Abstract
The androgen receptor (AR) has been found to be expressed in the majority of human breast cancer and AR antagonists, such as enzalutamide, have shown promising clinical activity in AR-positive (AR+) breast cancer. We have recently reported the discovery of a highly potent PROTAC AR degrader, ARD-61. In this study, we evaluated ARD-61 for its therapeutic potential and mechanism of action in breast cancer models in vitro and in vivo. ARD-61 potently and effectively induces AR degradation in AR+ breast cancer cell lines and is much more potent than enzalutamide in inhibition of cell growth and induction of cell cycle arrest and/or apoptosis. ARD-61 effectively induces complete AR degradation in xenograft tumor tissue and is more effective than enzalutamide in achieving tumor growth inhibition in the MDA-MB-453 xenograft model in mice. Our study provides strong preclinical rationale to develop AR degraders for the treatment of AR+ human breast cancer.
Abbreviations: AR, androgen receptor; ER, estrogen receptors; PR, progesterone receptors; GR, glucocorticoid receptor; TNBCs, triple negative breast cancer; VHL, Von Hippel-Lindau; CBR, clinical benefit ratio
Keywords: Androgen-receptor (AR) degraders, Breast cancer, New therapeutic strategy
Introduction
Despite tremendous progress made in the development of highly effective therapies, breast cancer remains the second cause of cancer death in women. While estrogen receptors (ER) and progesterone receptors (PR) are known to be prominent players in the progression and development of breast cancer, recent studies have suggested an important role played by the androgen receptor (AR), a male hormone, in breast cancer progression [1], [2]. The AR has been found to be expressed in the majority of all breast cancers [2], [3], [4], [5], [6]. For examples, AR is overexpressed in 70–90% of ER+ breast cancer, ∼60% of HER2+ breast cancers, and 10–35% of triple-negative breast cancers (TNBCs) [5], [6], [7].
Although the precise role of AR in breast cancer remains to be fully elucidated, it has been proposed that AR represents a potential therapeutic target in breast cancer [1], [2] and that AR antagonists may have a therapeutic potential for the treatment of AR+ breast cancers [1], [2], [8], [9]. Bicalutamide, an AR antagonist, was evaluated in AR+/ER−/PR− breast cancer [8] and demonstrated a clinical benefit ratio (CBR) of 19% with minimal toxicity. More recently, enzalutamide, a second-generation AR antagonist was evaluated in a phase 2 clinical trial with 75 patients with metastatic AR+, TNBCs. Enzalutamide showed a CBR of 35% and 29% at 16- and 24-weeks, respectively, and achieved a significant improvement of progression-free survival [9]. These clinical studies have provided the important proof-of-concept data that targeting AR may indeed provide clinical benefits for patients with AR+/ER−/PR− breast cancer.
While AR antagonists such as enzalutamide have demonstrated clinical benefits in AR+ breast cancer patients, resistance can develop. For example, the emergence of a F876L point mutation in AR causes resistance to enzalutamide and apalutamide [10]. In fact, this point mutation changes the conformation of the AR ligand binding domain in such a way that enzalutamide becomes an AR agonist [10], [11], [12]. Therefore, there is a clear need to develop more effective therapeutic strategies to target AR for the treatment of AR+ human breast cancer.
The proteolysis-targeting chimera (PROTAC) strategy has gained tremendous momentum for its promise for the development of completely new types of therapies through induced protein degradation [13], [14], [15]. Recently, we reported the discovery of ARD-61 as a highly potent PROTAC AR degrader [16], [17]. In our previous studies [16], we have shown that ARD-61 effectively induces AR degradation in AR+ prostate cancer cells in vitro and in prostate cancer xenograft tumor tissues in vivo. ARD-61 is a potent and effective inhibitor of AR signaling and cell growth in AR+ prostate cancer models. Significantly, ARD-61 is effective in enzalutamide-resistant prostate cancer models in vitro and in vivo [16].
In the present study, we investigated the therapeutic potential and mechanism of action of ARD-61 in human breast cancer models in vitro and in vivo. Our data demonstrate that ARD-61 potently and effectively induces AR degradation in human breast cancer cell lines in vitro and in xenograft tumor tissue in mice. ARD-61 is very effective in inhibition of tumor growth in the MDA-MB-453 AR+/ER−/PR− breast cancer xenograft model in mice at well tolerated dose-schedules. Our study provides strong preclinical data to support the clinical development of AR degraders for the treatment of AR+ human breast cancer.
Materials and methods
Synthesis
ARD-61, ARi-16 and VHL ligand were synthesized as previously described [16], [18], [17]. All compounds have purity of >95% based upon HPLC analysis. Enzalutamide (915087-33-1) was purchased from 1 Click Chemistry with >95% purity.
Cell lines
LNCaP (CRL-1740), MDA-MB-453 (HTB-131), HCC1428 (CRL-2327), MCF-7 (HTB-22), BT549 (HTB-122), T47D (HTB-133), BT20 (HTB-19), HCC1395 (SC-CRL-2324), MDA-MB-468 (HTB-132), HCC1806 (CRL-2335), MDA-MB-436 (HTB-130) and MDA-MB-231 (HTB-26) cancer cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines used in this study were cultured according to the manufacturer’s instructions and cells were maintained in culture for a maximum of 7–15 passages.
Western blot
Cells were lysed in RIPA lysis and extraction buffer (Thermo Fisher Scientific, 89901) supplemented with protease inhibitor cocktail (Roche, 11697498001) and phosphatase inhibitor cocktail (Roche, 4906837001) for 30 min on ice. Lysates were centrifuged at 15,000 rpm for 10 min and supernatants were analyzed by SDS/PAGE. Samples were then transferred onto PVDF membrane and incubated in 5% milk in TBST (Tris-buffered Saline with Tween 20) at room temperature for 1 h, followed by incubation with indicated primary antibodies overnight at 4 °C. Membranes were then incubated with HRP conjugated second antibodies for 1 h at room temperature. Membranes were visualized using the ECL western blotting detection reagent (BIO-RAD, 170506) and finally, films were developed using an X-ray film developer. PR A/B (#3176), GR (#3660), AKT (#4691), Phospho-AKT (#4060), P21 (#2947), β-catenin (#8480), FoxA1 (#53528), Phospho-HER3 (#4791), HER3 (#12708), Phospho-HER2 (#2247), HER2 (#4290), Cleaved caspase 3/7/8/9 (#9661, #8438, #9496, #9505), Cleaved PARP (#5625), and GAPDH (#8884) antibodies were all purchased from Cell Signaling Technology. AR antibody (#06-680) was purchased from Millipore Sigma. ER (Ab75635) antibody was purchased from Abcam. WNT7B (OAAN02407), c-Myc (NB600-302), and VHL (PA5-13488) antibodies were purchased form Aviva Systems Biology, Novus Biologicals and Thermo Fisher Scientific, respectively. MAD1 (sc-47746), Topo1 (sc-32736), anti-rabbit IgG (sc-2357) and anti-mouse IgG (sc-516102) antibodies were purchased from Santa Cruz Biotechnology.
Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
RNA was isolated using the RNeasy Mini Kit (Qiagen #74104). Reverse transcriptase reaction (RT) was performed with 1 mg of total RNA using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, 4387406), followed by polymerase chain reaction (PCR) using TaqMan Gene Expression Master Mix (Thermo Fisher Scientific, 4444557) on a QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific). The relative abundance of gene expression was calculated using the comparative CT method which compares the Ct value of target gene to that of GAPDH. GAPDH (Hs02786624-g1), AR (Hs00171172-m1), MYC (Hs00153408-m1), WNT7B (Hs00536497-m1), CDKN1A (Hs00355782-m1) and AQP3 (Hs00185020-m1) were all purchased from Thermo Fisher Scientific.
