Abstract
The hypoxic microenvironment, an important feature of solid tumors, promotes tumor cells to release exosomes and enhances tumor angiogenesis. However, the detailed functions of hypoxic exosomes and the mechanisms underlying their effects in pancreatic cancer (PC) remain mysterious. Here, we observed that hypoxic exosomes derived from PC cells promoted cell migration and tube formation of human umbilical vein endothelial cells (HUVECs). The long noncoding RNA (lncRNA) UCA1, a key factor, was highly expressed in exosomes derived from hypoxic PC cells and could be transferred to HUVECs through the exosomes. In addition, the expression levels of UCA1 in exosomes derived from PC patients’ serum were higher than in healthy controls and were associated with poor survival of PC patients. Moreover, hypoxic exosomal UCA1 could promote angiogenesis and tumor growth both in vitro and in vivo. With respect to the functional mechanism, UCA1 acted as a sponge of microRNA (miR)-96-5p, relieving the repressive effects of miR-96-5p on the expression of its target gene AMOTL2. Collectively, these results indicate that hypoxic exosomal UCA1 could promote angiogenesis and tumor growth through the miR-96-5p/AMOTL2/ERK1/2 axis and therefore, serve as a novel target for PC treatment.
Keywords: hypoxia, exosomes, lncRNA-UCA1, ceRNA, angiogenesis, pancreatic cancer
Graphical Abstract

Qiu, Guo, and other colleagues reported that lncRNA UCA1 was upregulated in exosomes secreted by hypoxic pancreatic cancer cells and could be transferred to HUVECs, promoting angiogenesis via mediating the miR-96-5p/AMOTL2/ERK1/2 axis. Additionally, exosomal UCA1 levels in human serum were associated with a late TNM stage and poor survival in patients with pancreatic cancer.
Introduction
Pancreatic cancer (PC) is a highly lethal malignancy and is notorious for its extremely poor prognosis worldwide. The 5-year relative survival rate of PC patients is about 8% and decreases to 3% when diagnosed at a distant stage.1 Angiogenesis, which is involved in tumor growth, invasion, and metastasis, comprises several complex steps and plays an important role in PC and most malignant tumors.2 Although PC is not a hypervascular neoplasm, it often exhibits enhanced foci of endothelial cell proliferation in the tumor tissues.3 Several studies have also reported a positive correlation between blood vessel density and PC progression.3,4 Furthermore, anti-angiogenic treatment efficiently prevented tumor growth and metastasis and decreased the tumor burden in an orthotopic mouse model of PC.5,6
Hypoxia, or low oxygen tension, has been widely acknowledged as a specific feature of the tumor microenvironment and is involved in tumor aggressiveness and metastasis.7 Tumor hypoxia induces adaptive mechanisms that rely on the stabilization and activation of hypoxia-inducible factors (HIFs), which induce the transcription of various genes that contribute to angiogenesis, metabolic reprogramming, metastasis, and immune evasion.8,9 A recent report has demonstrated that the hypoxia-induced protein MUC1 enhances angiogenesis through the upregulation of several proangiogenic factors in PC.10 However, whether the hypoxic microenvironment plays a role in modulating angiogenesis in PC and the underlying mechanisms in PC needs to be further elucidated.
Exosomes, which are secreted by a variety of cell types and tumor cells in particular, are single-membrane vesicles with a diameter of 30∼200 nm.11 Typical exosomes contain various types of nucleic acids, including mRNAs, microRNAs (miRNAs), and long noncoding RNAs (lncRNAs), and are released into the extracellular space and enter the circulation. They play an important role in intercellular communication through transferring genetic material.12,13 Recent studies have reported that hypoxia might promote tumor progression via stimulating exosomal secretion or regulating the content of exosomes.14,15 Exosomes released from hypoxic tumor cells contribute to the development of cancer through promoting cancer cell invasion and metastasis, enhancing cancer angiogenesis, and influencing the cancer-immune system interactions.16,17 For example, the exosomal lncRNA CCAT2, derived from glioma cells, enhances angiogenesis and inhibits endothelial cell apoptosis.18 However, whether hypoxic exosomes secreted by PC cells can regulate angiogenesis remains unknown.
LncRNAs are defined as a class of noncoding RNAs that are generally longer than 200 nucleotides with no evident protein-coding capacity.19 An increasing body of evidence has suggested that lncRNAs are abnormally expressed in cancer cells and are reported to modulate tumor growth, invasion, and metastasis.20 Recently, numerous lncRNAs have been reported to be involved in tumor angiogenesis.21, 22, 23 Interestingly, to adapt to the hypoxic microenvironment, numerous lncRNAs are highly expressed by tumor cells, packed into exosomes, and transported to the corresponding recipient cells, thus playing an important role in intercellular communication. For example, tumor cells may generate UCA1-rich exosomes that are delivered to normoxic cells to promote cell proliferation, migration, and invasion under hypoxia.24
In this study, we demonstrated that hypoxia enhances PC cell-derived exosome-mediated transfer of UCA1 to human umbilical vein endothelial cells (HUVECs) and promotes angiogenesis in vitro and in vivo. Furthermore, hypoxic exosomal UCA1 promotes angiogenesis via regulating the microRNA (miR)-96-5p/AMOTL2/ERK1/2 signaling pathway in HUVECs. In addition, the expression levels of exosomal UCA1 are higher in PC patients’ serum than in healthy donors’ serum. Therefore, our study reveals a novel mechanism of angiogenesis in PC and provides a potential diagnostic biomarker and treatment target for PC.
Results
Exosomes Derived from Hypoxic PC Cells Promoted Migration and Tube Formation of HUVECs
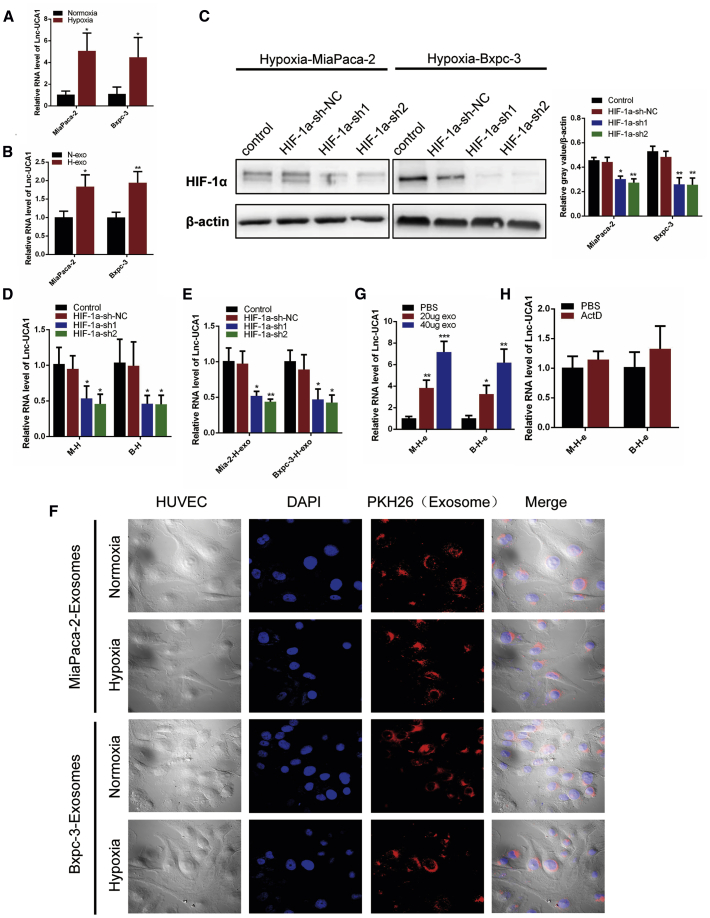
To examine the impact of exosomes derived from normoxic and hypoxic PC cells on the angiogenic ability of HUVECs, we first isolated and identified exosomes derived from normoxic and hypoxic PC cells. Equal number of MIA PaCa-2 cells were seeded under normoxia and hypoxia (1% O2) for 48 h. Compared with the cells cultured under normoxia, the protein levels of HIF-1α in MIA PaCa-2 cells increased under hypoxia (Figure 1A). Next, exosomes were isolated from the conditioned medium (CM) of MIA PaCa-2 cells under normoxic and hypoxic conditions after 48 h by ultracentrifugation and quantitated by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA). As shown in Figures 1B and 1C, TEM revealed typical rounded particles with a diameter of <200 nm, and NTA revealed a similar-size distribution of exosomes isolated from MIA PaCa-2 cells exposed to normoxic or hypoxic conditions. Moreover, western blot analysis confirmed the presence of the exosomal marker proteins CD63, TSG101, and ALIX, and the levels of all exosomal markers were higher in exosomes isolated from hypoxic MIA PaCa-2 cells than from normoxic cells (Figure 1D).
