Abstract
Antimicrobial peptides are a potential solution to the threat of multidrug-resistant bacterial pathogens. Recently, deep generative models including generative adversarial networks (GANs) have been shown to be capable of designing new antimicrobial peptides. Intuitively, a GAN controls the probability distribution of generated sequences to cover active peptides as much as possible. This paper presents a peptide-specialized model called PepGAN that takes the balance between covering active peptides and dodging nonactive peptides. As a result, PepGAN has superior statistical fidelity with respect to physicochemical descriptors including charge, hydrophobicity, and weight. Top six peptides were synthesized, and one of them was confirmed to be highly antimicrobial. The minimum inhibitory concentration was 3.1 μg/mL, indicating that the peptide is twice as strong as ampicillin.
1. Introduction
Antibiotic resistance is a serious and immediate threat against humanity as currently available antibiotics become increasingly obsolete. The annual deaths due to antimicrobial resistance are expected to exceed 10 million by 2050.1 Antimicrobial peptides (AMPs) are a possible solution to this problem.2 They are considered as less prone to resistance because microbes have been exposed to natural AMPs for millions of years, but widespread resistance against them has not been reported. Given the huge peptide space, however, it is very likely that numerous AMPs are yet to be found.
Deep generative models3 show encouraging results when applied to material and drug discovery.4−7 They are also one of the viable ways to boost the speed of AMP discovery, and several studies have been reported so far. Purely computational studies employing recurrent neural networks (RNNs),8 variational auto encoders,9 and generative adversarial networks (GANs)10 showed promising results in statistical terms, but experimental validation is yet to be done. Nagarajan et al.(11) were the first to show that a RNN can generate AMPs that works in vitro. They identified two peptides with a minimum inhibitory concentration (MIC) of 4 μg/mL against Escherichia coli. The potency of these peptides is at ampicillin-level because their MIC is comparable to that of ampicillin (6.25 μg/mL), a widely used antibiotic. Their neural network model has two parts. First, an RNN (i.e., generator) trained with known AMPs generates a large number of peptides. Next, a classifier neural network trained with peptide-MIC pairs ranks the generated sequences, and the top-ranked peptides are subject to experimental validation.
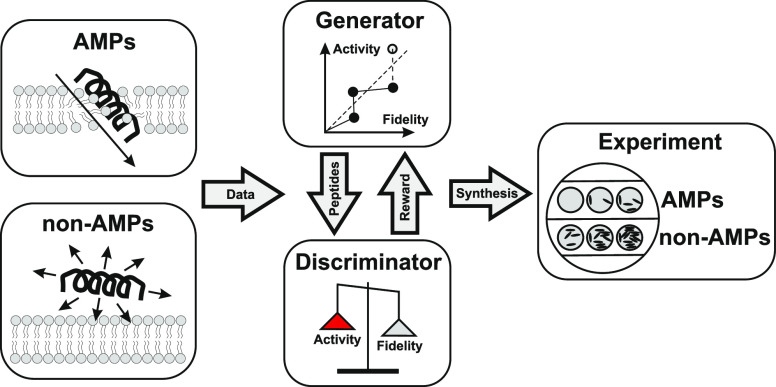
Drawbacks of the model by Nagarajan et al.(11) are as follows. (1) LSTM is an obsolete model that is often outperformed by GANs.12 (2) The generator is trained only with positive examples (i.e., AMPs) despite the fact that a plenty of negative examples (i.e., non-AMPs) are available. Aiming to solve the drawbacks, we develop a specialized model called PepGAN by engineering LeakGAN, one of the state-of-the-art sequence generators.13 PepGAN enhances the performance of LeakGAN with the help of activity predictor that is trained separately with positive and negative examples together (Figure 1).
Figure 1.
