Ant queen insemination induces brain anatomical and gene regulatory network changes that ants with reproductive workers co-opted.
Abstract
Caste differentiation happens early in development to produce gynes as future colony germlines and workers as present colony soma. However, gynes need insemination to become functional queens, a transition that initiates reproductive role differentiation relative to unmated gynes. Here, we analyze the anatomy and transcriptomes of brains during this differentiation process within the reproductive caste of Monomorium pharaonis. Insemination terminated brain growth, whereas unmated control gynes continued to increase brain volume. Transcriptomes revealed a specific gene regulatory network (GRN) mediating both brain anatomy changes and behavioral modifications. This reproductive role differentiation GRN hardly overlapped with the gyne-worker caste differentiation GRN, but appears to be also used by distantly related ants where workers became germline individuals after the queen caste was entirely or partially lost. The genes corazonin and neuroparsin A in the anterior neurosecretory cells were overexpressed in individuals with reduced or nonreproductive roles across all four ant species investigated.
INTRODUCTION
It is useful to distinguish between reproductive caste differentiation and reproductive role differentiation. The former represents irreversible differentiation in developmental pathways initiated well before adulthood to produce specialized breeder and unmated helper phenotypes, while the latter normally refers to differentiation among adult female breeders after insemination. Pioneers of the evolutionary study of social insects such as Darwin, Weismann, and Wheeler appropriately distinguished between perfect fertile female reproductives (gynes) and lifetime unmated “neuters” (1). However, it is less commonly appreciated that a gyne phenotype when it hatches from its pupal case is merely a necessary condition for becoming a functional queen—this reproductive role transition will be possible only after the sufficient condition of becoming inseminated is fulfilled. In that sense, gynes are functionally analogous to unfertilized metazoan eggs, and their insemination, followed by lifetime sperm storage, is comparable to the fertilization of such eggs to become zygotes. This analogy was aptly formulated more than a century ago by Wheeler (1), who defined colonies as higher-level (super)organisms, noting that “[t]he queen mother of the ant colony displays the generalized potentialities of all the individuals, just as the Metazoan egg contains in potentia all the other cells of the body.” Wheeler’s analogy was recently corroborated in a comparative study showing that the common ancestor of the ants had colonies with one singly inseminated queen (2) just like multicellular organismal development starts with a zygotic merger of a single egg and sperm cell (3). However, while individual caste-specific development is genetically hardwired, the colony-level process of caste differentiation is analogous to a higher-level form of ontogenetic development controlled by the social nurturing environment of the ant brood (4, 5).
After a number of decades with less precise definitions, the original version of colonial superorganismality was recently resurrected (1) because Wheeler’s original concept of germline sequestration at colony founding and subsequent somatic colony development is more fundamental than evolutionarily derived traits such as large colonies and complex division of labor (6). Taking a Wheeler perspective therefore emphasizes fundamental analogies that apply despite the obvious distinction between zygotic organismality and inseminated queen superorganismality. The organismal evo-devo principles apply to a clonal germline and soma, while a superorganism needs an extra meiotic level to realize colony-level development via the production of multiple cohorts of offspring zygotes. These will then become founding-stage sibling workers, later ergonomic colony growth workers, and lastly a mixture of gyne and male reproductives raised by mature worker cohorts (7). Metazoan organismal development is increasingly documented to unfold via differentiating gene regulatory networks (GRNs) where the first cell divisions may be controlled by the maternal gametic cytoplasm after which the zygotic genome takes control (8). Analogously, we hypothesized that the inseminated queen mother starts to unfold her superorganismal colonial potentialities because insemination triggers a GRN that decisively elaborates traits initiated during female virgin development.
To test the analogy drawn up above, one needs an ant model system where uninseminated gynes can continue to survive in their natal colony for substantial timespan so they can function as controls for inseminated gynes that complete reproductive role differentiation toward functional queens. The ant Monomorium pharaonis is unusual in offering this social colony structure because: (i) It has many inseminated queens per colony that are produced year around in captivity and can be freely combined with workers from other colonies in contrast to most other ants (9, 10). (ii) Gynes do not need dispersal flights and are inseminated in their colonies (9), allowing experimental control over the timing of insemination and the subsequent sampling of same age cohorts to monitor changes in brain anatomy and gene expression. (iii) Gynes that fail to become inseminated within a rather narrow time window after hatching assume somatic worker-like roles, although their phenotype developed for specialized reproductive functions, and they can survive in those roles for 4 (9) to 6 (this study) months.
M. pharaonis is convenient as a model ant for two other reasons as well. First, workers lack ovaries and haploid eggs laid by either gynes or queens are only reared as males when colonies are made queenless (9). We could thus assume that our experimental gynes and queens were unlikely to differ in their tendencies to, and incentives for, laying haploid eggs. Reproductive differences between gynes and queens must therefore be binary due to the production or nonproduction of fertilized eggs. Second, gyne-worker caste differentiation in M. pharaonis becomes irreversible already in the egg stage (9, 11), which is unusually early for ants but implies that the GRNs mediating immature gyne-worker caste differentiation and adult gyne-queen role differentiation are well separated in time (Fig. 1A).
Fig. 1. Experimental groups and time points.
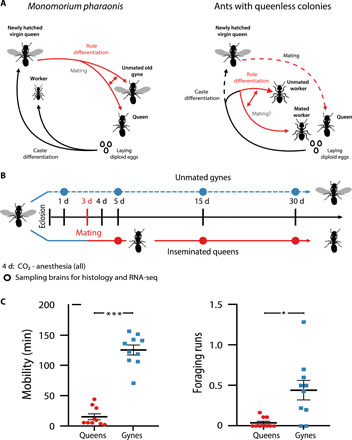
(A) Conceptual design showing ants have two fundamental developmental bifurcations: caste differentiation (virgin gynes versus lifetime unmated workers established before pupation) and adult role differentiation (virgin gynes versus inseminated queens). Left: The lifecycle and associated differentiation processes of M. pharaonis are unusual because caste is decided already in the egg stage and gynes failing insemination assume worker roles and survive alongside inseminated queens. Right: Several other ants secondarily lost the queen caste, partly or entirely (dashed arrows), and evolved role differentiation between reproductive and sterile workers via gamergate replacement queens or parthenogenesis. Queenless Dinoponera colonies always have a single gamergate, but Harpegnathos colonies have multiple gamergates forming a dominance hierarchy (fig. S1). (B) Cohorts of M. pharaonis gynes were divided, such that one group was allowed to mate on the third day to become inseminated queens, while the other group continued maturation without insemination. On day 4, all individuals were CO2-anesthetized. Gynes (blue) were collected at the same time points as queens (red) for neuroanatomical and transcriptomic investigations. (C) Behavioral observation experiments (n = 10, 3 hours per subcolony experiment) showed that the motility (average active time) and foraging activity (average number of foraging runs) of day 30 queens were significantly lower than that of same age gynes (t18 = 11.635, P < 0.001 and t18 = 3.311, P < 0.05, respectively). *P < 0.05, ***P < 0.001.
While cell differentiation during early metazoan zygote development relies on complex processes of cell signaling, role differentiation of newly inseminated gynes seems inconceivable without a prominent role of brains. We were therefore particularly interested in the GRNs expressed in the brains of M. pharaonis gynes versus queens because previous studies have shown that uninseminated gynes come to display worker-like behavior (9, 12), suggesting brain GRN changes in response to insemination. For the purpose of our study, we define a GRN as a cluster of genes with correlated expression in opposite direction for alternative phenotypes such as virgin gynes and inseminated queens as they diverge during reproductive role differentiation. We recently showed that a shared ancestral GRN for caste differentiation can be reconstructed from caste-specific biases in brain gene expression of adult gynes versus workers across different ant species (13). However, no attempt has yet been made to obtain a GRN for reproductive role differentiation in ants and to evaluate its degree of conservation.
While M. pharaonis has many social life history characteristics to allow the GRN for gyne-queen role differentiation to be identified, comparisons across ant lineages to explore whether that GRN appears to have been evolutionarily conserved were challenging because no highly polygynous ants with comparable gyne/queen characteristics have been targeted with advanced genomic techniques. However, several well-studied ant species secondarily evolved reproductive role differentiation within the worker caste after the original colony-founding queen caste had been partly or entirely lost (14–16). We therefore compared the obtained GRN for reproductive role differentiation in M. pharaonis gynes/queens (Fig. 1A) with published transcriptome data of three ant species with gamergate or parthenogenetic reproductive workers for which transcriptomes of nonreproductive control workers were also available (Fig. 1B and fig. S1): (i) Harpegnathos saltator where workers compete in dominance hierarchies to become coexisting gamergate reproductives after the founding queen has died (14, 17), (ii) Dinoponera quadriceps where the queen caste is completely lost and worker dominance hierarchies allow only a single gamergate to become inseminated (14), and (iii) the parthenogenetic clonal raider ant (Ooceraea biroi) where workers synchronously alternate between a reproductive and nonreproductive phase and where most workers become egg layers when their colony enters the reproductive phase (18).
In light of the above, the objectives of our study of reproductive role differentiation were to clarify: (i) How gyne/queen brain anatomy of M. pharaonis changes in response to insemination, (ii) whether and to what extent the insemination-induced GRN for reproductive role differentiation in M. pharaonis overlapped with the known GRN mediating gyne-worker caste differentiation (13), and (iii) whether the insemination-induced role differentiation GRN within the gyne/queen caste of M. pharaonis shares homologous modules with the reproductive role differentiation GRNs in ant species where worker caste individuals took over reproductive roles after the queen caste lost significance or disappeared.