Figure 1.
Both Hypoxic and Normoxic Exosomes Secreted from PC Cells Promoted Angiogenesis
(A) Western blot analysis of HIF-1α in MIA PaCa-2 cells under normoxic and hypoxic conditions. (B) Transmission electron microscopic image of exosomes. (C) The size distribution of exosomes was determined by NTA. (D) Western blot analysis of the exosomal markers CD63, Alix, and TSG101. Equal concentrations of HUVECs were treated with exosomes isolated from CM of MIA PaCa-2 and BxPC-3 cells cultured under normoxic and hypoxic conditions or PBS for 24 h. (E and F) The cell mobility (E) and migration (F) abilities of HUVECs were evaluated by wound healing and Transwell migration assays (n = 3). (G) The ability of tube formation in HUVECs was assessed by an in vitro Matrigel tube-formation assay (n = 3). Results are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Then we investigated the effects of normoxic and hypoxic exosomes on the angiogenic ability of HUVECs by a wound-healing assay, migration assay, and Matrigel tube-formation assay. HUVECs were pretreated with normoxic and hypoxic exosomes secreted by MIA PaCa-2 (M-N-e and M-H-e) or BxPC-3 (B-N-e and B-H-e) cells or PBS as a control for 24 h. Compared with normoxic exosomes or the PBS control, treatment with hypoxic exosomes significantly induced HUVEC migration in the wound-healing assay and the Transwell migration assay (Figures 1E and 1F) and tube-like structure formation in the Matrigel tube-formation assay (Figure 1G). These findings indicate that hypoxic exosomes secreted by PC cells enhance the angiogenic ability of HUVECs in vitro. The effects of hypoxia exosomes were more significant than those of normoxic exosomes.
UCA1 Was Highly Expressed in Exosomes Derived from Hypoxic PC Cells and Could be Transferred to HUVECs through Exosomes
It has been reported that noncoding RNAs are the predominant molecular constituents of tumor cell-derived exosomes. LncRNAs are also enriched in exosomes and can reflect the dysregulated noncoding RNA profiles in tumor cells.25 To identify hypoxic exosomal lncRNA candidates that may affect angiogenesis, next-generation sequencing was used. We have listed the top 20 lncRNAs with increased expression in hypoxic PANC-1-derived exosomes (Table S3). Then, we determined the expression levels of several lncRNAs that may be induced by hypoxia in several PC cells under normoxic and hypoxic conditions.26 Of these, UCA1 levels were increased in various PC cell lines under a hypoxic condition (Figures 2A and S1A). Consistently, analyses of The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEX) databases demonstrated that UCA1 was upregulated in human PC tissues and predicted poor survival of PC patients (Figures S1B and S1C). To further elucidate whether UCA1 is also highly expressed in exosomes derived from PC cells under hypoxia, we measured the expression of UCA1 in exosomes derived from normoxic and hypoxic MIA PaCa-2 and BxPC-3 cells by quantitative real-time PCR. The expression of UCA1 in both hypoxic MIA PaCa-2 and BxPC-3 cell-derived exosomes was markedly upregulated compared with normoxic exosomes (Figure 2B). To study whether hypoxia-induced UCA1 expression depends on HIF-1α, we knocked down the expression of HIF-1α in MIA PaCa-2 and BxPC-3 cells by lentivirus (Figure 2C). Under hypoxic conditions, knockdown of HIF-1α significantly decreased both cellular and exosomal UCA1 expression (Figures 2D and 2E). These results suggested that both cellular and exosomal UCA1 expression under hypoxia was, at least partly, dependent on HIF-1α.
Figure 2.
UCA1 Was Highly Expressed in Exosomes and Could be Transferred to HUVECs through Exosomes
(A and B) Quantitative real-time PCR analysis of UCA1 in MIA PaCa-2 and BxPC-3 cells (A) cultured under normoxic and hypoxic conditions and cell-derived exosomes (B). (C) Western blot analysis of HIF-1α in MIA PaCa-2 and BxPC-3 cells infected with indicated lentivirus under hypoxia. (D and E) Quantitative real-time PCR analysis of UCA1 in hypoxic exosomes (E) and corresponding cells (D) infected with indicated lentivirus. (F) Fluorescent image of the internalization of PKH26-labeled PC cell-derived exosomes (red) in HUVECs by confocal microscopy (60×). (G) Quantitative real-time PCR analysis of UCA1 in HUVECs after coculture with indicated hypoxic exosomes for 24 h and PBS as control (n = 3). (H) Quantitative real-time PCR analysis of UCA1 in HUVECs treated with actinomycin D followed by indicated hypoxic exosomes treatment for 24 h (n = 3). Results are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
To further confirm that hypoxic PC cell-secreted UCA1 can be transferred to HUVECs via exosomes, we first stained exosomes with PKH26. As shown in Figure 2F, the stained exosomes were found to rapidly enter HUVECs and were mainly distributed around the nucleus. Then, we determined UCA1 levels in HUVECs treated with exosomes isolated from MIA PaCa-2 and BxPC-3 cells under normoxic and hypoxic conditions. As expected, an increase in cellular UCA1 levels was observed in recipient HUVECs following treatment with exosomes of MIA PaCa-2 and BxPC-3 cells under hypoxic conditions (Figure 2G). In addition, the increase in UCA1 levels in recipient cells was not affected by the RNA polymerase II inhibitor, actinomycin D (ActD; 1 μg/mL) (Figure 2H). Collectively, these results indicated that hypoxic PC cell-derived exosomes containing UCA1 could be internalized by HUVECs, also transferring UCA1 to HUVECs.
UCA1 Expression Was Positively Correlated with Microvascular Density (MVD) and Negatively Correlated with Overall Survival in PC Patients
To further identify the correlation between UCA1 expression and the MVD in PC tissues, UCA1 was detected by in situ hybridization (ISH), and anti-CD31 was used to stain vascular endothelial cells, and then the MVD was calculated in clinical PC tissues. To further assess the expression of UCA1 in tissues, a “histo” score (H score) system was established. The MVD in tumor tissues with high UCA1 levels was significantly higher than in tumor tissues with low UCA1 levels (Figure 3A). The expression of UCA1 was statistically positively correlated with an increase in the MVD in PC tissues (Figure 3B). Collectively, these results suggested that UCA1 might play an important role in promoting angiogenesis in PC tissues.
Figure 3.
UCA1 Expression Was Positively Correlated with MVD and Negatively Correlated with Overall Survival in PC Patients
(A) Representative images of ISH staining for UCA1 and immunohistochemical (IHC) staining for CD31 in human PC tissue samples. (B) Correlation analysis of the H score of UCA1 and the MVD in PC tissues. (C) Representative transmission electron microscopy images of exosomes isolated from serum of PC patients and healthy individuals. (D and E) Western blot analysis (D) and relative gray value (E) of the serum exosomal markers CD63, Alix, and TSG101. (F) UCA1 expression in serum-derived exosomes was quantified by quantitative real-time PCR. (G) Kaplan-Meier survival curves (log-rank test) for the survival of all 46 PC patients in high and low UCA1 expression groups on the basis of the median UCA1 level. (H) The ROC curve for the circulating exosomal UCA1, where ACTB was used as an internal control. Results are presented as mean ± SD. ∗p < 0.05 and ∗∗p < 0.01.
