Abstract
Cilia assembly is under strict transcriptional control during animal development. In vertebrates, a hierarchy of transcription factors (TFs) are involved in controlling the specification, differentiation and function of multiciliated epithelia. RFX TFs play key functions in the control of ciliogenesis in animals. Whereas only one RFX factor regulates ciliogenesis in C. elegans, several distinct RFX factors have been implicated in this process in vertebrates. However, a clear understanding of the specific and redundant functions of different RFX factors in ciliated cells remains lacking. Using RNA-seq and ChIP-seq approaches we identified genes regulated directly and indirectly by RFX1, RFX2 and RFX3 in mouse ependymal cells. We show that these three TFs have both redundant and specific functions in ependymal cells. Whereas RFX1, RFX2 and RFX3 occupy many shared genomic loci, only RFX2 and RFX3 play a prominent and redundant function in the control of motile ciliogenesis in mice. Our results provide a valuable list of candidate ciliary genes. They also reveal stunning differences between compensatory processes operating in vivo and ex vivo.
INTRODUCTION
Cilia are highly conserved organelles serving fundamental functions in eukaryotes. Their assembly is tightly regulated during the cell cycle and tissue differentiation. For instance, primary cilia, which are present on many cells during vertebrate development, are disassembled at the onset of cell division and reassembled during subsequent cell cycle phases. During animal development, cilia assembly relies on strict transcriptional programs that coordinate cilia growth and tissue morphogenesis (reviewed in (1,2)). This transcriptional regulation has been thoroughly described for the differentiation of multi-ciliated epithelia in vertebrates (3,4). Several key transcription factors (TFs) are involved in this process. Upstream of the ciliogenesis pathway, two TFs of the Geminin family, Gemc1 and MCIDAS (or multicilin) (5,6), directly control centriole amplification, which is required for the assembly of multiple cilia. Downstream of these TFs, members of the FOXJ1 (7–10) and RFX families (11–16) have been shown to be direct regulators of core ciliogenic genes in multiciliated cells. In addition, members of the E2F and MYB families of TFs have been implicated in multiciliated cell differentiation (17–20). Recently, TAP73 was shown to regulate cilia assembly in mammalian airways (21–23). These different TFs engage in complex regulatory interactions, as revealed by specific cross regulatory loops, although most of these regulatory processes remain poorly understood.
Whereas, all of the aforementioned ciliogenic transcription factors play critical roles in the development of motile cilia, only RFX factors are also required for the assembly of non-motile cilia. Daf-19, the sole member of the RFX family in Caenorhabditis elegans, was initially shown to regulate cilia assembly in all ciliated cells of the nematode (24). In Drosophilamelanogaster, one of the two Rfx genes is required for sensory cilia assembly (25). In vertebrates and mammals, there are at least 8 different Rfx genes (2,26,27) several of which were shown to be required for ciliogenesis, either broadly, such as RFX3 in mice (11,12,28,29) and RFX2 in xenopus or zebrafish (13,30), or in specific tissues, such as RFX2 in multiciliated cells and RFX4 or RFX7 in the mouse neural tube (31–33).
In addition to their conserved roles in ciliogenesis, RFX proteins are involved in other processes ranging from the control of genome integrity and the cell cycle in yeast and fungi (34,35), to immune functions (36,37), and tissue and organ differentiation in the mouse or zebrafish (33,38–40). Several human diseases result from mutations in Rfx genes (37,38,41).
RFX proteins are characterized by a DNA binding domain of the winged-helix type (42) that specifically recognizes an inverted repeat element, the X-box, initially identified in Major Histocompatibility Complex Class II genes (36). Several of the RFX proteins also share a dimerization domain, allowing homo and heterodimerization (26,27,43).
Whereas many RFX target genes have been identified in various ciliated cell systems (13,44,45), little is known about how the functional specificity of different RFX factors is achieved and to what extent they have shared or redundant roles. To date, none of the studies aiming at identifying RFX targets has addressed the respective functions of several individual RFX proteins in the same cell type or tissue.
Here, we investigated the redundant and specific roles of three members of the RFX family—RFX1, RFX2 and RFX3—in the differentiation of multi-ciliated cells in mice, focusing on ependymal cells of the brain ventricle. These three transcription factors form a phylogenetic subgroup of highly related proteins, but only RFX2 and/or RFX3 have been implicated in cilia-associated functions in zebrafish, xenopus or mammals (2,26,27). Using high throughput RNA-seq and ChIP-seq approaches, we identified genes that are regulated directly or indirectly by these three TFs. We show that RFX1, RFX2 and RFX3 have complex overlapping functions and engage in potential regulatory interactions that cannot be explained solely by their binding-site specificities. Our study also provides valuable predictions of new ciliary genes. Finally, our results uncover unanticipated compensatory mechanisms operating in vivo compared to ex-vivo culture conditions, underlining the complexity of ciliary gene regulation in physiological settings.
MATERIALS AND METHODS
Mouse husbandry and genotyping
The generation of Rfx1−/− and Rfx3flox/flox mice was described in (11). The generation of Rfx2−/− and Rfx2flox/flox mice was described in (46). The generation of Rfx1flox/flox mice was described in (47). After deletion of exon 10 in Rfx1, splicing of exon 9 to exon 11 leads to a frame shift. For generation of Rfx1−/− mice, Rfx1flox/flox mice were crossed with a deleter strain expressing Cre ubiquitously under the control of a cytomegalovirus promoter (48). For generation of mice having conditional deletions of Rfx genes, mice carrying floxed Rfx alleles were crossed with FoxJ1-Cre mice. FoxJ1-Cre mice express Cre-recombinase in all multiciliated cells of the mouse (49). All mice were in a C57Bl/6 genetic background. Mice were genotyped using the following primers: Rfx1, TGTCTCCAGGGTAGGCACAAG and TTCTGTGACTGTGGGAGACTG; Rfx2, AGAATCTGCCCCTTGGCTAT and TGTCACCCACCTAGGCTTCT; Rfx3, GTCATGCTGGAAAATTTGAAG and AGTTGGCTTCTAACTTCTATG; FoxJ1, GACCGCCCCCTCGGAGAGTCCC and CCTGGCAATTTCGGCTATACG.
Scanning electron microscopy
Brains were dissected as previously described (50) and fixed in 2% glutaraldehyde (Sigma-Aldrich, France), 0.1 M cacodylate buffer (EMS, USA), pH 7.4. Brain samples were then washed several times in 0.2 M cacodylate buffer, pH 7.4 and postfixed for 60 min in 1% OsO4 (EMS, USA), 0.1 M cacodylate buffer. Fixed brain samples were washed extensively with distilled water and dehydrated in a graded series of ethanol solutions and finally in acetone. Brain samples were then prepared for scanning electron microscopy by the critical point freeze-dry procedure (Leica, EM CPD 300). Samples were surface-coated using a gold/palladium spattering device (Hummer 2, Technics) under optimal conditions for 5 min and observed with a scanning electron microscope (ZEISS Merlin Compact) at 5 keV. Observations were performed at the Centre for Microstructure Analysis of the University of Lyon.
Cell isolation and preparation
Brains of newborn pups (OF1 strain) were dissected and processed as described previously (12). After 4 days of serum deprivation, cells were fixed and processed for ChIP experiments as described (51). Briefly, 1/10 of fixation buffer (11% formaldehyde; 0.1 M NaCl; 50 mM HEPES pH 7.9) was added to cell dishes and cells were fixed at room temperature for 8 min with gentle rocking. Glycine was added to 0.18 M (final concentration) and cells were quickly scraped off on ice. After centrifugation, the cell pellet was washed once with cold PBS 1× and conserved at –80°C until further analysis. Before use, thawed cells were washed once with cold PBS and lysed in TE buffer (10 mM Tris–HCl at pH 8.0, 1 mM EDTA) containing protease inhibitors and 0.5% NP-40. Brains from E18.5 Rfx1−/− embryos (four independently processed embryos = four replicates), Rfx2−/− embryos (six replicates), Rfx3−/− embryos (six replicates) or wild type littermate controls (four, eight, six embryos, respectively) were dissected and processed as described (12). After 4 days of serum deprivation, total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific).
Immunofluorescence on tracheal and ependymal cells
Brains or tracheas were dissected and fixed in 4% paraformaldehyde (EMS, USA) overnight at 4°C. Tissues were saturated in 0.2% triton, 10% normal goat serum, 1% BSA in PBS for at least 1h at RT, before O/N incubation at 4°C with a monoclonal antibody (1/150, Sigma T6793) against mouse acetyl-tubulin in 0.1% triton, 10% normal goat serum, 1% BSA in PBS to visualize cilia. Samples were then washed several times in PBS before 2h incubation at room temperature with secondary goat anti-mouse antibodies labeled with Alexa 488 (Invitrogen, France) and for 5 min in Hoechst. Brains or trachea were mounted in Vectashield and visualized with a Confocal spectral Leica SP5. For ciliated cell enumeration: ten arbitrary pictures were taken of different areas of the trachea with a 40× water immersion Confocal spectral Leica SP5 objective. Images were stacked and ciliated cells were counted using ImageJ software.
For analysis of cell specific RFX2 or RFX3 deletion: brains were dissected and fixed in 4% paraformaldehyde overnight at 4°C. Tissues were embedded in sucrose. 12 μm cryostat sections were saturated in 0.1% triton, 10% normal goat serum, 1% BSA in PBS for at least 1h at RT, before O/N incubation at 4°C with RFX3 (Sigma HPA035689) and RFX2 (Sigma HPA048969) antibodies and against mouse acetyl-tubulin antibody (1/150, Sigma T6793) in 0.1% triton, 10% normal goat serum, 1% BSA in PBS. Samples were then washed several times in PBS before incubation with secondary goat anti-mouse Alexa 488 antibody and Alexa 555 anti-rabbit antibody (Thermo Fisher, France) and for 5 min in Hoechst. Brains or trachea were mounted in Vectashield and visualized with a Confocal spectral Leica SP5. Observations were performed at the PLATIM of the University of Lyon.
RNA-seq experiments
Four to eight replicates (see above) were processed for each condition. cDNA libraries were constructed by the Genomics platform of the University of Geneva using the Illumina TruSeq RNA Sample Preparation Kit according to the manufacturer's protocol. Libraries were sequenced using single-end 100nt-long reads on Illumina HiSeq2000. FastQ reads were mapped to the ENSEMBL reference genome (GRCm38.89) using STAR version 2.4.0j (52) with standard settings, except that any reads mapping to more than one location in the genome (ambiguous reads) were discarded (m = 1). Sequence data has been submitted to the GEO database under the accession number GSE145324. A unique gene model was used to quantify reads per gene. Briefly, the model considers all annotated exons of all annotated protein coding isoforms of a gene to create a unique gene where the genomic region of all exons are considered coming from the same RNA molecule and merged together. All reads overlapping with the exons of each unique gene model were reported using featureCounts version 1.4.6-p1 (53). Gene expression was reported as raw counts and normalized as reads per kb per million (RPKM) to filter out genes with low expression values (<1 RPKM) before identifying differentially expressed genes. Library size normalization and differential gene expression calculations were performed using the package edgeR (54) designed for the R software. Only genes having a significant fold-change (Benjamini–Hochberg corrected P-value < 0.001) were considered for the rest of the RNAseq analysis.
ChIP-seq experiments
Chromatin was purified from ex vivo derived ependymal cells from WT mice (OF1 strain). Chromatin immunoprecipitation was performed as previously described (46).