RESULTS
Insemination triggers behavioral changes and suppresses brain growth in M. pharaonis
It has been known for decades that newly inseminated queens of M. pharaonis become less active inside the nest, whereas gynes that remain unmated start to express explorative and foraging behavior similar to workers. Petersen-Braun (9) observed gynes that appeared outside the nest where they actively explored the surroundings and/or expressed foraging behavior, collected them, and investigated their flight muscles and ovaries by dissection. This showed that they all had degenerated flight muscles and ovaries, indicating that old gynes (possibly even old queens as insemination status was not checked) regress ovaries and perform worker-like behaviors (9). Petersen-Braun (9) also observed that most gynes that remained unmated after 2 to 4 weeks became active outside the nest near the food sources. To verify these observations, we quantified the differences in locomotion and foraging behavior between 30-day-old uninseminated gynes and queens of the same age that were inseminated on day 3 (see Fig. 1B and Materials and Methods for details). This confirmed that old gynes were significantly more active on both accounts (Fig. 1, B and C). However, nursing behavior of gynes remained highly variable (9), consistent with gyne phenotypes (irrespective of insemination status) not having been under selection for nursing tasks because M. pharaonis colonies multiply by budding so queens will never be without nursing workers. Our results combined with those of Petersen-Braun (9) thus indicate that old gynes display some fraction of the total worker behavioral syndrome. This allows old gynes to indirectly contribute to colony fitness and confirmed our confidence in these old gynes being proper controls in comparisons with same age inseminated queens.
To investigate the structural changes of the brain during gyne-queen reproductive role differentiation, we followed the experimental setup outlined in Fig. 1B to reconstruct the main neuropils of 64 individuals and to record the brain volume changes of newly eclosed gynes after insemination using confocal microscopy image stacks. We recorded the autofluorescence of brain tissues in whole head preparations for both the gyne and queen cohorts at specific ages, and we analyzed both the temporal effect of maturation and the age controlled effect of gyne/queen status on the volume of the antennal lobes (ALs), the optic lobes (OLs), the ocelli (OC), the mushroom bodies (MBs), and the supraesophageal zone (SPZ) (Fig. 2A). Overall, brain maturation differed significantly between queens and gynes but with a time lag as no divergence was yet apparent on day 5 (2 days after mating). However, the differences on day 15 clearly show that insemination had terminated further increase in total brain volume of queens while the volume of gyne brains continued to increase (Fig. 2B).
Fig. 2. Comparison of neuropil volumes of M. pharaonis gynes and queens over time.
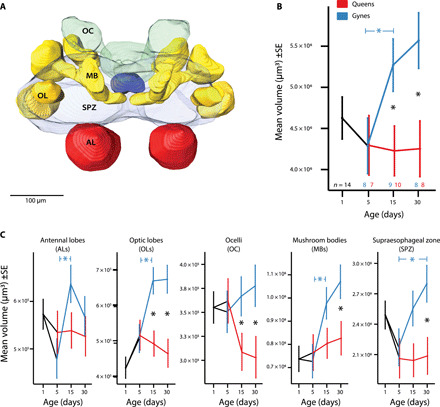
(A) Representative brain reconstruction of a day 5 gyne obtained from image stack of transparent head. In total, 64 brains were reconstructed in parallel series with AMIRA software 6.2.0 and enabled volume quantification based as sums of paired (ALs, OLs, and MBs) or triplet (OCs) parts. (B) Volumes of whole brains showed significant age or role differentiation effects in single comparisons (*P < 0.05) on days 15 and 30 and between days 5 and 15 gynes (horizontal *). An overall two-way ANOVA gave a stronger brain volume effect of gyne/queen status (F1,44 = 8.12, P = 0.007), adjusted for age and combining all sample sizes given above the x axis. (C) Volumes of separate brain parts also showed significances (*P < 0.05) in single t test comparisons for gyne/queen status (vertical *) or age (horizontal *). Capturing all data (days 5, 15, and 30) in a single three-way ANOVA again offered a more powerful test of overall differences (table S1). In particular, the age × gyne/queen status interaction term (F2,220 = 4.96, P = 0.0078) illustrated the generality of bifurcation across brain parts, while the brain-part × gyne/queen status interaction (F4,220 = 5.05, P = 6.54 × 10−4) illustrated that bifurcation also varied across brain parts (see table S1 for details and fig. S2 for relative volumes).
The patterns of change for the five brain parts separately revealed a modest but significant increase of the OLs and MBs in gynes over time (P < 0.05; Fig. 2C). These changes did not reach significance for the other brain parts, but a three-way analysis of variance (ANOVA) with considerably higher statistical power showed that patterns of change in all five parts of the brain across days 5, 15, and 30 were in fact very similar (table S1). This ANOVA approach also allowed statistical interactions to be identified. For example, the overall age × gyne/queen status interaction term was highly significant, confirming the increasing difference in brain volume with reproductive role differentiation over time (table S1) (Fig. 2C, fig. S2, and table S1). Functionally, the most pronounced volume increases of the OLs, the MBs, and the SPZ in gynes might indicate higher processing capacities for visual stimuli, better learning/memory performance, and behavioral control, consistent with the worker behaviors that old gynes express (Fig. 1C) (9, 19). In contrast, the smaller brain volumes in M. pharaonis queens might reflect predictable specialization because these individuals have achieved their adaptive reproductive roles, after which they only need to develop their reproductive organs to full potential and lay eggs in proportion to the amount and quality of nutrition provided by workers.
Divergence of brain gene expression during gyne-queen reproductive role differentiation
We obtained individual brain transcriptomes of freshly eclosed gynes (day 1) and of both unmated gynes and inseminated queens on days 5, 15, and 30 after eclosion (Fig. 1B), always with six to nine replicates at each time point, to quantify the divergence in brain gene expression over time and the involvement of GRNs. Principal components analysis (PCA) revealed that samples of the same age always grouped together on the first principal component (PC) axis, which explained 14.7% of the transcriptome variance. Samples from days 1 and 5 were separated from the samples taken at days 15 and 30 (Fig. 3A). Concordantly, the number of differentially expressed genes (DEGs) increased from around 350 across days 1 and 5 (372 in gynes and 327 in queens) to around 1000 across days 5 and 15 (817 in gynes and 1230 in queens) and decreased again between days 15 and 30 (104 in gynes and 410 in queens) (fig. S3). Focusing on age since hatching per se, rather than on age-specific gyne-queen comparison, we identified 1544 genes whose expression levels correlated significantly with age in both gynes and queens (partial Spearman’s correlation test, P < 0.05). These DEGs may represent candidate genes mediating the onset of sexual activity in Hymenoptera because haplodiploidy allows both gynes and queens to lay viable eggs (see Materials and Methods and table S2).
Fig. 3. Development of differential gene expression as gyne-queen role differentiation in M. pharaonis proceeds.
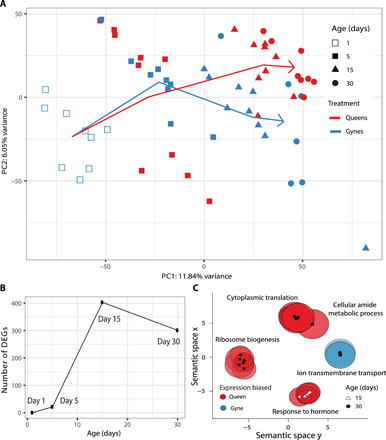
(A) PCA of the brain transcriptomes of gynes and queens of different ages, with each point representing the transcriptome from a single brain, colored according to binary reproductive role and with symbols representing different ages. The mean PC values for gynes and queens at days 1, 5, 15, and 30 were connected chronologically by solid lines (arrowed, young to old), showing that transcriptome profiles were similar on day 5 but had diverged on days 15 and 30. The larger scatter among the gyne dots may have contributed to the number of significant DEGs between gynes and queens on day 30 being less than on day 15. (B) The number of DEGs between gynes and queens of the same age increased drastically between days 5 and 15, which is the time period during which gynes started laying unfertilized eggs and queens fertilized (and possibly also some unfertilized) eggs, after which no further increase at day 30 was observed. (C) Multidimensional scaling clustering according to semantic similarities (gene membership) of Gene Ontology (GO) terms (62). Representative gene members were written next to GO clusters, showing that genes with queen-biased expression were involved in hormonal responses on day 15 and in biogenesis and metabolic processes on day 30, whereas genes with gyne-biased expression on day 30 were mainly involved in ion transport functions.
The brain transcriptomes of gynes and queens could only be separated along the second PC axis on days 15 and 30 (Fig. 3A). This reflects the cumulative effect of insemination, because the number of DEGs associated with reproductive role differentiation between gynes and queens increased from just 31 on day 5 (2 days after mating) to 414 on day 15 and 305 on day 30 (Fig. 3B). Of the total of 648 genes that were differentially expressed across all these age groups, 93 (14%) were significantly differentially expressed and showed a consistent direction of expression bias on both days 15 and 30 (table S3). Consistent with the changes in brain volume during the gyne-queen role differentiation process, we found that many genes involved in neuronal and glial activity were differentially expressed (table S3). Genes with gyne-biased expression on day 15 were significantly enriched for transmembrane ion transport functions (Fig. 3C), which might be associated with the activity of neural cells. Genes with queen-biased expression were significantly enriched for hormonal response functions on day 15 and for biosynthesis and metabolic process functions on day 30 (Fig. 3C).