It has been reported that exosomes can be secreted by cancer cells and enter the blood circulation. Therefore, exosomal UCA1 secreted by hypoxic PC cells can be detected in the blood. To determine the serum levels of exosomal UCA1 and its correlation with clinicopathological data, we first purified exosomes from serum samples obtained from 46 PC patients and 16 healthy donors. Purified serum exosomes were characterized by TEM analysis and western blot (Figures 3C–3E). We also determined the expression of UCA1 in the serum exosomes. The normalized expression levels of exosomal UCA1 in the serum of PC patients were markedly higher than in healthy controls (Figure 3F). Statistical analysis showed that elevated UCA1 expression in serum exosomes was significantly associated with tumor size (p = 0.038), lymphatic invasion (p = 0.018), and late TNM stage (p = 0.017) but not with other clinicopathological parameters, including sex, age, and tumor location (Table 1). Moreover, Kaplan-Meier survival analysis indicated that the survival rate of patients with high UCA1 expression was significantly lower than that of patients with low UCA1 expression (p = 0.0087; Figure 3G). In addition, a receiver operating characteristic (ROC) curve was drawn to evaluate the diagnostic value of serum exosomal UCA1. The results demonstrated that an area under curve (AUC) was of 0.7813 (95% confidence interval [CI] = 0.6497–0.9128, p = 0.00087) after normalization to β-actin (ACTB; Figure 3H). Altogether, these data revealed that serum exosomal UCA1 was negatively correlated with overall survival in PC patients and might serve as a promising diagnostic biomarker and treatment target for PC.
Table 1.
Relationship between lncRNA-UCA1 Expression and Clinicopathological Parameters of 46 Pancreatic Cancer Patients
| Parameters | Total | lncRNA-UCA1 Expression | p Value | |
|---|---|---|---|---|
| Low (n = 23) | High (n = 23) | |||
| Gender | ||||
| Male | 27 | 12 | 15 | 0.369 |
| Female | 19 | 11 | 8 | |
| Age (Years) | ||||
| ≤60 | 23 | 13 | 10 | 0.376 |
| >60 | 23 | 10 | 13 | |
| Tumor Location | ||||
| Head | 29 | 14 | 15 | 0.760 |
| Body or tail | 17 | 9 | 8 | |
| Tumor Size (cm) | ||||
| ≤2 | 21 | 14 | 7 | 0.038a |
| >2 | 25 | 9 | 16 | |
| Differentiation | ||||
| Well | 12 | 7 | 5 | 0.747 |
| Moderate | 18 | 9 | 9 | |
| Poor | 16 | 7 | 9 | |
| Lymph Node Metastasis | ||||
| Negative | 24 | 16 | 8 | 0.018a |
| Positive | 22 | 7 | 15 | |
| Neural Invasion | ||||
| No | 28 | 15 | 13 | 0.546 |
| Yes | 18 | 8 | 10 | |
| TNM Stage | ||||
| I + II | 20 | 14 | 6 | 0.017a |
| III + IV | 26 | 9 | 17 | |
p < 0.05.
Hypoxic Exosomal UCA1 Promoted Migration and Tube Formation of HUVECs In Vitro
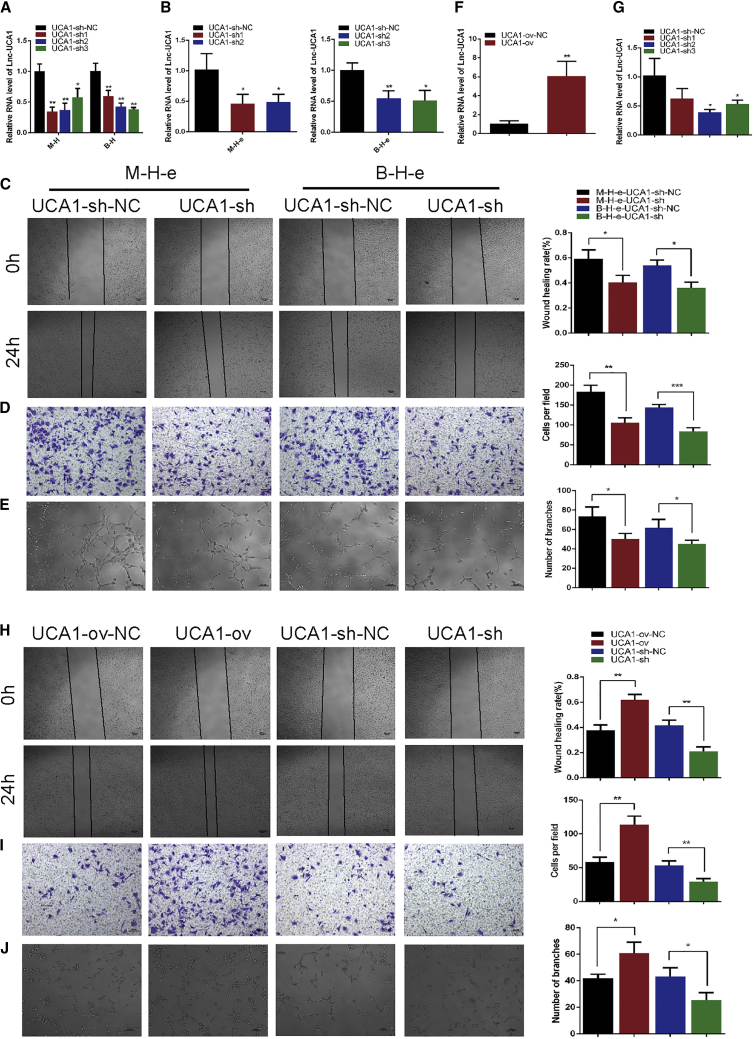
To investigate whether hypoxic exosome-mediated HUVEC migration and tube formation are directly dependent on exosomal UCA1, we first suppressed UCA1 expression in hypoxic MIA PaCa-2 and BxPC-3 cells by UCA1 short hairpin (sh)RNA. The suppression efficiency of shRNA in hypoxic MIA PaCa-2 and BxPC-3 cells was confirmed by quantitative real-time PCR (Figure 4A). In addition to affecting the expression levels of UCA1 in MIA PaCa-2 and BxPC-3 cells, UCA1 shRNA also reduced the expression levels of UCA1 in MIA PaCa-2 and BxPC-3 cell-derived hypoxic exosomes (Figure 4B). Then, we cocultured HUVECs with hypoxic UCA1-knockdown exosomes and hypoxic control shRNA exosomes and evaluated angiogenesis by examining the biological function of HUVECs. As expected, knockdown of hypoxic exosomal UCA1 resulted in the significant inhibition of the pro-angiogenic phenotype (Figures 4C–4E). Moreover, we directly transfected HUVECs with UCA1 overexpression plasmids and UCA1 shRNA to generate UCA1-ov and UCA1-sh cells, respectively. Quantitative real-time PCR analysis showed that UCA1 expression was upregulated in the UCA1-ov group and significantly downregulated in the UCA1-sh2 group (Figures 4F and 4G). Functional assays showed that HUVEC migration and tube-like structure formation were significantly enhanced in UCA1-ov cells and inhibited in UCA1-sh cells (Figures 4H–4J). These results suggested that hypoxic PC cell-derived exosomes promoted HUVEC angiogenesis partly by exosomal UCA1 in vitro.
Figure 4.
Hypoxic Exosomal UCA1 Promoted Migration and Tube Formation in HUVECs In Vitro
(A and B) Quantitative real-time PCR analysis of UCA1 in hypoxic PC cells (A) and exosomes (B) after the transfection of UCA1 shRNAs (n = 3). Equal numbers of HUVECs were pretreated with exosomes isolated from hypoxic PC cells transfected with UCA1 shRNAs for 24 h. (C and D) The cell mobility (C) and migration (D) abilities of HUVECs were evaluated by wound healing and Transwell migration assays (n = 3). (E) The ability of tube formation in HUVECs was assessed by an in vitro Matrigel tube-formation assay (n = 3). (F and G) Quantitative real-time PCR analysis of UCA1 in HUVECs directly transfected with UCA1 overexpression plasmids (F) or UCA1 shRNA (G) (n = 3). (H and I) The cell mobility (H) and migration (I) abilities of HUVECs were evaluated by wound healing and Transwell migration assays (n = 3). (J) The ability of tube formation in HUVECs was assessed by an in vitro Matrigel tube-formation assay (n = 3). Results are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
UCA1 Functioned As a Competing Endogenous RNA (ceRNA) for miR-96-5p in HUVECs
To explore the underlying mechanisms of UCA1-driven angiogenesis, we first determined the localization of UCA1 in HUVECs. After nuclear/cytoplasmic fractioning of HUVECs, we showed that UCA1 was predominantly localized in the cytoplasmic fraction (Figure 5A). Therefore, we hypothesized that it might function as a ceRNA. To confirm this, we performed an RNA immunoprecipitation (RIP) assay on Argonaute2 (Ago2), the core component of the RNA-induced mediating complex.27 The results showed that UCA1 overexpression in HUVECs led to increased enrichment of UCA1 on Ago2 (Figure 5B). These results indicated that UCA1 could competitively bind specific miRNAs, acting as a ceRNA, to block their downstream protein expression. Subsequently, we predicted the miRNAs that participated in the sponging of the UCA1 by the bioinformatics tools, miRcode (http://www.mircode.org/) and StarBase v.2.0 (http://starbase.sysu.edu.cn/) (Figure S2A). Four miRNA candidates fit the criteria. We validated the expression levels of these miRNAs in UCA1-ov and UCA1-sh HUVECs by quantitative real-time PCR. We found that miR-96-5p was downregulated in UCA1-ov HUVECs but upregulated in UCA1-sh HUVECs (Figure 5C), indicating that miR-96-5p was the most suitable candidate for further analysis. Subsequent detailed bioinformatics analysis predicted a potential binding site for miR-96-5p in UCA1 (Figure 5D). To confirm this hypothesis, we cloned wild-type and mutant UCA1 into the psiCHECK-2 luciferase reporter plasmid and performed a luciferase reporter assay to detect the interaction of miR-96-5p with UCA1. The luciferase activity of psiCHECK-2-UCA1 was significantly inhibited by the transfection of miR-96-5p mimics, but little effect was observed with mutant UCA1 (Figure 5E). We next determined whether miR-96-5p was functionally involved in UCA1-mediated angiogenesis of HUVECs. Restoration of miR-96-5p expression dampened the UCA1-mediated proangiogenic phenotype (Figures 5F–5H). Furthermore, transfection of miR-96-5p mimics also inhibited hypoxic exosome-mediated angiogenesis (Figures S2B–S2D). Taken together, these data showed that UCA1 functioned as a ceRNA for miR-96-5p in the regulation of angiogenesis.