RNA interference
ON-TARGETplus Human VHL and vector siRNAs were purchased from Dharmacon. MDA-MB-453 and MCF-7 cells were transfected with siRNAs against VHL (L-003936-00-0005) or vector and Lipofectamine™ RNAiMAX transfection reagent (Thermo Fisher, 13778150) according to manufacturer's instructions for 72 h. The expression of VHL was determined by immunoblotting.
Cell proliferation assay
Cells were seeded in 96-well plates in 200 μL of charcoal-stripped serum (CSS) contained medium and incubated at 37 °C for 2 days. MDA-MB-453 (4000 cells per 96-well), BT549 (2500 cells per 96-well), MDA-MB-415 (4000 cells per 96-well), HCC1428 (4000 cells per 96-well) and BT20 (3000 cells per 96-well) cells were seeded in RPMI 1640 medium supplemented with 10% charcoal-stripped fetal bovine serum (FBS). MCF-7 cells (3000 cells per 96-well) were seeded in DMEM medium supplemented with10% charcoal-stripped serum. Cells were treated with indicated concentrations of compounds. Treated cells were incubated at 37 °C for 7 days after which cell counting kit 8 reagent (DojinDo, CK04-11) was added to plates. Plates were then incubated at 37 °C for 1–4 h and the absorbance value was detected by microplate reader at 450 nm. Data were analyzed and plotted using Prism 8.0 software.
Colony formation assay
Cells were seeded in 12-well plates with 1000 cells per well in 1 ml of medium and incubated at 37 °C for 2 days. MDA-MB-453 and MDA-MB-415 cells were seeded in RPMI 1640 medium supplemented with 10% FBS. MCF-7 cells were seeded in DMEM medium supplemented with 10% FBS. Cells were then treated with indicated concentrations of compounds and incubated at 37 °C for 10 days. Colonies were then fixed with glutaraldehyde (6.0% v/v), stained with crystal violet (0.5% w/v).
Co-immunoprecipitation (Co-IP)
Co-IP was performed according to manufacturer’s instructions (Thermo Fisher Scientific, #88804). MDA-MB-453 cells were pretreated with charcoal-stripped FBS contained medium for 48 h. Cells were then collected after treatment with 1 nM R1881 alone or in combination with 1 µM ARD-61 or Enzalutamide for another 24 h, washed with PBS, and lysed with IP Lysis/Wash Buffer. Cell lysates were then centrifuged at 13,000 rpm for 10 min at 4 °C and supernatant was collected. For IP, 500 mg of total protein lysates was used for each IP condition, and 10% of the total lysates was used as the input. Cell lysates were mixed with 2–10 µg of IP antibody and incubated overnight at 4 °C to form the immune complex. 25 µl of Pierce protein A/G magnetic beads was added to lysates and incubated for 1 h at room temperature. The beads were then washed 3 times with IP Lysis/Wash Buffer and immunoprecipitated proteins were eluted by incubating with Elution Buffer for 10 min at room temperature and neutralized with Neutralization Buffer. Target proteins were detected by Western blot.
Nuclear and cytoplasmic extract preparation
MDA-MB-453 cells were treated with different compounds as indicated and cells were collected and washed twice with cold PBS. Cells were lysed with CREⅠbuffer plus PhoSTOP (Roche, 4906837001) and prostate inhibitor cocktail (Roche, 11697498001) on ice for 10 min and ice-cold CREⅡbuffer was added to the tube which was vortexed for 5 s on the highest setting, followed by incubation on ice for another 1 min. The supernatant after maximum speed centrifugation for 5 min is the cytoplasmic extract. The pellet fraction was then suspended in ice-cold NER on ice for 40 min and centrifuged at maximum speed for 10 min. The supernatant is the nuclear extract. The protein concentration was determined using the Bio-Rad protein assay and target proteins were determined by Western blot.
Cell cycle analysis by flow cytometry
MDA-MB-453, HCC1428 and MCF-7 cells were treated with different compounds as indicated and cells were collected, washed twice with cold PBS, fixed with 70% ethanol overnight at 4 °C and stained with PI (50 mg/ml, Sigma) plus 0.2 mg/ml DNase-free RNase A (Qiagen) for 30 min at rt. Cell cycle analysis was performed by the University of Michigan Flow Cytometry Core.
Apoptosis analysis by flow cytometry
MDA-MB-453, HCC1428 and MCF-7 cells were treated with compounds as indicated and cells were collected, washed twice with cold PBS, and stained with the cell apoptosis kit (Thermo Fisher Scientific, V13241) following manufacturer’s instructions. Apoptosis data were acquired on a flow cytometer and analyzed using Flowjo software.
In vivo PD and efficacy analysis
All in vivo studies were performed under an animal protocol (PRO00007499, PI: Shaomeng Wang) approved by the Institutional Animal Care & Use Committee (IACUC) of the University of Michigan, in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Male CB.17 SCID mice were injected subcutaneously with 5 × 106 MDA-MB-453 cells (ATCC) in 5 mg/ml Matrigel (Corning) for tumor growth. When tumors reached an average volume of 100 mm3, mice were randomized based upon their tumor sizes and assigned to different experimental groups. For PD study, drugs or vehicle control (20% PEG400 + 6% CremophorEL + 74% PBS) were given by intraperitoneal (IP) injection and tumor tissues were harvested at indicated time points for Western blotting analysis. For the in vivo efficacy experiment, drugs or vehicle control (20% PEG400 + 6% CremophorEL + 74% PBS) were administered daily by intraperitoneal (IP) injection. Tumor sizes and animal weights were measured 2 times per week. Tumor volume was calculated as: volume (mm3) = (length × width2)/2.
Statistical analyses
For tumor growth inhibition, cell cycle and apoptosis analyses, data were presented as mean ± SEM. For concentration–response cell growth, data were plotted as mean ± SD and fitted to a sigmoidal curve by nonlinear regression. When plotting dose–response fitting curves, DMSO controls were defined as the lowest doses of serial diluted compounds and set as 0 on x-axis. IC50 values were calculated using a nonlinear regression analysis of the mean ± SD from at least triplicates for each experiment. Differences in mean values between groups were analyzed by two-sided t test. All statistical analyses were performed using GraphPad Prism 8.0. The P values <0.05 are considered statistically significant.
Results
AR protein is expressed in both ER+ and ER− breast cancer cell lines
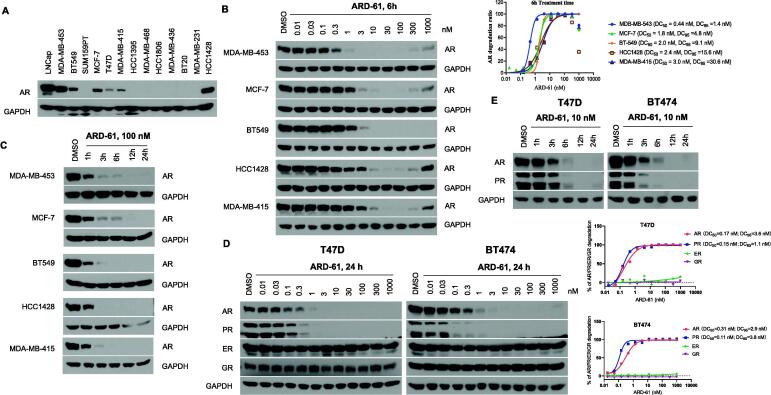
We first performed an immunoblotting analysis of AR protein levels in 13 representative breast cancer cell lines (Supplementary Table S1) with the AR+ LNCaP prostate cancer cell line included as a control, obtaining the data shown in Fig. 1A.