Insemination triggers the onset of genetic regulatory networks for neuroanatomical changes
Although only 31 genes were significantly differentially expressed between gynes and queens on day 5 (2 days after mating), these genes likely include early brain GRN responses to ovarian maturation after insemination while initiating the expression of the hundreds of other genes that we found to be differentially expressed on days 15 and 30. Nineteen of these 31 genes showed significantly higher expression in queens and 12 had enhanced expression in gynes. Many of these genes are involved in cell differentiation, proliferation processes or (neuro)trophic functions (table S3), such as neuroparsin A–like (NPA), SmD3/guf, and AdamTS-B, consistent with ongoing neuroanatomical changes in gynes and suppression of brain tissue expansion in queens (Fig. 2). NPA acts as a neurotrophic factor enhancing neurite outgrowth with an additional synergism in the presence of insulin (20), while SmD3/guf is required for neuron differentiation (21). AdamTS-B, on the other hand, negatively regulates binding between the epidermal growth factor receptor and its ligands (22). The neuropil-specific expansions in gyne brains that we observed (Fig. 2) might be coordinated by the early onset of differential expression of these morphogenic genes. The only morphogen with queen-biased expression was Krueppel-homolog 1 (Kr-h1) on day 5, a gene known to have higher expression associated with foraging behavior and increased neurite outgrowth in adult MB neurons of honeybees (23). On the other hand, Kr-h1 negatively modulates neuronal morphogenesis during larval and pupal development in Drosophila by interacting with ecdysone and the juvenile hormone signaling pathways (23, 24). Together, Kr-h1 might thus have an important function in mediating neuronal plasticity and inducing the regulatory reorganization of the brain when M. pharaonis gynes differentiate into queens after insemination.
Because only rather few DEGs had become established on day 5, we inspected the GRNs obtained for gynes and queens across the different sampling time points to look for early signatures of gyne-queen reproductive role differentiation. On the basis of the 648 genes that were differentially expressed between gynes and queens of any age cohort, we constructed temporal coexpression networks, aiming to infer the gene-gene interaction dynamics for both unmated gynes and inseminated queens. We found that the gene-level interaction activity for gyne brain transcriptomes remained relatively stable from days 1 to 15 but had slightly increased on day 30 (two-sided t test; P < 1 × 10−2) (fig. S4). This suggests that unmated gynes only experience GRN rewiring after day 15, consistent with their insemination time window having closed by that time (9) and their backup somatic worker-like roles becoming activated. In contrast, the gene-level interaction activity in queens had significantly increased already on day 5, i.e., 2 days after mating (P < 1 × 10−3) and gradually decreased afterward (fig. S4).
The gene-by-gene interactions that showed the highest connectivity are likely to be the key genes in the GRN as we defined it. Among the 648 candidate genes, we identified a cluster of 191 genes with a very high degree of interconnected expression particularly in 5-day-old queens (Fig. 4A). This gene cluster included neuropeptides represented by tachykinin and genes associated with the crucial neurotransmitter acetylcholine, represented by a subunit of the nicotinic acetylcholine (ACh) receptor (nAChRα2) and the choline O-acetyltransferase (ChAT) (Fig. 4B). In Drosophila, tachykinin-related proteins are required for multiple functions like odorant-specific sensitivity, inhibition of locomotor activity, aggression, and regulating the insulin/insulin-like peptide production (25). In the ant Acromyrmex echinatior, the expression level of tachykinin is also positively correlated with aggression levels, but only in workers and hardly in gynes (26). The gyne-queen reproductive role differentiation in M. pharaonis revealed a higher tachykinin expression in unmated gynes than in inseminated queens on both days 15 and 30 (fig. S5), suggesting a similar regulatory role in gyne-queen behavioral differentiation.
Fig. 4. Network analysis documenting effects of insemination in M. pharaonis queens.

(A) Coexpression profiling of DEG connectivity associated with gyne-queen role differentiation, calculated as the proportion of coexpressed genes among all 648 DEGs, ranging from 0 (no coexpression with other genes) to 1 (coexpression with all other genes). The dashed rectangle highlights 191 genes exclusively and highly activated in queens 5 days after eclosion (2 days after insemination). (B) Transitional coexpression networks for brain transcriptomes of gynes (day 1) and gynes and inseminated queens (day 5). Connected gene pairs had significantly correlated expression levels (P < 1 × 10−3). The 191 high-connectivity genes in young queens (red dots) clearly cluster relative to the remaining DEGs (gray dots), but the gyne coexpression network has hardly changed. Three genes (blue, yellow, and green for tachykinin, ChAT, and nAChRα2, respectively) are highlighted, showing that they were all closely connected (coexpressed) in inseminated queens, while tachykinin was isolated in gynes (see fig. S4 for coexpression networks at other time points). The coexpression networks are visualized with a force-directed graph drawing algorithm where edges are assumed to be of equal length (63), so that genes with higher interconnections (coexpression coefficient) cluster together.
Both nAChRα2 and ChAT were queen-biased DEGs on days 15 and 30 (fig. S5 and Fig. 4B). The ACh system was the only neurotransmitter system with queen-biased expression, because other neurotransmitters and neuropeptides exhibited gyne-biased expression patterns (table S3). To infer the putative function of the ACh neurotransmitter system in queens on day 5, we analyzed the closely related genes nAChRα2 and ChAT in the queen coexpression network. This showed that nAChRα2 was highly associated with genes involved in processes for transcription (fd68A), translation (EF2 and eIF5), protein modification (tun), neuron projection (fry), and signal transduction (CdGaPr). Expression of nAChRα2 was also highly correlated with expression of Gr28b, a gene expressed in the peripheral and central (brain) neurons of Drosophila melanogaster, where it can act as a detector to make flies avoid high temperatures (27). We hypothesize that this subnetwork of genes with correlated expression may mediate the negative phototactic behavior of newly mated queens toward solar radiation (28). ChAT expression was also highly correlated with the expression of genes such as nrv3, Glycogenin, and tachykinin, suggesting a shared function in both peptidergic and acetylcholinergic neurons. Because nAChRα2 is crucial for learning and memory processes in the honeybee and the fruit fly and ChAT is crucial for neural functions and behavioral control (29), these results suggest that nAChRα2 and ChAT activate the GRN for gyne-queen role differentiation triggered by insemination.
Shared GRN modules for gyne-queen and sterile-fertile worker reproductive role differentiation
We compared the GRN for gyne-queen role differentiation in M. pharaonis with the GRN for gyne-worker caste differentiation that we previously obtained (13), focusing on the 223 genes that were differentially expressed with consistent direction of bias between gynes and queens on day 15 and/or day 30. Assuming that these genes are core members of the GRN that mediates gyne-queen reproductive role differentiation in M. pharaonis, it was notable that only 21 of them were differentially expressed when comparing gyne-worker caste differentiation (table S3). This fraction (9%) is not significantly different from the background expectation (7%, P > 0.19 for a two-sided binomial test), indicating that the gyne-queen role differentiation GRN of our present study is distinctly different from the GRN for gyne-worker caste differentiation (13). This is an intriguing result because it suggests that the two major differentiation processes in M. pharaonis, one in the adult stage triggered by insemination and the other in the egg stage and likely triggered by both genetic and epigenetic mechanisms, have completely different evolutionary origins. We therefore looked for published data specifically targeting reproductive role differentiation in other ants that could shed light on whether the M. pharaonis GRN for reproductive role differentiation after insemination was also used in other ants.
As explained in Introduction, reproductive role differentiation within the worker caste has independently evolved in several ant lineages after the queen caste lost significance or disappeared (30). To elucidate possibly conserved elements in the M. pharaonis GRN for gyne-queen role differentiation after insemination, we made explicit transcriptomic comparisons with three cases of reproductive role differentiation within the worker caste, assuming that conservation would imply that this GRN could mediate differentiation independent of adult caste phenotype. Of the 223 reproductive role differentiation–related genes in M. pharaonis, 170 had one-to-one orthologs in all three other ant species (see Materials and Methods). To examine overall network-level regulation, i.e., whether these genes interacted similarly across species, we used singular value decomposition (SVD) to extract the first PC axis from the total expression profiles of the 170 orthologs in M. pharaonis. This PC axis, which explained 50.04% of the variance, was then assumed to represent the major GRN dimension mediating gyne-queen reproductive role differentiation in this ant species where the queen caste is present, as is normally the case in ants. We then projected the expression profiles between reproductive and sterile workers of the three other ant species onto this M. pharaonis PC axis (Materials and Methods and Fig. 5A) to assess the extent of match or mismatch. This approach calculated a new PC score for each sample by taking the sum of products of gene expression levels and their contributions (PC loadings) to reproductive role differentiation in M. pharaonis.
Fig. 5. Expression comparisons for genes involved in gyne-queen role differentiation for M. pharaonis and for three species from other ant subfamilies that secondarily evolved reproductive workers.
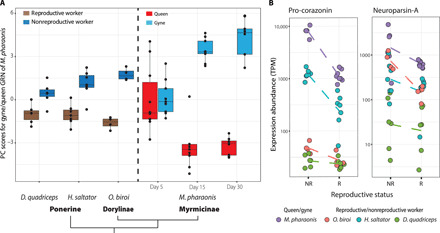
(A) Gene expression profiles (projected scores along the first PC axis) in the three ant species with reproductive role differentiation in the worker caste (left) and in the gyne caste for M. pharaonis differentiated by age (right) as illustrated in Fig. 1A. The reference PC axis for M. pharaonis was extracted from the combined data on days 15 and 30, using 170 DEGs with consistent differential expression on these 2 days and with one-to-one orthologs in all four ant species. The plotted values were the PC scores calculated from the expression profiles of these 170 genes, weighted by the gene-specific relative contributions to the overall gyne-queen DEG segregation in M. pharaonis. (B) Expression levels for two genes that were significantly differentially expressed between gynes and queens of M. pharaonis and exhibited the same direction of expression bias in nonreproductive (NR) versus reproductive (R) workers in the three other ant species. Dots represent the gene expression levels in each brain sample and solid lines connect the averages. Even shallow negative slopes represent considerable differences because expression abundances are plotted on a logarithmic scale. TPM, Transcripts Per Million.