Immunoprecipitated DNA was sequenced by the Genomics platform of the University of Geneva using the Illumina TruSeq ChIP Library Prep Kit according to the manufacturer's protocol. Libraries were sequenced using single-end 50nt-long reads on Illumina HiSeq2000. FastQ reads were mapped to the ENSEMBL reference genome (GRCm38.89) using bowtie version 0.12.7 (55) with standard settings, except that any reads mapping to more than one location in the genome (ambiguous reads) were discarded (m = 1). Duplicate reads were removed using Samtools version 0.1.18 (56). Sequence data has been submitted to the GEO database under the accession number n° GSE145324. Fragment length was estimated using cross-correlation (57). The Phantompeakqualtools R package (https: //www.encodeproject.org/software/phantompeakqualtools/) was used to measure the quality of the ChIPseq data, as assessed by the normalized ratio between the fragment-length cross-correlation and the background cross-correlation (normalized strand coefficient, NSC), the ratio between the fragment-length peak and the read-length peak (relative strand correlation, RSC) and the Qtag code. Peak calling was done with MACS2 version 2.0.10.20130520 (58) with no-model setting and shift-size parameter set to half of the estimated fragment length. Peak calling stringency was decreased by using P = 0.05 as threshold and applying the ’-to-large’ setting. Reproducible peaks were obtained by assessing the IDR for all pairs of replicates. The Fraction of Reads in Peaks (FRiP) (57) was found to be similar between replicates and correlated positively with the number of peaks. Antibodies used for ChIPseq are: RFX2 Antibody (C-15): sc-10657, RFX1, RFX3 mouse antibodies previously described in (28,59). All three were validated in vivo and in ChiP experiments (12,46,47,60).
Gene set enrichment analysis
Ciliary gene sets were generated from previously published data (61,62). Expressed genes included in the Rfx1, Rfx2 and Rfx3 RNA-seq data sets were ranked according to their fold change in knockout cells relative to WT cells. Gene set enrichment analysis was then done using the GSEA package Version 2.2 (63,64) from the Broad Institute (MIT, Cambridge, MA) for (a) different ciliary gene sets or (b) RFX1, RFX2 and RFX3 ChIP-seq peaks in promoters. Gene set permutations were performed 1000 times for each analysis. The Normalized Enrichment Score (NES) was calculated for each gene set. GSEA results with a nominal FDR < 0.05 and abs(NES) > 1 were considered significant.
De novo motif discovery and motif enrichment analysis
For analysis of differentially expressed genes, promoter sequences (−1000 bp to +500 bp relative to the TSS) were extracted, oriented according to gene orientation and used as input. For analysis of ChIP-seq data, peak sequences were used as input. De novo motif discovery was done using the cosmo package (65) designed for R (53). The probabilistic model used for the motif discovery was the zero-or-one-occurrence-per-sequence (ZOOPS), considering only the orientation of each promoter and motif lengths between 10nt and 20nt. Pscan (66) was used to identify overrepresented TF-binding-sites (TFBSs) using the JASPAR database (67) as matrices. TFBSs with P-values lower than 0.001 were considered to be significantly overrepresented. The motifs defined by the de novo analysis of ChIP-seq peaks (Figure 4C) was averaged and used to scan both strands of gene promoters (−1000/+500) to determine the presence of a X-box motifs. Results are reported in Figures 2C, 6C and 7 and Supplementary Figure S7.
Figure 4.
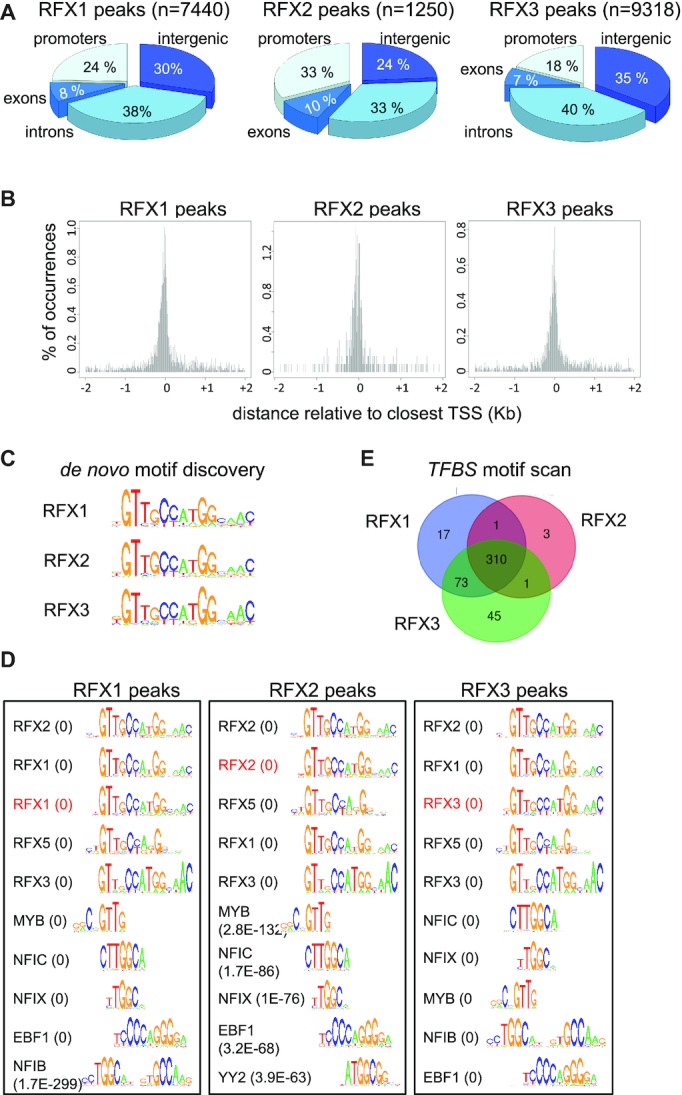
Identification of direct target genes and binding motifs for RFX1, RFX2 and RFX3. ChIP-seq was used to map genomic sites occupied by RFX1, RFX2 and RFX3 in mouse ependymal cells. (A) The pie charts illustrate the distribution of sites occupied by RFX1, RFX2 and RFX3 in the indicated portions of the genome. (B) The histograms represent the spatial distribution of RFX1, RFX2 and RFX3 occupied sites within promoter-proximal regions (−2kb to +2kb) centered on annotated TSSs. (C) The sequences logos show the top scoring DNA motifs identified by de novo motif discovery analysis (cosmo package) performed with the 100 most robust RFX1, RFX2 or RFX3 ChIP-seq peaks. All three motifs perfectly match the X-box motifs described in Figure 3B. (D) Sequence logos are shown for most enriched TFBS motifs identified using Pscan in ChIP-seq peaks for RFX1, RFX2 or RFX3. Only the 5 top ranking RFX motifs and non-RFX motifs are represented. The scan was done with all TFBS motifs from the JASPAR database. The de novo motifs (red) identified in (C) were included in the scan and are among the five best motifs. P-values are indicated in parentheses: values <1.0E–300 were rounded off to 0. The full list of motifs and P-values for their enrichment are provided in Supplementary Table S5. (E) The Venn diagrams show the overlap between sets of DNA motifs, distinct from the X-box, identified in regions bound by RFX1, RFX2 or RFX3 (see Supplementary Table S5). Certain motifs appear to be specifically enriched in RFX1 (17 motifs) or RFX3 (47 motifs) occupied regions. A significant number of other motifs are found in regions bound by both RFX1 and RFX3 (73 motifs) or by all three RFX factors (310 motifs).
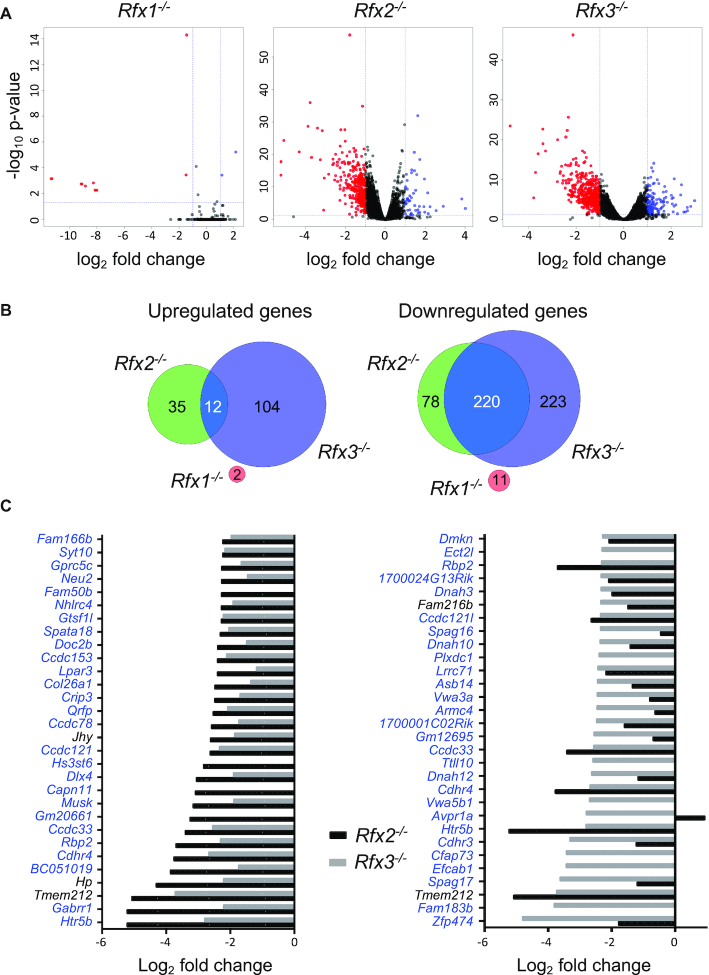
Figure 2.
Analysis of differentially expressed genes in ependymal cells from Rfx1, Rfx2 or Rfx3 deficient mice. (A) The volcano plots represent statistical significance (Y-axis, –log10P-value) and fold change (X-axis, log2 fold change) for alterations in gene expression observed in Rfx1−/−, Rfx2−/− or Rfx3−/− cells relative to WT cells. Whereas only few genes exhibited significantly altered expression in Rfx1−/− cells, numerous genes were differentially expressed in Rfx2−/− and Rfx3−/− cells. (B) The Venn diagrams summarize the overlaps observed between up (left) or down (right) regulated gene sets (fold change >2) in Rfx1−/−, Rfx2−/− or Rfx3−/− cells. Whereas there is little overlap between genes upregulated in Rfx2−/− and Rfx3−/− cells (left), most genes downregulated in Rfx2−/− cells are also downregulated in Rfx3−/− cells (right). There is no overlap between genes that are differentially expressed in Rfx1−/− cells and Rfx2−/− or Rfx3−/− cells. (C) The bar graphs show the fold changes in gene expression (log2 fold change) observed in Rfx2−/− (black bars) and Rfx3−/− (grey bars) cells for the 30 genes that are downregulated most strongly in Rfx2−/− (left) or Rfx3−/− (right) ependymal cells. Whereas nearly all genes that are strongly downregulated in Rfx2−/− cells are also strongly downregulated in Rfx3−/− cells (left), the reverse is not true (right). Genes highlighted in blue contain at least one X-box motif in their promoter region (−1000 to +500 bp).