Fig. 1.
ARD-61 potently induces AR degradation in breast cancer cells. (A) Western blotting of AR expression in different breast cancer cell lines. (B) AR+ breast cancer cells were treated with different concentrations of ARD-61 for 6 h and AR protein levels were assessed by Western blot. Protein levels were quantified using ImageJ and AR degradation percentage is plotted using Prism 8.0 and GAPDH was used as the loading control. One representative AR degradation dose–response curve for each cell line is shown. (C) AR+ breast cancer cells were treated with 100 nM of ARD-61 for indicated time points and whole protein lysates were used to determine AR protein levels by Western blot. (D) T47D and BT474 cells were treated by indicated concentrations of ARD-61 for 24 h. AR, PR, GR and ER protein levels were detected by Western blotting and quantified using ImageJ. Relative AR, PR, GR and ER levels in relation to the GAPDH protein level in percentage are plotted against drug concentrations. One representative dose–response curve is shown for each cell line. (E) T47D and BT474 breast cancer cells were treated with 10 nM of ARD-61 for indicated time points. AR and PR protein levels were determined by Western blotting.
Among these cell lines, MDA-MB-453 and HCC1428 have the highest AR protein level, followed by BT-549 and MCF-7. T47D and MDA-MB-415 have detectable levels of AR protein. In comparison, SUM-159PT, HCC1395, MDA-MB-468, HCC1806, MDA-MB-436, BT20 and MDA-MB-231 have undetectable levels of AR protein. Hence, MDA-MB-453, HCC1428, BT-549, MCF-7, T47D and MDA-MB-415 were classified as AR+ breast cancer cell lines in our study.
Among these 6 AR+ breast cancer cell lines, MDA-MB-453 is characterized as HER2+ subtype (ER−/PR−), BT-549 is characterized as triple negative (TNBC), HCC1428, MCF-7, T47D and MDA-MB-415 are classified into Luminal A subtype (ER+/PR+/HER2−) (Table S1) [19]. Therefore, AR is expressed in both ER+ and ER− breast cancer cell lines.
ARD-61 potently and efficiently degrades AR in breast cancer cell lines
We evaluated ARD-61 for its ability to induce AR protein degradation in the MDA-MB-453, MCF-7, BT549, MDA-MB-415 and HCC1428 cell lines by Western blotting, with obtained data shown in Fig. 1B.
ARD-61 is highly potent and effective in reducing AR protein levels in each of these 5 AR+ breast cancer cell lines. With 6 h treatment time, ARD-61 achieves DC50 values (concentration needed to reduce AR protein by 50% over the DMSO control) of 0.44 nM, 1.8 nM, 2.0 nM, 2.4 nM and 3.0 nM in the MDA-MB-453, MCF-7, BT-549, HCC1428 and MDA-MB-415 cell lines, respectively. ARD-61 is also capable of achieving near complete AR depletion and shows DC95 values of 1.4 nM, 4.8 nM, 9.1 nM, 15.6 nM and 30.6 nM in the MDA-MB-453, MCF-7, BT-549, HCC1428 and MDA-MB-415 cell lines, respectively. In some of these cell lines, at higher concentrations (≥300 nM), reduction of AR protein by ARD-61 was less than that observed at lower concentrations (3–100 nM), which is an example of the classical “hook” effect observed with PROTAC degraders in cells [20], [21].
We next investigated AR degradation kinetics by ARD-61 in these AR+ breast cancer cell lines (Fig. 1C). When treated with 100 nM of ARD-61, the AR protein level was reduced by >50% with 1 h treatment and >90% with 3 h treatment in each of these 5 AR+ breast cancer cell lines. These data showed that AR degradation by ARD-61 is rapid in these AR+ breast cancer cell lines.
The T47D cell line expresses not only AR and ER but also progesterone receptor (PR) and glucocorticoid receptor (GR). We evaluated ARD-61 for its effect on AR, ER, PR and GR in the T47D cell line (Fig. 1D). Consistent with the AR degradation data in other 5 AR+ breast cancer cell lines, with a 24 h treatment time, ARD-61 potently and effectively reduces the AR protein level in the T47D cells with a DC50 of 0.17 nM and DC95 of 3.6 nM. Interestingly, ARD-61 is also highly potent and effective in reducing the level of PR protein with a DC50 value of 0.15 nM and a DC95 of 1.1 nM in the T47D cells. ARD-61 has no obvious effect on ER and GR proteins at concentrations of 0.01–1000 nM. We also tested the degradation kinetics of AR and PR proteins by ARD-61 in the T47D cells and found that ARD-61 rapidly reduces both AR and PR proteins with near complete depletion with 6 h treatment at 10 nM (Fig. 1E).
To further confirm the depletion of PR protein by ARD-61, we tested ARD-61 in the BT474 breast cancer cell line (Fig. 1D-E), which also expresses AR, PR, ER and GR proteins. ARD-61 is also highly potent and effective in causing AR and PR depletion in the BT474 cell line with DC50 values of 0.31 nM and 0.11 nM respectively, and DC95 values of 2.9 nM and 3.8 nM, respectively. A kinetic experiment showed that ARD-61 at 10 nM markedly reduces AR and PR levels with a 3 h treatment time and achieves near complete depletion with a 6 h treatment time.
Taken together, these data show that ARD-61 is a highly potent and effective AR and PR degrader in breast cancer cell lines.
ARD-61 is a bona fide PROTAC degrader in breast cancer cell lines
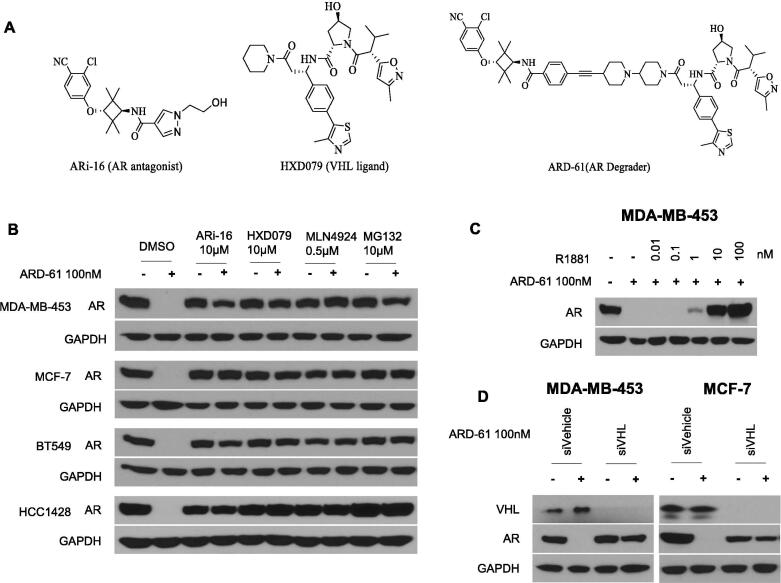
Based upon its PROTAC design, ARD-61 binds to AR protein through its AR antagonist portion and von Hippel-Lindau (VHL)/cullin 2 E3 ligase through its VHL ligand portion to recruit AR protein to cullin 2 for ubiquitination, followed by proteasome-dependent AR degradation. Accordingly, we examined the mechanism of AR degradation by ARD-61 in four representative AR+ breast cancer cell lines.
