in turn, more occupied than the average protein-coding gene, coincided well with their average mRNA production by RNA polymerase II (nascent transcription rates, nTR) (Jordán-Pla et al. 2015) and, as expected, with the occupancy profiles of RNA Pol II as measured by ChIP-seq of its major subunit, Rpb1, in the wild-type strain (Fig. 5C). So we wondered if Bud27 could be associated with chromatin in a transcription rate-dependent manner and if, in fact, as shown in Figure 5D, a clear positive correlation existed between Bud27 occupancy and transcription rates, nTR, by analyzing the average metagene profile of Bud27-TAP occupancy over the gene bodies and flanking regions of all the protein-coding genes represented by quartiles and decile 1 of the nascent transcription rate level, nTR (Jordán-Pla et al. 2015).
We also wondered whether the altered mRNA levels of genes RP and RiBi observed upon lack of Bud27 were caused by defects in RNA Pol II occupancy. As shown, the metagene analysis of the ChIP-seq data sets for Rpb1 evidenced alterations to the occupancy profiles in both groups of genes, with more severe effects noted in the RP genes (Fig. 5E). These results fall in line with the global mRNA accumulation analysis shown in Figure 4.
The FunSpec analysis (Robinson et al. 2002) of the GO categories for the RNA Pol II Bud27-occupied genes in our genome-wide analysis identified cellular components that corresponded mainly to the ribosome and biological functions related mainly with rRNA maturation, rRNA processing, ribosome biogenesis and assembly, and translation (Supplemental Fig. S9).
Altogether, these data reveal that the RP and RiBi mRNA level changes caused by lack of Bud27 are likely due to an alteration to RNA Pol II distribution along the gene bodies that depend on this protein being physically present in the chromatin of these genes.
Absence of Bud27 slightly impairs translation and ribosome biogenesis
As lack of Bud27 negatively impacts the synthesis of most ribosomal components, that is, rRNA transcription mediated by RNA Pol I and III, the expression of RP genes mediated by RNA Pol II and the processing of 7S pre-rRNAs, we explored in more detail whether lack of Bud27 could globally impair the ribosome biogenesis process. To test this, polysome profile analyses were performed in a bud27 null strain and its otherwise isogenic wild-type counterpart grown in liquid YPD at 30°C and shifted to 37°C for up to 6 h. Under these conditions, the bud27Δ mutant cells grew more slowly. As shown in Figure 6, when shifted to 37°C, the polysome profile from the bud27Δ mutant showed both a marked increase in the 80S peak and less polysome content compared to that of the wild-type strain. This result agrees with the previously reported role for Bud27 in the initiation of translation (Deplazes et al. 2009). Interestingly, a slight deficit of the free 60S versus the 40S r-subunits was also observed, which was not accompanied by the appearance of half-mer polysomes (Fig. 6). This result suggests a slight deficit in the 60S r-subunits in bud27Δ cells. Accordingly, the quantification of the total r-subunits by low Mg2+ sucrose gradients resulted in only an approximate 10% reduction in the 60S in relation to the 40S r-subunits ratio in the bud27Δ strain compared to the wild-type control after 6 or 12 h at 37°C. Altogether, these results indicate that Bud27 make a relevant contribution to ribosome production and function. Hence the deletion of BUD27 leads to a slight deficit in the 60S r-subunits in relation to the 40S r-subunits, which is compatible to the above-mentioned delay observed during the production of the 25S, 5.8S, and 5S mature rRNAs, and with a mild defect on 7S pre-rRNA processing.
FIGURE 6.
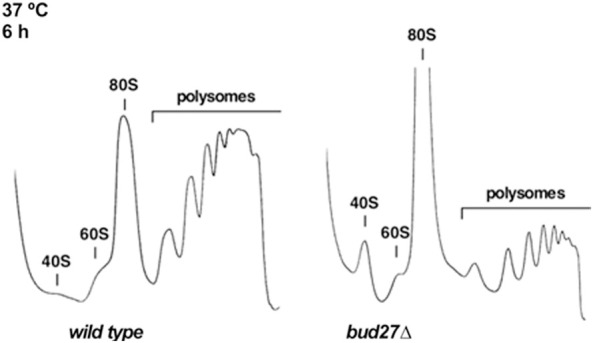
Absence of Bud27 impairs translation and 60S r-subunit metabolism. Polysomes profile analysis of the wild-type and bud27Δ cells grown to the mid-log phase in YPD medium at 30°C and then shifted to 37°C for 6 h. Cells were harvested at an OD600 of about 0.8. Whole-cell extracts were prepared and 10 A260 units of each extract were resolved in 7%–50% sucrose gradients. A254 was continuously measured. Sedimentation goes from left to right. The peaks of the free 40S and 60S r-subunits, 80S vacant ribosomes or monosomes and polysomes, are indicated.
Bud27 inactivation provokes a significant transcriptional response that partially mimics TOR pathway inactivation
Bud27 has been proposed as a TOR signaling cascade component by coordinating nutrient availability with gene expression driven by TORC1 kinase (Gstaiger et al. 2003). The TORC1 signaling pathway has also been shown to control ribosome biogenesis in yeast (Martin et al. 2006). To explore the molecular relation between Bud27 and the TOR pathway in more detail, the transcriptional responses achieved by inactivating Bud27 or the TOR cascade were compared. We first analyzed the RNA-seq data from the bud27Δ strain exponentially grown in YPD medium at 30°C versus its otherwise isogenic wild-type strain. In parallel, we compared the RNA-seq data for the wild-type yeast cells grown in YPD medium at 30°C in the absence and presence of rapamycin for 12 h to inactivate the TOR pathway (see Supplemental Fig. S1).
As a result, the expression of around 1000 genes changed by at least twofold in the bud27Δ strain compared to the wild-type control (406 up-regulated, 558 down-regulated). Notably, about 20%–30% of these genes (118 up-regulated, 127 down-regulated) were also altered in the rapamycin-treated cells (Fig. 7A). The comparison between the transcriptome profiles for the bud27Δ mutant versus its wild-type counterpart, and the rapamycin-treated cells versus the untreated control showed a mild positive correlation (r = 0.3) (Fig. 7B). When looking specifically at the RiBi and RP genes, the correlation increased to r = 0.33 and r = 0.42, respectively. This indicates that the effects of the experimental conditions were more specific for these two ribosome-related gene regulons than for the average protein coding gene (r = 0.27) (Fig. 7C).
We performed a functional analysis of the RNA-seq data and found that the commonly up-regulated or down-regulated genes under both experimental conditions were enriched in unique GO categories: biosynthetic processes-related categories among the up-regulated genes, and translation, ribosome biogenesis and RNA processing among the down-regulated genes. Similar results have been previously described for rapamycin-treated cells (Crespo and Hall 2002; Bandhakavi et al. 2008; Kumar et al. 2018).
All these data collectively suggest that lack of Bud27 leads to a significant transcriptional response that partially mimics the TOR inactivation transcriptional response, and indicate that Bud27 probably influences ribosome biogenesis via the TOR pathway. It is worth noting that while a major important alteration has been detected in our transcriptomic analysis of the bud27Δ null strain, one previous report has described differences only in 39 genes (Gstaiger et al. 2003). These significant disparities are probably due to differences in the technology used in each analysis and in their respective sensitivities.