Figure 5.
UCA1 Functioned as a ceRNA for miR-96-5p in HUVECs
(A) After nuclear/cytoplasmic fractionation, UCA1 was predominantly located in the cytoplasmic fraction. U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as controls (n = 3). (B) RIP assay of the enrichment of UCA1 on Ago2 relative to IgG in HUVECs transfected with UCA1 overexpression plasmids (n = 3). (C) Quantitative real-time PCR analysis of four miRNA candidates in HUVECs after transfection with UCA1 overexpression plasmids, UCA1-shRNA, or PBS (n = 3). (D) Schematic representation of the miR-96-5p binding site in UCA1 and the mutant site. (E) Luciferase reporter assays in HUVECs cotransfected with psiCHECK-2 luciferase plasmids with wild-type or mutant UCA1 and miR-96-5p mimics or negative control were performed (n = 3). (F and G) The cell mobility (F) and migration (G) abilities of HUVECs were evaluated by wound healing and Transwell migration assays (n = 3). (H) The ability of tube formation in HUVECs was assessed by an in vitro Matrigel tube-formation assay (n = 3). Results are presented as mean ± SD. ∗p < 0.05 and ∗∗p < 0.01.
UCA1 Promoted Angiogenesis via Regulating the miR-96-5p/AMOTL2/ERK1/2 Signaling Pathway in HUVECs
It is widely accepted that the regulatory effects of miRNAs on their target genes could be blocked by lncRNAs.28 In order to determine the genes that are regulated by UCA1 through sponging of miR-96-5p, we first predicted the potential target genes of miR-96-5p. With the use of TargetScan, PicTar, and miRanda databases, hundreds of genes were predicted (Figure S3A). We also used StarBase v.2.0 to predict the UCA1-regulated genes by the ceRNA mechanism, and 65 genes were predicted (Figure S3B). With the combination of these predictions, SLC7A8, NDRG1, and AMOTL2 were identified as potential downstream target genes of UCA1 and miR-96-5p. Then we measured the mRNA levels of these genes in UCA1-ov and UCA1-sh HUVECs by quantitative real-time PCR. We found that mRNA levels of AMOTL2 were upregulated in UCA1-ov HUVECs but downregulated in UCA1-sh HUVECs (Figure 6A). The RIP assay showed that overexpression of UCA1 in HUVECs led to a decrease in the enrichment of AMOTL2 on Ago2 (Figure 6B). Moreover, western blot analysis also indicated that the protein level of AMOTL2 was also upregulated in UCA1-ov HUVECs but downregulated in UCA1-sh HUVECs (Figure 6C). Additionally, we examined several classical signaling pathways that were reported to regulate the function of vascular endothelial cells in UCA1-ov and UCA1-sh HUVECs by western blot. The result showed that UCA1 could upregulate the phosphorylation level of ERK1/2 (p-ERK1/2) but had no effect on AKT or p38 mitogen-activated protein kinase (MAPK) (Figure 6D). As previously reported, AMOTL2 could mediate the activation of the ERK signaling pathway and promote proliferation, migration, and tube formation of HUVECs.29 These results suggested that UCA1 could regulate AMOTL2 expression and the downstream ERK1/2 signaling. Moreover, we examined the protein levels of AMOTL2 and p-ERK1/2 in HUVECs cocultured with hypoxic exosomes and found that hypoxic UCA1-knockdown exosomes derived from both MIA PaCa-2 and BxPC-3 cells decreased AMOTL2 and p-ERK1/2 protein levels (Figure S4A). Functionally, restoration of AMOTL2 expression restored the UCA1-mediated angiogenesis of HUVECs, whereas knockdown of AMOTL2 abolished UCA1-mediated angiogenesis of HUVECs (Figures 6E–6G). A similar phenomenon was observed after treatment with hypoxic exosomes (Figures S4B–S4D). In summary, the genetic and functional data indicated that AMOTL2 was responsible for UCA1-mediated angiogenesis.
Figure 6.
UCA1 Promoted Angiogenesis via the miR-96-5p/AMOTL2/ERK1/2 Signaling Pathway in HUVECs
(A) Quantitative real-time PCR analysis of three indicated mRNA candidates in UCA1-ov or UCA1-sh HUVECs. (B) RIP assay of the enrichment of AMOTL2 mRNA on Ago2 relative to IgG in HUVECs transfected with UCA1 overexpression plasmids (n = 3). (C) Western blot analysis was used to detect protein expression levels of AMOTL2 in UCA1-ov or UCA1-sh HUVECs. (D) Western blot analysis of indicated proteins in UCA1-ov or UCA1-sh HUVECs. (E and F) The effects of UCA1 and AMOTL2 on the mobility (E) and migration (F) were evaluated by wound healing and Transwell migration assays (n = 3). (G) The effects of UCA1 and AMOTL2 on the tube formation were assessed by an in vitro Matrigel tube-formation assay (n = 3). (H) Luciferase reporter assays were performed with HUVECs cotransfected with wild-type or mutant AMOTL2 3′ UTR with miR-96-5p mimics (n = 3). (I) Western blot analysis of indicated proteins in UCA1-ov HUVECs after transfection with miR-96-5p mimics, negative control, or PBS. Results are presented as mean ± SD. ∗p < 0.05 and ∗∗p < 0.01.
To further confirm that miR-96-5p functions as a bridge in the UCA1-mediated regulation of AMOTL2, we first cloned wild-type and mutant 3′ UTRs of AMOTL2 into the psiCHECK-2 luciferase reporter plasmid. The prediction of binding sites on the AMOTL2 3′ UTR with miR-96-5p was performed using the TargetScan database, a schematic representation of the miR-96-5p binding site in the AMOTL2 3′ UTR, and the mutant site was shown in Figures S3C and S3D. As expected, the luciferase reporter assay showed that the luciferase activity of psiCHECK-2-AMOTL2-3′ UTR was significantly inhibited by the transfection of miR-96-5p mimics, but little effect was observed with mutant AMOTL2-3′ UTR reporters (Figure 6H). Moreover, we examined the protein levels of AMOTL2 and p-ERK1/2 in UCA1-overexpressing HUVECs transfected with miR-96-5p mimics by western blot. As expected, transfection of miR-96-5p mimics significantly decreased the protein levels of AMOTL2 and p-ERK1/2 in the UCA1-overexpressing HUVECs (Figure 6I). In addition, transfection of miR-96-5p mimics decreased the protein levels of AMOTL2 and p-ERK1/2 in HUVECs cocultured with exosomes derived from both MIA PaCa-2 and BxPC-3 cells cultured under hypoxic conditions (Figure S4E). These data revealed that UCA1-mediated sequestration of miR-96-5p was responsible for the upregulation of AMOTL2 and p-ERK1/2.