Consistent with its PROTAC design, an excess of ARi-16 (a potent AR antagonist used to design ARD-61), HXD079 (a potent VHL ligand used to design ARD-61) (Fig. 2A), MLN4924 (a neddylation activating E1 enzyme inhibitor), and MG132 (a proteasome inhibitor) all effectively block AR degradation by ARD-61 in each of the AR+ breast cancer cell lines examined (Fig. 2B).
Fig. 2.
ARD-61 is a bona fide PROTAC degrader in human breast cancer cell lines. (A) Chemical structures of AR degrader ARD-61, AR antagonist ARi-16 and VHL ligand used for the design of ARD-61. (B) Cells were pretreated with AR antagonist ARi-16, VHL ligand (HXD079), neddylation activating E1 enzyme inhibitor MLN4924 and proteasome inhibitor MG132 for 2 h, followed by treatment with ARD-61 for another 6 h. AR protein levels were detected by Western blot and GAPDH was used as a loading control. (C) MDA-MB-543 cells were treated with ARD-61 or in combination with an AR agonist R1881 for 24 h. AR protein levels were detected by Western blot and GAPDH was used as a loading control. (D) MDA-MB-453 and MCF-7 cells were transfected with siVehicle (Control) or siVHL for 72 h and then treated with 100 nM of ARD-61 for another 24 h. AR protein levels were detected by Western blot and GAPDH was used as a loading control.
Using metribolone (R1881), a highly potent AR agonist, we further investigated if AR degradation induced by ARD-61 requires its binding to AR protein. R1881 is able to effectively block AR depletion induced by ARD-61 in the MDA-MB-453 cells (Fig. 2C), further supporting the requirement of AR binding for induced AR degradation by ARD-61.
To examine further the role of VHL in AR degradation by ARD-61, we knocked down VHL in the MDA-MB-453 and MCF-7 cell lines. Western blotting showed that efficient knock-down of VHL completely blocks AR degradation induced by ARD-61 in both MDA-MB-453 and MCF-7 cell lines (Fig. 2D).
Hence, AR degradation induced by ARD-61 in breast cancer cells requires its binding to AR and VHL and is neddylation- and proteasome-dependent. ARD-61 is therefore a bona fide PROTAC AR degrader in breast cancer cells.
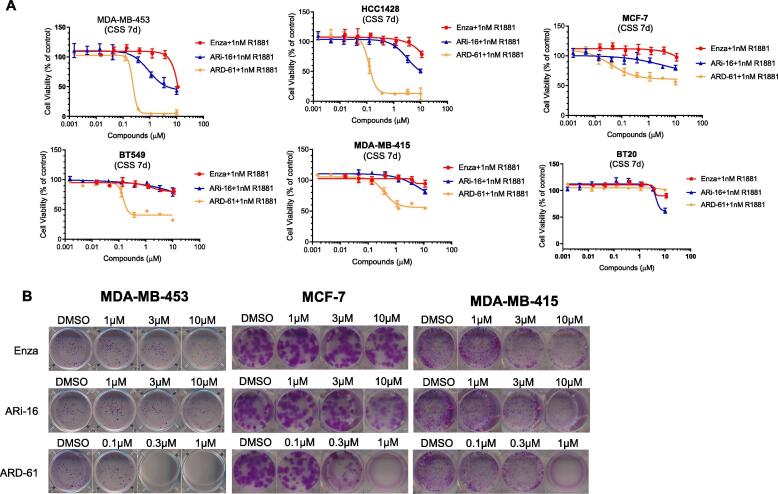
ARD-61 selectively inhibits cell growth in AR+ breast cancer cells
We evaluated the anti-proliferative activity of ARD-61 in a panel of AR+ breast cancer cell lines, with enzalutamide and ARi-16 included as the controls (Fig. 3A and Supplementary Table S2).
Fig. 3.
ARD-61 is much more potent than enzalutamide and ARi-16 in cell growth inhibition in AR+ breast cancer cell lines. (A) Breast cancer cells were pretreated in charcoal-stripped serum (CSS) contained medium for 72 h and then treated with ARD-61, enzalutamide (Enza) or ARi-16 for 7 days with the stimulation of 1 nM R1881. Cell growth was determined by a WST-8 assay. Data are represented as mean ± SEM (n = 3). (B) Breast cancer cells were seeded in 12-well plates and treated with ARD-61, enzalutamide (Enza) or ARi-16 for 10 days in regular medium. Cells were stained with purple crystal.
In the MDA-MB-453 and HCC1428 cell lines, which have the highest AR expression, ARD-61 achieves near complete inhibition of cell growth, and has IC50 values of 235 nM and 121 nM, respectively. In the MCF-7, BT-549 and MDA-MB-415 cell lines, which have a moderate level of AR protein, ARD-61 demonstrates partial cell growth inhibition, delivering IC50 values of 39, 147, and 380 nM, respectively. In comparison, enzalutamide and ARi-16 are much less potent than ARD-61 in each of these 5 AR+ breast cancer cell lines tested. Enzalutamide has a minimal activity in inhibition of cell growth in these cell lines at concentrations up to 10 µM and ARi-16 is >100-times less potent than ARD-61 in each of these 5 cell lines. These data show that the AR degrader ARD-61 is much more potent and effective in inhibition of cell growth in AR+ breast cancer cell lines than AR antagonists. In addition, ARD-61 also effectively decreased colony formation of MDA-MB-453, MCF-7 and MDA-MB-415 cells in full serum conditions at concentrations as low as 100 nM and is also much more effective than enzalutamide and ARi-16 (Fig. 3B).
To examine the specificity of ARD-61, we tested its cell growth inhibition in the BT-20 cell line which has an undetectable level of AR protein (Fig. 3A). Our data showed that ARD-61 has a minimal effect at concentrations up to 10 µM.
Taken together, these data show that ARD-61 potently and selectively inhibits cell growth in AR+ breast cancer cell lines and is much more potent and effective than AR antagonists.
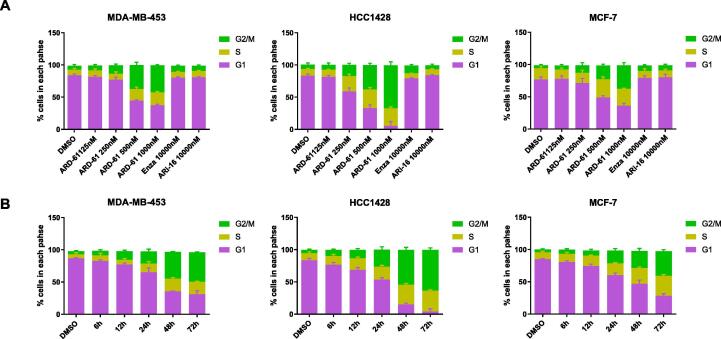
ARD-61 induces G2/M cell cycle arrest in AR+ Breast cancer cell lines
To shed light on the mode of action of cell growth inhibition by ARD-61, we performed cell cycle analysis in the MDA-MB-453, HCC1428 and MCF-7 cell lines by flow cytometry, with enzalutamide and ARi-16 included as controls (Fig. 4A-B and Supplementary Fig. S1).