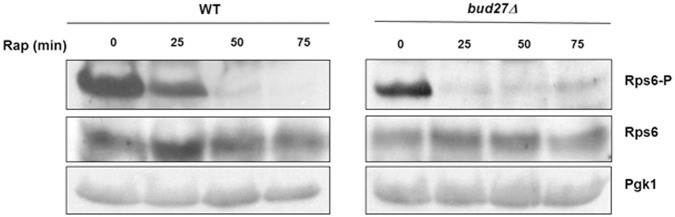
Bud27 influences S6 phosphorylation status and accumulation
In mammals, S6K kinase phosphorylates eS6, an RP of the 40S r-subunit, at several residues to promote the activation of the genes required for ribosome biogenesis (Chauvin et al. 2014). This phosphorylation is considered one of the most important targets of mTORC1 signaling and occurs in response to nutrients. Similarly, in S. cerevisiae, S6 (also named eS6 or Rps6) phosphorylation is also dependent on nutrients in a TORC1-dependent manner (González et al. 2015). To explore whether Bud27 functionally interacts with the TOR pathway and, therefore, provides a possible explanation for its impact on the ribosome biogenesis pathway, the phosphorylation/dephosphorylation status of yeast S6 protein was used as a readout of TOR activity upon Bud27 inactivation. A highly specific commercial antibody against human phospho-(Ser235/236) S6 was used, which cross-reacts with phosphorylated yeast S6. Wild-type and bud27Δ cells were firstly grown at 30°C in YPD medium before adding rapamycin to inactivate the TOR pathway. At different time points, cells were harvested, and whole cells extracts were performed, and later analyzed for the S6 phosphorylation/dephosphorylation status by western blot. As expected, S6 was strongly dephosphorylated upon rapamycin treatment in the wild-type cells (Fig. 8). This dephosphorylation was apparently maximal 50 min after adding rapamycin. Notably in bud27Δ cells, the amount of phosphorylated S6 was apparently less than in the wild-type cells, with a decrease of ∼25% noted in the untreated cells. Strikingly, the kinetics of the dephosphorylation of S6 also occurred apparently more quickly upon the addition of rapamycin, and was maximal at the 25-min time point, while residual phosphorylated S6 remained after this time point.
FIGURE 8.
The phosphorylation status of S6 is altered in the cells lacking Bud27. Western blot analysis of the phosphorylated (Rps6-P) and nonphosphorylated (Rps6) S6 RP in both the wild-type and bud27Δ mutant cells. Cells were grown in YPD medium at 30°C (time 0) or in YPD containing 400 ng/mL rapamycin for the indicated times. Anti-phospho (pSer235/236)-S6 RP and anti-S6 RP antibodies were used. Pgk1 was taken as the internal control.
Taken together, these results strongly suggest that Bud27 interplays with TORC1 activity kinase as lack of Bud27 accelerates S6 dephosphorylation and the inactivation of Bud27 and the TOR pathway leads to a partial transcriptional common response.
DISCUSSION
Ribosome biogenesis is a high-energy consumption process that requires stringent regulation to ensure proper ribosome production to deal with cell growth and protein synthesis in different environmental and metabolic situations. To support this statement, it is noteworthy that the synthesis of ribosomal components corresponds to ∼80% of total transcriptional activity in yeast, and to 50% in proliferating mammalian cells (Xiao and Grove 2009). One main regulation point of this process is performed in relation to the activity of the three RNA pols as these three complexes are required for the synthesis of the different ribosome components and the factors involved in all the steps during ribosome biogenesis, namely rRNA transcription, pre-rRNA processing, RP production and assembly, which are all temporally and spatially concomitant and interdependent processes (Loewith and Hall 2011; Jakob et al. 2012; de la Cruz et al. 2018; Vizoso-Vázquez et al. 2018). It has been reported that the TOR pathway is a master regulator of ribosomal components transcription, growth and ribosome biogenesis, and it performs its regulatory activity in different transcriptional and posttranscriptional steps of gene expression (Martin and Hall 2005). We herein evidence that, in yeast, the prefoldin-like Bud27 protein modulates the activities of all three RNA pols to properly influence the transcription of ribosomal components and ribosome biogenesis. The action of Bud27 differs and depends on each RNA pol. Thus, Bud27 modulates the synthesis of 35S rRNA by RNA Pol I, likely by influencing transcription elongation and pre-rRNA processing. Bud27 impacts 5S rRNA and tRNA transcription by modulating RNA Pol III occupancy to chromatin. Bud27 regulates the transcription of RP and RiBi genes by its association to these genes and by influencing both RNA Pol II transcription elongation in an transcription rate-dependent manner and mRNA splicing. Bud27 also seems to interplay with the TOR pathway to generally influence the ribosome biogenesis process.
One intriguing question remains: how Bud27 can control the three different transcriptional machinery activities. Our previous results suggested that this could be achieved by it coming into direct contact with a common subunit shared by the three RNA pols. In fact we have previously shown that Bud27 physically associates with Rpb5, which is indeed a universally shared subunit among the RNA pols of eukaryotes (Mita et al. 2011; Boulon et al. 2012; Mirón-García et al. 2013, 2014; Martínez-Fernández et al. 2015; Martínez-Fernández and Navarro 2018). Rpb5 is required for the transcription mediated by any of the three RNA pols (Zaros et al. 2007), although specific functions in RNA Pol II-mediated activity have also been reported (Miyao and Woychik 1998; Martínez-Fernández and Navarro 2018). The action of Rpb5 on the activity of the three RNA pols partly occurs through its physical and functional association with the RSC chromatin remodeler complex (Soutourina et al. 2006). Furthermore, the tripartite interaction of Rpb5 with RSC and Bud27 has been proven important for regulating transcription elongation by RNA Pol II (Mirón-García et al. 2014) and transcription by RNA Pol III (Vernekar and Bhargava 2015). Accordingly, this tripartite Bud27, Rpb5 and RSC interaction could also act to regulate RNA Pol I activity.
Regarding the functional interaction of Bud27 with RNA Pol I, our results indicated that lack of Bud27 resulted in low 35S pre-rRNA levels, as evidenced by the pulse-chase experiments. However, and strikingly, this defect was not accompanied by diminished RNA Pol I occupancy along the rDNA unit in relation to a wild-type strain, or the rDNA promoter, which also evidenced no defects in transcription initiation. This phenomenon seemed to differ from those that occur for RNA Pol II and RNA Pol III because, as shown by our results and previously reported ones, lack of Bud27 decreases RNA Pol II and RNA Pol III activities in concert with reduced RNA Pol II occupancy to chromatin, at least for some genes (Mirón-García et al. 2014; Vernekar and Bhargava 2015). Despite the fact that Bud27 mediates TOR-dependent transcription programs (Gstaiger et al. 2003; Martínez-Fernández et al. 2015, 2018) and that lack of Bud27 partially mimics TOR pathway inactivation (Gstaiger et al. 2003 and this work), the impact of BUD27 deletion in RNA Pol I transcription seemed specific and differed from that occurring under TOR inactivation by rapamycin addition or nutrient starvation. In these scenarios, RNA Pol I activity inhibition occurs as a consequence of diminished RNA Pol I occupancy to rDNA. This specific effect of Bud27 on RNA Pol I transcription does not seem atypical because, also in the post-log stationary phase, switching rRNA genes from an active to an inactive state has been also demonstrated to not be caused by the regulatory pathways involving TOR (Claypool et al. 2004). Notably, and despite the high RNA Pol I occupancy along the rDNA unit in the bud27Δ mutant, a drop between the 3′ and 5′ regions of the rDNA unit was observed, which suggests a defect in the relative processivity of the enzyme and in transcription elongation, as previously reported for other RNA Pol I mutants (Schneider et al. 2007). Concomitantly, the alteration of rRNA processing observed with BUD27 deletion (mild accumulation of 7S pre-rRNA, accompanied by a minor accumulation of 23S rRNA) could result from the transcriptional elongation defect. This would fall in line with the results from mutants of both RNA Pol I and elongation factor Spt4/5 showing that a defect in transcription elongation could perturb the processing of precursors for either 25S or 18S rRNAs. It would also evidence that the RNA Pol I transcription elongation rate and rRNA processing are functionally coupled (Schneider et al. 2006, 2007; Scull and Schneider 2019). In any case, our results do not discard a mild impact of exosome activity on 7S rRNA processing given that its Rrp6 subunit, which is needed for 7S rRNA processing (Guglielmi and Werner 2002; Cepeda et al. 2019), associates less efficiently with chromatin in the bud27Δ mutant (our unpublished data).