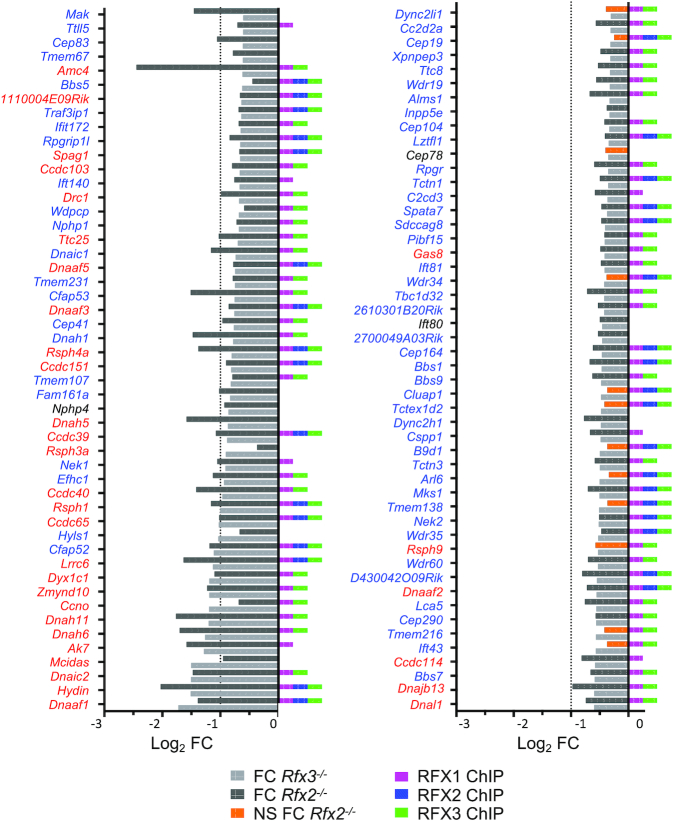
Figure 6.
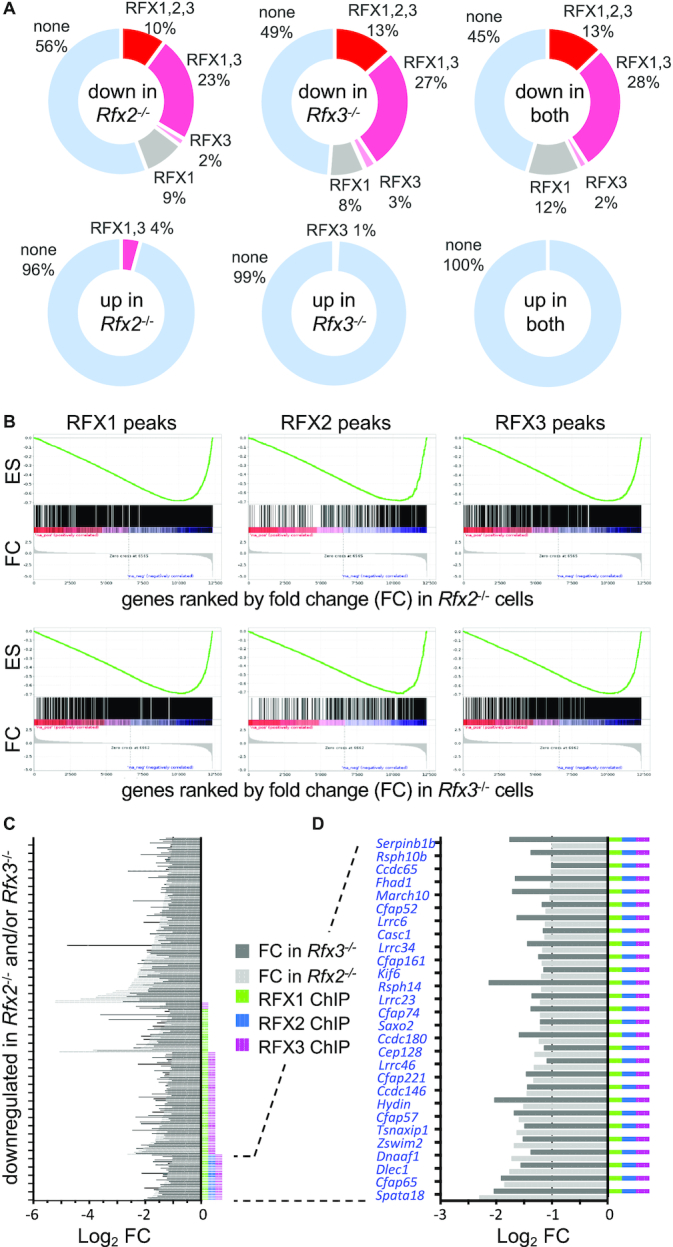
Identification of RFX target genes. (A) The pie charts show the fractions (%) of genes that are downregulated (top) or upregulated (bottom) in Rfx2−/− cells (left), Rfx3−/− cells (middle) or both (right) and have ChIP-seq peaks for RFX1, RFX2 and/or RFX3 within their promoters. Approximately one-third of downregulated genes have binding sites for either RFX1 plus RFX3 or all three RFX factors in their promoters. A minority of downregulated genes have binding sites for only RFX1 or RFX3. No downregulated genes are occupied by RFX2 alone. Most genes upregulated in Rfx2−/− or Rfx3−/− cells alone, and all genes upregulated in both Rfx2−/− and Rfx3−/− cells, lack RFX-occupied sites in their promoters. (B) Gene set enrichment analysis (GSEA) demonstrates that genes having RFX1, RFX2 or RFX3 ChIP-seq peaks in their promoters are strongly enriched in genes that are differentially expressed in Rfx2−/− or Rfx3−/− cells. (C, D) Histograms show the fold change (log2 FC) in expression for genes that are downregulated >2 fold in Rfx2−/− (light grey) or Rfx3−/− cells (dark grey). The presence of RFX1, RFX2 and/or RFX3 ChIP-seq peaks in the promoter is indicated for each gene at the right. (D) As in C, focusing on genes that are downregulated in both Rfx2−/− and Rfx3−/− cells, and have promoters occupied by all three RFX factors. All of these genes have at least one X-box motif in their promoter (−1000 to +500 bp), as highlighted by the blue font.
Figure 7.
Many ciliopathy genes are direct targets of RFX1, RFX2 and/or RFX3. The histograms show the fold change in expression (log2 FC) in Rfx2−/− and Rfx3−/− cells for 187 mouse orthologs of genes that are affected in human ciliopathies. The presence RFX1, RFX2 and/or RFX3 ChIP-seq peaks in their promoters is indicated at the right. Significant reductions in gene expression are highlighted in light (Rfx3−/− cells) or dark (Rfx2−/− cells) grey, whereas non-significant variations are indicated in orange (Rfx3−/− cells). Genes highlighted in red are orthologs of human genes implicated in Primary Ciliary Dyskinesia (PCD). Nearly all of these PCD genes are direct targets of RFX1, RFX2 and/or RFX3, and are downregulated significantly in Rfx2−/− and/or Rfx3−/− cells. All genes have at least one X-box motif in their promoter, except for those indicated in black font.
Gene ontology
GO term enrichment was performed using Panther classification tool (68).
RESULTS
Differential phenotypes of Rfx1−/−, Rfx2−/− and Rfx3−/− mice
We focused our study on three mouse RFX factors—RFX1, RFX2 and RFX3—exhibiting different patterns of expression in vivo. Expression of the Rfx1 gene is ubiquitous and does not exhibit selective enrichment in specific tissues (59,69). In contrast, expression data available in public databases reveal that the mouse Rfx2 and Rfx3 genes are both expressed preferentially in ciliated tissues: whereas Rfx2 expression is largely restricted to tissues harboring motile cilia, expression of Rfx3 is more widespread but enriched in tissues presenting motile cilia.
In agreement with their differential patterns of expression, germline deficiencies of the three RFX factors have different in vivo consequences. We previously reported that Rfx2−/− and Rfx3−/− mice exhibit different phenotypes (11,46). Whereas Rfx2−/− mice are viable and develop normally, showing only fertility defects, Rfx3−/− mice suffer from severe developmental defects and exhibit major hallmarks of ciliopathies (11,12,28).
Several different Rfx1−/− mouse models have been reported. In the first model, consisting of a gene trap insertion into the second exon, Rfx1−/− mice were found to die during embryogenesis (70). Two subsequent models were based on the excision of two exons encoding the DNA binding domain of RFX1 (37,71). These two independent models were used to demonstrate that Rfx1 inactivation in the testis leads to complete male sterility (71) and that Rfx1 inactivation in T cells leads to increased susceptibility to autoimmune disease (37). Finally, we previously reported the generation of mice harboring a LoxP-flanked (floxed) Rfx1 allele (Rfx1flox), which allows conditional Cre-mediated deletion of exons encoding the DNA binding domain. This deletion furthermore leads to a frame-shift in the protein coding sequence, thereby removing all downstream regions of the protein (Supplementary Figure S1A). This conditional Rfx1 allele was previously used to demonstrate that RFX1 and RFX3 have redundant functions in inner ear development (47).
To further investigate the function of RFX1 we investigated the phenotype of Rfx1−/− mice obtained by germline induced Cre-mediated recombination of the Rfx1flox allele. We observed that newborn (P0) Rfx1−/− pups arose at the expected Mendelian frequency from Rfx1+/− intercrosses and presented no overt growth or developmental defects. Homozygous Rfx1−/− pups were viable, reached adulthood and exhibited no obvious defects, although they did show an increased susceptibility to weaning compared to Rfx1+/+ and Rfx1+/− littermates (Supplementary Figure S1B). Adult Rfx1−/− males and females were fertile and produced litters of comparable size relative to heterozygous controls (Supplementary Figure S1C). These observations indicate that, in contrast to previously published observations, Rfx1 is in our hands dispensable for mouse development and survival. Several explanations could account for this discrepancy. First, acute inactivation induced by conditional deletion might lead to more severe phenotypes than those induced by constitutive deletion. This could explain the infertility observed after inactivating Rfx1 specifically in the testis (71). Second, variable phenotypes could result from differences in the genetic backgrounds of the mouse strains. Our model indicates that RFX1 is dispensable for mouse development in a C57Bl/6J background and under standard housing conditions.
In agreement with their good survival rates, Rfx1−/− mice exhibited normal ciliogenesis in the brain ventricles (Figure 1A) and trachea (Supplementary Figure S2A). As reported previously (46), a deficiency in RFX2 did not lead to specific alterations of motile cilia in the brain (Figure 1B). We had previously reported that Rfx3−/− mice exhibit severe hydrocephalus, which was not attributable to an observable defect in motile cilia but instead to differentiation defects in two major tissues required for cerebrospinal fluid homeostasis, namely the sub-commissural organ and the choroid plexus (28). As a consequence of these defects, all Rfx3−/− pups die at birth in a C57Bl/6 background (11). To overcome this embryonic lethality, and to analyze the brains of adult mice exhibiting a selective deficiency in RFX3, we used a conditional knockout approach based on cell-specific deletion of a floxed Rfx allele (Rfx3flox) by a Foxj1-Cre transgene (49), which drives Cre-mediated recombination in cells bearing motile cilia. Effective elimination of RFX3 expression was verified by immunofluorescence (Supplementary Figure S2D). RFX3-deficient ependymal cells did not show ciliated defects in vivo (Figure 1C). In summary, all three RFX factors are dispensable, individually, for the development of multiple cilia on ependymal cells in vivo.
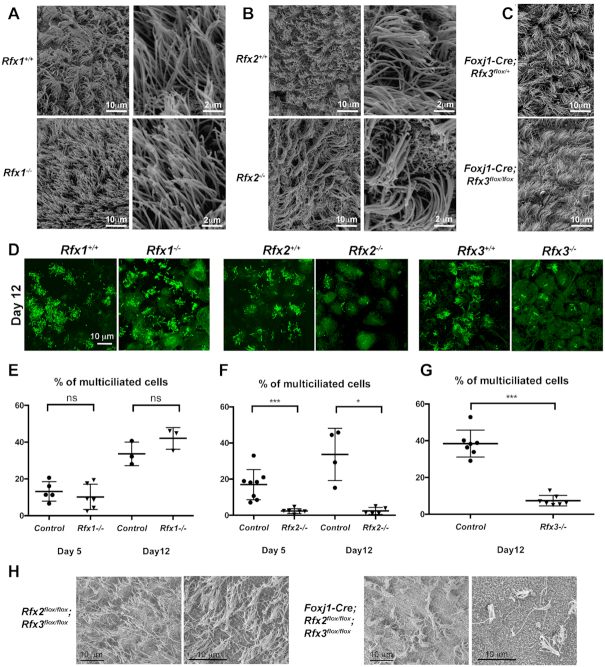
Figure 1.