Fig. 4.
ARD-61 effectively induces G2/M phase cell cycle arrest in AR+ breast cancer cell lines. (A) MDA-MB-453, HCC1428 and MCF-7 cells were treated with different concentrations of ARD-61, 10 µM enzalutamide (Enza) or 10 µM AR antagonist ARi-16 for 72 h. (B) MDA-MB-453, HCC1428 and MCF-7 cells were treated with 1 µM ARD-61 for indicated time points. Cell cycle data were acquired on a flow cytometer and analyzed using Flowjo software. Percentages of different cell cycle phases were plotted with Prism 8.0. Data are represented as mean ± SEM (n = 3).
Our cell cycle analysis showed that ARD-61 induces G2/M cell cycle arrest in a dose- and time-dependent manner in each of these three AR+ breast cancer cell lines. ARD-61 induces profound G2/M arrest at 0.5 µM in the MDA-MB-453 and HCC1428 cell lines and at 1 µM in the MCF-7 cell line, with a 3-day treatment time. In comparison, enzalutamide and ARi-16 have minimal effect in these three cell lines at 10 µM. Hence ARD-61 is much more potent and effective than enzalutamide and ARi-16 in inducing cell cycle arrest in these three representative AR+ cell lines.
ARD-61 induces apoptosis in some AR+ breast cancer cell lines
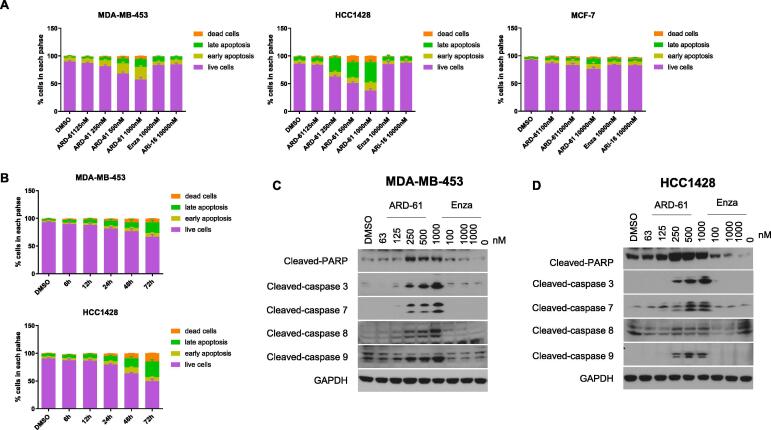
The near complete cell growth inhibition by ARD-61 in the MDA-MB-453 and HCC1428 cell lines suggested induction of cell death. We performed a flow cytometry analysis of apoptosis induction by ARD-61 in the MDA-MB-453, HCC1428 and MCF-7 cell lines with enzalutamide and ARi-16 included as controls. The results showed that ARD-61 induces apoptosis in the MDA-MB-453 and HCC1428 cell lines in a dose-dependent manner (Fig. 5A and Supplementary Fig. S2A). ARD-61 induces robust apoptosis at 0.5 µM in the MDA-MB-453 cell line and at 0.25 µM in the HCC1428 cell line with a 3-day treatment time. Consistent with its partial cell growth inhibition, ARD-61 induces minimal apoptosis at concentrations up to 10 µM in the MCF-7 cell line with a 3-day treatment time. In comparison, enzalutamide and ARi-16 induce minimal apoptosis in the MDA-MB-453, HCC1428 and MCF-7 cell lines at 10 µM with a 3-day treatment time. Therefore, ARD-61 is much more potent and effective in inducing apoptosis in the MDA-MB-453 and HCC1428 cell lines than AR antagonists.
Fig. 5.
ARD-61 induces apoptosis in AR+ breast cancer cell lines with high levels of AR expression. (A) MDA-MB-453, HCC1428 and MCF-7 cells were treated with different concentrations of ARD-61, 10 µM enzalutamide (Enza) or 10 µM AR antagonist ARi-16 for 72 h. (B) MDA-MB-453 and HCC1428 cells were treated with 1 µM ARD-61 for indicated time points. For apoptosis analysis, data were acquired on a flow cytometer and analyzed using Flowjo software. Percentages of live and apoptosis cells were plotted using Prism 8.0. Data were presented as mean ± SEM (n = 3). (C, D) MDA-MB-453 and HCC1428 cells were treated with different concentrations of ARD-61 or enzalutamide (Enza) for 48 h. Whole protein lysates were used for Western blot analysis.
We also examined the kinetics of apoptosis induction by ARD-61 in the MDA-MB-453 and HCC1428 cell lines by flow cytometry (Fig. 5B and Supplementary Fig. S2B). ARD-61 induces apoptosis in a time-dependent manner, with robust apoptosis observed with a 48 h treatment time in both cell lines.
Western blotting analysis (Fig. 5C-D) showed that ARD-61 induces cleavage of caspase-8, −9, −3, and −7 and PARP in a dose-dependent manner in the MDA-MB-453 and HCC1428 cell lines, with a clear effect at concentrations as low as 0.25 µM. These data indicated that both intrinsic and extrinsic apoptosis pathways are activated in the induction of apoptosis by ARD-61 in the MDA-MB-453 and HCC1428 cells. Consistent with its failure to induce apoptosis, enzalutamide has minimal effect on the cleavage of caspase-8, −9, −3, and −7 and PARP in both cell lines at 1 or 10 µM.
ARD-61 effectively inhibits Wnt/β-catenin and MYC signaling pathways
In AR+/ER− breast cancer cells, AR activation by androgens leads to upregulation of the WNT7B transcriptional factor, which in turns activates the Wnt/β-catenin signaling pathway and enhances c-Myc expression and its gene transcription activity [1], [3]. Activation of the Wnt/β-catenin signaling pathway or c-Myc stimulates the formation of HER2/HER3 heterodimers and activation of the down-stream PI3K/AKT pathway [1], [22], [23], promoting cell proliferation [3]. Because of the important roles of the Wnt/β-catenin and c-Myc signaling pathways in breast cancer, we investigated the effect of ARD-61 on Wnt/β-catenin and c-Myc in AR+/ER− MDA-MB-453 cells, with enzalutamide included as an AR antagonist control.
