Abstract
Background
Homologous recombination deficiencies (HRD) are present in approximately half of epithelial ovarian cancers, for which PARP inhibitors (PARPi) are becoming a preferred treatment option. However, a considerable proportion of these carcinomas acquire resistance or harbour de novo resistance, posing a significant challenge to treatment.
Methods
To identify new combinatorial therapeutics to overcome resistance to PARPi, we employed high-throughput conditional RNAi and drug screening of patient-derived ovarian cancer cells. To prioritise clinically relevant drug combinations, we integrated empirical validation with analysis of The Cancer Genome Atlas (TCGA) and Genomics of Drug Sensitivity in Cancer (GDSC) datasets to nominate candidate targets and drugs, reaching three main findings.
Findings
Firstly, we found that the PARPi rucaparib enhanced the effect of BET inhibitors (CPI-203 & CPI-0610) irrespective of clinical subtype or HRD status. Additional drug combination screens identified that dasatinib, a non-receptor tyrosine kinase inhibitor, augmented the effects of rucaparib and BET inhibitors, proposing a potential broadly applicable triple-drug combination for high-grade serous and clear cell ovarian carcinomas. Secondly, rucaparib synergised with the BCL2 family inhibitor navitoclax, with preferential activity in ovarian carcinomas that harbour alterations in BRCA1/2, BARD1, or MSH2/6. Thirdly, we identified potentially antagonistic drug combinations between the PARPi rucaparib and vinca alkaloids, anthracyclines, and antimetabolites, cautioning their use in the clinic.
Interpretation
These findings propose therapeutic strategies to address PARP inhibitor resistance using agents that are already approved or are in clinical development, with the potential for rapid translation to benefit a broad population of ovarian cancer patients.
Keywords: Ovarian cancer, Drug combinations, Poly(ADP-ribose) Polymerase Inhibitors, Rucaparib, Dasatinib, Navitoclax
Research in context.
Evidence before this study
Poly(ADP-ribose) polymerase (PARP) inhibitors (PARPi) demonstrate selective activity in ovarian cancer patients with germline or somatic BRCA1/2 mutations. Despite their success in these patients, many patients harbour de novo resistance or develop acquired resistance to PARP inhibitors. Although PARPi combinations including BRD4 or BCL2 inhibitors have been previously proposed, there have not been studies that systematically identify novel combinations by performing high-throughput conditional screening methods in the presence of PARPi using patient-relevant models of ovarian cancer. Aside from the recent FDA approval of olaparib in combination with bevacizumab for maintenance therapy in May 2020, there are currently no other clinically approved combinations with PARPi.
Added value of this study
Our study utilised a recent set of patient-derived ovarian cancer cells that closely recapitulate the genetic and molecular features of their original tumours. We performed high-throughput RNAi and drug screening in conjunction with genomic dataset analyses to reach three clinically relevant findings. Firstly, rucaparib synergised with BET inhibitors, irrespective of clinical subtype or homologous repair deficiency (HRD) status. Dasatinib, a non-receptor tyrosine kinase inhibitor, augmented the synergy of rucaparib and BET inhibitors, proposing a potential broadly applicable triple-drug combination for high-grade serous and clear cell ovarian carcinomas. Secondly, rucaparib synergised with the BCL2 family inhibitor navitoclax, with preferential activity in ovarian carcinomas that harbour alterations in BRCA1/2, BARD1, or MSH2/6. Thirdly, we identified potential antagonistic PARPi drug combinations with vinca alkaloids, anthracyclines, and antimetabolites, suggesting use of these combinations in the clinic may not be ideal.
Implications of all the available evidence
This study systematically identifies and proposes new combinations with PARP inhibitors using agents that are clinically available or in active development, with potential to be rapidly translated to the clinic. Currently, only subsets of high-grade serous ovarian cancer patients show clear benefit from PARP inhibitors. Here, we have identified PARP inhibitor combinations that could benefit broader populations of ovarian cancer patients, including those with high grade serous or clear cell carcinoma, irrespective of BRCA1/2 status.
Alt-text: Unlabelled box
1. Introduction
Ovarian cancer has the highest mortality of all gynaecologic cancers, with more than 22,000 new diagnoses and approximately 14,000 deaths per year in the United States alone [1]. The current standard of care consists of radical surgical resection of visible disease and treatment with platinum and taxane-based chemotherapies [2]. Although initial response rates are high, approximately 70% of patients recur [1]. Progress in identifying novel treatments for ovarian carcinoma has been relatively slow due to the complex genetic landscape, the lack of commonly occurring “druggable” targets, and scarcity of accurate model systems. A major breakthrough for precision medicine was provided by the discovery that carcinomas with deficiencies in homologous recombination pathway DNA repair genes such as BRCA1/2 utilise alternate PARP-dependent DNA repair pathways for survival, and are therefore more sensitive to PARP inhibition [3]. The implementation of PARP inhibitor (PARPi) therapy has transformed the standard care of treatment for many ovarian cancer patients.
Since this discovery, defects in other homologous recombination repair genes have also been demonstrated to cause sensitivity to PARPi [3], [4], [5], [6], [7]. Several clinical studies indicate that the broader category of homologous repair deficiency (HRD) status and/or genomic loss of heterozygosity predicts a more favourable response to PARPi in the maintenance setting (alone or in combination with bevacizumab) [8], [9], [10], which has led changes to clinical management of patients for PARPi therapy.
Unfortunately, resistance to PARPi is a clinical problem and disease progression following treatment is nearly inevitable [11]. BRCA1/2 mutated cancers can develop resistance to PARPi through somatic secondary BRCA1/2 (reversion) mutations that restore the open reading frame and functional homologous recombination repair [11], [12], [13], [14], [15]. Similarly, loss of BRCA1 methylation in BRCA1 methylated ovarian carcinoma provides another mechanism of PARPi resistance [16]. Other cancers are proficient in homologous recombination repair and are innately resistant to PARPi. Therefore, there is a pressing clinical need to develop therapeutic strategies that augment PARPi activity to overcome common resistance mechanisms and broaden the potential patient population that could benefit.
Previous PARPi sensitiser screens have identified candidate genes and drugs as possible combinations with PARPi; however, the use of long established cell lines raises concerns to the applicability of these results to larger patient populations [17], [18], [19], [20], [21], [22]. To discover and prioritise clinically relevant drug combinations to overcome acquired or innate PARPi resistance, we have combined conditional high-throughput siRNA and drug screening employing newer patient-derived ovarian cancer cells (PDCs) that include both high grade serous ovarian carcinoma (HGSC) and ovarian clear cell carcinoma (OCCC) subtypes [23]. These PDCs were derived using an optimised culture media and have been shown to genetically and phenotypically mirror the original patient's tumours, for example showing more than 95% identity with respect to genomic loss of heterozygosity pattern [23]. In addition, in vitro cisplatin response of PDCs correlated to the patient's clinical response [23]. Furthermore, the HGSC PDCs have consistent copy number variation pattern and proteomic profiles when compared to primary HGSC within the TCGA, indicating a potential applicability of our findings to larger patient populations [23,24].
In this study, we focused on rucaparib, a potent PARP1/2/3 inhibitor that was approved by the FDA in late 2016 for treatment of patients with deleterious BRCA1/2 mutation associated recurrent ovarian carcinoma and for maintenance treatment of recurrent ovarian carcinoma following response to platinum chemotherapy [8,9,25,26]. By focusing on druggable gene targets and independent cross-validation utilising clinically-focused oncology drug libraries, we generated a functional atlas to investigate novel strategies to overcome rucaparib resistance. The results were integrated with population-based genomics for target prioritisation and validated using orthogonal assays. This multifaceted approach identified the clinically relevant drug combinations of rucaparib and navitoclax in the context of HRD, and the three-drug combination of rucaparib, dasatinib, and BET inhibitors (BETi), irrespective of HRD status (Fig. 1a).
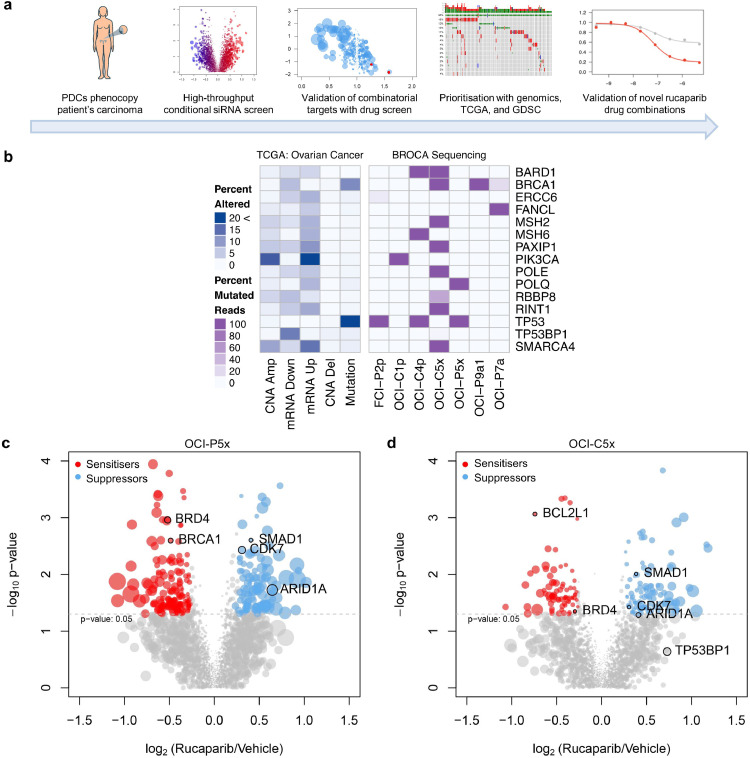
Fig. 1.