RFX2 and RFX3 play critical roles in ependymal cell ciliogenesis. (A–C) Lateral ventricles of P15 brains were analyzed by scanning electron microscopy for Rfx1−/− (A), Rfx2−/− (B) or conditional FoxJ1-Cre;Rfx3flox/flox mice (C). Brains from Rfx1+/+, Rfx2+/+ or FoxJ1-Cre; Rfx3flox/+ mice were used as controls, respectively. No alterations in the number of ciliated cells or in the number and size of cilia were detected in the mutants. (D–G) In vitro differentiated ependymal cells derived from lateral brain ventricular progenitors at E18.5. (D) Representative images of multiciliated ependymal cell cultures at 12 days post serum deprivation stained for acetylated tubulin (green). (E–G) Graphs represent the percentages of multiciliated cells observed after 5 or 12 days of serum deprivation. No significant differences were observed between control and Rfx1−/− cells (E), whereas strong reductions in the number of ciliated cells were observed for Rfx2−/− (F) and Rfx3−/− (G) cells. *P< 0.05; ***P< 0.001; ns, not significant. (H) Lateral ventricles of P15 brains were analyzed by scanning electron microscopy in conditional double-Rfx2-Rfx3 deficient mice generated using the Foxj1-Cre strain. Compared to control ependymal epithelia (left), a major reduction in the number of ciliated cells was observed in ependymal epithelia deficient for both Rfx2 and Rfx3 (right). Two representative images are shown for each genotype.
RFX2 and RFX3 are key regulators of ependymal cell ciliogenesis in vitro and in vivo
To further study the respective roles of RFX1–3 in the development of motile cilia, we differentiated ependymal cells in vitro from ventricular progenitors derived from E18.5 embryos. In agreement with our in vivo data (Figure 1A), ex vivo differentiated Rfx1−/− ependymal cells did not show major defects in ciliogenesis: neither the number of ciliated cells nor the number of cilia per cell were significantly affected compared to controls (Figure 1D, E). In contrast, Rfx3−/− ependymal cells cultivated in vitro exhibited impaired ciliogenesis (Figure 1D, G) (12), despite the fact that no ciliary defects were observed in vivo (28) (Figure 1C). Similarly, ex vivo Rfx2−/− ependymal cells exhibited completely defective ciliogenesis (Figure 1D, F), despite the presence of normal multiciliated cells in Rfx2−/− mice in vivo (Figure 1B). Thus, RFX2 and RFX3 play non-redundant roles in motile ciliogenesis under our in vitro cell culture conditions, whereas they appear to compensate for each-others absence in vivo.
To address the possibility that the roles of RFX2 and RFX3 in directing ependymal cell ciliogenesis might be redundant in vivo we studied the consequences of a combined deletion of Rfx2 and Rfx3 in these cells. Cell-specific deletion of floxed Rfx2 and Rfx3 alleles (Rfx2flox and Rfx3flox) was induced by means of the Foxj1-Cre transgene (49). Efficient Cre-mediated recombination was confirmed by immunofluorescence analysis, which demonstrated that the expression of RFX2 and RFX3 was effectively abolished in ependymal cells (Supplementary Figure S2D). We observed that the combined deletion of Rfx2 and Rfx3 led to a marked impairment of ependymal cell ciliogenesis (Figure 1H). These results confirm that RFX2 and RFX3 indeed have redundant functions in ependymal ciliogenesis in vivo. The same functional redundancy was also observed in vivo for airway ciliogenesis: whereas no defects in tracheal cilia were observed at birth in Rfx3−/− pups (11) or adult Rfx2−/− mice (46), we observed a significant reduction in the number of multiciliated tracheal cells at birth when both Rfx2 and Rfx3 were deleted (Supplementary Figure S2B and C).
RFX1, RFX2 and RFX3 regulate common and specific gene sets in ependymal cells
To understand how RFX1–3 exert their functions in ependymal cells, we performed RNA-seq experiments with ex vivo cultured ependymal cells obtained from E18.5 WT, Rfx1−/−, Rfx2−/− and Rfx3−/− embryos. Global analysis of differential gene expression was performed for each mutant cell type relative to WT cells (Figure 2A). Genes were considered to be differentially expressed if the fold-change was >2 with a P-value of <0.001 (Supplementary Table S1, genes indicated as down/up ‘high’; genes with a fold change between 2 and 1.5 were also included in the table, indicated as down/up ‘low’). Whereas 298 and 443 genes were significantly down regulated >2 fold, respectively, in Rfx2−/− or Rfx3−/− cells compared to WT cells, only 11 genes were down regulated in Rfx1−/− cells (Figure 2B). Markedly fewer genes were up-regulated: 47 for Rfx2−/−, 116 for Rfx3−/− and only 2 for Rfx1−/− cells (Figure 2B). There was no overlap between the differentially expressed (DE) gene sets in Rfx1−/− cells and DE gene sets in Rfx2−/− or Rfx3−/− cells (Figure 2B). Among upregulated genes, only a minority was shared by Rfx2−/− (12/47 genes) and Rfx3−/− (12/116 genes) cells (Figure 2B). In sharp contrast, there was strong overlap between downregulated gene sets in Rfx2−/− and Rfx3−/− cells: most genes downregulated in Rfx2−/− cells were also downregulated in Rfx3−/− cells (Figure 2B: 220/298 genes), and nearly half of the genes downregulated in Rfx3−/− cells were also downregulated in Rfx2−/− cells (220/443 genes). Interestingly, the genes that were downregulated most strongly in Rfx3−/− cells were nearly all downregulated as well in Rfx2−/− cells (Figure 2C, left), whereas the reverse was not true: only part of the most strongly downregulated genes in Rfx2−/− cells was also downregulated in Rfx3−/− cells (Figure 2C, right).
To determine if cross-regulation between different Rfx genes could contribute to the observed overlap in downregulated genes between Rfx2−/− and Rfx3−/− cells, we quantified the respective mRNA expression levels for Rfx1–3 genes in WT and the three Rfx-deficient ependymal cells. Rfx1−/− and Rfx2−/− cells exhibited no significant reductions in expression of the other two Rfx mRNAs. However, significant reductions in Rfx1 and Rfx2 expression were observed in Rfx3−/− cells (Supplementary Figure S3). These results suggest that deregulated expression in Rfx1−/− or Rfx2−/− cells is a direct consequence of the deficiency in RFX1 or RFX2, respectively. On the other hand, reduced RFX2 expression could account in part for the observed pattern of deregulated gene expression in Rfx3−/− cells. The latter mechanism is likely to be particularly relevant for genes that are downregulated in both Rfx2−/− and Rfx3−/− cells.
Gene Ontology (GO) analyses were performed for gene sets that were downregulated in Rfx2−/− and/or Rfx3−/− ependymal cells. Gene sets that were downregulated in Rfx3−/− cells alone or in both Rfx2−/− and Rfx3−/− cells were significantly enriched in genes encompassed by GO terms associated with cilium biogenesis or function (Supplementary Figure S4A, Supplementary Table S2). GO terms associated with microtubule-based processes and movement were also significantly enriched, which could be related to the regulation of ciliogenesis. No significant enrichment for specific GO terms was found in the set of genes that is downregulated only in Rfx2−/− cells (not shown).
To investigate further whether the downregulated gene sets could be used to predict novel ciliary genes, we performed gene set enrichment analyses (GSEA, (63)) for pathways associated with cilia assembly or function. The GSEA analyses were performed with several different sets of ciliary genes, including the previously defined Syscilia gold standard set of genes (set A) (62) and two sets comprising, respectively, human ciliopathy associated genes (set B) or potential ciliary genes identified on the basis of phenotypes documented in animal models (set C, Supplementary Table S3) (61). For all three gene sets, the distribution of ciliary genes was strikingly shifted towards the most down regulated genes in both Rfx2−/− and Rfx3−/− cells, indicating that genes exhibiting reduced expression in these cells are strongly enriched in functions associated with cilia assembly and function (Supplementary Figure S4B). Similar results were obtained using other ciliary gene sets (data not shown).
Genes regulated by RFX2 and RFX3 share indistinguishable X-Box motifs
Two complementary strategies were used to identify potential DNA-binding motifs for RFX factors in the promoter-proximal regions (defined here as −1000 to +500 bp relative to the transcription start site, TSS) of genes that are downregulated in Rfx2−/− and/or Rfx3−/− ependymal cells. As a first approach, Pscan software was used to quantify the enrichment of known transcription factor binding site (TFBS) motifs described in the JASPAR database. TFBS motifs were clustered on the basis of their enrichment scores in different downregulated gene sets, and results are presented as a heatmap (Figure 3A, Supplementary Figure S5). This analysis revealed that X-box motifs (TFBS motifs defined for RFXs are presented in Figure 3B) are highly enriched in the promoters of most subsets of genes downregulated in Rfx2−/− and Rfx3−/− cells, with the exception of genes regulated exclusively by RFX2 (Figure 3A, Supplementary Figure S5). As a second unbiased approach, a de novo motif discovery method (cosmo package for R) was used to identify motifs that are enriched in the promoters of different sets of genes downregulated in Rfx2−/− and/or Rfx3−/− cells (Figure 3C). Although applying this de novo search to −1000 to +500 promoter regions did not give significant results, using a more restricted promoter definition (−250 to +50) identified very similar motifs, all exhibiting nearly perfect matches to the previously defined X-box, in most subsets, with the exception of genes regulated by RFX2 alone. This motif discovery strategy revealed no differences between the X-box signatures for different downregulated gene subsets, as confirmed by TOMTOM (73) analysis of the identified motifs, which demonstrated that all show the best ranking value with the RFX1 MA0509.1 motif defined in Figure 3B (Supplementary Table S4). This indicates that differential target gene specificities of RFX2 and RFX3 cannot be accounted for simply by variations in the X-box motif itself, or that such variations are subtle and not detected by the analysis tools used here. For the set of genes downregulated only in Rfx2−/− cells, a motif very different from the X-box was identified. This motif is most closely related to UP00007_1, corresponding to the Egr1 motif. This suggests that most of the genes that are downregulated in Rfx2−/− cells are not regulated directly by RFX2, or that RFX2 is recruited to enhancer regions situated far away from their promoters.
Figure 3.
