Abstract
Host-associated bacteria and fungi, comprising the microbiota, are critical to host health. In the airways, the composition and diversity of the mucosal microbiota of patients are associated with airway health status. However, the relationship between airway microbiota and respiratory inflammation is not well-understood. Chronic rhinosinusitis (CRS) is a complex disease that affects up to 14% of the US population. Previous studies have shown decreased microbial diversity in CRS patients and enrichment of either Staphylococcus aureus or Pseudomonas aeruginosa. Although bacterial community composition is variable across CRS patients, Malassezia is a dominant fungal genus in the upper airways of the majority of healthy and CRS subjects. We hypothesize that distinct bacterial-fungal interactions differentially influence host mucosal immune response. Thus, we investigated in vitro and in vivo interactions between Malassezia sympodialis, P. aeruginosa, and S. aureus. The in vitro interactions were evaluated using the modified Kirby-Bauer Assay, Crystal Violet assay for biofilm, and FISH. A pilot murine model of acute sinusitis was used to investigate relationships with the host immune response. S. aureus and P. aeruginosa were intranasally instilled in the presence or absence of M. sympodialis (n = 66 total mice; 3–5/group). Changes in the microbiota were determined using 16S rRNA gene sequencing and host immune response was measured using quantitative real-time PCR (qRT-PCR). In vitro, only late stage planktonic P. aeruginosa and its biofilms inhibited M. sympodialis. Co-infection of mice with M. sympodialis and P. aeruginosa or S. aureus differently influenced the immune response. In co-infected mice, we demonstrate different expression of fungal sensing (Dectin-1), allergic responses (IL-5, and IL-13) and inflammation (IL-10, and IL-17) in murine sinus depending on the bacterial species that co-infected with M. sympodialis (p < 0.05). The pilot results suggest that species-specific interactions in airway-associated microbiota may be implicated driving immune responses. The understanding of the role of bacterial-fungal interactions in CRS will contribute to development of novel therapies toward manipulation of the airway microbiota.
Keywords: malassezia, sinus microbiome, bacterial-fungal interactions, interkingdom interactions, Staphylococcus aureus, Pseudomonas aeruginosa
Introduction
Emerging studies of the microbiota, defined as bacteria, fungi, viruses, and archaea that inhabit a niche, have profoundly altered our understanding of the role of the human microbiota in health and disease. The range of effects that these complex microbial assemblages have on human physiology is unimaginably broad. For example, many studies suggest that gastrointestinal microbiota are related to not only intestinal diseases but also obesity (Moise, 2017), cancers (Gopalakrishnan et al., 2018), allergies (Fujimura et al., 2016), and neurological disorders (Elizabeth and Zabielski, 2013). In the gut, Clostridium produces the short-chain fatty acid (SCFA) butyrate, which contributes to the polarization of FOXP3+CD4+ regulatory T-cells. In addition to butyrate, gut microbes produce propionate and acetic acid, which can act as ligands for G protein-coupled receptors (GPCRs) and affect neutrophil chemotaxis, regulation of regulatory T cells (Treg), and dendritic cell (DC) maturation (Kau et al., 2011; Thaiss et al., 2016). However, our understanding of the respiratory tract microbiota remains nascent, even though it is one of the first body sites to make contact with microorganisms after birth and has a larger surface area than the skin or GI tract (Fröhlich et al., 2016). The development of culture-independent methods has shown that complex, site-specific microbial communities inhabit the upper and lower airways (Brill et al., 2016; Pletcher et al., 2018).
Chronic rhinosinusitis (CRS) is a common upper respiratory inflammatory disease affecting ~14% of the Western population and costing about $65 billion each year in disease management (Caulley et al., 2015). CRS is diagnosed through a combination of patient history, nasal endoscopy, and radiography and is characterized by symptoms lasting at least 12 consecutive weeks (Sedaghat, 2017). The primary goal of CRS treatments is to manage the symptoms and improve patients' quality of life. Medical management for CRS includes nasal saline irrigation (Gupta and Singh, 2010), topical intranasal and oral corticosteroids (Epperson et al., 2019), and antibiotics if a pathogen is cultured (Mattila, 2012). If medicinal management is unsuccessful, endoscopic sinus surgery is performed. Even though these treatment methods can improve symptoms, the recurrence rate of CRS is as high as 78.9% (Sedaghat, 2017; Calus et al., 2019).
The sinuses harbor a complex bacterial and fungal microbiome. A recent study from our group demonstrated that the sinus bacterial microbiota exists in four different compositional states each dominated by one of four different bacterial families and low-abundance co-colonizers. Each bacterial community state was associated with a distinct host inflammatory response in CRS patients (Cope et al., 2017). In another study using targeted qPCR, we demonstrated that Malassezia was the predominant fungal taxa across all patient subgroups (Gelber et al., 2016). Recent sequence-based studies have confirmed that Malassezia species, including M. restricta and M. sympodialis, are dominant core members of the sinus fungal microbiome (Carter and Amedee, 2014; Cleland et al., 2014; Hoggard et al., 2019; Wagner Mackenzie et al., 2019). Malassezia is a dimorphic and lipophilic fungus that is commonly found on the skin as a commensal organism. An opportunistic pathogen, Malassezia produces virulence factors such as lipases, phospholipases, and allergens that can damage epithelial integrity and trigger predominantly TH2 immune responses (Blanco and Garcia, 2008; Boekhout et al., 2010). In the GI tract, a study showed that Malassezia was associated with the colonic mucosa of Crohn's disease patients and exacerbated colitis in mouse models (Limon et al., 2019). However, the role of Malassezia in the respiratory tract has not been studied.
Interactions between bacteria and fungi are critical mediators of microbial community composition and function. Several studies investigating the interactions between bacteria and fungi have focused predominantly on another dimorphic yeast, Candida albicans, and bacterial co-colonizers. Distinct types of interactions not only affect the survival of the bacteria but also affect both fungal and bacterial virulence and host immune responses. For example, the interaction between C. albicans and Staphylococcus aureus, a bacterial pathogen, results in increased C. albicans hyphal formation and higher invasiveness of S. aureus into mucosal membranes by attaching to the hyphae of C. albicans (Schlecht et al., 2015). These studies of fungal and bacterial interactions suggest that we need to consider bacterial-fungal interactions to advance our understanding of chronic airway diseases such as cystic fibrosis (CF) and chronic rhinosinusitis (CRS) in the context of microbial interactions and relationships with the host immune response.
In this study, we hypothesized that bacterial co-colonizers, selected from our prior studies of the CRS bacterial microbiota, differentially interact with Malassezia and that this drives specific sinonasal immune responses dependent on innate immune sensing of fungi. Here, we investigated the in vitro and in vivo interactions between Malassezia sympodialis and bacterial pathogens, Pseudomonas aeruginosa and Staphylococcus aureus. Malassezia is the predominant fungal genus in the sinuses (Gelber et al., 2016; Hoggard et al., 2019), and S. aureus and P. aeruginosa are predominant members of two of the four described bacterial community states in CRS (Cope et al., 2017), and they are commonly isolated bacteria in CRS patients (Zhang et al., 2015). We evaluated in vivo influence of bacterial-Malassezia co-infection using a murine model of sinonasal disease (Abreu et al., 2012). These pilot studies suggest that in vitro interactions between S. aureus or P. aeruginosa with M. sympodialis are highly dependent on the species of bacteria and co-infection of M. sympodialis with either S. aureus or P. aeruginosa yields distinct sinonasal immune responses.
Materials and Methods
Bacterial and Fungal Species
We used two bacterial species, which were previously demonstrated to be related to CRS endotypes, and one fungal genus that belongs to the dominant fungal genus in the nasal passage, Malassezia. The bacterial species used were isolated from CRS patients: Pseudomonas aeruginosa EC1 (Cope et al., 2016) and Staphylococcus aureus. Species IDs were confirmed by sequencing the full length 16S rRNA gene. The fungal species, Malassezia sympodialis, was gifted by Paal Anderson (University of Copenhagen).
Bacterial and Fungal Growth Media
P. aeruginosa and S. aureus were incubated in Sabouraud Dextrose Agar (SDA, HIMEDIA®) and SD broth. The lawn of M. sympodialis was grown on an SDA plate overlaid with ~300 μl sterile olive oil. Olive oil was not used in broth media for any organism as it inhibited some aspects of microbial growth. SDA with olive oil was not an optimal media because it inhibited aspects of microbial growth. The co-cultured biofilm experiment, the preparation for fluorescence in-situ hybridization (FISH) staining, and the in vivo mouse experiment were prepared using different media. The P. aeruginosa and S. aureus cultures were prepared in brain heart infusion broth (BHIB, BD DifcoTM). M. sympodialis were prepared in modified Dixon (mDixon) media. Briefly, the contents of the modified Dixon media are 36 g malt extract (MP Biomedicals), 10 g BactoTM peptone (Life Technologies), 10 ml Tween 40 (Sigma-Aldrich), and 2 ml glycerol (Sigma-Aldrich) added to 1 L dH2O before pH was adjusted to 6.0 (Guillot et al., 1998). The desiccated ox bile and antibiotics were omitted from the original Dixon media in order to facilitate use in co-culture experiments with other bacterial species.
In vitro Bacterial-Fungal Inhibitory Relationship Assay
We performed a modified Kirby-Bauer test to evaluate antagonistic relationships between M. sympodialis and S. aureus or P. aeruginosa. Each filter paper disc contained four different growth modes of P. aeruginosa or S. aureus culture (early phase planktonic: incubate for 3 h at 37°C with shaking; late phase planktonic: incubate for 24 h at 37°C with shaking). Growth discs containing biofilms were incubated for 24 h at 37°C in a 24-well plate. Biofilm discs were removed from the 24-well plate and immediately used in this assay. Finally, discs were inoculated with the cell-free supernatant fluid from late-phase planktonic bacterial growth media. The cell-free supernatant was prepared by filter-sterilizing the culture-supernatant after centrifugation at 8,000 × g for 1 min using a 0.22 μm syringe filter (MilliporeSigmaTM MillexTM). Discs were placed on the M. sympodialis lawn on Sabouraud Dextrose Agar (SDA) plates. The sterile Sabouraud Dextrose Broth (SDB) and 10% bleach solution on the discs were used for negative and positive growth inhibition controls, respectively. The plates were incubated at 30°C for 72 h, and the diameter of the inhibition zone around the discs was measured. This experiment was performed in triplicate.