This mechanism by which Bud27 regulates RNA Pol I activity is suggested to be dependent on the tripartite interaction between Bud27-Rpb5 and the chromatin remodeler complex RSC, which falls in line with Bud27 influencing chromatin conformation, and then transcription by RNA Pol II and Pol III through its association with RSC (Mirón-García et al. 2014; Vernekar and Bhargava 2015). Furthermore, the Rpb5-RSC interaction is necessary for the activity of the three yeast RNA polymerases (Soutourina et al. 2006). Accordingly, Bud27 could also act by influencing the chromatin structure of the rDNA unit via its interaction with RSC and RNA Pol I. Alternatively, Bud27 could influence the activity or association of other chromatin remodelers with rDNA, such as the SWI/SNF remodeler complex, by thereby modulating the RNA Pol I transcription process. Indeed an snf6Δ mutant (SWI/SNF) shows similar RNA Pol I transcriptional alterations to those observed for the bud27Δ mutant; that is, a defect in transcription elongation with no major alteration of RNA Pol I occupancy and a modest defect in pre-rRNA processing, which are consistent with a linkage between rRNA transcription and rRNA processing (Zhang et al. 2013). In agreement with an impact of Bud27 on the rDNA chromatin structure, our data from MNase-seq experiments in the bud27Δ mutant in relation to its otherwise isogenic wild-type strain showed clear differences for this DNA region (A Cuevas-Bermúdez, V Martí-nez-Fernández, AI Garrido-Godino et al., in prep.). Finally, our RNA-seq analysis suggested that the reduction in RNA Pol I activity provoked by lack of Bud27 was not due to a low expression level of transcription factors Rrn3 and Hmo1, both of which control RNA Pol I activity (Reiter et al. 2011).
Although no defect on 5S rRNA transcription due to BUD27 deletion has been previously reported (Vernekar and Bhargava 2015), our attempt to explore the impact of Bud27 on RNA Pol III activity more precisely by northern blot and pulse-chase experiments revealed that Bud27 mildly influenced 5S rRNA synthesis and then global rRNA cellular content, and in a way dependent on RNA Pol III occupancy to the corresponding DNA units. Similarly, and corroborating previous data, Bud27 also impacted tRNA synthesis and type 3 genes transcription (i.e., SCR1), which agrees with the major role proposed for Bud27 in regulating RNA Pol III-mediated transcription by influencing RNA Pol III occupancy and RSC association with the enzyme (Vernekar and Bhargava 2015).
Regarding RNA Pol II, our data remarkably showed that Bud27 could specifically contribute to the expression of RiBi and RP genes. In fact our results indicated a drop in RP and RiBi mRNA expressions in the bud27Δ strain, which could per se account for the defect in ribosome biogenesis that we observed in this strain. In line with this, a concerted expression of RP and RiBi genes has been described to ensure adequate ribosome biogenesis (Loewith and Hall 2011; Jakob et al. 2012; de la Cruz et al. 2018). The diminishing accumulation of RP and RiBi mRNAs in the bud27Δ strain might be explained by general reduced RNA Pol II activity as lack of Bud27 can negatively impact RNA Pol II-dependent transcription elongation (Mirón-García et al. 2014). However, we favor a scenario in which Bud27 plays a specific role in the expression of these genes. In line with this, our ChIP-seq data indicated that Bud27 specifically occupied RP and RiBi genes. Thus, by deleting the BUD27 gene, it could directly impact RP and the RiBi genes expression by altering Bud27 association with chromatin. Our data also evidenced that the Bud27 association with chromatin occurred in a transcription rate-dependent manner (this work) and influenced transcription elongation (Mirón-García et al. 2014), which agrees with the fact that the transcription regulation of RP and RiBi genes, which correspond to those with high average transcription rates (Jordán-Pla et al. 2015), occurs at the elongation level, as previously suggested (Pelechano et al. 2009). In addition, RP gene expression has been described to be dependent on the TFIIS transcription elongation factor (Gómez-Herreros et al. 2012). It has also been recurrently demonstrated that most RP and RiBi elements participate in rRNA processing (Choesmel et al. 2007; Reiter et al. 2011; Iadevaia et al. 2012; de la Cruz et al. 2018), a process that can occur in yeast either before or after 35S pre-rRNA completes its synthesis (Osheim et al. 2004; Kos and Tollervey 2010). Therefore, we cannot rule out that the lowered expression of the genes for RP and RiBi factors observed upon Bud27 inactivation could account for the rRNA maturation defect detected in the bud27Δ strain, at least in part. Not only RP and RiBi expression, but also global mRNA splicing, are affected upon Bud27 inactivation, which is likely a side consequence of its RNA Pol II transcription elongation defect by taking into account the concerted connection between RNA Pol II transcription elongation and mRNA splicing (Dujardin et al. 2014; Saldi et al. 2016). As ribosome biogenesis implies ∼90% of all cellular mRNA splicing, and many RP genes contain introns (Warner 1999; Metzl-Raz et al. 2017), this global and non-RP-specific defect could also influence ribosome biogenesis. The drop in the expression of some transcription factors directly involved in the up-regulation of RP and RiBi gene expression (Martin et al. 2004; Loewith and Hall 2011; Albert et al. 2016; Vizoso-Vázquez et al. 2018), which also occurs upon the deletion of BUD27, could also account for the lower detected RP and RiBi gene expression.
By considering together the influence of BUD27 deletion on the activity of the three RNA pols, the alteration to the ribosomal component transcription could also be coordinately impacted by the three enzymes, which would agree with the data that describe in S. cerevisiae cross-talk between tRNA and rRNA synthesis (Briand et al. 2001), cross-talk between RNA Pol I and RNA Pol II for RP genes transcription (Michels and Hernandez 2006), or with the concerted transcriptional activity of RNA Pol I and 5S rRNA, and RP mRNA synthesis (Laferte et al. 2006).
Bud27 is required for the cytoplasmic assembly of the three RNA pols in yeast (Mirón-García et al. 2013; Vernekar and Bhargava 2015). However, this circumstance does not seem to account for the different transcriptional defects that we observed in the absence of Bud27. Indeed, overcoming the RNA pols assembly defect shown by the bud27Δ mutant by RPB5 overexpression at either 30°C (Mirón-García et al. 2013) or 37°C (see Supplemental Fig. S5) did neither modify the pre-rRNA and pre-tRNA processing defects led by the bud27Δ mutation (data not shown) nor overcome the sensitivity of bud27Δ mutant cells to mycophenolic acid, a nucleotide triphosphate (NTP)-depleting drug used to detect S. cerevisiae strains defective in transcription elongation (Shaw et al. 2001). All these results did not seem to be the indirect consequence of the slow growth rate of the bud27Δ mutant strain at 37°C, because many of them were evident even at the permissive temperature of 30°C.