Integration of functional genomics with PDCs and big data for drug combination identification. (a) Schematic of the process used to identify rucaparib drug combinations, employing high-throughput conditional siRNA and drug screening on PDCs that genetically match the original tumour with prioritisation of drug targets using TCGA and GDSC. (b) Next generation sequencing of a targeted gene panel; only mutated genes within the PDCs are shown; darker purple indicates 100% mutated reads whereas lighter shades are a gradient of the percent mutated. (c) Volcano plots of the conditional siRNA screens on OCI-P5x cells. (d) Volcano plots of the conditional siRNA screens on OCI-C5x cells. Genes identified as rucaparib sensitisers are highlighted in red and genes identified as rucaparib suppressors in blue. Size of the dots represent the absolute difference in percent viability between siRNA alone and siRNA and rucaparib.
2. Materials and methods
2.1. Cell lines and culture
The patient derived Ovarian Cancer Ince (OCI) cell lines (OCI-P5x, OCI-C5x, FCI-P2p, OCI-P9a1, OCI-P7a, OCI-C1p, OCI-C2p, and OCI-C4p) were cultured in previously defined optimal media conditions [23]. These cells are available from the Ince Laboratory at Weill Cornell Medical College and OCMI medium will be available from US Biologicals. PE01, PE04, UWB1.289, UWB1.289 + Vector, and UWB1.289 + BRCA1 cell lines were provided by Dr. Swisher and grown in previously defined optimal media conditions [27,28]. OVCAR8 and CAOV3 cells were obtained from the American Type Culture Collection (ATCC) and cultured in RPMI 1640 and DMEM medium (Gibco) respectively, both supplemented with 10% fetal calf serum (FCS) and Primocin™ (Invivogen). KURAMOCHI cells were purchased from Sekisui Xenotech (Kansas City, KS) and cultured in RPMI 1640 medium supplemented with 10% FCS and Primocin™. Mycoplasma testing was performed either through IDEXX BioResearch (Columbia, MO) or using the MycoProbe® Mycoplasma Detection kit (R&D Systems). All cells were cultured at 37 °C in a humidified atmosphere with 5% CO2.
2.2. DNA sequencing
Genomic DNA was isolated using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer's protocol. The quantity and quality of DNA was analysed using a NanoDrop spectrophotometer. BROCA Cancer Risk Panel next generation sequencing was performed by the Department of Laboratory Medicine at the University of Washington. This targeted panel consists of ~60 DNA repair genes, covering 320 to >1,000 sequencing reads per bp [29] (Supp. Table 1).
2.3. Gene expression profiling
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol, and the quantity and quality was analysed by the NanoDrop spectrophotometer. RNA was analysed by gene expression profiling of 770 genes, using the nCounter® PanCancer Pathways panel (NanoString Technologies). Data was normalised to multiple endogenous control genes, calculating a Z-score for each gene.
2.4. siRNA screen
For the two PDCs chosen for screening, OCI-P5x and OCI-C5x, a preliminary feasibility assessment was conducted to identify optimal transfection conditions, rucaparib concentration, and readout timing. A matrix of 192 different conditions, including cell number, type of transfection reagent, siRNA ratio relative to transfection reagent, and final transfection concentrations were tested in triplicate. Viability was assessed with CellTiter-Glo® after ~1 h of incubation at room temperature. The readout timing was tested at 72 and 96 h. The conditions with the highest Z’ factor were then used for screening: Dharmafect 1 (Horizon Discovery) as the transfection reagent at a ratio of 1:1 volume with siRNAs; the siRNA amount at 1.25 pmol, followed by a 72 and 96 h incubation for OCI-C5x and OCI-P5x, respectively.
The custom siRNA library collection was designed to target all known human kinases, DNA damage repair genes, epigenetic factors, MYC-synthetic lethal genes, and a collection of genes that are commonly altered in ovarian cancer. siRNAs were purchased from Sigma or Qiagen. In total, the cells were transfected with 2400 different pooled siRNAs targeting 2187 unique genes in three technical replicates in the presence and absence of an IC30 concentration (20 μM) of rucaparib. This concentration of rucaparib was chosen because, while it did not dramatically decrease cellular viability, it has been shown to effectively inhibit PARP activity in other cell lines [30]. Cell viability was assessed after 72 and 96 h for OCI-C5x and OCI-P5x, respectively, using CellTiter-Glo® (Promega) and was quantified using an EnVision plate reader (Perkin-Elmer). We defined sensitiser genes as those that when knocked down in the presence of rucaparib, significantly decreased cell viability by >1.2 fold change or 20% of the vehicle. Suppressor genes were defined as those, that when knocked down in the presence of rucaparib, significantly increased cell viability by >1.2 fold or 20% of the vehicle.
2.5. Pathway analysis
Functional annotation of the suppressor and sensitiser genes was performed using DAVID Bioinformatics Resource 6.8 with the default threshold settings through the R package: gProfileR_0.6.6. DAVID integrates data from KEGG, REACTOME, BioCarta, among others. To account for potential siRNA library bias, the background genes used in this analysis were the genes probed by the siRNA library.
2.6. Drug screens
The 395-compound library was purchased from Selleck Chemicals and targets a broad range of cancer-related pathways. The drug library contains FDA-approved and tool compounds that target a broad range of oncogenic processes, including PI3K, HDAC, mTOR, CDK, JAK, and RTK. Drugs were diluted to an 8-point dose curve incorporating a 3-fold dilution step. The highest final drug concentrations were 5 μM; DMSO or PBS remained consistent across the wells at 0.05%. Cells were seeded at ~30% confluence in 384-well microtiter plates. The next day, drugs were added using a CyBio FeliX liquid handler (Analytik Jena AG), and cells were further incubated for an additional 144 h. Cell viability was determined using CellTiter-Glo® 2.0 and a BioTek H4 Synergy plate reader. Single agent responses were normalised to the solvent concentration (either 0.05% PBS or DMSO) while the combination drug responses were normalised to the solvent concentration and 10 μM of rucaparib. Z factors were calculated for each microtiter plate to ensure adequate screening quality.
The four-arm drug study was performed using an IC30 of CPI-0610 (1.18 μM) and rucaparib (10 μM). Cells were seeded at ~30% confluence in 384-well microtiter plates. The next day, drugs were added using a CyBio FeliX liquid handler (Analytik Jena AG), and cells were further incubated for an additional 144 h. Cell viability was determined using CellTiter-Glo® 2.0 and a BioTek H4 Synergy plate reader. Single agent responses were normalised to 0.1% DMSO. Combination responses were normalised to 0.1% DMSO + the respective combination of drugs (rucaparib, CPI-0610, or rucaparib + CPI-0610).
For all drug screens, IC50, area-under-the-curve (AUC), and Goodness of Fit (GOF) values were calculated with the R ‘nplr’ package version: 0.1–7. Drugs that showed an AUC fold change of ≥1.2 compared to the single agent and AUC combinations Z-score <-1 were further prioritised as top rucaparib drug combinations. The AUC Z-score was calculated by comparing each dose-response curve with a SEngine Precision Medicine internal database of drug responses across 54 different patient-derived cancer cell cultures on a per compound basis. This method of analysis allows identification of unique vulnerabilities across multiple samples [31].
2.7. Drug synergy assays
OCI-P5x, OCI-C5x, OCI-P5x, FCI-P2P, OVCAR8, KURAMOCHI and CAOV3 cells were seeded in 96-well plates at 1000–2000 cells/well, incubated overnight, and treated with test compounds or DMSO controls the next day. After 6 days, cell viability was determined by incubating with CellTiter-Glo® 2.0 and luminescence was read using a BioTek H4 Synergy plate reader. Drug synergy was calculated using the Bliss and Loewe methods and graphed using Combenefit software [32]. Both methods yielded similar results, and results from the Loewe method are shown in the figures.
2.8. Clonogenic assay
OCI-P5x and OCI-C5x cells were seeded in 12-well plates at 5000 cells/well, incubated overnight, and treated with test compounds or DMSO controls the next day. After 10–14 days, colonies were fixed with 25% methanol and stained with 0.5% crystal violet. Plates were scanned prior to quantitation by dissolving stain with 10% acetic acid for 15 min and then measuring absorbance at 590 nm. Measurements were normalised to the untreated control.