Crystal Violet Biofilm Assay
Primary broth cultures of P. aeruginosa or S. aureus were grown in brain heart infusion broth (BHIB, BD DifcoTM) for 24 h at 37°C with shaking. 100 μl of the primary cultures were transferred into fresh 10 ml of modified Dixon (mDixon) and incubated for 4 h at 37°C with shaking to reach the exponential growth phase. The exponential growth phase cultures were diluted 1:100 in 150 μl of mDixon broth in a sterile 96-well plate (Cafarchia et al., 2012). To grow the M. sympodialis biofilm, the primary culture was grown in mDixon for 48 h at 30°C with shaking. 100 μl of the primary cultures were transferred into fresh 10 ml of mDixon and incubated for 6 h at 30°C with shaking to reach the exponential growth phase. The exponential growth phase cultures diluted 1:50 in 150 μl of mDixon in a sterile 96 well plates. For the polymicrobial biofilms, 1.5 ml of diluted the exponential growth phase culture of M. sympodialis and P. aeruginosa or S. aureus in mDixon were combined in a sterile 96 well plates. All the biofilm plates were grown at 30°C for 48 hrs without agitation. Biofilms were quantified using the crystal violet biofilm assay, as previously described (O'Toole, 2011). Briefly, the media from the 96 wells were discarded, and the wells were rinsed with distilled water and dried. 200 μl of 1% crystal violet staining solution was added to the wells to stain the biofilms for 10 min. The crystal violet staining solution was discarded and then the wells were rinsed with distilled water 3 times. The crystal violet stained biofilms were dried in the air for 3 h and 200 μl of 70% ethanol was added to extract the crystal violet stain from the biofilms. The amount of crystal violet stain in the 70% ethanol was measured by observing the optical density at 600 nm wavelength with (BioTek SynergyTM HT). Uninoculated wells were used as negative controls. This experiment was repeated in quadruplicate.
In vivo Murine Challenge Experiment
The in vivo model was modified from a previous study by Abreu et al. (2012). This experiment was performed once, and we used the same number of animals in this study per group (n = 3 mice/group for molecular analysis, n = 2 mice/group for Fluorescent in situ Hybridization as applicable) as previously published (Abreu et al., 2012). These studies were approved under NAU IACUC approval number 16-008. Mice were acclimatized for 5 days prior to the experiments. Two groups of balb/c mice (n = 66 total) were used: an antimicrobial-naive group (n = 3 mice/group for molecular analysis and n = 2 mice for fluorescent in-situ hybridization), and an antimicrobial-treated group (n = 3 mice/group for molecular analysis). The antimicrobial-treated mice were intranasally administered with augmentin (15 mg/kg) and fluconazole (100 mg/kg) for 5 days after a 5 day acclimatization period. Each group was intranasally infected with one either (1) saline (control), (2–4) single species microorganisms (1.0 × 108 cells of P. aeruginosa, S. aureus, or M. sympodialis), or (5–6) dual-species microorganisms (1.0 × 108 cells of P. aeruginosa and M. sympodialis, or S. aureus and M. sympodialis). The antimicrobial-naive and -treated mice were infected with the six infection groups mentioned above for 3 days. One day after the third infection, the mice were euthanized using CO2, and the sinonasal tissue was harvested and stored in 700 μl of RNAlater for microbiome and immunological analysis (Figure S1).
Sinonasal Microbiome Analysis
The harvested sinonasal tissue samples in RNAlater® (Sigma-Aldrich) were stored at 4°C for 24 h before processing the samples. After 24 h in RNAlater®, DNA and RNA were extracted from the sinonasal tissue samples using the Allprep DNA/RNA mini kit (Qiagen). The V4 region of the 16S rRNA gene was amplified from DNA by polymerase chain reaction (PCR) using a 515f forward primer with an adapter sequence, AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTGCCAGCMGCCGCGGTAA, and the 806r indexed primers for reverse primers (Integrated DNA Technologies). The PCR reactions were carried out in 25 μL containing 1 μL of DNA template, 1x TaKaRa® Ex Taq PCR buffer (Mg2+ plus), 0.2 mM TaKaRa® dNTP mixture, 0.625 U TaKaRa® Ex Taq HS polymerase, 0.56 mg/μL BSA, 0.4 μM of each primer, and 16.375 μL of water. The PCRs were performed using a SimpliAmp® thermocycler (Applied Biosystems) under the following conditions: 98 °C for 2 min to release the polymerase antibody, followed by 30 cycles of 98°C for 20 s, 50°C for 30 s, and 72°C for 45 s. A final extension step of 72°C for 10 min was then conducted to ensure the completion of all fragments. All samples were triplicated and pooled in order to minimize the random amplification bias. The PCR amplicons were confirmed by agarose gel electrophoresis and quantified using Qubit 4 fluorometer (ThermoFisher Scientific). Amplicons were pooled at 50 ng/sample. The pooled amplicons were sequenced using Illumina MiSeq v3 (Illumina, inc.) at Translational Genomics North (TGen-North). The sequence data were analyzed using a bioinformatics platform, QIIME 2 (Bolyen et al., 2019).
Murine Immune Gene Expression Analysis
The immunological responses were analyzed by measuring gene expression of the key immune responses in CRS (Baba et al., 2015; Hamilos, 2015; Hulse, 2016), such as genes for interleukin (IL)-2, IL-5, IL-10, IL-13, IL-17, MUC5AC, Dectin-1, and Tumor Necrosis Factor (TNF)-α, by quantitative reverse transcription polymerase chain reaction (qRT-PCR) using the 1 ng/μl of cDNA, which was synthesized from the extracted mRNA. Gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (Table 1). qRT-PCR was done using QuantStudioTM 12k Flex Real-time PCR System (Applied BiosystemsTM) with Power SYBRTM Green PCR MasterMix (Applied BiosystemsTM). The qRT-PCR condition consists of hold (50°C for 2 min and 95°C for 10 min.), PCR (40 cycles of 95°C for 15 sec. and 60°C for 1 min.), and melt curve cycle (95°C for 15 sec. and decrease to 60°C by 1.6°C /s).
Table 1.
Primers used in this study.
| Primer | Sequence | Reference |
|---|---|---|
| IL-2F | GTCACATTGACACTTGTGCTCC | Telander et al., 1999 |
| IL-2R | AGTCAAATCCAGAACATGCCG | |
| IL-4F | TCGGCATTTTGAACGAGGTC | Kim et al., 2011 |
| IL-4R | GAAAAGCCCGAAAGAGTCTC | |
| IL-5F | ATGGAGATTCCCATGAGCAC | |
| IL-5R | GTCTCTCCTCGCCACACTTC | |
| IL-10F | GCGTCGTGATTAGCGATGATG | Trandem et al., 2011 |
| IL-10R | CTCGAGCAAGTCTTTCAGTCC | |
| IL-17F | GGACTCTCCACCGCAATGA | Atarashi et al., 2008 |
| IL-17R | GGCACTGAGCTTCCCAGATC | |
| IFNg-F | CTACCTTCTTCAGCAACAGC | Oestreich et al., 2012 |
| IFNg-R | GCTCATTGAATGCTTGGCGC | |
| GADPH-F | CCTCGTCCCGTAGACAAAATG | Ueda et al., 2010 |
| GADPH-R | TCTCCACTTTGCCACTGCAA | |
| MUC5AC-F | AGCTACAGTGCAACTGGACC | Lin et al., 2014 |
| MUC5AC-R | GGACACAGATGATGGTGACA | |
| IL13-F | AGGAGCTGAGCAACATCACAC | Kimura et al., 2014 |
| IL13R | CCATAGCGGAAAAGTTGCTT | |
| TNF-α-F | GTAGCCCACGTCGTAGCAA | |
| TNF-α-R | AAATGGCAAATCGGCTGAC | |
| Dectin-1_F | ATCAGCATTCTTCCCCAACTCG | |
| Dectin-1_R | CAGTTCCTTCTCACAGATACTGTATGA |
Fluorescence in situ Hybridization (FISH) of Polymicrobial Biofilms and Mouse Sinonasal Specimens
The bacterial pathogens, P. aeruginosa or S. aureus, and M. sympodialis co-developed biofilms were observed under the confocal laser scanning microscope (CLSM) using FISH. The biofilms were developed according to the protocol above. The biofilms were hybridized using the universal eubacterial probe, EUB 388, and Malassezia genus specific probe (Table 2). The biofilms were carefully transferred to glass slides and air-dried. The air-dried biofilms were fixed by dipping the slides in 50, 70, and 95% ETOH for 3 min each, and washed with DI water. The fixed biofilms were hybridized with 1 μl of the EUB 388 probe (10 nM) and 1 μl of the Malassezia_genus probe (10 nM) in 18 μl of the hybridization buffer for 90 min at 46°C in a hybridization chamber with damped paper with 5 ml of hybridization buffer for humidity. The hybridized biofilms were washed in a 48°C washing buffer for 30 min. The washed biofilms were rinsed briefly with DI water and mounted using VECTASHIELD® (VECTOR Laboratories). The hybridization buffer contained 0.9M NaCl, 20 mM Tris/HCl (pH 8.0), 0.01% SDS, and 35% formamide. The washing buffer contained 0.08M NaCl, 20 mM Tris/HCl (pH 8.0), 5 mM EDTA, and 0.01% SDS. Sinonasal specimens were prepared by fixing the skinned head of mice in methanol-Carnoy's fixative (60% methanol, 30% chloroform, 10% glacial acetic acid) for 24 h. Heads were decalcified in EDTA for 10 days, then embedded in a paraffin block. The micro-sectioned sinonasal tissue slide was prepared using the paraffin-embedded samples. The sinonasal tissue slides were deparaffinized by incubating the slide in xylene twice for 5 min each and washing it with 100% ethanol twice for 5 min each. The deparaffinized slides were hybridized using the same protocol with different probes. The probes used in the mouse sinonasal specimen FISH were 1 μl of 3 μM DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride), 1 μl of 10 nM P. aeruginosa specific probe (Paeru), 1 μl of 10 nM S. aureus specific probe (Saure), and 2 μl of 10 nM Malassezia_genus probe in 16 ul of the hybridization buffer (Table 2).
Table 2.