Hypoxic Exosomal UCA1 Promoted Angiogenesis and Tumor Growth in a Mouse Xenograft Model
To further determine whether hypoxic exosomal UCA1 also exerts proangiogenic effects in vivo, a mouse xenograft tumor model (BxPC-3 cell line) was established and treated with hypoxic MIA PaCa-2 cell-derived exosomes (hypoxic exosomes, hypoxic control shRNA exosomes, and hypoxic UCA1-knockdown exosomes) or PBS (Figure 7A). After five injections, mice were euthanized, and tumors were harvested on day 27. As shown in Figures 7B–7D, despite the administration of hypoxic exosomes or hypoxic control shRNA exosomes, tumors significantly increased in size and weight. Depletion of UCA1 in hypoxic exosomes, however, could abrogate tumor growth. Nevertheless, the body weight of nude mice was not significantly different between the groups (Figure 7E). We used anti-CD31 to stain vascular endothelial cells in tumors and then calculated the MVD. The MVD in tumor tissues treated with hypoxic exosomes was significantly higher than that in tumor tissues treated with PBS (Figure 7F). Moreover, compared with the hypoxic control shRNA exosome group, the MVD in tumor tissues treated with hypoxic UCA1-knockdown exosomes was markedly reduced (Figure 7F). In light of this, we surmise that hypoxic tumor-derived exosomes promote angiogenesis and tumor growth in a manner that is at least dependent on hypoxic exosomal UCA1 in vivo.
Figure 7.
Hypoxic Exosomal UCA1 Promoted Angiogenesis and Tumor Growth in a Mouse Xenograft Model
(A) Schematic flow chart of the mouse xenograft tumor model. (B) Images of tumors excised from tumor-implanted mice (n = 5). Scale bar, 10 mm. (C) Tumor volume curves of each group. Tumors were measured every 3 days. (D) Tumor weight of each group. (E) Mean body weight of nude mice (n = 5). (F) IHC staining against CD31 was performed to measure the MVD in tumor tissues. Scale bars, 200 μm. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Discussion
As a typical hallmark of most tumors, especially solid tumors, the hypoxic microenvironment induces adaptive modulation of cancer and stromal cells that promotes cancer development and aggressiveness.7 A large number of studies have shown that tumor cells release more exosomes in the hypoxic microenvironment, which are involved in the regulation of angiogenesis.15 Exosomes, which are cell-derived small particles with a stable bilayer membrane structure, can encapsulate multiple proteins and nucleic acids and play a significant role in the remodeling of the tumor microenvironment.30 In the present study, we extracted exosomes from the culture medium of hypoxic PC cells and investigated the impacts of hypoxic exosomal UCA1 on the progression of PC and the underlying mechanisms. Furthermore, we found that the hypoxic tumor microenvironment could enhance the secretion of exosomes from PC cells and promote exosomes to enter the peripheral blood circulation. The high expression of UCA1 in serum-derived exosomes was significantly associated with tumor size, lymphatic invasion, and late TNM stage and correlated with the progression and poor survival of patients with PC. Therefore, UCA1 might function as a prognostic oncogene in PC and be served as a potential treatment target for PC.
Angiogenesis is an important process in the growth and metastasis of PC and other solid tumors.31 Hypoxia is one of the main factors driving abnormal angiogenesis in tumors, mainly via regulating vascular endothelial growth factor (VEGF) signaling. In recent years, many tumor patients benefited from the combination treatment of VEGF targeting therapy and chemotherapy,32 but the overall outcomes are not optimistic in PC patients.33 However, several studies in animal models of PC have also found that the tumor burden could be efficiently decreased by treatment with anti-angiogenic drugs.5 Therefore, the study of VEGF-independent angiogenic pathways is important, and drugs targeting these pathways may provide possibilities to develop new anti-angiogenesis therapy for PC patients. It has recently been reported that the hypoxic microenvironment can promote cancer cells to release more exosomes and enhance angiogenesis.34 Hence, we determined whether exosomes secreted from hypoxic PC cells can promote angiogenesis. We purified the exosomes secreted by PC cells cultured under normoxic and hypoxic conditions and treated HUVECs with these exosomes. Exosomes secreted by PC cells under both hypoxic and normoxic conditions could promote the migration and tube formation of HUVECs in vitro, and the effects of hypoxic exosomes were more significant than those of normoxic exosomes.
LncRNAs are longer than 200 nucleotides in length and have highly diverse functions in diseases, including various cancers. Molecular mechanisms of lncRNAs are diverse and include chromatin interactions, transcriptional regulation, and RNA processing; and these mechanisms are mainly based on the cellular location of the lncRNAs.35 The levels of lncRNAs change in the hypoxic tumor microenvironment, and some of them can be encapsulated in exosomes.36 UCA1 is dysregulated in many malignant tumors, which affects the development and progression of tumors. Several studies have also reported that UCA1 was highly overexpressed in PC tissues and played an oncogenic role in tumor proliferation and metastasis.37,38 Moreover, UCA1 is induced by HIF-1α under hypoxic conditions and can be packed into exosomes.39 Here, we demonstrated not only that UCA1 levels were increased in hypoxic PC cells but also that UCA1 was enriched in hypoxic PC cell-derived exosomes and could be transferred to HUVECs through exosomes. The enhancement of UCA1 expression in both hypoxic PC cells and exosomes was regulated by HIF-1α. The enhancement of migration and tube formation in HUVECs by hypoxic exosomes was impaired upon knockdown of UCA1, and UCA1 knockdown also inhibited angiogenesis and tumor growth in vivo. These results suggested that hypoxic PC cell-derived exosome-mediated angiogenesis was dependent on UCA1.
It is well acknowledged that lncRNAs function via various mechanisms, which are mainly based on the cellular location. To further understand mechanisms by which UCA1 stimulates angiogenesis in HUVECs, we explored the location of UCA1 in HUVECs. UCA1 is mainly expressed in the cytoplasm. Cytoplasmic lncRNAs often competitively bind to certain miRNAs, referred to as ceRNAs, thus indirectly regulating the expression levels of targeted mRNA of the corresponding miRNA.28 For instance, long noncoding activated in renal cell carcinoma with sunitinib resistance (lncARSR) promotes sunitinib resistance in renal cancer by functioning as a ceRNA, modulating the expression of AXL/c-MET through regulating miR-34 and miR-449.40 UCA1 enhances cell proliferation through acting as a sponge for miR-204 in esophageal cancer.41 In this study, we also used RIP analysis to verify that UCA1 can be enriched by Ago2. Comprehensive bioinformatics analysis predicted that UCA1 and AMOTL2 share miRNA response elements (MREs) of miR-96-5p, implying the formation of the UCA1/miR-96-5p/AMOTL2 axis. Interestingly, a recent study has reported that platelet-derived miR-96 inhibited the wound healing and vascular network formation activities of HUVECs.42 Combined with our bioinformatics analysis and the luciferase reporter assay, we concluded that miR-96-5p functioned as a direct target of both UCA1 and AMOTL2. Western blot analysis also confirmed this hypothesis. In summary, our findings provided direct evidence that UCA1 functioned as a sponge of miR-96-5p, regulating AMOTL2 expression.
AMOTL2 belongs to the angiomotin (Amot) family of membrane-associated scaffold proteins, which includes three members.43 Intensive studies have revealed that Amot proteins play an irreplaceable role in tube formation and migration of endothelial cells.44,45 AMOTL2 has also been reported to play a significant role in promoting proliferation, polarity, and tube formation of vascular endothelial cells via positively regulating MAPK/ERK1/2 signaling.29 Additionally, a recent study demonstrated that AMOTL2 might link vascular endothelial (VE)-cadherin to contractile actin fibers, which is necessary for aortic lumen expansion.46 In the present study, we indicated that UCA1 could enhance the mRNA and protein levels of AMOTL2, as well as the p-ERK1/2 in HUVECs, and UCA1 promoted migration and tube formation of HUVECs. Hypoxic PC cell-derived exosomes could promote the expression of AMOTL2 and increase the p-ERK1/2 in HUVECs by transferring UCA1 to HUVECs. Knockdown of AMOTL2 abolished the effects of hypoxic exosomes on UCA1-mediated angiogenesis in HUVECs. These results indicated that hypoxic exosomal UCA1 played an important role in promoting angiogenesis via regulating the AMOTL2/ERK1/2 pathway.