2.9. Western blotting
Protein lysate preparation and SDS-PAGE were performed as previously described [33]. Immunoblot analysis was performed using the following primary antibodies from Cell Signaling Technology at 1:1000 dilution: p-SRC (Tyr416) (#6943, RRID:AB_10013641), SRC (#2123, RRID:AB_2106047), cyclin D1 (#55506), p-H2AX (Ser139) (#9718, RRID:AB_2118009), PARP (#9542, RRID:AB_2160739), cleaved PARP (Asp214) (#5625, RRID:AB_10699459), β-actin (#4970, RRID:AB_2223172). IRDye 680RD or 800CW secondary antibodies (Licor #926-68071, #926-68070, #926-32211, #926-32210; RRID: AB_10956166, AB_10956588, AB_621843, AB_621842) were purchased from Licor and used at 1:5,000 dilution. Images were analysed using the ChemiDoc MP imaging system with Image Lab software (BioRad Laboratories).
2.10. siRNA target validation
Dicer-substrate short interfering RNAs (DsiRNAs) to SRC, CDK9, BRD4, BRCA1, BCL2L1, and BCL2L2 were purchased from Integrated DNA Technologies. OCI-P5x and OCI-C5x cells were seeded in 96-well plates at 1000 cells/well and incubated overnight. The following day, siRNAs (25 nM) were transfected using Dharmafect 1 according to the manufacturer's recommendations. After 48 h, cells were treated with rucaparib to obtain a 10-point dose curve (0.0045–30 μM). After 6 days treatment with rucaparib, cell viability was determined by incubating with CellTiter-Glo® 2.0 and luminescence was read using a BioTek H4 Synergy plate reader.
2.11. In vivo mouse study
Xenograft studies were performed at Crown Bioscience. OCI-P5x cells were engrafted subcutaneously into female NOG mice. Each mouse was inoculated at the right flank with 5 × 106 OCI-P5x tumour cells in 0.1 ml of PBS mixed with Matrigel (1:1) for tumour development. Grouping was performed using StudyDirectorTM software (Studylog Systems). Treatments were initiated when the mean tumour size reached ~100 mm3. The drugs were administered to the tumour-bearing mice in 0.5% methylcellulose (Sigma-Aldrich) according to the predetermined regimen (Supp. Table 2). Tumour volumes were measured twice weekly using a caliper, and volumes were determined using the formula: V = 0.5 a x b2, where a and b are the length and width of the tumour, respectively. Tumour weight was measured at study termination. Establishment of the OCI-P5x cell line was previously described [23] under approved procedures by the Institutional Review Boards of the University of Miami, Brigham and Women's and Massachusetts General Hospitals with written consent from the patient. The protocol involving the care and use of animals in this study was approved by the Institutional Animal Care and Use Committee (IACUC) of Crown Bioscience (Pro. No. E3439-U1703). During the study, the care and use of animals was conducted in accordance with the regulations of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
2.12. Statistical analysis
For analyses of the siRNA screens and drug screens, a t-test p-value < 0.05 was considered statistically significant. For validation assays, statistical analyses were performed using Graphpad Prism. Significant differences between treatment groups were determined by one-way analysis of variance followed by Tukey's post-test. Differences were considered significant if p-value < 0.05.
2.13. Data deposition
BROCA sequencing data is available at the NCBI BioProject database (Accession No. PRJNA657639). Results from the siRNA and drug screens have been uploaded to PubChem (AID 1508597 & AID 1508598, respectively). Gene expression profiling data has been uploaded to GEO (GSE153440).
3. Results
3.1. Molecular characterisation of PDCs and functional genomic screening
To molecularly stratify seven of the ovarian cancer PDCs by DNA repair capabilities, we used a targeted-deep sequencing panel (BROCA test) (Fig. 1b, Supp. Table 1) [23,34]. Due to differences in mutational status and histology, optimal growth kinetics, and response to PARPi, we chose OCI-P5x, derived from a HGSC and OCI-C5x, derived from an OCCC for further investigation (Fig. 1b, Supp. Fig. 1a-c). OCI-P5x harbours mutations in TP53 (p.Y236N), while OCI-C5x exhibits numerous genomic alterations on the BROCA panel, including POLE and MSH2, suggesting a possible hypermutator phenotype (Fig. 1b) [23]. The hypermutator phenotype was recently shown to be present in ~6% of OCCC cases [35]. Although OCI-C5x harbours a deleterious BRCA1 (p.Q541X) mutation in 100% of its sequencing reads, its in vitro response to PARPi is more similar to that of a BRCA1/2 reverted or restored cell line (i.e., PE04 and UWB1.289 ++) than to cell lines carrying a known deleterious BRCA1/2 mutation (i.e. PE01 or UWB1.289) (Supp. Fig. 1b-c) [27,28]. Despite the presence of BRCA1 mutations in OCI-C5x cells, RAD51 foci were detected following DNA damage by ionising irradiation (Supp. Fig. 1d). OCI-C5x cells also have elevated levels of RAD51 protein compared to normal ovary epithelial cells (Supp. Fig. 1e), which could allow cells lacking BRCA1 function to circumvent its requirement in RAD51 subnuclear assembly and homologous repair [36]. The discrepancy between BRCA1 mutation status and PARPi response highlights the utility of functional testing in conjunction with genomic sequencing to accurately determine vulnerabilities in patient cells.
To interrogate the potential functional interactions with rucaparib in these two genetic settings, we employed a custom arrayed siRNA library targeting 2187 genes, including the kinome (712 genes), epigenome (1192 genes), DNA damage and repair (318 genes), commonly aberrated (mutated, copy number variation, or mRNA over/underexpressed) cancer-associated genes in TCGA HGSC cohort (123 genes), and previously identified MYC-synthetic lethal genes (48 genes) [37,38]. The siRNA library was selected based on potential druggability, novelty, and relevance to the mechanism of action of PARPi. To perform the rucaparib conditional siRNA screen, OCI-P5x and OCI-C5x cells were screened with the siRNA library alone or in the presence of the IC30 concentration of rucaparib. As a positive control, knockdown of BRCA1 sensitised OCI-P5x cells to rucaparib (Fig. 1c).
3.2. Inhibition of BRD4 and transcriptional machinery sensitises ovarian cancer to rucaparib in vitro and in vivo
The conditional siRNA screen identified a group of rucaparib sensitiser genes, i.e. knockdown of sensitiser genes increased cell death in the presence of rucaparib. These genes represent potential combinatorial targets with PARPi. Of the 2187 genes tested, 75 and 163 genes were identified as rucaparib sensitisers in OCI-C5x and OCI-P5x cells, respectively (Supp. Table 3). There were six genes in common to both PDCs, including the bromodomain-containing gene, BRD4 (Fig. 1c and d). Specific to OCI-P5x, the most represented gene families were members of the Mediator complex, MED4/8/10/16/23 (5 out of 27 in the siRNA library) as well as TBP-associated factors (TAFs), TAF1/1L/5L/7/12 (5 out of 11 in the siRNA library), indicating that, in addition to BRD4, other components of transcriptional complexes may modulate the response to rucaparib and potentially other PARPi (Supp. Table 3).
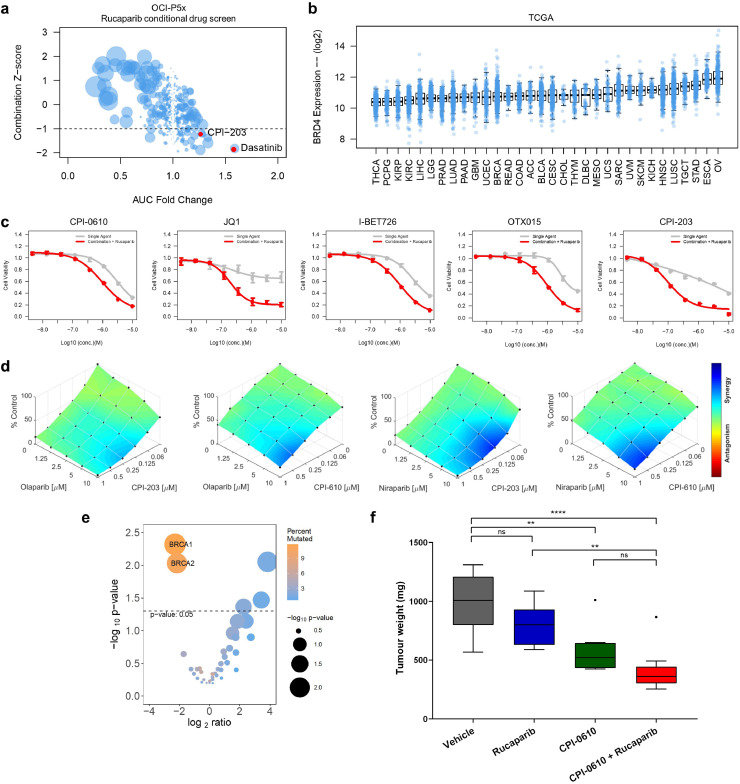
Consistent with the identification of BRD4 as a common rucaparib sensitiser in the siRNA screen, the BETi CPI-203 emerged as a top drug sensitiser in OCI-P5x cells in a rucaparib conditional drug screen (Fig. 2a), where cells were screened with a library of 395 oncology-focused compounds alone or in the presence of the IC30 concentration of rucaparib (Fig. 2a). Analysis of TCGA revealed that BRD4 mRNA expression levels are highest in HGSC when compared to all other cancer types (Fig. 2b), highlighting the potential clinical utility of BETi for ovarian cancer. To further validate rucaparib and BETi as an effective drug combination, we obtained a panel of clinically relevant and commonly used BETi for testing: CPI-0610, JQ1, I-BET-762, and OTX015 [39], [40], [41]. Confirming the findings of the siRNA and drug combination screen, the effect of each BETi was improved with the addition of rucaparib in OCI-P5x cells (Fig. 2c). Rucaparib and BETi synergy was also observed in 6 additional ovarian carcinoma PDCs and cell lines irrespective of HR status or clinical subtype, including OCI-C5x, FCI-P2p, OCI-C4p, OVCAR8, CAOV3, and KURAMOCHI (Supp. Fig. 2a). Further, this synergy was not restricted to rucaparib, as two other PARPi, olaparib and niraparib, also demonstrated synergistic activity with the BETi CPI-203 and CPI-0610 (Fig. 2d, Supp. Fig. 2b).