De novo AMP discovery with PepGAN. PepGAN receives sequence data consisting of AMPs and non-AMPs and creates new peptides with a generator and a discriminator. The generator samples a number of sequences stochastically. The reward for the sequences is evaluated by the discriminator and transmitted back to the generator to update the parameters. In normal GANs, the reward function represents fidelity, that is, how the sequences are similar to AMPs. In PepGAN, an activity predictor (shown in red) is incorporated in reward computation. Finally, top peptides are subject to experimental validation.
Another challenge in deep-learning-based AMP design is how to incorporate physicochemical properties such as charge, hydrophobicity, normalized van der Waals volume, and polarity. Deep learning models are essentially a language model, and it is not clear how to incorporate such information. To this aim, Nagarajan et al.(11) included several filtering steps in the model. Instead of complicating our model further, we simply chose to rerank PepGAN-generated peptides with an external AMP prediction tool (i.e., CAMP server14) trained with various physicochemical features. As a result of our experimental validation, the MIC of the best peptide was as low as 3.1 μg/mL, that is, twice as strong as ampicillin. We made a Python library of PepGAN publicly available at https://github.com/tsudalab/PepGAN to contribute in the developing open-source ecosystem of peptide design.
2. Results and Discussion
2.1. Generative Model
In various tasks including scheduling and maze solving, reinforcement learning has been used to generate a sequence of actions that maximizes a reward function.15 A reward function represents the quality of a generated sequence. In traditional settings, it is given a priori and stays unchanged during training. When optimizing multiple reward functions at once, a linear combination of them is used in many cases.16,17
Recently, Yu et al.(18) introduced SeqGAN that employs a machine-learned reward function for generating texts that resembles real sentences. A deep neural network called discriminator is trained to discriminate generated ones against real ones, and the training loss is adopted as the reward function. High reward implies high statistical fidelity: generated sequences are statistically indistinguishable from real ones. Later, SeqGAN is extended to LeakGAN13 by introducing the ideas of hierarchical reinforcement learning.19
In LeakGAN, the reward function for a sequence Y is designated as the output of the discriminator D(Y), that is, the probability of Y being real. The reward function of our model, PepGAN, is described as
where F(Y) is a separately trained activity predictor and λ denotes the mixing constant. The activity predictor has a gated recurrent unit20 with 256 hidden variables. Given a sequence Y, it computes a hidden vector at each position. The hidden vector is fed to a one-layer dense neural network to yield a partial score at each position. Finally, it is summarized to a final score by max-pooling. The output of the activity predictor represents the probability that Y is active and trained with both positive and negative examples.
Since LeakGAN is not informed of negative examples, it is likely to generate sequences that are similar to negative examples. By mixing the predicted activity into the reward function, we aim to shift the distribution of PepGAN-generated sequences away from the negative examples. In other words, PepGAN is more activity-aware than LeakGAN.
2.2. Statistical Fidelity
As training examples, we collect sequences not longer than 52 amino acids from the following databases: APD,21 CAMP,22 LAMP,23 and DBAASP.24 Redundant sequences are removed via multiple sequence alignment with a cutoff ratio of 0.35. The final dataset contains 16,648 positive sequences (i.e., AMPs) and 5583 negative sequences (i.e., non-AMPs). The activity predictor is first trained with all sequences, and later the rest of PepGAN is trained only with positive sequences. Statistical performance of the activity predictor is shown in Figure S3. PepGAN is used with three different parameter settings λ = 0, 0.5, and 1. Notice that LeakGAN corresponds to the case λ = 1. For each setting, 10,000 peptide sequences are generated.
We investigate the statistical fidelity of generated sequences from multiple viewpoints. The generated sequences are regarded as high-quality if their statistics match well with those of the positive sequence set. First, we investigate the following physicochemical properties: length, molar weight, charge, charge density, isoelectric point, aromaticity, global hydrophobicity, and hydrophobic moment. ModlAMP package8 was used to compute these properties. Obtained statistics are summarized in Table 1. In addition, the activity predictor scores of generated peptides are shown in Table S1. With respect to seven in eight properties, PepGAN with the activity predictor (λ = 0 and 0.5) was better than LeakGAN (λ = 1). This result shows that the activity predictor has a favorable impact in statistical fidelity. In the following experiments, λ = 0.5 is adopted because it achieved the best result here.