We found that the extracted PC axis could always adequately discriminate between the nonreproductive and reproductive workers of H. saltator, D. quadriceps, and O. biroi, irrespective of the reproductive workers being gamergates (H. saltator and D. quadriceps) or clonal reproductive phase workers (O. biroi). Furthermore, the gyne samples of M. pharaonis were always clustering with the nonreproductive workers in the three other ant species, and the M. pharaonis queen samples were always grouping with reproductive workers in the other species (Fig. 5A). This result is consistent with reproductive role differentiation within the reproductive gyne/queen caste being based on conserved ancestral GRN modules that have been retained across the ant family Formicidae (Fig. 5A) and that may derive from an older hymenopteran GRN than the one mediating the origin of caste differentiation in ants (13).
The projected separation between reproductive and nonreproductive individuals was more distinct in O. biroi, a species belonging to the formicoid ants that also M. pharaonis belongs to, than in the two other ant species that belong to the poneroid sister clade (Fig. 5A). Of the 170 orthologs involved in gyne-queen reproductive role differentiation in M. pharaonis, 56 (33%) were also significantly differentially expressed between nonreproductive and reproductive workers in O. biroi, and they exhibited the same direction of bias across caste phenotypes as in M. pharaonis. This ratio is significantly higher than background expectation (9%, P < 1 × 10−10; one-sided binomial test). Despite the data for the other two ants being a less accurate match (e.g., sampling was not time differentiated in D. quadriceps), 60 (35%) of the genes exhibited the same direction of biased across M. pharaonis and the two other ant species, which is once more significantly larger than background expectation (21%, P < 1 × 10−5). These results suggest that the gyne-queen reproductive role differentiation GRN in ants that we obtained directly for M. pharaonis shares gene modules with all independently modified GRNs that allowed the establishment of evolutionarily derived social systems based on reproductive workers.
Focusing on specific genes, our analyses took care to avoid false negatives. Such spurious results can easily emerge because statistical type II error increases multiplicatively when multiple datasets are compared, so the number of overlapping DEGs across species would quickly decrease. To identify orthologs that have conserved functions, we therefore calculated t-scores for each ortholog’s fold change in expression (reproductive versus nonreproductive) across all four ant species (see Materials and Methods for details). Our analysis thus considered the quantitative patterns of biased expression between reproductive role phenotypes across species and assigned a high absolute t-score to genes with a similarly high same direction fold change and a low t-score to genes with low same direction gene expression bias or with opposite direction fold changes.
We identified 45 specific genes that were significantly associated with reproductive role differentiations across all four species (P < 0.05, two-sided t test) (fig. S6 and table S4). Among these genes, 13 were consistently biased toward the nonreproductive individuals across all four species. These included NPA that is involved in cell growth and associated with the insulin/insulin-like receptor and the juvenile hormone pathways in locusts (20, 31), chrb that acts as a growth inhibitor and interacts with the Target of Rapamycin (TOR) pathway in Drosophila (31, 32), and pro-corazonin–like (crz) that was earlier shown to participate in differentiation between the nonreproductive and inseminated workers (gamergates) in the ant H. saltator (16). The remaining 32 genes exhibited consistent expression bias toward reproductive individuals in at least three of the four examined species and were either involved in neuron and glia cell proliferation and differentiation processes represented by Tctp, RpS8, repo, and elF-4a or associated with important pathways such as Tctp, CycG, and CG18619 that take part in the insulin/insulin-like pathway, CycG and CG18619 of the TOR pathway, and CycG of the Notch signalling pathway (fig. S6). The expression of vitellogenin 2, a neuroendocrine target of corazonin known to regulate foraging and reproductive behaviors (16), and insulin-like peptide 2 (ilp2), known to regulate adult ovarian activation in ants (33), were both consistently biased toward the reproductive individuals across all four species (table S4). These similarities suggest that these genes and pathways have conserved roles in differentiating between nonreproductive and reproductive roles of adult females across the examined ant species.
Crz is expressed in the lateral neurosecretory cells and NPA in the medial neurosecretory cells
Crz and NPA were among the conserved orthologs associated with adult reproductive role differentiation across the ant species that we investigated, showing high cross-species consistency for directionality in expression fold change between reproductive and nonreproductive phenotypes (Fig. 5B). In particular, crz had elevated expression levels in foraging workers across multiple ant species (16) and obtained a gyne-biased expression in M. pharaonis from day 15 onward relative to expression in same age inseminated queens (fig. S5). This suggests that the expression of crz might be correlated with worker-like behavior of old uninseminated gynes (9). Similarly, NPA, which we already showed above may be central for reproductive role differentiation, showed a consistent gyne-biased expression pattern at all three time points (days 5, 15, and 30) in the brains of M. pharaonis (fig. S5).
To better understand how molecular spatial gene expression patterns correlate with neuroanatomical or physiological changes in the brain, we used the hybridization chain reaction (HCR) technique (34) to localize crz and NPA in the brains of gynes and queens of different ages in M. pharaonis. Although in situ HCR would be suitable to locate several transcripts simultaneously (26), we only aimed for two transcripts, crz and NPA, to avoid driving the method to its limits (Fig. 6, A to D). We focused on these two genes not only because of their physiological importance but also because crz and NPA proteins have been located by antibody staining across different insects (35, 36). We detected crz-positive signals in the protocerebral lateral neurosecretory cells (LNSCs or PL) (Fig. 6E), the same location as recorded in other insects (35). As LNSCs are rich in neuropeptides that take part in oogenesis and diuresis across different insect species (37), the observed location of crz expression appears to be consistent with a functional role in the gyne-queen role differentiation process. We found that the NPA-positive cell cluster was positioned anterior-medial, next to or surrounding the optic stalk of the medial OC (Fig. 6, C and D). This localization of NPA-positive cells in the medial neurosecretory cells (MNSCs) appears to be conserved across the insects, as has been shown by antibody staining across 18 different species of six orders (36). The MNSCs are equipped with a diverse neuropeptide repertoire including the insulin/insulin-like peptide (37). Removal and transplantation experiments of the MNSCs have shown their direct and essential impact on oogenesis, ovarian development, and development of fat bodies in female insects (37), although it remains to be tested whether crz and NPA act as neurohormones or activate signaling cascades downstream.
Fig. 6. Localization of Crz- and NPA-positive cell clusters in the brain of a 5-day-old gyne.
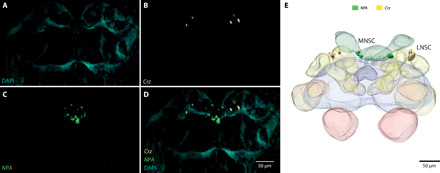
(A to D) Confocal microscopy image stacks for a gyne brain of M. pharaonis with (A) only nuclei staining [4′,6-diamidino-2-phenylindole (DAPI); blue], (B) mRNA in situ HCR probes of Crz-positive cells (yellow), (C) mRNA in situ HCR probes of NPA-positive cells (green), and (D) combined view after reducing all signals to a two-dimensional projection view. (E) Volume reconstructions of neuropils and crz-positive tissues enabled the localization of two mirrored crz-positive cell clusters between the medial and lateral calices and anterior to the respective hemispheres (LNSCs). The NPA-positive cells are part of the MNSCs surrounding the medial nerve fiber that connects the medial OC to the brain. No DAPI signal was detected in the LNSC or the MNSC, likely because the nuclei in these cell clusters are highly active. Scale bar, 50 μm.
DISCUSSION
Our study used several unique and evolutionary derived traits of ants to clarify how insemination induces changes in brain anatomy and behavior of queens relative to same age gynes that could be sampled simultaneously. This comparative approach to adult reproductive role differentiation was possible because gynes of our main model M. pharaonis do not need to disperse to be inseminated, an exceptional trait restricted to just a few ant species (38). This allowed us to combine analyses of behavior, brain anatomy, and brain transcriptomes to elucidate the mechanisms that drive reproductive role differentiation during the first month of adult life. After establishing that the GRN for gyne-queen role differentiation is fundamentally different from the conserved GRN mediating gyne-worker caste differentiation in ants (13), we reanalyzed transcriptomic data from three similarly idiosyncratic ant species where workers secondarily evolved to become fully functional reproductives that can replace the queen caste. This comparative analysis indicated that these ants used GRN modules that are derived from the same ancestral insemination-induced GRN that may have regulated reproductive role differentiation even before differentiated caste phenotypes evolved.
Insemination as female point of no return and its effect on brain functions
It is well known that the structure of a brain can be modified by influences from the social and physical environment even after brain development has been completed and that this process is normally accompanied by changes in memory and behavior (39). However, in comparison to the highly variable and largely unpredictable responses documented in these and other studies (39–42), the phenotypic changes in social insect queens following insemination are both profound and predictable. Queens are normally inseminated on a single day early in adult life, they will never re-mate again, and they will specialize on highly efficient reproductive functionality within a few weeks. The predictable adaptive syndrome of this reproductive role transition appears to be similarly fundamental as zygote differentiation toward the blastula stage under control of the biparental rather than the maternal genome. It is thus as expected that predictable changes in brain structure and function following sexual maturity and insemination have been reported in both honeybees and ants (28, 40). However, no previous study has covered the entire sequence of interconnected changes in neuroanatomy and brain gene expression induced by insemination.
Using the ant M. pharaonis as model species allowed us to investigate the transition from gyne to queen relative to a same age control group of gynes that remained uninseminated (Fig. 1A). A treatment control system like this would also apply in cooperative breeders without castes where females compete in dominance hierarchies that allow various degrees of reproductive monopolization and where subordinates become foragers and nurses. However, the obligate worker and queen castes in M. pharaonis have binary reproductive division of labor, so the gyne-queen reproductive role differentiation that we documented happens within the constraints of a specialized reproductive phenotype. This is very different from, for example, Polistes paper wasps where most if not all adult females remain reproductively totipotent so that insemination is associated with but not sufficient for reproductive status. Here, reproductive role differentiation remains plastic along a reproduction versus foraging and nursing axis (14, 43). Whether that differentiation is regulated by a brain GRN module remotely homologous to the one we found for ants remains to be investigated against the alternative hypothesis that premating dominance status induces ovary development with feedback via the brain to reinforce dominance, a causation pathway that is not expected in caste-differentiated ants. Our present analyses remained restricted to the ants, where we compared reproductive and nonreproductive individuals belonging to the same gyne or worker caste phenotype in each of the four species, spanning phylogenetic distances across the formicoid and poneroid ants. For brain anatomy, this produced the intriguing result that reproductive role bifurcation in M. pharaonis was consistent with changes in brain anatomy in the ponerine ant H. saltator where nonreproductive workers maintain their brain sizes, while the brains of inseminated gamergates become reduced (44).