Fig. 2.
Rucaparib synergises with BETi in HGSC. (a) Drug screen utilising a 395 compound library in combination with rucaparib on OCI-P5x cells. AUC fold change vs. AUC Z-score of the combination; the size of the dot corresponds to the absolute AUC difference between single agent and combination. Highlighted in red are the top hits: dasatinib and CPI-203. (b) Boxplot of BRD4 mRNA expression across TCGA published datasets sorted by median expression level. (c) Response of OCI-P5x cells to IC30 rucaparib alone and in combination with the BETi CPI-0610, JQ1, I-BET726, OTX015, CPI-203. (d) Dose-response plots of olaparib and niraparib in combination with the BETi CPI-203 or CPI-0610 demonstrating synergistic drug combination effects in OCI-P5x cells. (e) Mutation enrichment of BRD4 overexpression (Z-score > 2) or copy number amplification in TCGA HGSC, demonstrating that BRCA1 and BRCA2 mutations are mutually exclusive with BRD4 overexpression or amplification. (f) Tumour weights from in vivo study examining the rucaparib and CPI-0610 combination using OCI-P5x xenografts. Box-and-whisker plots are presented according to the Tukey method: **p < 0.01, ****p < 0.0001, ns=not significant (one- way ANOVA followed by Tukey's test).
Analysis of over 1000 cell lines within the Genomics of Drug Sensitivity in Cancer (GDSC) dataset revealed that ovarian cancer is the sixth most sensitive cancer type to the BETi, JQ1 (Supp. Fig. 3a) [42]. HGSC patients with high BRD4 expression (BRD4 mRNA overexpression (Z-score > 2) or copy number amplification) have a significantly decreased overall survival than those that do not (Supp. Fig. 3b). BRD4-high alterations are also mutually exclusive with BRCA1/2 mutations in the TCGA HGSC cohort (Fig. 2e). BRCA2-mutated colorectal cancer cell lines are significantly more sensitive to BET inhibition than their wild type counterparts (Supp. Fig. 3c) [43]. Additionally, when using a recently published aneuploidy score that reflects the total number of arm-level copy-number alterations in a sample, we found that BRD4-high HGSC have a significantly higher rate of aneuploidy than unaltered BRD4 HGSC (Supp. Fig. 3d–f) [44]. These analyses are consistent with the role BRD4 plays in NHEJ and copy number variation [45,46], and suggest the mechanism of observed synergy between rucaparib and BETi could involve perturbing BRD4-mediated adaptation of cancer cells to genomic instability.
For in vivo validation, OCI-P5x cells were engrafted into NOG female mice for a four-arm drug study (Supp. Table 2). CPI-0610 was chosen as the modifying agent due to its active clinical development (NCT02158858) and demonstrated synergistic in vitro effects with rucaparib (Supp. Fig. 2). The combination of rucaparib and CPI-0610 resulted in a significant 58% decrease in tumour weight when compared to vehicle (Fig. 2f). Single agent CPI-0610 treatment also significantly decreased tumour weight by 42% when compared to vehicle (Fig. 2f), supporting the potential for BETi to be used for ovarian cancer. The body weights of the mice did not decrease more than 10% compared to initial values over the course of the study, suggesting the regimens used were relatively well tolerated (Supp. Fig. 4a).
3.3. Dasatinib augments the synergy of rucaparib and BETi
While single agent CPI-0610 and the combination of CPI-0610 and rucaparib significantly decreased tumour weight in vivo, tumour volume was not significantly different between the single agent and combination treatments (Supp. Fig. 4b). Further immunohistochemical analysis of the tumours showed that the mitotic index was significantly reduced in each treatment group compared to the vehicle control, but not between the single agent or combination groups (Supp. Fig. 4c). The tumours also showed a high number of cells with aberrant mitotic shapes, which are hallmarks of mitotic catastrophe (Supp. Fig. 4d–e). Despite these observations, the discrepancy between tumour weight and volume responses is unclear. It is possible that the tumours were not effectively cleared using this immuno-compromised mouse model. Xue et al have previously reported that an innate immune response in vivo promotes tumour clearance [47]. Alternatively, the rucaparib and CPI-0610 combination was not effective enough to completely suppress the growth of OCI-P5x xenografts, and there are limitations to the subcutaneous xenograft model in modeling ovarian cancer.
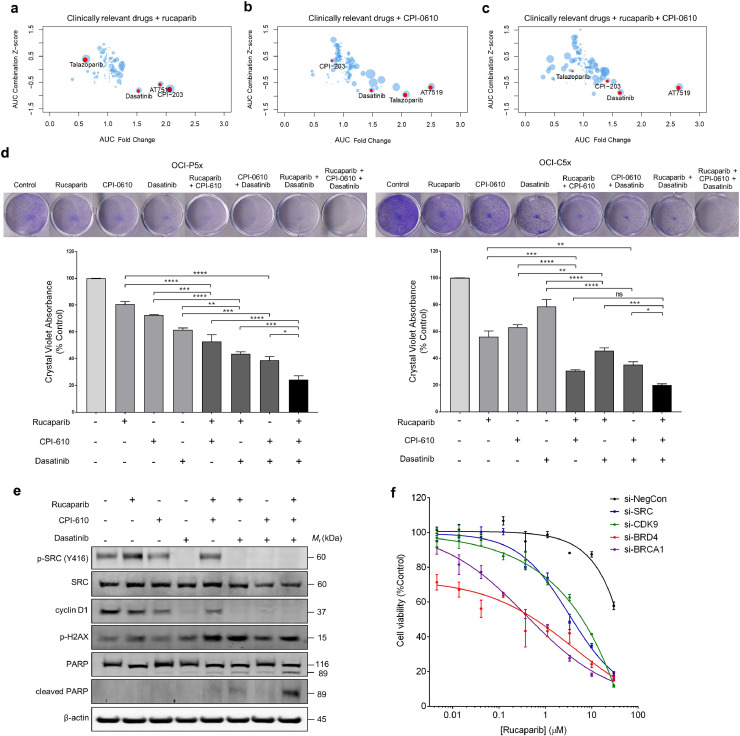
To explore the contribution of other tumour growth pathways in the presence of rucaparib and CPI-0610, we screened a clinical oncology drug library with this combination. This clinically focused drug library contains 131 FDA-approved oncology drugs and targeted agents in late-stage clinical development. This clinical drug library was tested on OCI-P5x cells as single agents, with rucaparib alone, CPI-0610 alone, and in combination with both rucaparib and CPI-0610. We compared the results of the drug screen to an internal database of drug responses across a panel of 54 PDCs, generating Z-scores for each compound. This method of analysis and internal PDC database has been shown to nominate drug combinations that are confirmed in patient-derived xenografts and led to tumour regression of recurrent chemo-resistant cancers [31]. Confirming the results of both the conditional siRNA and high throughput drug screens, the top drug combination for the rucaparib arm was the BETi, CPI-203 (Fig. 3A). Similarly, a top combination in the CPI-0610 arm was talazoparib, another recently approved PARPi (Fig. 3b, Supp. Fig. 5a) [48]. Dasatinib, a dual BCR/ABL and SRC family tyrosine kinase inhibitor, was a top hit in all three of the combinatorial arms (Fig. 3a–c). AT7519, a multi-CDK inhibitor, was also found to synergise with rucaparib and CPI-0610 (Fig. 3a–c). AT7519 potently inhibits CDK9, a kinase that phosphorylates BRD4 and RNA polymerase II, thereby regulating their activity [49], [50], [51].
Fig. 3.
Dasatinib augments the combination of rucaparib and BETi. Drug screen utilising a 131 compound clinically focused library in combination with (a) rucaparib, (b) CPI-0610, and (c) rucaparib and CPI-0610. Depicted results are fold change vs. AUC Z-score of the single agent plus rucaparib. Highlighted in red in the dot plot are confirmatory drug hits and drug hits common within all combination arms. (d) Clonogenic assays performed in OCI-P5x and OCI-C5x cells using rucaparib (10 μM), CPI-0610 (1 μM), and dasatinib (100 nM) as single agents, in double combinations, or in triple combination. Quantitation of crystal violet staining is representative of at least three independent experiments. Data is presented as mean ± SEM: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns=not significant (one- way ANOVA followed by Tukey's test). (e) Western blot analysis of phospho-SRC (Tyr 416), SRC, cyclin D1, phospho-H2AX, PARP, and cleaved PARP in response to treatment with rucaparib (10 μM), CPI-0610 (3 μM), and dasatinib (300 nM) for 48 h in OCI-P5x cells. Blots shown are representative of at least three independent experiments. (f) OCI-P5x cells were treated with siRNA specific to SRC, CDK9, BRD4 or BRCA1 for 48 h. Cells were subsequently treated with rucaparib for a further 6 days before cell viability was measured. Representative of 3 independent experiments, data is presented as mean ± SEM.