Table 1. Statistical Fidelity in Physicochemical Descriptorsa.
| descriptor | AMPs | λ = 0 | λ = 0.5 | λ = 1 |
|---|---|---|---|---|
| length | 28.01 ± 9.39 | 36.40 ± 10.66 | 33.78 ± 9.70 | 35.49 ± 10.16 |
| molar weight | 3125 ± 1049 | 4032 ± 1226 | 3682 ± 1062 | 3901 ± 1148 |
| charge | 3.27 ± 3.68 | 6.57 ± 6.00 | 5.88 ± 5.14 | 6.49 ± 5.69 |
| charge density | 0.0011 ± 0.0012 | 0.0016 ± 0.0013 | 0.0016 ± 0.0013 | 0.0017 ± 0.0013 |
| isoelectric point | 9.52 ± 2.47 | 10.30 ± 2.08 | 10.31 ± 1.96 | 10.39 ± 1.97 |
| aromaticity | 0.087 ± 0.069 | 0.064 ± 0.056 | 0.063 ± 0.055 | 0.060 ± 0.053 |
| global hydrophobicity | 0.004 ± 0.29 | –0.056 ± 0.37 | –0.019 ± 0.34 | –0.047 ± 0.35 |
| hydrophobic moment | 0.293 ± 0.19 | 0.298 ± 0.19 | 0.30 ± 0.19 | 0.32 ± 0.19 |
The mean and standard deviation of each descriptor are shown for the three variants of PepGAN (λ = 0, 0.5, and 1) and the positive sequence set (AMPs). The variant whose mean is the closest to that of AMPs is highlighted.
Next, k-gram counts are employed as the statistics to investigate the performance of PepGAN as a text generator. We employ BLEU (bilingual evaluation understudy) to measure the agreement of two distributions of k-gram counts.12Table 2 summarizes the BLEU scores at 2, 3, 4, and 5 g of PepGAN and the following three baseline models, MLE,18 CVAE,25 and SeqGAN.18 On average, PepGAN variants were better than the baseline models, and PepGAN (λ = 0.5) was slightly worse than LeakGAN (λ = 1). Since BLEU is concerned only with positive examples, the avoidance of negative examples with the activity predictor might have worked adversely to BLEU.
Table 2. BLEU Scores Based on k-grams (k = 2, 3, 4, and 5) for Three Baseline Models (MLE, CVAE, and SeqGAN) and Three Variants of PepGAN (λ = 0, 0.5, and 1)a.
| MLE | CVAE | SeqGAN | λ = 0 | λ = 0.5 | λ = 1.0 | |
|---|---|---|---|---|---|---|
| BLEU-2 | 0.939 | 0.975 | 0.947 | 0.922 | 0.938 | 0.930 |
| BLEU-3 | 0.711 | 0.714 | 0.729 | 0.759 | 0.778 | 0.776 |
| BLEU-4 | 0.374 | 0.331 | 0.388 | 0.469 | 0.487 | 0.493 |
| BLEU-5 | 0.182 | 0.149 | 0.188 | 0.246 | 0.257 | 0.264 |
| average | 0.551 | 0.542 | 0.563 | 0.599 | 0.615 | 0.616 |
The best method is highlighted.
It is reported that samples generated by GANs tend to lose diversity because of mode collapse.12 To check if mode collapse happened or not, the diversity of PepGAN-generated sequences is measured as follows. For each sequence, a BLEU score between that and all other sequences is computed. The diversity score called self-BLEU is then computed as the average of all BLEU scores. Table 3 shows the self-BLEU scores for the positive sequence set (i.e., AMPs) and the generated sequence sets of PepGAN variations. In all cases, the generated sequences were as diverse as the positive set, and mode collapse did not happen.
