Abstract
Precision medicine is an emerging paradigm that aims at maximizing the benefits and minimizing the adverse effects of drugs. Realistic mechanistic models are needed to understand and limit heterogeneity in drug responses. While pharmacokinetic models describe in detail a drug's absorption and metabolism, they generally do not account for individual variations in response to environmental influences, in addition to genetic variation. For instance, the human gut microbiota metabolizes drugs and is modulated by diet, and it exhibits significant variation among individuals. However, the influence of the gut microbiota on drug failure or drug side effects is under-researched. Here, we review recent advances in computational modeling approaches that could contribute to a better, mechanism-based understanding of drug–microbiota–diet interactions and their contribution to individual drug responses. By integrating systems biology and quantitative systems pharmacology with microbiology and nutrition, the conceptually and technologically demand for novel approaches could be met to enable the study of individual variability, thereby providing breakthrough support for progress in precision medicine.
Keywords: Precision medicine, Pharmacokinetic modeling, Constraint-based modeling, Drug metabolism, Gut microbiota
Highlights
-
•
The response to drug treatment is highly variable among individuals.
-
•
Pharmacokinetic models have been used to accelerate drug discovery but do not account for a person's diet and gut microbiota.
-
•
Here, we propose combining constraint-based and pharmacokinetic modeling to capture also dietary and gut microbial metabolism.
-
•
Such integrated models will enable the individual-specific prediction of drug response.
Introduction
The effect of drug treatment varies significantly among individuals, and genetic differences alone are insufficient to explain the observed inter-individual differences in drug response [1]. Human gut microbes metabolize many drugs [2]; however, their contribution to an individual's drug response and safety is poorly understood. Diet also modulates the microbiota composition and biochemical functions and alters drug bioavailability. Recent technological advances have led to a greater understanding of the diversity and abundance of gut microbial species. Consequently, research focus is shifting toward exploring the effects of a person's microbiota on metabolism and drug metabolism. Accordingly, constraint-based computational models have been applied to investigate how the gut microbiota can modulate the human metabolic phenotype [3]. In parallel, pharmacokinetic models are used to predict drug responses at the whole-body level [4].
Despite these advances, computational modeling efforts have yet not considered the joint effects of human gut microbiota metabolism, drug metabolism and diet. Consequently, neither the pharmaceutical industry nor academic researchers can properly exploit the increasing knowledge on the human gut microbiota as well as microbiota- and diet-related interpersonal variability for drug development and clinical trial design. The application of statistical methods to genomic or clinical data in pharmacogenomics has been of limited use for patient and therapeutic stratification [5] and does not provide a mechanistic system-level understanding of the targeted biological systems. Another limitation of pharmacogenomics is its failure to integrate exogenous factors that alter drug bioavailability, such as the human gut microbiota or diet [6].
Human and microbial drug metabolism
Individual drug response. Variations in individual treatment responses pose a major challenge to health professionals and patients as well as to drug development and clinical trial design [7]. Front-line physicians must therefore adapt a pragmatic or empirical prescription decision tree until an effective therapy for each patient is identified. Furthermore, the adverse drug reactions that may ensue are ranked among the top 10 causes of morbidity and mortality in the developed world [8]. Certain adverse effects are related to the production of toxic drug metabolites [9], and both the duration and extent of pharmacological action are related to the rate of drug metabolism [10]. Pharmacogenomic studies have greatly improved our understanding of individual variations in drug metabolism caused by genetic individuality [11]. However, these studies cannot explain the large observed individual variability in drug response as they only focus on the genetic variability of drug-metabolism related genes. Consequently, variation in a person's physiology, such as gender, ethnicity, body mass index, should be considered. Moreover, environmental factors, such as diet, gut microbiota composition, exercise, and stress, can modulate a person's metabolic phenotype and drug metabolism. As these factors can significantly alter drug efficacy and safety profiles [12], they must be accounted for in drug development and treatment.
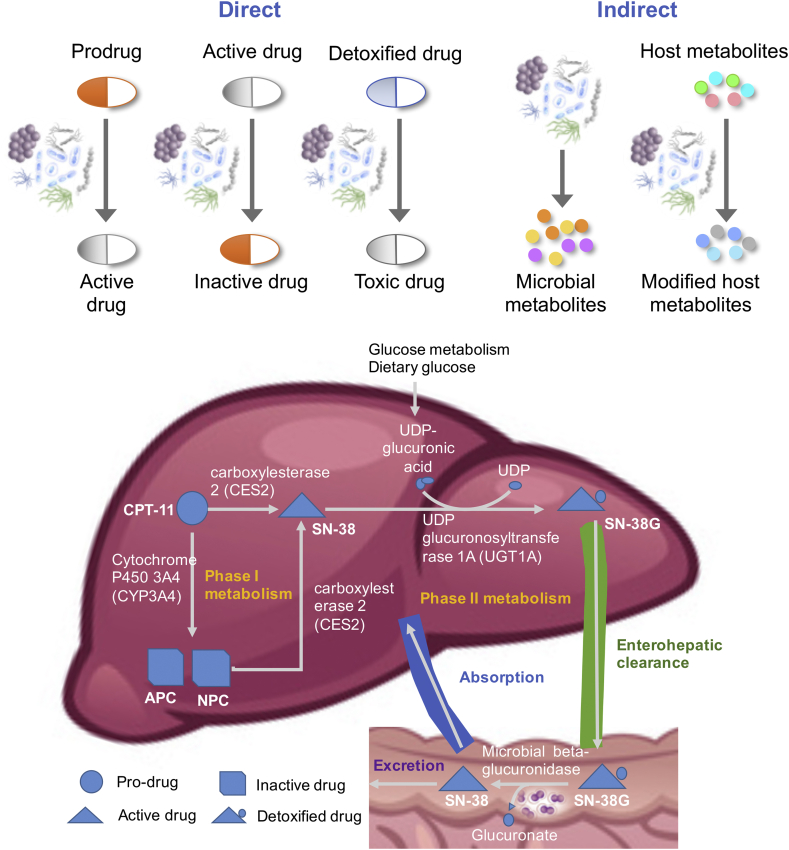
Gut microbial drug metabolism. The human gut microbiota is a metabolically active community of 10–100 trillion commensal, pathogenic, and symbiotic organisms composed of 500–1000 species and including two to four million different genes 13, 14. The gut microbiota contributes to the essential functions of the human host, such as food digestion, essential amino acids and vitamin synthesis, pathogen protection, and host immune system maturation [15]. The gut microbiota has also emerged as a significant factor influencing drug response [2]. Gut microbes affect drug efficiency both directly and indirectly. In turn, exposure to antibiotics and host-targeted drugs induced changes in gut microbiota gene expression across several phyla [16]. This xenobiotic modulation of microbial gene expression varies between human individuals suggesting a gut microbiota-dependent personalized drug response [16]. At least 30 host-targeted drugs are directly affected by gut microbial activity 17, 18 (Figure 1), yet mechanistic insight in the effects on drug efficiency and safety is often lacking [19].