Using OCI-P5x, OCI-C5x, and OCI-C4p cells, we demonstrated that the double combinations (rucaparib and CPI-0610, rucaparib and dasatinib, CPI-0610 and dasatinib) were significantly more effective at inhibiting colony formation compared to the respective single agent treatments (Fig. 3d, Supp. Fig. 5b). The rucaparib and dasatinib combination also demonstrated synergy in these cells (Supp. Fig. 5c). However, the triple combination of rucaparib, CPI-0610, and dasatinib was significantly more effective at inhibiting colony formation compared to each double combination, except for the rucaparib and CPI-0610 combination in OCI-C5x cells, which was not significantly different to the triple combination (Fig. 3d, Supp. Fig. 5b). Rucaparib treatment alone increased phosphorylation of the proto-oncogene SRC at Tyr416 in both OCI-P5x and OCI-C5x cells (Fig. 3e, Supp. Fig. 5d), suggesting activation of the pro-survival SRC pathway by PARPi which could have cytoprotective effects and/or mediate the development of PARPi resistance. Dasatinib treatment alone or in combination with rucaparib and CPI-0610 resulted in potent reduction in phospho-SRC levels, as well as downstream cyclin D1 levels (Fig. 3e). The double and triple combinations also resulted in increased levels of p-H2AX, indicating increased DNA damage. The triple combination treatment induced significantly higher levels of PARP cleavage, indicating greater induction of apoptosis compared to the single or double agent treatments (Fig. 3e).
To verify the molecular targets of CPI-0610 and dasatinib in sensitising OCI-P5x and OCI-C5x cells to rucaparib, we knocked down expression of BRD4 and SRC using siRNA for 48 h, then treated the cells with rucaparib for 6 days. Knockdown of SRC and BRD4 enhanced sensitivity of these cells to rucaparib (Fig. 3f, Supp. Fig. 5e). Knockdown of CDK9, a target of AT7519 that also regulates BRD4 activity, also sensitised cells to rucaparib. As a positive control, knockdown of BRCA1 also sensitised OCI-P5x cells to rucaparib. Collectively, these results suggest that the three-drug combination of dasatinib, rucaparib, and BETi could be a novel and clinically relevant therapeutic strategy for ovarian cancer, regardless of HRD status and clinical subtype, by collective suppression of the SRC pathway, enhanced DNA damage and induction of apoptosis.
3.4. Rucaparib sensitises cells with alterations in DNA repair to navitoclax
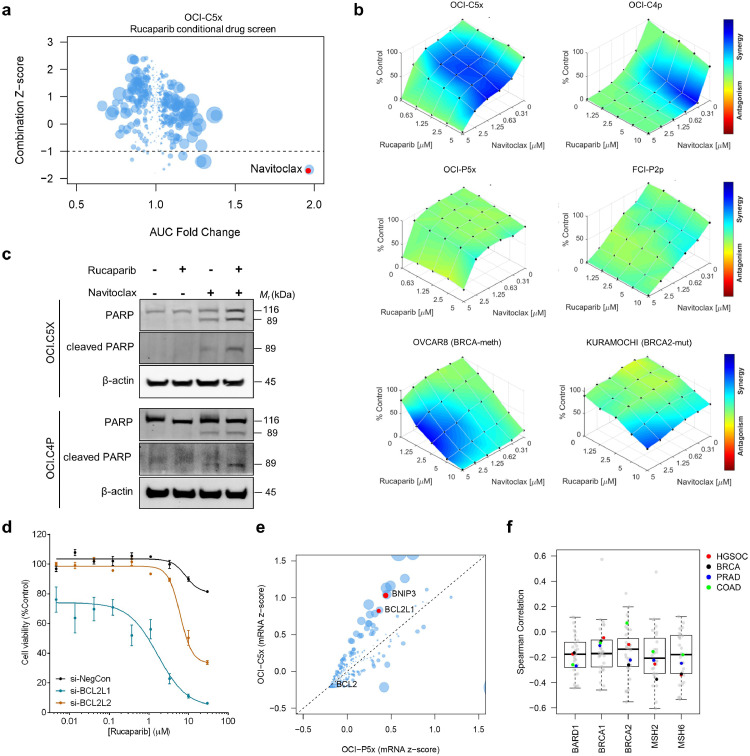
Among the 75 genes that were identified as rucaparib sensitisers in the BRCA1-mutant clear cell carcinoma OCI-C5x cells, BCL2L1, an anti-apoptotic gene, was one of the most statistically significant (Fig. 1d). Consistent with the siRNA screen results, the rucaparib conditional drug screen performed in OCI-C5x cells identified navitoclax, a BCL2, BCL2L1, and BCL2L2 inhibitor [52] as the strongest synergistic drug combination with rucaparib (Fig. 4a). Separate assays confirmed the synergy between rucaparib and navitoclax in the OCCC OCI-C5x and OCI-C4p cells, while little to no synergy was observed in HGSC OCI-P5x and FCI-P2p cells (Fig. 4b). To assess the generalisability of the synergy of navitoclax and rucaparib, we tested this combination in a total of 12 ovarian carcinoma PDCs and cell lines. We observed that the synergistic effect of rucaparib and navitoclax was more prominent for the PDCs and cell lines that harbour alterations in BRCA1/2, BARD1 and/or MSH2/6, which are genes involved in homologous recombination and mismatch repair, irrespective of the ovarian carcinoma subtype (Fig. 4b, Supp. Fig. 6a) [53,54]. Treatment of OCI-C5x and OCI-C4p cells with navitoclax resulted in cleavage of PARP that was further enhanced in the presence of rucaparib (Fig. 4c), indicating that enhanced induction of apoptosis contributed to the synergistic effect of this drug combination.
Fig. 4.
Navitoclax and rucaparib synergise in cancers with DNA repair mutations. (a) Drug screen utilising a 395-compound library in combination with rucaparib on OCI-C5x cells. AUC fold change vs. AUC Z-score of the combination; the size of the dot corresponds to the absolute AUC difference between single agent and combination. Highlighted in red is the top hit, navitoclax. (b) Dose-response plots of rucaparib in combination with navitoclax demonstrating synergistic drug combination effects in OCI-C5x, OCI-C4p, OVCAR8, and KURAMOCHI cells, but little synergy in OCI-P5x and FCI-P2P cells. (c) Western blot analysis of PARP and cleaved PARP levels in OCI-C5x and OCI-C4p cells in response to rucaparib (10 μM) and navitoclax (1 μM) for 48 h. Blots shown are representative of at least three independent experiments. (d) OCI-C5x cells were treated with siRNA specific to BCL2L1 and BCL2L2 for 48 h. Cells were subsequently treated with rucaparib for a further 6 days before cell viability was measured. Representative of 3 independent experiments, data is presented as mean ± SEM. (e) mRNA expression levels of 770 genes in OCI-C5x and OCI-P5x cells. Highlighted in red are genes involved in apoptosis. OCI-C5x has higher BCL2L1 mRNA expression compared to OCI-P5x. (f) Correlation of BCL2L1 expression with BRCA1/2, MSH2/6, and BARD1 expression in The Cancer Genome Atlas (TCGA): high grade serous ovarian carcinoma (HGSC), breast invasive carcinoma (BRCA), prostate adenocarcinoma (PRAD), and colon adenocarcinoma (COAD).
We did not observe a sensitising effect of rucaparib to venetoclax, the BCL2-specific inhibitor (Supp. Fig. 6b), suggesting that the navitoclax sensitisation is due to the inhibition of BCL2L1 and/or BCL2L2 rather than BCL2. To verify this, we silenced expression of BCL2L1 and BCL2L2 in OCI-C5x cells for 48h, then treated with rucaparib for 6 days. BCL2L1 knockdown dramatically sensitised cells to rucaparib, with a less potent effect observed for BCL2L2 knockdown (Fig. 4d). To assess the expression of BCL2 family genes in OCI-C5x and OCI-P5x cells, we analysed mRNA expression of 730 cancer pathway-related genes, which revealed that the pro-survival genes, BCL2L1 and BNIP3, were among the genes that were more highly expressed in OCI-C5x when compared to OCI-P5x cells (Fig. 4e). To investigate the application of the experimental findings to larger populations, we performed a pan-cancer TCGA analysis of BCL2L1 and BCL2L2. We observed significant negative correlation of mRNA expression levels between BCL2L1 or BCL2L2 with BRCA1, BRCA2, MSH2 or MSH6 in ovarian cancer and a variety of cancer types (Fig. 4f, Supp. Fig. 6c). These results suggest that carcinomas that harbour defects in DNA repair may have a reliance on the BCL2 family of anti-apoptotic proteins for cellular survival, suggesting a possible biomarker of response for this combination to investigate in future preclinical and clinical studies.