It has been previously reported that yeast Bud27 and human URI are components of the TOR signaling pathway (Gstaiger et al. 2003). As our results showed that Bud27 could regulate the transcriptional activities of all the elements related to ribosome biogenesis, it would likely be involved in a sort of coordination with the TOR pathway for proper ribosome biogenesis. One main question is to discern how Bud27 can indeed act in the TOR cascade. Our transcriptomic analyses of a bud27Δ strain provided results that resembled, but were not identical to, those upon TOR pathway inactivation by rapamycin (Hardwick et al. 1999; Crespo and Hall 2002; Bandhakavi et al. 2008; Kumar et al. 2018), which suggests a possible functional connection between the functions of Bud27 and those of the TOR pathway. Some other pieces of evidence have been described and agree with the action of Bud27 in ribosome biogenesis via the TOR pathway: diminished rRNA processing involving reduced RPs synthesis has already been shown upon rapamycin-dependent TOR pathway inactivation (Powers and Walter 1999; Reiter et al. 2011); the RSC complex, which associates with Bud27 (Mirón-García et al. 2014; Vernekar and Bhargava 2015), has been reported to play a role in the transcription of the genes related to ribosome biogenesis (Angus-Hill et al. 2001; Yu et al. 2015; Vizoso-Vázquez et al. 2018); the transcription elongation regulation of RP and RiBi genes depends on PKA kinase, which participates in the TOR signaling cascade (Pelechano et al. 2009). Hence, by taking into account the effects of inactivating Bud27 on the transcription mediated by the three RNA pols and its parallelism with TOR pathway inactivation, it is possible to speculate that the transcriptional coregulation needed to ensure proper ribosome biogenesis can be influenced by Bud27 as one possible coordinator. Obviously, further experiments are required to demonstrate this. Yet consistently with this hypothesis, our results revealed that Bud27 influenced the phosphorylation status of RP S6, a read-out of TOR activity, which depends on TORC1 and is necessary for ribosome biogenesis in both yeast and mammals (Du et al. 2012; González et al. 2015; Kos-Braun et al. 2017).
Despite the role of Bud27 in modulating transcription by the three RNA pols, its influence on pre-rRNA processing and its impact on ribosome synthesis, it is necessary to further experimentally explain the precise role of Bud27 in ribosome biogenesis. In line with this, and by considering the role of Bud27 as a cochaperone and its association with ribosomes and other chaperones (Deplazes et al. 2009; Martínez-Fernández et al. 2015, 2018), we cannot exclude the fact that lack of Bud27 might also account for a defect in the assembly of the elements required for ribosome synthesis. It is tempting to speculate that Bud27 may influence not only ribosome assembly in the nucleus, but also the association between the small and large r-subunits to be engaged in translation in the cytoplasm, which agrees with previously reported data (Deplazes et al. 2009). This function could be mediated by the PFD-binding domain of Bud27 as this domain is necessary for Bud27 to interact with the prefoldin complex, and probably with the R2TP module, which occurs for the human URI protein (Boulon et al. 2012; Martínez-Fernández et al. 2018).
Finally, further experiments are strictly required to decipher the functional relation between Bud27 and the TOR pathway, and to more precisely understand the impact of Bud27 on chromatin structure and transcription. It is worth mentioning that our data envisage that the impact of Bud27 inactivation on transcription, and then on cellular physiology, must be stronger than initially thought. Hence, this suggests that Bud27 must be a key element in many other cellular processes and, accordingly, human URI has been reported to be crucial for cancer development (Theurillat et al. 2011; Tummala et al. 2014; Buren et al. 2016). We assume that the ribosome biogenesis defects caused by Bud27 inactivation may influence cell growth not only in yeast, but also in humans and other eukaryotes, if we consider the close relation between Bud27 and URI (Chaves-Pérez et al. 2018; Martínez-Fernández et al. 2018). This phenomenon, and the major gene expression alterations caused by Bud27/URI deficiency, could impact many cellular processes, including some that might lead to different human diseases, including cancer (Theurillat et al. 2011; Teng et al. 2013; Tummala et al. 2014; Buren et al. 2016).
MATERIALS AND METHODS
Yeast strains and growth conditions
The Saccharomyces cerevisiae strains used in this study are listed in Table 1.
TABLE 1.
S. cerevisiae strains
The common yeast media, growth conditions and genetic techniques were used as described elsewhere (García-López et al. 2011). Yeast strains were cultured in liquid YPD or YPGal media containing 2% glucose or 2% galactose, respectively, at 30°C, unless otherwise specified. Synthetic SD medium supplemented with the specific nutrients required for auxotrophic deficiencies was also used. Rapamycin and mycophenolic acid (MPA) were used at the indicated concentrations.
In liquid cultures, cell growth was constantly monitored to maintain them in exponential phase, especially after the temperature shift to 37°C.
Chromatin immunoprecipitation and sequencing analyses
The chromatin immunoprecipitation experiments were performed using Dynabeads Pan Mouse IgG (Invitrogen) for Rpb8-TAP, as previously described (Garrido-Godino et al. 2013). For real-time PCR, a 1:100 dilution was used for input DNA and a 1:4 dilution was used for the DNA of immunoprecipitated samples. Genes were analyzed by quantitative real-time PCR in triplicate with at least three independent biological replicates using SYBR premix EX Taq (Takara). Quantification was performed as indicated in the Figure legends. The oligonucleotides utilized for the different PCRs are listed in Supplemental Table S1.
The chromatin immunoprecipitation and sequencing (ChIP-seq) of Bud27-TAP and Rpb1 were performed in biological duplicates. For the ChIP-seq experiments, 200 mL of each yeast culture were collected at an OD600 of about 0.6 and crosslinked by adding 1% formaldehyde. After a 15-min incubation, 10 mL of 2.5 M glycine were added and cultures were incubated for 5 min in the presence of this solution. Cells were harvested and washed four times with 150 mM NaCl, 20 mM Tris-HCl, pH 7.5 (by centrifugation). Then, they were resuspended in 300 µL of lysis buffer (50 mM HEPES, 140 mM NaCl, 1 mM EDTA, pH 8.0, 1% [v/v] Triton-X-100, 0.1% [w/v] sodium deoxycholate, 1 mM PMSF, 0.15% [w/v] benzamidine and protease inhibitor cocktail [Complete, Roche]). Cell disruption was carried out by vigorously vortexing these samples in the presence of 200 µL of glass beads for 15 min. After disruption, lysis buffer was added to cell extracts to adjust to 1.5 mL. Then samples were subsequently sonicated by a Bioruptor sonicator (Diagenode) for 45 min at 30-sec on/off intervals.