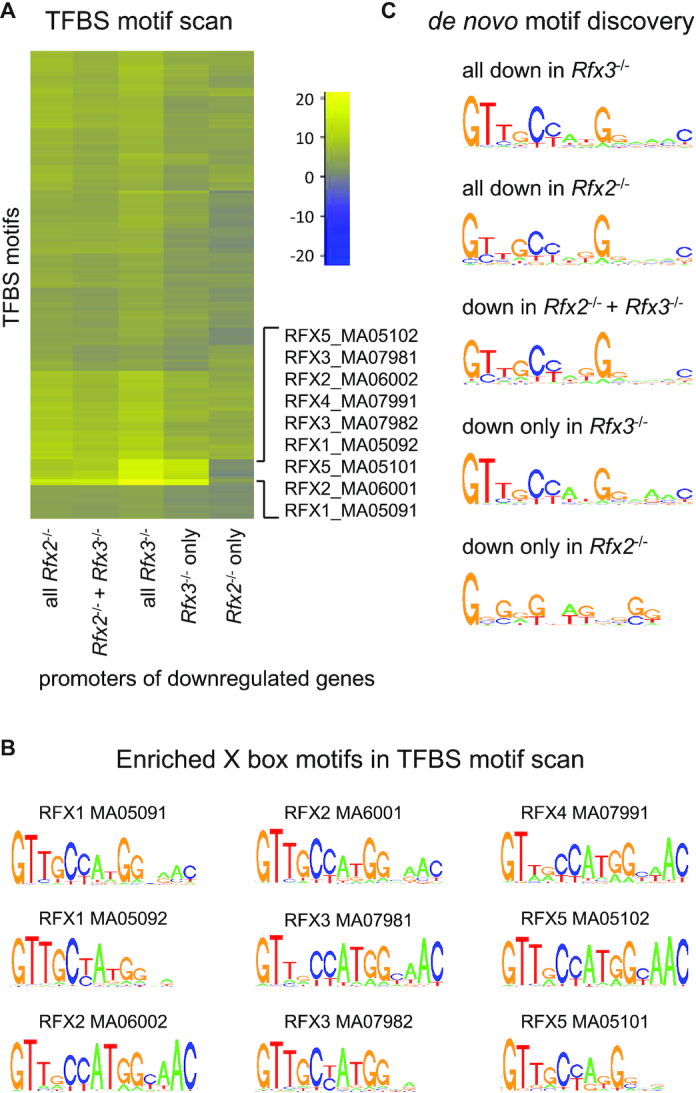
Identification of TFBS motifs in the promoters of genes that are downregulated in Rfx2 and/or Rfx3 KO cells. The promoters (−1000 to +500 pb relative to the TSS) of different sets of downregulated genes were examined by two unbiased approaches for the presence of specific TFBS motifs. (A) Selected region of a heatmap focusing on the most enriched motifs, determined using Pscan for the complete set of TFBS motifs from the JASPAR database, in the promoters of the indicated sets of downregulated genes. The full heatmap is presented in Supplementary Figure S5. Colour key: blue, under-represented motifs; yellow, over-represented motifs. In most gene sets examined, the most strongly enriched TFBS motifs were the X-box motifs previously defined for RFX factors (the corresponding sequence logos are provided in (B)). Only promoters of genes that are downregulated exclusively in Rfx2−/− cells did not exhibit an enrichment in these X-box motifs. (B) Sequence logos of RFX motifs from JASPAR database enriched in RFX-regulated genes. (C) A de novo motif discovery approach (cosmo package) was applied to the promoters (−250 to +50 pb relative to the TSS) of the same sets of downregulated genes. Typical X-box motifs were again revealed in the promoters of most sets of downregulated genes. As in (A) no motif resembling the X-box was revealed in the promoters of genes downregulated only in Rfx2−/− cells. In the latter gene set, a G-rich motif was instead identified, which could be related to the Egr1 motif (UP00007_1).
The Pscan analysis was also performed for the promoters of up-regulated genes, but this did not reveal any significantly enriched RFX binding sites, suggesting that these up-regulated genes could be indirect RFX targets. Interestingly, several TFBS motifs were significantly enriched in genes upregulated in the absence of RFX3 including, ZNF263, MAZ, ZNF148, E2F6, SP1, E2F3, TFAP (Supplemental Figure S5).
Altogether, these observations indicate that a significant fraction of downregulated genes in Rfx2−/− and Rfx3−/− ependymal cells are likely to be direct targets of either RFX3 alone or both RFX2 and RFX3.
RFX1-3 share numerous binding sites in mouse ependymal cells
To identify genes that are regulated directly by RFX1, RFX2 and/or RFX3 in ependymal cells, we performed chromatin immunoprecipitation sequencing (ChIP-seq) experiments for each transcription factor using ex-vivo cultured ependymal cells from WT mice. Total numbers of RFX occupied sites (ChIP-seq peaks) in the genome were 7440 for RFX1, 1250 for RFX2 and 9318 for RFX3. Sites occupied by the three factors exhibited similar distributions in promoter regions (defined here as −1000 to + 500 bp relative to the TSS), intergenic regions, introns and exons (Figure 4A). The peak distribution for each factor was centered just upstream of the TSS (Figure 4B).
TFBS motif analysis was performed as described above. The de novo motif discovery approach (cosmo package for R) was performed for the 100 most robust peaks (based on the number of sequence reads) for each RFX factor. For each factor a clear X-box motif was identified (Figure 4C, Supplementary Table S5). These motifs exhibited only minor differences between the three RFX factors, indicating that the best 100 peaks for each RFX harbor a similar RFX recognition sequence. It should be noted however, a significant portion of the 100 most robust peak sets defined for the three factors is shared between more than one RFX factor (Supplementary Figure S6A). Furthermore, peak overlaps are even greater when the top 100 peaks for each individual RFX factor are compared with all peaks for the remaining two factors (Supplementary Figure S6B). Therefore, to favor the identification of potential differences in sequence preference between RFX factors we performed the de novo motif analysis on the 100 most robust peaks that are unique to each RFX, using the least stringent window to identify coinciding peaks, i.e. <500 bp spacing between peaks. This strategy revealed nearly identical X-box motifs for RFX1 and RFX3 (Supplementary Figure S6C), indicating that no RFX1 and RFX3 motif preference could be discriminated. This analysis was not possible for RFX2, as only 10 of its most robust peaks did not overlap with any RFX1 or RFX3 peaks, a number that is too low to perform a reliable de novo motif analysis. A Pscan analysis was next performed on all peaks using the newly identified X-box motifs and all TFBS motifs described in the JASPAR database. For each RFX factor the ten most enriched motifs included the newly identified RFX binding sites and TFBS motifs previously defined for RFX2, RFX1 and RFX5 (Figure 4D, Supplementary Table S5), again indicating that the three RFX factors bind very similar motifs. Thus, RFX binding-site specificity or function does not seem to rely on readily detectable variations in their binding motifs.
Other predicted TFBS motifs were also found to be enriched in RFX ChIP-seq peaks. Close examination of these motifs indicated that several show partial overlap with the X-box (Figure 4D: MYB NFIC, NFIX, NFIB, YY2). However, a number of other significantly enriched motifs were identified as well. 17 and 47 TFBS motifs were found to be enriched specifically in ChIP-seq peak sets for RFX1 and RFX3, respectively (Figure 4E, Supplementary Table S5), suggesting that the corresponding factors could be RFX1 or RFX3 specific regulatory partners. Only three enriched TFBS motifs were identified in the RFX2-specific ChIP-seq peak set (FOSL1::JUND(VAR2); GMEB1; MYBL). A number of enriched motifs were shared by ChIP-seq peak sets defined for RFX1 and RFX3 (73 motifs) or all three RFX factors (310 motifs). The latter includes the Y-box, which is bound by NF-Y and is tightly associated with the X-box in the promoters of MHC class II genes (74). NFY is known to bind cooperatively with a transcription factor complex containing RFX5, a process that is essential for RFX5 function in antigen presenting cells (74). Thus, our results suggest that a similar functional association of X and Y-boxes could also be involved in the function of RFX1–3 in ependymal cells. The other enriched motifs found in RFX common peaks could represent TFBS motifs for regulatory partners of two or more RFX factors.
To identify genomic loci occupied by more than one RFX factor, we compared the sets of ChIP-seq peaks obtained for the three individual factors. Four different distance parameters were used to define coinciding peaks: two peaks were considered to overlap if their centers were less than 15, 50, 100 or 500 bp apart. The analysis was done separately for all peaks (Supplementary Figure S6D) or for peaks situated in promoter regions (Figure 5). Using all four distance parameters, a major proportion of the total number of RFX-bound loci were bound by more than one RFX factor. As the size of the X-box motif is only 15 bp, the overlap analysis done using this distance parameter likely identifies single binding sites bound by multiple factors. Increasing the distance parameter to 50, 100 and 500 bp led to relatively minor increases in the numbers of regions occupied by two or three factors, suggesting either technical issues leading to imprecise mapping of the same binding site or the presence of several closely spaced binding sites. As overlap numbers obtained with the 50, 100 and 500 bp distance parameters were very similar to each other, it is likely that the majority of regions occupied by multiple RFX factors contain either only a single binding site or 2–3 very closely spaced (<50 bp) binding sites. Lists of promoters occupied by single or multiple RFX factors are provided in Supplementary Table S6.
Figure 5.
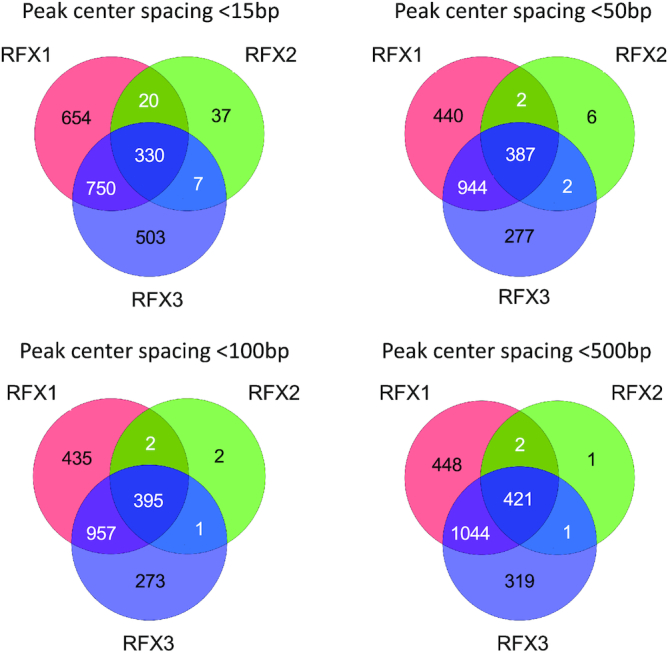
Overlap between sets of promoters occupied by RFX1, RFX2 and/or RFX3. The Venn diagrams illustrate the overlap between sites (situated between −1000 to +500 bp relative to the TSS) occupied by RFX1, RFX2 and/or RFX3. Sites were scored as being co-occupied by two or three RFX factors when the distance between their ChIP-seq peak centers were <15, <50, <100 or <500 bp. In most cases, the centers of ChIP-seq peaks at co-occupied promoters were situated <15 pb apart, which is less than the size of the X-box motif, indicating that RFX1, RFX2 and/or RFX3 are bound to the same binding site at these promoters. At a minority of co-occupied promoters, the centers of ChIP-seq peaks were separated by between 15 and 500 bp, suggesting the potential presence of 2–3 closely spaced binding sites.
Many RFX target genes are involved in cilia assembly or function in ependymal cells
A GO analysis was first performed using Panther (68) for gene sets having promoter regions bound by RFX1, RFX2 or RFX3 (Supplementary Table S7, Supplementary Figure S7). Most significantly enriched GO terms for biological functions defined for RFX1, RFX2 and RFX3 ChIP targets were found to be related to cilia, indicating that each factor binds preferentially to promoters of genes implicated in cilia assembly or function. Enriched GO terms also included microtubule or centrosome associated processes, which could be related to the regulation of ciliogenesis. Lastly, the enriched terms indicated an association with regulation of the cell cycle, in agreement with tight coupling between ciliogenesis and control of the cell cycle. Curiously, the set of RFX3 targets is also highly enriched in genes involved in mitochondrial respiratory chain assembly.
We also performed GO enrichment analyses for all ChIP-seq Peaks using GREAT analysis (75) to assign peaks to specific genes (Supplementary Table S8). Among the GO terms associated with biological processes (Supplementary Table S8), the top ranking GO terms are linked to cilia and cell cycle associated functions. Collectively, these analyses indicate that genes that are potentially regulated by RFX1–3 through promoters or distal enhancers are highly enriched in genes involved in cilia related functions. Lastly, RFX1 and RFX3 target genes are also associated with GO terms related to metabolic processes.