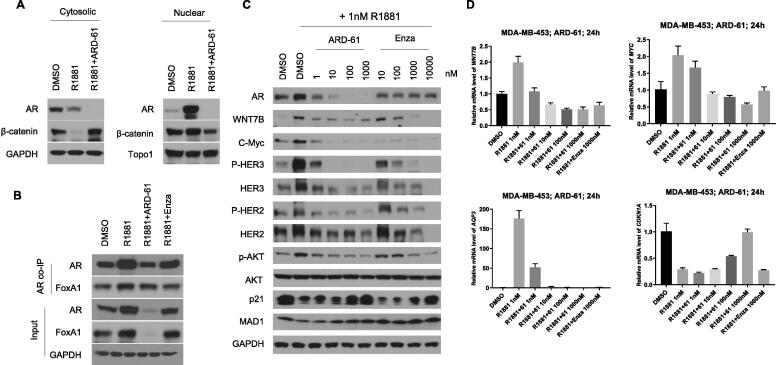
In MDA-MB-453 cells, R1881, an AR agonist, induces translocation of AR and β-catenin from cytoplasm to the nucleus and depletion of AR by ARD-61 effectively blocks translocation of β-catenin (Fig. 6A). R1881 also robustly induces upregulation of AR and FoxA1, which are effectively inhibited by ARD-61 (Fig. 6B). R1881 elevates the expression of WNT7B and MYC at both mRNA (Fig. 6D) and protein levels (Fig. 6C), which are effectively blocked by ARD-61. Interestingly, ARD-61 not only decreases both phosphorylated HER2 and HER3, but also un-phosphorylated HER2 and HER3 proteins. While the phosphorylated AKT level is down-regulated by ARD-61, the total AKT level is not affected. MAD1 was shown to be a negative regulator of the transcriptional activity of c-Myc by binding to MAX, a co-regulator of c-Myc. While R1881 reduces the level of MAD1 protein, ARD-61 restores the MDA1 protein level reduced by R1881 (Fig. 6C). AQP3, which is an AR target gene [1], is upregulated by R1881 but reduced to the control level by ARD-61. Furthermore, ARD-61 restores R1881-induced downregulation of p21, a cell cycle regulator, at both mRNA and protein levels (Fig. 6C–D). In comparison, while enzalutamide is similarly effective on the Wnt/β-catenin and c-Myc signaling pathways in the MDA-MB-453 cells as compared to ARD-61, it is at least 100-times less potent than ARD-61.
Fig. 6.
ARD-61 blocks AR signaling and represses AR-target genes expression in MDA-MB-453 cells. (A) MDA-MB-453 cells were pretreated with charcoal-stripped serum (CSS) contained medium for 72 h, and cells were then treated with 1 nM AR agonist R1881 alone or in combination with 1 µM ARD-61 for 24 h. Cytosolic and nuclear proteins were extracted and AR and β-catenin proteins were detected by Western blotting. GAPDH and Topo1 were used as the loading control for cytosolic and nuclear protein, respectively. (B) MDA-MB-453 cells were pretreated with charcoal-stripped serum (CSS) contained medium for 72 h, and cells were then treated with 1 nM AR agonist R1881 alone or in combination with 1 µM ARD-61 or 1 µM Enzalutamide (Enza) for 24 hr. Whole protein lysates were used for co-IP assay. (C) MDA-MB-453 cells were pretreated with charcoal-stripped serum (CSS) contained medium for 72 h. Cells were then treated with 1 nM AR agonist R1881 alone or in combination with different concentrations of ARD-61 or Enzalutamide (Enza) for 24 hr. Whole protein lysates were used to analyze the levels of AR signaling proteins by Western blot and GAPDH was used as the loading control. (D) MDA-MB-453 cells were pretreated with charcoal-stripped serum (CSS) contained medium for 72 h. Cells were then treated with 1 nM AR agonist R1881 alone or in combination with different concentrations of ARD-61 for 24 h. Expression of AR targeted genes were analyzed by qPCR and normalized to GAPDH. Data are mean ± SEM (n = 3).
Hence, ARD-61 is highly potent and effective in blocking activation of the Wnt/β-catenin and c-Myc signaling pathways and the activity of HER2/HER3 in MDA-MB-453 cells induced by an AR agonist.
ARD-61 effectively degrades AR protein in xenograft tumor tissue and inhibits AR signaling in vivo
We evaluated the ability of ARD-61 to degrade AR protein in the MDA-MB-453 xenograft tumor tissue in mice. Because MDA-MB-453 tumors grew very poorly in female mice, the xenograft tumors were grown in male SCID mice. Since ARD-61 was not designed as an oral bioavailable AR degrader, it was administered by intraperitoneal (IP) injection in this study.
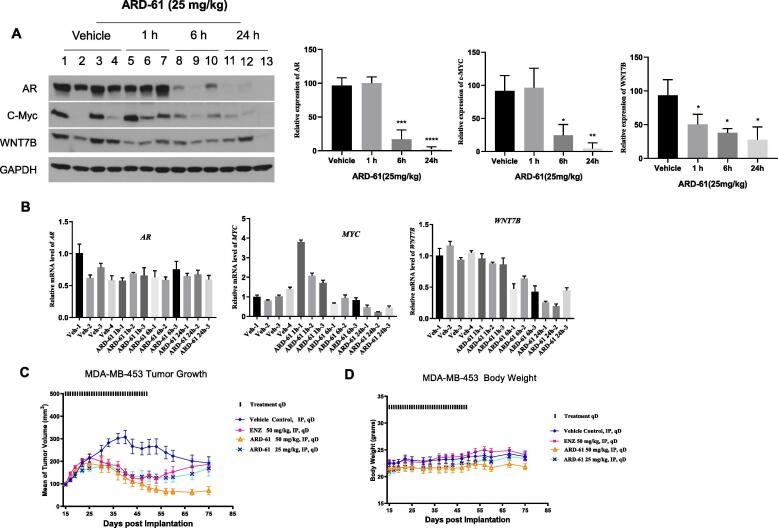
Western blotting (Fig. 7A) showed that a single dose of ARD-61 at 25 mg/kg effectively and rapidly reduces the AR protein in the MDA-MB-453 xenograft tissue, with the effect persisting for at least 24 h. qRT-PCR analysis of the tumor samples showed that ARD-61 has minimal effect on the mRNA level of AR (Fig. 7B), consistent with our in vitro data and its mode of action.
Fig. 7.
Pharmacodynamic (PD) analysis and antitumor activity of ARD-61 in the MDA-MB-453 xenograft tumor model. (A) Male SCID mice bearing MDA-MB-453 tumors were treated with a single dose of ARD-61 at 25 mg/kg by intraperitoneal injection. Mice were sacrificed and tumor tissues were collected at indicated time points for PD analysis. Data are mean ± SEM (n = 3). P values were calculated using Student’s t test. (B) Male SCID mice bearing MDA-MB-53 tumors were treated with a single dose of ARD-61 at 25 mg/kg by intraperitoneal injection. Mice were sacrificed and tumor tissues were collected at indicated time points. Total RNAs were isolated from tumor lysates for qPCR. Data are mean ± SEM (n = 3). (C, D) Male SCID mice bearing the MDA-MB-453 tumors were treated with different drugs at indicated dose-schedules. Data are mean ± SEM (n = 8).
In addition to AR, we analyzed the effect of ARD-61 on c-Myc and WNT7B, two transcriptional factors. Western blotting analysis (Fig. 7A) showed that ARD-61 effectively reduces the level of both c-Myc and WNT7B proteins at the 6 and 24 h time-points. qRT-PCR analysis (Fig. 7B) showed though the mRNA level of c-Myc was increased transiently by ARD-61 at the 1 h time-point, it is significantly down-regulated at the 24 h time-point. ARD-61 is very effective in reducing the mRNA level of WNT7B in a time-dependent manner, consistent with the kinetics of AR degradation in the tumor tissue.
Taken together, these data show that ARD-61 effectively depletes AR protein in the MDA-MB-453 tumor tissue, leading to transcriptional down-regulation of both c-Myc and WNT7B.
ARD-61 strongly inhibits growth of MDA-MB-453 tumors in mice
Next, we tested the antitumor activity of ARD-61 in the MDA-MB-453 xenograft tumor model in male SCID mice with enzalutamide included as a control.