Immunoprecipitation was performed in duplicate using 200 µL of magnetic beads Dynabeads Pan Mouse IgG (Invitrogen) for Bud27-TAP, or 200 µL of magnetic beads Dynabeads M-280 sheep anti-mouse IgG (Invitrogen) mixed with the 8WG16 anti-Rpb1 antibody (Santa Cruz) for Rpb1, and 300 µL of the sonicated samples. Washing and decrosslinking followed the conventional chromatin immunoprecipitation procedure; supernatants were collected in the same tube and DNA was purified with the Nucleospin Extract II Purification kit (Macherey-Nagel). The eluted DNA was concentrated by lyophilization. Immunoprecipitates and their corresponding input DNA samples were sequenced in an Illumina HiSeq 2000 instrument. The quality metrics of the Fastq sequencing data sets were obtained with FastQC and were visually inspected. Based on the first quality check, sequencing adapters were removed from raw reads with Fastx Clipper. A second inspection round with FastQC was carried out after the filtering steps to verify that filters were properly applied. The resulting high-quality sequences were mapped to the S. cerevisiae S288C genome (SGD R64) with Hisat2 using default parameters. The resulting BAM alignment files were sorted, indexed and visually inspected with the genome browser IGV (www.broadinstitute.org/igv/). Based on the good similarity of the average occupancy profiles from the two biological replicates (Supplemental Fig. S1), the two data sets from each sample were merged into one single data set with SamTools (Li et al. 2009).
Protein immunoprecipitation and TAP purification
For TAP purification, 400 mL of the appropriate cells growing exponentially (OD600 of about 0.6–0.8) in YPD medium were washed twice with ultrapure water and lysis buffer (50 mM HEPES pH 7.5, 120 mM NaCl, 1 mM EDTA, 0.3% CHAPS). Then, cells were resuspended in 1 mL of lysis buffer supplemented with 1× protease inhibitor cocktail (Complete, Roche), 0.5 mM PMSF, 2 mM sodium orthovanadate and 1 mM sodium fluoride. Whole-cell extracts were prepared using a MixerMill MM400 RETSCH (3 min 30Hz). Purifications were carried out as described elsewhere (Garrido-Godino et al. 2013) with some modifications: 150 µL of whole-cell extract (2 mg protein) per experiment and lysis buffer for all washes were used and 35 µL of Dynabeads Pan Mouse IgG (Invitrogen). The affinity-purified proteins were released from beads by boiling for 10 min and were analyzed by western blot with antibodies anti-Rpb1 (manufactured in our laboratory), anti-Rpa34.5 (a gift from P. Thuriaux), PAP (Sigma), and anti-phosphoglycerate kinase (459250, Invitrogen).
Chromatin-enriched fractions
The chromatin-enriched fractions were prepared by following the yChEFs procedure (Cuevas-Bermúdez et al. 2019) from 150 mL of yeast cells grown in YPD at 30°C until the mid-log phase. The western blots for the chromatin-associated proteins were performed with antibodies anti-Rpa34.5 (a gift from P. Thuriaux), 9E10 anti-c-Myc (Santa Cruz Biotechnology), anti-phosphoglycerate kinase (459250, Invitrogen), and anti-H3 (ab1791; Abcam).
RNA extraction for the global expression analyses and sequencing
For the global expression analyses, 50 mL cultures were grown in YPD until an OD600 of about 0.8. Cells were collected and used to extract total RNA. The rapamycin-treated cells corresponded to cells grown in YPD at 30°C that contained 50 ng/mL of rapamycin until an OD600 of about 0.8. RNA was prepared and quantified as previously described (Nonet et al. 1987).
cDNA libraries were performed and sequenced with the QuantSeq 3′ mRNA-seq Library Prep Kit from Illumina following the manufacturer's instructions; as above, samples were sequenced in an Illumina HiSeq 2000 instrument.
The data analysis run to reach the BAM alignment files was similar to those used for the ChIP-seq experiments. Briefly after obtaining the Fastq sequencing data sets with FastQC, adapters were removed with Fastx Clipper, and sequences were mapped to the S. cerevisiae S288C genome (SGD R64) with Hisat2. Subsequently, differential RNA abundance was obtained with the R/Bioconductor software based on an advanced version of the DESeq package, DESeq2 (Anders and Huber 2010). The gene ontology categories were analyzed by the FunSpec software (Robinson et al. 2002).
Reverse transcription and qRT-PCR
For the RNA analysis by real-time PCR, first-strand cDNA was synthesized by using 500 ng of RNA and the iScript cDNA synthesis kit (Bio-Rad) following the manufacturer's instructions. For the DNA contamination control, the same RNA samples were analyzed, but without performing the retrotranscription reaction.
Real-time PCR was performed in a CFX-384 Real-Time PCR instrument (BioRad) with the EvaGreen detection system “SsoFast EvaGreen Supermix” (Bio-Rad). Reactions were performed in a total 5 µL volume with 1:100 dilution of the synthesized cDNA. Each sample was analyzed in triplicate with at least three independent biological replicates per sample. Values were normalized to the steady-state 18S rRNA levels. The used oligonucleotides are listed in Supplemental Table S1.
Extraction of rRNA and northern blots
The RNA extraction and northern hybridization analyses of the pre- and mature rRNAs were carried out following standard procedures (García-Gómez et al. 2011). In all experiments, RNA was extracted from the samples corresponding to 10 OD600 units of the exponentially grown cells. Equal amounts of total RNA (5 µg) were loaded on either 1.2% agarose-6% formaldehyde or 7% polyacrylamide-8M urea gels. Specific oligonucleotides were 5′-end labeled with [γ-32P] ATP and used as probes.
The sequences of the oligonucleotides used for northern hybridization purposes are listed in Supplemental Table S2. The Phosphorimager analysis was performed in a FLA-5100 imaging system (Fujifilm) at the Biology Service (CITIUS) of the University of Seville (Spain).
Protein extraction and western blot analyses
Whole-cell protein extracts were prepared from 5 mL of the exponentially growing cells (OD600 of about 0.8) in appropriate media. Cells were harvested, washed twice with ultrapure water, and resuspended in 200 µL of 20% trichloroacetic acid (TCA) and 200 µL of glass beads (425–600 µm, SIGMA) before being subjected to vortexing for 1 min. The supernatant of each extract was transferred to a new tube, 400 µL of 5% TCA were added, and the mix was vortexed for 10 sec and centrifuged for 5 min at 14,000 rpm. The corresponding acid pellets were neutralized by adding 10 µL of 1.5 M Tris-HCl, pH 8.8, and 20 µL of loading buffer (50 mM Tris-HCl, pH 8.8, 2% [w/v] sodium dodecyl sulfate [SDS], 4% [w/v] bromophenol blue, 2% [v/v] 2-mercaptoethanol, 10% [v/v] glycerol). Finally, extracts were boiled for 10 min at 100°C and 3.5 µL were used for SDS-PAGE and western blot analyses with different primary antibodies: anti-phospho (pSer235/236)-S6 RP (#2211, Cell Signaling Technology); anti-S6 RP (ab40820, Abcam); anti-phosphoglycerate kinase (459250, Invitrogen).
Sucrose gradient centrifugation
Polyribosome and ribosomal subunit (r-subunit) preparations and analyses were done as previously described (Foiani et al. 1991; Kressler et al. 1997). Ten A260 units of cell extracts were loaded onto sucrose gradients, which were ultracentrifuged. Profiles were obtained by continuously monitoring A254 with a Teledyne-ISCO UA-6 system.
Ribosomal subunit analyses were also performed as previously described (Kressler et al. 1997). Briefly, cell extracts were prepared under polysome run-off conditions, by omitting cycloheximide, in a buffer lacking MgCl2 to dissociate ribosomes into the 40S and 60S r-subunits. Ten A260 units of each extract were resolved in 7%–50% sucrose gradients prepared in the same buffer. Profiles were obtained as above; the area under the curve of each peak was determined and the 60S/40S ratios were calculated. To do so, profiles were printed, and peak areas were cut out and weighed. Data are the average of three independent experiments. The relative standard deviation was <5% in each case.