Figure 1.
Top. The human gut microbiota modulates drug metabolism in a direct and indirect manner. Particularly, the indirect mechanism relies computational modeling approaches for identification. Adapted from Ref. [20]. Bottom. Human and microbial metabolism of irinotecan, a chemotherapeutic drug that is frequently used for treating solid tumors. Approximately 35% of irinotecan-treated patients experience severe life-threatening toxicity, commonly manifesting as neutropenia and severe diarrhea, and they require unplanned dose reductions or the discontinuation of the therapy, thus prohibiting effective treatment [72]. The liver enzyme uridine-diphosphate glucuronosyltransferase 1 (UDPGT1) catalyzes the inactivation of irinotecan. Polymorphisms in the corresponding gene have been reported [72] but are unlikely to be the only factor in the variability in drug toxicity. Inactivated (glucuronated) irinotecan is cleared via the enterohepatic route and may also be reabsorbed in the small intestine and excreted after passing the colon. Colonic microbial beta-glucuronidases can reactivate irinotecan by removing glucuronate, which can be used as a carbon source by certain microbes.
Direct microbial effects on drugs include chemically modifying drug structures, binding to drugs, and degrading drugs 17, 20. The gut microbiome encodes enzymes that perform drug transformations, including reduction, acetylation, deacetylation, and demethylation [17]. In certain cases, these transformations result in the desired conversion of a prodrug to an active drug. For example, the prodrug sulfasalazine, a treatment for inflammatory bowel disease, is cleaved into the active drug 5-aminosalicylic acid by intestinal microbial azoreductases [17]. However, in other cases, the drug is inactivated or converted into a more toxic form (Figure 1). For example, the cardiac drug digoxin is inactivated by the cardiac glycoside reductase of Eggerthella lenta [21]. This undesirable inactivation can be reduced by increasing the amount of dietary arginine, demonstrating that dietary interventions can influence drug–microbiota interactions [21]. Only E. lenta strains carrying the “cardiac glycoside reductase” (cgr) operon carry out this biotransformation. The abundance of cardiac glycoside reductase in stool samples has been shown to predict digoxin inactivation and the resulting reduction in drug activity 21, 22. This example clearly demonstrates that genomic and transcriptomic analyses of the gut microbiota can be useful for predicting drug responses 12, 16.
Additionally, gut microbes can indirectly affect the drug response and toxicity by the production of microbial metabolites. One example is the gut microbial production of p-cresol that competes with acetaminophen (paracetamol) for sulfonation by a liver enzyme and thus contributes to drug toxicity in certain individuals [10]. To date, few mechanistic insights have been provided for the effects of drug–microbiota interactions, including drug efficacy and safety, and the species capable of drug transformations are largely unknown [17]. Moreover, the gut microbiota is characterized by functional redundancy, i.e., the same functions can be performed by multiple bacteria that may be either closely or distantly related [23]. This redundancy also extends to drug-metabolizing genes in multiple species across phyla. Hence, a microbiota-wide systematic approach to exploiting and characterizing the capabilities of the gut microbiota to modulate drug metabolism is urgently required.
