Engineered spiral microvessels reveal unique contributions of curvature to EC responses to flow and support tissue perfusion.
Abstract
Vascularization remains a long-standing challenge in engineering complex tissues. Particularly needed is recapitulating 3D vascular features, including continuous geometries with defined diameter, curvature, and torsion. Here, we developed a spiral microvessel model that allows precise control of curvature and torsion and supports homogeneous tissue perfusion at the centimeter scale. Using this system, we showed proof-of-principle modeling of tumor progression and engineered cardiac tissue vascularization. We demonstrated that 3D curvature induced rotation and mixing under laminar flow, leading to unique phenotypic and transcriptional changes in endothelial cells (ECs). Bulk and single-cell RNA-seq identified specific EC gene clusters in spiral microvessels. These mark a proinflammatory phenotype associated with vascular development and remodeling, and a unique cell cluster expressing genes regulating vascular stability and development. Our results shed light on the role of heterogeneous vascular structures in differential development and pathogenesis and provide previously unavailable tools to potentially improve tissue vascularization and regeneration.
INTRODUCTION
The human vasculature forms a complex three-dimensional (3D) architecture spanning a large range of diameter, curvature, and torsion to provide efficient perfusion and tissue oxygenation. Its structure continues to adapt and remodel throughout life to meet changing energy demands in both regeneration and disease progression. The sources of vascular remodeling, however, remain elusive, partially due to the complex and heterogeneous vascular structure found in vivo and the lack of tools to recapitulate them in vitro. Vascularization remains a critical challenge in engineering complex tissue for potential tissue repair and disease modeling (1, 2).
Hemodynamic forces have been identified as one major driver of vascular development and remodeling (3, 4). By sensing the forces of blood flow, endothelial cells (ECs) lining the blood vessel wall regulate their morphology, phenotype, and cell behavior and further modify the structure of the vascular wall and vascular networks to support tissue growth (5–7). Extensive efforts have been made in understanding EC response to flow using flow chambers and microfluidic channels (8, 9), through which the EC response has been binned into one of two categories: laminar or disturbed (6, 10, 11). Laminar flow, defined as the parallel flow in devices such as parallel plate flow chambers, cone and plate viscometers, and straight tubes, applies uniform shear stress on ECs, and promotes EC alignment along the direction of flow and EC quiescence and maturation (12–14). Disturbed flow on the other hand has been described by distinct hemodynamic features like flow separation, flow reversal, or recirculation and modeled in step flow devices or specific pathological large vessel models in vitro (6, 10). Locations of disturbed flow are often associated with hotspots for atherosclerotic plaques in vivo, like in the aortic arch or at the bifurcations of vessels branching off the aorta (15, 16).
This dichotomous view of the endothelial response to flow has led to substantial advances in our understanding of EC biology, mechanotransduction, and vascular pathologies (7, 10, 17). Perfect axial flows and highly disturbed flows, however, represent the two extremes of a wide spectrum of flow profiles found throughout the vasculature. They do not reflect the extensive range of flow characteristics in the blood vessels that bend and twist through 3D space with varying levels of curvature and torsion (i.e., the deviation of curvature from its osculating plane) (17–21). For example, small vessels in vivo with low flow and high curvature and torsion have been associated with heterogeneous vascular remodeling (17, 18, 22, 23), but this phenomenon and the effect of complex nondisturbed flows have not been well studied.
The flow profile of curved vessels was first described by W. R. Dean, who found that curvature modifies the flow by introducing a secondary flow orthogonal to the primary flow axis, known as Dean flow. To characterize this phenomenon, the Dean number (De) was defined, combining the Reynolds number (; U, velocity; D, diameter; ρ, density; and μ, viscosity) with measures of the vessel’s curvature (; D, diameter; Rc, radius of curvature of the channel path). Additional research has extended Dean flows into 3D space and demonstrated theoretically that out-of-plane torsion modifies the secondary flow and also adds rotation into the primary flow at low Re (Re ≤ 1) (24, 25). In these curved vessels, ECs would experience not only the laminar shear force in the longitudinal direction but also curvature-dependent centrifugal and Coriolis forces and torsion-dependent forces that can be at normal, binormal (circumferential), or tangential (longitudinal) directions to the axial flow (26). In vivo, the values of curvature and torsion can range from 10−3 to 103 mm−1, with the largest values found in the microvasculature of complex vascular beds like the brain (27), where vessel diameter is very small, flow is very low, and therefore the Dean effect is very small. On the other extreme, a large vessel like the aortic arch has curvature and torsion on the order of 10−3 to 10−2 mm−1, but with high flow (and Reynolds number), which has been shown to lead to large Dean flows and a disturbed flow effect on ECs lining the curved walls (16, 28). At intermediate levels of curvature and torsion (i.e., of order ~100 mm−1) where Dean flows are not negligible, comparatively little is known despite the prevalence of these geometries in both normal and pathological vessels, like corkscrew collaterals and varicose veins (20).
This gap in the literature can be attributed to the difficulty of studying vessels with defined diameter, curvature, and torsion. Small curved vessels in vivo are often hard to image and measure flow, highly variable in geometry, and difficult to isolate for the study of cellular structure, function, and gene expression. In vitro, methods for engineering small vessels with 3D continuous curvature are limited. Microfluidic designs are typically confined to a single plane with zero torsion (29). Recent advances in bioprinting have made it possible to fabricate constant curvature conduits with torsion in biocompatible materials, but small vessel diameters are difficult to achieve and printing artifacts (e.g., layer lines) alter the shape of flow (30). A new method enabling the rapid fabrication of vessels with controllable diameter, curvature, and torsion would help fill this gap in understanding of the heterogeneous EC flow response and guide the design of vascularization in regenerative medicine.
Here, we have addressed these challenges by developing a spiral microvessel system with robust endothelium along the lumen and precisely controlled diameter, curvature, and torsion in 3D space. We showed that the spiral vessels supported rapid vascularization and homogeneous perfusion of thick (>5 mm) engineered tumor models and cardiac constructs. We show that small vessels of intermediate curvature and torsion (~100 mm−1) in laminar flow regimes induced distinct changes in EC phenotype and transcriptional profiles compared to straight vessels. Our results shed light on the role of vascular geometry in the heterogeneous vascular response to flow and show the promise of an easy-to-fabricate 3D platform for investigating the effect of curvature and torsion on vascular biology.
RESULTS
Fabrication and endothelialization of spiral tubes in PDMS and collagen gels
We exploited subtractive molding techniques to fabricate spiral tubes in both polydimethylsiloxane (PDMS) and collagen hydrogels and tested the fabrication limit and fidelity. In PDMS, stainless steel springs of various dimensions were molded in liquid-phase PDMS (10:1, base:curing agent) and manually removed after cross-linking. Robust perfusable spiral tubes with constant curvature were generated in PDMS with a diameter larger than 200 μm and a pitch greater than 1 mm per turn. The fabrication of smaller spiral tubes in PDMS is less consistent because of the distortion of the channel structure during spring removal. In collagen hydrogels (6 to 7.5 mg/ml), an automatic two-axis motion system was designed to retract the spring from the hydrogel after thermal gelation. Automatic retraction was critical to minimize distortion of the spiral pattern and maximize continuity of the luminal geometry in three dimensions in soft matrices (see Materials and Methods) (Fig. 1A). Spiral tubes of a wide range of wire diameter (dw = 120 to 400 μm), spiral diameter (ds = 1 to 3 mm), and pitch (p ≥ 400 μm) were formed in collagen hydrogels, corresponding to curvature κ in the range of 0.43 to 1.05 mm−1 and torsion τ in the range of 0.32 to 0.72 mm−1 (fig. S1A). Fluorescent beads were perfused to visualize the 3D structure of the spiral lumen [Fig. 1B (a); fig. S1, A and B; and movie S1], where loops of the spiral tubes were periodically spaced with distinct boundaries. Using off-the-shelf springs, we were able to achieve spacing between loops (δ = p − dw) as small as 210 μm. We further modified the spiral mold by adding a cylinder in the center of the spring to generate a second independently perfusable lumen in the hydrogel structure [Fig. 1B (b)]. We fabricated constructs with a central tube concentrically wrapped with a spiral tube and separated by a wall as thin as 200 μm.