Our brain transcriptome analyses revealed several neural genes that significantly changed their activity levels, but not necessarily in correlation with changes in brain volume, as for example, in the ACh neurotransmitter system where insemination had an almost immediate up-regulating effect at day 5 without a change in brain volume. As gynes are primarily adapted to be inseminated and proceed to develop full functionality as specialized egg layers, it is expected that the ACh system would mediate changes in synapses and neuropeptide or neurotransmitter levels to adjust information processing while, at the same time, terminating growth in brain volume now that phenotypic specialization is about to be completed. The NPA gene may be a marker of this stabilizing process, as its expression remained stable in queens while becoming up-regulated in gynes (fig. S5). This higher expression of NPA in gynes that remained uninseminated is associated with, and possibly causal to, continuing growth in brain volume. The higher levels of NPA transcripts in the MNSCs of gyne brains may also be responsible for inhibition of ovarian and egg development, as the experimental removal of these cells reduced oogenesis in other insect species [e.g., (37)]. The spatial separation of differential expression of the NPA and crz genes (see the next section) is consistent with distinctly different functions of the medial (MNSC) and lateral (LNSC) neurosecretory cells, with the former likely facilitating changes in ovarian and fat body physiology and the latter regulating the expression of worker-like behaviors. Thus, while ovary development may start as part of the normal maturation of adult gynes, insemination appears to establish a directional brain regulation network to modify ovary maturation toward full functionality.
Control over the time factor during our gyne-queen role differentiation experiment in M. pharaonis allowed us to investigate the expression dynamics of the DEGs even before most of them became significantly biased toward distinct overexpression in either unmated gynes or inseminated queens. This enabled us to putatively reconstruct how the GRN started with a limited number of highly coexpressed genes almost immediately after insemination (Fig. 4), so that a much more massive increase of DEGs could follow in the next 10 days. The identity of the initial DEGs suggests the unfolding of a highly coordinated network of molecular and cellular processes, which then cascade into recruiting many more DEGs to finally result in the distinct morphological, physiological, and behavioral syndromes typical for the functional queen phenotype. At the same time, the uninseminated gyne phenotype became rewired to provide useful somatic helper functions as far as its caste morphology and limited expected life span allowed. Uninseminated gynes behaving in a worker-like manner have been observed in several ant species (12, 26), but they remain severely understudied because almost no other ants would allow the controlled experiment that we did with M. pharaonis as gynes will normally embark on a dispersal flight to be inseminated and would thus no longer be available for comparative study.
Apart from consistent neuroanatomical and transcriptomic evidence for the operational significance and physical location of a brain GRN for role differentiation in the adult reproductive caste of M. pharaonis, our study also revealed some direct interactions between insemination and brain function at the specific gene level. In situ HCR staining showed that the crz gene is highly expressed in the central brain of M. pharaonis gynes and queens (Fig. 6). Injection of this protein in the brain is known to induce worker-like behavior and to suppress the expression of vitellogenin, which is required for reproductive functionality in an ant species where workers can become full reproductives (16) as we will elaborate in the next section. Crz thus appears to be important in regulating somatic helping behavior irrespective of whether we compared gyne/queen phenotypes with or without insemination or worker phenotypes with or without insemination (16). In either case, the expression of the crz gene became suppressed shortly after insemination to be maintained at low expression levels throughout further development of full reproductive potential. In contrast, crz expression continued to increase with age when insemination did not take place, consistent with older M. pharaonis gynes displaying worker-like behaviors. As we showed, also other genes are likely to be tightly associated with the expression of worker-like behaviors, as, for example, tachykinin (26).
Co-option of similar GRN modules for reproductive role differentiation in different ant lineages
Caste differentiation before pupal eclosion, resulting in permanent reproductive division of labor between adult gynes/queens and workers, appears to have originated only once in the last common ancestor of all ants and independently in the ancestors of the corbiculate bees and the vespine wasps. However, the workers of most lineages of social Hymenoptera have retained the ability to lay unfertilized eggs. M. pharaonis is exceptional in its workers having lost not only their sperm storage organ (spermatheca) but also their ovaries. At the other end of the scale, there are several lineages of poneroid ants that are exceptional in workers having retained both their ovaries and a sperm storage organ, although the latter is vestigial (45). It is in these lineages that selection could weaken or eliminate the queen caste when workers regained full reproductive potency, albeit with a smaller body size and lower fertility than the ancestral queen caste (46). We found two transcriptomic datasets from these ant lineages to test whether the ancestral M. pharaonis GRN for adult reproductive role differentiation has also been used in other ant lineages. A third test case of available transcriptome data concerned a special species of formicoid ants where workers do not have a spermatheca, but where parthenogenesis enabled cyclical reproduction by workers when the queen caste was lost. Although these three additional datasets were not collected for the type of test that we performed, and thus lacked the resolution of our M. pharaonis data, the match across two formicoid and two poneroid ants that we obtained for the role differentiation GRN was remarkable and encouraging.
In a previous study to elucidate the conserved GRN for gyne-worker caste differentiation, we primarily compared the brain transcriptomes of five formicoid ant species to establish the conservation of this GRN, after which we secondarily projected transcriptomic data from the literature to evaluate matches within the same gamergate and parthenogenetic ant as we used in the present study (13). These comparisons showed that the conserved GRN for gyne-worker caste differentiation during immature development was not used to mediate adult role differentiation between reproductive and nonreproductive workers—very few genes belonging to the GRN for gyne-worker caste differentiation had differential gene expression between reproductive and nonreproductive workers in colonies without a queen. This suggested that these scattered cases of secondarily evolved reproductive workers might have different, evolutionarily derived GRNs for mediating reproductive role differentiation among workers. Alternatively, these lineages might have co-opted elements of an existing GRN for reproductive role differentiation that is normally not expressed in worker phenotypes (6). Despite the heterogeneous nature of the transcriptome data, our comparison of reproductive role differentiation in M. pharaonis, the clonal formicoid ant (O. biroi), and in the two poneroid gamergate species (H. saltator and D. quadriceps) confirmed the latter hypothesis (Fig. 5A).
We lastly pursued a very first exploration of possible matches at the level of specific genes (table S4 and fig. S6). This revealed that corazonin and neuroparsin A, expressed in the anterior neurosecretory cells, were consistently overexpressed in individuals with “somatic” roles across all four investigated ant species, irrespective of their morphological caste phenotypes. On the basis of our present results and the ones obtained by Qiu et al. (13), it thus seems reasonable to conjecture that the GRNs for gyne-worker caste differentiation during immature development in the ants, derived corbiculate bees, and vespine wasps will be different both from each other and from the GRN for adult role differentiation that may go back to an ancestrally shared Hymenopteran version. Future studies testing this conjecture will also have bearing on the regulatory role of ovarian development per se but, as already highlighted above, without an expectation that lineage-specific GRNs for superorganismal gyne-worker caste differentiation are necessarily involved. Irreversible caste differentiation early in larval or embryonic development is expected to induce large qualitative differences between gynes and workers in overall morphology and ovariole number, while reproductive role differentiation within a single gyne (default) or worker caste (rare exceptions) is likely to have been ancestrally triggered by mere female insemination to realize full reproductive potential. If the adult reproductive role differentiation GRN would turn out to be older than the ant-specific GRN for gyne-worker caste differentiation, then we expect it to contain both highly conserved and ant-specific elements and to provide a rich platform for further research.
MATERIAL AND METHODS
Experimental ants and the creation of comparable gyne and queen cohorts
We used M. pharaonis as ant model system. The mature source colonies were established at the Copenhagen Center for Social Evolution since 2004, kept in a climate chamber at 26° ± 2°C and 50% relative humidity, and fed three times a week with frozen crickets, artificial protein-carbohydrate diet, and water provided ad libitum [see (47) for details on origin and history of the laboratory stock colonies]. From a stock of four closely related colonies, we created queenless colonies (Q− colonies) of similar size with brood and workers from all four main colonies, which produced gynes after 31 to 40 days. We daily collected newly eclosed gynes and isolated them with workers and worker pupae in smaller subcolonies to maintain eclosion day–specific cohorts of known age with an accuracy of ±12 hours as we collected newly hatched gynes once a day. Recognizable male pupae were removed daily from all Q− colonies to prevent uncontrolled mating of newly eclosed gynes. These pupae were allowed to hatch in separate subcolonies with workers, to obtain a stock for the later insemination trials. Some of the newly eclosed gynes of known age (day 1 gynes) were collected for parallel analysis of brain neuroanatomy and brain transcriptomes (14 replicates for neuroanatomy and 9 for brain transcriptomes).