FISH probes used in this study.
| Probe | Target | Fluor | Sequence 5′-3′ | Reference |
|---|---|---|---|---|
| EUB 388 | Bacteria | Alexa 594 | GCTGCCTCCCGTAGGAGT | Trebesius et al., 2000 |
| Malassezia_genus | Malassezia | Alexa 532 | CCGATATTTAGCTTTAGATGGAGTCTA | This study |
| Paeru | P. aeruginosa | Alexa 594 | GGTAACCGTCCCCCTTGC | Hogardt et al., 2000 |
| Saure | S. aureus | Alexa 660 | GAAGCAAGCTTCTCGTCCG | Hogardt et al., 2000 |
Bioinformatics and Statistical Analysis
For all comparisons, an α of 0.05 was considered significant. All p-values were corrected for false discovery using the Story method (Krzywinski and Altman, 2014). 16S rRNA gene sequences were analyzed using QIIME 2 version 2018.11 and 2019.1.0 (Bolyen et al., 2019); www.QIIME2.org). Sequences were demultiplexed and then truncated at 183 bases of the demultiplexed sequences to achieve a quality of >Q20 for all bases. Demultiplexed sequences were denoised and grouped into amplicon sequence variants (ASVs) using dada2 (Callahan et al., 2016). The feature table was rarified to a sampling depth of 1,134 sequences. A phylogenetic tree was built by aligning sequences with MAFFT and a phylogenetic tree was built using FastTree2 (Price et al., 2010; Katoh and Standley, 2013) which was subsequently rooted by midpoint rooting. Taxonomy was assigned to ASVs with a Naive Bayes classifier trained on the Greengenes 13_8 99% OTU database (DeSantis et al., 2006), using the q2-feature-classifier taxonomy classification plugin (Bokulich et al., 2018). Within-sample (α-diversity) was calculated using Pielou's Evenness index and a Kruskal Wallis test was used to compare across experimental groups. Between-sample (β-diversity) metrics were calculated using the UniFrac phylogenetic distance (Lozupone and Knight, 2005), Bray-Curtis, and Jaccard metrics (Goodrich et al., 2014). Permutational analysis of variance (PERMANOVA) between sinonasal communities of infection groups within antibiotic-treated and -naive groups was calculated on the Bray-Curtis dissimilarity matrix. The significance of the bacterial-fungal inhibitory relationship using the Kirby Bauer method was calculated using a paired t-test, and the Mann-Whitney U-test was used to compare the degree of biofilm development between the groups. For in vivo murine experiments, an unpaired t-test was used for comparisons of the murine immune gene expression.
Results
In vitro Bacterial and Fungal Interactions
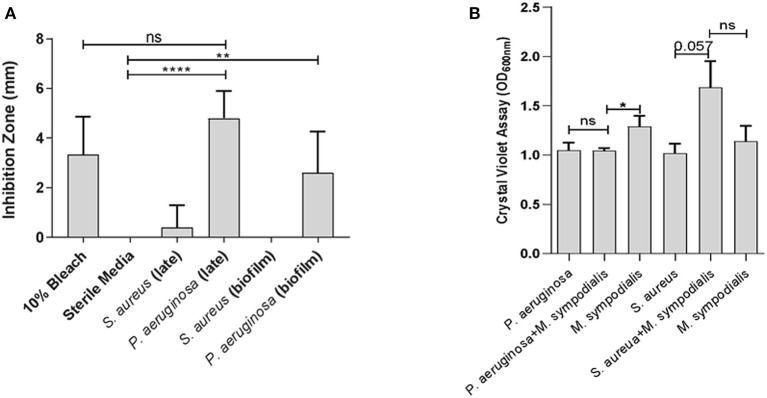
To assess whether differential interactions between M. sympodialis and S. aureus or P. aeruginosa exist in vitro, we used a modified Kirby-Bauer inhibition assay and a crystal violet biofilm quantification assay. Malassezia sympodialis lawn was plated and paper discs inoculated with early phase planktonic, late phase planktonic, 24 h biofilm, and cell-free supernatant of the late phase planktonic culture were placed on the M. sympodialis lawn. We observed significantly larger inhibition zones when P. aeruginosa late phase planktonic and biofilm were applied to the discs compared to the control discs (paired t-test p = 0.0006 and 0.0255, respectively). The late phase planktonic P. aeruginosa and 10% bleach solution were not significantly different (Figure 1A, paired t-test, p = 0.4226). S. aureus did not inhibit M. sympodialis in any treatment group (Figure 1A, paired t-test p > 0.05). Biofilm formation was measured using the crystal violet assay for biofilm development. Dual-species biofilms of P. aeruginosa and M. sympodialis were significantly reduced compared to the M. sympodialis single-species biofilm (Figure 1B, Mann-Whitney U-test p = 0.0286). Dual-species S. aureus and M. sympodialis dual-species biofilms were slightly increased compared to either single-species biofilms but this did not reach statistical significance (Figure 1B, Mann-Whitney U-test p = 0.0571). In order to determine spatial organization of bacteria and Malassezia in vitro, dual-species biofilms were grown for 48 h and viewed using Fluorescent in situ Hybridization (FISH) and CLSM. The co-cultured biofilms of M. sympodialis with S. aureus or P. aeruginosa were stained using FISH probes EUB 388 and Malassezia_genus. In vitro FISH of the M. sympodialis with P. aeruginosa showed inhibition of M. sympodialis as we were only able to observe P. aeruginosa (Figure S2). However, there was clear colocalization of S. aureus and M. sympodialis in vitro (Figure S2). These results are the first to demonstrate species-specific interactions with a clinically relevant species of Malassezia (M. sympodialis) and two common airway pathobionts, P. aeruginosa and S. aureus.
Figure 1.
In vitro interactions between Malassezia sympodialis and different growth stages of Pseudomonas aeruginosa or Staphylococcus aureus. Zones of inhibition indicating antagonistic interactions in the modified Kirby-Bauer assay (A), and quantification of biofilm development with P. aeruginosa and/or M. sympodialis or with S. aureus and/or M. sympodialis (B). (ns, not statistically significant; *P ≤ 0.05, **P ≤ 0.005, ****P ≤ 0.00005).
Sinonasal Microbiome Analysis
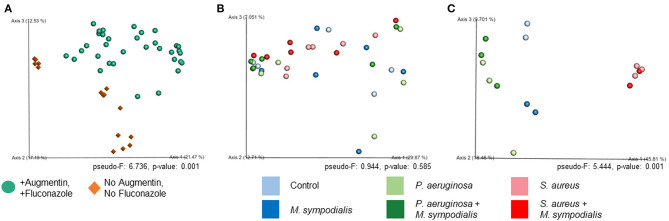
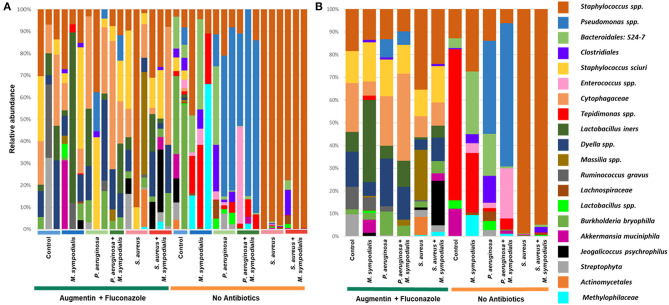
We next sought to understand whether nasal instillation of M. sympodialis or bacterial pathobionts altered the bacterial sinonasal microbiome composition or diversity (n = 3 mice per group were used for microbiome analysis). We were interested in whether the introduction of a potential fungal pathobiont would significantly change the surrounding bacterial community members. We anticipated that nasal administration of S. aureus or P. aeruginosa would result in dominance of the respective taxa in both antibiotic-treated and antibiotic-naïve mice. Indeed, we found that introduction of bacteria, but not fungi, significantly altered the composition of the murine sinonasal bacterial microbiome. Between-sample diversity (beta-diversity) was significantly different between antibiotic-naïve and antibiotic-treated mice (Figure 2A, PERMANOVA, p = 0.001, Bray-Curtis Dissimilarity). There was no distinct separation between the infection groups of the antibiotic-treated samples (Figure 2B, PERMANOVA, p = 0.585), whereas the antibiotic-naïve mice samples clustered by the bacterial species inoculated (Figure 2C, PERMANOVA, p = 0.001). The M. sympodialis infected mice clustered with control mice and the co-infected mice clustered with the singly infected groups (Figure 2C), indicating that introduction of M. sympodialis does not significantly alter the murine sinonasal bacterial microbiome. We also classified taxonomy to examine relative abundances of taxa across groups. We have generated taxonomic barplots of relative abundance of 20 most abundant taxa overall which covers 90.494% of total taxa observed in our study (Figure 3). The taxonomic barpolts represent relative abundance of the top 20 taxa in sinonasal samples (Figure 3A) or the averaged relative abundances of each treatment group (Figure 3B). Taxonomic barplots show that the sinonasal microbiota was dominated by the bacterial species introduced intranasally only in the antibiotic-naïve mice (Figure 3 and Supplemental Information). Surprisingly, the composition of the sinonasal bacterial microbiota in the antibiotic-treated mice did not show obvious changes by the infected bacterial species as shown in the antibiotic-naïve groups (Figure 3, PERMANOVA, p = 0.001). The antibiotic-treated groups possess high portions of Staphylococcus sciuri, Cytophagaceae, Lactobacillus iners, and Dyella species, which were not detected in the antibiotic-naïve samples, and Bacteriodalis: S24-7 (Muribaculaceae) was detected only in the antibiotic-naïve group (Figure 3, interactive visualization in the supplemental information and on https://github.com/e-cope/malassezia-ms and can be viewed on view.qiime2.org). These results suggest that presence of P. aeruginosa and S. aureus on the sinonasal mucosa in antibiotic-naïve mice 1 day after intranasal instillation, but also show that M. sympodialis has a negligible effect on the sinonasal bacterial microbiota in this experiment.
Figure 2.
Principal Coordinates Analysis of Bray-Curtis Dissimilarity of the Murine Sinonasal Microbiome. Two groups of mice, antibiotic-treated or -naive, were intranasally infected with either one species of bacteria, P. aeruginosa (light green), S. aureus (light red), or fungi, M. sympodialis (dark blue), or two species, M. sympodialis with P. aeruginosa (dark green) or S. aureus (dark red). (A) represents beta-diversity of antibiotic-treated (solid sphere) and antibiotic-naïve (diamond) mice groups. (B) represents beta-diversity of the infection groups in the antibiotic-treated mice, and (C) represents beta-diversity of the infection groups in the antibiotic-naïve mice.
Figure 3.