Fig. 1. Spiral vessel fabrication and engineered tissue models.
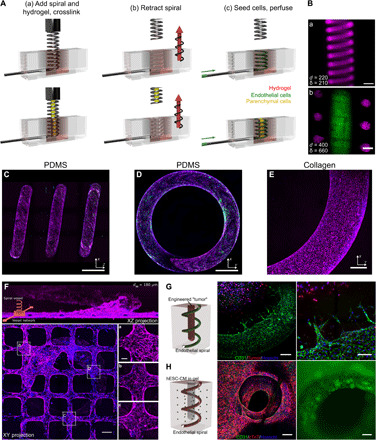
(A) Schematic of spiral vessel fabrication strategy. Top: A hydrogel is cross-linked around an off-the-shelf spring (a), the pattern is retracted from the gel via a two-axis motion system (b), and vessels were seeded with cells by perfusion (c). Bottom: An independent rod was introduced at the center of the spring to form an additional lumen for independent access and cell seeding. (B) (a) Maximum intensity projection (MIP) of a confocal z-stack of a spiral vessel in collagen perfused with fluorescent beads. (b) Optical section of a collagen spiral vessel (magenta) with an independently perfused center channel (green). Scale bars, 500 μm. (C to E) MIP of side (C) and top (D) views of an endothelialized spiral vessel in PDMS and top view of endothelialized collagen vessel (E). Scale bars, 750, 600, and 150 μm. (F) Integrated fabrication of spiral vessel (z directional flow) and planar microvessel (x and y directional flow) showing MIP of side and top views with magnified views of regions near (a) and distant from the connection of spiral to planar microvessels (b and c). Magenta, CD31; green, von Willebrand factor; blue, nuclei. Scale bars, 200 μm and 50 μm (inset). (G) Engineered vascularized tumor model with ECs from the spiral vessel sprouting toward avascular tumor cells embedded in the center lumen of the spiral. Green, CD31; red, KG1a cancer cells; blue, nuclei. Scale bars, 200 μm (left) and 100 μm (right). (H) Vascularized cardiac chamber model. Green, CD31; red, cTnT; blue, nuclei. Scale bars, 500 μm.
Next, we perfused human umbilical vein ECs (HUVECs) into the spiral tubes in either PDMS or collagen to allow cell attachment followed by culture under flow. Both materials supported the growth of a robust endothelium under steady flow for at least 1 week (Q = 1 μl/min; Fig. 1, C to E). PDMS spiral vessels with a lumen diameter of less than 200 μm often had sparse coverage of ECs on the vessel surface after seeding and were not used in experimental conditions. Collagen spiral vessels better supported endothelialization, and HUVECs were seeded and cultured under similar flow conditions for spiral vessels as small as 180 μm with high reproducibility (fig. S1C). ECs in PDMS vessels (lumen diameter > 200 μm) and all sized collagen vessels had robust junctions at cell-cell contacts and localized expression of CD31 to the plasma membrane (Fig. 1, C to E). Together, we successfully generated spiral microvessels with constant curvature and torsion at high fidelity and reproducibility and with robust endothelialization and perfusion.
Spiral vessels for 3D vascularization
The fabrication process for spiral vessels has the flexibility to integrate with existing vascularization approaches to further enhance tissue perfusion. By incorporating ECs into the bulk matrix, the endothelium in spiral tubes was readily anastomosed with self-assembled vessel networks and increased vascular density (fig. S1D). When combined with lithography and injection molding techniques (31), we successfully connected a spiral vessel with a microfabricated rectilinear vessel so that the spiral outflow was connected to the perfusion of microvessels in an orthogonal direction to the spiral. This integration allows the rotation of the spiral flow direction into another plane and mimics the architecture of the spiral artery to vascular bed connection found in vivo (Fig. 1F). We observed a continuous endothelium in the spiral microvessel connection [Fig. 1F (a)]. ECs in the planar microvessels near the spiral vessel outflow showed greater alignment with the direction of flow, likely due to higher flow stresses [Fig. 1F (a); average angle, 13.5° ± 10.2°] compared to cells in regions distant from the spiral microvessel interface [Fig. 1F (b and c); average angle, 39.3° ± 23.7° and 58.13° ± 29.25°, respectively]. These findings illustrate the potential of spiral vessels as a new strategy for rapidly generating long and high surface area vascular structures that may enhance tissue vascularization.
Using the concentric spiral platform in collagen gel, we further demonstrated the potential of spiral vessels in supporting 3D tissue function. By dispensing tumor cells (KG1a, a leukemia cell line; Materials and Methods) in a collagen gel (6 mg/ml) into the spatially defined center cylinder (1.3 mm diameter), we formed an artificial tumor surrounded by spiral vessels and monitored the sprouting of vessels from the spiral. This cell-remodelable system mimics the physiological origins of some tumors, where malignancies begin as an avascular cellular mass surrounded by host vasculature that it must recruit for expansion (32). When cultured under flow (Q = 1 μl/min) in normal growth medium, spiral vessels (dw = 400 μm, ds = 3.0 mm, and p = 1 mm) maintained patency throughout 7 days of culture and sprouted consistently by day 7, but not at day 3 (N = 4 for each time point) (Fig. 1G). These sprouts extended exclusively toward the tumor, with sprouts reaching as far as 220 μm from the vessel wall by day 7. No sprouts were observed when there were no tumor cells in the center.
We also created a thick cardiac chamber supported by a spiral vessel using the same concentric model (1.3-mm-diameter by 6-mm-long chamber surrounded by spiral vessel). GCaMP3-transduced human embryonic stem cell–derived cardiomyocytes (hESC-CMs) and stromal cells (HS27a) were added into the bulk collagen matrix (33) and ECs (HUVECs) into both the bulk matrix and the spiral lumen, while the center of the tissue was kept open (Fig. 1H). By day 12 of culture, organized calcium waves were observed and appeared to propagate in three dimensions along the spiral vessel wall (movie S2). The conduction velocity in engineered cardiac tissues was 2.7 ± 0.97 cm/s, as determined by analysis of the GCaMP3 signal (fig. S2). These proof-of-concept examples show that the spiral vessel platform can be used to support 3D vascularization and perfusion in large tissues, to study the vascular-tissue interaction in a spatially and temporally controlled manner, and to model complex tissue functions.
Flow characteristics of spiral tubes
We next examined the flow characteristics in these spiral microvessels and compared them with straight vessels of the same caliber. We visualized the flow characteristics by perfusing fluorescent bead solutions in two parallel streams through straight and spiral PDMS vessels of the same diameter and length (dvessel = 400 μm, dspiral = 3 mm, pspiral = 1 mm, κspiral = 0.46 mm−1, τspiral = 0.31 mm−1, and L = 6.5 cm) at three steady flow conditions (Q = 1, 50, and 100 μl/min, corresponding to Re = 0.01, 0.76, and 1.52, respectively). The 3D flow images were taken under confocal fluorescence microscopy at set distances (Lv = 5, 30, and 55 mm in straight vessels, or loops 3/4, 3 3/4, and 6 3/4 in spiral vessels) from the vessel inlet. Straight tubes displayed a classical parallel flow profile where the two streams of beads traveled to the outlet and maintained their position over the whole vessel length at both flow rates (Fig. 2, A and C). In spiral tubes, the two bead streams remained distinct and parallel at low flow (Q = 1 μl/min) but rotated over the vessel length without obvious mixing in the bulk [Fig. 2B (a to c)]. The orientation of the two parallel streams inverted after approximately four loops from the inlet [Fig. 2B (b)] and completed a full rotation at approximately loop 7 [Fig. 2B (c)]. At a higher flow rate (Q = 50 μl/min) in the same spiral geometry (De = 2.77), the two bead streams developed obvious bulk mixing with the leading edge of flow rotating 270° after three loops [Fig. 2B (e)] and completed another full rotation by loop 7 [Fig. 2B (f)]. At even higher flow (Q = 100 μl/min), a stronger mixing effect was observed in the same spiral geometry [Fig. 2B (g to i)], whereas the two streams remained parallel and unmixed in straight vessels under the same flow conditions.