Experimental design and specifications of the gyne and queen cohorts
Brain gene expression patterns and volumetric changes in the brain neuropils were compared across four time points throughout the maturation process (Fig. 1). The subcolonies with known-age gynes were randomly assigned to the mating and nonmating groups, which created the inseminated queen lines and unmated gyne control lines. Mating took place by moving gynes to “mating arena” petri dishes and exposing them to randomly collected males in a male-to-gyne ratio of 4:1. After 24 hours, the newly mated queens were reintroduced into their respective subcolonies. For further data acquisition of queens, we only used individuals that had shed their wings, which is a reliable marker of insemination. On day 4, all queens and gynes entering our further monitoring lines were CO2-anesthetized using a Flowbuddy Flow Regulator and Flypad (Genesee Scientific, USA) to synchronize their physiological status, after which we returned them to their respective age cohort–specific subcolonies. To investigate the effects of mating on the neuroanatomy and transcriptomes of brains, we started sampling subcolonies after 24-hour recovery time of gynes/queens in these subcolonies, i.e., on day 5 after eclosion (anatomy: 8 gynes, 7 queens; transcriptomes: 9 gynes, 10 queens). Because earlier studies have shown that both mated and unmated queens of M. pharaonis start egg laying 9 to 13 days after eclosion depending on the presence/absence of workers and brood (9), we collected our next set of samples 15 days after eclosion (12 days after insemination or not), ensuring that egg laying had started (anatomy: 9 gynes, 10 queens; transcriptomes: 9 gynes, 9 queens). Tissue degradation in the thorax and the abdomen and a lower receptiveness of unmated gynes to males have been documented 26 to 28 days after eclosion (9). Petersen-Braun (9) observed stagnation of egg laying among groups of unmated gynes, where each group contained five gynes with supporting workers and where the number of eggs per experimental group stopped increasing between days 16 and 20 after gyne eclosion. Taking these earlier histological and behavioral observations into account, we assumed that 30-day-old gynes would no longer be able to switch to reproductive functionality, an assumption that we checked in a separate experiment. We compared same age queens (ca. 35 days after eclosion) that were inseminated on day 4 (early mated queens) and day 30 (late mated queens) and counted the number of eggs laid after 24-hour isolation of single queens with ca. 10 workers each. We found that early mated queens laid ca. four eggs on average (n = 113), while late mated queens laid less than one egg on average (n = 56), a difference that was significant (P < 0.001, two-sided t test). This confirmed that old gynes have lost full reproductive potential. We therefore collected our final samples of brains for neuroanatomy and transcriptomics on day 30 after eclosion (anatomy: 8 gynes, 8 queens; transcriptomes: 6 gynes, 8 queens), at a point in developmental time at which queen egg laying was firmly established and gynes had become maximally similar to sterile workers.
Behavioral assays with 30-day-old gynes and queens
The extent of behavioral differentiation between adult gynes and queens depends on both the social (colony) environment and their age (9). We used 30-day-old unmated gynes and inseminated queens to verify these published results and to quantify general differences in activity, foraging behavior, and brood care. For each of these behavioral categories, we transferred 10 unlabeled individuals into a test arena, connected to a nest chamber with eggs and young larvae but no workers, and obtained 10 behavioral assay replicates for gynes and queens, respectively. After acclimatization for 1 hour, we added five larvae to monitor brood care behavior, and after an additional acclimatization hour, we also added food while continuing to record behaviors with video cameras for 3 hours. We calculated averages of gyne and queen behaviors per replicate trial by only considering time intervals during which at least 1 of 10 individuals was actively moving around in the arena and we always divided scores by the number of individuals (n = 10) to obtain average activity records per individual. To separate foraging and brood care activity from general activity, we recorded the number of encounters of an individual gyne or queen with either food items or larvae; also, these scores were divided by 10. We analyzed the data by independent sample t tests using IBM SPSS statistics 24.
Quantification of brain neuropil volumes
Our brain anatomy data of the inseminated queen cohort and the gyne control group allowed direct comparisons between neuronal structures and patterns of differential gene expression. At the same time, our experimental design allowed us to separate effects of insemination/nonmating and age, both known or suspected to affect histology, physiology, and behavior in M. pharaonis gynes and queens (9, 19, 48). The volumetric analyses comprised the major brain compartments shared by all insects: The ALs being anterior ventrally located in the brain and characterized by internal glomerular structures (49). The OLs being found laterally to the rest of the brain and extending from the compound eyes toward the central brain (49). The three OCs [simple photoreceptive organs (50, 51)] situated on the anterior part of the head capsule. The paired MBs located posterior dorsally to the rest of the brain, and the SPZ comprising the brain tissue above the level of the esophagus (49). Confocal image stacks were used to reconstruct the brain neuropils to assess changes in their volumes, which were obtained from whole head preparations to preserve the orientation of the brain in the head capsule. The preparation and imaging of these whole head samples were adapted from Smolla et al. (52) and imaged using a 20× objective (HCX PL APO CS 20.0x0.70 DRY UV) on a Leica SP5-X confocal laser scanning microscope and matching software (Leica, Germany). We thus reconstructed entire brains using volume information from AMIRA 6.2.0 (Mercury Computer Systems, Germany). Addressing maturation effects within and mating effects across the subsequent samples of gynes and queens on the absolute volume of the brain and the single neuropils, we used a two- or three-way ANOVA setup in addition to simple t test comparisons. The statistical analyses were conducted in R (53).
Sampling, RNA extraction, cDNA libraries, and RNA sequencing
Brains of individual samples were used to quantify transcriptome abundances of queens and gynes across age groups. We collected six to nine gyne and queen individuals from two colonies across all age categories and dissected the individual brains with the following procedure: (i) The head was removed and immersed in ice-cold 0.1 M phosphate-buffered saline (PBS)–diethyl pyrocarbonate (DEPC); (ii) the cuticle, muscles, and glands were removed from the brain tissue using a razor blade and forceps; and (iii) the whole brains were transferred into RNAlater (Invitrogen, Thermo Fisher Scientific, USA) and kept at 4°C overnight before transfer to a −24°C freezer for storage. Total RNA for each individual brain was extracted using a RNeasy Plus Micro Kit following the manufacturer’s instruction (catalog number 74034, Qiagen, Germany), and all RNA samples were stored at −80°C for maximally 1 month. RNA quality testing, library construction, and sequencing were performed at BGI, Shenzhen, China. The quality of RNA samples was tested with the Agilent 2100 Bioanalyzer, and Ambion ERCC RNA Spike-In Mix (catalog number 4456740) was added to each sample according to the manufacturer’s instructions before cDNA library construction. Extracted RNA was first reversed transcribed into cDNA following the Smart-seq2 protocol (54), which was then randomly fragmented, after which we added Tn5 enzymes to link with the sequencing adapter and obtain a complete cDNA library. Primers were then added to the cDNA libraries for polymerase chain reaction amplification, which gave us fragments of which those ranging from 150 to 350 bp were selected for further cDNA circularization to construct sequencing libraries. The samples were then sequenced on a BGISEQ-500 platform using a 100-nt paired-end sequencing protocol.
RNA sequencing quantification, quality control, and normalization of gene expression scores across samples
RNA expression levels were quantified using Salmon (v0.11.0) (55). We first trimmed adaptor sequences and filtered sequences with low read length from the RNA sequencing (RNA-seq) data using SOAPnuke (56). The filtered RNA-seq data were then quasi-mapped to an available brain transcriptome of M. pharaonis [in-house annotated; (13)] together with External RNA Controls Consortium (ERCC) spike-in sequences, using the default options of Salmon except that we turned on the bias-correction options to control for guanine-cytosine content bias and sequence-specific bias. RNA-seq quality was examined on the basis of transcriptome abundance similarity and ERCC spike-in abundance similarity with other samples. Pairwise inter-individual Spearman’s correlation coefficients for transcriptomes and ERCC spike-in abundances across all 64 samples were always higher than 0.9, but four showed reduced expression similarity and distinctly lower expression distribution similarity compared to all other samples. These four samples came from a single sequencing library, so we removed them as outliers from the downstream analyses—which thus worked with a data matrix where all Spearman’s correlation coefficients were above 0.92. Salmon output transcripts per million base-pairs values were used as raw transcriptome abundance data. They were log transformed [after adding a 1 × 10−5 pseudo count to avoid log(0)] and quantile normalized among samples to ensure that the overall distributions of gene expression levels remained the same across samples, which minimized the effects of any putative technical artifacts (57).
Identifying genes associated with gyne-queen reproductive role differentiation after insemination
DEGs between gynes and queens for each age group were identified with DESeq2 (58), using the raw count output of mapped reads from Salmon as input. Samples of the same age were first treated as subsets and then tested for DEGs using the generalized linear model
This model tested the effect of mating on gene expression levels while also taking any additive colony effects into account. Genes with adjusted P values < 5 × 10−2 [after false discovery rate (FDR) correction] were identified as DEGs between gynes and queens for each targeted age group. Because genes with low expression levels might suffer from high false-positive detection rates as DEGs, we removed genes whose mapped reads were less than five for over 60% of the individuals in the targeted age group. To identify genes associated with gyne-queen reproductive role differentiation on both days 15 and 30, we first identified 238 genes that were differentially expressed (using adjusted P values < 1 × 10−2) on either days 15 or 30 to obtain a comprehensive set of genes associated with reproductive role differentiation. We then filtered out some genes that exhibited different directions of differential expression in the two age groups and retained 223 genes that exhibited same direction expression bias on both days 15 and 30.
Ranking genes for their association with gyne or queen status or different age
To identify top ranking genes associated with gyne or queen phenotypes or developmental age, we calculated the partial correlation coefficient between each gene’s expression level (using quantile normalized transcriptome abundances) and the predictor variable of interest (gyne/queen status or age) for all samples while controlling for the other predictor variable. Calculations were performed with the Ppcor R package (59), using Spearman’s rank correlations to capture both linear and nonlinear associations. Genes were ranked on the basis of their significance (adjusted P values) for either gyne/queen status or age as predictor variable.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations for genes in the M. pharaonis genome were performed by using the annotation information for homologous genes between M. pharaonis and D. melanogaster, following the same procedure as in a previous study (13). To identify the potential functions for genes of interest (e.g., genes up-/down-regulated in gynes or queens), candidate genes were tested against a background gene set (genes with >3 read counts for at least five samples) using ClusterProfiler in R to examine their functional enrichment for both GO and KEGG gene sets (60). Gene sets were identified as being significantly enriched when FDRs were <0.05 in two-sided tests.