Computational modeling approaches
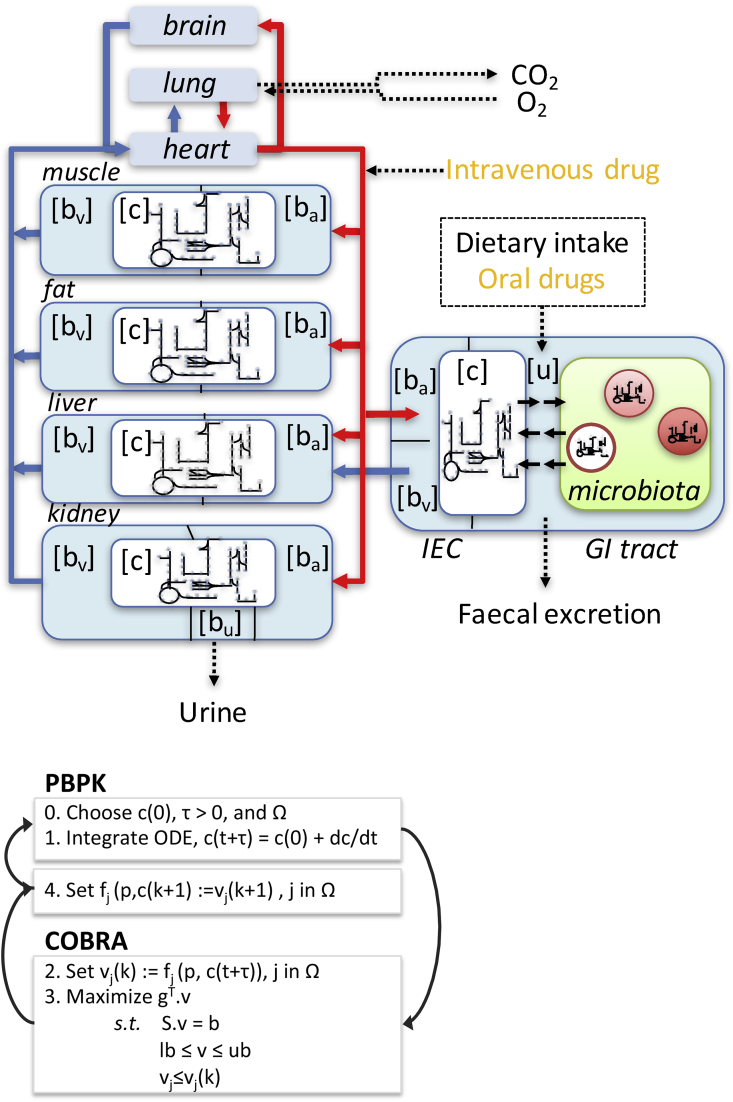
Pharmacokinetic models quantitatively describe the absorption, distribution, metabolism and elimination (ADME) of a drug to predict the time course of a drug's concentration in the body 24, 25. In particular, physiologically based pharmacokinetic (PBPK) models, which compose the core of quantitative systems pharmacology 26, 27, describe whole-body drug kinetics by using ordinary differential equations and an organ compartment structure 28, 29 (Figure 2). These models contain system-specific and drug-specific parameters. The system-specific parameters include blood flow, organ volumes, enzyme and transporter expression, and plasma protein concentrations [30]. The drug-specific parameters include intrinsic clearances, volume of distribution, solubility and physicochemical parameters, tissue partitioning, plasma protein binding affinity, and membrane permeability [30]. The drug-dependent parameters allow for the mechanistic extrapolation of human pharmacokinetics from in vitro and in silico data via a “bottom-up” approach [31]. Whole-body PBPK models have been published for at least 50 drugs 32, 33, ∗∗34, including 32 with an advanced compartment absorption and transit model (ACAT). The ACAT model [35] was based on a CAT model [36], which did not consider the dissolution of solid particles. The ACAT model considers nine gastrointestinal compartments, being the stomach, seven small intestinal segments, and the large intestine. It represents pH-dependent drug solubility, controlled release, drug absorption by the stomach and colon, metabolism in the gut or liver, degradation in the lumen, changes in absorption surface area, changes in drug transporter densities, and changes in efflux transporter densities. Drugs with low solubility or permeability may continue for absorption in the colon.
Figure 2.
Top: Schematic representation of a combined COBRA-PBPK multi-organ model. Each organ-specific model can be derived from the generic human metabolic reconstruction (e.g., [45]) and consists of seven intra-cellular compartments (cytosol [c], nucleus, mitochondria, peroxisome, lysosome, endoplasmatic reticulum, and the golgi apparatus). The microbial models consist of two compartments each, i.e., cytosol and extra-organismal space and could be individualized using metagenomic data. The arterial blood compartment is illustrated with red lines, while the venous blood is represented with blue lines. Each organ contains an exchange compartment with the respective blood compartment ([ba] and [bv]). GI tract = gastro-intestinal tract. IEC = Intestinal epithelial cell. Bottom: Pseudoalgorithm for solving iteratively the hybrid PBPK and COBRA model. Step 0 is initialization, and steps 1–4 are iteratively repeated for a user-specified duration. Symbols: c, concentration of a drug metabolite; t, time point; vj, jth reaction from the set of reactions that is present in both the PBPK and the COBRA model; v, flux vector containing a reaction rate for each reaction in the COBRA; model; lb, lower flux bound; and ub, upper flux bound.
PBPK models can be personalized by using system-specific parameters 37, 38. Physiological parameters from specific populations include specific parameters for infants 39, 40, pregnant women 41, 42, and elderly people [43]. Despite the mechanistic details captured in PBPK models, they do not account for microbial metabolism. Moreover, they do not permit personalization based on dietary, microbial, or genetic data. Also, current PBPK models do not yet connect with the underlying network of genes, proteins, and biochemical reactions of human metabolism.