Fig. 2. Flow in straight and spiral vessels.
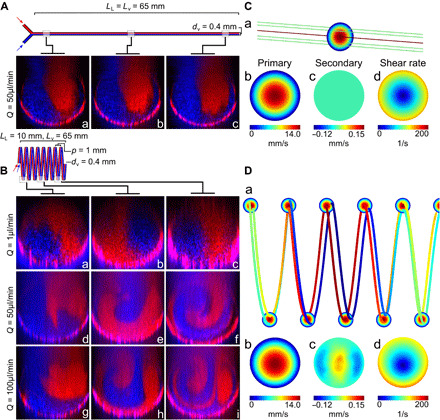
(A and B) Confocal cross sections of perfusion of two parallel streams of red and blue beads into a straight PDMS vessel (A) at a flow rate of Q = 50 μl/min and a spiral PDMS vessel (B) at three flow rates (Q = 1, 50, and 100 μl/min) at three distances from the vessel inlet (ΔLv,a ≈ 5 mm, ΔLv,b ≈ 30 mm, and ΔLv,c ≈ 55 mm), corresponding to the 3/4, 3 3/4, and 6 3/4 spiral loops (LL = linear length, Lv = vessel length, dv = vessel diameter, p = pitch). (C and D) Computational fluid dynamics plots of straight (C) and spiral (D) vessels at Q = 50 μl/min for (a) streamlines (color expressed with primary velocity magnitude), (b) primary velocity magnitude, (c) secondary flow velocity orthogonal to cross-sectional plane, and (d) shear rate at the cross-sectional views.
Using numerical simulation with COMSOL, we confirmed these flow characteristics: (i) Idealized parallel streamlines were present in fully developed flow in straight vessels [Fig. 2C (a)]; (ii) parallel streamlines in spiral vessels slightly rotate along circumferential direction at low flow (Q = 1 μl/min; fig. S3A); and (iii) streamline rotation was enhanced in spiral vessels and developed twists at higher flow [Q = 50 μl/min; Fig. 2D (a)] and had clear twists at Q = 100 μl/min (fig. S3B). The spiral geometry did not induce a significant change in the primary flow compared to straight vessels but did lead to the emergence of secondary flows with a peak magnitude of around 1% of the primary flow velocity [Q = 50 μl/min; Fig. 2, C (b and c) and D (b and c)]. This also led to the development of a shear stress gradient in 3D space and a change in the wall shear stress (WSS), with a maximum (10% increase over the straight tube) on the surface of the inner curvature and minimum on the outer bend, unlike in a straight tube where the WSS was constant across the lumen cross section with zero gradients [Fig. 2, C (d) and D (d)]. These data demonstrated that spiral vessels induced bulk flow mixing and heterogeneous hemodynamic forces on the endothelium lining the wall due to 3D curvature and torsion.
3D curvature alters endothelial morphology and mRNA expression under flow
To understand how the distinct hemodynamic features of flow in spiral vessels affected ECs, we cultured cells in both geometries under flow. In straight and spiral vessels, ECs formed robust junctions and a stable endothelium in low (Q = 1 μl/min and WSS = 0.1 dyne/cm2 in straight vessels) and high (Q = 50 μl/min and WSS = 4.6 dynes/cm2 in straight vessels) flow conditions. The increased flow appeared to change the EC morphology and enhance EC alignment in the direction of flow (Fig. 3A). Under low flow conditions (Q = 1 μl/min; Fig. 3B), fewer Ki67+ proliferating cells were observed in spiral vessels than in straight vessels. When exposed to higher flow, however, more proliferating cells were observed in the spiral geometry than the straight geometry, suggesting distinct roles for geometry and flow on the ECs. Previous literature has highlighted that very low laminar flows activate ECs, whereas high laminar flow enhances EC quiescence (11). Our data were consistent with this in straight vessels with significantly reduced cell proliferation at higher flow. In spiral vessels, however, the flow rotation in low flow may alter transport and promote quiescence at low flow. Given that the magnitude of flow forces is very low in the low flow conditions, it is also likely that differences in substrate curvature between straight and spiral geometries contribute to these observed differences (34).
Fig. 3. Endothelial response to flow in straight and spiral vessels.
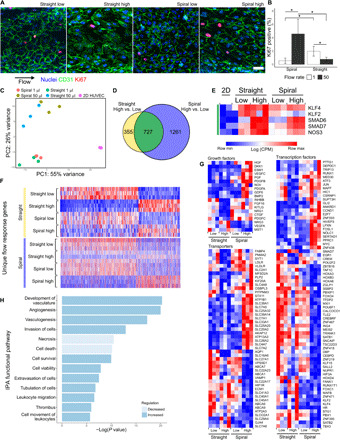
(A) MIP of EC cultured under flow (Q = 50 μl/min) for 24 hours. Blue, nuclei; green, CD31; red, Ki67. Scale bar, 50 μm. (B) Quantification of the percentage of Ki67-positive nuclei by counting 100+ cells per vessel in N = 3 vessels at two flow conditions (Q = 1 and 50 μl/min). Error bars represent 95% confidence interval of the mean. *P < 0.05 using a one-way analysis of variance (ANOVA) with Tukey’s pairwise comparisons. (C) PCA of RNA-seq data from cells cultured at static and at two flow conditions in two vessel geometries (N = 3). (D) Venn diagram showing the overlap of genes significantly changed by increasing flow in straight and spiral geometries. (E) Heatmap of log counts per million (CPM) values of known flow-responsive genes. All genes are present in the overlapping region of (D) (green). (F) Heatmaps of the CPM values of significantly regulated transcripts belonging to the nonoverlapping regions of (D). Three hundred fifty-five genes uniquely regulated in straight high versus low (top, yellow) and 1261 genes uniquely regulated in spiral high versus low (bottom, blue). (G) Heatmaps of the CPM values of selected growth factors, transporters, and transcription factors. (H) IPA functional pathways identified by comparing spiral to straight vessels under high flow.
We next examined the transcriptional changes in ECs in these conditions via RNA sequencing (RNA-seq) for ECs cultured under both flow conditions in straight and spiral vessels and under static conditions. Principal components analysis (PCA) of gene expression data showed clustering of individual groups, with the largest variance between static and all flow conditions (Fig. 3C). Activation of classical flow-dependent genes was confirmed in all flow conditions compared to static culture (Fig. 3, D and E). Among these genes, KLF2 and KLF4 appeared to only change with the onset of flow but were not sensitive to a further increase in flow, whereas SMAD6, SMAD7, and NOS3 increased further at higher flow conditions. Among the genes differentially expressed in straight vessels due to the increase of flow, 52% (533 of 1012) overlap with genes differentially expressed in the onset of flow (static versus low flow condition) (fig. S4, A and B). The genes unique to the increase of flow include up-regulation of many genes previously reported to regulate vascular development and flow sensing (35), such as Notch ligands JAG1 and JAG2; Notch target HEY2 and other transcription factors such as SNAI2; transmembrane proteins IL21R and EFNB2; transporter GJA5; peptidases MMP10, MMP1, and MT1F; growth factors and cytokines NOG, DKK2, WNT4, CXCL12, and TGFB1; and other molecules such as VCAN and CYP1B1 (fig. S4C). Gene Ontology (GO)–enriched terms for this group of genes showed up-regulation of cellular response to growth factors, vascular development, transmembrane receptor protein tyrosine kinase signaling pathway, blood vessel morphogenesis, cell migration and motility, and others (fig. S4D).