We next crossed our expression data for Rfx2−/− and Rfx3−/− cells with our RFX ChIP-seq data for the three RFX factors to identify genes that are regulated directly by RFX factors in ependymal cells (Figure 6, Supplementary Table S9). Approximately half (45–56%) of the down-regulated genes, and nearly all up-regulated genes, were found to lack RFX-bound sites in their promoters. When examining both distal enhancers and promoters using GREAT analysis, 15–21% of down-regulated genes and 22–48% of up-regulated genes were found to lack RFX-occupied sites in their regulatory sequences (Supplementary Figure S8). These results suggest that a substantial portion of differentially related genes are likely to be indirect targets of RFX factors in ependymal cells. Alternatively, they could be controlled by distal RFX-regulated enhancers that are difficult to correctly assign to a single gene. Among downregulated genes with RFX occupied sites in their promoters or putative distal enhancers, most were bound by at least two RFX factors—either all three RFX factors or RFX1 plus RFX3—whereas only a minority were bound by RFX3 or RFX1 alone, and none were bound by RFX2 alone or RFX2 plus RFX3 (Figure 6A, Supplementary Figure S8). We also performed a GSEA analysis of sets of genes containing RFX1, RFX2 or RFX3 occupied sites in their promoters in the expression data obtained for Rfx2−/− and Rfx3−/− cells (Figure 6B). We observed a strong enrichment of RFX-occupied promoters in downregulated genes, indicating a strong correlation between the occupancy of promoters by RFX proteins and their transcriptional regulation by RFX2 and/or RFX3.
A set of high-confidence RFX targets genes - namely the 28 genes that are downregulated in both Rfx2−/− and Rfx3−/− cells, and have promoters occupied by all three RFX factors - were examined for the presence of genes related to cilia (Figure 6C). Most (21/28) of these genes have been shown to be involved in ciliogenesis (Spata18, Rsph14, Hydin, Wdr65/Cfap57, Lrrc6, Lrrc34, Dnaaf1, Lrrc23, Saxo2/Fam154b, Dlec1, Cfap221, Wdr16/CFAP52, Cep128, Ccdc65, Rsph10b, march10) or to be highly expressed in ciliated tissues (Cfap161, Ccdc146, Ccdc108/Cfap65, Cfap74, Lrrc46). The remaining 7 genes (Fhad1, E230008N13Rik/ccdc180, Tsnaxip1, Casc1, Kif6, Zswim2, Serpinb1b) have not been previously associated with cilia. However, given their tight coregulation with other cilia-related genes, they are excellent candidates for having novel ciliary functions. A similar analysis done for downregulated genes exhibiting a fold change situated between –2 and –1.5 (Supplementary Figure S9) also highlighted a strong enrichment in genes associated with ciliary function in the set of direct RFX1–3 targets, thus providing further interesting ciliary candidates. Finally, we investigated whether genes linked to ciliopathies (187 mouse orthologs of genes from (61)) are direct RFX targets. Many of these genes are indeed bound by at least one RFX factor and/or are downregulated in Rfx2−/− and/or Rfx3−/− cells (Figure 7, Supplementary Table S9). In particular, the expression of all genes involved in primary ciliary dyskinesia are regulated by RFX2, RFX3 or both in ependymal cells, and most of these are direct RFX targets. In conclusion, our observations illustrate the critical role played by RFX2 and RFX3 in the motile ciliogenesis program in mice, and suggest that there are likely to be novel, previously unidentified, cilia-associated genes among the direct RFX target genes we have identified.
DISCUSSION
We show here that RFX2 and RFX3 have both redundant and specific functions in the biogenesis of motile cilia on mouse ependymal cells, whereas RFX1 does not seem to play a key regulatory role in this process. This is supported by the ependymal phenotypes of Rfx1, Rfx2 and Rfx3 single-knockout mice, and of conditional Rfx2:Rfx3 double-knockout mice, as well as by the analysis of direct and indirect targets of RFX1, RFX2 and RFX3 in ependymal cells. We demonstrate that many direct targets of RFX2 and RFX3 are known to be involved in cilia assembly or function, and furthermore provide valuable lists of new candidate ciliary genes. Our results also reveal complex compensatory mechanisms operating at the cellular level, as revealed by the phenotypic differences observed between in vivo ventricular ependymocytes and the corresponding in vitro differentiated cells. Compensatory mechanisms also appear to be operating at the molecular level, as revealed by the uncoupling between genome occupancy by RFX1 and genes effectively regulated by RFX1 versus the strong correlation observed between genome occupancy and gene expression for RFX2 and/or RFX3.
The promoters of numerous genes that are downregulated in Rfx2−/− or Rfx3−/− ependymal cells are actually occupied by these transcription factors in these cells, confirming that they are direct RFX targets. However, we found that a substantial portion of the deregulated genes do not show binding of RFX2 or RFX3 in their promoter regions or distal enhancers as assigned by GREAT analysis, suggesting that they could be indirect RFX targets. Alternatively, these genes could be regulated by binding of RFX2 or RFX3 at distant enhancers, which would be consistent with the finding that less than a third of all RFX ChIP-seq peaks are actually localized in promoter regions. The mechanisms by which RFX factors activate transcription is poorly understood. In C. elegans, most target genes are regulated by binding of RFX to sites situated close to the TSS (44,76). In mouse lymphocytes, RFX1 regulates chromatin modification at the IL17 promoter (37). The most thorough understanding comes from studies in xenopus, in which RFX2 was shown to act both at distant chromatin loci and at promoter sites, and to be involved in chromatin looping to stabilize FoxJ1 (14). Such long-range chromatin organizing functions could also be acting in ependymal cells for RFX2 and RFX3, and this could account for the regulation of specific subsets of genes lacking RFX-occupied sites in their promoters. Chromatin-conformation analysis studies will be necessary to understand further how RFX2 and RFX3 control target gene expression.
Motif discovery analyses of our ChIP-seq and RNA-seq data sets did not reveal any significant differences between the X-box motifs for RFX1, RFX2 and RFX3. Yet these transcription factors occupy distinct, albeit overlapping, sets of genomic loci. This may reflect inherent limitations in the motif discovery algorithms that were used, which could be insufficiently sensitive to reveal subtle differences. Alternatively, RFX1, RFX2 and RFX3 target site specificity could rely on associated promoter or enhancer binding sites for other transcription factors. Our de novo motif discovery approach did not reveal specific sequence motifs linked to the X-box in RFX1–3 target genes, indicating that potential associated motifs are more diverse or less well defined than the X-box motif. However, motif discovery analyses using Pscan did allow us to identify a number of transcription factor motifs enriched selectively in promoters occupied by RFX1–3 proteins. Certain of these motifs are specific for RFX1 or RFX3 occupied regions. TFs binding to these associated motifs could be responsible for determining differential occupancy by the RFX factors, and thus explain why, despite their identical X-box motifs, RFX1 and RFX3 occupy different sets of loci in ependymal cells. In addition, occupancy by RFX1 and RFX3 could rely on binding of factors to distant regions, or be sensitive to particular chromatin configurations that our study have not addressed.
We identified 310 transcription factor binding motifs that are enriched in promoters of genes bound by all three transcription factors. Many of these have binding sites that overlap with or resemble RFX motifs. However enriched motifs that are clearly distinct from RFX motifs include those for members of the E2F family of transcription factors (E2F4, E2F1, E2F3, E2F6). Interestingly, E2F4 and E2F5 have been shown to be involved in regulating ciliogenesis in the mouse, zebrafish or C. elegans, and E2F transcription factors associate with Geminin or multicilin to regulate ciliogenesis (20, 77–80). E2F transcription factors have also been shown to be involved in cell cycle regulation, which is in agreement with the tight link beween ciliogenesis and cell cycle control. Together with our observations, this indicates that E2F family members are likely to collaborate with RFX transcription factors to regulate ciliary gene expression. Two other transcription factors for which binding motifs are enriched in direct target genes of RFX1–3 are ZFP423/ZNF423 and ATOH (Supplementary Table S5). ZFP423 has been shown to regulate primary cilia formation and 12 genes involved in ciliogenesis were found to be regulated by ZFP423, including Tulp3, which was confirmed to be a direct target of ZFP423 (81). We show here that Tulp3 is also a direct target of RFX1–3, suggesting functional interactions between ZFP423 and RFX factors in the control of ciliogenesis. ATOH belongs to the family of proneural factors involved, in particular, in hair cell maintenance (14–16). The fly ortholog, atonal, has been shown to regulate Drosophila RFX genes as well as ciliary genes (82). This indicates that evolutionary conserved regulatory networks in ciliogenesis involve RFX and the atonal proneural factors. Collectively, these observations suggest that ciliary gene expression is regulated by complex transcription factor networks. Deciphering these complex networks will be a challenging endeavor.
Although RFX1 binds to numerous loci in ependymal cells, it does not appear to have a major impact on gene expression in these cells, and a deficiency in RFX1 does not lead to overt ciliary phenotypes in multiciliated epithelia. As each of the three RFX factors can form homodimers and heterodimers, it is likely that RFX1 homodimers as well as heterodimers with RFX2 and/or RFX3 can occupy regulatory regions containing X box motifs. However, our data suggests that RFX1 homodimers and heterodimers could be less efficient at activating transcription than RFX2 and RFX3 homodimers or heterodimers. Hence, removing RFX1 alone would not be sufficient to observe a major consequence on gene expression. These considerations further raise the question of whether or not RFX1 actually has any function in ciliogenesis. We have observed that the deletion of Rfx1 in conjunction with Rfx3 is highly deleterious during embryogenesis, leading to early embryonic lethality between E9 and E10 (data not shown). We have been unable to relate this lethality to evident defects in ciliogenesis in the early embryonic tissues that we have analyzed (primary cilia of the neural tube or mesenchymal cells, data not shown). We did also not observe clear defects in ciliogenesis in ependymal cells by a combined conditional deletion of RFX1 and RFX3 using the FoxJ1-cre deleter strain (data not shown). Thus, although we cannot fully exclude that RFX1 could have cilia-related functions during development, our data suggests that embryonic lethality induced by the combined deletion of Rfx1 and Rfx3 could well be due to the regulation of other pathways. In this respect, it should be mentioned that in yeast or fungi, RFX factors have been implicated in regulation of the cell cycle and genome repair (83–86). Defects in such pathways could also account for embryonic lethality observed in the double mutants. We identified >1400 genes which can potentially be regulated directly by RFX1 and RFX3 (Figure 2) and GO classification of these genes indicate that, in addition to ciliary genes, several could be essential for development, such as genes involved in assembly of the mitochondrial respiratory chain complex, in the assembly of ribonucleoprotein complexes, or in cell cycle progression. These target genes could explain the essential role played by RFX1 together with RFX3 in mouse embryonic development.
Here, we have identified RFX1, RFX2 and RFX3 target genes in ependymal cells. In a previous study, we analyzed RFX1 and RFX3 target genes by ChIP-seq (47) in a pancreatic β-cell line (Min6) because Rfx3 deficient mice showed impaired pancreatic development (60,72) associated with impaired primary cilia formation. We also previously identified RFX2 targets by Chip-seq and RNA-seq in the testis, where RFX2 is required for spermatid differentiation (46). Comparisons between the sets of target genes in these three different tissues demonstrated partial overlaps (Supplementary Table S10, Supplementary Figure S10). Common targets in the different cell types are significantly enriched in ciliogenic genes (Supplementary Table S11). Interestingly, GO analysis of tissue specific targets showed that genes regulated by RFX1 or RFX3 exclusively in Min6 cells are not associated with ciliogenesis, whereas target genes regulated directly by RFX1, 2 or 3 in ependymal cells are enriched in GO terms associated with cilia or microtubule-based processes. These observations indicate that RFX transcription factors regulate a core set of genes associated with ciliogenesis in all tissues, but that RFX1 or RFX3 could also play specific regulatory roles distinct from ciliogenesis in Min6 cells. By contrast, all sets of RFX2 target genes identified here are associated with ciliogenesis, indicating a focused function of RFX2 in this biological function. This is in agreement with observations made in other vertebrates, where RFX2 plays a prevalent role in the motile ciliogenesis program (13,14,30).