3.5. Suppressor genes and chemotherapies interfering with the effect of rucaparib
The conditional siRNA screen also identified rucaparib suppressor genes, i.e. knockdown of suppressor genes increased survival in the presence of rucaparib. Identification of rucaparib suppressor genes may inform mechanisms of action of rucaparib as well as mechanisms of resistance. The siRNA screen revealed 109 and 98 genes that suppressed the effect of rucaparib in OCI-P5x and OCI-C5x cells, respectively (Supp. Table 4). Of those, there were 12 genes in common, including SMAD1 and CDK7. SMAD1 and CDK7 are underexpressed (mRNA expression z-score < -2) in 15% and 26% of HGSC, respectively (Supp. Fig. 7a) [55]. Slightly outside the statistical cutoff for common rucaparib suppressors was ARID1A (Fig. 1c and d), a global transcriptional regulator that was recently shown to be mutated in ~67% of OCCC [35,56]. The low expression levels of SMAD1 and CDK7 in HGSC, and high mutational frequency of ARID1A within OCCC could contribute to innate resistance to PARPi.
DAVID pathway analysis identified an enrichment of genes involved with the ribosome, pyrimidine metabolism, nucleotide excision repair, DNA replication, spliceosome, and cell cycle among the rucaparib suppressors (Supp. Fig. 7b) [57]. These results suggest that agents that alter cell growth and division may interfere with the effect of rucaparib. Consistent with this idea, the rucaparib conditional drug screen in OCI-P5x cells (Fig. 2a), also found that addition of rucaparib suppressed the effect of many drugs that target the mitotic spindle and microtubules, including vinca alkaloids, AURKA inhibitors and PLK inhibitors (Supp. Fig. 7c). Also, DNA intercalators such as anthracyclines suppressed the effects of rucaparib. These findings suggest the use of certain chemotherapies in combination with PARPi could antagonise their activity in the clinic.
4. Discussion
In this study, we employed conditional high-throughput siRNA and drug screening of ovarian cancer PDCs that faithfully recapitulate patient tumour genetics and biology to identify and prioritise effective target and drug combinations with PARPi for HGSC and OCCC. The consistent validation of the siRNA gene hits through chemical functional screens and orthogonal assays demonstrates the accuracy of the automated arrayed siRNA platform to rapidly discover novel targets and drug combinations [58].
We identified the synergistic drug combination of rucaparib with the BETi CPI-203 and CPI0610 in ovarian cancer, irrespective of HRD status and histopathological subtype, potentially broadening the clinical reach of PARPi (Fig. 2). CPI-0610-mediated BET inhibition sensitised OCI-P5x cells to rucaparib-induced DNA damage and apoptosis (Fig. 3), consistent with previous reports using other BET inhibitors [59], [60], [61], [62]. While previous reports have focused on BRCA-mutant high grade serous ovarian cancers, our study demonstrates the broader potential of this combination to treat OCCC (Supp. Fig. 3), which generally shows poorer sensitivity to chemotherapeutics and remains difficult to treat in the clinic. Further, despite these studies reporting the potential for PARPi and BETi combinations, the mechanism of action remains unclear. We showed that knockdown of several components of the basic transcriptional machinery that directly or indirectly interact with BRD4 sensitise cells to rucaparib. AT7519, a multi-CDK inhibitor with high selectivity for CDK9, interacts with BRD4 in regulating transcription [49,50]. AT7519 synergises with both rucaparib and CPI-0610 (Fig. 3a–c), and silencing of CDK9 sensitises OCI-P5x cells to rucaparib (Fig. 3f). BRD4 may also have important roles in regulating the DNA damage response via NHEJ and the generation of copy number variations [45,46]. Collectively, these results suggest that the mechanism of synergy of rucaparib and BET inhibition may be due to attenuating the transcriptional functions of BRD4 that allow adaptation of cancer cells to genomic instability and/or its role in NHEJ.
We identified that double and triple combinations between rucaparib, CPI-0610, and/or dasatinib were effective in both OCI-P5x and OCI-C5x cells (Fig. 3), suggesting broad applicability for both HGSC and OCCC patients regardless of HRD status. The current clinical availability of these drugs points to the possibility of rapid clinical translation. Dasatinib, an FDA-approved inhibitor of SRC/ABL for the treatment of chronic myelogenous leukemia, demonstrated limited activity in a Phase II trial as a single agent in the treatment of recurrent epithelial ovarian cancer [63]. Combinations of dasatinib with carboplatin and paclitaxel have demonstrated some clinical activity, but cited concerns regarding myelosuppression and tolerability and the need for biomarker identification [64]. Future studies will seek to determine whether certain patient populations would benefit the most from rucaparib, CPI-0610 and dasatinib combinations. Assessing the level of SRC pathway activation as a biomarker for response to this combination is a possibility; phospho-SRC is elevated in OCI-P5x, FCI-P2p and OCI-C5x compared to other derived PDCs [23], and SRC amplification or high mRNA expression is observed in approximately 10% of serous ovarian cases in TCGA. A recent study also identified ARID1A-mutant OCCC cells as more sensitive to dasatinib through addiction to the SRC family kinase YES1 [65]. With regards to mechanism, our study identified that rucaparib treatment alone increased phosphorylation of the SRC proto-oncogene in both OCI-P5x and OCI-C5x cells, pointing to the oncogenic SRC signaling pathway as a potential mediator of acquired resistance to PARPi. Inhibition of SRC signaling with dasatinib treatment alone or in combination with rucaparib and/or CPI-0610 potently reduced p-SRC and downstream cyclin D1 levels that promote cell cycle progression (Fig. 3e). Thus, the potent activity of the triple combination of rucaparib, CPI-0610, and dasatinib involves dual mechanisms that include suppression of SRC pathway-mediated cell growth and proliferation, enhanced DNA damage and induction of apoptosis. SRC activation is also induced by the veliparib and carboplatin combination in triple-negative breast cancer [66], providing a mechanistic rationale to support the use of dasatinib in combination with other PARPi and across other cancers.
This study also identified the synergistic combination of rucaparib and navitoclax in both OCCC and HGSC cells, including OCI-C5x, OCI-C4p, OVCAR8, and KURAMOCHI cells. Navitoclax is a potent BCL2/BCL2L1/BCL2L2 inhibitor currently being investigated in ovarian cancer (NCT02591095) [67]. We observed rucaparib and navitoclax synergy in ovarian PDCs and cell lines with mutations in the HR pathway genes, BRCA1/2, BARD1, and/or MSH2/6, suggesting a patient population that could benefit from this combination. We observe a negative correlation of BRCA1/2 mRNA expression with BCL2L1, BCL2L2 mRNA expression in a variety of cancer types, including ovarian, breast, colon, and prostate, suggesting elevated BCL2L1 or BCL2L2 expression may compensate for BRCA1/2 deficiency. Studies utilising other PARPi have reported synergy between talazoparib and navitoclax [68], or olaparib and navitoclax [69,70], and BCL2L1/2 overexpression mediates ovarian cancer chemotherapy resistance [70], all in the context of HGSC. Our study demonstrates the feasibility of this combination to be extended to OCCC patients, particularly in those harbouring mutations in BRCA1/2, BARD1, and/or MSH2/6. PARP inhibition has been shown to induce DNA replication stress and genomic instability [71]. We hypothesize that BRCA1/2 mutated cancers, which harbour high levels of replication stress, are more reliant upon the anti-apoptotic proteins, BCL2L1 and/or BCL2L2, for cellular survival [72] and indicate a targetable weakness. Indeed, the rucaparib and navitoclax combination results in greater induction of apoptosis compared to navitoclax alone (Fig. 4c). Further, PARPi-induced senescent cells can re-initiate proliferation upon drug withdrawal, potentially explaining the requirement for sustained PARPi therapy in the clinic [73,74]. This renders cells susceptible to secondary synthetic lethal approaches targeting the senescent state using senolytic drugs, such as navitoclax, and providing further rationale for this combination to limit or overcome PARPi resistance.
Certain chemotherapies and targeted therapies were found to potentially antagonise the effect of rucaparib in OCI-P5x cells, including vinca alkaloids, anthracyclines, antimetabolites, AURKA inhibitors and PLK inhibitors. In contrast to our findings, there is evidence in the literature to suggest that combinations between PARPi and topoisomerase inhibitors, AURKA inhibitors and PLK inhibitors are promising therapeutic strategies [75], [76], [77]. However, these studies used different drugs than those used in the present study, different methods of inhibition, or different cell types. Numerous clinical trials involving PARPi in combination with chemotherapies have been conducted or are underway [78], although successes appear to be limited by toxicity and low overall response rates. Further mechanistic studies will be essential to examine other parameters, such as cell cycle status, to determine the implications of our findings for PARPi and chemotherapy combinations in ovarian cancer.