Approximately 66% (722 of 1136) of differentially expressed genes in straight vessels overlap with those in spiral vessels in response to increased flow (Fig. 3D). Almost all overlapping genes are changed in the same direction (99%), suggesting a conserved response to flow in both geometries (fig. S5A). MARC2, PTX3, and STX11 did not follow this trend and were up-regulated in spiral vessels with increased flow but down-regulated in straight vessels. PTX3 has been reported as a biomarker for endothelial dysfunction in preeclampsia, which is a disease caused by spiral artery dysfunction (36). Many genes down-regulated in straight vessels by the increase of flow did not show changes in spiral vessels, such as growth factors CTGF, FGF2, NRG1, and FGF16; transmembrane proteins CAV1, UNC5A, KIT, and SMAD4A; transcription regulators EGR1/2/3, MAF, MYRF, and MZF1; transporters such as LDLR; and cytokines TNFSF18, IL12A, CCL2, CCL16, and CCL28 (fig. S5B). This suggests that the EC response to flow in spiral vessels is a combination of both canonical flow pathways and a distinct response involving a wide range of other transcripts.
Increased flow also led to an additional 1294 genes significantly changed in spiral vessels that were not in straight ones (Fig. 3F). High flow in spiral vessels appeared to activate growth factors such as DKK1, ESM1, BMP2, PDGFA, OSGIN2, and VEGFC; many solute carrier (SLC) and adenosine triphosphate (ATP)–binding cassette (ABC) superfamily transporters; transcriptional regulators such as GLI2; cytokine CXCL1; peptidases TLL1, ADAMTS1, ADAMTS9, and TASP1; and kinases PODXL, EPHA5, HK2, PRKCA, CCT2, and MAP2K1 (Fig. 3G and fig. S6A). In addition, high flow in spiral vessels repressed growth factors such as MST, NRG2, GDF3, GAS6, and IGF2; transmembrane receptors CHRNA1, SELP, LRP1, ITGB3, and ROBO3; transporters including MAL2, ATP2A3, RBP1, and APOL1 and several members of SLC and ABC superfamilies; transcriptional regulators such as NOTCH3, CITED4, CAND2, FOXO4, DACH1, and EBF3; cytokines DKK3, CSF1, and FLT3LG; GPCR (G protein–coupled receptor) group SIPR4, OPRL1, and HTR2B; and kinases PDGFRB, CKB, and SBK1 (Fig. 3F and fig. S6A). GO term analysis showed the up-regulation of primarily ribosome biogenesis, which would be critical for cellular growth and proliferation (fig. S6B). These expression profiles show that spiral vessels share a common set of flow-responsive elements with straight vessels but have an additional response that appeared to promote vascular growth.
PCA analysis showed that the separation of straight and spiral geometries was enhanced under higher flow conditions (Fig. 3E). Under low flow conditions (Re << 1; inertia effect is negligible), ECs in the two geometries were largely similar, with only a handful of significantly regulated transcripts (fig. S7A). These included CYTL1, which is known to up-regulate proangiogenic function, but not proinflammatory pathways (37). HES2, a downstream Notch pathway gene, STK32B (serine/threonine kinase 32B), and CCND1, a cell cycle regulation gene, were also up-regulated in low flow spiral vessels. The up-regulation of these genes was further enhanced in high flow conditions. In addition, many genes that regulate vascular development were up-regulated when comparing spiral to straight vessels at high flow, for example, growth factors HGF, DKK1, ESM1, PGF, PDGFA, GDF6, PDGFB, CTGF, VEGFC, BMP2, and PDGFC; peptidases ADAMTS1, ADAMTS9, MME, and CTSS; kinases EPHA5, MPP4, PODXL, SPRY2, CDK7, and MAP2K1; transmembrane receptors KIT, SELE, ULBP2, PLXNA2, and LRP8; transcriptional regulators GLI2 (a hedgehog pathway mediator), ATF3 (required for endothelial regeneration) (38), and FOSL1 (required for vascular formation) (39); and many SLC and ATP family transporters (Fig. 3G). Ingenuity Pathway Analysis (IPA) showed that ECs in spiral vessels have activated upstream regulators including prosurvival factors HGF, PGF, EGF, VEGF, and HIF-1a. Up-regulated functional pathways included vascular development, angiogenesis, vasculogenesis, cell invasion, and cell survival, whereas cell death and necrosis were decreased compared to the straight vessel in high flow conditions (Fig. 3H and fig. S7B). Spiral vessels also showed the activation of antiapoptotic and proliferative pathways marked by cell cycle and mitotic genes. PDGF (platelet-derived growth factor) family members were relatively more abundant, as were molecules associated with IL-8 (interleukin-8) and HGF (hepatocyte growth factor) signaling (fig. S7B).
Together, the bulk RNA-seq showed that spiral vessels maintain a normal flow response to a certain extent, but curvature and torsion modified the response by up-regulating markers for transporters, cycling, and survival and down-regulating markers of cell death. These data suggest that flow in spiral vessels promoted vascular growth or development rather than inducing a common inflammatory response to disturbed flow.
Single-cell RNA-seq reveals heterogeneity in transcriptional responses to flow based on vessel geometry
We hypothesized that ECs exposed to flow within spiral vessels experienced a spatial variation in hemodynamic forces not present in straight vessels that would result in a heterogeneous transcriptional response to flow. To understand this heterogeneity at the single-cell level, we sequenced the transcriptomes of more than 2000 individual ECs pooled from three to four devices of each geometry cultured at high flow (Q = 50 μl/min). Dimensionality reduction by Uniform Manifold Approximation and Projection (UMAP) and cluster analysis was performed using Monocle (40–42). Projection in the top two UMAP dimensions shows overlapping contributions of ECs from spiral and straight vessels that form mostly contiguous clusters with nearly uniform expression of pan-endothelial markers such as CDH5 (VE-cadherin) (Fig. 4, A and B). Expression of classical flow-dependent genes, including KLF4 and NOS3, is distributed throughout the major clusters of ECs in this projection (Fig. 4C). We identified variation in gene expression across the first UMAP dimension driven largely by cell cycle genes that have been shown to be regulated, in part, by flow. Specifically, a large cluster of cells to the right in UMAP space (cluster 3) express genes such as MKI67, consistent with active cell cycle status, whereas cells clustered to the opposite pole (cluster 1) express genes implicated in cell cycle arrest and arterial phenotype shown to be regulated by the Notch pathway downstream of laminar shear stress (including CDKN1C, EFNB2, HEY1, GJA4, and IL33; Fig. 4C) (43–45).
Fig. 4. scRNA-seq comparison of cells cultured under high flow in spiral and straight vessels.
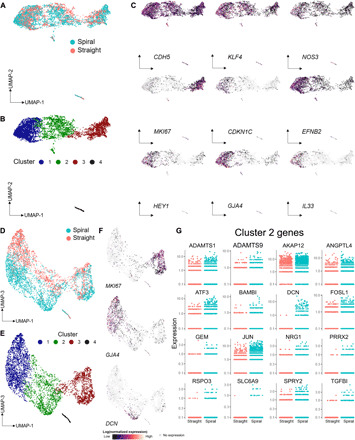
(A) UMAP plots of spiral high flow and straight high flow cells, computationally derived clusters (B), and the distribution of endothelial and cell cycling genes across cells (C). (D) UMAP plots of the first and third UMAP dimensions, the corresponding location of clusters in this dimensional space (E) with examples of cluster specific genes (F), as well as a selection of genes identified as significantly differentially expressed (G).