Conclusions
We demonstrated that UCA1 was upregulated in exosomes secreted by hypoxic PC cells and could be transferred to HUVECs, promoting angiogenesis. Moreover, hypoxic PC cell-derived exosomal UCA1 enhanced angiogenesis in vitro and in vivo by regulating the miR-96-5p/AMOTL2/ERK1/2 axis. Additionally, exosomal UCA1 levels in human serum were associated with a late TNM stage and poor survival in human PC and might function as a diagnostic biomarker and a novel therapeutic target in the treatment of PC.
Materials and Methods
Clinical Serum Samples and Ethical Approval
Forty-six PC blood samples were obtained from PC patients who underwent surgery in Shanghai General Hospital from 2013 to 2015, and healthy blood samples were collected from 16 volunteers without any malignancy. The clinical and pathological characteristics of the 46 PC patients were summarized in Table 1. Thirty-six PC tissue samples were obtained from PC patients who underwent surgery in Shanghai General Hospital from 2014 to 2015. The study was conducted with the approval of the Ethics Committee of Shanghai General Hospital, Shanghai Jiaotong University School of Medicine. Formal informed consent was signed by all patients before collecting blood samples.
Cell Lines and Culture Conditions
Human PC cell lines PANC-1, MIA PaCa-2, BxPC-3, Aspc-1, Sw1990, HUVECs, and human embryonic kidney 293T (HEK293T) cells were obtained from the American Type Culture Collection. The cell lines were maintained in DMEM or RPMI-1640 medium, supplemented with 10% fetal bovine serum (FBS) (normal medium), with 1% penicillin-streptomycin in a cell incubator with 5% CO2 at 37°C. To purify exosomes from hypoxic condition, PC cell lines were cultured in a hypoxia cell incubator with 1% O2.
Isolation and Identification of Exosomes from Cell Supernatants and Human Serum
The PC cell lines were cultured in the normal medium and normoxic condition until 70%–80% confluency, thereafter, cultured in DMEM or RPMI 1640 supplemented with 10% exosome-depleted FBS (exosome-free medium) under normoxic or hypoxic conditions for 48 h. Exosomes were isolated by differential centrifugation, as previously described. Briefly, CM was harvested and centrifuged at 300 × g for 10 min, 2,000 × g for 10 min, and 10,000 × g for 30 min and thereafter, ultracentrifuged at 100,000 × g for 70 min (Beckman Coulter, CA, USA), and the sediment was resuspended with PBS. ExoQuick Reagent (System Biosciences [SBI], CA, USA) was also used to isolate exosomes according to the manufacturer’s instruction. Additionally, the isolation of serum exosomes was conducted by a total exosome isolation reagent (Magen, Shanghai, China) according to the manufacturer’s instruction. The concentration of the exosome was detected by the Bicinchoninic Acid (BCA) Protein Assay Kit (Pierce, USA).
As previously reported, the images of exosomes were obtained by TEM.47 The diameter of exosomes was further analyzed by NTA.48 Moreover, exosomal marker proteins, including CD63, TSG101, and ALIX, were measured by western blot analysis.
Exosome Labeling and Uptake Assay
Indicated exosomes were isolated from the CM and labeled with PKH26, a red fluorescent membrane linker dye (Sigma-Aldrich), following the manufacturer’s protocol. Then, the labeled exosomes were isolated with ExoQuick Reagent and resuspended in PBS and added to the HUVECs at a concentrate of 10 μg exosomes/1 × 105 cells. After incubation for 10 h at 37°C, cells were harvested, fixed with 4% paraformaldehyde, and stained with 4,6-diamidino-2-phenylindole (DAPI; Solarbio, USA), according to the manufacturer’s instruction. The images were acquired with a confocal microscope (Nikon, Japan).
ISH
The relative expression of UCA1 in PC tissues was detected by ISH. The specific digoxin (DIG)-labeled probe (5′-DIG-GCAGATGGACGGCAGTTGGTGTGCTATA-DIG-3′) (Servicebio, China) was used. Briefly, paraffin sections were dewaxed in xylene and rehydrated through ethanol. After that, paraffin sections were digested with proteinase K, fixed in 4% paraformaldehyde, and hybridized with the DIG-labeled UCA1 synthetic oligonucleotide probe overnight and then incubated with anti-DIG-labeled peroxidase (anti-DIG-horseradish peroxidase [HRP]) (Jackson ImmunoResearch Laboratories, USA) at 4°C overnight. Subsequently, paraffin sections were stained with 3,3-diaminobenzidine and counterstained with hematoxylin. The staining scores were scored with an immunoreactivity H score.49 In a fixed field, the staining intensity (0, 1 +, 2+, or 3+) was determined for each cell. Then, the percentage of cells at each staining intensity level was counted, and an H score was calculated using the following formula: [1 × (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3 +)].
RNA Extraction and Quantitative Real-Time PCR Assay
According to the manufacturer’s protocol, total RNA was extracted from cells and exosomes by using Trizol reagent (Invitrogen). For lncRNA and mRNA, complementary DNA (cDNA) was obtained with the Reverse Transcription System (Promega, Madison, WI, USA), whereas for miRNA, cDNA was synthesized by the miDETECT A Track RT Reagent Kit (RiboBio, China). Quantitative lncRNA, mRNA, and miRNA expression was measured with triplicate samples by using SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) on the ABI 7300 PCR system (Applied Biosystems), according to the manufacturer’s instructions. ACTB and U6 were chosen as normalized controls for lncRNA, mRNA, and miRNA, respectively. The 2−ΔΔCt analytic method was employed to calculate the folding change of genes. Primers were shown in Table S1.
Plasmid Construction and Transfection
To knock down or increase the expression of genes, indicated plasmids were constructed. The shRNAs for UCA1 (shRNA-UCA1) were designed and cloned into the pGpU6/GFP/Neo vector by GenePharma (Shanghai, China). The targeting sequences of UCA1 were shown as follows: sh1 target: 5′-GGAGGATTCCCAGCCATATGA-3′; sh2 target: 5′-GGGTTCACCATTCCAGAATAA-3′; sh3 target: 5′-GGACAACAGTACACGCATATG-3′; and short hairpin negative control (shNC) target: 5′-TTCTCCGAACGTGTCACGT-3′. The full length of UCA1 was cloned into the EcoRI and XhoI sites of the pcDNA3.1 vector and named as pcDNA3.1-UCA1. The shRNA-HIF-1α-targeted sequences were shown as follows: sh1 target: 5′-TCCAGCAGACTCAAATACA-3′ and sh2 target: 5′-AGCTATTCACCAAAGTTGA-3′. The shRNA-AMOTL2-targeted sequences were shown as follows: sh1 target: 5′-GGAGATGTCTTGTTAGCAT-3′ and sh2 target: 5′-TGGTTCATGTGCATTGTTT-3′. The shRNA-HIF-1α and shRNA-AMOTL2 oligos were cloned into a pGIPZ lentivirus vector. The human AMOTL2 open reading frame (ORF) sequences were amplified and subcloned into a pLVX-IRES-Puro lentivirus vector. HEK293T cells were cotransfected with the lentivirus vector described above and packaging vectors psPAX2 and pMD2.G with Lipofectamine 2000 transfection reagent (Invitrogen) for producing lentivirus. The mimics and negative control for hsa-miR-96-5p were obtained from RiboBio (Guangzhou, China). The other plasmids were transfected into cells with Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions.
Protein Extraction and Western Blot Analysis
Cells and exosomes were decomposed by using 2 × SDS-lysis buffer. The protein samples were loaded into the SDS-PAGE gel and shifted onto a 0.22-μm nitrocellulose membrane (Axygen, USA). Membranes were blocked with 5% skim milk and then embraced with the primary antibodies overnight at 4°C. The next day, the membranes were washed and incubated with a second antibody for 1 h at room temperature. Finally, the membranes were visualized with an Enhanced Chemiluminescence (ECL) Detection Kit (Millipore, Billerica, USA) and by using ImageQuant LAS 4000 Mini (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). The primary antibodies used were listed as follows: anti-HIF-1α (1:1,000; Cell Signaling Technology, USA); anti-ACTB (1:5,000; Cell Signaling Technology, USA); anti-TSG101 (1:1,000; Proteintech, USA); anti-ALIX (1:500; Santa Cruz, USA); anti-CD63 (1:1,000; Abcam, UK); anti-AKT, anti-p-AKT, anti-ERK1/2, anti-p-ERK1/2, anti-p38MAPK, and anti-p-p38MAPK (1:1,000; Cell Signaling Technology, USA); and anti-AMOTL2 (1:500; Proteintech, USA).