In constraint-based reconstruction and analysis (COBRA), a metabolic reconstruction of an organism is assembled in a bottom-up manner on the basis of reaction stoichiometry and physicochemical properties obtained from genome annotations and biochemical and physiological data [44]. The conversion of a metabolic reconstruction into a condition-specific model includes the transformation of the biochemical reaction list into a mathematical format (a stoichiometric matrix, ). It also requires the imposition of physico-chemical constraints (e.g., mass conservation) and designation of reactions for exchange of mass across systems [44]. The COBRA approach assumes that the modeled system is in a steady state . These constraints result in an underdetermined system of linear equations that includes fewer equations (mass-balances) than variables (reaction fluxes, ); thus, a polyhedral convex steady-state solution space contains all of the feasible steady-state solutions [44]. Adding further constraints (e.g., nutrient uptake rates, enzyme reaction rates) to the model can restrict the solution space to solutions that are biologically relevant under the given condition. Despite incomplete knowledge of many reaction rates, kinetic parameters, and metabolites and enzyme concentrations, COBRA permits the prediction of feasible phenotypic properties of the modeled system. Comprehensive models of human metabolism (http://vmh.life) 45, 46, 47 exist. Additionally, we have created a stoichiometric reaction module, compatible with the human metabolic reconstruction [45] describing the metabolic transformation of the 18 most highly prescribed drugs, including statins, anti-hypertensions, immunosuppressants, and analgesics [48]. Such drug metabolic reactions are required to allow for the integration of the pharmacokinetic parameters with the COBRA models (see next section).
The COBRA approach has been applied to numerous biomedical questions, including the phenotypic consequences of single nucleotide polymorphisms [49] and enzyme deficiencies 45, 50, 51, and predictions of side and off-target effects of drugs 52, 53. With these resources in place, attention is now turning to their integration.
Integration of PBPK and COBRA modeling
To overcome the limitations associated with the steady-state assumption in COBRA models and the lack of biochemical details of PBPK models, hybrid COBRA-PBPK modeling approaches have been recently explored ∗54, ∗∗55, 56 (Figure 2). For instance, Krauss et al. [55] integrated a COBRA model of cellular liver metabolism, which consisted of 777 metabolites and 2539 reactions [57], with a PBPK model of an human adult to demonstrate an increase in predictive accuracy and mechanistic understanding for allopurinol treatment, ammonia detoxification, and paracetamol toxication. In a subsequent study, we combined seven copies of a COBRA model for a small intestinal epithelial cell [18], each consisting of 433 metabolites and 1318 reactions, with a physiology-based pharmacokinetic ACAT model [34]. We used this model to investigate the role of intestinal absorption on the bioavailability of levodopa, the predominant drug administered to patients with Parkinson's disease. Investigating the different model parameters, we identified that plasma-level of levodopa were most sensitive to the gastric emptying rate and the loss due to microbial activity. For instance, Helicobacter pylori, which is frequently found in the stomach, binds levodopa and thereby reduces its bioavailability.
Accounting for microbial metabolism and dietary information
As a next step, more organs in PBPK models need to be represented at a molecular level. To this end, numerous cell- and tissue-specific metabolic reconstructions have been published 57, 58, 59, 60, 61. To capture the gut microbial metabolism, we have recently published a most comprehensive collection of semi-manually curated metabolic reconstructions of 773 human gut microbes [62], named AGORA. This resource enables modeling of gut microbial communities and their interactions with the human host [63]. However, these microbial metabolic models do not yet capture xenobiotic metabolism [64], which will require the use using context-based comparative genomics techniques [65], to identify microbial enzymes known to modify drugs. The AGORA models can be combined into a microbiota community model [63] and parameterized using metagenomics data. This can be achieved by formulation of a microbial community biomass reaction summing all of the individual microbial biomass reaction fluxes. The stoichiometric coefficients of this community biomass reaction can be adapted on the basis of the relative microbial abundance reported in metagenomics data. The microbial community biomass reaction can be constrained such that its rate corresponds to the fecal excretion rate of an average human (e.g., once every 12–24 h). Finally, the effect of diet as a modulator of human health and microbial composition has been studied using COBRA modeling 59, 63, ∗66, 67. One excellent example has used the molecular composition of 24 defined food items for predicting their effect on human and microbial metabolism [66]. However, drug–diet interactions have not yet been computationally modeled using a COBRA modeling approach.
Development of efficient computational approaches for hybrid COBRA-PBPK modeling
To reliably and efficiently integrate PBPK and COBRA at large scale, using the available resources (Table1, Figure 2), the available hybrid modeling approaches needs to be refined. For instance, to ensure that the changes in calculated flux vectors between two consecutive time steps respond as well-behaved functions (single-valued and Lipschitz continuous) of perturbations in the input data, the flux balance analysis problem can be regularized to ensure that it is a strictly convex optimization problem 68, 69 by minimizing the Euclidean distance between the current and next optimal flux vector. In addition, one could mathematically reformulate the problem by adapting techniques from discrete mechanics to pose the dynamic trajectory of an integrated pharmacokinetic and metabolic system as the optimal solution to a variational integration problem [70]. Moreover, a new quad-precision linear programming solver has been recently developed to allow for solving optimization problems ranging multiple scales [71], e.g., due to macro – and micronutrients in the diet. High-precision solvers are slower but guarantee return of an optimal flux vector to 16 digits of numerical precision, which is far beyond the precision of the biological measurements that are applied as modeling constraints or used for the non-integer stoichiometric coefficients (e.g., microbial abundance in the community biomass reaction).
Table 1.
List of resources and tools for building and simulating with metabolic and pharmacokinetic models.
