Highlights
-
•
This is the first described case of isolation of Sars-CoV-2 in Vero E6 cell-culture from a 10-day-old newborn in Italy.
-
•
The infection was detected in the mother after the child was born and therefore a vertical transmission can be excluded.
-
•
Despite the presence of the virus, the newborn showed mild symptoms of the disease.
Keywords: Sars-CoV-2, COVID-19, Newborn, Cell-culture, Isolation
Abstract
This report describes the evolution of COVID-19 in a 10 day-old-baby. The mother developed the disease immediately after childbirth and therefore a vertical transmission can be excluded. The isolation of the virus in cell culture with a cytopathic effect already visible after 48 h, indicates that the viral load of the newborn was quite high, but not serious course of the disease was observed. This paper wants to highlight the possible role of newborns and children in the spread of the disease.
Introduction
It is not yet clear why children, and especially newborns, are less susceptible to COVID-19 than adults [1], despite viral infections being much more frequent from childhood onwards. One hypothesis is related to vaccinations that are carried out in the first weeks of life and which would favor an increase of a non-specific immunity related to an increase in inteleukin-2 (IL-2) or other factors not yet known [2]. Probably some interactions between the newborn and the most common respiratory viruses of childhood would stimulate an innate resistance to RNA viruses [3]. Another hypothesis concerns angiotensin-converting enzyme 2 (ACE2), a membrane-bound aminopeptidase which is highly expressed in pulmonary alveolar epithelial cells and small intestine enterocytes. Although ACE2 has been identified as a receptor for Sars-CoV-2, it is plausible that ACE2 tissue distribution differs between adults and children or that ACE2 binding capacity in children is lower than in adults [4].
One reason why infant do not develop severe forms of the disease could be due to the fact that the immune system of infants is not mature at all and therefore probably is not yet able to start the cytokine storm that occurs in COVID-19 infection of the adults [5]. In fact, from a clinical point of view, COVID-19 generally occurs in children with moderate or low fever accompanied by mild and non-specific symptoms. In some cases, gastrointestinal symptoms such as diarrhea, abdominal distension and aversion to food may occur in newborns [6].
Hypoxia and increased respiratory rate were recorded only in a small subgroup of children, which in severe cases does not respond to standard oxygen therapy and requires nasal cannula or mask. Based on the reported cases, most of these children have a benign course which resolves within 1–2 weeks after the onset of the disease. Infants with COVID-19 need more careful and cautious observation because they are more likely to present with non-specific symptoms such as lethargy and dehydration [4,7,8].
Case report
On 09 August 2020, a 34-year-old pregnant woman was admitted to the "Mater Dei" hospital (Bari, Apulia region, Italy) and following the standard procedures before the entrance was subjected to the nasopharyngeal swab for Sars-CoV-2 detection, resulting negative despite manifesting cold symptoms, but without fever. Also an internal reaction control was used in order to exclude a false negativity. On the same day the woman gave birth to a child weighing 3.100 Kg. Three days later mother and son came back home and after 10 days from the birth, the newborn developed low-grade fever and mild respiratory symptoms. For this reason he was transferred to the pediatric hospital “Giovanni XXIII” (Bari, Apulia region, Italy), but he couldn’t be hospitalized because he was less than 30 days old. On 20 August 2020 he was transferred to the neonatology department of “Miulli” hospital (Acquaviva delle Fonti, Apulia region, Italy) for suspected sepsis since he manifested fever with a temperature of 37.8 °C. Immediately he was subjected to the following tests: complete blood count, C reactive protein, film array for the detection of Adenovirus, Coronavirus 229E-HKQ1-NL63-OC43, Metapneumovirus human, Rhinovirus, Enterovirus, Influenza A-B, Parainfluenza 1-2-3-4, RSV, Pertussis, Chlamydia and Mycoplasma pneumonia. No lombar puncture was performed at the beginning, because the newborn showed mild respiratory symptoms and therefore a respiratory infection was suspected at first instance.
The values of the complete blood count were normal e C reactive protein resulted negative. Moreover, was not revealed the presence of respiratory pathogens. Finally, it was decided to perform also the canonical nasopharyngeal swab to verify an eventual Sars-CoV-2 infection.
The real time PCR specific for Sars-CoV-2 was performed by Allplex 2019-nCoV Assay Kit (Seegene, Seul, Korea) on the CFX96 Real Time System C1000 Thermal Cycler (Bio-Rad, Hercules, California-USA). The test resulted positive showing the amplification of the E gene with CT 9.7; RdRp gene with CT 13.3; and N gene with CT 14.5. After the results, the newborn was transferred to the infectious disease department of the pediatric hospital “Giovanni XXIII” (Bari). The day after, on 21 August, all his four family members (mother, father and two young brothers) were also tested for Sars-CoV-2, because the father complained of headache and sore throat and one of the children had 375 °C fever. Mother, father and the older brother resulted positive to the test, while the youngest one resulted negative, probable because in those days he had not been in strict contact with his family, but he was living temporarily with his maternal grandparents. It’s important to underline that the paternal grandmother during the days following the birth of the child complained a strong headache and cold flu symptoms and she went to visit the family at their home. The old woman was subjected to Sars-CoV-2 swab in the same day of the other relatives and she resulted to be positive. Probably she was the person who infected the others. Since that time all the family members were subjected to mandatory quarantine under the direct responsibility of the local health authorities.