Relative bacterial abundances in sinonasal microbiota of mice. Two groups of mice, antibiotic-treated or -naive, were intranasally infected with either one species of bacteria, P. aeruginosa or S. aureus, or fungi, M. sympodialis, or two species, M. sympodialis with P. aeruginosa or S. aureus. The 20 most abundant taxa covers 90.494% of total sequence hits. (A) represents relative abundance of each sinonasal samples, and (B) represents the averaged relative abundances of each treatment group samples. The legend is an orderly representation of 20 most abundant bacterial taxa, top taxa being the most abundant.
Analysis of the Sinonasal Immune Response
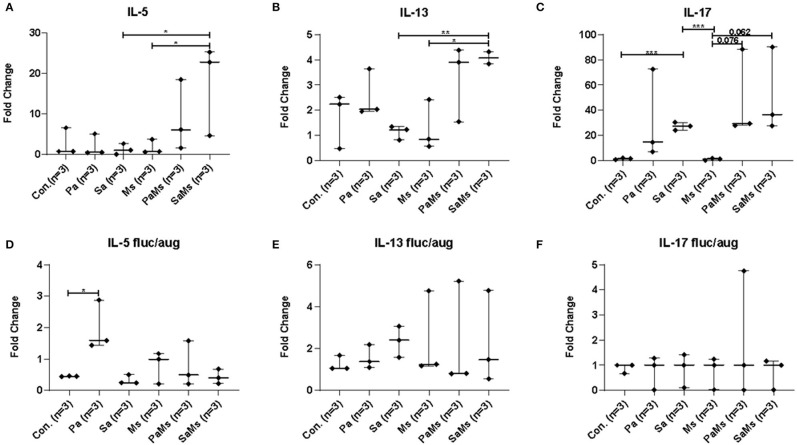
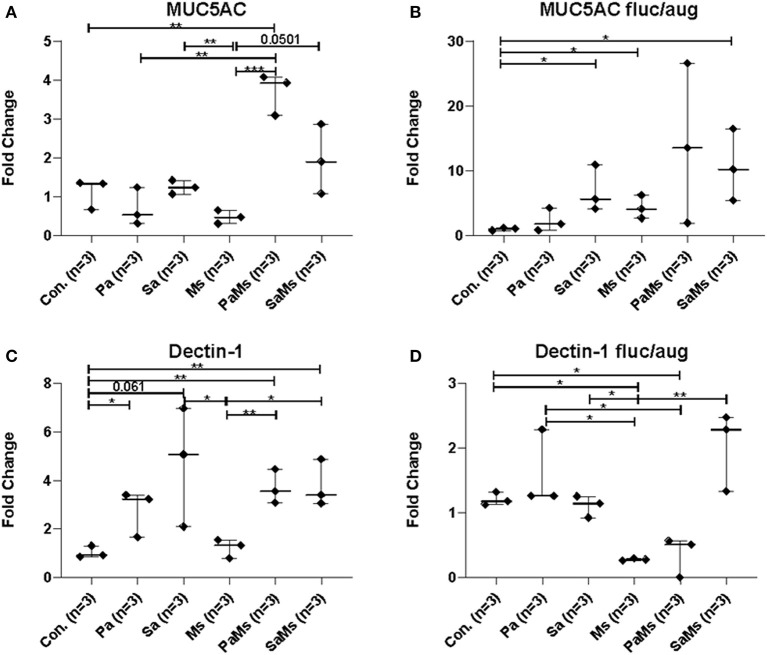
We next sought to determine whether co-infection with different combinations of P. aeruginosa, S. aureus, and M. sympodialis leads to distinct sinonasal immune responses. Gene expression of IL-5, IL-13, IL-17, MUC5AC, and Dectin-1 were measured using reverse transcriptase quantitative PCR (n = 3 mice/group). IL-5 gene expression significantly increased in S. aureus and M. sympodialis co-infected mice in antibiotic-naïve mice compared to control, S. aureus singly or M. sympodialis singly infected mice (Figure 4A, unpaired t-test, p = 0.0111, and p = 0.0188, respectively). However, IL-5 gene expression was not elevated in antibiotic-treated mice co-infected with S. aureus and M. sympodialis (Figure 4D). IL-5 expression significantly increased in the antibiotic-treated mice singly infected with P. aeruginosa (Figure 4D). IL-13 gene expression had the same pattern as the IL-5 gene expression. IL-13 gene expression increased significantly in S. aureus and M. sympodialis co-infected, antibiotic-naïve mice compared to the S. aureus singly, or M. sympodialis singly infected mice (Figure 4B, unpaired t-test, p = 0.035, p = 0.0017, respectively). In antibiotic-treated mice, none of the infection groups had significant changes in IL-13 gene expression (Figure 4E, unpaired t-test, p > 0.05).
Figure 4.
Relative gene expression of interleukin-5, interleukin-13, and interleukin-17 in the murine sinonasal cavity. The relative gene expression of IL-5 (A,D), IL-13 (B,E) and IL-17 (C,F) in antibiotic-naïve (A–C) and antibiotic-treated (D–F) mice in different infection groups, saline treated group (Con.) P. aeruginosa infected group (Pa), S. aureus infected group (Sa), M. sympodialis infected group (Ms), P. aeruginosa and M. sympodialis co-infected group (PaMs), and S. aureus and M. sympodialis co-infected group (SaMs), were measured by qRT-PCR. Gene expression was normalized with GAPDH, a housekeeping gene. Between-group comparisons without p-values were not statistically significant. (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005).
IL-17 gene expression significantly increased in S. aureus singly infected mice compared to controls (unpaired t-test, p = 0.0001) and M. sympodialis singly infected mice (unpaired t-test, p = 0.0001) in the antibiotic-naïve group. We observed a slight increase in increased in both co-infected mice, P. aeruginosa + M. sympodialis and S. aureus + M. sympodialis compared to the M. sympodialis singly infected mice in the antibiotic-naïve group but this was not statistically significant (Figure 4C, unpaired t-test, p = 0.061, p = 0.075). IL-17 gene expression did not change in any of the infection groups when mice were treated with antibiotics prior to infection (Figure 4F, unpaired t-test, p > 0.05).
MUC5AC gene expression significantly increased in antibiotic-naïve mice co-infected with P. aeruginosa and M. sympodialis when compared to control mice, M. sympodialis singly, and P. aeruginosa singly infected mice. We observed a slight but non-significant increase in MUC5AC gene expression in mice co-infected with S. aureus and M. sympodialis compared to mice singly infected with S. aureus or M. sympodialis (Figure 5A, unpaired t-test > 0.05). Of interest, MUC5AC gene expression was significantly decreased in M. sympodialis singly infected mice compared to S. aureus infected antibiotic-naïve mice (Figure 5A, unpaired t-test, p = 0.0059). In antibiotic-treated mice, MUC5AC was significantly increased in S. aureus singly, M. sympodialis singly infected mice, and S. aureus and M. sympodialis co-infected mice compared to control mice (Figure 5B, unpaired t-test, p < 0.05). We also evaluated the expression of dectin-1, a c-type lectin receptor involved in the recognition of Malassezia (Kistowska et al., 2014). Dectin-1 expression significantly increased in P. aeruginosa singly infected, P. aeruginosa and M. sympodialis co-infected, and S. aureus and M. sympodialis co-infected mice compared to controls in the antibiotic-naïve group (Figure 5C, unpaired t-test, p < 0.05). M. sympodialis infection did not induce dectin-1 gene expression, and dectin-1 expression was not different between either co-infection group compared to the corresponding single bacterial infected mice (Figure 5C). In antibiotic-treated mice, dectin-1 gene expression was not significantly different between control, P. aeruginosa singly infected, S. aureus singly infected, and S. aureus and M. sympodialis co-infected mice but it significantly decreased in M. sympodialis singly infected and P. aeruginosa and M. sympodialis co-infected mice compared to all other groups (Figure 5D, unpaired t-test, p < 0.05). In this pilot experiment, antibiotic-treated mice appeared to have a suppressive immune phenotype compared to antibiotic-naïve mice when infected with P. aeruginosa, S. aureus, or M. sympodialis. Future studies will confirm these findings in replicate experiments with additional mice.
Figure 5.
Relative gene expression of MUC5AC and Dectin-1 in the murine sinonasal cavity. Relative gene expressions of MUC5AC (A,B) and Dectin-1 (C,D) in antibiotic-naïve (A,C) and antibiotic-treated (B,D) mice on different infection groups, saline treated group (Con.) P. aeruginosa infected group (Pa), S. aureus infected group (Sa), M. sympodialis infected group (Ms), P. aeruginosa and M. sympodialis co-infected group (PaMs), and S. aureus and M. sympodialis co-infected group (SaMs), were measured by qRT-PCR. The gene expression was normalized with GAPDH, a housekeeping gene. Between group comparisons without p-value were not statistically significant. (* P ≤ 0.05, ** P ≤ 0.005, *** P ≤ 0.0005).
Ex vivo FISH on Murine Sinonasal Tissue
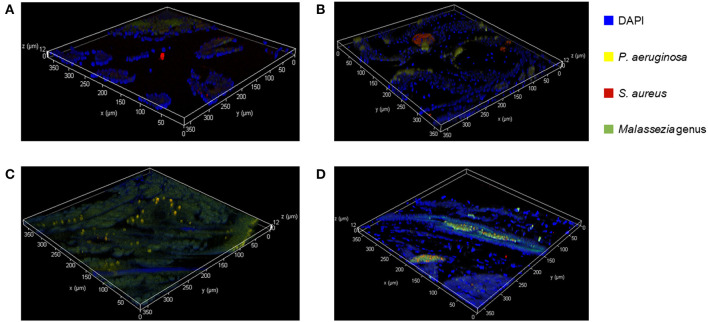
We used FISH to determine the spatial organization and confirm colonization of M. sympodialis, P. aeruginosa, and S. aureus ex vivo in the sinonasal cavity of mice that exhibited differential immune responses. In vitro FISH of the M. sympodialis with P. aeruginosa showed inhibition of M. sympodialis but there was clear colocalization of S. aureus and M. sympodialis (Figure S2). These results were confirmed in the sinonasal cavity of antibiotic-naïve mice after co-infection of each bacterial pathobiont with M. sympodialis. In control antibiotic-naïve mice, we observed Malassezia colonization and S. aureus biofilms, supporting our microbiome results (Figure 6A), and demonstrating the presence of Malassezia in the sinonasal microbiota. As expected, we observed increased Malassezia staining in mice singly infected with M. sympodialis. Endogenous S. aureus biofilms were also present, but we didn't observe strong colocalization of these two taxa in mice singly infected with M. sympodialis (Figure 6B). Notably, in mice co-instilled with M. sympodialis and S. aureus we demonstrate strong co-colonization of both microorganisms (Figure 6D). We also demonstrate potential intramucosal colonization of M. sympodialis and S. aureus, which may contribute to the differential immune response we observed in these mice. Intramucosal colonies of bacteria, most often Staphylococcus spp., have been observed in CRS patients, but the implications are unclear (Wood et al., 2012; Kim et al., 2013). Mice co-infected with M. sympodialis and P. aeruginosa had notably increased P. aeruginosa and less endogenous S. aureus colonies. We did not observe strong colocalization of P. aeruginosa with Malassezia, nor did we show a decrease in Malassezia genus (Figure 6C). These results show, for the first time, the spatial organization of Malassezia with bacterial pathobionts both in vitro and ex vivo.