Our work reveals redundant roles of RFX2 and RFX3 in the mouse, as well as surprising differences between compensatory mechanisms operating in the in vivo and in vitro differentiation processes of ependymal cells. Whereas removal of either RFX2 or RFX3 alone leads to multiciliated cell differentiation defects in vitro, only removal of both factors leads to ependymal defects in vivo. Although we do not as yet have any molecular explanations for this observation, a number of potential mechanisms could be at play. One major difference between the in vivo setting and the ex-vivo model is the spatio-temporal regulation of cell differentiation in a complex tissue in vivo. Ependymal cell differentiation is governed in vivo by finely tuned developmental cues, which are communicated to specific cells in a well-defined spatially and temporally controlled pattern. In sharp contrast, the ex vivo differentiation program is triggered by serum deprivation, which affects all cells in the culture at the same time. These differences could allow for compensatory regulatory pathways to be recruited in vivo but not in vitro. One might consequently question the biological relevance of the RNA-seq and ChiP-seq experiments, which were both performed using the in vitro differentiation model. However, as redundancy between RFX2 and RFX3 was observed at the levels of both their biochemical properties in vitro and their developmental functions in vivo, the in vitro data reported here are consistent with the in vivo roles of RFX3 and RFX2.
Similar redundancies between RFX factors need to be considered in other organisms, particularly in humans, where the functions of RFX1–3 remain unknown. Together with our observations, analyses of RFX expression patterns and the presence X-box motifs in the human genome should provide a valuable basis for furthering our understanding of the functions of RFX1–3 in man (87).
Our observations reveal a central role of RFX2 and RFX3 in the biogenesis of motile cilia and we therefore anticipate that the target gene sets we have identified are highly likely to include novel candidates implicated in ciliogenesis. In particular, we expect to find key candidates among genes that exhibit robustly reduced expression in Rfx2 and Rfx3 deficient ependymal cells and contain RFX2 and/or RFX3 occupied sites in their promoters and/or distal enhancers.
DATA AVAILABILITY
Sequence data has been submitted to the GEO database under the accession number n° GSE145324.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Sophie Mohamed Sidi from the mouse facility and Aurélie Mongope and Jean-Michel Vicat from the ALEC SPF platform for help with mouse husbandry. We would like to thank I. Kimura, E. Cortier, S. Ben Larbi for help with mouse genotyping. Electron Microscopy observations were performed at the CTMu from the University of Lyon-1. We are grateful to the iGE3 genomics platform of the University of Geneva for performing the high throughput sequencing.
Notes
Present address: Fabien Soulavie, IBDM – Institut de Biologie du Développement de Marseille, UMR 7288 Parc Scientifique de Luminy, France.
Present address: Julie Jerber, MRC Metabolic Diseases Unit, University of Cambridge Metabolic Research Laboratories, WT-MRC Institute of Metabolic Science, University of Cambridge, Cambridge CB2 OQQ, UK.
Contributor Information
Sylvain Lemeille, Department of Pathology and Immunology, University of Geneva Medical School, CMU, Geneva, Switzerland.
Marie Paschaki, Univ Lyon, Université Claude Bernard Lyon-1, CNRS UMR-5310, INSERM U-1217, Institut NeuroMyoGène, F-69008, Lyon, France.
Dominique Baas, Univ Lyon, Université Claude Bernard Lyon-1, CNRS UMR-5310, INSERM U-1217, Institut NeuroMyoGène, F-69008, Lyon, France.
Laurette Morlé, Univ Lyon, Université Claude Bernard Lyon-1, CNRS UMR-5310, INSERM U-1217, Institut NeuroMyoGène, F-69008, Lyon, France.
Jean-Luc Duteyrat, Univ Lyon, Université Claude Bernard Lyon-1, CNRS UMR-5310, INSERM U-1217, Institut NeuroMyoGène, F-69008, Lyon, France.
Aouatef Ait-Lounis, Department of Pathology and Immunology, University of Geneva Medical School, CMU, Geneva, Switzerland.
Emmanuèle Barras, Department of Pathology and Immunology, University of Geneva Medical School, CMU, Geneva, Switzerland.
Fabien Soulavie, Univ Lyon, Université Claude Bernard Lyon-1, CNRS UMR-5310, INSERM U-1217, Institut NeuroMyoGène, F-69008, Lyon, France.
Julie Jerber, Univ Lyon, Université Claude Bernard Lyon-1, CNRS UMR-5310, INSERM U-1217, Institut NeuroMyoGène, F-69008, Lyon, France.
Joëlle Thomas, Univ Lyon, Université Claude Bernard Lyon-1, CNRS UMR-5310, INSERM U-1217, Institut NeuroMyoGène, F-69008, Lyon, France.
Yong Zhang, Department of Medicine and Department of Cell Biology, Washington University School of Medicine, St. Louis, Missouri.
Michael J Holtzman, Department of Medicine and Department of Cell Biology, Washington University School of Medicine, St. Louis, Missouri.
W Stephen Kistler, Department of Chemistry and Biochemistry, University of South Carolina, Columbia, South Carolina, United States of America.
Walter Reith, Department of Pathology and Immunology, University of Geneva Medical School, CMU, Geneva, Switzerland.
Bénédicte Durand, Univ Lyon, Université Claude Bernard Lyon-1, CNRS UMR-5310, INSERM U-1217, Institut NeuroMyoGène, F-69008, Lyon, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondation pour la Recherche Médicale (FRM) [DEQ20131029168 to B.D.]; ANR (Ciliopath-X); F.S. was supported by a doctoral fellowship from the Région Rhône-Alpes; J.J. was supported by a doctoral fellowship from the Fondation pour la Recherche Médicale (Prix Line Pomaret-Delalande); Work in the team of W. Reith was supported by multiple grants from the Swiss National Science Foundation. Funding for open access charge: Research grants.
Conflict of interest statement. None declared.
REFERENCES
- 1. Thomas J., Morlé L., Soulavie F., Laurençon A., Sagnol S., Durand B.. Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol. Cell. 2010; 102:499–513. [DOI] [PubMed] [Google Scholar]
- 2. Choksi S.P., Lauter G., Swoboda P., Roy S.. Switching on cilia: transcriptional networks regulating ciliogenesis. Development. 2014; 141:1427–1441. [DOI] [PubMed] [Google Scholar]
- 3. Meunier A., Azimzadeh J.. Multiciliated cells in animals. Cold Spring Harb Perspect. Biol. 2016; 8:a028233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spassky N., Meunier A.. The development and functions of multiciliated epithelia. Nature. 2017; 18:423–436. [DOI] [PubMed] [Google Scholar]
- 5. Terré B., Piergiovanni G., Segura Bayona S., Gil Gómez G., Youssef S.A., Attolini C.S.O., Wilsch-Bräuninger M., Jung C., Rojas A.M., Marjanović M. et al.. GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J. 2016; 35:942–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou F., Narasimhan V., Shboul M., Chong Y.L., Reversade B., Roy S.. Gmnc is a master regulator of the multiciliated cell differentiation program. Curr. Biol. 2015; 25:3267–3273. [DOI] [PubMed] [Google Scholar]
- 7. Brody S., Yan X., Wuerffel M., Song S., Shapiro S.. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am. J. Respir. Cell Mol. Biol. 2000; 23:45–51. [DOI] [PubMed] [Google Scholar]
- 8. Huang T., You Y., Spoor M., Richer E., Kudva V., Paige R., Seiler M., Liebler J., Zabner J., Plopper C. et al.. Foxj1 is required for apical localization of ezrin in airway epithelial cells. J. Cell Sci. 2003; 116:4935–4945. [DOI] [PubMed] [Google Scholar]
- 9. Gomperts B., Gong-Cooper X., Hackett B.. Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. J. Cell Sci. 2004; 117:1329–1337. [DOI] [PubMed] [Google Scholar]
- 10. Yu X., Ng C., Habacher H., Roy S.. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 2008; 40:1445–1453. [DOI] [PubMed] [Google Scholar]
- 11. Bonnafe E., Touka M., AitLounis A., Baas D., Barras E., Ucla C., Moreau A., Flamant F., Dubruille R., Couble P. et al.. The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol. Cell Biol. 2004; 24:4417–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zein El,L., Ait-Lounis A., Morlé L., Thomas J., Chhin B., Spassky N., Reith W., Durand B.. RFX3 governs growth and beating efficiency of motile cilia in mouse and controls the expression of genes involved in human ciliopathies. J. Cell Sci. 2009; 122:3180–3189. [DOI] [PubMed] [Google Scholar]
- 13. Chung M.-I., Peyrot S.M., LeBoeuf S., Park T.J., McGary K.L., Marcotte E.M., Wallingford J.B.. RFX2 is broadly required for ciliogenesis during vertebrate development. Dev. Biol. 2012; 363:155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quigley I.K., Kintner C.. Rfx2 stabilizes Foxj1 binding at chromatin loops to enable multiciliated cell gene expression. PLoS Genet. 2017; 13:e1006538-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chung M.I., Kwon T., Tu F., Brooks E.R., Gupta R., Meyer M., Baker J.C., Marcotte E.M., Wallingford J.B.. Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife. 2014; 3:e01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kwon T., Chung M.-I., Gupta R., Baker J.C., Wallingford J.B., Marcotte E.M.. Identifying direct targets of transcription factor Rfx2 that coordinate ciliogenesis and cell movement. Genom. Data. 2014; 2:192–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Danielian P.S., Bender Kim C.F., Caron A.M., Vasile E., Bronson R.T., Lees J.A.. E2f4 is required for normal development of the airway epithelium. Dev. Biol. 2007; 305:564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tan F.E., Vladar E.K., Ma L., Fuentealba L.C., Hoh R., Espinoza F.H., Axelrod J.D., Alvarez-Buylla A., Stearns T., Kintner C. et al.. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development. 2013; 140:4277–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pan J.-H., Adair-Kirk T.L., Patel A.C., Huang T., Yozamp N.S., Xu J., Reddy E.P., Byers D.E., Pierce R.A., Holtzman M.J. et al.. Myb permits multilineage airway epithelial cell differentiation. Stem Cells. 2014; 32:3245–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma L., Quigley I., Omran H., Kintner C.. Multicilin drives centriole biogenesis via E2f proteins. Genes Dev. 2014; 28:1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nemajerova A., Kramer D., Siller S.S., Herr C., Shomroni O., Pena T., Gallinas Suazo C., Glaser K., Wildung M., Steffen H. et al.. TAp73 is a central transcriptional regulator of airway multiciliogenesis. Genes Dev. 2016; 30:1300–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marshall C.B., Mays D.J., Beeler J.S., Rosenbluth J.M., Boyd K.L., Guasch G.L.S., Shaver T.M., Tang L.J., Liu Q., Shyr Y. et al.. p73 is required for multiciliogenesis and regulates the Foxj1-Associated gene network. CellReports. 2016; 14:2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jackson P.K., Attardi L.D.. p73 and FoxJ1: programming multiciliated epithelia. Trends in Cell Biology. 2016; 26:239–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swoboda P., Adler H.T., Thomas J.H.. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell. 2000; 5:411–421. [DOI] [PubMed] [Google Scholar]
- 25. Dubruille R., Laurençon A., Vandaele C., Shishido E., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B.. Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development. 2002; 129:5487–5498. [DOI] [PubMed] [Google Scholar]
- 26. Emery P., Durand B., Mach B., Reith W.. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 1996; 24:803–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piasecki B.P., Burghoorn J., Swoboda P.. Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc. Natl Acad. Sci. USA. 2010; 107:12969–12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baas D., Meiniel A., Benadiba C., Bonnafe E., Meiniel O., Reith W., Durand B.. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur. J. Neurosci. 2006; 24:1020–1030. [DOI] [PubMed] [Google Scholar]