To evaluate heterogeneity in the transcriptional response of ECs resulting from vessel geometry, we next identified differentially expressed genes on the basis of single-cell RNA-seq (scRNA-seq) data of EC from straight versus spiral vessels. Examination of the third UMAP dimension revealed separation in transcriptional space between EC from straight versus spiral vessels, with many of the identified differentially expressed genes polarized in this dimension (Fig. 4, D to F). Among the genes up-regulated in EC from spiral vessels are many that were also identified as differentially expressed in bulk RNA-seq analysis, including ATF3, SPRY2, IL8, JUN, AKAP12, ANGPTL4, FOSL1, ADAMTS1, and ADAMTS9 (Fig. 4G and fig. S8A). Most of these genes are expressed in a common pattern, with increased expression in cells localized in UMAP space to the lower (spiral) portion of cluster 2. This suggests a distinct transcriptional program among the primarily spiral ECs in this region that may correspond with their transcriptional response to specific hemodynamic conditions unique to spiral vessels under high flow. In further support of this hypothesis, analysis of the scRNA-seq data also identified differentially expressed genes not detected in bulk RNA-seq, including DCN, SLC6A9, GEM, NRG1, RSPO3, BAMBI, TGFBI, and PRRX2, that were highly specific to EC from spiral vessels localized in cluster 2 (Fig. 4G and fig. S8B). These data suggest that flow in spiral vessels induced a population of ECs with unique gene expression profiles that are not present in straight vessels, with potential roles in processes such as angiogenesis, vascular growth, and inflammatory and stress responses.
DISCUSSION
3D vascular networks have been engineered to support complex tissues, but most designs have been geometrically limited to rectilinear structures with a constrained range of hemodynamic profiles (31, 46). Reproducing the structure of complex networks with continuous curvature was only made possible by recent advances in bioprinting (30) and is still restricted by printable materials. Printing artifacts create discontinuity and stepwise changes in wall geometry that disturbs the flow in vessels. Building perfusable and endothelialized vessels with continuous curvature and torsion as in vivo have not been achieved. Here, we developed a spiral vessel platform that allows the systematic control of 3D vessel geometry (diameter, curvature, and torsion) and is easy to fabricate rapidly and reproducibly. In one step, this approach creates a versatile vascular conduit that spans several orders of magnitude. Vessel diameters approach 100 μm and span spiral diameters in the millimeters, and the total vessel length stretches easily into many centimeters. Under perfusion, these vessels support a healthy endothelium with robust junctions and rich granules and appeared to guide the architecture and functional organization of parenchymal cells. This large vessel surface area can be tuned to fit into tissues of various shapes and sizes and can serve as a foundational vascular structure in engineering vascularized tissues.
The precise control of 3D geometries in this spiral vessel platform provides opportunities to study the contribution of individual geometric parameters to blood flow and corresponding vascular response that are otherwise hard to dissect in vivo or in other in vitro models. In this work, we took the first step to compare the 3D flow in spiral and straight vessels of same diameter at the same flow rate and the corresponding EC response. We showed that 3D curvature had little change in the primary flow but led to bulk flow rotation in spiral vessels in a creeping flow regime (Re < 0.1 and De < 0.1) and mixing at a slightly higher flow rate (Re < 1 and De < 2). The spiral curvature led to changes of WSS along circumferential direction and generated a stress gradient in 3D space. This additional stress gradient, distinct from the pressure gradient driving axial flow, likely induced flow mixing in the high flow condition. By culturing ECs in spiral and straight vessels with the same vessel diameter and length, we were able to distinguish the effect of curvature from the conventionally recognized flow response in ECs to laminar flow using both bulk and scRNA-seq.
The effects of laminar and disturbed flows have been documented through a variety of in vivo and in vitro systems to characterize a generalized response to disturbed flow conditions (10). Comparison of the responses of spiral vessels to known disturbed flow responses by bulk RNA-seq gives important context to the effect of secondary flows. A common marker of disturbed flows is the up-regulation of cell adhesion–associated transcripts (10). In this group, both E-selectin (SELE) and MCP-1 were significantly up-regulated in higher spiral flow, but neither VCAM-1 nor ICAM-1 was significantly changed relative to the straight vessel. This inflammatory axis also commonly correlates with the up-regulation of HuR (47), TGFβ1 (48), and BMP4 (49), but these were not significantly different between straight and spiral vessels. Some smooth muscle signaling–associated transcripts were activated in a disturbed flow pattern, with endothelial nitric oxide synthase decreased (NOS3; logFC = −0.60) and PDGFA (logFC = 1.09) and PDGFB (logFC = 0.98) mildly increased. Endothelin-1 transcripts, however, were not unchanged between spiral and straight vessels. Proangiogenic transcripts were mixed, with a mild disturbed-like increase in CTGF (logFC = 0.94) but a laminar-like decrease in VEGF transcripts (logFC = −1.03) and no significant changes in angiopoietin-2. Oxidative stress pathways are also commonly altered in disturbed flow through changes to genes like NOX4, NQO1, SOD1, HO-1, PRX, and GSH (50), but none of these transcripts were differentially regulated between straight and spiral vessels. Together, the bulk RNA-seq analysis revealed that spiral vessels display features of both laminar and disturbed flows. When disturbed flow–like responses were observed as in some proinflammatory and proangiogenic targets, however, the magnitude of change was generally small (logFC < 1.8). This mixed laminar-disturbed expression profile highlights the unique response of ECs in the presence of secondary flows.
Most studies on curvature have focused on large vessels, and cells located in the inner or outer curves have shown altered phenotypes corresponding to the different shear stress levels in these regions (15, 16, 26). Our spiral microvessels showed little change in the primary flow but introduced a weak secondary flow with no “disturbed flow” features when compared to the straight vessels at the same flow condition. Using scRNA-seq, we did not identify two distinct cell groups in spiral vessels deviating from the straight vessels; rather, cell clusters overlap significantly in the two geometries, confirming that ECs in spiral vessels share significant gene regulation with straight vessels in response to laminar flow. In addition, there is a group of ECs in spiral vessels separated from the main flow-responsive clusters, suggesting a different EC response mechanism than WSS. It is possible that the combined WSS gradient and bulk flow mixing provide a previously unidentified set of perturbations on ECs. This suggests that the twists and turns found throughout the vascular system contribute to a more heterogeneous EC response to flow than is currently appreciated.
During development and tissue regeneration, curved vessels often appear. Our data indicate that the transient appearance of complex vascular curvature may benefit vascular growth and remodeling. ECs in small curved vasculature may provide angiocrine or growth signals to surrounding tissue and promote tissue organization and regeneration. The spiral vessel system we developed here provides opportunity to systemically define and evaluate the role of heterogeneous vessel geometries in the EC response to flow. Future studies would be critical to understand how these transcriptional programs translate to EC behavior and modify vascular remodeling. This understanding will provide important insights into how heterogeneous vascular structures contribute to differential development and pathogenesis, as well as informing new engineering strategies to improve tissue vascularization and support tissue regeneration.
MATERIALS AND METHODS
Fabrication of endothelialized spiral vessels in PDMS
Spiral PDMS devices were formed by casting a mixture of Sylgard 184 (10:1, base:curing agent) around springs of varying pitch, wire diameter, and spiral diameter. The mixture was cured at 60°C for 2 to 3 hours, and the spring was removed from the polymerized PDMS. Before seeding cells, PDMS spiral devices were exposed to oxygen plasma for ~90 s and then immediately incubated with a 0.1% gelatin solution at 37°C for 20 min. HUVECs were trypsinized and resuspended to 10 million to 12 million cells/ml. Spiral vessels were seeded by perfusion of 20 to 40 μl of this suspension at 20 μl/min. Devices were rotated every 15 min for 1 hour to ensure even coverage of seeded cells on the vessel wall. Vessels were subject to constant flow perfusion between 4 and 12 hours after seeding. Unless noted otherwise, PDMS vessels were cultured under a flow of 1 μl/min, driven by syringe pump (KD Scientific KDS 220) with endothelial growth media (EGM) supplemented with 3.5% dextran.