Our efficacy experiment showed that ARD-61 effectively inhibits tumor growth at both 25 and 50 mg/kg doses (Fig. 7C). While ARD-61 completely inhibits tumor growth at 25 mg/kg, it is capable of inducing partial tumor regression at 50 mg/kg. Importantly, after the treatment of ARD-61 was stopped, persistent tumor growth inhibition was observed with ARD-61 at 50 mg/kg. In comparison, enzalutamide at 50 mg/kg IP dose had an antitumor activity which is very similar with that of ARD-61 at 25 mg/kg. No animal weight loss or other signs of toxicity was observed during the entire experiment for both ARD-61 and enzalutamide (Fig. 7D).
Hence, our efficacy experiment showed that while both ARD-61 and enzalutamide inhibit tumor growth in the AR+ MDA-MB-453 model, ARD-61 is more effective than enzalutamide in achieving long-term tumor growth inhibition.
Discussion
Induced protein degradation by bi-functional PROTAC molecules is being pursued as a new therapeutic strategy [13], [14]. A PROTAC small-molecule degrader has a number of important advantages over traditional small-molecule inhibitors. First, by depleting a protein in cells and tissues, a PROTAC degrader can inhibit all the functions or activity associated with the target protein. Second, a PROTAC small-molecule degrader can achieve an exceedingly high potency in inducing degradation of the target protein through its catalytic nature. Third, a PROTAC degrader can achieve a very high selectivity by inducing degradation of a single protein or very fewer target proteins.
We have recently reported the design of ARD-61 as a PROTAC degrader of AR. In our previous study [16], we demonstrated that ARD-61 is highly potent, effective and specific in inducing degradation of AR protein in human prostate cancer cell lines in vitro [16]. Furthermore, ARD-61 is much more potent than AR antagonists in suppressing the AR function in AR+ prostate cancer cells and in inhibition of cell growth [16]. Our previous data also demonstrated that ARD-61 is effective in inducing AR degradation in tumor tissue and in inhibition of tumor growth in AR+ prostate cancer xenograft models in vivo, including prostate cancer models resistant to enzalutamide [16].
Although AR antagonists have been developed primarily for the treatment of AR+ metastatic and non-metastatic prostate cancer, previous preclinical and clinical studies have provided evidence that AR antagonists may also have the therapeutic potential for the treatment of a subset of AR+ breast cancer [1], [3], [8], [24], [25]. In the present study, we evaluated ARD-61 for its therapeutic potential and mechanism of action in AR+ breast cancer cell lines in vitro and in vivo.
Consistent with our previous study in AR+ prostate cancer models [16], ARD-61 was highly potent and effective in inducing degradation of AR protein in all of the AR+ breast cancer cell lines tested. ARD-61 achieves DC50 values of 0.44–3.0 nM and DC95 values of 1.4–31 nM in a total of 7 AR+ breast cancer cell lines evaluated. Induction of AR degradation by ARD-61 is rapid with nearly complete AR degradation achieved within 3–6 h treatment time in all these AR+ breast cancer cell lines. Our mechanistic investigation showed that ARD-61 functions as a bona fide PROTAC degrader, through AR- and VHL-binding and proteasome- and neddylation dependent mechanisms.
ARD-61 potently and effectively inhibits cell growth in each of these AR+ breast cancer cell lines but with some notable differences among these AR+ cell lines. In the MDA-MB-453 and HCC1428 cell lines with the highest levels of AR expression, ARD-61 demonstrates nearly complete cell growth inhibition. In comparison, in three other AR+ cell lines (MCF-7, BT-549 and MDA-MB-415), ARD-61 only shows partial cell growth inhibition. Our flow cytometry analysis showed that while ARD-61 effectively induces G2/M cell cycle arrest in all these AR+ breast cancer cell lines, it induces robust apoptosis in the MDA-MB-453 and HCC1428 cell lines but not in the MCF-7 cell line. ARD-61 induces cleavage of caspase-8, −9, −3 and −7, suggesting that both intrinsic and extrinsic apoptosis pathways are activated by ARD-61. Further studies will be performed to define the precise mechanism of apoptosis induction in these AR+ breast cancer cell lines.
AR activation has been shown to induce expression of c-Myc and WNT proteins, leading to activation of HER2/HER3 and AKT in AR+ breast cancer cells [1], [3], [22], [26]. Our initial mechanistic investigation confirmed that AR activation by the AR agonist R1881 induces robust upregulation of c-Myc and WNT7B proteins in the MDA-MB-453 cell line. ARD-61 is highly effective and potent in blocking upregulation of c-Myc and WNT7B proteins induced by R1881. Of interest, ARD-61 not only inhibits phosphorylation of HER2 and HER3 but also down-regulates their protein levels. ARD-61 is also effective in reducing phosphorylation of AKT. While R1881 induces AR translocation from cytoplasm to nucleus, ARD-61 completely blocks AR translocation. Although enzalutamide is also effective in blocking AR signaling in the MDA-MB-453 cells, ARD-61 is >100-times much more potent than enzalutamide.
While ARD-61 achieves DC95 values in low nanomolar concentrations in these AR+ breast cancer cell lines, it has IC50 values in sub-micromolar concentrations in inhibition of cell growth. Because AR is a transcriptional factor, AR regulates cell growth, cell cycle and apoptosis through its targeted genes. Because AR functions as a transcriptional factor by binding to targeted DNAs in the nucleus, we examined the degradation of AR protein in both cytoplasm and nucleus by ARD-61. Our data showed that ARD-61 is very effective in reducing the levels of AR protein in both cytoplasm and nucleus in the MDA-MB-453 cells (Fig. 6A). We further evaluated the effect of ARD-61 on a number of genes known to be regulated by AR such as C-MYC, WNT7B and CDKN1A (Fig. 6D). Our data showed that while it only took 1 nM of ARD-61 to inhibit the increase of C-MYC induced by R1881, 10 nM of ARD-61 is needed to effectively suppress the increase of WNT7B. Moreover, it took at least 100 nM of ARD-61 to partially restore the decrease of CDKN1A (p21), a key cell cycle regulator, induced by R1881 and 1000 nM of ARD-61 to completely restore the decrease of CDKN1A. The potency of ARD-61 on CDKN1A (p21) is consistent with its potency in inhibition of cell growth.
Our in vivo PD experiments showed that a single dose of ARD-61 effectively depletes AR protein and suppresses the protein levels of c-Myc and WNT7B in the MDA-MB-453 xenograft tumor tissue in mice. Significantly, ARD-61 strongly inhibits tumor growth and is capable of achieving long-term tumor growth inhibition and partial tumor regression. In direct comparison, ARD-61 is more effective than enzalutamide in inhibition of tumor growth at the same dose. Consistent with our previous study [16], ARD-61 is well tolerated in mice.
Although our current study focuses on testing the ability of ARD-61 to degrade AR protein in breast cancer models, we also found that ARD-61 is equally potent and effective in inducing degradation of PR in the BT474 and T47D breast cancer cell lines, with DC95 values of 3.8 nM and 1.0 nM, respectively. ARD-61 fails to induce degradation of ER and GR in the BT474 and T47D breast cancer cell lines, highlighting its specificity. The potent and effective degradation of PR by ARD-61 provides an exciting opportunity to investigate the therapeutic potential of PR degradation in human breast cancer.
Currently, ARV-110, a PROTAC AR degrader discovered by scientists from Arvinas, is being evaluated in Phase 1 human clinical trials (ClinicalTrials.gov Identifier: NCT03888612) as a new therapy for the treatment of patients with castration-resistant AR+ prostate cancer after failed current AR targeted therapies such as AR antagonist enzalutamide and abiraterone, a CYP17A1 inhibitor blocking androgen synthesis. Our present study suggests that AR degraders should also be evaluated in the clinic as a new therapeutic strategy for the treatment of AR+ breast cancer.