Figure 6.
Fluorescent in-situ Hybridization (FISH) of the mice sinonasal tissues. Murine sinonasal tissues are hybridized with DAPI (blue) for tissue cells, and with FISH probes for P. aeruginosa (Alexa 594, pseudo-colored in yellow), S. aureus (Alexa 660, pseudo-colored in red) or genus Malassezia (Alexa 488, pseudo-colored in green). Hybridized specimens were observed using a confocal laser scanning microscope (CLSM) for 3 dimensional analysis. (A) Control, (B) M. sympodialis infected mouse, (C), M. sympodialis and P. aeruginosa infected mouse, and (D) M. sympodialis and S. aureus infected mouse.
Discussion
Physical and chemical interactions between fungi and bacteria play important roles in various ecosystems, such as in biofilm infections (Schlecht et al., 2015), in the oral cavity (Janus et al., 2016), in soil (Warmink and van Elsas, 2009), or in food products (Kastman et al., 2016). These cross-kingdom interactions result in changes to microbial fitness and virulence levels (Shirtliff et al., 2009; Schlecht et al., 2015). In this study, we investigated the interaction between predominant sinonasal bacterial pathogens, P. aeruginosa and S. aureus, and a dominant fungus present in the upper respiratory tract, M. sympodialis (Findley et al., 2013; Cope et al., 2017). Our goal was to assess the influence of microbial interactions in the upper airway on bacterial microbiome composition and immune response.
Here, we present two main conclusions. First, we show that there are species-specific interactions in vitro between a predominant member of sinonasal mycobiota, Malassezia, and major pathobionts associated with CRS (P. aeruginosa or S. aureus), and that co-infections with M. sympodialis and P. aeruginosa or S. aureus leads to distinct immune responses. This conclusion is supported by the in vitro studies and the increased immune response in co-infected mice. The second conclusion is that an intact sinonasal microbiota may be required for inducing distinct immune responses mediated by bacterial-fungal interactions. This is also inferred from the immune response analyses which shows that mice with depleted sinonasal microbiota by antimicrobial treatment had a suppressed immune response to intranasal pathobiont challenge. These pilot results are limited by a small sample size in mice and represent a single experiment. Taken together, these results seem to indicate that resident microbiota, and not just the single bacterial pathobionts, may be critical in inducing an immune response. Future studies using a larger sample size, longitudinal, and multiple-'omics analyses are necessary to identify the directionality and mechanism driving interactions.
In order to understand the interactions between S. aureus, P. aeruginosa, and M. sympodialis, we performed a modified Kirby-Bauer inhibition assay, a crystal violet biofilm quantification assay, and in vitro Fluorescent in situ Hybridization (FISH). The results from the Kirby-Bauer inhibition assay indicated that the late growth phase and biofilm-mode of P. aeruginosa inhibit M. sympodialis growth. The early growth phase and the cell-free supernatant of late growth phase P. aeruginosa did not inhibit M. sympodialis (data not shown), suggesting that inhibition of M. sympodialis by P. aeruginosa may require direct cell-cell contact and the inhibition mechanism may be dependent on the density of P. aeruginosa. In addition, quorum sensing (QS) molecules, which can trigger changes in biofilm physiology and gene expression and require high microbial density, may influence the observed negative interactions between P. aeruginosa and M. sympodialis. One possible inhibitory mechanism between P. aeruginosa and M. sympodialis is the type VI secretion system (T6SS). T6SS is a QS-regulated nanomachinery system in bacterial membranes that allow the bacteria to inject toxins into other cell membranes or cytoplasm (Gallique et al., 2017). P. aeruginosa possesses 3 genes for T6SS, H1-, H2-, and H3-T6SS, and H2- and H3-T6SS act not only against prokaryotes but also against eukaryotes (Hood et al., 2010; Trunk et al., 2018). A recently published study showed that programmed inhibitor cells can target bacterial species in a complex microbial community using T6SS (Ting et al., 2020), indicating a potential role for T6SS in altering microbiota composition.
We demonstrated a commensal or synergistic interaction between S. aureus and M. sympodialis. We observed a small positive effect on biofilm development when S. aureus was co-cultured with M. sympodialis in contrast to decreased biofilm growth when P. aeruginosa and M. sympodialis were cultured together. The in vitro FISH images also demonstrated colocalization of M. sympodialis and S. aureus in a biofilm but not with P. aeruginosa. This indicates that there may be a positive interaction between M. sympodialis and S. aureus. Other studies have shown positive interactions between the dimorphic yeast, Candida albicans, and S. aureus (Shirtliff et al., 2009; Schlecht et al., 2015). In these studies, co-existence of S. aureus and C. albicans induced fungal hyphae formation, and S. aureus formed biofilm specifically on fungal hyphae. As a result of this interaction, the hyphal form of C. albicans penetrated into the host tissue to obtain nutrients, and S. aureus secured a niche to invade host tissue, which increased the virulence of both microbes (Schlecht et al., 2015). There also are studies that reported a high incidence of co-existence of S. aureus and Malassezia spp in dermatological diseases, such as seborrheic dermatitis, atopic dermatitis, and erythroceruminous otitis (Buda and Miedzobrodzki, 2016; Nuttall, 2016; Tamer et al., 2018). Thus, the interaction between M. sympodialis and S. aureus may also have an influence on the host immune response related to chronic rhinosinusitis and should be studied further. Furthermore, we performed FISH on sinonasal specimens in order to confirm how co-infection impacts the abundance and localization of Malassezia in mice. We demonstrated an increase in Malassezia genus abundance in sinonasal tissue of mice singly infected with M. sympodialis, but we did not observe colocalization of Malassezia with endogenous Staphylococci. Of interest, in mice co-infected with exogenous Malassezia and S. aureus, we observed colocalization of both taxa in the submucosa. Submucosal S. aureus has been observed in chronic rhinosinusitis, but the implications are unclear. In one study, epithelial invasion of S. aureus was associated with an elevated immune response (Sachse et al., 2010), and in other studies, submucosal S. aureus is associated with reduced eosinophilia (Wood et al., 2012). Overall, these pilot results support that the interactions we observed in vitro are maintained in an acute sinusitis model in vivo. Future studies will evaluate the spatial organization of the microbiota in murine sinus tissue using Combinatorial Labeling and Spectral (CLASI)-FISH in a larger sample size.
The sinonasal microbiome was altered by antibiotic use and infection. The bacterial composition of the antibiotic-naïve mice was driven by the infected bacterial species, though we did not observe the same effect in antibiotic-treated mice. For the microbiome analyses, we grouped sequences into Amplicon Sequence Variants (ASVs), or 100% OTUs using DADA2. This is a relatively new approach that allows higher resolution than clustering into 97% OTUs, as was the previous gold standard. This allows us to differentiate some native Staphylococcus taxa from those we instilled into the murine nasal cavity if they differ by a single nucleotide. However, There are still limitations to 16S rRNA amplicon sequencing, including an inability to distinguish between species (Staphylococcus aureus and Staphylococcus epidermidis), particularly using the V4 region of the 16S rRNA gene. We did not detect any endogenous Pseudomonas in any control sample, though other studies have detected Pseudomonas in the murine airways (Cope et al., 2016). This could be due to different strains and vivarium conditions across studies. Our microbiome results show that the sinonasal microbiota of mice infected with P. aeruginosa or S. aureus were dominated by the infected bacterial species regardless of the co-infection with M. sympodialis. Interestingly, M. sympodialis infection alone did not significantly alter the composition of the sinonasal bacterial communities. These results indicated that the murine sinonasal microbiota was altered by antibiotic treatments, and these changes impaired colonization by the newly infected bacteria. The pathobionts, P. aeruginosa and S. aureus, were introduced to the undisturbed sinonasal microbiota and successfully colonized, becoming the predominant bacterial genera present in the sinonasal microbiota. Future studies should include additional animals in replicate experiments to confirm these results.
Antibiotic treatment affected the sinonasal immune response after single- and co-infection. Augmentin and Fluconazole were used to examine the effect of single or co-infections without the bias of an intact sinonasal microbiota. In a prior study of acute sinusitis, Abreu et al. demonstrated that the depletion of native microbiota using Augmentin resulted in increased goblet cell hyperplasia after nasal instillation of the pathobiont Corynebacterium tuberculostearicum compared to antibiotic-naïve mice with an intact sinonasal microbiome (Abreu et al., 2012). For this reason, we decided to include an antibiotic-treated group and antibiotic-naïve group to compare immune responses when the sinonasal microbiota are intact or depleted. We observed no statistically significant changes between different infection groups in the antibiotic-treated mice with one exception of increased IL-5 gene expression in P. aeruginosa-infected mice. However, there were several statistically significant immune responses in the antibiotic-naïve mice, in which we also observed major changes to the sinonasal microbiome. The interaction between S. aureus and M. sympodialis may contribute to the induction of Th2 and Th17 immune responses. IL-5 and IL-13 were elevated in S. aureus and M. sympodialis co-infected mice. These cytokines are involved in Th2 immune response, leading to increased mucus production and allergic response (Pope et al., 2001). In CRS, IL-5 expression is a signature immune endotype, often associated with concurrent asthma or nasal polyps (Tomassen et al., 2016; Hoggard et al., 2018). MUC5AC expression was also elevated in co-infected mice, which is downstream of IL-13 gene expression. Interestingly, we observed the greatest increase in M. sympodialis and P. aeruginosa-infected mice in the presence of an intact sinonasal microbiome. IL-17 expression was elevated in S. aureus infected mice. Th17 is another signature of CRS inflammatory endotypes, and may contribute to polyp generation in human disease (Tomassen et al., 2016; Miljkovic et al., 2017). IL-17 is required for protection against S. aureus cutaneous infection, so the induction of IL-17 in the sinonasal cavity may be protective (Cho et al., 2010). Pulmonary infection with S. aureus also induces a prominent Th17 response in mice (Frank et al., 2012). Interestingly, IL-17 was repressed in antibiotic-treated mice, suggesting a role for the sinonasal microbiome in Th17 induction in the airways as has been demonstrated in the GI tract, dependent on colonization with Segmented Filamentous Bacteria (Ivanov et al., 2009). However, we did not observe a significant immune response in the antibiotic-naive mice singly infected with M. sympodialis, indicating that the endogenous S. aureus strain is either at a low abundance and thus insufficiently interacts with M. sympodialis, or that the endogenous Staphylococci have a different interaction with Malassezia compared to the pathobiont that we co-infected mice with. Importantly, we actually observed a suppressive effect when M. sympodialis was singly inoculated into the mice.