- 29. Chu J.S.C., Baillie D.L., Chen N.. Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans. BMC Evol. Biol. 2010; 10:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bisgrove B.W., Makova S., Yost H.J., Brueckner M.. RFX2 is essential in the ciliated organ of asymmetry and an RFX2 transgene identifies a population of ciliated cells sufficient for fluid flow. Dev. Biol. 2012; 363:166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ashique A.M., Choe Y., Karlen M., May S.R., Phamluong K., Solloway M.J., Ericson J., Peterson A.S.. The Rfx4 transcription factor modulates Shh signaling by regional control of ciliogenesis. Science signaling. 2009; 2:ra70. [DOI] [PubMed] [Google Scholar]
- 32. Manojlovic Z., Earwood R., Kato A., Stefanovic B., Kato Y.. RFX7 is required for the formation of cilia in the neural tube. Mech. Dev. 2014; 132:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sedykh I., Keller A.N., Yoon B., Roberson L., Moskvin O.V., Grinblat Y.. Zebrafish Rfx4 controls dorsal and ventral midline formation in the neural tube. Dev. Dyn. 2018; 247:650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang M., Zhou Z., Elledge S.J.. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998; 94:595–605. [DOI] [PubMed] [Google Scholar]
- 35. Wu S., McLeod M.. The sak1+ gene of Schizosaccharomyces pombe encodes an RFX family DNA-binding protein thaht positively regulates cyclic AMP-dependant protein kinase mediated exit from the mitotic cycle. Mol. Cell Biol. 1995; 15:1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reith W., Herrero-Sanchez C., Kobr M., Silacci P., Berte C., Barras E., Fey S., Mach B.. MHC class II regulatory factor RFX has a novel DNA binding domain and functionally independant dimerization domain. Genes Dev. 1990; 4:1528–1540. [DOI] [PubMed] [Google Scholar]
- 37. Zhao M., Tan Y., Peng Q., Huang C., Guo Y., Liang G., Zhu B., Huang Y., Liu A., Wang Z. et al.. Nat. Commun. 2018; 9:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith S.B., Qu H.-Q., Taleb N., Kishimoto N.Y., Scheel D.W., Lu Y., Patch A.-M., Grabs R., Wang J., Lynn F.C. et al.. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010; 463:775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soyer J., Flasse L., Raffelsberger W., Beucher A., Orvain C., Peers B., Ravassard P., Vermot J., Voz M.L., Mellitzer G. et al.. Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development. 2010; 137:203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blackshear P.J., Graves J.P., Stumpo D.J., Cobos I., Rubenstein J.L.R., Zeldin D.C.. Graded phenotypic response to partial and complete deficiency of a brain-specific transcript variant of the winged helix transcription factor RFX4. Development. 2003; 130:4539–4552. [DOI] [PubMed] [Google Scholar]
- 41. Reith W., Satola S., Sanchez C., Amaldi I., Lisowska-Grospierre B., Griscelli C., Hadam M., Mach B.. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988; 53:897–906. [DOI] [PubMed] [Google Scholar]
- 42. Gajiwala K.S., Chen H., Cornille F., Roques B.P., Reith W., Mach B., Burley S.K.. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000; 403:916–921. [DOI] [PubMed] [Google Scholar]
- 43. Herrero Sanchez C., Reith W., Silacci P., Mach B.. The DNA-binding defect observed in major histocompatibility complex class II regulatory mutants concerns only one member of a family of complexes binding to the X boxes of class II promoters. Mol. Cell Biol. 1992; 12:4076–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Efimenko E., Bubb K., Mak H.Y., Holzman T., Leroux M.R., Ruvkun G., Thomas J.H., Swoboda P.. Analysis of xbx genes in C. elegans. Development. 2005; 132:1923–1934. [DOI] [PubMed] [Google Scholar]
- 45. Laurençon A., Dubruille R., Efimenko E., Grenier G., Bissett R., Cortier E., Rolland V., Swoboda P., Durand B.. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 2007; 8:R195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kistler W.S., Baas D., Lemeille S., Paschaki M., Seguín-Estévez Q., Barras E., Ma W., Duteyrat J.-L., Morlé L., Durand B. et al.. RFX2 is a major transcriptional regulator of spermiogenesis. PLoS Genet. 2015; 11:e1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elkon R., Milon B., Morrison L., Shah M., Vijayakumar S., Racherla M., Leitch C.C., Silipino L., Hadi S., Weiss-Gayet M. et al.. RFX transcription factors are essential for hearing in mice. Nat. Commun. 2015; 6:8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwenk F., Baron U., Rajewsky K.. A cre-transgenic mouse strain for the ubiquitous deletion of loxP- flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995; 23:5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y., Huang G., Shornick L., Roswit W., Shipley J., Brody S., Holtzman M.. A transgenic FOXJ1-Cre system for gene inactivation in ciliated epithelial cells. Am. J. Respir Cell Mol. Biol. 2007; 36:515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Delgehyr N., Meunier A., Faucourt M., Bosch Grau M., Strehl L., Janke C., Spassky N.. Ependymal cell differentiation, from monociliated to multiciliated cells. Methods Cell Biol. 2015; 127:19–35. [DOI] [PubMed] [Google Scholar]
- 51. Masternak K., Barras E., Zufferey M., Conrad B., Corthals G., Aebersold R., Sanchez J., Hochstrasser D., Mach B., Reith W.. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat. Genet. 1998; 20:273–277. [DOI] [PubMed] [Google Scholar]
- 52. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup . The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. et al.. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012; 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu T. Use model-based analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 2014; 1150:81–95. [DOI] [PubMed] [Google Scholar]
- 59. Reith W., Ucla C., Barras E., Gaud A., Durand B., Herrero-Sanchez C., Kobr M., Mach B.. RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins. Mol. Cell Biol. 1994; 14:1230–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ait-Lounis A., Bonal C., Seguín-Estévez Q., Schmid C.D., Bucher P., Herrera P.L., Durand B., Meda P., Reith W.. The transcription factor Rfx3 regulates beta-cell differentiation, function, and glucokinase expression. Diabetes. 2010; 59:1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reiter J.F., Leroux M.R.. Genes and molecular pathways underpinning ciliopathies. Nature. 2017; 18:533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van Dam T.J., Wheway G., Slaats G.G., Study Group S., Huynen M.A., Giles R.H.. The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia. 2013; 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E. et al.. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003; 34:267–273. [DOI] [PubMed] [Google Scholar]
- 65. Bembom O., Keles S., van der Laan M.J.. Supervised detection of conserved motifs in DNA sequences with cosmo. Stat. Appl. Genet. Mol. Biol. 2007; 6:8. [DOI] [PubMed] [Google Scholar]
- 66. Zambelli F., Pesole G., Pavesi G.. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009; 37:W247–W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sandelin A., Alkema W., Engström P., Wasserman W.W., Lenhard B.. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004; 32:D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D.. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017; 45:D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Horvath G.C., Kistler W.S., Kistler M.K.. RFX2 is a potential transcriptional regulatory factor for histone H1t and other genes expressed during the meiotic phase of spermatogenesis. Biol. Reprod. 2004; 71:1551–1559. [DOI] [PubMed] [Google Scholar]
- 70. Feng C., Xu W., Zuo Z.. Knockout of the regulatory factor X1 gene leads to early embryonic lethality. Biochem. Biophys. Res. Commun. 2009; 386:715–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang B., Qi T., Chen S.-Q., Ye L., Huang Z.-S., Li H.. RFX1 maintains testis cord integrity by regulating the expression of Itga6in male mouse embryos. Mol. Reprod. Dev. 2016; 83:606–614. [DOI] [PubMed] [Google Scholar]
- 72. Ait-Lounis A., Baas D., Barras E., Benadiba C., Charollais A., Nlend Nlend R., Liègeois D., Meda P., Durand B., Reith W.. Novel function of the ciliogenic transcription factor RFX3 in development of the endocrine pancreas. Diabetes. 2007; 56:950–959. [DOI] [PubMed] [Google Scholar]
- 73. Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S.. Quantifying similarity between motifs. Genome Biol. 2007; 8:R24–R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Reith W., Mach B.. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 2001; 19:331–373. [DOI] [PubMed] [Google Scholar]
- 75. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Henriksson J., Piasecki B.P., Lend K., Burglin T.R., Swoboda P.. Finding ciliary genes: a computational approach. Meth. Enzymol. 2013; 525:327–350. [DOI] [PubMed] [Google Scholar]
- 77. Miller J.G., Liu Y., Williams C.W., Smith H.E., O’Connell K.F.. The E2F-DP1 transcription factor complex regulates centriole duplication in caenorhabditis elegans. G3 (Bethesda). 2016; 6:709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chong Y.L., Zhang Y., Zhou F., Roy S.. Distinct requirements of E2f4 versus E2f5 activity for multiciliated cell development in the zebrafish embryo. Dev. Biol. 2018; 443:165–172. [DOI] [PubMed] [Google Scholar]
- 79. Kim S., Ma L., Shokhirev M.N., Quigley I., Kintner C.. Multicilin and activated E2f4 induce multiciliated cell differentiation in primary fibroblasts. Sci. Rep. 2018; 8:12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Arbi M., Pefani D.E., Kyrousi C., Lalioti M.E., Kalogeropoulou A., Papanastasiou A.D., Taraviras S., Lygerou Z.. GemC1 controls multiciliogenesis in the airway epithelium. EMBO Rep. 2016; 17:400–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hong C.-J., Hamilton B.A.. Zfp423 regulates sonic hedgehog signaling via primary cilium function. PLoS Genet. 2016; 12:e1006357-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cachero S., Simpson T.I., Lage Zur, P.I., Ma L., Newton F.G., Holohan E.E., Armstrong J.D., Jarman A.P.. The gene regulatory cascade linking proneural specification with differentiation in Drosophila sensory neurons. PLoS Biol. 2011; 9:e1000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wu S.Y., McLeod M.. The sak1+ gene of Schizosaccharomyces pombe encodes an RFX family DNA-binding protein that positively regulates cyclic AMP-dependent protein kinase-mediated exit from the mitotic cell cycle. Mol. Cell Biol. 1995; 15:1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Garg A., Futcher B., Leatherwood J.. A new transcription factor for mitosis: in Schizosaccharomyces pombe, the RFX transcription factor Sak1 works with forkhead factors to regulate mitotic expression. Nucleic Acids Res. 2015; 43:6874–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zaim J., Speina E., Kierzek A.M.. Identification of new genes regulated by the Crt1 transcription factor, an effector of the DNA damage checkpoint pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2005; 280:28–37. [DOI] [PubMed] [Google Scholar]
- 86. Min K., Son H., Lim J.Y., Choi G.J., Kim J.-C., Harris S.D., Lee Y.-W.. Transcription factor RFX1 is crucial for maintenance of genome integrity in Fusarium graminearum. Eukaryotic Cell. 2014; 13:427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sugiaman-Trapman D., Vitezic M., Jouhilahti E.-M., Mathelier A., Lauter G., Misra S., Daub C.O., Kere J., Swoboda P.. Characterization of the human RFX transcription factor family by regulatory and target gene analysis. BMC Genomics. 2018; 19:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data has been submitted to the GEO database under the accession number n° GSE145324.