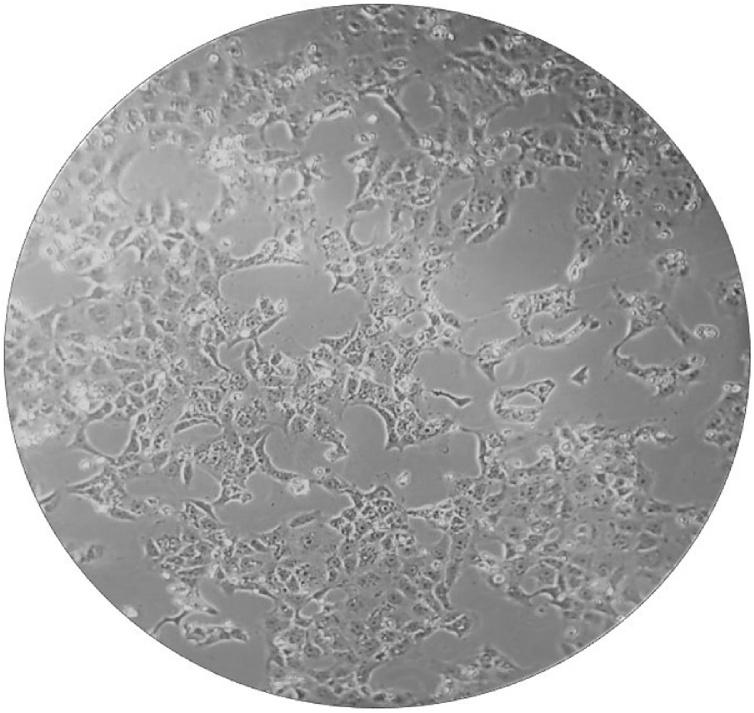
The nasopharyngeal swab of the newborn was sent to the BSL-3 laboratory of the Istituto Zooprofilattico Sperimentale della Puglia e della Basilicata (Foggia, Apulia region, Italy) for the isolation test. Briefly, 500 μL of the swab medium was incubated with 250 μL of an antibiotic solution for 1 h at room temperature. The suspension was then inoculated in a T25 Cell Culture Flask containing a monolayer of VeroE6 cells. The flask was incubated in a thermostat at 37 °C for 1 h. After incubation, 5.5 mL of EMEM with 6 % fetal bovine serum were added and incubated again at 37 °C. After 48 h of incubation a quite good cytopathic effect was appreciable. After further 24 h, an impressive cytopathic effect was highlighted consisting of rounding and detachment of most of the cells (Fig. 1). The replication of the virus was confirmed by the specific biomolecular test for Sars-CoV-2 carried out on 200 μL of the culture medium.
Fig. 1.
Cytopathic effects consisting of rounding and detachment of cells in Vero E 6 cell cultures, 72 h after the inoculation of the virus from the swab medium.
The newborn manifested fever for 3 days following the admission and then the heath conditions resulted stable.
Discussion
The newborn almost certainly contracted Sars-CoV-2 from one member of the family, presumably the paternal grandmother who, most likely, was infective in the days following the birth, when she went to visit the newborn at his home. We do not know if the newborn or another family member infected the others, but it’s very interesting that the viral load of the newborn was very high as demonstrated either by PCR or by isolation of the virus. The cytopathic effect on the Vero E6 monolayer was already evident after 48 h post inoculation and was much clearer after 72 h (Fig. 1). Despite the high viral load, the newborn never showed serious symptoms except mild respiratory symptoms and a not high fever for 3 days following the admission to the pediatric hospital. This case confirms what has already been reported in other similar works, even in presence of high viral loads, newborns usually show relatively mild symptoms [9].
This case requires a reflection on the potentiality of Sars-CoV-2 transmission from newborns and children that, maybe, often is underestimated. Moreover, some traditional behaviors of the Italian population and in particular of Southern Italy, where it is customary to visit newborn children in the days immediately following the return home, can amplify the transmission of Sars-CoV-2 from unknown infected children. In fact, newborns living inside home environment can spreadlarge quantities of virus and lead to high levels of contamination, so as to be dangerous for anyone who comes into contact with them. It’s unlikely that that infant would generate effective cough aerosols for infection, but however they can have infectious secretions such as stool or saliva as reported in literature [10,11].
Therefore, the aim of this paper is to report that also from Sars-CoV-2 positive newborns is possible to isolate alive virus even in presence of moderate symptoms, as showed by the isolation on cell culture of this study, indicating that they could be dangerous for the dissemination of the virus.
Ethical considerations
Written informed consent was obtained from the patient for the publication of this case report and accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal on request. All authors have made substantial contributions to thefollowing: (1) the conception and design of the study, oracquisition of data, or analysis and interpretation of data, (2) drafting the article or revising it critically for important intellectualcontent, (3) final approval of the version to be submitted.