We were surprised that depletion of the sinonasal microbiota generally suppressed the immune response; this result was in contradiction to our hypothesis and to prior studies (Abreu et al., 2012; Cope et al., 2016). In the current study, we administered antibiotics intranasally, while prior studies have used oral antibiotics. Intranasal antibiotics may have had a more robust effect on the sinonasal mucosal microbiome. In addition, we depleted both the bacterial and fungal microbiota using augmentin and fluconazole, while other studies have only used antibiotics targeting bacteria. It's possible that depleting the sinonasal fungal microbiota along with the bacterial microbiota, we have inhibited the potential for colonization, or that the remaining microbiota (e.g., Lactobacilli) are inhibitory toward exogenous M. sympodialis, S. aureus, or P. aeruginosa. We don't think that there are residual antibiotics after 24 h, prior studies have shown that when Augmentin and Fluconazole are cleared from the system ~1 day after the oral or intravenous administration (Foulstone and Reading, 1982; Ripa et al., 1993). However, this is a possibility that we will address in future studies.
Malassezia has the ability to bind C-type lectin, a protein that recognizes fungal carbohydrates and can induce an immune response by binding to ligand (Graham and Brown, 2009). Studies of atopic eczema have shown that the cellular component of Malassezia affects the expression of a C-type lectin receptor gene, dectin-1, which results in aggravation of the disease, as measured by increased IL-6/IL-8 gene expression and β-hexosaminidase, histamine, and tryptase release (Ribbing et al., 2011). In both antibiotic-treated and antibiotic-naïve mice infected with M. sympodialis alone, we observed no increase in dectin-1 gene expression. This was a surprising finding, but we also observed differential regulation of dectin-1 in mice infected with M. sympodialis with bacterial co-colonizers. When mice were co-infected with M. sympodialis and S. aureus, dectin-1 expression increased compared to M. sympodialis singly infected group, which may be due to increased M. sympodialis virulence mediated by the synergistic interaction with S. aureus. These results support our hypothesis that the interaction between S. aureus and M. sympodialis leads to a change in microbial community behavior, leading to increased fungal sensing. Future studies will seek to define the mechanisms underlying these findings. Dectin-1 activation can lead to increased Th2-type CD4+ T cell responses elicited by plasmacytoid dendritic cells, which could be one mechanism leading to elevated IL-5 and IL-13 gene expression in our study (Upchurch et al., 2016). Dectin-1 expression is also increased in CRS polyp tissue; future studies could evaluate whether this is correlated with co-colonization of S. aureus and Malassezia species (Gong et al., 2013). Interestingly, dectin-1 expression also increased in antibiotic-naïve mice that were singly infected with bacteria (S. aureus or P. aeruginosa), but this was not observed in antibiotic-naïve mice singly infected with M. sympodialis. This could be due to bacterial interactions with endogenous Malassezia present in the murine sinonasal cavity or due to the sensing of bacterial ligands by Dectin-1, as has been demonstrated in the lungs with Dectin-1 sensing of non-typeable Haemophilus influenzae (Heyl et al., 2015).
This study has limitations. First, we are presenting findings based on a single pilot murine experiment of 3–5 animals per group. These numbers and replicates were based on published studies of acute sinusitis (Abreu et al., 2012; Cope et al., 2016) but we acknowledge that future studies should aim to replicate these findings in a larger sample size across different strains of mice. Thus, while exciting, these findings should be considered in the context of a small samples size in a single experiment. We plan to address these pitfalls in future studies that are based off these initial findings.
In conclusion, we demonstrate that interkingdom, species-specific interactions between a predominant member of sinonasal mycobiota, Malassezia, and major pathobionts associated with CRS (P. aeruginosa or S. aureus) leads to distinct immune responses at the sinonasal mucosa. Malassezia is an understudied fungal taxa with potential importance in a myriad of gut, skin, and respiratory disease (Hoggard et al., 2019; Limon et al., 2019; Ianiri et al., 2020). We also demonstrated that an endogenous sinonasal microbiota may be required for inducing distinct immune responses mediated by bacterial-fungal interactions. Further studies of the mechanisms contributing to these interactions, such as quorum sensing or dectin-1 mediated activation of CD4+ Th2 cells, is necessary for understanding the pathophysiology of CRS and developing microbiome-mediated therapeutics.
Data Availability Statement
Sequence data for this study have been deposited in the NCBI. Short Read Archive (SRA) under BioProject ID PRJNA610287.
Ethics Statement
The animal study was reviewed and approved by Northern Arizona University Institutional Animal Care and Use Committee (IACUC).
Author Contributions
KL was the main investigator of the project and wrote the most part of the manuscript. OK contributed in animal experiments, processing samples, and analyzing the bioinformatics data. IZ and SK contributed in in-vitro experiments, in-vivo experiments, and processing samples. EC was the corresponding author who contributed in designing the research project, funding, and writing the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was partially supported by the Technology Research Initiative Fund (TRIF).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00472/full#supplementary-material
In vivo mouse co-infection experimental scheme. (A) acclimatized mice were intranasally infected with designated microbes for each group. Antibiotics were treated with Fluconazole (15 mg/kg) and Augmentin (100 mg/kg) for 5 days for the antibiotic-treated group. All mice were intranasally infected with designated microbes for each group. (B) The representation of infected microbes for each group.
Fluorescent in-situ hybridization (FISH) observed using confocal laser scanning microscopy (CLSM) confirmed the inhibition of P. aeruginosa on M. sympodialis and shows the coexistence of S. aureus and M. sympodialis.
Principal Coordinates Analysis of Unweighted UniFrac distances of the sinonasal microbiota. Two groups of mice, antibiotic-treated or -naive, were intranasally infected with either one species of bacteria, P. aeruginosa (light green), S. aureus (light red), or fungi, M. sympodialis (dark blue), or two species, M. sympodialis with P. aeruginosa (dark green) or S. aureus (dark red). (A) represents beta-diversity of antibiotic-treated (solid sphere) and antibiotic-naïve (diamond) mice groups. (B) represents beta-diversity of the infection groups in the antibiotic-treated mice, and (C) represents phylogenetically weighted beta-diversity of the infection groups in the antibiotic-naïve mice.
Principal Coordinates Analysis of Weighted UniFrac distances of the sinonasal microbiota. Two groups of mice, antibiotic-treated or -naive, were intranasally infected with either one species of bacteria, P. aeruginosa (light green), S. aureus (light red), or fungi, M. sympodialis (dark blue), or two species, M. sympodialis with P. aeruginosa (dark green) or S. aureus (dark red). (A) represents beta-diversity of antibiotic-treated (solid sphere) and antibiotic-naïve (diamond) mice groups. (B) represents beta-diversity of the infection groups in the antibiotic-treated mice, and (C) represents beta-diversity of the infection groups in the antibiotic-naïve mice.
Principal Coordinates Analysis of Jaccard distances of the sinonasal microbiota. Two groups of mice, antibiotic-treated or -naive, were intranasally infected with either one species of bacteria, P. aeruginosa (light green), S. aureus (light red), or fungi, M. sympodialis (dark blue), or two species, M. sympodialis with P. aeruginosa (dark green) or S. aureus (dark red). (A) represents beta-diversity of antibiotic-treated (solid sphere) and antibiotic-naïve (diamond) mice groups. (B) represents beta-diversity of the infection groups in the antibiotic-treated mice, and (C) represents beta-diversity of the infection groups in the antibiotic-naïve mice.
Relative bacterial abundances in sinonasal microbiota of intranasally infected mice with either bacteria or fungus or both. Available at this link (https://view.qiime2.org/visualization/?type=html&src=https%3A%2F%2Fdl.dropbox.com%2Fs%2F7r7k1c4ridwv8qv%2FDay1-sinus_taxa-barplot.qzv%3Fdl%3D1) or on github at https://github.com/e-cope/malassezia-ms. (Taxonomic level 7, Sort samples by Description and TreatmentGroup). The interactive QIIME 2 analysis file can be viewed at https://view.QIIME2.org/.