Pharmacokinetic simulations frequently focus on recurrent dynamics over a finite time interval, such as a regular 24 h-dosing regime. In this case, time can be explicitly discretized, thus producing a set of coupled metabolic and pharmacokinetic equations with a variable for each time point. Additional constraints enforce the dynamic continuity of each feasible trajectory. The optimal dynamic time course can be obtained by solving an optimization problem over this set of possible trajectories. The key to the tractability of the optimization problem is to couple the pharmacokinetic model variables to linear kinetic reactions within the metabolic model. Therefore, the rate of each drug diffusion reaction in a metabolic model is a linear function of the drug concentration, and with a specified diffusion coefficient, a rate variable can be linearly coupled in a COBRA model to a concentration variable in a discrete dynamical system. This discretized system is high-dimensional but amenable to parallel computing because only the constraints enforcing dynamic continuity share variables representing different time points.
Conclusion
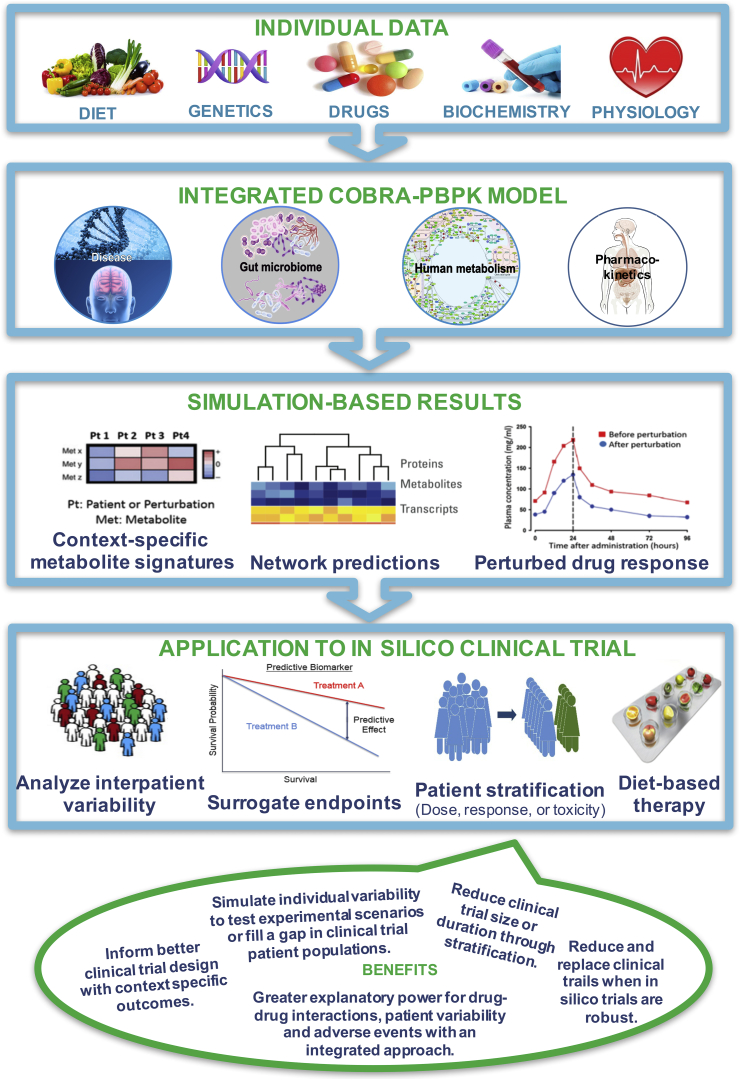
Despite the importance of diet and gut microbiota in drug metabolism, none of the current computational modeling approaches have integrated diet–microbiota–drug interactions. Hybrid COBRA-PBPK modeling leverages their individual strengths and overcomes some of their intrinsic weaknesses, e.g., steady-state assumption in COBRA and limited molecular details in PBPK modeling. Building on the growing knowledge of the gut microbiota and using cutting-edge computational modeling approaches will enable an unprecedented level of mechanistic understanding of personalized drug–microbiota–diet interactions. This new generation COBRA-PBPK models will combine a multi-level in silico description of human, microbial, and drug metabolism, which accurately represents the underlying network of genes, proteins, and biochemical reactions, as well as physiological processes and drug pharmacokinetics. Consequently, individual physiological parameters as well as exogenous factors that alter drug bioavailability can be used to generate personalized predictive models (Figure 3). More individualized treatment strategies, developed in silico yet based on individualized in vivo data, have great potential to contribute to personalized medicine and will enable in silico clinical trials and thereby directly contribute to the design of in vivo clinical trials, by enabling patient stratification to reduce the heterogeneity in treatment responses. (See Table 1)
Figure 3.
Overview of the proposed integrated computational approach for modeling host-diet-microbe-drug interactions. Different types of data, including genomic, biochemical and physiological data, drug administration, and dietary information are integrated into a model consisting of genome-scale reconstructions of human and gut microbial metabolism with pharmacokinetic models of drug absorption and metabolism. Such an integrated, personalized model may yield individual-specific predictions of drug metabolic pathway fluxes, end products of drug metabolism, and patients' drug responses. Taken together, the proposed computational pipeline enables the simulation of clinical trials through patient-specific prediction of drug response, drug toxicity, the effects of dietary interventions.
Acknowledgements
This work was supported by the Luxembourg National Research Fund (FNR) through the ATTRACT programme grant (FNR/A12/01), the National Centre of Excellence in Research (NCER) on Parkinson's disease, and the European Union's Horizon 2020 research and innovation programme under grant agreement #668738.