Pulse-chase analysis
The pulse-chase labeling of pre-rRNAs was performed exactly as previously described (Kressler et al. 1998) using 100 µCi of [5,6-3H]uracil (Perkin-Elmer; 45–50 Ci/mmol) per 40 OD600 of yeast cells. Cells were grown in SD-Ura medium at 30°C to the mid-log phase before being shifted to 37°C for 12 h. Then, cells were pulse-labeled for 2 min and chased for different times with the SD medium, which contained excess cold uracil. Total RNA was extracted by the hot phenol procedure (Ausubel et al. 1994). Radioactive incorporation was measured by scintillation counting (General Radioisotope Service-CITIUS of the University of Seville) and about 20,000 c.p.m. per RNA sample were loaded and resolved on 1.2% agarose-6% formaldehyde and 7% polyacrylamide-8M urea gels (Venema et al. 1995). RNA was transferred to nylon membranes and visualized by fluorography (Kressler et al. 1998).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. J.E. Pérez-Ortín for his helpful discussion and the “Servicios Centrales de Apoyo a la Investigación (SCAI)” of the University of Jaen for technical support. We also thank M. Carballo of the Biology Service (CITIUS) from the University of Seville for helping with the phosphorimager analysis. This work has been supported by grants from the Spanish Ministry of Economy and Competitiveness (MINECO) and ERDF (BFU2016-77728-C3-2-P to F.N. and BFU2016-75352-P to J.d.l.C.), Spanish Ministry of Science and Innovation (MICINN) and ERDF (RED2018-102467-T to F.N. and J.d.l.C.), the Junta de Andalucía (BIO258 to F.N. and BIO271 to J.d.l.C.), and FEDER-UJA (1260360 to F.N.). A.C.-B. is a recipient of the FPI predoctoral and a postdoctoral contract from MINECO. A.I.G.-G was financed by the University of Jaen, MINECO and ERDF funds (BFU2016-77728-C3-2-P to F.N.). F.G.-S is a recipient of a fellowship from the University of Jaen. A.J.-P. is funded by BFU2016-77728-C3-3-P to J.E. Pérez-Ortín.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.075507.120.
REFERENCES
- Albert B, Knight B, Merwin J, Martin V, Ottoz D, Gloor Y, Bruzzone MJ, Rudner A, Shore D. 2016. A molecular titration system coordinates ribosomal protein gene transcription with ribosomal RNA synthesis. Mol Cell 64: 720–733. 10.1016/j.molcel.2016.10.003 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SJ, Sikes ML, Zhang Y, French SL, Salgia S, Beyer AL, Nomura M, Schneider DA. 2011. The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J Biol Chem 286: 18816–18824. 10.1074/jbc.M110.202101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell 7: 741–751. 10.1016/S1097-2765(01)00219-2 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1994. Current protocols in molecular biology, pp. 13.10.11–13.14.17. Wiley, New York. [Google Scholar]
- Bandhakavi S, Xie H, O'Callaghan B, Sakurai H, Kim DH, Griffin TJ. 2008. Hsf1 activation inhibits rapamycin resistance and TOR signaling in yeast revealed by combined proteomic and genetic analysis. PLoS ONE 3: e1598 10.1371/journal.pone.0001598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum S, Bittins M, Frey S, Seedorf M. 2004. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem J 380: 823–830. 10.1042/bj20031962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, et al. 2010. HSP90 and its R2TP/prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell 39: 912–924. 10.1016/j.molcel.2010.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Bertrand E, Pradet-Balade B. 2012. HSP90 and the R2TP co-chaperone complex: building multi-protein machineries essential for cell growth and gene expression. RNA Biol 9: 148–154. 10.4161/rna.18494 [DOI] [PubMed] [Google Scholar]
- Briand JF, Navarro F, Gadal O, Thuriaux P. 2001. Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol 21: 189–195. 10.1128/MCB.21.1.189-195.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buren S, Gomes AL, Teijeiro A, Fawal MA, Yilmaz M, Tummala KS, Perez M, Rodriguez-Justo M, Campos-Olivas R, Megías D, et al. 2016. Regulation of OGT by URI in response to glucose confers c-MYC-dependent survival mechanisms. Cancer Cell 30: 290–307. 10.1016/j.ccell.2016.06.023 [DOI] [PubMed] [Google Scholar]
- Cepeda LPP, Bagatelli FFM, Santos RM, Santos MDM, Nogueira FCS, Oliveira CC. 2019. The ribosome assembly factor Nop53 controls association of the RNA exosome with pre-60S particles in yeast. J Biol Chem 294: 19365–19380. 10.1074/jbc.RA119.010193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, et al. 2014. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33: 474–483. 10.1038/onc.2012.606 [DOI] [PubMed] [Google Scholar]
- Chaves-Pérez A, Thompson S, Djouder N. 2018. Roles and functions of the unconventional prefoldin URI. Adv Exp Med Biol 1106: 95–108. 10.1007/978-3-030-00737-9_7 [DOI] [PubMed] [Google Scholar]
- Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. 2007. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood 109: 1275–1283. 10.1182/blood-2006-07-038372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool JA, French SL, Johzuka K, Eliason K, Vu L, Dodd JA, Beyer AL, Nomura M. 2004. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell 15: 946–956. 10.1091/mbc.e03-08-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P, Coulombe B. 2010. New insights into the biogenesis of nuclear RNA polymerases? Biochem Cell Biol 88: 211–221. 10.1139/O09-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo JL, Hall MN. 2002. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol Mol Biol Rev 66: 579–591. 10.1128/MMBR.66.4.579-591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Bermúdez A, Martínez-Fernández V, Garrido-Godino AI, Navarro F. 2017. Subunits common to RNA polymerases. In The yeast role in medical applications, Vol. 1 (ed. Abdulkhair WMH), pp. 151–165. IntechOpen, Rijeka, Croatia. [Google Scholar]
- Cuevas-Bermúdez A, Garrido-Godino A, Navarro F. 2019. A novel yeast chromatin-enriched fractions purification approach, yChEFs, for the chromatin-associated protein analysis used for chromatin-associated and RNA-dependent chromatin-associated proteome studies from Saccharomyces cerevisiae. Gene Rep 16: 100450 10.1016/j.genrep.2019.100450 [DOI] [Google Scholar]
- de la Cruz J, Gómez-Herreros F, Rodríguez-Galán O, Begley V, Muñoz-Centeno M, Chávez S. 2018. Feedback regulation of ribosome assembly. Curr Genet 64: 393–404. 10.1007/s00294-017-0764-x [DOI] [PubMed] [Google Scholar]
- Deplazes A, Mockli N, Luke B, Auerbach D, Peter M. 2009. Yeast Uri1p promotes translation initiation and may provide a link to cotranslational quality control. EMBO J 28: 1429–1441. 10.1038/emboj.2009.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Halova L, Kirkham S, Atkin J, Petersen J. 2012. TORC2 and the AGC kinase Gad8 regulate phosphorylation of the ribosomal protein S6 in fission yeast. Biol Open 1: 884–888. 10.1242/bio.20122022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, de la Mata M, Marasco LE, Muñoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR. 2014. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell 54: 683–690. 10.1016/j.molcel.2014.03.044 [DOI] [PubMed] [Google Scholar]
- Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. 1991. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol 11: 3203–3216. 10.1128/MCB.11.6.3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget D, Lacombe AA, Cloutier P, Al-Khoury R, Bouchard A, Lavallee-Adam M, Faubert D, Jeronimo C, Blanchette M, Coulombe B. 2010. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol Cell Proteomics 9: 2827–2839. 10.1074/mcp.M110.003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gómez JJ, Babiano R, Lebaron S, Froment C, Monsarrat B, Henry Y, de la Cruz J. 2011. Nop6, a component of 90S pre-ribosomal particles, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. RNA Biol 8: 112–124. 10.4161/rna.8.1.14143 [DOI] [PubMed] [Google Scholar]
- García-López MC, Pelechano V, Mirón-García MC, Garrido-Godino AI, García A, Calvo O, Werner M, Pérez-Ortín JE, Navarro F. 2011. The conserved foot domain of RNA Pol II associates with proteins involved in transcriptional initiation and/or early elongation. Genetics 189: 1235–1248. 10.1534/genetics.111.133215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Godino AI, García-López MC, Navarro F. 2013. Correct assembly of RNA polymerase II depends on the foot domain and Is required for multiple steps of transcription in Saccharomyces cerevisiae. Mol Cell Biol 33: 3611–3626. 10.1128/MCB.00262-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Herreros F, de Miguel-Jiménez L, Morillo-Huesca M, Delgado-Ramos L, Muñoz-Centeno MC, Chávez S. 2012. TFIIS is required for the balanced expression of the genes encoding ribosomal components under transcriptional stress. Nucleic Acids Res 40: 6508–6519. 10.1093/nar/gks340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A, Hall MN. 2017. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J 36: 397–408. 10.15252/embj.201696010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A, Shimobayashi M, Eisenberg T, Merle DA, Pendl T, Hall MN, Moustafa T. 2015. TORC1 promotes phosphorylation of ribosomal protein S6 via the AGC kinase Ypk3 in Saccharomyces cerevisiae. PLoS ONE 10: e0120250 10.1371/journal.pone.0120250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W. 2003. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302: 1208–1212. 10.1126/science.1088401 [DOI] [PubMed] [Google Scholar]
- Guglielmi B, Werner M. 2002. The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J Biol Chem 277: 35712–35719. 10.1074/jbc.M205526200 [DOI] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. 2011. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12: 483–492. 10.1038/nrm3152 [DOI] [PubMed] [Google Scholar]
- Haag JR, Brower-Toland B, Krieger EK, Sidorenko L, Nicora CD, Norbeck AD, Irsigler A, LaRue H, Brzeski J, McGinnis K, et al. 2014. Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Rep 9: 378–390. 10.1016/j.celrep.2014.08.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci 96: 14866–14870. 10.1073/pnas.96.26.14866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V, Zhang Z, Jan E, Proud CG. 2012. mTOR signaling regulates the processing of pre-rRNA in human cells. Nucleic Acids Res 40: 2527–2539. 10.1093/nar/gkr1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V, Liu R, Proud CG. 2014. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin Cell Dev Biol 36: 113–120. 10.1016/j.semcdb.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Jakob S, Ohmayer U, Neueder A, Hierlmeier T, Pérez-Fernández J, Hochmuth E, Deutzmann R, Griesenbeck J, Tschochner H, Milkereit P. 2012. Interrelationships between yeast ribosomal protein assembly events and transient ribosome biogenesis factors interactions in early pre-ribosomes. PLoS One 7: e32552 10.1371/journal.pone.0032552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordán-Pla A, Gupta I, de Miguel-Jiménez L, Steinmetz LM, Chávez S, Pelechano V, Pérez-Ortín JE. 2015. Chromatin-dependent regulation of RNA polymerases II and III activity throughout the transcription cycle. Nucleic Acids Res 43: 787–802. 10.1093/nar/gku1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M, Tollervey D. 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820. 10.1016/j.molcel.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos-Braun IC, Jung I, Kos M. 2017. Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. PLoS Biol 15: e2000245 10.1371/journal.pbio.2000245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, de la Cruz J, Rojo M, Linder P. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 17: 7283–7294. 10.1128/MCB.17.12.7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, de la Cruz J, Rojo M, Linder P. 1998. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol Cell Biol 18: 1855–1865. 10.1128/MCB.18.4.1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Awasthi A, Nain V, Issac B, Puria R. 2018. Novel insights into TOR signalling in Saccharomyces cerevisiae through Torin2. Gene 669: 15–27. 10.1016/j.gene.2018.05.081 [DOI] [PubMed] [Google Scholar]
- Kwapisz M, Beckouet F, Thuriaux P. 2008. Early evolution of eukaryotic DNA-dependent RNA polymerases. Trends Genet 24: 211–215. 10.1016/j.tig.2008.02.002 [DOI] [PubMed] [Google Scholar]
- Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev 20: 2030–2040. 10.1101/gad.386106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Tollervey D. 2000. Like attracts like: getting RNA processing together in the nucleus. Science 288: 1385–1389. 10.1126/science.288.5470.1385 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201. 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Hall MN. 2005. The expanding TOR signaling network. Curr Opin Cell Biol 17: 158–166. 10.1016/j.ceb.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979. 10.1016/j.cell.2004.11.047 [DOI] [PubMed] [Google Scholar]
- Martin DE, Powers T, Hall MN. 2006. Regulation of ribosome biogenesis: where is TOR? Cell Metab 4: 259–260. 10.1016/j.cmet.2006.09.002 [DOI] [PubMed] [Google Scholar]
- Martínez-Fernández V, Navarro F. 2018. Rpb5, a subunit shared by eukaryotic RNA polymerases, cooperates with prefoldin-like Bud27/URI. AIMS Genet 5: 63–74. 10.3934/genet.2018.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Fernández V, Garrido-Godino AI, Cuevas-Bermúdez A, Navarro F. 2015. Cytoplasmic and nuclear functions for the prefoldin-like URI/Bud27. In New research on molecular chaperones (ed. Chaperones NROM), pp. 57–73. Nova Science Publishers, Inc., NY. [Google Scholar]
- Martínez-Fernández V, Garrido-Godino AI, Cuevas-Bermúdez A, Navarro F. 2018. The yeast prefoldin Bud27. Adv Exp Med Biol 1106: 109–118. 10.1007/978-3-030-00737-9_8 [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391. 10.1038/sj.onc.1209883 [DOI] [PubMed] [Google Scholar]
- Metzl-Raz E, Kafri M, Yaakov G, Soifer I, Gurvich Y, Barkai N. 2017. Principles of cellular resource allocation revealed by condition-dependent proteome profiling. Elife 6: e28034 10.7554/eLife.28034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels AA, Hernandez N. 2006. Does Pol I talk to Pol II? Coordination of RNA polymerases in ribosome biogenesis. Genes Dev 20: 1982–1985. 10.1101/gad.1460706 [DOI] [PubMed] [Google Scholar]