Design and fabrication of spiral retractor system
An automated retraction device was designed to reproducibly create spiral features in cross-linked hydrogels. A custom two-axis machine was fabricated from a combination of 3D-printed and machined components. Linear motion in ±Z and rotation about Z were controlled through two stepper motors connected to an Arduino microcontroller. The microcontroller was programmed to “unscrew” a spring from a cross-linked hydrogel when given the spring pitch (mm/turn) as an input. This controlled movement is independent of the other features of the spring (i.e., wire diameter and spring diameter). While a purely mechanical device can be fabricated to accomplish the same task, it would require a unique set of components for every spring pitch of interest.
A set of spiral vessel holders was also designed and milled out of polysulfone. Each holder consisted of a small rectangular reservoir (~10 mm × 5 mm × 5 mm), where the spiral vessel was formed in collagen gel. For visualization, two of the walls of this reservoir were formed by using PDMS to bond glass microscope coverslips to the polysulfone device. To connect spirals to a perfusion system, a hole was drilled in the side of each holder to provide a slip-fit interface for connecting 23-gauge hypodermic tubing.
Fabrication of endothelialized straight round vessels in PDMS
Straight round vessels in PDMS were formed by suspending lengths of 28-gauge stainless steel hypodermic tube in a custom-machined acrylic mold. PDMS was poured over this mold and cured as for spiral PDMS devices. The hypodermic tubing was removed from the cured PDMS, leaving round straight channels in PDMS with a single inlet per channel and an outlet that drained into the tissue culture dish. Before seeding cells, round straight PDMS devices were exposed to oxygen plasma for ~30 s and then immediately incubated with a 0.1% gelatin solution at 37°C for 20 min. HUVECs were seeded into this lumen via perfusion, as described in spiral PDMS devices.
Fabrication of endothelialized spiral vessels in collagen
Collagen gels were prepared by mixing an acidic collagen stock solution (at 15 mg/ml) with 1 N NaOH, 10× Medium M199, and endothelial growth medium (Lonza) to reach neutralization and targeted concentration (6 or 7.5 mg/ml). After homogenizing, the collagen mixture was degassed by placing under vacuum for 30 to 60 min. While the gel solution was under vacuum, spiral vessel holders were prepped by brief corona treatment (<30 s) and subsequent coating of polyethylenimine (1%; Sigma-Aldrich) with glutaraldehyde (0.1%; Sigma-Aldrich) cross-linking. After five washes with autoclaved water, vessel holders were dried and placed in the spiral retractor system where the spiral pattern was brought into contact with a 25 mm length of 23-gauge hypodermic tubing within the holder. The degassed gel mixture was then pipetted into the reservoir and allowed to cross-link at room temperature for 1 hour. After cross-linking, the spiral pattern was removed from the gel using the spiral retraction system and submerged in warm medium. Each spiral vessel was seeded with cells following the same protocol as in PDMS vessels.
Fabrication of combination microvessel-spiral vessel devices in collagen
Planar vascular networks in collagen were formed by modification of a previously described protocol (31). Briefly, microvessels are fabricated by sandwiching a bottom flat layer of collagen gel with an injection-molded top layer of collagen gel bearing a microfabricated vessel pattern. To form combination vessels, the spiral pattern was introduced during the molding step for the top layer of collagen and the spiral vessel connected directly to the microfabricated pattern. Microvessels were otherwise assembled normally, and devices were seeded by gravity perfusion of a cell suspension from the top of the spiral vessel. Devices were cultured under gravity-driven flow (~6-mm H2O pressure head) that was replenished every 12 hours.
Anastomosis of spiral vessels with self-assembled connective vascular tubes in collagen
The spiral microvessels were formed following the same general protocol as collagen spiral vessels described above, but with the addition of ECs within the bulk matrix. Collagen gel was prepared and degassed, followed by the addition of HUVECs into the gel to reach a final concentration of 2.5 million cells/ml in collagen (7.5 mg/ml). The mixture was then added into spiral vessel holders for thermal gelation. Additional HUVECs were then seeded into the lumen of spiral vessels, as described above in other devices. The culture was then maintained under gravity-driven flow (~6-mm H2O pressure head) and replenished every 12 hours.
Fabrication of tumor spiral vessels to support tumor angiogenesis in collagen
An 18-gauge needle was added to the center of the spiral pattern before gelation of collagen gel to form tumor spiral vessels. After gelation at 37°C for 30 min, both the spiral and the 18-gauge needle were retracted simultaneously, leaving a spiral vessel and an open center channel for the final collagen structure. A separate collagen gel (6 mg/ml) was prepared with a suspension of KG1a cancer cells (final concentration of 5 million cells/ml). This KG1a-collagen gel was then pipetted into the central space in the collagen spiral device and polymerized at 37°C for 30 min. After this second step of polymerization, the spiral vessel was seeded with HUVEC as described previously and cultured under 1 μl/min flow with EGM supplemented with 3.5% dextran for up to 7 days.
Fabrication of thick cardiac tissue perfused with spiral vessels (cardiac spirals) in collagen
Cardiac spirals were formed using the same spiral and 18-gauge needle pattern as tumor spirals described as above. The collagen cell–gel mixture (6 mg/ml) for these devices consisted of hESC-derived cardiomyocytes (20 million cells/ml), HS27a stromal cells (10 million cells/ml), and HUVEC (2 million cells/ml) (33). Transgenic RUES2 hESCs expressing the fluorescent calcium reporter GCaMP3 were differentiated to cardiomyocytes by modulation of activin A, BMP4, and Wnt signaling using a previously reported protocol (51). The void space in the center of these devices was left unfilled but seeded with HUVEC in the process of seeding the spiral vessel lumen.
Computational fluid dynamic analysis in spiral vessels compared to straight vessels
The fluid flow characteristics in the 3D straight tubes and spiral tubes were simulated with COMSOL Multiphysics software, package version 5.0. The Navier-Stokes equation was used as predefined in COMSOL and solved with the stationary solver for laminar flow. The fluid properties were defined as follows: viscosity of 3 × 10−3 Pa/s (mimicking that of endothelial growth medium with 3% 70-kDa dextran) and density of 3 × 10−3 kg/m3. The inlet boundary conditions are laminar flow with a constant flow rate at Q = 1, 50, or 100 μl/min, corresponding to the average inlet velocity at 0.14, 7, and 14 mm/s, and the outlet boundary conditions are laminar flow with zero pressure. The channel walls were set to no-slip boundary condition. We decreased the length of the modeled channel (~40 mm and 6.5 loops) to enhance computation speed. Approximately 3 × 105 mesh elements were used in each simulation, and the simulation results were demonstrated mesh independent when changing the mesh size. The average secondary flow velocity was calculated as the velocity at the direction orthogonal to the primary flow direction. The primary flow, secondary flow velocity, and shear rate were exhibited at the cross-sectional plane that is orthogonal to the primary flow direction.
Immunofluorescent staining and imaging
Spiral and straight vessels, after culture for the designated time, were fixed by treatment with 4% formaldehyde for 15 min (for PDMS devices) or 60 min (for collagen devices). Devices were permeabilized by incubation in phosphate-buffered saline (PBS) containing Triton X-100 (0.5%) and bovine serum albumin (BSA; 2%) for 30 min. Unconjugated primary antibodies were added into the spirals (and onto collagen gels), incubated overnight at 4°C, followed by washing with PBS for three times (5 min for each time), and incubated with secondary antibodies and Hoechst 33342 (40 μg/ml) for 1 hour at room temperature before a final round of washing. Devices were stained for factors including CD31 (1:30; Abcam ab28364), VE-cadherin (1:100; Abcam ab33168), von Willebrand factor (1:100; Abcam ab8822), cTnT (cardiac troponin T) (1:100; Thermo Fisher Scientific MS-295-P1), and F-actin (1:100; Thermo Fisher Scientific A12379). Stained devices were imaged using a Nikon A1R confocal microscope or a Yokogawa W1 spinning disc confocal system.