This review comes from a themed issue on Pharmacology and drug discovery (2017)
Edited by Lars Kuepfer and Tobias Bollenbach
References
- 1.Nebert D.W., Zhang G., Vesell E.S. From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab Rev. 2008;40:187–224. doi: 10.1080/03602530801952864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haiser H.J., Turnbaugh P.J. Developing a metagenomic view of xenobiotic metabolism. Pharmacol Res: Off J Ital Pharmacol Soc. 2013;69:21–31. doi: 10.1016/j.phrs.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mardinoglu A., Shoaie S., Bergentall M., Ghaffari P., Zhang C., Larsson E., Backhed F., Nielsen J. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol Syst Biol. 2015;11:834. doi: 10.15252/msb.20156487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jamei M. Recent advances in development and application of physiologically-based pharmacokinetic (PBPK) models: a transition from academic curiosity to regulatory acceptance. Curr Pharmacol Rep. 2016;2:161–169. doi: 10.1007/s40495-016-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitano H. World Health Summit Yearbook; 2013. The grand challenge of systems biomedicine. [Google Scholar]
- 6.Wilson I.D. Drugs, bugs, and personalized medicine: pharmacometabonomics enters the ring. Proc Natl Acad Sci U. S. A. 2009;106:14187–14188. doi: 10.1073/pnas.0907721106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schork N.J. Personalized medicine: time for one-person trials. Nature. 2015;520:609–611. doi: 10.1038/520609a. [DOI] [PubMed] [Google Scholar]
- 8.Campbell J.E., Gossell-Williams M., Lee M.G. A review of pharmacovigilance. West Indian Med J. 2014;63:771–774. doi: 10.7727/wimj.2013.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alomar M.J. Factors affecting the development of adverse drug reactions (Review article) Saudi Pharm J. 2014;22:83–94. doi: 10.1016/j.jsps.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clayton T.A., Baker D., Lindon J.C., Everett J.R., Nicholson J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U. S. A. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drew L. Pharmacogenetics: the right drug for you. Nature. 2016;537:S60–S62. doi: 10.1038/537S60a. [DOI] [PubMed] [Google Scholar]
- 12.Nayak R.R., Turnbaugh P.J. Mirror, mirror on the wall: which microbiomes will help heal them all? BMC Med. 2016;14:72. doi: 10.1186/s12916-016-0622-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falony G., Joossens M., Vieira-Silva S., Wang J., Darzi Y., Faust K., Kurilshikov A., Bonder M.J., Valles-Colomer M., Vandeputte D. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. New York, N. Y. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. New York, N. Y. [DOI] [PubMed] [Google Scholar]
- 16.Maurice C.F., Haiser H.J., Turnbaugh P.J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klaassen C.D., Cui J.Y. Review: mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab Dispos. 2015;43:1505–1521. doi: 10.1124/dmd.115.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tralau T., Sowada J., Luch A. Insights on the human microbiome and its xenobiotic metabolism: what is known about its effects on human physiology? Expert Opin Drug Metab Toxicol. 2015;11:411–425. doi: 10.1517/17425255.2015.990437. [DOI] [PubMed] [Google Scholar]
- 19.Haiser H.J., Turnbaugh P.J. Is it time for a metagenomic basis of therapeutics? Science. 2012;336:1253–1255. doi: 10.1126/science.1224396. New York, N. Y. [DOI] [PubMed] [Google Scholar]
- 20.Spanogiannopoulos P., Bess E.N., Carmody R.N., Turnbaugh P.J. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev. 2016;14:273–287. doi: 10.1038/nrmicro.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haiser H.J., Gootenberg D.B., Chatman K., Sirasani G., Balskus E.P., Turnbaugh P.J. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. New York, N. Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haiser H.J., Seim K.L., Balskus E.P., Turnbaugh P.J. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes. 2014;5:233–238. doi: 10.4161/gmic.27915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moya A., Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016;24:402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Rowland M. Physiologically-based pharmacokinetic (PBPK) modeling and simulations principles, methods, and applications in the pharmaceutical industry. CPT Pharmacomet Syst Pharmacol. 2013;2:e55. doi: 10.1038/psp.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nestorov I. Whole body pharmacokinetic models. Clin Pharmacokinet. 2003;42:883–908. doi: 10.2165/00003088-200342100-00002. [DOI] [PubMed] [Google Scholar]
- 26.Knight-Schrijver V.R., Chelliah V., Cucurull-Sanchez L., Le Novere N. The promises of quantitative systems pharmacology modelling for drug development. Comput Struct Biotechnol J. 2017;14:363–370. doi: 10.1016/j.csbj.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kell D.B., Goodacre R. Metabolomics and systems pharmacology: why and how to model the human metabolic network for drug discovery. Drug Discov today. 2014;19:171–182. doi: 10.1016/j.drudis.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Upton R.N., Foster D.J., Abuhewla A.Y. An introduction to physiologically-based pharmacokinetic models. Paediatr Anaesth. 2016;26:1036–1046. doi: 10.1111/pan.12995. [DOI] [PubMed] [Google Scholar]
- 29.Nestorov I. Whole-body physiologically based pharmacokinetic models. Expert Opin Drug Metab Toxicol. 2007;3:235–249. doi: 10.1517/17425255.3.2.235. [DOI] [PubMed] [Google Scholar]
- 30.Jones H., Rowland-Yeo K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacomet Syst Pharmacol. 2013;2:e63. doi: 10.1038/psp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rostami-Hodjegan A. Physiologically based pharmacokinetics joined with in vitro-in vivo extrapolation of ADME: a marriage under the arch of systems pharmacology. Clin Pharmacol Ther. 2012;92:50–61. doi: 10.1038/clpt.2012.65. [DOI] [PubMed] [Google Scholar]
- 32.Sager J.E., Yu J., Ragueneau-Majlessi I., Isoherranen N. Physiologically based pharmacokinetic (PBPK) modeling and simulation approaches: a systematic review of published models, applications, and model verification. Drug Metab Dispos. 2015;43:1823–1837. doi: 10.1124/dmd.115.065920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida K., Maeda K., Kusuhara H., Konagaya A. Estimation of feasible solution space using Cluster Newton Method: application to pharmacokinetic analysis of irinotecan with physiologically-based pharmacokinetic models. BMC Syst Biol. 2013;7(Suppl 3):S3. doi: 10.1186/1752-0509-7-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guebila M.B., Thiele I. Model-based dietary optimization for late-stage, levodopa-treated, Parkinson's disease patients. Npj Syst Biol Appl. 2016;2:16013. doi: 10.1038/npjsba.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, a combined model of small intestinal metabolism and levodopa pharmacokinetics is used to predict the influence of dietary amino acids on the bioavailability of levodopa for Parkinson's disease patients.