Network analyses for identifying the regulatory network for gyne-queen reproductive role differentiation
GRNs were constructed independently for each age group of gynes and queens, based on the coexpression correlation matrix. We first identified genes associated with reproductive role differentiation by finding the genes that were differentially expressed in queens and gynes (adjusted P value < 5 × 10−2) for at least one age group (day 5, 15, or 30) to obtain a gross GRN that included 648 genes. We then subdivided the quantile normalized expression levels by age group for both gynes and queens and filtered out genes without expression variation (in any of the subset group) that could thus not be used for calculating coexpression levels. This retained 629 genes for which we calculated the pair-wise Spearman’s correlation coefficients among their pair-wise associations in expression. For each age group of gyne and queen individuals, these association scores between gene pairs were expressed as absolute values of correlation coefficients, to capture both positive and negative gene regulations, ranging from 1 (for 100% associated expression) to 0 (for the absence of association). We then replaced all nonsignificant association scores (P value > 0.05) by zeros in the matrix to remove noise and focus on the positive evidence for membership of the age group–specific GRN for gynes and queens.
Last, we identified hub genes for the entire developmental trajectory of gynes and queens across the age groups by evaluating the mean association scores for each gene with all other 628 genes, obtaining once more a matrix with association scores ranging from 1 to 0, but this time referring to genes’ connectivity (mean association score) within corresponding age phenotype–specific GRNs.
Identifying orthologous genes and quantifying brain transcriptomes in ant species with reproductive workers
Orthologous genes among M. pharaonis and the three other ants that have colonies without queens in which workers reproduce were based on reannotated genomes of these species. These were obtained in a previous study (13), where one-to-one orthologs were identified with reciprocal BLASTP combined with genome-wide synteny information [see (13) for details]. To examine the gene expression patterns associated with reproductive role differentiations of adult workers in these ant species with queenless colonies, we downloaded the previously published RNA-seq data of O. biroi, H. saltator, and D. quadriceps from National Center for Biotechnology Information (14–16). To reduce bias in transcriptome comparisons related to age effects, we used the reproductive workers and the brood nursing worker samples from day 12 in the O. biroi dataset, which is an age relatively close to our M. pharaonis samples on days 5, 15, and 30. For the gamergate and sterile worker samples in the H. saltator dataset, we had to work with older age groups of 120 days, and for the D. quadriceps dataset, we did not know age because the ant samples were collected from wild colonies. Transcriptome quantification and DEG detection for each of the ant species with queenless colonies were performed with the same procedure as in M. pharaonis, using the reannotated genomes for each of these species (13).
Comparative analysis of the role differentiation GRN in reproductive and sterile workers
Transcriptomes for one-to-one orthologs (n = 7776) were normalized across species, following the same procedure as in a previous study (13). Briefly, transcriptomes for each species were first quantile normalized to account for technical biased within datasets and then normalized across species with Combat (61) using species identity as a batch covariate and setting the mean.only option as “false.” This normalization method removed all unwanted interspecies differences in gene expression profiles, so final analyses could be based on means and variances of total expression levels being the same across species (13). The remaining residuals for individual genes thus came to reflect sample differences within species so that individuals with similar residuals now represented gene expression values with similar DEG scores [see (13) for details].
We focused on 170 genes that were both associated with the gyne-queen reproductive role differentiation in M. pharaonis and had one-to-one orthologs across the three ant species with queenless colonies. We used SVD to extract the first PC from the normalized expression profiles of these 170 genes using the combined days 15 and 30 datasets of M. pharaonis. The coefficients along the first PC axis were assumed to represent the reconstructed GRN for gyne-queen reproductive role differentiation. We then calculated the PC scores for the available samples of each of the ant species with queenless colonies by multiplying the coefficients of the first M. pharaonis PC with the normalized expression profiles for each of the samples of the other three ant species, to obtain an estimate of overall expression similarity between these ant species with queenless colonies and the gyne and queen samples of M. pharaonis (13). For example, samples of reproductive workers in an ant species with queenless colonies were expected to have a similar PC score as the queen samples of M. pharaonis because both should have an overall expression profile biased to reproductive functionality.
To identify genes with conserved roles in reproductive role differentiation between nonreproductive and reproductive adults, we used the log2 fold changes between reproductive and nonreproductive phenotypes for the expression levels of the 170 target genes in M. pharaonis at days 15 and 30 and the comparable samples of the three ant species with queenless colonies. To avoid overall fold change levels being biased by one or two species, we set the upper limit for the absolute value of 2log fold change to 1. We then performed two-sided t tests for each gene using their 2log fold changes expression levels in all four species. This procedure tested whether the overall means of gene-specific fold changes across the four species were significantly different from 0. Thus, genes with high absolute t-score values (i.e., low P values) were inferred to have overall similar expression biases across species, consistent with the hypothesized conserved GRN for adult role differentiation between sterile foragers and fertile egg layers to be operational in a similar way as for gyne-queen role differentiation in M. pharaonis. The null hypothesis of no correspondence between the gene expression profiles across the four ant species would produce absolute t-scores of around 0, i.e., high and nonsignificant P values, arising from either different directions of expression fold change, i.e., opposite across different species, or small and insignificant absolute values of expression fold change in the same direction.
In situ HCR
Spatial information about where DEGs are expressed is important to understand their impact on neuroanatomical and behavioral plasticity. We used the in situ HCR (34) to obtain whole brain in situ hybridizations for NPA and crz, two genes of major interest based on our transcriptomic results. We opened the head capsule from the posterior side to allow reagents to penetrate from the opposite site of where we expected the NPA- and crz-positive cells (35, 36). The brains were then partially dissected under 4% para-formaldehyde (PFA) in DEPC-treated 0.1 M PBS and incubated for 1 hour in glass vials on ice. After fixation, the brains were washed five times for 10 min in 0.2% Triton X-100 in PBS-DEPC, after which we used an HCR Kit for NPA and crz in M. pharaonis provided by Molecular Instruments (www.molecularinstruments.com). We used Alexa Fluor 488 for the detection of NPA and Alexa Fluor 546 for detecting crz and otherwise strictly followed the protocol for HCR in Drosophila embryos available at the company’s web site and publication (34). We added 4′,6-diamidino-2-phenylindole (DAPI) (1:500) to 5× SSC-Tx and 0.2% Tx-PBS-DEPC for nucleus staining in the final washings. The whole head preparations were imaged similar to the procedure used for the volume assessments. We recorded the signals for DAPI (excitation, 405 nm), NPA (excitation, 488 nm), and crz (excitation, 546 nm) and processed images using Fiji/ImageJ, after which we obtained the volume information using AMIRA 6.2.0 (Mercury Computer Systems, Germany).
Supplementary Material
Acknowledgments
We thank the Center for Advanced Bioimaging (CAB) Denmark at the University of Copenhagen for support and access to the technical equipment needed to collect our imaging data, L. Pontieri and D. R. Nash for discussion on statistics, and E. Abouheif and S. Kocher for constructive discussion of a previous version of the manuscript. Funding: This research was supported by a grant from the Lundbeck Foundation (R190-2014-2827) to G.Z. Author contributions: G.Z. directed the research. G.Z., M.N., and L.E.B. conceived and designed the research. L.E.B. and M.N. carried out the research and participated in all experiments, except for the behavioral assays, which were performed by R.S.L. and D.N. B.Q. performed the transcriptome analyses and analyzed the neuroanatomy data together with M.N. M.N., B.Q., J.J.B., and G.Z. discussed the results and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in this paper are present in the paper and/or the Supplementary Materials. The RNA-Seq data have been deposited under BioProject accession number PRJNA564022 and in the CNGB Nucleotide Sequence Archive (CNSA) with accession number CNP0001177. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eaaz5772/DC1
REFERENCES AND NOTES
- 1.W. M. Wheeler, Ants: Their Structure, Development and Behavior (Columbia Univ. Press, 1910), vol. 9. [Google Scholar]
- 2.Hughes W. O. H., Oldroyd B. P., Beekman M., Ratnieks F. L. W., Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science 320, 1213–1216 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Fisher R. M., Cornwallis C. K., West S. A., Group formation, relatedness, and the evolution of multicellularity. Curr. Biol. 23, 1120–1125 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Alvarado S., Rajakumar R., Abouheif E., Szyf M., Epigenetic variation in the Egfr gene generates quantitative variation in a complex trait in ants. Nat. Commun. 6, 6513 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Johnson B. R., Linksvayer T. A., Deconstructing the superorganism: Social physiology, groundplans, and sociogenomics. Q. Rev. Biol. 85, 57–79 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Boomsma J. J., Gawne R., Superorganismality and caste differentiation as points of no return: How the major evolutionary transitions were lost in translation. Biol. Rev. Camb. Philos. Soc. 93, 28–54 (2018). [DOI] [PubMed] [Google Scholar]
- 7.G. F. Oster, E. O. Wilson, Caste and Ecology in the Social Insects (Princeton Univ. Press, 1978). [PubMed] [Google Scholar]