The siRNA screen in the present study showed that knockdown of ARID1A in both HGSC and OCCC PDCs could suppress the effect of rucaparib, identifying additional potential mechanisms of PARPi resistance. This holds potential clinical significance as PARPi are currently being investigated in OCCC and uterine endometrioid carcinoma – cancer types that harbour mutations in ARID1A ~67% and ~58% of the time, respectively [35,79]. In corroboration with our results, in the ARIEL2 rucaparib clinical trial, patients harbouring ARID1A-mutated carcinomas showed significantly decreased progression free survival [80]. The siRNA screen also identified novel PARPi suppressor genes, such as SMAD1 and CDK7, which are often underexpressed in HGSC and when knocked down significantly protect against the effect of rucaparib. CDK7 has been shown to be a negative regulator of BRD4, indicating a potential rationale for the suppressing function [51]. The high mutational frequency of ARID1A across cancer types and low expression levels of SMAD1 and CDK7 in HGSC constitute potential means of innate PARPi resistance.
In summary, high-throughput functional genomic and combinatorial drug screens in ovarian PDCs revealed mechanisms of innate and acquired resistance to PARPi, highlighting potential antagonistic combinations. In both HGSC and OCCC, combinations with BETi and/or dasatinib present a novel strategy to overcome PARPi resistance through suppression of intrinsic or PARPi-induced SRC pathway signaling, enhanced DNA damage and induction of apoptosis. For patients with HR pathway mutations in BRCA1/2, BARD1 and/or MSH2/6, combinations with navitoclax and PARPi may also be a promising therapeutic approach. The studies herein provide a rationale for future efforts to assess the efficacy, tolerability, and toxicity of the proposed combinations in additional preclinical models and clinical trials. Careful assessment of dose escalation for each drug combination (both double and triple combinations) will be necessary, in addition to identification of optimal dosing regimens, to determine the translational feasibility of these findings to patients.
Author contributions
Study design: GYLL, RS, FXS, TAI, TCH, VKG, BAG, CJK, EMS, CG
Data collection: GYLL, RS, IS, KG, CB, RLD, RR, HAS, TCH
Data analysis and interpretation: GYLL, RS, FXS, IS, KG, CB, RLD, RR, HAS, TAI, TCH, VKG, BAG, CJK, EMS, CG
Manuscript writing: GYLL, RS, FXS, CJK, EMS, CG
Declaration of Competing Interest
RS, FXS, CB, RLD, RR, HAS, CG were employees of SEngine Precision Medicine during this study. FXS holds stock options in SEngine Precision Medicine. VKG, CJK, CG are cofounders of and have equity positions in SEngine Precision Medicine. TAI holds patents related to Ovarian Carcinoma Modified Ince (OCMI) medium formulation (#20190071634, #20160319239, #20140170693) licensed to US Biological/GLG, and has received grants from US Biological/GLG outside the submitted work. TCH is an employee and stockholder of Clovis Oncology, a company involved in the clinical development of PARP inhibitors. The remaining authors declare no competing interests with relevance to this study.
Funding sources
Support for this study was contributed by grants from: the NIH/NCI (U01 CA217883, U54 CA132381, and R01 CA214428) and Rivkin Center for Ovarian Cancer to Christopher Kemp; the Breast Cancer Research Foundation and NIH/NCI R33 CA214310 to Tan Ince. Elizabeth Swisher was supported by a Stand Up To Cancer-Ovarian Cancer Research Fund Alliance-National Ovarian Cancer Coalition Dream Team Translational Cancer Research Grant (SU2C-AACR-DT16-15). The siRNA screen was supported by Cure First, a 501(c)(3) public charity as part of “Project Sweetheart.” The funders had no role in study design, data collection, data analysis, interpretation, or writing of the manuscript.
Footnotes
Funding: National Institute of Health, Rivkin Center for Ovarian Cancer, Breast Cancer Research Foundation, American Association for Cancer Research, Cure First.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102988.
Contributor Information
Goldie Y.L. Lui, Email: glui@fredhutch.org.
Carla Grandori, Email: cgrandori@senginemedicine.com.
Appendix. Supplementary materials
References
- 1.Howlader N, Noone A, Krapcho M, Miller D, Brest A, Yu M. National Cancer Institute; Bethesda, MD: 2019. SEER cancer statistics review, 1975–2016. [Google Scholar]
- 2.Orr B, Edwards RP. Diagnosis and treatment of ovarian cancer. Hematol Oncol Clin N Am. 2018;32(6):943–964. doi: 10.1016/j.hoc.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16(2):110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 4.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108(44):18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stover EH, Konstantinopoulos PA, Matulonis UA, Swisher EM. Biomarkers of response and resistance to DNA repair targeted therapies. Clin Cancer Res. 2016;22(23):5651–5660. doi: 10.1158/1078-0432.CCR-16-0247. [DOI] [PubMed] [Google Scholar]
- 6.Yap TA, Plummer R, Azad NS, Helleday T. The DNA damaging revolution: PARP inhibitors and beyond. Am Soc Clin Oncol Educ Book. 2019;39:185–195. doi: 10.1200/EDBK_238473. [DOI] [PubMed] [Google Scholar]
- 7.Lui GYL, Grandori C, Kemp CJ. CDK12: an emerging therapeutic target for cancer. J Clin Pathol. 2018;71(11):957–962. doi: 10.1136/jclinpath-2018-205356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18(1):75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 9.Coleman RL, Oza AM, Lorusso D, Aghajanian C, Oaknin A, Dean A. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10106):1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray-Coquard I, Pautier P, Pignata S, Perol D, Gonzalez-Martin A, Berger R. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381(25):2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 11.Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68(8):2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29(22):3008–3015. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451(7182):1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451(7182):1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 15.Kondrashova O, Nguyen M, Shield-Artin K, Tinker AV, Teng NNH, Harrell MI. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2017;7(9):984–998. doi: 10.1158/2159-8290.CD-17-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondrashova O, Topp M, Nesic K, Lieschke E, Ho GY, Harrell MI. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun. 2018;9(1):3970. doi: 10.1038/s41467-018-05564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajrami I, Frankum JR, Konde A, Miller RE, Rehman FL, Brough R. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014;74(1):287–297. doi: 10.1158/0008-5472.CAN-13-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann M, Murina O, Reijns MAM, Agathanggelou A, Challis R, Tarnauskaite Z. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature. 2018;559(7713):285–289. doi: 10.1038/s41586-018-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettitt SJ, Krastev DB, Brandsma I, Drean A, Song F, Aleksandrov R. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun. 2018;9(1):1849. doi: 10.1038/s41467-018-03917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105(7):452–458. doi: 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillet JP, Calcagno AM, Varma S, Marino M, Green LJ, Vora MI. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci USA. 2011;108(46):18708–18713. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grandori C, Kemp CJ. Personalized cancer models for target discovery and precision medicine. Trends Cancer. 2018;4(9):634–642. doi: 10.1016/j.trecan.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ince TA, Sousa AD, Jones MA, Harrell JC, Agoston ES, Krohn M. Characterization of twenty-five ovarian tumour cell lines that phenocopy primary tumours. Nat Commun. 2015;6:7419. doi: 10.1038/ncomms8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coscia F, Watters KM, Curtis M, Eckert MA, Chiang CY, Tyanova S. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat Commun. 2016;7:12645. doi: 10.1038/ncomms12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin KY, Kraus WL. PARP inhibitors for cancer therapy. Cell. 2017;169(2):183. doi: 10.1016/j.cell.2017.03.034. [DOI] [PubMed] [Google Scholar]
- 26.Musella A, Bardhi E, Marchetti C, Vertechy L, Santangelo G, Sassu C. Rucaparib: an emerging parp inhibitor for treatment of recurrent ovarian cancer. Cancer Treat Rev. 2018;66:7–14. doi: 10.1016/j.ctrv.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 27.DelloRusso C, Welcsh PL, Wang W, Garcia RL, King MC, Swisher EM. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol Cancer Res. 2007;5(1):35–45. doi: 10.1158/1541-7786.MCR-06-0234. [DOI] [PubMed] [Google Scholar]
- 28.Langdon SP, Lawrie SS, Hay FG, Hawkes MM, McDonald A, Hayward IP. Characterization and properties of nine human ovarian adenocarcinoma cell lines. Cancer Res. 1988;48(21):6166–6172. [PubMed] [Google Scholar]
- 29.Norquist BM, Brady MF, Harrell MI, Walsh T, Lee MK, Gulsuner S. Mutations in homologous recombination genes and outcomes in ovarian carcinoma patients in GOG 218: an NRG oncology/gynecologic oncology group study. Clin Cancer Res. 2018;24(4):777–783. doi: 10.1158/1078-0432.CCR-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas HD, Calabrese CR, Batey MA, Canan S, Hostomsky Z, Kyle S. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther. 2007;6(3):945–956. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- 31.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7(5):462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics. 2016;32(18):2866–2868. doi: 10.1093/bioinformatics/btw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao J, Zhu Z, Wang H, Nichols TC, Lui GYL, Deng S. Combining CDK4/6 inhibition with taxanes enhances anti-tumor efficacy by sustained impairment of pRB-E2F pathways in squamous cell lung cancer. Oncogene. 2019;38(21):4125–4141. doi: 10.1038/s41388-019-0708-7. [DOI] [PubMed] [Google Scholar]