- Mirón-García MC, Garrido-Godino AI, García-Molinero V, Hernández-Torres F, Rodríguez-Navarro S, Navarro F. 2013. The prefoldin Bud27 mediates the assembly of the eukaryotic RNA polymerases in an Rpb5-dependent manner. PLoS Genet 9: e1003297 10.1371/journal.pgen.1003297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirón-García MC, Garrido-Godino AI, Martínez-Fernández V, Fernández-Pevida A, Cuevas-Bermúdez A, Martín-Expósito M, Chávez S, de la Cruz J, Navarro F. 2014. The yeast prefoldin-like URI-orthologue Bud27 associates with the RSC nucleosome remodeler and modulates transcription. Nucleic Acids Res 42: 9666–9676. 10.1093/nar/gku685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P, Savas JN, Djouder N, Yates JR, Ha S, Ruoff R, Schafler ED, Nwachukwu JC, Tanese N, Cowan NJ, et al. 2011. Regulation of androgen receptor mediated transcription by Rpb5 binding protein URI/RMP. Mol Cell Biol 31: 3639–3652. 10.1128/MCB.05429-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P, Savas JN, Ha S, Djouder N, Yates JR III, Logan SK. 2013. Analysis of URI nuclear interaction with RPB5 and components of the R2TP/Prefoldin-Like complex. PLoS One 8: e63879 10.1371/journal.pone.0063879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao T, Woychik NA. 1998. RNA polymerase subunit RPB5 plays a role in transcriptional activation. Proc Natl Acad Sci 95: 15281–15286. 10.1073/pnas.95.26.15281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Galván S, Jimeno S, Rothstein R, Aguilera A. 2013. Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet 9: e1003237 10.1371/journal.pgen.1003237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R. 1987. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol 7: 1602–1611. 10.1128/MCB.7.5.1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16: 943–954. 10.1016/j.molcel.2004.11.031 [DOI] [PubMed] [Google Scholar]
- Pelechano V, Jimeno-González S, Rodríguez-Gil A, García-Martínez J, Pérez-Ortín JE, Chávez S. 2009. Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet 5: e1000614 10.1371/journal.pgen.1000614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petibon C, Parenteau J, Catala M, Elela SA. 2016. Introns regulate the production of ribosomal proteins by modulating splicing of duplicated ribosomal protein genes. Nucleic Acids Res 44: 3878–3891. 10.1093/nar/gkw140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Walter P. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell 10: 987–1000. 10.1091/mbc.10.4.987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter A, Steinbauer R, Philippi A, Gerber J, Tschochner H, Milkereit P, Griesenbeck J. 2011. Reduction in ribosomal protein synthesis is sufficient to explain major effects on ribosome production after short-term TOR inactivation in Saccharomyces cerevisiae. Mol Cell Biol 31: 803–817. 10.1128/MCB.01227-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Grigull J, Mohammad N, Hughes TR. 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35 10.1186/1471-2105-3-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Zhao Y, Warner JR. 2005. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J 24: 533–542. 10.1038/sj.emboj.7600553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T, Cortazar MA, Sheridan RM, Bentley DL. 2016. Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J Mol Biol 428: 2623–2635. 10.1016/j.jmb.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. 2006. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci 103: 12707–12712. 10.1073/pnas.0605686103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M. 2007. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell 26: 217–229. 10.1016/j.molcel.2007.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scull CE, Schneider DA. 2019. Coordinated control of rRNA processing by RNA polymerase I. Trends Genet 35: 724–733. 10.1016/j.tig.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Wilson JL, Smith KT, Reines D. 2001. Regulation of an IMP dehydrogenase gene and its overexpression in drug-sensitive transcription elongation mutants of yeast. J Biol Chem 276: 32905–32916. 10.1074/jbc.M105075200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpakovski GV, Acker J, Wintzerith M, Lacroix JF, Thuriaux P, Vigneron M. 1995. Four subunits that are shared by the three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol Cell Biol 15: 4702–4710. 10.1128/MCB.15.9.4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Bhargava P. 2018. Regulation of tRNA gene transcription by the chromatin structure and nucleosome dynamics. Biochim Biophys Acta 1861: 295–309. 10.1016/j.bbagrm.2017.11.008 [DOI] [PubMed] [Google Scholar]
- Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, et al. 2006. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol 26: 4920–4933. 10.1128/MCB.00415-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T, Thomas G, Mercer CA. 2013. Growth control and ribosomopathies. Curr Opin Genet Dev 23: 63–71. 10.1016/j.gde.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Theurillat JP, Metzler SC, Henzi N, Djouder N, Helbling M, Zimmermann AK, Jacob F, Soltermann A, Caduff R, Heinzelmann-Schwarz V, et al. 2011. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer Cell 19: 317–332. 10.1016/j.ccr.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Torreira E, Louro JA, Pazos I, González-Polo N, Gil-Carton D, Duran AG, Tosi S, Gallego O, Calvo O, Fernández-Tornero C. 2017. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. Elife 6: e20832 10.7554/eLife.20832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala KS, Gomes AL, Yilmaz M, Grana O, Bakiri L, Ruppen I, Ximénez-Embun P, Sheshappanavar V, Rodriguez-Justo M, Pisano DG, et al. 2014. Inhibition of de novo NAD synthesis by oncogenic URI causes liver tumorigenesis through DNA damage. Cancer Cell 26: 826–839. 10.1016/j.ccell.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Venema J, Dirks-Mulder A, Faber AW, Raué HA. 1995. Development and application of an in vivo system to study yeast ribosomal RNA biogenesis and function. Yeast 11: 145–156. 10.1002/yea.320110206 [DOI] [PubMed] [Google Scholar]
- Vernekar DV, Bhargava P. 2015. Yeast Bud27 modulates the biogenesis of Rpc128 and Rpc160 subunits and the assembly of RNA polymerase III. Biochim Biophys Acta 1849: 1340–1353. 10.1016/j.bbagrm.2015.09.010 [DOI] [PubMed] [Google Scholar]
- Viktorovskaya OV, Appling FD, Schneider DA. 2011. Yeast transcription elongation factor SPT5 associates with RNA polymerase I and RNA polymerase II directly. J Biol Chem 286: 18825–18833. 10.1074/jbc.M110.202119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizoso-Vázquez A, Lamas-Maceiras M, González-Siso MI, Cerdán ME. 2018. Ixr1 regulates ribosomal gene transcription and yeast response to cisplatin. Sci Rep 8: 3090 10.1038/s41598-018-21439-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. 1999. The economics of ribosome byosynthesis in yeast. Trends Biochem Sci 24: 437–440. 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- Xiao L, Grove A. 2009. Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr Genomics 10: 198–205. 10.2174/138920209788185261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Imamura Y, Ueno M, Suzuki SW, Ohsumi Y, Yukawa M, Tsuchiya E. 2015. The yeast chromatin remodeler Rsc1-RSC complex is required for transcriptional activation of autophagy-related genes and inhibition of the TORC1 pathway in response to nitrogen starvation. Biochem Biophys Res Commun 464: 1248–1253. 10.1016/j.bbrc.2015.07.114 [DOI] [PubMed] [Google Scholar]
- Zaros C, Briand JF, Boulard Y, Labarre-Mariotte S, García-López MC, Thuriaux P, Navarro F. 2007. Functional organization of the Rpb5 subunit shared by the three yeast RNA polymerases. Nucleic Acids Res 35: 634–647. 10.1093/nar/gkl686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Anderson SJ, French SL, Sikes ML, Viktorovskaya OV, Huband J, Holcomb K, Hartman JL IV, Beyer AL, Schneider DA. 2013. The SWI/SNF chromatin remodeling complex influences transcription by RNA polymerase I in Saccharomyces cerevisiae. PLoS One 8: e56793 10.1371/journal.pone.0056793 [DOI] [PMC free article] [PubMed] [Google Scholar]