- 8.Lee M. T., Bonneau A. R., Giraldez A. J., Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30, 581–613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. D. Petersen-Braun, Untersuchungen zur Socialen Organisation der Pharaoameise Monomorium pharaonis (L.) (Hymenoptera, Formicidae), thesis, Rheinischen Friedrich-Wilhelms-Universität, Hohen Mathematish-Naturwissenschaftlichen Fakultät (1973). [Google Scholar]
- 10.Schmidt A. M., d’Ettorre P., Pedersen J. S., Low levels of nestmate discrimination despite high genetic differentiation in the invasive pharaoh ant. Front. Zool. 7, 20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khila A., Abouheif E., Evaluating the role of reproductive constraints in ant social evolution. Philos. Trans. R. Soc. B Biol. Sci. 365, 617–630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nehring V., Boomsma J. J., D’Ettorre P., Wingless virgin queens assume helper roles in Acromyrmex leaf-cutting ants. Curr. Biol. 22, R671–R673 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Qiu B., Larsen R. S., Chang N.-C., Wang J., Boomsma J. J., Zhang G., Towards reconstructing the ancestral brain gene-network regulating caste differentiation in ants. Nat. Ecol. Evol. 2, 1782–1791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patalano S., Vlasova A., Wyatt C., Ewels P., Camara F., Ferreira P. G., Asher C. L., Jurkowski T. P., Segonds-Pichon A., Bachman M., González-Navarrete I., Minoche A. E., Krueger F., Lowy E., Marcet-Houben M., Rodriguez-Ales J. L., Nascimento F. S., Balasubramanian S., Gabaldon T., Tarver J. E., Andrews S., Himmelbauer H., Hughes W. O. H., Guigó R., Reik W., Sumner S., Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc. Natl. Acad. Sci. U.S.A. 112, 13970–13975 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Libbrecht R., Oxley P. R., Kronauer D. J. C., Clonal raider ant brain transcriptomics identifies candidate molecular mechanisms for reproductive division of labor. BMC Biol. 16, 89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gospocic J., Shields E. J., Glastad K. M., Lin Y., Penick C. A., Yan H., Mikheyev A. S., Linksvayer T. A., Garcia B. A., Berger S. L., Liebig J., Reinberg D., Bonasio R., The neuropeptide corazonin controls social behavior and caste identity in ants. Cell 170, 748–759.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peeters C., Liebig J., Hölldobler B., Sexual reproduction by both queens and workers in the ponerine ant Harpegnathos saltator. Insectes Soc. 47, 325–332 (2000). [Google Scholar]
- 18.Tsuji K., Yamauchi K., Production of females by parthenogenesis in the ant, Cerapachys biroi. Insectes Soc. 42, 333–336 (1995). [Google Scholar]
- 19.Petersen-Braun M., Untersuchungen zur Sozialen Organisation der Pharaoameise Monomorium pharaonis (L.) (Hymenoptera, Formicidae) I. Der Brutzyklus und seine Steuerung durch Populationseigene Faktoren. Insectes Soc. 22, 269–291 (1975). [Google Scholar]
- 20.Vanhems E., Delbos M., Girardie J., Insulin and neuroparsin promote neurite outgrowth in cultured Locust CNS. Eur. J. Neurosci. 2, 776–782 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Salzberg A., Golden K., Bodmer R., Bellen H. J., gutfeeling, a Drosophila gene encoding an antizyme-like protein, is required for late differentiation of neurons and muscles. Genetics 144, 183–196 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butchar J. P., Cain D., Manivannan S. N., McCue A. D., Bonanno L., Halula S., Truesdell S., Austin C. L., Jacobsen T. L., Simcox A., New negative feedback regulators of Egfr signaling in Drosophila. Genetics 191, 1213–1226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grozinger C. M., Robinson G. E., Endocrine modulation of a pheromone-responsive gene in the honey bee brain. J. Comp. Physiol. A 193, 461–470 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Minakuchi C., Zhou X., Riddiford L. M., Krüppel homolog 1 (Kr-h1) mediates juvenile hormone action during metamorphosis of Drosophila melanogaster. Mech. Dev. 125, 91–105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoopfer E. D., Neural control of aggression in Drosophila. Curr. Opin. Neurobiol. 38, 109–118 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Howe J., Schiøtt M., Boomsma J. J., Tachykinin expression levels correlate with caste-specific aggression in workers of the leaf-cutting ant Acromyrmex echinatior. Front. Ecol. Evol. 4, 55 (2016). [Google Scholar]
- 27.Ni L., Bronk P., Chang E. C., Lowell A. M., Flam J. O., Panzano V. C., Theobald D. L., Griffith L. C., Garrity P. A., A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature 500, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julian G. E., Gronenberg W., Reduction of brain volume correlates with behavioral changes in queen ants. Brain Behav. Evol. 60, 152–164 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Dupuis J., Louis T., Gauthier M., Raymond V., Insights from honeybee (Apis mellifera) and fly (Drosophila melanogaster) nicotinic acetylcholine receptors: From genes to behavioral functions. Neurosci. Biobehav. Rev. 36, 1553–1564 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Schmidt C., Molecular phylogenetics of ponerine ants (Hymenoptera: Formicidae: Ponerinae). Zootaxa 3647, 201–250 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Girardie J., Boureme D., Couillaud F., Tamarelle M., Girardie A., Anti-juvenile effect of neuroparsin A, a neuroprotein isolated from locust corpora cardiaca. Insect Biochem. 17, 977–983 (1987). [Google Scholar]
- 32.Reiling J. H., Hafen E., The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 18, 2879–2892 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandra V., Fetter-Pruneda I., Oxley P. R., Ritger A. L., McKenzie S. K., Libbrecht R., Kronauer D. J. C., Social regulation of insulin signaling and the evolution of eusociality in ants. Science 361, 398–402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi H. M. T., Schwarzkopf M., Fornace M. E., Acharya A., Artavanis G., Stegmaier J., Cunha A., Pierce N. A., Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roller L., Tanaka Y., Tanaka S., Corazonin and corazonin-like substances in the central nervous system of the Pterygote and Apterygote insects. Cell Tissue Res. 312, 393–406 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Tamarelle M., Girardie J., Immunohistochemical investigation of locust neuroparsin-like substances in several insects, in some other invertebrates, and vertebrates. Histochemistry 91, 431–435 (1989). [DOI] [PubMed] [Google Scholar]
- 37.Shiga S., Anatomy and functions of brain neurosecretory cells in Diptera. Microsc. Res. Tech. 62, 114–131 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Hakala S. M., Perttu S., Helanterä H., Evolution of dispersal in ants (Hymenoptera: Formicidae): A review on the dispersal strategies of sessile superorganisms. Myrmecol. News. 29, 35–55 (2019). [Google Scholar]
- 39.Groh C., Meinertzhagen I. A., Brain plasticity in diptera and hymenoptera. Front. Biosci. (Schol. Ed.) 2, 268–288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groh C., Ahrens D., Rossler W., Environment- and age-dependent plasticity of synaptic complexes in the mushroom bodies of honeybee queens. Brain Behav. Evol. 68, 1–14 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Groh C., Röessler W., Comparison of microglomerular structures in the mushroom body calyx of neopteran insects. Arthropod Struct. Dev. 40, 358–367 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Kolb B., Whishaw I. Q., Brain plasticity and behavior. Annu. Rev. Psychol. 49, 43–64 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Toth A. L., Robinson G. E., Evo-devo and the evolution of social behavior. Trends Genet. 23, 334–341 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Gronenberg W., Liebig J., Smaller brains and optic lobes in reproductive workers of the ant Harpegnathos. Naturwissenschaften 86, 343–345 (1999). [Google Scholar]
- 45.Gobin B., Ito F., Billen J., Peeters C., Degeneration of sperm reservoir and the loss of mating ability in worker ants. Naturwissenschaften 95, 1041–1048 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Peeters C., Convergent evolution of wingless reproductives across all subfamilies of ants, and sporadic loss of winged queens (Hymenoptera: Formicidae). Myrmecol. News 16, 75–91 (2012). [Google Scholar]
- 47.Pontieri L., Schmidt A. M., Singh R., Pedersen J. S., Linksvayer T. A., Artificial selection on ant female caste ratio uncovers a link between female-biased sex ratios and infection by Wolbachia endosymbionts. J. Evol. Biol. 30, 225–234 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Berndt K.-P., Eichler W., Die Pharaoameise, Monomorium pharaonis (L.) (Hym., Myrmicidae). Mitteilungen aus dem Museum für Naturkd. Berlin. Zool. Museum und Inst. für Spez. Zool. 63, 3–186 (1987). [Google Scholar]
- 49.Ito K., Shinomiya K., Ito M., Armstrong J. D., Boyan G., Hartenstein V., Harzsch S., Heisenberg M., Homberg U., Jenett A., Keshishian H., Restifo L. L., Rössler W., Simpson J. H., Strausfeld N. J., Strauss R., Vosshall L. B.; Insect Brain Name Working Group , A systematic nomenclature for the insect brain. Neuron 81, 755–765 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Taylor C. P., Contribution of compound eyes and ocelli to steering of locusts in flight: I. Behavioural Analysis. J. Exp. Biol. 93, 1–18 (1981). [Google Scholar]
- 51.M. Mizunami, in Advances in Insect Physiology (Elsevier, 1995), vol. 25, pp. 151–265. [Google Scholar]
- 52.Smolla M., Ruchty M., Nagel M., Kleineidam C. J., Clearing pigmented insect cuticle to investigate small insects’ organs in situ using confocal laser-scanning microscopy (CLSM). Arthropod Struct. Dev. 43, 175–181 (2014). [DOI] [PubMed] [Google Scholar]
- 53.RStudio Team, RStudio: Integrated Development Environment for R (2016); www.rstudio.com/.
- 54.Picelli S., Faridani O. R., Björklund Å. K., Winberg G., Sagasser S., Sandberg R., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y., Chen Y., Shi C., Huang Z., Zhang Y., Li S., Li Y., Ye J., Yu C., Li Z., Zhang X., Wang J., Yang H., Fang L., Chen Q., SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 7, gix120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolstad B. M., Irizarry R. A., Åstrand M., Speed T. P., A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim S., ppcor: An R package for a fast calculation to semi-partial correlation coefficients. Commun. Stat. Appl. Methods 22, 665–674 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E., Storey J. D., The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE. 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Csárdi G., Nepusz T., The igraph software package for complex network research. InterJournal. 2006. Complex Syst. 1695, 1 (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eaaz5772/DC1