Wound-Healing Assay and Transwell Migration Assay
To assess the migration of HUVECs, we conducted a wound-healing assay and Transwell migration assay. First, indicated HUVECs were pretreated with corresponding exosomes for 24 h or transfected with plasmids. For the wound-healing assay, a certain number of HUVECs were seeded in 6-well plates and cultured to full confluence. After that, the wells were scratched with a 200-μL micropipette tip to create two separate boundaries that prevent cells from touching each other, and cells were washed with PBS to remove the floating cells and cultured with FBS-free medium. At 0 and 24 h after the scratch, cells were observed, and representative images were taken. The rate of wound healing was calculated according to the total healing area analyzed with ImageJ. All experiments were repeated independently in triplicate.
For the Transwell migration assay, 2 × 104 HUVECs resuspended in 200 μL serum-free medium were seeded in the upper chamber (8.0 μm pore; Corning, USA). About 600 μL normal medium supplemented with 10% FBS was added into the lower chamber as a chemoattractant. After 10 h of culture, cells on the inside of the membranes were wiped out gently, and outside invading cells were fixed with 4% paraformaldehyde and stained with Crystal Violet (Beyotime, China). The membranes were dried at room temperature and then observed under a light microscope, and representative images were taken. All experiments were repeated independently in triplicate.
Matrigel Tube-Formation Assay
As previously mentioned,50 in vitro Matrigel tube-formation analysis was performed. Briefly, indicated HUVECs were pretreated with corresponding exosomes for 24 h or transfected with plasmids, and then HUVECs were harvested. In advance, 50 μL growth factor-reduced Matrigel (BD Biosciences) was added to 96-well plates and cultured at 37°C for 30 min, and then, 2 × 104 HUVECs were resuspended in 100 μL serum-free DMEM and added to 96-well plates precoated with Matrigel. After incubation at 37°C for 6 to 8 h, the formation of tube structures was observed under the inverted microscope, and representative images were taken. The number of branches in each well was calculated by ImageJ software. All experiments were repeated independently in triplicate.
RIP
As previously mentioned,51 RIP assay was performed. Briefly, indicated HUVECs were transfected with pcDNA3.1-UCA1 or control for 48 h and then resuspended in radioimmunoprecipitation assay (RIPA) buffer and incubated on ice for 30 min. Human anti-Ago2 antibody (Proteintech, USA) was added into the lysate obtained by centrifugation and incubated for 4 h at 4°C, whereas normal mouse immunoglobulin G (IgG; Santa Cruz, USA) was chosen as a negative control. Meanwhile, the beads were resuspended in RIPA buffer and treated with proteinase K at 45°C for 45 min. Immunoprecipitated RNA samples were extracted with Trizol and tested by quantitative real-time PCR analysis. Proteins was isolated and detected by western blot analysis.
Luciferase Reporter Assay
The full length of human UCA1 and AMOTL2 3′ UTR sequence and mutant type for miR-96-5p were subcloned into the XhoI and NotI sites of the psiCHECK-2 vector by using ClonExpress II and Mut Express II Fast Mutagenesis Kit, v.2 (Vazyme, China), respectively. The primers used were shown in Table S2. The luciferase reporter plasmids psiCHECK-2 containing UCA1, AMOTL2 3′ UTR, and each mutant type were cotransfected into HUVECs with miR-96-5p mimics or negative control using Lipofectamine 2000 transfection reagent. After transfection for 48 h, the cells were harvested, and then firefly and Renilla luciferase activity were detected by using a Dual-Luciferase Kit (Promega), according to the manufacturer’s protocol. The activity of firefly luciferase was used for normalization. All experiments were independently repeated in triplicate.
Mouse Xenograft Model and Analysis of MVD in Tumor Tissues
All animal experiments were approved by the Ethics Committee for animal research of Shanghai Jiaotong University School of Medicine (Shanghai, China). As previously reported, PC cells (BxPC-3, 5 × 106 cells/animal) were transplanted subcutaneously into the right flank of 4-week-old female nude mice. Tumor size was measured every 3 days. On the 12th day, when the tumor volume reached approximately 100 mm2, the mice were randomly divided into four groups, and corresponding exosomes (10 μg) were intravenously injected into the center of tumors of mice every 3 days. After five injections, the mice were sacrificed, primary tumors were removed, and the volumes and weights were recorded. After that, the tumors were excised for immunohistochemical staining for CD31, which was always used as a marker of endothelial cells. The method of quantifying the blood vessels was described previously.52
Statistical Analysis
The IBM SPSS Statistics software was used for statistical analysis. Experiments were performed in triplicate, data are presented as the mean ± standard deviation (SD), and the graphs and diagrams were generated by GraphPad Prism. Student’s t test and one-way analysis of variance (ANOVA) were used to analyze statistics between two groups or among multiple groups, respectively. A p value <0.05 was considered statistically significant.
Author Contributions
Z.Q., Q.Z., C.H., and Z.G. designed the experiments, analyzed the data, and revised the manuscript. Z.G. wrote the manuscript. Z.G., X.W., and Y.Y. performed most of the experiments. W.C., K.Z., and B.T. conducted part of the experiments. All of the authors discussed the results and reviewed the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The work was done in Shanghai, People’s Republic of China. Thank you to all involved in this study. This work was supported by grants from the National Natural Science Foundation of China (81372640) received by Z.Q. and Shanghai Sailing Program Xiaofeng Wang (20YF1440100) received by X.W..
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.08.021.
Contributor Information
Chen Huang, Email: richard-hc@sohu.com.
Qian Zhao, Email: qzhao@shsmu.edu.cn.
Zhengjun Qiu, Email: qiuzjdoctor@sina.com.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Sunamura M., Duda D.G., Ghattas M.H., Lozonschi L., Motoi F., Yamauchi J., Matsuno S., Shibahara S., Abraham N.G. Heme oxygenase-1 accelerates tumor angiogenesis of human pancreatic cancer. Angiogenesis. 2003;6:15–24. doi: 10.1023/a:1025803600840. [DOI] [PubMed] [Google Scholar]
- 3.Huang C., Li Z., Li N., Li Y., Chang A., Zhao T., Wang X., Wang H., Gao S., Yang S. Interleukin 35 Expression Correlates With Microvessel Density in Pancreatic Ductal Adenocarcinoma, Recruits Monocytes, and Promotes Growth and Angiogenesis of Xenograft Tumors in Mice. Gastroenterology. 2018;154:675–688. doi: 10.1053/j.gastro.2017.09.039. [DOI] [PubMed] [Google Scholar]
- 4.Linder S., Blåsjö M., von Rosen A., Parrado C., Falkmer U.G., Falkmer S. Pattern of distribution and prognostic value of angiogenesis in pancreatic duct carcinoma: a semiquantitative immunohistochemical study of 45 patients. Pancreas. 2001;22:240–247. doi: 10.1097/00006676-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Dineen S.P., Sullivan L.A., Beck A.W., Miller A.F., Carbon J.G., Mamluk R., Wong H., Brekken R.A. The Adnectin CT-322 is a novel VEGF receptor 2 inhibitor that decreases tumor burden in an orthotopic mouse model of pancreatic cancer. BMC Cancer. 2008;8:352. doi: 10.1186/1471-2407-8-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M.-D., Liu L., Wu M.-Y., Jiang M., Shou L.-M., Wang W.-J., Wu J., Zhang Y., Gong F.R., Chen K. The combination of cantharidin and antiangiogenic therapeutics presents additive antitumor effects against pancreatic cancer. Oncogenesis. 2018;7:94. doi: 10.1038/s41389-018-0102-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rankin E.B., Giaccia A.J. Hypoxic control of metastasis. Science. 2016;352:175–180. doi: 10.1126/science.aaf4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu X., Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clin. Cancer Res. 2010;16:5928–5935. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih J.-W., Chiang W.-F., Wu A.T.H., Wu M.-H., Wang L.-Y., Yu Y.-L., Hung Y.W., Wang W.C., Chu C.Y., Hung C.L. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1α co-activator driving oral cancer progression. Nat. Commun. 2017;8:15874. doi: 10.1038/ncomms15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitamoto S., Yokoyama S., Higashi M., Yamada N., Takao S., Yonezawa S. MUC1 enhances hypoxia-driven angiogenesis through the regulation of multiple proangiogenic factors. Oncogene. 2013;32:4614–4621. doi: 10.1038/onc.2012.478. [DOI] [PubMed] [Google Scholar]