RNA sequencing
For RNA-seq studies, PDMS spirals, PDMS round straight channels, PDMS square straight channels, and collagen spiral devices were used. All PDMS devices were fabricated to have either a 400-μm lumen diameter or 400-μm square cross section and equal total vessel length (~68 mm). All devices were perfused at either 1 μl/min (“Low”) or 50 μl/min (“High”) for 3 days. On day 3, vessels were perfused with RLT lysis buffer and the lysate was collected. For PDMS devices, 350 μl of RLT was perfused per device and RNA was isolated using an RNeasy Mini kit (Qiagen). For collagen spirals, 175 μl of RLT was perfused per device. Before RNA isolation, the lysate from two collagen spirals was pooled to form one sample replicate.
Isolated RNA samples were then processed for RNA-seq (Illumina NGS, paired-end 50 cycles, low-input library prep). RNA-seq data were aligned using TopHat (52). Aligned count data were normalized and evaluated using EdgeR (53), where genes with adjusted P values and false discovery rates of less than 0.05 and log(fold change) ≥ 0.585 were considered differentially expressed. Gene set enrichment analysis was performed using clusterProfiler (54) to probe KEGG (55), WikiPathways (56), and GO (57) gene set libraries. IPA (Qiagen) was also used to probe for upstream regulators of differentially expressed genes.
scRNA sequencing
For scRNA-seq studies, spiral or straight vessels containing ECs were perfused under high flow conditions (50 μl/min). ECs were harvested from three to four vessels of each geometry by trypsinization and pooled. Cells were washed with PBS and resuspended in 0.04% ultrapure BSA (Thermo Fisher Scientific) in PBS on ice. Approximately 3500 cells were loaded on the 10X Genomics Chromium platform in one lane per sample. The 10X Genomics Version 2 Single Cell 3′ Kit was used to prepare single-cell mRNA libraries. Sequencing was performed for pooled libraries from each sample on Illumina NextSeq 500 using the 75-cycle, high-output kit. The Cell Ranger 2.1.1 pipeline (10X Genomics) was used to determine cell barcodes and unique molecular identifiers (UMIs) per cell and to exclude low-quality cells, using default parameters. The Monocle (version 2.99.3) platform (41) was used for downstream analysis of scRNA-seq data, combining read-depth normalized data from spiral and straight vessel samples. Dimensionality reduction with UMAP (42) was used to project cells in three dimensions, using the top 20 principal components, with the reduceDimension function (reduction_method = ‘UMAP’, max_components = 3). Clustering was performed by Louvain method with the clusterCells function, using default parameters with the exception of resolution set to 8 × 10−4. The principalGraphTest and differentialGeneTest functions in Monocle were used to determine genes differentially expressed between clusters in the UMAP projection and between ECs in spiral versus straight vessel geometries, respectively, selecting genes based on specificity and expression level (for cluster-specific genes) and significance (q < 0.01) (for genes differentially expressed between samples).
Supplementary Material
Acknowledgments
We acknowledge the Lynn and Mike Garvey Imaging Core in the Institute of Stem Cell and Regenerative Medicine, the Washington Nanofabrication Facility, and the Flow Cytometry Core, all at the University of Washington, and the Next Generation Sequencing facility at the Fred Hutchinson Cancer Research Institute. Funding: This work is supported by the NIH: R01HL141570, UG3TR002158, and UH2/UH3 DK107343 (to Y.Z.); NHLBI K08HL140143 (to B.H.); and Gree Foundation (to C.M.). Author contributions: Y.Z. conceived the project. C.M. and Y.Z. designed the experiments. C.M. performed the experiments and bulk RNA-seq analysis. B.H. performed single-cell RNA-seq and analysis. Y.Z. performed computational analysis. All authors interpreted the data and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in this paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the corresponding author.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eabb3629/DC1
REFERENCES AND NOTES
- 1.Atala A., Kasper F. K., Mikos A. G., Engineering complex tissues. Sci. Transl. Med. 4, 160rv12 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Ogle B. M., Bursac N., Domian I., Huang N. F., Menasché P., Murry C. E., Pruitt B., Radisic M., Wu J. C., Wu S. M., Zhang J., Zimmermann W.-H., Vunjak-Novakovic G., Distilling complexity to advance cardiac tissue engineering. Sci. Transl. Med. 8, 342ps13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucitti J. L., Jones E. A. V., Huang C., Chen J., Fraser S. E., Dickinson M. E., Vascular remodeling of the mouse yolk sac requires hemodynamic force. Development 134, 3317–3326 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Culver J. C., Dickinson M. E., The effects of hemodynamic force on embryonic development. Microcirculation 17, 164–178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gimbrone M. A. Jr., Anderson K. R., Topper J. N., The critical role of mechanical forces in blood vessel development, physiology and pathology. J. Vasc. Surg. 29, 1104–1151 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Hahn C., Schwartz M. A., Mechanotransduction in vascular physiology and atherogenesis. Nat. Rev. Mol. Cell Biol. 10, 53–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baeyens N., Bandyopadhyay C., Coon B. G., Yun S., Schwartz M. A., Endothelial fluid shear stress sensing in vascular health and disease. J. Clin. Investig. 126, 821–828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzima E., Irani-Tehrani M., Kiosses W. B., Dejana E., Schultz D. A., Engelhardt B., Cao G., DeLisser H., Schwartz M. A., A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature 437, 426–431 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Song J. W., Gu W., Futai N., Warner K. A., Nor J. E., Takayama S., Computer-controlled microcirculatory support system for endothelial cell culture and shearing. Anal. Chem. 77, 3993–3999 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Chiu J.-J., Chien S., Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol. Rev. 91, 327–387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien S., Mechanotransduction and endothelial cell homeostasis: The wisdom of the cell. Am. J. Physiol. Heart Circ. Physiol. 292, H1209–H1224 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Lin K., Hsu P.-P., Chen B. P., Yuan S., Usami S., Shyy J. Y.-J., Li Y.-S., Chien S., Molecular mechanism of endothelial growth arrest by laminar shear stress. Proc. Natl. Acad. Sci. U.S.A. 97, 9385–9389 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merna N., Wong A. K., Barahona V., Llanos P., Kunar B., Palikuqi B., Ginsberg M., Rafii S., Rabbany S. Y., Laminar shear stress modulates endothelial luminal surface stiffness in a tissue-specific manner. Microcirculation 25, e12455 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traub O., Berk B. C., Laminar shear stress: Mechanisms by which endothelial cells transduce an atheroprotective force. Arterioscler. Thromb. Vasc. Biol. 18, 677–685 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Davies P. F., Hemodynamic shear stress and the endothelium in cardiovascular pathophysiology. Nat. Rev. Cardiol. 6, 16–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent P. E., Plata A. M., Hunt A. A. E., Weinberg P. D., Sherwin S. J., Blood flow in the rabbit aortic arch and descending thoracic aorta. J. R. Soc. Interface 8, 1708–1719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong D. C., Yu Z., Brighton H. E., Bear J. E., Bautch V. L., Tortuous microvessels contribute to wound healing via sprouting angiogenesis. Arterioscler. Thromb. Vasc. Biol. 37, 1903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helisch A., Schaper W., Arteriogenesis: The development and growth of collateral arteries. Microcirculation 10, 83–97 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Ghaffari S., Leask R. L., Jones E. A. V., Flow dynamics control the location of sprouting and direct elongation during developmental angiogenesis. Development 142, 4151–4157 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Han H.-C., Twisted blood vessels: Symptoms, etiology and biomechanical mechanisms. J. Vasc. Res. 49, 185–197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pijnenborg R., Vercruysse L., Hanssens M., The uterine spiral arteries in human pregnancy: Facts and controversies. Placenta 27, 939–958 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Folkman J., Angiogenesis. Annu. Rev. Med. 57, 1–18 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Geudens I., Gerhardt H., Coordinating cell behaviour during blood vessel formation. Development 138, 4569–4583 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Wang C. Y., On the low-Reynolds-number flow in a helical pipe. J. Fluid Mech. 108, 185–194 (1981). [Google Scholar]