Table 3. Self-BLEU Scores Based on k-grams (k = 2, 3, 4, and 5) for Three Variants of PepGAN (λ = 0, 0.5, and 1) and the Positive Sequence Set (AMPs).
| AMPs | λ = 0 | λ = 0.5 | λ = 1 | |
|---|---|---|---|---|
| self-BLEU-2 | 0.965 | 0.969 | 0.970 | 0.970 |
| self-BLEU-3 | 0.802 | 0.835 | 0.842 | 0.846 |
| self-BLEU-4 | 0.550 | 0.592 | 0.608 | 0.621 |
| self-BLEU-5 | 0.393 | 0.372 | 0.381 | 0.405 |
2.3. Experimental Validation
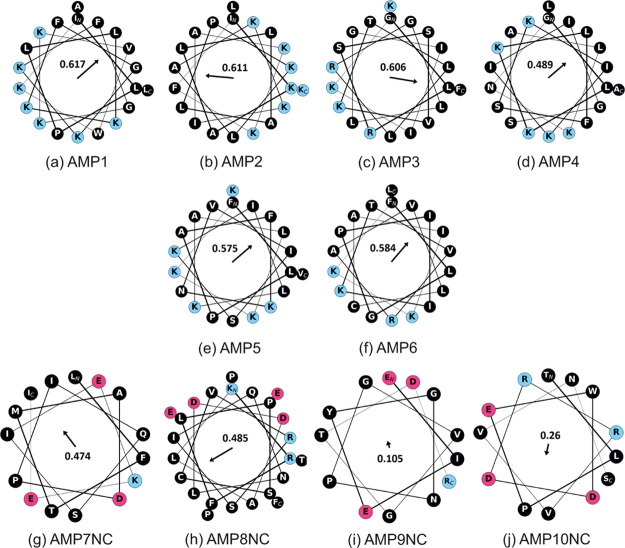
For experimental validation, generated peptides are prioritized according to the AMP likelihood computed by the CAMP server.14 Top six peptides are shown in Table 4. In addition, the worst four sequences are chosen as negative controls. Figure 2 shows the helical wheel plots of these peptides26 together with their hydrophobic moments.27 Cell-penetrating peptides tend to have a high relative abundance of positively charged amino acids and contain an alternating pattern of polar and hydrophobic amino acids (i.e., amphiphilicity).2 Our top peptides are observed as highly cationic and amphiphilic because they contain a large number of positively charged amino acids and no negatively charged ones, and their hydrophobic moments are high (0.58 ± 0.048). In comparison, negative controls are neither cationic nor amphiphilic.
Table 4. Results of Experimental Validationa.
| ID | sequence | MIC (μg/mL) | net charge at pH 7 |
|---|---|---|---|
| AMP1 | ILPLLKKFGKKFGKKVWKAL | 25 | 8 |
| AMP2 | IKALLALPKLAKKIAKKFLK | 50 | 8 |
| AMP3 | GLRSSVKTLLRGLLGIIKKF | >100 | 6 |
| AMP4 | GLKKLFSKIKIIGSALKNLA | 3.1 | 6 |
| AMP5 | FLPAFKNVISKILKALKKKV | 12.5 | 7 |
| AMP6 | FLGPIIKTVRAVLCAIKKL | 25 | 4.9 |
| AMP7NC | LFTMADPIQSIEKEI | >100 | –1 |
| AMP8NC | KRFLPSCVRSIQNLDDALPTPEEF | >100 | –0.1 |
| AMP9NC | EIEYGNPGVGTDR | >100 | –1 |
| AMP10NC | TLPEWDDRRVVNS | >100 | 0 |
| ampicillin | 6.25 |
AMP1–AMP6 are the top-ranked peptides chosen from PepGAN-generated sequences. AMP7NC–AMP10NC are negative controls.
Figure 2.