- 11.Pegtel D.M., Gould S.J. Exosomes. Annu. Rev. Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 12.Sun Z., Yang S., Zhou Q., Wang G., Song J., Li Z., Zhang Z., Xu J., Xia K., Chang Y. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol. Cancer. 2018;17:82. doi: 10.1186/s12943-018-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu R., Rai A., Chen M., Suwakulsiri W., Greening D.W., Simpson R.J. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 14.King H.W., Michael M.Z., Gleadle J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kucharzewska P., Christianson H.C., Welch J.E., Svensson K.J., Fredlund E., Ringnér M., Mörgelin M., Bourseau-Guilmain E., Bengzon J., Belting M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA. 2013;110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L., Li C., Wang S., Wang Z., Jiang J., Wang W., Li X., Chen J., Liu K., Li C., Zhu G. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016;76:1770–1780. doi: 10.1158/0008-5472.CAN-15-1625. [DOI] [PubMed] [Google Scholar]
- 17.Wang X., Luo G., Zhang K., Cao J., Huang C., Jiang T., Liu B., Su L., Qiu Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018;78:4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 18.Lang H.-L., Hu G.-W., Zhang B., Kuang W., Chen Y., Wu L., Xu G.H. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol. Rep. 2017;38:785–798. doi: 10.3892/or.2017.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau E. Non-coding RNA: Zooming in on lncRNA functions. Nat. Rev. Genet. 2014;15:574–575. doi: 10.1038/nrg3795. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai H., Liu X., Zheng J., Xue Y., Ma J., Li Z., Xi Z., Li Z., Bao M., Liu Y. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36:318–331. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Li L., Zhao K., Lin Q., Li H., Xue X., Ge W., He H., Liu D., Xie H. A novel LncRNA HITT forms a regulatory loop with HIF-1α to modulate angiogenesis and tumor growth. Cell Death Differ. 2020;27:1431–1446. doi: 10.1038/s41418-019-0449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar M.M., Goyal R. LncRNA as a Therapeutic Target for Angiogenesis. Curr. Top. Med. Chem. 2017;17:1750–1757. doi: 10.2174/156802661766616111644744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue M., Chen W., Xiang A., Wang R., Chen H., Pan J., Pang H., An H., Wang X., Hou H., Li X. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol. Cancer. 2017;16:143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhry H., Harris A.L. Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 2018;27:281–298. doi: 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Li Z., Xu P., Huang L., Tong J., Huang H., Meng A. Angiomotin-like2 gene (amotl2) is required for migration and proliferation of endothelial cells during angiogenesis. J. Biol. Chem. 2011;286:41095–41104. doi: 10.1074/jbc.M111.296806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whiteside T.L. Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Semin. Immunol. 2018;35:69–79. doi: 10.1016/j.smim.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarte-Waldhoff I., Volpert O.V., Bouck N.P., Sipos B., Hahn S.A., Klein-Scory S., Lüttges J., Klöppel G., Graeven U., Eilert-Micus C. Smad4/DPC4-mediated tumor suppression through suppression of angiogenesis. Proc. Natl. Acad. Sci. USA. 2000;97:9624–9629. doi: 10.1073/pnas.97.17.9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurwitz H., Saini S. Bevacizumab in the treatment of metastatic colorectal cancer: safety profile and management of adverse events. Semin. Oncol. 2006;33(Suppl 10):S26–S34. doi: 10.1053/j.seminoncol.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Kindler H.L., Niedzwiecki D., Hollis D., Sutherland S., Schrag D., Hurwitz H., Innocenti F., Mulcahy M.F., O’Reilly E., Wozniak T.F. Gemcitabine plus bevacizumab compared with gemcitabine plus placebo in patients with advanced pancreatic cancer: phase III trial of the Cancer and Leukemia Group B (CALGB 80303) J. Clin. Oncol. 2010;28:3617–3622. doi: 10.1200/JCO.2010.28.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu Y.L., Hung J.Y., Chang W.A., Lin Y.S., Pan Y.C., Tsai P.H., Wu C.Y., Kuo P.L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36:4929–4942. doi: 10.1038/onc.2017.105. [DOI] [PubMed] [Google Scholar]
- 35.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih J.-W., Kung H.-J. Long non-coding RNA and tumor hypoxia: new players ushered toward an old arena. J. Biomed. Sci. 2017;24:53. doi: 10.1186/s12929-017-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu X., Lin Y., Sui W., Zou Y., Lv Z. Analysis of distinct long noncoding RNA transcriptional fingerprints in pancreatic ductal adenocarcinoma. Cancer Med. 2017;6:673–680. doi: 10.1002/cam4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Gao F., Zhou L., Wang H., Shi G., Tan X. UCA1 Regulates the Growth and Metastasis of Pancreatic Cancer by Sponging miR-135a. Oncol. Res. 2017;25:1529–1541. doi: 10.3727/096504017X14888987683152. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., Yu Z., Yang J., Wang B., Sun H. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu L., Ding J., Chen C., Wu Z.-J., Liu B., Gao Y., Chen W., Liu F., Sun W., Li X.F. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Jiao C., Song Z., Chen J., Zhong J., Cai W., Tian S., Chen S., Yi Y., Xiao Y. lncRNA-UCA1 enhances cell proliferation through functioning as a ceRNA of Sox4 in esophageal cancer. Oncol. Rep. 2016;36:2960–2966. doi: 10.3892/or.2016.5121. [DOI] [PubMed] [Google Scholar]
- 42.Zhang M., Zhao Y., Zhang Y., Wang D., Gu S., Feng W., Peng W., Gong A., Xu M. LncRNA UCA1 promotes migration and invasion in pancreatic cancer cells via the Hippo pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864(5 Pt A):1770–1782. doi: 10.1016/j.bbadis.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Bratt A., Wilson W.J., Troyanovsky B., Aase K., Kessler R., Van Meir E.G., Holmgren L. Angiomotin belongs to a novel protein family with conserved coiled-coil and PDZ binding domains. Gene. 2002;298:69–77. doi: 10.1016/s0378-1119(02)00928-9. [DOI] [PubMed] [Google Scholar]
- 44.Levchenko T., Bratt A., Arbiser J.L., Holmgren L. Angiomotin expression promotes hemangioendothelioma invasion. Oncogene. 2004;23:1469–1473. doi: 10.1038/sj.onc.1207264. [DOI] [PubMed] [Google Scholar]
- 45.Zheng Y., Vertuani S., Nyström S., Audebert S., Meijer I., Tegnebratt T., Borg J.P., Uhlén P., Majumdar A., Holmgren L. Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ. Res. 2009;105:260–270. doi: 10.1161/CIRCRESAHA.109.195156. [DOI] [PubMed] [Google Scholar]
- 46.Hultin S., Zheng Y., Mojallal M., Vertuani S., Gentili C., Balland M., Milloud R., Belting H.G., Affolter M., Helker C.S. AmotL2 links VE-cadherin to contractile actin fibres necessary for aortic lumen expansion. Nat. Commun. 2014;5:3743. doi: 10.1038/ncomms4743. [DOI] [PubMed] [Google Scholar]
- 47.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;Chapter 3:Unit 3.22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 48.Soo C.Y., Song Y., Zheng Y., Campbell E.C., Riches A.C., Gunn-Moore F., Powis S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu X.G., Zhou C.F., Zhang Y.M., Yan R.M., Wei W.F., Chen X.J., Yi H.Y., Liang L.J., Fan L.S., Liang L. Cancer-derived exosomal miR-221-3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis. 2019;22:397–410. doi: 10.1007/s10456-019-09665-1. [DOI] [PubMed] [Google Scholar]
- 50.Nowak-Sliwinska P., Alitalo K., Allen E., Anisimov A., Aplin A.C., Auerbach R., Augustin H.G., Bates D.O., van Beijnum J.R., Bender R.H.F. Consensus guidelines for the use and interpretation of angiogenesis assays. Angiogenesis. 2018;21:425–532. doi: 10.1007/s10456-018-9613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai M.-C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olive K.P., Jacobetz M.A., Davidson C.J., Gopinathan A., McIntyre D., Honess D., Madhu B., Goldgraben M.A., Caldwell M.E., Allard D. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.