- 25.Germano M., On the effect of torsion on a helical pipe flow. J. Fluid Mech. 125, 1–8 (1982). [Google Scholar]
- 26.Alastruey J., Siggers J. H., Peiffer V., Doorly D. J., Sherwin S. J., Reducing the data: Analysis of the role of vascular geometry on blood flow patterns in curved vessels. Phys. Fluids 24, 031902 (2012). [Google Scholar]
- 27.Lorthois S., Lauwers F., Cassot F., Tortuosity and other vessel attributes for arterioles and venules of the human cerebral cortex. Microvasc. Res. 91, 99–109 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Yearwood T. L., Chandran K. B., Experimental investigation of steady flow through a model of the human aortic arch. J. Biomech. 13, 1075–1088 (198X). [DOI] [PubMed] [Google Scholar]
- 29.Nivedita N., Ligrani P., Papautsky I., Dean flow dynamics in low-aspect ratio spiral microchannels. Sci. Rep. 7, 44072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigoryan B., Paulsen S. J., Corbett D. C., Sazer D. W., Fortin C. L., Zaita A. J., Greenfield P. T., Calafat N. J., Gounley J. P., Ta A. H., Johansson F., Randles A., Rosenkrantz J. E., Louis-Rosenberg J. D., Galie P. A., Stevens K. R., Miller J. S., Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 364, 458–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y., Chen J., Craven M., Choi N. W., Totorica S., Diaz-Santana A., Kermani P., Hempstead B., Fischbach-Teschl C., López J. A., Stroock A. D., In vitro microvessels for the study of angiogenesis and thrombosis. Proc. Natl. Acad. Sci. U.S.A. 109, 9342–9347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillen F., Griffioen A. W., Tumour vascularization: Sprouting angiogenesis and beyond. Cancer Metastasis Rev. 26, 489–502 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts M. A., Tran D., Coulombe K. L. K., Razumova M., Regnier M., Murry C. E., Zheng Y., Stromal cells in dense collagen promote cardiomyocyte and microvascular patterning in engineered human heart tissue. Tissue Eng. Part A 22, 633–644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callens S. J. P., Uyttendaele R. J. C., Fratila-Apachitei L. E., Zadpoor A. A., Substrate curvature as a cue to guide spatiotemporal cell and tissue organization. Biomaterials 232, 119739 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Simons M., Eichmann A., Molecular controls of arterial morphogenesis. Circ. Res. 116, 1712–1724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cozzi V., Garlanda C., Nebuloni M., Maina V., Martinelli A., Calabrese S., Cetin I., PTX3 as a potential endothelial dysfunction biomarker for severity of preeclampsia and IUGR. Placenta 33, 1039–1044 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Schneller D., Hofer-Warbinek R., Sturtzel C., Lipnik K., Gencelli B., Seltenhammer M., Wen M., Testori J., Bilban M., Borowski A., Windwarder M., Kapel S. S., Besemfelder E., Cejka P., Habertheuer A., Schlechta B., Majdic O., Altmann F., Kocher A., Augustin H. G., Luttmann W., Hofer E., Cytokine-like 1 is a novel proangiogenic factor secreted by and mediating functions of endothelial progenitor cells. Circ. Res. 124, 243–255 (2019). [DOI] [PubMed] [Google Scholar]
- 38.McDonald A. I., Shirali A. S., Aragón R., Ma F., Hernandez G., Vaughn D. A., Mack J. J., Lim T. Y., Sunshine H., Zhao P., Kalinichenko V., Hai T., Pelegrini M., Ardehali R., Iruela-Arispe M. L., Endothelial regeneration of large vessels is a biphasic process driven by local cells with distinct proliferative capacities. Cell Stem Cell 23, 210–225.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evellin S., Galvagni F., Zippo A., Neri F., Orlandini M., Incarnato D., Dettori D., Neubauer S., Kessler H., Wagner E. F., Oliviero S., FOSL1 controls the assembly of endothelial cells into capillary tubes by direct repression of αv and β3 integrin transcription. Mol. Cell. Biol. 33, 1198–1209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S., Rinn J. L., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H. A., Trapnell C., Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I. W. H., Ng L. G., Ginhoux F., Newell E. W., Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Fang J. S., Coon B. G., Gillis N., Chen Z., Qiu J., Chittenden T. W., Burt J. M., Schwartz M. A., Hirschi K. K., Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat. Commun. 8, 2149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mack J. J., Mosqueiro T. S., Archer B. J., Jones W. M., Sunshine H., Faas G. C., Briot A., Aragón R. L., Su T., Romay M. C., McDonald A. I., Kuo C.-H., Lizama C. O., Lane T. F., Zovein A. C., Fang Y., Tarling E. J., de Aguiar Vallim T. Q., Navab M., Fogelman A. M., Bouchard L. S., Iruela-Arispe M. L., NOTCH1 is a mechanosensor in adult arteries. Nat. Commun. 8, 1620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundlisaeter E., Edelmann R. J., Hol J., Sponheim J., Küchler A. M., Weiss M., Udalova I. A., Midwood K. S., Kasprzycka M., Haraldsen G., The alarmin IL-33 is a notch target in quiescent endothelial cells. Am. J. Pathol. 181, 1099–1111 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Zhang B., Montgomery M., Chamberlain M. D., Ogawa S., Korolj A., Pahnke A., Wells L. A., Massé S., Kim J., Reis L., Momen A., Nunes S. S., Wheeler A. R., Nanthakumar K., Keller G., Sefton M. V., Radisic M., Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis. Nat. Mater. 15, 669–678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhee W. J., Ni C.-W., Zheng Z., Chang K., Jo H., Bao G., HuR regulates the expression of stress-sensitive genes and mediates inflammatory response in human umbilical vein endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 107, 6858–6863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen P.-Y., Qin L., Li G., Wang Z., Dahlman J. E., Malagon-Lopez J., Gujja S., Cilfone N. A., Kauffman K. J., Sun L., Sun H., Zhang X., Aryal B., Canfran-Duque A., Liu R., Kusters P., Sehgal A., Jiao Y., Anderson D. G., Gulcher J., Fernandez-Hernando C., Lutgens E., Schwartz M. A., Pober J. S., Chittenden T. W., Tellides G., Simons M., Endothelial TGF-β signalling drives vascular inflammation and atherosclerosis. Nat. Metab. 1, 912–926 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim C. W., Song H., Kumar S., Nam D., Kwon H. S., Chang K. H., Son D. J., Kang D.-W., Brodie S. A., Weiss D., Vega J. D., Alberts-Grill N., Griendling K., Taylor W. R., Jo H., Anti-inflammatory and antiatherogenic role of bmp receptor II in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 33, 1350–1359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chistiakov D. A., Orekhov A. N., Bobryshev Y. V., Effects of shear stress on endothelial cells: Go with the flow. Acta Physiol. 219, 382–408 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Shiba Y., Fernandes S., Zhu W.-Z., Filice D., Muskheli V., Kim J., Palpant N. J., Gantz J., Moyes K. W., Reinecke H., Van Biber B., Dardas T., Mignone J. L., Izawa A., Hanna R., Viswanathan M., Gold J. D., Kotlikoff M. I., Sarvazyan N., Kay M. W., Murry C. E., Laflamme M. A., Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature 489, 322–325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K., KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slenter D. N., Kutmon M., Hanspers K., Riutta A., Windsor J., Nunes N., Mélius J., Cirillo E., Coort S. L., Digles D., Ehrhart F., Giesbertz P., Kalafati M., Martens M., Miller R., Nishida K., Rieswijk L., Waagmeester A., Eijssen L. M. T., Evelo C. T., Pico A. R., Willighagen E. L., WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 46, D661–D667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young M. D., Wakefield M. J., Smyth G. K., Oshlack A., Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11, R14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/38/eabb3629/DC1