CRediT authorship contribution statement
Giuseppe Lenoci: Conceptualization, Project administration, Supervision, Visualization, Writing - original draft. Domenico Galante: Conceptualization, Supervision, Visualization, Writing - original draft. Edmondo Ceci: Conceptualization, Supervision, Visualization, Writing - original draft. Viviana Manzulli: Data curation, Formal analysis, Investigation. Angela Maria Moramarco: Investigation, Formal analysis, Investigation, Methodology, Software, Validation. Anna Chiaromonte: Formal analysis, Investigation, Methodology, Software, Validation. Giuseppina Labarile: Formal analysis, Investigation, Methodology, Software, Validation. Simone Lattarulo: Formal analysis, Investigation, Methodology, Software, Validation. Annalisa Resta: Formal analysis, Investigation, Methodology, Software, Validation. Lorenzo Pace: Data curation, Formal analysis, Investigation, Methodology. Valeria Rondinone: Data curation, Formal analysis, Investigation, Methodology. Antonio Parisi: Conceptualization, Formal analysis, Investigation, Methodology, Visualization. Dora Cipolletta: Data curation, Formal analysis, Investigation, Methodology. Leonardo Marino: Formal analysis, Investigation, Methodology. Iolanda Padalino: Formal analysis, Investigation, Methodology. Luigina Serrecchia: Formal analysis, Investigation, Methodology. Angela Aceti: Formal analysis, Formal analysis, Investigation, Methodology. Michela Iatarola: Formal analysis, Investigation, Methodology. Francesco Tolve: Formal analysis, Investigation, Methodology. Antonio Fasanella: Project administration, Supervision, Visualization, Writing - original draft.
Declaration of Competing Interest
The authors report no declaration of interests.
Acknowledgements
We thank Elena Poppa and Guido Asturio for their technical support.
Contributor Information
Giuseppe Lenoci, Email: g3.lenoci@miulli.it.
Domenico Galante, Email: domenico.galante@izspb.it.
Edmondo Ceci, Email: e.ceci@miulli.it.
Viviana Manzulli, Email: viviana.manzulli@izspb.it.
Angela Maria Moramarco, Email: a.moramarco@miulli.it.
Anna Chiaromonte, Email: annachiaromonte88@gmail.com.
Giuseppina Labarile, Email: g.labarile@miulli.it.
Simone Lattarulo, Email: lattarulo45@gmail.com.
Annalisa Resta, Email: a.resta@miulli.it.
Lorenzo Pace, Email: lorenzo.pace@izspb.it.
Valeria Rondinone, Email: valeria.rondinone@izspb.it.
Antonio Parisi, Email: antonio.parisi@izspb.it.
Dora Cipolletta, Email: dora.cipolletta@izspb.it.
Leonardo Marino, Email: leonardo.marino@izspb.it.
Iolanda Padalino, Email: iolanda.padalino@izspb.it.
Luigina Serrecchia, Email: luigina.serrecchia@izspb.it.
Angela Aceti, Email: angela.aceti@izspb.it.
Michela Iatarola, Email: michela.iatarola@izspb.it.
Francesco Tolve, Email: francesco.tolve@izspb.it.
Antonio Fasanella, Email: antonio.fasanella@izspb.it.
References
- 1.Caselli D., Aricò M. 2019-nCoV: polite with children! Pediatr Rep. 2020;12(1):8495. doi: 10.4081/pr.2020.8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Otto S., Mahner B., Kadow I., Beck J.F., Wiersbitzky S.K.W., Bruns R. General non-specific morbidity is reduced after vaccination within the third month of life – the Greifswald study. J InfSecur. 2000;41:172–175. doi: 10.1053/jinf.2000.0718. [DOI] [PubMed] [Google Scholar]
- 3.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J Innate Immun. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong Y., Mo X., Hu Y., Qi X., Jiang F., Jiang Z. Epidemiological characteristics of 2143 pediatric patients with 2019 coronavirus disease in China epidemiology of COVID-19 among children in China. Pediatrics. 2020;145(6) doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Q., Chen Y., Chen C., Chiu C. SARS-CoV-2 infection in children: transmission dynamics and clinical characteristics. J Formos Med Assoc. 2020;119(3):670–673. doi: 10.1016/j.jfma.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z.-M., Fu J.-F., Shu Q., Chen Y.-H., Hua C.-Z., Li F.-B. Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World J Pediatr. 2020;16:240–246. doi: 10.1007/s12519-020-00345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Rose D.U., Piersigilli F., Ronchetti M.P., Santisi A., Bersani I., Dotta A. Novel Coronavirus disease (COVID-19) in newborns and infants: what we know so far. Ital J Pediatr. 2020;46(1):56. doi: 10.1186/s13052-020-0820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.To K.-W., Tsang O.-Y., Yip C.-Y., Chan K.-H., Wu T.-C., Chan J.-C. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;(February (12)) doi: 10.1093/cid/ciaa14. [DOI] [PMC free article] [PubMed] [Google Scholar]