- 35.Agoram B., Woltosz W.S., Bolger M.B. Predicting the impact of physiological and biochemical processes on oral drug bioavailability. Adv Drug Deliv Rev. 2001;50(Suppl 1):S41–S67. doi: 10.1016/s0169-409x(01)00179-x. [DOI] [PubMed] [Google Scholar]
- 36.Yu L.X., Amidon G.L. Characterization of small intestinal transit time distribution in humans. Int J Pharm. 1998;171:157–163. [Google Scholar]
- 37.Price P.S., Conolly R.B., Chaisson C.F., Gross E.A., Young J.S., Mathis E.T., Tedder D.R. Modeling interindividual variation in physiological factors used in PBPK models of humans. Crit Rev Toxicol. 2003;33:469–503. [PubMed] [Google Scholar]
- 38.McNally K., Cotton R., Hogg A., Loizou G. PopGen: a virtual human population generator. Toxicology. 2014;315:70–85. doi: 10.1016/j.tox.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Strolin Benedetti M., Whomsley R., Baltes E.L. Differences in absorption, distribution, metabolism and excretion of xenobiotics between the paediatric and adult populations. Expert Opin Drug Metab Toxicol. 2005;1:447–471. doi: 10.1517/17425255.1.3.447. [DOI] [PubMed] [Google Scholar]
- 40.Barrett J.S., Della Casa Alberighi O., Laer S., Meibohm B. Physiologically based pharmacokinetic (PBPK) modeling in children. Clin Pharmacol Ther. 2012;92:40–49. doi: 10.1038/clpt.2012.64. [DOI] [PubMed] [Google Scholar]
- 41.Abduljalil K., Furness P., Johnson T.N., Rostami-Hodjegan A., Soltani H. Anatomical, physiological and metabolic changes with gestational age during normal pregnancy: a database for parameters required in physiologically based pharmacokinetic modelling. Clin Pharmacokinet. 2012;51:365–396. doi: 10.2165/11597440-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 42.Young J.F., Branham W.S., Sheehan D.M., Baker M.E., Wosilait W.D., Luecke R.H. Physiological “constants” for PBPK models for pregnancy. J Toxicol Environ Health. 1997;52:385–401. doi: 10.1080/00984109708984072. [DOI] [PubMed] [Google Scholar]
- 43.Thompson C.M., Johns D.O., Sonawane B., Barton H.A., Hattis D., Tardif R., Krishnan K. Database for physiologically based pharmacokinetic (PBPK) modeling: physiological data for healthy and health-impaired elderly. J Toxicol Environ Health B Crit Rev. 2009;12:1–24. doi: 10.1080/10937400802545060. [DOI] [PubMed] [Google Scholar]
- 44.Palsson B. Cambridge University Press; Cambridge: 2006. Systems biology: properties of reconstructed networks. [Google Scholar]
- 45.Thiele I., Swainston N., Fleming R.M., Hoppe A., Sahoo S., Aurich M.K., Haraldsdottir H., Mo M.L., Rolfsson O., Stobbe M.D. A community-driven global reconstruction of human metabolism. Nat Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agren R., Mardinoglu A., Asplund A., Kampf C., Uhlen M., Nielsen J. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol Syst Biol. 2014;10:721. doi: 10.1002/msb.145122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swainston N., Smallbone K., Hefzi H., Dobson P.D., Brewer J., Hanscho M., Zielinski D.C., Ang K.S., Gardiner N.J., Gutierrez J.M. Recon 2.2: from reconstruction to model of human metabolism. Metab: Off J Metab Soc. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S., Haraldsdottir H.S., Fleming R.M., Thiele I. Modeling the effects of commonly used drugs on human metabolism. FEBS J. 2015;282:297–317. doi: 10.1111/febs.13128. [DOI] [PubMed] [Google Scholar]; First paper presenting a metabolic, stoichiometrically accurate reaction module of 18 highly prescribed drugs, which is compatible with the human metabolic reconstruction, Recon 2.