References
- Abreu N. A., Nagalingam N. A., Song Y., Roediger F. C., Pletcher S. D., Goldberg A. N., et al. (2012). Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci. Transl. Med. 4:151ra124. 10.1126/scitranslmed.3003783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Nishimura J., Shima T., Umesaki Y., Yamamoto M., Onoue M., et al. (2008). ATP drives lamina propria T(H)17 cell differentiation. Nature 455, 808–812. 10.1038/nature07240 [DOI] [PubMed] [Google Scholar]
- Baba S., Kagoya R., Kondo K., Suzukawa M., Ohta K., Yamasoba T. (2015). T-cell phenotypes in chronic rhinosinusitis with nasal polyps in Japanese patients. Allergy Asthma Clin. Immunol. 11:33. 10.1186/s13223-015-0100-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J. L., Garcia M. E. (2008). Immune response to fungal infections. Vet. Immunol. Immunopathol. 125, 47–70. 10.1016/j.vetimm.2008.04.020 [DOI] [PubMed] [Google Scholar]
- Boekhout T., Guého-Kellermann E., Mayser P., Velegraki A. (2010). Malassezia and the Skin: Science and Clinical Practice. Berlin; Heidelberg: Springer Science & Business Media. [Google Scholar]
- Bokulich N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome 6:90. 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill S., James P., Cuthbertson L., Cox M., Cookson W., Wedzicha J., et al. (2016). Profiling the COPD airway microbiome using quantitative culture and 16S rRNA gene sequencing. Eur. Respir. J. 48(Suppl. 60):OA1787. 10.1183/13993003.congress-2016.OA1787 [DOI] [Google Scholar]
- Buda A., Miedzobrodzki J. (2016). The role of Staphylococcus aureus in secondary infections in patients with atopic dermatitis (AD). Pol. J. Microbiol. 65, 253–259. 10.5604/17331331.1215600 [DOI] [PubMed] [Google Scholar]
- Cafarchia C., Figueredo L. A., Favuzzi V., Surico M. R., Colao V., Iatta R., et al. (2012). Assessment of the antifungal susceptibility of Malassezia pachydermatis in various media using a CLSI protocol. Vet. Microbiol. 159, 536–540. 10.1016/j.vetmic.2012.04.034 [DOI] [PubMed] [Google Scholar]
- Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calus L., Van Bruaene N., Bosteels C., Dejonckheere S., Van Zele T., Holtappels G., et al. (2019). Twelve-year follow-up study after endoscopic sinus surgery in patients with chronic rhinosinusitis with nasal polyposis. Clin. Transl. Allergy 9:30. 10.1186/s13601-019-0269-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter J. M., Amedee R. G. (2014). Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. Am. J. Rhinol. Allergy 28:182. 10.2500/194589214810960481 [DOI] [PubMed] [Google Scholar]
- Caulley L., Thavorn K., Rudmik L., Cameron C., Kilty S. J. (2015). Direct costs of adult chronic rhinosinusitis by using 4 methods of estimation: results of the US medical expenditure panel survey. J. Allergy Clin. Immunol. 136, 1517–1522. 10.1016/j.jaci.2015.08.037 [DOI] [PubMed] [Google Scholar]
- Cho J. S., Pietras E. M., Garcia N. C., Ramos R. I., Farzam D. M., Monroe H. R., et al. (2010). IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J. Clin. Investig. 120, 1762–1773. 10.1172/jci40891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland E. J., Bassiouni A., Boase S., Dowd S., Vreugde S., Wormald P.-J. (2014). The fungal microbiome in chronic rhinosinusitis: richness, diversity, postoperative changes and patient outcomes. Int. Forum Allergy Rhinol. 4, 259–265. 10.1002/alr.21297 [DOI] [PubMed] [Google Scholar]
- Cope E. K., Goldberg A. N., Pletcher S. D., Lynch S. V. (2016). A chronic rhinosinusitis-derived isolate of Pseudomonas aeruginosa induces acute and pervasive effects on the murine upper airway microbiome and host immune response. Int. Forum Allergy Rhinol. 6, 1229–1237. 10.1002/alr.21819 [DOI] [PubMed] [Google Scholar]
- Cope E. K., Goldberg A. N., Pletcher S. D., Lynch S. V. (2017). Compositionally and functionally distinct sinus microbiota in chronic rhinosinusitis patients have immunological and clinically divergent consequences. Microbiome 5:53. 10.1186/s40168-017-0266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizabeth M., Zabielski R. (2013). Gut microbiome and brain-gut axis in autism — aberrant development of gut-brain communication and reward circuitry, in Recent Advances in Autism Spectrum Disorders - Volume I, ed M. Fitzgerald (InTech). [Google Scholar]
- Epperson M. V., Phillips K. M., Caradonna D. S., Gray S. T., Sedaghat A. R. (2019). Predictors of efficacy for combination oral and topical corticosteroids to treat patients with chronic rhinosinusitis with nasal polyps. Int. Forum Allergy Rhinol. 9, 1436–1442. 10.1002/alr.22431 [DOI] [PubMed] [Google Scholar]
- Findley K., Oh J., Yang J., Conlan S., Deming C., Meyer J. A., et al. (2013). Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370. 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulstone M., Reading C. (1982). Assay of amoxicillin and clavulanic acid, the components of Augmentin, in biological fluids with high-performance liquid chromatography. Antimicrob. Agents Chemother. 22, 753–762. 10.1128/aac.22.5.753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank K. M., Zhou T., Moreno-Vinasco L., Hollett B., Garcia J. G. N., Bubeck Wardenburg J. (2012). Host response signature to Staphylococcus aureus alpha-hemolysin implicates pulmonary Th17 response. Infect. Immun. 80, 3161–3169. 10.1128/IAI.00191-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich E., Mercuri A., Wu S., Salar-Behzadi S. (2016). Measurements of deposition, lung surface area and lung fluid for simulation of inhaled compounds. Front. Pharmacol. 7:181. 10.3389/fphar.2016.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K. E., Sitarik A. R., Havstad S., Lin D. L., Levan S., Fadrosh D., et al. (2016). Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191. 10.1038/nm.4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallique M., Bouteiller M., Merieau A. (2017). The type VI secretion system: a dynamic system for bacterial communication? Front. Microbiol. 8:1454. 10.3389/fmicb.2017.01454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelber J. T., Cope E. K., Goldberg A. N., Pletcher S. D. (2016). Evaluation of malassezia and common fungal pathogens in subtypes of chronic rhinosinusitis. Int. Forum Allergy Rhinol. 6, 950–955. 10.1002/alr.21777 [DOI] [PubMed] [Google Scholar]
- Gong J., Wang P., Qiu Z.-H., Chen Q.-J. (2013). Increased expression of dectin-1 in nasal polyps. Am. J. Otolaryngol. 34, 183–187. 10.1016/j.amjoto.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Goodrich J. K., Di Rienzi S. C., Poole A. C., Koren O., Walters W. A., Caporaso J. G., et al. (2014). Conducting a microbiome study. Cell 158, 250–262. 10.1016/j.cell.2014.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Helmink B. A., Spencer C. N., Reuben A., Wargo J. A. (2018). The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 33, 570–580. 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham L. M., Brown G. D. (2009). The Dectin-2 family of C-type lectins in immunity and homeostasis. Cytokine 48, 148–155. 10.1016/j.cyto.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot J., Breugnot C., de Barros M., Chermette R. (1998). Usefulness of modified Dixon's medium for quantitative culture of Malassezia species from canine skin. J. Vet. Diagn. Invest. 10, 384–386. 10.1177/104063879801000418 [DOI] [PubMed] [Google Scholar]
- Gupta V., Singh S. (2010). Nasal saline irrigation for chronic rhinosinusitis. Int. J. Clin. Rhinol. 3, 145–147. 10.5005/jp-journals-10013-1050 [DOI] [Google Scholar]
- Hamilos D. L. (2015). Drivers of chronic rhinosinusitis: Inflammation versus infection. J. Allergy Clin. Immunol. 136, 1454–1459. 10.1016/j.jaci.2015.10.011 [DOI] [PubMed] [Google Scholar]
- Heyl K. A., Klassert T. E., Heinrich A., Müller M. M., Klaile E., Dienemann H., et al. (2015). Dectin-1 is expressed in human lung and mediates the proinflammatory immune response to nontypeable haemophilus influenzae. Pneumologie 69:e01492–14. 10.1055/s-0035-1544861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogardt M., Trebesius K., Geiger A. M., Hornef M., Rosenecker J., Heesemann J. (2000). Specific and rapid detection by fluorescent in situ hybridization of bacteria in clinical samples obtained from cystic fibrosis patients. J. Clin. Microbiol. 38, 818–825. 10.1128/JCM.38.2.818-825.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard M., Waldvogel-Thurlow S., Zoing M., Chang K., Radcliff F. J., Mackenzie B. W., et al. (2018). Inflammatory endotypes and microbial associations in chronic rhinosinusitis. Front. Immunol. 9:2065. 10.3389/fimmu.2018.02065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard M., Zoing M., Biswas K., Taylor M. W., Douglas R. G. (2019). The sinonasal mycobiota in chronic rhinosinusitis and control patients. Rhinology 57, 190–199. 10.4193/Rhin18.256 [DOI] [PubMed] [Google Scholar]
- Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R. S., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse K. E. (2016). Immune mechanisms of chronic rhinosinusitis. Curr. Allergy Asthma Rep. 16:1. 10.1007/s11882-015-0579-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianiri G., Coelho M. A., Ruchti F., Sparber F., McMahon T. J., Fu C., et al. (2020). HGT in the human and skin commensal Malassezia: A bacterially derived flavohemoglobin is required for NO resistance and host interaction. Proc. Natl. Acad. Sci. U.S.A. 117:202003473. 10.1073/pnas.2003473117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Atarashi K., Manel N., Brodie E. L., Shima T., Karaoz U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janus M. M., Willems H. M. E., Krom B. P. (2016). Candida albicans in multispecies oral communities; a keystone commensal? Adv. Exp. Med. Biol. 931, 13–20. 10.1007/5584_2016_5 [DOI] [PubMed] [Google Scholar]
- Kastman E. K., Kamelamela N., Norville J. W., Cosetta C. M., Dutton R. J., Wolfe B. E. (2016). Biotic interactions shape the ecological distributions of Staphylococcus species. MBio 7:e01157-16. 10.1128/mBio.01157-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau A. L., Ahern P. P., Griffin N. W., Goodman A. L., Gordon J. I. (2011). Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336. 10.1038/nature10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Park B. S., Hong G. U., Lee B. J., Park J. W., Kim S. Y., et al. (2011). Anti-inflammatory effects of the R2 peptide, an inhibitor of transglutaminase 2, in a mouse model of allergic asthma, induced by ovalbumin. Br. J. Pharmacol. 162, 210–225. 10.1111/j.1476-5381.2010.01033.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R., Freeman J., Waldvogel-Thurlow S., Roberts S., Douglas R. (2013). The characteristics of intramucosal bacteria in chronic rhinosinusitis: a prospective cross-sectional analysis. Int. Forum Allergy Rhinol. 3, 349–354. 10.1002/alr.21117 [DOI] [PubMed] [Google Scholar]