Helical wheel plots of top-ranked peptides (AMP1–AMP6) and negative controls (AMP7NC–AMP10NC). Positively and negatively charged residues are shown in blue and red, respectively. The numbers and the corresponding arrows show Eisenberg’s hydrophobic moment.
The potency of the peptides is evaluated based on MIC against E. coli. MIC is determined as the minimum concentration of an antimicrobial at which the growth of a target microbe is suppressed. We found that as many as five out of six AMP peptides exhibited effective antimicrobial activity. Among them, AMP4 exhibited the best antimicrobial performance, 3.1 μg/mL, which is better than a well-known antimicrobial, ampicillin (6.25 μg/mL). The high production ratio, 5/6, and the sufficiently low MIC of AMP4 validate PepGAN’s ability to generate industry-level peptides. In contrast, all four negative control peptides did not exhibit effective antimicrobial activity.
We presented PepGAN, a generative model for designing peptides, and demonstrated its statistical and in vitro success in AMP design. AMP-specific tricks are intentionally left out of our Python library. Thus, our library can directly be applicable in development of other kinds of peptides such as drug-delivery peptides28 and anti-cancer peptides.29 To achieve our goal of boosting the speed of peptide development, experimental researchers, who are not necessarily familiar with machine learning, should be able to use computational tools such as PepGAN. Although we made our code public, we have not reached this level of utility. Open-source ecosystems in machine translation and computer vision are well-developed to the point that nonexperts can use them without difficulty. In the future work, we will continue to develop PepGAN with an aim to make it a core of the emerging ecosystem of peptide design tools.
3. Methods
3.1. Peptide Synthesis
We synthesized all peptides on rink amide resin (ProTide, CEM Corporation, NC, USA) using an automated microwave peptide synthesizer (Liberty Blue, CEM Corporation). Each peptide was cleaved from resin and purified by reversed-phase high-performance liquid chromatography using a C18 column (COSMOSIL 5C18-AR-II, 10 mm I.D. × 250 mm; Nacalai Tesque, Japan). In the purification, a mixture of solvent A [0.1% v/v trifluoroacetic acid (TFA) in water] and B (0.1% v/v TFA in acetonitrile) at 25 °C was used as the mobile phase, and a linear gradient from 20 to 80% B for 50 min at a flow rate of 2.5 mL/min was applied. The masses of the 10 purified peptides were verified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (microflex LT, Bruker Daltonics, USA); AMP1, m/z: 2340.377 [M + H]+, calcd m/z: 2341.537; AMP2, m/z: 2234.103 [M + H]+, calcd m/z: 2234.577; AMP3, m/z: 2198.801 [M + H]+, calcd m/z: 2198.423; AMP4, m/z: 2141.044 [M + H]+, calcd m/z: 2141.390; AMP5, m/z: 2284.861 [M + H]+, calcd m/z: 2284.500; AMP6, m/z: 2082.569 [M + H]+, calcd m/z: 2082.335; AMP7NC, m/z: 1733.915 [M + H]+, calcd m/z: 1733.856; AMP8NC, m/z: 2774.047 [M + H]+, calcd m/z: 2774.430; AMP9NC, m/z: 1406.266 [M + H]+, calcd m/z: 1405.671; AMP10NC, m/z: 1585.632 [M + H]+, calcd m/z: 1585.809 (monoisotopic mass for all). We quantified the purities of the peptides using another C18 column (COSMOSIL 5C18-AR-II, 4.6 mm × 250 mm; Nacalai Tesque, Japan) (Figure S1). The ellipticities of the peptides were recorded using a circular dichroism (CD) spectrometer (J1500; JASCO, Japan) to clarify the secondary structures of the peptides (Figure S2).