Acknowledgments
Acknowledgement
This study is supported in part by a research contract from Oncopia Therapeutics Inc. and from NIH P30 CA046592, the University of Michigan Comprehensive Cancer Center (now Rogel Cancer Center) Core grant from the National Cancer Institute (NCI), NIH.
Author contributions
L. Zhao conceived, designed and performed cell biology experiments, analyzed the data and wrote the paper; X. Han designed and synthesized ARD-61, ARi-16 and HXD079 used in this study and wrote the paper; J. Lu performed PD experiments and analyzed the data and wrote the paper; D. McEachern designed performed the in vivo PD and efficacy experiments and analyzed the data and wrote the paper; S. Wang conceived and designed experiments and supervised all the studies and wrote the paper.
Competing interests
The authors declare the following competing financial interest(s): The University of Michigan has filed a patent application on AR-61 and its analogues, which has been licensed by Oncopia Therapeutics Inc. S. Wang, and X. Han are co-inventors on the patent application. The University of Michigan has received a research contract from Oncopia Therapeutics. S.W. is a co-founder of Oncopia, owns shares in Oncopia and is a paid consultant to Oncopia. The University of Michigan also owns shares in Oncopia.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.07.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ni M., Chen Y., Lim E., Wimberly H., Bailey S.T., Imai Y. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20:119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higgins M.J., Wolff A.C. The androgen receptor in breast cancer: learning from the past. Breast Cancer Res Treat. 2010;124:619–621. doi: 10.1007/s10549-010-0864-5. [DOI] [PubMed] [Google Scholar]
- 3.Pietri E., Conteduca V., Andreis D., Massa I., Rocca A. Androgen receptor signaling pathways as a target for breast cancer treatment. Endocr-Relat Cancer. 2016;23:485–498. doi: 10.1530/ERC-16-0190. [DOI] [PubMed] [Google Scholar]
- 4.Collins L.C., Cole K.S., Marotti J.D., Hu R., Schnitt S.J., Tamimi R.M. Androgen receptor expression in breast cancer in relation to molecular phenotype: Results from the Nurses' Health Study(Article) Modern Pathol. 2011:924–931. doi: 10.1038/modpathol.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niemeier L.A., Dabbs D.J., Beriwal S., Striebel J.M., Bhargava R. Androgen receptor in breast cancer: Expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Modern Pathol. 2010;23:205–212. doi: 10.1038/modpathol.2009.159. [DOI] [PubMed] [Google Scholar]
- 6.Bilal R., Ruth O.R. AR signaling in breast Cancer. Cancers. 2017;9:21. doi: 10.3390/cancers9030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsang J.Y.S., Ni Y.B., Chan S.K., Shao M.M., Law B.K.B., Tan P.H. Androgen receptor expression shows distinctive significance in ER positive and negative breast cancers. Ann Surg Oncol. 2014;21:2218–2228. doi: 10.1245/s10434-014-3629-2. [DOI] [PubMed] [Google Scholar]
- 8.Gucalp A., Tolaney S., Isakoff S.J., Ingle J.N., Liu M.C., Carey L.A. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic. Breast Cancer Clin Cancer Res. 2013;19:5505–5512. doi: 10.1158/1078-0432.CCR-12-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traina T.A., O'Shaughnessy J., Nanda R., Schwartzberg L., Abramson V., Cortes J. Preliminary results from a phase 2 single-arm study of enzalutamide, an androgen receptor (AR) inhibitor, in advanced AR+ triple-negative breast cancer (TNBC) Cancer Res. 2015;75(9 Suppl) [Google Scholar]
- 10.Joseph J.D., Lu N., Qian J., Sensintaffar J., Shao G., Brigham D. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov. 2013;3:1020–1029. doi: 10.1158/2159-8290.CD-13-0226. [DOI] [PubMed] [Google Scholar]
- 11.Balbas M.D., Evans M.J., Hosfield D.J., Wongvipat J., Arora V.K., Watson P.A. Overcoming mutation-based resistance to antiandrogens with rational drug design. eLife. 2013;2 doi: 10.7554/eLife.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neklesa T.K., Winkler J.D., Crews C.M. Targeted protein degradation by PROTACs. Pharmacol Therapeut. 2017;174:138–144. doi: 10.1016/j.pharmthera.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 13.Burslem G.M., Crews C.M. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell. 2020;181:102–114. doi: 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schapira M., Calabrese M.F., Bullock A.N., Crews C.M. Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov. 2019;18:949–963. doi: 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- 15.Toure M., Crews C.M. Small-molecule PROTACS: new approaches to protein degradation. Angew Chem Int Ed. 2016;55:1966–1973. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- 16.Kregel S., Wang C., Han X., Xiao L., Fernandez-Salas E., Bawa P. Androgen receptor degraders overcome common resistance mechanisms developed during prostate cancer treatment. Neoplasia. 2020;22:111–119. doi: 10.1016/j.neo.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X., Wang C., Qin C., Xiang W., Fernandez-Salas E., Yang C.Y. Discovery of ARD-69 as a Highly Potent Proteolysis Targeting Chimera (PROTAC) Degrader of Androgen Receptor (AR) for the Treatment of Prostate Cancer. J Med Chem. 2019;62:941–964. doi: 10.1021/acs.jmedchem.8b01631. [DOI] [PubMed] [Google Scholar]
- 18.Han X., Zhao L., Xiang W., Qin C., Miao B., Xu T., Wang M., Yang C.Y., Chinnaswamy K., Stuckey J. Discovery of highly potent and efficient PROTAC degraders of androgen receptor (AR) by employing weak binding affinity VHL E3 ligase ligands. J Med Chem. 2019;62:11218–11231. doi: 10.1021/acs.jmedchem.9b01393. [DOI] [PubMed] [Google Scholar]
- 19.Dai X., Cheng H., Bai Z., Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer. 2017;8:3131–3141. doi: 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X., Dixit V.M. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 2016;26:484–498. doi: 10.1038/cr.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglass E.F., Jr., Miller C.J., Sparer G., Shapiro H., Spiegel D.A. A comprehensive mathematical model for three-body binding equilibria. J Am Chem Soc. 2013;135:6092–6099. doi: 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J., Blenis J., Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci. 2008;105:6584–6589. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou C.K., Lee D.F., Sun H.L., Li L.Y., Lin C.Y., Huang W.C. The suppression of MAD1 by AKT-mediated phosphorylation activates MAD1 target genes transcription. Mol Carcinogenesis. 2009;48:1048–1058. doi: 10.1002/mc.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traina T.A., Miller K., Yardley D.A., Eakle J., Schwartzberg L.S., O'Shaughnessy J. Enzalutamide for the Treatment of Androgen Receptor-Expressing Triple-Negative Breast Cancer. J Clin Oncol. 2018;36:884–890. doi: 10.1200/JCO.2016.71.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alain M., Rachel Y., Priyanka S. Targeting the androgen receptor in triple-negative breast cancer: current perspectives. Oncotargets Therapy. 2017;10:4675–4685. doi: 10.2147/OTT.S126051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni M., Chen Y., Fei T., Li D., Lim E., Liu X.S., Brown M. Amplitude modulation of androgen signaling by c-MYC. Genes Dev. 2013;27:734–748. doi: 10.1101/gad.209569.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.