- 34.Shirts BH, Casadei S, Jacobson AL, Lee MK, Gulsuner S, Bennett RL. Improving performance of multigene panels for genomic analysis of cancer predisposition. Genet Med. 2016;18(10):974–981. doi: 10.1038/gim.2015.212. [DOI] [PubMed] [Google Scholar]
- 35.Shibuya Y, Tokunaga H, Saito S, Shimokawa K, Katsuoka F, Bin L. Identification of somatic genetic alterations in ovarian clear cell carcinoma with next generation sequencing. Genes Chromosomes Cancer. 2018;57(2):51–60. doi: 10.1002/gcc.22507. [DOI] [PubMed] [Google Scholar]
- 36.Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007;67(20):9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- 37.Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A. 2012;109(24):9545–9550. doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 2016;3(4):e186–e195. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 40.Albrecht BK, Gehling VS, Hewitt MC, Vaswani RG, Cote A, Leblanc Y. Identification of a benzoisoxazoloazepine inhibitor (CPI-0610) of the Bromodomain and Extra-Terminal (BET) family as a candidate for human clinical trials. J Med Chem. 2016;59(4):1330–1339. doi: 10.1021/acs.jmedchem.5b01882. [DOI] [PubMed] [Google Scholar]
- 41.Mirguet O, Gosmini R, Toum J, Clement CA, Barnathan M, Brusq JM. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem. 2013;56(19):7501–7515. doi: 10.1021/jm401088k. [DOI] [PubMed] [Google Scholar]
- 42.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498(7453):246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33(4):676–689. doi: 10.1016/j.ccell.2018.03.007. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanlie A, Yousif AS, Akiyama H, Honjo T, Begum NA. Chromatin reader Brd4 functions in Ig class switching as a repair complex adaptor of nonhomologous end-joining. Mol Cell. 2014;55(1):97–110. doi: 10.1016/j.molcel.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 47.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Litton JK, Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379(8):753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santo L, Vallet S, Hideshima T, Cirstea D, Ikeda H, Pozzi S. AT7519, A novel small molecule multi-cyclin-dependent kinase inhibitor, induces apoptosis in multiple myeloma via GSK-3beta activation and RNA polymerase II inhibition. Oncogene. 2010;29(16):2325–2336. doi: 10.1038/onc.2009.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 51.Devaiah BN, Singer DS. Cross-talk among RNA polymerase II kinases modulates C-terminal domain phosphorylation. J Biol Chem. 2012;287(46):38755–38766. doi: 10.1074/jbc.M112.412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435(7042):677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 53.Tarsounas M, Sung P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat Rev Mol Cell Biol. 2020;21(5):284–299. doi: 10.1038/s41580-020-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edelbrock MA, Kaliyaperumal S, Williams KJ. Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat Res. 2013;743-744:53–66. doi: 10.1016/j.mrfmmm.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.TCGA Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trizzino M, Barbieri E, Petracovici A, Wu S, Welsh SA, Owens TA. The tumor suppressor ARID1A controls global transcription via pausing of RNA Polymerase II. Cell Rep. 2018;23(13):3933–3945. doi: 10.1016/j.celrep.2018.05.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 58.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 59.Yang L, Zhang Y, Shan W, Hu Z, Yuan J, Pi J. Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci Transl Med. 2017;9(400) doi: 10.1126/scitranslmed.aal1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun C, Yin J, Fang Y, Chen J, Jeong KJ, Chen X. BRD4 inhibition is synthetic lethal with PARP inhibitors through the induction of homologous recombination deficiency. Cancer Cell. 2018;33(3):401–416. doi: 10.1016/j.ccell.2018.01.019. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karakashev S, Zhu H, Yokoyama Y, Zhao B, Fatkhutdinov N, Kossenkov AV. BET bromodomain inhibition synergizes with PARP inhibitor in epithelial ovarian cancer. Cell Rep. 2017;21(12):3398–3405. doi: 10.1016/j.celrep.2017.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson AJ, Stubbs M, Liu P, Ruggeri B, Khabele D. The BET inhibitor INCB054329 reduces homologous recombination efficiency and augments PARP inhibitor activity in ovarian cancer. Gynecol Oncol. 2018;149(3):575–584. doi: 10.1016/j.ygyno.2018.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schilder RJ, Brady WE, Lankes HA, Fiorica JV, Shahin MS, Zhou XC. Phase II evaluation of dasatinib in the treatment of recurrent or persistent epithelial ovarian or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2012;127(1):70–74. doi: 10.1016/j.ygyno.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Secord AA, Teoh DK, Barry WT, Yu M, Broadwater G, Havrilesky LJ. A phase I trial of dasatinib, an SRC-family kinase inhibitor, in combination with paclitaxel and carboplatin in patients with advanced or recurrent ovarian cancer. Clin Cancer Res. 2012;18(19):5489–5498. doi: 10.1158/1078-0432.CCR-12-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller RE, Brough R, Bajrami I, Williamson CT, McDade S, Campbell J. Synthetic lethal targeting of ARID1A-mutant ovarian clear cell tumors with Dasatinib. Mol Cancer Ther. 2016;15(7):1472–1484. doi: 10.1158/1535-7163.MCT-15-0554. [DOI] [PubMed] [Google Scholar]
- 66.Sun Y, Lin X, Aske JC, Ye P, Williams C, Abramovitz M. Dasatinib attenuates overexpression of Src signaling induced by the combination treatment of veliparib plus carboplatin in triple-negative breast cancer. Cancer Chemother Pharmacol. 2019;84(6):1241–1256. doi: 10.1007/s00280-019-03962-8. [DOI] [PubMed] [Google Scholar]
- 67.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68(9):3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 68.Yokoyama T, Kohn EC, Brill E, Lee JM. Apoptosis is augmented in high-grade serous ovarian cancer by the combined inhibition of Bcl-2/Bcl-xL and PARP. Int J Oncol. 2017 doi: 10.3892/ijo.2017.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fleury H, Malaquin N, Tu V, Gilbert S, Martinez A, Olivier M. Exploiting interconnected synthetic lethal interactions between PARP inhibition and cancer cell reversible senescence. Nature Comm. 2019;10 doi: 10.1038/s41467-019-10460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stover EH, Baco MB, Cohen O, Li YY, Christie EL, Bagul M. Pooled genomic screens identify anti-apoptotic genes as targetable mediators of chemotherapy resistance in ovarian cancer. Mol Cancer Res. 2019;17(11):2281–2293. doi: 10.1158/1541-7786.MCR-18-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maya-Mendoza A, Moudry P, Merchut-Maya JM, Lee M, Strauss R, Bartek J. High speed of fork progression induces DNA replication stress and genomic instability. Nature. 2018;559(7713):279–284. doi: 10.1038/s41586-018-0261-5. [DOI] [PubMed] [Google Scholar]
- 72.Feng W, Jasin M. BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat Commun. 2017;8(1):525. doi: 10.1038/s41467-017-00634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, Gao J, Zhou J, Liu H, Xu C. Olaparib induced senescence under P16 or P53 dependent manner in ovarian cancer. J Gynecol Oncol. 2019;30(2):e26. doi: 10.3802/jgo.2019.30.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fleury H, Malaquin N, Tu V, Gilbert S, Martinez A, Olivier MA. Exploiting interconnected synthetic lethal interactions between PARP inhibition and cancer cell reversible senescence. Nat Commun. 2019;10(1):2556. doi: 10.1038/s41467-019-10460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.LoRusso PM, Li J, Burger A, Heilbrun LK, Sausville EA, Boerner SA. Phase I safety, pharmacokinetic, and pharmacodynamic study of the poly(ADP-ribose) polymerase (PARP) inhibitor veliparib (ABT-888) in combination with irinotecan in patients with advanced solid tumors. Clin Cancer Res. 2016;22(13):3227–3237. doi: 10.1158/1078-0432.CCR-15-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Do TV, Hirst J, Hyter S, Roby KF, Godwin AK. Aurora A kinase regulates non-homologous end-joining and poly(ADP-ribose) polymerase function in ovarian carcinoma cells. Oncotarget. 2017;8(31):50376–50392. doi: 10.18632/oncotarget.18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J, Wang R, Kong Y, Broman MM, Carlock C, Chen L. Targeting Plk1 to enhance efficacy of olaparib in castration-resistant prostate cancer. Mol Cancer Ther. 2017;16(3):469–479. doi: 10.1158/1535-7163.MCT-16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Veneris JT, Matulonis UA, Liu JF, Konstantinopoulos PA. Choosing wisely: selecting PARP inhibitor combinations to promote anti-tumor immune responses beyond BRCA mutations. Gynecol Oncol. 2020;156(2):488–497. doi: 10.1016/j.ygyno.2019.09.021. [DOI] [PubMed] [Google Scholar]
- 79.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu HM, Zhao X, Kaushik S, Robillard L, Barthelet A, Lin KK. A quantitative chemotherapy genetic interaction map reveals factors associated with PARP inhibitor resistance. Cell Rep. 2018;23(3):918–929. doi: 10.1016/j.celrep.2018.03.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.