3.2. MIC Determination
The MICs of the peptides were determined using the microdilution test with some modifications. We used E. coli TOP10 (Thermo Scientific, MA, USA) in the late-log phase for this test. The colony forming unit (cfu) of E. coli was determined using optical density at 600 nm. For each peptide, we prepared 11 wells containing 150 μL of 5 × 105 cfu/mL of E. coli and the series of concentration of each peptide from 10 to 0.01 μg/mL (2-fold dilution for 10 times). We also prepared one well containing only 150 μL of 5 × 105 cfu/mL of E. coli without the peptide. Similarly, we prepared 11 wells for ampicillin in the same well-plate. We incubate the plates for 26 h at 37 °C for the growth of E. coli. To read the optical density for each well, we set the plates in a plate reader (EnSpire 2300, PerkinElmer). After shaking the plate for 10 s at 300 rpm in double-orbital motion (diameter 1 mm), we measured the optical density of each solution (600 nm).
Acknowledgments
We thank Kei Terayama for helpful comments and suggestions. This work is supported by RIKEN engineering network fund, NEDO P15009, SIP (Technologies for Smart Bio-industry and Agriculture), JST CREST JPMJCR1502, and JST ERATO JPMJER1903. CD spectral measurements were supported by Molecular Structure Characterization Unit, RIKEN Center for Sustainable Resource Science (CSRS).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c02088.
Table of activity predictor scores, purities of the peptides, CD results, and performance of the activity predictor (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- O’Neill J.Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance, 2014; Vol. 20, pp 1–16. [Google Scholar]
- Fjell C. D.; Hiss J. A.; Hancock R. E. W.; Schneider G. Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discovery 2012, 11, 37–51. 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- LeCun Y.; Bengio Y.; Hinton G. Deep learning. Nature 2015, 521, 436–444. 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Segler M. H. S.; Preuss M.; Waller M. P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 2018, 555, 604–610. 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- Sanchez-Lengeling B.; Aspuru-Guzik A. Inverse molecular design using machine learning: Generative models for matter engineering. Science 2018, 361, 360–365. 10.1126/science.aat2663. [DOI] [PubMed] [Google Scholar]
- Sumita M.; Yang X.; Ishihara S.; Tamura R.; Tsuda K. Hunting for organic molecules with artificial intelligence: molecules optimized for desired excitation energies. ACS Cent. Sci. 2018, 4, 1126–1133. 10.1021/acscentsci.8b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhavoronkov A.; Ivanenkov Y. A.; Aliper A.; Veselov M. S.; Aladinskiy V. A.; Aladinskaya A. V.; Terentiev V. A.; Polykovskiy D. A.; Kuznetsov M. D.; Asadulaev A.; et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019, 37, 1038–1040. 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- Müller A. T.; Gabernet G.; Hiss J. A.; Schneider G. modlAMP: Python for Antimicrobial Peptides. Bioinformatics 2017, 33, 2753–2755. 10.1093/bioinformatics/btx285. [DOI] [PubMed] [Google Scholar]
- Kingma D. P.; Welling M.. PepCVAE: Semi-Supervised Targeted Design of Antimicrobial Peptide Sequences. 2018, arXiv:1810.07743. arXiv preprint. [Google Scholar]
- Gupta A.; Zou J. Feedback GAN for DNA optimizes protein functions. Nat. Mach. Intell. 2019, 1, 105–111. 10.1038/s42256-019-0017-4. [DOI] [Google Scholar]
- Nagarajan D.; Nagarajan T.; Roy N.; Kulkarni O.; Ravichandran S.; Mishra M.; Chakravortty D.; Chandra N. Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria. J. Biol. Chem. 2018, 293, 3492–3509. 10.1074/jbc.m117.805499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.; Lu S.; Zheng L.; Guo J.; Zhang W.; Wang J.; Yu Y.. Texygen: A benchmarking platform for text generation models. The 41st International ACM SIGIR Conference on Research & Development in Information Retrieval; SIGIR, 2018; pp 1097–1100.
- Guo J.; Lu S.; Cai H.; Zhang W.; Yu Y.; Wang J.. Long text generation via adversarial training with leaked information. Thirty-Second AAAI Conference on Artificial Intelligence; AAAI, 2018; pp 5141–5148.