- 49.Jamshidi N., Palsson B.O. Systems biology of SNPs. Mol Syst Biol. 2006;2:38. doi: 10.1038/msb4100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahoo S., Franzson L., Jonsson J.J., Thiele I. A compendium of inborn errors of metabolism mapped onto the human metabolic network. Mol Biosyst. 2012;8:2545–2558. doi: 10.1039/c2mb25075f. [DOI] [PubMed] [Google Scholar]
- 51.Pagliarini R., Castello R., Napolitano F., Borzone R., Annunziata P., Mandrile G., De Marchi M., Brunetti-Pierri N., di Bernardo D. In silico modeling of liver metabolism in a human disease reveals a key enzyme for histidine and histamine homeostasis. Cell Rep. 2016;15:2292–2300. doi: 10.1016/j.celrep.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaked I., Oberhardt M.A., Atias N., Sharan R., Ruppin E. Metabolic network prediction of drug side effects. Cell Syst. 2016;2:209–213. doi: 10.1016/j.cels.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Chang R.L., Xie L., Bourne P.E., Palsson B.O. Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadehn F.S.S., Eissing T., Krauss M., Kuepfer L. EMBC. 2016. A multiscale, model-based analysis of the multi-tissue interplay underlying blood glucose regulation in diabetes. (Orlando) [DOI] [PubMed] [Google Scholar]; Large-scale physiology-based pharmacokinetic model of whole-body glucose-insulin-metabolism.
- Krauss M., Schaller S., Borchers S., Findeisen R., Lippert J., Kuepfer L. Integrating cellular metabolism into a multiscale whole-body model. PLoS Comput Biol. 2012;8:e1002750. doi: 10.1371/journal.pcbi.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]; First paper demonstrating the hybrid modeling of constraint-based modeling and physiology-based pharmacokinetic modeling.
- 56.Toroghi M.K., Cluett W.R., Mahadevan R. A multi-scale model of the whole human body based on dynamic parsimonious flux balance analysis. IFAC-PapersOnLine. 2016;49:937–942. [Google Scholar]
- 57.Gille C., Bolling C., Hoppe A., Bulik S., Hoffmann S., Hubner K., Karlstadt A., Ganeshan R., Konig M., Rother K. HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol Syst Biol. 2010;6:411. doi: 10.1038/msb.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bordbar A., Feist A.M., Usaite-Black R., Woodcock J., Palsson B.O., Famili I. A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology. BMC Syst Biol. 2011;5:180. doi: 10.1186/1752-0509-5-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sahoo S., Thiele I. Predicting the impact of diet and enzymopathies on human small intestinal epithelial cells. Hum Mol Genet. 2013;22:2705–2722. doi: 10.1093/hmg/ddt119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. New York, N. Y. [DOI] [PubMed] [Google Scholar]
- 61.Aurich M.K., Thiele I. Computational modeling of human metabolism and its application to systems biomedicine. Methods Mol Biol. 2016;1386:253–281. doi: 10.1007/978-1-4939-3283-2_12. Clifton, N. J. [DOI] [PubMed] [Google Scholar]
- 62.Magnusdottir S., Heinken A., Kutt L., Ravcheev D.A., Bauer E., Noronha A., Greenhalgh K., Jager C., Baginska J., Wilmes P. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017;35:81–89. doi: 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- 63.Heinken A., Thiele I. Systematic prediction of health-relevant human-microbial co-metabolism through a computational framework. Gut Microbes. 2015;6:120–130. doi: 10.1080/19490976.2015.1023494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunemann M., Schmid M., Patil K.R. Computational tools for modeling xenometabolism of the human gut microbiota. Trends Biotechnol. 2014;32:157–165. doi: 10.1016/j.tibtech.2014.01.005. [DOI] [PubMed] [Google Scholar]; A conceptual overview of gut microbial xenometabolism and computational tools is provided.
- 65.Ravcheev D.A., Thiele I. Genomic analysis of the human gut microbiome suggests novel enzymes involved in quinone biosynthesis. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaie S., Ghaffari P., Kovatcheva-Datchary P., Mardinoglu A., Sen P., Pujos-Guillot E., de Wouters T., Juste C., Rizkalla S., Chilloux J. Quantifying diet-induced metabolic changes of the human gut microbiome. Cell metab. 2015;22:320–331. doi: 10.1016/j.cmet.2015.07.001. [DOI] [PubMed] [Google Scholar]; In this study, the authors showed how in silico diet variation may influence the gut microbial composition in silico.
- 67.Nogiec C.D., Kasif S. To supplement or not to supplement: a metabolic network framework for human nutritional supplements. PLoS One. 2013;8:e68751. doi: 10.1371/journal.pone.0068751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dontchev A.L., Rockafellar R.T. Primal-dual solution perturbations in convex optimization. Set-Valued Anal. 2001;9:49–65. [Google Scholar]
- 69.Fleming R.M., Maes C.M., Saunders M.A., Ye Y., Palsson B.O. A variational principle for computing nonequilibrium fluxes and potentials in genome-scale biochemical networks. J Theor Biol. 2012;292:71–77. doi: 10.1016/j.jtbi.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 70.Mardsen J.E., West M. Discrete mechanics and variational integrators. Acta Numer. 2001;10:357–514. [Google Scholar]
- 71.Ma, D.Y.L.; Fleming, R.M.T.; Thiele, I.; Palsson, B.O.; Saunders, M.A. (2016). Reliable and efficient solution of genome-scale models of Metabolism and macromolecular Expression. [DOI] [PMC free article] [PubMed]
- 72.Campbell J.M., Stephenson M.D., Bateman E., Peters M.D., Keefe D.M., Bowen J.M. Irinotecan-induced toxicity pharmacogenetics: an umbrella review of systematic reviews and meta-analyses. Pharmacogen J. 2016;17:21–28. doi: 10.2217/pgs.15.180. [DOI] [PubMed] [Google Scholar]