- Kimura Y., Chihara K., Honjoh C., Takeuchi K., Yamauchi S., Yoshiki H., et al. (2014). Dectin-1-mediated signaling leads to characteristic gene expressions and cytokine secretion via spleen tyrosine kinase (Syk) in rat mast cells. J. Biol. Chem. 289, 31565–31575. 10.1074/jbc.M114.581322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistowska M., Fenini G., Jankovic D., Feldmeyer L., Kerl K., Bosshard P., et al. (2014). Malassezia yeasts activate the NLRP 3 inflammasome in antigen-presenting cells via S yk-kinase signalling. Exp. Dermatol. 23, 884–889. 10.1111/exd.12552 [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Altman N. (2014). Comparing samples—part II. Nat. Methods 11, 355–356. 10.1038/nmeth.2900 [DOI] [Google Scholar]
- Limon J. J., Tang J., Li D., Wolf A. J., Michelsen K. S., Funari V., et al. (2019). Malassezia Is associated with crohn's disease and exacerbates colitis in mouse models. Cell Host Microbe 25, 377–388.e6. 10.1016/j.chom.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-H., Shen M.-L., Zhou N., Lee C.-C., Kao S.-T., Wu D. C. (2014). Protective effects of the polyphenol sesamin on allergen-induced T(H)2 responses and airway inflammation in mice. PLoS One 9:e96091. 10.1371/journal.pone.0096091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P. S. (2012). Treatment of chronic rhinosinusitis with antibiotics. Clin. Infect. Dis. 54, 69–70. 10.1093/cid/cir757 [DOI] [PubMed] [Google Scholar]
- Miljkovic D., Psaltis A. J., Wormald P. J., Vreugde S. (2017). Chronic rhinosinusitis with polyps is characterized by increased mucosal and blood Th17 effector cytokine producing cells. Front. Physiol. 8:898. 10.3389/fphys.2017.00898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moise A. M. R. (2017). The Gut Microbiome: Exploring the Connection between Microbes, Diet, and Health. ABC-CLIO Available online at: https://market.android.com/details?id=book-0oE5DwAAQBAJ
- Nuttall T. (2016). Successful management of otitis externa. In Pract. 38, 17–21. 10.1136/inp.i1951 [DOI] [Google Scholar]
- Oestreich K. J., Mohn S. E., Weinmann A. S. (2012). Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol. 13, 405–411. 10.1038/ni.2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. 47:2437. 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher S. D., Goldberg A. N., Cope E. K. (2018). Loss of microbial niche specificity between the upper and lower airways in patients with cystic fibrosis. Laryngoscope 129, 544–550. 10.1002/lary.27454 [DOI] [PubMed] [Google Scholar]
- Pope S. M., Brandt E. B., Mishra A., Hogan S. P., Zimmermann N., Matthaei K. I., et al. (2001). IL-13 induces eosinophil recruitment into the lung by an IL-5- and eotaxin-dependent mechanism. J. Allergy Clin. Immunol. 108, 594–601. 10.1067/mai.2001.118600 [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbing C., Engblom C., Lappalainen J., Lindstedt K., Kovanen P. T., Karlsson M. A., et al. (2011). Mast cells generated from patients with atopic eczema have enhanced levels of granule mediators and an impaired Dectin-1 expression. Allergy 66, 110–119. 10.1111/j.1398-9995.2010.02437.x [DOI] [PubMed] [Google Scholar]
- Ripa S., Ferrante L., Prenna M. (1993). Pharmacokinetics of fluconazole in normal volunteers. Chemotherapy 39, 6–12. 10.1159/000238967 [DOI] [PubMed] [Google Scholar]
- Sachse F., Becker K., Von Eiff C., Metze D., Rudack C. (2010). Staphylococcus aureus invades the epithelium in nasal polyposis and induces IL-6 in nasal epithelial cells in vitro. Allergy 65, 1430–1437. 10.1111/j.1398-9995.2010.02381.x [DOI] [PubMed] [Google Scholar]
- Schlecht L. M., Peters B. M., Krom B. P., Freiberg J. A., Hänsch G. M., Filler S. G., et al. (2015). Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology 161, 168–181. 10.1099/mic.0.083485-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedaghat A. R. (2017). Chronic rhinosinusitis. Am. Fam. Phys. 96, 500–506. [PubMed] [Google Scholar]
- Shirtliff M. E., Peters B. M., Jabra-Rizk M. A. (2009). Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 299, 1–8. 10.1111/j.1574-6968.2009.01668.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamer F., Yuksel M. E., Sarifakioglu E., Karabag Y. (2018). Staphylococcus aureus is the most common bacterial agent of the skin flora of patients with seborrheic dermatitis. Dermatol Pract Concept 8, 80–84. 10.5826/dpc.0802a04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telander D. G., Malvey E. N., Mueller D. L. (1999). Evidence for repression of IL-2 gene activation in anergic T cells. J. Immunol. 162, 1460–1465. [PubMed] [Google Scholar]
- Thaiss C. A., Zmora N., Levy M., Elinav E. (2016). The microbiome and innate immunity. Nature 535, 65–74. 10.1038/nature18847 [DOI] [PubMed] [Google Scholar]
- Ting S.-Y., Martínez-García E., Huang S., Bertolli S. K., Kelly K. A., Cutler K. J., et al. (2020). Targeted depletion of bacteria from mixed populations by programmable adhesion with antagonistic competitor cells. Cell Host Microbe. 28:313–321.e6. 10.1016/j.chom.2020.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassen P., Vandeplas G., Van Zele T., Cardell L.-O., Arebro J., Olze H., et al. (2016). Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J. Allergy Clin. Immunol. 137, 1449–1456.e4. 10.1016/j.jaci.2015.12.1324 [DOI] [PubMed] [Google Scholar]
- Trandem K., Jin Q., Weiss K. A., James B. R., Zhao J., Perlman S. (2011). Virally expressed interleukin-10 ameliorates acute encephalomyelitis and chronic demyelination in coronavirus-infected mice. J. Virol. 85, 6822–6831. 10.1128/JVI.00510-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebesius K., Leitritz L., Adler K., Schubert S., Autenrieth I. B., Heesemann J. (2000). Culture independent and rapid identification of bacterial pathogens in necrotising fasciitis and streptococcal toxic shock syndrome by fluorescence in situ hybridisation. Med. Microbiol. Immunol. 188, 169–175. 10.1007/s004300000035 [DOI] [PubMed] [Google Scholar]
- Trunk K., Peltier J., Liu Y.-C., Dill B. D., Walker L., Gow N. A. R., et al. (2018). The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol 3, 920–931. 10.1038/s41564-018-0191-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y., Kayama H., Jeon S. G., Kusu T., Isaka Y., Rakugi H., et al. (2010). Commensal microbiota induce LPS hyporesponsiveness in colonic macrophages via the production of IL-10. Int. Immunol. 22, 953–962. 10.1093/intimm/dxq449 [DOI] [PubMed] [Google Scholar]
- Upchurch K., Oh S., Joo H. (2016). Dectin-1 in the control of Th2-type T cell responses. Recep. Clin. Investig. 3:e1094. 10.14800/rci.1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner Mackenzie B., Chang K., Zoing M., Jain R., Hoggard M., Biswas K., et al. (2019). Longitudinal study of the bacterial and fungal microbiota in the human sinuses reveals seasonal and annual changes in diversity. Sci. Rep. 9:17416. 10.1038/s41598-019-53975-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmink J. A., van Elsas J. D. (2009). Migratory response of soil bacteria to Lyophyllum sp. strain karsten in soil microcosms. Appl. Environ. Microbiol. 75, 2820–2830. 10.1128/aem.02110-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. J., Fraser J. D., Swift S., Patterson-Emanuelson E. A. C., Amirapu S., Douglas R. G. (2012). Intramucosal bacterial microcolonies exist in chronic rhinosinusitis without inducing a local immune response. Am. J. Rhinol. Allergy 26, 265–270. 10.2500/ajra.2012.26.3779 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Adappa N. D., Doghramji L. J., Chiu A. G., Cohen N. A., Palmer J. N. (2015). Different clinical factors associated with Staphylococcus aureus and Pseudomonas aeruginosa in chronic rhinosinusitis. Int. Forum Allergy Rhinol. 5, 724–733. 10.1002/alr.21532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vivo mouse co-infection experimental scheme. (A) acclimatized mice were intranasally infected with designated microbes for each group. Antibiotics were treated with Fluconazole (15 mg/kg) and Augmentin (100 mg/kg) for 5 days for the antibiotic-treated group. All mice were intranasally infected with designated microbes for each group. (B) The representation of infected microbes for each group.
Fluorescent in-situ hybridization (FISH) observed using confocal laser scanning microscopy (CLSM) confirmed the inhibition of P. aeruginosa on M. sympodialis and shows the coexistence of S. aureus and M. sympodialis.
Principal Coordinates Analysis of Unweighted UniFrac distances of the sinonasal microbiota. Two groups of mice, antibiotic-treated or -naive, were intranasally infected with either one species of bacteria, P. aeruginosa (light green), S. aureus (light red), or fungi, M. sympodialis (dark blue), or two species, M. sympodialis with P. aeruginosa (dark green) or S. aureus (dark red). (A) represents beta-diversity of antibiotic-treated (solid sphere) and antibiotic-naïve (diamond) mice groups. (B) represents beta-diversity of the infection groups in the antibiotic-treated mice, and (C) represents phylogenetically weighted beta-diversity of the infection groups in the antibiotic-naïve mice.
Principal Coordinates Analysis of Weighted UniFrac distances of the sinonasal microbiota. Two groups of mice, antibiotic-treated or -naive, were intranasally infected with either one species of bacteria, P. aeruginosa (light green), S. aureus (light red), or fungi, M. sympodialis (dark blue), or two species, M. sympodialis with P. aeruginosa (dark green) or S. aureus (dark red). (A) represents beta-diversity of antibiotic-treated (solid sphere) and antibiotic-naïve (diamond) mice groups. (B) represents beta-diversity of the infection groups in the antibiotic-treated mice, and (C) represents beta-diversity of the infection groups in the antibiotic-naïve mice.
Principal Coordinates Analysis of Jaccard distances of the sinonasal microbiota. Two groups of mice, antibiotic-treated or -naive, were intranasally infected with either one species of bacteria, P. aeruginosa (light green), S. aureus (light red), or fungi, M. sympodialis (dark blue), or two species, M. sympodialis with P. aeruginosa (dark green) or S. aureus (dark red). (A) represents beta-diversity of antibiotic-treated (solid sphere) and antibiotic-naïve (diamond) mice groups. (B) represents beta-diversity of the infection groups in the antibiotic-treated mice, and (C) represents beta-diversity of the infection groups in the antibiotic-naïve mice.
Relative bacterial abundances in sinonasal microbiota of intranasally infected mice with either bacteria or fungus or both. Available at this link (https://view.qiime2.org/visualization/?type=html&src=https%3A%2F%2Fdl.dropbox.com%2Fs%2F7r7k1c4ridwv8qv%2FDay1-sinus_taxa-barplot.qzv%3Fdl%3D1) or on github at https://github.com/e-cope/malassezia-ms. (Taxonomic level 7, Sort samples by Description and TreatmentGroup). The interactive QIIME 2 analysis file can be viewed at https://view.QIIME2.org/.
Data Availability Statement
Sequence data for this study have been deposited in the NCBI. Short Read Archive (SRA) under BioProject ID PRJNA610287.