- Waghu F. H.; Barai R. S.; Gurung P.; Idicula-Thomas S. CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. 10.1093/nar/gkv1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R. S.; Barto A. G.; et al. Introduction to Reinforcement Learning; MIT Press Cambridge, 1998; Vol. 135. [Google Scholar]
- Van Moffaert K.; Drugan M. M.; Nowé A.. Scalarized multi-objective reinforcement learning: Novel design techniques. 2013 IEEE Symposium on Adaptive Dynamic Programming and Reinforcement Learning (ADPRL); IEEE, 2013; pp 191–199.
- Guimaraes G. L.; Sanchez-Lengeling B.; Outeiral C.; Farias P. L. C.; Aspuru-Guzik A.. Objective-Reinforced Generative Adversarial Networks (ORGAN) for Sequence Generation Models. 2017, arXiv:1705.10843. arXiv preprint. [Google Scholar]
- Yu L.; Zhang W.; Wang J.; Yu Y.. Seqgan: Sequence generative adversarial nets with policy gradient. Thirty-First AAAI Conference on Artificial Intelligence; AAAI, 2017; pp 2852–2858.
- Vezhnevets A. S.; Osindero S.; Schaul T.; Heess N.; Jaderberg M.; Silver D.; Kavukcuoglu K.. Feudal networks for hierarchical reinforcement learning. Proceedings of the 34th International Conference on Machine Learning, 2017; pp 3540–3549.
- Cho K.; van Merriënboer B.; Gulcehre C.; Bahdanau D.; Bougares F.; Schwenk H.; Bengio Y.. Learning Phrase Representations using RNN Encoder–Decoder for Statistical Machine Translation. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2014; pp 1724–1734.
- Wang Z.; Wang G. APD: the antimicrobial peptide database. Nucleic Acids Res. 2004, 32, D590–D592. 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghu F. H.; Gopi L.; Barai R. S.; Ramteke P.; Nizami B.; Idicula-Thomas S. CAMP: Collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2014, 42, D1154–D1158. 10.1093/nar/gkt1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.; Wu H.; Lu H.; Li G.; Huang Q. LAMP: a database linking antimicrobial peptides. PLoS One 2013, 8, e66557 10.1371/journal.pone.0066557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirtskhalava M.; Gabrielian A.; Cruz P.; Griggs H. L.; Squires R. B.; Hurt D. E.; Grigolava M.; Chubinidze M.; Gogoladze G.; Vishnepolsky B.; et al. DBAASP v.2: an enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res. 2016, 44, D1104–D1112. 10.1093/nar/gkv1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Bombarelli R.; Wei J. N.; Duvenaud D.; Hernández-Lobato J. M.; Sánchez-Lengeling B.; Sheberla D.; Aguilera-Iparraguirre J.; Hirzel T. D.; Adams R. P.; Aspuru-Guzik A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 2018, 4, 268–276. 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A. T.; Hiss J. A.; Schneider G. Recurrent Neural Network Model for Constructive Peptide Design. J. Chem. Inf. Model. 2018, 58, 472–479. 10.1021/acs.jcim.7b00414. [DOI] [PubMed] [Google Scholar]
- Eisenberg D.; Weiss R. M.; Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 1982, 299, 371–374. 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Wolfe J. M.; Fadzen C. M.; Choo Z.-N.; Holden R. L.; Yao M.; Hanson G. J.; Pentelute B. L. Machine learning to predict cell-penetrating peptides for antisense delivery. ACS Cent. Sci. 2018, 4, 512–520. 10.1021/acscentsci.8b00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisoni F.; Neuhaus C. S.; Hishinuma M.; Gabernet G.; Hiss J. A.; Kotera M.; Schneider G. De novo design of anticancer peptides by ensemble artificial neural networks. J. Mol. Model. 2019, 25, 112. 10.1007/s00894-019-4007-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.